Highlights
-
•
Serological studies on SARS- and MERS-coronavirus (CoV) diagnostics were reviewed.
-
•
Different types of serological assays and variable antigens were compared.
-
•
Immunogenic epitopes of CoV spike proteins were less conserved than nucleocapsid proteins.
-
•
Use of spike proteins was found to be superior over nucleocapsid proteins.
-
•
Applicability of serological assays for analysis of animal sera was reviewed.
Keywords: SARS-Coronavirus, MERS-Coronavirus, Reservoir, Serology, Antibody, Cross-reactivity
Abstract
More than a decade after the emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002/2003 the occurrence of a novel CoV termed Middle East respiratory syndrome (MERS) CoV challenges researchers and public health authorities. To control spread and finally contain novel viruses, rapid identification and subsequent isolation of infected individuals and their contacts is of utmost importance. Next to methods for nucleic acid detection, validated serological assays are particularly important as the timeframe for antibody detection is less restricted. During the SARS-CoV epidemic a wide variety of serological diagnostic assays were established using multiple methods as well as different viral antigens. Even though the majority of the developed assays showed high sensitivity and specificity, numerous studies reported on cross-reactive antibodies to antigens from wide-spread common cold associated CoVs. In order to improve preparedness and responsiveness during future outbreaks of novel CoVs, information and problems regarding serological diagnosis that occurred during the SARS-CoV should be acknowledged.
In this review we summarize the performance of different serological assays as well as the applicability of the two main applied antigens (spike and nucleocapsid protein) used during the SARS-CoV outbreak. We highlight challenges and potential pitfalls that occur when dealing with a novel emerging coronavirus like MERS-CoV. In addition we describe problems that might occur when animal sera are tested in serological assays for the identification of putative reservoirs. Finally, we give a recommendation for a serological testing scheme and outline necessary improvements that should be implemented for a better preparedness.
1. Introduction
In 2002/2003, a new disease emerged in Southeast Asia that was subsequently termed severe acute respiratory syndrome (SARS). A previously unknown coronavirus (CoV) was identified as the etiological agent (Drosten et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003). It was only the concerted efforts of public health authorities that made it possible to break the chain of transmission. No new cases have been reported since May 2004. Overall, SARS-CoV infected 8422 people, of whom 916 died, giving a case fatality ratio of about 11% (Chan-Yeung and Xu, 2003). Before the emergence of SARS-CoV, human pathogenic CoVs (HCoVs) such as HCoV-OC43 and HCoV-229E were known to cause mild upper respiratory diseases contributing to 5–30% of the seasonal common cold cases (Isaacs et al., 1983, Larson et al., 1980, Monto, 1974). This explains why, globally, more than 90% of the population has antibodies against the common cold CoV (Gorse et al., 2010).
SARS-CoV belongs to the Coronavirdae family within the order of Nidovirales. It harbors one of the largest known positive-strand RNA genomes comprising about 29 kb (Rota et al., 2003). The first two-thirds of the genome contain nonstructural proteins (NSP) that are well conserved among different CoV species (Rota et al., 2003). The NSPs include the RNA-dependent RNA polymerase and form the main part of the transcription/replication machinery. The last third of the genome encodes mainly the four structural proteins: spike (S), membrane (M), envelope (E) and nucleocapsid (N) (Rota et al., 2003). Interspersed between the structural proteins are group-specific open reading frames (ORFs) encoding a subset of accessory proteins with mostly unknown function (Narayanan et al., 2008). In the case of SARS-CoV, it was shown that, apart from the four structural proteins, some of the NSPs as well as the accessory proteins p3a and p7a, are incorporated into virions and may elicit an immune response in infected patients as it was shown for NSP13 (Leung et al., 2004, Neuman et al., 2008, Schaecher et al., 2007).
Diagnostic assays for SARS-CoV detection were rapidly developed after the identification of the virus. Testing of suspected cases helped considerably to contain the outbreak and understand the rapid disease progression that was observed in some of the patients. SARS patients had detectable viral RNA between three and 30 days after the first symptoms appeared, with high viral loads in lower respiratory tract and fecal samples. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) assays were the first assays that helped identify and subsequently isolate patients who were actively shedding the virus (Drosten et al., 2003). It was reported that viral RNA could be detected by qRT-PCR up to 30 days post onset of illness (dpoi) (Chan et al., 2004a). However, detection rates by these early qRT-PCR assays were often low as they relied on sampling during virus shedding and proper handling of samples (Yam et al., 2003). The specificity of the qRT-PCR assays has also been questioned because of the potential for nucleic acid contamination in the laboratories where many SARS-CoV samples were processed (Patrick et al., 2006).
As a consequence, the development of serological assays became crucial at the time. Many laboratories worldwide generated in-house assays using either virus-derived antigen or recombinant structural CoV proteins. For all laboratories, proper validation of serological assays was highly challenging because of its reliance on numerous well-characterized positive and negative serum samples. Furthermore, the exchange of patient serum samples was a major challenge during the outbreak for logistic and ethical reasons. Assay sensitivity (the number of positive samples that could be determined correctly) and specificity (measured by the number of negative samples that could be identified correctly) were therefore difficult to determine. Consequently, an external quality assurance study revealed that many laboratories had difficulties with SARS-CoV serodiagnostics (Niedrig et al., 2005). In particular, the high seroprevalence in the population of antibodies against the common cold CoV combined with the presence of cross-reactive antibodies against conserved parts of the immunogenic CoV proteins could have contributed to false positive results.
Nevertheless, as antibodies can be detected over a long period, developing reliable post-infection serological assays for SARS-CoV became a high priority. Serodiagnostic assays were applied to address epidemiological questions about transmission patterns, to observe silent infections, to analyze disease progression and to identify the origin of SARS-CoV. Patients usually developed IgM and IgG antibodies within 17–21 dpoi (Woo et al., 2004b). To identify infected contacts, including those that are asymptomatic, it is recommendable to analyze paired serum samples. Ideally, samples should be taken on day 0 and day 42 post exposure since it was shown that seroconversion of IgG or IgM occurred during that period (Chen et al., 2004a). According to the WHO criteria for serological diagnosis, a patient was considered to have seroconverted if one of the following statements were true: “Negative antibody test on acute-phase serum followed by positive antibody test on convalescent-phase serum tested in parallel” or a “Fourfold or greater rise in antibody titer between acute- and convalescent-phase sera tested in parallel” (WHO, 2004). Of note, WHO further recommended using a virus neutralization test (VNT) to exclude serological cross-reactions with other human or animal CoVs (WHO, 2004).
In this review, we summarize the various types of serological assays that were developed during the SARS-CoV outbreak and analyze the challenges and potential pitfalls of serodiagnostics. It should be acknowledged that the multitude of developed assays and the lack of standardized procedures and assay validation make it difficult to directly compare all studies. Finally, we discuss the implications and challenges facing serodiagnosis of the novel Middle East respiratory syndrome (MERS-CoV).
2. Testing parameters: IgM vs. IgG subclasses
Testing for different immunoglobulin (Ig) subclasses is common in serodiagnostics. Being pentameric with ten antigen-binding sites, IgM is characterized by a higher antigen avidity but lower antigen affinity than IgG (Murphy et al., 2008). As antibodies of IgM subclass are usually the first to develop following a primary challenge (Murphy et al., 2008), IgM is considered a parameter of the early phase of infection. However, Woo et al. (2004b) found that IgM could not be detected earlier than IgA and IgG subclass antibodies in SARS-CoV serodiagnostic assays. The low affinity of IgM antibodies also carries the increased risk of cross-reactivity with antigenically related epitopes, which are common in CoVs. IgM detection has an added value for CoV serodiagnostics as it is only present in very recently infected patients, but does not allow differential diagnosis. In contrast IgG antibodies comprise a higher specificity than IgM and can be detected even years after the infection. In this review, we therefore focus on studies describing assays for the detection of IgG antibodies.
3. Virus-based serological assays
During the outbreak of SARS-CoV, a wide variety of serological assays were established, including immunofluorescence assays (IFAs), enzyme-linked immunosorbent assays (ELISAs) and Western blot (WB) analysis.
Conventional IFAs (cIFAs) using virus-infected African green monkey kidney cells (VERO E6) spotted on glass slides and ELISAs using extracts or supernatant of infected cells were among the first assays used in serological diagnosis of SARS-CoV (Chan et al., 2004b, Chen et al., 2004a, Hsueh et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003, Wu et al., 2004). Both cIFAs and ELISAs for SARS-CoV were relatively easy to set up for experienced laboratories, as they primarily only required susceptible cell cultures and the virus. An obvious disadvantage of virus-based serological assays is the need for a biosafety level (BSL) 3 facility. In addition, IFAs can neither be properly standardized because the interpretation of fluorescence staining patterns is subjective, nor are they appropriate for high-throughput screening. In the case of ELISAs, antigen production can be an obstacle if it necessitates ultracentrifugation under BSL3 conditions. Ensuring that the antigen is properly inactivated can also be problematic. Ultimately, validation of the ELISA is dependent on access to a well-characterized serum collection for the determination of the assay-specific cut-off value.
A detailed summary of studies applying SARS virus-based cIFAs and ELISAs is given in Table 1 . Since SARS-CoV seroconversion generally occurred during the second week of illness (Hsueh et al., 2003, Ksiazek et al., 2003), only those studies in which sera was taken at least 14 dpoi are included in this overview. In the majority of cases, patient serum was tested in assays using virus-derived antigens. Most of the studies found that serum from between 85% and 100% of previously diagnosed SARS patients tested positive (Table 1, column 3), suggesting that the cIFA and ELISA used were highly sensitive. Of note, the studies with the most reliable results were those that used a large number of patient serum samples (n = 90; Chan et al. (2004b) and n = 224; Wu et al. (2004)), which found that 98.2% and 99.1%, respectively, were positive.
Table 1.
| Antigen | Assay | SARS sera (≥14 dpoi) | Control sera from respiratory patients or SARS contacts | Control sera from healthy donors | Reference | Comment |
|---|---|---|---|---|---|---|
| No. positive/no. tested (%) | No. positive/no. tested (%) | No. positive/no. tested (%) | ||||
| Whole Virus | IFA | 89/90 (98.8)a | 0/540 (0)a | 0/635 (0)a | Chan (2004, #6) | 24 control sera were excluded due to unspecific reaction |
| Whole Virus | IFA | 34/34 (100)a | n.d. | 1/100 (1)a | Che (2005, #9) | |
| Whole Virus | IFA | 6/7 (85.7)a | n.d. | 0/10 (0)a | Hsueh (2003, #22) | |
| Whole Virus | IFA | 10/10 (100)a | 0/27 (0)a | 0/384 (0)a | Ksiazek (2003, #66) | |
| Whole Virus | IFA | 46/46 (100)a | 0/40 (0)a | 0/16 (0)a | Leung (2004, #29) | |
| Whole Virus | IFA | 37/37 (100)a | 0/80 (0)a | 0/200 (0)a | Peiris (2003, #86) | |
| Whole Virus | IFA | 222/224 (99.1)b | 30/245 (12.2)a, b | n.d. | Wu (2004, #60) | Using VNT as gold standard |
| Whole Virus | ELISA | 10/10 (100)a | 0/27 (0)a | 1/384 (0.3)a | Ksiazek (2003, #66) | |
| Whole Virus | ELISA | 42/46 (91.3)a | 3/40 (7.5)a | 1/38 (2.6)a | Leung (2004, #29) | |
| Whole Virus | ELISA | 17/20 (85) | 0/20 (0) | 2/40 (5)a | Wang et al. (2004, #54) | |
| Whole Virus | ELISA | 220/224 (98.2)b | 3/245 (1.2)a, b | n.d. | Wu (2004, #60) | Using VNT as gold standard |
dpoi, days post onset of illness; IFA, immunofluorescence assay; n.d., not determined; ELISA, enzyme-linked immunosorbent assay; VNT, virus neutralization test.
Calculated from information given in the paper.
No differentiation between acute and convalescent sera.
Specificity of serological assays is vital to avoid false positive diagnoses. Critically, some of the HCoVs are antigenically closely related (Bradburne, 1970). This means, in combination with the observed high prevalence of HCoVs, such as HCoV-OC43, HCoV-229E and HCoV-NL63, in the population (Che et al., 2005, Dijkman et al., 2012), that the specificity of the SARS-CoV assay is of particular concern. We therefore analyzed and summarized the rate of false positive results found in serological studies and differentiated between the two types of control groups used; sera from individuals with non-SARS respiratory infection or in contact with SARS patients, or from healthy donors (Table 1, columns 4 and 5). One limitation of this analysis is that, in most of the studies, negative control sera were obtained from respiratory infection patients or blood donors, and therefore the inclusion of unidentified SARS cases cannot be completely discounted. Nevertheless, the detection rate of false positive samples gives a rough estimate of the specificity and allows the performance of these assays to be evaluated. In the various case control studies that were conducted during or after the SARS outbreak, cIFAs and viral antigen-based ELISAs detected between 0% and 12.2% false positives (Table 1, columns 4 and 5). Assay outcomes were highly dependent on the study design and the negative cohort selected. Notably, in one study, the rate of false positive results of cIFA increased to 12.2% when a virus neutralization assay was used as a gold standard (Wu et al., 2004). This could be explained by a lack of neutralizing capacity of certain cIFA-positive sera or indeed by cross-reactivity of sera with antigens from related HCoV in the cIFA.
A study by Guan and colleagues clearly emphasized the problems encountered when using whole virus-derived antigen. They applied whole viral lysates as antigen in WB analysis and observed a marked difference in reactivity to individual CoV proteins. N protein reactivity was detected in all SARS patient sera, indicating a sensitivity of 100%; however, it also gave false positive results in 17–66% of cases depending on the type of negative control subset tested. By contrast, serum from only 78% of SARS patients reacted with the S protein, but with no false positives, indicating that S protein detection by WB has a higher specificity. Interestingly, when, in one study, a recombinant glutathione S transferase (GST)-tagged N protein lacking a highly conserved part was used as the target there were no false positives. This clearly demonstrates that serological assays applying SARS-CoV-derived antigens should be interpreted with caution.
4. Recombinant protein-based assays
Serological assays that rely on recombinant proteins as antigen are widely used in laboratory diagnostics (Ecker et al., 1996, Farber et al., 1993). As a prerequisite, single immunogenic viral genes have to be cloned into prokaryotic or eukaryotic expression plasmids to produce proteins in bacterial or mammalian cell cultures. They have a major advantage as they do not require BSL3 containment and are appropriate for assay standardization if protocols are closely followed. In SARS-CoV, the N and S are the major immunogenic proteins (Qiu et al., 2005). Recombinant versions of both proteins have been used in IFAs, ELISAs and WB analysis (Table 2 ). Nevertheless, development and validation of assays using recombinant proteins may take longer and is also technically demanding as the necessary molecular cloning, transfection and protein purification techniques must first be optimized.
Table 2.
| Antigen | Assay | SARS sera (≥14 dpoi unless otherwise indicated) | Control sera from respiratory patients or SARS contacts | Control sera from healthy donors | Reference | Comment |
|---|---|---|---|---|---|---|
| No. positive/no. tested (%) | No. positive/no. tested (%) | No. positive/no. tested (%) | ||||
| N | ELISA | 6/6 (100)a | 0/73 (0)a | 0/20 (0)a | Carattoli (2005, #3) | Conflicting information: in the text two control sera mentioned as positive |
| N | ELISA | 56–61/76 (73.6–80.2)a | 1–4/100 (1–4)a | 8–23/1451 (0.6–1.6)a | Guan (2004a, #14) | Values depend on assay settings |
| N partial | ELISA | 74/74 (100)a | n.d. | 1/210 (0.5)a | Guan (2004b, #15) | Highly conserved motif deleted in N protein |
| N partial | ELISA | 41/46 (89.1)a | 2/40 (5)a | 2/35 (5.7)a | Leung (2004, #29) | N-terminal half |
| N | ELISA | 301/327 (92)a | 4/2049 (0.2)a | 3/1700 (0.2)a | Liu (2004, #73) | Antigen capture ELISA |
| N | ELISA | 121/135 (89.6)a, b | 0/64 (0)a | 1/940 (0.1)a | Shi (2003, #44) | Antigen capture ELISA |
| N | ELISA | 100/106 (94.3)a | 33/828 (4.0)a, c | 7/149 (4.7)a | Woo (2004, #87) | 29/33 false positive sera confirmed as false positives |
| N | WB | 34/34 (100)a | n.d. | 2/100 (2)a | Che (2005, #9) | |
| N partial | WB | 40/40 (100)a | 0/100 (0)a | 0/50 (0)a | Guan (2004, #16) | Highly conserved motif deleted |
| N partial | WB | 73/77 (94.8)a, d | 4/134 (3)a | 0/160 (0)a | He (2004a, #18) | 195 C-terminal aa |
| N | WB | 30/30 (100)a | n.d. | 48/48 (100)a | Maache (2006, #33) | |
| N | WB | 74/74 (100)a | n.d. | 0/99 (0)a | Tan (2004, #49) | |
| N | WB | 10/10 (100)a | n.d. | n.d. | Wang et al. (2005, #53) | |
| S partial | IFA | 15/15 (100)a | 0/42 (0)a | 0/100 (0)a | Manopo (2005, #36) | aa 441–700 |
| S | IFA | 74/74 (100)a | n.d. | 0/100 (0)a | Tan (2004, #49) | |
| S partial | ELISA | 6/46 (13)a | 5/40 (12.5)a | 2/38 (5.3)a | Leung (2004, #29) | app. 315 C-terminal aa |
| S partial | ELISA | 56/93 (60.2) | n.d. | 2/148 (1.4)a | Woo (2005, #59) | aa 14–667 |
| S1 | WB | 30/30 (100)a | n.d. | 0/48 (0)a | Maache (2006, #33) | aa 14–760 |
| S2 | WB | 26/30 (86.7)a | n.d. | 0/48 (0)a | Maache (2006, #33) | aa 761–1190 |
| S1 subunit | WB | 15/20 (75) | 6/20 (30) | 6/40 (15)a | Wang et al. (2004, #54) | |
| S2 subunit | WB | 17/20 (85) | 3/20 (15) | 0/40 (0)a | Wang et al. (2004, #54) | |
| S1 | WB | 5/10 (50)a | n.d. | n.d. | Wang et al. (2005, #53) | aa 14–403 |
| S2 | WB | 3/10 (30)a | n.d. | n.d. | Wang et al. (2005, #53) | aa 370–770 |
| S3 | WB | 7/10 (70)a | n.d. | n.d. | Wang et al. (2005, #53) | aa 738–1196 |
dpoi, days post onset of illness; N, SARS-CoV nucleocapsid protein; ELISA, enzyme-linked immunosorbent assay; n.d., not determined, WB, Western blot analysis; S, SARS-CoV spike protein; IFA, immunofluorescence assay; app., approximately.
Calculated from information given in the paper.
10–61 dpoi.
Includes 400 healthy blood donations from March–May 2003 since contact to SARS patients could not be excluded.
4–71 dpoi.
The N protein, as a major immunogenic protein of SARS-CoV (Qiu et al., 2005), was widely used in recombinant serological assays. Its small size and lack of glycosylation sites make it easy to clone and purify in vast quantities from bacteria. Serum from between 73% and 100% of SARS patients reacted with the complete or partial N protein in ELISAs or WB analyses indicating that these assays had high sensitivity (Table 2). No marked difference could be found between studies using a partial or complete N protein as antigen. By contrast, the rate of false positive results differed grossly between the individual studies. Most studies claimed that N protein-based ELISA and WB analysis detected only between 0% and 5.7% false positives in the sera of the negative control groups (Table 2), suggesting a low to moderate specificity. Of note, Woo et al. (2004a) were able to show that 29 of 33 sera that were false positive in an N protein-based ELISA were not reactive in a confirmatory WB assay using a SARS spike polypeptide. Therefore these sera were confirmed to be truly false positives, making this study more reliable than others. Conversely there are several reports showing that cross-reactivity with other human pathogenic CoVs occurs when using N protein-based assays (Che et al., 2005, Maache et al., 2006, Woo et al., 2004c). Of particular note is one study in which the N protein completely failed to differentiate serum from SARS patients and healthy individuals (Maache et al., 2006). These findings further highlight the complexity of serum diagnostics for CoVs and emphasize that a valid diagnosis cannot rely on reactivity to a single antigenic protein.
The other major immunodominant protein of SARS-CoV is the S protein (Qiu et al., 2005) which has been used in IFAs, ELISAs and WB analysis as well as for the generation of pseudotyped particles to develop a biosafe VNT (Fukushi et al., 2008, Hofmann et al., 2004). The majority of neutralizing antibodies were found to be directed against the S protein (Buchholz et al., 2004). For IFA, it was possible to use both full-length S, stably expressed in mammalian cells (Tan et al., 2004), and a fragment (amino acids (aa) 441–700) expressed in insect cells using baculovirus (Manopo et al., 2005). By contrast, because of the technical difficulties in expressing the full-length protein in bacteria, only fragments were used in ELISAs and WB analysis.
In general, antibodies directed against the S protein appeared later in infection than those directed against the N protein (Tan et al., 2004, Woo et al., 2005), indicating that S protein-based assays may be preferable for use with convalescent sera. Overall, reactivity of SARS patient sera with S protein differed grossly between different studies and types of assays, ranging from very low (13%) in one study (Leung et al., 2004) to 100% in several other studies (Table 2). There was a marked difference between detection rates with IFAs (100%), ELISAs (13–96%) and WB analysis (40–100%) (Table 2). The advantage of an IFA can be explained by its use of transfected cells that facilitate correct folding and post-translational modifications (e.g., glycosylation) of the S protein, whereas an ELISA and WB analysis use only denatured linear forms without proper glycosylation. For glycoprotein 120 (gp120) of the human immunodeficiency virus (HIV), it was shown that altered glycosylation patterns influence the host immune response (Grundner et al., 2004). Therefore patients that produce mainly antibodies recognizing conformational epitopes or glycosylation-dependent epitopes of the S protein could be tested as false negatives in an ELISA or WB analysis. That ELISAs and WB analysis are only able to use S protein fragments, as mentioned above, provides a plausible explanation for the variable results found between different assays.
Reactivity of negative control sera to the S protein varied widely between individual studies. Two studies using an IFA to test a sizeable control group of 100 and 142 sera did not detect any false positives (Table 2), which is indicative of the high specificity of the assays. By contrast, ELISAs and WB assays reported between 0% and 30% false positive results, although most studies used control groups of less than 60 sera making them less reliable to estimate the true performance of the assays. Interestingly, the only ELISA-based study that used a rather large control group (148 healthy donors), found only two false positive serum samples (Woo et al., 2005). These results suggest that IFAs using the complete S protein have a specificity advantage over ELISAs or WB assays.
5. Cross-reactivity and cross-neutralization among coronaviruses
Immunogenic proteins of closely related viruses that share common structural or linear epitopes can elicit cross-reactive and cross-neutralizing antibodies in the host. That both occur with CoVs has been recognized for many decades (Bradburne, 1970), and was reflected by the former classification of CoVs into serogroups (McIntosh et al., 1969, Monto, 1974).
As a result assays that used full virus particles or extracts of infected cells as antigen encountered several problems. Two studies reported the appearance of bands at molecular weights of 60 and/or 97 kDa in WB assays using crude viral lysates that could not be attributed to structural proteins (Guan et al., 2004c, Leung et al., 2004). It was speculated that these bands might represent NSPs recognized by SARS patient sera. In fact, it was shown that some NSPs and accessory ORF-encoded proteins can be incorporated into viral particles (Neuman et al., 2008) and are possibly able to elicit an immune response. Indeed, reactive antibodies against SARS-CoV proteins 3a and 9b have been demonstrated in SARS patients by protein micro array (Qiu et al., 2005). Antibodies directed against conserved NSPs or ORF proteins could contribute to false positive results in assays using whole virus as antigen. In addition, conserved structural proteins such as the N protein are produced at high levels in infected cells and viral particles, further increasing the potential for cross-reactivity between different CoVs. The influence of the N protein in serological assays will be outlined in detail below.
Particularly when using denatured antigens in serodiagnostic assays, cross-reactivity is more likely to occur when the applied target proteins are conserved among related viruses. To estimate the rate of conservation of the immunogenic proteins N and S, we analyzed the percentage of aa identity between all HCoV comprising of the genera alpha- and beta-CoV (Table 3 ; first column). Pairwise analysis of complete N sequences revealed that the SARS-CoV N protein had 25–29% aa identity with Alpha-CoV and 33–47% with the more closely related Beta-CoV. By comparison, the complete S protein sequence showed a lower degree of conservation with 23–25% aa identity with Alpha-CoV and around 29% with Beta-CoV. However, as antibodies are produced against the exposed domains of three-dimensional proteins, the overall degree of aa conservation alone cannot predict the occurrence of cross-reactivity. To gain a fuller picture, we therefore analyzed the available information on conserved aa sequences within known immunodominant regions of SARS-CoV N and S protein (Table 3). A study by Chen and colleagues revealed four immunodominant regions in the N protein termed EP1 aa 51–71, EP2 aa 134–208, EP3 aa 249–273 and EP4 aa 349–422 (Chen et al., 2004b). Upon comparison with other human pathogenic Beta-CoVs, one immunodominant region of the N protein showed a low degree of conservation (11–28%) and three regions showed a high degree (40–67%) (Table 3). In addition, Rota et al. found a very conserved motif of eight amino acids present in the N protein of all CoVs, including animal CoVs (Rota et al., 2003).
Table 3.
% AA Pairwise-Identity to SARS-CoV in immunogenic regions.
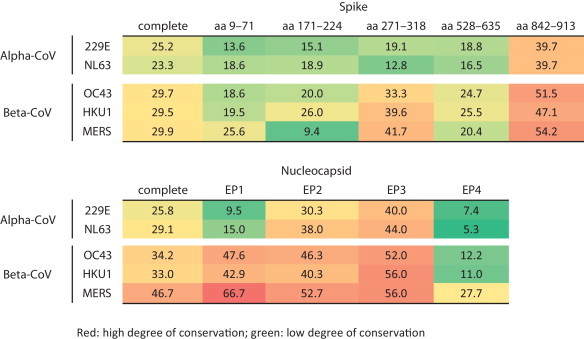
The S protein harbors five different immunogenic sites at aa positions 9–71, 171–224, 271–318, 528–635 and 842–913 (He et al., 2004b). Compared to human pathogenic Beta-CoVs, three immunodominant regions of the S protein show a low degree of conservation (9–26%), one region shows a medium degree (33–42%), and one region shows a high degree (47–55%). Taken together, these findings may explain why N protein-based serological assays are more often associated with cross-reactivity than S protein-based assays.
There are several studies that address the question of whether there is cross-neutralization between related CoVs. Cross-neutralization has been shown between closely related animal CoVs, as in transmissible gastroenteritis virus (TGEV) and feline infectious peritonitis virus type 2 (Horzinek et al., 1982) that have around 95% identity in the C-terminal 1150 aa of the S protein, and also HCoV-OC43 and bovine CoV (BCoV) (Gerna et al., 1981). Interestingly, even HCoV-HKU1, which is closely related to HCoV-OC43, is neither neutralized by HCoV-OC43-reactive sera, despite an aa identity of 64.5% in the S protein, nor by sera reactive with the more distantly related HCoV-229E or SARS-CoV (Chan et al., 2009). Similar results were found for the closely related CoVs HCoV-NL63 and HCoV-229E (Hofmann et al., 2005). To our knowledge, there is only one study describing cross-neutralization between HCoVs, namely of MERS-CoV using SARS-CoV reactive sera. However, it should be noted that neutralizing titers were very low (1:10), indicating that an assay-specific cut-off should be determined with well-defined reference sera to avoid false positive results (Chan et al., 2013). In conclusion, these data clearly show the value of VNTs as a tool to differentiate even between closely related CoVs and reaffirms their status as the gold standard in serological diagnostics.
To provide a valid serological diagnosis of infection with CoVs, it is vital to rule out cross-reactivity and cross-neutralization by using a second confirmatory assay. Widely accepted confirmatory assays like the VNT, which was recommended by WHO during the SARS outbreak (WHO, 2004), are labor-intensive leading to a high workload in diagnostic laboratories and subsequently to slow sample processing. The latter, in particular, is unfavorable in an outbreak situation when rapid identification of infected individuals and their contacts is necessary to contain the infectious agent. Establishing highly specific primary serological screening assays that avoid false positive results, and thus the need for further confirmation, is therefore an important goal.
6. Serological assays for animal screening
Serological assays have been used successfully as a pre-screening tool for virus discovery (Burbelo et al., 2012, Drexler et al., 2013, Muller et al., 2007, Peel et al., 2013). Antibody detection assays are particularly valuable because short phases of virus shedding or poor sample quality decrease the chances of detecting nucleic acids. In addition, they allow the distribution of pathogens in various species and the antigenic relatedness with already known pathogens to be defined (Drexler et al., 2013, Drexler et al., 2012). Importantly, testing of livestock and wild animals can help to trace the source of infection during an outbreak of a zoonotic virus (Li et al., 2005). The identification of the animal reservoir allows the pathogen to be contained by interrupting the chain of transmission; for example, by shutting down animal markets or culling infected animals. This has been a successful approach with outbreaks of avian Influenza and SARS-CoV (Kan et al., 2005, Tam, 2002, Tu et al., 2004, Watts, 2004).
A zoonotic origin of SARS-CoV was suspected early on as the first cases occurred among people handling wild exotic animals in Guangdong Province, China (Zhong et al., 2003). Shortly afterwards Guan and colleagues acquired the first evidence, using RT-PCR and a VNT, that Himalayan palm civets (Paguma larvata) and raccoon dogs (Nyctereutes procyanoides) harbor a SARS-CoV strain that is 99.8% identical to the human SARS-CoV strain (Guan et al., 2003). The lack of viral genetic diversity in positive animals, along with the fact that wild and other farmed palm civets were SARS-CoV seronegative (Tu et al., 2004), indicated that palm civets were not the natural reservoir of SARS-CoV. The search for the source of SARS-CoV continued by screening hundreds of animals from a multitude of species (Poon et al., 2005). A decade later the natural reservoir of SARS-CoV was eventually confirmed to be Rhinolophus sinicus bats in China (Ge et al., 2013). Valuable hints that facilitated the identification of the SARS-CoV reservoir were already provided in late 2005. Two studies found closely related SARS-CoV in Chinese rhinolophid bats in Asia by RT-PCR and serological methods (Lau et al., 2005, Li et al., 2005) while another study found more distantly related CoVs in other bat families (Poon et al., 2005). Li et al. (2005) used two different ELISAs, recombinant SARS-CoV N protein- and whole virus antigen-based, and detected a seroprevalence between 28% and 71% depending on the bat species. The second study by Lau et al. (2005) used an ELISA and WB analysis based on recombinant N protein derived from a bat SARS-like CoV and found a seroprevalence of 84% (ELISA) and 67% (WB) in bats. A marked difference between these two studies was observed in the neutralizing activity of bat sera toward human SARS-CoV. While Lau et al. could detect neutralizing antibodies in some bat sera (42%), Li et al. found no neutralizing activity despite positive serum titers by ELISA of up to 1:6400. Despite these differences, both studies indicated that there was a high level of cross-reactive antibodies when complete virus antigen- and recombinant N protein-based assays were used. VNTs showed a higher specificity, even for these closely related CoVs.
Several follow-up studies in bats were conducted after these findings. The first evidence of CoV in African bats came from the detection of antibodies that reacted against SARS-CoV proteins (Muller et al., 2007). Different serological assays were employed including a SARS-CoV protein-based ELISA, a cIFA using SARS-CoV infected cells and WB assays using either recombinant N or S protein or whole virus antigen (Muller et al., 2007). In this case, VNT revealed that African bats harbored antibodies that cross-reacted with different SARS-CoV antigens but were not able to neutralize SARS-CoV. In subsequent studies, we and others found that both Alpha- and Betacoronaviruses are highly diverse in African bats (Pfefferle et al., 2009, Quan et al., 2010), including close ancestral relatives of HCoV-229E and distant relatives of SARS-CoV. This example shows how the detection of cross-reactive bat serum antibodies against SARS-CoV antigen resulted in the discovery of highly diverse bat CoVs including relatives of SARS-CoV.
In conclusion, most assays performed well with animal sera. However, outcomes have to be evaluated cautiously as testing of animal sera adds another level of complexity. Animals may harbor antibodies against unidentified CoVs that can cross-react with the target antigen especially without knowing the degree of epitope conservation between test antigen and the unidentified CoV. In many cases, secondary detection antibodies are not always available or their performance in serological assays has not been validated. This is particularly true when human serodiagnostic assays are modified for use in animal screening. In the absence of viral nucleic acids, outcomes of serological animal screening should be interpreted cautiously and should only be used as an initial indicator for targeted follow-up studies.
7. Diagnostics for newly emerged MERS-CoV
In June 2012, a previously unknown CoV, now termed MERS-CoV, emerged in the Middle East (Zaki et al., 2012). To date, 180 people have been infected, of whom 77 have died (WHO, 2014), resulting in a case fatality ratio of 43%. So far, the virus is only endemic in the Middle East with only a few cases imported into countries outside this region (Bermingham et al., 2012, Buchholz et al., 2013, Drosten et al., 2013, Guery et al., 2013, HPA, 2013, Puzelli et al., 2013).
Similar to the SARS outbreak, the first diagnostic assays, including PCR and serological, were developed rapidly (Corman et al., 2012a, Corman et al., 2012b). Different serological assays were established including cIFAs using virus-infected cells, IFAs using VERO cells expressing recombinant N or S protein of MERS-CoV, as well as a WB analysis of lysates from cells expressing recombinant N or S protein (Corman et al., 2012b). A cell-free protein microarray was developed that uses the correctly folded and glycosylated S1 fragment of the MERS-CoV S protein as antigen (Reusken et al., 2013a). Finally, MERS-CoV- and S protein-pseudotyped viruses were used in neutralization assays (Buchholz et al., 2013, Drosten et al., 2013, Gierer et al., 2013a, Perera et al., 2013).
A major problem that faced all groups at the time of assay development was the need for a well-characterized serum sample collection to enable thorough validation. This should include defined positive sera for all known human pathogenic CoVs and MERS-CoV convalescent sera along with negative control groups. As only a very small number of positive sera were available at the time of testing, determination of assay sensitivity currently requires more rigorous validation. Assay specificity has been addressed in all studies. Similar to the SARS-CoV serological assays, using whole virus or recombinant N protein gave false positive results for IgM detection (Buchholz et al., 2013). In another study, 356 serum samples including those from 226 slaughterhouse workers in Saudi Arabia were screened using a cIFA followed by a recombinant S protein-based IFA and a VNT for confirmation. The cIFA in this study produced a significant number of false positives that required exclusion by confirmatory neutralization assay (Aburizaiza et al., 2013). Another study conducted in Saudi Arabia made use of MERS-S pseudotyped lentiviruses to test for virus-neutralizing antibodies (Gierer et al., 2013b). HCoV-NL63 S pseudotyped viruses were used as controls. All 268 tested serum samples were negative for MERS-CoV neutralizing antibodies. There was no unspecific cross-neutralization of MERS-S pseudotyped viruses by NL63-positive sera. Another study compared conventional VNT with the S pseudotyped lentivirus-based NT. In total, 1343 human serum samples collected from healthy donors in Egypt and, as controls, in Hong Kong were negative in both NT formats (Perera et al., 2013). Both studies emphasize that NT are less prone to unspecific reactivity and that the use of pseudotyped viruses has the added advantage of not requiring a BSL3 laboratory. Large-scale serological studies of the population in affected countries are urgently needed to further examine spread, prevalence and transmission of MERS-CoV.
Despite several attempts to find the source of the outbreak, the origin of MERS-CoV remains enigmatic. Several studies, based on nucleic acid detection assays, found closely related CoVs in different bat species in Africa, Saudi Arabia and Northern America (Annan et al., 2013, Anthony et al., 2013, Ithete et al., 2013, Memish et al., 2013). In parallel with PCR-based methods we took a serological approach and screened livestock animals from MERS-CoV-linked (Oman) and unlinked regions (Spain, The Netherlands). We showed that all tested dromedary camels from Oman as well as a few camels from the Canary Islands harbored MERS-CoV neutralizing antibodies (Reusken et al., 2013b). As the Canary Islands are not linked to any MERS cases this finding was surprising. Additionally we were able to identify MERS-CoV specific antibodies in camel sera from the United Arab Emirates (UAE) collected more than 10 years ago (Meyer et al., 2014). Importantly, since BCoV (a Beta-CoV of the A lineage) is commonly distributed among livestock, cross-neutralization of MERS-CoV by BCoV antibodies had to be excluded. Recently a study by Haagmans and colleagues could verify MERS-CoV infection in camels on a farm where human cases occurred by using RT-PCR detection methods (Haagmans et al., 2013). Still, since the genetic distance between the human and the camels MERS-CoV from that cluster was very small, it was not possible to differentiate whether camels were infected by humans or vice versa. The value of a MERS-S pseudotyped lentivirus-based NT has been shown in an independent camel study conducted in Egypt, which also found a high percentage of neutralizing antibodies in those animals (Perera et al., 2013). In conclusion, these data highlight the problem of interpreting CoV serological assays, especially if whole virus-derived antigen is used, and further stresses that demonstrating neutralization is indispensable to ultimately confirm seroconversion to a specific CoV.
8. Conclusion
Serological assays have proven valuable for a multitude of investigations, particularly as a complement to nucleic acid detection assays. Serology can help with the identification of the source of infection, the clarification of the epidemiology including transmission pattern analysis, patient contact studies and the identification of asymptomatic cases. All of these tasks can only be achieved with specific and sensitive serological assays. However, for properly validated assays, well-characterized serum samples from patients are necessary and they are not available in an outbreak scenario.
Despite the large number of assays developed in the aftermath of the outbreak of SARS- and MERS-CoV, serological differentiation of HCoVs remains challenging. Overall, it can be concluded that problems with cross-reactivity are more likely to arise with a more conserved antigen, and will often manifest as false positive results. Whether IF, ELISA or WB assays are the preferred screening tool depends on availability of molecular biological techniques, the exact study design as well as the number of samples. Compared to whole virus antigen, assays using recombinant proteins are in general more reliable and can be more easily standardized. Preferably, a recombinant serological assay should use an antigen which is highly immunogenic and provides epitopes specific for its virus species. In addition it is of importance that the protein is expressed in its natural conformation, i.e., displaying proper folding and glycosylation. As discussed above in case of coronaviruses only the S proteins combines all these features. Regardless of assay or antigen used, we recommend a thorough validation using a defined set of sera reactive with other HCoVs as well as a large number of negative serum samples from patients with other respiratory diseases. In any case, great care should be taken in selecting the most appropriate assay and the limitations of each assay should be considered when interpreting results. In our opinion to ultimately confirm serological results it is indispensable to perform a VNT as it is the most specific assay available.
To improve and coordinate the development of serological assays during future outbreaks of novel coronaviruses, a well-characterized subset of sera would be highly recommendable in order to implement a standardized protocol for validating new assays.
Acknowledgments
The work was funded by a FP7-European research project on emerging diseases detection and response, EMPERIE, http://www.emperie.eu/emp/, contract number 223498 and ANTIGONE (contract number 278976). Author C.D. received infrastructural support as the German National Consulting Laboratory for Coronavirus. The Consulting Laboratory is appointed by the Robert Koch-Institut (RKI), Berlin, Germany.
References
- Aburizaiza A.S., Mattes F.M., Azhar E.I., Hassan A.M., Memish Z.A., Muth D., Meyer B., Lattwein E., Muller M.A., Drosten C. Investigation of anti-middle East respiratory syndrome antibodies in blood donors and slaughterhouse workers in Jeddah and Makkah, Saudi Arabia, fall 2012. The Journal of infectious diseases. 2014;209(2):243–246. doi: 10.1093/infdis/jit589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K., Lina P.H., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19(3):456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., Ojeda-Flores R., Rico-Chavez O., Navarrete-Macias I., Zambrana-Torrelio C.M., Rostal M.K., Epstein J.H., Tipps T., Liang E., Sanchez-Leon M., Sotomayor-Bonilla J., Aguirre A.A., Avila-Flores R., Medellin R.A., Goldstein T., Suzan G., Daszak P., Lipkin W.I. Coronaviruses in bats from Mexico. J. Gen. Virol. 2013;94(Pt 5):1028–1038. doi: 10.1099/vir.0.049759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R., Pebody R.G., Green H.K., Boddington N.L., Gopal R., Price N., Newsholme W., Drosten C., Fouchier R.A., Zambon M. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. European Communicable Disease BulletinEuro surveillance: bulletin Europeen sur les maladies transmissibles. 2012;17(40):20290. [PubMed] [Google Scholar]
- Bradburne A.F. Antigenic relationships amongst coronaviruses. Arch. Gesamte Virusforsch. 1970;31(3):352–364. doi: 10.1007/BF01253769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U., Muller M.A., Nitsche A., Sanewski A., Wevering N., Bauer-Balci T., Bonin F., Drosten C., Schweiger B., Wolff T., Muth D., Meyer B., Buda S., Krause G., Schaade L., Haas W. Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October–November 2012. European Communicable Disease BulletinEuro surveillance: bulletin Europeen sur les maladies transmissibles. 2013;18(8) pii: 20406. [PubMed] [Google Scholar]
- Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. U.S.A. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo P.D., Dubovi E.J., Simmonds P., Medina J.L., Henriquez J.A., Mishra N., Wagner J., Tokarz R., Cullen J.M., Iadarola M.J., Rice C.M., Lipkin W.I., Kapoor A. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 2012;86(11):6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Di Bonito P., Grasso F., Giorgi C., Blasi F., Niedrig M., Cassone A. Recombinant protein-based ELISA and immuno-cytochemical assay for the diagnosis of SARS. J. Med. Virol. 2005;76(2):137–142. doi: 10.1002/jmv.20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Yeung M., Xu R.H. SARS: epidemiology. Respirology. 2003;8:S9–S14. doi: 10.1046/j.1440-1843.2003.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.M., Tse H., Wong S.S., Woo P.C., Lau S.K., Chen L., Zheng B.J., Huang J.D., Yuen K.Y. Examination of seroprevalence of coronavirus HKU1 infection with S protein-based ELISA and neutralization assay against viral spike pseudotyped virus. J. Clin. Virol.: Off. Publ. Pan Am. Soc. Clin. Virol. 2009;45(1):54–60. doi: 10.1016/j.jcv.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.H., Chan J.F., Tse H., Chen H., Lau C.C., Cai J.P., Tsang A.K., Xiao X., To K.K., Lau S.K., Woo P.C., Zheng B.J., Wang M., Yuen K.Y. Cross-reactive antibodies in convalescent SARS patients’ sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J. Infect. 2013;67(2):130–140. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.H., Poon L.L., Cheng V.C., Guan Y., Hung I.F., Kong J., Yam L.Y., Seto W.H., Yuen K.Y., Peiris J.S. Detection of SARS coronavirus in patients with suspected SARS. Emerg. Infect. Dis. 2004;10(2):294–299. doi: 10.3201/eid1002.030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.K., Ng K.C., Chan R.C., Lam R.K., Chow V.C., Hui M., Wu A., Lee N., Yap F.H., Cheng F.W., Sung J.J., Tam J.S. Immunofluorescence assay for serologic diagnosis of SARS. Emerg. Infect. Dis. 2004;10(3):530–532. doi: 10.3201/eid1003.030493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X.Y., Qiu L.W., Liao Z.Y., Wang Y.D., Wen K., Pan Y.X., Hao W., Mei Y.B., Cheng V.C., Yuen K.Y. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 2005;191(12):2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhou B., Li M., Liang X., Wang H., Yang G., Le X. Serology of severe acute respiratory syndrome: implications for surveillance and outcome. J. Infect. Dis. 2004;189(7):1158–1163. doi: 10.1086/380397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Pei D., Jiang L., Song Y., Wang J., Wang H., Zhou D., Zhai J., Du Z., Li B., Qiu M., Han Y., Guo Z., Yang R. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Chem. 2004;50(6):988–995. doi: 10.1373/clinchem.2004.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M., Bestebroer T., Muth D., Muller M., Drexler J., Zambon M., Osterhaus A., Fouchier R., Drosten C. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. European Communicable Disease BulletinEuro surveillance: bulletin Europeen sur les maladies transmissibles. 2012;17(39) doi: 10.2807/ese.17.39.20285-en. pii: 20285. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Muller M.A., Costabel U., Timm J., Binger T., Meyer B., Kreher P., Lattwein E., Eschbach-Bludau M., Nitsche A., Bleicker T., Landt O., Schweiger B., Drexler J.F., Osterhaus A.D., Haagmans B.L., Dittmer U., Bonin F., Wolff T., Drosten C. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. European Communicable Disease BulletinEuro surveillance: bulletin Europeen sur les maladies transmissibles. 2012;17(49) doi: 10.2807/ese.17.49.20334-en. pii: 20334. [DOI] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Gaunt E., Rossen J.W., Templeton K.E., Kuijpers T.W., van der Hoek L. The dominance of human coronavirus OC43 and NL63 infections in infants. J. Clin. Virol.: Off. Publ. Pan Am. Soc. Clin. Virol. 2012;53(2):135–139. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Muller M.A., Lukashev A.N., Gmyl A., Coutard B., Adam A., Ritz D., Leijten L.M., van Riel D., Kallies R., Klose S.M., Gloza-Rausch F., Binger T., Annan A., Adu-Sarkodie Y., Oppong S., Bourgarel M., Rupp D., Hoffmann B., Schlegel M., Kummerer B.M., Kruger D.H., Schmidt-Chanasit J., Setien A.A., Cottontail V.M., Hemachudha T., Wacharapluesadee S., Osterrieder K., Bartenschlager R., Matthee S., Beer M., Kuiken T., Reusken C., Leroy E.M., Ulrich R.G., Drosten C. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013;9(6):e1003438. doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Muller M.A., Maganga G.D., Vallo P., Binger T., Gloza-Rausch F., Rasche A., Yordanov S., Seebens A., Oppong S., Sarkodie Y.A., Pongombo C., Lukashev A.N., Schmidt-Chanasit J., Stocker A., Carneiro A.J., Erbar S., Maisner A., Fronhoffs F., Buettner R., Kalko E.K., Kruppa T., Franke C.R., Kallies R., Yandoko E.R., Herrler G., Reusken C., Hassanin A., Kruger D.H., Matthee S., Ulrich R.G., Leroy E.M., Drosten C. Bats host major mammalian paramyxoviruses. Nature Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Drosten C., Seilmaier M., Corman V.M., Hartmann W., Scheible G., Sack S., Guggemos W., Kallies R., Muth D., Junglen S., Muller M.A., Haas W., Guberina H., Rohnisch T., Schmid-Wendtner M., Aldabbagh S., Dittmer U., Gold H., Graf P., Bonin F., Rambaut A., Wendtner C.M. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 2013;13(9):745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker B., Vollenhofer S., Bares T., Schalkhammer T., Schinkinger M., Pittner F. Overexpression and purification of a recombinant chimeric HIV type 2/HIV type 1 envelope peptide and application in an accelerated immunobased HIV type 1/2 antibody detection system (AIBS): a new rapid serological screening assay. AIDS Res. Hum. Retrovir. 1996;12(12):1081–1091. doi: 10.1089/aid.1996.12.1081. [DOI] [PubMed] [Google Scholar]
- Farber I., Wutzler P., Wohlrabe P., Wolf H., Hinderer W., Sonneborn H.H. Serological diagnosis of infectious mononucleosis using three anti-Epstein-Barr virus recombinant ELISAs. J. Virol. Methods. 1993;42(2–3):301–307. doi: 10.1016/0166-0934(93)90041-o. [DOI] [PubMed] [Google Scholar]
- Fukushi S., Watanabe R., Taguchi F. Pseudotyped vesicular stomatitis virus for analysis of virus entry mediated by SARS coronavirus spike proteins. Methods Mol. Biol. 2008;454:331–338. doi: 10.1007/978-1-59745-181-9_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.Y., Wang L.F., Daszak P., Shi Z.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G., Cereda P.M., Revello M.G., Cattaneo E., Battaglia M., Gerna M.T. Antigenic and biological relationships between human coronavirus Oc43 and neonatal calf diarrhea coronavirus. J. Gen. Virol. 1981;54(May):91–102. doi: 10.1099/0022-1317-54-1-91. [DOI] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kramer-Kuhl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von Hahn T., Simmons G., Hofmann H., Pohlmann S. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87(10):5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Hofmann-Winkler H., Albuali W.H., Bertram S., Al-Rubaish A.M., Yousef A.A., Al-Nafaie A.N., Al-Ali A.K., Obeid O.E., Alkharsah K.R., Pohlmann S. Lack of MERS coronavirus neutralizing antibodies in humans, eastern province, Saudi Arabia. Emerg. Infect. Dis. 2013;19(12):2034–2036. doi: 10.3201/eid1912.130701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorse G.J., Patel G.B., Vitale J.N., O’Connor T.Z. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin. Vaccine Immunol. 2010;17(12):1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundner C., Pancera M., Kang J.M., Koch M., Sodroski J., Wyatt R. Factors limiting the immunogenicity of HIV-1 gp120 envelope glycoproteins. Virology. 2004;330(1):233–248. doi: 10.1016/j.virol.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Guan M., Chan K.H., Peiris J.S., Kwan S.W., Lam S.Y., Pang C.M., Chu K.W., Chan K.M., Chen H.Y., Phuah E.B., Wong C.J. Evaluation and validation of an enzyme-linked immunosorbent assay and an immunochromatographic test for serological diagnosis of severe acute respiratory syndrome. Clin. Diagn. Lab. Immunol. 2004;11(4):699–703. doi: 10.1128/CDLI.11.4.699-703.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan M., Chen H.Y., Foo S.Y., Tan Y.J., Goh P.Y., Wee S.H. Recombinant protein-based enzyme-linked immunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diagn. Lab. Immunol. 2004;11(2):287–291. doi: 10.1128/CDLI.11.2.287-291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan M., Chen H.Y., Tan P.H., Shen S., Goh P.Y., Tan Y.J., Pang P.H., Lu Y., Fong P.Y., Chin D. Use of viral lysate antigen combined with recombinant protein in Western immunoblot assay as confirmatory test for serodiagnosis of severe acute respiratory syndrome. Clin. Diagn. Lab. Immunol. 2004;11(6):1148–1153. doi: 10.1128/CDLI.11.6.1148-1153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guery B., Poissy J., el Mansouf L., Sejourne C., Ettahar N., Lemaire X., Vuotto F., Goffard A., Behillil S., Enouf V., Caro V., Mailles A., Che D., Manuguerra J.C., Mathieu D., Fontanet A., van der Werf S. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381(9885):2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Al Dhahiry S.H., Reusken C.B., Raj V.S., Galiano M., Myers R., Godeke G.J., Jonges M., Farag E., Diab A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., Al Romaihi H.E., Al Khal A., Bermingham A., Osterhaus A.D., Alhajri M.M., Koopmans M.P. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2013;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Chong K.H., Chng H.H., Leung B., Ling A.E., Wei T., Chan S.W., Ooi E.E., Kwang J. Development of a Western blot assay for detection of antibodies against coronavirus causing severe acute respiratory syndrome. Clin. Diagn. Lab. Immunol. 2004;11(2):417–422. doi: 10.1128/CDLI.11.2.417-422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Wu H., Luo B., Chen J., Li W., Jiang S. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173(6):4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M., Kuate S., Uberla K., Niedrig M., Pohlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78(12):6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102(22):7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzinek M.C., Lutz H., Pedersen N.C. Antigenic relationships among homologous structural polypeptides of porcine, feline, and canine coronaviruses. Infect. Immun. 1982;37(3):1148–1155. doi: 10.1128/iai.37.3.1148-1155.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hpa H.P.A. Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. European Communicable Disease BulletinEuro surveillance: bulletin Europeen sur les maladies transmissibles. 2013;18(11):20427. doi: 10.2807/ese.18.11.20427-en. [DOI] [PubMed] [Google Scholar]
- Hsueh P.R., Hsiao C.H., Yeh S.H., Wang W.K., Chen P.J., Wang J.T., Chang S.C., Kao C.L., Yang P.C. Microbiologic characteristics, serologic responses, and clinical manifestations in severe acute respiratory syndrome, Taiwan. Emerg. Infect. Dis. 2003;9(9):1163–1167. doi: 10.3201/eid0909.030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs D., Flowers D., Clarke J.R., Valman H.B., MacNaughton M.R. Epidemiology of coronavirus respiratory infections. Arch. Dis. Child. 1983;58(7):500–503. doi: 10.1136/adc.58.7.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithete N.L., Stoffberg S., Corman V.M., Cottontail V.M., Richards L.R., Schoeman M.C., Drosten C., Drexler J.F., Preiser W. Close relative of human middle East respiratory syndrome coronavirus in bat, South Africa. Emerg. Infect. Dis. 2013;19(10):1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Cui B., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Qi X., Chen K., Du L., Gao K., Zhao Y.T., Zou X.Z., Feng Y.J., Gao Y.F., Hai R., Yu D., Guan Y., Xu J. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79(18):11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. New Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Larson H.E., Reed S.E., Tyrrell D.A. Isolation of rhinoviruses and coronaviruses from 38 colds in adults. J. Med. Virol. 1980;5(3):221–229. doi: 10.1002/jmv.1890050306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.T., Tam F.C., Ma C.H., Chan P.K., Cheung J.L., Niu H., Tam J.S., Lim P.L. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190(2):379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Liu X., Shi Y., Li P., Li L., Yi Y., Ma Q., Cao C. Profile of antibodies to the nucleocapsid protein of the severe acute respiratory syndrome (SARS)-associated coronavirus in probable SARS patients. Clin. Diagn. Lab. Immunol. 2004;11(1):227–228. doi: 10.1128/CDLI.11.1.227-228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maache M., Komurian-Pradel F., Rajoharison A., Perret M., Berland J.L., Pouzol S., Bagnaud A., Duverger B., Xu J., Osuna A., Paranhos-Baccala G. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid-based western blot assay were rectified by the use of two subunits (S1 and S2) of spike for detection of antibody to SARS-CoV. Clin. Vaccine Immunol. 2006;13(3):409–414. doi: 10.1128/CVI.13.3.409-414.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manopo I., Lu L., He Q., Chee L.L., Chan S.W., Kwang J. Evaluation of a safe and sensitive Spike protein-based immunofluorescence assay for the detection of antibody responses to SARS-CoV. J. Immunol. Methods. 2005;296(1–2):37–44. doi: 10.1016/j.jim.2004.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K., Kapikian A.Z., Hardison K.A., Hartley J.W., Chanock R.M. Antigenic relationships among the coronaviruses of man and between human and animal coronaviruses. J. Immunol. 1969;102(5):1109–1118. [PubMed] [Google Scholar]
- Memish Z.A., Mishra N., Olival K.J., Fagbo S.F., Kapoor V., Epstein J.H., Alhakeem R., Durosinloun A., Al Asmari M., Islam A., Kapoor A., Briese T., Daszak P., Al Rabeeah A.A., Lipkin W.I. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19(11) doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Muller M.A., Corman V.M., Reusken C.B., Ritz D., Godeke G.J., Lattwein E., Kallies S., Siemens A., van Beek J., Drexler J.F., Muth D., Bosch B.J., Wernery U., Koopmans M.P., Wernery R., Drosten C. Antibodies against MERS Coronavirus in Dromedary Camels. United Arab Emirates, 2003 and 2013. Emerging infectious diseases. 2014;20(4):552–559. doi: 10.3201/eid2004.131746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto A.S. Medical reviews. Coronaviruses. Yale J. Biol. Med. 1974;47(4):234–251. [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., Paweska J.T., Leman P.A., Drosten C., Grywna K., Kemp A., Braack L., Sonnenberg K., Niedrig M., Swanepoel R. Coronavirus antibodies in African bat species. Emerg. Infect. Dis. 2007;13(9):1367–1370. doi: 10.3201/eid1309.070342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K., Travers P., Walport M. 7th ed. Garland Science; New York: 2008. The Distribution and Functions of Immunoglobulin Classes. Janeway's Immunobiology; pp. 400–401. [Google Scholar]
- Narayanan K., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133(1):113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., 3rd, Wuthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82(11):5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedrig M., Leitmeyer K., Lim W., Peiris M., Mackenzie J.S., Zambon M. First external quality assurance of antibody diagnostic for SARS-new coronavirus. J. Clin. Virol.: Off. Publ. Pan Am. Soc. Clin. Virol. 2005;34(1):22–25. doi: 10.1016/j.jcv.2005.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick D.M., Petric M., Skowronski D.M., Guasparini R., Booth T.F., Krajden M., McGeer P., Bastien N., Gustafson L., Dubord J., Macdonald D., David S.T., Srour L.F., Parker R., Andonov A., Isaac-Renton J., Loewen N., McNabb G., McNabb A., Goh S.H., Henwick S., Astell C., Guo J.P., Drebot M., Tellier R., Plummer F., Brunham R.C. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. The Canadian Journal of Infectious Diseases & Medical MicrobiologyJournal canadien des maladies infectieuses et de la microbiologie medicale/AMMI Canada. 2006;17(6):330–336. doi: 10.1155/2006/152612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel A.J., McKinley T.J., Baker K.S., Barr J.A., Crameri G., Hayman D.T., Feng Y.R., Broder C.C., Wang L.F., Cunningham A.A., Wood J.L. Use of cross-reactive serological assays for detecting novel pathogens in wildlife: assessing an appropriate cutoff for henipavirus assays in African bats. J. Virol. Methods. 2013;193(2):295–303. doi: 10.1016/j.jviromet.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R., Wang P., Gomaa M., El-Shesheny R., Kandeil A., Bagato O., Siu L., Shehata M., Kayed A., Moatasim Y., Li M., Poon L., Guan Y., Webby R., Ali M., Peiris J., Kayali G. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. European Communicable Disease BulletinEuro surveillance: bulletin Europeen sur les maladies transmissibles. 2013;18(36) doi: 10.2807/1560-7917.es2013.18.36.20574. pii=20574. [DOI] [PubMed] [Google Scholar]
- Pfefferle S., Oppong S., Drexler J.F., Gloza-Rausch F., Ipsen A., Seebens A., Muller M.A., Annan A., Vallo P., Adu-Sarkodie Y., Kruppa T.F., Drosten C. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 2009;15(9):1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J. Virol. 2005;79(4):2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzelli S., Azzi A., Santini M.G., Di Martino A., Facchini M., Castrucci M.R., Meola M., Arvia R., Corcioli F., Pierucci F., Baretti S., Bartoloni A., Bartolozzi D., de Martino M., Galli L., Pompa M.G., Rezza G., Balocchini E., Donatelli I. Investigation of an imported case of Middle East respiratory syndrome coronavirus (MERS-CoV) infection in Florence, Italy, May to June 2013. European Communicable Disease BulletinEuro surveillance: bulletin Europeen sur les maladies transmissibles. 2013;18(34) doi: 10.2807/1560-7917.es2013.18.34.20564. pii: 20564. [DOI] [PubMed] [Google Scholar]
- Qiu M., Shi Y., Guo Z., Chen Z., He R., Chen R., Zhou D., Dai E., Wang X., Si B., Song Y., Li J., Yang L., Wang J., Wang H., Pang X., Zhai J., Du Z., Liu Y., Zhang Y., Li L., Sun B., Yang R. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect./Institut Pasteur. 2005;7(5–6):882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan P.L., Firth C., Street C., Henriquez J.A., Petrosov A., Tashmukhamedova A., Hutchison S.K., Egholm M., Osinubi M.O., Niezgoda M., Ogunkoya A.B., Briese T., Rupprecht C.E., Lipkin W.I. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. mBio. 2010;1(4) doi: 10.1128/mBio.00208-10. pii: e00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C., Mou H., Godeke G.J., van der Hoek L., Meyer B., Muller M.A., Haagmans B., de Sousa R., Schuurman N., Dittmer U., Rottier P., Osterhaus A., Drosten C., Bosch B.J., Koopmans M. Specific serology for emerging human coronaviruses by protein microarray. European Communicable Disease BulletinEuro surveillance: bulletin Europeen sur les maladies transmissibles. 2013;18(14):20441. doi: 10.2807/1560-7917.es2013.18.14.20441. [DOI] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Vries L.S., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortazar-Schmidt C., Drosten C., Koopmans M.P. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13(10):859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81(2):718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Yi Y., Li P., Kuang T., Li L., Dong M., Ma Q., Cao C. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2003;41(12):5781–5782. doi: 10.1128/JCM.41.12.5781-5782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J.S. Influenza A (H5N1) in Hong Kong: an overview. Vaccine. 2002;20(Suppl. 2):S77L 81. doi: 10.1016/s0264-410x(02)00137-8. [DOI] [PubMed] [Google Scholar]
- Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11(2):362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Crameri G., Kong X., Chen J., Sun Y., Yu M., Xiang H., Xia X., Liu S., Ren T., Yu Y., Eaton B.T., Xuan H., Wang L.F. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 2004;10(12):2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chang Z., Ouyang J., Wei H., Yang R., Chao Y., Qu J., Wang J., Hung T. Profiles of IgG antibodies to nucleocapsid and spike proteins of the SARS-associated coronavirus in SARS patients. DNA Cell Biol. 2005;24(8):521–527. doi: 10.1089/dna.2005.24.521. [DOI] [PubMed] [Google Scholar]
- Wang Y.D., Li Y., Xu G.B., Dong X.Y., Yang X.A., Feng Z.R., Tian C., Chen W.F. Detection of antibodies against SARS-CoV in serum from SARS-infected donors with ELISA and Western blot. Clin. Immunol. 2004;113(2):145–150. doi: 10.1016/j.clim.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. China culls wild animals to prevent new SARS threat. Lancet. 2004;363(9403):134. doi: 10.1016/S0140-6736(03)15313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . vol. 2013. 2004. (WHO Guidelines for the Global Surveillance of Severe Acute Respiratory Syndrome (SARS)). [Google Scholar]
- WHO . 2014. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) – update. [Google Scholar]
- Woo P.C., Lau S.K., Tsoi H.W., Chan K.H., Wong B.H., Che X.Y., Tam V.K., Tam S.C., Cheng V.C., Hung I.F., Wong S.S., Zheng B.J., Guan Y., Yuen K.Y. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363(9412):841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Wong B.H., Chan K.H., Chu C.M., Tsoi H.W., Huang Y., Peiris J.S., Yuen K.Y. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin. Diagn. Lab. Immunol. 2004;11(4):665–668. doi: 10.1128/CDLI.11.4.665-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Wong B.H., Chan K.H., Hui W.T., Kwan G.S., Peiris J.S., Couch R.B., Yuen K.Y. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid enzyme-linked immunosorbent assay due to HCoV-OC43 and HCoV-229E rectified by Western blotting with recombinant SARS-CoV spike polypeptide. J. Clin. Microbiol. 2004;42(12):5885–5888. doi: 10.1128/JCM.42.12.5885-5888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Woo P.C., Lau S.K., Wong B.H., Tsoi H.W., Fung A.M., Kao R.Y., Chan K.H., Peiris J.S., Yuen K.Y. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 2005;43(7):3054–3058. doi: 10.1128/JCM.43.7.3054-3058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.S., Chiu S.C., Tseng T.C., Lin S.F., Lin J.H., Hsu Y.H., Wang M.C., Lin T.L., Yang W.Z., Ferng T.L., Huang K.H., Hsu L.C., Lee L.L., Yang J.Y., Chen H.Y., Su S.P., Yang S.Y., Lin S.Y., Lin T.H., Su I.S. Serologic and molecular biologic methods for SARS-associated coronavirus infection, Taiwan. Emerg. Infect. Dis. 2004;10(2):304–310. doi: 10.3201/eid1002.030731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam W.C., Chan K.H., Poon L.L., Guan Y., Yuen K.Y., Seto W.H., Peiris J.S. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 2003;41(10):4521–4524. doi: 10.1128/JCM.41.10.4521-4524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


