Highlights
-
•
MERS-CoV is a novel coronavirus causing SARS-like disease with high fatality rate.
-
•
Its S protein is necessary and sufficient to mediate binding, fusion and cell entry.
-
•
We and others identified neutralizing antibodies targeting RBD in S1 subunit.
-
•
We identified MERS-CoV fusion inhibitory peptides targeting HR1 in S2 subunit.
-
•
Combining MERS-CoV entry and fusion inhibitors have synergistic antiviral effect.
Keywords: Middle East respiratory syndrome coronavirus, MERS-CoV, Receptor-binding domain, Fusion inhibitor, Entry inhibitor, Six-helix bundle
Abstract
The recent outbreak of Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) infection has led to more than 800 laboratory-confirmed MERS cases with a high case fatality rate (∼35%), posing a serious threat to global public health and calling for the development of effective and safe therapeutic and prophylactic strategies to treat and prevent MERS-CoV infection. Here we discuss the most recent studies on the structure of the MERS-CoV spike protein and its role in virus binding and entry, and the development of MERS-CoV entry/fusion inhibitors targeting the S1 subunit, particularly the receptor-binding domain (RBD), and the S2 subunit, especially the HR1 region, of the MERS-CoV spike protein. We then look ahead to future applications of these viral entry/fusion inhibitors, either alone or in combination with specific and nonspecific MERS-CoV replication inhibitors, for the treatment and prevention of MERS-CoV infection.
1. Introduction
On June 13, 2012, a 60-year-old Saudi man suffering with a SARS-like disease characterized by fever, cough and shortness of breath was admitted to a hospital in Jeddah; eleven days later, he died from respiratory and kidney failure. Dr. Ali Moh Zaki, a Saudi Arabia virologist, isolated a new coronavirus (nCoV) from the patient's specimens. It was later verified as a betacoronavirus, named as hCoV-EMC (Zaki et al., 2012). In May of 2013, this virus was renamed as “Middle East respiratory syndrome coronavirus (MERS-CoV) by the International Committee on Taxonomy of Viruses (ICTV) (de Groot et al., 2013). As of 23 July 2014, WHO had been informed of 837 confirmed cases of Middle East respiratory syndrome (MERS), including 291 deaths, a higher case fatality rate (∼35%) than SARS (∼10%) (Drosten et al., 2003, Lu et al., 2013b, WHO, 2014). The case numbers in Saudi Arabia spiked in April and May of 2014, with more than 25% of the reported cases being health care workers (WHO, 2014), raising a concern about the pandemic potential of MERS.
The major clinical manifestation of MERS-CoV is rapidly progressive acute pneumonia, which closely resembles SARS. In some patients, other severe complications may occur, including acute renal failure, diarrhea and abdominal pain, consumptive coagulopathy, and pericarditis (Assiri et al., 2013). Most previously reported patients have been adults, and the average age is around 47 (WHO, 2014). It should be noted that over three-quarters of MERS-CoV cases have occurred in patients with comorbidities (Research Group, 2013). The most common comorbidities for MERS-CoV cases include some chronic diseases, such as diabetes, hypertension, obesity, cancer, and chronic kidney, heart, and lung disease (Assiri et al., 2013). From the viewpoint of clinical manifestation and treatment options, these chronic diseases are likely impact disease progression, but the biased correlation between chronic disease and MERS-CoV infection is still unknown.
MERS-CoV is an enveloped virus with a positive-sense single-stranded RNA genome belonging to lineage C betacoronaviruses (Zaki et al., 2012). The genome contains 30 kb nucleotides and 10 plus open reading frames (ORFs), which encode the nonstructural replicase polyprotein and structural proteins, including a number of enzyme proteins, S protein, E protein, M protein and N protein (Zaki et al., 2012, van Boheemen et al., 2012). MERS-CoV is the sixth human coronavirus known to infect humans. Phylogenetic analysis demonstrated that MERS-CoV is more closely related to the bat coronaviruses HKU4 and HKU5 than it is to SARS-CoV (van Boheemen et al., 2012, Lau et al., 2013, Woo et al., 2014), suggesting that MERS-CoV originated from bats, which may serve as the natural reservoir (Annan et al., 2013, Anthony et al., 2013). Indeed, the coronaviruses isolated from bats in South Africa were found to be genetically related to MERS-CoV and a coronavirus identified in a bat from Saudi Arabia with 100% nucleotide identity to virus from the human index case-patient (Memish et al., 2013, Milne-Price et al., 2014). Therefore, the possibility of direct transmission of MERS-CoV or MERS-CoV-like virus from bats to humans cannot be excluded.
Dromedary camels in the Middle East and Africa are now thought to be the intermediate hosts for transmission of MERS-CoV from bats to humans because MERS-CoV-specific antibodies and RNA fragments are widespread in camels from Saudi Arabia, Egypt, Tunisia, Nigeria, and Kenya (Matsuyama et al., 2005, Hemida et al., 2013, Lau et al., 2013, Reusken et al., 2013a, Reusken et al., 2013b, Barlan et al., 2014). Human infection of MERS-CoV from the infected camels was believed to occur through consumption of the animal's meat or milk. However, recent study suggests that MERS-CoV may also be airborne, since the genetic fragments of MERS-CoV were detected in an air sample from a barn that housed an infected camel (Azhar et al., 2014). Although further investigations are required to confirm this claim, the results suggest that people working with camels should take every precaution to stop the spread of MERS-CoV. For example, face masks should be worn by persons who handle camels, particularly those that appear sick.
The cellular receptor for MERS-CoV has been identified as dipeptidyl peptidase-4 (DPP4, also named CD26) (Raj et al., 2014). DPP4 is a type-II transmembrane glycoprotein which is widely expressed on nonciliated bronchial epithelium and the epithelial cells in kidney, small intestine, liver, parotid gland, even in testis and prostate (Lu et al., 2013a). The wide distribution is in good accordance with the diversity of clinical manifestations. Our recent studies have demonstrated that the bat coronavirus HKU4 could also use DPP4 for its entry into the target cell. However, HKU4 prefers to use bat DPP4, while human MERS-CoV prefers to utilize human DPP4 as its receptor. In the absence of exogenous proteases, both MERS-CoV and HKU4 pseudoviruses can enter the DPP4-expressing bat cells, while only MERS-CoV pseudovirus, but not HKU4 pseudovirus, is able to enter human DPP4-expressing human cells (Yang et al., 2014). As suggested by these findings, MERS-CoV, unlike HKU4, has adapted to human DPP4 and human cellular proteases to gain entry into host cells.
Like other coronaviruses, MERS-CoV enters the target cell through two pathways, either endocytosis or plasma membrane fusion, in a cathepsin L- and low pH-dependent or -independent manner, respectively (Qian et al., 2013, Shirato et al., 2013). For pH-dependent endocytosis, virus entry occurs after internalization with fusion taking place in the acidic environment of endosomal compartment (Belouzard et al., 2012). For pH-independent plasma membrane fusion, the virus is able to fuse directly at the cell surface after binding to the receptor, which is about 100- to 1000-fold more efficient than pH-dependent endocytosis (Matsuyama et al., 2005, Belouzard et al., 2012). After MERS-CoV S protein is cleaved by serine proteases during virus maturation, MERS-CoV enters the cell via fusion between viral envelope and plasma membrane at neutral pH, causing massive syncytia formation (Qian et al., 2013, Gierer et al., 2013). MERS-CoV seems to enter cells mainly through the plasma membrane fusion pathway since several MERS-CoV-infected cell lines, such as Calu-3 (lower airway) and Huh-7 (liver) cells, form syncytia (Chan et al., 2013a, Lu et al., 2014). MERS-CoV pseudovirus treated with serine proteases, such as TMPRSS2, becomes fusion-activated, suggesting that the plasma membrane fusion is mediated by the cleaved S1 and S2 subunits (Qian et al., 2013, Gierer et al., 2013).
The emergence of MERS-CoV as a cause of severe respiratory disease highlights an urgent need for the development of effective therapeutic and prophylactic agents for treatment and prevention of MERS-CoV infection. Currently, no specific anti-MERS-CoV drug is available. Some in vitro studies have shown that a nonspecific antiviral drug, Ribavirin, a nucleoside analog, could inhibit MERS-CoV replication (Chan et al., 2013b). Mycophenolic acid (MPA), which is commonly used in clinics as an immunosuppressant drug to prevent rejection in organ transplantation, is reported to be effective against a number of viruses, including hepatitis E virus (HEV) (Wang et al., 2014), HCV (Pan et al., 2012, Ye et al., 2012), influenza virus H1N1 (Chan et al., 2013b), West Nile virus (Morrey et al., 2002), Chikungunya virus (Khan et al., 2011), yellow fever virus (Leyssen et al., 2005), and MERS-CoV in cell culture (Chan et al., 2013b, Hart et al., 2014). Poly I:C, an immunostimulatory double-stranded RNA analog, can induce antiviral responses in vitro and in vivo (Leyssen et al., 2003, Kumar et al., 2006). Perlman and colleagues have shown that treatment of mice expressing human DPP4 with poly I:C before or after MERS-CoV challenge significantly accelerated virus clearance (Zhao et al., 2014). Type I interferons (IFNs), including IFN-β1b, IFN-β1a and IFN-α2b, were reported to be effective in inhibiting MERS-CoV replication in in vitro cell culture (Chan et al., 2013b). Recently, a number of compounds with inhibitory activity at low micromolar levels on MERS-CoV replication in cell cultures in vitro have been identified from the FDA-approved drug libraries (Dyall et al., 2014, de Wilde et al., 2014), but their mechanisms of action have not been well defined.
Entry inhibitors are a class of antiretroviral drugs that prevent the virus from entering the cell (Baldwin et al., 2003, Liu et al., 2007, Este and Telenti, 2007). As exemplified by the anti-HIV peptide T20 (enfuvirtide), entry inhibitors usually have good compatibility and complementarity with other kinds of drugs in cocktail therapies by increasing curative effect, while decreasing side effects (Lalezari et al., 2003, Lazzarin et al., 2003). Hence, virus entry inhibitors could serve as promising candidate drugs against MERS-CoV infection with good prospects in clinical application.
In this review, we will summarize recent advances in studies reporting on the structure and function of the MERS-CoV spike (S) protein, leading to a better understanding of MERS-CoV entry and fusion mechanisms. Following this, we will address the development of MERS-CoV entry/fusion inhibitors targeting the S1 and S2 subunits of the MERS-CoV spike protein, particularly focusing on the viral fusion inhibitors interacting with the HR2 region in the spike protein and their future application for treatment and prevention of MERS.
2. Structure of the MERS-CoV S protein and its role in MERS-CoV entry into the target cell
Similar to SARS-CoV, MERS-CoV spike (S) protein is also a type I transmembrane glycoprotein which is located at the viral envelope surface in a trimer state. MERS-CoV S protein contains 1353 amino acids and can be cleaved into two subunits, S1 and S2 (Fig. 1 ). The S1 subunit is responsible for binding to the cellular receptor (DPP4), and the S2 subunit mediates membrane fusion (Lu et al., 2013a, Raj et al., 2013, Mou et al., 2013, Lu et al., 2014).
Fig. 1.
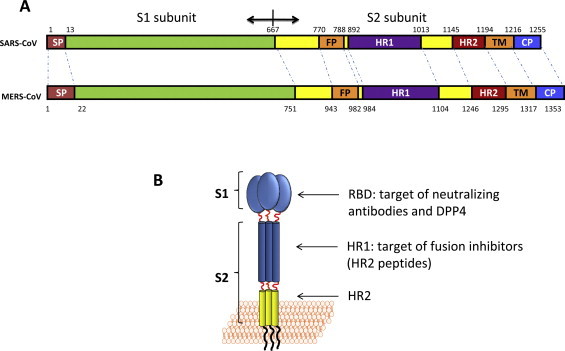
Structure of MERS-CoV S protein. (A) The schematic representation of MERS-CoV S protein compared with SARS-CoV S protein. SP, signal peptide; FP, fusion peptide; HR1, heptad repeat 1 domain; HR2, heptad repeat 2 domain; TM, transmembrane domain; CP, cytoplasmic domain. Residue numbers correspond to their positions in S protein of MERS-CoV. (B) The S1 subunit containing the receptor binding domain, the target of mAbs which could block MERS-CoV by binding to its receptor, DPP4. The S2 subunit contains the HR1 domain, the target of fusion inhibitors.
MERS-CoV S1 protein is located within the N-terminal 14–751 amino acids of S protein (Lu et al., 2013a, Chen et al., 2013, Wang et al., 2013), containing the receptor binding domain (RBD) (Fig. 1). Although the crystal structure of the DPP4/RBD complex had not previously been solved, we used multiple sequence alignment with SARS-CoV S1 subunit to predict that the RBD of MERS-CoV might be located at residues 377–662 (Du et al., 2013b). Later, three independent groups did solve the crystal structure of DPP4/RBD complex and determined that the RBD of MERS-CoV is located at residues 367–606 (Lu et al., 2013a, Chen et al., 2013, Wang et al., 2013). They also showed that the core structure of MERS-CoV S protein RBD is a five-stranded antiparallel β-sheet with several short α-helices, harboring three disulfide bonds that connect C383 to C407, C425 to C478, and C437 to C585, in order to stabilize the core structure (Lu et al., 2013a). The RBD also contains 2 glycans (Lu et al., 2013a, Chen et al., 2013, Wang et al., 2013). The receptor binding motif in RBD domain binds to the side-surface of the DPP4 β-propeller (Lu et al., 2013a). This interaction is very similar to the interaction between adenosine deaminase (ADA) and DPP4 (Weihofen et al., 2004). Therefore, ADA can compete with DPP4 for virus binding, acting as a natural antagonist for MERS-CoV infection (Raj et al., 2014). As an important domain to mediate the binding between MERS-CoV and its receptor DPP4, RBD serves as a critical target for the development of vaccines and therapeutics (Jiang et al., 2012, Hotez et al., 2014). We and others have previously demonstrated that the MERS-CoV RBD could induce significant neutralizing antibody responses in mice (Du et al., 2013a, Du et al., 2013b, Ma et al., 2014, Ying et al., 2014), providing a solid rationale for using the RBD as an attractive target to develop MERS-CoV entry inhibitors, such as soluble DPP4 (sDPP4) and RBD-specific neutralizing monoclonal antibodies (mAbs). By multiple sequence alignment with SARS-CoV S protein S2 subunit, we found that the S2 subunit of MERS-CoV harbors an fusion peptide (FP, residues 943–982), an HR1 domain (residues 984–1104), an HR2 domain (residues 1246–1295), a transmembrane domain (residues 1296–1317) and an intracellular domain (residues1318–1353) (Fig. 1A). Like HIV-1 gp41 and SARS-CoV S2 subunit, MERS-CoV S2 subunit is responsible for mediating viral fusion with the target cell membrane. After MERS-CoV S1 binding to DPP4, we hypothesize that the S2 subunit changes its conformation by inserting its FP into the target cell membrane. Its HR1 helices form a homotrimer with exposure of three highly conserved hydrophobic grooves on the surface. Its HR2 molecules then bind to the HR1 trimer to form a six-helix bundle (6-HB) core structure, which brings the viral and cell membranes into close proximity to facilitate fusion (Fig. 2A). Finally, the genetic materials of MERS-CoV enter the host cell via the fusion pore for replication in the cytoplasm (Lu et al., 2014). Alternatively, MERS-CoV can also enter cells via endocytosis in a cathepsin L- and low pH-dependent manner (Qian et al., 2013, Shirato et al., 2013).
Fig. 2.
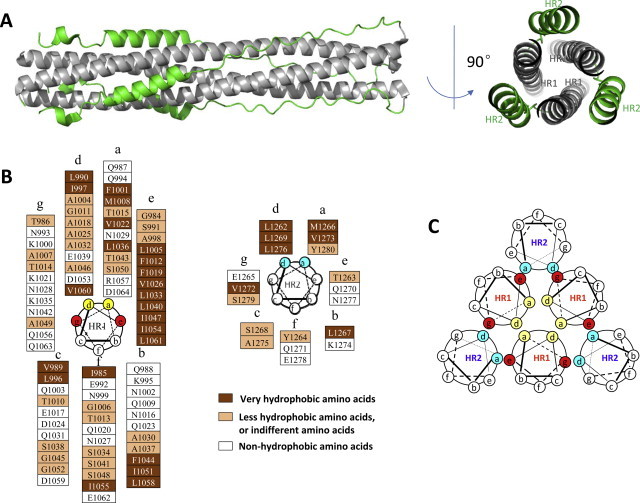
The structure of MERS-CoV 6-HB. (A) The cartoon of the MERS-CoV fusion core structure, with HR1 colored in gray and HR2 colored in green, respectively. (B) The helical wheel of HR1 and HR2. Most of the residues located at the “a” and “d” positions of the helical wheels are hydrophobic and more conserved than those in the “b”, “c”, and “f” positions, which are hydrophilic. (C) The interaction between HR1 and HR2. Through the interaction of the residues located at the “a” and “d” positions (yellow), three residues of HR1 form the internal trimer. The residues of HR1 located at the “e” and “g” positions (red) interact with residues of HR2 at the “a” and “d” positions (blue), finally forming the 6-HB structure.
Through further analysis, we have found that MERS-CoV uses a fusogenic mechanism to enter the target cell, which is similar to that of HIV or SARS-CoV to enter the target cell (Chan and Kim, 1998, Liu et al., 2003, Liu et al., 2004, Liu et al., 2007, Yu et al., 2012). The residues at the “a” and “d” positions in one of the HR1 helices interact with the residues at the “d” and “a” positions in the adjacent HR1 helices, respectively, to form the internal HR1 trimer. Those located at “e” and “g” positions in the HR1 helices interact with the residues at “e” and “g” positions in the HR2 helices to form 6-HB (Fig. 2B). The residues at the “b”, “c”, and “f” positions in HR2 helices form a hydrophilic face toward the solution to maintain the water-solubility of the oligomeric bundles (Fig. 2C).
In the crystal structure of MERS-CoV 6-HB, a notable difference in length is observed between the HR1 and HR2 helices (Fig. 2A). The N- and C-terminal tails of the HR2 region pack in an orderly manner against hydrophobic grooves of the HR1 trimer with several hydrophobic interactions, mainly distributed at the N-terminal portion and C-terminal portion of the HR2. One HR2 helix formed 11 hydrogen bonds to interact with its two neighboring HR1 domains. These hydrogen bonds located at N-terminal or C-terminal portion of the HR2 helix constitute two anchoring points, the N-Cap and C-Cap conformation, further stabilizing 6-HB construction. Compared with the 6-HB fusion core structure of SARS-CoV S protein (Liu et al., 2004, Xu et al., 2004), the MERS-CoV S2 subunit contains stronger hydrogen bonds, suggesting that the 6-HB of MERS-CoV S protein is more stable than that of SARS-CoV S protein. For this reason, the HR2 peptide derived from MERS-CoV S2 is expected to be more effective against MERS-CoV fusion than the HR2 peptide derived from SARS-CoV S2 against SARS-CoV fusion. These findings confirm that the HR1 region in MERS-CoV S protein S2 subunit is a promising target of HR2 peptides-based MERS-CoV fusion inhibitors.
3. MERS-CoV entry inhibitors targeting RBD in the S protein S1 subunit
MERS-CoV infection is initiated by binding of the viral particle via the RBD in S protein on viral surface with the cellular receptor DPP4 on the cell surface (Raj et al., 2013). Targeting of the binding site between the RBD and the receptor could block the initial step of virus entry and, thus, might provide superior pharmacological action to suppress MERS-CoV infection. In this regard, sDPP4 could serve as an excellent blocker for the attachment and entry of MERS-CoV to cells. Indeed, sDPP4 has been found to bind MERS-CoV RBD with high affinity (Du et al., 2013b, Ying et al., 2014) and prevent infection by a pseudotyped MERS-CoV with IC50 of about 10 μg/ml (Raj et al., 2013) (Table 1 ). The addition of some protein fragments to sDPP4 or chemical modification of sDPP4 may improve its antiviral activity and pharmacological properties.
Table 1.
MERS-CoV entry inhibitors targeting different parts in spike proteins.
| Inhibitors | Property | Target | Assay | IC50 (μg/ml) | Reference |
|---|---|---|---|---|---|
| sDPP4 | Protein | RBD in S1 | Pseudovirus | ∼10 | Raj et al. (2014) |
| Mersmab1 IgG | Mouse mAb | RBD in S1 | Pseudovirus | ∼0.10 | Du et al. (2014) |
| Mersmab1 IgG | Mouse mAb | RBD in S1 | Live virus (CPE) | ∼1.20 | Du et al. (2014) |
| m336 IgG | Human mAb | RBD in S1 | Pseudovirus | 0.005 | Ying et al. (2014) |
| m336 IgG | Human mAb | RBD in S1 | Live virus (CPE) | 0.07 | Ying et al. (2014) |
| MERS-4 IgG | Human mAb | RBD in S1 | Pseudovirus | 0.056 | Jiang et al. (2014) |
| MERS-4 IgG | Human mAb | RBD in S1 | Live virus (CPE) | 0.50 | Jiang et al. (2014) |
| 3B11 scFvFc | Human mAb | RBD in S1 | Live virus (CPE) | 1.83 | Tang et al. (2014) |
| 3B11 IgG | Human mAb | RBD in S1 | Live virus (CPE) | 3.5 | Tang et al. (2014) |
| HR2P | Peptide | HR1 in S2 | Cell–cell fusion | 3.314 | Lu et al. (2014) |
| HR2P | Peptide | HR1 in S2 | Live virus (CPE) | 2.485 | Lu et al. (2014) |
| HR2P-M2 | Peptide | HR1 in S2 | Cell–cell fusion | 2.278 | Lu et al. (2014) |
Alternatively, anti-DPP4 antibodies could also block the interaction between cellular DPP4 and viral RBD, thus inhibiting MERS-CoV infection (Ohnuma et al., 2013, Raj et al., 2013). However, it is impractical to apply this approach in vivo because DPP4 itself plays important roles in several distinct signaling pathways and in the regulation of many peptides (Zhong et al., 2013). As such, binding of the antibody to DPP4 on the cell surface may have pleiotropic effects on the host. Furthermore, because about 6 μg/ml of sDPP4 presents in serum (Ohnuma et al., 2013), a significant amount of antibody will be neutralized by sDPP4 before the antibody can bind to cellular DPP4 for inhibition of virus binding and infection. However, intranasal application of sDPP4 or anti-DPP4 antibodies for prevention of MERS-CoV is expected to circumvent the problems mentioned above.
We have previously found that the MERS-CoV RBD could induce significant neutralizing antibody responses in mice. This finding resulted in the development of a neutralizing mouse monoclonal antibody (mAb) which potently blocks MERS-CoV entry into human cells (Du et al., 2014). The neutralizing mAb was generated by immunizing mice with recombinant MERS-CoV S1 fused to a C-terminal IgG1 Fc tag. Stable hybridoma cell lines were then generated to screen positive clones. Subsequently, the selected anti-MERS-CoV mAbs were tested for their inhibition of pseudovirus entry mediated by MERS-CoV spike protein and for their neutralizing activity against live MERS-CoV infection in DPP4-expressing Vero E6 cells. The mAb Mersmab1, which had the most potent anti-MERS-CoV activity, was selected. Mersmab1 was not only highly effective in blocking the entry of MERS-CoV spike-mediated pseudoviruses into DPP4-expressing Huh-7 cells, but also potently neutralized live MERS-CoV infection of permissive Vero E6 cells by inhibiting the formation of MERS-CoV-induced CPE. In addition, Mersmab1 efficiently attenuated the formation of MERS-CoV-induced CPE in permissive Calu-3 cells, confirming its potent anti-MERS-CoV neutralizing activity (Table 1). To investigate the mechanism of Mersmab1 in neutralizing MERS-CoV cell entry, RBD/receptor-binding assays were performed in the presence of Mersmab1. It was found that the binding between MERS-CoV RBD and DPP4-expressing Huh-7 cells was blocked by Mersmab1 in a dose-dependent manner, suggesting that Mersmab1 neutralizes MERS-CoV entry into host cells by blocking the binding of MERS-CoV RBD to its host receptor, DPP4. We also found that Mersmab1 specifically bound to MERS-CoV S1-Fc and RBD-Fc, but not to MERS-CoV S2-Fc or human Fc (hFc). Furthermore, Mersmab1 lost most of its binding affinity for MERS-CoV S1 or RBD in the presence of DTT. These results suggest that Mersmab1 specifically binds to MERS-CoV RBD through recognizing conformational epitopes on the RBD. Thus, Mersmab1 can be humanized to serve as a potent passive immunotherapeutic agent for preventing and treating MERS-CoV infections.
Recently, neutralizing human mAbs of MERS-CoV have also been reported. Ying et al. constructed a very large phage-displayed antibody Fab library by using B cells from the blood of 40 healthy donors (Ying et al., 2014). By screening this library against recombinant MERS-CoV RBD, a panel of 12 human mAbs was identified. Three mAbs, m336, m337 and m338, which bound to the RBD with picomolar affinity, were selected for further characterization. Remarkably, all three RBD-specific mAbs exhibited exceptionally potent neutralizing activity. The mAbs neutralized pseudotyped MERS-CoV with 50% inhibitory concentration (IC50), ranging from 0.005 to 0.017 μg/ml (Table 1). Particularly noteworthy, the most potent mAb, m336, inhibited >90% MERS-CoV pseudovirus infection (IC90) at a concentration of 0.039 μg/ml. Similarly, m336 showed the most potent live virus neutralizing activity, with an IC50 of 0.07 μg/ml (Table 1). The three mAbs competed with each other for binding to the MERS-CoV RBD, and all the IgG1s potently inhibited the binding of RBD to the sDPP4 receptor. Consistent with the neutralization results, m336 IgG1 was slightly more potent than either m337 or m338 in blocking the binding of RBD to the receptor. The IC50s of m336, m337, and m338 were 0.034, 0.044, and 0.041 μg/ml, respectively. These results suggest that the mAbs neutralize MERS-CoV by competing with the host receptor DPP4 for binding to the virus and that they have overlapping epitopes. To further localize the mAb epitopes, a panel of RBD alanine-scanning mutants was developed, and some key residues on RBD involved in the binding of mAbs were identified. Interestingly, the three RBD-specific mAbs possess overlapping, but distinct, epitopes, suggesting the possibility of a synergistic effect by using three mAbs in combination. Molecular modeling and docking of these RBD-specific mAbs with the MERS-CoV RBD further confirmed the epitopes identified from RBD alanine-scanning mutants and also indicated a possible dominant role of the heavy chain in the mAb paratopes. The extensive overlapping between the mAb epitopes and the receptor binding sites on RBD explains the exceptional neutralizing potency of these mAbs.
Two other groups also identified several potent human mAbs. Jiang et al. used a nonimmune yeast-displayed scFv library to screen against the recombinant MERS-CoV RBD, and identified two potent RBD-specific neutralizing mAbs, MERS-4 and MERS-27 (Jiang et al., 2014). The most potent mAb, MERS-4, neutralized the pseudotyped MERS-CoV with an IC50 of 0.056 μg/ml and neutralized live MERS-CoV with an IC50 of 0.5 μg/ml (Table 1). mAbs could inhibit the binding of soluble RBD to DPP4-expressing susceptible Huh7 cells, suggesting that the neutralizing mechanism of MERS-4 and MERS-27 also occurred through the blocking of RBD binding to cellular receptor DPP4. Epitope mapping by mutagenesis of the key residues of RBD was also performed, and both MERS-4 and MERS-27 were found to recognize different epitopes on RBD. Indeed, the combination of MERS-4 and MERS-27 demonstrated a synergistic neutralizing effect against pseudotyped MERS-CoV. Meanwhile, Tang et al. also identified neutralizing mAbs from a nonimmune phage-displayed scFv library (Tang et al., 2014). The panning was performed by sequentially using MERS-CoV spike-containing paramagnetic proteoliposomes and 293T cells which expressed full-length S protein as antigens. A panel of 7 anti-S1 scFvs was identified and expressed in both scFv-Fc and IgG1 formats. Epitope mapping using different S1 fragments suggested that the epitopes of all seven mAbs also lie within the RBD domain. The binding competition assays indicated that the seven mAbs could recognize at least three different epitopes on MERS-CoV RBD. The most potent antibody, 3B11, neutralized MERS-CoV virus with an IC50 of 1.83 μg/ml in scFv-Fc format and 3.50 μg/ml in IgG format (Table 1). The antibodies could block DPP4 binding to MERS-CoV RBD and inhibit the attachment of pseudovirus to DPP4-expressing cells. DPP4 could also block the antibodies from RBD binding. These results confirmed that the neutralizing mechanism also occurred through the blocking of MERS-CoV RBD binding to cellular receptor DPP4.
Monoclonal antibodies are enjoying significant clinical success and have been used for the effective treatment of a number of diseases, in particular, cancer and immune disorders. Although more than 40 mAbs have been approved for clinical use, the humanized mAb palivizumab (Synagis) (Scott and Lamb, 1999) remains the only mAb approved by the FDA for use against a viral disease. However, an increasing number of mAbs against emerging viruses, including SARS-CoV and henipaviruses, as well as against HIV-1 and other viruses, have been developed, and many of them have exhibited high potency in vitro and in animal models of infection (Casadevall et al., 2004, Prabakaran et al., 2009). Interestingly, all the identified highly potent MERS-CoV mAbs targeting the RBD domain of MERS-CoV S glycoprotein have most likely utilized a competitive mechanism of virus neutralization, confirming the RBD as a critical target for the development of MERS-CoV vaccines and therapeutics. The high neutralization activity of these newly identified mAbs, especially m336, suggests that they show exceptional promise for prophylaxis and therapy of MERS-CoV infection in humans (Jiang et al., 2014, Tang et al., 2014, Ying et al., 2014).
4. MERS-CoV entry inhibitors targeting HR1 in the S protein S2 subunit
Two decades ago, we and others identified several highly potent HIV-1 fusion inhibitors derived from the HIV-1 gp41 HR2 region (Jiang et al., 1993, Wild et al., 1994). One of the HR2 peptides, T20, was approved by the U.S. FDA as the first HIV fusion/entry inhibitor for treatment of patients who failed to respond to the then current antiretroviral drugs (Kilby and Eron, 2003, Lalezari et al., 2003, Lazzarin et al., 2003). Since then, a series of HR2-peptides with increased anti-HIV activity or improved pharmacological properties or resistant profiles, such as SFT (Chong et al., 2014), T1249 (Qi et al., 2008), T1144 (Pan et al., 2009) and CP32M (He et al., 2008), have been identified.
The mechanisms of action of these HR2 peptides have been well documented. Binding of gp120 to CD4 and then a coreceptor, CCR5 or CXCR4, triggers a series of conformation changes of gp41, including the insertion of the fusion peptide (FP) into the target cell membrane, exposure of the pre-hairpin coiled coil of gp41 HR1 domain, and association of the viral HR2 domain with the HR1 trimer, resulting in the formation of the six-helical bundle (6-HB), finally bringing the viral envelope and cell membrane into close proximity for fusion. At the prefusion intermediate state, the synthetic HR2 peptide added can also bind to the viral HR1-trimer and form heterologous 6-HB to block virus-cell fusion (Liu et al., 2007).
Discovery of the HR2-based HIV fusion inhibitors has opened a new avenue to identify and develop peptidic viral entry inhibitors against enveloped viruses with class I membrane fusion proteins, such as Ebola virus, Nipahvirus, Hendravirus, and other paramyxoviruses, Newcastle disease virus, simian immunodeficiency virus (SIV), feline immunodeficiency virus (FIV), and respiratory syncytial virus (RSV) (Blacklow et al., 1995, Lombardi et al., 1996, Watanabe et al., 2000, Yu et al., 2002, Wang et al., 2003).
Ten years ago, we designed a peptide derived from the SARS-CoV S protein S2 subunit HR2 domain, named CP-1, and found that it could interact with the peptide derived from the HR1 domain of SARS-CoV S protein S2 subunit to form a stable 6-HB. CP-1 inhibited SARS-CoV infection in Vero E6 cells with an IC50 of about 19 μM (Liu et al., 2004). Later, several other groups also identified SARS-CoV fusion inhibitory peptides from the HR2 region with anti-SARS-CoV activity similar to that of CP-1 (Bosch et al., 2004, Yuan et al., 2004, Zheng et al., 2005).
By sequence alignment analysis of both SARS-CoV and MERS-CoV S protein S2 subunits, we predicted that the HR1 and HR2 domains of MERS-CoV S protein were located in regions overlapping residues 984–1104 and 1246–1295, respectively. The crystal structure of a canonical 6-HB formed by the HR1 and HR2 domains was solved. We noticed that the helical region of residues 1004–1023 in the HR1 domain closely interacted with the helical region of residues 1262–1280 in the HR2 domain (Fig. 2A) (Lu et al., 2014). Based on structure information, we designed and synthesized two peptides, HR1P (residues 998–1039, containing residues 1004–1023) and HR2P (residues 1251–1286, containing residues 1262–1280) (Fig. 3A).
Fig. 3.
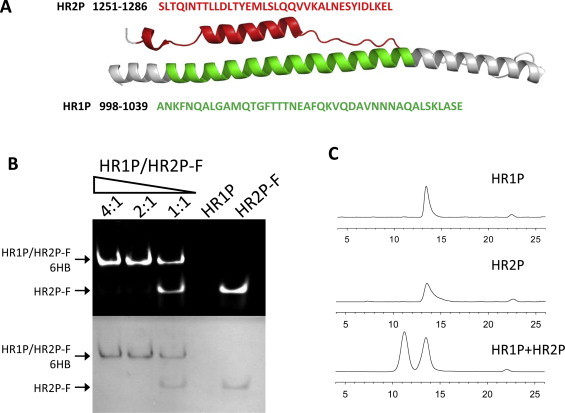
Design of S2-derived peptides (A) and determination of 6-HB formation between HR1P and HR2P-FITC by FN-PAGE (B), or by SE-HPLC (C). (A) The corresponding areas of the HR1P and HR2P in the cartoon of MERS-CoV 6-HB. Residue numbers correspond to their positions in S protein of MERS-CoV. (B) The association of HR2P-FITC and HR1P, as determined by N-PAGE. (C) The SE-HPLC profiles of HR1P, HR2P and their combination, HR1P/HR2P. The final concentration of peptides was 50 μM.
To determine the ability of the designed HR1P and HR2P peptides to form 6-HB, we first developed a fluorescence native polyacrylamide gel electrophoresis (FN-PAGE), which was based on the FN-PAGE we used to test 6-HB formation between the HIV-1 gp41 HR1 and HR2 peptides (Chan and Kim, 1998, Liu et al., 2003, Liu et al., 2004, Liu et al., 2007). HR2P was labeled with FITC. XCell SureLock® Mini-Cell (Invitrogen, Carlsbad, CA) was used for N-PAGE. Low molecular weight (LMW) markers were obtained from Amersham Pharmacia Biotech Inc. (Piscataway, NJ). Peptide HR1P at 40 μM in PBS (control: PBS) was incubated with HR2P-FITC 40 μM in PBS (control: PBS) at 37 °C for 30 min. The sample was then mixed with Tris-glycine native sample buffer (Invitrogen, Carlsbad, CA) at a ratio of 1:1 and loaded to a precast gel with 10 wells and 1.0 mm of thickness (25 μl each well). Gel electrophoresis was carried out with 125 V constant voltage at room temperature for 2 h. Immediately after electrophoresis, fluorescence bands in the gel were imaged by the FluorChem 8800 Imaging System using a transillumination UV light source with excitation wavelength at 302 nm and a fluorescence filter with emission wavelength at 520 nm (Fig. 3B, upper panel). The gel was then stained with Coomassie blue to explore the distribution of peptides (Fig. 3B, lower panel). Similar to N-PAGE, proteins or peptides were kept in their natural state under the condition of FN-PAGE, and their migration speed and direction depended on either their molecular weight or net charge. Under native electrophoresis condition, HR1P showed no band because it carries net positive charges, thus migrating up and off the gel. HR2P-FITC showed only one band at a lower position, while the combination of HR1P and HR2P-FITC showed two bands: a lower one with the same position of HR2P-FITC and a new upper band. By increasing HR1P concentration, the intensity of the upper band increased, while the intensity of the lower band decreased, suggesting that the upper band was the complex formed by HR1P and HR2P-FITC, while the lower band was the isolated HR2P-FITC.
We then used size-exclusion HPLC to assess the interaction between MERS-CoV HR1P and HR2P, as previously described for analyzing 6-HB formation between the HR1 and HR2 peptides of HIV-1 gp41 (Chan and Kim, 1998, Liu et al., 2003, Liu et al., 2004, Liu et al., 2007, Yu et al., 2012). HR1P or HR2P alone exhibited a single peak at 13.5 ml, while the combination of HR1P/HR2P showed a new peak at 11.5 ml (Fig. 3C). This result confirms that HR1P and HR2P could interact with each other to form an oligomeric complex.
We subsequently analyzed the complex using 15% SDS polyacrylamide protein gel without heating samples (Lu et al., 2014). Both HR1P and HR2P alone showed bands at molecular weights of about 3∼4 kDa, while the HR1P/HR2P complex exhibited a band of about 26 kDa, corresponding to the predicted mass of the 6-HB formed by HR1P and HR2P (25.8 kDa).
Finally, we used circular-dichroism (CD) spectroscopy to determine the secondary structures and assess the thermal stability of HR1P/HR2P complex. The mixture of HR1P/HR2P in equimolar concentration showed a typical helical complex. The thermal stability of the HR1P/HR2P complex of MERS-CoV S protein in phosphate buffer (Tm: ∼87 °C) (Lu et al., 2014) was much higher than that of complexes formed by the HR1 and HR2 peptides derived from the HIV-1 gp41 and SARS-CoV S protein S2 subunit, respectively (Chan and Kim, 1998, Liu et al., 2003, Liu et al., 2004, Liu et al., 2007).
By combining the above results from different biophysical and biochemical experiments, we concluded that the peptides derived from the HR1 and HR2 domains of MERS-CoV S protein S2 subunit could interact with each other to form highly stable 6-HB, mimicking the viral fusion core structure formed by the entire HR1 and HR2 domains of MERS-CoV S protein, as shown by crystallographic analysis (Lu et al., 2014).
To develop a MERS-CoV S protein-mediated cell–cell fusion model to test the membrane fusion inhibitory activity of the HR1 and HR2 peptides, we used Huh-7 cells that express DPP4 on the membrane surface, as the target cells (Raj et al., 2013), and 293T cells that instantaneously co-express MERS-CoV S protein and EGFP (293T/MERS/EGFP), as the effector cells (Lu et al., 2014). The 293T/MERS/EGFP cells were cocultured with Huh-7 cells at 37 °C for 2 to 4 h, using 293T cells expressing EGFP only (293T/EGFP) as the control cells. Fusion between 293T/MERS/EGFP and Huh-7 cells was visible under fluorescence microscopy. As shown in Fig. 4 , the fused cells showed larger size and weaker fluorescence than unfused cells because the fluorescence in one cell is diffused into two or more cells that fuse together. After staining the cell nucleus with DAPI, we could clearly visualize the presence of two or more nuclei in one fused cell with larger size and weaker fluorescence. The unfused cells had only one nucleus. This finding confirmed that MERS-CoV S protein was able to mediate membrane fusion between effector and target cells.
Fig. 4.
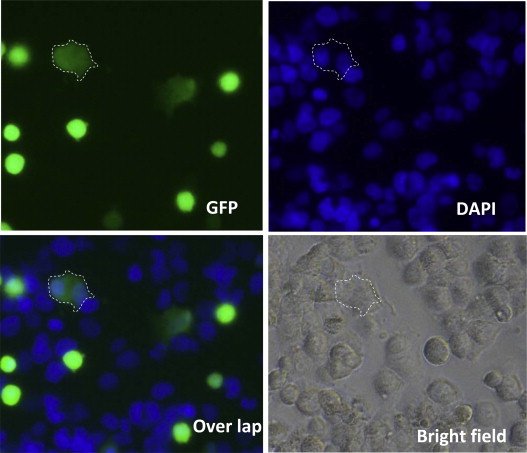
Images of MERS-CoV S protein-mediated cell–cell fusion. After 293T/MERS/EGFP cells were cocultured with Huh-7 cells at 37 °C for 4 h, cell–cell fusion was photographed under fluorescence microscopy (upper left). At the same time, cell nuclei were stained by DAPI (upper right). The overlap picture (bottom left) shows the fusion cell containing two or more nuclei. The bottom right is the image under visible light. The area encircled by the white dotted line is the fused cell.
Using this MERS-CoV S protein-mediated cell–cell fusion model, we tested the inhibitory activity of HR1P and HR2P derived from the HR1 and HR2 domains, respectively, of the MERS-CoV S protein S2 subunit. The result indicated that HR2P exhibited potent inhibitory effect (IC50 at 0.7–1.0 μM), while HR1P exhibited no appreciable activity. The HR2 peptides derived from the HIV-1 gp41 (T20) and SARS-CoV S protein (SC-1) exhibited no inhibitory activity on MERS-CoV S-mediated cell–cell fusion, suggesting that the HR2 peptide derived from different enveloped viruses cannot cross-inhibit viral fusion, even though these peptides have a similar mechanism of action. HR2P also showed potent inhibitory activity against pseudotyped and live MERS-CoV infection in DPP4-expressing cells, such as Huh-7 and Vero cells (Table 1) (Lu et al., 2014).
To improve the stability, solubility, and antiviral activity of HR2P, we adapted a approach described by Otaka et al. who modified the HIV-1 gp41 HR2 peptides by introducing EK mutations to form new intra-molecular salt-bridges (Otaka et al., 2002). We designed and synthesized two HR2P mutants, HR2P-M1 with 2 point mutations (T1263E and L1267R) and HR2P-M2 with 7 point mutations (T1263E, L1267K, S1268K, Q1270E, Q1271E, A1275K and N1277E) (Lu et al., 2014) in order to increase the salt-bridges and hydrophilicity of HR2P. We found that both HR2P-M1 and HR2P-M2 exhibited increased peptide solubility by about 67–72-fold and 1692–1879-fold in water and around 11.4–11.8-fold and 9.2–1.3-fold in PBS, respectively. Their inhibitory activity on MERS-CoV S-mediated cell–cell fusion was also increased (Lu et al., 2014). These results suggest that the antiviral activity and pharmacological properties of HR2-derived peptides can be further improved by using different approaches for optimization.
5. Conclusion and prospects
Like other enveloped viruses with class I membrane fusion proteins (e.g., HIV and SARS-CoV), MERS-CoV also utilizes its spike protein to enter into the target cells. The RBD in the S protein S1 subunit is responsible for virus binding to the receptor on the target cells, while the HR2 domain in the S protein S2 subunit is responsible for mediating fusion between the viral envelope and the target cell membrane for entry. As such, both may serve as targets for the development of MERS-CoV entry/fusion inhibitors, such as RBD-specific neutralizing mAbs and HR1-specific fusion inhibitory peptides.
Based on our previous experience in developing peptidic fusion inhibitors against HIV (Jiang et al., 1993, Wild et al., 1994) and SARS-CoV (Liu et al., 2004), we have successfully identified several specific MERS-CoV peptide fusion inhibitors, such as HR2P and HR2P-M2 (Lu et al., 2014). Like the HIV fusion inhibitor T20 (enfuvirutide), these peptides derived from the HR2 domain in S2 subunit of MERS-CoV spike protein can specifically bind to the HR1 domain in the S2 subunit of the viral MERS-CoV spike protein to form stable 6-HB and block viral fusion core formation, resulting in the inhibition of fusion between viral envelope and target cell membrane (Lu et al., 2014). These peptides show exceptional promise for further development as effective and safe therapeutic and prophylactic agents for treatment and prevention of MERS.
The clinical application of HIV fusion inhibitors has several limitations because of long-term repeated use, including serious local injection reaction, induction of anti-peptide drug antibodies or drug-resistant virus mutants. The peptide drugs against viruses that cause acute, often fatal, infectious diseases, such as MERS-CoV, are generally administered by injection for short periods of time, making these limitations moot. Furthermore, MERS-CoV fusion inhibitory peptide may also be intranasally used, avoiding local injection reaction.
Previous studies have shown that the addition of a fatty acid or cholesterol to the anti-HIV peptide drug T20 results in significant increase of its antiviral activity (Wexler-Cohen and Shai, 2007, Ingallinella et al., 2009, Lee et al., 2011, Ashkenazi et al., 2012). Conjugation of PEG to a peptide drug is expected to have the following advantages: (1) prolonged half-life in vivo, (2) decreased degradation by metabolic enzymes, and (3) reduction or elimination of protein immunogenicity (Veronese, 2001, Veronese and Pasut, 2005, Pasut and Veronese, 2009). Therefore, the above strategies can also be applied to modify the MERS-CoV fusion inhibitory peptides to improve their antiviral potency and pharmacological properties.
The RBD in S1 subunit and HR1 domain in S2 subunit can also be used as targets for screening small-molecule MERS-CoV entry inhibitors. The small molecules that bind to MERS-CoV RBD or HR1 domain with reasonable affinity and specificity can be considered alternatives to mAbs or inhibitory peptides. The use of small molecules could be advantageous under certain circumstances because of their easier and less expensive production, compared to biological molecules. Moreover, the high-resolution structures of the RBD-DPP4 complex, as well as the six-helix bundle fusion core of MERS-CoV spike protein S2 subunit, offer a significant opportunity for the structure-based rational design of entry inhibitors that target RBD or HR1 domain in the MERS-CoV spike protein.
Because of the long in vivo half-life and the superior stability of mAbs, they can be further developed as immunotherapeutics to both treat and prevent MERS-CoV infection. Although both RBD-specific neutralizing mAbs and fusion inhibitory HR2 peptides displayed high potency in inhibiting viral entry of MERS-CoV, they target different steps of viral entry: viral attachment to the receptor on the cell surface and viral fusion with the target cell membranes, respectively. Therefore, it is reasonable to speculate that the combination of neutralizing mAbs with HR2 peptides may have synergistic effect against MERS-CoV infection. It is well known that the combinatorial use of HIV reverse transcriptase (RT) inhibitors and protease inhibitors, known as highly active anti-retrovirus therapy (HAART) (Perelson et al., 1997, Louie et al., 2003, Di et al., 2004) has shown significant synergism in inhibiting HIV-1 infection, reducing adverse effects and delaying the emergence of drug resistance (Hogg et al., 1998), thus extending the lifespan of millions of HIV/AIDS patients (Simon et al., 2006, Richman et al., 2009).
One may question whether the use of HR2 peptides or RBD-specific neutralizing antibodies in controlling MERS-CoV infections may drive virus evolution, resulting in emergence of viral mutants with resistance to the fusion inhibitors or neutralizing antibodies. Unlike the long-term treatment of HIV infection and other chronic infectious diseases, the short-term treatment of MERS, SARS, and other acute infectious diseases may provide less opportunity and time to allow the virus to make drug-resistance mutations. So far, only one natural mutation at the position of 1020 in the HR1 domain of MERS-CoV spike protein was found (Cotten et al., 2014). However, no report has shown that this mutation affects the sensitivity of the mutant virus to MERS-CoV fusion inhibitors. The RBD in the MERS-CoV spike protein is relatively conservative (Ying et al., 2014, Cotten et al., 2014). The combinational use of an HR2 peptide and a RBD-specific neutralizing antibody or a conjugate consisting of an HR2 peptide and a RBD-binding molecule, like the bifunctional anti-HIV protein 2DLT (Lu et al., 2012, Xu et al., 2014) are expected to be effective against MERS-CoV with mutations in its spike protein HR1 domain or RBD, or to diminish the chance to induce drug-resistant viral mutants.
Considering that MERS-CoV entry into the target cell may occur through dual pathways, i.e., plasma membrane fusion and endocytosis, it may be necessary to use both membrane fusion inhibitors and endocytosis inhibitors in combination. Some clinically used drugs, such as chlorpromazine, were reported to function as endocytosis inhibitors against SARS-CoV (Inoue et al., 2007) and MERS-CoV (de Wilde et al., 2014). Falzarano et al. (2013) demonstrated the efficacy of combining ribavirin and IFN-α2b to reduce MERS-CoV replication in vitro. When this combination was given to rhesus macaques within 8 hours of inoculation with MERS-CoV, this treatment resulted in beneficial effects in reducing virus replication, lung injury and inflammation, improving clinical outcome. Therefore, this approach should not be limited to the combination of MERS-CoV entry inhibitors targeting different sites in spike protein, but rather include the combination of MERS-CoV entry inhibitors with the specific anti-MERS-CoV agents targeting other steps of MERS-CoV replication, such as protease inhibitors. Furthermore, combining specific anti-MERS-CoV agents with nonspecific MERS-CoV replication inhibitors identified from the FDA-approved drug libraries (Dyall et al., 2014, de Wilde et al., 2014) may also have synergistic effect. Overall, we anticipate that these endeavors will lead to a highly effective therapeutic approach for treatment of MERS patients and prevention of MERS-CoV infection in high-risk populations, including healthcare workers and family members.
Acknowledgement
This work was supported by grants from the National Science Fund of China (81173098 and 812111406 to SJ, 81102476 and 81373456 to LL).
References
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K.V., Lina P.H.C., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., Ojeda-Flores R., Rico-Chavez O., Navarrete-Macias I., Zambrana-Torrelio C.M., Rostal M.K., Epstein J.H., Tipps T., Liang E., Sanchez-Leon M., Sotomayor-Bonilla J., Aguirre A.A., vila-Flores R., Medellin R.A., Goldstein T., Suzan G., Daszak P., Lipkin W.I. Coronaviruses in bats from Mexico. J. Biol. Chem. 2013;94:1028–1038. doi: 10.1099/vir.0.049759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A., Viard M., Unger L., Blumenthal R., Shai Y. Sphingopeptides: dihydrosphingosine-based fusion inhibitors against wild-type and enfuvirtide-resistant HIV-1. FASEB J. 2012;26:4628–4636. doi: 10.1096/fj.12-215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F., Makhdoom H.Q., Zumla A.I., Memish Z.A. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., Hashem A.M., El-Kafrawy S.A., Sohrab S.S., Aburizaiza A.S., Farraj S.A., Hassan A.M., Al-Saeed M.S., Jamjoom G.A., Madani T.A. Detection of the Middle East respiratory syndrome coronavirus genome in an air sample originating from a camel barn owned by an infected patient. MBio. 2014;5:e01450–e1514. doi: 10.1128/mBio.01450-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin C.E., Sanders R.W., Berkhout B. Inhibiting HIV-1 entry with fusion inhibitors. Curr. Med. Chem. 2003;10:1633–1642. doi: 10.2174/0929867033457124. [DOI] [PubMed] [Google Scholar]
- Barlan A., Zhao J.C., Sarkar M.K., Li K., Mccray P.B., Perlman S., Gallagher T. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J. Virol. 2014;88:4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklow S.C., Lu M., Kim P. Trimeric subdomain of the simian immunodeficiency virus glycoprotein. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., Martina B.E.E., van der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J.R., de Groot R., Osterhaus A.D.M.E., Rottier P.J.M. Severe acute respiratory syndrome coroavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Dadachova E., Pirofski L.A. Passive antibody therapy for infectious diseases. Nat. Rev. Microbiol. 2004;2:695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- Chan D.C., Kim P.S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Chan K.H., Choi G.K., To K.K., Tse H., Cai J.P., Yeung M.L., Cheng V.C., Chen H., Che X.Y., Lau S.K., Woo P.C., Yuen K.Y. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013;207:1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Chan K.H., Kao R.Y., To K.K., Zheng B.J., Li C.P., Li P.T., Dai J., Mok F.K., Chen H., Hayden F.G., Yuen K.Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.Q., Rajashankar K.R., Yang Y., Agnihothram S.S., Liu C., Lin Y.L., Baric R.S., Li F. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:10777–10783. doi: 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong H., Yao X., Qiu Z., Sun J., Qiao Y., Zhang M., Wang M., Cui S., He Y. The M-T hook structure increases the potency of HIV-1 fusion inhibitor sifuvirtide and overcomes drug resistance. J. Antimicrob. Chemother. 2014;69:2759–2769. doi: 10.1093/jac/dku183. [DOI] [PubMed] [Google Scholar]
- Cotten M., Watson S.J., Zumla A.I., Makhdoom H.Q., Palser A.L., Ong S.H., Al Rabeeah A.A., Alhakeem R.F., Assiri A., Al-Tawfiq J.A., Albarrak A., Barry M., Shibl A., Alrabiah F.A., Hajjar S., Balkhy H.H., Flemban H., Rambaut A., Kellam P., Memish Z.A. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio. 2014;5:e01062–e1113. doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van N.S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di M.M., Markowitz M., Louie M., Hurley A., Hogan C., Simon V., Follmann D., Ho D.D., Perelson A.S. Dynamics of intermittent viremia during highly active antiretroviral therapy in patients who initiate therapy during chronic versus acute and early human immunodeficiency virus type 1 infection. J. Virol. 2004;78:10566–10573. doi: 10.1128/JVI.78.19.10566-10573.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der W.S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G., Chen Y., Yu F., Tseng C.T., Zhou Y., Jiang S. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLOS ONE. 2013;8:e81587. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Yang Y., Qiu H., Wang L., Kou Z., Tao X., Yu H., Sun S., Tseng C.T., Jiang S., Li F., Zhou Y. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L.Y., Zhao G.Y., Kou Z.H., Ma C.Q., Sun S.H., Poon V.K.M., Lu L., Wang L.L., Debnath A.K., Zheng B.J., Zhou Y.S., Jiang S.B. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 2013;87:9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jr., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Este J.A., Telenti A. HIV entry inhibitors. Lancet. 2007;370:81–88. doi: 10.1016/S0140-6736(07)61052-6. [DOI] [PubMed] [Google Scholar]
- Falzarano D., de W.E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P., Brining D., Bushmaker T., Martellaro C., Baseler L., Benecke A.G., Katze M.G., Munster V.J., Feldmann H. Treatment with interferon-alpha2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat. Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kramer-Kuhl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von H.T., Simmons G., Hofmann H., Pohlmann S. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart B.J., Dyall J., Postnikova E., Zhou H., Kindrachuk J., Johnson R.F., Olinger G.G., Jr., Frieman M.B., Holbrook M.R., Jahrling P.B., Hensley L. Interferon-beta and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2014;95:571–577. doi: 10.1099/vir.0.061911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Cheng J., Lu H., Li J., Hu J., Qi Z., Liu Z., Jiang S., Dai Q. Potent HIV fusion inhibitors against Enfuvirtide-resistant HIV-1 strains. Proc. Natl. Acad. Sci. USA. 2008;105:16332–16337. doi: 10.1073/pnas.0807335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida M.G., Perera R.A., Wang P., Alhammadi M.A., Siu L.Y., Li M., Poon L.L., Saif L., Alnaeem A., Peiris M. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro. Surveill. 2013;18:20659. doi: 10.2807/1560-7917.es2013.18.50.20659. [DOI] [PubMed] [Google Scholar]
- Hogg R.S., Rhone S.A., Yip B., Sherlock C., Conway B., Schechter M.T., O'Shaughnessy M.V., Montaner J.S. Antiviral effect of double and triple drug combinations amongst HIV-infected adults: lessons from the implementation of viral load-driven antiretroviral therapy. AIDS. 1998;12:279–284. doi: 10.1097/00002030-199803000-00005. [DOI] [PubMed] [Google Scholar]
- Hotez P.J., Bottazzi M.E., Tseng C.T., Zhan B., Lustigman S., Du L., Jiang S. Calling for rapid development of a safe and effective MERS vaccine. Microbes Infect. 2014;16:529–531. doi: 10.1016/j.micinf.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingallinella P., Bianchi E., Ladwa N.A., Wang Y.J., Hrin R., Veneziano M., Bonelli F., Ketas T.J., Moore J.P., Miller M.D., Pessi A. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc. Natl. Acad. Sci. USA. 2009;106:5801–5806. doi: 10.1073/pnas.0901007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L.W., Wang N.S., Zuo T., Shi X.L., Poon K.M.V., Wu Y.K., Gao F., Li D.Y., Wang R.K., Guo J.Y., Fu L.L., Yuen K.Y., Zheng B.J., Wang X.Q., Zhang L.Q. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014;6:239–259. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- Jiang S., Lin K., Strick N., Neurath A.R. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- Jiang S., Lu L., Liu Q., Xu W., Du L. Receptor-binding domains of spike proteins of emerging or re-emerging viruses as targets for development of antiviral vaccines. Emerg. Infect. Dis. 2012;1:e13. doi: 10.1038/emi.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Dhanwani R., Patro I.K., Rao P.V., Parida M.M. Cellular IMPDH enzyme activity is a potential target for the inhibition of Chikungunya virus replication and virus induced apoptosis in cultured mammalian cells. Antiviral Res. 2011;89:1–8. doi: 10.1016/j.antiviral.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Kilby J.M., Eron J.J. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 2003;348:2228–2238. doi: 10.1056/NEJMra022812. [DOI] [PubMed] [Google Scholar]
- Kumar A., Zhang J., Yu F.S. Toll-like receptor 3 agonist poly(I:C)-induced antiviral response in human corneal epithelial cells. Immunology. 2006;117:11–21. doi: 10.1111/j.1365-2567.2005.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalezari J.P., Henry K., O’Hearn M., Montaner J.S., Piliero P.J., Trottier B., Walmsley S., Cohen C., Kuritzkes D.R., Eron J.J., Jr., Chung J., DeMasi R., Donatacci L., Drobnes C., Delehanty J., Salgo M. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 2003;348:2175–2185. doi: 10.1056/NEJMoa035026. [DOI] [PubMed] [Google Scholar]
- Lau S.K.P., Li K.S.M., Tsang A.K.L., Lam C.S.F., Ahmed S., Chen H.L., Chan K.H., Woo P.C.Y., Yuen K.Y. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese Pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarin A., Clotet B., Cooper D., Reynes J., Arasteh K., Nelson M., Katlama C., Stellbrink H.J., Delfraissy J.F., Lange J., Huson L., DeMasi R., Wat C., Delehanty J., Drobnes C., Salgo M. Efficacy of enfuvirtide in patients infected with drug-resistant HIV-1 in Europe and Australia. N. Engl. J. Med. 2003;348:2186–2195. doi: 10.1056/NEJMoa035211. [DOI] [PubMed] [Google Scholar]
- Lee K.K., Pessi A., Gui L., Santoprete A., Talekar A., Moscona A., Porotto M. Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol-conjugated peptide, a new antiviral strategy for influenza virus. J. Biol. Chem. 2011;286:42141–42149. doi: 10.1074/jbc.M111.254243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyssen P., Balzarini J., De C.E., Neyts J. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J. Virol. 2005;79:1943–1947. doi: 10.1128/JVI.79.3.1943-1947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyssen P., Drosten C., Paning M., Charlier N., Paeshuyse J., De C.E., Neyts J. Interferons, interferon inducers, and interferon-ribavirin in treatment of flavivirus-induced encephalitis in mice. Antimicrob. Agents Chemother. 2003;47:777–782. doi: 10.1128/AAC.47.2.777-782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Wu S., Jiang S. HIV entry inhibitors targeting gp41: from polypeptides to small-molecule compounds. Curr. Pharm. Des. 2007;13:143–162. doi: 10.2174/138161207779313722. [DOI] [PubMed] [Google Scholar]
- Liu S., Xiao G.F., Chen Y.B., He Y.X., Niu J.K., Escalante C.R., Xiong H.B., Farmar J., Debnath A.K., Tien P., Jiang S.B. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhao Q., Jiang S. Determination of the HIV-1 gp41 fusogenic core conformation modeled by synthetic peptides: applicable for identification of HIV-1 fusion inhibitors. Peptides. 2003;24:1303–1313. doi: 10.1016/j.peptides.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Lombardi S., Massi C., Indino E., LaRosa C., Mazzetti P., Falcone M.L., Rovero P., Fissi A., Pieroni O., Bandecchi P., Esposito F., Tozzini F., Bendinelli M., Garzelli C. Inhibition of feline immunodeficiency virus infection in vitro by envelope glycoprotein synthetic peptides. Virology. 1996;220:274–284. doi: 10.1006/viro.1996.0315. [DOI] [PubMed] [Google Scholar]
- Louie M., Hogan C., Di M.M., Hurley A., Simon V., Rooney J., Ruiz N., Brun S., Sun E., Perelson A.S., Ho D.D., Markowitz M. Determining the relative efficacy of highly active antiretroviral therapy. J. Infect. Dis. 2003;187:896–900. doi: 10.1086/368164. [DOI] [PubMed] [Google Scholar]
- Lu G.W., Hu Y.W., Wang Q.H., Qi J.X., Gao F., Li Y., Zhang Y.F., Zhang W., Yuan Y., Bao J.K., Zhang B.C., Shi Y., Yan J.H., Gao G.F. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Du L., Jiang S. Middle East respiratory syndrome coronavirus (MERS-CoV): challenges in identifying its source and controlling its spread. Microbes Infect. 2013;15:625–629. doi: 10.1016/j.micinf.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhu Y., Chan K.H., Qin L.L., Li Y., Wang Q., Chan J.F.W., Du L.Y., Yu F., Ma C.Q., Ye S., Yuen K.Y., Zhang R.G., Jiang S.B. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Pan C., Li Y., Lu H., He W., Jiang S. A bivalent recombinant protein inactivates HIV-1 by targeting the gp41 prehairpin fusion intermediate induced by CD4 D1D2 domains. Retrovirology. 2012;9:104. doi: 10.1186/1742-4690-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Li Y., Wang L., Zhao G., Tao X., Tseng C.T., Zhou Y., Du L., Jiang S. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: Implication for designing novel mucosal MERS vaccines. Vaccine. 2014;32:2100–2108. doi: 10.1016/j.vaccine.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA. 2005;102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Mishra N., Olival K.J., Fagbo S.F., Kapoor V., Epstein J.H., Alhakeem R., Durosinloun A., Al A.M., Islam A., Kapoor A., Briese T., Daszak P., Al Rabeeah A.A., Lipkin W.I. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne-Price S., Miazgowicz K.L., Munster V.J. The emergence of the Middle East respiratory syndrome coronavirus. Pathog. Dis. 2014;71:119–134. doi: 10.1111/2049-632X.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrey J.D., Smee D.F., Sidwell R.W., Tseng C. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antiviral Res. 2002;55:107–116. doi: 10.1016/s0166-3542(02)00013-x. [DOI] [PubMed] [Google Scholar]
- Mou H., Raj V.S., van Kuppeveld F.J.M., Rottier P.J.M., Haagmans B.L., Bosch B.J. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma K., Haagmans B.L., Hatano R., Raj V.S., Mou H., Iwata S., Dang N.H., Bosch B.J., Morimoto C. Inhibition of Middle East respiratory syndrome coronavirus infection by anti-CD26 monoclonal antibody. J. Virol. 2013;87:13892–13899. doi: 10.1128/JVI.02448-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaka A., Nakamura M., Nameki D., Kodama E., Uchiyama S., Nakamura S., Nakano H., Tamamura H., Kobayashi Y., Matsuoka M., Fujii N. Remodeling of gp41-C34 peptide leads to highly effective inhibitors of the fusion of HIV-1 with target cells. Angew. Chem. Int. Ed. Engl. 2002;41:2937–2940. doi: 10.1002/1521-3773(20020816)41:16<2937::AID-ANIE2937>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Pan C., Cai L., Lu H., Qi Z., Jiang S. Combinations of the first and next generations of human immunodeficiency virus (HIV) fusion inhibitors exhibit a highly potent synergistic effect against enfuvirtide-sensitive and -resistant HIV type 1 strains. J. Virol. 2009;83:7862–7872. doi: 10.1128/JVI.00168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., de Ruiter P.E., Metselaar H.J., Kwekkeboom J., de J.J., Tilanus H.W., Janssen H.L., van der Laan L.J. Mycophenolic acid augments interferon-stimulated gene expression and inhibits hepatitis C Virus infection in vitro and in vivo. Hematology. 2012;55:1673–1683. doi: 10.1002/hep.25562. [DOI] [PubMed] [Google Scholar]
- Pasut G., Veronese F.M. PEGylation for improving the effectiveness of therapeutic biomolecules. Drugs Today (Barc.) 2009;45:687–695. doi: 10.1358/dot.2009.45.9.1416421. [DOI] [PubMed] [Google Scholar]
- Perelson A.S., Essunger P., Cao Y., Vesanen M., Hurley A., Saksela K., Markowitz M., Ho D.D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- Prabakaran P., Zhu Z., Xiao X., Biragyn A., Dimitrov A.S., Broder C.C., Dimitrov D.S. Potent human monoclonal antibodies against SARS CoV, Nipah and Hendra viruses. Expert Opin. Biol. Ther. 2009;9:355–368. doi: 10.1517/14712590902763755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Shi W., Xue N., Pan C., Jing W., Liu K., Jiang S. Rationally designed anti-HIV peptides containing multifunctional domains as molecule probes for studying the mechanisms of action of the first and second generation HIV fusion inhibitors. J. Biol. Chem. 2008;283:30376–30384. doi: 10.1074/jbc.M804672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Dominguez S.R., Holmes K.V. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLOS. ONE. 2013;8:e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Smits S.L., Provacia L.B., van den Brand J.M.A., Wiersma L., Ouwendijk W.J.D., Bestebroer T.M., Spronken M.I., van Amerongen G., Rottier P.J.M., Fouchier R.A.M., Bosch B.J., Osterhaus A.D.M.E., Haagmans B.L. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4-mediated entry of the Middle East respiratory syndrome coronavirus. J. Virol. 2014;88:1834–1838. doi: 10.1128/JVI.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Research Group State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans. PLoS Curr. 2013 doi: 10.1371/currents.outbreaks.0bf719e352e7478f8ad85fa30127ddb8. 0bf719e352e7478f8ad85fa30127ddb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Ababneh M., Raj V.S., Meyer B., Eljarah A., Abutarbush S., Godeke G.J., Bestebroer T.M., Zutt I., Muller M.A., Bosch B.J., Rottier P.J., Osterhaus A.D., Drosten C., Haagmans B.L., Koopmans M.P. Middle East Respiratory Syndrome coronavirus (MERS-CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Euro. Surveill. 2013;18:14–20. doi: 10.2807/1560-7917.es2013.18.50.20662. [DOI] [PubMed] [Google Scholar]
- Reusken C.B.E.M., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B., Muth D., Raj V.S., Smits-De Vries L., Corman V.M., Drexler J.F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortazar-Schmidt C., Drosten C., Koopmans M.P.G. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D.D., Margolis D.M., Delaney M., Greene W.C., Hazuda D., Pomerantz R.J. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- Scott L.J., Lamb H.M. Palivizumab. Drugs. 1999;58:305–311. doi: 10.2165/00003495-199958020-00009. [DOI] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87:12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V., Ho D.D., Abdool K.Q. HIV/AIDS epidemiology, pathogenesis, prevention, and treatment. Lancet. 2006;368:489–504. doi: 10.1016/S0140-6736(06)69157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.C., Agnihothram S.S., Jiao Y., Stanhope J., Graham R.L., Peterson E.C., Avnir Y., Tallarico A.S., Sheehan J., Zhu Q., Baric R.S., Marasco W.A. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. USA. 2014;111:2018–2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D.M.E., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A.M. A newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473–e512. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese F.M. Peptide and protein PEGylation: a review of problems and solutions. Biomaterials. 2001;22:405–417. doi: 10.1016/s0142-9612(00)00193-9. [DOI] [PubMed] [Google Scholar]
- Veronese F.M., Pasut G. PEGylation, successful approach to drug delivery. Drug Discovery Today. 2005;10:1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- Wang E.X., Sun X., Qian Y., Zhao L.Q., Tien P., Gao G.F. Both heptad repeats of human respiratory syncytial virus fusion protein are potent inhibitors of viral fusion. Biochem. Biophys. Res. Commun. 2003;302:469–475. doi: 10.1016/s0006-291x(03)00197-9. [DOI] [PubMed] [Google Scholar]
- Wang N.S., Shi X.L., Jiang L.W., Zhang S.Y., Wang D.L., Tong P., Guo D.X., Fu L.L., Cui Y., Liu X., Arledge K.C., Chen Y.H., Zhang L.Q., Wang X.Q. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhou X., Debing Y., Chen K., Van Der Laan L.J., Neyts J., Janssen H.L., Metselaar H.J., Peppelenbosch M.P., Pan Q. Calcineurin inhibitors stimulate and mycophenolic acid inhibits replication of hepatitis E virus. Gastroenterology. 2014;146:1775–1783. doi: 10.1053/j.gastro.2014.02.036. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Takada A., Watanabe T., Ito H., Kida H., Kawaoka Y. Functional importance of the coiled-coil of the Ebola virus glycoprotein. J. Virol. 2000;74:10194–10201. doi: 10.1128/jvi.74.21.10194-10201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihofen W.A., Liu J.G., Reutter W., Saenger W., Fan H. Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface. J. Biol. Chem. 2004;279:43330–43335. doi: 10.1074/jbc.M405001200. [DOI] [PubMed] [Google Scholar]
- Wexler-Cohen Y., Shai Y. Demonstrating the C-terminal boundary of the HIV 1 fusion conformation in a dynamic ongoing fusion process and implication for fusion inhibition. FASEB J. 2007;21:3677–3684. doi: 10.1096/fj.07-8582com. [DOI] [PubMed] [Google Scholar]
- WHO . 2014. Middle East respiratory syndrome coronavirus (MERS-CoV)-update.http://www.who.int/csr/don/2014_07_23_mers/en/ [Google Scholar]
- Wild C.T., Shugars D.C., Greenwell T.K., McDanal C.B., Matthews T.J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Li K.S., Tsang A.K.Y.K.Y. Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Pathog. Dis. 2014;71:119–134. doi: 10.1038/emi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Wang Q., Yu F., Lu L., Jiang S.B. Synergistic effect resulting from combinations of a bifunctional HIV-1 antagonist with antiretroviral drugs. J. Acquir. Immune Defic. Syndr. 2014;67:1–6. doi: 10.1097/QAI.0000000000000265. [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhu J., Liu Y., Lou Z., Yuan F., Liu Y., Cole D.K., Ni L., Su N., Qin L., Li X., Bai Z., Bell J.I., Pang H., Tien P., Gao G.F., Rao Z. Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus. Biochemistry. 2004;43:14064–14071. doi: 10.1021/bi049101q. [DOI] [PubMed] [Google Scholar]
- Yang Y., Du L., Liu C., Wang L., Ma C., Tang J., Baric R.S., Jiang S., Li F. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. USA. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Li J., Zhang T., Wang X., Wang Y., Zhou Y., Liu J., Parekh H.K., Ho W. Mycophenolate mofetil inhibits hepatitis C virus replication in human hepatic cells. Virus Res. 2012;168:33–40. doi: 10.1016/j.virusres.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Du L., Ju T.W., Prabakaran P., Lau C.C., Lu L., Liu Q., Wang L., Feng Y., Wang Y., Zheng B.J., Yuen K.Y., Jiang S., Dimitrov D.S. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J. Virol. 2014;88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Wang E.X., Liu Y.F., Cao D.J., Jin N.Y., Zhang C.W.H., Bartlam M., Rao Z.H., Tien P., Gao G.F. Six-helix bundle assembly and characterization of heptad repeat regions from the F protein of Newcastle disease virus. J. Gen. Virol. 2002;83:623–629. doi: 10.1099/0022-1317-83-3-623. [DOI] [PubMed] [Google Scholar]
- Yu X., Lu L., Cai L., Tong P., Tan S., Zou P., Meng F., Chen Y.H., Jiang S. Mutations of Gln64 in the HIV-1 gp41 N-terminal heptad repeat render viruses resistant to peptide HIV fusion inhibitors targeting the gp41 pocket. J. Virol. 2012;86:589–593. doi: 10.1128/JVI.05066-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Yi L., Chen J., Qu X., Qing T., Rao X., Jiang P., Hu J., Xiong Z., Nie Y., Shi X., Wang W., Ling C., Yin X., Fan K., Lai L., Ding M., Deng H. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem. Biophys. Res. Commun. 2004;319:746–752. doi: 10.1016/j.bbrc.2004.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale M.J., Jr., Baric R.S., Enjuanes L., Gallagher T., McCray P.B., Jr., Perlman S. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. USA. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B.J., Guan Y., He M.L., Sun H.Z., Du L.Y., Zheng Y., Wong K.L., Chen H.L., Chen Y., Lu L.Y., Tanner J.A., Watt R.M., Niccolai N., Bernini A., Spiga O., Woo P.C.Y., Kung H.F., Yuen K.Y., Huang J.D. Synthetic peptides outside the spike protein heptad repeat regions as potent inhibitors of SARS-associated coronavirus. Antivir. Ther. 2005;10:393–403. [PubMed] [Google Scholar]
- Zhong J., Rao X., Rajagopalan S. An emerging role of dipeptidyl peptidase 4 (DPP4) beyond glucose control: potential implications in cardiovascular disease. Atherosclerosis. 2013;226:305–314. doi: 10.1016/j.atherosclerosis.2012.09.012. [DOI] [PubMed] [Google Scholar]


