Abstract
Infectious salmon anaemia virus (ISAV) is a piscine orthomyxovirus causing a serious disease in farmed Atlantic salmon (Salmo salar L.). The virus surface glycoprotein hemagglutinin-esterase (HE) is responsible for both viral attachment and release. Similarity to bovine and porcine torovirus hemagglutinin-esterase (BToV HE, PToV HE), bovine coronavirus HE (BCoV HE) and influenza C hemagglutinin-esterase-fusion (InfC HEF) proteins were exploited in a computational homology-based structure analysis of ISAV HE. The analysis resolved structural aspects of the protein and identified important features of relevance to ISAV HE activity. By recombinant expression and purification of secretory HE (recHE) proteins, receptor-binding and quantitative analyses of enzymatic activities displayed by ISAV HE molecules are presented for the first time. Three different recHE molecules were constructed: one representing a high virulent isolate, one a low virulent, while in the third a Ser32 to Ala32 amino acid substitution was introduced in the enzymatic catalytic site as inferred from the model. The three amino acid differences between the high and low virulent variants, of which two localized to the putative receptor-binding domain and one in the esterase domain, had no impact on receptor-binding or -release activities. In contrast, the Ser32 amino acid substitution totally abolished enzymatic activity while receptor binding increased, as observed by agglutination of Atlantic salmon red blood cells. This demonstrates the essential role of a serine in the enzyme's catalytic site. In conclusion, structural analysis of ISAV HE in combination with selected recHE proteins gave insights into structure–function relationships and opens up for further studies aiming at dissecting molecular determinants of ISAV virulence.
Keywords: Infectious salmon anaemia virus (ISAV), Hemagglutinin-esterase (HE), Receptor binding, Receptor-destroying enzyme (RDE), Protein structure, Recombinant secretory HE (recHE)
1. Introduction
Infectious salmon anaemia virus (ISAV) is a piscine orthomyxovirus, genus Isavirus (Kawaoka et al., 2005), causing a systemic disease in farmed Atlantic salmon (Salmo salar L.). The ongoing devastating ISA disease outbreaks in Chile, threatening the country's entire Atlantic salmon farming industry (Mardones et al., 2009, Godoy et al., 2008), is a reminder of the importance of this emerging disease. Similar to influenza A and B viruses, the ISAV is an enveloped virus with a genome consisting of eight negative sense single-stranded RNA segments encoding at least 10 proteins (Mjaaland et al., 1997, Clouthier et al., 2002, Falk et al., 2004). ISAV has two major surface proteins; a 50 kDa fusion (F) protein which is encoded by gene segment 5 and responsible for fusion of viral and cellular membranes (Aspehaug et al., 2005) and the 38–43 kDa hemagglutinin-esterase (HE) protein encoded by gene segment 6, providing both the receptor-binding and the receptor-destroying enzyme (RDE) activities (Falk et al., 2004, Krossøy et al., 2001, Rimstad et al., 2001). This functional organisation differs from influenza A and B viruses where the hemagglutinin (HA) has receptor-binding and fusion activities, while RDE activity is performed by the neuraminidase (NA) (Colman et al., 1983, Palese et al., 1974, Wiley and Skehel, 1977, Wiley and Skehel, 1987). In fact, ISAV HE shows higher functional similarity to the bovine and porcine torovirus and bovine coronavirus hemagglutinin-esterase proteins (BToV HE, PToV HE, BCoV HE), as well as to the influenza C virus hemagglutinin-esterase-fusion protein (InfC HEF) where all three functions are incorporated in one single surface protein (Herrler et al., 1988). The X-ray crystallographic structures of BToV HE, PToV HE, BCoV HE and InfC HEF have been determined (Langereis et al., 2009, Zeng et al., 2008, Rosenthal et al., 1998).
The RDE activity prevents the viruses from self-aggregation and promotes release of viral progenies from the infected cell (Höfling et al., 1996, Liu et al., 1995). In addition, the RDE may also play a role early during infection (Brossmer et al., 1993, Huang et al., 1985, Matrosovich et al., 2004, Ohuchi et al., 1995, Strobl and Vlasak, 1993). For influenza A viruses, a functional balance between the antagonistic functions of its receptor-binding- and RDE activities seems to be required for viral replicative fitness and virulence (Wagner et al., 2002, Lu et al., 2005, Kaverin et al., 1998, Chen et al., 2007, Gulati et al., 2005, Mitnaul et al., 2000, Baigent and McCauley, 2001, Shtyrya et al., 2009). The function of the ISAV RDE and its role in virulence remains to be clarified. Enzyme inhibition studies indicate that the acetylesterase of the ISAV HE contains a serine in its active site, probably constituting a part of a Ser-His-Asp catalytic triad, similar to the RDE of the InfC HEF protein (Falk et al., 2004, Kristiansen et al., 2002, Muchmore and Varki, 1987, Pleschka et al., 1995). Furthermore, data from multiple sequence alignment including HE sequences from ISAV and other viruses suggest that these residues are located at position 32 for Ser, 261 for Asp and 264 for His (Falk et al., 2004). This remains to be confirmed by functional studies to provide conclusive evidence. The cellular receptor for ISAV has not yet been defined. While InfC HEF specifically binds to and deacetylates 5-N-acetyl-9-O-acetylsialic acid residues (Herrler et al., 1985, Rogers et al., 1986), the most likely substrate for ISAV HE is 5-N-acetyl-4-O-acetylsialic acid (Hellebø et al., 2004).
ISAV binds to erythrocytes from several fish (Atlantic salmon, rainbow trout, Atlantic cod, crucian carp and wolffish) and mammalian species (horse and rabbit) (Falk et al., 2004, Falk et al., 1997). The acetylesterase responsible for the RDE activity allows the virus to detach (elute) from these erythrocytes. One general exception is Atlantic salmon erythrocytes. Most experiments point to the inability of the ISAV to elute from these cells (Falk et al., 2004, Falk et al., 1997, Hellebø et al., 2004). Recent results, however, suggest that inability to elute from Atlantic salmon erythrocytes in some way can be related to virulence, as low virulent strains may elute (K. Falk, O.B. Dale, personal communication). This contradicts the situation in influenza A and B viruses where an inhibition of the RDE hampers release of viral particles from the cell surface and is used as treatment against influenza infections in humans (Moscona, 2005, von Itzstein et al., 1993).
Despite large differences in virulence between isolates, the ISAV genome is remarkably conserved (Markussen et al., 2008, Mjaaland et al., 2005, Mjaaland et al., 2002). The ISAV protein with highest sequence variation is the HE containing a highly polymorphic region (HPR) in the stalk of the protein, near the transmembrane domain (Mjaaland et al., 2002, Krossøy et al., 2001, Rimstad et al., 2001, Markussen et al., 2008). It has been suggested that the HPR results from differential deletions of the full-length HPR (HPR0), possibly as a consequence of transmission from a viral reservoir to densely populated farmed Atlantic salmon (Mjaaland et al., 2002). The presence of HPR0 has been confirmed in healthy wild and farmed Atlantic salmon (Cunningham et al., 2002, Cook-Versloot et al., 2004, Anonymous, 2005, Nylund et al., 2007, Markussen et al., 2008). Moreover, ISAV isolated from fish with clinical ISA always contain a deletion in this region (Mjaaland et al., 2002, Nylund et al., 2007). It has therefore been hypothesized that variations in the length of the HPR may affect the structure/plasticity of the molecule, which again may affect the receptor-binding affinities and RDE activities, as well as potential interactions with the F protein. The role of amino acid differences outside the HPR region on structure and/or function has not yet been addressed. The aim of this study was to identify different domains of the HE protein, including the active site of the RDE, by comparison to known HE structures. Functional testing of these predictions through mutagenesis and functional/quantitative analyses were made possible by the development of a recombinant secreted form of HE (recHE). In addition, the analysis was used to select different HE variants for quantitative comparisons of their functions.
2. Materials and methods
2.1. Sequence analysis and modeling
Sequences were retrieved from the NCBI nr (Sayers et al., 2010) protein sequence database. The sequence analysis and fold recognition was performed on the ISAV HE sequence Q911Y4 (AJ276859, Scottish) from strain 390/98. Complementary analysis was also done on the ISAV HE sequence Q9JOYO (AF220607) from the strain Glesvær/2/90 (ISAV4) (Rimstad et al., 2001). These two HE sequences are 95% identical at the amino acid sequence level. Fold recognition was performed with the GeneSilico (Kurowski and Bujnicki, 2003) and BioInfo (Bujnicki et al., 2001) meta-servers. These servers collect data from a large number of prediction methods and compare the results on a unified scale using a jury system (Ginalski et al., 2003). Secondary structures for experimental structure data were classified with DSSP (Kabsch and Sander, 1983) and extracted using local software. Experimental structures were downloaded from RCSB/PDB (Berman et al., 2000). Structure-based sequence alignment for the experimental structures was generated with FatCat (Ye and Godzik, 2003), allowing for structural flexibility. Identification of receptor–ligand interactions in the experimental structures was done with the Ligand Explorer at RCSB (Berman et al., 2000). Visualization and analysis of the alignment with ISAV HE was done with DeepView (SwissPDBViewer) 4.0 (Guex and Peitsch, 1997). The final alignment (Fig. 1 ) was drawn with Alscript (Barton, 1993).
Fig. 1.
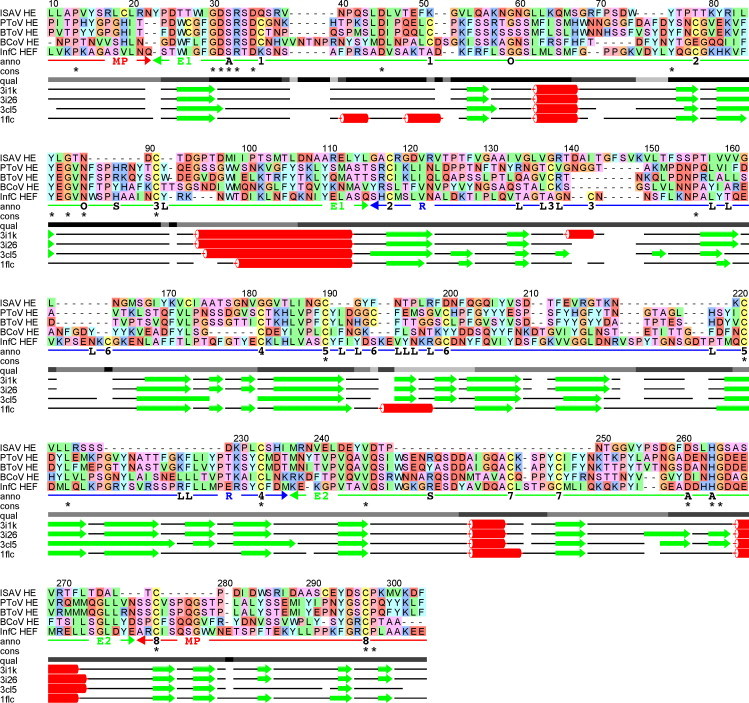
Multiple alignment of hemagglutinin sequences, showing the FFAS (Rychlewski et al., 2000) alignment of ISAV HE (Q911Y4) against the structure-based alignment of porcine torovirus HE (PToV HE; PDB id code 3I1K), bovine torovirus HE (BToV HE; 3I26), bovine coronavirus HE (BCoV HE; 3CL5) and influenza C virus HEF (InfC HEF; 1FLC). The upper part shows the sequence alignment, the lower part shows the corresponding alignment of secondary structure elements determined from the experimental protein structures (β-strands: green arrows, α-helices: red coils). The qual line indicates alignment quality based on the degree of consensus between FFAS alignments against each of the structural templates, ranging from full consensus in black to no consensus in light grey. The cons line shows completely conserved positions in the alignment. The anno line shows relevant annotation of the alignment, including residues involved in active site (A), oxyanion hole (O), ligand binding (L), substrate coordination (S) and Cys–Cys pairing (1–8). The anno line also shows domain organisation (MP: membrane-proximal (red); E1, E2: esterase (green); R: receptor (blue)) taken from Langereis et al. (2009). The receptor domain is frequently annotated as a lectin domain in other publications, e.g. in Langereis et al. (2009). The numbering of the alignment is according to the full-length ISAV HE sequence. The alignment includes part of the signal peptide for ISAV HE (LLAPVYS from position 10 to 16), but no part of the signal peptide for any of the other sequences. This shows a clear sequence similarity between part of the ISAV HE signal peptide and the structural templates.
2.2. Cells and recombinant baculoviruses
Spodoptera frugiperda (Sf9) cells were maintained in suspension culture at 28 °C using serum-free insect cell medium (Sf-900 II SFM) (Invitrogen). Recombinant baculoviruses were generated by homologous recombination in Sf9 cells of a transfer plasmid containing a HE DNA sequence insert or without an insert (negative control) and linearized DNA of the baculovirus Autographa californica nuclear polyhedrosis virus (AcNPV) (BacVector™-2000 Triple Cut Virus DNA) (Novagen). Except for minor modifications (described in Müller et al., 2008), the procedure as well as subsequent amplification and determination of the viral titres were performed according to the instructions given by the manufacturer.
2.3. Construction of transfer plasmids and site-directed mutagenesis
Isolation of RNA from ISAV isolates, TOPO-cloning and sequencing was performed as previously described (Markussen et al., 2008). The two pCR®2.1-TOPO plasmids containing the full-length HE genes from the isolates ISAV4 (Glesvær/2/90, AF220607) (HE4) and ISAV7 (AF427053) (HE7) were used as PCR templates. Primers were designed to produce truncated forms of the HEs (Table 1 ), without its putative transmembrane and cytoplasmic regions (amino acids 354–392 relative to the start codon). To enable secretion, all HE constructs maintained their own signal peptide (amino acids 1–16). The PCR cycling conditions were; 95 °C/2 min, followed by 35 cycles of 95 °C/1 min, 63 °C/1 min and 72 °C/2 min, and a final extension step at 72 °C/10 min. The PCR products were gel-purified using the QIAquick® Gel Extraction Kit (Qiagen), cloned into the pCR®2.1-TOPO vector (Invitrogen), subcloned into the SacI/XhoI sites of the pBACgus-1 transfer plasmid (Novagen), and transformed into Escherichia coli XL10-Gold® Ultracompetent cells (Stratagene), according to protocols provided by the manufacturers. Plasmids were isolated using the QIAprep Spin Miniprep Kit (Qiagen). The recombinant transfer plasmids with the correct inserts were named pBACgus-1-sHE4 and pBACgus-1-sHE7. The mutated plasmid pBACgus-1-recHE4S32A was made by site-directed mutagenesis using the QuikChange® XL Site-Directed Mutagenesis Kit (Stratagene) with pBACgus-1-sHE4 as template, and primers designed according to the manufacturer's guidelines (Table 1). A C-terminal His6-tag present in all three constructs facilitated identification and purification of the expressed recHE proteins. All plasmid DNAs were sequenced on both strands at GATC-Biotech AG using the sequencing primers given in Table 1.
Table 1.
Oligonucleotide primers used for PCR amplification, site-directed mutagenesis and sequencing.
| Primer | Oligonucleotide sequence (5′ → 3′) |
|---|---|
| HE fwd (PCR) | ATTATTGAGCTCATAAATATGGCACGATTCATAATTTTATTCC |
| HE4 rev (PCR) | TAATTACTCGAGACCCATAGTTTGGTTCAGCTGAGG |
| HE7 rev (PCR) | TAATTACTCGAGACCCATAGTTTGGTTCAGCTG |
| HE4(S32A) fwd (mutagenesis) | CCTGGATAGGTGACgCTCGAAGCGATCAGTC |
| HE4(S32A) rev (mutagenesis) | GACTGATCGCTTCGAGcGTCACCTATCCAGG |
| pBACgus-1 fwd (sequencing) | ATAACCATCTCGCAAATAAATAAGTAT |
| pBACgus-1 rev (sequencing) | CTGTAAATCAACAACGCACAG |
Sequences representing restriction enzyme recognition sites are underlined. Start codon (ATG) is shown in bold, while additional nucleotides necessary to provide translation initiation signals are in italics. Substituted nucleotides designed to introduce the single amino acid mutation in pBACgus-1-recHE4 are shown in small bold letters. fwd = forward, rev = reverse.
2.4. Protein expression and purification
All recHEs were expressed and purified as previously described (Müller et al., 2008). The purified recombinant proteins were named recHE4, recHE7 and recHE4S32A, while protein purified from Sf9 cells infected with the negative control baculovirus was called rec empty-1. The amount of soluble protein was determined by the DC protein assay (BioRad Laboratories Inc.) with bovine serum albumin (BSA) as the protein standard. The purity and identity of the recombinant proteins were determined by SDS-PAGE and Western blot analysis using anti-His antibody and anti-HE peptide serum as previously described (Müller et al., 2008). Purified proteins were stored in 20 mM HEPES, 150 mM NaCl at pH 8.0 with glycerol added to a final concentration of 50% before storage at −20 °C.
2.5. Immobilisation of recHEs to magnetic cobalt beads
Purified recHE4, recHE7 and recHE4S32A were coupled to magnetic Dynabeads®TALON™ (Invitrogen) (2 μg protein/2.5 μl beads) in 700 μl 20 mM HEPES, 500 mM NaCl, pH 7.5 at 20 °C for 1 h with slow rotation. The coupled beads were washed twice with buffer, once with L-15 (Leibovitz) medium, and then resuspended in 200 μl L-15 medium. Negative controls included rec empty-1 (described above) and drNimA, an unrelated His6-tagged protein (NimA from Deinococcus radiodurans) (kindly provided by Dr. Hanna-Kirsti S. Leiros at The Norwegian Structural Biology Centre, Tromsø, Norway) (Leiros et al., 2004, Müller et al., 2008). To compare the amounts of protein conjugated to the beads, the four His6-tagged proteins were eluted using 20 mM HEPES, 500 mM NaCl, 500 mM imidazole, pH 7.5 followed by analysis on SDS-PAGE under reducing conditions, and Coomassie blue staining. The intensities of the protein bands (three parallels of each sample) were quantified and compared using a Gene Genius Bioimage analyzer (Syngene).
2.6. Immunostaining
Dynabeads®TALON™, coated with proteins as described above, were incubated with a mixture of two mouse monoclonal antibodies (3H6F8/10C9F5) directed against ISAV HE (kindly provided by Dr. K. Falk at The Norwegian Veterinary Institute, Oslo, Norway) (Falk et al., 1998) at a dilution of 1:100 for 1 h at room temperature (RT). Incubation with secondary antibody Alexa405® goat anti-mouse IgG (Invitrogen), used at 2 μg/ml, was for 2 h at RT. RecHE4-coupled beads incubated with secondary antibody only served as a negative control. Fluorescence microscopy was performed and pictures taken at 40× magnification using an Olympus IX81 microscope.
2.7. Hemagglutination assay
Blood from Atlantic salmon and rabbit were collected using VENOJECT Lithium Heparin (LH) tubes (Terumo Europe NV) and centrifuged at 400 × g at 4 °C for 10 min. The Atlantic salmon- and rabbit red blood cells (AsRBCs and rRBCs) were washed three times in L-15 medium, centrifuged at 400 × g at 4 °C for 10 min and diluted in L-15 medium to 0.25% (v/v) AsRBC or 0.5% (v/v) rRBC suspension. To reduce non-specific interactions with tube walls, 1.5 ml Eppendorf tubes (Eppendorf) were pre-treated with 20 mg/ml BSA in PBS, pH 7.4, supplemented with 1.5% NaCl, and washed once with the same buffer. The hemagglutination reaction was performed by gently mixing 200 μl 0.25% AsRBC, or 200 μl 0.5% rRBCs, with 100 μl of recHE-coated Dynabeads®TALON™ (described above). The effect of the serine esterase inhibitor 3,4-dichloroisocoumarin (DCIC) (Sigma–Aldrich Inc.) on hemagglutination was studied by pre-incubating the recHE-coated beads with 2 μl of 5 mM DCIC solubilized in DMSO at 16 °C for 30 min before the addition of AsRBCs or rRBCs. Control samples were added 2 μl of DMSO only. Aliquots of the hemagglutination samples, with and without inhibitor, were collected at two time intervals, 1–3 h and 5–7 h for AsRBCs, and at 30 min and 1 h for rRBCs. Quantification of agglutination was performed by counting unbound beads in a Bürker chamber. The experiments were repeated three times, and data from one representative experiment are presented. Aliquots of the bead-agglutination mixtures were also transferred onto chamber slides (Lab-Tek®) containing L-15 medium and photographed at 40× magnification in a light microscope.
2.8. Enzyme activity assay
The acetylesterase activities of the recHE4, recHE7 and recHE4S32A proteins were determined using synthetic substrates as described previously (Falk et al., 1997, Kristiansen et al., 2002), with minor modifications. Various concentrations of 4-methylumbelliferyl acetate (4MUAc) (Sigma–Aldrich Inc.) (0–500 μM) in PBS pH 7.4 at RT were incubated with 4 μl recHE-coated Dynabeads®TALON™ (described above). Fluorescence was measured in a time-dependent manner in a Perkin Elmer plate reader at 360 nm excitation/465 nm emission. Pre-incubation with the serine esterase inhibitor DCIC was conducted as described above, and end-point data were collected following 40 min incubation with substrate. Enzyme activities of soluble proteins were determined by adding 200 μl p-nitrophenyl acetate (pNPA) (Sigma–Aldrich Inc.) at concentrations ranging from 50 μM to 4000 μM to 5 μl (0.7–1 mg/ml) of the soluble purified recHE protein sample diluted in 20 mM HEPES, 150 mM NaCl, pH 8.0. Enzyme activity was measured at OD405 in a time-dependent manner. For the two assay types, spontaneous hydrolysis of 4MUAc and pNPA in the buffer was quantified and subtracted from sample values. The rec empty-1 sample and/or the drNimA protein served as negative controls. Data on enzyme kinetics are presented as Michaelis–Menten and Lineweaver–Burk plots. The assays were performed in 96-well plates (Nunc™, Thermo Fisher Scientific) at RT, and run in minimum three parallels. All experiments were repeated minimum two times for verification of results, and in each case the data from one such experiment are presented.
3. Results
3.1. Fold recognition
The ISAV HE sequence was subjected to fold recognition and 3D structure analysis to identify the localisation of the ISAV receptor-destroying acetylesterase function. This analysis also enables identification and visualization of additional structural domains of this protein, such as the receptor-binding domain. The fold recognition servers unambiguously identified ISAV HE as a member of the SGNH hydrolase family, originally characterised by four conserved blocks of residues, with Ser (S), Gly (G), Asn (N) and His (H) completely conserved (Mølgaard et al., 2000). In particular the fold and function assignment system (FFAS) fold recognition software (Rychlewski et al., 2000) identified ISAV HE as significantly similar to PToV HE (PDB id code 3I1K) and BToV HE (PDB 3I26), and clearly similar to BCoV HE (PDB 3CL5) and InfC HEF (PDB 1FLC), with a sequence identity between 16 and 21%. These predictions were used for alignment and structure prediction for ISAV HE.
3.2. Sequence alignment
The sequence alignment of ISAV HE against the known experimental structures identified by fold recognition is shown in Fig. 1. This alignment is based on the FFAS predicted alignment of the ISAV HE sequence against each of the four structural templates, as well as the structure-based alignment of the experimental structures against each other. The structure-based alignments are taken directly from the FatCat (Ye and Godzik, 2003) pairwise alignments, adding the necessary gaps to get a consensus alignment of all four sequences. The structural similarity in the alignment is indicated with secondary structure elements of the experimental structures. The ISAV HE sequence was added to the structure-based alignment by using the consensus of FFAS alignments against each of the individual experimental structures. This turned out to be identical to the alignment against PToV HE, which has the most significant similarity to ISAV HE according to the FFAS fold recognition method. The alignment has been annotated with domain organisation according to PToV HE (Langereis et al., 2009), indicated as receptor domain, esterase domain and membrane-proximal domain. The receptor domain is frequently annotated as a lectin domain in other publications, e.g. in Langereis et al. (2009). Also indicated is the active site residues of the catalytic triad (Ser-His-Asp), residues forming the oxyanion hole of the active site (Langereis et al., 2009) and the pattern of disulphide bridges. Residues with side chains involved in ligand binding were identified in all four structures, either by computational analysis or from the original paper (InfC HEF), and annotated in the alignment. The alignment does not include sequence regions outside of the domains identified by FFAS, like the coiled-coil helix of InfC HEF, the transmembrane region and part of the signal peptide.
3.3. Structural visualization
The annotated alignment was used to visualize predicted structural properties of ISAV HE, using PToV HE (PDB 3I1L) as a template (Fig. 2A). This structure is identical to 3I1K used in the alignment, but with a ligand in the receptor-binding site. The ligand is Neu4,5,9Ac3α2Me, a synthetic analog of sialic acid. The overlap in alignment between ISAV and PToV HE is indicated on the structure, as well as active site residues (Fig. 2B), ligand binding residues in the receptor domain (Fig. 2C) and ISAV4 HE versus ISAV7 HE sequence differences (Fig. 2C). This enabled us to use the experimental structure of PToV HE as an approximate model of the ISAV HE structure.
Fig. 2.
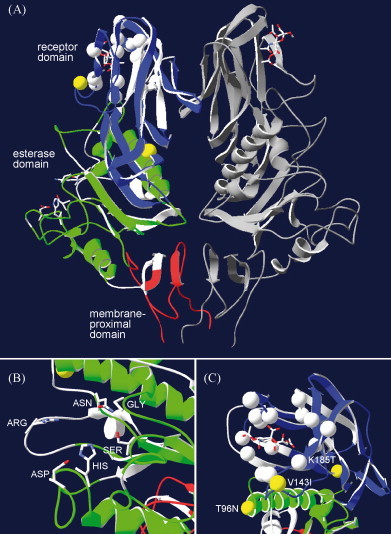
(A) Visualization of ISAV HE based on the experimental structure of porcine torovirus HE (PToV HE; PDB id code 3I1L). The structure includes an analog of the natural ligand in the receptor-binding pocket. The right monomer of the dimer structure is shown in grey. The left monomer shows the sequence alignment overlap with ISAV HE, color coded according to domain (membrane-proximal: red; esterase: green; receptor: blue). Seven residues of the first membrane-proximal sequence overlap with the ISAV HE signal peptide (see Fig. 1 caption). Non-overlapping regions are shown in white. Also shown are the active site residues in the esterase domain (see B), ligand binding residues in the receptor domain (white spheres) and the three amino acid residues differing between ISAV4 HE and ISAV7 HE (yellow spheres; see C). In (B) the active site region is magnified. The side chains for the active site residues, Ser32-His264-Asp261, are shown, as well as the oxyanion hole residues Gly59 and Asn89, and the substrate coordinating Arg (not conserved in ISAV HE). In (C) the ligand binding region of HE is magnified, showing the three amino acid differences between ISAV4 HE and ISAV7 HE (yellow spheres) as well as the residues involved in ligand binding in any of the known 3D structures (white spheres). The ligand of the 3I1L PToV HE structure is also shown.
3.4. Expression, purification and folding of recHEs
By using the information extracted from the model, two wild-type ISAV HE variants differing at three positions, recHE4 and recHE7 (Fig. 2C), and one mutated recHE4 form, recHE4S32A, containing a Ser to Ala amino acid substitution at position 32 (relative to the start codon) in the catalytic triad (Ser32-His264-Asp261), were expressed along with the negative control rec empty-1 in a secreted form. All three recHEs were obtained at a high purity in the range of 300–600 μg per liter cell culture (Fig. 3A).
Fig. 3.
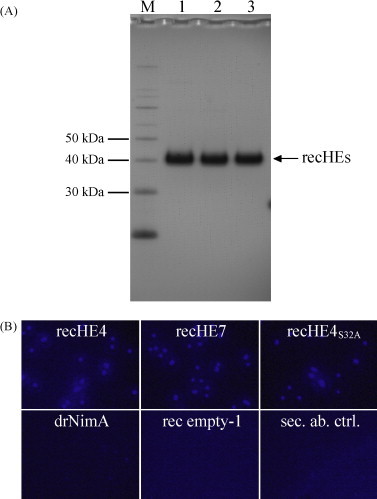
(A) Recombinant (rec) protein samples (4 μg each) were separated by SDS-PAGE and stained with Coomassie blue; lane 1: recHE4, lane 2: recHE7, lane 3: recHE4S32A; M: MagicMark™ XP protein standard (5 μl). (B) Immunostaining of recHEs coupled to Dynabeads®TALON™ using a mixture of two monoclonal anti-ISAV HE antibodies as a primary antibody and Alexa405® goat anti-mouse IgG as a secondary antibody. The negative controls included the unrelated His6-tagged protein drNimA, rec empty-1 and recHE4-coupled beads stained with secondary antibody (sec. ab. ctrl.) only.
A mixture of two mouse anti-ISAV HE monoclonal antibodies recognised all three recHEs bound by their C-terminal His6-tag to magnetic cobalt beads. No staining was observed with beads coupled with drNimA, rec empty-1 or when recHE4-bound beads were incubated with secondary antibody only (Fig. 3B). This indicates that the beads are coated with recHEs. The binding of recHE proteins to the beads was also validated by elution of the proteins using imidazole, followed by quantitative analysis of the eluates on a Coomassie blue-stained SDS gel. The recHE protein band intensities (each in three parallels) were only slightly, but not significant, different from each other as calculated by single factor ANOVA analysis (data not shown). Also, binding of the His6-tagged protein drNimA to the beads was verified by analyses of the eluate on both Coomassie blue-stained SDS gel and by Western blot using an anti-His-antibody (data not shown), indicating the specificity of the assay.
3.5. Hemagglutination by recHEs
The ability of the three recHEs to agglutinate Atlantic salmon- and rabbit red blood cells (AsRBCs and rRBCs), and the effect of a serine esterase inhibitor on hemagglutination were investigated using proteins immobilized to magnetic cobalt beads. Large aggregates of bead-bound recHE4S32A and AsRBCs were formed after 1–3 h, while smaller aggregates were observed with beads coupled with recHE4 and recHE7. The degree of hemagglutination was indistinguishable for the latter two proteins, and no hemagglutination was observed with any of the negative controls (Fig. 4A). During the incubation period (1–3 h), aggregates formed by recHE4- and recHE7-coated beads got smaller as opposed to the recHE4S32A sample (not shown). Following incubation for 5–7 h, the aggregates of AsRBCs generated by recHE4 and recHE7 disappeared more or less completely, while hemagglutination by recHE4S32A was practically unchanged (Fig. 4A), indicating a continuous elution facilitated by the RDE. To test this possibility hemagglutination was studied in the presence of a specific esterase inhibitor (DCIC). By adding DCIC a stronger agglutination was observed after 1–3 h incubation, in particular with the recHE4- and recHE7-coupled beads (Fig. 4A). Although smaller aggregates were generated following 5–7 h incubation with recHE4 or recHE7 and the inhibitor, agglutination to AsRBCs was still apparent for both proteins. For recHE4S32A, the agglutination pattern with DCIC was indistinguishable from that without the inhibitor (Fig. 4A). This suggests that elution of aggregates indeed is the result of the esterase activity, although the inhibition may not have been 100% efficient. The results also suggest that Ser32 is a key residue in the elution process.
Fig. 4.
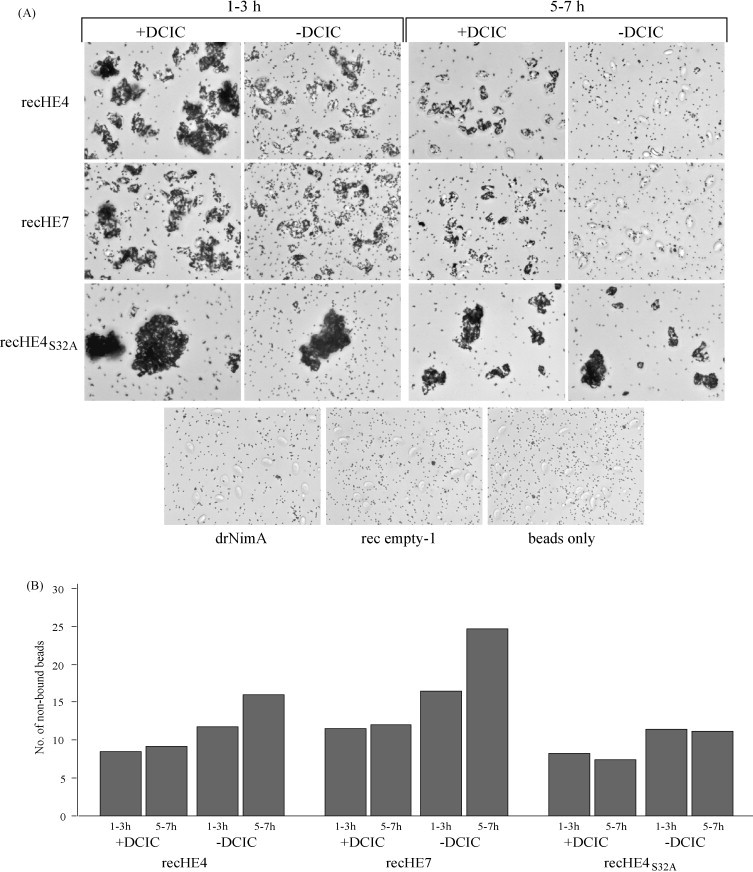
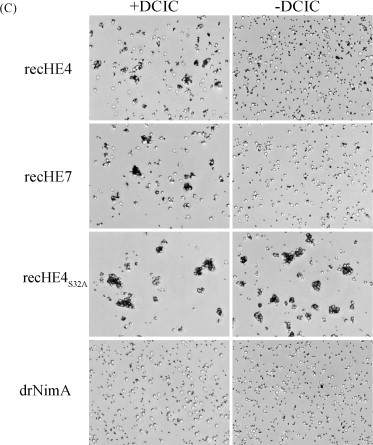
Agglutination of red blood cells (RBCs) with recHEs. RecHE4, recHE7 and recHE4S32A coupled to Dynabeads®TALON™ were pre-treated with DCIC in DMSO, or DMSO only, and mixed with RBCs. (A) The mixtures of Atlantic salmon RBCs (AsRBCs) and beads were observed under the microscope between 1–3 h and 5–7 h after adding AsRBCs. Negative controls included beads coupled with the His6-tagged protein drNimA, rec empty-1 and beads only. (B) Aliquots from the AsRBC-bead mixtures in (A) were taken out between 1–3 h and 5–7 h after addition of AsRBCs, and free beads (not associated with AsRBCs) were counted using a Bürker chamber. These experiments were repeated three times, and data from one representative experiment is presented. (C) The mixtures of rabbit RBCs (rRBCs) and beads were observed under the microscope 1 h after adding of rRBCs. Beads coupled with the His6-tagged protein drNimA served as a negative control. All photographs presented were taken at 40× magnification.
To quantify the hemagglutination level the number of non-aggregated beads was counted in the presence or absence of DCIC. The data from these assays provide quantitative support for the observations described above. For the recHE4 and recHE7 samples, in absence of DCIC, the number of free beads was higher following 5–7 h incubation, as compared to 1–3 h (Fig. 4B). In the presence of DCIC the amounts of free beads were similar after 1–3 h and 5–7 h incubation (Fig. 4B), although the aggregates were smaller after the prolonged incubation (Fig. 4A). It was not possible to identify any differences between recHE4 and recHE7 with respect to the level of hemagglutination. For recHE4S32A, the numbers of free beads were unaffected by both incubation time and presence of DCIC (Fig. 4B). For the negative controls, drNimA, rec empty-1 and beads only, the numbers of free beads (50–55) were unchanged regardless of incubation time or presence of DCIC (not shown). Proteins from two separate batches were tested and similar results obtained, supporting the reliability of the data. Together with the visual observations (Fig. 4A), these results suggest that all three recHEs have an intact receptor-binding domain. Substitution of Ser32 abolished elution of recHE from erythrocytes indicating the importance of this amino acid for the RDE and elution, but not for receptor binding. The three amino acid differences between recHE4 and recHE7 have no or minimal effect on hemagglutination, suggesting that they do not have a major impact on receptor binding.
To determine whether the recHEs have the ability also to agglutinate RBCs from a species other than Atlantic salmon, rRBCs were tested as above. Following 0.5 or 1 h incubation only a limited number of beads coupled with recHE4 and recHE7 were associated to rRBCs (Fig. 4C). A shorter incubation time did not change this result, while incubation more than 1 h abolished agglutination completely (not shown). RecHE4S32A-coated beads, on the other hand, clearly aggregated rRBCs. While pre-incubating with DCIC did not change the hemagglutination activity of the recHE4S32A, the presence of DCIC resulted in formation of small aggregates using the recHE4 and recHE7 samples (Fig. 4C). Agglutination of rRBCs was not observed with any of the negative controls (only drNimA shown in Fig. 4C). This is consistent with the results obtained with AsRBC, but the level of hemagglutination induced by the three recHEs was generally weaker for rRBC as compared to AsRBCs (Fig. 4A and C). This indicates that the recHEs use a different receptor determinant and/or substrate for the RDE on rRBCs, or that the receptor density on rRBCs is low relative to AsRBCs.
3.6. Acetylesterase activities of the recHEs
To compare the acetylesterase activities of the three recHEs, both bead-bound and soluble form of the proteins were analyzed. Although data from single experiments are presented, repeating the experiments produced similar results. The recHE4 and recHE7 proteins exhibit similar acetylesterase activities with all substrate concentrations tested, indicating a similar affinity for the synthetic substrates 4MUAc and pNPA (Fig. 5A and C). In contrast, recHE4S32A showed no acetylesterase activity, comparable to that of the negative control (Fig. 5A and C). The rec empty-1 and beads (not shown) gave a similar result as the drNimA sample.
Fig. 5.
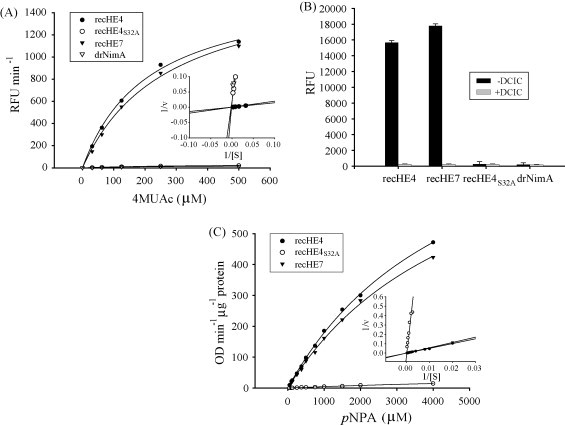
Acetylesterase activities of recHEs. (A) The acetylesterase activities of recHEs coupled to Dynabeads®TALON™ were determined using increasing concentrations of the substrate 4MUAc ([S]). The data (each data point is the average of at least three parallels) are presented by Michaelis–Menten and Lineweaver–Burk plots. The experiment was repeated twice, but only one is shown. Beads coupled to the His6-tagged protein drNimA served as negative control. RFU: relative fluorescence units; 1/v: reciprocal values of RFU min−1. (B) The influence of DCIC on acetylesterase activities of the recHEs coupled to Dynabeads®TALON™ using 4MUAc was studied. End-point data following 40 min incubation with substrate are shown. Beads coupled with the His6-tagged protein drNimA served as negative control. RFU: relative fluorescence units. (C) The acetylesterase activities of recHEs in solution were determined using increasing concentrations of the substrate pNPA ([S]). Rec empty-1 and drNimA served as negative controls (not shown). The data (each data point is the average of at least three parallels) are presented by Michaelis–Menten and Lineweaver–Burk plots. The experiment was repeated twice, but only one is shown. 1/v: reciprocal values of OD min−1 μg−1 protein.
The effect of the serine esterase inhibitor DCIC was tested using 4MUAc as a substrate. Pre-treatment of bead-coupled recHE4 and recHE7 with DCIC abolished acetylesterase activity completely, while recHE4S32A as well as drNimA remained unaffected (Fig. 5B). These results indicate that Ser32 is vital for the acetylesterase activity and that the three amino acid differences between recHE4 and recHE7 do not have any effect on the affinities of the enzymes to the used substrates.
4. Discussion
The HE is one of the two major ISAV surface glycoproteins, and is responsible for both viral attachment and release (Falk et al., 2004, Krossøy et al., 2001, Rimstad et al., 2001). Several reports indicate an important role of this protein in virulence. In this study, a structural model of the ISAV HE molecule was constructed, using the PToV HE protein as a template, and exploited to study the structure and functions of the HE protein. The model is consistent with and supports the hypothesis that ISAV HE is a structural homologue of torovirus and coronavirus HEs as well as InfC HEF, including the two characteristic active site sequence patterns in the putative enzyme domain, separated by the putative receptor domain. The GDSRsD subsequence includes the active site Ser32 (S) residue, and a DslHG motif holds the Asp261 (D) and His264 (H) active site residues (lower case residue codes indicate less conserved positions) (Fig. 1). Together this constitutes a Ser-His-Asp active site triad similar to the one found in traditional esterases, although the topology of this fold is atypical compared to more traditional lipases and esterases (Mølgaard et al., 2000). In particular the important strand–turn–helix motif for the active site Ser (the nucleophilic elbow motif) is less well defined in esterases that are similar to InfC HEF (Mølgaard et al., 2000). Still, the fact that these motifs are conserved is consistent with the experimental results showing that the ISAV contains an acetylesterase with a serine in the active site (Falk et al., 2004, Kristiansen et al., 2002).
It is however important to realize that significant fold recognition does not necessarily guarantee an optimal sequence alignment. In particular the ISAV HE receptor domain seems to be less similar to the template sequences than the enzyme domain, and the receptor domain is also approximately 40 residues shorter than the corresponding template domains. Virus sequences show a high degree of reassortment (Lai, 1992, Brown, 2000), meaning that alternative domains (and potentially alternative folds) can be combined and recombined from different sources. The weak sequence similarity may reflect the fact that for example InfC HEF specifically deacetylates 5-N-acetyl-9-O-acetylsialic acid residues, which is a high-affinity receptor determinant for binding of influenza C virus to erythrocytes, while the enzyme substrate for ISAV HE is 5-N-acetyl-4-O-acetylsialic acid (Hellebø et al., 2004, Herrler et al., 1985, Rogers et al., 1986). The cellular receptor for ISAV has not yet been defined but solid phase binding assays have shown that HE specifically binds 4-O-acetylated sialic acids, and 5-N-acetyl-4-O-acetylsialic acids probably represent a major receptor determinant for the virus (Hellebø et al., 2004). In the elucidation of the receptor domain structure it was assumed that the general fold of this domain is conserved, based on similarity in secondary structure content between ISAV HE and the other sequences (not shown). The alignment in Fig. 1 and the visualization in Fig. 2A does not necessarily represent an optimal solution, which is indicated, e.g. by the pattern of Cys–Cys bridges. These bridges are not completely conserved in the known structures (shown in Fig. 1), indicating a certain flexibility. This is consistent with previous studies showing that disulphide bonds are only moderately conserved between distant homologues (Thangudu et al., 2008). However, the current alignment leads to some unmatched Cys pairs, indicating that some shifts in alignment and probably also in tertiary structure may be needed for an optimal model, in particular in the receptor domain. The very low sequence similarity in this region makes it difficult to judge the relative merit of alternative alignments, and in general the current alignment seems reasonable. In addition to the conserved active site residues also the oxyanion hole residues (Gly59 and Asn89), involved in stabilizing the transition state, seem to be conserved. The majority of the receptor domain ligand binding residues identified from the experimental structures are found in regions with alignment between ISAV HE and the template structures. There may be an important difference in esterase ligand binding, where a loop with an Arg implied in substrate coordination seems to be missing in ISAV HE (indicated in Fig. 2B). It is known that alternative sequence positions may be involved in substrate coordination, e.g. in BToV HE (Langereis et al., 2009), but no clear alternative candidate has been found in ISAV HE. This may indicate important differences in substrate binding in ISAV HE, possibly consistent with different substrate specificity.
To test the features visualized by the model, ISAV HE variants were analyzed with respect to functions. The HE proteins from the isolates ISAV4 (recHE4) and ISAV7 (recHE7), in addition to a HE4 variant containing a single Ser to Ala substitution at position 32 (recHE4S32A) were therefore expressed, secreted and purified. ISAV4 and ISAV7 represents a high (ISAV4) and a low (ISAV7) virulent strain (Mjaaland et al., 2005, Mjaaland et al., 2002). The HE4 and HE7 proteins differ at three amino acid positions only, and share an identical HE-HPR (Mjaaland et al., 2002, Markussen et al., 2008). Therefore, comparing recHE4 and recHE7 is appropriate to test the influence of the three amino acid differences on the HE's function and hence their role in virulence, while analysis of the recHE4S32A focuses on the role of the Ser32 on the protein's acetylesterase activity.
Functional analyses can only be performed on correctly folded proteins, especially analyses of domains that have a function depending on the tertiary structure. The hemagglutination and acetylesterase activities (Fig. 4, Fig. 5) of the recHE proteins suggest that they adopt a conformation akin to the viral protein (Falk et al., 2004, Falk et al., 1997, Kristiansen et al., 2002). Coupled onto magnetic beads through their C-terminal His6-tag, recHE4 and recHE7 both displayed receptor-binding abilities as observed by agglutination of AsRBCs. The presence of an intact RDE activity for these two proteins was indicated by the observed elution of recHE-coupled beads from AsRBCs at later time points, with further evidence provided by the absence of elution in the presence of the serine esterase inhibitor DCIC, which attacks a serine in the enzyme's active site. Such elution caused by the RDE activity is not observed in agglutination assays using AsRBCs and purified ISAV4 particles, even after a period of 6–24 h incubation (Falk et al., 2004, Falk et al., 1997). The apparent contradiction may be explained by conformational differences between the native HE incorporated into the viral membrane versus the recombinant and bead-bound HE. In fact, Orlova et al. (2003) showed for horseradish peroxidase that differences in enzymatic properties are due to conformational variations between the recombinant and the native horseradish peroxidase. Also, the oligomerization status may vary between the native and the recombinant protein and can thus affect both structure as well as functional properties.
Our results clearly show that recHE4 elutes from AsRBCs, as does recHE7. This function may vary between different isolates, but apparently does not do so between the recHEs from these two isolates, as measured by the present analyses. Thus, the three amino acid substitutions (Fig. 2C), two of which are located in the putative receptor-binding domain (V143I and K185T), and one located in the proposed esterase domain, but close to the receptor-binding domain (T96N), do apparently not affect the ability to agglutinate AsRBCs. This seems consistent with the model, where none of these amino acid differences are located in the actual ligand binding site. Only V143I is close to this region, but still outside of the most likely binding site. However, it cannot be excluded that the ligand binding site is located elsewhere in the structure.
In the present study, the slight reduction in agglutination observed at later time points in presence of DCIC, may be due to either degradation of protein and/or of the AsRBCs, or that the inhibition is not 100% efficient. In any case it is clear that the esterase activity is the mechanism behind the elution. The mutant protein recHE4S32A has a stronger ability to agglutinate AsRBCs as compared to both recHE4 and recHE7. The mutated protein has no detectable RDE/acetylesterase activity as shown by lack of elution and enzyme activity in the esterase assay. Hemagglutination is thus a function related to the RDE activity which may serve to regulate the level of receptor binding by the virus. Moreover the mutant protein was not affected by DCIC. This provides functional evidence for the requirement of Ser32 in HE function. As suggested by the model (Fig. 2A and B) the residue is part of a catalytic triad, Ser32-His264-Asp261, constituting the enzyme's active site, which is also consistent with the assumptions made by Falk and co-workers (Falk et al., 2004). However, the roles of His264 and Asp261 remain to be proven. Of course, the lack of esterase activity displayed by the recHE4S32A mutant may be a consequence of an improperly folded enzyme domain. However, the residues in the catalytic triad are in their modelled positions all located to loops, and the Ser to Ala mutation will thus probably not affect the framework structure.
Falk and co-workers (Falk et al., 2004) have shown that ISAV particles can agglutinate RBCs from several different species, including those from rabbit (rRBCs), and subsequently elute from them within 6 h. Here, it was found that rRBCs were associated to beads coupled with recHE4 and recHE7, but considerable agglutination occurred only in the presence of DCIC. This may reflect a difference in receptor density, in affinity to the receptor of the two RBC types by the two recombinant proteins, and/or a stronger RDE activity towards the receptor determinant on rRBCs. RecHE4S32A agglutination of rRBCs was both obvious and independent of DCIC, as observed with AsRBCs, suggesting that the esterase activity regulates the binding to the rRBC receptor as well.
The acetylesterase assays (Fig. 5) clearly show an enzyme activity expressed by recHE4 and recHE7. The mutated protein has no activity with either substrate, verifying again the importance of Ser32 in the esterase activity of recHEs, and indicates that the enzyme activity is the cause for elution of the recHEs from RBCs. The affinities of bead-bound recHE4 and recHE7 to both synthetic substrates were similar (Fig. 5A and C). Also, using a natural substrate for the acetylesterase, as represented by the surface molecules on erythrocytes, no difference in elution could be observed between the two proteins. Thus, the three amino acid substitutions (Fig. 2C) probably do not affect the RDE activity.
Taken together, no differences in receptor-binding or RDE activities were observed between recHE4 and recHE7. Hence, the differences in virulence between ISAV4 and ISAV7 are most likely not linked to variations in these two activities. However, Aspehaug and co-workers have shown that co-expression of HE and the fusion protein increased cell–cell fusion (Aspehaug et al., 2005), opening up for the possibility that the three amino acid differences between HE4 and HE7 may affect the fusion process and in this way contribute to the differences in virulence between ISAV4 and ISAV7. In addition, the possibility that that the ISAV F protein may be involved in receptor binding similar to what has been reported for the paramyxovirus respiratory syncytial virus (RSV), should not be excluded (Karron et al., 1997, Karger et al., 2001, Techaarpornkul et al., 2001). Moreover, sequence differences in the remaining gene segments of these virus isolates may also account for the virulence differences (Markussen et al., 2008).
In conclusion, a structural model of ISAV HE, visualizing putative receptor-binding and receptor-destroying domains, has been developed. The two functions were studied using purified secreted forms of recombinant HEs from two different virus isolates. The differences in virulence displayed by these two isolates could not be linked to the functional activities of recombinant HEs. Moreover, using information from sequence analyses and related structures, a HE variant containing a single amino acid mutation (Ser32 to Ala32) at the putative catalytic site of the enzyme was generated. This mutated protein displayed no enzymatic activity but intact receptor-binding ability. This provides experimental evidence for the importance of Ser32 in enzyme activity.
Acknowledgments
This work was supported by The Research Council of Norway (project no. 151939/150). Part of the bioinformatics analysis was performed with the FUGE Bioinformatics platform funded by the Functional Genomics (FUGE) programme of the Research Council of Norway.
Contributor Information
Anita Müller, Email: anita.mueller@nfh.uit.no.
Turhan Markussen, Email: turhan.markussen@vetinst.no.
Finn Drabløs, Email: finn.drablos@ntnu.no.
Tor Gjøen, Email: tor.gjoen@farmasi.uio.no.
Trond Ø. Jørgensen, Email: trondj@nfh.uit.no.
Stein Tore Solem, Email: stein.tore.solem@biotec.no.
Siri Mjaaland, Email: siri.mjaaland@fhi.no.
References
- Anonymous Epizootiological investigation into a case of suspicion of infectious salmon anaemia (ISA) in Scotland in November 2004. Report by FRS Marine Laboratory; Aberdeen; 2005. [Google Scholar]
- Aspehaug V., Mikalsen A.B., Snow M., Biering E., Villoing S. Characterization of the infectious salmon anemia virus fusion protein. J. Virol. 2005;79(19):12544–12553. doi: 10.1128/JVI.79.19.12544-12553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigent S.J., McCauley J.W. Glycosylation of haemagglutinin and stalk-length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus Res. 2001;79(1–2):177–185. doi: 10.1016/s0168-1702(01)00272-6. [DOI] [PubMed] [Google Scholar]
- Barton G.J. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6(1):37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brossmer R., Isecke R., Herrler G. A sialic acid analogue acting as a receptor determinant for binding but not for infection by influenza C virus. FEBS Lett. 1993;323(1–2):96–98. doi: 10.1016/0014-5793(93)81456-a. [DOI] [PubMed] [Google Scholar]
- Brown E.G. Influenza virus genetics. Biomed. Pharmacother. 2000;54(4):196–209. doi: 10.1016/S0753-3322(00)89026-5. [DOI] [PubMed] [Google Scholar]
- Bujnicki J.M., Elofsson A., Fischer D., Rychlewski L. Structure prediction meta server. Bioinformatics. 2001;17(8):750–751. doi: 10.1093/bioinformatics/17.8.750. [DOI] [PubMed] [Google Scholar]
- Chen H., Bright R.A., Subbarao K., Smith C., Cox N.J., Katz J.M., Matsuoka Y. Polygenic virulence factors involved in pathogenesis of 1997 Hong Kong H5N1 influenza viruses in mice. Virus Res. 2007;128(1–2):159–163. doi: 10.1016/j.virusres.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Clouthier S.C., Rector T., Brown N.E., Anderson E.D. Genomic organization of infectious salmon anaemia virus. J. Gen. Virol. 2002;83(Pt 2):421–428. doi: 10.1099/0022-1317-83-2-421. [DOI] [PubMed] [Google Scholar]
- Colman P.M., Varghese J.N., Laver W.G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303(5912):41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- Cook-Versloot M., Griffith S., Cusack R., McGeachy S., Richie R. Identification and characterization of infectious salmon anaemia virus (ISAV) haemagglutinin gene highly polymorphic region (HPR) type 0 in North America. Bull. Eur. Assoc. Fish Pathol. 2004;24(4):203–208. [Google Scholar]
- Cunningham C.O., Gregory A., Black J., Simpson I., Raynard R.S. A novel variant of the infectious salmon anemia virus (ISAV) haemagglutinin gene suggests mechanisms for virus diversity. Bull. Eur. Assoc. Fish Pathol. 2002;22(6):366–374. [Google Scholar]
- Falk K., Aspehaug V., Vlasak R., Endresen C. Identification and characterization of viral structural proteins of infectious salmon anemia virus. J. Virol. 2004;78(6):3063–3071. doi: 10.1128/JVI.78.6.3063-3071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk K., Namork E., Dannevig B.H. Characterization and applications of a monoclonal antibody against infectious salmon anaemia virus. Dis. Aquat. Org. 1998;34(2):77–85. doi: 10.3354/dao034077. [DOI] [PubMed] [Google Scholar]
- Falk K., Namork E., Rimstad E., Mjaaland S., Dannevig B.H. Characterization of infectious salmon anemia virus, an orthomyxo-like virus isolated from Atlantic salmon (Salmo salar L.) J. Virol. 1997;71(12):9016–9023. doi: 10.1128/jvi.71.12.9016-9023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K., Elofsson A., Fischer D., Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19(8):1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Godoy M.G., Aedo A., Kibenge M.J., Groman D.B., Yason C.V., Grothusen H., Lisperguer A., Calbucura M., Avendano F., Imilan M., Jarpa M., Kibenge F.S. First detection, isolation and molecular characterization of infectious salmon anaemia virus associated with clinical disease in farmed Atlantic salmon (Salmo salar) in Chile. BMC Vet. Res. 2008;4(28):1–13. doi: 10.1186/1746-6148-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Gulati U., Wu W., Gulati S., Kumari K., Waner J.L., Air G.M. Mismatched hemagglutinin and neuraminidase specificities in recent human H3N2 influenza viruses. Virology. 2005;339(1):12–20. doi: 10.1016/j.virol.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Hellebø A., Vilas U., Falk K., Vlasak R. Infectious salmon anemia virus specifically binds to and hydrolyzes 4-O-acetylated sialic acids. J. Virol. 2004;78(6):3055–3062. doi: 10.1128/JVI.78.6.3055-3062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrler G., Durkop I., Becht H., Klenk H.D. The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J. Gen. Virol. 1988;69(Pt 4):839–846. doi: 10.1099/0022-1317-69-4-839. [DOI] [PubMed] [Google Scholar]
- Herrler G., Rott R., Klenk H.D., Müller H.P., Shukla A.K., Schauer R. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. EMBO J. 1985;4(6):1503–1506. doi: 10.1002/j.1460-2075.1985.tb03809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfling K., Brossmer R., Klenk H., Herrler G. Transfer of an esterase-resistant receptor analog to the surface of influenza C virions results in reduced infectivity due to aggregate formation. Virology. 1996;218(1):127–133. doi: 10.1006/viro.1996.0172. [DOI] [PubMed] [Google Scholar]
- Huang R.T., Dietsch E., Rott R. Further studies on the role of neuraminidase and the mechanism of low pH dependence in influenza virus-induced membrane fusion. J. Gen. Virol. 1985;66(Pt 2):295–301. doi: 10.1099/0022-1317-66-2-295. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Karger A., Schmidt U., Buchholz U.J. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J. Gen. Virol. 2001;82(Pt 3):631–640. doi: 10.1099/0022-1317-82-3-631. [DOI] [PubMed] [Google Scholar]
- Karron R.A., Buonagurio D.A., Georgiu A.F., Whitehead S.S., Adamus J.E., Clements-Mann M.L., Harris D.O., Randolph V.B., Udem S.A., Murphy B.R., Sidhu M.S. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. U.S.A. 1997;94(25):13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverin N.V., Gambaryan A.S., Bovin N.V., Rudneva I.A., Shilov A.A., Khodova O.M., Varich N.L., Sinitsin B.V., Makarova N.V., Kropotkina E.A. Postreassortment changes in influenza A virus hemagglutinin restoring HA-NA functional match. Virology. 1998;244(2):315–321. doi: 10.1006/viro.1998.9119. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Cox N.J., Haller O., Hongo S., Kaverin N., Klenk H.D., Lamb R.A., McCauley J., Palese P., Rimstad E., Webster R.G. Infectious salmon anaemia virus. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy—Eighth Report of the International Committee on Taxonomy Viruses. Elsevier Academic Press; New York: 2005. pp. 681–693. [Google Scholar]
- Kristiansen M., Frøystad M.K., Rishovd A.L., Gjøen T. Characterization of the receptor-destroying enzyme activity from infectious salmon anaemia virus. J. Gen. Virol. 2002;83(Pt 11):2693–2697. doi: 10.1099/0022-1317-83-11-2693. [DOI] [PubMed] [Google Scholar]
- Krossøy B., Devold M., Sanders L., Knappskog P.M., Aspehaug V., Falk K., Nylund A., Koumans S., Endresen C., Biering E. Cloning and identification of the infectious salmon anaemia virus haemagglutinin. J. Gen. Virol. 2001;82(Pt 7):1757–1765. doi: 10.1099/0022-1317-82-7-1757. [DOI] [PubMed] [Google Scholar]
- Kurowski M.A., Bujnicki J.M. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31(13):3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56(1):61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis M.A., Zeng Q., Gerwig G.J., Frey B., von Itzstein M., Kamerling J.P., de Groot R.J., Huizinga E.G. Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases. Proc. Natl. Acad. Sci. U.S.A. 2009;106(37):15897–15902. doi: 10.1073/pnas.0904266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiros H.K., Kozielski-Stuhrmann S., Kapp U., Terradot L., Leonard G.A., McSweeney S.M. Structural basis of 5-nitroimidazole antibiotic resistance: the crystal structure of NimA from Deinococcus radiodurans. J. Biol. Chem. 2004;279(53):55840–55849. doi: 10.1074/jbc.M408044200. [DOI] [PubMed] [Google Scholar]
- Liu C., Eichelberger M.C., Compans R.W., Air G.M. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol. 1995;69(2):1099–1106. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Zhou H., Ye D., Kemble G., Jin H. Improvement of influenza A/Fujian/411/02 (H3N2) virus growth in embryonated chicken eggs by balancing the hemagglutinin and neuraminidase activities, using reverse genetics. J. Virol. 2005;79(11):6763–6771. doi: 10.1128/JVI.79.11.6763-6771.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardones F.O., Perez A.M., Carpenter T.E. Epidemiologic investigation of the re-emergence of infectious salmon anemia virus in Chile. Dis. Aquat. Org. 2009;84(2):105–114. doi: 10.3354/dao02040. [DOI] [PubMed] [Google Scholar]
- Markussen T., Jonassen C.M., Numanovic S., Braaen S., Hjortaas M., Nilsen H., Mjaaland S. Evolutionary mechanisms involved in the virulence of infectious salmon anaemia virus (ISAV), a piscine orthomyxovirus. Virology. 2008;374(2):515–527. doi: 10.1016/j.virol.2008.01.019. [DOI] [PubMed] [Google Scholar]
- Matrosovich M.N., Matrosovich T.Y., Gray T., Roberts N.A., Klenk H.D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004;78(22):12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitnaul L.J., Matrosovich M.N., Castrucci M.R., Tuzikov A.B., Bovin N.V., Kobasa D., Kawaoka Y. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 2000;74(13):6015–6020. doi: 10.1128/jvi.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjaaland S., Hungnes O., Teig A., Dannevig B.H., Thorud K., Rimstad E. Polymorphism in the infectious salmon anemia virus hemagglutinin gene: importance and possible implications for evolution and ecology of infectious salmon anemia disease. Virology. 2002;304(2):379–391. doi: 10.1006/viro.2002.1658. [DOI] [PubMed] [Google Scholar]
- Mjaaland S., Markussen T., Sindre H., Kjøglum S., Dannevig B.H., Larsen S., Grimholt U. Susceptibility and immune responses following experimental infection of MHC compatible Atlantic salmon (Salmo salar L.) with different infectious salmon anaemia virus isolates. Arch. Virol. 2005;150(11):2195–2216. doi: 10.1007/s00705-005-0588-8. [DOI] [PubMed] [Google Scholar]
- Mjaaland S., Rimstad E., Falk K., Dannevig B.H. Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): an orthomyxo-like virus in a teleost. J. Virol. 1997;71(10):7681–7686. doi: 10.1128/jvi.71.10.7681-7686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscona A. Neuraminidase inhibitors for influenza. N. Engl. J. Med. 2005;353(13):1363–1373. doi: 10.1056/NEJMra050740. [DOI] [PubMed] [Google Scholar]
- Muchmore E.A., Varki A. Selective inactivation of influenza C esterase: a probe for detecting 9-O-acetylated sialic acids. Science. 1987;236(4806):1293–1295. doi: 10.1126/science.3589663. [DOI] [PubMed] [Google Scholar]
- Müller A., Solem S.T., Karlsen C.R., Jørgensen T.O. Heterologous expression and purification of the infectious salmon anemia virus hemagglutinin esterase. Protein Expr. Purif. 2008;62(2):206–215. doi: 10.1016/j.pep.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Mølgaard A., Kauppinen S., Larsen S. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure. 2000;8(4):373–383. doi: 10.1016/s0969-2126(00)00118-0. [DOI] [PubMed] [Google Scholar]
- Nylund A., Plarre H., Karlsen M., Fridell F., Ottem K.F., Bratland A., Saether P.A. Transmission of infectious salmon anaemia virus (ISAV) in farmed populations of Atlantic salmon (Salmo salar) Arch. Virol. 2007;152(1):151–179. doi: 10.1007/s00705-006-0825-9. [DOI] [PubMed] [Google Scholar]
- Ohuchi M., Feldmann A., Ohuchi R., Klenk H.D. Neuraminidase is essential for fowl plague virus hemagglutinin to show hemagglutinating activity. Virology. 1995;212(1):77–83. doi: 10.1006/viro.1995.1455. [DOI] [PubMed] [Google Scholar]
- Orlova M.A., Chubar T.A., Fechina V.A., Ignatenko O.V., Badun G.A., Ksenofontov A.L., Uporov I.V., Gazaryan I.G. Conformational differences between native and recombinant horseradish peroxidase revealed by tritium planigraphy. Biochem. Mosc. 2003;68(11):1225–1230. doi: 10.1023/b:biry.0000009137.75375.d2. [DOI] [PubMed] [Google Scholar]
- Palese P., Tobita K., Ueda M., Compans R.W. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61(2):397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- Pleschka S., Klenk H.D., Herrler G. The catalytic triad of the influenza C virus glycoprotein HEF esterase: characterization by site-directed mutagenesis and functional analysis. J. Gen. Virol. 1995;76(Pt 10):2529–2537. doi: 10.1099/0022-1317-76-10-2529. [DOI] [PubMed] [Google Scholar]
- Rimstad E., Mjaaland S., Snow M., Mikalsen A.B., Cunningham C.O. Characterization of the infectious salmon anemia virus genomic segment that encodes the putative hemagglutinin. J. Virol. 2001;75(11):5352–5356. doi: 10.1128/JVI.75.11.5352-5356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.N., Herrler G., Paulson J.C., Klenk H.D. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J. Biol. Chem. 1986;261(13):5947–5951. [PubMed] [Google Scholar]
- Rosenthal P.B., Zhang X., Formanowski F., Fitz W., Wong C.H., Meier-Ewert H., Skehel J.J., Wiley D.C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396(6706):92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlewski L., Jaroszewski L., Li W., Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9(2):232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E.W., Barrett T., Benson D.A., Bolton E., Bryant S.H., Canese K., Chetvernin V., Church D.M., Dicuccio M., Federhen S., Feolo M., Geer L.Y., Helmberg W., Kapustin Y., Landsman D., Lipman D.J., Lu Z., Madden T.L., Madej T., Maglott D.R., Marchler-Bauer A., Miller V., Mizrachi I., Ostell J., Panchenko A., Pruitt K.D., Schuler G.D., Sequeira E., Sherry S.T., Shumway M., Sirotkin K., Slotta D., Souvorov A., Starchenko G., Tatusova T.A., Wagner L., Wang Y., John Wilbur W., Yaschenko E., Ye J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38(Database issue):D5–16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtyrya Y., Mochalova L., Voznova G., Rudneva I., Shilov A., Kaverin N., Bovin N. Adjustment of receptor-binding and neuraminidase substrate specificities in avian-human reassortant influenza viruses. Glycoconj. J. 2009;26(1):99–109. doi: 10.1007/s10719-008-9169-x. [DOI] [PubMed] [Google Scholar]
- Strobl B., Vlasak R. The receptor-destroying enzyme of influenza C virus is required for entry into target cells. Virology. 1993;192(2):679–682. doi: 10.1006/viro.1993.1087. [DOI] [PubMed] [Google Scholar]
- Techaarpornkul S., Barretto N., Peeples M.E. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 2001;75(15):6825–6834. doi: 10.1128/JVI.75.15.6825-6834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangudu R.R., Manoharan M., Srinivasan N., Cadet F., Sowdhamini R., Offmann B. Analysis on conservation of disulphide bonds and their structural features in homologous protein domain families. BMC Struct. Biol. 2008;8:55. doi: 10.1186/1472-6807-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Itzstein M., Wu W.Y., Kok G.B., Pegg M.S., Dyason J.C., Jin B., Van P.T., Smythe M.L., White H.F., Oliver S.W., Colman P.M., Varghese J.N., Ryan D.M., Woods J.M., Bethell R.C., Hotham V.J., Cameron J.M., Penn C.R. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363(6428):418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- Wagner R., Matrosovich M., Klenk H.D. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 2002;12(3):159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- Wiley D.C., Skehel J.J. Crystallization and X-ray diffraction studies on the haemagglutinin glycoprotein from the membrane of influenza virus. J. Mol. Biol. 1977;112(2):343–347. doi: 10.1016/s0022-2836(77)80149-6. [DOI] [PubMed] [Google Scholar]
- Wiley D.C., Skehel J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Ye Y., Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl. 2):ii246–255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- Zeng Q., Langereis M.A., van Vliet A.L.W., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. U.S.A. 2008;105(26):9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]


