Highlights
-
•
Porcine epidemic diarrhea virus (PEDV) is a causative agent of porcine epidemic diarrhea; consequently, the small intestine was believed to be its only target organ.
-
•
We found that PEDV infected not only the small intestines, but also the respiratory tract.
-
•
Infection and replication of PEDV in the respiratory tract from naturally PEDV-infected piglets were examined by reverse transcription polymerase chain reaction, immunohistochemistry, and virus re-isolation.
-
•
Our observations were confirmed by experimental inoculation, and we found that PEDV infection in the respiratory tract was specifically associated with alveolar macrophages in the lung.
-
•
The discovery that PEDV infects and replicates in alveolar macrophages provides new insights into its pathogenesis.
Keywords: Porcine epidemic diarrhea, Coronavirus, Tropism, Respiratory tract, Alveolar macrophages, Pathogenesis
Abstract
Porcine epidemic diarrhea virus (PEDV) is a causative agent of porcine epidemic diarrhea; consequently, the small intestine was believed to be its only target organ. In this study, we found that PEDV infected not only the small intestines, but also the respiratory tract. Infection and replication of PEDV in the respiratory tract from naturally PEDV-infected piglets were examined by reverse transcription polymerase chain reaction, immunohistochemistry, and virus re-isolation. Our observations were confirmed by experimental inoculation, and we found that PEDV infection in the respiratory tract was specifically associated with alveolar macrophages in the lung. Vero cell-adapted PEDV was able to replicate in both primary alveolar macrophages and continuous porcine alveolar macrophage cells. Sequencing analysis of the spike (S) glycoprotein revealed that mutations in S might be a potential determinant of auxiliary targets for PEDV. The discovery that PEDV infects and replicates in alveolar macrophages provides new insights into its pathogenesis.
1. Introduction
Coronaviruses infect many animals, including humans, in a species-specific manner. Most coronaviruses in farm animals cause respiratory or gastrointestinal tract infections in young animals, resulting in huge economic losses (Saif, 2004a, Saif, 2004b, Stoddart et al., 1988, Weiss and Navas-Martin, 2005). Several viruses in the family coronaviridae infect pigs, including transmissible gastroenteritis virus (TGEV), porcine respiratory coronavirus (PRCoV), porcine hemagglutinating encephalitis virus, and porcine epidemic diarrhea virus (PEDV) (Cartwright et al., 1969, Greig et al., 1962). TGEV mainly infects epithelial cells in the small intestine and causes fatal enteritis in piglets (Enjuanes et al., 1995). Additionally, TGEV replicates in porcine respiratory tract tissues but does not induce primary respiratory disease (Kemeny et al., 1975, Saif, 2004a, Saif, 2004b). PRCoV, a spontaneously occurring variant of TGEV, replicates more extensively in the respiratory tract without causing clinical signs (Wesley et al., 1991). It has been reported that PRCoV replicates in different types of lung cells, including alveolar, bronchiolar and bronchial epithelial cells and in bronchoalveolar macrophages, but not in enterocytes (Cox et al., 1990).
PEDV is one of the major etiological agents of seasonal acute diarrhea in piglets (Debouck and Pensaert, 1980). The pathology following PEDV infection resembles that of TGEV, which is mainly characterized by acute watery diarrhea and dehydration in suckling piglets (Hess et al., 1980, Pensaert et al., 1981, Takahashi et al., 1983). Based on these symptoms, the enterocytes have largely been thought of as the target cells of PEDV infection. Indeed, experimental infection with the prototype strain CV777 showed that replication in epithelial cells covering small and large intestinal villi resulted in villous atrophy, malabsorption and diarrhea (Debouck and Pensaert, 1980). Similarly, Korean strains of PEDV caused severe villous atrophy in the duodenum, jejunum and ileum of experimentally infected pigs (Kim and Chae, 2000, Kim and Chae, 2003). Replication was mainly localized in the cytoplasm of villous epithelial cells of the small intestine and colon (Sueyoshi et al., 1995). Microscopically, marked cytoplasmic vacuolation and exfoliation of enterocytes with subsequent considerable shortening and fusion of villi were observed (Coussement et al., 1982, Pospischil et al., 1981). However, according to immunohistochemistry (IHC) results reported by Lee et al., PEDV antigens were detected not only in the cytoplasm of the villous enterocytes but also in macrophages that had infiltrated the lamina propria (Lee et al., 2000). PEDV infection in macrophages may reflect the possibility of widespread pathology or the presence of extra-gastrointestinal infection and replication, as observed in other species infected with coronaviruses (Gu et al., 2005). Most recent studies on the pathogenesis of PEDV have been limited to partial autopsies of the gastrointestinal tract. Therefore, a comprehensive approach to identify pathogenesis in different organs or cell types was needed.
In this study, we used histological and molecular approaches to acquire a better understanding of PEDV pathogenesis. The presence of PEDV genome and antigens in both the small intestines and respiratory tracts of PEDV-infected piglets suggests that, in addition to the small intestine, the porcine respiratory tract could also be a replication site for PEDV.
2. Materials and methods
2.1. Cells and viruses
African green monkey kidney cells (Vero, CCL-81) were prepared in minimum essential medium (MEM, Gibco-BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco-BRL). 3D4 cells are a porcine monomyeloid cell line, which was established following transfection of primary porcine alveolar macrophage cultures with plasmid pSV3neo carrying genes for neomycin resistance and SV40 large T antigen (Weingartl et al., 2002). 3D4 cells were maintained in RPMI 1640 (Gibco-BRL) supplemented with 10% FBS, 10 mM HEPES (Gibco-BRL), 1.0 mM sodium pyruvate (Gibco-BRL) and 0.1 mM nonessential amino acids (Gibco-BRL) (Weingartl et al., 2002). PK-15 and MARC-145 cells were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco-BRL) supplemented with 10% FBS. All cells were incubated in a humidified incubator at 37 °C with 5% CO2.
The Korean Vero cell-adapted PEDV isolates, KPEDV-9 and SM98LVec, were propagated as described previously (Hofmann and Wyler, 1988). Briefly, Vero cells were inoculated with virus at a multiplicity of infection (m.o.i.) of 1 and cultured in serum-free MEM for 48 h at 37 °C with 5% CO2. The progeny virions were titrated using the TCID50 method. CSFV (LOM strain) and PRRSV (VR-2332 strain) were propagated in PK-15 cells and MARC-145 cells, respectively.
2.2. RT-PCR and sequence comparison
Clinical samples were obtained from piglets in the Chungman province that were exhibiting watery diarrhea and were screened by RT-PCR. Viral RNA was extracted from tissue homogenates using the Viral gene-spin viral DNA/RNA extraction kit (iNtRON Biotechnology, Sungnam, Korea) following the manufacturer's instructions. For first-strand cDNA synthesis, M-MLV reverse transcriptase (iNtRON Biotechnology, Korea) was used along with 10 μl of extracted viral RNA in a 50 μl randomly primed reaction. To exclude possible infection by other swine enteric viruses, specific primer sets were used for the detection of PEDV, TGEV and rotavirus (Song et al., 2006).
The specific primers used for the sequencing analysis of spike (S) protein were PEDV-S forward (GTGATGTTGTGTTAGGCTTGTTGAAG, 20541–20566) and PEDV-S reverse (CATCACTGCACGTGGACCTTTTC, 24769–24791). RT-PCR was performed at 94 °C for 2 min, followed by 40 cycles of 94 °C for 20 s, 54 °C for 10 s, 72 °C for 4 min, and a final extension was performed at 72 °C for 10 min. Amplified products were purified using the PCR clean-up kit following the manufacturer's instructions (GeneAll, Seoul, Korea) and sequenced. The nucleotide and deduced amino acid sequences of S protein were compared with those from published reference strains; CV777 (NC003436), DR13 (DQ462404), JS-2004-2 (AY654204), Chinju99 (AY167585), and KNU-0901 (GU180144). The multiple-sequencing alignments were generated using the MultAlin (http://multalin.toulouse.inra.fr) program, and similarities between nucleotide sequences were further assessed using the same software.
2.3. Experimental inoculation
With permission (permission number 20110345) and following the protocols approved by the Institutional Animal Care and Ethnic in Chungnam National University (CNU), Korea, all animals were cared for and experiments were performed in the animal facility at CNU. Healthy 3-day old colostrum-free piglets were inoculated orally with 2 ml of tissue homogenates from PVED infected animals or KPEDV-9. Controls were inoculated with phosphate buffered saline (PBS, pH 7.2). All piglets were examined daily for clinical signs such as diarrhea, anorexia, weight loss, vomiting, and dehydration. At 72 hpi, all piglets were euthanized and tissue samples processed for histological and molecular analysis.
2.4. Tissue sampling and histopathology
After the piglets were euthanized, necropsies were performed. Small intestines, lungs, livers, kidneys, and spleens were collected and each divided into two parts. For histological studies, tissue specimens were fixed in 10% (w/v) buffered formaldehyde for 48 h, and embedded in paraffin according to standard laboratory procedures. For virus re-isolation, tissue specimens were homogenized in PBS containing protease inhibitors (Sigma) using Tissue Lyser II homogenizer (Qiagen).
For histopathologic studies, sections were prepared from each of the formalin-fixed, paraffin-embedded tissues. These sections were deparaffinized in xylene and rehydrated through graded alcohols. Antigen retrieval was performed in retrieval buffer (pH 6) using a pressure boiler as a heat source. The endogenous peroxidase was quenched by treating sections with 3% H2O2 in methanol for 30 min. All slides were incubated with 2% skim milk for 30 min at room temperature (RT) to saturate nonspecific protein-binding sites and then incubated with primary antibodies overnight at 4 °C. Mouse polyclonal anti-PEDV, which was obtained by immunizing mice with KPEDV-9, was used as the primary antibody. After three washes with PBS, sections were incubated for 30 min with biotinylated horse anti-mouse IgG diluted 1:200 in PBS. These were washed 4 times with PBS and incubated for 30 min with alkaline phosphatase-conjugated streptavidin. The slides were then washed 4 times with PBS and processed by immersing sections in 3,3′-diaminobenzidine substrate solution (Vector Laboratories, Burlingame, CA) at RT. Sections from all tissues were also stained with hematoxylin and eosin for histopathological examination.
2.5. Virus re-isolation
Tissue homogenates from PEDV-infected piglets were centrifuged and the resulting supernatant was used for virus isolation. Each sample resulting from these tissue homogenates was diluted (1:100 for small intestinal samples, 1:5 for other organ samples) in MEM containing 10 μg/ml of trypsin and added to a Vero cell monolayer and incubated for 1 h. After adsorption, cells were washed twice with PBS and then incubated in MEM containing 10 μg/ml of trypsin at 37 °C for 48 h. PEDV-infected cells were confirmed by an indirect immunofluorescence assay using mouse polyclonal anti-PEDV antibody. Cells were stained with Texas red-conjugated goat anti-mouse IgG and observed under a fluorescence microscope.
2.6. Susceptibility of porcine alveolar macrophages to PEDV
3D4 cells were seeded in 96-well Plates 24 h prior to infection with PEDV (KPEDV-9 strain), CSFV (LOM strain) or PRRSV (VR-2332 strain) at a MOI of 1. After virus adsorption, the inoculums were removed and the cells washed twice with RPMI and incubated for 5 days at 37 °C. At the indicated time points, cells were fixed for immunostaining and progeny virions in the supernatants were titrated using the TCID50 method.
Porcine primary alveolar macrophages were isolated from healthy piglets by bronchoalveolar lavage (BAL). Briefly, the trachea was cannulated and the airways were filled with PBS. The fluid was collected from the airways using gentle suction. The procedure was repeated twice and the lavages were combined. Primary porcine alveolar macrophages were separated from BAL fluid via centrifuge at 1000 × g for 10 min. The isolated cells were cultured in RPMI 1640 containing 20% FBS for 24 h. After cells stabilized and became confluent, they were inoculated with PEDV. After virus adsorption, the inoculums were removed and the cells washed twice with PBS and incubated for 5 days at 37 °C in RPMI 1640. At 48 hpi, cells were collected for viral RNA extraction and virus infection was confirmed by RT-PCR.
For a kinetic assay, Vero and 3D4 cells were inoculated with different Vero cell-adapted strains of PEDV (KPEDV-9 and SM98LVec with m.o.i. 1). Two hours later, the cells were washed twice with PBS and incubated at 37 °C. At the indicated time points, supernatants were collected and titrated for progeny virions using the TCID50 method. At 24 hpi, infected cells were fixed with 5% paraformaldehyde and stained with mouse polyclonal anti-PEDV.
2.7. Immunocytochemistry
Cells were fixed with 5% paraformaldehyde for 5 min and permeabilized with 1% NP40. Following three washes with PBS, cells were incubated with antisera specific for PEDV, CSFV, or PRRSV for 1 h. Cells were washed 3 times with PBS and then incubated with a 1:1000 dilution of goat anti-mouse IgG conjugated with Texas red. Immunostained cells were observed under a fluorescence microscope (Zeiss, Oberkochen, Germany).
3. Results
3.1. Confirmation of wild-type PEDV infection in piglets
Piglets were scarified upon the development of watery diarrhea, which is a major clinical sign for PEDV infection. For PEDV screening, RNA was extracted from fecal samples and reverse transcription polymerase chain reaction (RT-PCR) targeting the nucleocapsid gene of PEDV was performed. To exclude possible infection by other swine enteric viruses, we also performed PCR targeting TGEV and porcine rotavirus. Of the four piglets tested, all four were positive for PEDV, but were negative for the other porcine enteric pathogens (data not shown).
It has been reported that Vero cell-adapted PEDV only infects villous epithelial cells in the small intestine in vivo (Kim and Chae, 2000, Kim and Chae, 2003). To evaluate which organs are affected by natural wild-type PEDV infection, we collected various organs (lungs, livers, kidneys, and spleens) including the small intestines and confirmed PEDV by RT-PCR. Surprisingly, PEDV infection was detected not only in the small intestine, but also in the lung, liver, and kidney from one of the four piglets we tested (Table 1 ). To evaluate tissue damage, we stained sections with hematoxylin and eosin, and to detect PEDV antigens we performed IHC on these tissue samples. As shown in Fig. 1A, severe histopathological changes were observed in the small intestine, including villous atrophy, extensive vacuolation, and flattening of the villous enterocytes. Since the small intestines were severely damaged, most PEDV-positive cells were already detached and found in the lumen while PEDV was detected in a few enterocytes in the intestinal villi (Fig. 1F). Interestingly, PEDV was also detected in lung tissue by IHC, mostly in the mixed gland and epithelial cells, but not in the alveolar respiratory bronchiole (Fig. 1G). Despite this, we were unable to find significant pathological changes in the lung (Fig. 1B and G). Samples from the liver, kidney, and spleen were negative for PEDV as evaluated by IHC, and we were unable to find significant pathological changes (Fig. 1C, D, E, H, I, and J). Together, the results from RT-PCR and histology suggest that PEDV can replicate not only in the intestine but also in the lung.
Table 1.
RT-PCR analysis of different organ samples obtained from naturally PEDV-infected piglets.
| Intestines | Lungs | Liver | Kidneys | Spleen | |
|---|---|---|---|---|---|
| CNU-091222-01 | + | + | + | + | + |
| CNU-091222-02 | + | − | − | + | + |
| CNU-091222-03 | + | − | − | − | − |
| CNU-091222-04 | + | − | − | − | − |
Fig. 1.
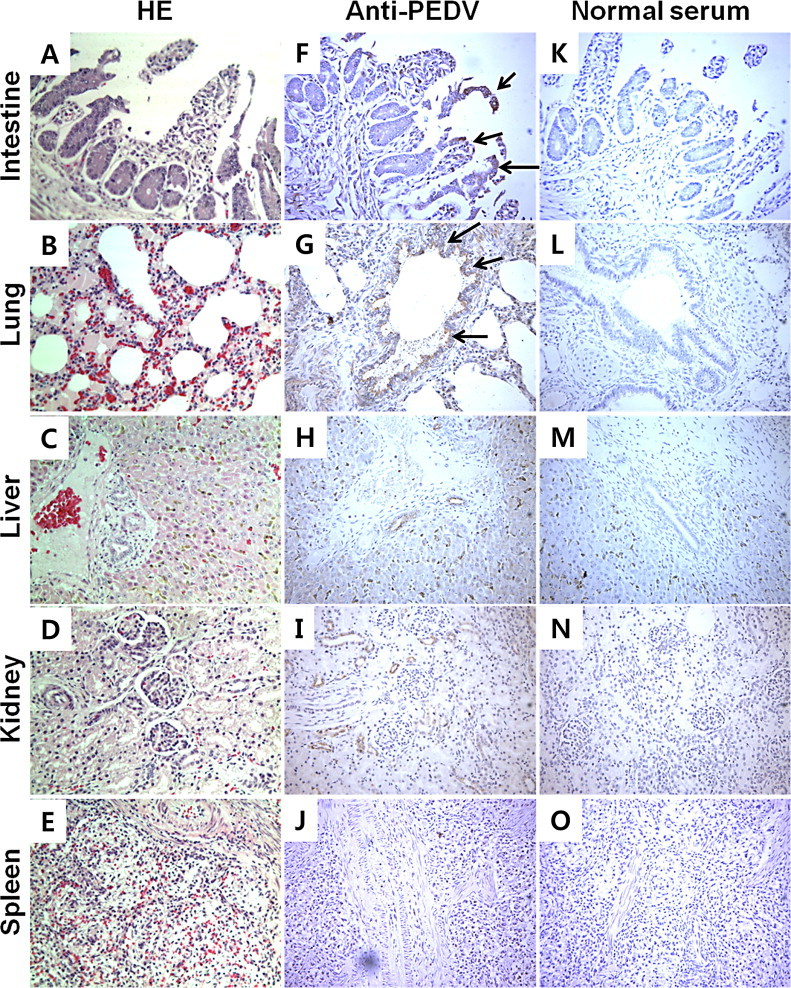
Pathological examination and IHC of samples from piglets naturally infected with PEDV. Samples from the small intestine, lung (bronchus and alveoli), liver, kidney and spleen stained with hematoxylin and eosin (HE) are shown in the left panel (A–E). Images from IHC with mouse polyclonal anti-PEDV antibody (F–J) or normal mouse serum (K–O) are shown in the middle and right panel, respectively. Arrows indicate PEDV-infected cells. Magnification: 100×.
3.2. PEDV isolation from the small intestine and alveolar macrophages
To confirm our observations from piglets that had been naturally infected with wild-type PEDV, we experimentally inoculated colostrum-free piglets with wild-type PEDV and monitored their pathology. Since it is difficult to isolate wild-type PEDV from infected piglets, we used intestinal tissue homogenates from infected piglets as inoculums. To compare pathologic differences between wild-type PEDV and Vero cell-adapted PEDV, one group was inoculated with tissue homogenates from those naturally infected piglets, another with the Vero cell-adapted KPEDV-9 strain as the positive control, and the last with PBS as the negative control. Typical clinical signs of PEDV-infected piglets, such as watery diarrhea, were observed in both the homogenate and KPEDV-9 inoculated groups at 3 days post-inoculation. As shown in Table 2 , the RT-PCR results testing for PEDV were positive in the small intestines, kidneys, and spleens from both KPEDV-9 and homogenate inoculated piglets. Lung samples obtained from homogenate inoculated piglets, but not from KPEDV-9 or PBS inoculated piglets, were positive when tested by RT-PCR.
Table 2.
RT-PCR analysis of different organ samples obtained from CNU-091222-01-, KPEDV9- or PBS-inoculated piglets.
| Inoculum | Intestine | Lung | Liver | Kidney | Spleen |
|---|---|---|---|---|---|
| PBS | − | − | − | − | − |
| KPEDV-9a | + | − | − | + | + |
| CNU-091222-01b | + | + | − | + | + |
Vero cell-adapted PEDV.
PEDV-infected tissue homogenates.
To confirm the RT-PCR results, we performed an immunohistological analysis of the small intestines and lungs (Fig. 2 ). In the small intestine, the highest level of PEDV was found in entire intestinal villi epithelial cells (Fig. 2B and C). Highly increased cellularity was observed in the lung, possibly owing to the infiltration of inflammatory cells (Fig. 2E). As shown in Fig. 2H, morphologically identical lymphocytes and macrophages were found in the alveolar interstitium, and PEDV was detected in mixed glands in the bronchus and alveoli. This pattern was similar to our findings from naturally infected piglets. With higher magnification, we were able to detect PEDV in lung cells, predominantly in alveolar macrophages (arrows in Fig. 2H). We were unable to detect PEDV in any of the liver, kidney or spleen samples (Data not shown).
Fig. 2.
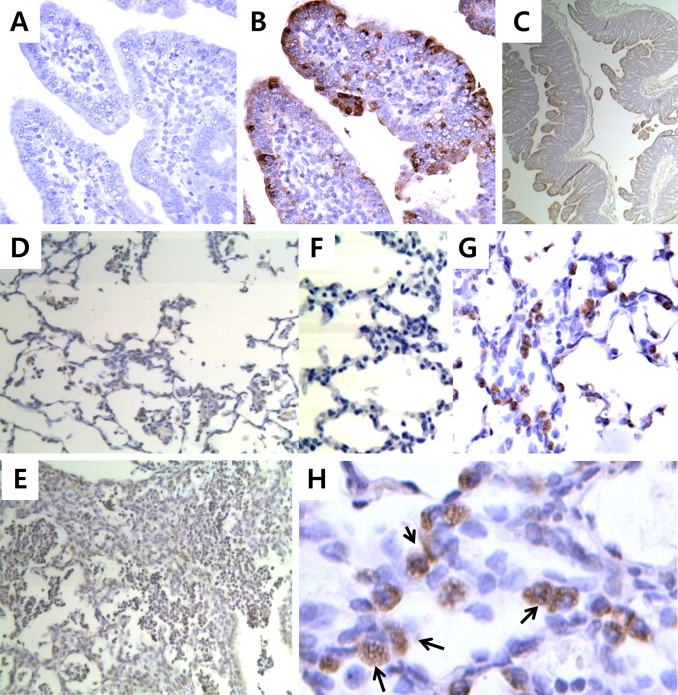
IHC with mouse polyclonal anti-PEDV of samples from piglets inoculated with PEDV-infected tissue homogenates. Intestinal sections from the PBS-inoculated (A) and homogenate-inoculated piglets (B and C) at 96 hpi. Lung sections from PBS-inoculated (D and F) and homogenate-inoculated piglets (G, E and H) at 96 hpi. Arrows indicate PEDV-infected porcine alveolar macrophages. Magnification: C–E, 40×; A, B, F and G, 400×; H, 1000×.
We re-isolated the virus from tissue homogenates from the experimentally infected piglets. As shown in Fig. 3 , wild-type PEDV was isolated from Vero cells infected with homogenates made from the small intestines and from the alveolar macrophages of PEDV-infected piglets, indicating that PEDV had infected and replicated in both small intestine and alveolar macrophages. To the best of our knowledge, this is the first time replication in alveolar macrophages has been observed for PEDV and this may provide important new insights into its pathogenesis.
Fig. 3.
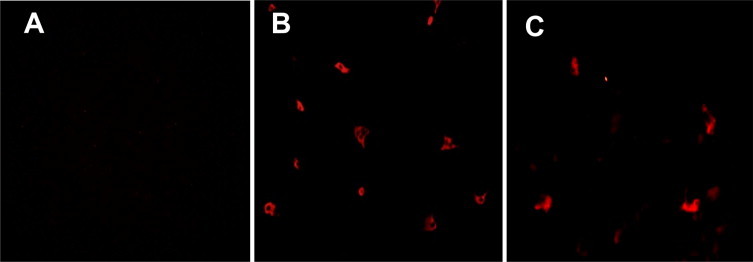
PEDV isolation from intestines and alveolar macrophages. Vero cells were inoculated with small intestine homogenates (A) from PBS-inoculated piglets, or small intestine homogenates (B) or alveolar macrophage homogenates (C) from wild-type PEDV-inoculated piglets. At 24 hpi, infected Vero cells were fixed and stained with mouse polyclonal anti-PEDV. Scale bar = 100 μm.
3.3. Replication of PEDV in porcine alveolar macrophages
Previous data suggested that alveolar macrophages might be a target cell type for PEDV infection in the respiratory tract. For further confirmation of the susceptibility of porcine alveolar macrophages to PEDV, 3D4 cells (continuous porcine alveolar macrophages) were inoculated with KPEDV-9, classical swine fever virus (CSFV, as positive control), or porcine respiratory and reproductive syndrome virus (PRRSV, as negative control). At 24 h post-infection (hpi), progeny virions in cell-free supernatants were titrated in susceptible cell lines (Vero cells for PEDV, PK15 cells for CSFV, and MARC145 cells for PRRSV). We found that 3D4 cells supported KPEDV-9 replication; however, the susceptibility of 3D4 cells was much lower compared with that of Vero cells, which showed a titer about 10-fold higher than 3D4 cells (Fig. 4A). To determine whether primary alveolar macrophages are susceptible to PEDV, primary porcine alveolar macrophages were collected from piglets by lung lavage and inoculated with KPEDV-9 or SM98LVec. At 24 hpi, virus infection was assessed by RT-PCR. As shown in Fig. 4B, both KPEDV-9 and SM98LVec inoculated primary porcine alveolar macrophages were positive for PEDV.
Fig. 4.
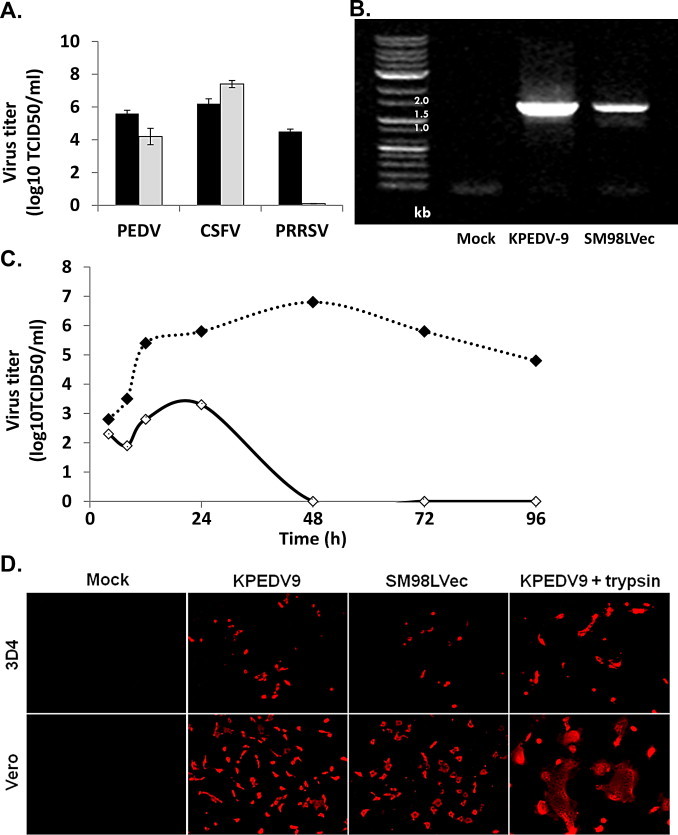
PEDV infection in porcine alveolar macrophages. (A) The susceptibility of 3D4 cells to PEDV. The 3D4 cells were infected with PEDV, CSFV (LOM strain), and PRRSV (VR-2332 strain), and progeny viruses were titrated in appropriate cell lines. Titers of 3 different swine viruses in the 3D4 cells (gray bar) were compared. For better understanding, viral titer was also compared with those in known susceptible cells (black bar); Vero cells for PEDV, PK15 cells for CSFV, and MARC145 cells for PRRSV, respectively. (B) The susceptibility of porcine primary alveolar macrophages to PEDV. Primary alveolar macrophages were collected and infected with Vero cell-adapted strains of PEDV and then virus infectivity was evaluated by RT-PCR. (C) Viral growth kinetics in alveolar macrophage cell line. Vero and 3D4 cells were inoculated with KPEDV9 and progeny viruses were titrated at the indicated time points. Vero (black circles) and 3D4 (white circles) cells were inoculated with Vero cell-adapted strains of PEDV and progeny viruses were titrated at the indicated time points. In Vero cells, the amount of KPEDV-9 increased continuously and gradually until reaching a titer of 107 TCID50 around 48 hpi, which slowly reduced afterwards. In 3D4 cells, the titer of KPEDV-9 increased slightly, reaching 103.4 TCID50 by 24 hpi. The titer was slowly decreased by 48 hpi, and we were unable to detect any infectious viruses after 48 hpi. (D) Vero and 3D4 cells were inoculated with Vero cell adapted PEDVs (KPEDV9 or SM98LVec) in supplemented with 10 μg/ml trypsin as indicated, and the cells were stained with mouse polyclonal anti-PEDV at 24 h after infection. Scale bar = 100 μm. A reduced infection in 3D4 cells was also supported by the results of an immunofluorescence assay. The percentage of PEDV-infected cells was lower in 3D4 cells than in Vero cells. We also compared CPE formation with trypsin condition.
Furthermore, we examined the growth kinetics of KPEDV-9 in Vero cells and 3D4 cells. In Vero cells, the amount of KPEDV-9 increased continuously and gradually until reaching a titer of 107 TCID50 (50% tissue culture infectious dose) around 48 hpi, which slowly reduced afterwards (Fig. 4C). We were still able to detect PEDV after 96 hpi. In 3D4 cells, the titer of KPEDV-9 increased slightly, reaching 103.4 TCID50 by 24 hpi. The titer was slowly decreased by 48 hpi, and we were unable to detect any infectious viruses after 48 hpi. A reduced infection in 3D4 cells was also supported by the results of an immunofluorescence assay. The percentage of PEDV-infected cells was lower in 3D4 cells than in Vero cells (Fig. 4D). Although the growth of PEDV is slower and peaks at lower levels in 3D4 cell lines than in Vero cells, these findings confirm the susceptibility of porcine alveolar macrophages to PEDV, and show that PEDV can successfully replicate in porcine alveolar macrophages.
3.4. Comparison of PEDV spike (S) protein
The S protein of PEDV consists of an N-terminal domain and a C-terminal domain. The N-terminal domain is responsible for viral attachment. It is also a major determinant that recognizes receptors and affects tissue tropism. To determine the impact of S sequences on PEDV tropism, S genes were amplified and compared with reference sequences. The sequence analysis showed that the S gene consisted of 4161 nucleotides and 1386 amino acids. The similarities among S proteins are described in Table 3 . The S protein in our sample shared high levels of identity with KNU-0901, which was previously reported as Korean wild-type strain (95.0 and 94.2% homology of nucleotide and amino acid sequences, respectively). However, the similarity with other strains, including the reference strain CV777, was relatively low and showed similarities ranging from 93.9 to 94.0% and 91.9 to 92.9% identity in nucleotide and amino acid sequences, respectively. As shown in Fig. 5 , more mutations were observed in the N-terminal portions of the S proteins. New PEDV isolates were in different cluster from those published as shown in Fig. 6 .
Table 3.
Comparisons of the amino acid sequences on S protein from newly isolated PEDV with other reference strains.
| Strains | Amino acid homology (%) |
||||
|---|---|---|---|---|---|
| CV777 | DR13 | Chinju99 | KNU-0901 | CNU-091222-01 | |
| CV777a | – | 99.7 | 92.8 | 93.5 | 92.9 |
| DR13b | 99.9 | – | 92.6 | 93.4 | 92.8 |
| Chinju99c | 94.4 | 94.3 | – | 95.2 | 91.9 |
| KNU-0901d | 94.2 | 94.2 | 96.3 | – | 94.2 |
| CNU-091222-01e | 93.9 | 93.9 | 94.0 | 95.0 | – |
The prototype strain (Belgium, 1988, AF353511).
Korean PEDV vaccine strain (Korea, 2006, DQ862099).
Korean PEDV field isolate (Korea, 1999, AY167585).
Sequenced S protein of Korean field sample (Korea, 2009, GU180145).
Sequenced S protein of Korean field sample in the paper (Korea, 2009, JN184634).
Fig. 5.
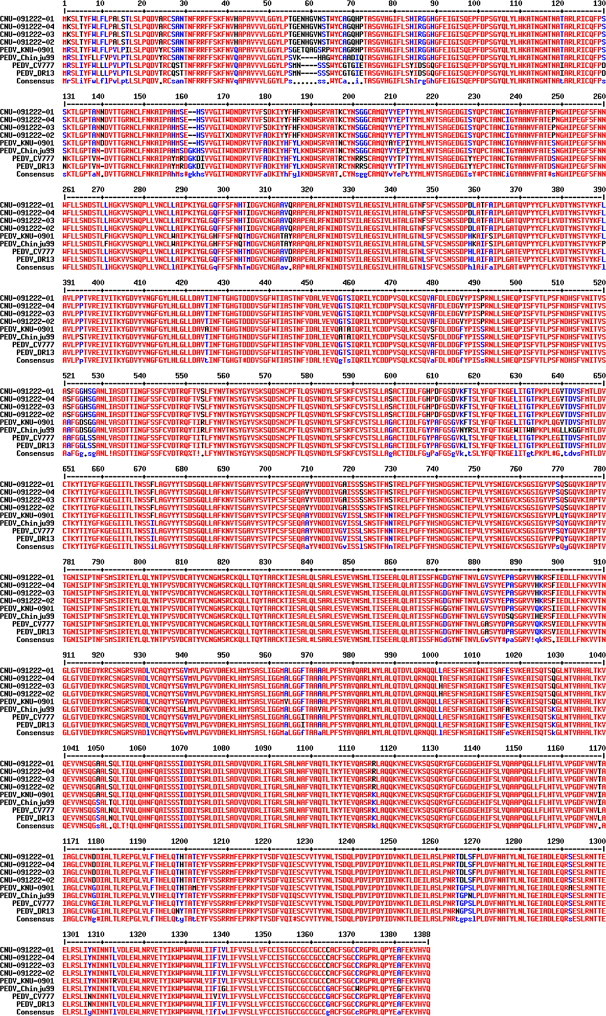
Sequence comparison and alignment report on S proteins. The deduced amino acid sequences of the S protein of different PEDV strains were aligned using the MultiAlign program. Numbers indicate amino acid position.
Fig. 6.
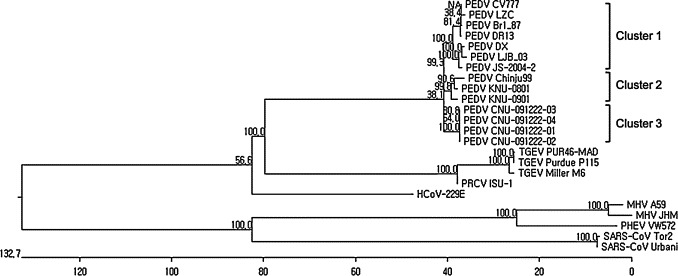
Phylogenetic analysis based on the amino acid sequences. The phylogenetic tree was generated using the Clustal W method following multiple alignments of amino acid sequences. Putatively similar regions of spike proteins of other distantly related coronaviruses were also included in this tree. The numbers represent bootstrap values greater than 50% of 1000 replicates. New PEDV isolates were in different cluster from those published.
Interestingly, all four sequenced S proteins showed similar sequences although their RT-PCR results on different tissues were similar but still bit different. These data indicate that the recent Korean field isolate is genetically different from previously published strains. It also suggests that mutations on S might affect cellular tropism of wild-type PEDV, allowing a respiratory infection in addition to infection of the small intestines. We still do not know which mutation(s) are involved in changes in tropism. However, further examination is required to confirm this hypothesis.
4. Discussion
The most obvious clinical symptom of PEDV infection in piglets is watery diarrhea, which has led to the belief that the target cells for PEDV were limited to porcine enterocytes (Sueyoshi et al., 1995). Until now, PEDV replication has not been reported in cells other than those of the intestinal tract, with the exception of a few studies that have reported PEDV infection in macrophages as well as enterocytes from the gastrointestinal tract (Lee et al., 2000). Here, we evaluated the pathogenicity of new wild-type PEDV in comparison with Vero cell-adapted KPEDV-9. We found that new wild-type PEDV, but not Vero cell adapted KPEDV-9 strain, can infect not only small intestines but also the respiratory tract, especially porcine alveolar macrophages. We are not so sure whether all the new PEDV wild type has same activity or not and this findings could be limited to this specific isolate. Further studies required with other isolates, but there is big limitation on new isolation. Genetic analyses suggest that the extra-intestinal tropism of wild-type PEDV might be obtained by natural mutations on PEDV S.
As the author's knowledge, this study is the first to demonstrate the extra-intestinal tropism of wild-type PEDV in piglets. In piglets naturally infected with wild-type PEDV, we observed that PEDV is able to infect not only in the small intestine but also other organs, including the lung. We further investigated this observation in piglets, which were experimentally inoculated with infected tissue homogenates and compared with piglets inoculated with Vero cell-adapted KPEDV-9. As with the piglets that were naturally infected with wild-type PEDV, the virus was detected in both small intestines and lungs of experimentally inoculated animals. Additionally, we found that lung infection of PEDV was associated with alveolar macrophages. These results indicate that PEDV replication in the lungs might be related to increased cellularity and partially owing to infiltration of macrophages into the alveolar and interalveolar septa.
For further confirmation of PEDV tropism in alveolar macrophages, we tested whether porcine alveolar macrophages were susceptible to PEDV. We found that both primary porcine alveolar macrophages and 3D4 cells were susceptible to PEDV and able to support PEDV replication. However, growth of Vero cell-adapted PEDV in 3D4 cells was relatively slow and inefficient compared with its growth in Vero cells. It is not clear why PEDV was unable to replicate as successfully in 3D4 cells as it does in Vero cells. Our sequencing results indicate that wild-type PEDV has differences in the N-terminal domain of its S protein sequences compared with other reference and Vero cell-adapted strains of PEDV. We propose that the changes in amino acids in the N-terminal domain may be involved in the observed tropism changes, and thus Vero cell-adapted PEDV strains may have lost their ability to infect alveolar macrophages. Similarly, PRCoV, a deletion mutant of TGEV with deletion of nucleotides 621–681 in the S gene, is non-enteropathogenic and has a tropism for respiratory epithelium and alveolar macrophages (Sanchez et al., 1999). Although the growth curves and titers were different in 3D4 cells from those produced by infected Vero cells, the fact that the virus was able to grow at all suggests that macrophages may be another possible target for PEDV replication. It seems likely that the ability to induce pathogenesis might be largely dependent on the ability of the S protein to mediate entry into target cells.
The inefficient infection in alveolar macrophages also suggests the possibility of a persistent infection or presence of PEDV in pig farms. The same farms experienced repeated outbreaks and some farms were found to be positive for PEDV infection most of the time. Based on our findings, we hypothesize that alveolar macrophages may be susceptible to PEDV although they are probably not a primary replication site. Perhaps, when protective antibodies are generated by vaccination or infection, PEDV might escape into alveolar macrophages. As such, these cells might make a good target for persistence. We found many sows tested that confirmed positive for PEDV, as measured by RT-PCR of their lung samples, although they have never shown any of clinical signs of infection (personal observations). We also found many PEDV-positive cases from day-old piglet samples, as measured by RT-PCR. We can only explain these positive results from day-old piglet samples if the infection spread from sows during pregnancy or delivery. In summary, we hypothesize that alveolar macrophages are not the primary target or a common replication site for PEDV, but they are likely important for its survival and pathogenesis.
To the best of our knowledge, this is the first report on PEDV lung infection. Our results clearly showed that (i) wild-type PEDV exhibits heterogeneity in the S protein with other PEDV strains that induce only enteric diseases, (ii) PEDV can replicate in multiple organs other than the small intestine, and (iii) PEDV is able to replicate and infect porcine macrophages in vivo and in vitro. However, further studies are still required to fully understand the mechanism of PEDV infection since our findings conflict with the previous understanding of PEDV replication sites and pathogenesis. Although we found some variations in sequence with other isolates, it remains unknown exactly where and what the critical factors are which cause these changes in its pathogenesis. Additional studies with other stains will be required to resolve these questions. In conclusion, our confirmation of PEDV lung infection provides a new perspective on the tropism of this virus and could be an important finding for the management of PEDV pathogenesis in pig farms.
5. Conclusions
Our observations were confirmed by experimental inoculation, and we found that PEDV infection in the respiratory tract was specifically associated with alveolar macrophages in the lung. Vero cell-adapted PEDV was able to replicate in both primary alveolar macrophages and continuous porcine alveolar macrophage cells. The discovery that PEDV infects and replicates in alveolar macrophages provides new insights into its pathogenesis.
Acknowledgments
This research was supported by a grant from the Technology Development Program for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries, and the Korea Research Foundation Grant funded by the Korean Government (MOEHRD No. 2011-0023942).
References
- Cartwright S.F., Lucas M., Cavill J.P., Gush A.F., Blandford T.B. Vomiting and wasting disease of piglets. Vet. Rec. 1969;84:175–176. doi: 10.1136/vr.84.7.175. [DOI] [PubMed] [Google Scholar]
- Coussement W., Ducatelle R., Debouck P., Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. I. Histological and histochemical study. Vet. Pathol. 1982;19:46–56. doi: 10.1177/030098588201900108. [DOI] [PubMed] [Google Scholar]
- Cox E., Hooyberghs J., Pensaert M.B. Sites of replication of a porcine respiratory coronavirus related to transmissible gastroenteritis virus. Res. Vet. Sci. 1990;48:165–169. doi: 10.1016/S0034-5288(18)30984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck P., Pensaert M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am. J. Vet. Res. 1980;41:219–223. [PubMed] [Google Scholar]
- Enjuanes L., Smerdou C., Castilla J., Anton I.M., Torres J.M., Sola I., Golvano J., Sanchez J.M., Pintado B. Development of protection against coronavirus induced diseases. A review. Adv. Exp. Med. Biol. 1995;380:197–211. doi: 10.1007/978-1-4615-1899-0_34. [DOI] [PubMed] [Google Scholar]
- Greig A.S., Mitchell D., Corner A.H., Bannister G.L., Meads E.B., Julian R.J. A hemagglutinating virus producing encephalomyelitis in baby pigs. Can. J. Comp. Med. Vet. Sci. 1962;26:49–56. [PMC free article] [PubMed] [Google Scholar]
- Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z., Zhuang H., Wu B., Zhong H., Shao H., Fang W., Gao D., Pei F., Li X., He Z., Xu D., Shi X., Anderson V.M., Leong A.S. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess R.G., Bollwahn W., Pospischil A., Heinritzi K., Bachmann P.A. Current aspects in the etiology of viral diarrheas of swine: occurrence of infections with the epizootic viral diarrhea (EVD) virus. Berlin. Munch. Tierarztl. Wochenschr. 1980;93:445–449. [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemeny L.J., Wiltsey V.L., Riley J.L. Upper respiratory infection of lactating sows with transmissible gastroenteritis virus following contact exposure to infected piglets. Cornell Vet. 1975;65:352–362. [PubMed] [Google Scholar]
- Kim O., Chae C. Experimental infection of piglets with a Korean strain of porcine epidemic diarrhoea virus. J. Comp. Pathol. 2003;129:55–60. doi: 10.1016/S0021-9975(02)00170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O., Chae C. In situ hybridization for the detection and localization of porcine epidemic diarrhea virus in the intestinal tissues from naturally infected piglets. Vet. Pathol. 2000;37:62–67. doi: 10.1354/vp.37-1-62. [DOI] [PubMed] [Google Scholar]
- Lee H.M., Lee B.J., Tae J.H., Kweon C.H., Lee Y.S., Park J.H. Detection of porcine epidemic diarrhea virus by immunohistochemistry with recombinant antibody produced in phages. J. Vet. Med. Sci. 2000;62:333–337. doi: 10.1292/jvms.62.333. [DOI] [PubMed] [Google Scholar]
- Pensaert M.B., Debouck P., Reynolds D.J. An immunoelectron microscopic and immunofluorescent study on the antigenic relationship between the coronavirus-like agent, CV 777, and several coronaviruses. Arch. Virol. 1981;68:45–52. doi: 10.1007/BF01315166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospischil A., Hess R.G., Bachmann P.A. Light microscopy and ultrahistology of intestinal changes in pigs infected with epizootic diarrhoea virus (EVD): comparison with transmissible gastroenteritis (TGE) virus and porcine rotavirus infections. Zentralbl. Veterinarmed. B. 1981;28:564–577. doi: 10.1111/j.1439-0450.1981.tb01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J. Animal coronavirus vaccines: lessons for SARS. Dev. Biol. (Basel) 2004;119:129–140. [PubMed] [Google Scholar]
- Saif L.J. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev. Sci. Tech. 2004;23:643–660. doi: 10.20506/rst.23.2.1513. [DOI] [PubMed] [Google Scholar]
- Sanchez C.M., Izeta A., Sanchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Duran J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Kang B.K., Oh J.S., Ha G.W., Yang J.S., Moon H.J., Jang Y.S., Park B.K. Multiplex reverse transcription-PCR for rapid differential detection of porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine group A rotavirus. J. Vet. Diagn. Invest. 2006;18:278–281. doi: 10.1177/104063870601800309. [DOI] [PubMed] [Google Scholar]
- Stoddart M.E., Gaskell R.M., Harbour D.A., Pearson G.R. The sites of early viral replication in feline infectious peritonitis. Vet. Microbiol. 1988;18:259–271. doi: 10.1016/0378-1135(88)90092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi M., Tsuda T., Yamazaki K., Yoshida K., Nakazawa M., Sato K., Minami T., Iwashita K., Watanabe M., Suzuki Y. An immunohistochemical investigation of porcine epidemic diarrhoea. J. Comp. Pathol. 1995;113:59–67. doi: 10.1016/S0021-9975(05)80069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Okada K., Ohshima K. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Nihon Juigaku Zasshi. 1983;45:829–832. doi: 10.1292/jvms1939.45.829. [DOI] [PubMed] [Google Scholar]
- Weingartl H.M., Sabara M., Pasick J., van Moorlehem E., Babiuk L. Continuous porcine cell lines developed from alveolar macrophages: partial characterization and virus susceptibility. J. Virol. Methods. 2002;104:203–216. doi: 10.1016/S0166-0934(02)00085-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R.D., Woods R.D., Cheung A.K. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. J. Virol. 1991;65:3369–3373. doi: 10.1128/jvi.65.6.3369-3373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


