Highlights
-
•
Complete and up to date coverage of replicase protein structures for SARS-CoV.
-
•
Discusses SARS-CoV structure in the context of other coronavirus structures.
-
•
Summarizes data from a variety of structural methods to illuminate protein function.
-
•
Uses models and predictions to fill gaps in the SARS-CoV structure.
-
•
Discusses the high percentage of novel protein folds among SARS-CoV proteins.
Keywords: SARS coronavirus, Nonstructural protein, Protein structure, NMR, X-ray crystallography, Cryoelectron microscopy, Small angle X-ray scattering
Abstract
The international response to SARS-CoV has produced an outstanding number of protein structures in a very short time. This review summarizes the findings of functional and structural studies including those derived from cryoelectron microscopy, small angle X-ray scattering, NMR spectroscopy, and X-ray crystallography, and incorporates bioinformatics predictions where no structural data is available. Structures that shed light on the function and biological roles of the proteins in viral replication and pathogenesis are highlighted. The high percentage of novel protein folds identified among SARS-CoV proteins is discussed.
1. Introduction
In the wake of the SARS crisis, a wave of structural proteomics swept the coronavirus research community. The focus of this effort was to understand the interplay between structure and function in what had been, until that time, a somewhat neglected branch of the positive-stranded viruses. The unusual aspect of the SARS proteomics at the time was its evenhandedness – rather than focusing exclusively on proteins with well-defined roles in pathogenesis, competing international teams attempted to solve structures and assign functions across the entire viral proteome.
This effort brought fresh attention to several little-known replicase cofactors, such as the European group's structure of the obscure but important RNA binding protein nsp9 (Egloff et al., 2004), the Chinese group's barrel-shaped 16-protein structure of nsp7 + 8 primase complex (Zhai et al., 2005) and the American group's long crawl through the giant multi-domain, multi-enzymatic protein nsp3 which found the first of three SARS-CoV macrodomain folds (Saikatendu et al., 2005).
Shortly after the outbreak, the sequence of the genome was completed and the 3-D structure of Mpro, the main protease essential for viral replication, was deposited in the Protein Data Bank (PDB). By 2007, 100 entries in the PDB were on 14 of the 28 SARS CoV proteins, and at present count there are 99 structures of coronavirus Mpro available in the PDB alone, providing an unprecedented database for investigators working on this and related viruses. This review summarizes the findings of functional and structural studies including those derived from cryoelectron microscopy, small angle X-ray scattering, NMR spectroscopy, and X-ray crystallography in an attempt to understand the function and biological roles of the proteins in viral replication and pathogenesis.
2. A note on functional organization
The new wealth of structural and functional information revealed that the coronavirus replicase, which is but one biologically successful example of the conserved nidovirus replicative machinery (Lauber et al., 2013), is not a patchwork amalgam of evolutionary jetsam, but an organized piece of biological machinery where proteins are generally organized into units with related functions (Fig. 1 ). The first two parts of the replicase, nsp1 and nsp2 are somewhat enigmatic, but appear to work by interfering with host defenses rather than by directly supporting virus replication. Subunits nsp3–6 contain all the viral factors that are necessary to form viral replicative organelles (Angelini et al., 2013), as well as two proteinases that are responsible for processing all of the viral replicase proteins (Ziebuhr et al., 2000). The small subunits nsp7–11 comprise the viral primer-making activities and provide other essential support for replication (Donaldson et al., 2007b, Imbert et al., 2006, Miknis et al., 2009). The final part of the replicase from nsp12–16 contains the remaining RNA-modifying enzymes needed for replication, RNA capping and proofreading.
Fig. 1.
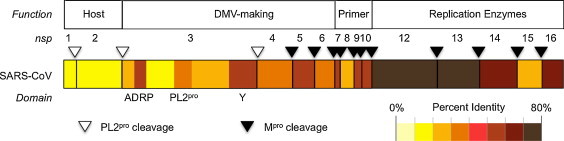
Conservation of the SARS-CoV replicase. Replicase subunits, or domains for nsp3, were color-coded according to percent identity between homologous proteins of SARS-CoV and MERS-CoV. Alignments and identity calculations were performed using Clustal Omega (Sievers et al., 2011).
The organization of replicase has a sort of chronological logic to it. Nsp1–2 help to colonize the host, followed by Nsp3–6 which lay a foundation to organize and protect the replicative machinery. This is followed by the primer-making activities of nsp7–11 which also interact with downstream capping and RNA synthesis factors (Bouvet et al., 2010). Finally, in the proper framework, the RNA-synthesizing enzymes from the C-terminus of the replicase are able to function. While this may be an appealing way to think of the replicase, the reality is probably much more complex. The replicase proteins are all processed from large polyproteins, and therefore are produced at the same time. Because of this, the order in which different proteins are active during the viral replication cycle remains poorly understood.
The organization of the replicase also roughly follows a gradient of primary sequence conservation. Levels of sequence conservation among the different coronaviruses are highest at the 3′ end of the replicase gene, and the sequences are very divergent at the 5′ end, especially in nsp1–3, which are products of nsp3 PLpro cleavage. The DMV-making proteins and the primase group of proteins show intermediate levels of conservation with the exception of the well-conserved nsp5Mpro. Fig. 1 illustrates amino acid conservation across the replicase using the comparison between SARS-CoV and MERS-CoV as an example.
3. The Atlas
3.1. Nsp1
See Box 1 .
Box 1. Key nsp1 and nsp2 structures.
| Virus | Protein | Method | Accession | Reference |
|---|---|---|---|---|
| SARS-CoV | nsp1 | NMR | 2HSX | Almeida et al. (2007) |
| TGEV | nsp1 | X-ray (1.49 Å) | 3ZBD | Jansson (2013) |
| IBV | nsp2 | X-Ray (2.5 Å) | 3LD1 | Yu et al. (2012) |
3.1.1. Structure
Nsp1 is the N-terminal cleavage product of the replicase polyprotein and is produced by the action of PLpro. Nsp1 is not found in the gammacoronavirus or deltacoronavirus lineages, which encode a distant homolog of SARS-CoV nsp2 at the N-terminus of the replicase po lyprotein. This has led to a suggestion that nsp1 is useful as a group-specific marker (Snijder et al., 2003). SARS-CoV nsp1 is 179 residues long.
In the alphacoronaviruses, nsp1 (also known as p9) is a protein of about 110 residues, with 20–50% sequence identity among all alphacoronaviruses. The betacoronaviruses of subgroup A, such as murine hepatitis virus (MHV) and human coronavirus OC43, encode an nsp1 protein of about 245 residues, also known as p28 (Brockway and Denison, 2005). The nsp1 of SARS-CoV and its bat equivalents, which have been classified as the only members to date of the betacoronavirus subgroup B (Gorbalenya et al., 2006, Gorbalenya et al., 2004, Snijder et al., 2003), have 179 residues. Nsp1 sequences are divergent between subgroups of betacoronavirus, and no sequence similarity between SARS-CoV nsp1 and betacoronavirus subgroup A nsp1 proteins could be identified using standard searching tools such as PSI-BLAST.
Almeida et al., 2006, Almeida et al., 2007 determined the NMR structure of the nsp1 segment from residue 13 to 128 and also showed that the polypeptide segments of residues 1–12 and 129–179 are flexibly disordered (PDB ID 2GDT; 2HSX) (Almeida et al., 2007). Residues 13–128 of nsp1 represents a novel α/β-fold formed by a mixed parallel/antiparallel 6-stranded β-barrel, an α-helix covering one opening of the barrel, and a 310-helix alongside the barrel (Fig. 2 ). NMR data indicate that full-length nsp1 has the same globular fold as the truncated nsp1, but with additional flexibly disordered regions that correspond to the N-terminal region (residues 1–12) and the long C-terminal tail (residues 129–179).
Fig. 2.
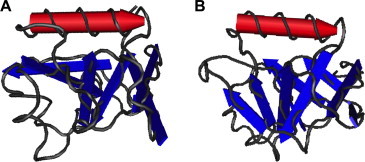
Comparison of nsp1 structure in alpha- and betacoronavirus lineages. The SARS-CoV nsp1 structure comes from PDB entry 2HSX, and the TGEV nsp1 structure comes from PDB entry 3ZBD.
The C-terminal portion of SARS-CoV nsp1 is flexibly disordered. Interestingly, it has been determined that the C-terminal half of MHV nsp1 (Lys124–Leu241) is dispensable for viral replication in culture but is important for efficient proteolytic cleavage at the nsp1–2 peptide linkage by the papain-like protease and optimal viral replication (Brockway and Denison, 2005). Likewise, the long disordered terminus of SARS-CoV nsp1 are probably important for the efficient proteolytic processing of this protein from the nascent viral polyprotein chain.
The nsp1 of transmissible gastroenteritis virus (TGEV) was recently solved, and was found to contain a similar fold to SARS-CoV nsp1 (Jansson, 2013). This was surprising as there is no detectable homology between alphacoronavirus nsp1 proteins and betacoronavirus nsp1 proteins. However, the relationship of the structures suggests that coronavirus nsp1 proteins share a common evolutionary origin.
3.1.2. Function
In several coronaviruses, nsp1 suppresses host gene expression (Huang et al., 2011, Kamitani et al., 2006, Narayanan et al., 2008, Zust et al., 2007). The mechanism of nsp1-mediated host suppression remains a topic of active research. SARS-CoV nsp1 binds to 40S subunits and exerts suppression of host gene expression (Kamitani et al., 2009). It has also been shown that SARS-CoV nsp1 promotes host mRNA degradation but coronavirus RNA species are protected from degradation (Kamitani et al., 2009, Tanaka et al., 2012).
In MHV, nsp1 arrests the cell cycle of transfected cells in G0/G1 phase (Chen et al., 2004). MHV mutants that are incapable of liberating nsp1 from the nascent polyprotein exhibit delayed replication, diminished peak titers, and reduced RNA synthesis compared to wild-type controls (Denison et al., 2004). A point mutation in the proteolytic cleavage site between nsp1 and nsp2 in full-length TGEV genome blocks the release of nsp1 from the nascent polyprotein and causes a dramatic reduction in virus recovery (Galan et al., 2005).
Kamitani et al., have shown that plasmid-driven expression of nsp1 (driven by SV40, CMV and IFN-β promoters) sharply reduces protein expression (Kamitani et al., 2006). This correlates with reduction in the specific mRNA, whereas rRNA remained unaffected. More generally, transfected nsp1 mRNA that was capped and polyadenylated decreased host protein synthesis, and the inclusion of actinomycin D (to block new transcription) showed a much stronger inhibition of protein synthesis in the presence of nsp1, demonstrating that that while translation of new transcripts was proceeding (in cells not treated with actinomycin D), translation from pre-existing transcripts was blocked by nsp1. Decreased mRNA levels and decreased translation of pre-existing mRNA, presumably as a result of degradation, were also seen during infection with SARS-CoV.
SARS-CoV nsp1 has also been shown to be a potent inducer of CCL5, CXCL10 and CCL3 expression in human lung epithelial cells via the activation of NF-κB (Law et al., 2007). The pathogenesis of SARS-CoV infection is characterized by a hyperimmune response and the massive elevation of chemokine levels. In contrast, HCoV-229E, HCoV-OC43, and MHV did not significantly induce chemokine expression, perhaps because these only cause mild upper respiratory tract diseases.
The kinetics of nsp1 expression suggests that it might have an early regulatory role during the viral life cycle. Nsp1 is the first mature protein processed from the gene 1 polyprotein and is likely cleaved quickly following translation of PL1pro within nsp3 (Baker et al., 1989, Baker et al., 1993, Denison and Perlman, 1987, Denison et al., 1992, Denison et al., 1995, Denison and Perlman, 1986). MHV mutants that are incapable of liberating nsp1 from the nascent polyprotein exhibit delayed replication, diminished peak titers, small plaques, and reduced RNA synthesis compared to wild-type controls (Denison et al., 2004). These results emphasize the importance of nsp1 cleavage for optimal viral RNA synthesis and suggest that nsp1 might play an important role at MHV replication complexes. However, later in infection, nsp1 is distinct from replication complexes and instead co-localizes with MHV structural proteins at virion assembly sites (Brockway et al., 2004).
In MHV, nsp1 interacts with p10 and p15 (counterparts of SARS nsp7 and nsp10, respectively; (Brockway et al., 2004)). Previous immunolocalization and interaction studies in MHV have also indicated that in vivo, nsp1 may act in concert with numerous other viral proteins – counterparts of SARS nsp2, 5, 8, 9, 12, 13 and sars9a (Bost et al., 2000, Brockway et al., 2004). However, at some stages in the MHV life cycle, nsp1 has spatially different membrane localization from p65, the SARS nsp2 counterpart. It appears that later in infection, MHV nsp1 co-localizes with structural proteins at virion assembly sites rather than with the replication complexes (Brockway et al., 2004). Y2H and co-immunoprecipitation studies indicate that nsp1 interacts with E, and sars3a (von Brunn et al., 2007).
3.2. Nsp2
3.2.1. Structure
SARS-CoV nsp2 is a counterpart of the p65 protein (Denison et al., 1995) of MHV. As with nsp1, sequence homology is very low and does not permit confident sequence alignment. However, as with nsp1, bioinformatics gives an indication that coronavirus nsp2 proteins share a common fold and origin (Fig. 3 ). The structure of the N-terminal 359 amino acids of the IBV equivalent of nsp2 has been released, but is currently awaiting full publication, though a crystallization report is available (Yang et al., 2009, Yu et al., 2012). Crystallization of part of the SARS-CoV nsp2 (Li et al., 2011) has also been reported, but the structure is not currently available pending full publication. The solved region of IBV nsp2 comprises about half the protein. It represents a novel multi-domain fold, though further structural and functional details await full publication.
Fig. 3.
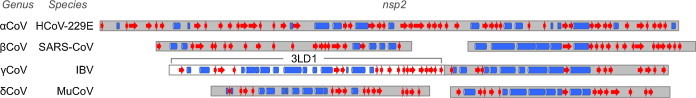
Bioinformatics detection of homology and domain organization in nsp2. The structure of IBV nsp2 is taken from PDB entry 3LD1. Predicted secondary structures for representative coronavirus lineages were made by PsiPRED 3.0 (McGuffin et al., 2000). Rectangles represent α-helices and arrows represent beta-strands.
Interestingly, secondary structure prediction suggests that coronavirus nsp2 proteins consist of a duplicated fold, with a second, more conserved fold similar to the structure 3LD1 immediately following the region solved in 3LD1 (Fig. 3). This is seen most clearly in the gammacoronaviruses and deltacoronaviruses. This would fit the context of domain and fold duplication at the N-terminal part of the replicase polyprotein which has been observed across the coronaviridae, which includes duplicated ubiquitin-like, papain-like, and macrodomain folds (Neuman et al., 2008).
3.2.2. Function
The functions of nsp2 remain unknown. In MHV p65 has spatially different membrane localization from nsp1 and co-localizes with the MHV homologue of SARS nsp8 (Sims et al., 2000). In MHV, p65 plays an important role in the viral life cycle (Hughes et al., 1993) that appears to be distinct from that of its counterparts in other coronaviruses (Bost et al., 2001, Denison et al., 2004, Sims et al., 2000). Based on immunolocalization studies in MHV, p65 may function in concert with counterparts of SARS nsp1, 5, 7, 8, 9, 10, 12, 13 and sars9a (Bost et al., 2000, Brockway et al., 2004). Deletion mutagenesis with infectious clones of SARS and MHV indicated that nsp2 is dispensable for viral replication in cell culture; however, deletion of the nsp2 coding sequence attenuates viral growth and RNA synthesis.
The exact nature of the role of nsp2 in viral growth and RNA synthesis is still not clear. However, IBV nsp2 has a weak PKR antagonist activity, which may hint at a role complementary to that of nsp1 in interfering with intracellular immunity. A proteomics study with full-length SARS-CoV nsp2 also found that nsp2 bound prohibitin 1 and prohibitin 2, which could contribute to the hypothetical role of nsp2 in counteracting intracellular immunity (Cornillez-Ty et al., 2009). Based on immunolocalization studies in MHV, p65 may function in concert with counterparts of SARS nsp1, 5, 7, 8, 9, 10, 12, 13 and sars9a (Bost et al., 2000, Brockway et al., 2004).
3.3. Nsp3
3.3.1. Structure
SARS nsp3 is a large multidomain protein with 1922 residues (Snijder et al., 2003, Thiel et al., 2003). Nsp3, with nsp4, nsp5 and nsp6 forms a conserved block of proteins that are involved in forming the double-membrane vesicles that are the site of viral RNA synthesis (Fig. 4 ). Every completely sequenced coronavirus has an nsp3-related protein. All nsp3s are ∼200 kDa, cleaved from the polyprotein 1a or 1ab by PLpro.
Fig. 4.
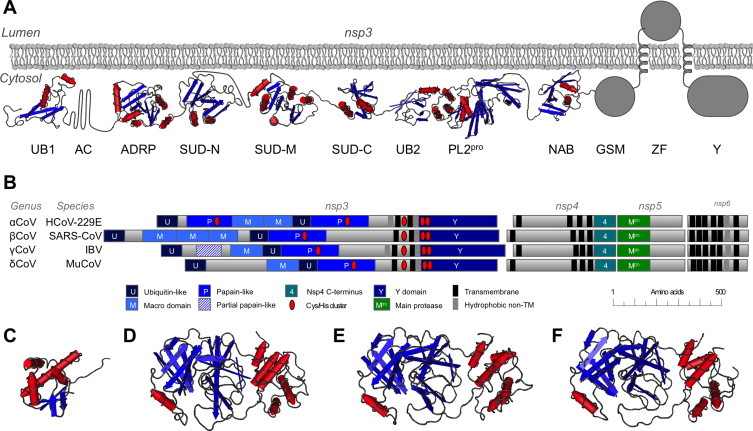
Domain architecture and structure of the conserved DMV-making proteins nsp3 through nsp6. A partially schematic view combining the known domain structures of SARS-CoV nsp3 is shown in panel A, combining the PDB entries 2IDY, 2ACF, 2FE8, 2KAF, 2JZF, 2W2G and 2K87. Conservation of structural domains and other important sequence features is shown for four representative coronaviruses in panel B. Structures of the C-terminal domain of FCoV nsp4 (3GZF), SARS-CoV nsp5 (1UJ1), TGEV nsp5 (1LVO) and IBV nsp5 (2Q6F) are shown in panels C–F, respectively.
We have compiled a higher-resolution analysis of nsp3 domain architecture as a tool for novel structural and functional characterization (Neuman et al., 2008). Based on phylogenetic analysis of coronavirus and torovirus nsp3 homologues, results from previously published studies (Gorbalenya et al., 2006, Ratia et al., 2006, Saikatendu et al., 2005, Serrano et al., 2007, Thiel et al., 2003, Ziebuhr et al., 2001) and de novo domain prediction software (Jaroszewski et al., 2005), we estimate that SARS-CoV nsp3 has about 14 domains – UB1, AC (it is missing PL1pro found in several other CoVs), ADRP, SUD-N, SUD-M, SUDC, UB2, PL2pro, NAB, G2M, TM1, ZF, TM2, and Y, which may contain three structural domains. A partially schematic model of the nsp3 structure is shown (Fig. 3). Inferring from the presence of PL2pro cleavage sites at both termini of nsp3, the observed glycosylation at positions 1431 and 1434 in the ZF domain of SARS-CoV (Harcourt et al., 2004) and the homologous region of MHV (Kanjanahaluethai et al., 2007). SARS-CoV nsp3 contains two transmembrane spans, placing the first 1395 residues (including the PL2pro domain), and the last 377 residues (the Y domain) on the same face of the membrane (Oostra et al., 2008). The two TM helices probably consist of residues 1396–1418 and 1523–1545. This transmembrane topology is similar to that proposed for MHV nsp3 (Kanjanahaluethai et al., 2007). Between helices two and three, there is a central, absolutely conserved tetrad of cysteines (CX14–18C4–5C2C) – which may represent a Zn finger – which is likely on the same side of the membrane as the domains N- and C-terminal to the TM region.
3.3.2. Function
Although the function of the N-terminal region of polyprotein 1a/polyprotein 1ab is not known, both the transcription-negative phenotypeof an alphavirus X domain mutant (von Brunn et al., 2007) and the conservation of a transcription factor-like zinc finger in coronavirus PLpro domains (Culver et al., 1993) indicated that nsp3 might be involved in coronavirus RNA synthesis. This hypothesis is strongly supported by a report in which the equine arteritis virus nonstructural protein 1, which, most probably, is a distant homolog of the coronavirus PLpro, is shown to be a transcriptional factor that is indispensable for sg mRNA synthesis (Phizicky and Greer, 1993).
3.4. UB1 and AC
3.4.1. Structure
The sequence of the N-terminal domain of nsp3 (1–183) is highly conserved in different SARS coronavirus isolates but shows less than 25% of sequence identity with other known proteins. This region exhibits two well defined regions with different physico-chemical and structural properties. NMR was used to determine the structure of the N-terminal domain (residues 1–110); this exhibits a ubiquitin-like fold with two additional helices which make the overall structure of this domain (UB1 domain) more elongated than other ubiquitin-like proteins (Serrano et al., 2007). NMR studies revealed that the highly acidic (51% Glu/Asp residues) C-terminal domain (residues 111–183; AC domain) is structurally disordered (Serrano et al., 2007).
3.4.2. Function
UB1 has high structural homology to Ras-interacting proteins such as the Ras-interacting domain (RID) of RALGDS, a member of the RA family and conservation of residues important for the interaction with Ras. Ras family proteins (RFPs) act as molecular switches that cycle between inactive GDP- and active GTP-bound states. RFPs control cell growth, motility, intracellular transport and differentiation. Ras plays a fundamental role in cell progression from phase G0 to G1 (Dobrowolski et al., 1994, Peeper et al., 1997) Molecular interactions that result in a Ras inactivation avoid cell progression to G1 phase. SARS-CoV and other coronaviruses such as MHV are able to induce cell arrest in G0/G1 phase during the lytic infection cycles for their own replication advantage (Chen and Makino, 2004, Yuan et al., 2005). Sars3b plays a role in this process (Yuan et al., 2005) and nsp3 may also be involved in arresting the cell cycle arrest in the G0 phase.
Additionally, UB1 has structural homology with ISG15, an interferon-induced protein constitutively present in higher eukaryotes. This protein conjugates with cellular targets as a primary response to interferon-α/β induction and other markers of viral or parasitic infection. High levels of this protein are essential for cellular antiviral response. It is known that ISG15 is able to inhibit virus replication by abrogating nuclear processing of unspliced viral RNA precursors. However, some viruses have developed a mechanism to avoid the expression of ISG15. For example, influenza B virus blocks its expression by means of NS1 protein in order to overcome the immune response. It is possible that the PL2pro domain of nsp3 may bind ISG15 and subvert the antiviral response of the cell.
The functional significance of RNA-binding by this domain is unknown. It possesses a ubiquitin fold, as does the domain N-terminal to the PLpro domain. We also do not know if this fact has any functional significance. Also, the predicted function of this domain based on its similarity to Ras-binding proteins and ISG15 remains to be experimentally validated.
NMR experiments indicated a ligand bound to UB1, which was identified as a small RNA fragment by mass spectrometry. NMR studies have identified the interacting molecular interfaces. UB1 of MHV has recently been shown to interact with the nucleoprotein, effectively tethering nsp3 to viral RNA during the replication process (Hurst et al., 2013). This activity does not require the AC domain that follows UB1 and is hypervariable.
3.5. ADRP/Macro1
3.5.1. Structure
The crystal structure of a construct consisting of residues 184–365 has been determined for SARS-CoV (Saikatendu et al., 2005), and the corresponding region has since been solved for several other coronaviruses (see Box 2 ). This region of nsp3 adopts a macro H2A domain fold. The putative active site and substrate-binding residues were conserved in its three structural homologues yeast Ymx7, Archaeoglobus fulgidus AF1521 and Er58 from E. coli, and its sequence homologue, yeast YBR022W, a known phosphatase that acts on ADP ribose-1”-phosphate (Appr-1”-p or ADRP). The notable exception is that proposed active site residue Asp90 in YMX7 is an alanine in both the SARS-CoV ADRP (Ala50) and AF1521 (Ala44). Histidine residues in both enzymes proximal to the terminal 1″ phosphate of the substrate (His45 in ADRP and His39 in AF1521) might therefore be involved in catalysis (Saikatendu et al., 2005). Alternatively, the predominant nucleophile in the catalytic site may actually be an Asp or Glu in the conformationally flexible loop 101NAGEDIQ107 in SARS-CoV and the corresponding region in other coronaviral ADRPs (Saikatendu et al., 2005). The former proposition was verified by site directed mutagenesis data in the HCoV-229E ADRP, which showed that residues Asn37, Asn40, His45, Gly44 and Gly48 are part of the active site in the SARS-CoV ADRP (Putics et al., 2005).
Box 2. Key nsp3 and nsp4 structures.
| Virus | Domain | Method | Accession | Reference |
|---|---|---|---|---|
| SARS-CoV | UB1 and Ac | NMR | 2IDY | Serrano et al. (2007) |
| HCoV-229E | ADRP | X-ray (2.0 Å) | 3EWR | Xu et al. (2009) |
| FCoV | ADRP | X-ray (3.9 Å) | 3JZT | Wojdyla et al. (2009) |
| IBV | ADRP | X-ray (2.0 Å) | 3EWP | Xu et al. (2009) |
| HCoV-NL63 | ADRP | X-ray (1.9 Å) | 2VRI | Awaiting publication |
| SARS-CoV | ADRP | X-ray (1.4 Å) | 2ACF | Saikatendu et al. (2005) |
| TGEV | PL1pro | X-ray (2.5 Å) | 3MP2 | Wojdyla et al. (2010) |
| SARS-CoV | UB2-PL2pro | X-ray (1.9 Å) | 2FE8 | Ratia et al. (2006) |
| SARS-CoV | SUD-N-M | X-ray (2.2 Å) | 2W2G | Tan et al. (2009) |
| SARS-CoV | SUD-M | NMR | 2JZF | Chatterjee et al. (2009) |
| SARS-CoV | SUD-C | NMR | 2KAF | Johnson et al. (2010) |
| SARS-CoV | NAB | NMR | 2K87 | Serrano et al. (2009) |
| FCoV | nsp4-CTD | X-ray (2.8 Å) | 3GZF | Manolaridis et al. (2009) |
3.5.2. Function
The SARS ADRP readily hydrolyzes the 1” phosphate group from Appr-1”-p in vitro demonstrating that it is an active enzyme (Saikatendu et al., 2005). Another group validated this finding: both the SARS ADRP and the human coronavirus HCoV-229E counterpart were shown to dephosphorylate Appr-1”-p to ADP-ribose in a highly specific manner, the enzyme having no detectable activity on several other nucleoside phosphates (Putics et al., 2005).
The role of an ADRP in the coronavirus life cycle may closely parallel that in the eukaryotic tRNA splicing pathway (Culver et al., 1993, Phizicky and Greer, 1993, Saikatendu et al., 2005, Snijder et al., 2003). In coronaviruses, an early post infection event is the transcription of a nested set of sub-genomic mRNAs. Each sub-genomic mRNA contains a short 5′-terminal ‘leader’ sequence derived from the 5′ end of the genome (Lai and Holmes, 2001a, Lai and Holmes, 2001b, Thiel et al., 2003). The fusion of the two noncontiguous RNA segments is a poorly understood process. It is thought to be achieved by a discontinuous step in the synthesis of the minus-strand and involves transcription regulatory sequences (Pasternak et al., 2001, Thiel et al., 2003). In eukaryotes, pre-tRNA splicing is initiated by cleavage at the splice site by an endonuclease. The resulting tRNA fragments are then ligated to yield mature tRNA that retains the 2′ phosphomonoester group at the splice site (Phizicky and Greer, 1993). Using NAD as an acceptor, a phosphotransferase removes the 2′ phosphate to yield ADP-ribose-1″-2″ cyclic phosphate (Culver et al., 1994). A cyclic phosphodiesterase then hydrolyzes Appr>p to yield Appr-1″-p (Culver et al., 1994) (Martzen et al., 1999). Finally, a phosphatase converts Appr-1″-p into ADP-ribose and releasing inorganic phosphate. While the equivalent for the cyclic phosphodiesterase appears absent in the SARS proteome, the Appr-1″-p phosphatase (SARS ADRP) and an endonuclease (nsp15) are present. Characterization of an Appr-1″-phosphatase-deficient HCoV-229E mutant revealed no significant effects on viral RNA synthesis and virus titer (Putics et al., 2005).
Egloff et al. (2006) suggested that ADRP may primarily be a poly-ADP-ribose binding (PAR-binding) module. PARylation occurs in compromised cells to trigger apoptosis. PAR polymerases (PARPs) are responsible for so tagging proteins. PARP is activated on recognizing nicked DNA, and it helps in DNA repair. It auto-PARylates itself, and in case of extreme DNA damage, gets overactivated and depletes the cell of its nucleotide pool. If ADRP binds PAR, then it can bind proteins that are PARylated, including PARP. Indeed, binding the latter may be most beneficial, since it can tether down this protein, slow down apoptosis, and prevent nucleotide depletion, prolonging viral replication and transcription in the infected cell.
The existence of the ADRP domain in all CoV nsp3s (as well as in several other viruses) argues for its critical role in the viral life cycle. Its function as an ADRP in recycling organic phosphate appears to be a dispensable function and does not correlate with the conservation of this domain. It appears that its role as an ADRP may be secondary to a more important role – such as its proposed role as a PAR-binding module. If so, the role of PAR-binding in the viral life cycle needs to be delineated experimentally.
3.6. SARS-unique domain
3.6.1. Structure
The region corresponding to residues 366 to 722 has been considered a domain unique to SARS-CoV and is called the SUD (SARS Unique Domain). The corresponding regions which are located just downstream of the ADRP domain in HCoV-NL63, PEDV, HCoV-229E (among alphacoronaviruses) and all Betacoronaviruses have no assigned domain prediction, but secondary structure prediction suggests the presence of an additional macrodomain fold (Chatterjee et al., 2009).
It has been demonstrated that the SUD may actually consist of three domains, termed by position: the N-terminal SUD-N, the middle SUD-M and the C-terminal SUD-C Deuterium exchange mass spectrometry data on a construct nsp3:451–651 initially appeared to support this notion, indicating that a well ordered domain exists from residues 523–651. Constructs representing nsp3:365–722 have been shown to be particularly susceptible to proteolysis (Stefanie Tech et al., 2004). Size exclusion chromatography and PFO-PAGE indicates that it forms a dimer in solution and 1D-NMR spectra show that is well-folded.
The structure of SUD-N and SUD-M has been solved and surprisingly each domain was found to contain a macrodomain fold that was a close structural match for the SARS-CoV ADRP/Macro1 domain despite a lack of detectable amino acid homology between these proteins (Tan et al., 2009). The presence of these additional macrodomain folds has also been confirmed by the NMR structure of the complete SUD (Johnson et al., 2010) and the NMR structure of SUD-M (Chatterjee et al., 2009). The SUD-C domain contained a novel fold that consisted of an antiparallel beta sheet (Johnson et al., 2010).
3.6.2. Function
All three of the domains that make up the SUD have been demonstrated to interact with nucleic acid in some way. The SUD-NM has a high affinity for G-rich sequences and G-quadruplexes (Tan et al., 2009), while the SUD-MC showed a general preference for purine nucleotides (Johnson et al., 2010). While the SUD-N and SUD-M domains bear a close structural resemblance to the SARS-CoV ADRP domain, neither domain has any demonstrable affinity for ADP-ribose (Tan et al., 2009). The amino acids responsible for SUD-M and SUD-C RNA binding have been mapped, and appear to fall near the region of SUD-M that corresponds to the active site in the structurally similar ADRP domain (Chatterjee et al., 2009). Together this suggests that the cluster of three macrodomains in SARS-CoV nsp3 arose through gene duplication and that the SUD may contribute to the function of nsp3 as an accessory to the viral replication process (Neuman et al., 2008).
3.7. UB2 and PL2pro
3.7.1. Structure
Unlike many coronaviruses that encode two papain-like protease, SARS-CoV has a single copy of papain-like cysteine protei ase (PL2pro) that cleaves polyprotein 1a at three sites at the N-terminus (177LNGG↓AVT183, 815LKGG↓API821, and 2737LKGG↓KIV2743) to release nsp1, nsp2, and nsp3, respectively (Harcourt et al., 2004, Thiel et al., 2003). SARS-CoV PL2pro is also a deubiquitinating enzyme; it efficiently disassembles diubiquitin and branched polyubiquitin chains, cleaves ubiquitin-AMC substrates, and has de-ISGylating activity (Chen et al., 2007b, Lindner et al., 2005). Thus, PL2pro may have critical roles not only in proteolytic processing of the replicase complex but also in subverting cellular ubiquitination machinery to facilitate viral replication. Thestructure of a PL2pro construct nsp3:723–1037 revealed a ubiquitin fold (residues 723–783; UB2) and a well-ordered papain-like protease catalytic domain (residues 784–1036; PL2pro) (Ratia et al., 2006). The catalytic domain adopts the canonical “thumb, palm and fingers” domain architecture. The thumb domain is formed by four prominent helices, the palm is made up of a six-stranded β-sheet that slopes into the active site, which is housed in a solvent-exposed cleft between the thumb and palm domains, and a four-stranded, twisted, anti-parallel β-sheet makes up the “fingers” domain. Two β-hairpins at the fingertips region contain four cysteine residues, which coordinate a zinc ion with tetrahedral geometry. Mutational analysis of the zinc-coordinating cysteines of SARS-CoV PLpro, that zinc-binding ability is essential for structural integrity and protease activity (Barretto et al., 2005). PL2pro has several structural homologues from the cysteine protease superfamily, the most significant being USP14 and HAUSP, both of which are cellular DUBs. The active site of PLpro consists of a catalytic triad of cysteine, histidine, and aspartic acid residues, consistent with catalytic triads found in many PLpro domains. The recent structure of the TGEV PL1pro demonstrates that the coronavirus-like PLpro folds have a common architecture (Wojdyla et al., 2010), and likely arose through gene duplication.
3.7.2. Function
It has been demonstrated that an LXGG motif at the P4–P1 positions of the substrate is essential for recognition and cleavage by PL2pro (Barretto et al., 2005, Han et al., 2005). There appear to be no preferences for the P′ positions or for residues N-terminal to P4. It is not surprising then that PLpro is able to cleave after the four C-terminal residues of ubiquitin, LRGG. As predicted by Sulea et al. (2005) SARS-CoV PL2pro (nsp3 residues 1507–1858) does possess de-ubiquitinating activity (Barretto et al., 2006, Lindner et al., 2005) in addition to its better-known cysteine protease activity. The specific deubiquitinating enzyme inhibitor, ubiquitin aldehyde, inhibited its activity at a Ki of 210 nM.
Interestingly, a number of cellular deubiquitinases, including full-length USP14 and Ubp6, possess an N-terminal ubiquitin-like domain. Although the significance of this domain in these proteins is not well established, it has been demonstrated that the presence of the ubiquitin-like domain in USP14 and Ubp6 serves a regulatory function by mediating interactions between these deubiquitinases and specific components of the proteasome (Hu et al., 2005, Leggett et al., 2002). Comparisons of deubiquitinase activities between wild-type and mutant Ubp6 lacking the Ubl domain reveal that these associations are responsible for a 300-fold increase in catalytic rate and serve to activate the enzyme (Leggett et al., 2002). It is intriguing to consider that the Ubl-like domain of PLpro may instead act as a sort of “decoy” or “lure” to detract cellular ubiquitinating enzymes from other viral proteins, or it may mediate protein–protein interactions between the replicase components.
While the role of PL2pro in polyprotein processing is well understood, the physiological significance of its deubiquitinating activity in the viral replication cycle is still not completely clear. However the conserved structural protein E is readily ubiquitinated in infected cells, suggesting that deubiquitination may be important in the assembly process (Alvarez et al., 2010). There is now mounting evidence that PL2pro interferes with interferon transcriptional activation pathways by inactivating TBK1, blocking NF-kappaB signaling and preventing translocation of IRF3 to the nucleus (Frieman et al., 2009, Wang et al., 2011, Zheng et al., 2008).
3.8. NAB and GSM
3.8.1. Structure
The region between the PL2pro domain and the transmembrane region of nsp3 does not show sequence similarity to any known domain. Disorder prediction programs Disembl 1.4, FoldIndex and RONN predict a long disordered stretch of residues in the central region of the segment in SARS-CoV, suggesting that it may be a consist of two domains with the second domain beginning from around residue 1226. A NMR structural study confirmed that the NAB is an independently folded functionally active unit. The solved region comprised residues 1066 to 1181 and the constructs nsp3(1066–1203) and nsp3(1035–1181). This globular domain represents a new fold, with a parallel four-strand β-sheet holding two α-helices of three and four turns that are oriented antiparallel to the beta-strands. Two antiparallel two-strand β-sheets and two 310-helices are anchored against the surface of this barrel-like molecular core. A positively charged patch on the molecule surface was identified by NMR ascontaining the nucleic acid binding activity.
3.8.2. Function
The NAB has been demonstrated to form homodimers upon incubation at 37 °C (Neuman et al., 2008), and displayed a high affinity for nucleic acid. While the NAB was able to interact with both single-stranded and double-stranded nucleic acids, cooling the protein-nucleic acid complex released single-stranded RNA, demonstrating that the NAB may function as a ssRNA binding protein with RNA chaperone-like activity (Neuman et al., 2008). Little else is known about the function of NAB in the viral replication cycle, or about the structure and function of the GSM domain that follows or the conserved hydrophobic, non-transmembrane region that immediately precedes the first transmembrane region of nsp3.
3.9. TM, ZF and Y
3.9.1. Structure
The region of nsp3 after the TM domain is highly conserved in all CoVs, but this region has not been structurally characterized yet. An Fold and Function Annotation System search (FFAS; Jaroszewski et al., 2005) using the sequence from the SARS-CoV RBD to the end of nsp3 domain, TM domain and Y domain reveals three of seven significant hits (with expect values of −8 or better) to viral RdRp proteins, which may hint at the evolutionary origin of nsp3, which comprises nearly one fifth of most coronavirus genomes. The level of conservation in the Y domain in particular approaches levels consistent with the other enzymatic domains of nsp3, and exceeds the conservation of other domains that are believed to be non-enzymatic (Neuman et al., 2008).
3.9.2. Function
It appears that domains from PL2pro to the Y domain have not undergone significant deletion or rearrangement during coronavirus evolution, while other nsps like nsp1, nsp2, and the N-terminal regions of nsp3 clearly have evolved by duplication and deletion of domains (Neuman et al., 2008). Therefore nsp3 is more likely to confer a basic and important function in a variety of hosts. UB1, SUD and RBD bind RNA, and ADRP is part of the RNA-processing machinery. If not for the proteinase(s), nsp3 would be classified exclusively as an RNA binding/modifying protein. These regions have been shown to change the localization of nsp4 (Hagemeijer et al., 2011), and cause a membrane proliferation phenotype in transfected cells (Angelini et al., 2013).
The topology of nsp3 leaves only one domain, ZF, on the lumenal side of the membrane. If nsp3 participates directly in the membrane pairing exhibited in cells transfected with SARS-CoV nsp3 and nsp4 (Angelini et al., 2013), then the ZF domain likely participates in this interaction.
3.10. Nsp4
3.10.1. Structure
Nsp4 is a transmembrane protein with four transmembrane helices and an internal C-terminal domain (Oostra et al., 2007). Coronavirus nsp4 is approximately 500 amino acids in length, and is the only part of the viral polyprotein that is released after processing by both the PLpro and Mpro. The location and topology of the four transmembrane regions has been mapped (Oostra et al., 2007).
The C-terminal domain of nsp4 is conserved in all known coronaviruses, but deletion of this domain from a MHV infectious clone resulted in only slightly attenuated virus growth, consistent with a non-essential function virus (Sparks et al., 2007). Mutation of the two glycosylation sites in nsp4, however, led to defective DMV formation and attenuation (Gadlage et al., 2010). The structure of the C-terminal domain of FCoV nsp4 has been reported (Fig. 4). It consists of two small antiparallel β-sheets and four α-helices (Manolaridis et al., 2009).
3.10.2. Function
SARS-CoV Nsp4 is an essential component for the formation of viral double-membrane vesicles (Angelini et al., 2013). Intracellular expression studies have demonstrated a biological interaction between the carboxyl-terminal region of MHV nsp3 (Hagemeijer et al., 2011), and co-expression of full-length SARS-CoV nsp3 and nsp4 results in extensive membrane pairing, in which the paired membranes are held at the same distance as observed in authentic DMVs (Angelini et al., 2013). Nsp4 has also been shown to interact with nsp2 in a yeast two-hybrid screen (von Brunn et al., 2007), and to interact with other nsp4 molecules in cells (Hagemeijer et al., 2011). Nsp4 has been shown to cause aberrant DMV formation upon mutation, leading to a loss of nsp4 glycosylation (Gadlage et al., 2010, Sparks et al., 2007)
3.11. Nsp5
3.11.1. Structure
Mpro, a chymotrypsin like protease is encoded within the mature polypeptide nsp5. It emerges by self trans-cleavage at nsp4/5 and 5/6 boundaries at residues 3238VLQ↓SGF3243 and 3544TFQ↓GKF3549 of polyprotein polyprotein 1a/1ab. It belongs to the C30 family of endopeptidases and is responsible for cleavage at 11 sequence specific sites within polyprotein 1a/1ab. The resultant “mature” protein products (nsp4–16) assemble into components of the replication complexes. Given its paramount importance in replicase processing and therefore its role in viral replication, this protein has been extensively studied both from structural and functional perspectives (reviewed in Hilgenfeld et al., 2006, Ziebuhr et al., 2000).
Based on both structure and sequence characteristics, nsp5 can be divided into three domains. This domain prediction has been confirmed by the numerous crystal structures. It is conserved in all coronaviruses, indeed in all three nidoviral groups and several other RNA viruses that share common polyprotein processing scheme (Ziebuhr et al., 2000). The sequence is related to chymotrypsin-like protease superfamily of endopeptidases.
Structures of Mpro from five different coronaviruses have been reported (Box 3 ; Fig. 4), and structures for HCoV-NL63 and HKU4 viruses have been released prior to publication. All these structures show stringent conservation of a three-domain tertiary architecture and a partially surface exposed catalytic core.
Box 3. Key nsp5 structures.
| Virus | Domain | Method | Accession | Reference |
|---|---|---|---|---|
| HCoV-229E | Mpro | X-ray (2.5 Å) | 1P9S | Anand et al. (2003) |
| HCoV-HKU1 | Mpro | X-ray (2.5 Å) | 3D23 | Zhao et al. (2008) |
| HCoV-HKU4 | Mpro | X-ray (1.6 Å) | 2YNB | Awaiting publication |
| IBV | Mpro | X-ray (2.0 Å) | 2Q6F | Xue et al. (2008) |
| HCoV-NL63 | Mpro | X-ray (1.6 Å) | 3TLO | Awaiting publication |
| SARS-CoV | Mpro | X-ray (1.9 Å) | 1UJ1 | Yang et al. (2003) |
| TGEV | Mpro | X-ray (2.0 Å) | 1LVO | Anand et al. (2002) |
The first two domains (N terminal 8–101 amino acids of domain 1 and 102–184 form domain 2; (Yang et al., 2003)) are duplicated closed β-barrels of type with n = 6 and S = 8 (Murzin et al., 1995) in which the strands are arranged in a greek key motif – all hallmarks of trypsin-like serine protease fold as defined in SCOP structure classification database and has been placed under “viral cysteine protease of the trypsin fold” family. Close homologs of this family include picornvirus-like 3C cysteine proteases.
The critical role of the first seven residues at the N terminus in dimerization and its close proximity to the active site results in this enzyme to be an obligate dimer, although modification of the termini appears to modulate higher order oligomerization (Zhang et al., 2010). Deletion of the first five amino acids results in complete inactivation of this enzyme. The helical C-terminal domain III mediates homodimerization of coronaviral Mpro proteases. This interaction is believed to be important for its trans-proteolytic activity. The active site is located at the interface of the two β-barrels with the catalytic residues H41 and C144 being contributed by domains 1 and 2 respectively.
The active site is located in a substantially solvent exposed cleft that is located between the two β-barrels. Structures of Mpro complexed with peptide/peptidomimetic substrates reveal that the substrate peptides occupy the S1 and S2 pockets in an anti-parallel β-sheet orientation to the two interacting β-strands of the enzyme active site – a feature seen in subtilisin and related serine proteases. Sequence specificity is mainly determined by the S1 binding pocket. All coronaviral Mpro recognize a glutamine as the P1 residue, a feature that is largely determined because of structural complimentarity. The wall of P1 pocket is lined by residues His 163, Phe 140 (that contribute sidechains) and Met-165, Glu-166, and His-172 contributing main chain atoms (Anand et al., 2003, Yang et al., 2003). Comparison of ligand bound and apo structures have shown that unlike the S1 pocket (which houses the P1 residue) that largely remains unchanged, the S2 pocket undergoes significant conformational changes upon ligand binding. The specificity for leucine being the most common residue at P2 position (see Table 1 ) was structurally explained by a ligand fit-induced structural ordering of the S2 pocket by Yin and coworkers (Yin et al., 2007).
Table 1.
Cleavage sites of SARS Mpro using Tor2 as the reference SARS strain.
| …P3,P2,P1↓P-1,P-2,P-3…. | …P3,P2,P1↓P-1,P-2,P-3… | ||
|---|---|---|---|
| Nsp4/5 | 3238VLQ↓SGF3243 | Nsp11/12 | 4367LMQ↓SAD4372 |
| Nsp5/6 | 3544TFQ↓GKF3549 | Nsp12/13 | 5299VLQ↓AVG5304 |
| Nsp6/7 | 3834TVQ↓SKM3839 | Nsp13/14 | 5900TLQ↓AEN5905 |
| Nsp7/8 | 3917TLQ↓AIA3922 | Nsp14/15 | 6427RLQ↓SLE6432 |
| Nsp8/9 | 4115KLQ↓NNE4120 | Nsp15/16 | 6773KLQ↓ASQ6778 |
| Nsp9/10 | 4228RLQ↓AGN4233 |
3.11.2. Function
The most well-studied of all SARS proteins, nsp5, also known as the main protease (Mpro) or in older literature as the chymotrypsin-3C like proteast (3CLpro) is the primary molecule responsible for cleaving and maturation of SARS polyprotein Polyprotein 1a and Polyprotein 1ab. As part of its proteolytic activity, it is destined to interact with all the non-structural proteins from nsp4 to 16, presumably near its catalytic site.
It cleaves the polyproteins at 11 sequence specific cleavage sites (Table 1). The other three (nsp1/2, 2/3 and 3/4) are cleaved by the PLpro. It is specific for a glutamine followed by a small hydrophobic residue, usually an alanine or a glycine, sometimes a serine.
Hsu and others (Hsu et al., 2005) have described a four step process by which a mature catalytically competent Mpro head-to-head dimer is formed from multiple copies of polyprotein Polyprotein 1a. (Lin et al., 2004) have dissected its trans cleavage (by ELISA based assays) and cis peptide cleavage activities by cell based assays.
Unlike the canonical chymotrypsin-like proteases, Mpro houses a conserved catalytic dyad–residues His 41 and Cys 45 and not the more common catalytic triad of cysteine proteases. Instead, from the structures, it is apparent that the role of the third catalytic residue has been taken over by a conserved water molecule that often lies within hydrogen bonding distance of H41. The role of a conserved “catalytic” water molecule has long been recognized in many proteolytic cleavage schemes, especially in serine proteases (Perona et al., 1993). The mechanism of substrate hydrolysis is however similar to its cysteine protease cousins, in which the acylation (the first step) is performed by His 41, which acts as a general base and helps the sulfur atom of the catalytic cysteine residue's sidechain to carry out the nucleophilic attack on the backbone C O group of the peptide bond to be cleaved (Yin et al., 2007). The first transition state is a tetrahedral intermediate (TI-1). The next step of the cleavage cycle is the implosion (collapse) of this transition state and leaving of the C-terminal half of the peptide product from the active site. At this stage, the other portion of the peptide substrate is covalently bound to the enzyme via a thio-ester linkage. In the other half of the reaction cycle (the de-acylation step), a water molecule that is activated by His41 acts as a nucleophilic hydroxyl ion (OH−), attacks the carbonyl atom of the thioester and releases the N-terminal half of the peptide product, thus regenerating the cysteine. Many excellent and exhaustive studies have based this mechanism of catalysis and the structure of the catalytic site architecture to design several different classes of peptidomimetic inhibitors targeted against Mpro of coronaviruses (including SARS) and other pathogenic viruses. Several Mpro inhibitors have also been structurally characterized (Akaji et al., 2011, Bacha et al., 2008, Chu et al., 2006, Chuck et al., 2013, Grum-Tokars et al., 2008, Lee et al., 2007, Lee et al., 2009, Lee et al., 2005, Shan and Xu, 2005, Shao et al., 2007, Turlington et al., 2013, Verschueren et al., 2008, Wei et al., 2006, Yang et al., 2006, Yang et al., 2003, Yang et al., 2007, Zhang et al., 2010, Zhu et al., 2011).
3.12. Nsp6
3.12.1. Structure and function
The membrane topology of nsp6 has been determined (Oostra et al., 2008). Although SARS-CoV nsp6 is predicted by TMHMM2.0 (Krogh et al., 2001) to contain seven transmembrane regions, only six of these function as membrane-spanning helices. The presence of additional non-transmembrane hydrophobic domains near authentic transmembrane domains is a common theme running through the DMV making proteins nsp3, nsp4 and nsp6. IBV and SARS-CoV nsp6 have been shown to activate autophagy, inducing vesicles containing Atg5 and LC3-II (Cottam et al., 2011). MHV Nsp6 is relocalized when it is co-expressed with nsp4 (Hagemeijer et al., 2012), suggesting that the two proteins interact. Nsp6 has also been shown to interact with nsp2, nsp8, nsp9 and sars9b via yeast two-hybrid assays (von Brunn et al., 2007).
3.13. Nsp7 and Nsp8
See Box 4 .
Box 4. Key nsp7 and nsp8 structures.
| Virus | Domain | Method | Accession | Reference |
|---|---|---|---|---|
| SARS-CoV | nsp7 | NMR | 1YSY | Peti et al. (2005) |
| SARS-CoV | nsp7 + nsp8 | X-ray (2.6 Å) | 2AHM | Zhai et al. (2005) |
| FCoV | nsp7 + nsp8 | X-ray (2.4 Å) | 3UB0 | Xiao et al. (2012) |
3.13.1. Structure
Nsp7 and nsp8 are two mature proteins that emerge due to cleavage of polyprotein Polyprotein 1a at 3834TVQ↓SKM3839 (nsp6/7), 3917TLQ↓AIA3922 (nsp7/8) and 4115KLQ↓NNE4120 (nsp8/9) boundaries. SARS-CoV Nsp7 and nsp8 self associate and form a large sixteen-subunit supercomplex that has been directly implicated in replication (Bartlam et al., 2005, Zhai et al., 2005).
The structure of nsp7 was first determined by NMR (Peti et al., 2005) which revealed the presence of a four helical bundle arranged in a novel sheet-like arrangement, with three of the helices arranged anti-parallel to each other while the fourth oriented at an angle to the bundle. Much of the structure derived functional information about nsp7 and nsp8 came from the study by Rao and co-workers (Bartlam et al., 2005, Zhai et al., 2005) who determined the 2.4 Å resolution crystal structure of the nsp7/8 supercomplex (Fig. 5 ). Eight subunits of nsp7 and nsp8 each form a tight hexadecameric complex. In this complex, nsp7 reveals a tertiary structure that is similar to its solution structure with the minor deviation in that the fourth helix is oriented at a slightly different angle and is more ordered. This is possibly due to its existence as a complex with nsp8 in crystals. SARS-CoV Nsp8 adopts two major conformations described as the “golf club” and the “bent golf club” fold, which has an extended long shaft domain with three helices (one of which is very long) and a globular core at the C terminus (Zhai et al., 2005).
Fig. 5.
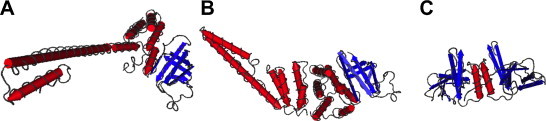
Structures of the coronavirus replicase proteins nsp7, nsp8 and nsp9. Structures for the nsp7 and nsp8 heterodimers from SARS-CoV (2AHM; panel A) and FCoV (3UB0; panel B) are shown to illustrate the distinctive structures of these proteins. The structure of homodimeric SARS-CoV nsp9 is taken from PDB entry 1QZ8 and is shown in panel C.
The supercomplex, which is formed by a stoichiometric association of eight subunits each of nsp7 and nsp8, is a hollow cylindrical structure with a central channel, and two handles (one on either side of the structure) has a very distinct bimodal distribution of electrostatic charge on its surface in which the outer skin of the complex is composed of predominantly negatively charged residues while the inner core channel is lined with positively charged sidechains. RNA binding studies using gel mobility shift assays suggest that the function of the central positively charged channelis to preferentially guide dsRNA through the supercomplex either toward the polymerase (nsp12) or away from it during replication. Mutagenesis experiments indicate that residues R26 and K32 of nsp7 and K77, R80, K63, R84 and R85 are among those that line the channel and are primarily responsible for this translocation (Zhai et al., 2005).
The FCoV nsp7 and nsp8 proteins were recently shown to adopt similar structures to the SARS-CoV equivalents (Fig. 5), but in a distinctive 2:1 protein complex (Xiao et al., 2012). No known homologs exist for either of these proteins outside of coronaviridae lineage within statistical limits of significance.
SCOP places nsp7 as a member of the “immunoglobulin/albumin-binding domain-like” fold, with three of its helices arranged as a bundle and having an overall topology that mirrors spectrin-like fold. The globular core domain of Nsp8 golf-club has been defined as a new fold.
3.13.2. Function
The most striking functional insight on the supercomplex has been obtained by Canard and co-workers who have shown that coronaviral nsp8 encodes a second non-canonical RNA polymerase activity (Imbert et al., 2006). This template-dependent oligonucleotide-synthesizing activity, which is dependent on Mn2+ or Mg2+ cations, was found to be preferentially enhanced by internal 5′-(G/U)CC-3′ trinucleotides that are present on RNA templates and were used to initiate the synthesis of complementary oligonucleotides. Typical extension products were found to be <6 residues long. Nsp8 effectively polymerized poly(rC) and oligo(rC15) templates and poly(rU) to a weaker extent but not poly(rA). This accessory polymerase, which is both catalytically weaker and has a lesser fidelity than the main viral RdRp (nsp12), was potently inhibited with 3′-dGTP and to a lesser extent by ddGTP and 2′-O-methyl-GTP suggesting an avenue for possible therapeutic inhibition. The primase activity of nsp8 was blocked by N-terminal extension of nsp8 with peptides other than nsp7 (te Velthuis et al., 2012).
Initial mutagenesis experiments on nsp8 implicated four residues K58, R75, K82 and S85 to be essential for polymerization, but a more recent study also identified a magnesium ion binding site at D50 and D52 corresponding to functional a D/E–x–D/E motif (te Velthuis et al., 2012). All of these residues localize on the long α-helix (the stem of the golf club) and map onto one of several dimer interface regions of the supercomplex. The main function of nsp8 overall appears to be to catalyze the synthesis of short stretches of RNA primers that can be utilized by the primer-dependent main SARS polymerase nsp12. Using dual labeling immunofluorescence microscopic studies, Prentice and co-workers have shown that nsp8 co-localizes along with nsp2 and nsp3 in cytoplasmic complexes, which also contain the protein LC3, which is a general marker for autophagic vacuole (Prentice et al., 2004).
Astriking observation by Masters and co-workers (Züst et al., 2007) provided compelling evidence that nsp8 specifically interacts with a molecular switch composed of a bulged stem-loop and an RNA pseudoknot that exists in the 3ʹ untranslated region of MHV, indeed most coronaviral genomes. These studies are aiding in developing a model that explains the origins and initiation of negative strand genomic RNA synthesis (te Velthuis et al., 2012).
Yeast two-hybrid screening and co-immunoprecipitation experiments, which were subsequently confirmed by in vivo co-localization studies by Lal and co-workers have shown that nsp8 interacts with sars6 gene product as well, thereby implicating this accessory protein in the replication complex (Kumar et al., 2007). In a proteome-wide yeast two-hybrid screening study nsp8 was found to be one of the most promiscuous non-structural protein, which interacted with no less than 13 out of 29 SARS proteins tested (von Brunn et al., 2007).
3.14. Nsp9
See Box 5 .
Box 5. Key nsp9, nsp10 and nsp11 structures.
| Virus | Domain | Method | Accession | Reference |
|---|---|---|---|---|
| HCoV-229E | nsp9 | NMR | 2J97 | Ponnusamy et al. (2008) |
| SARS-CoV | nsp9 | X-ray (2.6 Å) | 1QZ8 | Egloff et al. (2004) |
| SARS-CoV | nsp10 | X-ray (1.8 Å) | 2FYG | Joseph et al. (2006) |
| SARS-CoV | nsp10 + nsp11 | X-ray (2.1 Å) | 2G9T | Su et al. (2006) |
3.14.1. Structure
Two groups have independently determined the structure of nsp9 (Egloff et al., 2004, Sutton et al., 2004). It adopts a β-barrel fold with a C-terminal α-helix (Fig. 5). The structure of HCoV-229E nsp9 was subsequently solved, and found to contain a similar fold (Ponnusamy et al., 2008). Nsp9 of both viruses was found to be dimeric, although the dimer interface was stabilized by an additional disulfide bond in HCoV-229E nsp9.
3.14.2. Function
Nsp9 binds ssRNA and dsDNA in a concentration dependent manner (Egloff et al., 2004, Sutton et al., 2004). Optimal binding occurs with 45-mer oligonucleotides, consistent with binding occurring by the nsp9 dimer wrapping the DNA fragment around itself once (Egloff et al., 2004). Since RNA-binding is not sequence specific, nsp9 may protect nascent ssRNA from nucleases during viral RNA synthesis, given its natural abundance in the infected cell (Egloff et al., 2004). Nsp9 colocalizes in the perinuclear region along with other components of the replication complex (Bost et al., 2000). While the precise role of nsp9 in viral replication is not yet clear, Minkis and co-workers investigated the role of the dimer interface demonstrated that SARS-CoV nsp9 is essential for efficient viral growth (Miknis et al., 2009).
3.15. Nsp10–11
3.15.1. Structure
The tenth coronavirus nonstructural protein constitutes the carboxyl-terminal conserved domain of replicase polyprotein polyprotein 1a, and a region of this polyprotein homologous to nsp10 can be readily identified in all coronaviruses (Joseph et al., 2006). Originally described as a growth factor-like protein on the basis of high cysteine content and sequence homology (Gorbalenya et al., 1989), nsp10 is a highly conserved component of the coronavirus replicase machinery. Reciprocal BLAST searches using various nsp10 homologs identify a region of homology near the carboxyl terminus of polyprotein 1a in more distantly related nidoviruses such as torovirus and the newly identified white bream virus (Schutze et al., 2006). However, no region of nsp10 homology has been noted to date in the ronivirus or arterivirus lineages of the Nidovirales. Bioinformatic analysis of nsp10 does not yield any consistent matches to conserved enzymatic signatures. Thus, the profile of nsp10 more likely fits a role as an auxiliary replicase component, rather than an essential replicase enzyme.
X-ray crystallography structures (Bhardwaj et al., 2006, Joseph et al., 2006) have revealed that nsp10 is a single domain protein consisting of a pair of antiparallel N-terminal helices stacked against an irregular β-sheet, a coil-rich C terminus, and two Zn fingers. As such, nsp10 represents a novel fold, as might be expected from the lack of protein or domain homology to other known proteins. Bacterially expressed nsp10 binds generic single-stranded and double-stranded nucleic acids with micromolar affinity (Joseph et al., 2006).
Within the polyprotein, coronavirus nsp10 is followed by a short peptide of highly variable sequence that maps to the region of the genomic RNA where the ribosomal frameshift signal leading to the translation of the replicase enzyme cluster in open reading frame 1b is located. In SARS-CoV, nsp11 is a 13-residue peptide which can theoretically be processed from the C-terminus of polyprotein 1a, however processing of nsp11 has not been demonstrated in infected cells. The structure of the uncleaved nsp10–11 polypeptide showed some differences in oligomerization and crystal packing, but little difference in the core nsp10 structure (Bhardwaj et al., 2006). In that study the nsp11 density was flexibly disordered (Bhardwaj et al., 2006). Thus, nsp11 more likely forms part of an essential translation reading frame shift mechanism, and is unlikely to significantly influence the function of nsp10. Synthesized nsp11 peptide is fairly insoluble in aqueous buffers (J. Joseph, unpublished data).
3.15.2. Function
The first assignment of a function to nsp10 was noted from a study of MHV strains that contained temperature-sensitive lesions affecting viral RNA synthesis (Sawicki et al., 2005). It was further noted that this defect in nsp10 could not be compensated in cells by co-infection with viruses harboring temperature-sensitive lesions in nsp4 or nsp5, suggesting that coronavirus polyprotein 1a (at least from nsp4 onward) forms a single functional unit important for coronavirus discontinuous negative-strand RNA synthesis (Sawicki et al., 2005). Mutagenesis studies have confirmed the importance of nsp10 for general RNA synthesis and for controlling the ratio of subgenomic to genomic RNA (Donaldson et al., 2007b). Deletion of nsp10 or rearrangement of the genes encoding nsp7–10 completely inhibited virus growth, while alteration of the Mpro cleavage site between nsp9 and nsp10 reduced viral growth (Deming et al., 2007). An unexpected finding was that the temperature sensitive lesion in nsp10 correlates with a severe inhibition of Mpro activity at the non-permissive temperature (Donaldson et al., 2007a). From these results it appears clear that the function of nsp10 is closely tied to viral RNA synthesis. Nsp10 is now known to form part of the viral mRNA cap methylation complex (Bouvet et al., 2010) which is discussed below with the viral methyltransferase subunits.
3.16. Nsp12
3.16.1. Structure
Nsp12 is the cleavage product of the replicase polyprotein polyprotein 1ab and is produced by the action of Mpro. It is 932 residues long in the SARS-CoV. Immunoblotting and immunofluorescence analyses indicated that full length, 106-kDa, RdRp protein is present in infected Vero cells and is part of the viral replication cycle of (Prentice et al., 2004).
Cheng et al. (Cheng et al., 2005) expressed nsp12 in E. coli and found that during purification, the protein was cleaved into three stable fragments (1–110, 111–368, 369–932), which may correspond to separate domains.
Nsp12 has high sequence identity with other coronavirus RdRps, but very low similarity to other viral polymerases. Based on manual sequence alignments with other the RdRps of poliovirus, rabbit hemorrhagic disease virus, hepatitis C virus, reovirus and bacteriophage Φ6 polymerases, as well as HIV-1 reverse transcriptase, Xu et al. (2003) were able to identify conserved sequence motifs and, by homology, to assign functions to these regions in the catalytic domain of the polymerase.
Xu et al. (2003) built a three-dimensional model of the catalytic domain of nsp12 (PDB ID 1O5S), based on alignments of conserved motifs with other viral polymerase proteins. Based on the model of SARS-CoV nsp12, the catalytic domain forms the canonical “palm and fingers” domain. The fingers subdomain is predicted to span residues 376–584 and 626–679 and is predicted to consist of α-helices in the base and β-strands and coils at the tip (Xu et al., 2003). Similar to the HCV and RHDV RdRps, its fingers subdomain also contains an N-terminal portion (residues 405–444) that forms a long loop starting from the fingertip that bridges the fingers and thumb subdomains. The palm subdomain of SARS-CoV nsp12 (residues 585–625 and 680–807) forms the catalytic core and contains the four highly conserved sequence motifs (A–D) found in all polymerases and a fifth motif (E) unique to RdRps and RTs (Poch et al., 1989). The core structure of the palm subdomain is well conserved across all classes of polymerases. It consists of a central three-stranded β-sheet flanked by two α-helices on one side and a β-sheet and an α-helix on the other. Residues forming the catalytic active site are found within motifs A and C.
The structures of RdRps of a few non-CoVs have been determined – for example, those of hepatitis C virus, poliovirus, rabbit hemorrhagic disease virus, reovirus, bacteriophage Φ6 and HIV-1 (see Xu et al., 2003). However, there is very low sequence similarity between these structures and CoV RdRps. Xu et al. (2003) constructed a comparative molecular model for SARS-CoV RdRp based on these structures, using manual sequence alignments anchored by conserved sequence motifs shared by all RdRps and reverse transcriptases.
3.16.2. Function
The RdRp is the central enzyme in the multi-component viral replicase complex that replicates the viral RNA genome (Bost et al., 2000, Brockway et al., 2003) would contain several other viral proteins as well. The replicase transcribes (i) full-length negative and positive strand RNAs; (ii) a 3′-co-terminal set of nested subgenomic mRNAs that have a common 5′ ‘leader’ sequence derived from the 5′ end of the genome; and (iii) subgenomic negative strand RNAs with common 5′ ends and leader complementary sequences at their 3′ ends (Thiel et al., 2003). Full-length nsp12 has RdRp activity. The “catalytic” 64 kDa domain and the N-terminal 12 kDa domain form a complex that possesses comparable RdRp activity. However, the 64 kDa domain in isolation has no activity. Cheng and coworkers suggest that the N-terminal domain is required for polymerase activity possibly via involvement in template-primer binding (Cheng et al., 2005). Snijder and co-workers were able to confirm that the full-length nsp12 has robust, primer-dependent RNA polymerase activity (te Velthuis et al., 2010), a finding generally confirmed by the later study of Ahn et al. (2012).
There has been some success in the use of inhibitors of viral polymerases as therapeutics. Hence, the RdRp is an attractive drug target. Lu et al. used a short RNAi targeting the RdRp and found that it significantly reduced plaque formation of SARS-CoV in Vero-E6 cells (Brockway et al., 2004). However, such an approach would affect expression of the entire polyprotein 1a/ab and would not be specific to the RdRp. He et al. (2004) showed that aurintricarboxylic acid could potently reduce viral titer by more than 1000-fold when added to cells in culture. The same group subsequently suggested, by analogy to other RNA polymerases, that this compound may act on nsp12, and performed docking studies to predict the site of binding (Yap et al., 2005).
The main RdRp would be predicted to interact either directly or indirectly with several other viral proteins, including the nsp3–6 scaffold proteins, the nsp10–16 methylation complex and the nsp 7–8 primase. Adedeji and coworkers showed that SARS-CoV nsp12 enhances the helicase activity of nsp13 by two-fold (Adedeji et al., 2012). Nsp12 interacts with nsp8, nsp13, sars3a and sars9b according to yeast two-hybrid experiments and with nsp8 by co-immunoprecipitation experiments. Previous immunolocalization and interaction studies in MHV have also indicated that in vivo, nsp12 may act in concert with numerous other viral proteins – counterparts of SARS nsp1, 2, 5, 8, 9, 13 and sars9a (Bost et al., 2000, Brockway et al., 2003, von Brunn et al., 2007).
3.17. Nsp13
3.17.1. Structure and function
Nsp13 is a helicase capable of unwinding both RNA and DNA duplexes in a 5’-to-3’ direction with high processivity(Ivanov et al., 2004, Tanner et al., 2003). It possesses deoxynucleoside triphosphatase (dNTPase) activity against all standard nucleotides and deoxynucleotides, and also RNA 5′-triphosphatase activity which may be involved in the first step of formation of the 5′ cap structure of the viral mRNAs (Ivanov et al., 2004, Tanner et al., 2003). The two hydrolase activities likely have a common active site, which contains a canonical Walker A NTPase-like motif (Ivanov et al., 2004). Since NTPase/helicase proteins are considered essential for viral viability (Kadare and Haenni, 1997), they are potential drug targets (Anand et al., 2003, Holmes, 2003). Promising inhibitors are in trials for herpes simplex virus (Kleymann, 2003) and hepatitis C viral infections (Borowski et al., 2002). Several SARS-CoV helicase inhibitors – bananin derivatives – have been identified (Tanner et al., 2005).
While the structure of nsp13 has not yet been determined, the protein has been modeled based on the E. coli Rep ATP-dependent DNA helicase (PDB accession 1UAA). The model of the helicase domain at the position 80–568 of SARS-CoV nsp13 has been deposited (PDB accession 2G1F; Bernini et al., 2006). The N-terminus of nsp13 contains conserved cysteine and histidine residues that are probably homologous with the metal binding domains at the N-terminus of arterivirus helicases, which coordinate up to four Zn2+ (van Dinten et al., 2000).
IBV Nsp13 also has a proposed role in modulating the host response, although it is not yet clear whether this role is conserved in other coronaviruses (Xu et al., 2011). Overexpression of nsp13 led to cell cycle arrest by interfering with DNA polymerase delta, though the report did not determine whether this effect occurs normally during viral infection.
3.18. Nsp14
3.18.1. Structure
SARS-CoV nsp14 is 527 residues long and is multifunctional. The structure of nsp14 has not yet been solved. All coronaviruses have homologues of nsp14, all containing an N-terminal domain with 3′ to 5′ exonuclease motifs I (DE), II (D) and III (D) within the first ∼280 residues of the protein (Moser et al., 1997, Thiel et al., 2003, Zuo and Deutscher, 2001) and a C-terminal cap N7-methyltransferase domain (Chen et al., 2013, Minskaia et al., 2006). Compared to other RNA ExoNs, CoV and torovirus ExoNs have an additional putative Zn finger between Exo I and II motifs (Thiel et al., 2003).
The coronaviral N7-methyltransferase is unusual in that it is physically and functionally linked with the exoribonuclease domain (Chen et al., 2013). Most of the residues known to be essential for methylation are located around the sequence motif DxGxPxA at positions 331–338 of nsp14, which is predicted to form the S-adenosyl-l-methionine binding pocket of the methyltransferase domain.
3.18.2. Function
Arteriviruses, which are related to nidoviruses except that they are about two-fold smaller, do not have an ExoN homologue or a homologue of either of the viral methyltransferases. This seems to indicate that this enzyme in CoVs is required for stable synthesis of exceptionally large RNA templates (Minskaia et al., 2006).
SARS-CoV nsp14 has 3′→5′ exonuclease activity on both ssRNA and dsRNA (Minskaia et al., 2006). Recombinant nsp14 (as a maltose binding protein fusion) hydrolyzed ssRNA to a ∼12 nucleotide product. When the DE-D-D residues are substituted with alanine, this activity was abolished or greatly impaired; D90A/E92A andH268A mutants had very low activity while N238A, D243A and D273A had undetectable activity (Minskaia et al., 2006). DsRNA also significantly enhanced exonuclease activity of the enzyme (Minskaia et al., 2006). DNA and ribose-2′-O-methylated RNA are resistant to cleavage (Minskaia et al., 2006). The activity of nsp14 is strictly dependent on divalent cations (Chen et al., 2007a). Its activity was highest in the presence of Mg2+ or Mn2+, lower in the presence of low amounts of Zn2+ (0.5 mM) and undetectable with Ca2+ or higher concentrations of Zn2+. With Mn2+, the size of the product is slightly smaller than that obtained with Mg2+ or Zn2+, indicating that the metal ions may modulate the configuration of the active site differently (Chen et al., 2007a, Minskaia et al., 2006).
In MHV, nsp14 greatly enhances replication fidelity, essential for the replication and stability of the unusually large CoV genome (Eckerle et al., 2007). Recombinant viruses with mutations in the nsp14 active site were defective in growth and RNA synthesis and possessed 15-fold more mutations than wild-type viruses. Nsp14 therefore appears to play a role in error prevention or repair of nucleotide incorporation during RNA synthesis (Eckerle et al., 2007). Recombinant HCoV-229E containing mutations in the active site of nsp14 had severe defects in RNA synthesis and no viable virus could be recovered. Besides strongly reduced genome replication, specific defects in sg RNA synthesis, such as aberrant sizes of specific sg RNAs and changes in the molar ratios between individual sg RNA species, were observed (Minskaia et al., 2006). Sperry et al. (Eckerle et al., 2006, Sperry et al., 2005) have shown that a Tyr→His mutation (equivalent to SARS-nsp14 Tyr420His) in an infectious clone of MHV-A59 shows attenuated virus replication and virulence in mice, also arguing for the importance of this protein as a proof-reading component of the viral replication machinery. Based on temperature-sensitive mutants of MHV, Sawicki et al. (2005) showed that nsp14 is essential for the assembly of a functional replicase-transcriptase complex and appears to affect the positive-strand synthesis, as would be expected for a protein involved in both capping and mismatch repair.
Nsp14 interacts with nsp10 and nsp16 to form the viral cap methylation complex, as described in more detail under nsp16 below. Y2H and co-immunoprecipitation studies suggest that nsp14 may also interact with nsp8 and sars9b (von Brunn et al., 2007).
3.19. Nsp15
See Box 6 .
Box 6. Key nsp15 and 16 structures.
| Virus | Domain | Method | Accession | Reference |
|---|---|---|---|---|
| MHV | nsp15 | X-ray (2.7 Å) | 2GTH | Xu et al. (2006) |
| SARS-CoV | nsp15 | X-ray (2.6 Å) | 2H85 | Ricagno et al. (2006b) |
| SARS-CoV | nsp10 + nsp16 | X-ray (2.0 Å) | 2XYQ | Decroly et al. (2011) |
3.19.1. Structure
Nsp15 of SARS-CoV is a 346-residue polypeptide that results from the cleavage of polyprotein 1ab at sites 6427RLQ↓SLE6432 and 6773KLQ↓ASQ6778 by Mpro. It is one of the most well studied RNA processing enzyme of the coronaviral replicase with several recent studies focusing on its structural and functional characterization due to its potential importance as a drug target. Studies on HCoV-229E and equine arteritis virus have shown that inactivating this enzyme by site-directed mutagenesis renders these viruses nonviable. This enzyme is a specific marker for coronaviruses as no known homologs of nsp15 exists among other RNA viruses outside of nidovirales. Nsp15 preferentially cleaves the 3′ end of uridylates of RNA at GUU or GU sequences to produce molecules with 2′–3′ cyclic phosphate ends (Bhardwaj et al., 2004). It acts on both double-stranded RNA and single-stranded RNA (ssRNA) and its activity is dependent on the presence of Mn2+ ions (Bhardwaj et al., 2004, Guarino et al., 2005). The ion binds only weakly but nonetheless produces substantial conformational changes in the active site loops (Bhardwaj et al., 2004, Bhardwaj et al., 2006).
Several groups have characterized the structure of nsp15, both by cryoEM (Guarino et al., 2005) and X-ray crystallography from SARS-CoV (Bhardwaj et al., 2008, Guarino et al., 2005, Joseph et al., 2007, Ricagno et al., 2006a, Ricagno et al., 2006b, Xu et al., 2006), and MHV (Xu et al., 2006) and its eukaryotic homolog, XendoU from Xenopus laevis (Renzi et al., 2006). The coronaviral structures have revealed a three-domain architecture (Fig. 6 ). Again not surprisingly, the catalytic C-terminal domain contains a novel fold. The first two domains (residues 1–190) have a topological similarity to methyltransferases forming a ‘spitting image’ of the SAM-dependent methyltransferase fold as defined in SCOP database (Murzin A, personal communication; Fig. 6). The full length MHV and SARS nsp15 enzymes were shown to be packed as hexamers, their biologically relevant oligomeric state, forming a hollow, toroid shaped structure. Hexamerization is absolutely essential for both metal ion binding and catalytic activity (Guarino et al., 2005). The eukaryotic homolog XendoU from X. levis is much shorter (missing the first two domains) and shares only the conserved catalytic domain. In fact, it was the first structure of this endoribonuclease fold to be structurally characterized. It is a functional monomer in solution. The catalytic center of nsp15 retains features that resemble the active site of an unrelated nuclease, RNase A (Cuchillo et al., 2011).
Fig. 6.
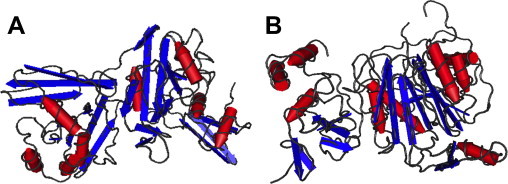
Structures of the replicase proteins nsp10, 15 and 16. The hexameric structure of SARS-CoV nsp15 comes from PDB entry 2H85 and is shown in panel A. The structure of the SARS-CoV nsp10–16 shown in panel B complex is taken from PDB entry 2XYQ (nsp10 is shown on the left).
3.19.2. Function
While the enzymatic activity of nsp15 is now fairly well understood, the role of nsp15 in the coronavirus replication cycle is not. Nsp15 cleaves at uridylates preceded by cytidylate or adenylate residues. When model RNA substrates were 2′-O-ribose methylated, it blocked endonucleolytic activity, indicating a possible functional link between nsp15 and the 2′-O-ribose methyltransferase nsp16. A structure based catalytic mechanism of endonucleolytic activity of nsp15 was proposed by Ricagno et al. (2006b), based on active site similarities with RNAse A. In this model, Lys-289, His-249, and His-234 residues act as the main catalytic triad while Ser-293 and Tyr-342 provide the supporting role by stabilizing the aromatic ring of the nucleotide. Despite structural uniqueness of nsp15, the actual mode of sessile bond cleavage is thought to be very similar to those of several RNAses of this class that share the same catalytic triad e.g., RNAse T1, RNAse A and others. The six active sites of the hexamer are spatially segregated and are thought to function independent of one another. The actual electrostatic contribution of the Mn2+ ion in catalysis is unclear. In XendoU, Mn2+ does not impede either RNA substrate binding of cleavage (Renzi et al., 2006). However, fluorescence experiments indicate that upon metal ion binding, the protein undergoes large structural transitions suggesting an indirect, possibly structural role for metal in either stabilizing the enzyme in a catalytically competent “on” state from an otherwise inactive “off” state (Renzi et al., 2006). Drawing analogies with endonuclease EndN, a Zn dependent enzyme, Ricagno and co-workers have hypothesized that, given its proximity to the catalytic site, Y342 might be the residue involved in Mn2+ ion binding by forming a cation-II interactions, assisted by the histidine H249 and the 2′-O-ribose moiety of the substrate. Molecular modeling and docking studies led Renzi et al. (2006) to propose a similar mechanism for endonucleolytic cleavage by the eukaryotic homolog XendoU, wherein the O4 of UMP nucleotide forms a potential hydrogen bond with the catalytic histidine H178 and the pyrimidine ring of the nucleotide involved in stacking interaction with the aromatic ring of tyrosine Y280.
The structure of a truncated form of nsp15 from SARS-CoV, that was lacking the N terminal hexamerization domain, revealed striking changes in the active site loops in the catalytic domain – suggesting allosteric control of endonucleolytic activity and providing a direct link between oligomerization and function (Joseph et al., 2007). In this structure, which lacked the first 27 amino acids of nsp15, a dramatic shift was noticed in the active site loop (residues 234–249, referred to as the “active site loop” spanning the two active site histidines H234 and H249) that was flipped by as much as ∼120° into the active site cleft. In the full-length nsp15 hexamer, the “active site loop” and the “supporting loop” are packed against each other and are stabilized by intimate interactions with residues contributed by the adjacent monomer.
3.20. Nsp16
3.20.1. Structure
nsp16 lies at the C-terminal end of polyprotein Polyprotein 1ab and results when Mpro cleaves the polyprotein at nsl15/16 junction (Snijder et al., 2003). Although first identified in flavi- and reoviruses (Koonin, 1993) about two decades ago, the role of viral methyltransferases in viral replication has been only now begun to be explored systematically (Gorbalenya et al., 2006). After rigorous sequence and structure analysis using 3D-jury based metaserver prediction methods, Richlewski and co-workers noticed a strong but remote homology between SARS nsp16 and an ancient family of S-adenosyl-L-methionine (SAM) dependent 2′-O-ribose methyltransferases enzymes (von Grotthuss et al., 2003, Ferron et al., 2002). The sequence of SARS MTase has features that place it in the viscinity of the RrmJ/fibrillarin superfamily of 2′-O-ribose methyltransferases (Feder et al., 2003).
The structure of SARS-CoV nsp16 has been determined as part of an nsp16-nsp10 complex by two groups independently (Chen et al., 2011, Decroly et al., 2011). Nsp16 adopts a canonical S-adenosyl-l-methionine dependent methyltransferase fold, with a central beta sheet framed by a helical clamp and a conserved catalytic KDKE tetrad (Martin and McMillan, 2002). The nsp16 topology matches those of the dengue virus NS5 methyltransferase (Egloff et al., 2002) and vaccinia virus VP39 O-methyltransferase (Hodel et al., 1996). The structure of the nsp16/nsp10 interaction interface shows that nsp10 interacts with and probably helps to stabilize the S-adenosyl-l-methionine binding pocket. This has the effect of making the putative RNA-binding groove of nsp16 longer. The study by Decroly et al. (2011) also demonstrated that the methyltransferase inhibitor sinefungin interacts with the nsp16 active site, and could therefore form the basis of a new generation of inhibitors that attack the coronavirus methylation process. The structure of nsp10 was found to be virtually identical when solved in the presence and absence of nsp16 (Chen et al., 2011, Decroly et al., 2011, Joseph et al., 2006, Su et al., 2006).
3.20.2. Function
Nsp16 has been shown to interact with nsp10 and nsp14 to form a viral cap methylation complex (Bouvet et al., 2010). All eukaryotic mRNAs posses this modified guanosine at the 5′ terminus, a feature that confers protection against degradation by host nucleases. First reported in the early 70s (Gingras, 2009), the “cap” structure and has been found to be present in almost all eukaryotic viral RNAs. The generic nomenclature that's been widely adopted is m7G(5′)ppp(5′)X(m)pY(m) where m7G corresponds to the modified 7-methylguanosine nucleotide. O-methyltransferases such as nsp16 perform the final step of cap synthesis, which involves adding a methyl group to the first nucleotide following the m7G, and sometimes adding a methyl group at the same position on subsequent nucleotides. While the m7G cap is essential for efficient translation splicing, nuclear export, translation and stability of eukaryotic mRNA, O-methylation is not (Cougot et al., 2004, Lewis and Izaurralde, 1997, Schwer et al., 1998).
Zust et al. (2011) explored the role of nsp16 methylation in the viral replication cycle and found that O-methylation acts as a recognition marker that helps the host cell to recognize its own RNA species, and respond to incompletely methylated cap structures. Nsp16 makes replication possible by camoflauging newly synthesized viral RNA to resemble host mRNA, which therefore blocks the induction of an interferon response. This suggests that drugs that act on nsp16 have the potential to interfere with viral replication both at the level of inhibition of the replication process, and in promoting intracellular recognition and response to viral RNA species.
4. Conclusion – a Galapagos of new folds
New coronavirus protein structures continue to have important implications for virus biology and antiviral design. An unexpected benefit of the coronavirus structure boom is a wealth of previously undiscovered protein folds. The protein data bank keeps records of the discovery of new protein structures over time, and currently classifies all known protein structures into fewer than 1500 folds.
Since 2003, 18 out of 28 coronavirus proteins encompassing a total of 27 domains have been determined experimentally. Several of these have been described as “new folds” commonly defined as one with sufficiently different fold/topology based on comparison methods (DALI), fold classification schemes (CATH and SCOP) and family assignment schema of PFAM. By these criteria 16 out of the 27 domains are indeed new folds – a striking rate of fold discovery, when compared to the ∼10% for model pro- and eukaryotes being reported by structural genomics centers.
Why do coronaviruses possess an abundance of new folds? One obvious reason might be that these structures have been relatively unexplored, and therefore under-represented in PDB. This is the first proteome-scale structural characterization of a coronavirus, and one with a disproportionately large number of singletons. The new folds are significantly contributed by the 16 nonstructural proteins of the replicase machinery, several of which do not have counterparts outside Nidovirales. Ideally, new folds enable us to model sequence homologues, thereby filling out the immediate neighborhood in structure space. This is non-trivial for SARS-CoV proteins, since the new folds are (so far) either true sequence singletons (nsp1, nsp2 nsp3a, sars9b) or found exclusively in Coronaviridae (nsp4, 7, 8, 9, 10, 15, spike RBD, sars9a).
Fast mutation rates in viruses may encourage divergent sampling of fold space (Andreeva and Murzin, 2006). This, along with oligomerization has been proposed to be major facilitators of fold evolution (Andreeva and Murzin, 2006) allowing a protein to morph to a new fold analogous to structural drift (Krishna and Grishin, 2005) Elucidation of new folds, especially for isolated groups of divergent homologues should help in improving fold recognition and comparative modeling algorithms.
These observations also have ramifications in evolution of new viral strains, a phenomenon which is the result of two antagonistic forces: greater adaptability within an ecological niche (because of intrinsically fast mutation rates) and increased evolutionary constraints due to their small genomes (Holmes and Rambaut, 2004). Overall, we are left with the piquant notion that proteins in viral proteomes may probably occupy a unique niche in fold space and coronaviruses, a peculiar island in this niche. Viruses are the most diverse biological entities on this planet and second only to prokaryotes in terms of sheer biomass. While the diversity of protein structures they represent certainly defies imagination, our understanding of protein folds and their migration in tertiary fold space may well be locked up in them.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2013.12.004.
Appendix A. Supplementary data
References
- Adedeji A.O., Marchand B., Te Velthuis A.J., Snijder E.J., Weiss S., Eoff R.L., Singh K., Sarafianos S.G. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE. 2012;7(5):e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D.G., Choi J.K., Taylor D.R., Oh J.W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Archives of Virology. 2012;157(11):2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaji K., Konno H., Mitsui H., Teruya K., Shimamoto Y., Hattori Y., Ozaki T., Kusunoki M., Sanjoh A. Structure-based design, synthesis, and evaluation of peptide-mimetic SARS 3CL protease inhibitors. Journal of Medicinal Chemistry. 2011;54(23):7962–7973. doi: 10.1021/jm200870n. [DOI] [PubMed] [Google Scholar]
- Almeida M.S., Johnson M.A., Herrmann T., Geralt M., Wuthrich K. Novel β-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. Journal of Virology. 2007;81(7):3151–3161. doi: 10.1128/JVI.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida M.S., Johnson M.A., Wuthrich K. NMR assignment of the SARS-CoV protein nsp1. Journal of Biomolecular NMR. 2006;36(Suppl. 1):46. doi: 10.1007/s10858-006-9018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez E., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Marcos-Villar L., Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology. 2010;402(2):281–291. doi: 10.1016/j.virol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. The EMBO Journal. 2002;21(13):3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Andreeva A., Murzin A.G. Evolution of protein fold in the presence of functional constraints. Current Opinion in Structural Biology. 2006;16(3):399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. MBio. 2013;4(4.) doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacha U., Barrila J., Gabelli S.B., Kiso Y., Mario Amzel L., Freire E. Development of broad-spectrum halomethyl ketone inhibitors against coronavirus main protease 3CL(pro) Chemical Biology & Drug Design. 2008;72(1):34–49. doi: 10.1111/j.1747-0285.2008.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.C., Shieh C.K., Soe L.H., Chang M.F., Vannier D.M., Lai M.M. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. Journal of Virology. 1989;63(9):3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.C., Yokomori K., Dong S., Carlisle R., Gorbalenya A.E., Koonin E.V., Lai M.M. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. Journal of Virology. 1993;67(10):6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. Journal of Virology. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. Deubiquitinating activity of the SARS-CoV papain-like protease. Advances in Experimental Medicine and Biology. 2006;581:37–41. doi: 10.1007/978-0-387-33012-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlam M., Yang H., Rao Z. Structural insights into SARS coronavirus proteins. Current Opinion in Structural Biology. 2005;15(6):664–672. doi: 10.1016/j.sbi.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernini A., Spiga O., Venditti V., Prischi F., Bracci L., Huang J., Tanner J.A., Niccolai N. Tertiary structure prediction of SARS coronavirus helicase. Biochemical and Biophysical Research Communications. 2006;343(4):1101–1104. doi: 10.1016/j.bbrc.2006.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. Journal of Virology. 2004;78(22):12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Palaninathan S., Alcantara J.M., Yi L.L., Guarino L., Sacchettini J.C., Kao C.C. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem. 2008;283(6):3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Sun J., Holzenburg A., Guarino L.A., Kao C.C. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. Journal of Molecular Biology. 2006;361(2):243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski P., Schalinski S., Schmitz H. Nucleotide triphosphatase/helicase of hepatitis C virus as a target for antiviral therapy. Antiviral Research. 2002;55(3):397–412. doi: 10.1016/s0166-3542(02)00096-7. [DOI] [PubMed] [Google Scholar]
- Bost A.G., Carnahan R.H., Lu X.T., Denison M.R. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. Journal of Virology. 2000;74(7):3379–3387. doi: 10.1128/jvi.74.7.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost A.G., Prentice E., Denison M.R. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology. 2001;285(1):21–29. doi: 10.1006/viro.2001.0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathogens. 2010;6(4):e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. Journal of Virology. 2003;77(19):10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Denison M.R. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology. 2005;340(2):209–223. doi: 10.1016/j.virol.2005.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockway S.M., Lu X.T., Peters T.R., Dermody T.S., Denison M.R. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. Journal of Virology. 2004;78(21):11551–11562. doi: 10.1128/JVI.78.21.11551-11562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Johnson M.A., Serrano P., Pedrini B., Joseph J.S., Neuman B.W., Saikatendu K., Buchmeier M.J., Kuhn P., Wuthrich K. Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. Journal of Virology. 2009;83(4):1823–1836. doi: 10.1128/JVI.01781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Makino S. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. Journal of Virology. 2004;78(11):5658–5669. doi: 10.1128/JVI.78.11.5658-5669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Sugiyama K., Kubo H., Huang C., Makino S. Murine coronavirus nonstructural protein p28 arrests cell cycle in G0/G1 phase. Journal of Virology. 2004;78(19):10410–10419. doi: 10.1128/JVI.78.19.10410-10419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Jiang M., Hu T., Liu Q., Chen X.S., Guo D. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. Journal of Biochemistry and Molecular Biology. 2007;40(5):649–655. doi: 10.5483/bmbrep.2007.40.5.649. [DOI] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., Ahola T., Liang Y., Liu X., Guo D. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathogens. 2011;7(10):e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tao J., Sun Y., Wu A., Su C., Gao G., Cai H., Qiu S., Wu Y., Ahola T., Guo D. Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase. Journal of Virology. 2013;87(11):6296–6305. doi: 10.1128/JVI.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wang Y., Ratia K., Mesecar A.D., Wilkinson K.D., Baker S.C. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. Journal of Virology. 2007;81(11):6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Zhang W., Xie Y., Jiang W., Arnold E., Sarafianos S.G., Ding J. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology. 2005;335(2):165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L.H., Choy W.Y., Tsai S.N., Rao Z., Ngai S.M. Rapid peptide-based screening on the substrate specificity of severe acute respiratory syndrome (SARS) coronavirus 3C-like protease by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Protein Sciene. 2006;15(4):699–709. doi: 10.1110/ps.052007306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck C.P., Chen C., Ke Z., Wan D.C., Chow H.F., Wong K.B. Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases. European Journal of Medicinal Chemistry. 2013;59:1–6. doi: 10.1016/j.ejmech.2012.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillez-Ty C.T., Liao L., Yates J.R., 3rd, Kuhn P., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. Journal of Virology. 2009;83(19):10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7(11):1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., van Dijk E., Babajko S., Seraphin B. Cap-tabolism. Trends in Biochemical Sciences. 2004;29(8):436–444. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Cuchillo C.M., Nogues M.V., Raines R.T. Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry. 2011;50(37):7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver G.M., Consaul S.A., Tycowski K.T., Filipowicz W., Phizicky E.M. tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose 1,2-cyclic phosphate. Journal of Biological Chemistry. 1994;269(40):24928–24934. [PubMed] [Google Scholar]
- Culver G.M., McCraith S.M., Zillmann M., Kierzek R., Michaud N., LaReau R.D., Turner D.H., Phizicky E.M. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1–2 cyclic phosphate. Science. 1993;261(5118):206–208. doi: 10.1126/science.8392224. [DOI] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathogens. 2011;7(5):e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D.J., Graham R.L., Denison M.R., Baric R.S. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. Journal of Virology. 2007;81(19):10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M., Perlman S. Identification of putative polymerase gene product in cells infected with murine coronavirus A59. Virology. 1987;157(2):565–568. doi: 10.1016/0042-6822(87)90303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Hughes S.A., Weiss S.R. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology. 1995;207(1):316–320. doi: 10.1006/viro.1995.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Perlman S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. Journal of Virology. 1986;60(1):12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Yount B., Brockway S.M., Graham R.L., Sims A.C., Lu X., Baric R.S. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. Journal of Virology. 2004;78(11):5957–5965. doi: 10.1128/JVI.78.11.5957-5965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Zoltick P.W., Hughes S.A., Giangreco B., Olson A.L., Perlman S., Leibowitz J.L., Weiss S.R. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189(1):274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolski S., Harter M., Stacey D.W. Cellular ras activity is required for passage through multiple points of the G0/G1 phase in BALB/c 3T3 cells. Molecular and Cellular Biology. 1994;14(8):5441–5449. doi: 10.1128/mcb.14.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Graham R.L., Sims A.C., Denison M.R., Baric R.S. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. Journal of Virology. 2007;81(13):7086–7098. doi: 10.1128/JVI.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Sims A.C., Graham R.L., Denison M.R., Baric R.S. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. Journal of Virology. 2007;81(12):6356–6368. doi: 10.1128/JVI.02805-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Brockway S.M., Sperry S.M., Lu X., Denison M.R. Effects of mutagenesis of murine hepatitis virus nsp1 and nsp14 on replication in culture. Adv Exp Med Biol. 2006;581:55–60. doi: 10.1007/978-0-387-33012-9_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14-exoribonuclease mutants. Journal of Virology. 2007 doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Benarroch D., Selisko B., Romette J.L., Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. The EMBO Journal. 2002;21(11):2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(11):3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. Journal of Virology. 2006;80(17):8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder M., Pas J., Wyrwicz L.S., Bujnicki J.M. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene. 2003;302(1–2):129–138. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- Ferron F., Longhi S., Henrissat B., Canard B. Viral RNA-polymerases – a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends in Biochemical Sciences. 2002;27(5):222–224. doi: 10.1016/s0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. Journal of Virology. 2009;83(13):6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadlage M.J., Sparks J.S., Beachboard D.C., Cox R.G., Doyle J.D., Stobart C.C., Denison M.R. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. Journal of Virology. 2010;84(1):280–290. doi: 10.1128/JVI.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan C., Enjuanes L., Almazan F. A point mutation within the replicase gene differentially affects coronavirus genome versus minigenome replication. Journal of Virology. 2005;79(24):15016–15026. doi: 10.1128/JVI.79.24.15016-15026.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C. Journal Club: 35 years later, mRNA caps still matter. Nature Reviews. Molecular Cell Biology. 2009;10(11):734. doi: 10.1038/nrm2789. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Research. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Research. 1989;17(12):4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Snijder E.J., Spaan W.J. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. Journal of Virology. 2004;78(15):7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grum-Tokars V., Ratia K., Begaye A., Baker S.C., Mesecar A.D. Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drugdiscovery. Virus Res. 2008;133(1):63–73. doi: 10.1016/j.virusres.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. Journal of Molecular Biology. 2005;353(5):1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Rottier P.J., de Haan C.A. Biogenesis and dynamics of the coronavirus replicative structures. Viruses. 2012;4(11):3245–3269. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Ulasli M., Vonk A.M., Reggiori F., Rottier P.J., de Haan C.A. Mobility and interactions of coronavirus nonstructural protein 4. Journal of Virology. 2011;85(9):4572–4577. doi: 10.1128/JVI.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.S., Chang G.G., Juo C.G., Lee H.J., Yeh S.H., Hsu J.T., Chen X. Papain-like protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS-CoV): expression, purification, characterization, and inhibition. Biochemistry. 2005;44(30):10349–10359. doi: 10.1021/bi0504761. [DOI] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. Journal of Virology. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Adonov A., Traykova-Adonova M., Cao J., Cutts T., Grudesky E., Deschambaul Y., Berry J., Drebot M., Li X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochemical and Biophysical Research Communications. 2004;320(4):1199–1203. doi: 10.1016/j.bbrc.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R., Anand K., Mesters J.R., Rao Z., Shen X., Jiang H., Tan J., Verschueren K.H. Structure and dynamics of SARS coronavirus main proteinase (Mpro) Advances in Experimental Medicine and Biology. 2006;581:585–591. doi: 10.1007/978-0-387-33012-9_106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel A.E., Gershon P.D., Shi X., Quiocho F.A. The 1.85 A structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell. 1996;85(2):247–256. doi: 10.1016/s0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- Holmes E.C., Rambaut A. Viral evolution and the emergence of SARS coronavirus. Philosophical Transactions of the Royal Society of London. 2004;359(1447):1059–1065. doi: 10.1098/rstb.2004.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. The Journal of Clinical Investigation. 2003;111(11):1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M.F., Kuo C.J., Chang K.T., Chang H.C., Chou C.C., Ko T.P., Shr H.L., Chang G.G., Wang A.H., Liang P.H. Mechanism of the maturation process of SARS-CoV 3CL protease. Journal of Biological Chemistry. 2005;280(35):31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Li P., Song L., Jeffrey P.D., Chenova T.A., Wilkinson K.D., Cohen R.E., Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. The EMBO Journal. 2005;24(21):3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. Alphacoronavirus transmissible gastroenteritis virus nsp1 protein suppresses protein translation in mammalian cells and in cell-free HeLa cell extracts but not in rabbit reticulocyte lysate. Journal of Virology. 2011;85(1):638–643. doi: 10.1128/JVI.01806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S.A., Denison M.R., Bonilla P., Leibowitz J.L., Baric R.S., Weiss S.R. A newly identified MHV-A59 ORF1a polypeptide p65 is temperature sensitive in two RNA negative mutants. Advances in Experimental Medicine and Biology. 1993;342:221–226. doi: 10.1007/978-1-4615-2996-5_35. [DOI] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. Journal of Virology. 2013;87(16):9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. The EMBO Journal. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. Journal of Virology. 2004;78(11):5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson A.M. Structure of alphacoronavirus transmissible gastroenteritis virus nsp1 has implications for coronavirus nsp1 function and evolution. Journal of Virology. 2013;87(5):2949–2955. doi: 10.1128/JVI.03163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Research. 2005;33:W284–W288. doi: 10.1093/nar/gki418. (Web Server Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., Chatterjee A., Neuman B.W., Wuthrich K. SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding. Journal of Molecular Biology. 2010;400(4):724–742. doi: 10.1016/j.jmb.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Brooun A., Griffith M., Moy K., Yadav M.K., Velasquez J., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. Journal of Virology. 2006;80(16):7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. Journal of Virology. 2007;81(12):6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadare G., Haenni A.L. Virus-encoded RNA helicases. Journal of Virology. 1997;71(4):2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nature Structural & Molecular Biology. 2009;16(11):1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(34):12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanahaluethai A., Chen Z., Jukneliene D., Baker S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361(2):391–401. doi: 10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleymann G. Novel agents and strategies to treat herpes simplex virus infections. Expert Opinion on Investigational Drugs. 2003;12(2):165–183. doi: 10.1517/13543784.12.2.165. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. The Journal of General Virology. 1993;74(Pt 4):733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- Krishna S.S., Grishin N.V. Structural drift: a possible path to protein fold change. Bioinformatics (Oxford, England) 2005;21(8):1308–1310. doi: 10.1093/bioinformatics/bti227. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kumar P., Gunalan V., Liu B., Chow V.T., Druce J., Birch C., Catton M., Fielding B.C., Tan Y.J., Lal S.K. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366(2):293–303. doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M., Holmes K. In: Fundamental Virology. Knipe D.M,H.P., editor. Lippincott Williams & Wilkins; Philadelphia: 2001. Coronaviridae: the viruses and their replication. [Google Scholar]
- Lai M.M.C., Holmes K.V. In: Fields Virology. Knipe D.M., Howley P.M., editors. Lippincott Williams and Wilkins; Philadelphia: 2001. Coronaviridae: the viruses and their replication; pp. 1163–1185. [Google Scholar]
- Lauber C., Goeman J.J., Parquet Mdel C., Nga P.T., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathogens. 2013;9(7):e1003500. doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law A.H., Lee D.C., Cheung B.K., Yim H.C., Lau A.S. Role for nonstructural protein 1 of severe acute respiratory syndrome coronavirus in chemokine dysregulation. Journal of Virology. 2007;81(1):416–422. doi: 10.1128/JVI.02336-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.C., Kuo C.J., Hsu M.F., Liang P.H., Fang J.M., Shie J.J., Wang A.H. Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors. FEBS Lett. 2007;581(28):5454–5458. doi: 10.1016/j.febslet.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.C., Kuo C.J., Ko T.P., Hsu M.F., Tsui Y.C., Chang S.C., Yang S., Chen S.J., Chen H.C., Hsu M.C., Shih S.R., Liang P.H., Wang A.H. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. Journal of Biological Chemistry. 2009;284(12):7646–7655. doi: 10.1074/jbc.M807947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.W., Cherney M.M., Huitema C., Liu J., James K.E., Powers J.C., Eltis L.D., James M.N. Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate-like aza-peptide epoxide. Journal of Molecular Biology. 2005;353(5):1137–1151. doi: 10.1016/j.jmb.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett D.S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R.T., Walz T., Ploegh H., Finley D. Multiple associated proteins regulate proteasome structure and function. Molecular Cell. 2002;10(3):495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Izaurralde E. The role of the cap structure in RNA processing and nuclear export. European Journal of Biochemistry/FEBS. 1997;247(2):461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- Li Y., Ren Z., Bao Z., Ming Z., Li X. Expression, crystallization and preliminary crystallographic study of the C-terminal half of nsp2 from SARS coronavirus. Acta Crystallographica Section F: Structural Biology and Crystallization Communications. 2011;67(Pt 7):790–793. doi: 10.1107/S1744309111017829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.W., Tsai C.H., Tsai F.J., Chen P.J., Lai C.C., Wan L., Chiu H.H., Lin K.H. Characterization of trans- and cis-cleavage activity of the SARS coronavirus 3CLpro protease: basis for the in vitro screening of anti-SARS drugs. FEBS Letters. 2004;574(1–3):131–137. doi: 10.1016/j.febslet.2004.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Menard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. Journal of Virology. 2005;79(24):15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolaridis I., Wojdyla J.A., Panjikar S., Snijder E.J., Gorbalenya A.E., Berglind H., Nordlund P., Coutard B., Tucker P.A. Structure of the C-terminal domain of nsp4 from feline coronavirus. Acta Crystallographica. Section D, Biological crystallography. 2009;65(Pt 8):839–846. doi: 10.1107/S0907444909018253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.L., McMillan F.M. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Current Opinion in Structural Biology. 2002;12(6):783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Martzen M.R., McCraith S.M., Spinelli S.L., Torres F.M., Fields S., Grayhack E.J., Phizicky E.M. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286(5442):1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics (Oxford, England) 2000;16(4):404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Miknis Z.J., Donaldson E.F., Umland T.C., Rimmer R.A., Baric R.S., Schultz L.W. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. Journal of Virology. 2009;83(7):3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M.J., Holley W.R., Chatterjee A., Mian I.S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Research. 1997;25(24):5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. Journal of Molecular Biology. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T., Makino S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. Journal of Virology. 2008;82(9):4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., 3rd, Wuthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. Journal of Virology. 2008;82(11):5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P., te Lintelo E.G., Rottier P.J., de Haan C.A. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. Journal of Virology. 2008;82(24):12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., te Lintelo E.G., Deijs M., Verheije M.H., Rottier P.J., de Haan C.A. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. Journal of Virology. 2007;81(22):12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born E., Spaan W.J., Snijder E.J. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. The EMBO Journal. 2001;20(24):7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeper D.S., Upton T.M., Ladha M.H., Neuman E., Zalvide J., Bernards R., DeCaprio J.A., Ewen M.E. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386(6621):177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- Perona J.J., Craik C.S., Fletterick R.J. Locating the catalytic water molecule in serine proteases. Science. 1993;261(5121):620–622. doi: 10.1126/science.8342029. [DOI] [PubMed] [Google Scholar]
- Peti W., Johnson M.A., Herrmann T., Neuman B.W., Buchmeier M.J., Nelson M., Joseph J., Page R., Stevens R.C., Kuhn P., Wuthrich K. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. Journal of Virology. 2005;79(20):12905–12913. doi: 10.1128/JVI.79.20.12905-12913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E.M., Greer C.L. Pre-tRNA splicing: variation on a theme or exception to the rule? Trends in Biochemical Sciences. 1993;18(1):31–34. doi: 10.1016/0968-0004(93)90085-2. [DOI] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. The EMBO Journal. 1989;8(12):3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy R., Moll R., Weimar T., Mesters J.R., Hilgenfeld R. Variable oligomerization modes in coronavirus non-structural protein 9. Journal of Molecular Biology. 2008;383(5):1081–1096. doi: 10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. Journal of Virology. 2004;78(18):9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A., Filipowicz W., Hall J., Gorbalenya A.E., Ziebuhr J. ADP-ribose-1-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. Journal of Virology. 2005;79(20):12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(15):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzi F., Caffarelli E., Laneve P., Bozzoni I., Brunori M., Vallone B. The structure of the endoribonuclease XendoU: from small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12365–12370. doi: 10.1073/pnas.0602426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Coutard B., Grisel S., Bremond N., Dalle K., Tocque F., Campanacci V., Lichiere J., Lantez V., Debarnot C., Cambillau C., Canard B., Egloff M.P. Crystallization and preliminary X-ray diffraction analysis of Nsp15 from SARS coronavirus. Acta Crystallographica Section F: Structural Biology and Crystallization Communications. 2006;62(Pt 4):409–411. doi: 10.1107/S1744309106009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(32):11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13(11):1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathogens. 2005;1(4):e39. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze H., Ulferts R., Schelle B., Bayer S., Granzow H., Hoffmann B., Mettenleiter T.C., Ziebuhr J. Characterization of White bream virus reveals a novel genetic cluster of nidoviruses. Journal of Virology. 2006;80(23):11598–11609. doi: 10.1128/JVI.01758-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Mao X., Shuman S. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic Acids Research. 1998;26(9):2050–2057. doi: 10.1093/nar/26.9.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Johnson M.A., Almeida M.S., Horst R., Herrmann T., Joseph J.S., Neuman B.W., Subramanian V., Saikatendu K.S., Buchmeier M.J., Stevens R.C., Kuhn P., Wuthrich K. Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. Journal of Virology. 2007;81(21):12049–12060. doi: 10.1128/JVI.00969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P., Johnson M.A., Chatterjee A., Neuman B.W., Joseph J.S., Buchmeier M.J., Kuhn P., Wuthrich K. Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3. Journal of Virology. 2009;83(24):12998–13008. doi: 10.1128/JVI.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y.F., Xu G.J. Study on substrate specificity at subsites for severe acute respiratory syndrome coronavirus 3CL protease. Acta Biochimica et Biophysica Sinica (Shanghai) 2005;37(12):807–813. doi: 10.1111/j.1745-7270.2005.00114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y.M., Yang W.B., Peng H.P., Hsu M.F., Tsai K.C., Kuo T.H., Wang A.H., Liang P.H., Lin C.H., Yang A.S., Wong C.H. Structure-based design and synthesis of highly potent SARS-CoV 3CL protease inhibitors. Chembiochem: A European Journal of Chemical Biology. 2007;8(14):1654–1657. doi: 10.1002/cbic.200700254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims A.C., Ostermann J., Denison M.R. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. Journal of Virology. 2000;74(12):5647–5654. doi: 10.1128/jvi.74.12.5647-5654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. Journal of Molecular Biology. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks J.S., Lu X., Denison M.R. Genetic analysis of Murine hepatitis virus nsp4 in virus replication. Journal of Virology. 2007;81(22):12554–12563. doi: 10.1128/JVI.01257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry S.M., Kazi L., Graham R.L., Baric R.S., Weiss S.R., Denison M.R. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. Journal of Virology. 2005;79(6):3391–3400. doi: 10.1128/JVI.79.6.3391-3400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanie Tech, C.L.S., Mutschall, D., Schmidtke, S., Moll, R., Ziebuhr, J., Wadhwani, P., Ulrich, A.S., Hilgenfeld, R., 2004. Evidence for protease activity of the SARS Unique Domain (SUD). International Conference on SARS – One Year After the (First) Outbreak, Lübeck, Germany. http://www.egms.de/static/en/meetings/sars2004/04sars130.shtml.
- Su D., Lou Z., Sun F., Zhai Y., Yang H., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. Journal of Virology. 2006;80(16):7902–7908. doi: 10.1128/JVI.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Purisima E.O., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? Journal of Virology. 2005;79(7):4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12(2):341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathogens. 2009;5(5):e1000428. doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kamitani W., DeDiego M.L., Enjuanes L., Matsuura Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. Journal of Virology. 2012;86(20):11128–11137. doi: 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. Journal of Biological Chemistry. 2003;278(41):39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.A., Zheng B.J., Zhou J., Watt R.M., Jiang J.Q., Wong K.L., Lin Y.P., Lu L.Y., He M.L., Kung H.F., Kesel A.J., Huang J.D. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chemistry & Biology. 2005;12(3):303–311. doi: 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Research. 2010;38(1):203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J., van den Worm S.H., Snijder E.J. The SARS-coronavirus nsp7 + nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Research. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. The Journal of General Virology. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Turlington M., Chun A., Tomar S., Eggler A., Grum-Tokars V., Jacobs J., Daniels J.S., Dawson E., Saldanha A., Chase P., Baez-Santos Y.M., Lindsley C.W., Hodder P., Mesecar A.D., Stauffer S.R. Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: Identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorganic & Medicinal Chemistry Letters. 2013;23(22):6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dinten L.C., van Tol H., Gorbalenya A.E., Snijder E.J. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. Journal of Virology. 2000;74(11):5213–5223. doi: 10.1128/jvi.74.11.5213-5223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren K.H., Pumpor K., Anemuller S., Chen S., Mesters J.R., Hilgenfeld R. A structural view of the inactivation of the SARS coronavirus main proteinase by benzotriazole esters. Chemistry & Biology. 2008;15(6):597–606. doi: 10.1016/j.chembiol.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS ONE. 2007;2(5):e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113(6):701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Chen G., Zheng D., Cheng G., Tang H. PLP2 of mouse hepatitis virus A59 (MHV-A59) targets TBK1 to negatively regulate cellular type I interferon signaling pathway. PLoS ONE. 2011;6(2):e17192. doi: 10.1371/journal.pone.0017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P., Fan K., Chen H., Ma L., Huang C., Tan L., Xi D., Li C., Liu Y., Cao A., Lai L. The N-terminal octapeptide acts as a dimerization inhibitor of SARS coronavirus 3C-like proteinase. Biochemical and Biophysical Research Communications. 2006;339(3):865–872. doi: 10.1016/j.bbrc.2005.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyla J.A., Manolaridis I., Snijder E.J., Gorbalenya A.E., Coutard B., Piotrowski Y., Hilgenfeld R., Tucker P.A. Structure of the X (ADRP) domain of nsp3 from feline coronavirus. Acta Crystallographica. Section D, Biological Crystallography. 2009;65(Pt 12):1292–1300. doi: 10.1107/S0907444909040074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyla J.A., Manolaridis I., van Kasteren P.B., Kikkert M., Snijder E.J., Gorbalenya A.E., Tucker P.A. Papain-like protease 1 from transmissible gastroenteritis virus: crystal structure and enzymatic activity toward viral and cellular substrates. Journal of Virology. 2010;84(19):10063–10073. doi: 10.1128/JVI.00898-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Ma Q., Restle T., Shang W., Svergun D.I., Ponnusamy R., Sczakiel G., Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. Journal of Virology. 2012;86(8):4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.H., Huang M., Fang S.G., Liu D.X. Coronavirus infection induces DNA replication stress partly through interaction of its nonstructural protein 13 with the p125 subunit of DNA polymerase delta. Journal of Biological Chemistry. 2011;286(45):39546–39559. doi: 10.1074/jbc.M111.242206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Liu Y., Weiss S., Arnold E., Sarafianos S.G., Ding J. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Research. 2003;31(24):7117–7130. doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. Journal of Virology. 2006;80(16):7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Cong L., Chen C., Wei L., Zhao Q., Xu X., Ma Y., Bartlam M., Rao Z. Crystal structures of two coronavirus ADP-ribose-1-monophosphatases and their complexes with ADP-Ribose: a systematic structural analysis of the viral ADRP domain. Journal of Virology. 2009;83(2):1083–1092. doi: 10.1128/JVI.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. Journal of Virology. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Wei L., Zhao W., Xu Y., Rao Z. Expression, crystallization and preliminary X-ray diffraction analysis of the N-terminal domain of nsp2 from avian infectious bronchitis virus. Acta Crystallographica Section F: Structural Biology and Crystallization Communications. 2009;65(Pt 8):788–790. doi: 10.1107/S1744309109024749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Bartlam M., Rao Z. Drug design targeting the main protease, the Achilles’ heel of coronaviruses. Current Pharmaceutical Design. 2006;12(35):4573–4590. doi: 10.2174/138161206779010369. [DOI] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Tanner J.A., Zheng B.J., Watt R.M., He M.L., Lu L.Y., Jiang J.Q., Shum K.T., Lin Y.P., Wong K.L., Lin M.C., Kung H.F., Sun H., Huang J.D. Bismuth complexes inhibit the SARS coronavirus. Angewandte Chemie (International ed. In English) 2007;46(34):6464–6468. doi: 10.1002/anie.200701021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap Y., Zhang X., Andonov A., He R. Structural analysis of inhibition mechanisms of aurintricarboxylic acid on SARS-CoV polymerase and other proteins. Computational Biology and Chemistry. 2005;29(3):212–219. doi: 10.1016/j.compbiolchem.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Niu C., Cherney M.M., Zhang J., Huitema C., Eltis L.D., Vederas J.C., James M.N. A mechanistic view of enzyme inhibition and peptide hydrolysis in the active site of the SARS-CoV 3C-like peptidase. Journal of Molecular Biology. 2007;371(4):1060–1074. doi: 10.1016/j.jmb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Ming Z., Li Y., Chen C., Bao Z., Ren Z., Liu B., Tao W., Rao Z., Lou Z. Purification, crystallization and preliminary X-ray analysis of nonstructural protein 2 (nsp2) from avian infectious bronchitis virus. Acta Crystallographica Section F: Structural Biology and Crystallization Communications. 2012;68(Pt 6):716–719. doi: 10.1107/S1744309112018623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Shan Y., Zhao Z., Chen J., Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virology Journal. 2005;2:66. doi: 10.1186/1743-422X-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer. Nature Structural & Molecular Biology. 2005;12(11):980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zhong N., Xue F., Kang X., Ren X., Chen J., Jin C., Lou Z., Xia B. Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease. Protein & Cell. 2010;1(4):371–383. doi: 10.1007/s13238-010-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Li S., Xue F., Zou Y., Chen C., Bartlam M., Rao Z. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. Journal of Virology. 2008;82(17):8647–8655. doi: 10.1128/JVI.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Chen G., Guo B., Cheng G., Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Research. 2008;18(11):1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., George S., Schmidt M.F., Al-Gharabli S.I., Rademann J., Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Research. 2011;92(2):204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. The Journal of General Virology. 2000;81(Pt 4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Thiel V., Gorbalenya A.E. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. Journal of Biological Chemistry. 2001;276(35):33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y., Deutscher M.P. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Research. 2001;29(5):1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B., Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nature Immunology. 2011;12(2):137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathogens. 2007;3(8):e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Miller T.B., Goebel S.J., Thiel V., Masters P.S. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J. Virol. 2007;82(3):1214–1228. doi: 10.1128/JVI.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


