Abstract
RNA molecules are functionally diverse in part due to their extreme structural flexibility that allows rapid regulation by refolding. RNA folding could be a difficult process as often molecules adopt a spatial conformation that is very stable but not biologically functional, named a kinetic trap. RNA chaperones are non-specific RNA binding proteins that help RNA folding by resolving misfolded structures or preventing their formation. There is a large number of viruses whose genome is RNA that allows some evolutionary advantages, such as rapid genome mutation. On the other hand, regions of the viral RNA genomes can adopt different structural conformations, some of them lacking functional relevance and acting as misfolded intermediates. In fact, for an efficient replication, they often require RNA chaperone activities. There is a growing list of RNA chaperones encoded by viruses involved in different steps of the viral cycle. Also, cellular RNA chaperones have been involved in replication of RNA viruses. This review briefly describes RNA chaperone activities and is focused in the roles that viral or cellular nucleic acid chaperones have in RNA virus replication, particularly in those viruses that require discontinuous RNA synthesis.
Keywords: RNA viruses, Template switch
1. Introduction
Some RNA molecules easily become trapped in inactive conformations (kinetic traps) because of their structural and functional flexibility. These unproductive folds can be very stable and persistent. RNA chaperones existence was postulated to solve this RNA folding problem (Herschlag, 1995). Although RNA chaperone activity has been shown using different in vitro assays (see below), their in vivo activity was only recently demonstrated (Lorsch, 2002). RNA chaperones are non-specific nucleic acid binding proteins that rescue RNAs trapped in unproductive folding states (Cristofari and Darlix, 2002, Schroeder et al., 2004). One of the most important characteristics of RNA chaperones is that, once the RNA has been folded, they can be removed without alteration in the RNA conformation (Cristofari and Darlix, 2002, Rajkowitsch et al., 2005). RNA chaperone activity cannot be predicted based on the protein domain structure or the existence of discrete motifs. Nevertheless, RNA chaperones have the highest frequency of disordered regions, and it has been proposed that they act according to an entropy transfer model, allowing correct RNA folding by successive cycles of protein–substrate order–disorder (Fig. 1 ) (Dyson and Wright, 2005, Ivanyi-Nagy et al., 2005, Tompa and Csermely, 2004). In fact, viral proteins with large disordered regions, postulated as RNA chaperones based on this criteria, act as nucleic acid chaperones, at least in vitro (Ivanyi-Nagy et al., 2008, Zuñiga et al., 2007). The number of proteins with RNA chaperone activity is steadily growing [(Rajkowitsch et al., 2005), RCA website: http://www.projects.mfpl.ac.at/rnachaperones], but there is no consensus on the definition of RNA chaperone activity or the minimum assays required to establish the nucleic acid activity of a protein. Furthermore, the mode of action of RNA chaperones remains to be determined and a careful mechanistic comparison of several nucleic acid chaperones should be essential to achieve this objective (Rajkowitsch et al., 2005).
Fig. 1.
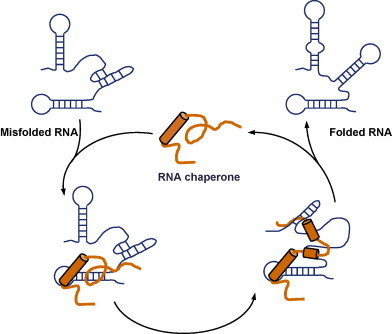
Model of RNA chaperone mechanism. Entropy transfer model of the action of RNA chaperones, adapted from Tompa and Csermely (2004). RNA chaperones (orange) have long intrinsically disordered regions. In the first step of the model, this kind of proteins bind misfolded RNAs, in general, non-specifically. Disordered domains of the protein become ordered while domains of the substrate are unfolded (entropy transfer from protein to substrate). The RNA is correctly folded while the protein is disordered again (entropy transfer from RNA substrate to protein). Finally, after several cycles of order–disorder, the RNA substrate, correctly folded, is released, and the RNA chaperone is free to act again.
2. RNA chaperone activities
A number of in vitro assays, with different technical difficulty, have been used through the years to analyze the RNA chaperone activity of a protein [for a review see Cristofari and Darlix, 2002, Rajkowitsch et al., 2005]. In vitro conditions differ from intracellular conditions, and very few in vivo RNA chaperone assays have been reported. This could be due, at least in part, to the pleiotropic effects that RNA chaperones can cause, and to the difficulty in analyzing their activity in the context of a whole cell.
2.1. In vitro activities
The in vitro assays to analyze the nucleic acid chaperone activity of a protein could be divided into: simple assays, with relatively low technical difficulty as they are focused on a single aspect of chaperone activity (i.e., annealing and strand transfer assays), and advanced assays, in which several chaperone activities are analyzed simultaneously (i.e., ribozyme or intron cleavage) (Cristofari and Darlix, 2002, Rajkowitsch and Schroeder, 2007, Rajkowitsch et al., 2005). Although there is no consensus, at least two different positive assays should be required to establish the RNA chaperone activity of a protein. In the simple assays, it should be at least desirable to show the absence of protein requirement after it has acted as nucleic acid chaperone, as this is the main difference between an RNA chaperone and an RNA binding protein that stabilizes a determined RNA folding while remaining bound. In some cases, biophysical technologies, such as fluorescence resonance energy transfer (FRET), NMR spectroscopy, fluorescence correlation spectroscopy (FCS), or single-molecule spectroscopy (SMS), have been applied to investigate the chaperone mechanism of action in in vitro assays providing precise information on the RNA chaperone mode of action (Égelé et al., 2005, Hong et al., 2003, Liu et al., 2007, Rajkowitsch and Schroeder, 2007, Ramalanjaona et al., 2007, Tisne et al., 2004, Zeng et al., 2007).
2.1.1. Nucleic acid annealing and strand displacement assays
The effect of proteins on RNA or DNA hybridization has been widely used to determine their nucleic acid chaperone activity (Cristofari et al., 2004, DeStefano and Titilope, 2006, Henriet et al., 2007, Huang et al., 2003, Ivanyi-Nagy et al., 2008, Pontius and Berg, 1992, Tsuchihashi and Brown, 1994, Vo et al., 2006, Zuñiga et al., 2007). It has been shown that RNA chaperones facilitate nucleic acid annealing. Nevertheless, annealing experiments must be carefully planned, as RNA annealing may occur in the absence of RNA chaperone activity. It is necessary to study activities including more than one step, and to perform the reactions in stringent conditions, such that only proteins with RNA chaperone activity could overcome all the requirements. To test activities that only RNA chaperones, but no other kind of proteins, could display, two strategies have been developed: (i) use of highly structured nucleic acid molecules, which requires a previous unwinding step by the RNA chaperone prior to the annealing, or (ii) protein elimination after base pairing has taken place (i.e., by treatment with proteinase K).
Nucleic acid chaperones also facilitate nucleic acid unwinding and strand displacement, and RNA chaperone activity could be clearly monitored within this kind of in vitro reactions (Cristofari et al., 2004, DeStefano and Titilope, 2006, Huang et al., 2003, Mir and Panganiban, 2006a, Mir and Panganiban, 2006b, Pontius and Berg, 1992, Tsuchihashi and Brown, 1994). Recently, an experimental approach consisting on detection of double-stranded molecules by FRET has been described. This technique includes a first annealing step and a second strand displacement reaction, mediated by the addition of an excess of non-labeled nucleic acid competitor. Therefore, this process combines real-time analysis of both reactions within the same experiment, measuring annealing and strand displacement before and after the addition of the competitor RNA, respectively (Rajkowitsch and Schroeder, 2007). These type of experiments should be more informative about RNA chaperone activity and could avoid the problems of simple assays that could lead to a misinterpretation of the results.
2.1.2. Ribozyme cleavage
A more complex assay is the study of the cleavage, in cis or in trans, of a ribozyme. Hammerhead ribozymes have been widely used to monitor RNA chaperone activity, as nucleic acid chaperones enhance ribozyme cleavage (Bertrand and Rossi, 1994, Daros and Flores, 2002, Herschlag et al., 1994, Ivanyi-Nagy et al., 2008, Zuñiga et al., 2007). This assay implies more restrictive conditions for chaperone activity, as it requires unwinding and annealing steps that must finally render an RNA conformation capable of self-cleavage.
2.1.3. Group I intron splicing
Group I intron splicing is a special case of splicing that may occur within the introns of some organisms, in the absence of proteins. It depends on the folding of the RNA in a defined three-dimensional structure, as these type of introns are able of self-splice. This reaction could be very inefficient and slow in vitro, unless an RNA chaperone is present. Both cis and trans splicing assays have been used to demonstrate RNA chaperone activity of several proteins, especially bacterial proteins (Belisova et al., 2005, Coetzee et al., 1994, Grohman et al., 2007, Mayer et al., 2002, Semrad et al., 2004, Zheng et al., 1995).
2.1.4. Template switch
This assay has been used to study the RNA chaperone activity of human immunodeficiency virus (HIV)-1 nucleocapsid (NC) protein (Guo et al., 1997, Guo et al., 2002, Heilman-Miller et al., 2004, Henriet et al., 2007, Hong et al., 2003, Wu et al., 2007) and, more recently, to analyze the RNA chaperone activity of TGEV N protein (S. Zuñiga, I. Sola, L. Enjuanes, unpublished results). The synthesis of different cDNA species is analyzed, including a cDNA generated by a switch from the donor RNA template to an acceptor RNA. This assay involves a second level of complexity, as unwinding of very stable secondary RNA structures, efficient RNase H activity, inhibition of self-priming, and annealing of separate RNAs are required. This type of assay is considered below in detail.
2.2. In vivo activities
Evaluation of nucleic acid chaperone activity in vivo, using a physiological system, almost requires a “customized” assay, depending on the RNA chaperone tested. Therefore, as previously indicated, just a few in vivo RNA chaperone activity assays have been reported to date. The first system described was based on an in vivo RNA folding trap existing in the bacteriophage T4 thymidylate synthase (td) group I intron (Clodi et al., 1999). Several RNA chaperones, overexpressed in Escherichia coli, are able to resolve the kinetic trap or impede the formation of misfolded structures, therefore promoting intron splicing. This system has been successfully used to test nucleic acid chaperone activity in vivo (Belisova et al., 2005, Clodi et al., 1999). Nevertheless, its physiological relevance remains uncertain (Lorsch, 2002) and, it also has a great limitation when overexpression of the RNA chaperone is toxic for E. coli.
Other reported assays, with more physiological relevance are based on: (i) transcription antitermination in vivo, for E. coli CspA-family nucleic acid chaperone proteins (Bae et al., 2000); (ii) Neurospora crassa group I intron splicing by CYT-19 protein, data that were also supported by the splicing phenotype of several mutants (Mohr et al., 2002); (iii) in vivo hepatitis delta ribozyme activity (Jeng et al., 1996); and (iv) in vivo reverse transcriptase template switching assay, using a MLV-based retroviral vector (Zhang et al., 2002).
An ideal assay for RNA chaperone activity is the in vivo demonstration of function associated with the natural RNA partner. Nevertheless, the analysis of RNA chaperone activity in vivo is quite difficult mainly because of the pleiotropic effects that RNA chaperones could have in the cell, and also because they act in different steps of a single biological process. All these reasons are in part responsible for the lack of appropriate in vivo systems for the analysis of RNA chaperone activity, and for a tendency to use complex in vitro systems with defined components.
3. Virus-encoded RNA chaperones and their roles in viral life cycle
The first viral RNA chaperone activities were reported in 1988, and were associated with the retrovirus nucleocapsid proteins. These activities have been extensively studied, and their nucleic acid chaperone activity was clearly established using different types of assays. Ten years have passed until another virus-encoded RNA chaperone was described, the HDV delta antigen. Then the list of virus-encoded RNA chaperones has been quickly growing, starting with the identification in 2004 of another viral RNA chaperone, the HCV core protein. Thus, in 2007, five new viral nucleic acid chaperones were described (Table 1 ). Nevertheless, the role of some of these proteins on viral life cycle, as RNA chaperones, is still unclear. This is mostly due to the difficulty in analyzing in an in vivo viral context the chaperone activity of these proteins.
Table 1.
RNA chaperones involved in virus life cycle
| Proteina | Virusb | Function | Reference |
|---|---|---|---|
| Virus encoded | |||
| NC | Retrovirus | Reverse transcription, template switch | Levin et al. (2005) |
| SdAg | HDV | Replication | Huang et al. (2003) |
| Core | Flaviviridae | Replication | Ivanyi-Nagy et al. (2008) |
| N | Coronavirus | RNA synthesis | Zuñiga et al. (2007) |
| N | Hantavirus | Replication | Mir and Panganiban (2006a) |
| 3AB | Poliovirus | Replication, recombination | DeStefano and Titilope (2006) |
| Vif | HIV-1 | Temporal regulation of reverse transcription | Henriet et al. (2007) |
| Tat | HIV-1 | Viral DNA transcription | Kuciak et al. (2008) |
| Host factors | |||
| PTB | Picornavirus, HCV, coronavirus, calicivirus | Translation, RNA synthesis | Anderson et al. (2007); Domitrovich et al. (2005); Karakasiliotis et al. (2006); Shi and Lai (2005) |
| hnRNP A1 | Coronavirus | RNA synthesis | Shi and Lai (2005) |
| La | HCV, poliovirus | Translation, replication | Domitrovich et al. (2005) |
| Unr | HRV-2 | Translation | Hunt et al. (1999) |
| PARBP33 | ASBVd | Replication | Daros and Flores (2002) |
NC, nucleocapsid protein; SdAg, small delta antigen; N, nucleocapsid protein; Vif, viral infectivity factor; Tat, transcription activation factor; PTB, polypyrimidine tract-binding protein; hnRNP A1, heterogeneous nuclear ribonucleoprotein A1; Unr, upstream of N-ras.
HDV, hepatitis delta virus; HIV-1, human immunodeficiency virus-1; HCV, hepatitis C virus; HRV-2, human rhinovirus-2; ASBVd, avocado sunblotch viroid.
Virus-encoded RNA chaperones have no common motif features (Fig. 2 ) and, sometimes, even RNA chaperone proteins of the same viral genus have little sequence similarity. Nevertheless, they fulfill the rule of having long disordered regions, with more than 50% of the protein being disordered in some cases. In fact, protein disorder has been the selected criterion to investigate the chaperone activity of several candidate viral RNA chaperone proteins.
Fig. 2.
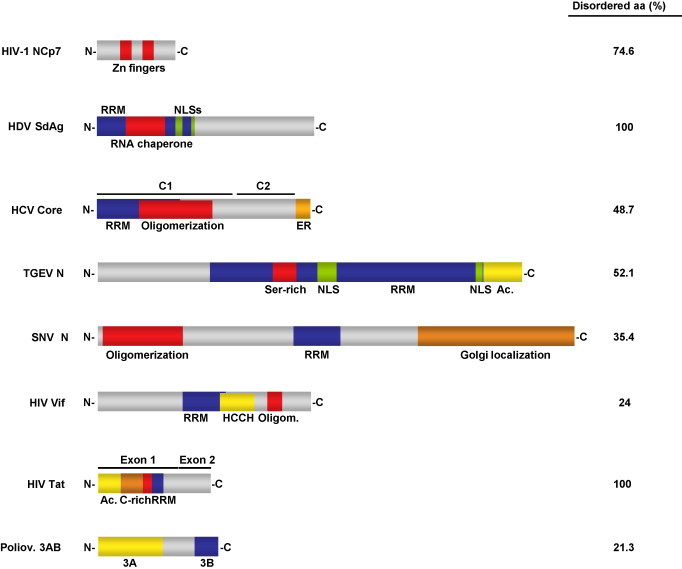
Virus-encoded RNA chaperones. The left part of the figure schematically represents domain organization of viral RNA chaperones described to date. Numbers in the right column show the percentage of disordered aminoacids in each case, calculated using DisProt Predictor VL3H (Obradovic et al., 2003, Peng et al., 2005). Sequences used and GeneBank accession numbers: human immunodeficiency virus (HIV)-1 nucleocapsid protein (NCp7) (AAB21888), hepatitis delta virus (HDV) small delta antigen (SdAg) (AAQ09794), hepatitis C virus (HCV) core protein (AAX11912), transmissible gastroenteritis virus (TGEV) N protein (AJ271965), hantavirus Sin Nombre (SNV) N protein (M14626), HIV-1 viral infectivity factor (Vif ) (AAC05239), HIV-1 transcriptional activator (Tat) (AAL01567) and human poliovirus 1 Mahoney 3AB polypeptide (P03300). RRM, RNA recognition or binding motif; NLS, nuclear localization signal; ER, endoplasmic reticulum retention signal; Ac., acidic domain; HCCH, HCCH motif; C-rich, cysteine rich domain.
3.1. Retrovirus-encoded RNA chaperones
Retroviruses are enveloped viruses infecting avian and mammalian species. Their genome is a dimer of two positive-sense single-stranded (ss) RNAs of around 10 kb, with a capped 5′-end and a poly-A at the 3′-end (Frankel and Young, 1998; ICTVdB-The Universal Virus Database, version 4; http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/). The viral genome can also be integrated into a host cell chromosome, and viral replication and transcription occurs in the host cell nucleus. Subsequent steps of the viral life cycle take place in the cell cytoplasm, i.e., viral protein translation and production of infectious viral particles. HIV-1 could be taken as a retrovirus model. Its genome encodes nine open reading frames (ORFs) (Fig. 3A). Three of these, Gag, Pol and Env polyproteins, are proteolyzed to generate mature structural proteins (from Gag and Env) and proteins with enzymatic functions (from Pol) such as protease, reverse transcriptase (RT) and integrase. The other six ORFs, encode accessory proteins, some of them found in the viral particle (Vif, Vpr and Nef) (Frankel and Young, 1998).
Fig. 3.
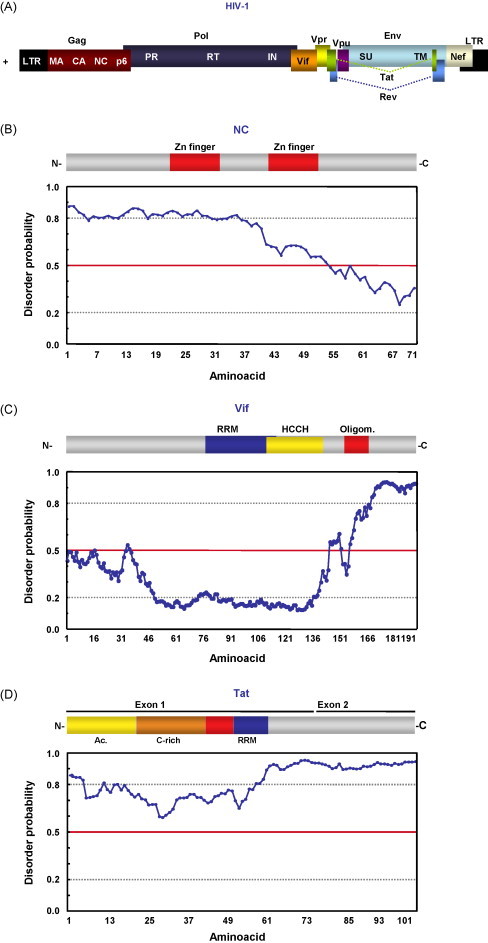
Retrovirus-encoded RNA chaperones. (A) Schematic representation of HIV-1 genome. Gag, Pol and Env polyproteins, and their processing products are represented. LTR, long terminal repeat; MA, matrix protein; CA, capsid protein; NC, nucleocapsid; PR, protease; RT, reverse transcriptase; IN, integrase; Vif, viral infectivity factor; Vpr, viral protein R; Vpu, viral protein U; Tat, transactivator protein; SU, surface glycoprotein gp120; TM, transmembrane protein gp41; Nef, negative replication factor. Domain organization and disorder prediction, calculated using DisProt Predictor VL3H (Obradovic et al., 2003, Peng et al., 2005), for HIV NC (B), Vif (C) and Tat (D) proteins. Aminoacids with a disorder score equal or above 0.5 are considered to be in a disordered environment, while a value below 0.5 is considered ordered. GeneBank accession numbers and acronyms are as in Fig. 2.
Avian and murine retrovirus NC proteins were the first virus-encoded RNA chaperones described (Prats et al., 1988). Later, the chaperone activity of HIV-1 NC was reported (Bertrand and Rossi, 1994, Tsuchihashi and Brown, 1994) and most of the current data on NC chaperone function came from HIV [for comprehensive reviews see Darlix et al., 2002, Levin et al., 2005]. The scenario has recently been complicated, as HIV-1 viral infectivity factor (Vif) and transcription activator (Tat) also have RNA chaperone activity (Henriet et al., 2007, Kuciak et al., 2008).
3.1.1. NC protein
The mature HIV-1 NC protein (usually named NCp7) is a short (55 aa), basic protein, produced by the proteolytic cleavage of the Gag precursor (Fig. 3B). It is found in the interior of the viral particle, bound to the genomic RNA. NC contains two conserved zinc-finger domains with the CCHC Zn binding motif. Both Zn fingers are required for virus replication, but mutations on the first CCHC motif had a greater impact on RNA chaperone activity and virus replication (Guo et al., 2002).
NC plays a role in almost all the steps of retrovirus replication cycle, from reverse transcription to RNA packaging (Levin et al., 2005). As an RNA chaperone, NC facilitates dimerization of genomic RNA (Darlix et al., 1990), initiation of the reverse transcription and transfer events during reverse transcription. The effect of NC in these processes will be discussed below in more detail. Also, NC improves the processivity of the RT by reducing polymerase pausing during transcription (Ji et al., 1996, Wu et al., 1996) and stimulates RNase H activity of the RT (Levin et al., 2005).
3.1.1.1. Initiation of reverse transcription
Reverse transcription is initiated by annealing of a cellular tRNA with the primer binding site (PBS), located in the 5′ untranslated region (UTR) of the viral genome (Fig. 4A). The tRNA, specific for each retrovirus, will act as primer for the reverse transcriptase. These tRNAs are selectively incorporated into the viral particle where they are already placed onto the viral RNA. NC protein is required for the formation of an initiation-competent complex by facilitating tRNA–PBS annealing and unwinding of local domains. It was shown that NC induces conformational changes in the tRNA structure, allowing annealing with the viral genome and also additional interactions between the RNA genome and the tRNA (Tisne et al., 2004).
Fig. 4.
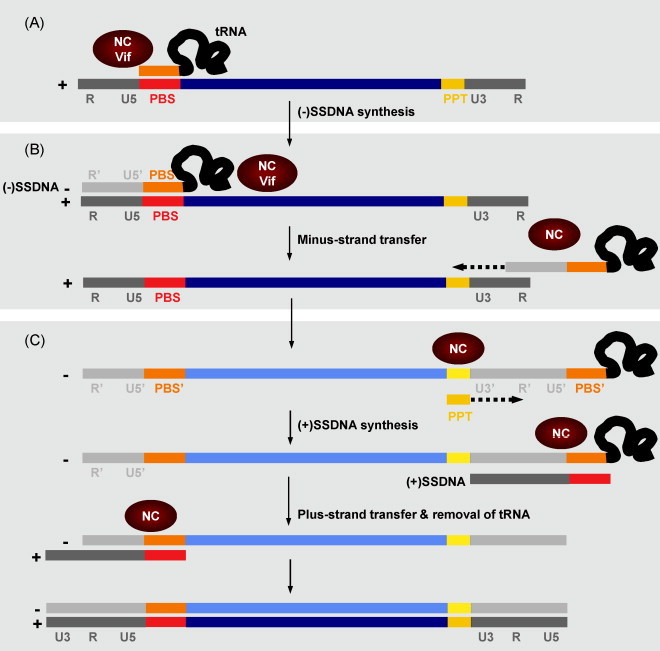
Retrovirus replication. Representation of the key steps in retrovirus replication. Negative polarity DNA synthesis is initiated using a partially unwound tRNA annealed to the primer-binding site (PBS, in red) at the 5′-end of the viral genomic RNA. The reverse transcriptase elongates the 3′-end of tRNA, synthesizing the (−) strong-stop DNA (SSDNA), complementary to U5 and R. RNase H activity of RT degrades R and U5 sequences in the retroviral genomic RNA. The first RT template switch occurs, and the minus strand DNA anneals with R sequences at the 3′-end of the genomic RNA. RT elongates the negative-strand DNA to synthesize a (−) DNA strand complementary to the viral genome. RNase H degrades all viral genomic RNA except the polypurine tract (PPT, in yellow) sequence. The RT begins the synthesis of positive strand DNA using PPT RNA as a primer. The RNA primer is degraded by RNase H, and the second RT template switch occurs, annealing the (+) DNA PBS (in red) with its complementary sequence on the (−) DNA strand. Finally, RT elongates both strands to complete synthesis of a double-stranded proviral DNA. Positive polarity RNA and DNA strands are represented in dark colors. Negative polarity DNA strands are shown in light colors. Long terminal repeat (LTR) sequences are shown in grey. Steps in which RNA chaperones have a role are indicated by the red ellipses.
3.1.1.2. Minus strand transfer
Following initiation of the reverse transcription, RT synthesizes the negative strong-stop DNA [(−) SSDNA], that must be translocated to the 3′-end of the viral genome (Fig. 4B). This step is guided by the annealing between the complementary repeat regions (R) in each case, that contain the highly structured trans-activation response (TAR) element sequence. This step is highly efficient in vivo, as (−) SSDNA is not accumulated in infected cells. NC promotes the minus strand transfer by facilitating the annealing of the complementary TAR sequences in the (−) SSDNA and the acceptor RNA (Liu et al., 2007, Vo et al., 2006, Zeng et al., 2007), inhibiting self-priming of (−) SSDNA (Guo et al., 1997, Hong et al., 2003), and promoting RNase H cleavage of the donor RNA (Levin et al., 2005). After strand transfer, (−) SSDNA is elongated by the RT (Fig. 4B). NC increases the elongation efficiency by reducing RT pausing at genomic RNA secondary structures (Ji et al., 1996, Wu et al., 1996).
3.1.1.3. Plus strand transfer
While synthesis of minus strand DNA occurs, RT initiates the plus strand DNA synthesis. Similarly to minus strand DNA synthesis, a short DNA product, termed positive strong-stop DNA [(+) SSDNA] is produced by elongation of a polypurine tract (PPT), used as a primer (Fig. 4C). The synthesis of (+) SSDNA is followed by a plus strand transfer event, required for the subsequent elongation of plus strand DNA. During this transfer process, the complementary PBS sequences, present in the minus strand DNA and (+) SSDNA, anneal to form a circular intermediate. As an RNA chaperone, NC facilitates plus strand transfer by contributing to tRNA primer removal from the minus strand DNA donor, and enhancing the PBS sequences annealing (Égelé et al., 2005, Ramalanjaona et al., 2007, Wu et al., 1999).
3.1.2. Vif protein
HIV-1 Vif is an accessory 192 aa protein (Fig. 3C), important for the efficient viral DNA synthesis in certain cell types, named “non-permissive” (Frankel and Young, 1998, Sakai et al., 1993, Sova and Volsky, 1993). Vif is required in non-permissive cells to counteract cellular inhibitors of viral infection, such as APOBEC3G and APOBEC3F (Opi et al., 2006, Wiegand et al., 2004). It has been recently shown that Vif has RNA chaperone activity, and that it is involved in the early steps of reverse transcription. Vif enhances tRNA primer–PBS annealing, decreases RT pausing and enhances SSDNA synthesis. Nevertheless, it has modest effects in NC-mediated strand transfer events that are different depending on the maturation product of NC used in the assays. It has been proposed that Vif could act as a negative temporal regulator preventing premature initiation of reverse transcription (Henriet et al., 2007).
3.1.3. Tat protein
HIV-1 Tat is a small nuclear regulatory protein that acts as a transcriptional activator (Fig. 3D). It is required for the transcription of integrated proviral DNA, driven by the 5′ long terminal repeat (LTR) sequence and the interaction of Tat with TAR element. Interaction of Tat with TAR recruits several transcription factors such as the TATA-binding protein, transcription factor TFIIB and positive transcription elongation factor B (P-TEFB). These interactions favor the hyperphosphorylation of the carboxy terminus domain (CTD) of the RNA polymerase II by the cyclin-dependent kinase 9 (CDK9) component of P-TEFB, allowing the elongation of the nascent RNA (Gatignol, 2007). Tat is a highly unstructured protein, and its RNA chaperone activity has been recently demonstrated (Kuciak et al., 2008). As an RNA chaperone, Tat enhances TAR DNA annealing, RNA–DNA exchange, ribozyme cleavage and RNA trans-splicing. It has been proposed that, as an RNA chaperone, Tat may act at the levels of proviral DNA transcription, viral RNA splicing and suppression of silencing (Kuciak et al., 2008).
3.2. Hepatitis delta virus delta antigen
Hepatitis delta virus (HDV) is an enveloped subviral satellite agent with hepatitis B virus (HBV) as its natural helper virus. Viral genome is a 1679 nt negative-sense, ssRNA with a circular rod-like conformation (Kos et al., 1986, Wang et al., 1986). The genome contains several ORFs, but only two proteins are produced from a single ORF: the small (195 aa) and large (214 aa) delta antigens. The large delta antigen isoform is generated later in the infection by post-transcriptional RNA editing, acts as an inhibitor of virus replication, and is required for viral assembly (Cornillez-Ty and Lazinski, 2003).
The small delta antigen (SdAg) is required for viral replication (Kuo et al., 1989). The RNA chaperone activity of SdAg was shown in different in vitro assays. This activity was proportional to the RNA binding activity of the protein, and was located in a domain from aa 24 to 59 (Huang et al., 2003, Huang and Wu, 1998, Wang et al., 2003).
During HDV replication, monomeric size RNA molecules must be generated. The ribozyme activity present in the viral RNA allows cis cleavage of both genome and antigenome RNAs. It was shown that, although delta antigen is not essential for the in vivo cleavage of HDV RNA, it enhances HDV ribozyme activity both in vitro and in vivo (Huang et al., 2003, Jeng et al., 1996).
3.3. Flaviviridae core proteins
Flaviviruses are enveloped viruses that infect vertebrates and are transmitted by an arthropod vector. Their genome is a positive-sense ssRNA close to 11 kb. The family comprises three genera: flavivirus, pestivirus and hepacivirus (Lindenbach and Rice, 2001). RNA chaperone activity was reported for hepacivirus (hepatitis C virus, HCV) core protein (Cristofari et al., 2004). Recently, based on disorder conservation in Flaviviridae core proteins, the RNA chaperone activity of core proteins from different members of each genera has been analyzed. It has been found that, although core proteins from different Flaviviridae genera have low sequence similarity, all of them have RNA chaperone activity (Ivanyi-Nagy et al., 2008). Furthermore, data strongly suggest that nucleic acid chaperone activity resides in disordered regions of Flaviviridae core proteins as their RNA chaperone activity is resistant to heat denaturation of the protein (Ivanyi-Nagy et al., 2008).
HCV has a 9.6 kb RNA genome, with a unique ORF encoding a long polyprotein (Fig. 5A). Structural and non-structural proteins are generated by co- and post-translational processing of the polyprotein. The 5′ and 3′ UTR regions contain RNA domains involved in viral replication. The 5′ UTR is folded with an IRES structure that promotes cap-independent translation. The 3′ UTR consists on a short variable region, a U/(UC) motif, and a X tail with highly conserved stem loop structures (Moradpour et al., 2007). The mature 178 aa core protein is produced by cleavage of the polyprotein and further signal peptide processing. Core protein could be divided in two different domains: C1, involved in RNA binding and oligomerization, and C2, involved in association with lipid droplets (Moradpour et al., 2007) (Fig. 5B).
Fig. 5.
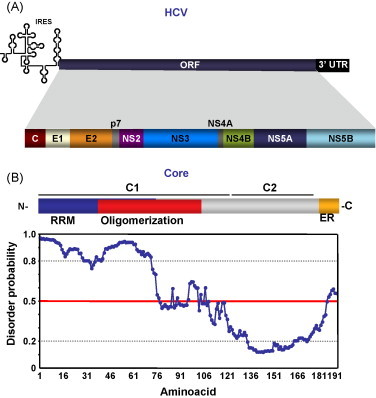
Hepacivirus-encoded RNA chaperone. (A) Representation of HCV genome. C, core protein; E1, envelope protein 1; E2, envelope protein 2; NS, non-structural protein. (B) Domain organization and disorder prediction plot for HCV core protein. Plot obtention and interpretation as in Fig. 3. GeneBank accession number and acronyms are as in Fig. 2.
HCV core protein has RNA chaperone activity that has been demonstrated using different approaches (Cristofari et al., 2004). It was also shown that, acting as a nucleic acid chaperone, core protein allows dimerization of the HCV RNA 3′ UTR (Cristofari et al., 2004, Ivanyi-Nagy et al., 2006). This HCV RNA region could adopt different conformations, with unknown role on viral life cycle. It has been suggested that HCV RNA 3′ UTR interconversion between different structures could act by regulating transitions between translation–replication and replication–packaging of the genomic RNA. In this context, the RNA chaperone activity of the core protein could regulate these riboswitches by facilitating interconversion of the different RNA structures (Ivanyi-Nagy et al., 2006).
3.4. Coronavirus nucleocapsid protein
Coronaviruses are enveloped viruses, of the Coronaviridae family, included in the Nidovirales order (Enjuanes et al., 2008, Gorbalenya et al., 2006). Their genomes are positive-sense, ssRNAs of around 30 kb, the largest known viral RNA genomes. The 5′ two-thirds of the genomic RNA encode the replicase genes (rep1a and rep1ab). The 3′ one-third of the genome encodes structural and non-structural proteins (Fig. 6A). Coronavirus replication and transcription occur in the cytoplasm of infected cells, and are based on RNA-dependent RNA synthesis. Transcription leads to a nested set of subgenomic (sg) mRNAs that are 5′- and 3′-coterminal with the viral genome. These sg mRNAs are generated by a discontinuous process (Fig. 7 ) that implies base pairing of nascent RNAs of negative polarity, driven by the transcription-regulating sequences (TRSs) preceeding each gene, and sequences located at the 3′-end of the leader within the genomic RNA (Sawicki and Sawicki, 2005, Sawicki et al., 2007, Sola et al., 2005, Zuñiga et al., 2004).
Fig. 6.
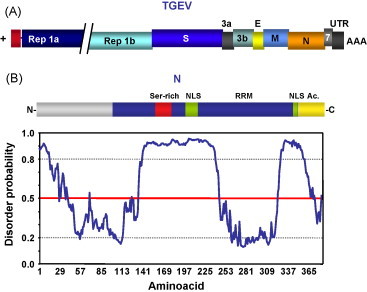
Coronavirus-encoded RNA chaperone. (A) Schematic representation of TGEV genome, as a model for coronavirus genomes. Rep, replicase polyproteins; S, spike protein; E, envelope protein; M, membrane protein; N, nucleocapsid protein. (B) Domain organization and disorder prediction for TGEV N protein. Plot obtention and interpretation as in Fig. 3. GeneBank accession number and acronyms are as in Fig. 2.
Fig. 7.
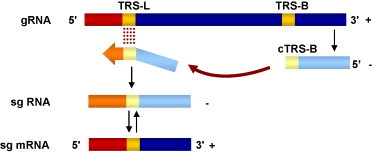
Coronavirus transcription. Representation of the coronavirus discontinuous transcription during negative-strand synthesis. Transcription regulating sequences from the leader (TRS-L) and preceding a gene (TRS-B) are shown in yellow. Negative polarity RNA synthesis begins from 3′-end of the coronavirus genome. Once the transcription complex copies the TRS-B template switch of the nascent RNA to the leader sequence (red) takes place and the sequence complementary to the TRS-B (cTRS-B) hybridizes with the TRS-L. This process is indicated by the dark red arrow. Nascent RNA synthesis continues with the copy of the leader sequence (red), leading to a negative polarity subgenomic RNA (sg RNA) that will be used as a template for the generation of the sg mRNAs of positive polarity. Genomic RNA and sg mRNA strands are shown in dark colors. Negative polarity RNA strands are shown in light colors.
It has been shown that the free energy of duplex formation between nascent RNA sequences complementary to the body (donor) TRS and the leader (acceptor) TRS is the main factor driving coronavirus discontinuous transcription (Sola et al., 2005, Zuñiga et al., 2004) (Fig. 7). Nevertheless, on top of this mechanism operating in the synthesis of all sg mRNAs, the transcription of specific genes, such as N gene, is regulated by a recently described enchancer mechanism (Moreno et al., 2008).
Coronavirus nucleocapsid (N) protein has a structural role and has also been involved in RNA synthesis (Almazan et al., 2004, Enjuanes et al., 2006). The N proteins from different coronaviruses are variable in length and primary sequence. Nevertheless, some motifs with functional relevance are conserved, and the proteins share a common domain organization. A three-domain structure has been proposed for N protein (Parker and Masters, 1990). Recently, based on disorder predictions, a modular organization including two structured domains separated by a long disordered region has been proposed for coronavirus N protein (Chang et al., 2006, Zuñiga et al., 2007) (Fig. 6B).
Using transmissible gastroenteritis virus (TGEV) as a model, RNA chaperone activity has been demonstrated for N protein (Zuñiga et al., 2007). RNA chaperone activity was also reported for severe and acute respiratory syndrome virus (SARS-CoV) (Zuñiga et al., 2007). Based on these results and on the high conservation of disorder patterns for different coronavirus N proteins, RNA chaperone was postulated as a general activity of all coronavirus nucleoproteins. In fact, it has been recently shown that the central disordered domain of TGEV N protein includes the nucleic acid chaperone activity of the protein (S. Zuñiga, I. Sola, L. Enjuanes, unpublished results).
The template switch involved during the discontinuous RNA synthesis in coronavirus transcription is a complex process that includes numerous steps (see below). To accomplish some of these steps we have proposed that RNA chaperones, such as the N protein, could decrease the energy barrier needed to dissociate the nascent minus RNA chain from the genomic RNA template, in order to perform a template switch leading to the hybridization with the TRS of the leader sequence during discontinuous transcription (Zuñiga et al., 2007). In fact, we have recently shown that TGEV N protein enhanced template switch in vitro using the HIV-derived system (S. Zuñiga, I. Sola, L. Enjuanes, unpublished results).
It is worth noting that the number of coronavirus-encoded RNA chaperones could grow with the addition of some non-structural proteins, such as SARS-CoV nsp3 (Neuman et al., 2008).
3.5. Hantavirus nucleocapsid protein
Hantaviruses are enveloped viruses included within the Bunyaviridae family. Their genome is composed of three segments of negative-sense, ssRNA (Khaiboullina et al., 2005). The large (L), medium (M) and small (S) segments, encode RNA dependent RNA polymerase, the glycoprotein precursor yielding surface glycoproteins G1 and G2, and the nucleocapsid (N) protein, respectively (Plyusnin et al., 1996). These RNA segments have complementary 5′ and 3′ termini that are base-paired, forming a panhandle structure leading to circular, supercoiled RNAs. Formation of the panhandle structure is important for bunyavirus replication, but the precise role in viral RNA synthesis is unclear (Barr and Wertz, 2005).
Hantavirus N proteins have around 433 aa, and interact with viral RNA, plus strand cRNA, and mRNAs (Mir and Panganiban, 2005, Plyusnin et al., 1996). Hantavirus N protein has been found to be involved in viral RNA packaging and virus replication (Mir and Panganiban, 2006b). The RNA chaperone activity of the hantavirus Sin Nombre (SNV) N protein was recently reported (Mir and Panganiban, 2006b). It has been proposed that N protein facilitates panhandle formation and mediates panhandle dissociation during replication initiation, acting as an RNA chaperone (Mir and Panganiban, 2006a, Mir and Panganiban, 2006b).
Hantavirus N protein is the first RNA chaperone described encoded by a negative-stranded RNA virus. Genomic RNA panhandle formation and dissociation during virus replication could be a common feature of other negative strand RNA viruses. Nevertheless, influenza A virus nucleocapsid protein has no RNA chaperone activity, at least in in vivo assays based on an RNA folding trap (Clodi et al., 1999).
3.6. Poliovirus 3AB protein
Polioviruses belong to the Picornaviridae family. Their genome is a positive-sense ssRNA of around 8 kb, encoding a single polypeptide. This polyprotein is processed leading, at least, to 10 different protein products (Agol, 2006). Processing intermediates of the polyprotein often have specific functions. RNA chaperone activity was recently reported for one of these products, the 3AB precursor, that has annealing and helix-destabilizing activities (DeStefano and Titilope, 2006). 3B protein, also named as VPg, is linked to the 5′-end of the viral genome and acts as primer for viral RNA synthesis. 3A protein was proposed as the protein that anchors the replication complexes to membranes. 3AB precursor stimulates RNA synthesis by the RNA-dependent RNA polymerase, and has RNA chaperone activity. Nevertheless, its role as RNA chaperone in the viral life cycle is unclear.
4. Cellular RNA chaperones involved in viral replication
A large collection of cellular factors has been found to be involved in virus replication (Table 1). Most of them were first identified using viral RNA–protein binding assays, followed by functional assays, i.e., silencing of cell protein expression and relevance on virus replication. Most of the cellular proteins identified using these approaches are common for unrelated viruses. RNA chaperone activity has been associated to several proteins identified using these strategies. Nevertheless, some of these proteins do not act as real RNA chaperones, as they need to remain bound to the RNA to achieve the expected functional activity. In fact, their relevance in viral replication acting as canonical RNA chaperones has been clearly demonstrated only in a reduced number of cases. Therefore, the exact role of these proteins, as RNA chaperones, on viral life cycle still needs to be elucidated. The activity of some of these proteins is described below.
4.1. Heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1)
hnRNP A1 is a cellular RNA binding protein involved in different aspects of RNA metabolism. It is predominantly nuclear, but also shuttles to the cytoplasm (Dreyfuss, 1986, Dreyfuss et al., 1993). hnRNP A1 is a well-characterized RNA chaperone (Herschlag et al., 1994, Pontius and Berg, 1992, Portman and Dreyfuss, 1994) that promotes RNA annealing and enhances ribozyme cleavage.
Mouse hepatitis coronavirus (MHV) RNAs and N protein bind hnRNP A1 (Zhang et al., 1999). It has been found that the binding of hnRNP A1 to viral TRSs correlates with the sg mRNA level transcribed from these TRSs (Zhang and Lai, 1995). hnRNP A1 has also been involved in 5′–3′-end cross-talk (Huang and Lai, 2001). All data reported indicated that hnRNP A1 plays a role in vivo in coronavirus RNA synthesis, as over-expression of the protein facilitates MHV replication and dominant-negative mutants of hnRNP A1 reduce virus replication (Shi and Lai, 2005). Nevertheless, its role, as nucleic acid chaperone, in viral replication has not been fully established. That was mostly due to the fact that other hnRNP proteins could substitute for hnRNP A1 in vivo (Shen and Masters, 2001, Shi and Lai, 2005), making it very difficult to evaluate the precise role of hnRNP A1 even in its cellular functions.
4.2. Polypyrimidine tract-binding protein (PTB)
PTB is an RNA binding protein involved in regulation of alternative splicing and translation of cellular RNAs. Similarly to hnRNP A1, it is mainly located in the nucleus, although it can shuttle to cell cytoplasm under different conditions (i.e., viral infections). There are at least three different PTB isoforms: PTB1, PTB2 and PTB3 (Gil et al., 1991), and differences in the activity of these proteins in cellular RNA splicing have been reported (Wollerton et al., 2001). Only PTB3, also known as hnRNP I (Ghetti et al., 1992), has a well-established RNA chaperone activity based on group I splicing assays (Belisova et al., 2005). Nevertheless, the RNA chaperone activity of other PTB isoforms is not clear. In fact, when analyzed, PTB1 did not enhance nucleic acids annealing and hammerhead ribozyme cleavage (Zuñiga et al., 2007).
PTBs are the most recurrent proteins reported to bind viral RNAs. It is difficult to identify the specific PTB isoform involved in each case, due to the small aa difference between them, and because in most cases, it has been identified by proteomic approaches that do not distinguish PTB isoforms. PTBs bind picornavirus IRES, both entero-rhinovirus group (including poliovirus and HRV-2) and cardio-aphthovirus (including encephalomyocarditis virus [ECMV] and foot-and-mouth disease virus [FMDV]) (Florez et al., 2005, Hunt et al., 1999, Song et al., 2005). PTBs also bind HCV UTR regions (Domitrovich et al., 2005), calicivirus RNA (Karakasiliotis et al., 2006) and coronavirus RNAs (Shi and Lai, 2005).
In viral life cycle, PTBs mainly act by facilitating translation. For picornavirus and HCV, it has been proposed that PTBs, acting as RNA chaperones, may help IRES folding into a translation-competent conformation (Anderson et al., 2007, Domitrovich et al., 2005, Gosert et al., 2000, Hunt and Jackson, 1999, Song et al., 2005). Nevertheless, in these cases, it has not been shown whether PTBs could be eliminated after the IRES has folded properly. Therefore, a key property of RNA chaperones has not been demonstrated.
The role of PTBs in viral RNA synthesis seems to depend on virus taxonomy. It was reported that PTBs modulate coronavirus RNA synthesis and also could facilitate 5′–3′-end interactions (Shi and Lai, 2005). A positive effect on feline calicivirus RNA synthesis was observed, but only at temperatures proposed to difficult functional folding of calicivirus RNA (Karakasiliotis et al., 2006). In HCV RNA synthesis the role of PTBs as RNA chaperones needs additional work. It has been shown that PTBs have an inhibitory effect on HCV RNA synthesis (Domitrovich et al., 2005) and it has been proposed that they block replication initiation while enhancing IRES-promoted translation, facilitating the replication–translation switch. At the same time, it has also been reported that PTBs are required for an efficient HCV RNA replication (Aizaki et al., 2006, Chang and Luo, 2006).
4.3. La autoantigen
La is a nuclear RNA-binding phosphoprotein that is associated with RNA polymerase III transcripts (Maraia and Intine, 2001, Pannone et al., 1998). La has RNA chaperone activity, as it facilitates group I intron splicing both in vitro and in vivo (Belisova et al., 2005). La interacts with the UTR regions of HCV and poliovirus, and enhances translation from the IRES of these viruses (Domitrovich et al., 2005, Meerovitch et al., 1993). In addition, La autoantigen is also required for efficient replication of HCV viral RNA (Domitrovich et al., 2005). Nevertheless, the relevance of La autoantigen RNA chaperone activity in these processes has not been clarified.
4.4. Upstream of N-ras (Unr)
Unr is a cytoplasmic RNA binding protein essential for transcriptional regulation and mRNA metabolism. Unr contains five cold-shock domains and, therefore, is predicted to act as RNA chaperone by similarity with prokaryotic cold-shock domain containing proteins (Rajkowitsch et al., 2007). Unr binds and stimulates translation from human rhinovirus-2 (HRV-2) IRES (Brown and Jackson, 2004, Hunt et al., 1999). It has been proposed that Unr could act as an RNA chaperone to allow folding of HRV-2 IRES into a translation-competent structure that could bind PTB and ribosome (Anderson et al., 2007), similarly to cellular Apaf-1 IRES (Mitchell et al., 2003).
4.5. PARBP33
PARBP33 is a chloroplast RNA binding protein identified by UV-crosslinking of avocado sunblotch viroid (ASBVd) infected leaves and purification of the RNA–protein complexes obtained. It has been reported that PARBP33 acts as an RNA chaperone in vitro, enhancing ASBVd hammerhead ribozyme cleavage (Daros and Flores, 2002). According to this report, PARBP33 should act in vivo facilitating ribozyme-mediated cleavage of oligomeric ASBVd RNA, a key step in the replication mechanism of this viroid.
5. RNA chaperones, template switch and transcription
Template switch is a complex process in which different steps could be differentiated: (i) slow down or stop of nucleic acid synthesis; (ii) template switch itself; (iii) reassociation of the nascent nucleic acid strand with the acceptor sequence; and (iv) elongation of the nascent strand using the acceptor nucleic acid as template (Enjuanes et al., 2007). Some aspects of these steps will be expanded below, specially focused on the two viral systems where template switch is an essential process of their life cycle: coronavirus discontinuous transcription and retrovirus reverse transcription.
5.1. Polymerase pausing
Two different slow-down or stop signals could be acting, both in coronavirus transcription and retrovirus reverse transcription. First, the termination of nascent strand synthesis might be mediated by secondary RNA structures located either in the template or in the nascent strand, as described for prokaryotic (Wilson and von Hippel, 1995) or eukaryotic (Reeder and Lang, 1997) RNA polymerases. Extensive studies have been made on HIV RT pausing and on secondary RNA structures influencing this process. It has been shown that the RT pausing could be promoted by a highly structured hairpin. This RT slow-down should increase RNase H cleavages at the site of pausing, increasing template switch at this location (Basu et al., 2008, Roda et al., 2003). In coronavirus little is known about secondary structures and pausing sites, although 5′ TRS sequence secondary structures are good candidates for stopping signals (Enjuanes et al., 2007). TRS secondary structures could be specially relevant in the case of torovirus non-discontinuous transcription (Smits et al., 2005), that resembles the premature transcription termination mechanism of tombusviruses (Lin and White, 2004, White, 2002, White and Nagy, 2004). In these viruses, it has been proposed that RNA secondary structures formed in the genome, upstream to the transcription initiation sites, could facilitate transcription termination (White and Nagy, 2004).
Alternatively, sequence complementarity of nascent nucleic acid with the acceptor molecule could have a role on pausing decision. According to this model, in coronavirus, both the CS and the TRS 3′-flanking sequences combine their complementarity to decide whether template switch would take place. This model fits with the results obtained for bovine coronavirus (BCoV) (Ozdarendeli et al., 2001) and TGEV (Sola et al., 2005, Zuñiga et al., 2004) showing that sequences downstream the CS exert a dominant influence on the template-switching decision. Similar results were obtained in HIV systems, in which increased complementarity before the pausing site facilitates acceptor invasion (Basu et al., 2008, Gao et al., 2007). In HIV it was also reported that a pause-independent strand transfer occurs in low structured RNA regions (Basu et al., 2008, Derebail and DeStefano, 2004), although the relevance of sequence complementarity on this process has not been established.
The role of RNA chaperones on the RNA secondary structure mediated pausing is not obvious, as this kind of proteins are expected to unwind secondary structures. In fact, reduction of polymerase pausing has been shown for NC (Levin et al., 2005), and proposed for coronavirus N protein (Sawicki et al., 2007). Nevertheless, in HIV it has been shown that NC enhances RNase H cleavage, that is a key step of RT hairpin-mediated pausing, as mentioned above (Levin et al., 2005). The possible role of RNA chaperones in sequence complementarity mediated pausing seems a probable event, as RNA chaperones enhance nucleic acids annealing.
5.2. Template switch
Template switch both in corona and retrovirus resembles a similarity-assisted high-frequency copy-choice recombination (Negroni and Buc, 2000, Sawicki et al., 2007). A copy-choice mechanism implies that the polymerase leaves the first template (the donor) and continues synthesis on a second template (the acceptor) (Kim and Kao, 2001). In similarity-assisted recombination, sequence similarity between nucleic acids influences the frequency and site of the recombination event, but additional structure determinants, generally only present in one of the nucleic acids, are essential for efficient recombination (Nagy and Simon, 1997). For instance, in turnip crinkle virus (TCV), efficient recombination needs RNA sequence similarity and a hairpin structure present only in the acceptor RNA (Nagy et al., 1998).
In retroviruses, a secondary DNA structure present in the donor strand, that allows acceptor invasion, has been postulated in the recombination process (Negroni and Buc, 2000). Similar increase in recombination levels has been found when only the acceptor or both the donor and acceptor molecules are coated with NC protein, indicating that this protein could display its chaperone activity when coating a nucleic acid molecule (Negroni and Buc, 2000). In contrast, a secondary structure on one of the nucleic acid molecules is not required for either the minus or the plus strand transfer events. In fact, for an efficient template switch, the unwinding of a stable secondary structure must occur, both in the minus and plus strand transfer (Basu et al., 2008, Levin et al., 2005). NC protein plays a key role in this process, acting as an RNA chaperone (Hong et al., 2003, Wu et al., 1999). One of the determinant steps in HIV minus strand transfer is the annealing of the two highly structured TAR sequences (Basu et al., 2008, Levin et al., 2005). Without the nucleic acid chaperone activity of NC, the annealing reactions would occur much more slowly, as NC increases the annealing of structured sequences around 3000-fold, depending on the assays (Basu et al., 2008, Levin et al., 2005, Vo et al., 2006, Zeng et al., 2007).
According to the current model of coronavirus transcription there is a structural requirement for the leader TRS, that should be in an adequate context to serve as the acceptor for the nascent minus RNA strand transfer (Enjuanes et al., 2007, Sawicki et al., 2007). In fact, a leader stem-loop structure was predicted for several nidovirus, and its existence and relevance on RNA synthesis has been shown for equine arteritis arterivirus (EAV) (van den Born et al., 2004, van den Born et al., 2005) and for TGEV coronavirus (J.L. Moreno, D. Dufour, I. Sola, J. Gallego, L. Enjuanes, unpublished results). In coronavirus template switch, the annealing of the leader and nascent minus RNA sequences complementary to the body TRSs is one of the main factors regulating transcription. It has been demonstrated that the free-energy of the duplex formation between those sequences is the driving force of template switch, regulating the amount of each sg mRNA, and the places where template switch takes place (Sola et al., 2005, Zuñiga et al., 2004). Acting as an RNA chaperone, N protein could be involved in the correct folding of the leader structure. In fact, it has been proposed that N protein could mediate the annealing of TRSs by decreasing the energy barrier required for this process (Zuñiga et al., 2007). Nowadays, our group is working on the design of an in vivo system to analyze template switch step during coronavirus transcription.
5.3. Experimental systems to analyze coronavirus template switch
In the case of retroviruses, there are several experimental systems concerning just one step of the transfer events. These kind of approaches have been essential to develop extensive knowledge on the role of retrovirus NC RNA chaperone on strand transfer (Levin et al., 2005). In contrast, the experimental system to study coronavirus template switch at the molecular level needs to be developed.
Coronaviruses have the longest viral RNA genome known, but it is possible to perform reverse genetics on these viruses, as infectious cDNA clones are available for several coronaviruses (Enjuanes, 2005). Nevertheless, the analysis of RNA chaperone activity in the context of the whole infectious virus represents a challenge. An alternative approach is the use of coronavirus replicons. In the case of TGEV, a set of replicons has been generated (Almazan et al., 2004), that may be used for in vivo testing the chaperone activity of the candidate protein.
In vitro systems are required to precisely define the role that RNA chaperones have on the different steps of coronavirus template switch. The purification of in vitro active coronavirus replication–transcription complexes has been recently described (van Hemert et al., 2008). These systems could be used to analyze the role of chaperones on template switch. The main limitation of these in vitro systems is that the precise components required to reproduce the activity still need to be identified and, very likely, in addition to viral RNA chaperones, cellular ones could be part of the purified active complexes. An alternative is to develop in vitro systems based on defined components, similarly to the current retrovirus in vitro systems. Acceptor and donor RNAs must be carefully selected, attending to the features to be assayed. Probably, the key difficulty in the development of these in vitro systems is the precise number of protein components required. The RNA-dependent RNA-polymerase (RdRp) from SARS-CoV has been purified and its polymerization activity has been shown (Cheng et al., 2005). In addition RNA chaperones and other still unidentified proteins will be required for a fully functional in vitro system.
6. Perspectives
There is little evidence of the in vivo folding mechanism of RNA molecules, but it is generally accepted that RNAs exist bound to proteins within the cells. In 1995, based on some in vitro observations, and from the point of view of the existence of a functional association between RNA and proteins, RNA chaperone activity was postulated (Herschlag, 1995). Since then, several RNA chaperones have been identified using diverse in vitro assays, or postulated, based on the effects observed when these proteins are mutated. A helpful attempt to provide a precise and restricted definition of RNA chaperone activity has recently been reported (Rajkowitsch et al., 2007). According to this definition, RNA chaperones are proteins that resolve misfolded RNAs, and that also must fulfill several requirements: (i) transient interaction with the nucleic acid, mainly due to a weak and low specificity RNA–protein binding; (ii) lack of requirement of an external energy input, such as ATP hydrolysis; and (iii) lack of RNA–protein interaction requirement to maintain the functional RNA conformation.
Previous confusion in the precise identification of RNA chaperones has arised in large extent due to the experimental difficulty in analyzing RNA chaperone activity. As mentioned above, simple studies such as annealing or strand displacement assays could involve other non-chaperone activities (i.e., RNA crowding or matchmaker activities) (Rajkowitsch et al., 2007). New technologies that monitor single molecules have been used to clarify the mechanism of RNA chaperone activity by measuring RNA folding in real-time. These approaches include force spectroscopy and single-molecule stretching using optical tweezers, and time-resolved NMR (Furtig et al., 2007, Williams et al., 2002, Williams et al., 2001). Nevertheless, performance and interpretation of the results provided by these technologies require highly specialized personnel and equipment.
Complex refolding events, such as ribozyme cleavage or intron splicing, are in general suited for the assessment of candidate proteins and to evaluate RNA chaperone mutant activity. Nevertheless, several assays are needed to avoid misinterpretation of the results, such as combination of ribozyme cleavage and transcription activity. These assays must be concentrated on measurements performed using RNA, it also must be shown that after the initial steps protein can be removed, and that the process is independent of external energy sources (Rajkowitsch et al., 2007). In more advanced systems, RNA chaperone should be also tested in vivo, using its natural RNA partner. The analysis of RNA chaperone activity in vivo has the additional difficulty of the pleiotropic effects that RNA chaperones could have in the cell. Nevertheless, the development of in vivo systems is possible. In coronaviruses progress is being made to develop a homologous template switch system that could be easily evaluated.
Acknowledgements
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT, BIO2007-60978), the Community of Madrid (S-SAL-0185/06), the Ministry of Science and Innovation (MICINN, CIT-010000-2007-8) and the European Union (Frame VI, DISSECT Project, SP22-CT-2004-511060). S.Z. and I.S. received contracts from Highest Council of Scientific Research (CSIC). J.L.G.C. received contract from Community of Madrid.
References
- Agol V.I. Molecular mechanisms of poliovirus variation and evolution. Curr. Top. Microbiol. Immunol. 2006;299:211–259. doi: 10.1007/3-540-26397-7_8. [DOI] [PubMed] [Google Scholar]
- Aizaki H., Choi K.S., Liu M., Li Y.J., Lai M.M. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J. Biomed. Sci. 2006;13:469–480. doi: 10.1007/s11373-006-9088-4. [DOI] [PubMed] [Google Scholar]
- Almazan F., Galan C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.C., Hunt S.L., Jackson R.J. Internal initiation of translation from the human rhinovirus-2 internal ribosome entry site requires the binding of Unr to two distinct sites on the 5′ untranslated region. J. Gen. Virol. 2007;88:3043–3052. doi: 10.1099/vir.0.82463-0. [DOI] [PubMed] [Google Scholar]
- Bae W., Xia B., Inouye M., Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. U.S.A. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J.N., Wertz G.W. Role of the conserved nucleotide mismatch within 3′- and 5′-terminal regions of Bunyamwera virus in signaling transcription. J. Virol. 2005;79:3586–3594. doi: 10.1128/JVI.79.6.3586-3594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu V.P., Song M., Gao L., Rigby S.T., Hanson M.N., Bambara R.A. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Belisova A., Semrad K., Mayer O., Kocian G., Waigmann E., Schroeder R., Steiner G. RNA chaperone activity of protein components of human Ro RNPs. RNA. 2005;11:1084–1094. doi: 10.1261/rna.7263905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E.L., Rossi J.J. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.C., Jackson R.J. All five cold-shock domains of unr (upstream of N-ras) are required for stimulation of human rhinovirus RNA translation. J. Gen. Virol. 2004;85:2279–2287. doi: 10.1099/vir.0.80045-0. [DOI] [PubMed] [Google Scholar]
- Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C., Lee S.J., Hsiao H.H., Wu W.J., Chang W.L., Lin C.H., Huang T.H. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.S., Luo G. The polypyrimidine tract-binding protein (PTB) is required for efficient replication of hepatitis C virus (HCV) RNA. Virus Res. 2006;115:1–8. doi: 10.1016/j.virusres.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Cheng A., Zhang W., Xie Y., Jiang W., Arnold E., Sarafianos S.G., Ding J. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology. 2005;335:165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clodi E., Semrad K., Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 1999;18:3776–3782. doi: 10.1093/emboj/18.13.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee T., Herschlag D., Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 1994;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- Cornillez-Ty C.T., Lazinski D.W. Determination of the multimerization state of the hepatitis delta virus antigens in vivo. J. Virol. 2003;77:10314–10326. doi: 10.1128/JVI.77.19.10314-10326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G., Darlix J.L. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Ivanyi-Nagy R., Gabus C., Boulant S., Lavergne J.P., Penin F., Darlix J.L. The hepatitis C virus core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004;32:2623–2631. doi: 10.1093/nar/gkh579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J.L., Gabus C., Nugeyre M.T., Clavel F., Barré-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- Darlix, J.L., Lastra, M.L., Mély, Y., Roques, B., 2002. Nucleocapsid protein chaperoning of nucleic acids at the heart of HIV structure, assembly and cDNA synthesis. In: Kuiken, C., Foley, B., Freed, E., Hahn, B., Marx, P., McCutchan, F., Mellors, J.W., Wolinsky, S., Korber, B. (Eds.), HIV Sequence Compendium. Los Alamos, NM, pp. 69–88.
- Daros J.A., Flores R. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 2002;21:749–759. doi: 10.1093/emboj/21.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derebail S.S., DeStefano J.J. Mechanistic analysis of pause site-dependent and -independent recombinogenic strand transfer from structurally diverse regions of the HIV genome. J. Biol. Chem. 2004;279:47446–47454. doi: 10.1074/jbc.M408927200. [DOI] [PubMed] [Google Scholar]
- DeStefano J.J., Titilope O. Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities. J. Virol. 2006;80:1662–1671. doi: 10.1128/JVI.80.4.1662-1671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domitrovich A.M., Diebel K.W., Ali N., Sarker S., Siddiqui A. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology. 2005;335:72–86. doi: 10.1016/j.virol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G. Structure and function of nuclear and cytoplasmic ribonucleoprotein particles. Annu. Rev. Cell Biol. 1986;2:459–498. doi: 10.1146/annurev.cb.02.110186.002331. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Égelé C., Schaub E., Piémont E., Rocquigny H., Mély Y. Investigation by fluorescence correlation spectroscopy of the chaperoning interactions of HIV-1 nucleocapsid protein with the viral DNA initiation sequences. C. R. Biol. 2005;328:1041–1051. doi: 10.1016/j.crvi.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Enjuanes, L. (Ed.), 2005. Coronavirus replication and reverse genetics, vol. 287. Curr. Top. Microbiol. Immunol. Springer, Berlin. [DOI] [PMC free article] [PubMed]
- Enjuanes L., Almazan F., Sola I., Zuñiga S. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 2006;60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- Enjuanes L., Gorbalenya A.E., de Groot R.J., Cowley J.A., Ziebuhr J., Snijder E.J. The Nidovirales. In: Mahy B.W.J., Van Regenmortel M., Walker P., Majumder-Russell D., editors. Encyclopedia of Virology. 3rd ed. Elsevier Ltd; Oxford: 2008. pp. 419–430. [Google Scholar]
- Enjuanes L., Sola I., Zuñiga S., Moreno J.L. Coronavirus RNA synthesis: transcription. In: Thiel V., editor. Coronaviruses: Molecular and Cellular Biology. Caister Academic Press; Norfolk: 2007. pp. 81–107. [Google Scholar]
- Florez P.M., Sessions O.M., Wagner E.J., Gromeier M., Garcia-Blanco M.A. The polypyrimidine tract binding protein is required for efficient picornavirus gene expression and propagation. J. Virol. 2005;79:6172–6179. doi: 10.1128/JVI.79.10.6172-6179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A.D., Young J.A.T. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- Furtig B., Buck J., Manoharan V., Bermel W., Jaschke A., Wenter P., Pitsch S., Schwalbe H. Time-resolved NMR studies of RNA folding. Biopolymers. 2007;86:360–383. doi: 10.1002/bip.20761. [DOI] [PubMed] [Google Scholar]
- Gao L., Balakrishnan M., Roques B.P., Bambara R.A. Insights into the multiple roles of pausing in HIV-1 reverse transcriptase-promoted strand transfers. J. Biol. Chem. 2007;282:6222–6231. doi: 10.1074/jbc.M610056200. [DOI] [PubMed] [Google Scholar]
- Gatignol A. Transcription of HIV: Tat and cellular chromatin. Adv. Pharmacol. 2007;55:137–159. doi: 10.1016/S1054-3589(07)55004-0. [DOI] [PubMed] [Google Scholar]
- Ghetti A., Piñol-Roma S., Michael W.M., Morandi C., Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Sharp P.A., Jamison S.F., Garcia-Blanco M.A. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Chang K.H., Rijnbrand R., Yi M., Sangar D.V., Lemon S.M. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 2000;20:1583–1595. doi: 10.1128/mcb.20.5.1583-1595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohman J.K., Del Campo M., Bhaskaran H., Tijerina P., Lambowitz A.M., Russell R. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Henderson L.E., Bess J., Kane B., Levin J.G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Wu T., Kane B.F., Johnson D.G., Henderson L.E., Gorelick R.J., Levin J.G. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 2002;76:4370–4378. doi: 10.1128/JVI.76.9.4370-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilman-Miller S.L., Wu T., Levin J.G. Alteration of nucleic acid structure and stability modulates the efficiency of minus-strand transfer mediated by the HIV-1 nucleocapsid protein. J. Biol. Chem. 2004;279:44154–44165. doi: 10.1074/jbc.M401646200. [DOI] [PubMed] [Google Scholar]
- Henriet S., Sinck L., Bec G., Gorelick R.J., Marquet R., Paillart J.C. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007;35:5141–5153. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Khosla M., Tsuchihashi Z., Karpel R.L. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M.K., Harbron E.J., O’Connor D.B., Guo J., Barbara P.F., Levin J.G., Musier-Forsyth K. Nucleic acid conformational changes essential for HIV-I nucleocapsid protein-mediated inhibition of self-priming in minus-strand transfer. J. Mol. Biol. 2003;325:1–10. doi: 10.1016/s0022-2836(02)01177-4. [DOI] [PubMed] [Google Scholar]
- Huang P., Lai M.M.C. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′-untranslated region and mediates potential 5′–3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 2001;75:5009–5017. doi: 10.1128/JVI.75.11.5009-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.S., Su W.H., Wang J.L., Wu H.N. Selective strand annealing and selective strand exchange promoted by the N-terminal domain of hepatitis delta antigen. J. Biol. Chem. 2003;278:5685–5693. doi: 10.1074/jbc.M207938200. [DOI] [PubMed] [Google Scholar]
- Huang Z.S., Wu H.N. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J. Biol. Chem. 1998;273:26455–26461. doi: 10.1074/jbc.273.41.26455. [DOI] [PubMed] [Google Scholar]
- Hunt S.L., Hsuan J.J., Totty N., Jackson R.J. unr, a cellular cytoplasmic RNA-binding protein with five cold shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S.L., Jackson R.J. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Davidovic L., Khandjian E.W., Darlix J.L. Disordered RNA chaperone proteins: from functions to disease. Cell. Mol. Life Sci. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Kanevsky I., Gabus C., Lavergne J.P., Ficheux D., Penin F., Fossé P., Darlix J.L. Analysis of hepatitis C virus RNA dimerization and core–RNA interactions. Nucleic Acids Res. 2006;34:2618–2633. doi: 10.1093/nar/gkl240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Lavergne J., Gabus C., Ficheux D., Darlix J. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008;36:2618–2633. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng K.S., Su P.Y., Lai M.M.C. Hepatitis delta antigens enhance the ribozyme activities of hepatitis delta virus RNA in vivo. J. Virol. 1996;70:4205–4209. doi: 10.1128/jvi.70.7.4205-4209.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Klarmann G.J., Preston B.D. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- Karakasiliotis I., Chaudhry Y., Roberts L.O., Goodfellow I.G. Feline calicivirus replication: requirement for polypyrimidine tract-binding protein is temperature-dependent. J. Gen. Virol. 2006;87:3339–3347. doi: 10.1099/vir.0.82153-0. [DOI] [PubMed] [Google Scholar]
- Khaiboullina S.F., Morzunov S.P., St Jeor S.C. Hantaviruses: molecular biology, evolution and pathogenesis. Curr. Mol. Med. 2005;5:773–790. doi: 10.2174/156652405774962317. [DOI] [PubMed] [Google Scholar]
- Kim M.J., Kao C. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA–RNA recombination. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4972–4977. doi: 10.1073/pnas.081077198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos A., Dijkema R., Arnberg A.C., van der Meide P.H., Schellekens H. The hepatitis delta (d) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- Kuciak M., Gabus C., Ivanyi-Nagy R., Semrad K., Storchak R., Chaloin O., Muller S., Mély Y., Darlix J.L. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucleic Acids Res. 2008;36:3389–3400. doi: 10.1093/nar/gkn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.Y., Chao M., Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Lin H.X., White K.A. A complex network of RNA–RNA interactions controls subgenomic mRNA transcription in a tombusvirus. EMBO J. 2004;23:3365–3374. doi: 10.1038/sj.emboj.7600336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Rice C.M. Flaviviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields Virology. 4th ed. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 991–1041. [Google Scholar]
- Liu H.W., Zeng Y., Landes C.F., Kim Y.J., Zhu Y., Ma X., Vo M.N., Musier-Forsyth K., Barbara P.F. Insights on the role of nucleic acid/protein interactions in chaperoned nucleic acid rearrangements of HIV-1 reverse transcription. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5261–5267. doi: 10.1073/pnas.0700166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch J.R. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109:797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- Maraia R.J., Intine R.V.A. Recognition of nascent RNA by the human La antigen: conserved and divergent features of structure and function. Mol. Cell. Biol. 2001;21:367–379. doi: 10.1128/MCB.21.2.367-379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer O., Waldsich C., Grossberger R., Schoeder R. Folding of the td pre-RNA with the help of the RNA chaperone StpA. Biochem. Soc. Trans. 2002;30:1175–1180. doi: 10.1042/bst0301175. [DOI] [PubMed] [Google Scholar]
- Meerovitch K., Svitkin Y.V., Lee H.S., Lejbkowicz F., Kenan D.J., Chan E.K., Agol V.I., Keene J.D., Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J. Virol. 2005;79:1824–1835. doi: 10.1128/JVI.79.3.1824-1835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. Characterization of the RNA chaperone activity of hantavirus nucleocapsid protein. J. Virol. 2006;80:6276–6285. doi: 10.1128/JVI.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. RNA. 2006;12:272–282. doi: 10.1261/rna.2101906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.A., Spriggs K.A., Coldwell M.J., Jackson R.J., Willis A.E. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and Unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Mohr S., Stryker J.M., Lambowitz A.M. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109(6):769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- Moradpour D., Penin F., Rice C.M. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Moreno J.L., Zuñiga S., Enjuanes L., Sola I. Identification of a coronavirus transcription enhancer. J. Virol. 2008;82:3882–3893. doi: 10.1128/JVI.02622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Simon A.E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Zhang C., Simon A.E. Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J. 1998;17:2392–2403. doi: 10.1093/emboj/17.8.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negroni M., Buc H. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6385–6390. doi: 10.1073/pnas.120520497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., Wüthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obradovic Z., Peng K., Vucetic S., Radivojac P., Brown C., Dunker A.K. Predicting intrinsic disorder from aminoacid sequence. Proteins. 2003;53:566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- Opi S., Takeuchi H., Kao S., Khan M.A., Miyagi E., Goila-Gaur R., Iwatani Y., Levin J.G., Strebel K. Monomeric APOBEC3G is catalytically active and has antiviral activity. J. Virol. 2006;80:4673–4682. doi: 10.1128/JVI.80.10.4673-4682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdarendeli A., Ku S., Rochat S., Williams G.D., Senanayake S.D., Brian D.A. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 2001;75:7362–7374. doi: 10.1128/JVI.75.16.7362-7374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone B., Xue D., Wollin S.L. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.M., Masters P.S. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990;179:463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K., Obradovic Z. Optimizing intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- Plyusnin A., Vapalahti O., Vaheri A. Hantaviruses: genome structure, expression and evolution. J. Gen. Virol. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- Pontius B.W., Berg P. Rapid assembly and disassembly of complementary DNA strands through as equilibrium. J. Biol. Chem. 1992;267:13815–13818. [PubMed] [Google Scholar]
- Portman D.S., Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A.C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J.L. Small finger protein of avian and murine retrovirus has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988;7:1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L., Chen D., Stampfl S., Semrad K., Waldsich C., Mayer O., Jantsch M.F., Konrat R., Bläsi U., Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- Rajkowitsch L., Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:1–8. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L., Semrad K., Mayer O., Schroeder R. Assays for the RNA chaperone activity of proteins. Biochem. Soc. Trans. 2005;33:450–456. doi: 10.1042/BST0330450. [DOI] [PubMed] [Google Scholar]
- Ramalanjaona N., de Rocquigny H., Millet A., Ficheux D., Darlix J.L., Mély Y. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J. Mol. Biol. 2007;374:1041–1053. doi: 10.1016/j.jmb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Reeder R.H., Lang W.H. Terminating transcription in eukaryotes: lessons learned from RNA polymerase I. Trends Biochem. Sci. 1997;22:473–477. doi: 10.1016/s0968-0004(97)01133-x. [DOI] [PubMed] [Google Scholar]
- Roda R.H., Balakrishnan M., Hanson M.N., Wöhrl B.M., Le Grice S.F.J., Roques B.P., Gorelick R.J., Bambara R.A. Role of the reverse transcriptase, nucleocapsid protein, and template structure in the two-step transfer mechanism in retroviral recombination. J. Biol. Chem. 2003;278:31536–31546. doi: 10.1074/jbc.M304608200. [DOI] [PubMed] [Google Scholar]
- Sakai H., Shibata R., Sakuragi J., Kawamura M., Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. J. Virol. 1993;67:1663–1666. doi: 10.1128/jvi.67.3.1663-1666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 2005;287:31–55. doi: 10.1007/3-540-26765-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R., Barta A., Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell. Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- Semrad K., Green R., Schoeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA. 2004;10:1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Masters P.S. Evaluation of the role of heterogeneous nuclear ribonucleoprotein A1 as a host factor in murine coronavirus discontinuous transcription and genome replication. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2717–2722. doi: 10.1073/pnas.031424298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, S.T., Lai, M.M., 2005. Viral and cellular proteins involved in coronavirus replication. In: Enjuanes, L. (Ed.), Curr. Top. Microbiol. Immunol., vol. 287, pp. 95–131. [DOI] [PMC free article] [PubMed]
- Smits S.L., van Vliet A.L., Segeren K., el Azzouzi H., van Essen M., de Groot R.J. Torovirus non-discontinuous transcription: mutational analysis of a subgenomic mRNA promoter. J. Virol. 2005;79:8275–8281. doi: 10.1128/JVI.79.13.8275-8281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Moreno J.L., Zuñiga S., Alonso S., Enjuanes L. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 2005;79:2506–2516. doi: 10.1128/JVI.79.4.2506-2516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Tzima E., Ochs K., Bassili G., Trusheim H., Linder M., Preissner K.T., Niepmann M. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sova P., Volsky D.J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisne C., Roques B.P., Dardel F. The annealing mechanism of HIV-1 reverse transcription primer onto the viral genome. J. Biol. Chem. 2004;279:3588–3595. doi: 10.1074/jbc.M310368200. [DOI] [PubMed] [Google Scholar]
- Tompa P., Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z., Brown P.O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type I nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E., Gultyaev A.P., Snijder E.J. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA. 2004;10:424–437. doi: 10.1261/rna.5174804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E., Posthuma C.C., Gultyaev A.P., Snijder E.J. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 2005;79:6312–6324. doi: 10.1128/JVI.79.10.6312-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert M.J., van den Worm S.H., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS–coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo M.N., Barany G., Rouzina I., Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Wang C.C., Chang T.C., Lin C.W., Tsui H.L., Chu P.B., Chen B.S., Huang Z.S., Wu H.N. Nucleic acid binding properties of the nucleic acid chaperone domain of hepatitis delta antigen. Nucleic Acids Res. 2003;31:6481–6492. doi: 10.1093/nar/gkg857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.S., Choo Q.L., Weiner A.J., Ou J.H., Najarian R.C., Thayer R.M., Mullenbach G.T., Denniston K.J., Gerin J.L., Houghton M. Structure, sequence and expression of the hepatitis delta (d) viral genome. Nature. 1986;323:508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- White K.A. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology. 2002;304:147–154. doi: 10.1006/viro.2002.1732. [DOI] [PubMed] [Google Scholar]
- White K.A., Nagy P.D. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- Wiegand H.L., Doehle B.P., Bogerd H.P., Cullen B.R. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.C., Gorelick R.J., Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone functions. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M.C., Rouzina I., Wenner J.R., Gorelick R.J., Musier-Forsyth K. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.S., von Hippel P.H. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton M.C., Gooding C., Robinson F., Brown E.C., Jackson R.J., Smith C.W. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–832. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Guo J., Bess J., Henderson L.E., Levin J.G. Molecular requirements for human immunodeficiency virus type 1 plus-strand transfer: analysis in reconstituted and endogenous reverse transcription systems. J. Virol. 1999;73:4794–4805. doi: 10.1128/jvi.73.6.4794-4805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Heilman-Miller S.L., Levin J.G. Effects of nucleic acid local structure and magnesium ions on minus-strand transfer mediated by the nucleic acid chaperone activity of HIV-1 nucleocapsid protein. Nucleic Acids Res. 2007;35:3974–3987. doi: 10.1093/nar/gkm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Henderson L.E., Copeland T.D., Gorelick R.J., Bosche W.J., Rein A., Levin J.G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Liu H.W., Landes C.F., Kim Y.J., Ma X., Zhu Y., Musier-Forsyth K., Barbara P.F. Probing nucleation, reverse annealing, and chaperone function along the reaction path of HIV-1 single-strand transfer. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12651–12656. doi: 10.1073/pnas.0700350104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Hwang C.K., Hu W.S., Gorelick R.J., Pathnak V. Zinc finger domain of murine leukemia virus nucleocapsid protein enhances the rate of viral DNA synthesis in vivo. J. Virol. 2002;76:7473–7484. doi: 10.1128/JVI.76.15.7473-7484.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lai M.M.C. Interactions between the cytoplasmic proteins and the intergenic (promoter) sequence of mouse hepatitis virus RNA: correlation with the amounts of subgenomic mRNA transcribed. J. Virol. 1995;69:1637–1644. doi: 10.1128/jvi.69.3.1637-1644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li H.P., Xue W., Lai M.C.C. Formation of a ribonucleoprotein complex of mouse hepatitis virus involving heterogeneous nuclear ribonucleoprotein A1 and transcription-regulatory elements of viral RNA. Virology. 1999;264:115–124. doi: 10.1006/viro.1999.9970. [DOI] [PubMed] [Google Scholar]
- Zheng A., Derbyshire V., Salvo J.L., Belfort M. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- Zuñiga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuñiga S., Sola I., Moreno J.L., Sabella P., Plana-Duran J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]


