Highlights
-
•
We expressed and purified the cytoplasmic domain of the 3a protein.
-
•
Cyto3a domain binds calcium.
-
•
Calcium binding causes a conformational change.
-
•
3a protein in vivo to have significant role in viral pathogenesis.
Abbreviations: SARS-CoV, Severe Acute Respiratory Syndrome Coronavirus; orf, open reading frame
Keywords: SARS-CoV, Cyto3a protein, Calcium, Ion channel, Protein conformation
Abstract
The Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) is a positive stranded RNA virus with ∼30 kb genome. Among all open reading frames (orfs) of this virus, the orf3a is the largest, and encodes a protein of 274 amino acids, named as 3a protein. Sequence analysis suggests that the orf3a aligned to one calcium pump present in Plasmodium falciparum and the enzyme glutamine synthetase found in Leptospira interrogans. This sequence similarity was found to be limited only to amino acid residues 209–264 which form the cytoplasmic domain of the orf3a. Furthermore, this region was predicted to be involved in the calcium binding. Owing to this hypothesis, we were driven to establish its calcium binding property in vitro. Here, we expressed and purified the cytoplasmic domain of the 3a protein, called Cyto3a, as a recombinant His-tagged protein in the E. coli. The calcium binding nature was established by performing various staining methods such as ruthenium red and stains-all. 45Ca overlay method was also done to further support the data. Since the 3a protein forms ion channels, we were interested to see any conformational changes occurring in the Cyot3a upon calcium binding, using fluorescence spectroscopy and circular dichroism. These studies clearly indicate a significant change in the conformation of the Cyto3a protein after binding with calcium. Our results strongly suggest that the cytoplasmic domain of the 3a protein of SARS-CoV binds calcium in vitro, causing a change in protein conformation.
With the news pouring in about advances in infection by influenza viruses, the name of a closely related candidate called the coronavirus cannot be overlooked. Severe Acute Respiratory Syndrome (SARS) virus is one such member of the coronavirus family that spread viral respiratory illness in 2003 which attracted attention globally. After April 2004, no new cases of the SARS virus infection have been reported until September 2012, when a fresh case of viral respiratory illness got noticed with the causative agent belonging to the coronavirus family (Martina et al., 2003). SARS-CoV is a newly identified coronavirus in humans that leads to a dangerous acute inflammation and is more lethal than other human coronaviruses (Drosten et al., 2003). Among 14 orfs of this virus, the largest unique orf is the orf 3a which codes for 3a protein. This protein has been proposed to contribute to increased virulence of the virus in humans (Pacheco et al., 2003). It is an important protein of this virus that needs to be studied extensively owing to its unique existence and multi-tasking behavior.
The 3a protein of SARS-CoV forms homo- as well as hetero-tetramers in transfected and possibly in infected cells (Yuan et al., 2005, Yuan et al., 2007). This multi-meric pattern is commonly observed in most of the viral proteins that show ion channel activity commonly known as viroporins (Lu et al., 2006). Indeed, 3a protein was shown to form ion channels which are selective to potassium ions (Lu et al., 2006). However, till date, only a few ion channel proteins for viruses have been identified, and some of the important ones include Kcv protein of Paramecium bursaria chlorella virus (Plugge et al., 2000), M2 protein of influenza virus (Pinto et al., 1992) and Vpu and Vpr of HIV (Ewart et al., 1996, Piller et al., 1996). The functions of these ion channels vary. The Kcv is associated with virus replication (Plugge et al., 2000), and M2 protein is responsible for influenza A virus uncoating, which is a critical step in viral infection (Ciampor et al., 1995). The 3a protein shows similar function to those of Vpuprotein in HIV-1 (Lu et al., 2006). The formation of a pore structure in the virus-infected cell membrane makes the cell more permeable which could be an important factor in the SARS-CoV life cycle. The ion channel activity of the 3a protein is also linked to its pro-apoptotic function (Chan et al., 2009).
The 3a protein is predicted to contain three transmembrane regions (34–56, 77–99 and 103–125), and a C-terminal cytoplasmic domain of 149 amino acid residues (Rota et al., 2003). The 3a protein has been studied to form ion channel sensitive to potassium ions (Lu et al., 2006). Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Moreover, in case of bacterial potassium channels, a portion of the protein functions as gate to the channel (Doyle et al., 1998, Jiang et al., 2002a, Jiang et al., 2002b). The binding of calcium to the cytoplasmic domain of these channels lead to conformational change that is shown to effect the pore lining (Schumacher and Adelman, 2002). Based on these evidences we speculated that calcium might be playing same kind of role as a ligand to the gated channel. It has been reported on the basis of bioinformatics analysis that the cytoplasmic domain of SARS 3a protein is a calcium-binding protein (Singh et al., 2004). Calcium binding motifs present in viral proteins are known to play an important role in virion assembly and stability (Gorbalenya et al., 1991, Garbutt et al., 1999, Ruiz et al., 2000). The calcium-binding pockets in 3a cytoplasmic domain shows a putative ‘EF hand’ motif commonly found in many calcium-binding proteins. We focused our study on the cytoplasmic domain, the Cyto3a. We expressed and purified the recombinant Cyto3a protein with His-tag and subjected it to various staining methods that specifically stain calcium binding proteins to prove its calcium binding nature. Fluorescence emission helps in studying the interaction of a protein with various ligands and presents adequate information about the magnitude and the microenvironment of the protein residues (Hirshfield et al., 1996, Deepa and Mishra, 2005, Zhao et al., 2006, Zhang et al., 2008). Hence, fluorescence spectroscopic study was done to monitor the changes in the intrinsic fluorescence of the Cyto3a protein upon its interaction with calcium followed by Circular dichroism (CD) study.
The orf3a gene (nucleotides 25,268–26,092) of the SARS-CoV genome (GenBank NC_004718) was provided by Dr. Vincent Chow (National University of Singapore). From this clone,the Cyto3awas PCR amplified using primers, X1C1, GAATTCATGAGATGTTGGCTTTGTTGG and X1R1,GTCGACTTACAAAGGCACGCTAGTAGT and cloned into pGEMT-Easy, sequence verified, and an EcoRI-SalI fragment was sub-cloned into the EcoRI-SalI digested vector pET28a+ in frame with the hexa-histidine tag. The Cyto3a was expressed in E. coli BL21 DE3 strain containing pET28 a+-Cyto3a by induction with IPTG; as ∼17 kDa protein with His tag (Fig. 1A). We did not remove the His tag because it is not known to bind calcium (Ishitani et al., 2000). The purification was done using Ni-NTA affinity chromatography (Qiagen). All the working buffers like Tris–Cl buffer (10 mM Tris–Cl, pH 8.0) and purified Cyto3a were passed through Chelex 100 resin to remove calcium. The protein samples after passing through Chelex 100 gave calcium free Cyto3a (called here apoCyto3a).
Fig. 1.
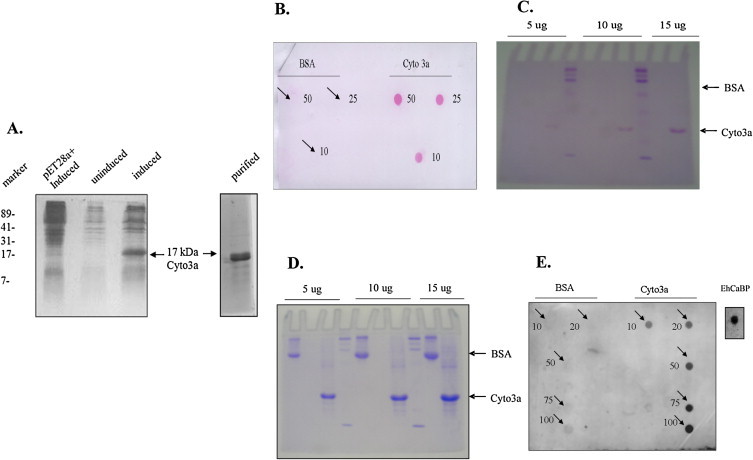
(A) SDS-PAGE of un-induced and induced sample of E. coli BL21 DE3 cells expressing pET28a+-Cyto3a. The cytoplasmic domain of 3a was expressed as a protein of approximately 17 kDa. (B) Ruthenium red staining showing red stained blots of Cyto3a while no color with BSA. (C) Stains all staining shows purple band of Cyto3a protein. (D) Commassie Brilliant blue stains both BSA and Cyto3a. (E) 45Ca Overlay method shows signals for Cyto3a and a positive control EhCaBP3 whereas BSA does not give any signal. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The first hypothesis that the Cyto3a binds calcium was proven by the various staining methods. The purified protein was subjected to stains specific for calcium binding proteins. At first ruthenium red staining was performed in which different concentrations (10, 25 and 50 μg) of the purified recombinant Cyto3a protein were spotted onto the nitrocellulose membrane. The membrane was washed with the Chelex-100 (Sigma–Aldrich) treated water to remove any residual calcium and then subjected to ruthenium red (Sigma–Aldrich) staining (25 μg ml−1) for 10 min at room temperature (Charuk et al., 1990). The negative control used here was BSA which is a non-calcium binding protein. Result showed thatCyto3adid stain red whereas BSA did not show any staining (Fig. 1B). However, ruthenium red does not bind exclusively to calcium-binding proteins; it can also stain polyanionic substrates (Charuk et al., 1990). So cationic carbocyanine dye “Stains-all” was used which preferentially stains calcium-binding proteins as dark blue or purple (Campbell et al., 1983). Different concentrations of BSA and purified recombinant Cyto3a protein (5, 10 and 15 μg) were separated by 12% SDS-PAGE. After separation, the gel was fixed and stained with “Stains-all” (Sigma–Aldrich) for 48 h. in the dark. BSA did not show purple staining whereas Cyto3a did (Fig. 1C). After this the same gel was stained with coomassie brilliant blue (Campbell et al., 1983) and both proteins showed blue bands (Fig. 1D). We clearly observed that stains-all stained only Cyto3a while there was no staining of BSA. We finally performed 45Ca overlay method in which different concentrations (10, 25 and 50 μg) of the purified protein were dot blotted onto the nitrocellulose membrane. Then the membrane was subjected to 45CaCl2 binding by overlay method as described by Maruyama et al. (1984) and Rajini et al. (2001). BSA was the negative control whereas CaBP3 of E. histolytica, EhCaBP3 was the positive control (Kumar et al., 2007). The overlay gave strong signals for both Cyto3a and EhCaBP3 whereas for BSA, there was no signal. The 45Ca signals were proportional to the amounts of the Cyto3a protein blotted on the membrane (Fig. 1E). These results unequivocally demonstrated the presence of calcium binding feature in the Cyto3a protein.
It is established that the intrinsic fluorescence of a protein is well dominated by the contribution of the Trp and Tyr residues (Lakowicz et al., 1983). The emission is highly sensitive to changes in environment of the chromophore, such as binding to a ligand or metal ion, solvent and pH change, etc. The intrinsic fluorescence measurement is more sensitive and features much larger spectral changes during conformational transitions. The interaction of ligand to a protein causes an alteration in the secondary and tertiary structures due to the conformational change in the protein (Khan et al., 2002). The primary protein sequence of Cyto3a shows aromatic amino acids (Fig. 2A) and the fluorescence spectrum of Cyto3a is due to the presence of aromatic residues. So to see if interaction of calcium would confer structural changes in the Cyto3a protein we monitored changes in fluorescence intensity at 350 nm on the successive addition of different concentrations of CaCl2 to the purified Cyto3a. The titration of apo Cyto3a protein against increasing calcium concentrations showed sharp inflection in the curve (Fig. 2B). This suggested that the intrinsic fluorescence of Cyto3a protein was quenched by calcium binding indicating a perturbation in the microenvironment around the tryptophan residues in the protein sequence. When a tryptophan residue is buried inside the protein, quenching is not expected to occur, but if the residue is on the protein surface quenching is observed. This leads to exposure of hydrophobic regions of the protein as is reported in many Ca2+ sensor proteins (Valcarce et al., 1993). The observed result suggests that the interaction of Cyto3a with calcium indeed changes the microenvironment of Trp and Tyr residues in the protein.
Fig. 2.
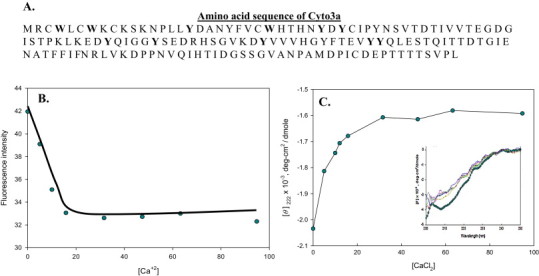
(A) The amino acid sequence of Cyto3a. (B) The intrinsic fluorescence intensity of Cyto3aat 350 nm as a function of increasing calcium concentration (μM). (C) Plot of the far-UV CD of Cyto3a at 222 nm against increasing calcium concentration (μM). The inset shows the full CD spectra at different [Ca2+].
Furthermore, the far-UV CD data can also detect large changes in the secondary structure of a protein upon its interaction with the ligand. Here we employed it as a complementary tool to understand the structural changes in the Cyto3a protein upon calcium binding. The binding of Cyto3a to Ca2+ was associated with very small change in secondary structure of Cyto3a (Fig. 2C). In the cases of calmodulin and troponin C, it has been shown that the conformational change in response to Ca2+ binding may involve reorientation of pre-existing helices with no changes in the overall content or distribution of regular secondary structures (Nelson and Chazin, 1998). The phenomenon may be caused by rearrangements in the tertiary structure (Nelson and Chazin, 1998). Results herein reported also indicate that the cytoplasmic domain of 3a protein displays some properties that are common in typical Ca2+ sensor proteins (Nelson and Chazin, 1998). The fluorescence data showed that binding of Ca2+ would promote the exposure of a hydrophobic cleft due to a conformational change, as observed in regulatory proteins triggered by calcium (James et al., 1995).
The SARS-CoV 3a protein is unique to the virus and till now, there are very few reports for potential homologues in other coronaviruses, which may explain the unexpectedly high virulence of the virus (Law et al., 2005, Wang et al., 2011). The 3a protein has multitasking behavior. So we emphasize the need of further studies on the calcium binding nature of full length 3a protein which would help in a deeper understanding of the viral pathogenesis.
Funding
This work was supported by a grant from the Department of Biotechnology (DBT), Government of India. The funding agency had no role in designing and execution of the experiments, data analysis and preparation of the manuscript.
Acknowledgements
We thank Dr. Shahid Jameel for generously providing us the lab space and reagents for carrying out some experiments. We thank Dr. Narendra Padhan for providing us the purified EhCBP3.
References
- Campbell K.P., MacLennan D.H., Jorgensen A.O. Staining of the Ca2+-binding proteins, calsequestrin, calmodulin, troponin C, and S-100, with the cationic carbocyanine dye “Stains-all”. J. Biol. Chem. 1983;258:11267–11273. [PubMed] [Google Scholar]
- Chan C.M., Tsoi H., Chan W.M., Zhai S., Wong C.O., Yao X., Chan W.Y., Tsui S.K., Chan H.Y. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charuk J.H., Pirraglia C.A., Reithmeier R.A. Interaction of ruthenium red with Ca2(+)-binding proteins. Anal. Biochem. 1990;188:123–131. doi: 10.1016/0003-2697(90)90539-l. [DOI] [PubMed] [Google Scholar]
- Ciampor F., Cmarko D., Cmarkova J., Zavodska E. Influenza virus M2 protein and haemagglutinin conformation changes during intracellular transport. Acta Virol. 1995;39:171–181. [PubMed] [Google Scholar]
- Deepa S., Mishra A.K. Fluorescence spectroscopic study of serum albumin–bromadiolone interaction: fluorimetric determination of bromadiolone. J. Pharm. Biomed. Anal. 2005;38:556–563. doi: 10.1016/j.jpba.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Drosten C., Preiser W., Gunther S., Schmitz H., Doerr H.W. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol. Med. 2003;9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart G.D., Sutherland T., Gage P.W., Cox G.B. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J. Virol. 1996;70:7108–7115. doi: 10.1128/jvi.70.10.7108-7115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt M., Law L.M., Chan H., Hobman T.C. Role of rubella virus glycoprotein domains in assembly of virus-like particles. J. Virol. 1999;73:3524–3533. doi: 10.1128/jvi.73.5.3524-3533.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Lai M.M. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield K.M., Toptygin D., Grandhige G., Kim H., Packard B.Z., Brand L. Steady-state and time-resolved fluorescence measurements for studying molecular interactions: interaction of a calcium-binding probe with proteins. Biophys. Chem. 1996;62:25–38. doi: 10.1016/s0301-4622(96)00027-0. [DOI] [PubMed] [Google Scholar]
- Ishitani M., Liu J., Halfter U., Kim C.S., Shi W., Zhu J.K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000;12:1667–1678. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Vorherr T., Carafoli E. Calmodulin-binding domains: just two faced or multi-faceted? Trends Biochem. Sci. 1995;20:38–42. doi: 10.1016/s0968-0004(00)88949-5. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., MacKinnon R.A. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., MacKinnon R.B. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- Khan M.A., Muzammil S., Musarrat J. Differential binding of tetracyclines with serum albumin and induced structural alterations in drug-bound protein. Int. J. Biol. Macromol. 2002;30:243–249. doi: 10.1016/s0141-8130(02)00038-7. [DOI] [PubMed] [Google Scholar]
- Kumar S., Padhan N., Alam N., Gourinath S. Crystal structure of calcium binding protein-1 from Entamoeba histolytica: a novel arrangement of EF hand motifs. Proteins. 2007;68:990–998. doi: 10.1002/prot.21455. [DOI] [PubMed] [Google Scholar]
- Lakowicz J.R., Maliwal B.P., Cherek H., Balter A. Rotational freedom of tryptophan residues in proteins and peptides. Biochemistry. 1983;22:1741–1752. doi: 10.1021/bi00277a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law P.T., Wong C.H., Au T.C., Chuck C.P., Kong S.K., Chan P.K., To K.F., Lo A.W., Chan J.Y., Suen Y.K., Chan H.Y., Fung K.P., Waye M.M., Sung J.J., Lo Y.M., Tsui S.K. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Mikawa T., Ebashi S. Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J. Biochem. 1984;95:511–519. doi: 10.1093/oxfordjournals.jbchem.a134633. [DOI] [PubMed] [Google Scholar]
- Nelson M.R., Chazin W.J. An interaction-based analysis of calcium-induced conformational changes in Ca2+ sensor proteins. Protein Sci. 1998;7:270–282. doi: 10.1002/pro.5560070206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco J.M., Henry T.M., O’Donnell V.K., Gregory J.B., Mason P.W. Role of nonstructural proteins 3A and 3B in host range and pathogenicity of foot-and-mouth disease virus. J. Virol. 2003;77:13017–13027. doi: 10.1128/JVI.77.24.13017-13027.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piller S.C., Ewart G.D., Premkumar A., Cox G.B., Gage P.W. Vpr protein of human immunodeficiency virus type 1 forms cation-selective channels in planar lipid bilayers. Proc. Natl. Acad. Sci. U.S.A. 1996;93:111–115. doi: 10.1073/pnas.93.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L.H., Holsinger L.J., Lamb R.A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- Plugge B., Gazzarrini S., Nelson M., Cerana R., Van Etten J.L., Derst C., DiFrancesco D., Moroni A., Thiel G. A potassium channel protein encoded by chlorella virus PBCV-1. Science. 2000;287:1641–1644. doi: 10.1126/science.287.5458.1641. [DOI] [PubMed] [Google Scholar]
- Rajini B., Shridas P., Sundari C.S., Muralidhar D., Chandani S., Thomas F., Sharma Y. Calcium binding properties of gamma-crystallin: calcium ion binds at the Greek key beta gamma-crystallin fold. J. Biol. Chem. 2001;276:38464–38471. doi: 10.1074/jbc.M102164200. [DOI] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Ruiz M.C., Cohen J., Michelangeli F. Role of Ca2+ in the replication and pathogenesis of rotavirus and other viral infections. Cell Calcium. 2000;28:137–149. doi: 10.1054/ceca.2000.0142. [DOI] [PubMed] [Google Scholar]
- Schumacher M., Adelman J.P. Ion channels: an open and shut case. Nature. 2002;417:501–502. doi: 10.1038/417501a. [DOI] [PubMed] [Google Scholar]
- Singh A.N., Gupta D., Jameel S. Bioinformatic analysis of the SARS virus X1 protein shows it to be a calcium-binding protein. Curr. Sci. 2004;86:842–844. [Google Scholar]
- Valcarce C., Selander-Sunnerhagen M., Tamlitz A.M., Drakenberg T., Bjork I., Stenflo J. Calcium affinity of the NH2-terminal epidermal growth factor-like module of factor X. Effect of the gamma-carboxyglutamic acid-containing module. J. Biol. Chem. 1993;268:26673–26678. [PubMed] [Google Scholar]
- Wang K., Xie S., Sun B. Viral proteins function as ion channels. Biochim. Biophys. Acta (BBA) Biomembr. 2011;1808:510–515. doi: 10.1016/j.bbamem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Li J., Shan Y., Yang Z., Zhao Z., Chen B., Yao Z., Dong B., Wang S., Chen J., Cong Y. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Yao Z., Wu J., Zhou Y., Shan Y., Dong B., Zhao Z., Hua P., Chen J., Cong Y. G1 phase cell cycle arrest induced by SARS-CoV 3a protein via the cyclin D3/pRb pathway. Am. J. Respir. Cell Mol. Biol. 2007;37:9–19. doi: 10.1165/rcmb.2005-0345RC. [DOI] [PubMed] [Google Scholar]
- Zhang G., Wang A., Jiang T., Guo J. Interaction of the irisflorentin with bovine serum albumin: a fluorescence quenching study. J. Mol. Struct. 2008;891:93–97. [Google Scholar]
- Zhao H., Su W., Luo Y., Ji Y., Li Z., Jiu H., Liang H., Chen B., Zhang Q. Rectification of excitation with bathochromic shift induced by intense absorption of organic ligands during emission measurement of Eu(III) complex. Spectrochim. Acta A: Mol. Biomol. Spectrosc. 2006;65:846–851. doi: 10.1016/j.saa.2006.01.018. [DOI] [PubMed] [Google Scholar]


