Abstract
The capsid (C) protein of Classical swine fever virus (CSFV) is proposed to play an essential role in the replication and translation of the viral RNA. In this study, a monoclonal antibody (mAb) directed against the C protein was generated with the recombinant C protein expressed in Escherichia coli as immunogen. IFA and IPMA analysis showed that the native C protein of CSFV virions was reactive to the mAb. By truncating the C protein, we identified a linear epitope recognized by the mAb, corresponding to amino acids 61TQDGLYHNKN70 of the CSFV C protein, which is well conserved among pestiviruses. Laser confocal analysis showed that the C protein mainly locates in the cellular nucleoplasm and nucleolus of PK-15 cells. The results have implications for further study of CSFV replication.
Keywords: Classical swine fever virus, Capsid protein, Monoclonal antibody, Epitope, Subcellular localization
1. Introduction
Classical swine fever virus (CSFV) is the causative agent of classical swine fever (CSF), a disease of pigs characterized by fever, leukopenia, hemorrhage, and widespread apoptosis of lymphocytes (Summerfield et al., 1998, Summerfield et al., 2000). The disease causes significant economic loss worldwide and is one of the Office International des Epizooties listed notifiable diseases (Paton and Greiser-Wilke, 2003). CSFV is a small, enveloped virus with a single-stranded, positive-sense RNA genome, which is classified in the genus Pestivirus within the family Flaviviridae, along with bovine viral diarrhea virus 1 (BVDV-1), BVDV-2, and border disease virus (BDV) (Becher et al., 2003). The 12.5-kb CSFV genome contains a single open reading frame that encodes a 3898-amino-acid polyprotein and yields 12 final cleavage products: NH2-Npro-C-Erns-E1-E2-p7-NS2-NS3-NS4ANS4B-NS5A-NS5B-COOH through co- and post-translational processing of the polyprotein by cellular and viral proteases (Meyers et al., 1989, Meyers and Thiel, 1996, Thiel et al., 1991). The virion consists of four structural proteins, the capsid (C) protein and the glycoproteins Erns, E1, and E2 (Thiel et al., 1991).
The C protein of CSFV is located between the N-terminal protease Npro and the glycoprotein Erns in the polyprotein. The C protein is a small protein and rich in basic amino acids (lysine and arginine) (Meyers et al., 1989, Thiel et al., 1991). Mature C protein has 86 amino acids and its apparent molecular weight (MW) is 14 kDa (Heimann et al., 2006). The N terminus (169Ser) of the C protein is cleaved by autocatalysis of the Npro protein (Rumenapf et al., 1998, Stark et al., 1993). The C protein is followed by the Erns protein, of which N terminus (268Asp) is generated by the signal peptidase (SP) (Heimann et al., 2006, Stark et al., 1993). The C protein acts as a transcriptional regulator and plays an important role in the CSFV virion maturation (Liu et al., 1998a). Formation of a C protein–RNA complex inside the virion suggests a protective function of the C protein (Meyers and Thiel, 1996, Thiel et al., 1991). Furthermore, C proteins may function in RNA structural rearrangements taking place during virus replication (Ivanyi-Nagy et al., 2008) and RNA packaging and virion morphogenesis (Murray et al., 2008). To our knowledge, other potential functions of the C protein remain unknown.
The C protein of viruses is important for virus infection and assembly (Matsumoto et al., 1996). Also, the C protein may also play an important role in the pathogenesis of viral infection (Major et al., 1995). To understand the functions of the C protein of CSFV and directly elucidate the mechanisms involved in CSFV replication, the monoclonal antibody (mAb) tool is needed. Availability of specific antibodies against the C protein of CSFV may facilitate further studies on viral replication and biosynthesis. To date, however, there is no report about the mAb against C protein. In this study, we expressed the C protein of CSFV in Escherichia coli and prepared a mAb against the C protein, and identified a linear epitope targeted by the mAb. In addition, the cellular localization of the C protein was analyzed.
2. Materials and methods
2.1. Viruses and cells
The CSFV Shimen strain used in this study was maintained in the Harbin Veterinary Research Institute (Li et al., 2009). PK-15 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, Grand Island, USA) supplemented with 10% fetal bovine serum (FBS) at 37 °C in a 5% CO2 incubator.
2.2. Construction of prokaryotic expression vectors
The 297-bp DNA fragment encoding the C protein of CSFV was amplified with the specific primers F-C and R-C (Table 1 ). PCR was performed by pre-denaturing at 94 °C for 5 min, denaturing at 94 °C for 45 s, annealing at 52 °C for 45 s, and extending at 72 °C for 1.5 min for 30 cycles, with a final elongation step for 10 min at 72 °C. PCR products were cloned into the pMD18-T vector (TaKaRa). The resulting expression plasmid, designated as pMD18-T-C, was then verified by restriction digestion and sequencing. Subsequently, the C gene was subcloned into the prokaryotic expression vector pET-32a (Novagen). The resulting recombinant expression plasmid was designated as pET-CSFV-C.
Table 1.
The primers used in this study.
| Primers | Nucleotide sequences (5′ → 3′)\ | Restriction sites |
|---|---|---|
| F-C | CTGAATTCATGTCTGATGATGGCGCAAGTGG | EcoRI |
| R-C | CGCTCGAGCTAGGCTTCAACTGGTTGATACA | XhoI |
| F-GFP-C | CTCTCGAGATGTCTGATGATGGCGCAAGTGG | XhoI |
| R-GFP-C | CGGGATCCCTAGGCTTCAACTGGTTGATACA | BamHI |
| F-91 | TACTCGAGGACAGCAGAACTAAGCCACCCG | XhoI |
| R-120 | CTGGATCCCGTTACGTAGCGTCGGGTGGCTTAG | BamHI |
| F-181 | TACTCGAGACCCAAGACGGCCTGTACCAC | XhoI |
| R-210 | CAGGATCCCGTTAATTCTTGTTGTGGTACAGG | BamHI |
2.3. Expression and purification of the recombinant protein
The E. coli BL21 (DE3) cells containing pET-CSFV-C was propagated at 37 °C for 4 h under induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for the expression of the His-C protein. The His-C protein was purified by Ni-NTA affinity chromatograph column (Novagen, Madison, WI, USA) according to the manufacturer's protocol. The purified protein was detected by Western blot using mouse anti-His antibody (Santa Cruz). The pET-32a plasmid encodes downstream the T7 promotor 105-aa thioredoxin and various additional sequences that account for the higher MW of the expression product (MW of the tag is about 20 kDa). Total protein concentration was determined by the Bradford method (Bradford, 1976) using bovine serum albumin (BSA) as a reference.
2.4. Production and characterization of the mAb against the C protein
Three 8-week-old SPF BALB/c mice were immunized intraperitoneally (i.p.) and subcutaneously (s.c.) with the entire fusion protein (His-tagged C protein) (50 μg per mouse) emulsified in complete Freund's adjuvant (Sigma–Aldrich, St. Louis, MO). The mice were boosted with the protein (50 μg per mouse) emulsified in incomplete Freund's adjuvant (Sigma–Aldrich) two weeks after the first immunization. Two more weeks later, the mice were injected i.p. with 100 μg of the purified His-C. Three days after the final immunization, the spleen cells were harvested to prepare cell suspension. The spleen cells were fused with SP2/0 cells as described essentially by Kohler and Milstein (1975). The hybridoma cells were cultured in RPMI-1640 containing HAT and HT for 10–14 days. Further selection and characterization were carried out by the indirect ELISA based on the His-C protein as described previously (Peng et al., 2008). After three times of limiting dilution, the hybridoma cells of interest were propagated, suspended in serum-free medium, and then inoculated into the pristine-primed mice, from which ascetic fluids containing the mAb 3E8 (see below) against the C protein was collected 7–10 days post-inoculation (dpi). Reactivity and titer of the mAb were tested by Western blot and ELISA using the His-C protein. The IgG subtype analysis of the mAb was performed using a SBA Clonotyping System/horseradish peroxidase (HRP) (Southern Biotechnology Associates, Inc., Birmingham, AL, USA).
2.5. Western blot
The purified His-C protein was subjected to 12% SDS-PAGE gels and then transferred to 0.22 μm nitrocellulose membranes (Hybond-C Extra, Amersham Biosciences). After blocking, the membranes were incubated with the mAb 3E8 at 37 °C for 60 min. After washing three times with PBS containing 0.5% Tween-20 (PBST), the membranes were inoculated with HRP-conjugated goat anti-mouse IgG (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) at 37 °C for 60 min and visualized using DAB substrate (Zhongshan Goldenbridge, Beijing, China).
2.6. Construction of eukaryotic expression vectors
To map the epitope on the CSFV C protein, four eukaryotic expression vectors containing overlapped truncated C protein fragments as well as the full-length C gene were constructed. Briefly, the full-length C gene and four partial C genes, corresponding to amino acids (aa) 1–99 (nt 1–297), aa 1–40 (nt 1–120), aa 31–70 (nt 91–210), aa 61–99 (nt 181–297), and aa 61–70 (nt 181–210) of the CSFV C protein, were amplified with a panel of primers containing XhoI and BamHI sites (Table 1) as described above. The purified PCR products were digested with XhoI and BamHI and inserted into the eukaryotic expression vector pEGFP-N1 (Clontech, Mountain View, CA, USA), in which EGFP is a C-terminal fusion. So, five recombinant plasmids pEGFP-C1–99, pEGFP-C1–40, pEGFP-C31–70, pEGFP-C61–99, and pEGFP-C61–70 were constructed. The recombinant plasmids were identified by DNA sequencing.
2.7. Transfection
The recombinant plasmids pEGFP-C1–99, pEGFP-C1–40, pEGFP-C31–70, pEGFP-C61–99, pEGFP-C61–70 and pEGFP-N1 were purified using a high-purity plasmid purification kit (Qiagen) in accordance with the manufacturer's instructions. The transfection was performed using Lipofectamine™ 2000 reagent (Invitrogen) in 96-well plates according to the instruction manual as described previously (Zhang et al., 2009). Briefly, PK-15 cells of 95% confluence were transfected with 0.2 μg of plasmids and 0.6 mL of Lipofectamine™ 2000 reagent diluted in 25 mL of DMEM without serum and antibiotics.
2.8. Immunoperoxidase monolayer assay (IPMA)
PK-15 cells, transfected with pEGFP-C1–99, pEGFP-C1–40, pEGFP-C31–70, pEGFP-C61–99, pEGFP-C61–70 or pEGFP-N1, were fixed with cold acetone–methanol (1:1) for 20 min at −20 °C, and then allowed to air dry. After blocking with 5% skimmed milk, the fixed cells were incubated with the mAb 3E8 for 60 min at 37 °C in a humidified chamber. After washing three times with PBST, the fixed cells were incubated with HRP-labeled goat anti-mouse IgG (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD). The cells were visualized using DAB substrate (Zhongshan Goldenbridge, Beijing, China) and examined using microscope.
2.9. Immunofluorescence assay (IFA)
PK-15 cells infected with 100 TCID50 CSFV Shimen strain were cultured for 48 h. The cells were digested with trypsin and fixed as mentioned above. After blocking with 5% skimmed milk, the fixed cells were incubated with the mAb 3E8 and rabbit anti-E2 sera (Sun et al., 2010) for 90 min at 37 °C in a humidified chamber. After washing three times with PBST, the fixed cells were incubated with FITC-labeled goat anti-mouse IgG (Kirkegaard & Perry, Gaithersburg, MD, USA) and TRITC-conjugated goat anti-rabbit IgG (Sigma, Saint Louis, MO, USA). The cells were washed three times with PBST and examined using fluorescence microscope.
3. Results
3.1. Identification of the recombinant C protein
After induction with 1 mM IPTG, the recombinant protein His-C, with the MW of 34 kDa, was expressed in pET-CSFV-C-transformed cells. The purified His-C protein could be recognized by anti-His mAb in Western blot (Fig. 1 ).
Fig. 1.
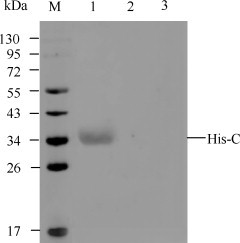
Western blot analysis of the purified His-C protein. The purified His-C recombinant protein (lane 1) was recognized by anti-His mAb (Novagen). Lysates of E. coli BL21 (DE3) containing pET-32a (lane 2) and lysates of E. coli BL21 (DE3) (lane 3) were used as controls. Lane M, PageRuler™ Prestained Protein Ladder (Fermentas, Hanover, MD, Catalog SM0671).
3.2. Characterization of the mAb against the C protein
A mouse mAb designated 3E8 against the C protein were produced using the purified C protein. The mAb 3E8 was identified to belong to IgG1 isotype with a κ light chain. The ascite titer of the mAb 3E8 was up to 1:512,000. The mAb specifically recognized the CSFV-infected PK-15 cells and CSFV virions, but not mock-treated PK-15 cells, in Western blot assay (Fig. 2 ).
Fig. 2.
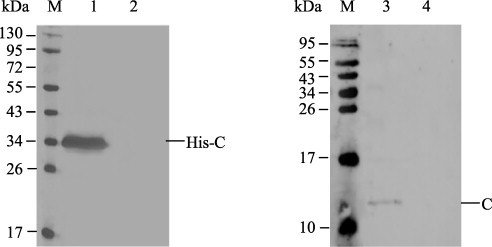
Reactivity of the mAb 3E8 with the CSFV C protein. Lane 1, cell lysates of E. coli BL21 (DE3) harboring pET-CSFV-C; lane 2, cell lysates of E. coli BL21 (DE3) harboring pET-32a; lane 3, CSFV virions purified from CSFV-infected PK-15 cells; lane 4, cell lysates of uninfected PK-15 cells; lane M, protein marker.
3.3. Reactivity of the mAb 3E8 with the C protein in CSFV-infected cells
IFA was used to verify the reactivity of the mAb 3E8 with the C protein in CSFV-infected and uninfected PK-15 cells. As shown in Fig. 3 , the mAb 3E8 showed reactivity with the PK-15 cells infected with CSFV, but not with the uninfected PK-15 cells.
Fig. 3.
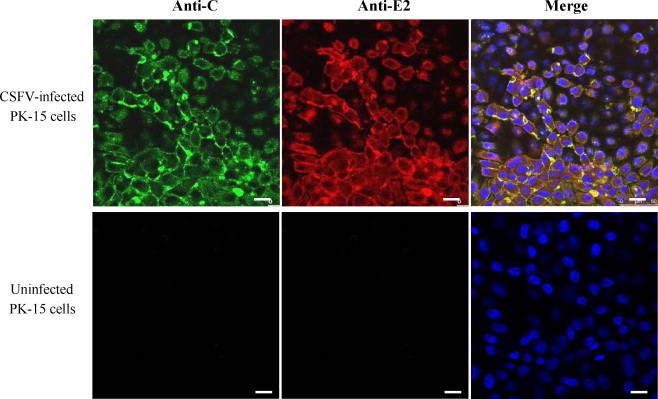
Binding of the mAb 3E8 to the C protein of CSFV in IFA. CSFV-infected PK-15 cells were tested using 3E8 and rabbit anti-E2 sera. Uninfected PK-15 cells were used as control. The nucleus was labeled with the DAPI dye. Bar = 10 μm.
3.4. Epitope mapping of the mAb 3E8
To identify the epitope of the mAb 3E8, the truncated C genes were expressed in transfected PK-15 cells (Fig. 4A). The IPMA showed that the mAb 3E8 was reactive with the complete C protein expressed in PK-15 cells, and the mAb 3E8 was able to recognize aa 31–70 and aa 61–99, but not aa 1–40 (Fig. 4B). Subsequently, the fragment encoding aa 61–70 of the C protein was cloned and expressed in PK-15 cells, and was shown to be reactive with the mAb 3E8 (Fig. 4B). This indicates that the epitope recognized by the mAb 3E8 was 61TQDGLYHNKN70 on the CSFV C protein. This epitope is well conserved among pestiviruses, as shown by the sequence alignment of 35 different pesitivirus species and genotypes (Fig. 5 ).
Fig. 4.
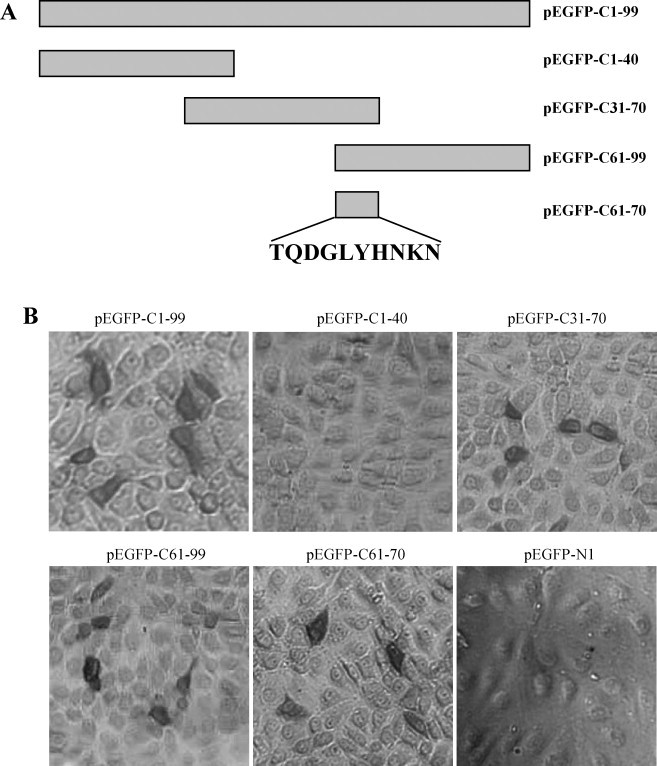
Epitope mapping of the mAb 3E8. (A) The schematic representations of the constructs used to map the epitope recognized by 3E8. (B) Reactivity of the mAb 3E8 with truncated C proteins in IPMA. PK-15 cells transfected with pEGFP-C1–99, pEGFP-C1–40, pEGFP-C31–70, pEGFP-C61–99, pEGFP-C61–70 or pEGFP-N1 were tested using the mAb 3E8 against the C protein of CSFV (200×).
Fig. 5.
Alignment of the sequences around the epitope 61TQDGLYHNKN70 among pestiviruses. A total of 35 pestiviruses from GenBank database are included. GenBank accession numbers are shown in parentheses. Dots indicate identical residues. The epitope is boxed.
3.5. Subcellular localization of the C protein of CSFV
The kinetic subcellular localization of the C protein in PK-15 cells transfected with the eukaryotic expression vector containing the C gene of CSFV was investigated. As shown in Fig. 6 , the C protein of CSFV was distributed in the nucleolus and cytoplasm in PK-15 cells. A pat 7 pattern nuclear localization signal (NLS) (PESRKKL) on the CSFV C protein was predicted by PSORT II (Horton and Nakai, 1997).
Fig. 6.
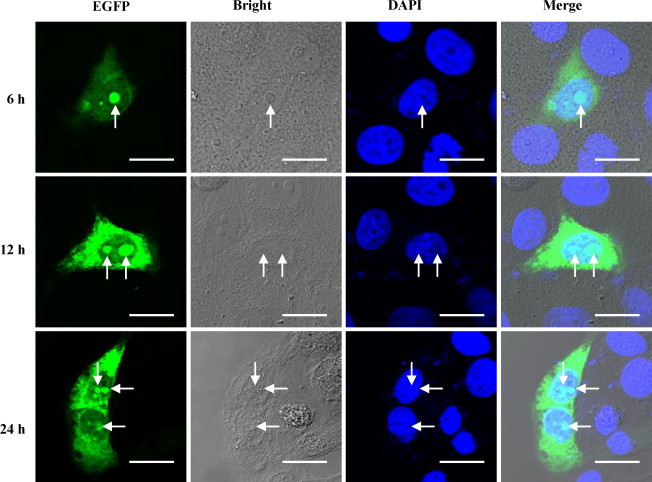
Subcellular localization of the CSFV C protein. The PK-15 cells transfected with the pEGFP-C1–99 containing the C gene of CSFV. The nucleus was labeled with the DAPI dye. Bar = 10 μm.
4. Discussion
There is very limited knowledge about the structure and function of the CSFV C protein because of the lack of tools such as mAb against the C protein. In this study, we generated the mAb 3E8 recognizing the native CSFV C protein, using the recombinant protein His-C expressed in E. coli as immunogens. Consequently, the mAb 3E8 provides a tool to analyze the function of the C protein in CSFV infection. Then, using the truncated C proteins, an epitope recognized by the mAb 3E8 corresponding to aa 61–70 (TQDGLYHNKN) on the CSFV C protein was identified (Fig. 4). To our knowledge, this epitope is the first one identified on the C protein of pestiviruses. Fine mapping of the epitope needs to be done to determine the minimum motif and the contribution of each residue.
The C protein of many viruses is an important target of humoral and cellular immune responses to virus infection, and the primary function of viral core proteins is to provide a protective shell for the viral genome inside the virion (Matsumoto et al., 1996). In the case of CSFV, the C protein of CSFV was suggested to represent an important target for the host's immune response (Liu et al., 1998b). Using the expressed and purified the CSFV C protein, we prepared an mAb against the C protein, which will be beneficial to uncover the function of the C protein in CSFV infection and replication.
Up to now, there is no report about the distribution of the C protein of CSFV in host cells. In this study, using the recombinant expression vector pEGFP-C1–99, the distribution of the C protein was investigated. We demonstrated that the C protein localized in the cytoplasm and nucleolus of transfected PK-15 cells (Fig. 6). This subcellular distribution of the C protein of CSFV is reasonable, for that as a structural protein inside the virion, the major population of the C protein should reside in the cytoplasm. On the other hand, in order to carry out the transcriptional regulation, the C protein may enter into the nucleus including the nucleolus, just as the localization of the nucleoprotein of avian coronavirus and bovine herpesvirus-1 infected cell protein 27 (BICP27) (Chen et al., 2002, Guo et al., 2009). Meanwhile, we detected the nucleolar localization of the C protein of CSFV using 3E8 in CSFV-infected PK-15 cells. However, no nucleolar localization of the C protein was observed in CSFV-infected cells. Possible explanations may be that successful detection of nucleolar proteins using antibodies can be related to the concentration of the protein within the nucleolus and the nucleolus is not always refractive to antibody staining (Sheval et al., 2005). In addition, charged proteins can also migrate through cells post-fixation and become localized to the nucleus (Lundberg and Johansson, 2001, Lundberg and Johansson, 2002). In a previous report, no nucleolar localization of SARS-CoV N protein was observed using anti-N antibody in infected cells (You et al., 2005).
The mechanism how the C protein of CSFV localizes to the nucleolus has not been determined in this study. However, nuclear pore complexes allow the passive transport in both directions between the cytoplasm and nucleoplasm of ions, small molecules, and proteins with MW ranging from 40 to 60 kDa (Peters, 1986). Then the transport of larger proteins through the pore is an active process requiring ATP, and such proteins contain a NLS (Nigg et al., 1991, Richardson et al., 1988). The C protein of CSFV meets both criteria in that the C protein is less than 40 kDa in size and is predicated to contain a NLS (PESRKKL). Therefore, the C protein of CSFV might enter the nucleus via both passive and active pathways.
In summary, we prepared the mAb 3E8 directed against the C protein of CSFV, and identified a linear epitope of C protein of CSFV. In addition, for the first time, we found that the nucleolar localization of the C protein. The results of this study have implications for further analysis of the replication and pathogenicity of CSFV.
Acknowledgements
This study was supported by the National 973 Program (No. 2005CB523202) funded by the Ministry of Science and Technology of China.
References
- Becher P., Avalos Ramirez R., Orlich M., Cedillo Rosales S., Konig M., Schweizer M., Stalder H., Schirrmeier H., Thiel H.J. Genetic and antigenic characterization of novel pestivirus genotypes: implications for classification. Virology. 2003;311:96–104. doi: 10.1016/s0042-6822(03)00192-2. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:154–248. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen H., Wurm T., Britton P., Brooks G., Hiscox J.A. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 2002;76:5233–5250. doi: 10.1128/JVI.76.10.5233-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ding Q., Lin F., Pan W., Lin J., Zheng A.C. Characterization of the nuclear and nucleolar localization signals of bovine herpesvirus-1 infected cell protein 27. Virus Res. 2009;145:312–320. doi: 10.1016/j.virusres.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimann M., Roman-Sosa G., Martoglio B., Thiel H.J., Rumenapf T. Core protein of pestiviruses is processed at the C terminus by signal peptide peptidase. J. Virol. 2006;80:1915–1921. doi: 10.1128/JVI.80.4.1915-1921.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Lavergne J., Gabus C., Ficheux D., Darlix J. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008;36:712–715. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Li M., Wang Y.F., Wang Y., Gao H., Li N., Sun Y., Liang B.B., Qiu H.J. Immune responses induced by a BacMam virus expressing the E2 protein of classical swine fever virus in mice. Immunol. Lett. 2009;125:145–150. doi: 10.1016/j.imlet.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Liu J.J., Wong M.L., Chang T.J. The recombinant nucleocapsid protein of classical swine fever virus can act as a transcriptional regulator. Virus Res. 1998;53:75–80. doi: 10.1016/s0168-1702(97)00132-9. [DOI] [PubMed] [Google Scholar]
- Liu J.J., Wong M.L., Chen P.F., Chang T.J. Cloning, expression and sequence analysis of the classical swine fever virus nucleocapsid protein. Virus Genes. 1998;16:225–234. doi: 10.1023/a:1007976208935. [DOI] [PubMed] [Google Scholar]
- Lundberg M., Johansson M. Is VP22 nuclear homing an artifact? Nat. Biotechnol. 2001;19:713. doi: 10.1038/90741. [DOI] [PubMed] [Google Scholar]
- Lundberg M., Johansson M. Positively charged DNA binding proteins cause apparent cell membrane translocation. Biochem. Biophys. Res. Commun. 2002;291:367–371. doi: 10.1006/bbrc.2002.6450. [DOI] [PubMed] [Google Scholar]
- Major M.E., Vitvitski L., Mink M.A., Schleef M., Whalen R.G., Trepo C., Inchauspe G. DNA-based immunization with chimeric vectors for the induction of immune responses against the hepatitis C virus nucleocapsid. J. Virol. 1995;69:5798–5805. doi: 10.1128/jvi.69.9.5798-5805.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M., Hwang S.B., Jeng K.S., Zhu N., Lai M.M. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology. 1996;218:43–51. doi: 10.1006/viro.1996.0164. [DOI] [PubMed] [Google Scholar]
- Meyers G., Rumenapf T., Thiel H.J. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology. 1989;171:555–567. doi: 10.1016/0042-6822(89)90625-9. [DOI] [PubMed] [Google Scholar]
- Meyers G., Thiel H.J. Molecular characterization of pestiviruses. Adv. Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- Murray C.L., Marcotrigiano J., Rice C.M. Bovine viral diarrhea virus core is an intrinsically disordered protein that binds RNA. J. Virol. 2008;82:1294–1304. doi: 10.1128/JVI.01815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., Baeuerle P.A., Luhrmann R. Nuclear import–export: in search of signals and mechanisms. Cell. 1991;66:15–22. doi: 10.1016/0092-8674(91)90135-l. [DOI] [PubMed] [Google Scholar]
- Paton D.J., Greiser-Wilke I. Classical swine fever—an update. Res. Vet. Sci. 2003;75:169–178. doi: 10.1016/s0034-5288(03)00076-6. [DOI] [PubMed] [Google Scholar]
- Peng W.P., Hou Q., Xia Z.H., Chen D., Li N., Sun Y., Qiu H.J. Identification of a conserved linear B-cell epitope at the N-terminus of the E2 glycoprotein of classical swine fever virus by phage-displayed random peptide library. Virus Res. 2008;135:267–272. doi: 10.1016/j.virusres.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Peters R. Fluorescence microphotolysis to measure nucleocytoplasmic transport and intracellular mobility. Biochim. Biophys. Acta. 1986;864:305–359. doi: 10.1016/0304-4157(86)90003-1. [DOI] [PubMed] [Google Scholar]
- Richardson W.D., Mills A.D., Dilworth S.M., Laskey R.A., Dingwall C. Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell. 1988;52:655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- Rumenapf T., Stark R., Heimann M., Thiel H.J. N-terminal protease of pestiviruses: identification of putative catalytic residues by site-directed mutagenesis. J. Virol. 1998;72:2544–2547. doi: 10.1128/jvi.72.3.2544-2547.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheval E.V., Polzikov M.A., Olson M.O., Zatsepina O.V. A higher concentration of an antigen within the nucleolus may prevent its proper recognition by specific antibodies. Eur. J. Histochem. 2005;49:117–123. [PubMed] [Google Scholar]
- Stark R., Meyers G., Rumenapf T., Thiel H.J. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J. Virol. 1993;67:7088–7095. doi: 10.1128/jvi.67.12.7088-7095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield A., Knoetig S.M., Tschudin R., McCullough K.C. Pathogenesis of granulocytopenia and bone marrow atrophy during classical swine fever involves apoptosis and necrosis of uninfected cells. Virology. 2000;272:50–60. doi: 10.1006/viro.2000.0361. [DOI] [PubMed] [Google Scholar]
- Summerfield A., Knotig S.M., McCullough K.C. Lymphocyte apoptosis during classical swine fever: implication of activation-induced cell death. J. Virol. 1998;72:1853–1861. doi: 10.1128/jvi.72.3.1853-1861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Liu D.F., Wang Y.F., Liang B.B., Cheng D., Li N., Qi Q.F., Zhu Q.H., Qiu H.J. Generation and efficacy evaluation of a recombinant adenovirus expressing the E2 protein of classical swine fever virus. Res. Vet. Sci. 2010;88:77–82. doi: 10.1016/j.rvsc.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Thiel H.J., Stark R., Weiland E., Rumenapf T., Meyers G. Hog cholera virus: molecular composition of virions from a pestivirus. J. Virol. 1991;65:4705–4712. doi: 10.1128/jvi.65.9.4705-4712.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J., Dove B.K., Enjuanes L., Dediego M.L., Alvarez E., Howell G., Heinen P., Zambon M., Hiscox J.A. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Gen. Virol. 2005;86:3303–3310. doi: 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- Zhang X., Ma G., Li Y., Jiang X., He J., Zhou J. Characterization of monoclonal antibody against replication-associated protein of porcine circovirus. DNA Cell Biol. 2009;28:23–29. doi: 10.1089/dna.2008.0800. [DOI] [PubMed] [Google Scholar]


