Abstract
The immune surveillance system protects host cells from viral infection, and viruses have evolved to escape this system for efficient proliferation in the host. Host cells produce cytokines and chemokines in response to viral infection, and among such effector molecules, type I interferons are the principal antiviral cytokines and therefore effective targets for viruses to disarm host surveillance. Porcine reproductive and respiratory syndrome virus (PRRSV) expresses proteins that circumvent the IFN response and other cellular processes, and to compensate the small coding capacity of PRRSV, these proteins are multifunctional. To date, at least four viral proteins have been identified and studied as viral antagonists of host defenses: N as a structural protein and three non-structural proteins, Nsp1 (Nsp1α and Nsp1β), Nsp2, and Nsp11. Among these, N and Nsp1 are nuclear-cytoplasmic proteins distributed in both the nucleus and cytoplasm of cells. Nsp1 and Nsp2 are viral proteases while Nsp11 is an endoribonuclease. This review describes the current understanding of the role of these proteins in modulating the host innate immune responses. Blocking against virus-mediated inhibition of the innate response may lead to the future development of effective vaccines. The understanding of viral mechanisms modulating the normal cellular processes will be a key to the design of an effective control strategy for PRRS.
Keywords: PRRSV, Interferons, Immune evasion, Immune modulation, NF-κB, CBP degradation, IRF3, PIAS, Nucleocapsid, non-structural proteins, Arterivirus
1. Introduction
1.1. Recognition of non-self and innate immunity
The existence of a virus in a host requires a balance between replication of the virus and the antiviral defense of the host. The antiviral defense of the host is armed with a variety of immune surveillance mechanisms to eliminate invading viruses, and viruses in turn have evolved to subvert these barriers. Recognition of the invading virus is initiated by innate immunity mediated by an intricate system of pattern recognition receptors (PRRs) that specifically recognize pathogen-associated molecular patterns (PAMPs). The sensing of PAMPs initiates the induction of an appropriate immune response to virus infection. Toll-like receptors (TLRs) represent an essential family of PRRs, and several TLRs may be triggered by single-stranded RNA, double-stranded RNA, and a number of viral proteins activating signal transduction. Thus, TLR signaling is determined by the repertoire of PAMPs presented to TLRs by invading viruses (for reviews, see Akira et al., 2006, Kawai and Akira, 2010).
The events that occur following TLR recognition by invading viruses include induction of interferon regulatory factor 3 (IRF3) and nuclear factor-κB (NF-κB), which then switches on the expression of proinflammatory cytokines and chemokines that include interleukin (IL)-6, IL-8, tumor necrosis factor (TNF)-α, RANTES, and type I interferons (IFN-α/β). Production of type I IFNs mediates antiviral host response through four main effector pathways; the Mx1 (myxovirus resistance 1) GTPase pathway, the dsRNA-dependent kinase PKR pathway, the 2′,5′-oligoadenylate synthetase-directed RNase L pathway, and the ISG15 (IFN-stimulated gene product of 15 kDa) uniquitin-like pathway (for review, see Katze et al., 2002). Cytotoxic T lymphocytes (CTLs) are also activated leading to a full range induction of viral-specific adaptive immune surveillance. The CTL activation is dependent especially on TLR3, -4, -7, and -9, and therefore the crosstalk between innate immunity and adaptive immunity is also triggered by TLRs.
TLRs are not the only family of PRRs that sense viral PAMPs. Retinoic acid-inducible gene I (RIG-I)-like RNA helicases (RLHs) also constitute an important family of PRRs that comprise RIG-I and melanoma differentiation-associated gene 5 (MDA-5) (Fig. 1 ) (Takeuchi and Akira, 2008). RIG-I and MDA-5 are expressed in the cytosol where no TLR is expressed, and thus constitute an alternative defense mechanism, especially for RNA viruses that replicate exclusively in the cytoplasm. RLHs possess dsRNA unwinding helicase activity that is coupled to caspase activation and recruitment domains (CARDs) and that allow interaction with another CARD-containing protein, CARD adaptor inducing IFN-β (Cardif; also known as MAVS, IPS-1, or VISA), to stimulate activation of the antiviral transcription factors NF-κB and IRF3. A key difference between RIG-I and TLRs is that RIG-I functions mainly in fibroblasts, macrophages, and conventional dendritic cells, whereas in plasmacytoid dendritic cells (pDCs), RIG-I has no role and type I IFN production is solely dependent on TLRs 7/8 and 9.
Fig. 1.
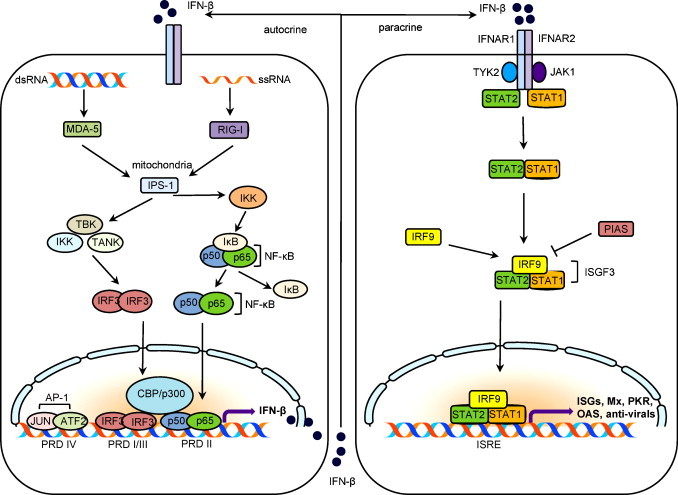
Production of type I interferon via RIG-I dependent pathway and IFN-mdediated JAK-STAT signalling pathway. For explanation, see the text (adopted from Aaronson and Horvath, 2002, Bowie and Unterholzner, 2008, Sadler and Williams, 2008).
NF-κB is another family of inducible transcription factors activated by TLRs and virus-induced RIG-I/MDA-5 pathways (Vallabhapurapu and Karin, 2009). The NF-κB transcriptional activators are comprised of five member proteins (RelA/p65, cRel, RelB, p100/p52, and p105/p50), and they share a structurally conserved Rel homology domain that contains a functional region responsible for dimerization and DNA binding. NF-κB proteins are sequestered in the cytoplasm as a complex with inhibitory IkB proteins that are activated by TLR ligands. Activating signals for NF-κB trigger phosphorylation of IkB by IkB kinase B (IKK-B), and phosphorylated IkB becomes ubiquitinated and degraded in the proteasome. As a consequence, NF-κB is released from IkB, and translocates to the nucleus to bind specific DNA sequences, which activates targeted gene expression (for a review, see Vallabhapurapu and Karin, 2009).
1.2. IFN production and signaling pathway
Among the various factors involved in type I IFN production, IRF3 plays a major role. IRF3 is constitutively expressed in most cell types and resides in the cytoplasm in its inactive form. When stimulated, IRF3 becomes phosphorylated and undergoes conformational changes, leading to dimerization and unveiling of the nuclear localization signal (NLS). Dimerized IRF3 translocates to the nucleus where it forms a complex with the transcription co-activator CREB (cAMP responsive element binding)-binding protein (CBP)/p300 (Lin et al., 1998, Dragan et al., 2007). The IRF3-CBP/p300 complex then binds to target DNA sequences, including the positive regulatory domain (PRD) I–III regions of the IFN-β promoter, and assembles basal transcription machinery and RNA polymerase II for IFN gene expression (Panne et al., 2007). Therefore, the transcriptional activity of IRF3 is entirely dependent on its association with CBP/p300 co-activators. Once IFN is expressed, it is secreted extracellularly and binds to IFN-aR (IFNAR) receptors on the plasma membrane of its own cells (autocrine) or neighbor cells (paracrine) (Fig. 1) Immediately after IFNs bind to the IFNAR2 subunit, the IFNAR1 subunit is recruited to form the receptor–ligand complex. Janus kinase 1 (JAK1) and tyrosine kinase 2 (Tyk2) then trans-phosphorylate one another, resulting in phosphorylation of the cytoplasmic tails of receptors and recruitment of STAT (signal transducers and activator of transcription) proteins. STAT1 and STAT2 are tyrosine-phosphorylated and activated to form the STAT1/STAT2/IRF9 heterotrimeric complex designated interferon-stimulated gene factor 3 (ISGF3). STAT-bound ISGF3 then translocates to the nucleus where it binds to IFN-stimulated regulatory response elements (ISRE) in the promoter regions of IFN-α/β-regulated genes and upregulates their transcription. Thus, secreted IFN triggers the IFN-signaling cascade through the JAK-mediated STAT pathway and achieves transcriptional upregulation of IFN-stimulated genes (ISGs) (Aaronson and Horvath, 2002).
1.3. Viral strategies for IFN evasion
More than 300 ISGs have been identified so far and they are mostly related to innate immunity, thus specifying the antiviral state of the host. In turn, many viruses have evolved to evade the host immune system by expressing antagonistic proteins that interfere with host signaling pathways needed for expression of Type I IFN (reviewed in Takeuchi and Akira, 2008, Koyama et al., 2008, Pichlmair and Sousa, 2007, Bowie and Unterholzner, 2008, Sadler and Williams, 2008, Randall and Goodbourn, 2008, De Clercq, 2006). Some RNA viruses express proteins which function in the cytoplasm while other proteins are localized in the nucleus to intercept the antiviral defense despite the fact that RNA viruses generally replicate in the cytoplasm. Various strategies are employed for viral evasion from IFN and some examples are as follow.
1.3.1. Inhibition of biomolecular transports
The poliovirus 3D protein, which localizes to the nucleus with 3Cpro in the form of precursor 3BCD, degrades the nuclear pore complex (NPC) structure, thereby blocking nucleo-cytoplasmic transport of host cell mRNA subsequent IFN induction (for a review, see Gustin, 2003). The VSV (vesicular stomatitis virus) M protein targets another component of NPC and changes the nuclear transport channel (Enninga et al., 2002, Glodowski et al., 2002). The picornavirus proteases (poliovirus and FMDV [foot and mouth disease virus]) and pestivirus proteases (CSFV [classical swine fever virus] and BVDV [bovine viral diarrhea virus]) cause a shut-down of host cell metabolism to interfere with the IFN response (Bauhofer et al., 2007, La Rocca et al., 2005, Ruggli et al., 2005, Weidman et al., 2003).
1.3.2. Inhibition of IRF3 activation
The hepatitis C virus NS3-4A protease specifically cleaves IKK to inactivate IRF3 (Luquin et al., 2007), and the P protein of rabies virus and G1 protein of Hantavirus prevent IRF3 phosphorylation (Brzózka et al., 2005, Prescott et al., 2007). IRF3 itself is degraded by the NPro protein CSFV and BVDV (La Rocca et al., 2005, Baigent et al., 2002, Gil et al., 2006). Recently, the SARS coronavirus Nsp1 protein has been shown to regulate IRF3-dependent innate immunity for IFN production (Wathelet et al., 2007, Devaraj et al., 2007). The NS1 protein of influenza A virus binds host RNA and then forms complexes with RIG-I that antagonize IFN induction (Min and Krug, 2006). The V proteins of paramyxoviruses bind MDA-5 and inhibit the IFN promoter activation (Andrejeva et al., 2004).
1.3.3. Inhibition of JAK-STAT pathway
Paramyxovirus V protein is of particular interest as it participates in the JAK-STAT pathway (Young et al., 2000, Rodriguez et al., 2002, Rodriguez et al., 2003, Didcock et al., 1999, Palosaari et al., 2003). The SV5V protein induces STAT1 degradation while the paramyxovirus type 2 (PIV2) V protein induces STAT2 degradation, which in turn antagonizes IFN-α and -β signal transductions, or ISGF3 (interferon stimulated gene factor 3) function. As described above, IFN-α/-β induces an antiviral state in cells as an end point of signal transduction through the JAK-STAT pathway, and therefore evasion from IFNs by direct inhibition of STAT proteins is of a considerable benefit to the survival of the virus. The V protein-induced IFN antagonism is species-specific and is linked to successful host range determination for measles virus, Nipha virus, Hendra viruses, PIV2, and Newcastle disease virus (NDV) (Palosaari et al., 2003, Rodriguez et al., 2002; for a review, see Horvath, 2004).
1.3.4. Modulation of NF-κB activity
The role of NF-κB during viral infection is more complex than that of IRF3. Some viruses activate NF-κB to prevent the cells from apoptosis (Lee et al., 2005), and some viruses block NF-κB activity to delay the innate immune response until an infection is established.
2. PRRS and host response to PRRS
Porcine reproductive and respiratory syndrome (PRRS) is an emerged and re-emerging disease in swine. PRRS was first recognized in 1987 in the U.S. and in Germany in 1990, independently (Keffaber, 1989, Benfield et al., 1992). PRRS quickly spread globally to most pig-producing countries and has become one of the most economically important diseases to the pork industry worldwide. Since its emergence, the virus has continued to evolve. A new type of PRRSV has been identified in the US (Ropp et al., 2004, Fang et al., 2007), and a highly virulent strain has appeared in China (Tian et al., 2007, Tong et al., 2007, Zhou et al., 2008) and has spread to neighboring countries (Normile, 2007, Feng et al., 2008, Kukushkin et al., 2008). Commercial vaccines are available against PRRS but their safety and efficacy are deemed unsatisfactory and the disease continues to plague the pork industry. A major obstacle in the development of a successful PRRS vaccine is an unconventional immune response of the pig to the virus. Infection of pigs with PRRSV instigates abundant production of antibodies in the infected animal (Lemke et al., 2004, Loemba et al., 1996). However, the PRRSV antibodies are mostly non-neutralizing, and neutralizing antibody response is vague and weak with a delayed appearance (Mateu and Diaz, 2008). Infection of pig with PRRSV may lead to a poor lung defense and predisposes the pig to secondary bacterial infections that may increase clinical severity. Viremia eventually ceases but the pigs become persistently infected in the tonsils and lymphoid tissues for 6 months or longer (Allende et al., 2000, Batista et al., 2002, Wills et al., 1997) and may spread the virus to naïve animals, which may serve as an important means of transmission to susceptible herds (Wills et al., 2003, Christopher-Hennings et al., 1995, Albina et al., 1994, Lamontagne et al., 2003, Bierk et al., 2001).
The effects of cytokines on PRRSV have been extensively studied. A transient T cell response is detected at one month and lasts for two to three additional months post-infection (Bautista and Molitor, 1999, López Fuertes et al., 1999). During this period, cytokine response is mainly composed of IFN-γ secreted largely from CD4+ or CD8+ T cells (Diaz et al., 2005, Diaz et al., 2006, Meier et al., 2004) and to a lesser extent for IL-2 (Meier et al., 2003, Meier et al., 2004, Xiao et al., 2004). TNF-α and IL-12 are also detectable in alveolar macrophages and dendritic cells, and the IL-10 response is unusually strong and high. During PRRSV infection, IL-10 expression increases in PBMC and also in the bronchoalveolar lavage fluids in infected pigs (Aasted et al., 2002, Suradhat and Thanawongnuwech, 2003, Suradhat et al., 2003). IL-10 levels seem to inversely correlate with IFN-γ response, indicating an involvement of T-cell, IL-10, and IFN-γ responses against PRRSV. PRRSV seems to be highly susceptible to both IFN-α and -β (Albina et al., 1998, Overend et al., 2007), but PRRSV-affected pigs do not elicit adequate induction of IFNs. IFN-α is not detected in the lungs of pigs where PRRSV is actively replicating. Both IFN-α and INF-β production are low and clearly down-regulated in pulmonary alveolar macrophages (PAMs) and in PRRSV-permissive monkey kidney cells (MARC-145) (Albina et al., 1998, Buddaert et al., 1998, Van Reeth et al., 1999, Royaee et al., 2004). Accumulating data suggests that PRRSV may inhibit IFN-α response (Lee et al., 2004, Miller et al., 2004), and that this inhibition is RIG-I mediated (Luo et al., 2008). Such inadequate innate responses may result in the delayed clearance and persistence of virus. Impairment of IFN-α production by PRRSV would also affect the development of an effective T helper type 1 (Th1) response and lead to a weakly induced cellular response and viral persistence in infected pigs, which would be of significant benefit to the virus. Viral persistence is generally related to the ability of the virus to evade host antiviral defense combined with a viral capacity to establish persistent infection within cells. Virus-infected animals activate both innate and adaptive immune responses, and when antiviral defense is unsuccessful, viruses may still persist. Different PRRSV isolates appear to have different abilities to induce or inhibit IFN-α (Lee et al., 2004), and this difference may be attributed to the genetic diversity of the virus. The mechanism for virulence and persistence of PRRSV is yet unclear, and the molecular basis for IFN suppression by PRRSV has not been studied until recently. It is postulated that at least one of the mechanisms for viral persistence may be related to the modulation of type I IFN production by PRRSV. This series of interactive processes between virus and cell trigger differential expression of cellular genes, which has been of interest in understanding how altered gene expression plays a role during PRRSV infection. PRRSV seems to possess an ability to disarm the host antiviral defense. As a consequence, the virus readily survives in the pig without being eliminated and is spread to naïve animals. Counter-acting the ability of PRRSV to disarm the host defense would be the key to controlling PRRSV infection and blocking transmission.
3. PRRSV proteins and their role in host response modulation
PRRSV is a porcine arterivirus belonging to the family Arteriviridae in the order Nidovirales (http://www.ictvonline.org/virusTaxonomy.asp?version=2009). PRRSVs in North America and Europe share only 55–80% of sequence identity in different viral genes and form two distinct genotypes (for a review, see Music and Gagnon, 2010). The viral genome is a positive-sense RNA of 15 kb in size and contains 9 ORFs; ORF1a, ORF1b, ORF2a, ORF2b, and ORF3 through ORF7. The viral genome is translated to produce two polyproteins from ORF1a and ORF1b, and ORF1a alone can produce PP1a of 2503 amino acids. ORF1b does not contain an initiating codon and therefore is expressed as PP1a/1b in the form of a fusion protein of 3960 amino acids with PP1a by ribosomal frame-shifting (den Boon et al., 1991, Wootton et al., 2000). PP1a and PP1a/1b are co-translationally processed to 14 proteolytic cleavage products. These proteins are only found in PRRSV-infected cells and thus are non-structural proteins (Nsps) designated Nsp1 through Nsp12 in order from the N-terminus (van Dinten et al., 1996, Snijder et al., 1992, Snijder et al., 1994, Snijder et al., 1995, Snijder et al., 1996, Wassenaar et al., 1997). Nsp1 and Nsp7 are both internally cleaved and produce Nsp1α and Nsp1β, and Nsp7α and Nsp7β, respectively. Similar to those of coronaviruses and equine arteritis virus (EAV), PRRSV Nsp's function in genome replication and mRNA synthesis. PP1a contains three protease motifs: papain-like cysteine protease (PCP) in Nsp1, poliovirus 3C-like cysteine protease (CP) in Nsp2, and serine protease (SP) in Nsp4. The polyprotein processing for PRRSV is similar to that of EAV such that PCP generates Nsp1α and Nsp1β, CP cleaves off Nsp2, and SP generates Nsp3 through Nsp12.
PRRSV structural proteins are encoded in the 3′-most 3 kb of the viral genome. This region contains ORF2a, ORF2b, and ORF3 through ORF7 and codes for seven structural proteins: glycoprotein 2 (GP2), small envelope (E), GP3, GP4, GP5, membrane (M), and nucleocapsid (N) proteins, respectively (Meulenberg et al., 1993, Wootton et al., 2000). The E protein is a small, hydrophobic, myristoylated membrane protein of 73 amino acids (Du et al., 2010). E is likely an ion-channel protein of the virus involved in the uncoating process during virus entry (Lee and Yoo, 2006). GP5 and M form a heterodimer with unknown function. GP2, GP3, and GP4 are minor components of the virion and form a heterotrimer (Wieringa et al., 2003) that may function as a viral ligand that recognizes CD163 as the cellular receptor to initiate infection (Das et al., 2010). N protein binds to viral genomic RNA and constitutes the viral capsid.
3.1. Nuclear-cytoplasmic proteins of PRRSV
As with many other RNA viruses, the entire life cycle of PRRSV occurs in the cytoplasm of infected cells, and thus viral-specific proteins are predominantly localized in the cytoplasm of infected cells, especially in the perinuclear region, endoplasmic reticulum (ER) and Golgi, and internal membranes. An unusual feature of PRRSV is that the viral N protein localizes to the nucleus and nucleolus in addition to its normal cytoplasmic distribution (Rowland et al., 1999). Since the N protein is a major structural component essential for viral assembly, and because PRRSV assembly and maturation takes place in the ER and Golgi of the cell, the N protein has been postulated to play dual roles - a structural role in the cytoplasm and a non-structural role in the nucleus. Recently, the nonstructural protein Nsp1 has been identified as an additional nuclear protein of PRRSV (Yoo et al., 2009; Fig. 3 ). Nsp1 is the first viral protein synthesized during infection, and both N and Nsp1 appear to modulate normal host cell process and immune surveillance.
Fig. 3.
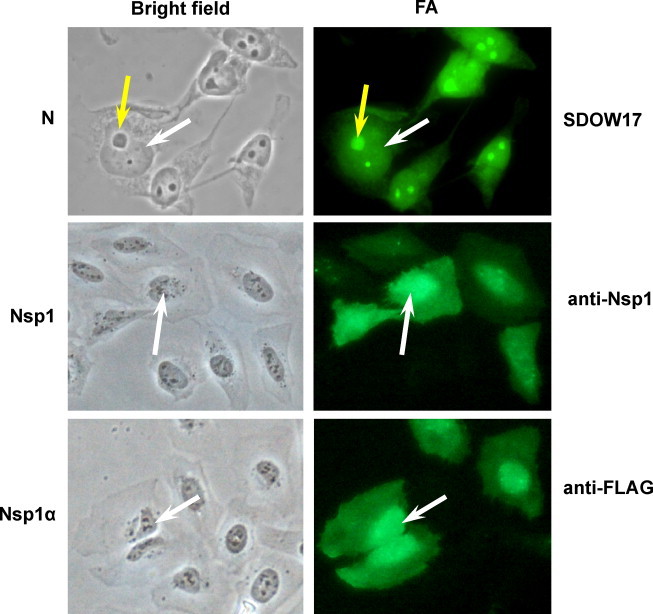
Nuclear cytoplasmic distribution of N, Nsp1, and Nsp1α subunit of Nsp1. HeLa cells were gene-transfected and stained at 24 h with N-specific monoclonal antibody SDOW17 (top panel), or with anti-FLAG antibody for Nsp1 (middle panel) and Nsp1α proteins (bottom panel). Nuclear distribution is indicated by white or black arrows. The N protein in particular is distributed in the nucleolus (yellow arrows), in addition to the nucleus and cytoplasm.
3.1.1. Nucleocapsid (N) protein
The N protein is a small basic protein of 123 amino acids. As for many other RNA viruses, N is the most abundant virion component and the most immunogenic protein in PRRSV-infected pigs. The PRRSV N protein is a serine-phosphoprotein with unknown function (Wootton et al., 2002). As the sole protein component of the viral capsid, N associates with itself by both covalent and non-covalent interactions, providing the basis for viral capsid assembly. The covalently-linked N-N homodimerization is formed in the lumen of the ER and the Golgi complex through disulfide linkages via a cysteine residue at position 23 (Wootton and Yoo, 2003). Cysteine 23 of N is essential for virus replication and infectivity, indicating the importance of N-N dimerization for virion assembly (Lee et al., 2005).
3.1.1.1. Nuclear localization of N and interaction with fibrillarin
Since N protein nuclear localization has been reported for PRRSV, N proteins of EAV (Tijms et al., 2002), lactate dehydrogenase elevating virus of mouse (LDV; Mohammadi et al., 2009), simian hemorrhagic fever virus (SHFV; Mohammadi, 2010), and coronaviruses including avian infectious bronchitis virus, transmissible gastroenteritis virus, and murine hepatitis virus (Hiscox et al., 2001, Wurm et al., 2001) have been studied for nuclear localization which seems to be a common feature in nidoviruses. A functional motif for the nuclear localization signal (NLS) in PRRSV N is 41PGKKNKK which interacts with the nuclear transporters importin-α and importin-β (Rowland et al., 2003). Therefore, the N protein seems to localize in the nucleus through the NLS-dependent and importin α/β-mediated nuclear transport pathway. Once in the nucleus, the N protein co-localizes and specifically interacts with a set of cellular proteins. Cellular proteins that have been identified as interacting partners of N include fibrillarin (Yoo et al., 2003), I-mfa domain containing protein (HIC) (Song et al., 2009), protein inhibitor of activated STAT1 (PIAS1), and other proteins including nucleolin, B23, poly-A binding protein (Song, 2003), importin-α and -β, and exportin (Rowland et al., 2003). The nuclear import of N protein is faster than its nuclear export, and thus the N protein is apparently localized in greater amounts in the nucleolus (You et al., 2008).
The nucleolus, where PRRSV N localizes, is a subcellular structure involved in cellular processes such as ribosomal RNA synthesis, modulation of cell growth, and response to cell stress (Sirri et al., 2008). Fibrillarin is a major cellular protein in the nucleolus, and thus was the first protein studied for its interaction with PRRSV N. Fibrillarin is a component of the small nucleolar ribonucleoprotein (snoRNP) complex that participates in the precursor ribosomal RNA processing and cell cycle progression (Reichow et al., 2007). The fibrillarin and N protein complex appears to be restricted to the dense fibrillar component and nearby regions of the nucleolus and associated with box C/D snoRNAs to form snoRNP complexes (Yoo et al., 2003). The small nucleolar RNAs contain an antisense element complementary to rRNAs, and the antisense snoRNAs function as the guide for snoRNPs in the dense fibrillar component. The snoRNPs then participate in the site-specific 2′-O-ribose methylation of the pre-rRNA and the precise pre-rRNA cleavage processing for assembly of ribosomal subunits (Tollervery and Kiss, 1997). Since both fibrillarin and N protein function as RNA binding proteins, co-localization to the same site may reflect a common RNA binding function, and in fact, the N protein and fibrillarin interaction requires RNA. Since replication of PRRSV genomic RNA is restricted to the cytoplasm and because fibrillarin exists mainly in the nucleus and nucleolus, it is likely that precursor rRNA, which is abundantly present in the dense fibrillar component where the N and fibrillarin co-localization is detected, may be the RNA species participating in this interaction in the nucleolus. Indeed, the PRRSV N protein has been shown to be able to bind to both 28S and 18S rRNAs (Yoo et al., 2003). Therefore, it is possible that N and fibrillarin may compete for the same rRNA substrate in the dense fibrillar component of the nucleolus, which may result in the modification of fibrillarin function in virus-infected cells. Alternatively, the binding of nucleolar RNAs in the fibrillar region may direct a conformational change of N sufficient to facilitate a stable interaction with other nucleolar proteins, such as fibrillarin, and subsequently allow the N protein to counter-regulate the fibrillarin function such as site-specific methylation, leading to uncontrolled cleavages of pre-rRNA. This hypothesis is supported by the observation that the co-localization of N and fibrillarin does not occur in the Cajal body (Yoo et al., 2003), where unlike the nucleolus, rRNA is absent. Additional supporting evidence includes the cleavage of 28S rRNA in PRRSV-infected MARC-145 cells (unpublished data). These studies implicate the involvement of N in the ribosome biogenesis and cell cycle progression. Hijacking the nucleolar function and cell cycle arrest are considerable benefits to the virus since the available pool of cellular machineries can be maximally utilized towards the production of progeny virus at the early stage of infection.
3.1.1.2. Interaction of N with HIC
Using N as bait in yeast-2 hybrid screening, an additional cellular protein interacting with N has been identified, I-mfa (inhibitor of MyoD family) domain-containing protein (HIC) (Song et al., 2009). HIC is a transcription factor that has been identified as a binding partner of CD4 in humans (Thébault et al., 2000), and it differentially regulates transcriptional activities of human immunodeficiency virus (HIV) and human T-cell leukemia virus (HTLV) long terminal repeats (LTR). I-mfa is a myogenic repressor protein and is an inhibitor of MyoD family (Chen et al., 1996). MyoD is a family of transcription factors regulating somitogenesis. I-mfa inhibits MyoD-mediated muscle differentiation (Kraut et al., 1998). The physical interaction between the cysteine-rich domain at the C-terminus of I-mfa and the myogenic basic helix–loop–helix protein contribute to the inhibitory effect of I-mfa. The porcine HIC contains a cysteine-rich domain analogous to that of I-mfa and contains a sequence that resembles a nucleolar localization signal. HIC is a zinc-binding protein, and confocal microscopy shows co-localization of PRRSV N with the HIC-p40 isomer in the nucleus and nucleolus, and in the cytoplasm with the HIC-p32 isomer, which is an N-terminal truncation of HIC-p40. Porcine HIC is universally expressed in tissues of pig including alveolar macrophages. The interaction of viral capsid protein with the cellular transcription factor implicates a possible regulation of host cell gene expression by N during PRRSV infection. The I-mfa signature domain is found in porcine HIC. The N protein binds HIC-p32 effectively, and co-expression of N with HIC-p32 or HIC-p40 does not change their subcellular distributions, suggesting that N binding to HIC likely results in the functional modification of HIC in association with the I-mfa signature domain. Although the biological function of HIC is largely unknown, recent studies show that HIC interacts with cyclin T1 in cells and stimulates positive transcription elongation factor (P-TEFb)-dependent transcription. Cyclin T1 is a component of the P-TEFb complex, which is known to participate in the control of cell cycle progression. Since PRRSV N protein can undergo physical interaction with fibrillarin and co-localizes with fibrillarin in the nucleolus (Yoo et al., 2003), it is possible that the N-HIC interaction in conjunction with N-fibrillarin interaction participates in the regulation of cell cycle progression. During the expression of N, cells accumulate at the G2/M phase (Mohammadi, 2010), indicating that the N protein is responsible for delay of cell cycle progression. The arrest of cell cycle progression is beneficial for the virus to subvert host cell function and increases the efficiency of virus production (Hiscox, 2007, Emmott and Hiscox, 2009).
3.1.1.3. Upregulation of IL-10 by N
Accumulating evidence shows that PRRSV strongly induces interleukin (IL)-10 production in pigs during an early phase of infection (Suradhat and Thanawongnuwech, 2003, Suradhat et al., 2003, Thanawongnuwech and Thacker, 2003). IL-10 induction has been observed in both North American and European types of PRRSV. IL-10 is a potent cytokine that functions to suppress innate and adaptive immune responses. Because protection against viral infection relies on the induction of viral-specific cell-mediated immunity, interference and delay of the crosstalk between antigen presenting cells and T cells are significant benefits for the virus to survive in the host. The induction of IL-10 and other inhibitory factors by PRRSV is mediated by interaction between PRRSV proteins and macrophages/dendritic cells as these cells are susceptible targets for PRRSV infection and play important roles in the induction of anti-viral innate and adaptive immunity. We have sought to identify a viral protein responsible for IL-10 induction using individual PRRSV genes, and have recently obtained convincing evidence that the N protein plays a significant role (unpublished data). The PRRSV N protein induces IL-10 production in peripheral blood mononuclear cells (PBMCs) of pig and in cultivated PAMs as the primary target cells of PRRSV (Wongyanin et al., 2009). The expression of PRRSV N results in significant induction of IL-10 expression in PAMs. PRRSV N stimulated IL-10 production is similar to the observation for human hepatitis C virus infection (Barrett et al., 2008). For HCV, the core protein is linked to the increase of IL10 production in PBMCs. A study is in progress in my laboratory to elicit the mechanism of IL-10 induction by PRRSV N.
3.1.1.4. In vivo role of N in PRRSV-infected pigs
The in vivo role of N protein nuclear localization in pigs has been examined using an NLS-null mutant PRRSV (Lee et al., 2006, Pei et al., 2008). Two lysine residues at the NLS motif of 41PGKKNKK were substituted to glycine, and the modified NLS to 41PGGGNKK restricted the N protein to the cytoplasm. Then using an infectious cDNA clone, the NLS-null mutation was introduced into the viral genome, and an NLS-null mutant virus was rescued, showing that the nuclear localization of N is not essential for PRRSV infectivity. When the host response to the NLS-null virus was examined in the piglet model, NLS-null virus-infected pigs showed milder clinical signs with a significantly shorter mean duration of viremia compared to wild-type PRRSV-infected pigs, and developed higher titers of neutralizing antibodies. Mutations occurred at the NLS locus in one pig during viremia, and both wild-type and NLS-null viruses persisted in the tonsils. The NLS-null virus persisting in the tonsils was found to be mutated in the NLS locus. No other mutation was found in the N gene. All types of reversions that occurred during viremia and persistence were able to translocate the mutated N proteins to the nucleus, indicating strong selection pressure for reversion at the NLS locus of the N protein in vivo. Reversions from NLS-null to functional NLS in the tonsils suggest a possible correlation of viral persistence with N protein nuclear localization. To prevent the reversion to functional NLS, a series of small deletions were additionally made in NLS of N, and the resulting mutant viruses were tested in pigs (Pei et al., 2008). Similar results were obtained. These studies show that N protein nuclear localization is non-essential for PRRSV multiplication but does play an important role in viral attenuation and in pathogenesis in pigs.
3.1.2. Nsp1 as a modulator for IFN response
3.1.2.1. Processing and structure of Nsp1
Nsp1 is the first viral protein synthesized during PRRSV infection. Unlike EAV Nsp1 that has a single proteinase motif, two proteinase motifs are found for PRRSV Nsp1. PRRSV Nsp1 is a 382-amino acid protein and contains two papain-like cysteine proteinase (PCP) domains, designated PCPα and PCPβ. Active sites for PCPα and PCPβ have been mapped to C76-H146 and C271-H340, respectively (den Boon et al., 1995). The protease activity of PCPα is responsible for generation of Nsp1α, whereas the PCPβ activity generates Nsp1β by cleaving Y382-A383 from the nascent polypeptide chain of the PP1a. After cleavage of Nsp1β from Nsp2, no trans-cleavage protease activity is observed for Nsp1α and Nsp1β and therefore the enzymatic activity of PCPα and PCPβ seems to be no longer functional, which is probably due to conformational changes after cleavage. The PCPα activity is required for subgenomic mRNA synthesis but is not involved in genomic RNA replication (Kroese et al., 2008). The internal cleavage of Nsp1 to generate Nsp1α and Nsp1β was initially predicted to occur at Q166-R167, and this prediction had long been the structural basis for two subunits: 166 amino acids for Nsp1α and 216 amino acids for Nsp1β (den Boon et al., 1995). A zinc finger motif (ZF1) of C8–C10–C25–C28 is found at the N-terminal region of Nsp1α, and this motif along with PCPα activity is required for transactivation of subgenomic RNA synthesis (Tijms and Snijder, 2003, Tijms et al., 2001, Tijms et al., 2007). Recently, an X-ray crystallographic study was conducted for Nsp1α and mapped the precise internal cleavage site to M180/A181, yielding Nsp1α subunit of 180 amino acids and Nsp1β subunit of 202 amino acids (Sun et al., 2009b) (Fig. 2B). In parallel with the crystallographic study, Chen et al. (2010a) has also determined by peptide sequencing the precise internal cleavage site of Nsp1 to M180/A181. These findings overturn the previous prediction for Nsp1 cleavage and show that the naturally cleaved authentic Nsp1α subunit is 14 amino acids extended at the C-terminus, and therefore Nsp1β is 14 amino acids shorter at its N-terminus when compared to the previous prediction. Most importantly, the presence of a second zinc-finger (ZF2) configuration of C70–C76–H146–M180 has been identified in the C-terminal half of Nsp1α and shown to hold an additional Zn++ ion in addition to the ZF1 motif in the N-terminal region. The ZF2 configuration includes M180 which is the terminal residue of Nsp1α, suggesting that Nsp1α may contain a previously unidentified biological function that may be associated with the ZF2 structure.
Fig. 2.
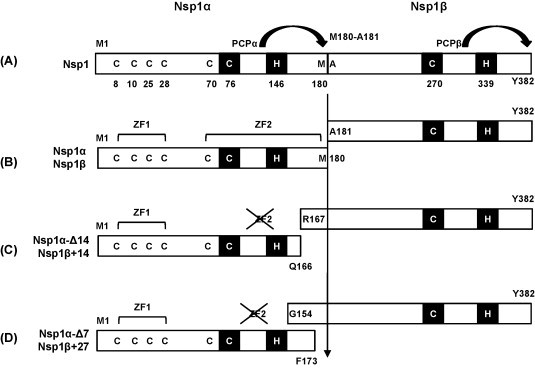
Processing of PRRSV Nsp1 protein and primary structure of Nsp1α and Nsp1β subunits of Nsp1. Numbers indicate amino acid positions. ZF1 represents the zinc-finger motif composed of C8–C10–C25–C28 in the N-terminal region, and ZF2 represents the second zinc-finger motif of C70–C76–C146–C180 in the C-terminal region of Nsp1α identified by X-ray cystallography (Sun et al., 2009b). Nsp1 is autocleaved internally at M180-A181 by PCPα to release Nsp1α, and Nsp1β is released from Nsp2 (not shown here) by PCPβ (arrows). PCPα and PCPβ motifs are indicated by black areas. C76 and H46 participate in both PCPα activity and ZF2 formation. Panel (B) represents the authentic structures of the Nsp1α and Nsp1β subunits used in our study (Sun et al., 2009b, Yoo et al., 2009). Panel (C) represents the structure of two subunits based on the previous prediction (den Boon et al., 1995), and panel (D) represents the constructs used in the study by Beura et al. (2010). ZF2 is destroyed in the Nsp1α constructs in (C) and (D) due to the loss of M180.
3.1.2.2. Downregulation of type I IFN by Nsp1
3.1.2.2.1. Nsp1
The Nsp1 protein of EAV has been reported to localize in the nucleus (Tijms et al., 2002), and recently, PRRSV Nsp1 has also been observed in the nucleus in addition to its cytoplasmic distribution (Fig. 3). It is noteworthy that unlike N which contains a distinct NLS motif for its nuclear localization, no known NLS motif is present in either of the Nsp1 cleavage products, suggesting that Nsp1 nuclear localization is not importin-dependent but that an intermediary is involved. The nuclear localization of PRRSV Nsp1 has prompted us to hypothesize that Nsp1 may play an accessory role in the nucleus of PRRSV-infected cells. Recent studies from my laboratory and others show that PRRSV Nsp1 modulates type I IFN response in cultured cells (Beura et al., 2010, Chen et al., 2010a, Kim et al., 2010). Nsp1 appears to block dsRNA-induced IRF3 and IFN promoter activities, but IRF3 phosphorylation and its nuclear translocation occur normally in the presence of Nsp1. Degradation of IRF3 was initially hypothesized as the mechanism for IFN down-regulation by PRRSV. It is striking however, that PRRSV Nsp1 degrades the CREB (cyclic AMP response element binding)-binding protein (CBP) in the nucleus and thus down-regulates the recruitment of CBP by IRF3 in the nucleus (Kim et al., 2010). For full activation, IRF3 requires phosphorylation and dimerization, and when in the nucleus recruits co-activator CBP/p300, which directs the complex to bind IRF3 responsive elements for transcription initiation (Yang et al., 2004, Suhara et al., 2002). By degrading CBP, Nsp1 seems to block the full activation of IRF3 and subsequently inhibits the assembly of the IFN enhanceosome, leading to the inhibition of IFN production. IRF3 is stable in the presence of Nsp1, and CBP degradation is proteasome-dependent, indicating that CBP degradation is not due to Nsp1 protease activity but that an intermediary is involved. CBP degradation may be induced via polyubiquitinylation.
CBP possesses histone acetyltransferase activity and dissociates histones for chromatin relaxation in the DNA promoter region. The CBP/p300 co-activators function in concert with a variety of transcription factors including c-Myc, p53, STATs, NF-κB, PIAS1, and the IRF family (Barlev et al., 2001, Lin et al., 2000, Ma et al., 2005, Oelgeschläger et al., 1996, Yin et al., 2005, Zhong et al., 2002). Therefore, CBP acts as an important regulator for many cellular processes. Since PRRSV Nsp1 degrades CBP, a question arises whether PRRSV Nsp1 will interfere with general cell transcription activities. The Thogoto virus ML protein, which is a homolog of the influenza virus NS1 protein, inhibits association of IRF3 with CBP and also interacts with RNA polymerase II transcription factor IIB (TFIIB; Jennings et al., 2005, Vogt et al., 2008). However, the ML protein–TFIIB interaction has surprisingly little effects on host cell gene expression in general, but shows a strong negative impact on the IRF3-regulated and NF-κB-regulated promoters. Thus, it is conceivable that PRRSV Nsp1-mediated CBP degradation may also be negligible for general cellular transcription but have a more specific and potent role for IRF3-regulated IFN induction. CBP degradation is a novel strategy for viral evasion from the host response, and PRRSV Nsp1 may form a new class of viral antagonists for IFN modulation. For EAV, Nsp1 is interactive with p100, a transcription co-activator that also interacts with regulatory proteins of other viruses (Tijms and Snijder, 2003). The significance of EAV Nsp1 binding to p100 has not yet been explored and remains to be determined.
3.1.2.2.2. Nsp1α and Nsp1β
Since PRRSV Nsp1 is autocleaved into two subunits, we have sought to determine the role of the individual subunits for IFN regulation. Two constructs were made for Nsp1α and Nsp1β according to the authentic cleavage of Nsp1 and each subunit was FLAG-tagged for detection. It appears that Nsp1α contains potent activity for IFN down-regulation even though Nsp1β also harbors a moderate level of regulatory activity (Yoo et al., 2009). Recent data from my laboratory reveals that Nsp1α has the ability to suppress NF-κB activation when stimulated with dsRNA or TNF-α. Nsp1α substantially suppressed NF-κB activation, and the suppressive activity was RIG-I dependent. Since IFNβ production requires the coordinated activation of transcription factors to result in the formation of a transcription competent enhanceosome, we have also examined whether NF-κB suppression by Nsp1α is associated with the suppression of IFN-β production. Besides IRF3, which binds at PRD I/III, the IFNβ promoter region also contains PRD II for NF-κB binding and PRD IV for AP-1 binding, and thus NF-κB binding for PRD II has specifically been examined. It appears that Nsp1α indeed inhibited the binding of NF-κB to the PRD II region of IFN promoters, and this inhibition resulted from the blocking of NF-κB nuclear translocation from the cytoplasm. The C-terminal extension of fourteen amino acids in the Nsp1α subunit was critical in maintaining the biological activity of Nsp1α for IFN-β and NF-κB, suggesting again that the newly identified ZF2 in Nsp1α may participate in modulating host response. Nsp1α inhibited IκB phosphorylation and as a consequence NF-κB translocation to the nucleus was blocked, and subsequently NF-κB stimulated gene expression was inhibited. Since Nsp1 degrades CBP in the nucleus (Kim et al., 2010) and blocks NF-κB activation in the cytoplasm, and because Nsp1 is distributed to both the nucleus and cytoplasm of the cell, it is conceivable that the nuclear form of Nsp1α is responsible for CBP degradation in the nucleus and the cytoplasmic form of Nsp1α participates in NF-κB inactivation in the cytoplasm, which both lead to a potent suppression of IFN-β production. Thus PRRSV Nsp1 may be considered a multifunctional protein modulating the IFN surveillance system of the host.
In our study, neither Nsp1α nor Nsp1β blocked the nuclear localization of IRF3 (Yoo et al., 2009), and this finding is contradictory to a previous report (Beura et al., 2010). Beura reported that IRF3 nuclear localization was inhibited by Nsp1β, and as a consequence, the IFNβ promoter activity was down-regulated in the presence of Nsp1β. In their study however, both Nsp1α and Nsp1β constructs were modified forms from the authentic cleavage products, such that Nsp1α was shorter by 7 residues at the C-terminus and therefore ZF2 was destroyed, while their Nsp1β had 26 additional residues at the N-terminus, when compared to the authentic subunits (Fig. 2D). Such unusual features of their constructs may have attributed to the contradictory observations compared to our data. Pestivirus classical swine fever virus (CSFV) and bovine viral diarrhea virus (BVDV) Npro protein down-regulates IFN production via IRF3 degradation (La Rocca et al., 2005, Bauhofer et al., 2007). For BVDV, both cytopathic (cp) and non-cytopathic (ncp) biotypes can cause acute infections, but only ncpBVDV establishes a persistent infection in infected bovine fetus, and subsequently causes mucosal disease in the calf. NcpBVDV inhibits IFN-β production through binding of Npro to IRF3 (Baigent et al., 2002, Chen et al., 2007), and Npro also contains a zinc-binding TRASH motif (Ettema et al., 2003). This zinc-finger motif is essential to IRF3 degradation and IFN regulation by BVDV (Szymanski et al., 2009). PRRSV Nsp1α contains two zinc finger motifs, ZF1 and ZF2, and a study is in progress using mutant PRRSVs generated from an infectious cDNA clone to examine whether any of these motifs in Nsp1α also regulates CBP degradation, NF-κB suppression, and/or IFN induction. Such studies will warrant the functional contribution of the newly identified zinc-finger structure in Nsp1α to biological significance.
The downregulation of NF-κB by Nsp1α is striking. Lee and Kleiboeker (2005) previously reported the activation of the NF-κB pathway by PRRSV in MARC-145 cells and suggested the generation of reactive oxygen species (ROS) as the cause for NF-κB activation. This report initially led us to exclude the possibility of NF-κB downregulation by Nsp1. Further studies in my laboratory however found that PRRSV Nsp1 and Nsp1α possessed the ability to downregulate NF-κB in both MARC-145 and HeLa cells. The NF-κB response is generally instant upon stimulation, and NF-κB downregulation by PRRSV Nsp1α is observed within minutes upon stimulation. The previous report by Lee and Kleiboeker (2005) examined NF-κB activation at one and two days post-infection, and it is possible that the timing of determination may have caused the differences between our results. Various confirmatory experiments are underway.
3.1.2.3. Interaction of Nsp1 with PIAS1
By yeast 2-hybrid screening, PRRSV Nsp1 has also been found to interact with the protein inhibitor of activated STAT1 (PIAS1) (unpublished data). PIAS1 is a member of the PIAS protein family known to modulate the activity of many transcription factors including STATs and NF-κB (Shuai and Liu, 2005). PIAS1 is a negative regulator for STAT1 in the IFN-mediated JAK-STAT pathway and thus regulates the IFN response (Starr and Hilton, 1999, Coccia et al., 2002). The specific binding of PIAS with Nsp1 has been confirmed independently in another laboratory (personal communication, Dr. H.C. Liu, North Carolina State University, Raleigh, NC). This finding supports a previous report that Nsp1 also plays a role in the JAK-STAT IFN signaling pathway (Chen et al., 2010a). In that study, PRRSV Nsp1 was shown to inhibit ISRE (IFN-stimulated regulatory response element) promoter activity and also inhibited STAT1 nuclear translocation. The PIAS1 protein also possesses SUMO (small ubiquitin-like modifier) E3 ligase activity. Many proteins are post-translationally modified by SUMO, known as sumoylation (Wilkinson and Henley, 2010). Sumoylation occurs through a unique pathway but analogous to ubiquitinylation during proteosome-dependent protein degradation, and for this process, a SUMO molecule is conjugated to the lysine residue of the target protein within the consensus sequence of ψK×E (ψ represents a hydrophobic amino acid) by E3 ligase. Since PIAS1 may function as an E3 ligase required for ubiquitinylation of a protein, Nsp1 may bring CBP to PIAS1, facilitating the ubiquitinylation of CBP and subsequent degradation. Alternatively, it is possible that the binding of Nsp1 to PIAS1 triggers sumoylation of Nsp1 and facilitates Nsp1 transport to the nucleus.
3.2. Cytoplasmic proteins of PRRSV and their modulatory role for innate immunity
3.2.1. Immune modulation by Nsp11
The core enzymes required for PRRSV RNA synthesis include RNA-dependent RNA polymerase (RdRp) and RNA helicase in Nsp9 and Nsp10, respectively, and Nsp11 is a recently discovered additional viral enzyme. Nsp11 contains endoribonuclease activity and is highly conserved in nidoviruses. The domain for this enzyme activity is unique and designated ‘NendoU’. A distant relationship has been identified between NendoU and the XendoU family, an endoribonuclease derived from Xenopus laevis. For arteriviruses, the NendoU domain is located in the C-terminal region of Nsp11 (Posthuma et al., 2006), and for coronaviruses, it is mapped to Nsp15 and contains uridylate-specific cleavage activity of RNA (Bhardwaj et al., 2004, Ivanov et al., 2004). Coronavirus Nsp15 forms hexamers to achieve optimal enzymatic activity (Xu et al., 2006). The role of NendoU in viral RNA synthesis has been demonstrated for EAV and SARS-CoV, and partially for PRRSV (Nedialkova et al., 2009). NendoU is able to process RNA independently from Mn2+ with the preference of uridylate using a cleavage mechanism resembling that of RNase A. At least one other viral protein containing RNase activity has been shown to inhibit type I IFN induction. Pestivirus BVDV Erns is a glycoprotein found on the surface of the viral membrane, and the Erns protein contains exoribonuclease activity that targets viral RNA. Cleavage of extracellular viral RNA prevents activation of the RNA-binding receptor TLR3, blocking the dsRNA-mediated signaling pathways (Magkcuras et al., 2008), and thus BVDV Erns acts as an inhibitor of dsRNA-induced IFN response of cells.
In our laboratory, PRRSV Nsp11 has been examined for its role for IFNβ production (Sun et al., 2009a; unpublished data). In Nsp11 expressing cells, the IFNβ promoter and IRF3 reporter activities were significantly suppressed, and the inhibitory activity was mediated through the RIG-I signaling pathway. The Nsp11-mediated IFN inhibition was triggered by either dsRNA, a constitutively active form of RIG-I and IPS-1, or IRF3 as an inducer. Subsequent experiments showed that PRRSV Nsp11 functions to block IRF3 activation. Nsp11 inhibited IRF3 phosphorylation and thus blocked the nuclear translocation of IRF3 in stimulated cells. It is hypothesized that the NendoU activity of Nsp11 is associated with the inhibition of IRF3 activation. The amino acids at positions H3735, H3750, and H3779 of PP1a/1b are catalytic sites for PRRSV NendoU activity, and mutation of these amino acids to H3735A, H3750A, and H3779A resulted in the loss of IFN inhibitory function of Nsp11.
Since RNA binds to pattern recognition receptors such as PKR, RIG-I/MAD5, TLR-3, and TLR7, and because Nsp11 is an RNase, it is possible that Nsp11 cleaves mRNAs non-specifically leading to suppression of IFN response. We have only examined the IRF3-mediated IFN production pathway, but Nsp11 may also function in other signaling branches in the production pathway and the JAK-STAT signaling pathway. A recent report indicates that PRRSV Nsp11 is able to reduce NF-κB promoter activation (Beura et al., 2010). Although this report did not specify whether the reduction of NF-κB promoter activity was associated with suppression of type I IFN production or other cellular processes, it is in support of our data that Nsp11 functions in IFN down-regulation. NendoU activity in Nsp11 may have multiple functions in the modulation of host cell response.
We have constructed a series of NendoU mutant PRRSVs such that the association of NendoU activity with IFN response may be determined using infectious viruses (Sun et al., 2009a, unpublished data). Of 7 NendoU mutant viruses generated using an infectious clone, catalytic site mutant viruses were non-viable. Mutation of D3786A and D3810A completely blocked viral RNA synthesis and virus production, which is consistent with the previous observation for EAV and coronaviruses (Ivanov et al., 2004, Kang et al., 2007, Posthuma et al., 2006). It is believed that in PRRSV, both D3786 and D3810 play a pivotal role in stabilizing the secondary structure of Nsp11, similar to Nsp15 in SARS-CoV. Thus, mutation of D3786A and D3810A abolishes their NendoU activity and therefore the IFN inhibitory activity. Our results provide first evidence that the NendoU domain of PRRSV Nsp11 plays an important role associated with the modulation of type I IFN induction.
3.2.2. Immune modulation by Nsp2
Nsp2 is the largest protein in PRRSV. It is highly heterogenous and extensive genetic and antigenic variations exist with deletions and mutations. The highly virulent PRRSV reported in the recent outbreaks in China contains two non-contiguous deletions in the Nsp2 hypervariable region (Tian et al., 2007, Zhou et al., 2008), and these deletions serve as genetic markers for differentiation from other PRRSV. Recent studies suggest the involvement of Nsp2 in host immune modulation. A deletion of 12 amino acids at position 456–469 in the cysteine proteinase (CP) domain of Nsp2 did not result in the production of viable virus. A deletion of 32 amino acids at positions 691–722, designated ES3, downstream of CP rendered a viable virus production and the ES3 mutant virus showed enhanced cytolytic activity and increased growth kinetics. When infected with the ES3 mutant virus, pigs produced a higher viral load and decreased production of IL-1β and TNF-α in PBMCs and macrophages compared with parental virus (Chen et al., 2010b), suggesting that Nsp2 play a role in the modulation of host innate immune response during infection. An independent study (Frias-Staheli et al., 2007) shows that the CP of Nsp2 belongs to the ovarian-tumor (OTU) domain deubiquitinases superfamily and may deconjugate ubiquitin (Ub) and ISG15 from cellular proteins thereby inhibiting Ub- and ISG15-dependent innate immune responses. ISG15 is a 15-kDa protein that is able to covalently modify target proteins in much the same way as ubiquitin to induce antiviral state. The addition of ISG15 targets RIG-I and JAK1 and plays an important role in IFN response. A further study shows that the PRRSV Nsp2 OTU domain antagonizes the type I interferon induction by interfering with the NF-κB signaling pathway (Sun et al., 2010). Inhibition of NF-κB activation is mediated by interfering with the poly-ubiquitination process of IKKα, which subsequently prevents IKKα degradation. The OTU domain in PRRSV Nsp2 acts as broad spectrum deISGylases and deubuquitinases. The dual deconjugating activity provides an additional example that Nsp2 is a multifunctional IFN antagonist. Protease activity by the Nsp2 OTU domain has the capacity to evade cytokine pathways that are important for antiviral immunity. Deletion of the OTU domain from the viral genome generates a nonviable virus.
4. Conclusions
The immune surveillance system protects host cells from viral infection, and viruses have evolved to escape this system for efficient proliferation in the host. Host cells produce cytokines and chemokines in response to viral infection and among such effector molecules, type I IFNs are principal antiviral cytokines and therefore are effective targets for viruses to disarm host surveillance. Many viruses are known to express proteins that counteract the production of type I IFN. RNA viruses are generally small, and to compensate for their small coding capacity, their proteins are often multifunctional. Ample evidence is available for different evasion strategies for different viruses, and viral proteins function to circumvent the IFN response by interfering with host cell gene expression, blocking the IFN induction cascade, and inhibiting the IFN signaling pathways (Koyama et al., 2008, Randall and Goodbourn, 2008, Pichlmair and Sousa, 2007, Kawai and Akira, 2006, Takaoka and Yanai, 2006).
The PRRSV genome is duplicated into dsRNA during replication, which can trigger a cascade of cellular events, and PRRSV has evolved to subvert host IFN surveillance for effective survival. To date, at least four viral proteins involved in the modulation of innate immunity have been studied: N as a structural protein and three non-structural proteins, Nsp1, Nsp2, and Nsp11. Among these, N and Nsp1 are nuclear-cytoplasmic proteins distributed in the both nucleus and cytoplasm of cell. The N protein in the nucleolus appears to participate in the cell cycle progression as well as immune modulation, whereas Nsp1 inhibits IFN production by degrading CREB-binding protein in the nucleus. Specifically, the Nsp1α subunit of Nsp1 blocks phosphorylation of IκB and thus prevents IκB degradation to retain NF-κB in the cytoplasm in the silent form. It is probable that CBP degradation is mediated by Nsp1α in the nucleus while NF-κB activation is blocked by the cytoplasmic form of Nsp1α. While the N protein nuclear localization is NLS-dependent, the basis for Nsp1α nuclear transport is unknown, and the structural differences between the nuclear and cytoplasm forms of Nsp1α are also unknown. It is intriguing that PIAS protein appears to interact with both N protein and Nsp1α subunit, and their biological consequences need to be further studied. Both N and Nsp1α may be sumoylated by PIAS1 to function in cellular processes, or binding of N and Nsp1α to PIAS1 may alter the normal PIAS function to gain benefits for favorable replication of the virus. Nsp2 is a cysteine protease, and deconjugation of ubiquitin and ISG15 by Nsp2 from key signaling molecules is an emerging mechanism of immune evasion by PRRSV to silence the inflammatory and IFN responses. Nsp4 is a serine protease whereas Nsp11 is an endoribonuclease, and both Nsp's have also been suggested to participate in the immune evasion strategy of PRRSV. Blocking against PRRSV-induced inhibition of the innate immune response may lead to the future development of effective vaccines or the discovery of novel antiviral compounds. The understanding of viral mechanisms modulating the normal cellular processes during infection will be a key to developing an effective future control strategy for PRRS.
Acknowledgments
This study was supported by funding awarded to DY by the National Research Initiatives of the US Department of Agriculture Cooperative State Research Education and Extension Service, grant number 2008-35204-04634.
References
- Aaronson D.S., Horvath C.M. A road map for those who don’t know JAK-STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- Aasted B., Bach P., Nielsen J., Lind P. Cytokine profiles in peripheral blood mononuclear cells and lymph node cells from piglets infected in utero with porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 2002;9:1229–1234. doi: 10.1128/CDLI.9.6.1229-1234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Tacheuchi O. Pathogenic recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Albina E., Carrat C., Charley B. Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J. Interferon Cytokine Res. 1998;18:485–490. doi: 10.1089/jir.1998.18.485. [DOI] [PubMed] [Google Scholar]
- Albina E., Madec F., Cariolet R., Torrison J. Immune response and persistence of the porcine reproductive and respiratory syndrome virus in infected pigs and farm units. Vet. Rec. 1994;134:567–573. doi: 10.1136/vr.134.22.567. [DOI] [PubMed] [Google Scholar]
- Andrejeva J., Childs K.S., Young D.F., Carlos T.S., Stock N., Goodbourn S., Randall R.E. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende R., Laegreid W.W., Kutish G.F., Galeota J.A., Wills R.W., Osorio F.A. Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J. Virol. 2000;74:10834–10837. doi: 10.1128/jvi.74.22.10834-10837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigent S.J., Zhang G., Fray M.D., Flick-Smith H., Goodbourn S., McCauley J.W. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 2002;76:8979–8988. doi: 10.1128/JVI.76.18.8979-8988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev N.A., Liu L., Chehab N.H., Mansfield K., Harris K.G., Halazonetis T.D., Berger S.L. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- Barrett L., Gallant M., Howley C., Bowmer M.I., Hirsch G., Peltekian K., Grant M. Enhanced IL-10 production in response to hepatitis C virus proteins by peripheral blood mononuclear cells from human immunodeficiency virus-monoinfected individuals. BMC Immunol. 2008;9:28. doi: 10.1186/1471-2172-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista L., Dee S.A., Rossow K.D., Deen J., Pijoan C. Assessing the duration of persistence and shedding of PRRSV in a large population of breeding-age gilts. Can. J. Vet. Res. 2002;66:196–200. [PMC free article] [PubMed] [Google Scholar]
- Bauhofer O., Summerfield A., Sakoda Y., Tratschin J.D., Hofmann M.A., Ruggli N. Classical swine fever virus Npro interacts with interferon regulatory factor 3 and induces its proteasomal degradation. J. Virol. 2007;81:3087–3096. doi: 10.1128/JVI.02032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista E.M., Molitor T.W. IFN gamma inhibits PRRSV replication in macrophages. Arch. Virol. 1999;144:1191–1200. doi: 10.1007/s007050050578. [DOI] [PubMed] [Google Scholar]
- Benfield D.A., Nelson E., Collins J.E., Harris L., Goyal S.M., Robison D., Christianson W.T., Morrison R.B., Gorcyca D., Chladek D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332) J. Vet. Diagn. Invest. 1992;4:127–133. doi: 10.1177/104063879200400202. [DOI] [PubMed] [Google Scholar]
- Beura L.K., Sarkar S.N., Kwon B., Subramaniam S., Jones C., Pattnaik A.K., Osorio F.A. Porcine reproductive and respiratory syndrome virus non structural protein 1-beta modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 2010;84:1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierk M.D., Dee S.A., Rossow K.D., Otake S., Collins J.E., Molitor T.W. Transmission of PRRSV from persistently-infected sows to contact controls. Can. J. Vet. Res. 2001;65:261–266. [PMC free article] [PubMed] [Google Scholar]
- Bowie A.G., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzózka K., Finke S., Conzelmann K.K. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 2005;79:7673–7681. doi: 10.1128/JVI.79.12.7673-7681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddaert W., Van Reeth K., Pensaert M. In vivo and in vitro interferon (IFN) studies with the porcine reproductive and respiratory syndrome virus (PRRSV) Adv. Exp. Med. Biol. 1998;440:461–467. doi: 10.1007/978-1-4615-5331-1_59. [DOI] [PubMed] [Google Scholar]
- Chen C.M., Kraut N., Groudine M., Weintraub H. I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell. 1996;86:731–741. doi: 10.1016/s0092-8674(00)80148-8. [DOI] [PubMed] [Google Scholar]
- Chen Z., Lawson S., Sun Z., Zhou X., Guan X., Christopher-Hennings J., Nelson E.A., Fang Y. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp function as interferon antagonist. Virology. 2010;398:87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhou X., Lunney J.K., Lawson S., Sun Z., Brown E., Christopher-Hennings J., Knudsen D., Nelson E., Fang Y. Immunodominant epitopes in nsp2 of porcine reproductive and respiratory syndrome virus are dispensable for replication, but play an important role in modulation of the host immune response. J. Gen. Virol. 2010;91:1047–1057. doi: 10.1099/vir.0.016212-0. [DOI] [PubMed] [Google Scholar]
- Chen Z., Rijnbrand R., Jangra R.K., Devaraj S.J., Qu L., Ma Y., Lemon S.M., Li K. Ubiquitination and proteasomal degradation of interferon regulatory factor-3 induced by Npro from a cytopathic bovine viral diarrhea virus. Virology. 2007;366:277–292. doi: 10.1016/j.virol.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher-Hennings J., Nelson E.A., Hines R.J., Nelson J.K., Swenson S.L., Zimmerman J., Chase C.L., Yaeger M.J., Benfield D.A. Persistence of porcine reproductive and respiratory syndrome virus in serum and semen of adult boars. J. Vet. Diagn. Invest. 1995;7:456–464. doi: 10.1177/104063879500700406. [DOI] [PubMed] [Google Scholar]
- Coccia E.M., Stellacci E., Orsatti R., Benedetti E., Giacomini E., Marziali G., Valdez B.C., Battistini A. Protein inhibitor of activated signal transducer and activator of transcription (STAT)-1 (PIAS-1) regulates the IFN-gamma response in macrophage cell lines. Cell Signal. 2002;14:537–545. doi: 10.1016/s0898-6568(01)00272-8. [DOI] [PubMed] [Google Scholar]
- Das P.B., Dinh P.X., Ansari I.H., de Lima M., Osorio F.A., Pattnaik A.K. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 2010;84:1731–1740. doi: 10.1128/JVI.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Interferon and its inducers—a never-ending story: “old” and “new” data in a new perspective. J. Infect. Dis. 2006;194:S19–S26. doi: 10.1086/505351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A.F., Horzinek M.C., Spaan W.J.M. Equine arteritis virus is not a togavirus but belongs to the coronavirus-like superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Faaberg K.S., Meulenberg J.J., Wassenaar A.L., Plagemann P.J., Gorbalenya A.E., Snijder E.J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papain-like cysteine proteases. J. Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T., Baker S.C., Li K. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz I., Darwich L., Pappaterra G., Pujols J., Mateu E. Immune responses of pigs after experimental infection with a European strain of PRRSV. J. Gen. Virol. 2005;86:1943–1951. doi: 10.1099/vir.0.80959-0. [DOI] [PubMed] [Google Scholar]
- Diaz I., Darwich L., Pappaterra G., Pujols J., Mateu E. Different European-type vaccines against PRRSV have different immunological properties and confer different protection to pigs. Virology. 2006;351:249–259. doi: 10.1016/j.virol.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Didcock L., Young D.F., Goodbourn S., Randall R.E. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 1999;73:3125–3133. doi: 10.1128/jvi.73.4.3125-3133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragan A.I., Hargreaves V.V., Makeyeva E.N., Privalov P.L. Mechanisms of activation of interferon regulator factor 3: the role of C-terminal domain phosphorylation in IRF-3 dimerization and DNA binding. Nucl. Acid Res. 2007;35:3525–3534. doi: 10.1093/nar/gkm142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Zuckermann F.A., Yoo D. Myristoylation of the small envelope protein of porcine reproductive and respiratory syndrome virus is non-essential for virus infectivity but promotes its growth. Virus Res. 2010;147:294–299. doi: 10.1016/j.virusres.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enninga J., Levy D.E., Blobel G., Fontoura B.M. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science. 2002;295:1523–1525. doi: 10.1126/science.1067861. [DOI] [PubMed] [Google Scholar]
- Emmott E., Hiscox J.A. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema T.J.G., Huynen M.A., de Vos W.M., van der Oost J. TRASH: a novel metal-binding domain predicted to be involved in heavy-metal sensing, trafficking and resistance. Trends Biochem. Sci. 2003;28:170–173. doi: 10.1016/S0968-0004(03)00037-9. [DOI] [PubMed] [Google Scholar]
- Fang Y., Schneider P., Zhang W.P., Faaberg K.S., Nelson E.A., Rowland R.R. Diversity and evolution of a newly emerged North American Type 1 porcine arterivirus: analysis of isolates collected between 1999 and 2004. Arch. Virol. 2007;152:1009–1017. doi: 10.1007/s00705-007-0936-y. [DOI] [PubMed] [Google Scholar]
- Feng Y., Zhao T., Nguyen T., Inui K., Ma Y., Nguyen T.H., Nguyen V.C., Liu D., Bui Q.A., To L.T., Wang C., Tian K., Gao G.F. Porcine respiratory and reproductive syndrome virus variants, Vietnam and China, 2007. Emerg. Infect. Dis. 2008;14:1774–1776. doi: 10.3201/eid1411.071676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N., Glannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J., Richt J.A., Rowland R.R.R., Schmaljohn C.S. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil L.H., Ansari I.H., Vassilev V., Liang D., Lai V.C., Zhong W., Hong Z., Dubovi E.J., Donis R.O. The amino-terminal domain of bovine viral diarrhea virus Npro protein is necessary for alpha/beta interferon antagonism. J. Virol. 2006;80:900–911. doi: 10.1128/JVI.80.2.900-911.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glodowski D.R., Petersen J.M., Dahlberg J.E. Complex nuclear localization signals in the matrix protein of vesicular stomatitis virus. J. Biol. Chem. 2002;277:46864–46870. doi: 10.1074/jbc.M208576200. [DOI] [PubMed] [Google Scholar]
- Gustin K.E. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex. Virus Res. 2003;95:35–44. doi: 10.1016/S0168-1702(03)00165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C.M. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 2004;271:4621–4628. doi: 10.1111/j.1432-1033.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings S., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. Thogoto virus ML protein suppresses IRF3 function. Virology. 2005;331:63–72. doi: 10.1016/j.virol.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Kang H., Bhardwaj K., Li Y., Palaninathan S., Sacchettini J., Guarino L., Leibowitz J.L., Kao C.C. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze M.G., He Y., Gale Y., Jr. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Keffaber K.K. Reproductive failure of unknown etiology. Am. Assoc. Swine Pract. Newslett. 1989;1:1–9. [Google Scholar]
- Kim, O., Sun, Y., Lai, F., Yoo, D., 2010. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology. doi:10.1016/j.virol.2010.03.039. [DOI] [PMC free article] [PubMed]
- Koyama S., Ishii K.J., Coban C., Akira S. Innate immune response to viral infection. Cytokine. 2008;43:336–341. doi: 10.1016/j.cyto.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Kraut N., Snider L., Chen C.M., Tapscott S.J., Groudine M. Requirement of the mouse I-mfa gene for placental development and skeletal patterning. EMBO J. 1998;17:6276–6288. doi: 10.1093/emboj/17.21.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroese M.V., Zevenhoven-Dobbe J.C., Bos-de Ruijter J.N.A., Peeters B.P.H., Meulenberg J.J.M., Cornelissen L., Snijder E.J. The nsp1-alpha and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 2008;89:494–499. doi: 10.1099/vir.0.83253-0. [DOI] [PubMed] [Google Scholar]
- Kukushkin A.S., Baybikov T.Z., Scherbakov A.V., Timina A.M., Baborenko E.P., Puzankova O.S., Pronin I.A., Fomin A.E. First outbreak of atypical porcine reproductive and respiratory syndrome virus in Russia caused by highly pathogenic Chinese-like PRRS virus. 2008 International PRRS Symposium, Abstract; Chicago, IL, December 5–6; 2008. [Google Scholar]
- Lamontagne L., Pagé C., Larochelle R., Magar R. Porcine reproductive and respiratory syndrome virus persistence in blood, spleen, lymph nodes, and tonsils of experimentally infected pigs depends on the level of CD8 high T cells. Viral Immunol. 2003;16:395–406. doi: 10.1089/088282403322396181. [DOI] [PubMed] [Google Scholar]
- La Rocca S.A., Herbert R.J., Crooke H., Drew T.W., Wileman T.E., Powell P.P. Loss of interferon regulatory factor 3 in cells infected with classical swine fever virus involves the N-terminal protease Npro. J. Virol. 2005;79:7239–7247. doi: 10.1128/JVI.79.11.7239-7247.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Calvert J.G., Welch S.K., Yoo D. A DNA-launched reverse genetics system for porcine reproductive and respiratory syndrome virus reveals that homodimerization of the nucleocapsid protein is essential for virus infectivity. Virology. 2005;331:47–62. doi: 10.1016/j.virol.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Lee C., Hodgins D., Calvert J.G., Welch S.K., Jolie R., Yoo D. Mutations within the nuclear localization signal of the porcine reproductive and respiratory syndrome virus nucleocapsid protein attenuate virus replication. Virology. 2006;346:238–250. doi: 10.1016/j.virol.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Yoo D. The small envelope protein of porcine reproductive and respiratory syndrome virus possesses ion channel protein-like properties. Virology. 2006;355:30–43. doi: 10.1016/j.virol.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Kleiboeker S.B. Porcine arterivirus activates the NF-κB pathway through IκB degradation. Virology. 2005;342:47–59. doi: 10.1016/j.virol.2005.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Schommer S.K., Kleiboeker S.B. Porcine reproductive and respiratory syndrome virus field isolates differ in in vitro interferon phenotypes. Vet. Immunol. Immunopathol. 2004;102:217–231. doi: 10.1016/j.vetimm.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke C.D., Haynes J.S., Spaete R., Adolphson D., Vorwald A., Lager K., Butler J.E. Lymphoid hyperplasia resulting in immune dysregulation is caused by PRRSV infection in neonatal pigs. J. Immunol. 2004;172:1916–1925. doi: 10.4049/jimmunol.172.3.1916. [DOI] [PubMed] [Google Scholar]
- Loemba H.D., Mounir S., Mardassi H., Archambault D., Dea S. Kinetics of humoral immune response to the major structural proteins of the PRRSV. Arch. Virol. 1996;141:751–761. doi: 10.1007/BF01718333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Heylbroeck C., Pitha P.M., Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Genin P., Mamane Y., Hiscott J. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 2000;20:6342–6353. doi: 10.1128/mcb.20.17.6342-6353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Fuertes L., Doménech N., Alvarez B., Ezquerra A., Domínguez J., Castro J.M., Alonso F. Analysis of cellular immune response in pigs recovered from porcine respiratory and reproductive syndrome infection. Virus Res. 1999;64:33–42. doi: 10.1016/s0168-1702(99)00073-8. [DOI] [PubMed] [Google Scholar]
- Luo R., Xiao S., Jiang Y., Jin H., Wang D., Liu M., Chen H., Fang L. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-β production by interfering with the RIG-I signaling pathway. Mol. Immunol. 2008;45:2839–2846. doi: 10.1016/j.molimm.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquin E., Larrea E., Civeira M.P., Prieto J., Aldabe R. HCV structural proteins interfere with interferon-alpha Jak/STAT signalling pathway. Antiviral Res. 2007;76:194–197. doi: 10.1016/j.antiviral.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Ma Z., Chang M.J., Shah R.C., Benveniste E.N. Interferon-gamma-activated STAT-1a suppresses MMP-9 gene transcription by sequestration of the coactivators CBP/p300. J. Leukoc. Biol. 2005;78:515–523. doi: 10.1189/jlb.0205112. [DOI] [PubMed] [Google Scholar]
- Magkcuras I., Matzener P., Rumenapf T., Peterhans E., Schweizer M. RNase-dependent inhibition of extracellular, but not intracellular, dsRNA-induced interferon synthesis by Erns of pestiviruses. J. Gen. Virol. 2008;89:2501–2506. doi: 10.1099/vir.0.2008/003749-0. [DOI] [PubMed] [Google Scholar]
- Meier W.A., Galeota J., Osorio F.A., Husmann R.J., Schnitzlein W.M., Zuckermann F.A. Gradual development of the interferon-gamma response of swine to PRRSV infection or vaccination. Virology. 2003;309:18–31. doi: 10.1016/s0042-6822(03)00009-6. [DOI] [PubMed] [Google Scholar]
- Meier W.A., Husmann R.J., Schnitzlein W.M., Osorio F.A., Lunney J.K., Zuckermann F.A. The use of cytokines and synthetic double-stranded RNA to augment the T helper 1 immune response of swine to PRRSV. Vet. Immunol. Immunopathol. 2004;102:299–314. doi: 10.1016/j.vetimm.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu E., Diaz I. The challenge of PRRS immunology. Vet. J. 2008;177:345–351. doi: 10.1016/j.tvjl.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg J.J.M., Hulst M.M., de Meijer E.J., Moonen P.J.M., den Besten A., De Kluyver E.P., Wensvoort G., Moormann R.J.M. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.C., Laegreid W.W., Bono J.L., Chitko-McKown C.G., Fox J.M. Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch. Virol. 2004;149:2453–2463. doi: 10.1007/s00705-004-0377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.Y., Krug R.M. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′–5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi, H., 2010. Molecular characterization of the nucleocapsid protein of arteriviruses. Ph.D. Thesis. University of Guelph, Guelph, Canada.
- Mohammadi H., Sharif S., Rowland R.R., Yoo D. The lactate dehydrogenase-elevating virus capsid protein is a nuclear-cytoplasmic protein. Arch. Virol. 2009;154:1071–1080. doi: 10.1007/s00705-009-0410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Music N., Gagnon C.A. The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim. Health Res. Rev. 2010;14:1–29. doi: 10.1017/S1466252310000034. [DOI] [PubMed] [Google Scholar]
- Nedialkova D.D., Ulferts R., van den Born E., Lauber C., Gorbalenya A.E., Ziebuhr J., Snijder E.J. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J. Virol. 2009;83:5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile D. China, Vietnam grapple with ‘rapidly evolving’ pig virus. Science. 2007;317:1017. doi: 10.1126/science.317.5841.1017. [DOI] [PubMed] [Google Scholar]
- Oelgeschläger M., Janknecht R., Krieg J., Schreek S., Lüscher B. Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- Overend C., Mitchel R., He D., Rompato G., Grubman M.J., Garmendia A.E. Recombinant swine beta interferon protects swine alveolar macrophages and MARC-145 cells from infection with Porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2007;88:925–931. doi: 10.1099/vir.0.82585-0. [DOI] [PubMed] [Google Scholar]
- Palosaari H., Parisien J.P., Rodriguez J.J., Ulane C.M., Horvath C.M. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 2003;77:7635–7644. doi: 10.1128/JVI.77.13.7635-7644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D., Maniatis T., Harrison S.C. An atomic model of the interferon-β enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Hodgins D.C., Lee C., Calvert J.G., Welch S.K.W., Jolie R., Keith M., Yoo D. Functional mapping of the PRRSV capsid protein nuclear localization signal and its pathogenic association. Virus Res. 2008;135:107–114. doi: 10.1016/j.virusres.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Pichlmair A., Sousa C.R. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Posthuma C.C., Nedialkova D.D., Zevenhoven-Dobbe J.C., Blokhuis J.H., Gorbalenya A.E., Snijder E.J. Site-directed mutagenesis of the nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J. Virol. 2006;80:1653–1661. doi: 10.1128/JVI.80.4.1653-1661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J.B., Hall P.R., Bondu-Hawkins V.S., Ye C., Hjelle B. Early innate immune responses to Sin Nombre hantavirus occur independently of IFN regulatory factor 3, characterized pattern recognition receptors, and viral entry. J. Immunol. 2007;179:1796–1802. doi: 10.4049/jimmunol.179.3.1796. [DOI] [PubMed] [Google Scholar]
- Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Reichow S.L., Hamma T., Ferré-D’Amaré A.R., Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J.J., Parisien J.P., Horvath C.M. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 2002;76:11476–11483. doi: 10.1128/JVI.76.22.11476-11483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J.J., Wang L.F., Horvath C.M. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. J. Virol. 2003;77:11842–11845. doi: 10.1128/JVI.77.21.11842-11845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropp S.L., Wees C.E., Fang Y., Nelson E.A., Rossow K.D., Bien M., Arndt B., Preszler S., Steen P., Christopher-Hennings J., Collins J.E., Benfield D.A., Faaberg K.S. Characterization of emerging European-like porcine reproductive and respiratory syndrome virus isolates in the United States. J. Virol. 2004;78:3684–3703. doi: 10.1128/JVI.78.7.3684-3703.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland R.R., Kervin R., Kuckleburg C., Sperlich A., Benfield D.A. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 1999;64:1–12. doi: 10.1016/s0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- Rowland R.R., Schneider P., Fang Y., Wootton S., Yoo D., Benfield D.A. Peptide domains involved in the localization of the porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus. Virology. 2003;316:135–145. doi: 10.1016/S0042-6822(03)00482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royaee A.R., Husmann R.J., Dawson H.D., Calzada-Nova G., Schnitzlein W.M., Zuckermann F.A., Lunney J.K. Deciphering the involvement of innate immune factors in development of the host immune response to PRRSV vaccination. Vet. Immunol. Immunopathol. 2004;102:199–216. doi: 10.1016/j.vetimm.2004.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggli N., Bird B.H., Liu L., Bauhofer O., Tratschin J.D., Hofmann M.A. N(pro) of classical swine fever virus is an antagonist of double-stranded RNA-mediated apoptosis and IFN-alpha/beta induction. Virology. 2005;340:265–276. doi: 10.1016/j.virol.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem. Cell. Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K., Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L., Spaan W.J.M. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J. Virol. 1992;66:7040–7048. doi: 10.1128/jvi.66.12.7040-7048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L., Spaan W.J.M. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L., Spaan W.J., Gorbalenya A.E. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 1995;270:16671–16676. doi: 10.1074/jbc.270.28.16671. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L.M., van Dinten L.C., Spaan W.J.M., Gorbalenya A.E. The arterivirus Nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J. Biol. Chem. 1996;271:4864–4871. doi: 10.1074/jbc.271.9.4864. [DOI] [PubMed] [Google Scholar]
- Song, C., 2003. Identification and characterization of a novel cellular protein interacting with the porcine arterivirus capsid protein. PhD Thesis. University of Guelph, Guelph, Canada.
- Song C., Lu R., Bienzle D., Liu S., Yoo D. PRRSV nucleocapsid protein interacts with the human I-mfa domain containing protein HIC. Biol. Chem. 2009;390:215–223. doi: 10.1515/BC.2009.028. [DOI] [PubMed] [Google Scholar]
- Starr R., Hilton D.J. Negative regulation of the JAK/STAT pathway. Bioessays. 1999;21:47–52. doi: 10.1002/(SICI)1521-1878(199901)21:1<47::AID-BIES6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Suhara W., Yoneyama M., Kitabayashi I., Fujita T. Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J. Biol. Chem. 2002;277:22304–22313. doi: 10.1074/jbc.M200192200. [DOI] [PubMed] [Google Scholar]
- Sun Y., Chen, N., Yoo, D., 2009a. Inhibition of type I interferon signaling by Nsp11 of PRRSV. International PRRS Symposium, Chicago, IL, December 4–5, 2009, p. 24 (Abstract 9).
- Sun Y., Xue F., Guo Y., Ma M., Hao N., Zhang X.C., Lou Z., Li X., Rao Z. Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1-alpha. J. Virol. 2009;83:10931–10940. doi: 10.1128/JVI.02579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z., Chen, Z., Lawson, S., Fang, Y., 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism function. J. Virol. May 26 [Epub ahead of print] doi:10.1128/JVI.00217-10. [DOI] [PMC free article] [PubMed]
- Suradhat S., Thanawongnuwech R. Upregulation of interleukin-10 gene expression in the leukocytes of pigs infected with PRRSV. J. Gen. Virol. 2003;84:2755–2760. doi: 10.1099/vir.0.19230-0. [DOI] [PubMed] [Google Scholar]
- Suradhat S., Thanawongnuwech R., Poovorawan Y. Upregulation of IL-10 gene expression in porcine peripheral blood mononuclear cells by porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2003;84:453–459. doi: 10.1099/vir.0.18698-0. [DOI] [PubMed] [Google Scholar]
- Szymanski M.R., Fiebach A.R., Tratschin J.D., Gut M., Ramanujam V.M.S., Patel P., Ye M., Ruggli N., Choi K.H. Zinc-binding in pestivirus Npro is required for interferon regulatory factor 3 (IRF3) interaction and degradation. J. Mol. Biol. 2009;391:438–449. doi: 10.1016/j.jmb.2009.06.040. [DOI] [PubMed] [Google Scholar]
- Takaoka A., Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. MDA5/RIG-I and virus regonition. Curr. Opin. Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Thanawongnuwech R., Thacker E.L. Interleukin (IL)-10, IL-12 and interferon-gamma (IFN-γ) levels in the respiratory tract following Mycoplasma hyopneumoniae and PRRSV infection in pigs. Viral Immunol. 2003;16:357–367. doi: 10.1089/088282403322396154. [DOI] [PubMed] [Google Scholar]
- Thébault S., Gachon F., Lemasson I., Devaux C., Mesnard J.M. Molecular cloning of a novel human I-mfa domain-containing protein that differently regulates human T-cell leukemia virus type I and HIV-1 expression. J. Biol. Chem. 2000;275:4848–4857. doi: 10.1074/jbc.275.7.4848. [DOI] [PubMed] [Google Scholar]
- Tian K., Yu X., Zhao T., Feng Y., Cao Z., Wang C., Hu Y., Chen X., Hu D., Tian X., Liu D. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE. 2007;6:1–10. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms M.A., Nedialkova D.D., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Snijder E.J. Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease. J. Virol. 2007;81:10496–10505. doi: 10.1128/JVI.00683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms M.A., Snijder E.J. Equine arteritis virus non-structural protein 1, an essential factor for viral subgenomic mRNA synthesis, interacts with the cellular transcription co-factor p100. J. Gen. Virol. 2003;84:2317–2322. doi: 10.1099/vir.0.19297-0. [DOI] [PubMed] [Google Scholar]
- Tijms M.A., van Dinten L.C., Gorbalenya A.E., Snijder E.J. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms M.A., van der Meer Y., Snijder E.J. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 2002;83:795–800. doi: 10.1099/0022-1317-83-4-795. [DOI] [PubMed] [Google Scholar]
- Tollervery D., Kiss D. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Tong G.Z., Zhou Y.J., Hao X.F., Tian Z.J., An T.Q., Qiu H.J. Highly pathogenic porcine reproductive and respiratory syndrome in China. Emerg. Infect. Dis. 2007;13:1434–1436. doi: 10.3201/eid1309.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- van Dinten L.C., Wassenaar A.L., Gorbalenya A.E., Spaan W.J.M., Snijder E.J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reeth K., Abarque G., Nauwynck H., Pensaert M. Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res. Vet. Sci. 1999;67:47–52. doi: 10.1053/rvsc.1998.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt C., Preuss E., Mayer D., Weber F., Schwemmle M., Kochs G. The interferon antagonist ML protein of Thogoto virus targets general transcription factor IIB. J. Virol. 2008;82:11446–11453. doi: 10.1128/JVI.01284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar A.L., Spaan W.J., Gorbalenya A.E., Snijder E.J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J. Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 2007;81:16620–16632. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidman M.K., Sharma R., Raychaudhuri S., Kundu P., Tsai W., Dasgupta A. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 2003;95:75–85. doi: 10.1016/s0168-1702(03)00164-3. [DOI] [PubMed] [Google Scholar]
- Wieringa R., de Vries A.A.F., Rottier P.J.M. Formation of disulfide-linked complexes between the three minor envelope glycoproteins (GP2b, GP3, and GP4) of equine arteritis virus. J. Virol. 2003;77:6216–6226. doi: 10.1128/JVI.77.11.6216-6226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K.A., Henley J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills R.W., Doster A.R., Galeota J.A., Sur J.H., Osorio F.A. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J. Clin. Microbiol. 2003;41:58–62. doi: 10.1128/JCM.41.1.58-62.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills R.W., Zimmerman J.J., Yoon K.J., Swenson S.L., McGinley M.J., Hill H.T., Platt K.B., Christopher-Hennings J., Nelson E.A. PRRSV: a persistent infection. Vet. Microbiol. 1997;55:231–240. doi: 10.1016/s0378-1135(96)01337-5. [DOI] [PubMed] [Google Scholar]
- Wongyanin, P., Buranapraditkun, S., Thanawonguwech, R., Roth, J.A., Suradhat, S., 2009. Effect of nucleocapsid protein of PRRSV on the induction of interleuklin-10 producing lymphocytes. International PRRS Symposium, December 4–5, 2009, Chicago, IL (Abstract p95).
- Wootton S.K., Yoo D. Homo-oligomerization of the porcine reproductive and respiratory syndrome virus nucleocapsid protein and the role of disulfide linkages. J. Virol. 2003;77:4546–4557. doi: 10.1128/JVI.77.8.4546-4557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton S.K., Rowland R.R., Yoo D. Phosphorylation of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. J. Virol. 2002;76:10569–10576. doi: 10.1128/JVI.76.20.10569-10576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton S., Yoo D., Rogan D. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch. Virol. 2000;145:2297–2323. doi: 10.1007/s007050070022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm T., Chen H., Hodgson T., Britton P., Brooks G., Hiscox J.A. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Batista L., Dee S., Halbur P., Murtaugh M.P. The level of virus-specific T-Cell and macrophage recruitment in PRRSV infection in pigs is independent of virus load. J. Virol. 2004;78:5923–5933. doi: 10.1128/JVI.78.11.5923-5933.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 2006;80:7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Ma G., Lin C.H., Orr M., Wathelet M.G. Mechanism for transcriptional synergy between interferon regulatory factor (IRF)-3 and IRF-7 in activation of the interferon-β gene promoter. Eur. J. Biochem. 2004;271:3693–3703. doi: 10.1111/j.1432-1033.2004.04310.x. [DOI] [PubMed] [Google Scholar]
- Yin X., Warner D.R., Roberts E.A., Pisano M.M., Greene R.M. Novel interaction between nuclear coactivator CBP and the protein inhibitor of activated Stat1 (PIAS1) J. Interferon Cytokine Res. 2005;25:321–327. doi: 10.1089/jir.2005.25.321. [DOI] [PubMed] [Google Scholar]
- Yoo D., Wootton S.K., Li G., Song C., Rowland R.R. Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J. Virol. 2003;77:12173–12183. doi: 10.1128/JVI.77.22.12173-12183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, D., Kim, O., Song, C., Sun, Y., Du, Y., Liu, H.C., 2009. Modulation of type I interferon production and evasion strategies of PRRSV from the host defense. International PRRS Symposium, Chicago, IL, December 4–5, 2009, p. 24 (Abstract 9).
- You J.H., Howell G., Pattnaik A.K., Osorio F.A., Hiscox J.A. A model for the dynamic nuclear/nucleolar/cytoplasmic trafficking of the porcine reproductive and respiratory syndrome virus (PRRSV) nucleocapsid protein based on live cell imaging. Virology. 2008;378:34–47. doi: 10.1016/j.virol.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D.F., Didcock L., Goodbourn S., Randall R.E. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology. 2000;269:383–390. doi: 10.1006/viro.2000.0240. [DOI] [PubMed] [Google Scholar]
- Zhong H., May M.J., Jimi E., Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- Zhou Y.J., Hao X.F., Tian Z.J., Tong G.Z., Yoo D., An T.Q., Zhou T., Li G.X., Qiu H.J., Wei T.C., Yuan X.F. Highly virulent porcine reproductive and respiratory syndrome virus emerged in China. Transbound. Emerg. Dis. 2008;55:152–164. doi: 10.1111/j.1865-1682.2008.01020.x. [DOI] [PubMed] [Google Scholar]


