Graphical abstract
Keywords: Protein synthesis regulation, Translation initiation, Translation reinitiation, Translation recycling, Density regulated protein (DENR)
Abstract
The density regulated protein (DENR) forms a stable heterodimer with malignant T-cell-amplified sequence 1 (MCT-1). DENR-MCT-1 heterodimer then participates in regulation of non-canonical translation initiation and ribosomal recycling. The N-terminal domain of DENR interacts with MCT-1 and carries a classical tetrahedral zinc ion-binding site, which is crucial for the dimerization. DENR-MCT-1 binds the small (40S) ribosomal subunit through interactions between MCT-1 and helix h24 of the 18S rRNA, and through interactions between the C-terminal domain of DENR and helix h44 of the 18S rRNA. This later interaction occurs in the vicinity of the P site that is also the binding site for canonical translation initiation factor eIF1, which plays the key role in initiation codon selection and scanning. Sequence homology modeling and a low-resolution crystal structure of the DENR-MCT-1 complex with the human 40S subunit suggests that the C-terminal domain of DENR and eIF1 adopt a similar fold. Here we present the crystal structure of the C-terminal domain of DENR determined at 1.74 Å resolution, which confirms its resemblance to eIF1 and advances our understanding of the mechanism by which DENR-MCT-1 regulates non-canonical translation initiation and ribosomal recycling.
1. Introduction
The density regulated protein (DENR) was discovered as a protein, whose synthesis is increased at high cell density [1]. It is also overexpressed in breast and ovarian cancers [1], [2]. In translation, DENR functions as a stable heterodimer with malignant T-cell-amplified sequence 1 (MCT-1 or MCTS1), which interacts with the 40S ribosomal subunit during initiation, reinitiation and recycling stages [3], [4], [5].
Initiation of protein synthesis in eukaryotes is a complex process orchestrated by more than a dozen translation initiation factors (eIFs). It includes recruitment of the mRNA to the small ribosomal subunit, mRNA scanning to locate the initiation codon (usually the first AUG codon), selection of the initiator tRNA (Met-tRNAiMet) and the joining of the small (40S) and large (60S) ribosomal subunits, which results in the 80S ribosome becoming primed for protein synthesis. The key factor that controls fidelity of translation initiation is eIF1. eIF1 allows scanning of the mRNA and dissociation of aberrantly assembled ribosomal complexes, discrimination between an optimal and non-optimal context for AUG codons and promotes ejection of deacylated tRNA during recycling [6], [7], [8].
Recycling of the 40S subunit, which remains bound to mRNA along with deacylated tRNA in the P site, is promoted by the combined action of canonical initiation factors eIF1, eIF1A and eIF3, or by eIF2D or the heterodimer formed by DENR and the malignant T cell-amplified sequence 1 (MCT-1/MCTS1) [4], [9], [10], [11]. It also was shown that after translation of some protein-coding open reading frames (ORFs) the 40S subunit remains bound to the mRNA, leading to reinitiation of protein synthesis from the nearest AUG [12], [13], [14]. Reinitiation is used by viruses to express polycistronic RNAs, and in eukaryotes this process regulates translation of some ORFs [12], [15].
The molecular mechanism of translation reinitiation is not well understood. Reinitiation may require that the 40S subunit remain bound to mRNA after termination and that some eIFs (e.g., eIF4F and eIF3) remain bound to the 40S subunit even after completing translation of the first, usually short ORF. It is not clear if the same requirement can be applied for a long upstream ORF [6], [12], [16]. Reinitiation is promoted by canonical initiation factors eIF1, eIF1A, eIF3 and, as was shown recently, by eIF2D and DENR-MCT-1 [11], [17], [18], [19]. The later regulates translation reinitiation on some mRNAs with short upstream ORFs in Drosophila and in HeLa cells [17], [18]. In addition, ribosome profiling in mouse fibroblasts linked DENR-dependent translation reinitiation with regulation of the mammalian circadian clock [20].
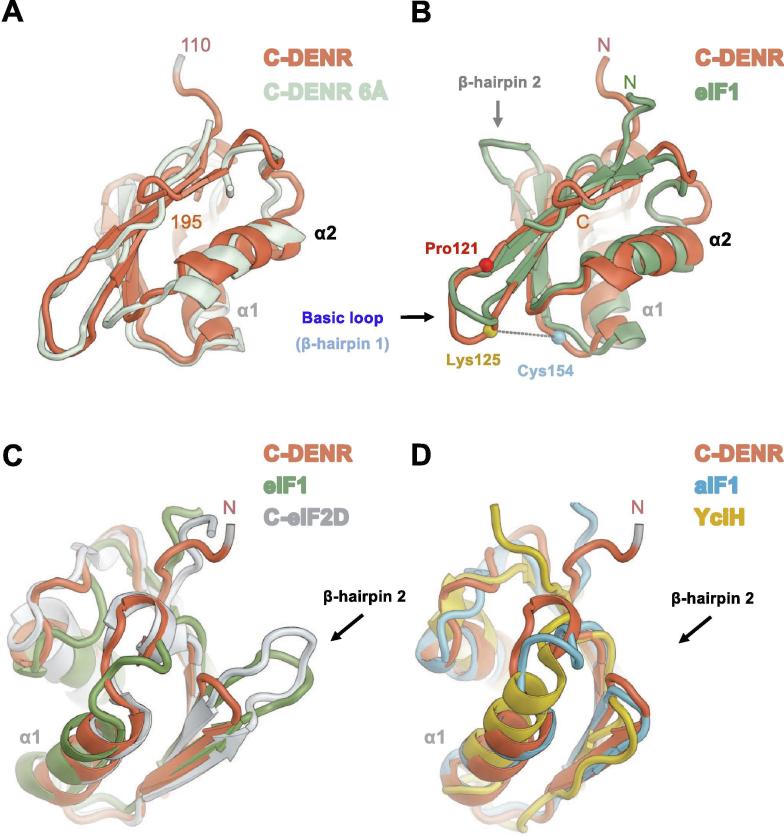
Noncanonical translation initiation factors eIF2D and DENR-MCT-1 share similar domains involved in RNA binding and start codon recognition (Fig. 1A). Recently, their interactions with the human 40S subunit were characterized by X-ray crystallography and cryoelectron microscopy (cryo-EM) [3], [5]. DENR-MCT-1 binds to the human 40S ribosomal subunit through interactions between the N-terminal domain of MCT-1 and helix h24 of 18S rRNA, and between the C-terminal domain of DENR and the top of helix h44 of 18S rRNA, which constitutes the eIF1 binding site as well. The C-terminal domains of both eIF2D (C-eIF2D) and DENR (C-DENR) have the same eIF1-like fold and the same binding site on the 40S ribosomal subunit, suggesting a similar mechanism for DENR-MCT-1, eIF2D and eIF1 in regulating binding of the Met-tRNAiMet in the P site of the 40S subunit, which is crucial for translation initiation, reinitiation, and ribosomal recycling [21]. In addition, DENR-MCT-1 may interact with and stabilize the CCA-end of the P-site bound initiator tRNA in a manner similar to the homologous initiation factor eIF2D [3], [5], [22], [23]. It was shown that DENR-MCT-1, as well as eIF2D, promotes translation initiation via an internal ribosome entry site (IRES) by recruiting Met-tRNAiMet to the start codon AUG that directly positioned in the P site of the 40S subunit. Their action does not require GTP and the assistance of canonical translation initiation factors (eIF1, eIF1A, eIF2, eIF5, eIF5B) [7], [11].
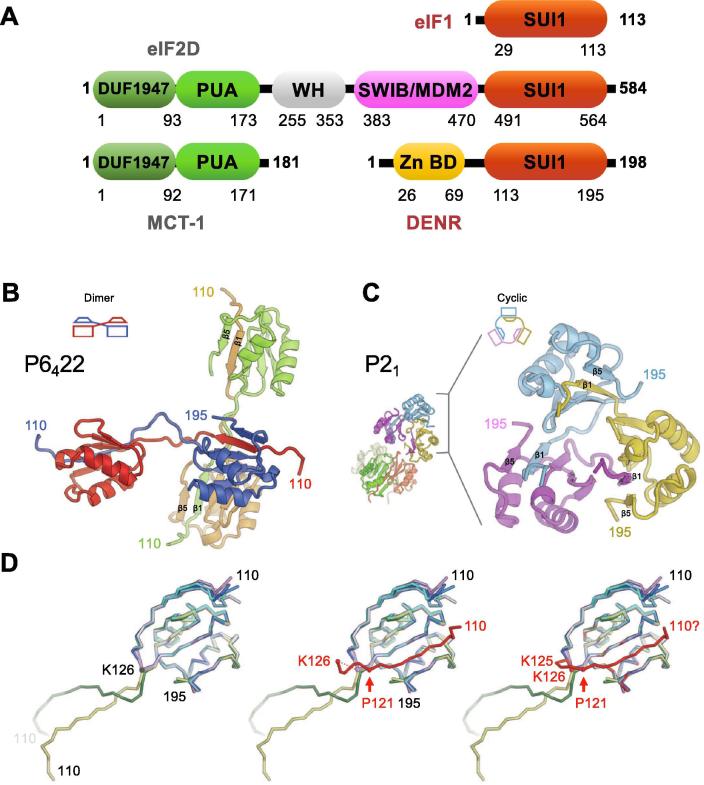
Fig. 1.
The structure of the C-terminal domain of human DENR110–195. A. Domain organization of human eIF1, eIF2D, MCT-1, and DENR (numbers showing amino acid residues at the borders). Domain swapping and packing of the C-DENR molecules in the: B. P6422 and C. P21 crystal form. D. Superposition of C-DENR molecules: from both crystal forms (left), with the β strand 1 swapped (red), present (middle), with the modeled basic loop (red, right). β strands 1 and 5, and positions of some amino acid residues are marked. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
High-resolution crystal structures of the N-terminal domain of human DENR (N-DENR) bound to MCT-1 revealed that N-DENR includes a zinc ion-binding site, which preserves the structure of the DENR’s MCT-1–binding interface that is essential for the dimerization [22], [23]. Recent X-ray crystallography and cryo-electron microscopy (cryo-EM) studies showed that the C-terminal domains of DENR and eIF2D have an eIF1-like fold (SUI1 in yeast) and share the binding site on the 40S ribosomal subunit. However, insufficient resolution of these data did not enable construction of an atomic-resolution model of C-DENR. Therefore, only a homology-based model is available, which limits our understanding of the mechanism by which C-DENR interacts with the 18S rRNA, tRNA and mRNA molecules in the P site of the 40S ribosomal subunit [3], [5], [24]. To overcome this limitation, we determined the crystal structure of the C-terminal domain of DENR at 1.74 Å resolution. The structure confirmed that C-DENR has eIF1-like fold. However, the β-hairpin loop 2 of C-DENR, which, in case of eIF1, interacts with the D-stem loop of the P-site tRNA, is much smaller than that of eIF1. A structural comparison between C-DENR and eIF1 allowed us to elucidate further the differences and similarities in the mechanism by which these factors regulate translation initiation, reinitiation and recycling.
2. Results and discussion
2.1. Design of deletion mutants and structure determination of the recombinant C-terminal domain of DENR
Our recent model of the C-terminal domain of DENR (amino acid residues 113–191) was build based on sequence and structural homology [3]. To confirm this model, we designed, expressed, purified and attempted to crystalize three C-DENR fragments: C-DENR106–198, C-DENR109–198, C-DENR110–195 (Materials and Methods). Only one of these constructs, C-DENR110–195, was successfully crystalized, yielding two types of crystals. The first one grew at 20 °C and belongs to the P6422 hexagonal space group. The structure was determined at 2.3 Å resolution by single-wavelength anomalous diffraction (SAD) using a mercury derivative as described in Materials and Methods (Fig. 1B). The asymmetric unit contains two C-DENR molecules with an evident eIF1-like fold of two α-helices on one side of a five-stranded parallel and antiparallel β-sheet [25]. However, continuous electron density in the 2Fo-Fc map shows that β-strand 1 of each molecule is flipped from its own β-sheet and replaced by β-strand 1 from the crystallographic symmetrically related molecule (Fig. 1A). The mechanism of formation of the protein dimer or higher oligomer by exchanging identical structural elements is known as “domain swapping”, which can be either biologically relevant or a crystallization artefact [26]. The second crystal type grew at 12 °C and belongs to the P21 monoclinic space group, wherein six C-DENR molecules form two trimers per asymmetric unit. Surprisingly, each C-DENR trimer is also formed by swapping of the same domain: β-strand 1 (Fig. 1C). A complete data set for P21 crystals was collected to 1.74 Å resolution (Table 1) and initial phases were determined by a molecular replacement method using as a search model the crystal structure of C-DENR with β-strand 1 swapped as determined by SAD (Materials and Methods). The quality of the maps allows us to build the entire region of C-DENR (amino acid residues 110–195). The final model was refined at 1.74 Å resolution to a Rfree of 24.7% (Table 1).
Table 1.
C-DENR110–195 data collection and refinement statistics.
| Hg absorption edge | Native | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.008 | 0.9792 |
| Space group | P6(4)22 | P2(1) |
| Cell dimensions | ||
| a, b, c (Å) | 116.60, 116.60, 75.23 | 64.67, 47.26, 95.50 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 106.29, 90.00 |
| Resolution (Å) | 100.00–2.2 (2.2) a | 91.67–1.74 (1.74) |
| Rmerge | 16.8 (1207.0) | 10.5 (659.6) |
| I/σ (I) | 11.7 (0.1) | 7.8 (0.2) |
| CC1/2b | 99.9 (2.7) | 99.7 (7.0) |
| Anomal. correlation (%) | 56 (30)c | |
| Completeness (%) | 100 (1 0 0) | 98.9 (99.4) |
| Redundancy | 20.6 (19.9) | 4.4 (4.4) |
| Refinement | ||
| Resolution (Å) | 100.0–2.2 (2.2) | 91.67–1.74 (1.74) |
| No. reflections | 15,752 | 51,222 |
| Rwork/Rfreed (%) | 22.4/28.8 | 17.8/24.7 |
| No. molecules/ASU | 2 | 6 |
| No. atoms | 1459 | 4926 |
| rmsd Bond length (Å) | 0.009 | 0.0072 |
| rmsd Bond angle (°) | 1.04 | 0.81 |
Values in parentheses are for highest-resolution shell.
CC1/2 is the correlation coefficient.
Anomalous correlation (30 at 3.1 Å)
Cross-validation dataset corresponds to 5% randomly selected reflections.
2.2. Overview of the C-DENR110–195 structure
Superposition of C-DENR structures built for P6422 and P21 crystal forms revealed that they are almost identical except for the positions of the β-strand 1, which were swapped (Fig. 1D). However, these β-sheets with the swapped β-strand 1 in place are identical and, together with the rest of the molecule, will represent the biologically relevant state of the C-DENR110–195 structure that will be discussed further in this paper (Figs. 1C, 2). This high-resolution structure of C-DENR superposed well with the structure of C-DENR bound to the human 40S ribosomal subunit, which we determined previously at 6.0 Å resolution (Fig. 2) [3]. This suggests that the C-terminal domain of DENR does not undergo major conformational rearrangements upon binding to the ribosome. Likewise, no such rearrangements were observed for eIF1 or eIF1-like domain of eIF2D [5], [24], [27], [28], [29]. Superposition of the structure of C-DENR with the structure of eIF1 determined by NMR (amino acid residues 27–113) confirmed that it resembles that of eIF1 (rmsd 1.78 Å), but revealed some differences in the β-hairpin loop 2 region (Fig. 2) [25].
Fig. 2.
Superposition of the C-DENR110–195 structure (coral) with: A. the structure of the human C-DENR (light gray) bound to the human 40S subunit; B. the NMR structure of the human eIF1 (green, PDB: 2IF1, amino acid residues 28–108); C. the structure of the human C-eIF2D (gray, PDB: 5W2F); D. structures of archaeal aIF1 (blue, PDB: 5ZCY) and bacterial YciH (yellow, PDB: 1D1R). The basic loop and β-hairpin 2 loop are marked by an arrow; N- and C-terminus of a protein are marked by N and C, respectively. Positions of amino acid residues of C-DENR are presented by spheres: Pro121 (red), Lys125 (gold), Cys154 (blue). A crosslink between residues Lys125 and Cys154 is marked by a dotted line. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
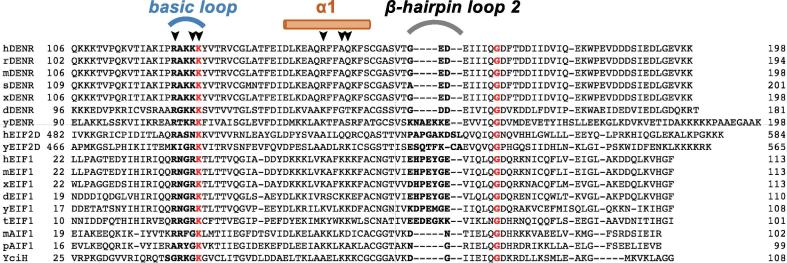
The β-hairpin loop 1, namely the basic loop (Arg122-Lys126), plays the main role in C-DENR interactions with the 40S subunit and the tRNA positioned in the P site [3], [5]. Structural studies of eukaryotic translation preinitiation complexes (PIC) predict that the basic loop of eIF1 interacts/clashes with the anticodon stem loop (ASL) of Met-tRNAiMet. During scanning, the basic loop displaces the ASL of Met-tRNAiMet and prevents the locking of Met-tRNAiMet in the P site. Base pairing of the anticodon loop of Met-tRNAiMet with the initiation codon, locks tRNA in the P site, displacing the basic loop of eIF1 out of the P site, which lead to dissociation of eIF1 from the ribosome [8], [27], [28], [30]. Only initiator Met-tRNAMet induces dissociation of eIF1 upon base pairing with AUG. Exclusivity of this function of initiator Met-tRNA is attributed to the presence of three universally conserved G-C base pairs in its ASL [30]. This allows eIF1 to promote ejection of deacylated tRNA from the P site during recycling, or be ejected itself at the end of translation initiation. We showed earlier that a potential clash of the basic loop of C-DENR with the ASL of Met-tRNAi is more severe than that in the case of eIF1. We also proposed that Pro121, which is conserved in all DENR from vertebrates, provides a structural kink and may impose local rigidity for the basic loop, thereby enhancing its activity in the dissociation of the P-site tRNA (Fig. 3, Fig. 4) [3]. The functional relevance of this is emphasized by the discovery that mutation Pro121Leu in human DENR is associated with Asperger syndrome and impairs neuronal development [31]. Amino acid residues Arg122, Lys125 and Lys126 of the basic loop of C-DENR are conserved among eIF1-like domains in eukaryotes (Fig. 3). In yeast eIF1, substitution of corresponding residues Arg33, Arg36 and Lys 37 with Glu weakened its binding to the 40S subunit in vitro and conferred a Sui¯ phenotype, mutation Lys37Glu is lethal [32]. The basic loop of C-DENR has an additional positively charged residue, Lys124 (Gly in eIF1), that may make the interaction of the basic loop with the phosphate backbone of the h24 and h44 of the 18S rRNA in the mRNA binding channel stronger than that of eIF1 and eIF2D (Fig. 3). In addition, comparison of the swapped β-strand 1 in C-DENR structures presented here defines Lys126 as a hinge when the loop Arg122-Lys125 adopts an extended conformation upon rotation and swapping (Fig. 1D). These suggest that the only flexible part of the β-hairpin 1 is the region Arg122-Lys125. Moreover, we observed a crosslink between Lys125 and oxidized Cys154. (Fig. 2, Fig. 7). This crosslink is present only in the P21 crystal form, and is likely an artefact of crystallization. Though, if this crosslink formed in vivo, it would strongly decrease both mobility of the basic loop and its positive charge (see also discussion below).
Fig. 3.
Sequence alignment of the eIF1-like proteins. Multiple sequence alignment using program Clustal Omega (www.clustal.org/omega) with default settings was performed for protein sequences (accession/organism) of DENR: h (NP_003668.2, Homo sapiens), r (NP_001108518.1, Rattus norvegicus), m (NP_080879.1, Mus musculus), s (NP_001134755.1, Salmo salar), x (NP_001086186.1, Xenopus laevis), d (NP_573176.1, Drosophila melanogaster), y (P47089.1, Saccharomyces cerevisiae); eIF2D: h (P41214.3), y (DAA11962.1); eIF1: h (P41567.1), m (P48024.2), x (NP_001079402.1), d (NP_001079402.1), y (NP_014155.1), t (DAA33997.1, Tetrahymena thermophila); aIF1: m (PDBID 4MO0, Methanocaldococcus jannaschii), p (PDBID 5ZCY, Pyrococcus horikoshii); YciH (VWQ02336.1, Escherichia coli). Conserved amino acid residues are marked in red, amino acid residues involved in rRNA binding are marked by arrows, regions are marked by lines: blue (basic loop), gray (β-hairpin 2 loop), brown (helix α1). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
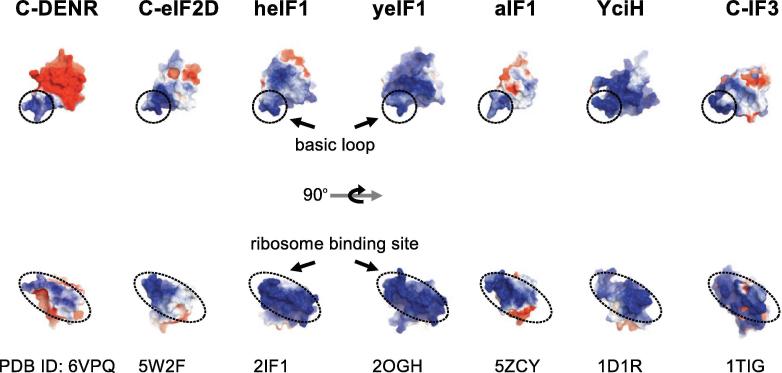
Fig. 4.
Electrostatic charge distribution of the human C-DENR110–195, C-eIF2 (PDB: 5W2F) and eIF1 (heIF1, PDB: 2IF1), Saccharomyces cerevisiae eIF1 (yeIF1, PDB: 2OGH), Pyrococcus horikoshii aIF1 (PDB: 5ZCY), Escherichia coli YciH (PDB: 1D1R) and Geobacillus stearothermophilus C-IF3 (PDB: 5ZCY). Surfaces are colored by potential on solvent accessible surface.
Fig. 7.
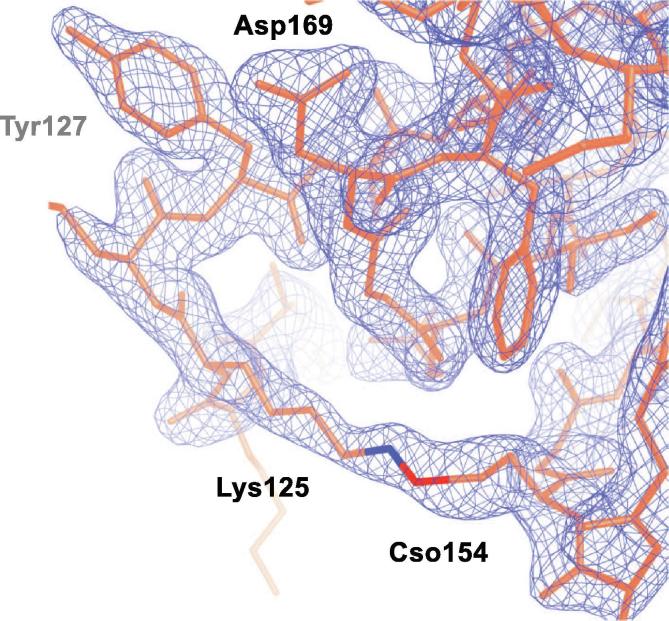
The electron density map 2Fo-Fc contoured at σ = 1.5 (blue mesh) calculated for the region of C-DENR were the crosslink between Lys125 and oxidized Cys154 (S-hydrohycysteine, Cso154) was observed in the P21 crystal form. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The β-hairpin loop 2 of C-DENR is small, in contrast to eIF1 and C-eIF2D, and similar in size to that of aIF1 and YciH (Fig. 2, Fig. 3). In the canonical PIC the β-hairpin loop 2 of eIF1, which is negatively charged, will clash with the D stem of Met-tRNAiMet that is base paired with the AUG in the ribosomal P site (Pin state of the tRNAi), providing an additional force to displace eIF1 from the P site when scanning is finished [28], [29], [33], [34]. In the PIC assembled on the HCV IRES the longer, but less negatively charged, β-hairpin loop 2 of eIF2D contacts the D stem of Met-tRNAiMet, likely stabilizing tRNAi in a hybrid P/E-like state, which is not possible in the context of a canonical PIC [5]. Superposition of PICs structures suggested that the short β-hairpin loop 2 of C-DENR does not interact with the Met-tRNAiMet in the Pin state, as also was seen for aIF1 [3], [35]. Interesting, the N-terminal domain of aIF1 of many archaeal organisms and N-terminal domain of DENR possess a zinc-binding site, which is essential for formation of the DENR-MCT-1 heterodimer. However, its role in aIF1 is not clear yet [22], [36].
The helix α1 of human C-DENR contains only three positively charged amino acid residues, Lys142, Arg146, Lys151, which correspond to Lys52, Lys56, Asp61 of the yeast eIF1, respectively. In addition, there are three more positively charged residues in the helix α1 of the yeast eIF1: Arg53, Lys59 and Lys60 (Fig. 3). It was shown that conserved Lys56, Lys59, Lys60 interact with the helix h45 of 18S rRNA and are essential for ribosome binding. Whereas substitutions of Lys56 and Lys60 confer the Sui− phenotype, substitution of Lys59 is lethal [27], [32].
The total number of positively charged residues in helix α1 of eIF1 may vary. However, it was suggested that the different combination of these residues keep overall affinity to the 40S subunit constant across phylogeny (Fig. 3) [37]. Indeed, a dramatic change in the charge distribution in helix α1 caused by double substitution of Lys56 and Lys60 with glutamic acid or substitution of all five basic residues with alanines are lethal [32], [38]. Essential for yeast 40S subunit binding, Lys59 and Lys60 of eIF1 are substituted in human C-DENR by alanine and glutamine, respectively (Fig. 3). Compared to eIF1s, these substitutions will decrease affinity of helix α1 of C-DENR to the ribosome. However, DENR is tightly bound to MCT-1, which binds on its own to helix h24 of the 18S rRNA and may preserve overall affinity of DENR-MCT-1 to the 40S subunit [3]. Alternatively, higher affinity between the 18S rRNA and the basic loop of C-DENR due to an additional lysine residue (Lys124), may compensate for the weaker binding of helix α1 (Fig. 3). The total number of positively charged residues in helix α1 is also reduced in the C-terminal domain of eIF2D, which is covalently attached to the MCT-1-like N-terminal domain (Fig. 1A). However, the basic loop of C-eIF2D also has less positively charged residues, compared to eIF1 (Fig. 3). These likely reflect not only differences in the kinetics of interaction of eIF1, eIF2D or DENR-MCT-1 with the rRNA, mRNA and P-site tRNA, but also the differences in the composition of the ribosomal complexes, with which these factors interact, i.e. complexes with or without eIF1A, eIF2, eIF3 or eIF5.
2.3. Surface charge distribution of C-DENR
We compared electrostatic potential surfaces of C-DENR and eIF1-like domains of eIF2D (C-eIF2D), eIF1, archaea IF1 (aIF1), bacterial YciH, and also the C-terminal domain of IF3 (C-IF3), the latter is a functional homologue of eIF1 (Fig. 4) [30]. All these proteins interact with the rRNA and the ASL of the tRNA in the P site of the small ribosomal subunit through their basic loop/β-hairpin loop 1 (β-hairpin loop in C-IF3), which constitutes a strong positive electrostatic potential surface. The ribosome interacting region of these proteins is also positively charged, as it is expected from the rRNA interacting proteins (Fig. 4). Interesting, the solvent-exposed surface of C-DENR is highly negatively charged, which is similar to C-eIF2D and aIF1 (Fig. 4). The corresponding area in eIF1 from yeast and most of this area in eIF1 from humans are positively charged. eIF1 is essential for the scanning ability of the PIC, whereas DENR-MCT-1, eIF2D or aIF1 are not. A striking difference in the charge distribution of the solvent-exposed surface of these two groups of translation factors may reflect differences in their interactions with other eIFs required for scanning, initiation codon selection or ribosomal recycling.
2.4. Stability and oligomerization state of C-DENR
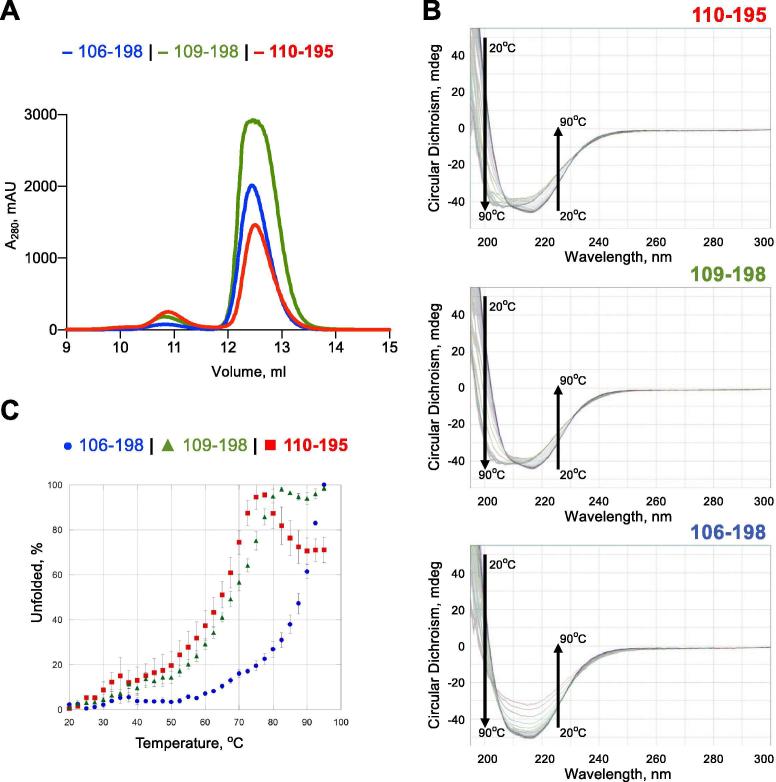
Domain swapping observed in both crystal forms raises some interesting questions: does it happen in vivo and, if yes, is it physiological? Size-exclusion chromatography shows that about 85% of C-DENR110–195 behaves as a monomer in solution at 4 °C (Fig. 5A). We also analyzed by circular dichroism (CD) a temperature dependence of the fold of all three C-DENR deletion mutants that we constructed (Fig. 5B and C). The data illustrate that the CD intensity of C-DENR110–195 and C-DENR109–198 decreases and the negative maxima at 218 nm is shifted to 208 nm when temperature is increased. These indicate that the folding of these mutants at room temperature converts to a more random structure at higher temperatures when β-strand 1 is flipped from the β-sheet and β-strands 1 and 5 became unstructured. The C-DENR106–198 mutant is more stable, which suggests that amino acid residues 106–109 (QKKK) are crucial for the stability of the C-DENR’s β-sheet.
Fig. 5.
Size-exclusion chromatography analysis and thermal unfolding of the C-DENR mutants using Circular Dichroism. A. Chromatogram of C-DENR mutants runs on the Superdex 75 10/300 column. B. Temperature-dependent CD spectra of C-DENR110–195 (top), C-DENR109–198 (middle), C-DENR106–199 (bottom). C. Temperature-dependent transitions from folded to unfolded state monitored at 220 nm.
In addition, conventional Molecular Dynamics (MD) simulations were performed on both the extended and semi-extended conformations observed in the C-DENR110–195 structures from P6422 and P21 crystals forms, respectively. Here, both conformations quickly relax into a non-native intermediate state and get trapped into this local minimum until the end of the observed simulation (data not shown). To overcome this local energy barrier and push the trajectories to explore more extensive and relevant conformation space, we next performed enhanced sampling metadynamics simulations along the two chosen collective variables (Materials and Methods). A free energy landscape (Fig. 6) constructed for the semi-extended conformation metadynamics trajectory shows that this starting crystal conformation is highly destabilized and thus quickly progresses downhill towards a more well-folded, compact ground state (eIF1-like fold). This folding pathway proceeds via two local minima or “intermediate states” where residues 116 to 118 in the N-terminus of C-DENR mutant form non-native contacts with residues 158–160 in the molecule. From intermediate state 2, the structures quickly converge to the ground state where the N-terminus forms a β-strand embedded in the C-DENR molecule to form the five-stranded parallel and antiparallel β-sheet. The metadynamics simulations of the extended conformation however show that it is more resistant to conformational changes and doesn’t fold back into a compact ground state. These results indicate that the semi-extended conformation is a non-native, destabilized alternative conformation of the C-DENR110–195 mutant and it folds into a compact ground state conformation with five-stranded parallel and antiparallel β-sheet by crossing an energy barrier equivalent to 14 kJ/mol. The extended conformation however remains stable due to the possibility of domain swapping of β-strand 1 between molecules to generate a C-DENR dimer, where each monomer is stabilised in the compact ground state due to the swapping.
Fig. 6.
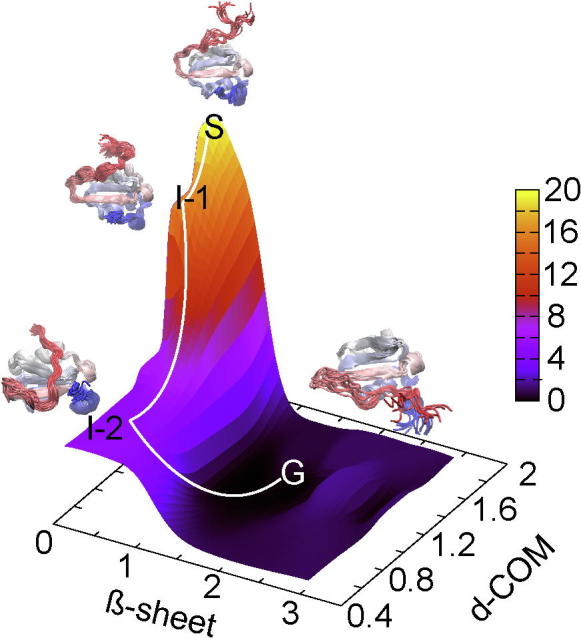
Free energy landscape of the C-DENR semi-extended conformation. S denotes the starting crystal structure, I-1 and I-2 are the intermediate states and G is the compact ground state. The two collective variables are: β-sheet, β-strand character of the N-terminal residue stretch 121–133 of the C-DENR110–195 mutant; d-COM, the distance of the center of mass of these residues with the rest of the molecule.
2.5. Crosslink between Lys125 (basic loop) and – Cso154 (S-hydroxycysteine) in P21 crystal
Upon building the model in the electron density map of the P21 crystal form, we discovered density connecting Lys125 and Cys154, which is not present in the P6422 crystals (Figs. 2B, 7). When the formation of a crosslink between Cys and Lys side chains in a crystal structure of a protein is only partial, it would be extremely difficult to distinguish whether the formed bridge is an O bridge or a CH2 bridge, solely based on the electron density map. However, our previous analysis of change in bond energies suggests that only an O bridge can be formed spontaneously between Cys and Lys side chains upon exposure to air, but not a methylene bridge [39]. We believe that a covalent bridge between Lys25 and Cys154 of side chains of C-DENR110–195 is likely to have occurred from air oxidation during the crystal growth, though, analysis of an electron density map alone cannot distinguish if oxidation occurred before or after crystallization, or from X-ray diffraction experiments [40], [41].
3. Conclusion
As expected, the crystal structure of the C-terminal domain of human DENR shows that it has an eIF1-like fold, though there are some differences in charge distribution and the size of the functionally important regions. Compared to eIF1, the basic loop of C-DENR contains an additional positively charged amino acid residue, Lys124 (Gly in eIF1), which may provide an additional binding affinity to the P site of the 40S subunit and reduce probability of the tRNA to displace C-DENR from that site. It was shown that eIF1 promotes dissociation of deacylated tRNA from the posttermination 40S subunits in the presence of eIF3, whereas eIF2D and DENR-MCT-1 can do it alone [4], [9], [10]. Our data may explain this difference. First, the position of DENR-MCT-1 or eIF2D on the 40S subunit is incompatible with eIF3. As a result, DENR-MCT-1 either displace eIF3 from the interface of the 40S subunit or acts at ribosomal complexes that do not have eIF3 bound [3], [5]. Second, increased rigidity and positive charge of the basic loop of C-DENR together with the enhanced binding to the 40S subunit through interaction with MCT-1, are sufficient to dissociate the deacylated tRNA by DENR-MCT-1 alone [21].
Though the observed crosslink between Lys125 and the oxidized Cys154 in the C-DENR structure derived from the P21 crystal form is a spontaneous event, it demonstrates an additional possibility to regulate stability/rigidity of the basic loop and the protein itself. In vivo, at least twelve protein lysine methyltransferases modify translation elongation factors and ribosomal proteins in yeast, which likely is important for fine tuning cell physiology [42], [43]. We speculate that Lys125 may be methylated or Cys154 may be oxidized in vivo under some conditions, providing possibility for the aforementioned crosslink. As a result, increased rigidity of the basic loop and eliminating Lys125 from the interaction with the rRNA may fine tune regulation of physiological activity of DENR-MCT-1 in translation or, contrary, affect translation of some genes that will results in malignancy or other aberrant phenotypes.
4. Materials and methods
4.1. Plasmid constructs, protein expression and purification
The QuikChange Lightning® site-directed mutagenesis kit (Agilent Technologies) and the plasmid pET28-CDENR (110–198) [3] were used to obtain C-DENR106–198, C-DENR109–198, C-DENR110–195 expression constructs. C-DENR mutants were expressed and purified as described previously [3]. The last purification step for all C-DENR mutants was size exclusion chromatography on the Superdex™ 75 (16/60) column (GE Healthcare, Marlborough, MA, USA) in the buffer: 0.01 M Tris-HCl pH 8.0, 0.1 M NaCl, 2 mM DTT. Concentrated proteins were then analyzed on the Superdex™ 75 (10/300) column equilibrated in the buffer 0.01 M Tris-HCl pH 8.0, 0.3 M NaCl, 2 mM DTT.
4.2. Crystals growth, heavy atoms soaks and freezing
Crystals were grown in 24-wells sitting-drop plates using the vapor diffusion technique. 3 μl of DENR110–195 (10 mg/ml) were mixed with 2 μl of reservoir solution 1 (0.2 M K/Na tartrate, 2.5 M Ammonium Sulfate, 0.1 M tri-Sodium citrate, pH 5.6) or 3 μl of reservoir solution 2 (0.1 M CaCl2, 15% PEG 400, 0.1 M Sodium acetate, pH 5.0). Plates with reservoir solution 1 were incubated at 20 °C for 21–31 days and with reservoir solution 2 for 42 days at 12 °C. Crystals growing at 20 °C were stabilized by soaking for 15 min in the following buffer: 0.2 M K/Na tartrate, 2.8 M Ammonium Sulfate, 0.1 M tri-Sodium citrate, pH 5.6 and 25% of glycerol, with or without 10 mM Hg(OAc)2. Crystals growing at 12 °C were stabilized by soaking for 20 min in the buffer: 0.1 M Ca Acetate, 40% PEG 400, 0.1 M Sodium acetate, pH 5.6. After stabilization, crystals were frozen in liquid nitrogen.
4.3. Data collection and processing
X-ray diffraction data were collected at the beamline 24ID-C, the Advanced Photon Source in the Argonne National Laboratory. A complete data set was collected from a single crystal to a 1.74 (native) and 2.3 (Hg) Å resolution. SAD data set was collected at the Hg absorption peak wavelengths of 1.008 Å. Diffraction data were processed and scaled using XDS (Table 1) [44]. Experimental phases were generated and initial model was built by AutoSol Wizard from PHENIX [45].
The final structure (P21 crystal form) was solved by molecular replacement using PHASER from the CCP4 program suite and the model of C-DENR built in the experimental map (P6422 crystal form). This model had the swapped β-strand 1 from the other molecule and did not include the basic loop. Program Coot was used for model building, Refmac 5 from the CCP4 suite and Phenix was used for the model refinement [45], [46], [47]. The final cross-validated Rfree after model refinement was 24.7%.
The electrostatic charge distribution of each protein was generated using PDB2PQR Server and the APBS plugin of the molecular visualization tool PyMOL [Delano Scientific, The PyMOL Molecular Graphics System, Version 1.8 Schrödinger (https://www.pymol.org)] [48], [49].
4.4. Circular dichroism (CD) measurements and analysis
C-DENR mutants (final concentration 3 mg/ml in 10 mM Phosphate buffer, pH 7.4 with 100 mM KF) were incubated at 20 °C for 10 min. The CD spectra for thermal unfolding were acquired on a Chirascan Circular Dichroism Spectrometer (Applied Photophysics, Inc., Beverly, MA, USA) from 20 °C to 90 °C with the 5 °C interval as the average of three scans. Thermal unfolding was performed after a 10-minute equilibration at the desired temperature and an integration time of 2.5 s. Thermal stability was assessed by monitoring the change in the CD signal at 220 nm as a function of temperature.
4.5. Molecular dynamics simulations
Molecular dynamics simulations were performed on the C-DENR110–195 mutant crystal structures showing both the extended and semi-extended conformations using GROMACS 5 [50] with the AMBER-03 forcefield [51], where the AMBER-99 potential has been revised based on QM-continuum solvent calculations. The crystal structure was first placed in the center of a 280 nm3 dodecahedron simulation box with each side 1.2 nm away from the protein. The box was solvated with TIP3P water model and the charge on the system was neutralized by adding seven Na+ atoms and an additional 100 mM NaCl was included to provide stability to the protein structure. The system was then energy minimized using steepest descent and conjugate gradient methods followed by 50 ps of position restraints on the solute (C-DENR) while the temperature of the box was raised to 200 K by coupling to an external bath. Next, the position restraints were removed, and the system was simulated under NVT (constant Number of particles, Volume and absolute Temperature) conditions for 100 ps. Finally, the box was heated to the final simulation temperature of 298.15 K and equilibrated under NPT (constant Number of particles, Pressure and absolute Temperature) conditions for a further 5 ns. The equilibration of the simulation was monitored using the pairwise root mean square distance (RMSD) of the solute configuration during the simulation with the starting crystal structure. Following equilibration, production runs under NPT conditions were continued for a further 300 ns.
4.6. Metadynamics
For each of the C-DENR110–195 extended and semi-extended conformations, two NPT simulations with different velocity seeds were initiated from the equilibrated trajectories generated above. These were continued for a further 5 ns, during which the Pearson Correlation Co-efficient was monitored for the pairwise RMSD between the two trajectories at each time point to ensure that the trajectory progressions are sufficiently un-correlated at the end of 5 ns. Subsequently, metadynamics was switched on in the bias-exchange mode to enhance the conformational sampling of the system along the chosen collective variables (CV). CV1 was chosen to be the beta-strand character of the N-terminal residue stretch 121–133 of the C-DENR110–195 mutant and CV2 was chosen to the distance of the center of mass of these residues with the rest of the molecule. In the bias-exchange mode, the evolution of the trajectory is changed stochastically with time according to rules of the replica-exchange method. Each CV was imposed on one replica, and the trajectories were simulated until they converged.
5. Accession numbers
PDB accession numbers for coordinates and structure factors reported in this paper are: 6VPQ and 6VPR.
CRediT authorship contribution statement
Ivan B. Lomakin: Conceptualization, Methodology, Investigation, Writing - original draft, Writing - review & editing, Supervision. Swastik De:Investigation, Writing - original draft. Jimin Wang: Formal analysis, Writing - original draft. Aditi N. Borkar: Investigation, Writing - original draft, Writing - review & editing. Thomas A. Steitz: Conceptualization, Funding acquisition.
Acknowledgements
We thank professor Ronald R. Breaker (Yale University) for invaluable help and support in keeping our research running after decease of professor Thomas A. Steitz and for critical reading of the manuscript; the staff of the Advanced Photon Source beamline NECAT 24-ID and the Richards Center facility at Yale University for support. This work was supported by NIH grant GM022778 to R.R.B. and the Sir Henry Wellcome Postdoctoral Fellowship, UK and Anne McLaren Fellowship, University of Nottingham, UK to A.N.B.
References
- 1.Deyo J.E., Chiao P.J., Tainsky M.A. drp, a novel protein expressed at high cell density but not during growth arrest. DNA Cell Biol. 1998;17(5):437–447. doi: 10.1089/dna.1998.17.437. [DOI] [PubMed] [Google Scholar]
- 2.Oh J.J. Identification of differentially expressed genes associated with HER-2/neu overexpression in human breast cancer cells. Nucleic Acids Res. 1999;27(20):4008–4017. doi: 10.1093/nar/27.20.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lomakin I.B. Crystal structure of the human ribosome in complex with DENR-MCT-1. Cell Rep. 2017;20(3):521–528. doi: 10.1016/j.celrep.2017.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skabkin M.A. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24(16):1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weisser, M., et al., Structural and functional insights into human re-initiation complexes. Mol Cell, 2017;67(3): p. 447–456 e7. [DOI] [PubMed]
- 6.Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weisser M., Ban N. Extensions, extra factors, and extreme complexity: ribosomal structures provide insights into eukaryotic translation. Cold Spring Harb Perspect Biol. 2019;11(9) doi: 10.1101/cshperspect.a032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinnebusch A.G. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci. 2017;42(8):589–611. doi: 10.1016/j.tibs.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Pisarev A.V., Hellen C.U., Pestova T.V. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131(2):286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pisarev A.V. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37(2):196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skabkin M.A. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell. 2013;51(2):249–264. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinnebusch A.G., Ivanov I.P., Sonenberg N. Translational control by 5'-untranslated regions of eukaryotic mRNAs. Science. 2016;352(6292):1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson R.J., Hellen C.U., Pestova T.V. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 14.Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29(24):5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryabova L.A., Pooggin M.M., Hohn T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 2006;119(1):52–62. doi: 10.1016/j.virusres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip Rev RNA. 2014;5(6):765–778. doi: 10.1002/wrna.1245. [DOI] [PubMed] [Google Scholar]
- 17.Schleich S. Identification of transcripts with short stuORFs as targets for DENR*MCTS1-dependent translation in human cells. Sci Rep. 2017;7(1):3722. doi: 10.1038/s41598-017-03949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schleich S. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014;512(7513):208–212. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zinoviev A., Hellen C.U., Pestova T.V. Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol Cell. 2015;57(6):1059–1073. doi: 10.1016/j.molcel.2015.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castelo-Szekely V. Charting DENR-dependent translation reinitiation uncovers predictive uORF features and links to circadian timekeeping via Clock. Nucleic Acids Res. 2019;47:5193–5209. doi: 10.1093/nar/gkz261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellen C.U.T. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol. 2018 doi: 10.1101/cshperspect.a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomakin I.B., Dmitriev S.E., Steitz T.A. Crystal structure of the DENR-MCT-1 complex revealed zinc-binding site essential for heterodimer formation. Proc Natl Acad Sci U S A. 2019;116(2):528–533. doi: 10.1073/pnas.1809688116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed Y.L. DENR-MCTS1 heterodimerization and tRNA recruitment are required for translation reinitiation. PLoS Biol. 2018;16(6) doi: 10.1371/journal.pbio.2005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaidya A.T. Crystal structure of the c-terminal domain of human eIF2D and its implications on eukaryotic translation initiation. J Mol Biol. 2017;429(18):2765–2771. doi: 10.1016/j.jmb.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fletcher C.M. Structure and interactions of the translation initiation factor eIF1. EMBO J. 1999;18(9):2631–2637. doi: 10.1093/emboj/18.9.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascarenhas N.M., Gosavi S. Understanding protein domain-swapping using structure-based models of protein folding. Prog Biophys Mol Biol. 2017;128:113–120. doi: 10.1016/j.pbiomolbio.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llacer J.L. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell. 2015;59(3):399–412. doi: 10.1016/j.molcel.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomakin I.B., Steitz T.A. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500(7462):307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisser M. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat Struct Mol Biol. 2013;20(8):1015–1017. doi: 10.1038/nsmb.2622. [DOI] [PubMed] [Google Scholar]
- 30.Lomakin I.B. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25(1):196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas M.A. De novo mutations in DENR disrupt neuronal development and link congenital neurological disorders to faulty mRNA translation re-initiation. Cell Rep. 2016;15(10):2251–2265. doi: 10.1016/j.celrep.2016.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Marcos P. beta-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNA(Met) recruitment and accuracy of AUG selection in vivo. J Biol Chem. 2013;288(38):27546–27562. doi: 10.1074/jbc.M113.498642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hussain T. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014;159(3):597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabl J. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331(6018):730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 35.Coureux P.D. Cryo-EM study of start codon selection during archaeal translation initiation. Nat Commun. 2016;7:13366. doi: 10.1038/ncomms13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monestier A. Role of aIF1 in Pyrococcus abyssi translation initiation. Nucleic Acids Res. 2018;46(20):11061–11074. doi: 10.1093/nar/gky850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Marcos P. Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy. RNA. 2014;20(2):150–167. doi: 10.1261/rna.042069.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin-Marcos P., Cheung Y.N., Hinnebusch A.G. Functional elements in initiation factors 1, 1A, and 2beta discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol. 2011;31(23):4814–4831. doi: 10.1128/MCB.05819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J. Crystallographic identification of spontaneous oxidation intermediates and products of protein sulfhydryl groups. Protein Sci. 2019;28(3):472–477. doi: 10.1002/pro.3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J. X-ray radiation-induced addition of oxygen atoms to protein residues. Protein Sci. 2016;25(8):1407–1419. doi: 10.1002/pro.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J. Oxygen additions in serial femtosecond crystallographic protein structures. Protein Sci. 2016;25(10):1797–1802. doi: 10.1002/pro.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polevoda B., Sherman F. Methylation of proteins involved in translation. Mol Microbiol. 2007;65(3):590–606. doi: 10.1111/j.1365-2958.2007.05831.x. [DOI] [PubMed] [Google Scholar]
- 43.White J.T. Protein methylation and translation: role of lysine modification on the function of yeast elongation factor 1A. Biochemistry. 2019;58(49):4997–5010. doi: 10.1021/acs.biochem.9b00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams P.D. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vagin A.A. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 47.Winn M.D. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolinsky T.J. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32(Web Server):W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schrodinger, LLC, The AxPyMOL Molecular Graphics Plugin for Microsoft PowerPoint, Version 1.8. 2015.
- 50.Pronk S. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duan Y. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24(16):1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]