Highlights
► We designed six small interfering (si)RNAs that target the RABV nucleoprotein gene. ► SiN796, siN580 and siN799 significantly inhibited CVS-11 replication in vitro. ► In vitro analysis indicated that the antiviral activity lasted for four days. ► All of the three could partially protect mice from a lethal dose of CVS-11 virus. ► Liposome facilitates optimal cellular uptake of transfected genetic-based reagents.
Keywords: Small interfering RNA, Rabies virus, Nucleoprotein gene
Abstract
Rabies virus (RABV) infection continues to be a global threat to human and animal health, yet no curative therapy has been developed. RNA interference (RNAi) therapy, which silences expression of specific target genes, represents a promising approach for treating viral infections in mammalian hosts. We designed six small interfering (si)RNAs (N473, N580, N783, N796, N799 and N1227) that target the conserved region of the RABV challenge virus standard (CVS)-11 strain nucleoprotein (N) gene. Using a plasmid-based transient expression model, we demonstrated that N796, N580 and N799 were capable of significantly inhibiting viral replication in vitro and in vivo. These three siRNAs effectively suppressed RABV expression in infected baby hamster kidney-21 (BHK-21) cells, as evidenced by direct immunofluorescence assay, viral titer measurements, real-time PCR, and Western blotting. In addition, liposome-mediated siRNA expression plasmid delivery to RABV-infected mice significantly increased survival, compared to a non-liposome-mediated delivery method. Collectively, our results showed that the three siRNAs, N796, N580 and N799, targeting the N gene could potently inhibit RABV CVS-11 reproduction. These siRNAs have the potential to be developed into new and effective prophylactic anti-RABV drugs.
1. Introduction
Infection with the rabies virus (RABV) manifests as a neurodegenerative disease known as rabies, which is fatal in the clinical stage. Affecting both animals and humans, rabies cases are reported worldwide but are most prevalent in undeveloped countries, specifically in rural areas of Africa and Asia. No definitive treatment exists to cure the disease once the clinical stage has been reached, and only a small portion of clinical cases responds to intensive therapeutic management and survives (Willoughby et al., 2005). Thus, the current strategy to control rabies is pre-exposure prophylaxis by vaccine injection, but research efforts are focused on developing an effective antiviral strategy against established RABV infection.
RNA interference (RNAi) is considered one of the most promising methods to achieve antiviral defense against RABV. The RNAi therapeutic approach is based on the ability of double-stranded small interfering (si)RNAs to specifically trigger mRNA degradation without causing significant cytotoxicity. siRNA-mediated gene silencing occurs through the activities of the RNA-induced silencing complex (RISC), a ubiquitous cytoplasmic protein complex that harbors dsRNA-binding domains and an exonuclease domain. The RNAi approach has already been successfully applied to several human pathogenic viruses, including hepatitis B virus (HBV) (Morrissey et al., 2005, Song et al., 2003), dengue virus (DNV) (Padwad et al., 2009), human immunodeficiency virus (HIV) (Boden et al., 2004, Kumar et al., 2008), poliovirus (Saleh et al., 2004), hepatitis C virus (Lupberger et al., 2008), human papilloma virus (Niu et al., 2006), respiratory syncytial virus (RSV) (Bitko et al., 2005), severe acute respiratory syndrome (SARS) coronavirus (Li et al., 2005), and influenza virus (Zhang et al., 2009). In addition, several researchers have investigated the potential of RNAi to treat RABV infections. Brandao et al. demonstrated that siRNA designed against the rabies virus nucleoprotein (N) mRNA was able to partially protect cultured cells (baby hamster kidney (BHK)-21) from rabies virus infection, as evidenced by direct fluorescent antibody test (Brandao et al., 2007); however, no other in vitro assays or in vivo data was provided to support the siRNA inhibitory effect. More recently, Gupta et al. and Sonwane et al. reported that adenoviral vector-mediated delivery of small hairpin (sh)RNAs targeting the RABV N or polymerase (L) mRNA led to a slight increase in survival of RABV-infected mice (Gupta et al., 2011, Sonwane et al., 2011).
While viral vectors are undoubtedly efficient tools for gene delivery, their use has been restricted by several inherent features that pose potentially significant safety risks to the recipients. For example, the viral vectors randomly integrate their vector DNA into the host chromosome. In addition, the vector itself may be immunogenic, cytotoxic, or have specific tissue tropism. There is also a possibility of the vector virus recombining with the wild-type viruses, generating a potentially virulent or toxic pathogen. Thus, researchers in gene therapy have turned to non-viral gene delivery systems in the hopes of creating a safe and effective delivery system. To date, the cationic lipids have proven to be one of the most efficient non-viral gene transfer methods. These amphiphilic molecules possess two elements that are crucial for gene delivery: a cationic head group that condenses DNA and a lipid moiety that acts as a fusogenic group to enhance penetration into cells.
In the present study, we aimed to develop a universal siRNA agent that is capable of targeting different RABV strains. The N gene was chosen for this study since it is the most highly conserved among the five RABV genes. We designed six siRNAs that were specific to the conserved region of the RABV nucleoprotein gene. An siRNA expression vector-based transient expression model was used to determine their abilities to inhibit RABV replication in vitro and in vivo.
2. Materials and methods
2.1. Cells, viruses and animals
BHK-21 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 2% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin (Hyclone, Logan, UT, USA) at 37 °C in an atmosphere of 5% CO2. The RABV challenge virus standard (CVS)-11 strain was obtained from the Changchun Institute of Veterinary Science (China). RABV CVS-11 was used at a multiplicity of infection (MOI) of 0.01 for in vitro BHK-21 infection experiments, and at 50% lethal dose (LD50) of 10 for in vivo mice challenge experiments. Seventy-two pathogen-free, female BALB/c mice (weighing 13–15 g; Changchun Institute of Biological Products, China) were used to assess the antiviral activities of the six siRNAs in vivo. All experiments with the RABV virus were conducted in a biosafety level three laboratory facility at the Changchun Institute of Veterinary Science.
2.2. Plasmid construction
The siRNAs targeting conserved sequences of the RABV N gene were designed by Dharmacon's siRNA design algorithm (http://www.dharmacon.com/DesignCenter/DesignCenterPage.aspx). A scrambled siRNA sequence was designed for use as a negative control. The siRNA target sequences, including the control, were verified as specific to RABV, and absent in either the human or Mus musculus genomes, by performing a BLAST search. Then, complementary single-stranded DNA oligos encoding the specific small-hairpin siRNAs (shRNAs) were synthesized, annealed, and cloned into the pSilence2.1-U6 Hygro siRNA expression vector (Ambion, Austin, TX, USA) to generate six ps-shRNA plasmids. The 67 oligonucleotide conserved sequence region encoding the specific shRNAs is shown in Fig. 1 .
Fig. 1.
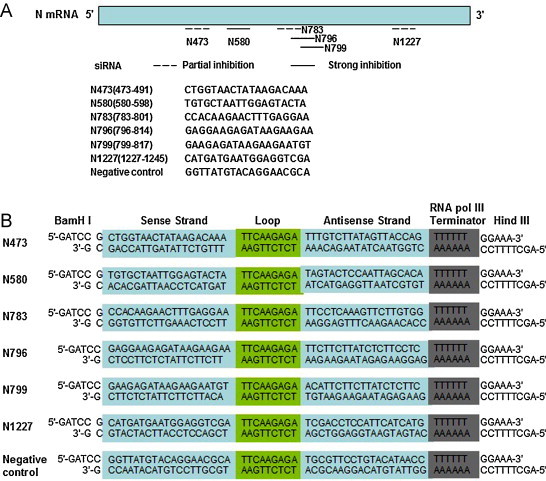
siRNAs were designed to target the conserved sequences of the N gene from the RABV CSV-11 strain. (A) Schematic diagram showing the location of the six siRNAs in the N mRNA and their relative potency in inhibiting rabies virus replication in BKH-21 cells (based on the data presented in Fig. 2, Fig. 3, Fig. 4). (B) Schematic diagram showing the coding sequences of shRNAs. The 5′-ends of the two oligonucleotides are noncomplementary and form the BamHI and HindIII restriction site overhangs that facilitated directional cloning into the pSilence2.1-U6 Hygro siRNA expression vector.
2.3. BHK-21 cell transfection with siRNA expression plasmid and virus infection
BHK-21 cells were plated at 2 × 105 cells/well in 24-well plates and incubated overnight. The various ps-shRNA plasmids were mixed respectively with Lipofectamine 2000 transfection reagent (1:2.5 ratio using 0.8 μg plasmids and 2 μL Lipofectamine 2000; Invitrogen, Carlsbad, CA, USA) in Opti-MEM medium (Invitrogen) and added to the wells. Scrambled siRNA plasmid was used as a negative control. After 6 h of transfection, the transfected BHK-21 cells were infected with 0.01 MOI of the RABV CVS-11 virus for 48 h. The non-transfected infected cells served as viral controls.
2.4. Direct immunofluorescence assay (DFA)
After transfection and 48 h of infection, the supernatants were removed from the BHK-21 cells. The cells were fixed with 80% cold acetone for 1 h and washed three times with phosphate buffered saline (PBS). A fluorescein isothiocyanate (FITC)-conjugated mouse anti-N monoclonal antibody (prepared by the Changchun Institute of Veterinary Science) was added to each well at 1:200 concentration and incubated at 37 °C for 1 h. Immunoreactivity was observed by fluorescence microscopy (BX51FL; Olympus, Japan).
2.5. Viral titer assay
Viral titer was measured using 50% tissue culture infective dose (TCID50) assays. Serial 10-fold dilutions of supernatants from the treated and viral samples were added onto a monolayer of BHK-21 cells in 96-well culture plates and incubated for two days. Virus concentrations were measured by DFA, as described above, and the viral titer for each sample was calculated by the Reed–Muench method (Pandiri et al., 2007).
2.6. Quantitative real-time reverse-transcription-polymerase chain reaction (qRT-PCR)
siRNA-transfected BHK-21 cells were harvested after 24 h of RABV infection and total RNA was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The RNA was reverse transcribed into cDNA using AMV reverse transcriptase (Promega, Madison, WI, USA) and poly-T oligonucleotide primer (5′-TTTTTTTTTTTTTTT-3′; Promega). The levels of N mRNA transcripts were determined by qPCR using the Brilliant II SYBR Green QPCR Master Mix (Stratagene, La Jolla, CA, USA) and the following gene-specific primers: forward 5′-TCAAGAATATGAGGCGGCTG-3′ and reverse 5′-TGGACGGGCTTGATGATTGG-3′ for CVS11-N (109 bp amplicon), and forward 5′-TGACAGGATGCAGAAGGAGA-3′ and reverse 5′-GCTGGAAGGTGGACAGTGAG-3′ for β-actin (86 bp amplicon). The PCR reactions (25 μL; in triplicate) were carried out on the Mx 3000P System (Stratagene) under the following thermal cycling conditions: 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s and 60 °C for 1 min. The β-actin normalized (relative) levels of N mRNA transcripts were calculated by the double standard curve method using the MxPro QPCR software.
2.7. Western blotting
siRNA-transfected BHK-21 cells were harvested after 48 h of RABV infection and total protein samples were obtained by incubating in cell lysis buffer (Beyotime Biotech Inc., Nantong, China). Protein concentration was measured by the bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL, USA). Fifty microgram aliquots of total protein were resolved by sodium SDS-PAGE and transferred to a nitrocellulose membrane (Amersham Biosciences, Uppsala, Sweden). The resultant blots were probed with a mouse monoclonal antibody to RABV N protein (1:200) or a mouse monoclonal antibody to β-actin (1:2000, Santa Cruz Biotechnology, Santa Cruz, CA, USA), followed by the horseradish peroxidase-conjugated goat anti-mouse IgG (1:2000, Santa Cruz Biotechnology, Santa Cruz, CA, USA). Immunoreactive bands were detected with the enhanced chemiluminescent reagent (Pierce) and the band area was quantified using Image J software (US National Institutes of Health).
2.8. Viral challenge in mice
Mice, in groups of nine, were inoculated intracerebrally with the liposome/ps-shRNA (40 μL of DMEM containing 50 μg ps-shRNA and 25 μL of liposome) or ps-shRNA alone (100 μg). After 6 h, the mice were infected with 10 LD50 of RABV CVS-11 at the same intracerebral site of inoculation. The mice were observed for 21 days post-infection for development of rabies-specific symptoms and death.
2.9. Statistical analysis
Intergroup differences in quantitative data were analyzed by t-test and one-way ANOVA. Intergroup comparison of the survival rate was carried out by the log-rank test. All statistical analyses were performed by SPSS ver 13.0 statistical software (SPSS, Inc., Chicago, IL, USA). A probability (p-value) of less than 0.05 was considered statistically significant.
3. Results
3.1. Screening of the antiviral effect of siRNA in BKH-21 cells
To evaluate the antiviral effects of siRNAs targeting the conserved sequences of the CVS11-N gene, the six siRNA expression plasmids (ps-N473, ps-N580, ps-N783, ps-N796, ps-N799 and ps-N1227) and a negative control were respectively transfected into BHK-21 cells. DFA results indicated that the ps-N796, ps-N580 and ps-N799 constructs mediated a significant reduction in the RABV N protein fluorescence signal. In contrast, the other three N gene targeting siRNAs and the negative control had no effect on the fluorescence signals, compared to the non-transfected viral-infected control cells (Fig. 2 ). In addition, the three effective constructs significantly lowered the viral titer and relative mRNA levels in infected cells. Specifically, ps-N796, ps-N580 and ps-N799 transfected RABV-infected cells had TCID50/mL of 6.25, 6.75 and 6.92, respectively (vs. 7.50 for the negative control, p < 0.01) (Fig. 3A). The viral relative mRNA levels in the ps-N796, ps-N580 and ps-N799 groups were 6.74, 5.68 and 8.19, respectively (vs. 100 for the negative control, p < 0.01) (Fig. 3B).
Fig. 2.
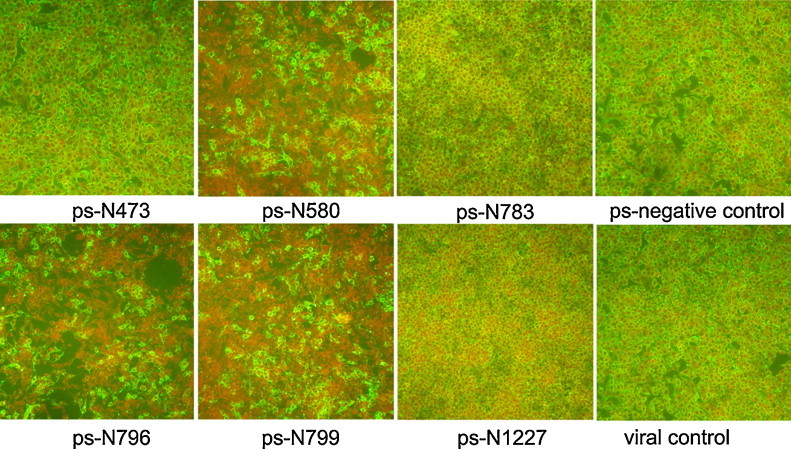
siRNA-mediated inhibition of the CVS-11 rabies virus presence in BKH-21 cells, as detected by DFA. Transfection of the ps-N796, ps-N580 and ps-N799 vectors led to significant reduction in the fluorescence signal (green: CVS-11 N protein), compared to the scrambled siRNA-transfected (ps-negative) control and the untransfected RABV-infected (viral) control.
Fig. 3.
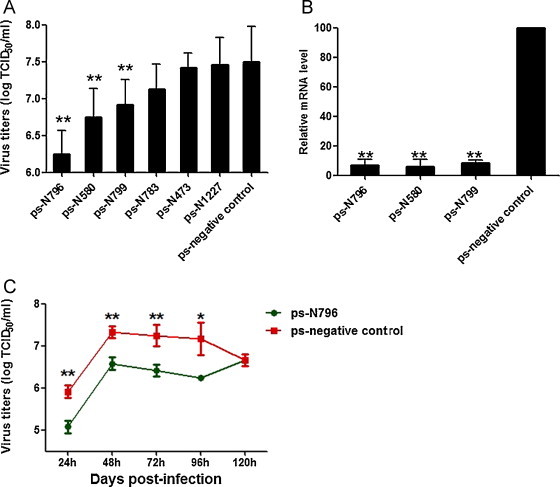
Transfection of ps-shRNA vectors inhibited the replication of CVS-11 rabies virus in BKH-21 cells and the effect lasted several days. (A) Viral titers in culture supernatants were determined by the TCID50 assay. Data are expressed as mean log TCID50/mL ± SD for each group from three separate experiments. **p < 0.01 vs. negative control, one-way ANOVA test. (B) qPCR detection of CSV11-N mRNA transcripts relative to β-actin transcripts. Data are shown as means ± SD from three separate experiments. **p < 0.01 vs. negative control, one-way ANOVA test. (C) Time duration of the ps-N796 inhibitive effect. Data are expressed as mean log TCID50/mL ± SD for each group from three separate experiments. *p < 0.05, **p < 0.01 vs. negative control, one-way ANOVA test.
To determine the duration of the siRNA-mediated antiviral activity, the siRNA with the most robust inhibitive property, ps-N796 was selected for further analysis. After transfection and infection, the cells’ supernatants were harvested for viral titer assays at 24, 48, 72, 96, and 120 h time points. The results showed that the significant inhibitive effect lasted for 96 h. At 120 h, the viral titer had dropped to the same amount as that of the negative control (Fig. 3C).
The N protein expression was analyzed in the transfected and infected cells by Western blotting. The ps-N796, ps-N580 and ps-N799 vectors led to significantly reduced CVS11-N expression, compared to the negative control cells (p < 0.01) (Fig. 4 ).
Fig. 4.
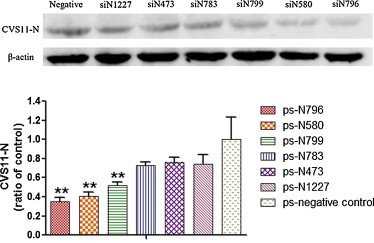
siRNAs targeting the RABV N gene suppressed N protein expression in RABV CSV-11-infected BKH-21 cells, as detected by Western blotting. The immunoreactive N protein bands were quantified using Image J Software and expressed as means ± SD relative to the corresponding β-actin protein. **p < 0.01 vs. negative control.
Collectively, these results demonstrated that the three siRNAs, N796, N580 and N799, could significantly inhibit replication of the CVS-11 virus in BKH-21 cells; however, the other three siRNAs, N783, N473 and N1227, had no significant inhibitive properties and were excluded from further analysis.
3.2. Antiviral activity of the ps-N796, ps-N580 and ps-N799 in RABV-infected mice
To test whether the inhibition of RABV replication observed in vitro for the ps-N796, ps-N580 and ps-N799 vectors was adequate for protection in vivo, we used an established BALB/c mouse model of the rabies virus. The mice were first treated with ps-N796, ps-N580 or ps-N799 using liposome-mediated delivery or direct delivery (no liposome), and challenged with a lethal dose (10 LD50) of RABV CVS-11 6 h later. Compared to untreated (no siRNA) RABV-infected control mice, all of the siRNA-treated mice showed fewer clinical signs of disease and lower morbidity. However, the mice in the siRNA/liposome-treated groups showed different survival rates than the mice treated with siRNA alone (ps-N796: 33.3% vs. 44.4%; ps-N580: 44.4% vs. 33.3%; and ps-N799: 55.6% vs. 33.3%; Fig. 5 ). The mice that survived did not develop symptoms of rabies and remained healthy until the end of the experiment (21 days post-challenge). All untreated control mice died from the RABV infection. These results suggested that all three of the siRNAs targeting the conserved regions of the RABV N gene were able to alleviate RABV-related morbidity and could partially protect mice that had been challenged with a lethal dose of the rabies virus.
Fig. 5.
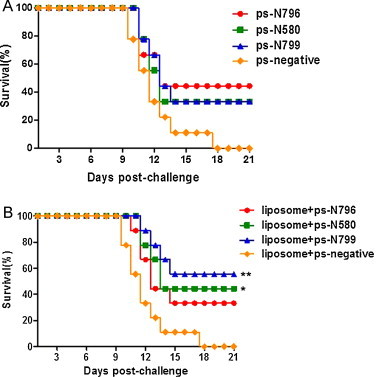
Liposome-mediated delivery of siRNAs increased survival of RABV CSV-11-infected mice. Mice were treated intracerebrally with ps-shRNA (A) or liposome + ps-shRNA (B) and intracerebrally challenged with 10 LD50 of lethal CVS-11 6 h later at the same site. The challenged mice were observed for 21 days, and percent survival was calculated. (A) The survival for groups of mice treated with ps-shRNA did not differ from those treated with a ps-negative control. (B) The survival of the liposome + ps-N799 and liposome + ps-N580 group was significantly different from that of the liposome + ps-negative group (*p < 0.05, **p < 0.01 vs. negative control; log-rank).
4. Discussion
RNA interference has emerged as a useful technique for developing nucleic acid-based gene silencing therapeutics for treatment of viral diseases (Anesti et al., 2008, Habayeb et al., 2009, Kim et al., 2008, Liu et al., 2009, Pacca et al., 2009). RNAi can be achieved by either endogenously encoded small RNAs, known as microRNAs (miRNAs), or exogenously introduced synthetic siRNAs or plasmid/viral vector constructs containing shRNAs. Results from a collection of in vivo studies (McCaffrey et al., 2003) and a phase I clinical trial (DeVincenzo et al., 2010) have provided the proof-of-concept for RNAi as an effective therapeutic agent in humans; however, some safety concerns remain.
Since an effective treatment for clinical rabies has yet to be developed (Hemachudha et al., 2006), RNAi is considered a promising therapeutic strategy. The RABV genome consists of a single negative-stranded RNA molecule of about 12 kb that encodes five viral proteins, in the order of 3′-N-P-M-G-L-5′. RNA transcription and RABV replication requires an intricate interplay between the nucleoprotein N, the RNA-dependent RNA polymerase (RdRp) L, the nonenzymatic polymerase cofactor P, and the RNA genome enwrapped by N, which is also known as the RABV nucleocapsid (N–RNA). The N-RNA complex acts as the template for the viral polymerase. During mRNA synthesis, the P and L complex bind to the N–RNA template through an N–P interaction that involves two adjacent N proteins in the nucleocapsid. The L–P binding to the N–RNA is believed to trigger conformational changes that allow the polymerase to access the RNA (Albertini et al., 2008, Albertini et al., 2011). The N, P and L proteins are thus important factors for both viral transcription and replication; therefore, they are considered as promising candidates for siRNA/miRNA inhibition strategies. In fact, studies that sought to identify genetic suppressor elements that inhibit RABV replication identified the N and P proteins as effective targets (Israsena et al., 2009, Wunner et al., 2004).
In our current study, we attempted to develop a universal siRNA agent that would recognize different RABV strains by using the N gene, which is the most highly conserved gene among the five RABV genes in the different RABV strains. Moreover, we selected the most conserved sequences of the N transcript for use as RNAi targets. Six siRNAs (N473, N580, N783, N796, N799 and N1227) designed, and three (N796, N580 and N799) were capable of significantly inhibiting CVS-11 rabies virus replication. Furthermore, in vitro analysis indicated that the antiviral activity lasted for four days. In vivo, all of the three could partially protect mice from a lethal dose of CVS-11 rabies virus. Interestingly, when ps-N580 and ps-N799 were delivered at lower concentration but together with a liposome reagent, the survival rates significantly increased with both, but to a remarkably high extent with the ps-N799. These results indicated that the cellular uptake of the siRNA therapy was not dependent on dosage alone, but was significantly influenced by the carrier.
There are some discrepancies in the animal experiments that should be noted. N796 worked best as naked RNA, and worse when delivered with liposomes. The protection rates of N580 and N799 increased when delivered with liposomes, which is consistent with our theoretical design. However, the result of N796 was opposite, with N796 working better as naked RNA than with liposomes. We remain perplexed by this finding but speculate that the reason may be due to the distinctive features of in vitro and in vivo environments. Moreover, the mouse grouping was random and this result may reflect individual physical differences in the tested mice. Nonetheless, the mechanism remains unclear and we intend to investigate it in detail in our subsequent studies. The purpose of the in vivo experiments was to verify whether the siRNAs that were selected by in vitro analysis were effective in vivo. The results showed a partial protective effect. However, to achieve complete protection, other siRNA delivery methods (targeted delivery), route (intravenous crossing of the blood–brain barrier), dose (single or multiple siRNAs), and timing of therapeutic administration should be assessed to generate an optimal treatment strategy. Since the siRNAs against RABV N gene sequences were capable of reducing RABV replication and protected mice against rabies, this strategy may represent the foundation for which each of the above-listed parameters may be optimized. Although it is difficult to reach absolute protection, the fact that there is currently no effective way to cure rabies makes these agents promising candidates for adjuvant therapy. For example, in RABV exposure vaccination the body generally needs at least seven days to produce antibodies, and siRNA adjuvant may exert suppressive effects on the virus in the early stages of exposure.
In conclusion, we developed three siRNAs (N796, N580 and N799) that successfully targeted the N gene of RABV. All three effectively inhibited RABV replication and protected mice from challenges with the CVS-11 strain. We believe these siRNAs have the potential to be developed into new and effective prophylactic anti-RABV drugs.
Acknowledgements
This study was supported by grants from the Special Fund for Agro-Scientific Research in the Public Interest: Animal rabies rapid diagnosis and effective vaccine research (201103032) and the Natural Science Foundation for Young Scholars (31101791).
We also thank Medjaden Bioscience Limited for assisting in the preparation of this manuscript.
Contributor Information
Song-Tao Yang, Email: yst10223@yahoo.com.cn.
Xian-Zhu Xia, Email: xiaxianzhu@gmail.com.
References
- Albertini A.A., Ruigrok R.W., Blondel D. Rabies virus transcription and replication. Advances in Virus Research. 2011;79:1–22. doi: 10.1016/B978-0-12-387040-7.00001-9. [DOI] [PubMed] [Google Scholar]
- Albertini A.A., Schoehn G., Weissenhorn W., Ruigrok R.W. Structural aspects of rabies virus replication. Cellular and Molecular Life Sciences. 2008;65(2):282–294. doi: 10.1007/s00018-007-7298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesti A.M., Peeters P.J., Royaux I., Coffin R.S. Efficient delivery of RNA interference to peripheral neurons in vivo using herpes simplex virus. Nucleic Acids Research. 2008;36(14):e86. doi: 10.1093/nar/gkn371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitko V., Musiyenko A., Shulyayeva O., Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nature Medicine. 2005;11(1):50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- Boden D., Pusch O., Silbermann R., Lee F., Tucker L., Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Research. 2004;32(3):1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandao P.E., Castilho J.G., Fahl W., Carnieli P., Jr., Oliveira Rde N., Macedo C.I., Carrieri M.L., Kotait I. Short-interfering RNAs as antivirals against rabies. Brazilian Journal of Infectious Diseases. 2007;11(2):224–225. doi: 10.1590/s1413-86702007000200011. [DOI] [PubMed] [Google Scholar]
- DeVincenzo J., Lambkin-Williams R., Wilkinson T., Cehelsky J., Nochur S., Walsh E., Meyers R., Gollob J., Vaishnaw A. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(19):8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.K., Sonwane A.A., Singh N.K., Meshram C.D., Dahiya S.S., Pawar S.S., Gupta S.P., Chaturvedi V.K., Saini M. Intracerebral delivery of small interfering RNAs (siRNAs) using adenoviral vector protects mice against lethal peripheral rabies challenge. Virus Research. 2011;163(1):11–18. doi: 10.1016/j.virusres.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habayeb M.S., Ekstrom J.O., Hultmark D. Nora virus persistent infections are not affected by the RNAi machinery. PLoS One. 2009;4(5):e5731. doi: 10.1371/journal.pone.0005731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemachudha T., Sunsaneewitayakul B., Desudchit T., Suankratay C., Sittipunt C., Wacharapluesadee S., Khawplod P., Wilde H., Jackson A.C. Failure of therapeutic coma and ketamine for therapy of human rabies. Journal of Neurovirology. 2006;12(5):407–409. doi: 10.1080/13550280600902295. [DOI] [PubMed] [Google Scholar]
- Israsena N., Supavonwong P., Ratanasetyuth N., Khawplod P., Hemachudha T. Inhibition of rabies virus replication by multiple artificial microRNAs. Antiviral Research. 2009;84(1):76–83. doi: 10.1016/j.antiviral.2009.07.012. [DOI] [PubMed] [Google Scholar]
- Kim S.M., Lee K.N., Park J.Y., Ko Y.J., Joo Y.S., Kim H.S., Park J.H. Therapeutic application of RNA interference against foot-and-mouth disease virus in vitro and in vivo. Antiviral Research. 2008;80(2):178–184. doi: 10.1016/j.antiviral.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Kumar P., Ban H.S., Kim S.S., Wu H., Pearson T., Greiner D.L., Laouar A., Yao J., Haridas V., Habiro K., Yang Y.G., Jeong J.H., Lee K.Y., Kim Y.H., Kim S.W., Peipp M., Fey G.H., Manjunath N., Shultz L.D., Lee S.K., Shankar P. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134(4):577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.J., Tang Q., Cheng D., Qin C., Xie F.Y., Wei Q., Xu J., Liu Y., Zheng B.J., Woodle M.C., Zhong N., Lu P.Y. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nature Medicine. 2005;11(9):944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.P., von Eije K.J., Schopman N.C., Westerink J.T., ter Brake O., Haasnoot J., Berkhout B. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Molecular Therapy. 2009;17(10):1712–1723. doi: 10.1038/mt.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupberger J., Brino L., Baumert T.F. RNAi: a powerful tool to unravel hepatitis C virus–host interactions within the infectious life cycle. Journal of Hepatology. 2008;48(3):523–525. doi: 10.1016/j.jhep.2007.12.007. [DOI] [PubMed] [Google Scholar]
- McCaffrey A.P., Nakai H., Pandey K., Huang Z., Salazar F.H., Xu H., Wieland S.F., Marion P.L., Kay M.A. Inhibition of hepatitis B virus in mice by RNA interference. Nature Biotechnology. 2003;21(6):639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Morrissey D.V., Lockridge J.A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V., Vaish N., Zinnen S., Vargeese C., Bowman K., Shaffer C.S., Jeffs L.B., Judge A., MacLachlan I., Polisky B. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nature Biotechnology. 2005;23(8):1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Niu X.Y., Peng Z.L., Duan W.Q., Wang H., Wang P. Inhibition of HPV 16 E6 oncogene expression by RNA interference in vitro and in vivo. International Journal of Gynecological Cancer. 2006;16(2):743–751. doi: 10.1111/j.1525-1438.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- Pacca C.C., Severino A.A., Mondini A., Rahal P., D’Avila S.G., Cordeiro J.A., Nogueira M.C., Bronzoni R.V., Nogueira M.L. RNA interference inhibits yellow fever virus replication in vitro and in vivo. Virus Genes. 2009;38(2):224–231. doi: 10.1007/s11262-009-0328-3. [DOI] [PubMed] [Google Scholar]
- Padwad Y.S., Mishra K.P., Jain M., Chanda S., Karan D., Ganju L. RNA interference mediated silencing of Hsp60 gene in human monocytic myeloma cell line U937 revealed decreased dengue virus multiplication. Immunobiology. 2009;214(6):422–429. doi: 10.1016/j.imbio.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Pandiri A.R., Reed W.M., Mays J.K., Fadly A.M. Influence of strain, dose of virus, and age at inoculation on subgroup J avian leukosis virus persistence, antibody response, and oncogenicity in commercial meat-type chickens. Avian Diseases. 2007;51(3):725–732. doi: 10.1637/0005-2086(2007)51[725:IOSDOV]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Saleh M.C., Van Rij R.P., Andino R. RNA silencing in viral infections: insights from poliovirus. Virus Research. 2004;102(1):11–17. doi: 10.1016/j.virusres.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Song E., Lee S.K., Wang J., Ince N., Ouyang N., Min J., Chen J., Shankar P., Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nature Medicine. 2003;9(3):347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Sonwane A.A., Dahiya S.S., Saini M., Chaturvedi V.K., Singh R.P., Gupta P.K. Inhibition of rabies virus multiplication by siRNA delivered through adenoviral vector in vitro in BHK-21 cells and in vivo in mice. Research in Veterinary Science. 2012;93(1):498–503. doi: 10.1016/j.rvsc.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Willoughby R.E., Jr., Tieves K.S., Hoffman G.M., Ghanayem N.S., Amlie-Lefond C.M., Schwabe M.J., Chusid M.J., Rupprecht C.E. Survival after treatment of rabies with induction of coma. New England Journal of Medicine. 2005;352(24):2508–2514. doi: 10.1056/NEJMoa050382. [DOI] [PubMed] [Google Scholar]
- Wunner W.H., Pallatroni C., Curtis P.J. Selection of genetic inhibitors of rabies virus. Archives of Virology. 2004;149(8):1653–1662. doi: 10.1007/s00705-004-0299-6. [DOI] [PubMed] [Google Scholar]
- Zhang W., Wang C.Y., Yang S.T., Qin C., Hu J.L., Xia X.Z. Inhibition of highly pathogenic avian influenza virus H5N1 replication by the small interfering RNA targeting polymerase A gene. Biochemical and Biophysical Research Communications. 2009;390(3):421–426. doi: 10.1016/j.bbrc.2009.09.039. [DOI] [PubMed] [Google Scholar]


