Highlights
-
•
Cyclophilin A (CypA) is a host factor for human coronavirus NL63 replication.
-
•
CypA is a target for anti-coronaviral therapy.
-
•
Non-immunosuppressive CsA derivatives (Alisporivir, NIM811) inhibit CoV replication.
-
•
New classes of non-immunosuppressive CsA/FK506 derivatives inhibit CoV replication.
Abbreviations: EC50, 50% effective inhibitory concentration; CsA/CsD, cyclosporine A/D; PPIase, peptidyl prolyl cis/trans isomerase; CypA/B, cyclophilin A/B; ALV, Alisporivir; FKBP, FK506-binding protein
Keywords: HCoV-NL63, Cyclosporine/FK506-non-immunosuppressive derivatives, Inhibition of viral replication, Cyclophilin A, FKBP
Abstract
Until recently, there were no effective drugs available blocking coronavirus (CoV) infection in humans and animals. We have shown before that CsA and FK506 inhibit coronavirus replication (Carbajo-Lozoya, J., Müller, M.A., Kallies, S., Thiel, V., Drosten, C., von Brunn, A. Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Res. 2012; Pfefferle, S., Schöpf, J., Kögl, M., Friedel, C., Müller, M.A., Stellberger, T., von Dall’Armi, E., Herzog, P., Kallies, S., Niemeyer, D., Ditt, V., Kuri, T., Züst, R., Schwarz, F., Zimmer, R., Steffen, I., Weber, F., Thiel, V., Herrler, G., Thiel, H.-J., Schwegmann-Weßels, C., Pöhlmann, S., Haas, J., Drosten, C. and von Brunn, A. The SARS-Coronavirus-host interactome: identification of cyclophilins as target for pan-Coronavirus inhibitors. PLoS Pathog., 2011). Here we demonstrate that CsD Alisporivir, NIM811 as well as novel non-immunosuppressive derivatives of CsA and FK506 strongly inhibit the growth of human coronavirus HCoV-NL63 at low micromolar, non-cytotoxic concentrations in cell culture. We show by qPCR analysis that virus replication is diminished up to four orders of magnitude to background levels. Knockdown of the cellular Cyclophilin A (CypA/PPIA) gene in Caco-2 cells prevents replication of HCoV-NL63, suggesting that CypA is required for virus replication. Collectively, our results uncover Cyclophilin A as a host target for CoV infection and provide new strategies for urgently needed therapeutic approaches.
1. Introduction
Coronaviruses cause severe diseases of the respiratory and gastrointestinal tract and the central nervous system in animals (Perlman and Netland, 2009). The infection of humans with HCoV-OC43 and HCoV-229E are known since the mid sixities to be associated with respiratory tract i.e. common cold-like diseases. SARS-CoV (severe acute respiratory syndrome-Corona Virus) is a highly aggressive human agent, causing the lung disease SARS, with often fatal outcome (Drosten et al., 2003). This virus appeared as an epidemic in 2003 after it had crossed the species barrier most likely from bats to civet cats and humans demonstrating the potential of coronaviruses to cause high morbidity and mortality in humans (Lau et al., 2005, Li et al., 2005). As no treatment was available, the epidemic could eventually be controlled by highly effective traditional public health measures of quarantine and case isolation. The strains HCoV-NL63 and HCoV-HKU1 were discovered in 2004 and 2005, respectively (van der Hoek et al., 2004, Woo et al., 2005). They cause more severe lower respiratory tract infections like bronchiolitis and pneumonia especially in young children (van der Hoek, 2007). In 2012, a new human CoV MERS (Middle East Respiratory Syndrome virus, previously called “EMC”) emerged from the Middle East with clinical outcomes such as renal failure and acute pneumonia, similar to those of SARS-CoV but with an even higher mortality rate of about 50% (de Groot et al., 2013, van Boheemen et al., 2012, Zaki et al., 2012).
Human coronaviruses cause approximately 10–15% of all upper and lower respiratory tract infections. They account for significant hospitalizations of children under 18 years of age, the elderly and immunocompromised individuals. According to a number of international studies 1- 10% of the acute respiratory diseases are caused by HCoV-NL63 (for review see Abdul-Rasool and Fielding, 2010). These numbers are probably an underestimation with regard to the general population since during routine diagnostic screening for respiratory viruses tests for HCoV are frequently not included. An important aspect of HCoV-NL63 infection is the co-infection with other human coronaviruses, influenza A, respiratory syncytial virus (RSV), parainfluenza virus or human metapneumovirus (Abdul-Rasool and Fielding, 2010). In children they are associated with acute respiratory tract illness, pneumonia and Croup leading in many cases to hospitalization. In a recent epidemiological study out of 1471 hospitalized children (<2years) 207 (14%) were HCoV-positive (Dijkman et al., 2012). Infection frequencies in children with mild symptoms and in hospitalized children occurred in the order HCoV-OC43 > HCoV-NL63 > HCoV-HKU1 > HCoV-229E. In a large-scale survey on 11,661 diagnostic respiratory samples collected in Edinburgh, UK, between 2006 and 2009, 267 (2.30%) were positive for at least one coronavirus accounting for 8.15% of all virus detections (Gaunt et al., 2010). 11% to 41% of coronaviruses detected were present in samples tested positive for other respiratory viruses (e.g. RSV).
Inhibitors of coronavirus enzymes (reviewed by Tong, 2009a, Tong, 2009b) and compounds inhibiting in vitro replication have been described (Kono et al., 2008, Milewska et al., 2013, Pyrc et al., 2006, te Velthuis et al., 2010, Vincent et al., 2005). The most instensely studied anti-viral compounds are directed against viral proteases not present in the mammalian host (Chaudhuri et al., 2011, Chuck et al., 2011, Chuck et al., 2013, Yang et al., 2005, Zhu et al., 2011). However, clinically licensed antivirals for coronavirus infection are absent. Coronaviruses represent the group of the largest single-stranded RNA viruses with plus strand orientation. In general, RNA viruses replicate at low fidelity and are thus prone to rapid evolutionary changes. Although coronaviruses encode a proofreading exoribonuclease (nsp14 ExoN) increasing replicative robustness of its large genomes, mutations within this domain increase mutation rates significantly (Smith and Denison, 2013). Virus replication depends on a variety of host factors (de Haan and Rottier, 2006, Vogels et al., 2011, Wang and Li, 2012) which represent potential antiviral targets. These might be more preferable targets than viral proteins as development of resistance is much less likely.
In a recent study we performed a genome-wide SARS-CoV yeast-two-hybrid interaction screen with human cDNA libraries identifying human immunophilins (including cyclophilins [Cyps] and FK506-binding proteins [FKBPs] as interaction partners of CoV non-structural protein 1 [Nsp1] (Pfefferle et al., 2011). A pronounced feature of most mammalian cyclophilins is their ability to bind the immunosuppressive drug cyclosporine A (CsA). We showed that the drug acts as a replication inhibitor of a number of human (SARS-CoV, HCoV-NL63 and HCoV-229E) and animal coronaviruses (Feline CoV [serotypes I and II], porcine transmissible gastroenteritis virus (TGEV), and avian infectious bronchitis virus [IBV]) suggesting host cyclophilins as targets for pan-coronavirus inhibition (Pfefferle et al., 2011). Inhibition of SARS-CoV, HCoV-229E and in addition of Mouse Hepatitis virus (MHV) was subsequently also confirmed by de Wilde et al. (2011). Inhibition of feline CoV replication was also found by Tanaka et al. (2012). Similarly, we showed that FK506 inhibits the replication of SARS-CoV, HCoV-NL63 and HCoV-229E and the dependence of HCoV-NL63 on FKBP1A/B (Carbajo-Lozoya et al., 2012).
Cyclophilins and FKBPs represent large, independent families of peptidyl-prolyl cis/trans isomerases (PPIases, EC number 5.2.1.8) thus exerting important functions on folding, maturation and trafficking of proteins within the eukaryotic cell (Blackburn and Walkinshaw, 2011, Davis et al., 2010). Both CsA and FK506 act as tight-binding, reversible and competitive inhibitors of the active site of these enzymes (Fischer et al., 1989). Physical interaction of cyclophilins with viral proteins, and thus replication sensitivity to CsA have been shown for several viruses, e.g. the capsid proteins of Human Immunodeficiency Virus (HIV-1) (Strebel et al., 2009, Ylinen et al., 2009) and Human Papilloma virus (HPV) types 16 (Bienkowska-Haba et al., 2009), the N protein of Vesicular stomatitis Virus (Bose et al., 2003), the NS5a of Hepatitis C Virus (HCV) (Fernandes et al., 2010, Fischer et al., 1989), the NS4A protein of the mosquito-borne Japanese encephalitis virus (Kambara et al., 2011), the NS5 protein of West Nile virus (Qing et al., 2009) and the M1 protein of influenza A virus (Liu et al., 2009). The most prominent cyclophilins thought to be involved are CypA and CypB. PPIase-independent activities of CsA and FK506 exerted by gain-of-function, result from the binary complexes formed by binding of the drugs to Cyps and FKBPs, respectively. Based on the inhibition of the protein phosphatase activity of calcineurin, these complexes block the cellular calcineurin (CaN)/NFAT pathway thereby interfering with T-cell activation and Il-2 production. Chemically changed derivatives covering specific side-chain modifications, the so-called non-immunosuppressive cyclosporine or FK506 analogues, can discriminate between alternative signalling pathways either based on PPIase- or CaN-inhibiting functions.
Identifying the interaction of the SARS-CoV Nsp1 protein with Cyps and FKBPs, and the sensitivity of CoV replication to both drugs, CsA and FK506, we suggested CsA as a potential pan-CoV inhibitor (Pfefferle et al., 2011). Here we demonstrate, by using Alisporivir, NIM811 and a series of newly synthesized CsA and FK506 derivatives, inhibition of HCoV-NL63 replication independent of the immunosuppressive character of the compounds. We further show that CypA but not CypB is required for virus replication.
2. Materials and methods
2.1. Antibodies and drugs
Mouse antibody 1H11 (1:20,000) recognizing HCoV-NL63 N-protein was obtained from INGENASA, Spain (Sastre et al., 2011). Anti-Lamin A (1:20,000) was purchased from Biomol, Hamburg, Germany. Goat-anti-Lamin B (1:400), rabbit anti-CypA (1:2000) and rabbit anti-CypB (1:1000) were obtained from Santa Cruz Biotechnology, Enzo Life Sciences and Abcam, respectively. Secondary antibodies were received from Dianova (goat anti-rabbit-Ig-horse radish peroxidase HRP, [1:3000] and rabbit-anti-goat-Ig-HRP [1:3000]) and Sigma (anti-mouse-Ig-HRP [1:40,000]).
Compounds 1, 2, 3, 4, and 5 were synthesized as previously described (Malesevic et al., 2013, Prell et al., 2013). The synthesis of 6 will be described elsewhere. Alisporivir and NIM811 were generously provided by Novartis (Switzerland). CsA, CsD and FK506 were obtained from Sigma-Aldrich, Santa Cruz (Germany) and Enzo Life Sciences (Germany), respectively.
2.2. NFAT reporter gene assay
The tests were performed as described (Prell et al., 2013). Briefly, Jurkat cells were transfected with NFAT reporter gene plasmid and incubated with 0.5 μM inhibitor or 0.5% DMSO (control) for 30 min. Ca2+ mobilization was initiated by phorbol 12-myristate 13-acetate/ionomycin or tumor necrosis factor-α and cultured for additional 5 h before harvesting and determining luciferase activity in cell lysates. NFAT activities are expressed as mean SD of triplicates in three independent experiments.
2.3. NFAT-GFP nuclear translocation assay
HeLa cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% FCS and 1% penicillin/streptomycin. NFAT3-GFP plasmid was transfected into HeLa cells by using Lipofectamine LTX and Plus Reagent (Life Technologies) when the cells were 70% confluent. Subsequently, drugs were added to the medium at a final concentration of 40 nM. 19 h later, ionomycin was added at a final concentration of 2 μM to induce NFAT3-GFP translocation. Pictures were taken using a fluorescence microscope (Leica DM4000 B) 10 min after ionomycin induction.
2.4. RNA isolation and real-time reverse transcription-PCR (RT-PCR)
CaCo-2 cells were infected with HCoV-NL63 at MOI 0.004 for 1 h. After removal of virus inoculum and two PBS washes, fresh medium supplemented with increasing inhibitor concentrations was added. After 48 h RNA was extracted from 10 μl culture supernatant using the High Pure Viral Nucleic Acid Kit (Roche) and eluted in 13 μl. Quantification was done by real-time PCR SensiFAST Probe Hi-ROX One-Step kit (Bioline GmbH, Germany) allowing reverse transcription, cDNA synthesis and PCR amplification in a single step. Samples were analyzed by ABI Prism 7000 Cycler I Sequencing Detection System. A standard curve was produced using serial dilutions of viral RNA of HCoV-NL63 virus stock with known virus titer.
PCR primers (Herzog et al., 2008) used were NL-63RF2for 5′-CTTCTGGTGACGCTAGTACAGCTTAT-3′ (genome position nt 14459–14484) and NL-63RR2rev 5′-AGACGTCGTTGTAGATCCCTAACAT-3′ (genome position nt 14573–14597) and NL-63 probe was 5′-FAMCAGGTTGCTTAGTGTCCCATCAGATTCAT-TAMRA-3′ (genome position nt 14532–14560).
2.5. Western blotting
To determine N-protein expression in the presence of inhibitors Caco-2 cells were infected at virus MOI 0.004 for one hour in six-well plates. Virus was washed off with PBS and inhibitors were added to the medium at the respective concentrations. After 48 h cells were harvested and lysed with 1% NP-40 in 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10 mM DTT and Protease Inhibitor Cocktail (Hoffmann La Roche) in 250 μl lysis buffer. Proteins were separated by 8% or 12.5% SDS-PAGE and electroblotted onto nitrocellulose membranes. Latter were blocked with 5% milk powder in TBST (150 mM NaCl, 20 mM Tris–HCl [pH 7.6], 0.1% Tween 20) buffer. Incubation with primary antibodies was usually carried out at 4 °C overnight. Secondary antibody incubation was performed at room temperature for 2 h. After each incubation step membranes were washed three times with TBST for 10 min. HRP was developed with Immobilon Western blot HRP chemiluminiscent substrate from Milipore. Membranes were exposed to X-ray film (Agfa).
2.6. Cyclophilin knockdown cell lines.
CaCo-2 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% FCS and 1% penicillin/streptomycin. Cyclophilin knockdown cell lines were generated using shRNA expression vectors (Sirion GmbH, Martinsried, Germany) as recently described for FKBP1A/B (Carbajo-Lozoya et al., 2012). Briefly, cells were transduced at MOI 30 with MISSIONTM lentiviral non-target control gene, PPIA from a target set SHVRS-NM_021130 (TRCN000049232) and PPIB from a target set SHVRSNM_000942 (TRCN0000049251). Sequences used for gene knockdown are listed in Table 1 . Stably shRNA-expressing cells were generated through 3 weeks of bulk-selection in 10–15 μg/ml puromycin-containing medium (DMEM + 10% FCS + 2 mM l-glutamine + 1 mM Na-pyruvate).
Table 1.
shRNA sequences used for lentiviral-based gene knockdown (A) and sequences of primers used for quantification of gene knockdown (B).
| Particle set | Target | shRNA sequence (5′ ≥ 3′) |
|---|---|---|
| (A) | ||
| Non-target control TRC1.5 Vector (pLKO.1-puro) | CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTG | |
| PPIA: SHVRS-_NM_021130 | TRCN000049232 | CCGGGTTCCTGCTTTCACAGAATTACTCGAGTAATTCTGTGAAAGCAGGAACTTTTTG |
| PPIB: SHVRS-NM_000942 | TRCN000049251 | CCGGGTTCTTCATCACGACAGTCAACTCGAGTTGACTGTCGTGATGAAGAACTTTTTG |
| (B) | |
| qRT-PCR primer | Sequence (5′ ≥ 3′) |
| hPPIA_F | CAGACAAGGTCCCAAAGACAG |
| hPPIA_R | TTGCCATCCAACCACTCAGTC |
| hPPIB_F | CTCTCCGAACGCAACATGAAG |
| hPPIB_R | ACCTTGACGGTGACTTTGGG |
3. Results
3.1. Characterization of non-immunosuppressive CsA- and FK506-derivatives
First CsA analogues were isolated in 1977 from Trichoderma polysporum (Traber et al., 1977). CsD was described in 1993 as a weak immunosuppressant in lymphocyte proliferation assays with about 10% of the CsA activity (Sadeg et al., 1993a, Sadeg et al., 1993b). In CsD a valine is located at position 2 instead of l-α-aminobutyric acid. The two prominent CsA derivatives NIM811 (contains a methyl-isoleucine at position 4 instead of the methyl–leucine) and Alisporivir (contains a methyl–alanine at position 3 instead of sarcosine and an N-ethyl valine at position 4, instead of N-methyl leucine) were intensively tested in clinical trials as anti- HIV-1 and anti-HCV drugs (Fischer et al., 2010, Gallay and Lin, 2013, Lin and Gallay, 2013, Membreno et al., 2013, Vermehren and Sarrazin, 2011).
A further set of CsA analogues was developed, fine-tuned by derivatization at MeBmt residue 1 of CsA and a FK506 derivative with different properties regarding the inhibition of drug-Cyp/FKBP complexes, CaN phosphatase-, NFAT activities (Fig. 1 , Table 2 ). Synthesis of drug derivatives compounds 1 and 2 were previously described (Prell et al., 2013). The CsA derivatives 3, 4, 5 were synthesized as recently published (Malesevic et al., 2013). The chemical synthesis of the non-immunosuppressive FK506 analogue 6 starting from the parent drug FK506 will be published elsewhere.
Fig. 1.
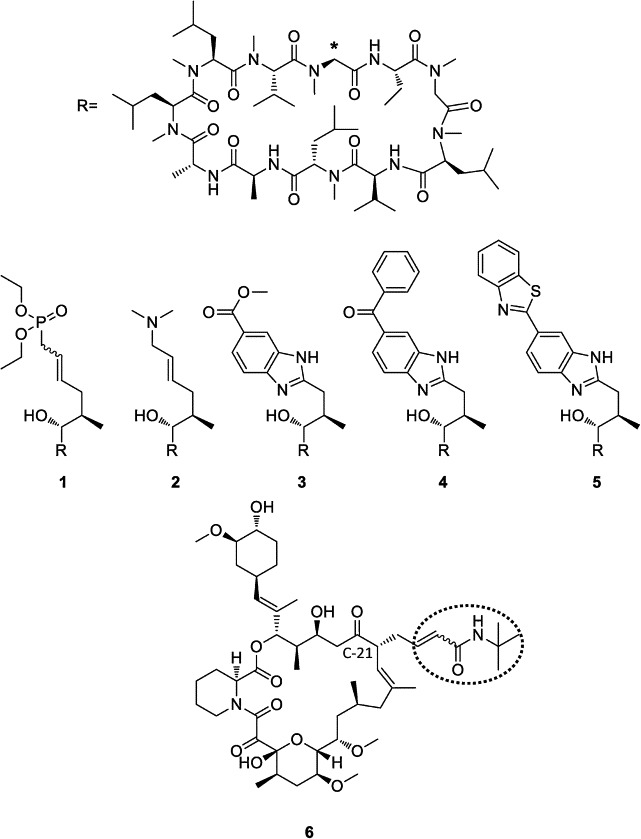
Chemical structures of novel CsA derivatives (compounds 1–5) and the FK506 derivative (compound 6) used in this work. The position where the R substituents are attached to the CsA ring core at position 1 is marked with an asterisk. Parent FK506 is substituted at position C-21 (dotted circle).
Table 2.
Biochemical characterization of PPIase inhibitor compounds.
| Name | In vitro Inhibition human CypA IC50 [nM] | In vitro Inhibition CnA IC50 [μM] | In vitro inhibition NFAT reporter gene assay IC50 [μM] | NFAT-GFP Translocation in HEK- 293 cells | Inhibition of HCoV-NL63 in CaCo-2 EC50 [μM] |
|---|---|---|---|---|---|
| CsA | 9.1 ± 0.8 | 0.05 | 1.6 ± 0.0003 | no | 0.9–2.0 |
| CsD | nd | nd | nd | no | 2.5 |
| ALV | nd | nd | nd | yes | 0.8 |
| NIM811 | nd | nd | nd | yes | 0.8 |
| 1 | 9.1 ± 1.4 | no | no | yes | 1.6 |
| 2 | 57.5 ± 6.9 | no | no | yes | 7.9 |
| 3 | 11.9 ± 0.9 | 6.9 | 45% at 10 μM | yes | 1.1 |
| 4 | 14.1 ± 1.9 | ∼10 | 1.3 | yes | 8.1 |
| 5 | 16.6 ± 2.3 | >10 | 1.2 | yes | 2.3 |
| FK506 | nd | nd | nd | no | 6.6 |
| 6 | 10.2 ± 1.9* | no | no | yes | 4.2 |
Properties of CsA (compounds 1–5) and FK506 (compound 6) analogues with respect to inhibition of PPIA in a protease-coupled PPIase assay, FKBP12 inhibition (*), CaN, NFAT, and NFAT-GFP nuclear translocation. Methods are described in Prell et al. (2013). The last column represents the EC50 inhibitory values of the peptides on HCoV-NL63 infection of Caco-2 cells.
These PPIase inhibitors were biochemically characterized by determining their inhibitory potency in a standard PPIase assay. Inhibition of the CaN phosphatase, the influence on cell-based NFAT reporter gene activity and NFAT translocation by the drugs (Table 2) have been performed using published procedures (Pfefferle et al., 2011, Prell et al., 2013). While the IC50 values for PPIase inhibition were similar to that of CsA, compound 2 (57.5 ± 6.9 nM) showed a higher IC50 value indicating lower binding affinity, but inhibition was still found in the low nanomolar range. CnA activity could not be inhibited at all by binary PPIase/drug complexes of 1, 2, 6. In contrast, CnA activity was weakly inhibited at very high concentrations of the drug/CypA complexes as indicated by IC50 values in the range of 10 μM for compounds 3 (IC50 6.9 μM), 4 (IC50 ∼ 10 μM) and 5 (IC50 > 10 μM)). In addition, these derivatives also demonstrated low ability (IC50 10 μM, 1.3 μM and 1.2 μM, respectively), compared with CsA (IC50 1.6 nM), to reduce the NFAT-driven reporter gene expression in a luciferase-coupled NFAT reporter gene assay indicating a greatly diminished immunosuppressive activity in a cellular assay. Although, peptides 4 and 5 showed higher CnA inhibition in vitro NFAT inhibition was remarkable indicating a gain of CaN effects in vivo. The 45% NFAT inhibitory activity at 10 μM of 3 still represents a 5000-fold lower influence of the CsA derivative on NFAT-regulated signaling pathways as compared to CsA.
The drug derivatives were further characterized by a NFAT-GFP nuclear translocation assay. HeLa 93 cells were transfected with a plasmid encoding a NFAT-GFP fusion construct under the control of the CMV promoter (Fig. 2 ). Upon Ca2+ mobilization by ionomycin the cellular phosphatase CaN dehosphorylates the NFAT transcription factor which is subsequently translocated to the nucleus. As the cells are not synchronized NFAT-GFP translocation is not expected to occur simultaneously explaining minor transfer efficiencies in some cells. The Cyp-binding immunosuppressants CsA, CsD and the FKBP-binding FK506 induce the binding as protein complexes to CaN thus inactivating its phosphatase activity. As a result NFAT is not transferred to the nucleus. Complexes of the modified drugs 1–6 with both types of PPIases, i.e. Cyp/modified CsA or FKBP/modified FK506 derivative complexes, have a greatly reduced potency to inhibit CaN and thus might allow the translocation of NFAT to the nucleus and the transcriptional regulation of immune genes. Consequently, ALV, NIM811 and the whole series of newly synthesized drug derivatives 1–6 clearly allow NFAT-GFP translocation to the nucleus and can be thus considered as non-immunosuppressive under these assay conditions (Table 2). In conclusion, in three different assays all synthetic drug derivatives proved to be inert or nearly inert in the suppression of the cellular immune response at an almost unchanged binding and inhibitory potency against the PPIase activity of the respective drug receptor.
Fig. 2.
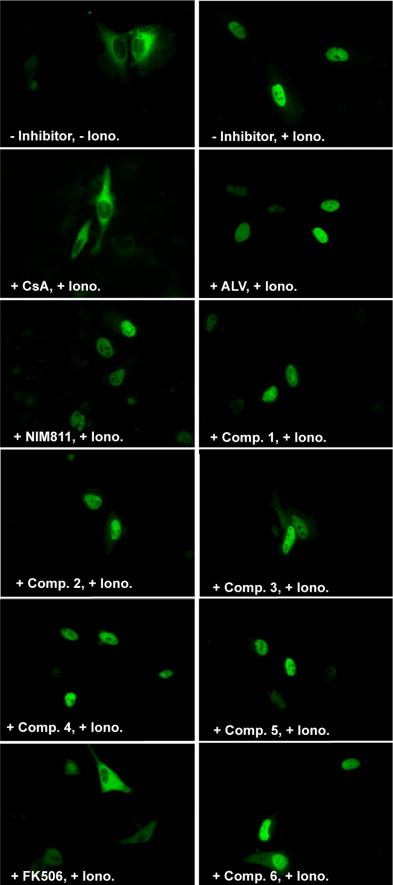
Effect of CsA, FK506 and derivatives on NFAT-GFP nuclear translocation as a measure of immunosuppressive activity. NFAT3-GFP expression plasmid was transfected into HeLa cells. Subsequently, inhibitor compounds (comp.) were added to the medium at a final concentration of 40 nM. 19 h later, ionomycin was added at a final concentration of 2 μM to induce NFAT3-GFP translocation. Pictures were taken 10 min after Ionomycin (Iono.) induction. CsD (not depicted here) showed the same behavior as CsA.
3.2. Non-immunosuppressive CsA- and FK506- derivatives inhibit replication of HCoV-NL63
To examine the inhibitory effects of the described non-immunosuppressive CsA and FK506 derivatives on the replication of HCoV-NL63, Caco-2 cells were infected with HCoV-NL63 as described (Carbajo-Lozoya et al., 2012). Fig. 3 shows the effects of CsA and seven non-immunosuppressive derivatives (ALV, NIM811, 1, 2, 3, 4, 5) and of FK506 and its descendant 6 on HCoV-NL63 replication in Caco-2 cells. Left panels represent the percentage reduction of virus replication (left Y-axes) and cell viabilities (right Y-axes). The right panels indicate the respective log10 titer reduction. The figure shows two independent sets of experiments (Fig. 3A and B) carried out at different times using different batches of CsA. When performing individual virus inhibition experiments CsA was always included as an internal control. Minor variations are explained by the use of different compound batches. Fig. 3A summarizes results obtained with peptides CsA, 3, ALV and NIM811. Fig. 3B delineates results with peptides CsD, 1, 2, 4, 5, and of FK506 and 6. The combination of the two peptides in each individual graph was chosen arbitrarily. Only FK506 and its derivative 6 were grouped together as they belong to the same compound family.
Fig. 3.
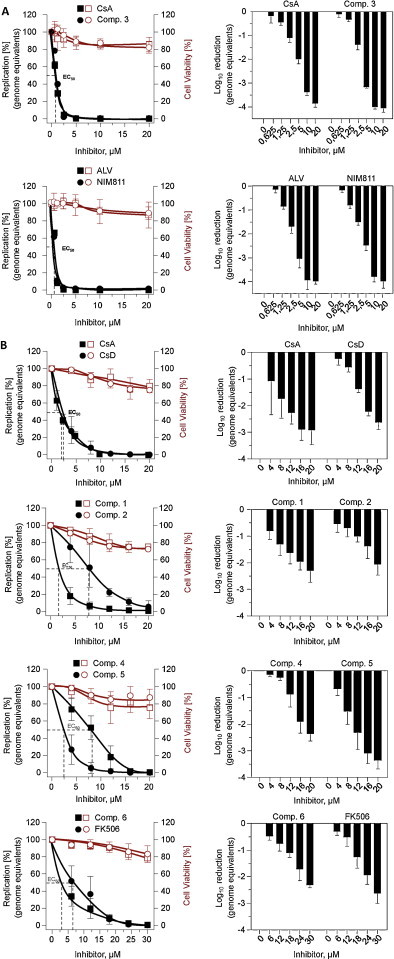
Effect of CsA and FK506 derivatives on HCoV-NL63 replication in Caco-2 cells. (A) and >(B) show independent sets of experiments using compounds CsA, 3, ALV, NIM811 and CsD, CsA, 1, 2, 4, 5, and FK506, 6, respectively. CsA represents an internal control included in each set.
Genome equivalents were determined by qPCR and cell viabilities by Cell Titer Glow kit (Promega). Data shown are mean values of a representative experiment performed in at least triplicates. Left Y-axes represent the percentage of reduction of virus replication in linear scale (left panel column) and in log scale (right panel column). Right Y-axes indicate the percentage of the cell viabilities. X-axes indicate increasing inhibitor concentrations at which virus replication was determined. Closed/open squares and closed/open circles represent the reduction of genome equivalents and cell viability at the indicated inhibitor concentrations (X-axes), respectively. The graphs were plotted using Prism 5 (GraphPad Software, Inc.) and by a non-linear regression with a variable slope algorithm, the curve was fitted for each respective inhibitor and the EC50 calculated.
From the inhibition curves it can be concluded that all peptides tested, i.e. CsA, CsD and FK506 as well as ALV, NIM811 and the new derivatives, clearly inhibit the replication of HCoV-NL63 in Caco-2 cells at low micromolar concentration levels. The EC50 inhibitory scores (Table 2) reside in a range between 0.8 μM and 8.1 μM with ALV/NIM811 and 4 showing the lowest and highest concentrations, respectively. 1 and 3 acted similarly. Also, the FK506 derivative behaved very similar to its ancestor molecule. There was no clear correlation of the inhibitory effect to in vitro inhibition of CypA, CnA or NFAT activity.
3.3. Effects of inhibitory drugs on HCoV-NL63 N protein expression
To study the effect of CsA, ALV, NIM-811 and substance 3 on viral protein expression cells were incubated with concentrations of 0 to 20 μM of the respective inhibitors for 48 h. Western blot analysis of HCoV-NL63-infected CaCo-2 cells was performed utilizing an anti-N–protein antibody. Fig. 4 clearly shows a significant decrease of the N-protein between 1.25 μM and 5 μM. It is not detectable any more at 20 μM of the respective inhibitor. This suggests that the drugs inhibit an important step in the viral replicative cycle.
Fig. 4.
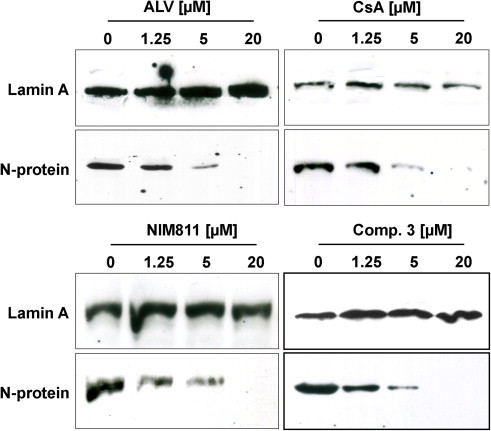
Western blot analysis of N protein expression in HCoV-NL63- infected CaCo-2 cells upon treatment with increasing concentrations of CsA, ALV, NIM811 and compound 3. N- Protein was detected with a mouse mab against N-protein. A rabbit anti-Lamin A antibody was used to detect Lamin A as a loading control.
3.4. HCoV-NL63 replication depends on cyclophilin A
In order to examine whether cellular CypA or CypB, encoded by the PPIA and PPIB genes, respectively, are required for HCoV-NL63 replication, cyclophilin Caco-2 knockdown cell lines were established using lentiviral shRNA expression vectors as recently described for FKBP1A/B (Carbajo-Lozoya et al., 2012). Rather high puromycin concentrations (10–15 μg/ml) were needed for selection as Caco-2 cells are very efficiently depleted from inhibitory drug molecules because of enhanced expression of multi drug resistant protein 1 gene (Takara et al., 2002).
To characterize Cyp A and CypB knockdown quality, mRNA expression was quantified initially from bulk-selected knockdown and control cells by real-time RT-PCR. For reverse transcription, 1 μg of total RNA was used. Amplification products were detected by SYBR I, and amplicon integrity was verified by melting point analysis. Human topoisomerase 1 gene (hTOP1) served as a reference gene for non-target control, PPIA and PPIB to determine the specificity of the knockdown. mRNA expression levels of PPIA and PPIB were then determined by real-time RT-PCR with primers listed in >Table 1. In the case of PPIA the knockdown in the puromycin bulk-selected Caco-2 cells was incomplete >as determined by Western blot (about 79%) and qPCR analysis. Here HCoV-NL63 virus growth was detectable by qPCR (not shown). This was not a surprising result as CypA comprises 0.1–0.4% of total cytosolic protein of most eukaryotic tissues (Harding et al., 1986). We therefore reasoned that these cells were not appropriate for testing the effect of CypA on virus replication and that it was necessary to search for better quality knockdown clones. Individual cell clones were then selected from the Caco-2-PPIA bulk KD cells by limited dilution in the presence of puromycin, and checked for knockdown quality.
Individual clones were expanded and tested for expression by Western blot and qPCR (Fig. 5 ). Fig. 5A shows Western blot analyses of the PPIA KD clone (polyclonal rabbit anti-CypA) used for further infection experiments. The CypB KD was demonstrated (Fig. 5A) using a polyclonal rabbit anti-CypB antibody. For control the housekeeping gene Lamin B was probed with a polyclonal goat anti-Lamin B antibody. As Lamin B levels showed no differences in Caco-2wt, Caco-2sh control and both knockdown cells, CypA was barely detectable while CypB was completely reduced. By qPCR comparison to non-target controls the CypA and CypB knockdown cell lines were determined to be 97% and 98%, respectively (Fig. 5B).
Fig. 5.
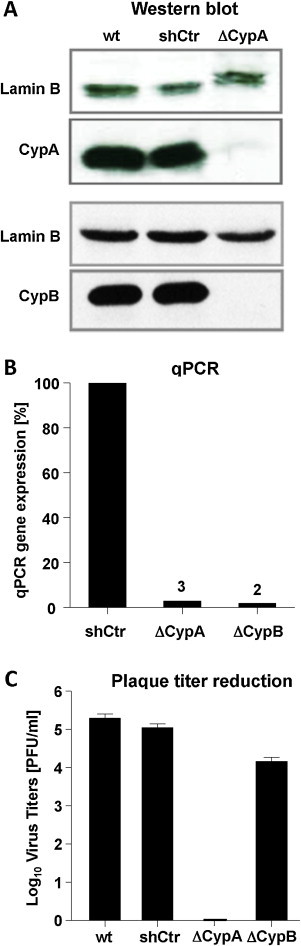
Characterization of CypA and CypB in Caco-2 knockdown cells and growth analysis of HCoV-NL63. CypA and CypB expression was determined by Western blot using an anti-CypA and anti-CypB (A) antibody and by qPCR (B). Lamin B was detected with an anti-lamin B antibody as a loading control. hTOP1 was used in qPCR to standardize cyclophilin expression. Growth of HCoV-NL63 on Caco-2 wt, Caco-2 shCtr non-target cells, Caco-2 ΔCypA (CypA KD) and Caco-2 ΔCypB (CypB KD) knockdown mutants was analyzed by plaque titration assay. Virus titers are depicted in (C).
To examine virus propagation on the knockdown cell lines >Caco-2wt, Caco-2sh control, Caco-2-PPIA KD and Caco-2-PPIB KD cells were infected with serial dilutions of HCoV-NL63. Plaque titration clearly showed comparable virus titers (Fig. 5C) in both control and in the Caco-2-PPIB-KD cell lines. The virus did not grow in the Caco-2-PPIA KD cells at all indicating the dependence of virus replication on CypA. Similar results were obtained by qPCR (not shown).
4. Discussion
Common and in many cases very successful antiviral therapeutic approaches adhere to the use of drugs that inhibit viral replication by directly acting on viral components important for the viral life cycle like reverse transcriptases, proteases, integrases etc. A drawback of these drugs is the high mutational susceptibility of the viral genomes due to the poor proofreading activities of viral replicative enzymes, especially of RNA viruses. These difficulties might be circumvented by new, promising therapeutic drugs targeting host factors involved in viral entry, replication, assembly and release, or in general cellular functions such as protein translation and folding. A well characterized example is inhibition of hepatitis C virus infection in cell culture models and humans by host-targeting agents (Zeisel et al., 2013). Cyclophilins are a class of mostly intracellular enzymes required for the correct folding and function of a various cellular and also viral proteins (von Hahn et al., 2011). CypA has been shown to bind to several HIV-1 and HCV proteins and being essential for replication of several viruses which can be inhibited by CsA and non-immunosuppressive derivatives thereof (Baugh and Gallay, 2012, Daelemans et al., 2010, Fassati, 2012, Frausto et al., 2013, Gallay, 2012, Membreno et al., 2013). In HCV patients safety and efficacy has been shown for the three Cyp inhibitors ALV, NIM811 (both from Novartis) and SCY-635 (SCYNEXIS) in phase I and phase II trials. ALV is the first-in-class and the most advanced cyclophilin inhibitor with clinical proof-of-concept including phase III randomized clinical trials (Flisiak et al., 2013, Gallay and Lin, 2013, Membreno et al., 2013).
By unbiased yeast-two-hybrid screening we have found that Nsp1 proteins of SARS-CoV (Pfefferle et al., 2011, Schöpf et al., 2010) and of other CoVs (unpublished) bind to cyclophilins including CypA and that CsA inhibits coronaviruses including human SARS-CoV, HCoV-NL63, HCoV-229E and animal CoVs FCoV (two serotypes), IBV, TGEV. We further showed inhibitory action of FK506 on the three human CoVs. The main goal of our study was to test and compare the inhibitory effect of the chemically difficult-to-synthesize cyclosporines ALV and NIM811 with a set of drug derivatives 1–5 which result from the chemically well-tractable side chain in position 1 of CsA. In addition, the FK506 derivative 6 should provide a first indication of feasibility of expanding the concept of antiviral non-immunosuppressive CypA inhibitors into the field of FKBP inhibitors. Data on the inhibition of HCoV-NL63 replication were compared with the immunosuppressive immunophilin binders CsA, CsD and FK506. All derivatives inhibit the PPIase activity of their respective binding proteins thus preventing their catalytic function in assisting client proteins to fold correctly. When compared with already known non-immunosuppressive drugs, the new compounds were not only more easily accessible by chemical synthesis but also showed a favorable ratio of PPIase inhibition to cellular toxicity. All compounds were tested in an NFAT-GFP nuclear translocation assay (Fig. 2). Upon mobilization of Ca2+ by ionomycin NFAT-GFP remained in the cytoplasm in the presence of CsA, CsD and FK506. This indicates that the CypA/CsA, CypA/CsD or FKBP/FK506 complexes inactivate the CaN phosphatase thus preventing NFAT dephosphorylation and nuclear translocation which constitutes the basis for immunosuppression. For CsD it is clear that it exerts a rather strong immunosuppressive activity as opposed to earlier reports ascribing only about 10% of the CsA activity to the molecule (Sadeg et al., 1993a). In the presence of ALV, NIM811 and all the newly synthesized drug derivatives 1 to 6 NFAT migrated to the nucleus within minutes confirming their non-immunosuppressive activity. The new set of drugs was also characterized in vitro with respect to inhibition of CypA in a protease-coupled PPIase assay, CnA and NFAT assay and cell permeability. The potency of PPIAse inhibition was comparable to CsA. Only in the case of compound 2 it was increased by about sixfold. The IC50 values of CnA inhibition was achieved for CsA at 0.05 μM whereas for three of the new compounds 138-fold to 200-fold higher concentrations were needed.
Virus inhibition experiments (Fig. 3, Table 2) clearly show the highly effective inhibition of HCoV-NL63 by ALV (EC50: 0.8 μM) and NIM811 (EC50: 0.8 μM) which closely resembles the patterns of CsA (EC50: 0.9–2.0 μM) and CsD (EC50: 2.5 μM). The EC50 values of the CsA derivatives range between 1.1 μM and 8.1 μM. EC50 values of FK506 and its derivative 6 were 6.6 μM and 4.2 μM, respectively. Taken together, all the derivatives inhibited virus replication in the low micromolar range similar to our previous report on the inhibition of various human and animal CoVs with CsA (Pfefferle et al., 2011). Interestingly, inhibition of HCV with CsA, ALV and NIM811 is commonly observed at nanomolar concentrations with ALV as the most effective compound. We have no explanation for this but it can be speculated that in the case of HCV the interaction of viral proteins and cyclophilin A is more sensitive to the inhibitors. The micromolar ranges of CoV inhibition was nicely confirmed by another group (de Wilde et al., 2011).
The HCoV-NL63-N protein plays a crucial role in the viral life cycle. It was analyzed as a representative of viral protein expression in the presence of increasing concentrations of CsA, ALV, NIM-811 and substance 3. The protein decreased at 1.25 μM and it was not detectable any more at concentrations above 5 μM. Thus, there is a clear inhibitory effect of the different peptides on N protein expression and virus replication. Whether inhibition is a result of lacking cyclophilin interaction with Nsp1 or another viral protein cannot be decided at the current stage. As the N protein of SARS-CoV was reported to bind to CypA (Luo et al., 2004) and CypA is incorporated into SARS-CoV particles (Neuman et al., 2008) the inhibitors might also act directly on CypA/N-protein complexes, if these also exist in HCoV-NL63. It can also not be ruled out that further viral proteins require CypA functions.
An important question was whether CypA is the crucial cyclophilin required for CoV replication. For HCV there were some discrepancies on the necessity of CypA and CypB for virus growth. Recent studies demonstrate that CypA is the key host factor for HCV replication (Baugh and Gallay, 2012, Kaul et al., 2009). In order to address the role of the two members of the Cyp family for HCoV-NL63 replication, we developed CaCo-2 cell lines with individual knockdowns for CypA and CypB on a lentiviral basis (Fig. 5). The characterization at protein (Fig. 5A) and RNA (Fig. 5B) levels indicated efficient KD of the two Cyps. In plaque titration experiments performed on CaCo-2wt, CaCo-2sh (non-target) and CaCo-2ΔCypB the virus grew at comparable titers (Fig. 5C). In contrast, CaCo-2ΔCypA cells did not support virus growth at all indicating the dependence on functional CypA.
Regarding Cyp requirement for CoV replication contradictory results were reported by de Wilde et al. (2011). While our CoV data on CsA inhibition (Schöpf et al., 2010) were basically confirmed, the report claims that neither CypA nor CypB are required for replication of SARS-CoV and Mouse Hepatitis Virus (MHV) on the basis of siRNA PPIA and PPIB knockdown experiments. However, both siRNA knockdowns were rather incomplete in terms of the residual CypA or CypB protein level leaving enough PPIase activity in the infected cells to support viral replication. At least for HCoV-NL63 we clearly demonstrate that in lentivirally produced knockdown cells CypA but not CypB is the required molecule. Interestingly, showing a similarly poor knockdown of PPIA the same authors claim in a follow-up study on arteriviruses that PPIA is the required cyclophilin for nidovirus replication (de Wilde et al., 2013). Coronaviridae and Arteriviridae are both families within the order of Nidovirales. They are both positive-stranded with similar genome organization and genome length of 27 to 32 kb and 13 to 16 kb, respectively, and they share a number of enzymatic functions. Since both families are sensitive to CsA the involvement of cyclophilins in nidovirus replication as a general principle is reasonable.
From our HCoV-NL63 infectivity studies in Caco-2-PPIA/PPIB KD cells we conclude that the most abundant CypA of the viral host is a prerequisite of CoV replication. The interaction of SARS-CoV Nsp1 in Y2H and in mammalian protein-binding assays with several cyclophilin isoforms suggests the potential involvement of additional cyclophilins in the infection process. It has to be examined systematically whether CypA and further cyclophilins bind to other viral proteins. Furthermore, knowledge about the regulatory role of the catalytic activity of the PPIase subfamilies of cyclophilins and FK506-binding proteins, both of which were shown here to be critical in the viral replication process requires identification of their protein substrates in the coronaviral background.
The four non-SARS-related HCoVs (HCoV-OC43/229E/NL63 and/HKU1) are major causes of relatively mild respiratory tract infections in immunocompetent hosts. However, clinical manifestations like bronchiolitis and pneumonia can be severe especially in young children, the elderly and immunocompromised patients (van der Hoek, 2007). Infection occurs in early childhood and the detection of anti-S IgG antibodies in >70% of the general human population demonstrates their high prevalence (Zhou et al., 2013). Furthermore, the zoonotic transmission potential of HCoV-NL63 (Huynh et al., 2012) and of the highly aggressive SARS-CoV and MERS-CoV (Gallagher and Perlman, 2013) demand the development of effective drugs preventing or alleviating virus growth and pathogenicity.
The marked inhibition of HCoV-NL63 replication by the non-immunosuppressive derivatives of CsA and of FK506 highlights the functional relevance of host cell cyclophilins and FKBPs as antiviral targets.
Acknowledgment
This work was supported by the “Bundesministerium fuer Bildung und Forschung” of the German Government (Zoonosis Network, Consortium on ecology and pathogenesis of SARS, project code 01KI1005A,F; http://www.gesundheitsforschungbmbf.de/de/1721.php#SARS) to AvB. We thank Julia Schöpf for technical help. We are grateful to Novartis (Switzerland) for the provision of Alisporivir and NIM811. Special thanks to Drs. C. Thirion and M. Salomon for help with the Caco-2 KD cell lines using lentiviral technology.
References
- Abdul-Rasool S., Fielding B.C. Understanding human coroonavirus HCoV-NL63. Open Virol. J. 2010;4:76–84. doi: 10.2174/1874357901004010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh J., Gallay P. Cyclophilin involvement in the replication of hepatitis C virus and other viruses. Biol. Chem. 2012;393(7):579–587. doi: 10.1515/hsz-2012-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska-Haba M., Patel H.D., Sapp M. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog. 2009;5(7):e1000524. doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E.A., Walkinshaw M.D. Targeting FKBP isoforms with small-molecule ligands. Curr. Opin. Pharmacol. 2011;11(4):365–371. doi: 10.1016/j.coph.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Bose S., Mathur M., Bates P., Joshi N., Banerjee A.K. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J. Gen. Virol. 2003;84(Pt 7):1687–1699. doi: 10.1099/vir.0.19074-0. [DOI] [PubMed] [Google Scholar]
- Carbajo-Lozoya J., Müller M.A., Kallies S., Thiel V., Drosten C., von Brunn A. Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Res. 2012;165:112–117. doi: 10.1016/j.virusres.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R., Tang S., Zhao G., Lu H., Case D.A., Johnson M.E. Comparison of SARS and NL63 papain-like protease binding sites and binding site dynamics: inhibitor design implications. J. Mol. Biol. 2011;414(2):272–288. doi: 10.1016/j.jmb.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck C.-P., Chen C., Ke Z., Chi-Cheong Wan D., Chow H.-F., Wong K.-B. Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases. Eur. J. Med. Chem. 2013;59(0):1–6. doi: 10.1016/j.ejmech.2012.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck C.-P., Chow H.-F., Wan D.C.-C., Wong K.-B. Profiling of substrate specificities of 3C-like proteases from group 1, 2a, 2b, and 3 coronaviruses. PLoS One. 2011;6(11):e27228. doi: 10.1371/journal.pone.0027228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daelemans D., Dumont J.-M., Rosenwirth B., De Clercq E., Pannecouque C. Debio-025 inhibits HIV-1 by interfering with an early event in the replication cycle. Antiviral Res. 2010;85(2):418–421. doi: 10.1016/j.antiviral.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Davis T.L., Walker J.R., Campagna-Slater V., Finerty P.J., Paramanathan R., Bernstein G., MacKenzie F., Tempel W., Ouyang H., Lee W.H., Eisenmesser E.Z., Dhe-Paganon S. Structural and biochemical characterization of the human cyclophilin family of peptidyl–prolyl isomerases. PLoS Biol. 2010;8(7):e1000439. doi: 10.1371/journal.pbio.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV); announcement of the Coronavirus Study Group. J. Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Rottier P.J. Hosting the severe acute respiratory syndrome coronavirus: specific cell factors required for infection. Cell. Microbiol. 2006;8(8):1211–1218. doi: 10.1111/j.1462-5822.2006.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Li Y., van der Meer Y., Vuagniaux G., Lysek R., Fang Y., Snijder E.J., van Hemert M.J. Cyclophilin inhibitors block arterivirus replication by interfering with viral RNA synthesis. J. Virol. 2013;87(3):1454–1464. doi: 10.1128/JVI.02078-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Zevenhoven-Dobbe J.C., van der Meer Y., Thiel V., Narayanan K., Makino S., Snijder E.J., van Hemert M.J. Cyclosporin A inhibits the replication of diverse coronaviruses. J. Gen. Virol. 2011;92(Pt 11):2542–2548. doi: 10.1099/vir.0.034983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Gaunt E., Rossen J.W., Templeton K.E., Kuijpers T.W., van der Hoek L. The dominance of human coronavirus OC43 and NL63 infections in infants. J. Clin. Virol. 2012;53(2):135–139. doi: 10.1016/j.jcv.2011.11.011. (the official publication of the Pan American Society for Clinical Virology) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der W.S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Fassati A. Multiple roles of the capsid protein in the early steps of HIV-1 infection. Virus Res. 2012;170(1–2):15–24. doi: 10.1016/j.virusres.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Fernandes F., Ansari I.U., Striker R. Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis C NS5A. PLoS One. 2010;5(3):e9815. doi: 10.1371/journal.pone.0009815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G., Gallay P., Hopkins S. Cyclophilin inhibitors for the treatment of HCV infection. Curr. Opin. Invest. Drugs. 2010;11(8):911–918. [PubMed] [Google Scholar]
- Fischer G., Wittmann-Liebold B., Lang K., Kiefhaber T., Schmid F.X. Cyclophilin and peptidyl–prolyl cis-trans isomerase are probably identical proteins. Nature. 1989;337(6206):476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- Flisiak R., Jaroszewicz J., Parfieniuk-Kowerda A. Emerging treatments for hepatitis C. Expert Opin. Emerg. Drugs. 2013;18(4):461–475. doi: 10.1517/14728214.2013.847089. [DOI] [PubMed] [Google Scholar]
- Frausto S.D., Lee E., Tang H. Cyclophilins as modulators of viral replication. Viruses. 2013;5(7):1684–1701. doi: 10.3390/v5071684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T., Perlman S. Public health: broad reception for coronavirus. Nature. 2013;495(7440):176–177. doi: 10.1038/495176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay P.A. Cyclophilin inhibitors: a novel class of promising host-targeting anti-HCV agents. Immunol. Res. 2012;52(3):200–210. doi: 10.1007/s12026-011-8263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay P.A., Lin K. Profile of alisporivir and its potential in the treatment of hepatitis C. Drug Des. Dev. Ther. 2013;7:105–115. doi: 10.2147/DDDT.S30946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48(8):2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding M.W., Handschumacher R.E., Speicher D.W. Isolation and amino acid sequence of cyclophilin. J. Biol. Chem. 1986;261(18):8547–8555. [PubMed] [Google Scholar]
- Herzog P., Drosten C., Muller M.A. Plaque assay for human coronavirus NL63 using human colon carcinoma cells. Virol. J. 2008;5:138. doi: 10.1186/1743-422X-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C., Nagel J., Johnson J.B., Agnihothram S., Gates J.E., Frieman M.B., Baric R.S., Donaldson E.F. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86(23):12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H., Tani H., Mori Y., Abe T., Katoh H., Fukuhara T., Taguwa S., Moriishi K., Matsuura Y. Involvement of cyclophilin B in the replication of Japanese encephalitis virus. Virology. 2011;412(1):211–219. doi: 10.1016/j.virol.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Kaul A., Stauffer S., Berger C., Pertel T., Schmitt J., Kallis S., Zayas Lopez M., Lohmann V., Luban J., Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5(8):e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono M., Tatsumi K., Imai A.M., Saito K., Kuriyama T., Shirasawa H. Inhibition of human coronavirus 229E infection in human epithelial lung cells (L132) by chloroquine: involvement of p38 MAPK and ERK. Antiviral Res. 2008;77(2):150–152. doi: 10.1016/j.antiviral.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. PNAS. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lin K., Gallay P. Curing a viral infection by targeting the host: the example of cyclophilin inhibitors. Antiviral Res. 2013;99(1):68–77. doi: 10.1016/j.antiviral.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Sun L., Yu M., Wang Z., Xu C., Xue Q., Zhang K., Ye X., Kitamura Y., Liu W. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell. Microbiol. 2009;11(5):730–741. doi: 10.1111/j.1462-5822.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- Luo C., Luo H., Zheng S., Gui C., Yue L., Yu C., Sun T., He P., Chen J., Shen J., Luo X., Li Y., Liu H., Bai D., Yang Y., Li F., Zuo J., Hilgenfeld R., Pei G., Chen K., Shen X., Jiang H. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem. Biophys. Res. Commun. 2004;321(3):557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malesevic M., Gutknecht D., Prell E., Klein C., Schumann M., Nowak R.A., Simon J.C., Schiene-Fischer C., Saalbach A. Anti-inflammatory effects of extracellular cyclosporins are exclusively mediated by CD147. J. Med. Chem. 2013;56(18):7302–7311. doi: 10.1021/jm4007577. [DOI] [PubMed] [Google Scholar]
- Membreno F.E., Espinales J.C., Lawitz E.J. Cyclophilin inhibitors for hepatitis C therapy. Clin. Liver Dis. 2013;17(1):129–139. doi: 10.1016/j.cld.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Milewska A., Ciejka J., Kaminski K., Karewicz A., Bielska D., Zeglen S., Karolak W., Nowakowska M., Potempa J., Bosch B.J., Pyrc K., Szczubialka K. Novel polymeric inhibitors of HCoV-NL63. Antiviral Res. 2013;97(2):112–121. doi: 10.1016/j.antiviral.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., III, Wuthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3 2. J. Virol. 2008;82(11):5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Schöpf J., Kögl M., Friedel C., Müller M.A., Stellberger T., von Dall’Armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Züst R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.-J., Schwegmann-Weßels C., Pöhlmann S., Haas J., Drosten C., von Brunn A. The SARS-Coronavirus-host interactome: identification of cyclophilins as target for pan-Coronavirus inhibitors. PLoS Pathog. 2011;7(10):e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prell E., Kahlert V., Rucknagel K.P., Malesevic M., Fischer G. Fine tuning the inhibition profile of cyclosporine A by derivatization of the MeBmt residue. ChemBioChem. 2013;14(1):63–65. doi: 10.1002/cbic.201200621. [DOI] [PubMed] [Google Scholar]
- Pyrc K., Bosch B.J., Berkhout B., Jebbink M.F., Dijkman R., Rottier P., van der Hoek L. Inhibition of human coronavirus NL63 infection at early stages of the replication cycle. Antimicrob. Agents Chemother. 2006;50(6):2000–2008. doi: 10.1128/AAC.01598-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing M., Yang F., Zhang B., Zou G., Robida J.M., Yuan Z., Tang H., Shi P.Y. Cyclosporine inhibits flavivirus replication through blocking the interaction between host cyclophilins and viral NS5 protein. Antimicrob. Agents Chemother. 2009;53(8):3226–3235. doi: 10.1128/AAC.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeg N., Pham-Huy C., Rucay P., Righenzi S., Halle-Pannenko O., Claude J.R., Bismuth H., Duc H.T. In vitro and in vivo comparative studies on immunosuppressive properties of cyclosporines A, C, D and metabolites M1, M17 and M21. Immunopharmacol. Immunotoxicol. 1993;15(2-3):163–177. doi: 10.3109/08923979309025992. [DOI] [PubMed] [Google Scholar]
- Sadeg N., Pham Huy C., Martin C., Warnet J.M., Claude J.R. Effect of cyclosporin A and its metabolites and analogs on lipid peroxidation in rabbit renal microsomes. Drug Chem. Toxicol. 1993;16(2):165–174. doi: 10.3109/01480549309031994. [DOI] [PubMed] [Google Scholar]
- Sastre P., Dijkman R., Camunas A., Ruiz T., Jebbink M.F., van der Hoek L., Vela C., Rueda P. Differentiation between human coronaviruses NL63 and 229E using a novel double-antibody sandwich enzyme-linked immunosorbent assay based on specific monoclonal antibodies. Clin. Vaccine Immunol. 2011;18(1):113–118. doi: 10.1128/CVI.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöpf J., Pfefferle S., Kögl M., Herzog P., Müller M.A., Kallies S., D. Muth D., Kuri T., Ebel T., Friedel F., Zimmer C., Weber R., Haas F., Thiel J., Herrler H.-J., Schwegmannn-Wessels G., Thiel C., Drosten V., von Brunn C.A. Applying systems biology to severe acute respiratory coronavirus and its human host: identification of a cellular target and its inhibitor for antiviral intervention. Fourth European Congress of Virology; Cernobbio, Lake Como, Italy; 2010. [Google Scholar]
- Smith E.C., Denison M.R. Coronaviruses as DNA Wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013;9(12):e1003760. doi: 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K., Luban J., Jeang K.T. Human cellular restriction factors that target HIV-1 replication. BMC Med. 2009;7:48. doi: 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takara K., Tsujimoto M., Ohnishi N., Yokoyama T. Digoxin up-regulates MDR1 in human colon carcinoma Caco-2 cells. Biochem. Biophys. Res. Commun. 2002;292(1):190–194. doi: 10.1006/bbrc.2002.6619. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Sato Y., Osawa S., Inoue M., Tanaka S., Sasaki T. Suppression of feline coronavirus replication in vitro by cyclosporin A. Vet. Res. 2012;43(1):41. doi: 10.1186/1297-9716-43-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J., van den Worm S.H., Sims A.C., Baric R.S., Snijder E.J., van Hemert M.J. Zn(2+) inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6(11):e1001176. doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T.R. Therapies for coronaviruses. Part 2: Inhibitors of intracellular life cycle. Expert Opin. Ther. Pat. 2009;19(4):415–431. doi: 10.1517/13543770802600698. [DOI] [PubMed] [Google Scholar]
- Tong T.R. Therapies for coronaviruses. Part I of II—Viral entry inhibitors. Expert Opin. Ther. Pat. 2009;19(3):357–367. doi: 10.1517/13543770802609384. [DOI] [PubMed] [Google Scholar]
- Traber R., Kuhn M., Loosli H.R., Pache W., von Wartburg A. [New cyclopeptides from Trichoderma polysporum (Link ex Pers.) Rifai: cyclosporins B, D and E (author's transl)] Helv. Chim. Acta. 1977;60(5):1568–1578. doi: 10.1002/hlca.19770600513. [DOI] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6) doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L. Human coronaviruses: what do they cause? Antiviral Ther. 2007;12(4 Pt B):651–658. [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermehren J., Sarrazin C. New HCV therapies on the horizon. Clin. Microbiol. Infect. 2011;17(2):122–134. doi: 10.1111/j.1469-0691.2010.03430.x. [DOI] [PubMed] [Google Scholar]
- Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels M.W., van Balkom B.W., Kaloyanova D.V., Batenburg J.J., Heck A.J., Helms J.B., Rottier P.J., de Haan C.A. Identification of host factors involved in coronavirus replication by quantitative proteomics analysis. Proteomics. 2011;11(1):64–80. doi: 10.1002/pmic.201000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hahn T., Ciesek S., Manns M.P. Arrest all accessories—inhibition of hepatitis C virus by compounds that target host factors. Discovery Med. 2011;12(64):237–244. [PubMed] [Google Scholar]
- Wang R.Y., Li K. Host factors in the replication of positive-strand RNA viruses. Chang Gung Med. J. 2012;35(2):111–124. doi: 10.4103/2319-4170.106160. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., Poon L.L., Wong S.S., Guan Y., Peiris J.S., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10):e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylinen L.M., Schaller T., Price A., Fletcher A.J., Noursadeghi M., James L.C., Towers G.J. Cyclophilin A levels dictate infection efficiency of human immunodeficiency virus type 1 capsid escape mutants A92E and G94D. J. Virol. 2009;83(4):2044–2047. doi: 10.1128/JVI.01876-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zeisel M.B., Lupberger J., Fofana I., Baumert T.F. Host-targeting agents for prevention and treatment of chronic hepatitis C—perspectives and challenges. J. Hepatol. 2013;58(2):375–384. doi: 10.1016/j.jhep.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Zhou W., Wang W., Wang H., Lu R., Tan W. First infection by all four non-severe acute respiratory syndrome human coronaviruses takes place during childhood. BMC Infect. Dis. 2013;13:433. doi: 10.1186/1471-2334-13-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., George S., Schmidt M.F., Al-Gharabli S.I., Rademann J., Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Res. 2011;92(2):204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


