Highlights
-
•
This retrospective reviews 30 years of research on the retroviral nucleocapsid protein (NC) focusing on HIV-1 NC.
-
•
Originally considered as a non-specific nucleic-acid binding protein, NC has seminal functions in virus replication.
-
•
Indeed NC turns out to be a all-in-one viral protein that chaperones viral DNA synthesis and integration, and virus formation.
-
•
As a chaperone NC provides assistance to genetic recombination thus allowing the virus to escape the immune response and antiretroviral therapies against HIV-1.
Abbreviations: RV, retroviruses; CA, capsid; MA, matrix; NC, nucleocapsid; RT, reverse transcriptase; IN, integrase; PBS, primer binding site; gRNA, genomic RNA; DLS, dimer linkage structure; DIS, dimer initiation sequence; MuLV, murine leukemia virus; MoMuLV, Moloney MuLV; ASLV, avian sarcoma leukosis virus; BLV, bovine leukemia virus; HIV, SIV, human and simian immunodeficiency viruses; Ty3, retrotransposon 3 from yeast SC; ZF, zinc finger; CCHC motif, ZF of the form Cys-Cys-His-Cys; 3D, three dimensional; LTR, long terminal repeat; UV, ultraviolet light; PDB, Protein Data Bank; IDP, instrinsically disordered protein; NABP, nucleic acid binding protein
Keywords: Retrovirus, Nucleocapsid protein, Zinc fingers, Viral DNA synthesis, Virus formation, Intrinsically disordered protein
Abstract
This review aims at briefly presenting a retrospect on the retroviral nucleocapsid protein (NC), from an unspecific nucleic acid binding protein (NABP) to an all-in-one viral protein with multiple key functions in the early and late phases of the retrovirus replication cycle, notably reverse transcription of the genomic RNA and viral DNA integration into the host genome, and selection of the genomic RNA together with the initial steps of virus morphogenesis. In this context we will discuss the notion that NC protein has a flexible conformation and is thus a member of the growing family of intrinsically disordered proteins (IDPs) where disorder may account, at least in part, for its function as a nucleic acid (NA) chaperone and possibly as a protein chaperone vis-à-vis the viral DNA polymerase during reverse transcription. Lastly, we will briefly review the development of new anti-retroviral/AIDS compounds targeting HIV-1 NC because it represents an ideal target due to its multiple roles in the early and late phases of virus replication and its high degree of conservation.
1. Forewords
During the past hundred years or so investigations on retroviruses have triggered seminal discoveries in biology and medicine such as that of oncogenes and their implications in cancer (Bishop, 1983, Brugge and Erikson, 1977, Hanafusa, 1969, Hu et al., 2003a, Hu and Temin, 1990, Huebner and Todaro, 1969) and the fact that retroelements constitute a large fraction of all eukaryotic genomes acting as a driving force in genome plasticity and evolution (Baltimore, 1970, Garfinkel et al., 1985, Hansen et al., 1988).
In this context, our understanding of the structure and functions of the retroviral nucleocapsid protein (i.e. NC) has come a long way since the first descriptions of oncoretroviruses and the oncoviral nucleoprotein (Ellermann and Bang, 1908, Rous, 1910, Davis and Rueckert, 1972). Initially studies on the retroviral NC focused on that of alpharetroviruses and gammaretroviruses, namely avian sarcoma leukosis viruses (ASLV) and the murine leukemia viruses (MuLV), respectively. This has fueled a large number of multidisciplinary studies on HIV-1 NC structure and functions as well as on NC of ancient yeast retroelements (Gabus et al., 1998, Garfinkel et al., 1985, Hansen et al., 1988) and on NC-like proteins encoded by Flaviviruses as well as other RNA viruses (Cristofari et al., 2004, Mir and Panganiban, 2005).
The retroviral NC story began with the isolation of large ribonucleoprotein complexes (RNPs) (Davis and Rueckert, 1972) from purified viral particles of ASLV and MoMuLV, which were able to support viral DNA synthesis. These viral RNP complexes, consisting of the 60–70S genomic RNA dimer coated by about 2000 NC molecules, represent the most stable inner structure of the viral particle, with a circular chromatin-like conformation (Chen et al., 1980, Pager et al., 1994). These viral RNPs or nucleocores also contain molecules of the viral reverse transcriptase (RT) and integrase (IN) enzymes together with molecules of cellular tRNAs and ribosomal RNA (Chen et al., 1980, Darlix et al., 1995, Dickson et al., 1985).
The hallmark of all retroviral NC proteins, with the exception of Foamy viruses (Stenbak and Linial, 2004), is the presence of one or two small highly conserved CX2CX4HX4C (CCHC) motifs called zinc fingers (ZFs) or zinc knuckles that bind zinc ions with high affinity (Berg, 1986, Mely et al., 1991, Mely et al., 1996, Summers et al., 1990, Summers et al., 1992), and that are flanked by unstructured basic sequences. In retrospect, some 38 years ago NC of alpha- and gammaretroviruses was first isolated from purified virions and considered to be an unspecific nucleic acid binding protein (NABP) with some preference for single stranded sequences within structured RNA domains (Davis et al., 1976).
Thirty years of multidisciplinary research, notably on HIV-1, radically changed our views since NC was discovered to be a potent nucleic acid chaperoning factor exerting key functions throughout the virus replication cycle. In addition to being a viral structural protein, NC assists the viral RT enzyme throughout viral DNA synthesis (Allain et al., 1994, Barat et al., 1989), ensuring viral DNA completion and maintenance during the early phase of retrovirus (RV) replication (reviewed in Bampi et al., 2004, Darlix et al., 1995, Darlix et al., 2000, Darlix et al., 2011, Levin et al., 2005, Rein et al., 1998, Rein et al., 2011, Thomas et al., 2008, Wu et al., 2010). At the same time NC was found to foster recombination reactions during reverse transcription of the dimeric viral RNA template, generating a large genetic diversity that precludes a sustained antiviral immune response and causes the emergence of antiviral resistance (Brenner et al., 2002, Coffin, 1995, Galetto and Negroni, 2005, Gallant et al., 2003, Negroni and Buc, 2001, Onafuwa-Nuga and Telesnitsky, 2009). At the start of virus assembly during the late phase, NC initially as part of the structural polyprotein Gag drives specific interactions between the ZF region of NC and specific sequences in the 5′ leader of the genomic RNA causing its dimerization and forming a GagNC assembly plateform (Cimarelli and Darlix, 2002, D'Souza and Summers, 2005, Lu et al., 2011a, Mirambeau et al., 2010, Muriaux et al., 2001). Later in assembly, the mature NC molecules drive the maturation of the 60S RNA (Feng et al., 1996, Fu et al., 1994) and condensation of the core structure that ensures stability of the viral particle while exerting a negative control over the reverse transcription reaction (Mirambeau et al., 2006, Mougel et al., 2009).
This review aims at presenting a brief retrospect on the retroviral NC protein, now recognized as a central player during retrovirus replication (Fig. 1 ). In addition we will discuss the notion that NC protein is endowed with a high level of flexibility (Dunker et al., 2001, Wright and Dyson, 1999), which may account for its multiple functions as a viral DNA and RNA chaperone during virus replication. Due to its multiple implications in HIV replication NC represents an ideal target for anti-HIV drugs, which will also be reviewed here.
Fig. 1.
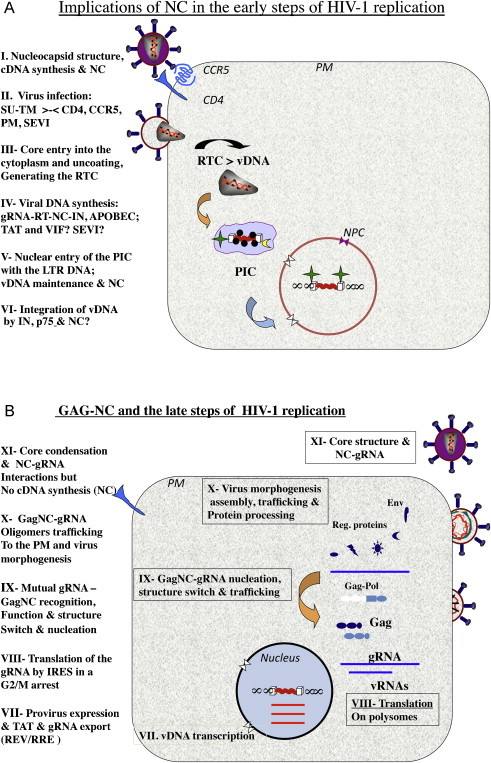
Schematic representation of HIV-1 replication. (a) The early steps of HIV-1 replication. The implications of mature NC protein (NCp7) are at the levels of (I) the structure of a stable, condensed nucleo-core where 1500–2000 NC molecules coat the dimeric RNA genome of 60S, providing protection and negatively regulating reverse transcription; (II) virus infection is mediated by molecular recognition and interactions between the virus trimeric SUgp120-TMgp41 proteins and the cellular CD4 receptor and CCR5 coreceptor at the plasma membrane (PM), that are believed to be augmented in vivo by peptides present in the semen (SEVI; see Section 4.1). Next the viral core (III) is delivered into the cytoplasm where two dynamic processes take place, namely partial core uncoating and viral DNA (vDNA) synthesis. (IV) Viral DNA synthesis by the viral RT enzyme, chaperoned by NC molecules from initiation to completion; this slow process (6–12 h in cells effectively targeted by HIV-1 in vivo (see Section 4.1)) is believed to be facilitated by the viral VIF and TAT proteins and by the cellular SEVI that are all nucleic acid chaperones. In the absence of VIF expression in virus producer cells, the restriction factor APOBEC 3G can be incorporated into the newly made viral particles by interacting with NC, which in turn causes deamination of C residues in nescent cDNA. (V) Once the viral DNA flanked by the LTRs is made it is found in a poorly defined molecular complex called the preintegration complex (PIC) (Arhel, 2010). Integrity of the vDNA is essential since the LTR ends called inverted repeats (ir) are required for the IN-mediated integration of the vDNA into the host genome. Protection of the LTR ends against cellular exonucleases such as TREX-1 (Yan et al., 2010) is thought to be mediated by the binding of molecules of NC and IN. In agreement with this notion, mutating the highly conserved CCHC zinc fingers results in partial trimming of the LTR DNA ends (Buckman et al., 2003, Tanchou et al., 1998), which impairs vDNA integration. (VI) Once the PIC is actively delivered into the nucleus via the NPC (nucleopore complex), the tetrameric IN enzyme in a complex with the cellular helper factor LEDGF/p75 mediates vDNA integration into the host genome. This reaction is believed to be chaperoned by NC but this is still a matter of debate. Also once in the nucleus and before its integration the vDNA can be expressed to produce the viral factors TAT, REV and NEF that are essential for the late steps of HIV-1 replication and the immune response (Wu and Marsh, 2001). (b) Schematic representation of the late steps of HIV-1 replication. (VII) The integrated proviral DNA is transcribed by the host cell RNA polymerase B machinery, with the assistance of the viral trans-acting factor TAT at the levels of initiation and elongation (Wu, 2004). The newly made full length (FL) vRNA is either exported from the nucleus which requires the viral protein REV bound to the RRE or spliced to generate the single and multiple spliced subgenomic RNAs. (VIII) The Gag and GagPol polyprotein precursors to the structural proteins and enzymes, respectively, of the virus are synthesized by the host cell translation machinery using the FL RNA as the messenger RNA template. This is carried out by an original mechanism whereby the host ribosomes directly bind to sequences within the multifunctional 5′ leader, involved in RNA synthesis, splicing and export, and translation, dimerization and packaging, and vDNA synthesis. Such a small domain is called internal ribosome entry site (IRES), which ensures a high level of viral translation throughout the cell cyle, notably in the G2M arrest caused by the viral protein VPR (Balvay et al., 2007). (IX) As soon as Gag accumulates in the cytosol several Gag molecules recognize and bind to the 5′ Packaging region of the FL viral RNA (such as SL2; see Fig. 2d). This in turn would render the 5′ leader inaccessible to translating ribosomes due to structural rearrangements (see text) together with the presence of bound Gag molecules (Huthoff and Berkhout, 2001a, Huthoff and Berkhout, 2001b, Huthoff and Berkhout, 2002). Thus the specific interactions between the NC domain of Gag, notably the ZFs, and the Psi packaging signal would cause a functional switch from translation to virus assembly, representing the nucleation event where the FL RNA acts as the initial platform or scaffold for Gag assembly. (X) Further to the nucleation event, another molecular switch takes place at the opposite end of Gag where the N-terminal myristate becomes accessible, and addresses the Gag–RNP complexes to cellular membranes to pursue and complete virus assembly via interactions with, and recruitment of viral and cellular proteins and NAs (Cimarelli and Darlix, 2002, Freed and Mouland, 2006, Hamard-Peron and Muriaux, 2011) and references therein. (XI) As virion formation progresses, GagPol and Gag become oligomeric which in turn activates the viral protease (PR). Subsequently the viral PR processes Gag and GagPol in a sophisticated manner, first giving rise to MAp17 and NCp15, and RT. Ultimately, upon and/or during virus budding the fully mature Gag proteins are generated, notably NCp7, leading to core condensation while the dimeric RNA genome becomes a mature 60S competent for reverse transcription.
2. NC from a simple nucleic acid binding protein (NABP) to a multifunctional all-in-one nucleic acid chaperone
2.1. Characterization of NC protein as the major viral RNA ligand
The interactions between NC protein and the genomic RNA in alpha- (RSV) and gamma-RV (MoMuLV) virions were initially examined using UV light at 252 nm for cross-linking since this approach allows the specific spatial relationship of certain amino acids and RNA bases to be characterized (see Burd and Dreyfuss, 1994). Results showed that NC was by far the major protein tightly bound to the genomic 70S RNA in the nucleocapsid (Darlix and Spahr, 1985, Meric et al., 1984, Prats et al., 1990, Tanchou et al., 1995). But these results were obscured and a matter of intense debate because MAp19 protein of RSV and pp12 protein of MoMuLV were originally reported to be the viral proteins tightly linked to the genome in virions (Darlix and Spahr, 1982, Sen et al., 1977).
The NC cross-linked RNA fragments were recovered and upon sequencing it was found that NC tightly binds a small number of genomic RNA sites rich in U and G residues, located in the multifunctional 5′ untranslated leader region (5′ UTR) (Dannull et al., 1994, Darlix and Spahr, 1982, Meric et al., 1984, Tanchou et al., 1995). This was of special interest since these sites are close to the dimer linkage structure (DLS) (Darlix and Spahr, 1982, Prats et al., 1990) that holds together the two genomic RNA in the virion nucleocapsid (Murti et al., 1981), and the DLS is part of the packaging psi signal (Pugatsch and Stacey, 1983). These findings suggested that NC interaction with high affinity sites presents in the 5′ leader region, close to the DLS, might be required for the selective packaging of the genomic RNA during virus morphogenesis (Aldovini and Young, 1990, Lever et al., 1989). In fact, mutations in the ZF or flanking basic residues of NC have a strong negative impact on genomic RNA packaging as initially reported for RSV and MoMuLV and later for HIV-1 (Aldovini and Young, 1990, Dorfman et al., 1993, Gorelick et al., 1990, Meric and Spahr, 1986) (see section on NC functions in the late phase of RV replication, Fig. 1b).
2.2. The original reports on NC functions
In the 1980s, two series of findings effectively kick-started the NC story. Firstly, NC was shown to be the driving force in genomic RNA selection, dimerization and packaging during ASLV and MoMuLV assembly (Fig. 1IX and X) (Gorelick et al., 1999, Meric et al., 1988). Secondly, NC was found to direct viral RNA dimerization in vitro and annealing of the replication primer tRNA to the genomic primer binding site (PBS) both in vitro and in ASLV and MuLV virions (Darlix et al., 1990, Prats et al., 1991, Prats et al., 1988). This second series of observations led to the discovery that NC was a viral protein having potent nucleic acid chaperoning activities that was later found to be necessary for bona fide proviral DNA synthesis (reviewed in Darlix et al., 1995, Rein et al., 1998). These findings have since been confirmed (Allain et al., 1998, Auxilien et al., 1999, Barat et al., 1989, Buckman et al., 2003, Darlix et al., 1993, Gao et al., 2003, Godet et al., 2006, Gorelick et al., 1990, Gorelick et al., 1999, Guo et al., 1997, You and McHenry, 1994, Yu and Darlix, 1996) paving the way for attempts to identify anti-HIV drugs specifically targeting NC (Breuer et al., 2012, de Rocquigny et al., 2008, Druillennec et al., 1999b, Goldschmidt et al., 2010, Grigorov et al., 2011, Jenkins et al., 2005, Mori et al., 2012, Rice et al., 1993, Rice et al., 1995, Schito et al., 2003, Tummino et al., 1996).
2.3. Large scale synthesis of RV NC proteins and zinc binding
Originally NC proteins were purified from RV virions (Davis et al., 1976, Henderson et al., 1981, Henderson et al., 1988, Henderson et al., 1990, Henderson et al., 1992, Hizi et al., 1987, Prats et al., 1988, Prats et al., 1990) and later from recombinant Escherichia coli or synthesized in vitro by chemical methods (see below) and found to bind a wide variety of nucleic acids (NA) molecules with a preference for the genomic 70S RNA (Darlix and Spahr, 1985, Wu et al., 1996). Accordingly, the relative affinity of in vitro synthesized NC protein for retroviral RNA, DNA and oligonucleotides is in the order ‘retroviral RNA > ssDNA > dsDNA > oligonucleotides’ where the apparent affinity constant for the genomic RNA is 5 × 107 M−1, about 25 times higher than that for poly(rA) (You and McHenry, 1993).
NC from various retroviruses such as NCp12 from ASLV, NCp10 of MoMuLV, NCp12 from BLV and NCp15 from HTLV have been sequenced and comparison of their primary sequences revealed the presence of a common highly conserved CCHC domain (Copeland et al., 1983a, Copeland et al., 1983b, Henderson et al., 1981), later called zinc fingers (ZFs). Up to the 90s NC protein was purified from virions or recombinant bacteria. Therefore, in addition to a series of affinity or mass exclusion chromatographies, the protein solutions were treated with reducing agents (β mercapto and DTT) in the presence of EDTA. As a consequence, the initial RNA binding experiments showed that the stoechiometry and affinity of NCs for various RNA/DNA substrates were independent from zinc ions (Casas-Finet et al., 1988, Karpel et al., 1987). These data were reinforced by results from the group of Leis who showed that the CCHC domain was essential for viral replication but not required as a zinc binding domain (Jentoft et al., 1988). However it remained that the CCHC sequence was a zinc binding domain as suggested by Berg (1986). In support of this notion, site directed mutageneses of zinc ligands namely the cysteine or histidine residues were found to render the virus replication defective (Gorelick et al., 1988, Meric and Goff, 1989). At last original biophysical studies notably by NMR demonstrated that one zinc atom was interacting per CCHC domain (Bess et al., 1992, Chance et al., 1992, Summers et al., 1992) which in turn was causing a structural organization of the primary CCHC sequence around the zinc ion (South et al., 1990, Summers et al., 1990).
Therefore it is likely that the lack of consensus on the role of zinc in NC activity and structure was due, at least in part, to the purification method. This is why it appeared necessary to in vitro synthesize NC proteins in order to tightly control the oxidation state of the cysteine residues. The first attempt was carried out on MoMuLV NCp10 which is 56 residues long with one zinc finger flanked by basic residues. NCp10 was obtained in a pure form by solid phase peptide synthesis (SPPS) using the tBoc strategy (Cornille et al., 1990). However the tBoc chemistry was found to exhibit a number of drawbacks that hamper the synthesis of large peptides. This is why in a next step the synthesis of the HIV-1 NC protein was performed using the Fmoc chemistry. HIV-1 NC contains two zinc fingers surrounded by a series of basic residues and the first NC obtained represented the 1–72 sequence corresponding to fully mature NCp7 (55 residues) and spacer peptide sp2 (56–72) (Dannull et al., 1994, de Rocquigny et al., 1991, de Rocquigny et al., 1992, de Rocquigny et al., 1997, Dong et al., 1997, Surovoy et al., 1993).
All together such a chemical-based challenging production of RV NCs and other NA chaperones turned out to have several advantages: (i) the quantity of peptide was easily adapted using various resin types and scales; (ii) the peptides were not contaminated by nucleic acid and cysteine residues were preserved from oxidation since purification was performed in acidic conditions and further handling carried out using oxygen free solutions in the presence of Zn2+; (iii) synthesis of NC fragments and/or replacement of key residues for structure activity analyses or substitution of natural amino acids for exotic ones (fluorescent derivatives, biotins, etc.) became routine procedures. This rendered possible a great deal of in vitro investigations on RV NCs ranging from structure analyses, NA binding, chaperoning and condensing activities to NC implications and functions in viral DNA synthesis and integration (see below).
Due to its merits this chemical protocol was also used to synthesize active SIV NCp8 (Morellet et al., 2006) together with many other NC proteins from FIV, ASLV and yeast Ty3 as well as HIV-1 TAT, and HCV core and HBV core peptides (Peloponese et al., 1999, Sharma et al., 2012) as well as putative NA chaperone domains of cellular proteins such as the SEVI (Munch et al., 2007) and the human prion protein (Gabus et al., 2001b). All these peptides were found to exhibit potent NA binding, chaperoning and condensing activities in vitro under physiological like conditions (see below and data not shown for the SEVI).
2.4. Brief description of HIV-1 NC and NC–nucleic acid complex structures
Three-dimensional structures of intact nucleocapsid proteins from HIV-1 have been determined using nuclear magnetic resonance (NMR) methods (Morellet et al., 1992, Summers et al., 1992). Additionally the dynamical behavior of the protein has also been studied using NMR methods (Lee et al., 1998, Ramboarina et al., 2002). Moreover, the structure of different NC–nucleic acid complexes has been solved by the same technique and data show that such nucleoprotein complexes are different while displaying some common features (Amarasinghe et al., 2000b, Bazzi et al., 2011, Bourbigot et al., 2008, De Guzman et al., 1998, Morellet et al., 1998, Spriggs et al., 2008). The contacts between NC and the NA partner involve electrostatic, hydrophobic (Fig. 2 ) and hydrogen bonds (not shown) components. The main features are that guanines bind deeply into the ZF1 and ZF2 hydrophobic pockets (Fig. 2). With respect to NC protein alone (Fig. 2a), the hydrophobic platform, formed of residues V13, F16, T24, A25, W37, Q45 and M46, undergoes important structure changes upon nucleic acid binding. In NC–RNA complexes, one guanine is inserted in the hydrophobic platform of each zinc finger and this insertion is stabilized by hydrogen bonding of the basic residues spread along the NC sequence (K14, K20, K26, R29, R32, K38) (not shown). In NC–DNA complexes only one guanine is inserted in ZF2 while a thymine partly interacts with ZF1 (Fig. 2d). Another key structural feature involved in NC–NA complex stabilization is the strong stacking interactions between the guanine nucleobases and the ZF-aromatic residues (F16 for ZF1 and W37 for ZF2 see Fig. 2b). There is an additional important difference between NC–DNA complexes (three complexes resolved (Bazzi et al., 2011, Bourbigot et al., 2008, Morellet et al., 1998)) and the NC–RNA complexes (three complexes (Amarasinghe et al., 2000a, De Guzman et al., 1998, Spriggs et al., 2008)) where the orientation of ZF1 and ZF2 along the nucleic acid chain is opposite in NC–RNA complexes as compared to NC–DNA complexes.
Fig. 2.
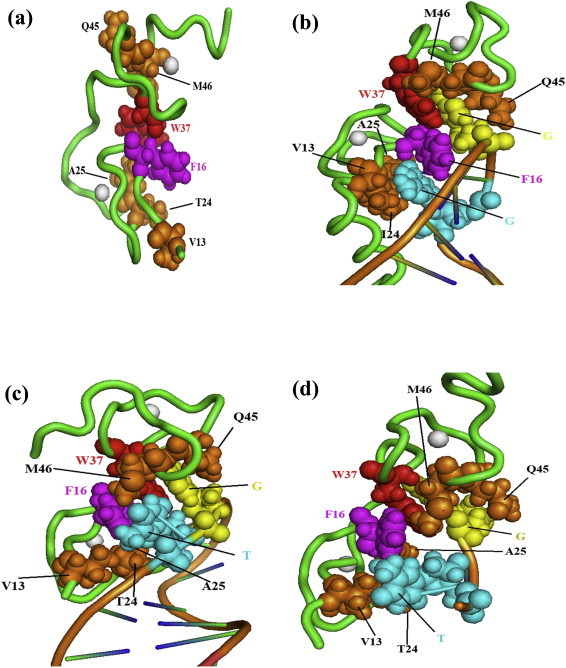
Three dimensional structure representations of free HIV-1 NC and bound to various RNA and DNA molecules. For the sake of clarity, only the major structural determinants of NC are shown here. The NC protein backbone is shown as a green tube and the two zinc atoms coordinated by the CCHC tetrad in each zinc finger are indicated as white spheres. Key hydrophobic interactions taking place between NC and the NA are highlighted here. In addition the hydrophobic residues contacting the nucleic acid are illustrated as spheres (with Van der Waals radius set at 0.75 to facilitate visualization). The F16 and W37 aromatic residues are in magenta and red, respectively, while other residues in ZF1 (V13, T24, A25) and ZF2 (Q45, M46) are in orange. The two nucleotides with the tightest hydrophobic interaction with the protein side chains are 2G residues in blue and yellow in (b) (NC–SL3 RNA complex (De Guzman et al., 1998), pdb1A1T), T and G in (c) (NC–PBS(−) DNA complex (Bourbigot et al., 2008), pdb2jzw) and (d) (NC–minicTAR DNA complex (Bazzi et al., 2011), pdb2L4L). Relative to free NC (a) (pdb1esk (Morellet et al., 1998)), note that the hydrophobic interface is reorganized in the NC–NA complexes (b–d). In (b) the two guanines in blue and yellow are deeply inserted within the hydrophobic pocket formed by the zinc fingers. In (c) and (d), one guanine (G in yellow) is inserted in the ZF2 hydrophobic pocket while the thymine (T in blue) is on the exterior of the hydrophobic pocket. Also note that the T24, V13 and A25 residues are in close contact with the double stranded part of PBS(−) DNA in (c), while there is no such contact in the NCp7–SL3 RNA complex in (b).
3. The NC network of molecular interactions
Our comprehension of the nature of NC profoundly changed once it was realized that the RV NC could interact in multiple ways with NA (see Fig. 2) and with distinct NA molecules at the same time and by so doing, NC has an impact on NA conformation. In fact the present view is that RV NCs are members of a growing family of nucleic acid binding proteins (NABPs) named RNA chaperones that are abundant in all living organisms and viruses where they perform essential functions ranging from gene transcription and regulation to mRNA translation and maintenance (Cristofari and Darlix, 2002, Herschlag, 1995, Schroeder et al., 2004, Tompa and Csermely, 2004). These chaperone proteins belong to a large class of intrinsically disordered proteins (IDPs) because they contain flexible/disordered domains (Dunker et al., 2001, Uversky, 2002, Wright and Dyson, 1999) rich in basic residues and bind nucleic acids with broad sequence specificity and different modes (see Fig. 2) (Herschlag, 1995, Schroeder et al., 2004).
Such RNA chaperone proteins are indispensable partners of NAs because they assist RNA folding by preventing misfolding or by resolving misfolded RNA species (Cristofari and Darlix, 2002, Herschlag, 1995, Schroeder et al., 2004, Tompa and Csermely, 2004). This RNA folding assistance is essential to ensure that RNAs reach their correct functional conformation in a rapid manner under physiological conditions (Schroeder et al., 2004). Examples of cellular RNA chaperones with essential functions include the tumor suppressor P53, hnRNP A1, the major mRNA binding protein YB1/P50, the fragile X mental retardation protein FMRP and the Prion protein (Cristofari and Darlix, 2002, Evdokimova et al., 2006, Gabus et al., 2001b, Ivanyi-Nagy et al., 2005). In this context RV NCs represent remarkable examples of multifunctional NA chaperones that drive the necessary structural rearrangements of the genomic RNA during the early and late phases of virus replication (see below; reviewed in Darlix et al., 1995, Levin et al., 2005). In addition RV NCs might well be endowed with protein chaperoning activity by providing assistance to the viral RT and IN enzymes during viral DNA synthesis and integration (Buckman et al., 2003, Carteau et al., 1997, Lapadat-Tapolsky et al., 1993, Poljak et al., 2003; reviewed in Darlix et al., 2011).
Based on simple and advanced in vitro assays to examine the properties of RNA chaperones that involve rearrangement of nucleic acid structures in physiological conditions (reviewed in Cristofari and Darlix, 2002, Godet and Mely, 2010, Schroeder et al., 2004) other RV proteins with NA chaperone activities – or with nucleic acid annealing activity – have been identified and include TAT (Boudier et al., 2010, Boudier et al., 2014, Godet et al., 2012, Kameoka et al., 2002b, Kuciak et al., 2008) and VIF (Batisse et al., 2012, Henriet et al., 2005) that perform essential functions in HIV replication.
In conclusion the NA chaperoning activities of NC indicate that this small viral protein can direct NA structure remodeling so that the most thermodynamically stable conformations are gained (Fig. 2). This remodeling reaction appears to rely on three essential NC properties, that are (i) its binding to NAs causing their condensation as large nucleoprotein complexes where molecular crowding can take place (Echols, 1990, Le Cam et al., 1998, Mirambeau et al., 2006, Williams et al., 2001), (ii) the partial destabilization of NA structures in an ATP-independent manner (Azoulay et al., 2003, Beltz et al., 2003, Beltz et al., 2005, Bernacchi et al., 2002, Bourbigot et al., 2008, Heilman-Miller et al., 2004) and (iii) a rapid on-and-off NA binding kinetics (Cruceanu et al., 2006a, Cruceanu et al., 2006b). The mechanism whereby RV NC protein can remodel NA structures in an ATP-independent manner is yet poorly understood but it might be a direct consequence of the multiple modes of NC binding to single and double stranded RNA and DNA molecules (Fig. 2, compare panels a with b–d) and the rapid on and off binding kinetics of NC possibly involving an entropy exchange between NC and the NA (see Section 6 on NC flexibility) (Ivanyi-Nagy et al., 2005).
4. The multiple roles of NC nucleic acid chaperoning in viral DNA synthesis by reverse transcriptase
4.1. NC as an essential cofactor of RT
A canonical hallmark of retroviruses, shared by LTR and non-LTR retroelements, is their replication strategy that necessitates the reverse transcription of the genomic RNA (gRNA) by the viral DNA polymerase, known as reverse transcriptase (RT) during the early phase of virus replication that spans virus-cell recognition and entry to viral DNA synthesis and integration into the host genome. The double stranded viral DNA is synthesized by the viral RT during a succession of specific reactions that take place in an RT-complex (RTC) most probably corresponding to the viral nucleocapsid (reviewed in Darlix et al., 1995, Levin et al., 2005, Rein et al., 1998). The overall process of reverse transcription necessitates profound structural rearrangements of the genomic RNA, the cellular primer tRNA and of the newly made viral DNA, which are all assisted by NC (Fig. 1III and IV). Subsequently the viral DNA is imported into the nucleus within the preintegration complex (PIC) and ultimately integrated into the host cell genome, then called the provirus (Fig. 1I–V).
It is not the purpose of this short review to provide once more a simplified scheme of the reverse transcription process which ultimately leads to the synthesis of a complete viral DNA flanked by the 5′ and 3′ LTR with the inverted repeats (ir) required for vDNA integration (see a number of reviews, i.e. Bampi et al., 2004, Levin et al., 2005).
The present view is that NC protein assists RT from the initiation to the completion of reverse transcription (RTion) to ensure fidelity and completeness of vDNA synthesis with the LTRs. This is achieved by means of complementary mechanisms. Firstly, NC chaperones a number of specific obligatory viral nucleic acid annealing reactions, namely primer tRNA annealing to the RTion initiation site (PBS), and the minus strand and the plus strand DNA strand transfers. Secondly NC protein possibly through direct interactions with both the NA and the RT enzyme (Druillennec et al., 1999a, Lener et al., 1998) appears to contribute to the fidelity of vDNA synthesis (i) by preventing minus strand and plus strand false initiations (Beltz et al., 2005, Guo et al., 1997, Lapadat-Tapolsky et al., 1997), (ii) by largely increasing the time of residence of active RT molecules on the genomic RNA template (Grohmann et al., 2008) and (iii) by eliciting a nucleotide excision-repair activity of RT (Bampi et al., 2006; reviewed in Darlix et al., 2007).
Thus NC can be considered as an essential cofactor of RT chaperoning both the viral NA annealing reactions and the RT enzyme in order to ensure fidelity and completeness of viral DNA synthesis. Hence NC could be viewed as a Janus chaperone assisting both nucleic acids and proteins to the benefit of their functional interactions (Kovacs et al., 2009).
At this point, it should be mentioned that two other viral factors, namely the viral trans-acting factor TAT and the viral infectivity factor VIF endowed with NA chaperoning activities (Henriet et al., 2005, Kuciak et al., 2008) can provide assistance to vDNA synthesis (Apolloni et al., 2003, Boudier et al., 2010, Boudier et al., 2014, Kameoka et al., 2002a). But this is still a debated issue since only low to very low amounts of TAT and VIF are found in virions. On the other hand, TAT and VIF could assist the initiation of vDNA synthesis which can already take place in the infected cell (Lori et al., 1992, Trono, 1992, Zhang et al., 1994). Along this line, a cellular factor called SEVI present in abundance in the seminal fluid was found to be a potent NA chaperone resembling NC in vitro (Darlix, unpublished data) and to augment the level of vDNA synthesized in vitro and in cell culture (Munch et al., 2007). This same component of the seminal fluid was found to drastically increase the virus titer on primary human blood cells (Munch et al., 2007, Zhang et al., 1996, Zhang et al., 1998).
The potent NC chaperoning activity is also believed to drive recombination reactions during reverse transcription of the dimeric RNA genome (Galetto et al., 2006, Galetto and Negroni, 2005, Onafuwa-Nuga and Telesnitsky, 2009). In fact the pseudiploidy of the RV genome constitutes the basis for forced and unforced recombinations (Hu et al., 2003b, Hu and Temin, 1990, Katz and Skalka, 1990, Zhang and Temin, 1993) in the course of minus strand DNA synthesis, which ultimately lead to the formation of a complete viral DNA despite nicks in each monomeric RNA. Moreover these NC-directed recombination reactions should cause the reassortment of specific genetic traits when the two RNAs are different as it is in heterozogous viruses, which constitutes the basis for a high level of viral variability as indicated by the presence of large quasispecies populations in HIV infected person (Wain-Hobson et al., 2003). This rather high level of virus variability as indicated by high throughput sequencing analyses (HTPS) allows the virus to escape the immune response and HAART (highly active antiretroviral therapies) (Barbaro et al., 2005, Gallant et al., 2003). Thus the emerging view is that NC is central to the network of dynamic interactions taking place between RT, the dimeric RNA template and the viral cDNA being synthesized. This NC network fulfills two functions, firstly by ensuring the synthesis of a complete functional vDNA flanked by the LTRs and secondly by fueling the genetic diversity of newly made virus by reassorting genetic traits through recombination reactions to the benefit of HIV-1 fitness in vivo (Barbour and Grant, 2005, Bocharov et al., 2005, Wain-Hobson et al., 2003).
4.2. Impacts of zinc finger mutations on viral DNA synthesis and integration
At this stage it should be mentioned that synthesis of a complete double stranded viral DNA flanked by the LTR is a slow process that takes 6–8 h in human T cell lines and up to 16–24 h in PBLs and macrophages that are the major cells infected by HIV-1 in vivo (Arfi et al., 2008). Once vDNA synthesis is completed, NC is thought to protect the vDNA product against nuclease attack (Krishnamoorthy et al., 2003), notably the ir ends required for integration. In fact mutating either one of the highly conserved CCHC residues of the zinc fingers (ZFs) caused a lower affinity of NC for DNA in vitro and rendered the LTR ends accessible to nucleases in an in vitro model system containing the IN enzyme and the LTR DNA (Bampi et al., 2004).
Along this line, early observations show that point mutations in the highly conserved CCHC ZFs resulted in the formation of incomplete LTR ends and in the production of replication defective viruses (Buckman et al., 2003, Tanchou et al., 1998) where viral DNA synthesis was partially impaired and at the same time being unstable (Demene et al., 1994, Gorelick et al., 1996, Gorelick et al., 1999). Additional results by Gorelick et al. also indicated that the integration reaction was impaired (Buckman et al., 2003, Thomas et al., 2006), supporting the view that functional interactions exist between NC and IN as it stands between NC and RT since mutating the ZF strongly diminish NC–RT interactions (Druillennec et al., 1999a, Lener et al., 1998). The existence of critical interactions between NC and IN is further supported by the fact that NC can strongly stimulate concerted LTR DNA integration by IN under physiological conditions in vitro (Buckman et al., 2003, Carteau et al., 1997, Carteau et al., 1999, Poljak et al., 2003, Thomas et al., 2006).
In conclusion, these findings support the view that NC and notably the CCHC ZFs play a central role in viral DNA synthesis, maintenance and integration possibly through chaperoning interactions with both the viral NAs and the viral enzymes.
4.3. NC and the timing of viral DNA synthesis
The interplay of NC with the viral replication machinery also extends into the late phase of the replication cycle corresponding to virus assembly and budding (Gottlinger, 2001) (see below). As said before, the highly conserved CCHC ZFs flanked by basic amino acids are a hallmark of RV NC proteins and were originally shown to drive the specific packaging of the genomic RNA, most probably in a dimeric form, during RSV and MoMuLV virion formation (Gorelick et al., 1988, Gorelick et al., 1996, Housset et al., 1993, Meric et al., 1988, Meric and Spahr, 1986).
Interestingly, active reverse transcription was shown to occur in newly made RSV, MuLV and HIV-1 virions (Lori et al., 1992, Trono, 1992, Zhang et al., 1994; reviewed in Darlix et al., 1995). More recently the impact of NC mutations on cDNA synthesis in newly made HIV-1 virions was investigated. In fact point mutations of the NC CCHC motif resulted in the formation of virions where the core was only partially condensed (Gelderblom et al., 1987, Grigorov et al., 2006, Grigorov et al., 2007, Tanchou et al., 1998). Moreover such virions were changed to DNA containing particles since they contained a rather large amount of vDNA including two LTR DNA molecules, and much less genomic RNA as compared to wild-type virions. This endogenous reverse transcription reaction was shown to occur in virus producer cells, most probably during morphogenesis of the ZF mutant virus (Thomas and Gorelick, 2008; see review Mougel et al., 2009 and references therein).
These results indicate that NC exerts a dual control over the timing of viral DNA synthesis by RT, a positive one after viral entry in target cells, and a negative one that delays the process in the course of virus production (Houzet et al., 2008; reviewed in Mougel et al., 2009). Therefore, NC should be viewed as an essential viral component of the replicative machinery, a all-in-one small chaperone impacting on the timing, kinetics and completeness of reverse transcription from the initiation up to the synthesis of the complete viral DNA with the two LTRs and its maintenance (Fig. 1).
As soon as the proviral DNA is imported into the cell nucleus and before its integration, viral genes can be expressed by the host cell transcription and translation machineries to produce the essential viral factors TAT, REV and NEF (Wu and Marsh, 2001). The very early and efficient synthesis of TAT is central to the late steps of HIV-1 replication since it boosts provirus transcription by the cellular RNA polymerase B machinery.
5. NC, the nucleation of retrovirus assembly and structure switches
Two distinct mechanisms of retrovirus assembly have been described so far and have been the subject of many reviews. In the first instance retroviral particles are preformed in the cytoplasm of infected cells and transported to the plasma membrane in the case of the betaretroviruses (MMTV, MPMV). In the second case, RV particles directly assemble underneath the plasma membrane (PM) as for alpha- and gammaretroviruses (ASLV and MLV, respectively) (Dickson et al., 1985). Until recently, HIV-1 assembly was thought to follow the second mode (Ivanchenko et al., 2009, Jouvenet et al., 2006, Jouvenet et al., 2008, Ono et al., 2004) but recent findings indicate that this may not be the sole mechanism. Indeed, according to several reports HIV-1 assembly can also take place in intracellular vesicles such as endosomes in human macrophages, and in T cells (Grigorov et al., 2006, Ono and Freed, 2004, Pelchen-Matthews et al., 2003, Raposo et al., 2002, Rudner et al., 2005).
It is not the purpose of the present report to review the choreography of RV assembly and budding as there are many excellent reviews on the subject (Adamson and Freed, 2007, Resh, 2005), notably on the role of each Gag domain, including NC, in Gag trafficking and Gag-vRNA oligomer formation. However much less is known on how and where the very first step of RV assembly is taking place and the possible role of NC in this nucleation reaction.
The Gag polyprotein precursor contains all the signals and domains required for viral particle assembly, and as such it orchestrates the particle assembly from the nucleation step to virus release by budding (Kutluay and Bieniasz, 2010, Muriaux and Darlix, 2010, Sundquist and Krausslich, 2012). In infected cells, the unspliced viral RNA acts both as the messenger coding for Gag and Gag-Pol and as the genome to be recruited, dimerized and packaged into assembling Gag particles (Butsch and Boris-Lawrie, 2000, Dorman and Lever, 2000, Griffin et al., 2001, Levin and Rosenak, 1976, Ni et al., 2011). But the full length (FL) viral RNA cannot fulfill both functions at the same time, thus raising a long standing issue on how the fate of the FL viral RNA, translation versus packaging, is regulated (Brasey et al., 2003, Lopez-Lastra et al., 2005). As originally reported for ASLV and MoMuLV and more recently for HIV and SIV (Attal et al., 1996, Berlioz and Darlix, 1995, Berlioz et al., 1995, Brasey et al., 2003, Buck et al., 2001, Deffaud and Darlix, 2000, Herbreteau et al., 2005, Lopez-Lastra et al., 2010, Ohlmann et al., 2000, Vagner et al., 1995), Gag is synthesized by an IRES dependent mechanism whereby the active ribosomes directly bind to sequences within the 5′ untranslated region (UTR or leader) of the FL RNA, upstream of the initiator AUG of Gag. At this stage it is believed that newly made Gag molecules accumulate on or in the vicinity of the translating polysomes. Next or concomitant with translation, the nucleation step would take place via specific interactions between GagNC and the high affinity binding sites present in the IRES/Packaging signals within the 5′ UTR (Aldovini and Young, 1990, Baudin et al., 1993, Berkhout, 1996, Clever et al., 1995, Clever et al., 2000, Clever et al., 2002, Hayashi et al., 1992, Lever et al., 1989, McBride and Panganiban, 1996, Sakaguchi et al., 1993, Shubsda et al., 2002) for reviews (Cimarelli and Darlix, 2002, Lever, 2007, Muriaux and Darlix, 2010).
This proposed mechanism stipulates that binding of several just-made GagNC molecules to the Packaging region of the FL viral RNA would render it inaccessible to translating ribosomes due to structural rearrangements of the 5′ UTR such as base pairing interactions involving the Gag AUG initiation codon (Abbink and Berkhout, 2003, Berkhout, 1996, D'Souza and Summers, 2004, Lu et al., 2011a, Lu et al., 2011b). This together with bound Gag molecules (Huthoff and Berkhout, 2001a, Huthoff and Berkhout, 2001b, Huthoff and Berkhout, 2002) would cause a functional reorientation of the FL RNA from translation to dimerization and packaging (Heng et al., 2012) effectively starting Gag assembly where the FL RNA acts as the initial platform or scaffold (Burniston et al., 1999, Cimarelli et al., 2000, Muriaux et al., 2001, Ott et al., 2009; reviewed in Butsch and Boris-Lawrie, 2000, Butsch and Boris-Lawrie, 2002, Darlix et al., 1995, Darlix et al., 2000). As a result of the nucleation event is the targeting of the viral Gag–RNP complex to cellular membranes due to another conformational switch at the opposite end of Gag. In fact the N-terminal matrix myristyl group undergoes a switch whereby the myristate becomes accessible, enabling the anchoring of the Gag–RNP complex to cellular membranes to pursue and complete virus assembly (Freed and Mouland, 2006, Hamard-Peron and Muriaux, 2011, Saad et al., 2006).
Taken together, these findings suggest that the NC domain of Gag plays a major role in determining the fate of the genomic RNA and that this contribution will depend on both NC-induced RNA structural rearrangements (Huthoff and Berkhout, 2001a, Huthoff and Berkhout, 2002) and molecular crowding brought about by Gag molecules bound to the 5′ UTR via the NC domain (Abbink and Berkhout, 2003, Brasey et al., 2003, Darlix et al., 1995, Heng et al., 2012). In support of this possible mechanism, we found HIV-1 Gag–viral RNA complexes together with unbound Gag molecules cosedimenting with a fraction of polyribosomes of high sedimentation values (250–350S) purified from HIV-1 Gag-Pol expressing 293 cells (unpublished data). Upon addition of EDTA to dissociate the ribosomes from the RNA template, Gag–RNA complexes and free Gag molecules were recovered by sucrose gradient centrifugation (unpublished data).
6. Viral NC–RNA RNPs, fuzziness and functions
The all-in-one retroviral NC performs multiple functions in the virus replication cycle, during both the early and late phases, which represents a significant body of scientific research since its original description as a non-specific simple NABP. But how could NC be endowed with so many essential functions in virus structure, vDNA synthesis and viral particle morphogenesis?
Before providing a possible clue to the above question, one should remember the nature of NC with its structured zinc fingers flanked by disordered basic sequences driving specific and nonspecific interactions, respectively, with NAs (Amarasinghe et al., 2000a, Amarasinghe et al., 2000b, Bazzi et al., 2011, Bourbigot et al., 2008, De Guzman et al., 1998, Morellet et al., 1998). This in turn can result in NA condensation (Krishnamoorthy et al., 2003, Lapadat-Tapolsky et al., 1995, Mirambeau et al., 2006, Stoylov et al., 1997, Tanchou et al., 1995) together with structural rearrangements as it occurs during reverse transcription. Thus NC–RNA and GagNC–RNA RNP complexes most probably represent the driving force behind the functions. Indeed NC-based oligomeric structures can be formed in vitro, and are easily seen by electron microscopy (Gabus et al., 2001a) as highly dense heterogeneous complexes exhibiting various sizes and forms. In these NC-based oligomeric structures multiple tight nucleoprotein interactions are holding together the NC and RNA molecules causing a molecular crowding phenomenon thus allowing many reactions to take place (Darlix et al., 1995, Darlix et al., 2000, Herschlag, 1995, Rein et al., 1998). But at the same time these NC-based oligomeric structures are accessible to small ODNs and to the RT enzyme which can gain access to the RNA and perform cDNA synthesis with high efficiency. Therefore such NC-based oligomers are either porous or else highly dynamic, the latter being probably the case due to the rapid on and off binding kinetics of NC to the nucleic acid (Cruceanu et al., 2006a) together with different modes of interactions driven by the ZFs and/or the disordered basic domains (reviewed in Darlix et al., 2011, Ivanyi-Nagy et al., 2012). Taken together, these findings favor the notion that NC–RNA ribonucleoprotein complexes can be regarded as fuzzy flexible molecular assemblages where multiple different interactions between the viral NC and the NA (Fig. 2) as well as with the RT enzyme and possibly IN are highly dynamic which is essential during the early steps of RV replication.
The functionality of such fuzzy nucleocapsid assemblages is also illustrated by the dimeric nature of the genomic RNA, which provides the basis for multiple forced and unforced recombination reactions during vDNA synthesis by RT (Hu et al., 2003a, Hu and Temin, 1990, Katz and Skalka, 1990, Onafuwa-Nuga and Telesnitsky, 2009). These recombinations are chaperoned by NC and fulfill two functions, formation of a complete viral DNA by RT in conditions where each RNA monomer contains nicks, and reassortments of specific genetic traits when the two monomers are different, which fuels a high level of genetic diversity resulting in a quasispecies population in HIV-1 infected persons (Wain-Hobson et al., 2003).
7. Anti-NC drugs: an exciting challenge
Since resistant HIV-1 strains have been identified for each of the clinically available antiretroviral drugs targeting the viral enzymes, namely RTi, PRi, INi (reviewed in Das and Arnold, 2013a, Das and Arnold, 2013b) there is an urgent need to identify new molecules preferably targeting viral proteins not yet targeted by available therapies. Due to its high degree of conservation and to its multiple fundamental roles in HIV-1 replication, NC constitutes a highly promising target. Since the NC properties and functions largely rely on its zinc fingers, the latter were selected as the primary target for developing zinc ejectors able to eject the zinc atoms from the NC fingers, resulting in NC and the concomitant total loss of HIV-1 infectivity. Starting from 3-nitrosobenzamide, the first identified zinc ejector (Rice et al., 1993) various classes of compounds were designed (reviewed in de Rocquigny et al., 2008, Goldschmidt et al., 2010, Musah, 2004, Turpin et al., 2008). These compounds were found to show strong antiviral activity against a large spectrum of HIV-1 strains and elicit little viral resistance. A few of them were tested in clinical assays (Goebel et al., 2001, Turpin, 2003, Vandevelde et al., 1996) that were unfortunately not conclusive. In fact, these compounds showed limited selectivity and thus were quite toxic, preventing their use in HAART protocols. Current efforts are directed toward the development of zinc ejectors to be used as topical microbicides in the prevention of sexually transmitted HIV-1 infection (Srivastava et al., 2004, Wallace et al., 2009). In this respect, S-acyl-2-mercaptobenzamide thioesters (SAMT) (Jenkins et al., 2005) and more recently 2-methyl-3-phenyl-2H-[1,2,4]thiazol-5-yideneamine (WDO-217) (Vercruysse et al., 2012) were found to constitute valuable candidates for topical microbicide formulations.
As an alternative to zinc ejectors, non-covalent NC inhibitors (NCis) were developed as possible antiretroviral therapeutics with greater specificity and thus less toxicity than zinc ejectors. Two main classes of compounds were identified, namely (i) non-covalent NCis binding to NC and (ii) non-covalent NCis binding to nucleic acid partners of NC. Within the first class, high throughput screenings were performed to identify small molecules able to inhibit the NA-binding (Breuer et al., 2012, Stephen et al., 2002) and chaperone properties (Shvadchak et al., 2009) of NC. Compounds able to bind NC with an affinity in the micromolar range, but with only modest anti-HIV activity were obtained, indicating that these compounds still need to be optimized. An important step toward the rational optimization of NCis has been recently achieved by the resolution of the first 3D structure of a complex between NC and an NCi (Goudreau et al., 2013). In this complex, the NCi was found to bind preferentially within the hydrophobic pocket of NCp7 performing a π–π stacking interaction with the side chain of Trp37, and thus, behaving as a guanosine mimetic. Moreover, a second NCi molecule was found to bind NC by connecting the hydrophobic pocket with the N-terminal region. Another key progress was achieved by the application of molecular dynamics simulations and molecular modeling studies on NMR structures of NC in complex with oligonucleotides to identify pharmacophoric hot-spots for NCis to compete with NAs for the NC binding sites (Mori et al., 2010, Mori et al., 2011). Based on this knowledge, a virtual screening was performed and NCis with antiviral activity in the low micromolar range were identified (Mori et al., 2012), indicating that this strategy constitutes a promising approach to develop and improve NCis. In parallel, methylated oligoribonucleotides (mODNs) able to inhibit HIV-1 replication in MT4 cells with an IC50 value of 0.3 nM were recently disclosed (Grigorov et al., 2011). These molecules were found to impair reverse transcription, both by sequestering NCp7 molecules (Avilov et al., 2012) and by direct interaction with reverse transcriptase. Unfortunately, HIV-1 mutants resistant to these mODNs were obtained, so that their development was stopped.
A second approach to inhibit NC functions is to target the NA sequences interacting with NC during HIV-1 replication. Of special interest are the stem-loops of the 5′ untranslated leader of HIV-1 RNA, which control viral translation and virus assembly through their interaction with NC. Various classes of molecules such as aminoglycosides, intercalators, anthraquinones and small peptides were designed or screened to target the stem-loops of the ψ packaging signal (Bernacchi et al., 2007, Dietz et al., 2008, Freisz et al., 2008, Pustowka et al., 2003, Turner et al., 2006, Turner et al., 2009, Warui and Baranger, 2009), as well as the PBS and/or the TAR sequence (Raja et al., 2006, Sosic et al., 2013). These compounds were able to prevent the interaction of NC with its NA targets or dissociate the already formed nucleoprotein complexes, but showed only limited anti-viral activity (Dietz et al., 2008, Ennifar et al., 2006, Raja et al., 2006, Sosic et al., 2013). As for NCi directly targeting NC, it is believed that the structural determinants and the in vivo mechanism of action of NCis targeting the NA targets of NC need to be further understood in order to rationally design the next generation of compounds.
8. Conclusion
As it stands now, RV NC oligomers resemble fuzzy RNP assemblages that probably also exist in simple RV such as ASLV and MuLV and in ancient retrotransposons of yeast such as TY3. NC appears to be central to the formation of such fuzzy assemblies, and by coordinating under a dynamic mode multiple NA:NA, NA:NC and NC:enzymes interactions it fulfills multiple functions from the inner structure of the viral particle to vDNA synthesis and integration (Ivanyi-Nagy et al., 2012). In addition the dimeric nature of the viral genome is very well accommodated in such vaguely ordered replication machinery, which ensures a genetic diversity of the virus fueled by multiple recombination events. In the case of HIV-1, this turns out to be sufficient for the virus to escape HAART therapies and specific immune responses.
From a general perspective with the aim of better understanding the functions of viral nucleoproteins, the core proteins of flaviviruses, hepadnaviruses (Ivanyi-Nagy and Darlix, 2010), paramyxoviruses (Longhi et al., 2010, Longhi and Oglesbee, 2010) and coronaviruses (Chang et al., 2006, Chang et al., 2009) were studied. In all cases examined so far, it turned out that such viral nucleoproteins, contained intrinsic disordered domains, were forming dense nucleoprotein complexes upon binding to NAs and were endowed with potent NA chaperoning activities (Ivanyi-Nagy et al., 2005, Longhi et al., 2010, Longhi and Oglesbee, 2010).
Therefore, the determination of the composition of such fuzzy viral functional complexes and of the multiple dynamic interactions in these complexes (Fig. 2) is central to the identification of possible druggable sites in viral nucleoprotein cores from highly pathogenic viruses such as HIV, HCV, HBV, MV and SARS coronavirus (Cheng et al., 2006, Csermely et al., 2005).
Acknowledgements
The authors acknowledge INSERM, CNRS, ANRS, SIDACTION and UDS.
References
- Abbink T.E.M., Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- Adamson C.S., Freed E.O. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. (San Diego, CA) 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- Aldovini A., Young R.A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain B., Lapadat-Tapolsky M., Berlioz C., Darlix J.L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain B., Rascle J.B., de Rocquigny H., Roques B., Darlix J.L. CIS elements and trans-acting factors required for minus strand DNA transfer during reverse transcription of the genomic RNA of murine leukemia virus. J. Mol. Biol. 1998;277:225–235. doi: 10.1006/jmbi.1997.1596. [DOI] [PubMed] [Google Scholar]
- Amarasinghe G.K., De Guzman R.N., Turner R.B., Chancellor K.J., Wu Z.R., Summers M.F. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- Amarasinghe G.K., De Guzman R.N., Turner R.B., Summers M.F. NMR structure of stem-loop SL2 of the HIV-1 psi RNA packaging signal reveals a novel A-U-A base-triple platform. J. Mol. Biol. 2000;299:145–156. doi: 10.1006/jmbi.2000.3710. [DOI] [PubMed] [Google Scholar]
- Apolloni A., Hooker C.W., Mak J., Harrich D. Human immunodeficiency virus type 1 protease regulation of tat activity is essential for efficient reverse transcription and replication. J. Virol. 2003;77:9912–9921. doi: 10.1128/JVI.77.18.9912-9921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfi V., Riviere L., Jarrosson-Wuilleme L., Goujon C., Rigal D., Darlix J.L., Cimarelli A. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. J. Virol. 2008;82:6557–6565. doi: 10.1128/JVI.02321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhel N. Revisiting HIV-1 uncoating. Retrovirology. 2010;7 doi: 10.1186/1742-4690-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attal J., Theron M.C., Taboit F., Cajero-Juarez M., Kann G., Bolifraud P., Houdebine L.M. The RU5 (‘R’) region from human leukaemia viruses (HTLV-1) contains an internal ribosome entry site (IRES)-like sequence. FEBS Lett. 1996;392:220–224. doi: 10.1016/0014-5793(96)00815-0. [DOI] [PubMed] [Google Scholar]
- Auxilien S., Keith G., Le Grice S.F.J., Darlix J.L. Role of post-transcriptional modifications of primer tRNA(Lys, 3) in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription. J. Biol. Chem. 1999;274:4412–4420. doi: 10.1074/jbc.274.7.4412. [DOI] [PubMed] [Google Scholar]
- Avilov S.V., Boudier C., Gottikh M., Darlix J.L., Mely Y. Characterization of the inhibition mechanism of HIV-1 nucleocapsid protein chaperone activities by methylated oligoribonucleotides. Antimicrob. Agents Chemother. 2012;56:1010–1018. doi: 10.1128/AAC.05614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoulay J., Clamme J.P., Darlix J.L., Roques B.P., Mely Y. Destabilization of the HIV-1 complementary sequence of TAR by the nucleocapsid protein through activation of conformational fluctuations. J. Mol. Biol. 2003;326:691–700. doi: 10.1016/s0022-2836(02)01430-4. [DOI] [PubMed] [Google Scholar]
- Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- Balvay L., Lopez Lastra M., Sargueil B., Darlix J.L., Ohlmann T. Translational control of retroviruses. Nat. Rev. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bampi C., Bibillo A., Wendeler M., Divita G., Gorelick R.J., Le Grice S.F., Darlix J.L. Nucleotide excision repair and template-independent addition by HIV-1 reverse transcriptase in the presence of nucleocapsid protein. J. Biol. Chem. 2006;281:11736–11743. doi: 10.1074/jbc.M600290200. [DOI] [PubMed] [Google Scholar]
- Bampi C., Jacquenet S., Lener D., Decimo D., Darlix J.L. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Int. J. Biochem. Cell Biol. 2004;36:1668–1686. doi: 10.1016/j.biocel.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M.T., Gruninger-Leitch F., Barre-Sinoussi F., LeGrice S.F., Darlix J.L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaro G., Scozzafava A., Mastrolorenzo A., Supuran C.T. Highly active antiretroviral therapy: current state of the art, new agents and their pharmacological interactions useful for improving therapeutic outcome. Curr. Pharm. Des. 2005;11:1805–1843. doi: 10.2174/1381612053764869. [DOI] [PubMed] [Google Scholar]
- Barbour J.D., Grant R.M. The role of viral fitness in HIV pathogenesis. Curr. HIV/AIDS Rep. 2005;2:29–34. doi: 10.1007/s11904-996-0006-1. [DOI] [PubMed] [Google Scholar]
- Batisse J., Guerrero S., Bernacchi S., Sleiman D., Gabus C., Darlix J.L., Marquet R., Tisne C., Paillart J.C. The role of Vif oligomerization and RNA chaperone activity in HIV-1 replication. Virus Res. 2012;169:361–376. doi: 10.1016/j.virusres.2012.06.018. [DOI] [PubMed] [Google Scholar]
- Baudin F., Marquet R., Isel C., Darlix J.L., Ehresmann B., Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- Bazzi A., Zargarian L., Chaminade F., Boudier C., De Rocquigny H., Rene B., Mely Y., Fosse P., Mauffret O. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011;39:3903–3916. doi: 10.1093/nar/gkq1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz H., Azoulay J., Bernacchi S., Clamme J.P., Ficheux D., Roques B., Darlix J.L., Mely Y. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- Beltz H., Clauss C., Piemont E., Ficheux D., Gorelick R.J., Roques B., Gabus C., Darlix J.L., de Rocquigny H., Mely Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J. Mol. Biol. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- Berg J.M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- Berlioz C., Darlix J.L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlioz C., Torrent C., Darlix J.L. An internal ribosomal entry signal in the rat VL30 region of the Harvey murine sarcoma virus leader and its use in dicistronic retroviral vectors. J. Virol. 1995;69:6400–6407. doi: 10.1128/jvi.69.10.6400-6407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S., Freisz S., Maechling C., Spiess B., Marquet R., Dumas P., Ennifar E. Aminoglycoside binding to the HIV-1 RNA dimerization initiation site: thermodynamics and effect on the kissing-loop to duplex conversion. Nucleic Acids Res. 2007;35:7128–7139. doi: 10.1093/nar/gkm856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S., Stoylov S., Piemont E., Ficheux D., Roques B.P., Darlix J.L., Mely Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol. 2002;317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- Bess J.W., Jr., Powell P.J., Issaq H.J., Schumack L.J., Grimes M.K., Henderson L.E., Arthur L.O. Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type I, and other retroviruses. J. Virol. 1992;66:840–847. doi: 10.1128/jvi.66.2.840-847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J.M. Oncogenes and proto-oncogenes. Hosp. Pract. (Off. Ed.) 1983;18:67–74. doi: 10.1080/21548331.1983.11702609. [DOI] [PubMed] [Google Scholar]
- Bocharov G., Ford N.J., Edwards J., Breinig T., Wain-Hobson S., Meyerhans A. A genetic-algorithm approach to simulating human immunodeficiency virus evolution reveals the strong impact of multiply infected cells and recombination. J. Gen. Virol. 2005;86:3109–3118. doi: 10.1099/vir.0.81138-0. [DOI] [PubMed] [Google Scholar]
- Boudier C., Humbert N., Chaminade F., Chen Y.Y., de Rocquigny H., Godet J., Mauffret O., Fosse P., Mely Y. Dynamic interactions of the HIV-1 Tat with nucleic acids are critical for Tat activity in reverse transcription. Nucleic Acids Res. 2014;42:1065–1078. doi: 10.1093/nar/gkt934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudier C., Storchak R., Sharma K.K., Didier P., Follenius-Wund A., Muller S., Darlix J.L., Mely Y. The mechanism of HIV-1 Tat-directed nucleic acid annealing supports its role in reverse transcription. J. Mol. Biol. 2010;400:487–501. doi: 10.1016/j.jmb.2010.05.033. [DOI] [PubMed] [Google Scholar]
- Bourbigot S., Ramalanjaona N., Boudier C., Salgado G.F., Roques B.P., Mely Y., Bouaziz S., Morellet N. How the HIV-1 nucleocapsid protein binds and destabilises the (−)primer binding site during reverse transcription. J. Mol. Biol. 2008;383:1112–1128. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- Brasey A., Lopez-Lastra M., Ohlmann T., Beerens N., Berkhout B., Darlix J.L., Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner B.G., Turner D., Wainberg M.A. HIV-1 drug resistance: can we overcome? Expert Opin. Biol. Ther. 2002;2:751–761. doi: 10.1517/14712598.2.7.751. [DOI] [PubMed] [Google Scholar]
- Breuer S., Chang M.W., Yuan J.Y., Torbett B.E. Identification of HIV-1 inhibitors targeting the nucleocapsid protein. J. Med. Chem. 2012;55:4968–4977. doi: 10.1021/jm201442t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugge J.S., Erikson R.L. Identification of a transformation-specific antigen induced by an avian sarcoma virus. Nature. 1977;269:346–348. doi: 10.1038/269346a0. [DOI] [PubMed] [Google Scholar]
- Buck C.B., Shen X., Egan M.A., Pierson T.C., Walker C.M., Siliciano R.F. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckman J.S., Bosche W.J., Gorelick R.J. Human immunodeficiency virus type 1 nucleocapsid Zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Burniston M.T., Cimarelli A., Colgan J., Curtis S.P., Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid–dimer interface and the basic region of matrix protein. J. Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M., Boris-Lawrie K. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J. Virol. 2000;74:11531–11537. doi: 10.1128/jvi.74.24.11531-11537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M., Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 2002;76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S., Batson S.C., Poljak L., Mouscadet J.F., de Rocquigny H., Darlix J.L., Roques B.P., Kas E., Auclair C. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J. Virol. 1997;71:6225–6229. doi: 10.1128/jvi.71.8.6225-6229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S., Gorelick R.J., Bushman F.D. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Finet J.R., Jhon N.I., Maki A.H. p10, a low molecular weight single-stranded nucleic acid binding protein of murine leukemia retroviruses, shows stacking interactions of its single tryptophan residue with nucleotide bases. Biochemistry. 1988;27:1172–1178. doi: 10.1021/bi00404a016. [DOI] [PubMed] [Google Scholar]
- Chance M.R., Sagi I., Wirt M.D., Frisbie S.M., Scheuring E., Chen E., Bess J.W., Jr., Henderson L.E., Arthur L.O., South T.L. Extended X-ray absorption fine structure studies of a retrovirus: equine infectious anemia virus cysteine arrays are coordinated to zinc. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10041–10045. doi: 10.1073/pnas.89.21.10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C., Lee S.J., Hsiao H.H., Wu W.J., Chang W.L. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Garon C.F., Papas T.S. Native ribonucleoprotein is an efficient transcriptional complex of avian myeloblastosis virus. Proc. Natl. Acad. Sci. U.S.A. 1980;77:1296–1300. doi: 10.1073/pnas.77.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., LeGall T., Oldfield C.J., Mueller J.P., Van Y.Y.J., Romero P., Cortese M.S., Uversky V.N., Dunker A.K. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–442. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Cimarelli A., Darlix J.L. Assembling the human immunodeficiency virus type 1. Cell. Mol. Life Sci. 2002;59:1166–1184. doi: 10.1007/s00018-002-8495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A., Sandin S., Hoglund S., Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever J., Sassetti C., Parslow T.G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever J.L., Mirandar D., Jr., Parslow T.G. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 2002;76:12381–12387. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever J.L., Taplitz R.A., Lochrie M.A., Polisky B., Parslow T.G. A heterologous, high-affinity RNA ligand for human immunodeficiency virus Gag protein has RNA packaging activity. J. Virol. 2000;74:541–546. doi: 10.1128/jvi.74.1.541-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J.M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Copeland T.D., Morgan M.A., Oroszlan S. Complete amino acid sequence of the nucleic acid-binding protein of bovine leukemia virus. FEBS Lett. 1983;156:37–40. doi: 10.1016/0014-5793(83)80243-9. [DOI] [PubMed] [Google Scholar]
- Copeland T.D., Oroszlan S., Kalyanaraman V.S., Sarngadharan M.G., Gallo R.C. Complete amino acid sequence of human T-cell leukemia virus structural protein p15. FEBS Lett. 1983;162:390–395. doi: 10.1016/0014-5793(83)80793-5. [DOI] [PubMed] [Google Scholar]
- Cornille F., Mely Y., Ficheux D., Savignol I., Gerard D., Darlix J.L., Fournie-Zaluski M.C., Roques B.P. Solid phase synthesis of the retroviral nucleocapsid protein NCp10 of Moloney murine leukaemia virus and related “zinc-fingers” in free SH forms. Influence of zinc chelation on structural and biochemical properties. Int. J. Pept. Protein Res. 1990;36:551–558. [PubMed] [Google Scholar]
- Cristofari G., Darlix J.L. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Ivanyi-Nagy R., Gabus C., Boulant S., Lavergne J.P., Penin F., Darlix J.L. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004;32:2623–2631. doi: 10.1093/nar/gkh579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruceanu M., Gorelick R.J., Musier-Forsyth K., Rouzina I., Williams M.C. Rapid kinetics of protein–nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J. Mol. Biol. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- Cruceanu M., Urbaneja M.A., Hixson C.V., Johnson D.G., Datta S.A., Fivash M.J., Stephen A.G., Fisher R.J., Gorelick R.J., Casas-Finet J.R. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P., Agoston V., Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol. Sci. 2005;26:178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- D'Souza V., Summers M.F. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- D'Souza V., Summers M.F. How retroviruses select their genomes. Nat. Rev. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- Dannull J., Surovoy A., Jung G., Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J.L., Cristofari G., Rau M., Pechoux C., Berthoux L., Roques B. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol. (San Diego, CA) 2000;48:345–372. doi: 10.1016/s1054-3589(00)48011-7. [DOI] [PubMed] [Google Scholar]
- Darlix J.L., Gabus C., Nugeyre M.T., Clavel F., Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- Darlix J.L., Garrido J.L., Morellet N., Mely Y., de Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. (San Diego, CA) 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- Darlix J.L., Godet J., Ivanyi-Nagy R., Fosse P., Mauffret O., Mely Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- Darlix J.L., Lapadat-Tapolsky M., de Rocquigny H., Roques B.P. First glimpses at structure–function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- Darlix J.L., Spahr P.F. Binding sites of viral protein P19 onto Rous sarcoma virus RNA and possible controls of viral functions. J. Mol. Biol. 1982;160:147–161. doi: 10.1016/0022-2836(82)90172-3. [DOI] [PubMed] [Google Scholar]
- Darlix J.L., Spahr P.F. Isolation, characterization, genome structure of a Rous-sarcoma virus derivative defective in replication and RNA packaging. Experientia. 1985;41:797–807. [Google Scholar]
- Darlix J.L., Vincent A., Gabus C., de Rocquigny H., Roques B. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C.R. Acad. Sci. III. 1993;316:763–771. [PubMed] [Google Scholar]
- Das K., Arnold E. HIV-1 reverse transcriptase and antiviral drug resistance, Part 1. Curr. Opin. Virol. 2013;3:111–118. doi: 10.1016/j.coviro.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K., Arnold E. HIV-1 reverse transcriptase and antiviral drug resistance, Part 2. Curr. Opin. Virol. 2013;3:119–128. doi: 10.1016/j.coviro.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J., Scherer M., Tsai W.P., Long C. Low-molecular-weight Rauscher leukemia virus protein with preferential binding for single-stranded RNA and DNA. J. Virol. 1976;18:709–718. doi: 10.1128/jvi.18.2.709-718.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N.L., Rueckert R.R. Properties of a ribonucleoprotein particle isolated from Nonidet P-40-treated Rous sarcoma virus. J. Virol. 1972;10:1010–1020. doi: 10.1128/jvi.10.5.1010-1020.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman R.N., Wu Z.R., Stalling C.C., Pappalardo L., Borer P.N., Summers M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- de Rocquigny H., Ficheux D., Gabus C., Fournie-Zaluski M.C., Darlix J.L., Roques B.P. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 1991;180:1010–1018. doi: 10.1016/s0006-291x(05)81166-0. [DOI] [PubMed] [Google Scholar]
- de Rocquigny H., Gabus C., Vincent A., Fournie-Zaluski M.C., Roques B., Darlix J.L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rocquigny H., Petitjean P., Tanchou V., Decimo D., Drouot L., Delaunay T., Darlix J.L., Roques B.P. The zinc fingers of HIV nucleocapsid protein NCp7 direct interactions with the viral regulatory protein Vpr. J. Biol. Chem. 1997;272:30753–30759. doi: 10.1074/jbc.272.49.30753. [DOI] [PubMed] [Google Scholar]
- de Rocquigny H., Shvadchak V., Avilov S., Dong C.Z., Dietrich U., Darlix J.L., Mely Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini Rev. Med. Chem. 2008;8:24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- Deffaud C., Darlix J.L. Characterization of an internal ribosomal entry segment in the 5′ leader of murine leukemia virus env RNA. J. Virol. 2000;74:846–850. doi: 10.1128/jvi.74.2.846-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demene H., Dong C.Z., Ottmann M., Rouyez M.C., Jullian N., Morellet N., Mely Y., Darlix J.L., Fournie-Zaluski M.C., Saragosti S. 1H NMR structure and biological studies of the His23 → Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for virus infectivity. Biochemistry. 1994;33:11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- Dickson C., Eisenman R., Fan H., Hunter E., Reich N. Protein biosynthesis and assembly. In: Weiss R., Teich N., Varmus H., Coffin J., editors. RNA Tumor Viruses, Part 2. 2nd ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, New York: 1985. pp. 513–648. [Google Scholar]
- Dietz J., Koch J., Kaur A., Raja C., Stein S., Grez M., Pustowka A., Mensch S., Ferner J., Moller L. Inhibition of HIV-1 by a peptide ligand of the genomic RNA packaging signal Psi. ChemMedChem. 2008;3:749–755. doi: 10.1002/cmdc.200700194. [DOI] [PubMed] [Google Scholar]
- Dong C.Z., De Rocquigny H., Remy E., Mellac S., Fournie-Zaluski M.C., Roques B.P. Synthesis and biological activities of fluorescent acridine-containing HIV-1 nucleocapsid proteins for investigation of nucleic acid–NCp7 interactions. J. Pept. Res. 1997;50:269–278. doi: 10.1111/j.1399-3011.1997.tb01468.x. [DOI] [PubMed] [Google Scholar]
- Dorfman T., Luban J., Goff S.P., Haseltine W.A., Gottlinger H.G. Mapping of functionally important residues of a cysteine–histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman N., Lever A. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J. Virol. 2000;74:11413–11417. doi: 10.1128/jvi.74.23.11413-11417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druillennec S., Caneparo A., de Rocquigny H., Roques B.P. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J. Biol. Chem. 1999;274:11283–11288. doi: 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- Druillennec S., Dong C.Z., Escaich S., Gresh N., Bousseau A., Roques B.P., Fournie-Zaluski M.C. A mimic of HIV-1 nucleocapsid protein impairs reverse transcription and displays antiviral activity. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4886–4891. doi: 10.1073/pnas.96.9.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., Lawson J.D., Brown C.J., Williams R.M., Romero P., Oh J.S., Oldfield C.J., Campen A.M., Ratliff C.M., Hipps K.W. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Echols H. Nucleoprotein structures initiating DNA replication, transcription, and site-specific recombination. J. Biol. Chem. 1990;265:14697–14700. [PubMed] [Google Scholar]
- Ellermann V., Bang O. Experimentelle Leukämie bei hühnern. Zentralbl. Backteriol. 1908;46:595–609. [Google Scholar]
- Ennifar E., Paillart J.C., Bodlenner A., Walter P., Weibel J.M., Aubertin A.M., Pale P., Dumas P., Marquet R. Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell. Nucleic Acids Res. 2006;34:2328–2339. doi: 10.1093/nar/gkl317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V., Ovchinnikov L.P., Sorensen P.H.B. Y-box binding protein 1 – providing a new angle on translational regulation. Cell Cycle (Georgetown, TX) 2006;5:1143–1147. doi: 10.4161/cc.5.11.2784. [DOI] [PubMed] [Google Scholar]
- Feng Y.X., Copeland T.D., Henderson L.E., Gorelick R.J., Bosche W.J., Levin J.G., Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E.O., Mouland A.J. The cell biology of HIV-1 and other retroviruses. Retrovirology. 2006;3:77. doi: 10.1186/1742-4690-3-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freisz S., Lang K., Micura R., Dumas P., Ennifar E. Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site. Angew. Chem. Int. Ed. 2008;47:4110–4113. doi: 10.1002/anie.200800726. [DOI] [PubMed] [Google Scholar]
- Fu W., Gorelick R.J., Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabus C., Auxilien S., Pechoux C., Dormont D., Swietnicki W., Morillas M., Surewicz W., Nandi P., Darlix J.L. The prion protein has DNA strand transfer properties similar to retroviral nucleocapsid protein. J. Mol. Biol. 2001;307:1011–1021. doi: 10.1006/jmbi.2001.4544. [DOI] [PubMed] [Google Scholar]
- Gabus C., Derrington E., Leblanc P., Chnaiderman J., Dormont D., Swietnicki W., Morillas M., Surewicz W.K., Marc D., Nandi P. The prion protein has RNA binding and chaperoning properties characteristic of nucleocapsid protein NCp7 of HIV-1. J. Biol. Chem. 2001;276:19301–19309. doi: 10.1074/jbc.M009754200. [DOI] [PubMed] [Google Scholar]
- Gabus C., Ficheux D., Rau M., Keith G., Sandmeyer S., Darlix J.L. The yeast Ty3 retrotransposon contains a 5′–3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J. 1998;17:4873–4880. doi: 10.1093/emboj/17.16.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galetto R., Giacomoni V., Veron M., Negroni M. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J. Biol. Chem. 2006;281:2711–2720. doi: 10.1074/jbc.M505457200. [DOI] [PubMed] [Google Scholar]
- Galetto R., Negroni M. Mechanistic features of recombination in HIV. AIDS Rev. 2005;7:92–102. [PubMed] [Google Scholar]
- Gallant J.E., Gerondelis P.Z., Wainberg M.A., Shulman N.S., Haubrich R.H., St Clair M., Lanier E.R., Hellmann N.S., Richman D.D. Nucleoside and nucleotide analogue reverse transcriptase inhibitors: a clinical review of antiretroviral resistance. Antivir. Ther. 2003;8:489–506. [PubMed] [Google Scholar]
- Gao K., Gorelick R.J., Johnson D.G., Bushman F. Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. J. Virol. 2003;77:1598–1603. doi: 10.1128/JVI.77.2.1598-1603.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D.J., Boeke J.D., Fink G.R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42:507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- Gelderblom H.R., Hausmann E.H., Ozel M., Pauli G., Koch M.A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- Godet J., Boudier C., Humbert N., Ivanyi-Nagy R., Darlix J.L., Mely Y. Comparative nucleic acid chaperone properties of the nucleocapsid protein NCp7 and Tat protein of HIV-1. Virus Res. 2012;169:349–360. doi: 10.1016/j.virusres.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet J., de Rocquigny H., Raja C., Glasser N., Ficheux D., Darlix J.L., Mely Y. During the early phase of HIV-1 DNA synthesis, nucleocapsid protein directs hybridization of the TAR complementary sequences via the ends of their double-stranded stem. J. Mol. Biol. 2006;356:1180–1192. doi: 10.1016/j.jmb.2005.12.038. [DOI] [PubMed] [Google Scholar]
- Godet J., Mely Y. Biophysical studies of the nucleic acid chaperone properties of the HIV-1 nucleocapsid protein. RNA Biol. 2010;7:687–699. doi: 10.4161/rna.7.6.13616. [DOI] [PubMed] [Google Scholar]
- Goebel F.D., Hemmer R., Schmit J.C., Bogner J.R., de Clercq E., Witvrouw M., Pannecouque C., Valeyev R., Vandevelde M., Margery H. Phase I/II dose escalation and randomized withdrawal study with add-on azodicarbonamide in patients failing on current antiretroviral therapy. AIDS (London, England) 2001;15:33–45. doi: 10.1097/00002030-200101050-00007. [DOI] [PubMed] [Google Scholar]
- Goldschmidt V., Miller Jenkins L.M., De Rocquigny H., Darlix J., Mély Y. The nucleocapsid protein of HIV-1 as a promising therapeutic target for antiviral drugs. HIV Ther. 2010;4:179–198. [Google Scholar]
- Gorelick R.J., Chabot D.J., Ott D.E., Gagliardi T.D., Rein A., Henderson L.E., Arthur L.O. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R.J., Fu W., Gagliardi T.D., Bosche W.J., Rein A., Henderson L.E., Arthur L.O. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from moloney murine leukemia virus. J. Virol. 1999;73:8185–8195. doi: 10.1128/jvi.73.10.8185-8195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R.J., Henderson L.E., Hanser J.P., Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R.J., Nigida S.M., Jr., Bess J.W., Jr., Arthur L.O., Henderson L.E., Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger H.G. The HIV-1 assembly machine. AIDS (London, England) 2001;15(Suppl. 5):S13–S20. doi: 10.1097/00002030-200100005-00003. [DOI] [PubMed] [Google Scholar]
- Goudreau N., Hucke O., Faucher A.M., Grand-Maitre C., Lepage O., Bonneau P.R., Mason S.W., Titolo S. Discovery and structural characterization of a new inhibitor series of HIV-1 nucleocapsid function: NMR solution structure determination of a ternary complex involving a 2:1 inhibitor/NC stoichiometry. J. Mol. Biol. 2013;425:1982–1998. doi: 10.1016/j.jmb.2013.02.022. [DOI] [PubMed] [Google Scholar]
- Griffin S.D., Allen J.F., Lever A.M. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 2001;75:12058–12069. doi: 10.1128/JVI.75.24.12058-12069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorov B., Arcanger F., Roingeard P., Darlix J.L., Muriaux D. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 2006;359:848–862. doi: 10.1016/j.jmb.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Grigorov B., Bocquin A., Gabus C., Avilov S., Mely Y., Agopian A., Divita G., Gottikh M., Witvrouw M., Darlix J.L. Identification of a methylated oligoribonucleotide as a potent inhibitor of HIV-1 reverse transcription complex. Nucleic Acids Res. 2011;39:5586–5596. doi: 10.1093/nar/gkr117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorov B., Decimo D., Smagulova F., Pechoux C., Mougel M., Muriaux D., Darlix J.L. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann D., Godet J., Mely Y., Darlix J.L., Restle T. HIV-1 nucleocapsid traps reverse transcriptase on nucleic acid substrates. Biochemistry. 2008;47:12230–12240. doi: 10.1021/bi801386r. [DOI] [PubMed] [Google Scholar]
- Guo J., Henderson L.E., Bess J., Kane B., Levin J.G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard-Peron E., Muriaux D. Retroviral matrix and lipids, the intimate interaction. Retrovirology. 2011;8:15. doi: 10.1186/1742-4690-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa H. Rapid transformation of cells by Rous sarcoma virus. Proc. Natl. Acad. Sci. U.S.A. 1969;63:318–325. doi: 10.1073/pnas.63.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L.J., Chalker D.L., Sandmeyer S.B. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol. Cell. Biol. 1988;8:5245–5256. doi: 10.1128/mcb.8.12.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Shioda T., Iwakura Y., Shibuta H. RNA packaging signal of human-immunodeficiency-virus type-1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- Heilman-Miller S.L., Wu T.Y., Levin J.G. Alteration of nucleic acid structure and stability modulates the efficiency of minus-strand transfer mediated by the HIV-1 nucleocapsid protein. J. Biol. Chem. 2004;279:44154–44165. doi: 10.1074/jbc.M401646200. [DOI] [PubMed] [Google Scholar]
- Henderson L.E., Benveniste R.E., Sowder R., Copeland T.D., Schultz A.M., Oroszlan S. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne) J. Virol. 1988;62:2587–2595. doi: 10.1128/jvi.62.8.2587-2595.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson L.E., Bowers M.A., Sowder R.C., 2nd, Serabyn S.A., Johnson D.G., Bess J.W., Jr., Arthur L.O., Bryant D.K., Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson L.E., Copeland T.D., Sowder R.C., Smythers G.W., Oroszlan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J. Biol. Chem. 1981;256:8400–8406. [PubMed] [Google Scholar]
- Henderson L.E., Sowder R.C., Copeland T.D., Oroszlan S., Benveniste R.E. Gag precursors of HIV and SIV are cleaved into six proteins found in the mature virions. J. Med. Primatol. 1990;19:411–419. [PubMed] [Google Scholar]
- Heng X., Kharytonchyk S., Garcia E.L., Lu K., Divakaruni S.S., LaCotti C., Edme K., Telesnitsky A., Summers M.F. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 2012;417:224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriet S., Richer D., Bernacchi S., Decroly E., Vigne R., Ehresmann B., Ehresmann C., Paillart J.C., Marquet R. Cooperative and specific binding of Vif to the 5′ region of HIV-1 genomic RNA. J. Mol. Biol. 2005;354:55–72. doi: 10.1016/j.jmb.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Herbreteau C.H., Weill L., Decimo D., Prevot D., Darlix J.L., Sargueil B., Ohlmann T. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Hizi A., Henderson L.E., Copeland T.D., Sowder R.C., Hixson C.V., Oroszlan S. Characterization of mouse mammary tumor virus gag-pro gene products and the ribosomal frameshift site by protein sequencing. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7041–7045. doi: 10.1073/pnas.84.20.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housset V., De Rocquigny H., Roques B.P., Darlix J.L. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J. Virol. 1993;67:2537–2545. doi: 10.1128/jvi.67.5.2537-2545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L., Morichaud Z., Didierlaurent L., Muriaux D., Darlix J.L., Mougel M. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 2008;36:2311–2319. doi: 10.1093/nar/gkn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.S., Rhodes T., Dang Q., Pathak V. Retroviral recombination: review of genetic analyses. Front. Biosci. 2003;8:D143–D155. doi: 10.2741/940. [DOI] [PubMed] [Google Scholar]
- Hu W.S., Rhodes T., Dang Q., Pathak V. Retroviral recombination: review of genetic analyses. Front. Biosci. 2003;8:d143–d155. doi: 10.2741/940. [DOI] [PubMed] [Google Scholar]
- Hu W.S., Temin H.M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Huebner R.J., Todaro G.J. Oncogenes of RNA tumor viruses as determinants of cancer. Proc. Natl. Acad. Sci. U.S.A. 1969;64:1087–1094. doi: 10.1073/pnas.64.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H., Berkhout B. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 2001;29:2594–2600. doi: 10.1093/nar/29.12.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H., Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H., Berkhout B. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry. 2002;41:10439–10445. doi: 10.1021/bi025993n. [DOI] [PubMed] [Google Scholar]
- Ivanchenko S., Godinez W.J., Lampe M., Krausslich H.G., Eils R., Rohr K., Brauchle C., Muller B., Lamb D.C. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Darlix J.L. Intrinsic disorder in the core proteins of flaviviruses. Protein Pept. Lett. 2010;17:1019–1025. doi: 10.2174/092986610791498911. [DOI] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Davidovic L., Khandjian E.W., Darlix J.L. Disordered RNA chaperone proteins: from functions to disease. Cell. Mol. Life Sci. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Makowska Z., Darlix J.L. Intrinsic flexibility of nucleic acid chaperone proteins from pathogenic RNA viruses. In: Schweitzer-Stenner R., editor. Protein and Peptide Folding, Mis-folding and Non-folding. Wiley and Sons Inc.; USA: 2012. pp. 279–306. (Chapter 10) [Google Scholar]
- Jenkins L.M., Byrd J.C., Hara T., Srivastava P., Mazur S.J., Stahl S.J., Inman J.K., Appella E., Omichinski J.G., Legault P. Studies on the mechanism of inactivation of the HIV-1 nucleocapsid protein NCp7 with 2-mercaptobenzamide thioesters. J. Med. Chem. 2005;48:2847–2858. doi: 10.1021/jm0492195. [DOI] [PubMed] [Google Scholar]
- Jentoft J.E., Smith L.M., Fu X.D., Johnson M., Leis J. Conserved cysteine and histidine residues of the avian myeloblastosis virus nucleocapsid protein are essential for viral replication but are not “zinc-binding fingers”. Proc. Natl. Acad. Sci. U.S.A. 1988;85:7094–7098. doi: 10.1073/pnas.85.19.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Bieniasz P.D., Simon S.M. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008:e435. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N., Neil S.J., Bess C., Johnson M.C., Virgen C.A., Simon S.M., Bieniasz P.D. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka M., Morgan M., Binette M., Russell R.S., Rong L., Guo X., Mouland A., Kleiman L., Liang C., Wainberg M.A. The Tat protein of human immunodeficiency virus type 1 (HIV-1) can promote placement of tRNA primer onto viral RNA and suppress later DNA polymerization in HIV-1 reverse transcription. J. Virol. 2002;76:3637–3645. doi: 10.1128/JVI.76.8.3637-3645.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka M., Morgan M., Binette M., Russell R.S., Rong L.W., Guo X.F., Mouland A., Kleiman L., Liang C., Wainberg M.A. The Tat protein of human immunodeficiency virus type 1 (HIV-1) can promote placement of tRNA primer onto viral RNA and suppress later DNA polymerization in HIV-1 reverse transcription. J. Virol. 2002;76:3637–3645. doi: 10.1128/JVI.76.8.3637-3645.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpel R.L., Henderson L.E., Oroszlan S. Interactions of retroviral structural proteins with single-stranded nucleic acids. J. Biol. Chem. 1987;262:4961–4967. [PubMed] [Google Scholar]
- Katz R.A., Skalka A.M. Generation of diversity in retroviruses. Annu. Rev. Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- Kovacs D., Rakacs M., Agoston B., Lenkey K., Semrad K., Schroeder R., Tompa P. Janus chaperones: assistance of both RNA- and protein-folding by ribosomal proteins. FEBS Lett. 2009;583:88–92. doi: 10.1016/j.febslet.2008.11.049. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy G., Roques B., Darlix J.L., Mely Y. DNA condensation by the nucleocapsid protein of HIV-1: a mechanism ensuring DNA protection. Nucleic Acids Res. 2003;31:5425–5432. doi: 10.1093/nar/gkg738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuciak M., Gabus C., Ivanyi-Nagy R., Semrad K., Storchak R., Chaloin O., Muller S., Mely Y., Darlix J.L. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucleic Acids Res. 2008;36:3389–3400. doi: 10.1093/nar/gkn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay S.B., Bieniasz P.D. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6:e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M., De Rocquigny H., Van Gent D., Roques B., Plasterk R., Darlix J.L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M., Gabus C., Rau M., Darlix J.L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J. Mol. Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M., Pernelle C., Borie C., Darlix J.L. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam E., Coulaud D., Delain E., Petitjean P., Roques B.P., Gerard D., Stoylova E., Vuilleumier C., Stoylov S.P., Mely Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45:217–229. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lee B.M., De Guzman R.N., Turner B.G., Tjandra N., Summers M.F. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- Lener D., Tanchou V., Roques B.P., Le Grice S.F., Darlix J.L. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 1998;273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- Lever A., Gottlinger H., Haseltine W., Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A.M. HIV-1 RNA packaging. Adv. Pharmacol. (San Diego, CA) 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Levin J.G., Rosenak M.J. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 1976;73:1154–1158. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi S., Habchi J., Mamelli L., Darbon H. Structural disorder within Henipavirus nucleoprotein and phosphoprotein. Biophys. J. 2010;98:256a. doi: 10.1371/journal.pone.0011684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi S., Oglesbee M. Structural disorder within the measles virus nucleoprotein and phosphoprotein. Protein Pept. Lett. 2010;17:961–978. doi: 10.2174/092986610791498894. [DOI] [PubMed] [Google Scholar]
- Lopez-Lastra M., Ramdohr P., Letelier A., Vallejos M., Vera-Otarola J., Valiente-Echeverria F. Translation initiation of viral mRNAs. Rev. Med. Virol. 2010;20:177–195. doi: 10.1002/rmv.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lastra M., Rivas A., Barria M.I. Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol. Res. 2005;38:121–146. doi: 10.4067/s0716-97602005000200003. [DOI] [PubMed] [Google Scholar]
- Lori F., di Marzo Veronese F., de Vico A.L., Lusso P., Reitz M.S., Jr., Gallo R.C. Viral DNA carried by human immunodeficiency virus type 1 virions. J. Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Heng X., Garyu L., Monti S., Garcia E.L., Kharytonchyk S., Dorjsuren B., Kulandaivel G., Jones S., Hiremath A. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Heng X., Summers M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride M.S., Panganiban A.T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mely Y., Cornille F., Fournie-Zaluski M.C., Darlix J.L., Roques B.P., Gerard D. Investigation of zinc-binding affinities of Moloney murine leukemia virus nucleocapsid protein and its related zinc finger and modified peptides. Biopolymers. 1991;31:899–906. doi: 10.1002/bip.360310709. [DOI] [PubMed] [Google Scholar]
- Mely Y., De Rocquigny H., Morellet N., Roques B.P., Gerad D. Zinc binding to the HIV-1 nucleocapsid protein: a thermodynamic investigation by fluorescence spectroscopy. Biochemistry. 1996;35:5175–5182. doi: 10.1021/bi952587d. [DOI] [PubMed] [Google Scholar]
- Meric A.L., 3rd, Purtell M.J., Levy C.C. Characterization of a p30 fraction from Rauscher leukemia virus which has an associated ATPase activity. J. Biol. Chem. 1984;259:12865–12872. [PubMed] [Google Scholar]
- Meric C., Goff S.P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J. Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric C., Gouilloud E., Spahr P.F. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J. Virol. 1988;62:3328–3333. doi: 10.1128/jvi.62.9.3328-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric C., Spahr P.F. Rous-sarcoma virus nucleic acid-binding protein-p12 is necessary for viral-70S RNA dimer formation and packaging. J. Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J. Virol. 2005;79:1824–1835. doi: 10.1128/JVI.79.3.1824-1835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G., Lyonnais S., Coulaud D., Hameau L., Lafosse S., Jeusset J., Justome A., Delain E., Gorelick R.J., Le Cam E. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;364:496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Mirambeau G., Lyonnais S., Gorelick R.J. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010;7:724–734. doi: 10.4161/rna.7.6.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morellet N., Demene H., Teilleux V., Huynh-Dinh T., de Rocquigny H., Fournie-Zaluski M.C., Roques B.P. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J. Mol. Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- Morellet N., Jullian N., De Rocquigny H., Maigret B., Darlix J.L., Roques B.P. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morellet N., Meudal H., Bouaziz S., Roques B.P. Structure of the zinc finger domain encompassing residues 13–51 of the nucleocapsid protein from simian immunodeficiency virus. Biochem. J. 2006;393:725–732. doi: 10.1042/BJ20051203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Dietrich U., Manetti F., Botta M. Molecular dynamics and DFT study on HIV-1 nucleocapsid protein-7 in complex with viral genome. J. Chem. Inform. Model. 2010;50:638–650. doi: 10.1021/ci100070m. [DOI] [PubMed] [Google Scholar]
- Mori M., Manetti F., Botta M. Targeting protein–protein and protein–nucleic acid interactions for anti-HIV therapy. Curr. Pharm. Des. 2011;17:3713–3728. doi: 10.2174/138161211798220972. [DOI] [PubMed] [Google Scholar]
- Mori M., Schult-Dietrich P., Szafarowicz B., Humbert N., Debaene F., Sanglier-Cianferani S., Dietrich U., Mely Y., Botta M. Use of virtual screening for discovering antiretroviral compounds interacting with the HIV-1 nucleocapsid protein. Virus Res. 2012;169:377–387. doi: 10.1016/j.virusres.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Mougel M., Houzet L., Darlix J.L. When is it time for reverse transcription to start and go? Retrovirology. 2009;6:24. doi: 10.1186/1742-4690-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch J., Rucker E., Standker L., Adermann K., Goffinet C., Schindler M., Wildum S., Chinnadurai R., Rajan D., Specht A. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell. 2007;131:1059–1071. doi: 10.1016/j.cell.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Muriaux D., Darlix J.L. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 2010;7:744–753. doi: 10.4161/rna.7.6.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D., Mirro J., Harvin D., Rein A. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murti K.G., Bondurant M., Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous-sarcoma viruses as observed by electron-microscopy. J. Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musah R.A. The HIV-1 nucleocapsid zinc finger protein as a target of antiretroviral therapy. Curr. Top. Med. Chem. 2004;4:1605–1622. doi: 10.2174/1568026043387331. [DOI] [PubMed] [Google Scholar]
- Negroni M., Buc H. Mechanisms of retroviral recombination. Annu. Rev. Genet. 2001;35:275–302. doi: 10.1146/annurev.genet.35.102401.090551. [DOI] [PubMed] [Google Scholar]
- Ni N., Nikolaitchik O.A., Dilley K.A., Chen J., Galli A., Fu W., Prasad V.V., Ptak R.G., Pathak V.K., Hu W.S. Mechanisms of human immunodeficiency virus type 2 RNA packaging: efficient trans packaging and selection of RNA copackaging partners. J. Virol. 2011;85:7603–7612. doi: 10.1128/JVI.00562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T., Lopez-Lastra M., Darlix J.L. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- Onafuwa-Nuga A., Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 2009;73:451–480. doi: 10.1128/MMBR.00012-09. (table of contents) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Ablan S.D., Lockett S.J., Nagashima K., Freed E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Freed E.O. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott D.E., Coren L.V., Shatzer T. The nucleocapsid region of human immunodeficiency virus type 1 Gag assists in the coordination of assembly and Gag processing: role for RNA–Gag binding in the early stages of assembly. J. Virol. 2009;83:7718–7727. doi: 10.1128/JVI.00099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pager J., Coulaud D., Delain E. Electron microscopy of the nucleocapsid from disrupted Moloney murine leukemia virus and of associated type VI collagen-like filaments. J. Virol. 1994;68:223–232. doi: 10.1128/jvi.68.1.223-232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchen-Matthews A., Kramer B., Marsh M. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 2003;162:443–455. doi: 10.1083/jcb.200304008. Epub 2003 Jul 2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloponese J.M., Collette Y., Gregoire C., Bailly C., Campese D., Meurs E.F., Olive D., Loret E.P. Full peptide synthesis, purification, and characterization of six Tat variants – differences observed between HIV-1 isolates from Africa and other continents. J. Biol. Chem. 1999;274:11473–11478. doi: 10.1074/jbc.274.17.11473. [DOI] [PubMed] [Google Scholar]
- Poljak L., Batson S.M., Ficheux D., Roques B.P., Darlix J.L., Kas E. Analysis of NCp7-dependent activation of HIV-1 cDNA integration and its conservation among retroviral nucleocapsid proteins. J. Mol. Biol. 2003;329:411–421. doi: 10.1016/s0022-2836(03)00472-8. [DOI] [PubMed] [Google Scholar]
- Prats A.C., Housset V., de Billy G., Cornille F., Prats H., Roques B., Darlix J.L. Viral RNA annealing activities of the nucleocapsid protein of Moloney murine leukemia virus are zinc independent. Nucleic Acids Res. 1991;19:3533–3541. doi: 10.1093/nar/19.13.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A.C., Roy C., Wang P.A., Erard M., Housset V., Gabus C., Paoletti C., Darlix J.L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J. Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A.C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J.L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugatsch T., Stacey D.W. Identification of a sequence likely to be required for avian retroviral packaging. Virology. 1983;128:505–511. doi: 10.1016/0042-6822(83)90279-9. [DOI] [PubMed] [Google Scholar]
- Pustowka A., Dietz J., Ferner J., Baumann M., Landersz M., Konigs C., Schwalbe H., Dietrich U. Identification of peptide ligands for target RNA structures derived from the HIV-1 packaging signal psi by screening phage-displayed peptide libraries. ChemBioChem. 2003;4:1093–1097. doi: 10.1002/cbic.200300681. [DOI] [PubMed] [Google Scholar]
- Raja C., Ferner J., Dietrich U., Avilov S., Ficheux D., Darlix J.L., de Rocquigny H., Schwalbe H., Mely Y. A tryptophan-rich hexapeptide inhibits nucleic acid destabilization chaperoned by the HIV-1 nucleocapsid protein. Biochemistry. 2006;45:9254–9265. doi: 10.1021/bi052560m. [DOI] [PubMed] [Google Scholar]
- Ramboarina S., Srividya N., Atkinson R.A., Morellet N., Roques B.P., Lefevre J.F., Mely Y., Kieffer B. Effects of temperature on the dynamic behaviour of the HIV-1 nucleocapsid NCp7 and its DNA complex. J. Mol. Biol. 2002;316:611–627. doi: 10.1006/jmbi.2001.5379. [DOI] [PubMed] [Google Scholar]
- Raposo G., Moore M., Innes D., Leijendekker R., Leigh-Brown A., Benaroch P., Geuze H. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic (Copenhagen, Denmark) 2002;3:718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- Rein A., Datta S.A., Jones C.P., Musier-Forsyth K. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 2011;36:373–380. doi: 10.1016/j.tibs.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A., Henderson L.E., Levin J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- Resh M.D. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev. 2005;7:84–91. [PubMed] [Google Scholar]
- Rice W.G., Schaeffer C.A., Harten B., Villinger F., South T.L., Summers M.F., Henderson L.E., Bess J.W., Jr., Arthur L.O., McDougal J.S. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature. 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- Rice W.G., Supko J.G., Malspeis L., Buckheit R.W., Jr., Clanton D., Bu M., Graham L., Schaeffer C.A., Turpin J.A., Domagala J. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science. 1995;270:1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]
- Rous P. A transmissible avian neoplasm:sarcoma for the common fowl. J. Exp. Med. 1910;12:696–705. doi: 10.1084/jem.12.5.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner L., Nydegger S., Coren L.V., Nagashima K., Thali M., Ott D.E. Dynamic fluorescent imaging of human immunodeficiency virus type 1 gag in live cells by biarsenical labeling. J. Virol. 2005;79:4055–4065. doi: 10.1128/JVI.79.7.4055-4065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J.S., Miller J., Tai J., Kim A., Ghanam R.H., Summers M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K., Zambrano N., Baldwin E.T., Shapiro B.A., Erickson J.W., Omichinski J.G., Clore G.M., Gronenborn A.M., Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schito M.L., Goel A., Song Y., Inman J.K., Fattah R.J., Rice W.G., Turpin J.A., Sher A., Appella E. In vivo antiviral activity of novel human immunodeficiency virus type 1 nucleocapsid p7 zinc finger inhibitors in a transgenic murine model. AIDS Res. Hum. Retroviruses. 2003;19:91–101. doi: 10.1089/088922203762688595. [DOI] [PubMed] [Google Scholar]
- Schroeder R., Barta A., Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- Sen A., Sherr C.J., Todaro G.J. Phosphorylation of murine type-C viral p12 proteins regulates their extent of binding to homologous viral-RNA. Cell. 1977;10:489–496. doi: 10.1016/0092-8674(77)90036-8. [DOI] [PubMed] [Google Scholar]
- Sharma K.K., de Rocquigny H., Darlix J.L., Lavergne J.P., Penin F., Lessinger J.M., Mely Y. Analysis of the RNA chaperoning activity of the hepatitis C virus core protein on the conserved 3′X region of the viral genome. Nucleic Acids Res. 2012;40:2540–2553. doi: 10.1093/nar/gkr1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubsda M.F., Paoletti A.C., Hudson B.S., Borer P.N. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry. 2002;41:5276–5282. doi: 10.1021/bi016045+. [DOI] [PubMed] [Google Scholar]
- Shvadchak V., Sanglier S., Rocle S., Villa P., Haiech J., Hibert M., Van Dorsselaer A., Mely Y., de Rocquigny H. Identification by high throughput screening of small compounds inhibiting the nucleic acid destabilization activity of the HIV-1 nucleocapsid protein. Biochimie. 2009;91:916–923. doi: 10.1016/j.biochi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Sosic A., Frecentese F., Perissutti E., Sinigaglia L., Santagada V., Caliendo G., Magli E., Ciano A., Zagotto G., Parolin C. Desig, synthesis and biological evaluation of TAR and cTAR binders as HIV-1 nucleocapsid inhibitors. MedChemComm. 2013;4:1388–1393. [Google Scholar]
- South T.L., Blake P.R., Sowder R.C., 3rd, Arthur L.O., Henderson L.E., Summers M.F. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry. 1990;29:7786–7789. doi: 10.1021/bi00486a002. [DOI] [PubMed] [Google Scholar]
- Spriggs S., Garyu L., Connor R., Summers M.F. Potential intra- and intermolecular interactions involving the unique-5′ region of the HIV-1 5′-UTR. Biochemistry. 2008;47:13064–13073. doi: 10.1021/bi8014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P., Schito M., Fattah R.J., Hara T., Hartman T., Buckheit R.W., Jr., Turpin J.A., Inman J.K., Appella E. Optimization of unique, uncharged thioesters as inhibitors of HIV replication. Bioorg. Med. Chem. 2004;12:6437–6450. doi: 10.1016/j.bmc.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Stenbak C.R., Linial M.L. Role of the C terminus of foamy virus Gag in RNA packaging and Pol expression. J. Virol. 2004;78:9423–9430. doi: 10.1128/JVI.78.17.9423-9430.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen A.G., Worthy K.M., Towler E., Mikovits J.A., Sei S., Roberts P., Yang Q.E., Akee R.K., Klausmeyer P., McCloud T.G. Identification of HIV-1 nucleocapsid protein: nucleic acid antagonists with cellular anti-HIV activity. Biochem. Biophys. Res. Commun. 2002;296:1228–1237. doi: 10.1016/s0006-291x(02)02063-6. [DOI] [PubMed] [Google Scholar]
- Stoylov S.P., Vuilleumier C., Stoylova E., De Rocquigny H., Roques B.P., Gerard D., Mely Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Summers M.F., Henderson L.E., Chance M.R., Bess J.W., Jr., South T.L., Blake P.R., Sagi I., Perez-Alvarado G., Sowder R.C., 3rd, Hare D.R. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers M.F., South T.L., Kim B., Hare D.R. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry. 1990;29:329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- Sundquist W.I., Krausslich H.G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovoy A., Dannull J., Moelling K., Jung G. Conformational and nucleic acid binding studies on the synthetic nucleocapsid protein of HIV-1. J. Mol. Biol. 1993;229:94–104. doi: 10.1006/jmbi.1993.1011. [DOI] [PubMed] [Google Scholar]
- Tanchou V., Decimo D., Pechoux C., Lener D., Rogemond V., Berthoux L., Ottmann M., Darlix J.L. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J. Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanchou V., Gabus C., Rogemond V., Darlix J.L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J. Mol. Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- Thomas J.A., Bosche W.J., Shatzer T.L., Johnson D.G., Gorelick R.J. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J. Virol. 2008;82:9318–9328. doi: 10.1128/JVI.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.A., Gorelick R.J. Nucleocapsid protein function in early infection processes. Virus Res. 2008;134:39–63. doi: 10.1016/j.virusres.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.A., Shulenin S., Coren L.V., Bosche W.J., Gagliardi T.D., Gorelick R.J., Oroszlan S. Characterization of human immunodeficiency virus type 1 (HIV-1) containing mutations in the nucleocapsid protein at a putative HIV-1 protease cleavage site. Virology. 2006;354:261–270. doi: 10.1016/j.virol.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Tompa P., Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J. Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummino P.J., Scholten J.D., Harvey P.J., Holler T.P., Maloney L., Gogliotti R., Domagala J., Hupe D. The in vitro ejection of zinc from human immunodeficiency virus (HIV) type 1 nucleocapsid protein by disulfide benzamides with cellular anti-HIV activity. Proc. Natl. Acad. Sci. U.S.A. 1996;93:969–973. doi: 10.1073/pnas.93.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K.B., Hagan N.A., Fabris D. Inhibitory effects of archetypical nucleic acid ligands on the interactions of HIV-1 nucleocapsid protein with elements of Psi-RNA. Nucleic Acids Res. 2006;34:1305–1316. doi: 10.1093/nar/gkl004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K.B., Kohlway A.S., Hagan N.A., Fabris D. Noncovalent probes for the investigation of structure and dynamics of protein–nucleic acid assemblies: the case of NC-mediated dimerization of genomic RNA in HIV-1. Biopolymers. 2009;91:283–296. doi: 10.1002/bip.21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin J.A. The next generation of HIV/AIDS drugs: novel and developmental antiHIV drugs and targets. Expert Rev. Anti Infect. Ther. 2003;1:97–128. doi: 10.1586/14787210.1.1.97. [DOI] [PubMed] [Google Scholar]
- Turpin J.A., Schito M.L., Jenkins L.M., Inman J.K., Appella E. Topical microbicides: a promising approach for controlling the AIDS pandemic via retroviral zinc finger inhibitors. Adv. Pharmacol. (San Diego, CA) 2008;56:229–256. doi: 10.1016/S1054-3589(07)56008-4. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S., Waysbort A., Marenda M., Gensac M.C., Amalric F., Prats A.C. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J. Biol. Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- Vandevelde M., Witvrouw M., Schmit J.C., Sprecher S., De Clercq E., Tassignon J.P. ADA, a potential anti-HIV drug. AIDS Res. Hum. Retroviruses. 1996;12:567–568. doi: 10.1089/aid.1996.12.567. [DOI] [PubMed] [Google Scholar]
- Vercruysse T., Basta B., Dehaen W., Humbert N., Balzarini J., Debaene F., Sanglier-Cianferani S., Pannecouque C., Mely Y., Daelemans D. A phenyl-thiadiazolylidene-amine derivative ejects zinc from retroviral nucleocapsid zinc fingers and inactivates HIV virions. Retrovirology. 2012;9:95. doi: 10.1186/1742-4690-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain-Hobson S., Renoux-Elbe C., Vartanian J.P., Meyerhans A. Network analysis of human and simian immunodeficiency virus sequence sets reveals massive recombination resulting in shorter pathways. J. Gen. Virol. 2003;84:885–895. doi: 10.1099/vir.0.18894-0. [DOI] [PubMed] [Google Scholar]
- Wallace G.S., Cheng-Mayer C., Schito M.L., Fletcher P., Miller Jenkins L.M., Hayashi R., Neurath A.R., Appella E., Shattock R.J. Human immunodeficiency virus type 1 nucleocapsid inhibitors impede trans infection in cellular and explant models and protect nonhuman primates from infection. J. Virol. 2009;83:9175–9182. doi: 10.1128/JVI.00820-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warui D.M., Baranger A.M. Identification of specific small molecule ligands for stem loop 3 ribonucleic acid of the packaging signal Psi of human immunodeficiency virus-1. J. Med. Chem. 2009;52:5462–5473. doi: 10.1021/jm900599v. [DOI] [PubMed] [Google Scholar]
- Williams M.C., Rouzina I., Wenner J.R., Gorelick R.J., Musier-Forsyth K., Bloomfield V.A. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. Intrinsically unstructured proteins: re-assessing the protein structure–function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Wu T., Datta S.A., Mitra M., Gorelick R.J., Rein A., Levin J.G. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: biological implications. Virology. 2010;405:556–567. doi: 10.1016/j.virol.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Henderson L.E., Copeland T.D., Gorelick R.J., Bosche W.J., Rein A., Levin J.G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. HIV-1 gene expression: lessons from provirus and non-integrated DNA. Retrovirology. 2004;1:13. doi: 10.1186/1742-4690-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.T., Marsh J.W. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science. 2001;293:1503–1506. doi: 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- Yan N., Regalado-Magdos A.D., Stiggelbout B., Lee-Kirsch M.A., Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J.C., McHenry C.S. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J. Biol. Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]
- You J.C., McHenry C.S. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- Yu Q., Darlix J.L. The zinc finger of nucleocapsid protein of Friend murine leukemia virus is critical for proviral DNA synthesis in vivo. J. Virol. 1996;70:5791–5798. doi: 10.1128/jvi.70.9.5791-5798.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Bagasra O., Niikura M., Poiesz B.J., Pomerantz R.J. Intravirion reverse transcripts in the peripheral-blood plasma of human-immunodeficiency-virus type 1-infected individuals. J. Virol. 1994;68:7591–7597. doi: 10.1128/jvi.68.11.7591-7597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Dornadula G., Beumont M., Livornese L., Van Uitert B., Henning K., Pomerantz R.J. Human immunodeficiency virus type I in the semen of men receiving highly active antiretroviral therapy. N. Engl. J. Med. 1998;339:1803–1809. doi: 10.1056/NEJM199812173392502. [DOI] [PubMed] [Google Scholar]
- Zhang H., Dornadula G., Pomerantz R.J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J. Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Temin H.M. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science. 1993;259:234–238. doi: 10.1126/science.8421784. [DOI] [PubMed] [Google Scholar]


