Abstract
The genetic diversity of the host is believed to be the key of the diversity in the clinical presentation of bronchiolitis. The aim of this study was to determine whether the known rs12979860 and rs8099917 single nucleotide polymorphisms (SNPs) in interleukin (IL)28B region, influence clinical features and natural history of bronchiolitis. Both SNPs showed no significant association with the risk of hospitalization for respiratory syncytial virus (RSV), viral load, disease severity, and other clinical features of patients. Interestingly infants carrying IL28B rs12979860 TT genotype had lower age at hospital admission than that of infants carrying CC/CT genotypes. Overall our results indicate that both IL28B SNPs had no impact on the clinical course of bronchiolitis with the only exception of the IL28B rs12979860 SNP which increased the risk of hospitalization for bronchiolitis at early age.
Keywords: RSV, IFN, IL28B, Bronchiolitis, Viral load, Respiratory virus
It is well established that the clinical spectrum of respiratory syncytial virus (RSV)-associated bronchiolitis in infants is extremely variable, ranging from mild upper respiratory symptoms to severe respiratory distress and, occasionally, death (Wainwright, 2010). There are several known risk factors associated with severe bronchiolitis, such as: young age (<6 months), premature birth (<35 weeks of gestation), immunodeficiency or immunosuppression status, congenital heart or chronic lung disease. However, most seriously affected infants have no evident risk factors (Vicencio, 2010). Thus it is likely that other factors, such as immune response and genetic heterogeneity of the host, together with well-known viral risk factors, contribute to RSV disease severity (Amanatidou et al., 2009). Recent genome-wide association studies have identified single nucleotide polymorphisms (SNPs) near the interleukin (IL) 28B gene [namely rs12979860 (C/T) and rs8099917 (T/G)], which are strongly associated with spontaneous and treatment-induced clearance in hepatitis C virus (HCV) infection (Ge et al., 2009, Suppiah et al., 2009, Tanaka et al., 2009, Thomas et al., 2009). The rs12979860 is 3 kb upstream of IL28B gene, whereas rs8099917 is located 8.9 kb downstream from IL28B and 16 kb upstream of IL28A (Ge et al., 2009, Suppiah et al., 2009).
The IL28B gene encodes the IL28B protein, also known as interferon lambda3 (IFN lambda3), an innate immune response cytokine with antiviral activity that is distantly related to type I IFN and the IL-10 family (Gad et al., 2010). The biological mechanisms involved in the association of these IL28B SNPs over HCV control are still unclear. Many papers have reported conflicting data on the correlation of these SNPs with IL28 mRNA levels in both PBMC and liver of HCV positive patients (Abe et al., 2011a, Honda et al., 2010, Suppiah et al., 2009, Tanaka et al., 2009, Urban et al., 2010). Furthermore it has been shown that IL28B rs12979860 genotype is strongly associated with intrahepatic interferon stimulated genes (ISGs) expression, with the poor-response (non-CC) IL28B genotypes exhibiting higher ISG levels compared to patients with the favorable CC IL28B genotype (Dill et al., 2011, Honda et al., 2010, Lagging et al., 2011, Urban et al., 2010). The clinical significance of these IL28B SNPs have not been characterized in RSV or other respiratory viral infections. Interestingly recent studies have highlighted the importance of IFN lambda in the protection against viral invaders in the respiratory tract (Mordstein et al., 2010, Jewell et al., 2010). In addition our previous studies have demonstrated that in infants with RSV-associated bronchiolitis there is a strong upregulation of ISGs and pattern recognition receptors and that the severity of illness is inversely related to the level of expression of ISGs (Scagnolari et al., 2007, Scagnolari et al., 2009).
Given the shared pathways for receptor signalling and biological effectors function of type I IFN and IFN lambda and the importance of innate immune response in RSV infections, we evaluated whether the IL28B rs12979860 and rs8099917 SNPs were associated with RSV susceptibility, RSV load, disease severity and other demographic and clinical parameters in infants suffering from bronchiolitis. Specifically 138 infants (69 tested positive only for RSV plus 69 tested negative for other respiratory viruses) were selected for IL28B SNPs genotyping from a total of 528 infants for whom both nasopharingeal washings (NPW) and peripheral blood mononuclear cells (PBMC) samples were collected. These infants were admitted because suffering from respiratory diseases over four successive winter seasons (2006–2010) to the Paediatric Department of Policlinico Umberto I Hospital, later clinically diagnosed as bronchiolitis
The study was approved by the institutional review board at Sapienza University of Rome and written informed consent was obtained from the children's parents. Bronchiolitis was diagnosed clinically according to the presence of a history of upper respiratory tract infection followed by acute onset of respiratory distress with cough, tachypnea, retractions and bilateral crackles on auscultation (having wheezing alone was not considered sufficient for inclusion in the study) (Midulla et al., 2010). Infants were grouped according to the worst severity of bronchiolitis experienced during their admission as previously described (Papoff et al., 2011). In particular the least severe group (gr.0) required only conservative treatment (no need for supplemental oxygen or intravenous fluids); the mild severity group (gr1) needed intravenous fluids or oxygen treatment or both for less than 12 h; the moderate severity group (gr.2) needed oxygen for more than 12 h without ventilatory support and intravenous fluids; and the severe group (gr.3) needed either mechanical ventilation or noninvasive respiratory support. In particular NPW and PBMC were collected from each infants in the first 24/48 h after admission to hospital and an aliquot of NPW was tested for the detection of fourteen respiratory viruses (RSV, influenza A and B, coronavirus OC43, 229 E, NL63, HKU1, metapneumovirus, adenovirus, rhinovirus and parainfluenza 1–3, Bocavirus) by using a panel of reverse transcription (RT) PCR or nested PCR assays, some in a multiplex format, as previously described (Pierangeli et al., 2007, Pierangeli et al., 2008). PBMCs were separated using Ficoll–Hypaque gradient sedimentation and frozen at–80 °C for subsequent IL28 SNPs analysis. The infants were divided into two groups on the basis of the detection of single RSV infection (n = 69) or the failure to detect any of the 14 tested respiratory viruses (n = 69) in their NPW and matched by age and gender. Demographic and clinical characteristics of the 2 patient groups are shown in Table 1 . McNemar's test and paired t-test were used to compare respectively the categorical and the continuous outcomes between patient groups. There were no differences in the age at hospital admission or gestational age, weight at birth or at hospital admission, numbers of neutrophils, lymphocytes, eosinophils or platelets, levels of C-reactive protein and duration of illness between the RSV positive and the virus-negative groups (p > 0.5). Number of infants with moderate or severe bronchiolitis (gr. 2 + 3) was not significantly different between RSV positive group and uninfected infants (Mcnemar test, p = 0.38).
Table 1.
Comparison of the clinical and demographic characteristics between virus-negative or RSV positive infants hospitalized for bronchiolitis.
| Item* | Virus-negative group*** | RSV single infection positive group |
|---|---|---|
| Gender | ||
| Female | 34/69 (49.3) | 34/69 (49.3) |
| Male | 35/69 (50.7) | 35/69 (50.7) |
| Gestational age (weeks) | 38.9 (0.2) | 38.4 (0.2) |
| Birth weight (kg) | 3.2 (0.1) | 3.1 (0.1) |
| Age at hospital admission (days) | 62.0 (4.9) | 63.0 (5.1) |
| Weight at hospital admission (kg) | 5.0 (0.2) | 4.8 (0.2) |
| Severity of bronchiolitis** | ||
| Group 0 + 1(%) | 55/69 (79.7) | 49/69 (71.0) |
| Group 2 + 3 (%) | 14/69 (20.3) | 20/69 (29.0) |
| Fever | 22/69 (31.9) | 22/69 (31.9) |
| Days in hospital (days) | 5.3 (0.3) | 5.8 (0.4) |
| C-reactive protein (mg/dl) | 1.1 (0.2) | 1.5 (0.4) |
| Neutrophils (cells/μl) | 4121 (321) | 3970 (373) |
| Lymphocytes (cells/μl) | 5124 (366) | 4859 (213) |
| Eosinophils (cells/μl) | 115 (15) | 95 (15) |
| Platelets (no./μl) | 452,815 (19350) | 462,708 (21678) |
The number of patients of each gender are shown, with percentages in parentheses, and for other variables the means are shown with SE in parentheses. McNemar's test and gender paired t-test was used to compare respectively the categorical or continuous variables between patient group.
Infants were grouped according to the worst severity of bronchiolitis as previously described (Papoff et al., 2011)
Infants who tested negative for the detection of fourteen respiratory viruses (RSV, influenza A and B, coronavirus OC43, 229 E, NL63, HKU1, metapneumovirus, adenovirus, rhinovirus and parainfluenza 1–3, Bocavirus) by using a panel of reverse transcription (RT) PCR or nested PCR assays, some in a multiplex format (Pierangeli et al., 2007, Pierangeli et al., 2008).
Next, the frequencies of the IL-28B SNPs were compared between RSV positive and uninfected infants by using a newly developed sensitive pyrosequencing screening assays. Briefly genomic DNA was isolated from PBMC using an EZ1® DNA blood mini kit (Qiagen, Milan, Italy) in an EZ1 automatic extractor, following the manufacturer's instructions. The rs12979860 and rs8099917 SNPs were analyzed by SNP mode pyrosequencing analysis in a PyroMark™ Q96 ID instrument (Qiagen, Italy). Specific amplification and sequencing primers for each SNP were created by PyroMark PSQ Assay Design v.1.0.6 (Biotage, Uppsala, Sweden). Amplification primers were designed to match a few bases upstream of the specific SNP in order to obtain an easy and reliable analysis by sequencing only a few bases. Results indicated that the frequency of TT (rs12979860) and GG (rs8099917) IL28B genotypes was 8.8% and 2.9% respectively in the RSV group, while that was 11.8%, and 2.9% respectively in the no virus-detected group (Fig. 1 ). There were not significant differences in the IL28B rs12979860 or IL28B rs8099917 genotype frequencies between the two groups (McNemar test, p > 0.05). Therefore our results indicate that genetic variations around IL28 are not associated with the risk of hospitalization for RSV infection during early life and suggest that the effect of these SNPs, though strongly associated to the outcome of HCV infection (Balagopal et al., 2010, McHutchison, 2011, Rauch et al., 2010, Thomas et al., 2009), cannot be extended to this specific respiratory viral infection. In addition, recent studies had failed to found such association also in other viral infection in which IFN play an important role. To this regards Rallon and co-authors have reported that IL28B SNP (rs12979860) had no role in the protection either against HIV acquisition or from HIV disease progression (Rallon et al., 2011). Furthermore no influence of IL28B polymorphisms (rs12979860) on the outcome of hepatitis B virus infection has been also demonstrated (Martin et al., 2010).
Fig. 1.
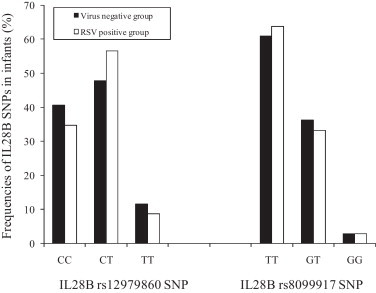
Comparison of the frequencies of interleukin (IL) 28B rs12979860 and rs8099917 single nucleotide polymorphisms (SNPs) between infants with single respiratory syncytial virus (RSV) infection and infants who tested negative for the detection of fourteen respiratory viruses (RSV, influenza A and B, coronavirus OC43, 229 E, NL63, HKU1, metapneumovirus, adenovirus, rhinovirus and parainfluenza 1–3, Bocavirus). RSV positive group vs virus negative group: IL28B rs12979860 SNP p = 0.597 (CC vs CT + TT); p = 0.754 (TT vs CT + CC); IL28B rs8099917 SNP p = 0.851 (TT vs TG + GG); p = 1.00 (GG vs GT + TT) by using McNemar test for paired samples.
Because controversy exists about the impact of rs12979860 and rs8099917 IL28B SNPs on HCV load (Del Campo et al., 2010, Labarga et al., 2011, Thompson et al., 2010), it was examined whether IL28B SNPs affects RSV load. A TaqMan-based real-time PCR technique for RSV RNA quantification was performed on all NPW specimens with positive RT-PCR results for RSV as previously described (Scagnolari et al., 2009). Viral load values were log10 transformed for analysis and data was expressed as the log10 number of RSV copies per ml of NPW. Titers of RSV in NPW varied widely, ranging from 1.7 to 8.9 RNA copies/ml. As show in Fig. 2 , none of these SNPs correlated with the RSV-RNA levels stratified in two groups according to the RSV-RNA levels below or above the 50th percentile of 5.8 RSV-RNA copies/ml. Unfortunately in this study we could not determined the impact of these IL28B SNPs on RSV load decline because we collected NPW samples from infants with bronchiolitis only at the time of the admission and not on hospital discharge. Having established that IL28B rs12979860 and rs8099917 SNPs had no affects on the RSV susceptibility and on the viral load in infants with bronchiolitis, we evaluated whether these SNPs were associated with specific demographic or clinical characteristics of all infants included in this study independently of the presence or absence of RSV infection. Chi square test or t-test was used to find such associations respectively for categorical or continuous variables. Results are shown in Table 2 . It can been seen that the distribution of demographic characteristics and clinical factors was comparable among infants with different IL28B rs12979860 or rs8099917 genotypes (chi square or t-test, p > 0.05). The only exception was represented by the significantly relation between the age of infants at hospital admission and IL28B rs12979860 SNP. In particular infants carrying IL28B rs12979860 TT genotype had lower age at hospital admission that the infants carrying CC/CT genotypes. On the contrary infants with IL28B rs12979860 CC genotype had higher age at hospital admission compared to those with CT/TT genotypes.
Fig. 2.
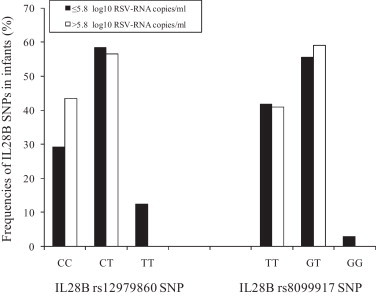
Analysis of the relationship between IL28B single nucleotide polymorphisms (SNPs) (rs12979860 and rs8099917) and levels of viral load in single respiratory syncytial virus (RSV) positive infants. RSV load (≤5.8 vs > 5.8 log10 copies of RSV-RNA/ml): IL28B rs12979860 SNP p = 0.51 (CC vs CT + TT); p = 0.41(TT vs CT + CC); IL28B rs8099917 SNP p = 0.89 (TT vs TG + GG); p = 0.82 (GG vs GT + TT) by using McNemar test for paired samples.
Table 2.
Frequencies of IL28B rs12979860 and rs8099917 SNP in 138 infants hospitalized for bronchiolitis.
| Gender* |
Gestational age (weeks) | Birth weight (kg) | Age at hospital admission (days) | Weight at hospital admission (kg) | Severity of bronchiolitis** |
Fever | Days in hospital (days) | CRP (mg/dl) | Neutrophils (cells/μl) | Lymphocytes (cells/μl) | Eosinophils (cells/μl) | Platelets (n̊/μl) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Group 0–1 | Group 2–3 | ||||||||||||
| IL28B rs12979860 | |||||||||||||||
| CC | 29 | 23 | 38.7 (0.2) | 3.3 (0.1) | 73.2 (6.9) | 5.3 (0.2) | 42 | 10 | 14 | 5.5 (0.4) | 1.5 (0.4) | 4019.9 (374.8) | 4882.2 (281.1) | 86.7 (17.3) | 476921.6 (24713.9) |
| CT + TT | 40 | 46 | 38.6 (0.2) | 3.1 (0.1) | 55.9 (3.6) | 4.7 (0.1) | 58 | 23 | 30 | 5.7 (0.3) | 1.2 (0.2) | 4104.3 (322.4) | 5037.1 (293.4) | 115.3 (13.9) | 446679 (17282.5) |
| p-value | 0.297 | 0.609 | 0.068 | 0.029 | 0.008 | 0.241 | 0.329 | 0.682 | 0.515 | 0.868 | 0.723 | 0.203 | 0.304 | ||
| TT | 5 | 9 | 39.1 (0.4) | 3.2 (0.1) | 45.7 (4.9) | 4.5 (0.3) | 10 | 4 | 7 | 6.1 (0.4) | 1.1 (0.4) | 4194.5 (915.3) | 4511.4 (765.2) | 128.1 (26.2) | 508769.2 (44466.8) |
| CT + CC | 64 | 60 | 38.6 (0.2) | 3.1 (0.1) | 64.4 (3.8) | 4.9 (0.1) | 90 | 29 | 37 | 5.5 (0.2) | 1.4 (0.2) | 4058.3 (253.1) | 5032.6 (218.4) | 101.8 (11.7) | 452857.1 (15040.4) |
| p-value | 0.277 | 0.326 | 0.773 | 0.005 | 0.197 | 0.719 | 0.165 | 0.391 | 0.734 | 0.866 | 0.453 | 0.461 | 0.245 | ||
| IL28B rs8099917 | |||||||||||||||
| GG | 2 | 2 | 39.3 (0.5) | 2.7 (0.2) | 50 (12) | 4.1 (0.5) | 3 | 1 | 2 | 6.3 (1.1) | 1.6 (1.1) | 2888.8 (550.1) | 5228.3 (2522) | 212.4 (36.9) | 386500 (61582.6) |
| GT + TT | 67 | 67 | 38.6 (0.2) | 3.2 (0) | 62.8 (3.6) | 4.9 (0.1) | 97 | 32 | 42 | 5.6 (0.2) | 1.3 (0.2) | 4108.5 (251.1) | 4970.9 (206.4) | 101.2 (11.0) | 460609 (14578.1) |
| p-value | 1.00 | 0.48 | 0.17 | 0.54 | 0.22 | 1.00 | 0.60 | 0.60 | 0.80 | 0.40 | 0.84 | 0.08 | 0.38 | ||
| TT | 48 | 38 | 38.6 (0.2) | 3.2 (0.1) | 63.9 (4.8) | 4.9 (0.1) | 66 | 20 | 26 | 5.6 (0.3) | 1.4 (0.3) | 3794.3 (264.9) | 4830.5 (218.3) | 101.4 (14.2) | 463141.2 (17741.6) |
| GT + GG | 21 | 31 | 38.6 (0.3) | 3.1 (0.1) | 60.1 (4.6) | 4.8 (0.2) | 34 | 13 | 18 | 5.5 (0.4) | 1.2 (0.3) | 4545.2 (480.0) | 5230.2 (432.0) | 109.6 (16.8) | 449723.4 (24224.2) |
| p-value | 0.11 | 0.95 | 0.76 | 0.61 | 0.72 | 0.67 | 0.57 | 0.82 | 0.72 | 0.14 | 0.36 | 0.72 | 0.65 | ||
The number of patients of each gender are shown and for other variables the means are shown with SE in parentheses. Chi square and t-test were used to compare respectively the categorical and the continuous outcome between patient group.
Infants were grouped according to the worst severity of bronchiolitis as previously described (Papoff et al., 2011). Significant correlations are highlighted in boldface.
The biological mechanism underlying the association of the IL28B SNPs with the age of hospitalized infants with bronchiolitis is unknown and was not investigated here. However it could be hypothesized that IL28 SNPs affects levels of expression of IL-28B or the activation of some IFN-related pathways, which indirectly increased in infants the risk of development of bronchiolitis. Our previous study showed an inverse correlation between the severity of illness and the airway levels of ISGs (Scagnolari et al., 2007); even so an influence of IL28BSNPs on the severity of bronchiolitis has not been found. Furthermore an association between levels of ISGs and IL28B SNPs (rs12979860 and rs8099917) in HCV infected has been reported in previous papers, whereas other studies have failed to detect such an association (Abe et al., 2011b, Balagopal et al., 2010, Urban et al., 2010). These findings make more evident the complexity in the understanding the biological significance of rs12979860 and rs8099917 IL28B SNPs and suggest the importance of evaluating the pathway regulated by type III IFN in infants suffering from bronchiolitis.
In conclusion this is the first study to our knowledge that has investigated the role of rs12979860 and rs8099917 IL-28B single nucleotide polymorphisms in one of the most important paediatric acute respiratory diseases. Our results clearly suggest that genetic variations around IL28 B region do not affect the risk of hospitalization either for RSV infection or for severe bronchiolitis. In addition this study indicates that rs12979860 and rs8099917 SNPs are also not associated with any of the demographical or clinical parameters of infants with bronchiolitis. The only exception was the age of appearance of respiratory disease which was lower for infants with TT rs12979860 genotype. Further larger studies are needed to evaluate the biological mechanisms underlying the association of the IL28B rs12979860 SNP with the age of hospitalized infants with bronchiolitis and to determine the association between both IL28B SNPs (rs12979860 and rs8099917) and RSV load decline.
Acknowledgements
This work was supported by grants to G.A from Pasteur Institute (Cenci Bolognetti Foundation; title of the project: “Molecular characterization of viruses causing bronchiolitis and study of viral and host factors affecting Type I IFN antiviral response induced by respiratory viruses”), and to A.P. from “Sapienza” Università di Roma (Fondi ricerche Universitarie) and Italian Ministry of Health (Ricerca Finalizzata Conv. No. 88).
References
- Abe H., Hayes C.N., Ochi H., Tsuge M., Miki D., Hiraga N., Imamura M., Takahashi S., Kubo M., Nakamura Y., Kamatani N., Chayama K. Inverse association of IL28B genotype and liver mRNA expression of genes promoting or suppressing antiviral state. J. Med. Virol. 2011;83:1597–1607. doi: 10.1002/jmv.22158. [DOI] [PubMed] [Google Scholar]
- Abe H., Hayes C.N., Ochi H., Maekawa T., Tsuge M., Miki D., Mitsui F., Hiraga N., Imamura M., Takahashi S., Kubo M., Nakamura Y., Chayama K. IL28 variation affects expression of interferon stimulated genes and peg-interferon and ribavirin therapy. J. Hepatol. 2011;54:1094–1101. doi: 10.1016/j.jhep.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Amanatidou V., Apostolakis S., Spandidos D.A. Genetic diversity of the host and severe respiratory syncytial virus-induced lower respiratory tract infection. Pediatr. Infect. Dis. J. 2009;28:135–140. doi: 10.1097/INF.0b013e31818c8d17. [DOI] [PubMed] [Google Scholar]
- Balagopal A., Thomas D.L., Thio C.L. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139:1865–1876. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo J.A., Maraver Zamora M., Ramýrez-Lorca R., Pardo-Yules B., Sáez M.E., Real L.M. IL28B polymorphism predicts sustained virological response in hepatitis C but is not associated with fibrosis or viral load. Hepatology. 2010;52:748A. [Google Scholar]
- Dill M.T., Duong F.H., Vogt J.E., Bibert S., Bochud P.Y., Terracciano L., Papassotiropoulos A., Roth V., Heim M.H. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology. 2011;140:1021–1031. doi: 10.1053/j.gastro.2010.11.039. [DOI] [PubMed] [Google Scholar]
- Gad H.H., Hamming O.J., Hartmann R. The structure of human interferon lambda and what it has taught us. J. Interferon Cytokine Res. 2010;30:565–571. doi: 10.1089/jir.2010.0062. [DOI] [PubMed] [Google Scholar]
- Ge D., Fellay J., Thompson A.J., Simon J.S., Shianna K.V., Urban T.J., Heinzen E.L., Qiu P., Bertelsen A.H., Muir A.J., Sulkowski M., McHutchison J.G., Goldstein D.B. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- Honda M., Sakai A., Yamashita T., Nakamoto Y., Mizukoshi E., Sakai Y., Yamashita T., Nakamura M., Shirasaki T., Horimoto K., Tanaka Y., Tokunaga K., Mizokami M., Kaneko S., Hokuriku Liver Study Group Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- Jewell N.A., Cline T., Mertz S.E., Smirnov S.V., Flaño E., Schindler C., Grieves J.L., Durbin R.K., Kotenko S.V., Durbin J.E. Lambda interferon is the predominant interferon induced by influenza A virus infection in vivo. J. Virol. 2010;84:11515–11522. doi: 10.1128/JVI.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarga P., Soriano V., Caruz A., Poveda E., Di Lello F., Hernandez-Quero J., Moreno S., Bernal E., Miró J.M., Leal M., Gutierrez F., Portilla J., Pineda J.A., CoRIS Association between IL28B gene polymorphisms and plasma HCV-RNA levels in HIV/HCV-co-infected patients. AIDS. 2011;25:761–766. doi: 10.1097/QAD.0b013e32834488e7. [DOI] [PubMed] [Google Scholar]
- Lagging M., Askarieh G., Negro F., Bibert S., Söderholm J., Westin J., Lindh M., Romero A., Missale G., Ferrari C., Neumann A.U., Pawlotsky J.M., Haagmans B.L., Zeuzem S., Bochud P.Y., Hellstrand K., DITTO-HCV Study Group Response prediction in chronic hepatitis C by assessment of IP-10 and IL28B-related single nucleotide polymorphisms. PLoS One. 2011;6:e17232. doi: 10.1371/journal.pone.0017232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.P., Qi Y., Goedert J.J., Hussain S.K., Kirk G.D., Hoots W.K., Buchbinder S., Carrington M., Thio C.L. IL28B polymorphism does not determine outcomes of hepatitis B virus or HIV infection. J. Infect. Dis. 2010;202:1749–1753. doi: 10.1086/657146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHutchison J.G. The role of genetic markers in hepatitis C virus therapy: a major step for individualized care. Liver Int. 2011;1:29–35. doi: 10.1111/j.1478-3231.2010.02389.x. [DOI] [PubMed] [Google Scholar]
- Midulla F., Scagnolari C., Bonci E., Pierangeli A., Antonelli G., De Angelis D., Berardi R., Moretti C. Respiratory syncytial virus, human bocavirus and rhinovirus bronchiolitis in infants. Arch. Dis. Child. 2010;95:35–41. doi: 10.1136/adc.2008.153361. [DOI] [PubMed] [Google Scholar]
- Mordstein M., Neugebauer E., Ditt V., Jessen B., Rieger T., Falcone V., Sorgeloos F., Ehl S., Mayer D., Kochs G., Schwemmle M., Günther S., Drosten C., Michiels T., Staeheli P. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoff P., Moretti C., Cangiano G., Bonci E., Roggini M., Pierangeli A., Scagnolari C., Antonelli G., Midulla F. Incidence and predisposing factors for severe disease in previously healthy term infants experiencing their first episode of bronchiolitis. Acta Paediatr. 2011;100:17–23. doi: 10.1111/j.1651-2227.2011.02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierangeli A., Gentile M., Di Marco P., Pagnotti P., Scagnolari C., Trombetti S., Lo Russo L., Tromba V., Moretti C., Midulla F., Antonelli G. Detection and typing by molecular techniques of respiratory viruses in children hospitalized for acute respiratory infection in Rome, Italy. J. Med. Virol. 2007;79:463–468. doi: 10.1002/jmv.20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierangeli A., Scagnolari C., Trombetti S., Grossi R., Battaglia M., Moretti C., Midulla F., Antonelli G. Human bocavirus infection in hospitalized children in Italy. Influenza Other Respir. Viruses. 2008;2:175–179. doi: 10.1111/j.1750-2659.2008.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallon N.I., Restrepo C., Naggie S., Lopez M., Del Romero J., Goldstein D., McHutchison J., Soriano V., Benito J.M. Interleukin-28B gene polymorphisms do not influence the susceptibility to HIV-infection or CD4 cell decline. AIDS. 2011;25:269–271. doi: 10.1097/QAD.0b013e328341b84e. [DOI] [PubMed] [Google Scholar]
- Rauch A., Kutalik Z., Descombes P., Cai T., Di Iulio J., Mueller T., Bochud M., Battegay M., Bernasconi E., Borovicka J., Colombo S., Cerny A., Dufour J.F., Furrer H., Günthard H.F., Heim M., Hirschel B., Malinverni R., Moradpour D., Müllhaupt B., Witteck A., Beckmann J.S., Berg T., Bergmann S., Negro F., Telenti A., Bochud P.Y., Swiss Hepatitis C., Cohort Study Swiss HIV Cohort Study Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- Scagnolari C., Midulla F., Pierangeli A., Moretti C., Bonci E., Berardi R., De Angelis D., Selvaggi C., Di Marco P., Girardi E., Antonelli G. Gene expression of nucleic acid-sensing pattern recognition receptors in children hospitalized for respiratory syncytial virus-associated acute bronchiolitis. Clin. Vaccine Immunol. 2009;16:816–823. doi: 10.1128/CVI.00445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scagnolari C., Midulla F., Trombetti S., Pierangeli A., Tromba V., Grossi R., Di Marco P., Dianzani C., Girardi E., Antonelli G. Upregulation of interferon-induced genes in infants with virus-associated acute bronchiolitis. Exp. Biol. Med. (Maywood) 2007;232:1355–1359. doi: 10.3181/0705-BC-124. [DOI] [PubMed] [Google Scholar]
- Suppiah V., Moldovan M., Ahlenstiel G., Berg T., Weltman M., Abate M.L., Bassendine M., Spengler U., Dore G.J., Powell E., Riordan S., Sheridan D., Smedile A., Fragomeli V., Müller T., Bahlo M., Stewart G.J., Booth D.R., George J. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., Hige S., Ito Y., Mita E., Tanaka E., Mochida S., Murawaki Y., Honda M., Sakai A., Hiasa Y., Nishiguchi S., Koike A., Sakaida I., Imamura M., Ito K., Yano K., Asaki N., Sugauchi F., Izumi N., Tokunaga K., Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- Thomas D.L., Thio C.L., Martin M.P., Qi Y., Ge D., O’Huigin C., Kidd J., Kidd K., Khakoo S.I., Alexander G., Goedert J.J., Kirk G.D., Donfield S.M., Rosen H.R., Tobler L.H., Busch M.P., McHutchison J.G., Goldstein D.B., Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.J., Muir A.J., Sulkowski M.S., Ge D., Fellay J., Shianna K.V., Urban T., Afdhal N.H., Jacobson I.M., Esteban R., Poordad F., Lawitz E.J., McCone J., Shiffman M.L., Galler G.W., Lee W.M., Reindollar R., King J.W., Kwo P.Y., Ghalib R.H., Freilich B., Nyberg L.M., Zeuzem S., Poynard T., Vock D.M., Pieper K.S., Patel K., Tillmann H.L., Noviello S., Koury K., Pedicone L.D., Brass C.A., Albrecht J.K., Goldstein D.B., McHutchison J.G. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–129. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Urban T.J., Thompson A.J., Bradrick S.S., Fellay J., Schuppan D., Cronin K.D., Hong L., McKenzie A., Patel K., Shianna K.V., McHutchison J.G., Goldstein D.B., Afdhal N. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicencio A.G. Susceptibility to bronchiolitis in infants. Curr. Opin. Pediatr. 2010;22:302–306. doi: 10.1097/MOP.0b013e32833797f9. [DOI] [PubMed] [Google Scholar]
- Wainwright C. Acute viral bronchiolitis in children – a very common condition with few therapeutic options. Paediatr. Respir. Rev. 2010;11:39–45. doi: 10.1016/j.prrv.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


