Abstract
Our knowledge about the structure and function of the nonstructural proteins (nsps) encoded by the arterivirus replicase gene has advanced in recent years. The continued characterization of the nsps of the arterivirus prototype equine arteritis virus has not only corroborated several important functional predictions, but also revealed various novel features of arteriviral replication. For porcine reproductive and respiratory syndrome virus (PRRSV), based on bioinformatics predictions and experimental studies, a processing map for the pp1a and pp1ab replicase polyproteins has been developed. Crystal structures have been resolved for two of the PRRSV nonstructural proteins that possess proteinase activity (nsp1α and nsp4). The functional characterization of the key enzymes for arterivirus RNA synthesis, the nsp9 RNA polymerase and nsp10 helicase, has been initiated. In addition, progress has been made on nsp functions relating to the regulation of subgenomic mRNAs synthesis (nsp1), the induction of replication-associated membrane rearrangements (nsp2 and nsp3), and an intriguing replicative endoribonuclease (nsp11) for which the natural substrate remains to be identified. The role of nsps in viral pathogenesis and host immunity is also being explored, and specific nsps (including nsp1α/β, nsp2, nsp4, nsp7, and nsp11) have been implicated in the modulation of host immune responses to PRRSV infection. The nsp3–8 region was identified as containing major virulence factors, although mechanistic information is scarce. The biological significance of PRRSV nsps in virus-host interactions and the technical advancements in engineering the PRRSV genome by reverse genetics are also reflected in recent developments in the area of vaccines and diagnostic assays.
Keywords: RNA synthesis, Viral enzymes, Nonstructural proteins, Vaccines, Diagnostic assays
1. Introduction
Porcine reproductive and respiratory syndrome (PRRS) continues to be a threat to the swine industry worldwide. The etiological agent, porcine reproductive and respiratory syndrome virus (PRRSV) belongs to the family Arteriviridae (order Nidovirales), which also includes equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV), and simian hemorrhagic fever virus (SHFV) (den Boon et al., 1991, Conzelmann et al., 1993, Meulenberg et al., 1993, Cavanagh, 1997, Snijder and Meulenberg, 1998, Gorbalenya et al., 2006). Comparative genome sequence analysis has revealed that PRRSV segregates into two distinct genotypes, European (Type I) and North American (Type II) PRRSV (Allende et al., 1999, Nelsen et al., 1999, Ropp et al., 2004).
Arteriviruses are small, enveloped viruses (diameter about 65 nm) containing a single-stranded RNA genome of positive polarity. This polycistronic RNA molecule is about 13–16 kb in length (∼15 kb in the case of PRRSV) and contains at least nine open reading frames (Fig. 1 ). The replicative enzymes of arteriviruses are encoded in ORF1a and ORF1b, which are situated in the 5′-proximal three quarters of the genome. ORF1a and ORF1b encode two long nonstructural polyproteins, pp1a and pp1ab, with expression of the latter depending on a ribosomal frameshift signal in the ORF1a/ORF1b overlap region (den Boon et al., 1991, Snijder and Meulenberg, 1998). Following their synthesis from the genomic mRNA template, the arterivirus pp1a and pp1ab replicase polyproteins are processed into functional nonstructural proteins (nsps) by a complex proteolytic cascade that is directed by four (PRRSV/LDV) or three (EAV) proteinase domains encoded in ORF1a. Over the past two decades, by using bioinformatics, nsp-specific antisera, and various experimental systems, a tentative processing scheme has been developed for EAV pp1a and pp1ab (Snijder and Meulenberg, 1998, Ziebuhr et al., 2000, van Aken et al., 2006a). For PRRSV, polyprotein processing of the nsp3–12 region by the nsp4 ‘main protease’ is thought to largely follow the same outline, in particular since this is the most conserved part of the replicase and both nsp4 and all of its cleavage sites identified for EAV are fully conserved in the sequences of PRRSV and other arteriviruses. However, a number of important differences are found in the nsp1–2 region, and these will be discussed below, in particular the presence of an additional autoprotease, cleaving the nsp1 region into nsp1α and nsp1β, and a remarkable size difference for nsp2.
Fig. 1.
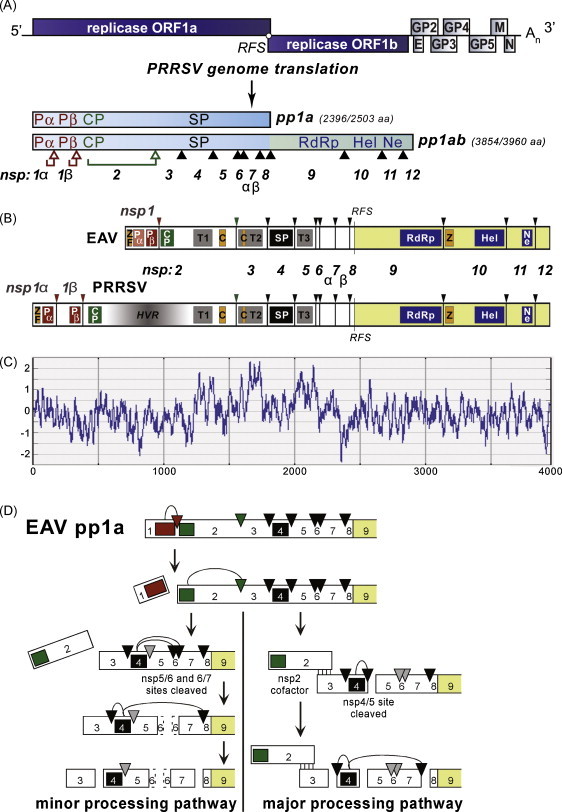
Processing and functional domains of the PRRSV replicase, and comparison to EAV. (A). Top: PRRSV genome organization, showing the 5′-proximal replicase open reading frames (ORFs) 1a and 1b, as well as the downstream ORFs encoding the viral structural proteins envelope (E), membrane (M), nucleocapsid (N), and glycoproteins (GP) 2–5 and the 3′ poly(A) tail (An). Bottom: overview of the pp1a and pp1ab replicase polyproteins that derive from genome translation, with pp1ab synthesis requiring a -1 ORF1a/1b ribosomal frameshift (RFS). Arrowheads represent sites cleaved by the four virus-encoded proteases (open for sites cleaved by the three N-proximal ‘accessory proteases’, the cysteine proteases PCPα [Pα], PCPβ [Pβ], and CP; closed for sites processed by the ‘main protease’, the nsp4 serine protease [SP]). The numbering/nomenclature of the resulting nonstructural proteins (nsp) is indicated. In addition to the four protease domains, three key enzymatic domains are depicted: the nsp9 RNA-dependent RNA polymerase (RdRp), the nsp10 helicase (Hel), and the nsp11 endoribonuclease (Ne). Adapted from Nedialkova et al. (2010). (B). Comparison of the replicase polyprotein processing maps of PRRSV and the arterivirus prototype EAV. Note the presence of an uncleaved nsp1 in EAV and nsp1α and nsp1β subunits in PRRSV, and the remarkable size difference between the nsp2 subunits of both viruses. ZF: zinc finger; HVR: hypervariable region; T: (putative) transmembrane domain; C: cluster of conserved Cys residues; Z: predicted zinc-binding domain. (C). Kyte & Doolittle hydrophobicity plot (window size 21; above the axis is hydrophobic) of replicase pp1ab of PRRSV (strain VR2332). Prominent hydrophobic domains are present in nsp2, nsp3, and nsp5, and are thought to mediate the membrane association of the replication and transcription complex. (D). Model for alternative processing of the EAV nsp3–8 region in pp1a (and, likely, also pp1ab), as proposed by Wassenaar et al. (1997). The association of cleaved nsp2 with nsp3–8 was shown to direct cleavage of the nsp4/5 site by the nsp4 SP (major pathway). Alternatively, in the absence of nsp2, the nsp5/6 and 6/7 sites were found to be processed and the nsp4/5 junction remained uncleaved. The status of the small nsp6 subunit (fully cleaved or partially associated with nsp5 or nsp7) remains to be elucidated.
The key enzymes for arterivirus RNA synthesis are encoded in ORF1b, in particular the viral RNA-dependent RNA polymerase (RdRp; nsp9) and RNA helicase (Hel; nsp10). Together with putative “accessory subunits”, these enzymes assemble into a membrane-associated viral replication and transcription complex (RTC; Pedersen et al., 1999), which mediates both genome replication and the synthesis of a nested set of subgenomic (sg) mRNAs. The latter, a hallmark of all nidoviruses, form a 5′- and 3′-coterminal nested set (de Vries et al., 1990) and is required to express the arterivirus structural protein genes (Snijder and Meulenberg, 1998, Pasternak et al., 2006). In addition to the specific viral nsps discussed below, regulatory RNA sequences are critical for sg mRNA synthesis, which is thought to rely on discontinuous minus-strand RNA synthesis to produce the templates for sg mRNA synthesis, a mechanism shared with coronaviruses. The mechanistic details of corona- and arterivirus sg mRNA synthesis have recently been summarized in the reviews of e.g. Pasternak et al. (2006), Snijder and Spaan (2006), and Sawicki et al. (2007), and will not be addressed in detail here, with the exception of arterivirus nsp1 function. In addition to their decisive role in viral RNA synthesis, it has become clear that certain arterivirus nsps or nsp domains may serve specific roles in viral pathogenesis and antagonizing host immunity.
In this review, we will discuss the current knowledge on the structure, function, and biochemistry of PRRSV nsps, obtained from studies in cell culture systems, reverse genetics, and animal models. For several aspects, this information will be placed against the background of the EAV replicase, which has been more extensively characterized in terms of nsp molecular biology and biochemistry. We will summarize (i) our current knowledge of the expression and molecular biology of PRRSV nsps and their functions in viral replication; (ii) recent studies of PRRSV nsps suggesting that they may modulate host immune responses and viral pathogenesis; (iii) the application of our knowledge on PRRSV nsps in the development of vaccines and diagnostic assays. Finally, we outline future perspectives concerning arterivirus replicase research and discuss how such information may lead to improved strategies for diagnosis and prevention of PRRS.
2. PRRSV replicase processing and nsp functions in viral RNA synthesis
2.1. Proteolytic processing of the replicase polyproteins
As in all nidoviruses, arterivirus nsps derive from the post-translational processing of two large precursor polyproteins, pp1a and pp1ab. The proteolytic cleavages yielding the individual PRRSV nsps (Fig. 1A–B; Table 1 ) were initially predicted from comparative sequence analysis (Ziebuhr et al., 2000) in combination with data from experimental studies on EAV, the arterivirus prototype (Fig. 1B). Likewise, the initial functional assignments of various individual arterivirus nsps were largely based on sequence analysis (within the arterivirus group and in the broader context of the nidovirus order) and the dissection of the EAV replicative cycle through molecular biological, biochemical, and (reverse) genetics approaches.
Table 1.
PRRSV polyprotein pp1ab cleavage products and their (potential) functions.
| Cleavage Product | (predicted) N- and C-terminal residues of each cleavage product in polyprotein pp1aba |
Known or predicted nsp properties/functionsb | |
|---|---|---|---|
| Type I PRRSV | Type II PRRSV | ||
| nsp1α | Met1–? (? aa) | Met1–Met180 (180 aa) | zinc finger-protein; accessory protease PCPα; regulator of sg mRNA synthesis; potential IFN antagonist |
| nsp1β | ?–Gly385 (? aa) | Ala181–Gly383 (203 aa) | accessory protease PCPβ; potential IFN antagonist |
| nsp2 | Ala386–Gly1463c (1078 aa) | Ala384–Gly1579c (1196 aa) | accessory protease CP; deubiquitinating (DUB) enzyme (OTU class); potential IFN antagonist; transmembrane protein, involved in membrane modification |
| nsp3 | Ala1464–Glu1693 (230 aa) | Gly1580–Glu1809 (230 aa) | transmembrane protein, involved in membrane modification |
| nsp4 | Gly1694–Glu1896 (203 aa) | Gly1810–Glu2013 (204 aa) | main protease SP |
| nsp5 | Gly1897–Glu2066 (170 aa) | Gly2014–Glu2183 (170 aa) | transmembrane protein, possibly involved in membrane modification |
| nsp6 | Gly2067–Glu2082 (16 aa) | Gly2184–Glu2199 (16 aa) | ? |
| nsp7α | Ser2083–Glu2231 (149 aa) | Ser2200–Glu2348 (149 aa) | ? |
| nsp7β | Asn2232–Glu2351 (120 aa) | Asn2349–Glu2458 (110 aa) | ? |
| nsp8 | Ala2352–Cys2396d (45 aa) | Ala2459–Cys2503d (45 aa) | ? |
| nsp9 | Ala2352–Glu3036 (685 aa) | Ala2459–Glu3143 (685 aa) | RNA-dependent RNA polymerase |
| nsp10 | Gly3037–Glu3478 (442 aa) | Gly3144–Glu3584 (441 aa) | NTPase; RNA helicase; contains putative zinc-binding domain |
| nsp11 | Gly3479–Glu3702 (224 aa) | Gly3585–Glu3807 (223 aa) | endoribonuclease (NendoU) |
| nsp12 | Gly3703–Pro3854 (152 aa) | Gly3808–Asn3960 (153 aa) | ? |
Amino acid numbers refer to the pp1ab sequence of Type I PRRSV strain Lelystad (GenBank accession #M96262) or Type II PRRSV strain VR2332 (GenBank accession #PRU87392).
PCP: Papain-like cysteine proteine protease; CP: cysteine protease; SP: serine protease;? unknown function or cleavage site.
Adjusted compared to earlier predictions to match the general LxGG↓ specificity of DUBs of the OTU class.
Due to the ORF1a/1b -1 ribosomal frameshift immediately upstream of its codon, this C-terminal Cys residue of pp1a is only present in nsp8, but not in nsp9, in which it is essentially replaced by Leu, the first ORF1b-encoded residue after the frameshift site.
The arterivirus replicase processing scheme (Fig. 1) involves the rapid autoproteolytic release of two or three N-terminal nsps [nsp1 (or nsp1α/1β) and nsp2] and the subsequent processing of the remaining polyproteins by the “main protease” residing in nsp4 (Snijder et al., 1996), together resulting in a set of 13 or 14 individual nsps. It should be noted, however, that many intermediate cleavage products of unknown function have also been detected (Snijder et al., 1994, van Dinten et al., 1996) and that alternative major and minor processing pathways have been defined for the EAV nsp3–8 region (Wassenaar et al., 1997).
According to the current processing scheme, PRRSV pp1a is processed into 10 end products (including nsp1α/nsp1β and nsp7α/nsp7β) as a result of 9 proteolytic cleavages (Snijder et al., 1994, den Boon et al., 1995, Ziebuhr et al., 2000, van Aken et al., 2006a). The same cleavages are presumed to occur in pp1ab, and in addition, the nsp4 main protease cleaves three sites in the ORF1b-encoded portion of pp1ab to produce nsp9 to nsp12 (van Dinten et al., 1996). Nsp8, the last subunit upstream of the ribosomal frameshift site, is fully colinear with the N-terminal domain of the RdRp-containing subunit nsp9, of which the N-terminus is released by cleavage of the nsp7/8 site (Fig. 1). Wassenaar et al. (1997) proposed a model for differential processing of the EAV nsp3–8 region, in which cleaved nsp2 acts as a co-factor for cleavage of the nsp4/5 site in the nsp3–8 processing intermediate by the nsp4 protease (Fig. 1D). This is thought to constitute the ‘major’ processing pathway, but when nsp2 does not associate with nsp3–8, a ‘minor’ pathway can be followed in which the nsp4/5 site remains uncleaved and the nsp5/6 and nsp6/7 sites are processed instead.
The arterivirus nsp1 region contains a tandem of papain-like autoprotease domains (PCPα and PCPβ), but in EAV PCPα has lost its enzymatic activity, resulting in the ‘merge’ of nsp1α and nsp1β into a single nsp1 subunit (den Boon et al., 1995). Thus, instead of three self-cleaving N-terminal replicase subunits, EAV has two: nsp1 and nsp2 (Fig. 1B). Recently, the exact PRRSV nsp1α/1β and nsp1β/2 cleavage sites were identified by direct N-terminal sequence analysis (Sun et al., 2009a, Chen et al., 2010a). The nsp1α/1β site of Type II PRRSV was found to be at 180M↓A181, which is 14 amino acids downstream of a previously proposed candidate (166Q↓R167). Based on a sequence alignment of this part of nsp1, the corresponding cleavage site in Type I PRRSV would be 180H↓S181 (Chen et al., 2010a), but this remains to be confirmed experimentally, in particular since it would be unusual for a PCP cleavage site to have a His residue at the P1 position (cleavage site nomenclature according to Schechter and Berger, 1967). Chen et al. (2010a) also confirmed that the nsp1β/2 cleavage site (383G↓A384 in Type II PRRSV) matches the previous prediction of Ziebuhr et al. (2000).
2.2. The nonstructural proteases
The two or three N-proximal ‘accessory proteinases’ in arterivirus replicase polyproteins are most similar to cellular papain-like enzymes. They are all predicted to employ a Cys-His tandem as active site residues, and this similarity, together with their sequential position in the genome (the three PRRSV papain-like domains are present in the first 500 residues of the replicase), suggests that duplications of accessory proteases are likely to have occurred in the course of arterivirus evolution (Ziebuhr et al., 2000).
The arterivirus PCPα and PCPβ domains were found to be active in reticulocyte lysates and E. coli, systems that were used to corroborate their functionality and first document the generation of the nsp1(α/β) cleavage products (Snijder et al., 1992, den Boon et al., 1995). Both PCP domains appear to act exclusively in cis, and it has been speculated that this property may be due to the post-cleavage retention of the C-terminal residues of nsp1α and nsp1β in the active sites of PCPα and PCPβ, respectively (Ziebuhr et al., 2000). A recent PRRSV research highlight, the crystal structure of nsp1α by Sun and co-workers (Sun et al., 2009a; Fig. 2A), indeed documented the presence of a number of nsp1α C-terminal residues in the PCPα substrate-binding pocket, in a well-stabilized form that would preclude further proteolytic reactions. The same study also corroborated the presence of a zinc-binding domain near the N-terminus of the arterivirus nsp1 region (see below).
Fig. 2.
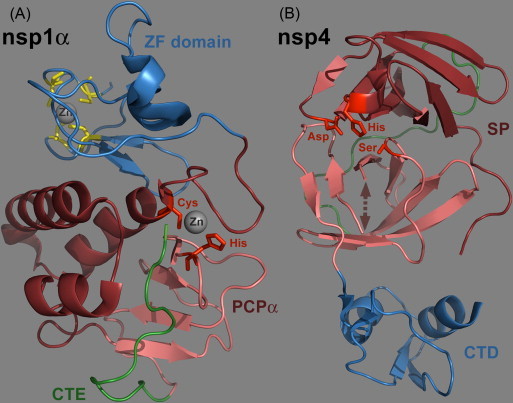
Ribbon diagrams of the PRRSV nsp1α and nsp4 crystal structures. Representations were made using the Pymol molecular graphics system (DeLano, 2002) and PDB entries 3IFU (Sun et al., 2009a) and 3FAN (Tian et al., 2009), respectively. (A). Crystal structure of an nsp1α monomer consisting of five β strains and five α helices (Sun et al., 2009a). The N-terminal ZF domain (Met1 to Glu65; Zn-binding residues highlighted in yellow) and C-terminal PCPα domain (Pro66 to Gln166) are colored as blue and dark red ribbons, respectively. The ‘C-terminal extension’ (CTE: Arg167 to Met 180) that resides inside the PCPα substrate-binding pocket is featured in green. (B). Crystal structure of PRRSV nsp4 (Tian et al., 2009). The protein consists of three domains of which the first two (in dark red and pink) form the typical chymotrypsin-like two-β-barrel fold. A long loop connecting domains I and II is highlighted in green, whereas the arrow represents a loop consisting of residues 136–140, which was devoid of visible electron density. The residues of the catalytic triad are highlighted in red. The C-terminal domain was shown to be dispensible for proteolytic activity in EAV nsp4 and is depicted in blue (Barrette-Ng et al., 2002, van Aken et al., 2006b). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
The nsp2 subunit has been shown to be highly variable in length among the sequenced isolates of different arteriviruses. In the PRRSV Type I prototypic strain, Lelystad virus, nsp2 is (predicted) to be 1077 amino acids in length, while the protein of the Type II prototypic strain, VR2332, has an additional 118 amino acids. The (predicted) EAV-, LDV-, and SHFV-encoded nsp2 proteins are 571, 906, and 757 amino acids in length, respectively (Meulenberg et al., 1993, Godeny et al., 1993, Allende et al., 1999, Nelsen et al., 1999, Ziebuhr et al., 2000). The arterivirus nsp2 contains a cysteine protease domain (CP) in its N-terminal domain, which mediates its rapid release from pp1a/pp1ab and possesses both cis- and trans-cleavage activities (Snijder et al., 1995, Han et al., 2009). The arterivirus nsp2 CP domain was also identified as a member of the ovarian tumor domain (OTU) family of deubiquitinating enzymes (Makarova et al., 2000, Frias-Staheli et al., 2007, Sun et al., 2010), a property that will be discussed in more detail below. Two candidate PRRSV nsp2/3 cleavage sites were predicted by comparative sequence analysis with EAV (Allende et al., 1999, Nelsen et al., 1999, Ziebuhr et al., 2000). The work of Han et al. (2009) indicated that the more downstream candidate, which flanks the highly conserved nsp3 subunit, most likely is the actual nsp2/3 cleavage site, although the classification of the CP as an OTU family member would suggest that the enzyme does not cleave inside but downstream of the proposed Gly–Gly doublet (Table 1). A recent analysis of the C-terminus of immunoprecipitated nsp2 from cells infected with a Type I PRRSV (SD01-08) further supports this notion. The nsp2/3 cleavage was found to most likely occur at 1445GG↓A1447 (corresponding to 1462GG↓A1464 in the prototypic Lelystad virus sequence), which is one amino acid downstream from the previously predicted 1445G↓G1446 site that equals 1462G↓G1463 in Lelystad virus (Fang et al., unpublished data).
The arterivirus nsp4 main proteinase is a member of a relatively rare group of proteolytic enzymes, the 3C-like serine proteases (Snijder et al., 1996), which combine the catalytic triad (His-Asp-Ser) of classical chymotrypsin-like proteases with the substrate specificity of the 3C-like cysteine proteases, a subgroup of chymotrypsin-like enzymes named after the picornavirus 3C proteases. Specific residues in the substrate-binding region of the arterivirus nsp4 SP are assumed to determine its specificity for cleavage sites containing a Glu residue at the P1 position and a small residue (commonly Gly, Ser or Ala) occupying the P1′ position. Crystal structures for EAV and PRRSV nsp4 (Barrette-Ng et al., 2002, Tian et al., 2009; Fig. 2B) revealed that the protein consists of three domains, with domains I and II forming the typical chymotrypsin-like two-β-barrel fold. The C-terminal domain III is dispensable for proteolytic activity and may be involved in fine-tuning replicase polyprotein proteolysis (van Aken et al., 2006b). Based on the analysis of EAV replicase processing, the PRRSV nsp4 SP was proposed to mediate 9 cleavages in the nsp3–12 region. While assessing the proteolytic activity of recombinant PRRSV nsp4 in vitro, the nsp3/4, nsp4/5, and nsp11/12 cleavage sites previously predicted by Ziebuhr et al. (2000) were confirmed as cleavable substrates (Tian et al., 2009).
2.3. Arterivirus nsp1: interconnecting various stages of the replicative cycle
Arterivirus RNA synthesis entails both genome amplification and the production of sg mRNAs, which serve as templates for the translation of viral structural protein genes (Fig. 3 ). Due to the fact that all arterivirus mRNAs are equipped with a common leader sequence, which is identical to the 5′-terminal sequence of the genome, they form a 5′- and 3′-coterminal nested set of transcripts. The majority of data on arteri- (and coronavirus) RNA synthesis supports a model proposed by Sawicki and Sawicki (1995), according to which the unique structure of sg mRNAs derives from discontinuous extension during minus-strand RNA synthesis. This process resembles copy-choice RNA recombination and is guided by specific RNA signals (transcription-regulatory sequences, TRS; van Marle et al., 1999a, Pasternak et al., 2001). In the genomic RNA, these RNA motifs are found upstream of each structural protein gene (body TRS) and in the 3′-end of the leader sequence (leader TRS) [recently reviewed in Pasternak et al. (2006) and Sawicki et al. (2007)]. Minus-strand RNA synthesis is invariably initiated at the genomic 3′ end and can presumably be “attenuated” at a body TRS motif. The genomic leader TRS then serves as a base-pairing target for the body TRS complement presented at 3′ end of the nascent minus strand, which is translocated to the 5′-proximal region of the genomic template, base pairs to the leader TRS, and is extended with the complement of the leader sequence. The subgenome-length minus-strand templates produced in this manner then serve as templates for the synthesis of the various sg mRNAs. Discontinuous minus-strand RNA synthesis likely ‘competes’ with the synthesis of the full-length minus strand, implying that regulatory mechanisms must exist to control the use of the genomic RNA template for full-length versus subgenome-length minus-strand production. In the EAV model, nsp1 appears to be a key regulatory factor in this context (Fig. 4 ) and was recently proposed to control the production of each of the viral mRNA by determining the accumulation level of their respective minus-strand templates (Nedialkova et al., 2010).
Fig. 3.
Overview of the PRRSV replicative cycle highlighting the various steps in viral RNA and protein synthesis. Genome translation yields the pp1a and pp1ab replicase polyproteins that are cleaved into the mature nonstructural proteins. These assemble into a replication and transcription complex (RTC) that recognizes the 3′ of the genome to initiate minus-strand RNA synthesis. Continuous minus-strand RNA synthesis yields the genome-length minus strand template for genome replication. Guided by regulatory RNA sequences (TRSs, in orange) and proteins (see Fig. 4), discontinuous minus-strand RNA synthesis produces a nested set of subgenome-length minus strands that serve as templates for sg mRNA synthesis. The ORFs expressed from the respective mRNAs are indicated. The common 5′ leader (L) sequence on the mRNAs and the common 3′ anti-leader (A) on the minus-strand RNAs are indicated. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Fig. 4.
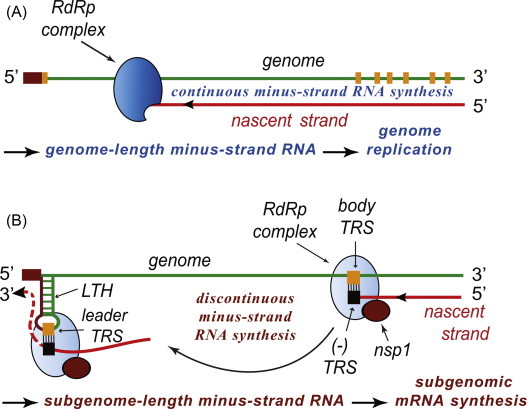
Arterivirus nsp1 modulates genome replication and transcription at the level of minus-strand RNA synthesis. (A). Continuous minus-strand RNA synthesis by the viral RdRp complex yields a genome-length minus strand template for genome replication, a process for which nsp1 is dispensable. (B) EAV nsp1 (presumably nsp1α in PRRSV) switches the RdRp complex into ‘discontinuous mode’ and modulates minus-strand RNA synthesis to control the relative abundance of both genomic and sg mRNAs (Nedialkova et al., 2010). Discontinuous RNA synthesis is guided by a base pairing interaction between the TRS complement [(−)TRS] at the 3′ end of the nascent minus-strand and the genomic leader TRS, which is present in an RNA hairpin structure (LTH). Following base pairing, minus-strand RNA synthesis resumes to add the anti-leader sequence to the nascent subgenome-length minus strand, which subsequently serves as a template for sg mRNA synthesis. The RNA and/or protein element(s) with which nsp1 interacts to control viral mRNA abundance remain to be identified.
Figures adapted from Nedialkova et al. (2010).
By reverse genetics, EAV nsp1 was first shown to be dispensable for genome amplification but essential for sg mRNA production (Tijms et al., 2001). A predicted zinc finger domain in its N-terminal region, whose existence was recently confirmed in the PRRSV nsp1α structure (Sun et al., 2009a), was implicated in the regulation of sg RNA synthesis. Subsequently, by extensive characterization of a set of mutants, nsp1 was identified as a multifunctional protein that interconnects three important steps of the EAV replicative cycle: replicase polyprotein cleavage, sg mRNA production, and virion biogenesis (Tijms et al., 2007, Nedialkova et al., 2010). Although replication can proceed in the absence of nsp1, its autoproteolytic release was found to be essential for RNA synthesis, suggesting that nsp2 functionality depends on cleavage of the nsp1/2 site. A similar observation was made for PRRSV nsp2 and the critical processing of the nsp1β/2 site (Kroese et al., 2008). The postulated key role for the nsp1/nsp1α zinc finger domain was supported by the complete and selective block of sg mRNA accumulation observed upon mutagenesis of zinc-coordinating residues in EAV nsp1 (Tijms et al., 2007). Subsequently, transcription-negative phenotypes were also obtained upon replacements of charged residues in the PCPα and PCPβ domains. This underlines the importance of all subdomains of nsp1, which was recently proposed to modulate the accumulation of both genome- and subgenome-length minus-strand RNA and thereby fine-tune the relative abundance of each of the viral mRNAs in the infected cell (Fig. 4; Nedialkova et al., 2010). In PRRSV, this function is likely performed by nsp1α, since inactivation of the nsp1α/nsp1β cleavage yielded a replication-positive, sg mRNA-negative phenotype similar to that observed for a variety of EAV nsp1 mutants (Kroese et al., 2008). Examining the interactions of nsp1/nsp1α with other viral proteins, like RdRp and helicase, and the RNA motifs directing viral RNA synthesis is an obvious starting point in dissecting the molecular basis of the regulatory activities of nsp1 proteins in arterivirus RNA synthesis.
Surprisingly, replacements of certain residues in the zinc finger and PCPα domains of EAV nsp1 had little effect on the accumulation of EAV RNAs, but greatly decreased the production of infectious progeny particles, suggesting that nsp1 is important for virion biogenesis as well (Nedialkova et al., 2010) and participates in the poorly characterized multi-step process of virion assembly and release. A final remarkable feature of EAV nsp1 and PRRSV nsp1α and nsp1β, their partially nuclear localization (Tijms et al., 2002, Tijms and Snijder, 2003, Chen et al., 2010a), will be discussed in more detail below.
2.4. Hydrophobic nsps inducing extensive reorganization of intracellular membranes
With the exception of nsp1(α/β) proteins, all arterivirus replicase subunits studied to date were found to exclusively localize to the perinuclear region of the infected cell (Fig. 5A–B; van der Meer et al., 1998, Kroese et al., 2008; Fang et al., unpublished data). They are associated with intracellular membranes, likely derived from the endoplasmic reticulum (ER), which are modified into vesicular double-membrane structures with which the viral replication and transcription complex (RTC) is thought to be associated (Pedersen et al., 1999). The formation of closely paired membranes and double membrane vesicles (DMVs; Fig. 5C–E) is a typical feature of infected cells that was described many years ago for all arteriviruses, including PRRSV (Weiland et al., 1995, Pol et al., 1997). Biochemical and electron microscopy studies have implicated the ORF1a-encoded subunits that contain (predicted) membrane-spanning domains (in particular nsp2, nsp3, and nsp5; Fig. 1C) in inducing these membrane modifications. They may thus serve to anchor nsps lacking such hydrophobic domains to the membranes that are postulated to form a scaffold for viral RNA synthesis. Expression of EAV nsp2–3 in a heterologous expression system induced strikingly similar structures, indicating that DMV formation does not depend on other viral proteins and/or EAV-specific RNA synthesis (Snijder et al., 2001). Nsp2 and nsp3 are known to interact (Snijder et al., 1994), and a lumenal nsp3 domain containing a cluster of 4 conserved Cys residues was recently implicated in DMV formation (Posthuma et al., 2008).
Fig. 5.
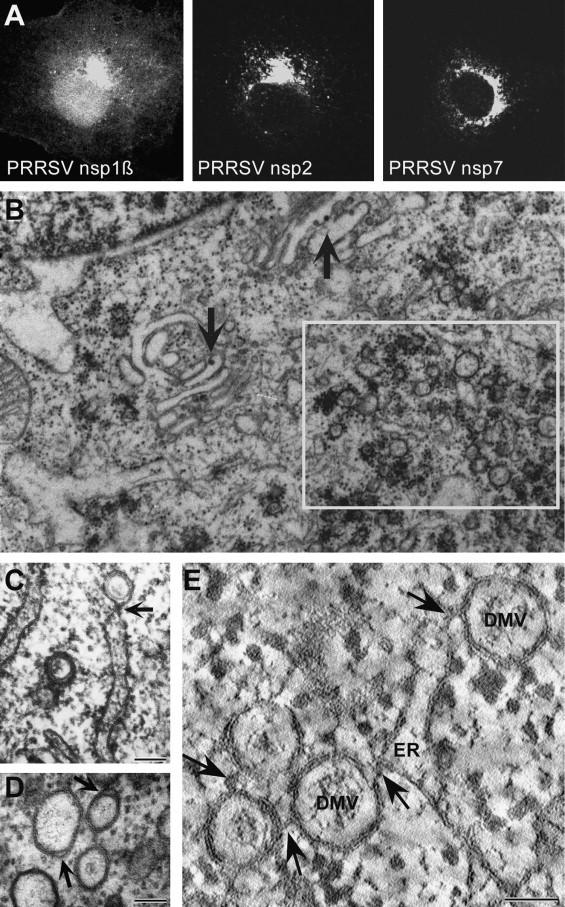
Subcellular localization of replicase subunits and virus-induced membrane structures. (A). Distribution of selected PRRSV nsps in MARC-145 cells infected with strain VR2332. At 12 h post infection, nsps were detected using FITC-labeled monoclonal antibodies. Images of representative infected cells were obtained with a 63 x oil objective, Zeiss LSM 510 confocal microscope. Note that both nsp2 and nsp7 map to the perinuclear region where extensive membrane changes can be observed (panels B–E), whereas part of nsp1β is present in the nucleus (see text for details). (B–E). Electron micrographs of EAV-infected BHK-21 cells. Electron micoscopy analysis of arterivirus-induced membrane changes in BHK-21 (B–D) or Vero-E6 (E) cells infected with EAV. Scale bars represent 100 nm. (B). The rectangle indicates a region containing many double membrane vesicles (DMVs), and also many ribosomes, with which viral RNA synthesis is presumably associated. The arrows point towards virus particles budding from smooth intracellular membranes in or close to the Golgi region. (C). Possible intermediate in DMV formation from endoplasmic reticulum (ER) membranes. Whereas the inner DMV membrane appears to be detached, the outer DMV membrane is continuous with an ER cisterna. An electron dense, neck-like connection is clearly visible (arrow). Adapted from Snijder et al. (2001). (D). Image of typical DMVs with apparently sealed inner and outer membranes. The membranes are generally tightly apposed; arrows point to regions which readily visualize the double-membrane nature of the vesicles. Adapted from Snijder and Meulenberg (1998). (E). Example of a recent electron tomography analysis revealing the interconnected nature of EAV–induced DMVs. Double-tilt electron tomography was applied to a semi-thick section of an EAV-infected Vero-E6 cell, cryofixed at 7 h p.i., and images were processed to compute tomograms. The image represents a tomographic slice with a thickness of 1.2 nm showing a rough ER cisterna and various DMVs that are connected with each other and with the ER by their outer membranes (arrows). Despite a major difference in the size of the DMVs, which are about twice smaller in arteriviruses, this membrane structure resembles the reticulovesicular network recently documented for SARS-coronavirus (Knoops et al., 2008).
Courtesy of Dr. Kèvin Knoops and co-workers, Leiden University Medical Center.
The formation of DMVs is a feature shared with coronaviruses, for which electron tomography analysis of infected cells revealed that these unusual structures are part of an extensive network of modified ER (Knoops et al., 2008). Although arterivirus-induced DMVs are substantially smaller than those in coronavirus-infected cells (Pedersen et al., 1999), a similar network of interconnected DMVs that is continuous with ER membranes has recently been uncovered for EAV (Knoops, Koster, Mommaas, and Snijder, unpublished data). Both SARS-coronavirus and EAV DMVs were found to contain double-stranded RNA, the presumed intermediate in viral RNA synthesis. However, it is unclear whether this reflects actual RNA production inside DMVs or merely ‘RNA accumulation’, for example to delay double-stranded RNA-triggered host innate immune responses. The fact that early RNA synthesis can apparently proceed in the absence of (large numbers of) DMVs raises the possibility that the active RTC may localize elsewhere in the cell and that DMVs may in fact be a ‘side effect’ of overproduction of membrane-spanning viral nsps during the later stages of infection. On the other hand, the general importance of an organelle like the ER certainly does not exclude that nidoviruses may induce its dramatic remodelling to manipulate specific host cell processes. Furthermore, membranes were found to be important for the activity of EAV RTCs isolated from infected cells, which were also found to require a cytosolic host cell protein for in vitro genome and sg mRNA synthesis (van Hemert et al., 2008).
2.5. The key enzymes in arterivirus RNA synthesis: the nsp9 RdRp and nsp10 helicase
ORF1b is the most conserved part of arterivirus (and nidovirus) genomes and encodes the key enzymes for RNA-templated RNA synthesis—the RdRp and helicase, originally identified by comparative sequence analysis (den Boon et al., 1991). The RdRp domain is found in the C-terminal portion of replicase subunit, nsp9, which contains an additional upstream domain of unknown function. A protocol to purify an enzymatically active recombinant form of the EAV nsp9-RdRp from E. coli was recently developed (Beerens et al., 2007), and its initial biochemical characterization revealed that the protein is able to initiate RNA synthesis de novo. Although RNA polymerization in vitro did not depend on the presence of other viral proteins, the recombinant EAV RdRp could not utilize short sequences representing the 3′ end of the viral genome as template, suggesting that additional factors are required for template selection and/or activity in vivo. Clearly, not only mechanistic studies into nidovirus RNA synthesis but also antiviral drug development may benefit from the availability of functional recombinant arterivirus RdRps.
RNA helicases are a diverse class of enzymes that use energy from ATP hydrolysis to unwind RNA duplexes. Much like the RdRp domain, the arterivirus helicase is part of a larger replicase subunit, nsp10, which also contains an N-terminal predicted zinc-binding domain. This domain, containing 13 conserved Cys and His residues (van Dinten et al., 2000), is conserved in all nidovirus helicases. Recombinant EAV and PRRSV nsp10 have ATPase activity and can unwind RNA and DNA duplexes in a 5′-to-3′ direction in vitro (Seybert et al., 2000, Bautista et al., 2002). Notably, this 5′-to-3′ polarity of the helicases of arteriviruses and other nidoviruses has so far not been reconciled with their presumed role in unwinding double-stranded RNA structures hindering the progress of the RdRp, which proceeds in the opposite direction during viral RNA synthesis.
The zinc-binding domain of arterivirus nsp10 is also critical for the in vitro ATPase and helicase activities of the protein (Seybert et al., 2005). Although residues from this domain are unlikely to be involved in catalysis, zinc coordination might assist the proper folding of nsp10 and/or mediate interactions with substrate RNA molecules. Already before the discovery of nsp1's function in sg RNA synthesis, the serendipitous introduction of a mutation in the EAV nsp10 ‘linker’ region connecting the zinc-binding and helicase domains revealed that arterivirus sg mRNA synthesis can be functionally uncoupled from genome replication (van Dinten et al., 1997). This Ser-2429 to Pro substitution in nsp10 did not interfere with replicase polyprotein processing, accumulation of genomic RNA, or in vitro helicase activity, but resulted in a more than 100-fold reduction of subgenome-length minus- and plus-strand RNA accumulation (van Marle et al., 1999b).
2.6. The nsp11 endoribonuclease: a potential ‘suicide enzyme’ found in all nidoviruses
In addition to the core viral enzymes described above, the replicative machinery of nidoviruses includes several subunits with rather unusual RNA-processing activities that have few or no known counterparts in the RNA virus world. The majority of these enzymes are encoded only by nidoviruses with large genomes, like coronaviruses, but one of them—the nidovirus endoribonuclease (NendoU) domain is conserved throughout the nidovirus order (Snijder et al., 2003, Gorbalenya et al., 2006). To date, no NendoU counterparts have been identified in other RNA viruses, and this domain is thus considered a genetic marker of Nidovirales. The arterivirus NendoU domain resides in nsp11 and is N-terminally fused to another domain of unknown function. Its critical role in the arterivirus replicative cycle was first established by reverse genetics (Posthuma et al., 2006). Remarkably, however, replacements of any of the three predicted active site residues of EAV NendoU were not lethal, although they yielded severely crippled mutants exhibiting a moderate but specific defect in sg mRNA synthesis and a dramatic reduction of infectious progeny titers by up to 5 logs. A recent account of a reverse genetics analysis of the PRRSV NendoU domain corroborated the observations made for EAV, including a specific effect on sg mRNA accumulation, and strengthened the notion that nsp11 is involved in multiple steps of the arterivirus replicative cycle (Yoo et al., personal communication).
NendoU enzymes share certain features with bovine pancreatic RNase A, such as the identity of the active site residues and the proposed mechanism of RNA hydrolysis. Recombinant EAV and PRRSV nsp11 were recently shown to exhibit broad substrate specificity in vitro (Nedialkova et al., 2009). Nsp11 cleaves both single-stranded and double-stranded RNA substrates 3′ of pyrimidines, with a modest preference for cleavage at single-stranded uridylates. Heterologous expression of enzymatically active arterivirus nsp11 in both prokaryotic and eukaryotic cells is extremely toxic (Nedialkova et al., 2009), suggesting that the ‘uncontrolled’ enzyme targets essential cellular RNA components. A similar broad activity towards viral RNA substrates in infected cells would make NendoU expression potentially ‘suicidal’ for a replicating RNA virus and strongly suggests that its access to RNA substrates must be strictly controlled in the context of natural infection. Its integration into the multisubunit RTC and a confined intracellular localization of NendoU-containing nsps likely contribute to a strategy to protect viral RNA from rapid degradation by NendoU. Integration of the currently available data from bioinformatics, in vitro assays, and reverse genetics into a coherent hypothesis on NendoU function clearly requires the identification of the in vivo substrate(s) of the enzyme. The fact that NendoU knockout mutants remain capable of RNA synthesis has opened the possibility that, opposite to earlier predictions, the enzyme may not function in a nidovirus-specific step of viral RNA synthesis but rather targets unknown cellular substrates. Their identification undoubtedly will be a technically challenging task, but also an inevitable step towards understanding the basis for the conservation of this enzymatic activity in nidoviruses. Apart from unique insights into the role of a replicative endoribonuclease in the life cycle of positive strain RNA viruses, this knowledge will also be an important prerequisite for assessing the suitability of NendoU as a target for the development of antiviral compounds.
2.7. Arterivirus mRNA capping and polyadenylation: unexplored territory
The arterivirus genomic and sg mRNAs are generally assumed to have a 5′-terminal cap structure and are known to be 3′ polyadenylated. The 5′ end of the SHFV genome was reported to contain a type I cap (Sagripanti et al., 1986) and, in addition, cap analogue was found to be an essential component of in vitro transcription reactions used to generate infectious PRRSV and EAV RNA from full-length cDNA templates (Meulenberg et al., 1998; Snijder et al., unpublished data). Three basic enzymatic activities are commonly required for mRNA capping: (i) an RNA triphosphatase removing the 5′ γ-phosphate group of the RNA substrate; (ii) a guanylyltransferase to catalyze the transfer of GMP to the remaining 5′-diphosphate terminus; and (iii) an N7-methyl transferase to methylate the cap guanine at the N7-position, a step preventing the pyrophosphorolytic reversal of the guanylyltransfer reaction. In higher eukaryotes, the ‘cap-0 structure’ (7MeGpppN) structure produced by these three consecutive reactions is commonly converted into cap-1 or cap-2 structures by 2′O methyl tranferases that can modify the ribose 2′O-position of the first or second nucleotide of the mRNA, respectively. Given their entire cytoplasmic replication cycle, positive-stranded RNA viruses cannot rely on the capping machinery of the host cell's nucleus and have developed specific mechanisms for cytoplasmic mRNA capping, or alternative mechanism to ensure the efficient translation of their mRNAs.
In the distantly related coronaviruses, essentially only the guanylyltransferase involved in the second step of the default mRNA capping pathway outlined above remains to be identified (summarized by Bouvet et al., 2010). In contrast, none of the arterivirus nsps has been shown or predicted to possess one of the activities required for capping thus far. The 5′ RNA triphosphatase activity reported for the coronavirus nsp13-helicase (Ivanov et al., 2004), which was proposed to initiate the capping reactions, remains to be investigated for its arterivirus homolog, the nsp10-helicase. The N7- and 2′O-methyl transferase domains found in the coronavirus replicase subunits nsp14 and nsp16, respectively, are conspicuously lacking in the arterivirus replicase. Consequently, the investigation of mRNA capping is one of the remaining challenges in arterivirus enzymology. In addition to asking whether cellular enzymes might be recruited to perform one or multiple of the required enzymatic reactions, the biochemical characterization of the domains that currently form the major ‘white spots’ on the arterivirus replicase map (the C-terminal domain of nsp2, nsp7, the N-terminal half of nsp9, and nsp12) could be a first step in the hunt for the enzymes directing this presumably critical function in arterivirus RNA processing. Likewise, the signals for and mechanism of 3′ polyadenylation of arterivirus mRNAs remain to be investigated, although it is commonly assumed that this process in RNA viruses is primarily directed by the viral RdRp subunit.
3. Function of PRRSV nonstructural proteins in host immunity
3.1. Adaptive immune responses against PRRSV nsps
PRRSV nsp1 α/β, nsp2, and nsp7 were found to be important players in the humoral immune response. Oleksiewicz et al. (2001a) used a phage display library to screen for linear oligopeptide epitopes that reacted with antisera from pigs infected with Type I PRRSV. Ten linear B-cell epitope sites (ES) were identified, which were scattered throughout the nonstructural and structural protein regions of the virus. Interestingly, six of the ten epitopes mapped to nsp2 and two others were located in nsp1β and nsp4. By using phage-displayed peptides (epitopes) from PRRSV nsps as ELISA antigen, Oleksiewicz and co-workers were able to detect specific serum antibody responses against nsps. Such antibodies were also consistently found in the semen of boars, beginning from 1–4 weeks post infection. A correlation between seminal IgA anti-nsp responses and the duration of PRRSV RNA detection in semen was also observed (Oleksiewicz et al., 2001b). A recent study by Brown et al. (2009) also demonstrated the antigenic nature of certain PRRSV nsps (Fig. 6 ). Serum antibody responses to Type II PRRSV ORF1a-encoded nsps were screened by ELISA, revealing that in particular nsp1α/β, nsp2, and nsp7 reacted strongly with pig immune sera. The antibodies specific to these proteins could be detected as early as 14 days post infection (dpi) and lasted at least until 202 dpi. Further comparison of these three antigens with nucleocapsid (N) protein showed that there was a decrease of antibody titers to the N protein after 126 dpi, while the antibody titers to nsp1α/β, nsp2, and nsp7 were maintained at high levels. Johnson et al. (2007) indicated that the antibody reactivity to nsp1α/β depended on refolding of the denatured protein, suggesting that the porcine antibody response is directed primarily to conformational epitopes. Sera from pigs infected with various PRRSV strains cross-reacted with nsp1α/β and nsp2 antigens, indicating that multiple epitopes are conserved. Strain-specific antibody responses to highly variable regions of nsp2 were also detected, which provide a basis for the development of immunoassays that can differentiate between serological responses against vaccines and field isolates (see Section 4.1 below). Johnson et al. (2007) postulated that the early and robust antibody responses induced by nsp1α/β and nsp2 may be related to their expression and rapid autoproteolytic release early in the viral replicative cycle. This might make these proteins available right away for degradation by the macrophage proteosome machinery and presentation to the immune system in the context of MHC classes I and II.
Fig. 6.
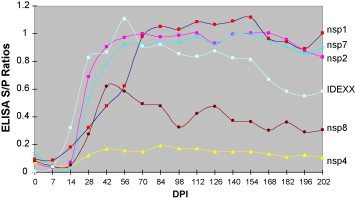
Kinetics of antibody response to PRRSV nsps. Pigs were experimentally infected with Type II PRRSV, VR2332. The serum samples were collected from 0 to 202 days post inoculation (DPI) as indicated. For nsp4 and nsp8, serum samples from 10 pigs were tested; for nsp1, nsp2, and nsp7, serum samples from 30 pigs were tested.
Figure adapted from Brown et al. (2009).
3.2. PRRSV nsps modulating innate immune responses
PRRSV infection appears to elicit poor innate immune responses, presumably resulting in a weak adaptive immune response. The latter is apparent from short-lasting cell-mediated immune responses and slow development of virus-specific interferon (IFN)-γ secreting cells, leading to a prolonged viremia (Meier et al., 2003, Royaee et al., 2004). Stimulation of MARC-145 cells by exogenous double-stranded RNA resulted in a significant increase in Type I IFN mRNA expression, but in the presence of PRRSV infection, Type I IFN activation was significantly inhibited (Miller et al., 2004). Results from Luo et al. (2008) showed that PRRSV infection significantly inhibits dsRNA-induced IFN-β production. Various PRRSV nonstructural proteins appear to be able to modulate Type I IFN activity in expression systems employing reporter gene-based read-outs. Expression of PRRSV nsp1α/β and nsp2 were found to exert strong inhibitory effects on IFN-β promoter activation (Beura et al., 2010, Chen et al., 2010a, Kim et al., 2010a, Sun et al., 2010). Nsp4 and nsp11 were also reported to have inhibitory effect on IFN-β promoter activation (Sun et al., 2009b, Beura et al., 2010), although this type of studies requires detailed follow-up experiments to exclude the possibility that the observed effects may be (partially) due to more general consequences of the expression of these potentially toxic viral enzymes. The deubiquitinating activity of the arterivirus nsp2 CP/OTU domain appears to be capable of suppressing IFN-β production and signaling. Frias-Staheli et al. (2007) reported that the EAV and PRRSV nsp2 CP domains are capable of deconjugating both ubiquitin and interferon stimulated gene (ISG) 15 from cellular proteins, which may thus inhibit ubiquitin- and ISG15-dependent innate immune responses. The mechanistic aspects of the role of PRRSV nsp1 α/β and nsp2 in counteracting host innate immune response are reviewed in more detail in Sections 3.3, 3.4.
3.3. Suppression of Type I interferon synthesis and signaling by PRRSV nsp1α and nsp1β
The host innate immune response is the first line of defense in counteracting virus infection. A key aspect of the antiviral innate immune response is the synthesis and secretion of Type I interferon such as IFN-α and IFN-β, which exhibit antiviral, anti-proliferative, and immunomodulatory functions (reviewed by Samuel, 2001, Haller and Weber, 2007, Randall and Goodbourn, 2008). The two N-terminal PRRSV replicase cleavage products, nsp1α and nsp1β, appear to affect Type I IFN synthesis and signaling in different manners. Individually expressed nsp1β has the ability to inhibit both IFN synthesis and signaling, while nsp1α strongly inhibits IFN synthesis only. Nsp1β expression strongly inhibited IFN regulatory factor 3 (IRF3)- dependent gene induction in the signaling pathway leading to IFN-β synthesis (Beura et al., 2010, Chen et al., 2010a). Specifically, when tested in a porcine myelomonocytic cell line, nsp1β inhibited Sendai virus-mediated activation of porcine IFN-β promoter activity (Beura et al., 2010). Chen et al. (2010a) tested each component in the IRF3 signaling pathway in response to dsRNA stimulation. The result suggests that the PRRSV nsp1α and nsp1β proteins act downstream of IRF-3 activation, which may have a direct effect on the formation of the transcription enhanceosome on the IFN-β promoter in the nucleus. Kim et al. (2010a) reported that nsp1α/β may interfere with IFN-β production through degradation of transcription factor CREB-binding protein (CBP300) in the cell nucleus. When expressed in HeLa or MARC-145 cells, nsp1α/β did not block IRF3 phosphorylation or its nuclear translocation, but inhibited IRF3 association with CBP300 to form the transcription enhanceosome in the nucleus. While IRF3 was stable, CBP300 was degraded in the presence of nsp1α/β. Although this effect needs to be further corroborated in infected, natural host cells, the result is consistent with the predominantly nuclear localization of nsp1α and nsp1β (Chen et al., 2010a) during the later stages of infection and the fact that IRF3 appears to be activated and translocated to the nucleus normally after SeV infection of cells expressing PRRSV nsp1α or nsp1β. These studies suggested that these proteins may affect enhanceosome assembly in the cell nucleus, which could be one of the mechanisms leading to a block of the IFN response during infection. Also part of EAV nsp1 was reported to localize into the nucleus of the infected cell (Tijms et al., 2002), likely mediated by its C-terminal region (amino acids 157–260) that roughly corresponds to PRRSV nsp1β. Interestingly, arterivirus nsp1 sequences do not contain any of the canonical nuclear localization signals, suggesting that nuclear import may depend on interactions with other proteins that shuttle between cytoplasm and nucleus. Tijms and Snijder (2003) further demonstrated an interaction between EAV nsp1 and a host cell transcriptional co-activator, p100, in the cell nucleus and suggested that the nuclear localization of nsp1 might be the basis for modulation of host cell transcription. Recent in vitro studies also showed that EAV nsp1 is a potent IFN antagonist (Chen et al., manuscript in preparation), suggesting that the protein may both modulate host transcription and interfere with host innate immunity.
After being secreted, Type I IFN binds to its receptor on adjacent cells to activate the so-called JAK-STAT signaling pathway, which in turn induces the transcription of interferon-stimulated genes (ISGs) enabling the host cell to fight infection and inhibit virus replication. PRRSV nsp1β was found to be able to inhibit the IFN-dependent signaling pathway leading to ISG expression (Beura et al., 2010). The protein strongly inhibited reporter gene expression from an IFN-stimulated response element (ISRE) promoter after IFN-β stimulation (Chen et al., 2010a). Its impact on this pathway appears to be based on a block of the phosphorylation of transcription factor STAT1 and prevention of STAT1's nuclear translocation. Yoo et al. (2009) identified a protein inhibitor of activated STAT1 (PIAS1) as a specific cellular interaction partner for nsp1α and nsp1β. Beura et al. (2010) reported that both nsp1α and nsp1β are able to inhibit upregulation of endogenous ISG56 expression. Further studies in this area are needed to unravel and understand PRRSV nsp1α/β effects on IFN signaling in more detail.
3.4. Multifunctional PRRSV nsp2 modulates the host immune response at different levels
Nsp2 is the largest (mature) PRRSV protein and contains at least four distinct domains: the N-terminal CP/OTU domain, a central hypervariable region, a putative transmembrane domain, and a C-terminal region of unknown function that is rich in conserved Cys residues (Snijder et al., 1994). The CP domain performs the critical cleavage of the nsp2/3 site, and in the EAV model, fully cleaved nsp2 was reported to be a co-factor for the nsp4 main protease during cleavage of the nsp4/5 site (Wassenaar et al., 1997). Subsequently, comparative sequence analysis revealed that the nsp2 CP belongs to the OTU family of deubiquitinating enzymes (Makarova et al., 2000), a prediction now confirmed for both EAV and PRRSV (Frias-Staheli et al., 2007, Sun et al., 2010). Frias-Staheli et al. (2007) also documented the ability of the nsp2 CP to deconjugate ISG15, an interferon-induced ubiquitin-like molecule exhibiting antiviral activity. The biological significance of this activity was supported by its capacity to inhibit NF-κB dependent signaling and antagonize the antiviral effects of ISG15 when expressed in combination with a Sindbis virus infection in vivo. Ongoing in-depth studies may elucidate the specific viral and/or host protein target(s) of the interferon-signaling network with which arterivirus nsp2 OTU domains interact.
As discussed in Section 3.1, PRRSV nsp2 is an immunodominant protein with the ability to induce a strong humoral antibody response (Oleksiewicz et al., 2001a). In Type II PRRSV nsp2, de Lima et al. (2006) demonstrated that nsp2 contains the highest frequency of B-cell epitopes, and Yan et al. (2007) identified six linear B-cells epitopes using a panel of monoclonal antibodies and swine antisera. Another important characteristic of nsp2 is that its central region is highly variable with deletions and insertions having been identified in various studies (Fig. 7 ; Shen et al., 2000, Gao et al., 2004, Fang et al., 2004, Fang et al., 2007, Ropp et al., 2004, Han et al., 2006, Tian et al., 2007, Yoshii et al., 2008, Kim et al., 2010b). Fang et al. (2004) reported the nsp2 sequences from a group of Type I PRRSV strains that had emerged in North America since 1999. A hallmark of this group is a17-aa deletion in nsp2, which is accompanied by additional deletions/insertions. Interestingly, most of the B-cell epitope sites identified in nsp2 are associated with these deletion/insertion regions. A search for potential T cell epitopes in the region of the 17-aa deletion revealed a MHC class I HLA-A2.1-restricted epitope motif, LINLVGGNL and a general T cell epitope motif, VSAGLINLVG (Rothbard and Taylor, 1988, Falk et al., 1991, Hunt et al., 1992). Deletions and insertions in nsp2 were also reported in Type II PRRSV (Shen et al., 2000, Gao et al., 2004, Han et al., 2006, Tian et al., 2007, Yoshii et al., 2008). For example, the highly pathogenic PRRSV (HP-PRRSV) associated with 2006 outbreaks of porcine high fever disease in Southeastern Asia was found to possess a discontinuous (29 and 1) 30-aa deletion in the nsp2 region (Tian et al., 2007), which contains previously identified B-cell epitopes (aa 536–560; de Lima et al., 2006) and potential T-cell epitopes (Falk et al., 1991, Sette et al., 1989).
Fig. 7.
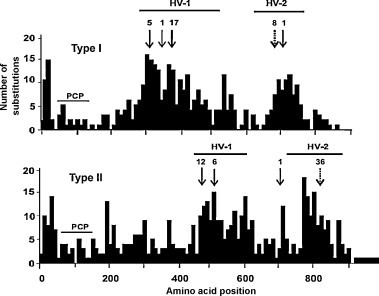
Hypervariability of PRRSV nsp2 region. Nsp2 peptide sequences of 18 Type I (top panel) and 15 Type II (lower panel) PRRSV were aligned using Clustal X. The histogram shows the number of amino acid substitutions along the nsp2 peptide sequence for all viruses. Each bar covers 10 amino acids along the nsp2 peptide sequence. The locations of amino acid insertions and deletions are identified by dashed and solid arrows, respectively. The number above each arrow identifies the length of the insertion/deletion. HV: hypervariable region.
Figure adapted from Fang et al. (2007).
Current research efforts are directed towards elucidating whether there is an association between PRRSV pathogenicity and nsp2 sequence variation. The 30-aa nsp2 deletion in Chinese HP-PRRSV was initially suspected to be a determinant of fatal disease, but a recent study from Zhou et al. (2009) concluded that this is unlikely to be the case. A chimeric virus in which the nsp2 region of a highly virulent strain (JXwn06) containing the 30-aa deletion was replaced by the corresponding region of a low virulent strain (HB-1/3.9) retained high virulence for piglets, although prolonged survival of infected animals was observed. Moreover, introduction of the same 30-aa deletion into strain HB1-1/3.9 did not alter the virulence phenotype of that virus. Therefore, it was concluded that this nsp2 deletion is not a primary determinant of virulence in HP-PRRSV. A recent study from Ellingson et al. (2010) showed that a virulent strain carrying a large deletion in the central region of nsp2 (rMN184Δ618) was neither sufficiently attenuated nor efficacious in protecting against a heterologous challenge when tested as a potential vaccine strain, although a lower replication rate was observed in the earlier stages of infection. In Type 1 PRRSV, each of the six identified B-cell epitopes (ES2 through ES7) were deleted by reverse genetics (Chen et al., 2010b). Deletion of ES3, ES4, or ES7 allowed the generation of viable recombinant viruses, in line with data from Han et al. (2007) demonstrating that the hypervariable central region of nsp2 is dispensable for virus replication. Further in vitro and in vivo characterization of these nsp2 epitope deletion mutants showed that mutants ΔES4 and ΔES7 are attenuated, whereas mutant ΔES3 produced higher peak viral loads in pigs as compared to the parental virus. The expression of innate and T helper 1 cytokines was measured in peripheral blood mononuclear cells and virus-infected macrophages and, compared to the parental virus and other nsp2 deletion mutants, IL-1β and TNF-α expression levels were found to be consistently down-regulated in the case of the ΔES3 mutant.
The studies summarized above make it clear that PRRSV nsp2 is a multi-functional replicase subunit, with different domains being involved in different functions in viral replication and pathogenesis. Certain nsp2 regions appear less- or non-essential for PRRSV replication, but may play important roles in the modulation of host immune response in vivo. Deletions and hypervariability in this region may represent a mechanism for the virus to escape from host immune surveillance.
4. The PRRSV replicase in vaccine and diagnostic test development
4.1. Development of genetic markers in PRRSV nsp2: Implications for recombinant marker vaccines
Modified-live attenuated vaccines against PRRSV are currently used to combat clinical disease. However, vaccinated pigs cannot be distinguished serologically from pigs that have recovered from a natural infection. A genetically marked vaccine that would allow the differentiation of naturally infected and vaccinated animals (DIVA) would be invaluable for PRRSV control and eradication programs. Modified-live vaccines containing such genetic markers have been developed for various animal RNA and DNA viruses, either by insertion of foreign antigens to create a positive marker or by deletion of an immunogenic domain (epitope) to create a negative marker. The antibody responses to the foreign insert or deleted epitope were used to differentiate vaccinated animals from naturally infected animals (Moormann et al., 1990, Kaashoek et al., 1994, van Oirschot et al., 1996, Walsh et al., 2000a, Walsh et al., 2000b, Widjojoatmodjo et al., 2000, Mebatsion et al., 2002, Castillo-Olivares et al., 2003).
Given the compact organization of the rest of the PRRSV genome and in view of the available data on nsp2 sequence heterogeneity (see above), the PRRSV nsp2 region is considered one of the primary candidates for engineering such vaccine markers. Another important property related to marker engineering is the presence of several immunodominant epitopes in this region, whose removal could be used as a negative marker. To obtain proof of principle, several laboratories have explored the insertion into nsp2 of green fluorescent protein (GFP) as a positive marker. Using an American Type I PRRSV infectious clone, pSD 01-08, the GFP gene was inserted into the central region of the nsp2-coding sequence (Fang et al., 2006). In order to obtain a negative marker virus, the B-cell epitope ES4 that is located downstream of the GFP insertion was deleted. Infectious progeny virus (GFP/ΔES4 marker virus) was recovered (Fang et al., 2008) and its in vivo characteristics were studied in a nursery pig disease model. GFP- and ES4 epitope-based ELISA assays were developed as accompanying tests for marker detection. Marker virus-vaccinated pigs could indeed be identified by GFP-positive ELISA results, whereas wild-type virus-exposed pigs were characterized by an ES4-positive ELISA outcome. Finally, animals exposed to both the vaccine and wild-type virus could be identified by producing positive ELISA results for both GFP and ES4. Thus, this study demonstrated that genetically engineered recombinant PRRSV and companion ELISA can be used to correctly identify the vaccination and/or infection status in vivo.
The genetic flexibility of nsp2 was also explored for marker engineering in Type II viruses by several laboratories. Using a VR2332 infectious cDNA clone as a backbone, Han et al. (2007) inserted the GFP gene into the site of an nsp2 deletion (V7-nsp2Δ324–434) located in the position where a natural deletion had been found. The recombinant virus was viable but exhibited slower growth kinetics. The GFP-coding insert was unstable, and the recombinant virus gradually gained parental growth kinetics by loss of most of the GFP gene. A similar study was reported by Kim et al. (2007), who modified the pCMV-129 infectious clone by replacing 131-aa in the C-terminal region of nsp2 by the GFP gene. Other oligo- and polypeptide tags, including FLAG and luciferase, were also inserted into this region. Their results showed that only the genomes containing the GFP-coding insert were replication competent and produced virus progeny. However, GFP fluorescence, but not GFP immunoreactivity, was lost during subsequent passaging of recombinant viruses in culture. In vivo studies showed that GFP was recognized by the sera from pigs infected with the deletion mutant, but not by sera from pigs infected with wild-type virus. In contrast, the 131-aa peptide was recognized by the sera from pigs infected with the wild-type virus, but not the sera from pigs infected with the deletion mutant (Kim et al., 2009).
Also another negative marker virus was constructed using the pFL12 infectious clone (Truong et al., 2004). By reverse genetics, de Lima et al. (2008) deleted epitope 44 and 45 from the nsp2 region and recovered viable viruses. In vivo analysis showed that pigs inoculated with the 44 epitope deletion mutant (FLdNSP2/44) did not develop antibodies against the deleted epitope, whereas a strong reaction was observed in the same peptide-based ELISA using sera from animals infected with the wild-type virus. Taken together, the studies summarized above provide proof of concept to support the concept of PRRSV marker vaccine development by deletion of an individual immunodominant epitope (negative marker) and/or inclusion of a foreign immunogenic domain (positive marker). Two major obstacles still need to be overcome for future marker vaccine development. The first is the limited stability of the marker-coding region, since markers need to be stable during passaging in cell culture and animals. Secondly, negative marker candidates with a high degree of sequence conservation need to be identified to ensure reaction with a broad array of field viruses. Thus, the central region of nsp2 may not be the best target for marker engineering due to its hypervariable nature. On the other hand, the N-terminal nsp2 CP domain region is highly conserved and immunogenic, suggesting it is worthwhile exploring. Also other PRRSV genome regions, including ORF5, ORF6 and ORF7 regions are currently being investigated for their potential in genetic marker engineering.
4.2. Identification of virulence determinants in the PRRSV replicase and their importance for PRRSV vaccine development
A recent development in the generation of improved live PRRSV vaccines is the search for genetic markers for virulence and the application of this knowledge in reverse genetics-based vaccine development. Wang et al. (2008) developed two genetically distinct recombinant viruses (pMLV, constructed from Ingelvac® PRRS MLV; pMN184, constructed from virulent strain MN184) to study the attenuation of the contemporary PRRSV strain MN184. Two reciprocal chimeric clones (pMLVORF1/MN184 and pMN184ORF1/MLV) were constructed, such that the 5′ UTR/replicase of one strain was linked to ORF2a–7/3′ UTR from the other strain. The parental and chimeric viruses were characterized in a nursery pig model, and a complete loss of acute pathogenicity was observed upon introduction of the 5′ four-fifths of the vaccine virus into strain MN184, suggesting that the replicase gene is an important player in viral virulence and attenuation. Their challenge study further showed improved protection in the groups infected with the chimeric viruses (no mortality), which appeared to be protected following the exposure to the virulent heterologous challenge virus. This study suggested that a new PRRSV vaccine could be developed by retaining the 5′UTR/replicase region of an early vaccine strain such as Ingelvac® PRRS MLV and replacing the structural protein-coding domain and 3′UTR with that of an emerging virulent virus.
Kwon et al. (2008) reported a more detailed analysis of virulence factors in the PRRSV replicase gene. A series of chimeric viruses were generated in which specific genomic regions in a highly virulent PRRSV infectious clone (pFL12) were replaced with their counterparts from an attenuated vaccine strain (Prime Pac). The in vivo virulence of the chimeric viruses was determined in a sow reproductive failure model. In the replicase gene, the most important region contributing to PRRSV virulence appeared to be the nsp3–8-coding region, as reflected in the sows inoculated with the chimeric virus cPNSP3.8, in which the part of nsp3–8 region of a virulent strain was replaced by the corresponding region from a vaccine strain. These animals exhibited a significantly improved piglet survival rate in comparison to the offspring of sows inoculated with the parental pFL12 virus. In addition, the nsp1–3 and nsp10–12 regions were proposed to contain potential virulence determinants.
Taken together, the studies summarized above establish model systems for dissecting the molecular basis of attenuation of PRRSV virulence during the future generation of more rationally attenuated PRRSV vaccines. As a general caveat, however, it has to be pointed out that the engineering of chimeric, truncated, or otherwise mutated RNA virus genome sequences, as done in various of the studies described above, can have unforeseen consequences. For example, known and unknown RNA signals underlying specific protein sequences, or overall viral RNA structure, may be affected when making such recombinant viruses. Likewise, the interplay between viral nsps could be influenced by point mutations present in swapped replicase domains. These effects could result in subtle or more pronounced changes in – for example – replicase polyprotein processing, general viral RNA synthesis properties, sg mRNA production in particular (e.g. due to changes in leader TRS-body TRS interplay), or interactions with the host's innate immune system. Clearly, this calls for an in-depth characterization of a variety of molecular biological properties of engineered recombinant viruses before solid conclusions about the role of specific nsps in virulence and pathogenesis can be drawn.
4.3. PRRSV nonstructural proteins as potential diagnostic targets
Nsp1α/β, nsp2, and nsp7 have been explored as potential antigens for the development of serological diagnostic assays. As discussed in Section 3.1, nsp1α/β, nsp2, and nsp7 react strongly with pig immune sera. Brown et al. (2009) have performed a detailed comparison of these three antigens in terms of their sensitivity upon early seroconversion and the correlation with the response to the N protein, which is the antigen in the IDEXX HerdChek® PRRS 2XR ELISA. Interestingly, there is a decrease of the antibody titer to the N protein after 126 dpi, while the antibody titers of nsp1, nsp2, and nsp7 continued to be high. Nsp7 was selected as the target in further diagnostic development based on a combination of characteristics: (i) nsp7 can be expressed as soluble recombinant protein in bacterial cultures, which is convenient for ELISA antigen preparation, especially when applied in large-scale diagnostic screening campaigns; (ii) the nsp7-coding region of both PRRSV genotypes is more conserved compared to the regions encoding nsp1α/β and nsp2; (iii) the use of nsp7 offers the possibility of reliable antibody detection beyond 126 dpi. Using recombinant nsp7 as antigen, a dual-ELISA for the simultaneous detection and differentiation of serum antibodies against Type I and Type II PRRSV was validated. The nsp7-based ELISAs showed good sensitivity and specificity for identification and differentiation of Type I and Type II PRRSV. The capability of the nsp7 dual-ELISA to detect serum antibody response from pigs infected with genetically different field strains was measured. The nsp7 dual-ELISA showed 97.6% agreement with the IDEXX HerdChek® PRRS 2XR ELISA. Upon further testing of suspected false positive IDEXX samples, the nsp7 dual-ELISA resolved 98% of these samples as negative. These results indicate that the nsp7 dual-ELISA can be used as a highly sensitive and specific differential test in PRRSV serology. This ELISA offers an additional tool for routine or follow-up diagnostics, as well as having substantial value in epidemiological surveys and outbreak investigations.
5. Future prospects
A principal reason for the inability of complete PRRSV control is the lack of knowledge on the mechanisms the virus employs to evade host innate and adaptive immune responses. We are at the initial stages of exploring the roles that PRRSV nonstructural proteins play in this key aspect of virus–host interplay, the development of improved vaccines being one of the driving forces behind most of this basic research. At the same time, our knowledge of the molecular biological and biochemical characteristics of arterivirus nonstructural proteins continues to develop, with important implications for our understanding of viral RNA synthesis. There is little need to emphasize the importance of this process, given the fact that – generally speaking – a ‘simple’ reduction of its replication efficiency is one of the most straightforward mechanisms of virus attenuation. Moreover, through their general low fidelity, RNA virus RdRp-complexes are key players in virus evolution, which is reflected in the gradual but continuous genetic changes observed in particular in globally spreading RNA viruses like PRRSV. A more detailed understanding of the functions of arterivirus nsps in sg mRNA synthesis is important, given the critical role of these transcripts in the production of progeny virions, a process in which nsps also may participate through functions not related to sg RNA synthesis. Finally, if the use of such compounds in veterinary settings would ever become a financially attractive and accepted supplement to vaccination strategies, the biochemical and structural characterization of key arterivirus enzymes, like the proteases, RdRp, and helicase, may provide important leads for the development of specific anti-PRRSV drugs.
One of the most important aspects will be to develop a strategy on how to appropriately manipulate PRRSV-encoded antagonists of the IFN system and virulence factors during the development of a new generation of modified live vaccines. The initial research efforts in this area have raised many important questions: (i) Are PRRSV nsps modulating host gene expression and/or immune responses in a virus strain-specific manner? (ii) Does the postulated IFN-antagonizing activity of various nsps play a significant role in PRRSV replication and/or pathogenesis in animal models? (iii) Could certain nsp mutations be used to generate viable recombinant viruses with stably altered replication and virulence properties in vivo? (iv) Do mutations and deletions in specific nsps alter the IFN and cytokine responses in infected cells and (or) animals? If so, how does this affect disease progression in animals? (v) Can an attenuated PRRSV mutant with changes in its replicase gene provide protective immunity from wild-type PRRSV infection? Addressing these questions will be crucial to advance our understanding of the interplay between PRRSV replication and pathogenesis in the host, and to provide valuable information for vaccine development.
Another important aspect of nsp structure/function studies is to develop more sensitive and specific diagnostic assays. Currently available PRRSV diagnostic assays provide good sensitivity and specificity in controlled settings. However, the worldwide experience with PRRSV outbreaks suggests that substantial improvement is needed in some of these areas. This diagnostic challenge is mainly due to characteristics shared with all RNA viruses: PRRSV undergoes rapid evolution, which creates genetically and antigenically heterogeneous populations. The functional importance of PRRSV nsps in viral replication and host immunity makes them attractive targets for the diagnostic test development. The use of the generally more conserved nsps may offer certain advantages, although a diagnostic test based on a single PRRSV strain may still not be effective in detecting the wide spectrum of PRRSV field strains.
Persistence is another significant factor impeding the successful elimination of PRRS. Current diagnostic methods are unable to differentiate persistently infected pigs from pigs that have developed protective immunity. The PRRSV nsp7 region has now been successfully tested as a potential antigen for the development of serological assays. An important property of this protein is that the antibody response against this target persists for at least 202 dpi, which could also be used as a potential indicator of persistence. This study represents an initial effort to explore replicase subunits as a diagnostic target. Future advancements in our understanding of PRRSV nsp structure and function in replication and host immunity will likely provide a basis for additional and improved approaches to the development of diagnostic assays.
Acknowledgements
We would like to acknowledge the contributions, over many years, of a large number of colleagues who worked with us in studies on arterivirus replicase structure and function.
References
- Allende R., Lewis T.L., Lu Z., Rock D.L., Kutish G.F., Ali A., Doster A.R., Osorio F.A. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 1999;80:307–315. doi: 10.1099/0022-1317-80-2-307. [DOI] [PubMed] [Google Scholar]
- Barrette-Ng I.H., Ng K.K.S., Mark B.L., van Aken D., Cherney M.M., Garen C., Kolodenko Y., Gorbalenya A.E., Snijder E.J., James M.N.G. Structure of arterivirus nsp4—The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J. Biol. Chem. 2002;277:39960–39966. doi: 10.1074/jbc.M206978200. [DOI] [PubMed] [Google Scholar]
- Bautista E.M., Faaberg K.S., Mickelson D., McGruder E.D. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology. 2002;298:258–270. doi: 10.1006/viro.2002.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N., Selisko B., Ricagno S., Imbert I., van der Zanden L., Snijder E.J., Canard B. De novo initiation of RNA synthesis by the arterivirus RNA-dependent RNA polymerase. J. Virol. 2007;81:8384–8395. doi: 10.1128/JVI.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Sarkar S.N., Kwon B., Subramaniam S., Jones C., Pattnaik A.K., Osorio F.A. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 2010;84:1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-Coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4):e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E., Lawson S., Welbon C., Murtaugh M.P., Nelson E.A., Zimmerman J.J., Rowland R.R.R., Fang Y. Antibody response of nonstructural proteins: implication for diagnostic detection and differentiation of Type I and Type II porcine reproductive and respiratory syndrome virus. Clin. Vac. Immunol. 2009;16:628–635. doi: 10.1128/CVI.00483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Olivares J., Wieringa R.T., Bakonyi A.A.F., de Vries N.J., Poyner D., Rottier P.J.M. Generation of a candidate live marker vaccine for equine arteritis virus by deletion of the major virus neutralization domain. J. Virol. 2003;77:8470–8480. doi: 10.1128/JVI.77.15.8470-8480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Chen Z., Lawson S., Sun Z., Zhou X., Guan X., Christopher-Hennings J., Nelson E.A., Fang Y. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology. 2010;398(1):87–97. doi: 10.1016/j.virol.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhou X., Lunney J.K., Lawson S., Sun Z., Brown E., Christopher-Hennings J., Knudsen D., Nelson E., Fang Y. Immunodominant epitopes in nsp2 of porcine reproductive and respiratory syndrome virus are dispensable for replication but play an important role in modulation of host immune response. J. Gen. Virol. 2010;91:1047–1057. doi: 10.1099/vir.0.016212-0. [DOI] [PubMed] [Google Scholar]
- Conzelmann K.K., Visser N., Van Woensel P., Thiel H.J. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 1993;193:329–339. doi: 10.1006/viro.1993.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. DeLano Scientific; San Carlos, CA, USA: 2002. The PyMOL Molecular Graphics System.http://www.pymol.org [Google Scholar]
- de Lima M., Pattnaik A.K., Flores E.F., Osorio F.A. Mapping of B-cell linear epitopes on Nsp2 and structural proteins of a North American strain of porcine reproductive and respiratory syndrome virus. Virology. 2006;353:410–421. doi: 10.1016/j.virol.2006.05.036. [DOI] [PubMed] [Google Scholar]
- de Lima M., Kwon B., Ansari I.H., Pattnaik A.K., Flores E.F., Osorio F.A. Development of a porcine reproductive and respiratory syndrome virus differentiable (DIVA) strain through deletion of specific immunodominant epitopes. Vaccine. 2008;26:3594–3600. doi: 10.1016/j.vaccine.2008.04.078. [DOI] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A.F., Horzinek M.C., Spaan W.J.M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Faaberg K.S., Meulenberg J.J.M., Wassenaar A.L.M., Plagemann P.G.W., Gorbelenya A.E., Snijder E.J. Processing and evolution of the N-Terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J. Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries A.A., Chirnside E.D., Bredenbeek P.J., Gravestein L.A., Horzinek M.C., Spaan W.J. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucl. Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingson J.S., Wang Y., Layton S., Ciacci-Zanella J., Roof M.B., Faaberg K.S. Vaccine efficacy of porcine reproductive and respiratory syndrome virus chimeras. Vaccine. 2010;28:2679–2686. doi: 10.1016/j.vaccine.2009.12.073. [DOI] [PubMed] [Google Scholar]
- Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H.G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- Fang Y., Kim D.Y., Ropp S., Steen P., Christopher-Hennings J., Nelson E.A., Rowland R.R.R. Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res. 2004;100:229–235. doi: 10.1016/j.virusres.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Fang Y., Rowland R.R.R., Roof M., Lunney J.K., Christopher-Hennings J., Nelson E.A. A full-length cDNA infectious clone of North American type 1 porcine reproductive and respiratory syndrome virus: expression of green fluorescent protein in the nsp2 region. J. Virol. 2006;80:11447–11455. doi: 10.1128/JVI.01032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Schneider P., Zhang W.P., Faaberg K., Nelson E.A., Rowland R.R.R. Diversity and evolution of a newly emerged North American Type 1 porcine arterivirus. Arch. Virol. 2007;152:1009–1017. doi: 10.1007/s00705-007-0936-y. [DOI] [PubMed] [Google Scholar]
- Fang Y., Christopher-Hennings J., Brown E., Liu H., Chen Z., Lawson S., Breen R., Clement T., Gao X., Bao J., Knudsen D., Daly R., Nelson E.A. Development of genetic markers in the nonstructural protein 2 region of a US type 1 porcine reproductive and respiratory syndrome virus: implications for future recombinant marker vaccine development. J. Gen. Virol. 2008;89:3086–3096. doi: 10.1099/vir.0.2008/003426-0. [DOI] [PubMed] [Google Scholar]
- Frias-Staheli N., Glannakopoulos N.V., Klkkert M., Taylor S.L., Bridgen A., Paragas J., Richt J.A., Rowland R.R.R., Schmaljohn C.S., Lenschow D.J., Snijder E.J., Garcia-Sastre A., Virgin H.W. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbes. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z.Q., Guo X., Yang H.C. Genomic characterization of two Chinese isolates of porcine respiratory and reproductive syndrome virus. Arch. Virol. 2004;149:1341–1351. doi: 10.1007/s00705-004-0292-0. [DOI] [PubMed] [Google Scholar]
- Godeny E.K., Chen L., Kumar S.N., Methven S.L., Koonin E.V., Brinton M.A. Complete genomic sequence and phylogenetic analysis of the lactate dehydrogenase-elevating virus (LDV) Virology. 1993;194:585–596. doi: 10.1006/viro.1993.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O., Weber F. Pathogenic viruses: smart manipulators of the interfeon system. Curr. Top. Microbiol. Immunol. 2007;316:315–334. doi: 10.1007/978-3-540-71329-6_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Wang Y., Faaberg K.S. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 2006;122:175–182. doi: 10.1016/j.virusres.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Han J., Liu G., Wang Y., Faaberg K.S. Identification of nonessential regions of the nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J. Virol. 2007;81:9878–9890. doi: 10.1128/JVI.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Rutherford M.S., Faaberg K.S. The porcine reproductive and respiratory syndrome virus nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities. J. Virol. 2009;83:9449–9463. doi: 10.1128/JVI.00834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D.F., Henderson R.A., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox A.L., Appella E., Engelhard V.H. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.R., Yu W., Murtaugh M. Cross-reactive antibody responses to nsp1 and nsp2 of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 2007;88:1184–1195. doi: 10.1099/vir.0.82587-0. [DOI] [PubMed] [Google Scholar]
- Kaashoek M.J., Moerman A., Madic J., Rijsewijk F.A., Quak J., Gielkens A.L., van Oirschot J.T. A conventionally attenuated glycoprotein E-negative strain of bovine herpesvirus type 1 is an efficacious and safe vaccine. Vaccine. 1994;12:439–444. doi: 10.1016/0264-410x(94)90122-8. [DOI] [PubMed] [Google Scholar]
- Kim D.Y., Calvert J.G., Chang K.O., Horlen K., Kerrigan M., Rowland R.R.R. Expression and stability of foreign tags inserted into nsp2 of porcine reproductive and respiratory syndrome virus (PRRSV) Virus Res. 2007;128:106–114. doi: 10.1016/j.virusres.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Kim D.Y., Kaiser T.J., Horlen K., Keith M.L., Taylor L.P., Jolie R., Calvert J.G., Rowland R.R.R. Insertion and deletion in a non-essential region of the nonstructural protein 2 (nsp2) of porcine reproductive and respiratory syndrome (PRRS) virus: effects on virulence and immunogenicity. Virus Genes. 2009;38:118–128. doi: 10.1007/s11262-008-0303-4. [DOI] [PubMed] [Google Scholar]
- Kim O., Sun Y., Lai F.W., Song C., Yoo D. Modulation of type I interferon induction by porcine reproductive and respiratory syndrome virus and degradation of CREB-binding protein by non-structural protein 1 in MARC-145 and HeLa cells. Virology. 2010;402(2):315–326. doi: 10.1016/j.virol.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Roh I.S., Choi E.J., Lee C., Lee C.H., Lee K.H., Lee K.K., Song Y.K., Lee O.S., Park C.K. A molecular analysis of European porcine reproductive and respiratory syndrome virus isolated in South Korea. Vet. Microbiol. 2010;143:394–400. doi: 10.1016/j.vetmic.2009.11.039. [DOI] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroese M.V., Zevenhoven-Dobbe J.C., Bos-de Ruijter J.N., Peeters B.P., Meulenberg J.J., Cornelissen L.A., Snijder E.J. The nsp1α and nsp1β papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 2008;89:494–499. doi: 10.1099/vir.0.83253-0. [DOI] [PubMed] [Google Scholar]
- Kwon B., Ansari I.H., Pattnaik A.K., Osorio F.A. Identification of virulence determinants of porcine reproductive and respiratory syndrome virus through construction of chimeric clones. Virology. 2008;380:371–378. doi: 10.1016/j.virol.2008.07.030. [DOI] [PubMed] [Google Scholar]
- Luo R., Xiao S., Jiang Y., Jin H., Wang D., Liu M., Chen H., Fang L. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-β production by interfering with the RIG-I signaling pathway. Mol. Immunol. 2008;45:2839–2846. doi: 10.1016/j.molimm.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Aravind L., Koonin E.V. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 2000;25:50–52. doi: 10.1016/s0968-0004(99)01530-3. [DOI] [PubMed] [Google Scholar]
- Mebatsion T., Koolen M.J.M., de Vaan L.T.C., de Haas N., Braber M., Romer-Oberdorfer A., van den Elzen P., van der Marel P. Newcastle disease virus marker vaccine: an immunodominant epitope on the nucleoprotein gene of NDV can be deleted or replaced by a foreign epitope. J. Virol. 2002;76:10138–10146. doi: 10.1128/JVI.76.20.10138-10146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier W.A., Galeota J., Osorio F.A., Husmann R.J., Schnitzlein W.M., Zuckermann F.A. Gradual development of the interferon-gamma response of swine to porcine reproductive and respiratory syndrome virus infection or vaccination. Virology. 2003;309:18–31. doi: 10.1016/s0042-6822(03)00009-6. [DOI] [PubMed] [Google Scholar]
- Meulenberg J.J., Hulst M.M., de Meijer E.J., Moonen P.L., den Besten A., de Kluyver E.P., Wensvoort G., Moormann R.J. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulenberg J.J.M., Bos-de Ruijter J.N.A., Wensvoort G., Moormann R.J.M. Infectious transcripts from cloned genome-length cDNA of porcine reproductive respiratory syndrome virus. J. Virol. 1998;72:380–387. doi: 10.1128/jvi.72.1.380-387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.C., Laegreid W.W., Bono J.L., Chitko-Mckown C.G., Fox J.M. Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch. Virol. 2004;149:2453–2463. doi: 10.1007/s00705-004-0377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moormann R.J., de Rover T., Briaire J., Peeters B.P., Gielkens A.L., van Oirschot J.T. Inactivation of the thymidine kinase gene of a g1 deletion mutant of pseudorabies virus generates a safe but still highly immunogenic vaccine strain. J. Gen. Virol. 1990;71:1591–1595. doi: 10.1099/0022-1317-71-7-1591. [DOI] [PubMed] [Google Scholar]
- Nedialkova D.D., Ulferts R., van den Born E., Lauber C., Gorbalenya A.E., Ziebuhr J., Snijder E.J. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J. Virol. 2009;83(11):5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova D.D., Gorbalenya A.E., Snijder E.J. Arterivirus Nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLoS Pathog. 2010;6(2):e1000772. doi: 10.1371/journal.ppat.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen C.J., Murtaugh M.P., Faaberg K.S. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiewicz M.B., Botner A., Toft P., Normann P., Storgaard T. Epitope mapping porcine reproductive and respiratory syndrome virus by phage display: the nsp2 fragment of the replicase polyprotein contains a cluster of B-cell epitopes. J. Virol. 2001;75:3277–3290. doi: 10.1128/JVI.75.7.3277-3290.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksiewicz M.B., Bøtner A., Normann P. Semen from boars infected with porcine reproductive and respiratory syndrome virus (PRRSV) contains antibodies against structural as well as nonstructural viral proteins. Vet. Microbiol. 2001;81:109–125. doi: 10.1016/s0378-1135(01)00341-8. [DOI] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born E., Spaan W.J., Snijder E.J. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J.M., Snijder E.J. Nidovirus transcription: how to make sense? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- Pedersen K.W., van der Meer Y., Roos N., Snijder E.J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol J.M.A., Wagenaar F., Reus J.E.G. Comparative morphogenesis of three PRRS virus strains. Vet. Microbiol. 1997;55:203–208. doi: 10.1016/s0378-1135(96)01329-6. [DOI] [PubMed] [Google Scholar]
- Posthuma C.C., Nedialkova D.D., Zevenhoven-Dobbe J.C., Blokhuis J.H., Gorbalenya A.E., Snijder E.J. Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J. Virol. 2006;80:1653–1661. doi: 10.1128/JVI.80.4.1653-1661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma C.C., Pedersen K.W., Lu Z., Joosten R.G., Roos N., Zevenhoven-Dobbe J.C., Snijder E.J. Formation of the arterivirus replication/transcription complex: a key role for nonstructural protein 3 in the remodeling of intracellular membranes. J. Virol. 2008;82:4480–4491. doi: 10.1128/JVI.02756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R.E., Goodbourn S. Interferons and viruses: an interplay between induction, signaling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Ropp S.L., Mahlum Wees C.E., Fang Y., Nelson E.A., Rossow K.D., Bien M., Arndt B., Preszler S., Steen P., Christopher-Hennings J., Collins J.E., Benfield D.A., Faaberg K.S. Characterization of emerging European-like PRRSV isolates in the United States. J. Virol. 2004;78:3684–3703. doi: 10.1128/JVI.78.7.3684-3703.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbard J.B., Taylor W.R. A sequence pattern common to T cell epitopes. EMBO J. 1988;7:93–100. doi: 10.1002/j.1460-2075.1988.tb02787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royaee A.R., Husmann R., Dawson H.D., Calzada-Nova G., Schnitzlein W.M., Zuckermann F., Lunney J.K. Deciphering the involvement of innate immune factors in the development of the host responses to PRRSV vaccination. Vet. Immunol. Immunopathol. 2004;102:199–216. doi: 10.1016/j.vetimm.2004.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagripanti J.L., Zandomeni R.O., Weinmann R. The cap structure of simian hemorrhagic fever virion RNA. Virology. 1986;151:146–150. doi: 10.1016/0042-6822(86)90113-3. [DOI] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I., Berger A. On the size of the active site in proteases. I: Papain. Biochem. Biophys. Res. Commun. 1967;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Sette A., Buus S., Appella E., Smith J.A., Chesnut R., Miles C., Colon S.M., Grey H.M. Prediction of major histocompatibility complex binding regions of protein antigens by sequence pattern analysis. Proc. Natl. Acad. Sci. 1989;86:3296–3300. doi: 10.1073/pnas.86.9.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., van Dinten L.C., Snijder E.J., Ziebuhr J. Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J. Virol. 2000;74:9586–9593. doi: 10.1128/jvi.74.20.9586-9593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A., Posthuma C.C., van Dinten L.C., Snijder E.J., Gorbalenya A.E., Ziebuhr J. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 2005;79:696–704. doi: 10.1128/JVI.79.2.696-704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Kwang J., Liu W., Liu D.X. Determination of the complete nucleotide sequence of a vaccine strain of porcine reproductive and respiratory syndrome virus and identification of the Nsp2 gene with a unique insertion. Arch. Virol. 2000;145:871–883. doi: 10.1007/s007050050680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L.M., Spaan W.J. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J. Virol. 1992;66:7040–7048. doi: 10.1128/jvi.66.12.7040-7048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L.M., Spaan W.J. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L.M., Spaan W.J., Gorbalenya A.E. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 1995;270:16671–16676. doi: 10.1074/jbc.270.28.16671. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Wassenaar A.L.M., van Dinten L.C., Spaan W.J.M., Gorbalenya A.E. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J. Biol. Chem. 1996;271:4864–4871. doi: 10.1074/jbc.271.9.4864. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Meulenberg J.J. The molecular biology of arteriviruses. J. Gen. Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., van Tol H., Roos N., Pedersen K.W. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 2001;82:985–994. doi: 10.1099/0022-1317-82-5-985. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E.J., Spaan, W.J., 2006. Arteriviruses. In: Knipe, D. M., and P. M. Howley, (Eds.), Fields Virology, vol. 1, fifth ed. Lippincott Williams & Wilkins, Philidelphia, pp. 1337–1355.
- Sun Y., Xue F., Guo Y., Ma M., Hao N., Zhang X.C., Lou Z., Li X., Rao Z. Crystal structure of porcine reproductive and respiratory syndrome virus (PRRSV) leader protease Nsp1{alpha} J. Virol. 2009;83:10931–10940. doi: 10.1128/JVI.02579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Chen, N., Yoo, D., 2009b. Inhibition of type 1 interferon signaling by nsp11 of PRRSV. In: The 2009 International PRRS Symposium, abstract no. 33, 4–5 December 2009, Chicago, IL, USA.
- Sun, Z., Chen, Z., Lawson, S., Fang, Y., 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism function. J. Virol. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Tian K., Yu X., Zhao T., Feng Y., Cao Z., Wang C. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE. 2007;2:526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Lu G., Gao F., Peng H., Feng Y., Ma G., Bartlam M., Tian K., Yan J., Hilgenfeld R., Gao G.F. Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV) J. Mol. Biol. 2009;392:977–993. doi: 10.1016/j.jmb.2009.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms M.A., van Dinten L.C., Gorbalenya A.E., Snijder E.J. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. 2001;98:1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms M.A., van der Meer Y., Snijder E.J. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 2002;83:795–800. doi: 10.1099/0022-1317-83-4-795. [DOI] [PubMed] [Google Scholar]
- Tijms M.A., Snijder E.J. Equine arteritis virus non-structural protein 1, an essential factor for viral subgenomic mRNA synthesis, interacts with the cellular transcription co-factor p100. J. Gen. Virol. 2003;84:2317–2322. doi: 10.1099/vir.0.19297-0. [DOI] [PubMed] [Google Scholar]
- Tijms M.A., Nedialkova D.D., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Snijder E.J. Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease. J. Virol. 2007;81:10496–10505. doi: 10.1128/JVI.00683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong H.M., Lu Z., Kutish G.F., Galeota J., Osorio F.A., Pattnaik A.K. A highly pathogenic porcine reproductive and respiratory syndrome virus generated from an infectious cDNA clone retains the in vivo virulence and transmissibility properties of the parental virus. Virology. 2004;325:308–319. doi: 10.1016/j.virol.2004.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aken D., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Snijder E.J. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 2006;87:3473–3482. doi: 10.1099/vir.0.82269-0. [DOI] [PubMed] [Google Scholar]
- van Aken D., Snijder E.J., Gorbalenya A.E. Mutagenesis analysis of the nsp4 main proteinase reveals determinants of arterivirus replicase polyprotein autoprocessing. J. Virol. 2006;80:3428–3437. doi: 10.1128/JVI.80.7.3428-3437.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer Y., van Tol H., Locker J.K., Snijder E.J. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dinten L.C., Wassenaar A.L., Gorbalenya A.E., Spaan W.J., Snijder E.J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J. Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dinten L.C., den Boon J.A., Wassenaar A.L., Spaan W.J., Snijder E.J. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc. Natl. Acad. Sci. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dinten L.C., van Tol H., Gorbalenya A.E., Snijder E.J. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J. Virol. 2000;74:5213–5223. doi: 10.1128/jvi.74.11.5213-5223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., de Wilde A.H., Gorbalenya A.E., Snijder E.J. The in vitro RNA synthesizing activity of the isolated arterivirus replication/transcription complex is dependent on a host factor. J. Biol. Chem. 2008;283:16525–16536. doi: 10.1074/jbc.M708136200. [DOI] [PubMed] [Google Scholar]
- van Marle G., Dobbe J.C., Gultyaev A.P., Luytjes W., Spaan W.J., Snijder E.J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., van Dinten L.C., Spaan W.J., Luytjes W., Snijder E.J. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J. Virol. 1999;73:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oirschot J.T., Kaashoek M.J., Rijsewijk F.A., Stegeman J.A. The use of marker vaccines in eradication of herpesviruses. J. Biotechnol. 1996;44:75–81. doi: 10.1016/0168-1656(95)00129-8. [DOI] [PubMed] [Google Scholar]
- Walsh E.P., Baron M.D., Rennie L.F., Anderson J., Barrett T. Development of a genetically marked recombinant rinderpest vaccine expressing green fluorescent protein. J. Gen. Virol. 2000;81:709–718. doi: 10.1099/0022-1317-81-3-709. [DOI] [PubMed] [Google Scholar]
- Walsh E.P., Baron M.D., Rennie L.F., Monaghan P., Anderson J., Barrett T. Recombinant rinderpest vaccines expressing membrane-anchored proteins as genetic markers: evidence of exclusion of marker protein from the virus envelope. J. Virol. 2000;74:10165–10175. doi: 10.1128/jvi.74.21.10165-10175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liang Y., Han J., Burkhart K.M., Vaughn E.M., Roof M.B., Faaberg K.S. Attenuation of porcine reproductive and respiratory syndrome virus strain MN184 using chimeric construction with vaccine sequence. Virology. 2008;371(2):418–429. doi: 10.1016/j.virol.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Wassenaar A.L., Spaan W.J., Gorbalenya A.E., Snijder E.J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that Nsp2 acts as a cofactor for the Nsp4 serine protease. J. Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland, F., Granzow, H., Wieczorek-Krohmer, M., Weiland, E., 1995. Electron microscopic studies on the morphogenesis of PRRSV in infected cells—comparative studies. In: Schwyzer, M., Ackermann, M., Bertoni, G., Kocherhans, R., McCullough, K., Engels, M., Wittek, R., Zanoni, R. (Eds.), Immunobiology of Viral Infections. Proceedings of the 3rd Congress of the European Society of Veterinary Virology, pp. 499–502.
- Widjojoatmodjo M.N., van Gennip H.G., Bouma A., van Rijn P.A., Moormann R.J. Classical swine fever virus E(rns) deletion mutants: trans-complementation and potential use as non transmissible, modified, live-attenuated marker vaccines. J. Virol. 2000;74:2973–2980. doi: 10.1128/jvi.74.7.2973-2980.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Guo X., Ge X., Chen Y., Cha Z., Yang H. Monoclonal antibody and porcine antisera recognized B-cell epitopes of Nsp2 protein of a Chinese strain of porcine reproductive and respiratory syndrome virus. Virus Res. 2007;126:207–215. doi: 10.1016/j.virusres.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Yoo, D., Kim, O., Song, C., Sun, Y., Du, Y., Liu, H.C., 2009. Modulation of type 1 interferon production and evasion strategies of PRRSV from the host defense. In: The 2009 International PRRS Symposium, 4–5 December 2009, Chicago, IL, USA (abstract no. 9).
- Yoshii M., Okinaga T., Miyazaki A., Kato K., Ikeda H., Tsunemitsu H. Genetic polymorphism of the nsp2 gene in North American type porcine reproductive and respiratory syndrome virus. Arch. Virol. 2008;153(7):1323–1334. doi: 10.1007/s00705-008-0098-6. [DOI] [PubMed] [Google Scholar]
- Zhou L., Zhang J., Zeng J., Yin S., Li Y., Zheng L., Guo X., Ge X., Yang H. The 30 amino acids deletion in Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J. Virol. 2009;83:5156–5167. doi: 10.1128/JVI.02678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]


