Abstract
In this review, we summarize the researches on animal reservoirs of the SARS coronavirus (SARS-CoV). Masked palm civets were suspected as the origin of the SARS outbreak in 2003 and was confirmed as the direct origin of SARS cases with mild symptom in 2004. Sequence analysis of the SARS-CoV-like virus in masked palm civets indicated that they were highly homologous to human SARS-CoV with nt identity over 99.6%, indicating the virus has not been circulating in the population of masked palm civets for a very long time. Alignment of 10 complete viral genome sequences from masked palm civets with those of human SARS-CoVs revealed 26 conserved single-nucleotide variations (SNVs) in the viruses from masked palm civets. These conserved SNVs were gradually lost from the genomes of viruses isolated from the early phase to late phase human patients of the 2003 SARS epidemic. In 2005, horseshoe bats were identified as the natural reservoir of a group of coronaviruses that are distantly related to SARS-CoV. The genome sequences of bat SARS-like coronavirus had about 88–92% nt identity with that of the SARS-CoV. The prevalence of antibodies and viral RNA in different bat species and the characteristics of the bat SARS-like coronavirus were elucidated. Apart from masked palm civets and bats, 29 other animal species had been tested for the SARS-CoV, and the results are summarized in this paper.
Keywords: SARS-CoV, Animal reservoirs, Bats, Masked palm civets
1. Introduction
Severe acute respiratory syndrome (SARS), a disease that first occurred in November 2002 in Guangdong Province of China (Xu et al., 2004a, Xu et al., 2004b), has spread to 28 regions around the world in 2003 causing 774 deaths in 8096 infected individuals (WHO, 2004). A novel coronavirus, SARS-CoV, was identified as the etiological agent of the disease (Drosten et al., 2003, Ksiazek et al., 2003, Peiris et al., 2003) and phylogenetic analysis indicated that the virus was different from previously known coronaviruses (Marra et al., 2003, Rota et al., 2003). Although the identification of the etiological agent and molecular studies on the SARS-CoV proceeded rather rapidly, the puzzle on the origin of the disease was still not fully resolved. From the very beginning, data suggested that SARS was originated from animals. For examples: (1) Animal food handlers who handle, kill, or butcher “exotic” animals for food were over represented among early-onset SARS cases (Xu et al., 2004a, Xu et al., 2004b, Zhong et al., 2003). (2) Serological studies of traders from several selected animal markets in Guangzhou, Guangdong Province in 2003 indicated that the animal traders had significantly higher prevalence of IgG antibodies against the SARS-CoV than vegetable traders or control groups (Xu et al., 2004a, Xu et al., 2004b, CDC, 2003). These data pointed to the fact that animals being sold in markets as a likely source of the virus. Therefore, after the outbreak of SARS, many scientists concentrated their efforts on investigating the animal origin of SARS-CoV.
A breakthrough came in May of 2003 when viruses related to SARS were identified in animals. A team led by Yi Guan (Hong Kong University) and colleagues from the Center for Disease Control (CDC) Shenzhen, studied 25 individuals from 8 different species including 6 masked palm civets (Paguma larvata), 1 raccoon dog (Nyctereutes procyonoides), 2 Chinese ferret-badgers (Melogale moschata), 3 hog-badgers (Arctonyx collaris), 3 beavers (Castor fiber), 4 domestic cats (Felis catus), 3 Chinese hares (Lepus sinensis) and 2 Chinese muntjac (Muntiacus reevesi) gathered in Dongmen Market, Shenzhen, Guangdong Province (Guan et al., 2003). Nasal and faecal swabs from the animals were tested for SARS-CoV nucleic acid by using reverse transcription-polymerase chain reaction (RT-PCR) on the N gene of the human SARS-CoV, and all the swaps were inoculated into FRhK-4 cells for virus isolation. All of the six masked palm civets and one raccoon dog were positive for SARS-CoV by either RT-PCR and/or virus isolation. Sera from five animals including three masked palm civets, a raccoon dog and a Chinese ferret badger, had neutralizing antibody against a SARS-CoV isolated from a masked palm civet (SZ16). The neutralizing antibody titer was ≥640, 40, 40 for the three masked palm civets, ≥640 for the raccoon dog and 160 for the Chinese ferret badger (Guan et al., 2003). It is interesting to note that one of the masked palm civet, which was tested positive for SARS-CoV nucleic acid both in nasal and faecal swabs was negative for neutralizing antibodies against its own SZ16 virus. It implied that this animal was either newly infected or it had developed mechanisms to avoid generating neutralizing antibodies. This study raised the initial alert that masked palm civets and other wildlife animals may play a role in the transmission of SARS-CoV and led to a temporary ban on the hunting, sale, transportation and export of all wild animals in Guangdong Province.
Later in 2003, the role of masked palm civets and other animals was questioned and a debate ensued on whether these animals are actually the origin of the SARS-CoV. A rather convincing evidence implicating masked palm civets came in approximately half a year later in the winter of 2003–2004. Between 16 December 2003 and 8 January 2004, a total of four patients were identified as SARS cases with mild flu-like syndromes and no secondary transmission (Liang et al., 2004). Epidemiological information collected by the Guangdong CDC and Guangzhou CDC indicated that three of the patients had direct or indirect contact to masked palm civets. The second patient was a waitress in a local restaurant (TDLR), where live masked palm civets were kept for exotic dishes, and she often passed by or stood in close proximity of the animal cages (Wang et al., 2005b). The fourth patient dined in the same restaurant and his dining table was within 5 m of civet cages (Wang et al., 2005b). The third patient dined in a neighboring restaurant (SJR), which shared the kitchen with TDLR (Song et al., 2005). The masked palm civets kept in TDLR restaurant were reported to have been originally purchased from the Xinyuan Live Animal Market (Wang et al., 2005b). Three research teams published their results on the SARS-CoV-like viruses in masked palm civets from the TDLR restaurant and from the Xinyuan Live Animal Market as well as the four patients (Kan et al., 2005, Song et al., 2005, Wang et al., 2005b). Almost identical sequences of the SARS-CoV spike genes were identified in samples from some of the patients and from masked palm civets in either the restaurant or the Xinyuan Live Animal Market (Kan et al., 2005, Song et al., 2005, Wang et al., 2005b). Based on virological and epidemiological findings, Guangdong provincial officials took an aggressive action on 5 January 2004 to destroy any animals that might harbor the SARS-CoV in farms and in animal food markets. Quarantine on all civets reared for human consumption in farms all over China was imposed. Since then, no additional community-acquired SARS cases have been reported.
Although masked palm civets were identified as direct origin of at least some of the SARS cases, the following data indicated that it might not necessarily be the natural animal reservoir of the SARS-CoV: (1) The so far sequenced SARS-CoVs from masked palm civets harbored a nucleic acid identity higher than 99.6%, implying that SARS-CoV-like viruses might have recently entered into the masked palm civet population. (2) Although masked palm civets in markets had a high prevalence for SARS-CoV during the epidemic (Guan et al., 2003, Kan et al., 2005, Tu et al., 2004), investigations on farmed masked palm civets were often negative for the SARS-CoV (Kan et al., 2005, Tu et al., 2004) with only a few exceptions (Hu et al., 2005, Tu et al., 2004). (3) An investigation of 21 wild masked palm civets trapped in Hong Kong indicated that those animals were free of SARS-CoV-like viruses suggesting the virus is not widely circulating in wild masked palm civets (Poon et al., 2005).
For a number of reasons, attention turned to bats as a likely reservoir of the SARS-CoV. Bats have long been known as an important reservoir for many zoonotic viruses including rabies virus, Hendra virus, Nipha virus, Ebola virus and St. Louis encephalitis viruses (Halpin et al., 2000, Leroy et al., 2005, Mackenzie et al., 2001, Mackenzie and Field, 2004). An early study by Poon et al. (2005) on 81 bats belonging to 12 different species identified a novel bat coronavirus (Bat-CoV) from three different bat species of the same genus, Miniopterus magnater, Miniopterus pusillus, and Miniopterus schreibersi. However, the Bat-CoV belonged to group 1 coronavirus and was not closely related to SARS-CoV since its RNA fragments shared only 41–62% nucleotide sequence identities to SARS-CoV (Poon et al., 2005). This initial failure to identify a SARS-CoV-like virus in bats was soon compensated by the successful detection, by two other independent groups, of several SARS-CoV-like viruses in different species of horseshoe bats (Lau et al., 2005, Li et al., 2005b).
This review summarizes the research aimed at investigating the origin in masked palm civets, bats, as well as other animals. We tried to cover investigations on ecology, epidemiology, as well as molecular virology. For readers interested in more detailed information on interaction of SARS-CoV spike protein with receptors of different species in relation to animal origin of SARS-CoV, we highly recommend the recent review paper by Li et al. (2006).
2. Masked palm civets and SARS-CoV
2.1. Masked palm civets
Masked palm civets (P. larvata) belong to Carnivora, Viverridae. In China, they are distributed mainly south of Yangzhi River, but are also found in other parts such as HeBei, Shaanxi and Shanxi Provinces and south of Tibet (Zhang, 1999, Gao et al., 1987). There are nine subspecises of P. larvata in China, including P. l. larvata, P. l. taivana, P. l. reevesi, P. l. hainana, P. l. intrudens, P. l. chichingensis, P. l. neglecta, P. l. nigriceps, and P. l. lanigera (Gao et al., 1987). The animals are mainly arboreal, nocturnal and largely solitary. They live in mountain and hill forests taking shelter in hollows of trees. It was reported that they live in a family based group of 2–15 individuals (Zhang et al., 1991b). They eat mostly fruits but also supplement their diet with rodents, birds, insects and roots.
Farming of masked palm civets in China started in late 1950s mainly for fur (Liu, 1959), but breeding became popular in the late 1980s with increasing demands for them as an exotic food. It was reported that in 2003 about 40,000 masked palm civets were raised in 660 farms all over China (Jiang et al., 2003). The farms started either from capture and breeding of local wild animals or from breeding stocks brought in from other farms. The feed for the farmed animals varied depending on the particular farm but usually contained grains such as maize, beans, rice and animal feed such as powdered products from fish or bones and often supplemented with fresh meat and/or parts from other animals (Zhou, 1994, Shao and Li, 1997). By studying body temperature and weight, food intake, shedding and living habits of 87 wild masked palm civets in farms, Zhang et al. (1991a) found that masked palm civets in Shanxi Province hibernated slightly from December to February for about 75–85 days. A similar study showed that the masked palm civets in Hunan Province (south of Yangzhi River) also hibernated slightly from December to February as their body temperature dropped 3–4 °C and other physiological indexes decreased significantly compared with those during normal period (Kang et al., 1997).
Masked palm civets are naturally timid. Farm breeding seemed to have been successful and only limited number diseases have been reported. Viral enteritis caused by a parvovirus was one of the common diseases in masked palm civets. An inactivated vaccine against this virus was developed to immunize the animals (Duan et al., 2001). An investigation of four apparently healthy masked palm civets in Taiwan has shown that they had antibodies against feline parvovirus (FPV) but not to feline herpesvirus type 1 or to feline calicivirus (Ikeda et al., 1999). Moreover, the titer of neutralizing antibodies against FPV ranged from 160 to >5120 (Ikeda et al., 1999). Bacterial and parasitic diseases have also been reported for masked palm civets (Yang et al., 2000, Zhao, 2003).
2.2. SARS-CoV-like viruses in masked palm civets
When SARS-CoV-like viruses were first identified in masked palm civets from a Shenzhen market (Guan et al., 2003), several investigators concentrated their efforts on exploring the prevalence of these viruses in this animal (Table 1 ).
Table 1.
Summary of investigations of SARS-CoV in masked palm civets from markets, farms and wilda
| Market/farm/wild | Samples |
Anitibody |
Nucleic acid/virus |
Reference | |||
|---|---|---|---|---|---|---|---|
| Location (Province) | Sample date | Methods | Prevalence | Method | Prevalence | ||
| Market | Shenzheng (Guangdong) | May, 2003 | VNT | 3/4 (75%) | RT-PCR, isolation | 6/6 (100%) | Guan et al. (2003) |
| Market | Xinyuan (Guangdong) | May, 2003 | ND | ND | RT-PCR | 2/7 (29%) | Tu et al. (2004) |
| Market | Xinyuan (Guangdong) | January, 2004 | ND | ND | RT-PCR | 91/91 (100%) | Kan et al. (2005) |
| Market | Xinyuan (Guangdong) | January, 2004 | VNT, IFA, WB | 14/18 (78%) | RT-PCR | 0/18 | Tu et al. (2004) |
| Market | Guangzhou (Guangdong) | November, 2004 | ND | ND | Real-time RT-PCR | 0/12 | Wang et al., 2005a, Wang et al., 2005b |
| Market | Shenzheng (Guangdong) | December, 2004 | ND | ND | Real-time RT-PCR | 0/12 | Wang et al., 2005a, Wang et al., 2005b |
| Farm | Luoning (Henan) | June, 2003 | VNT, IFA | 0/17 | ND | ND | Tu et al. (2004) |
| Farm | Changsha (Hunan) | June, 2003 | VNT, IFA | 0/30 | ND | ND | Tu et al. (2004) |
| Farm | Qianguan (Guangdong) | January, 2004 | VNT, IFA | 0/9 | RT-PCR | 0/9 | Tu et al. (2004) |
| Farm | Shanguan (Guangdong) | January, 2004 | VNT, IFA | 0/10 | RT-PCR | 0/10 | Tu et al., 2004 |
| Farm | Shanwei (Guangdong) | January, 2004 | VNT, IFA, WB | 4/10 (40%) | RT-PCR | 0/10 | Tu et al. (2004) |
| Farm | Zhuhai (Guangdong) | January, 2004 | VNT, IFA | 0/10 | RT-PCR | 0/10 | Tu et al. (2004) |
| Farm | Guangzhou (Guangdong) | May, 2003 | ND | ND | RT-PCR | 2/9 (22%) | Tu et al. (2004) |
| Farm | (Jiangxi) | May, 2003 | ND | ND | RT-PCR | 0/15 | Tu et al. (2004) |
| 25 Farms | (12 Provinces) | January–September, 2004 | ND | ND | RT-PCR | 0/1107 | Kan et al. (2005) |
| Farm | (Hubei) | April, 2004 | ND | ND | Real-time RT-PCR | 7/7 (100%) | Hu et al. (2005) |
| Wild | Hong Kong | 2003–2004 | VNT | 0/21 | RT-PCR | 0/21 | Poon et al. (2005) |
aND, not done; VNT, virus neutralization test; IFA, immunofluorescent antibody test; WB, Western blot test.
Tu et al. (2004) examined 56 masked palm civets including 38 from four farms in different regions of Guangdong Province and 18 from the Xinyuan Live Animal Market in Guangzhou, Guangdong Province in January 2004. They found anti-SARS-CoV antibodies in 78% of the market animals (14 out of 18), while only one out of the four farms contained animals with anti-SARS-CoV antibodies with a prevalence of 40% (4 out of 10). SARS-CoV antibody levels in farmed animals were lower than those from the market. Intestinal tissues collected from the 56 civets were tested by RT-PCR and none were positive. Analysis of 47 serum samples collected in June 2003 from two farms in Henan and Hunan Provinces were negative by a neutralization test and an immunofluoresence antibody assay. The authors also mentioned previously unpublished data on SARS-CoV in the Xinyuan Live Animal Market in Guangzhou, in Guangdong Centre for Rescue and Care of Wildlife Animals, and in a civet farm in Jiangxi Province. These data indicated a high percentage of infected civets in one particular market at a specific time, while no indication of civet infection was seen on most farms during the same period. The authors suggested that the high prevalence in market civets might be associated with trading activities under conditions of overcrowding and mixing of various animal species (Tu et al., 2004).
Kan et al. (2005) investigated 1107 palm civets from 25 farms in 12 Provinces from January to September of 2004 in China. All of the 1107 civets sampled tested negative for the virus by RT-PCR. While all of 91 animals from the Xinyuan Live Animal Market in January of 2004 were positive by RT-PCR (Kan et al., 2005). The authors suggested that the palm civets were infected at the market by other palm civets or by other animals harboring the virus rather than at the farm (Kan et al., 2005). In contrast, by using a real-time RT-PCR assay, Hu et al. (2005) reported seven out of seven masked palm civets in a farm in Hubei Province were positive, although the copy numbers of the nucleic acid of most of the samples were quite low.
Wang et al. (2005a) examined the prevalence of the virus in civets present in a Guangdong market in late 2004. Twelve and 10 masked palm civets were sampled from markets from Guangzhou and Shenzhen, respectively and none of them tested positive by real-time RT-PCR. Tests on 21 wild civets captured in Hong Kong did not reveal an infection by SARS-CoV-like virus (Poon et al., 2005).
Taken all together, the above data indicate that one cannot derive an unequivocal conclusion at the present time. It seems, however, that masked palm civets are not widely infected in the wild or when raised in farms. Therefore, investigation of larger number of masked palm civets in a wider distribution is needed in the future.
2.3. Viral sequence analysis from masked palm civets
To date, the genomes of 10 viruses from civets have been completely sequenced, including 2 (SZ3 and SZ16) from Shenzhen market in 2003 (Guan et al., 2003), 4 (civet010, civet007, civet020, PC4-13) from TDLR restaurant in 2004 (Wang et al., 2005b, Song et al., 2005) and 4 (PC4-136, PC4-227, A022, B039) from the Xinyuan Live Animal Market in 2004 (Song et al., 2005, Kan et al., 2005). When the genome sequences were compared with 92 human SARS-CoVs (Song et al., 2005), a total of 212 single-nucleotide variation (SNV) sites with multiple occurrences were identified, among which 209 sites were in the protein coding regions (data not shown). Among the 209 SNV sites, 73 have synonymous amino acid (aa) mutations and 136 have nonsynonymous aa mutations. There was only one site with multiple base substitutions: the original nucleotide at position 28,590 (Tor2) is C, while Civet020 and A022 have T instead, and B039 has the nucleotide G. All these mutations resulted in synonymous aa substitutions.
One hundred and twenty-nine SNV sites contain variations in viral sequences from masked palm civets, among which 26 SNVs were conserved in the civet viruses (Table 2 ). Twenty of the SNVs cause nonsynonymous aa substitution, including seven in ORF1ab (aa 1121, 1663, 2746, 2971, 3047, 3072, 5767), eight in the spike protein (aa 77, 244, 344, 360, 487, 665, 778, 1163), three in ORF3a (aa 7, 81, 121), one in M protein (aa 5) and one in N protein (aa 17) (Table 2). It is interesting to see that all but one of the 26 SNVs from masked palm civets were also conserved in sequences from the 2004 patients with mild symptoms (Table 2). Based on epidemiological and molecular data, the patients of the 2003 epidemic were classified as early, middle and late phases (Chinese SARS Molecular Epidemiology Consortium, 2004). Many of the 26 SNVs from masked palm civets were conserved in the genomes from early human cases in the 2003 epidemic. Some remained unchanged in the genomes of the middle phase patients but all were lost in the late phase patients. At least one of the mutations, aa 487 in the spike protein, was shown to be important in the adaptation of SARS-CoV to human cells (Li et al., 2005c, Qu et al., 2005). Analysis of the structures of the spike protein and its cellular receptor angiotensin-converting enzyme 2 (ACE2) (Li et al., 2003) indicated that aa 487 of spike is important for spike-ACE2 interaction and a mutation from Ser487 (civet) to Thr487 appeared to enhance human-to-human transmission of the SARS-CoV (Li et al., 2005c). Residues 344 and 360 of the spike protein are distant from the binding interface in the spike-ACE2 complex (Li et al., 2005a), and these two mutations do not affect binding affinity or infectivity of the SARS-CoV (Li et al., 2005c, Qu et al., 2005). Whether the rest of the aa residues play important roles during cross species transmission or human-to-human transmission shall need further investigations.
Table 2.
SNVs of SASR-CoVs from masked palm civets and human patientsa
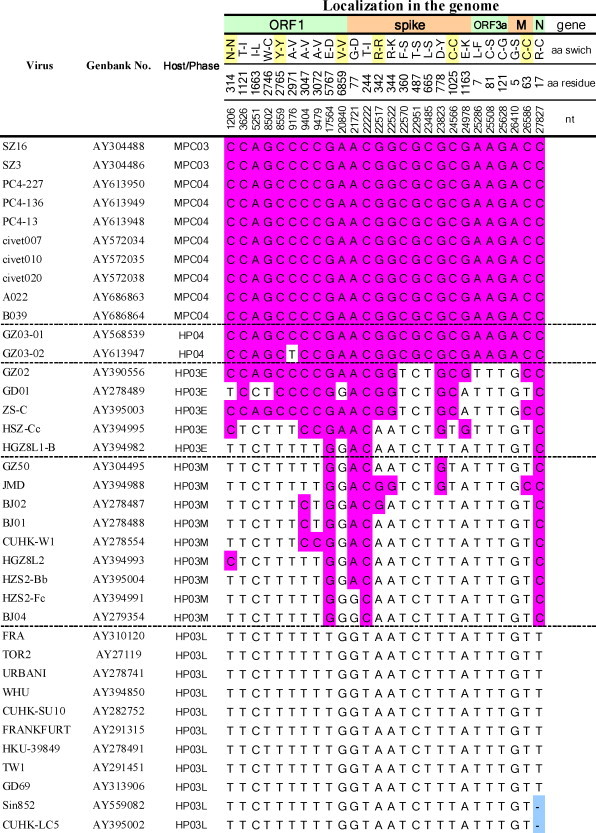
aMPC, masked palm civets; HP, human patients; 03, year 2003; 04, year 2004; E, M and L stands for early, middle and late phase of 2003 epidemic (according to the Chinese SARS Molecular Epidemiology Consortium, 2004).
All the so far identified masked palm civet SARS-CoV genomes except PC4-227 contain a 29-nt sequence which is missing in the epidemic strain of SARS-CoV. It is a 27-nt sequence in PC4-227 lacking the last two nts of the 29-nt. The 29-nt sequence was first identified in SZ3 and SZ16 (Guan et al., 2003) and was also found in some early epidemic human samples such as GD01, GZ02, HSZ-Bb, HSZ-Bc, HSZ-Cb, and HSZ-Cc (Chinese SARS Molecular Epidemiology Consortium, 2004). The 29-nt was suggested as a marker to distinguish the strain that originated from masked palm civet and the later epidemic strains (Guan et al., 2003, Chinese SARS Molecular Epidemiology Consortium, 2004). The 29-nt fuses ORF8a and ORF8b into a single ORF. A recent study by Yount et al. (2005) using recombinant virus has shown that the 29-nt had little if any impact on in vitro growth or RNA synthesis. It was predicted that the occurrences of deletions in ORF8a/8b region might be related to the RNA secondary structure of this region (Chinese SARS Molecular Epidemiology Consortium, 2004). Nevertheless, this 29-nt deletion reflected an important evolutionary event of SARS-CoV-like viruses in their adaptation from masked palm civets to humans. Interestingly, the bat SARS-CoV-like viruses also harbor a 29-nt sequence and encode a single ORF8, although the similarity of the ORF8 is very low to that of SARS-CoV-like viruses in masked palm civets (see below).
2.4. Evolutionary perspectives
The rate of evolution of a virus in a certain host could sometimes infer whether the host is a natural animal reservoir of the virus. Several studies have been conducted to estimate the substitution rate and to identify the most recent common ancestor of the SARS-CoV. By analyzing 10 genome sequences of the 2003 Chinese patient samples and their epidemiological information, it was determined that the synonymous substitution rate of SARS-CoV genome to be 8.26 × 10−6 (±2.16 × 10−6) nucleotides per site per day and the date for the most recent common ancestor was estimated to range from early June to the end of December 2002 (Chinese SARS Molecular Epidemiology Consortium, 2004). Zhao et al. (2004) analyzed 11 genomic sequences from different parts of the world during 2003 SARS epidemic and found that the mutation rate to be in the order of 0.80–2.38 × 10−3 nucleotide substitution per site per year (2.2–6.7 × 10−6 nucleotide substitutions per site per day) and the most recent common ancestor of the sequences was inferred to be present as early as the spring of 2002. Analysis on the Singapore isolates from the 2003 epidemic revealed that the mutation rate to be 5.7 × 10−6 nucleotide substitutions per site per day, with the rates for synonymous and non-synonymous mutations at 2.5 × 10−6 and 3.2 × 10−6 nucleotide substitutions per site per day, respectively (Vega et al., 2004). All the above studies used human samples with available clinical data from the 2003 outbreak and the substitution rates of SARS-CoV genome obtained from the various studies were in good agreement with each other. In comparison to other coronaviruses, the estimated substitution rate of SARS-CoV is lower than that in mouse hepatitis virus (Chen and Baric, 1995) and similar to that in the transmissible gastroenteritis virus (Enjuanes et al., 1992), but higher than that in the infectious bronchitis virus (Cavanagh et al., 1998).
Song et al. (2005) used the concatenated five major coding sequences (ORF1ab, S, E, M, and N) of SARS-CoV genomes to estimate the neutral mutation rate. The total length of the concatenated sequence accounted for 91.25% of the whole genome. By taking GZ02 as an outgroup, the number of synonymous substitutions per synonymous site, K s, was calculated for two 2003 masked palm civet SARS-CoVs (SZ16 and SZ3), three 2004 masked palm civet SARS-CoVs (PC4-136, PC4-227, PC4-13), and two 2004 human patient sequences (GZ03-01 and GZ03-02). Based on these K s values, the neutral mutation rate among this group of SARS-CoVs was estimated to be 8.00 × 10−6 nucleotides per site per day. This suggested that SARS-CoV evolves at a relatively constant neutral rate both in humans and in masked palm civet (Song et al., 2005).
The spike genes of late human patient isolates were undergoing negative selection during 2003 SARS outbreak (Hu et al., 2003). However, when spike sequences from masked palm civets were used as an outgroup to estimate the K a /K s (the ratios of the rates of nonsynonymous to synonymous changes) of the human isolates, the value was significantly over 1, indicating that the spike gene was under positive selection pressure when transmitted from civets to human (Yeh et al., 2004). Phylogenetic analysis showed that the viruses from masked palm civets collected in 2003 were different from those of 2004 indicating the viral transmission from animal-to-human occurred independently in these two instances (Song et al., 2005).
2.5. Isolation of SARS-CoV-like virus from masked palm civets
Isolation of virus is an important step for identifying the viral host. So far, propagation in cell culture of SARS-CoV-like viruses from masked palm civets seems to be difficult. Initially, it was reported that SZ16 replicated in FRhK-4 cells (Guan et al., 2003), but no further reports have been published. Wang et al. (2005b) reported that SARS-CoV-like virus from a rectal swab specimen of a masked palm civet replicated in FRhK-4 cells cultured but not in Vero E6 cells. Similarly, viruses from the four mild symptom patients of 2004 could not be propagated in cell lines (Liang et al., 2005). Because it has been reported that certain aa residues are important for the viral spike protein to interact with its specific receptor ACE2 (Li et al., 2005a, Li et al., 2005c, Li et al., 2006, Qu et al., 2005), selecting or engineering a cell line capable of supporting the interaction of receptor and S proteins of SARS-CoV-like viruses may be essential to overcome the obstacles in isolation of live viruses from civets and other animal sources.
3. Bats and SARS-CoV
Bats were recently identified as reservoirs of the SARS-like coronavirus (SL-CoV) or SARS-CoV-like virus by two independent research teams (Lau et al., 2005, Li et al., 2005b). Both teams reported their findings of coronaviruses, which are highly similar to SARS-CoV in several horseshoe bat species of the genus Rhinolophus within the family Rhinolophidae.
3.1. Prevalence of antiviral antibodies and viral nucleic acids in bats
The prevalence of anti-SARS-CoV nucleocapsid antibodies was examined in bats in the Provinces of Hubei, Guangdong Guangxi and Tianjing (Li et al., 2005b) and in the Hong Kong Special Administrative Region (Lau et al., 2005) (Table 3 ). A rather high prevalence of antibodies was observed in the lesser horseshoe bat Rhinolophus pussilus (2/6), the great-eared horseshoe bat Rhinolophus macrotis (5/7) and the Pearson's horseshoe bat Rhinolophus pearsoni (13/46) by a sandwich ELISA method (Li et al., 2005b). It was also seen in the Chinese horseshoe bat Rhinolophus sinicus (12/18 by Western blots, 31/37 positive by EIA with titer ≥1:400, and 8/19 by a neutralization assay for human SARS-CoV with titer ≥1:20) (Lau et al., 2005). Antiviral antisera from bats did not neutralize human SARS-CoV (Li et al., 2005b). A very low percentage of positive samples were detected in the fruit bat Rousettus leschenaultia (1.2%). No antibodies were detected in other bat species sampled, including the lesser dog-faced fruit bat Cynopterus sphinx, the mouse-eared bats Myotis ricketti, Myotis altarium and Myotis chinensis, the noctutule bats Nyctalus plancyi and N. noctula, the great round leaf bat Hipposideros armiger and the intermediate horseshoe Rhinolophus affinus (Table 3).
Table 3.
Detection of prevalence of antibodies and RNAs of SARS-CoV-like viruses in bats and genome identity between bat SL-CoVs and SARS-CoV
| Sampling location | Bat species | Antibody test positive/total (%) | RT-PCRa positive/total (%) | Genome size (nt) | Identity with SARS-CoV (%) |
|---|---|---|---|---|---|
| Guangxi | Rousettus leschenaulti | 1/142 (1.4%)b | 0/165 | ||
| Rhinolophus pearsoni | 13/46 (28.3%)b | 3/30 (10%) | 29,736 | 92 | |
| Rhinolophus pussilus | 2/6 (33.3%)b | 0/6 | |||
| Guangdong | Rousettus leschenaulti | 0/42b | 0/45 | ||
| Cynopterus sphinx | 0/17b | 0/27 | |||
| Tianjin | Myotis ricketti | ND | 0/21 | ||
| Hubei | R. pussilus | ND | 0/15 | ||
| Rhinolophus ferrumequinum | 0/4b | 1/8 (12.5%) | 29,743 | 87 | |
| Rhinolophus macrotis | 5/7 (71%)b | 1/8 (12.5%) | 29,749 | 87 | |
| Nyctalus plancyi | 0/1b | 0/1 | |||
| Miniopterus schreibersi | 0/1b | 0/1 | |||
| Myotis altarium | 0/1b | 0/1 | |||
| Hong Kong | Hipposideros armiger | ND | 0/12 | ||
| Miniopterus magnater | ND | 0/23 | |||
| Miniopterus pusillus | ND | 0/24 | |||
| Myotis chinensis | ND | 0/3 | |||
| M. ricketti | ND | 0/2 | |||
| Nyctalus noctula | ND | 0/2 | |||
| Rhinolophus affinus | ND | 0/2 | |||
| Rhinolophus sinicus | 12/18 (67%)c | 23/59 (39%) | 29,709 | 88 | |
Modified according to Lau et al. (2005) and Li et al. (2005b). ND, not done.
Sandwich ELISA based on SARS-CoV N protein (Li et al., 2005b).
Western blot with recombinant N protein of bat SL-CoV (Lau et al., 2005).
Faecal swabs were used for RT-PCR.
Both teams subsequently confirmed by RT-PCR with SARS specific primers the presence of SARS-CoV-like virus in some feacal swabs. A relatively low prevalence of viruses was detected in Rhinolophus ferrumequinum (1/8), R. macrotis (1/8) and R. pearsoni (3/30), compared with a high prevalence in R. sinicus (23/59) (Table 3). The authors pointed out that bats with neutralizing antibodies had a lower viral load in their anal swabs (Lau et al., 2005). The quantitative RT-PCR was more sensitive than routine RT-PCR method. No positive samples were found in 219 respiratory swabs collected in these studies (Table 3). Unfortunately, both teams failed in their attempt to isolate the viruses.
3.2. The sequences of the SARS-CoV-like viruses from bats
The complete genome sequences of three viruses from the R. sinicus bat were determined by direct sequencing of the RT-PCR products. The three genomes, termed HKU3-1, HKU3-2 and HKU3-3 were 29,709 bp in size (Lau et al., 2005). HUK3-1 and HUK3-3 are identical and HKU3-1 and HKU3-2 have only 18 nucleotide difference, indicating that these virus isolates belong to the same genotype. Three full-length genomes of bat SL-CoV were obtained from R. pearsoni (Rp3), R. ferrumequinum (Rf1) and R. macortis (Rm1) by Li et al. (2005b) and Ren et al. (2006). All full genomes have similar gene organization to the SARS-CoV and contained a 5′ leader sequence followed by non-structural proteins ORF1ab and the structural proteins S, E, M and N, and 3′ polyA tail. There are some SARS specific accessory proteins termed ORF3a, ORF6, ORF7a, ORF7b, ORF8, ORF9b and 9c between the S and N genes (the nomenclature of genes and ORFs followed the nomenclature used for other coronaviruses) (Spaan et al., 2005) (Fig. 1 ). All gene products of SARS-CoV and bat SL-CoV were similar in size except that bat SL-CoV HKU3-1 and Rf1 had a shorter ORF1a (4376 and 4377 aa respectively).
Fig. 1.
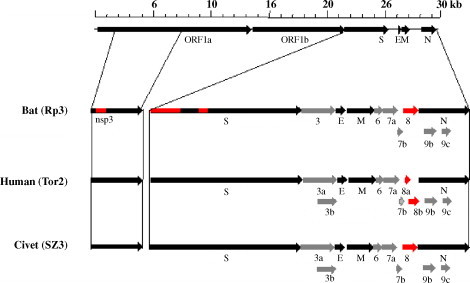
The genome organization of SARS-CoV in human and masked palm civet, and SL-CoV in bat. The black arrows indicate the conserved genes of the family Coronaviridae; the gray arrows indicate SARS-CoV unique genes; the red indicate the most variable regions in SARS-CoV and SL-CoV. The bar scale may not represent the real size of the genes.
The genome sequences of SL-CoVs isolated from horseshoe bat shared a sequence identity of 89–90%. Similar sequence identity, 87–92%, exists between bat and human or civet isolates. Except for the putative S1, ORF3, ORF8 and ORF9b and c, most of bat SL-CoVs gene products shared an aa identity of 90–100% with that of human SARS-CoVs or civet SARS-CoV-like viruses. The most conserved proteins were ORF1b (98–99% identity), E (98–100% identity), M (97–98% identity) and N (97–96% identity). The less conserved proteins were of ORF1a (94–96% identity), putative S2 (92–96% identity), ORF6 (92–93% identity), ORF7a (91–95% identity), 7b (90–93% identity), 9b (81–90% identity), 9c (80–91% identity) and ORF3a (81–86% identity). Moreover, the transcription regulatory sequences (TRS) and the stem-loop II motif (s2m) similar to SARS-CoV were identified in bat SL-CoVs. Phylogenetic analyses based on full sequences of bat SL-CoV revealed that bat SL-CoVs are closely related to SARS-CoVs in humans and civets and distantly related to other coronaviruses in group 2 (Fig. 2 ). Some authors suggested that these groups of viruses constitute a SARS coronavirus cluster, which are assembled into group 2 coronavirus within the genus Coronavirus (Li et al., 2005b). Others have suggested that these viruses form group 2b which are distinct from other group 2 coronavirus (group 2a) within the genus Coronavirus (Lau et al., 2005).
Fig. 2.
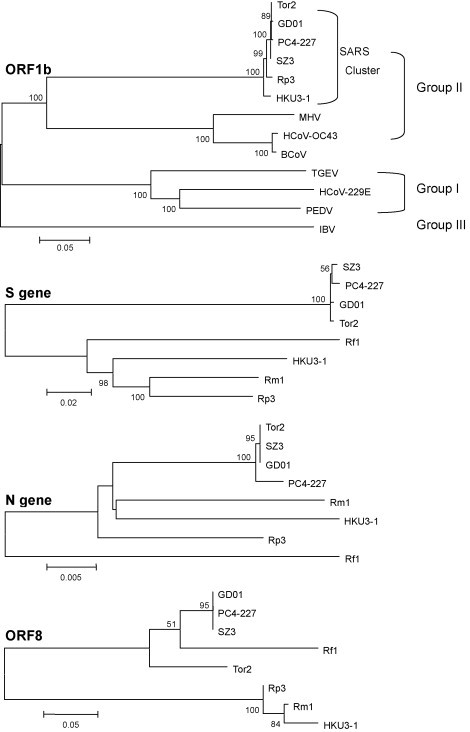
Phylogenetic trees based on aa sequence of ORF1b, nucleotide sequence of S, N and ORF8 gene. Sequence used for analysis in this study are as follows (GenBank accession number given in parenthesis): Tor2 (NC-004718), human isolate from late phase of the 2002/3 oubreaks; GD01 (AY278489), human isolate from early phase of the 2002/3 outbreaks; SZ3 (AY304486), masked palm civet isolate from 2003; PC4-227 (AY613950), masked palm civet isolate from 2004; HKU3-1 (DQ022305), isolate from Rhinolophus sinicus; Rp3 (DQ071615), isolate from Rhinolophus pearsoni; Rf1 partial sequence (DQ71611, DQ159956), isolate from Rhinolophus ferrumequinum; Rm1 partial sequence (DQ71612, DQ159957), isolate from Rhinolophus macrotis. The phylogenetic trees were constructed using the Neighbor-Joining algorithm in the MEGA3.1 software with a bootstrap of 1000 replicates. Other coronavirus sequences used in this study: HCoV-229E (AF304460), HCoV-OC43 (AY391777), PEDV (AF353511), MHV (AY700211), IBV (AY851295), and BCoV (AF391542) and TGEV (NC_002306). Genetic variation scales are indicated for each tree and different genetic scales are used for different trees.
Nucleotide variations are present throughout all bat SL-CoV genomes when compared with SARS-CoV. The most variable regions in bat SL-CoVs relative to SARS-CoVs are in the nonstructural protein nsp3, the spike protein (particularly in S1 domain, Fig. 3 ), ORF3 and ORF8. Most of deletion and insertion sites are present in nsp3, S and ORF8 (Fig. 1, Fig. 3), which result in a low similarity with SARS-CoV in the S protein (76–78%) and in ORF8 (35% for Rp3, HKU3-1, HKU3-2 and Rm1, 80% for Rf1). The 29-nt region, which is present only in SARS-CoV from masked palm civets and in the early phase human patients, also existed in all bat SL-CoVs and resulted in a fused ORF (ORF8). The low similarity in ORF8 between SARS-CoV from human/civet and bat SL-CoV is also seen between bat SL-CoV Rf1 and other three isolates. Analysis of ORF8 indicated that Rf1 was more closely related to SARS-CoV than to other bat SL-CoV isolates. The high mutated region in ORF8 further demonstrates that this locus is prone to deletions (Chiu et al., 2005, Lau et al., 2005, Li et al., 2005b, Song et al., 2005).
Fig. 3.
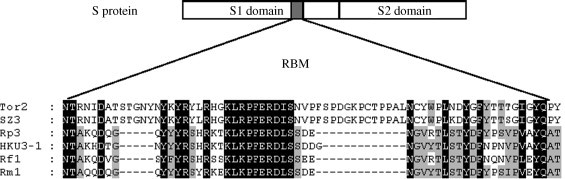
Comparison of the receptor binding motif of S protein of bat SL-CoV with that of SARS-CoV. Alignment between the receptor binding motif of S protein (RBM) of SARS-CoV Tor2, SZ3 (masked palm civet) and the corresponding regions of bat SL-CoVs S proteins is shown. The displayed RBM region represents aa 424–495 of the Tor2 spike protein sequence.
The key difference between bat SL-CoVs and SARS-CoVs in humans and masked palm civets is seen in the spike protein, which determines host range and tissue tropism in coronaviruses (Holmes, 2001). The putative S2 domain of all SL-CoVs has a high identity with that of SARS-CoVs (96%). All sites that are critical for SARS virus fusion to cell membranes are highly conserved in SL-CoVs putative S2 domain, suggesting that SL-CoVs possess the same fusion mechanism to host cells as SARS-CoVs. However, the putative S1 domain of SL-CoVs has low similarity to that of SARS-CoVs, especially in N-terminus and the receptor-binding domain (RBD, 318-510 aa of Tor2 S1 domain) (Wong et al., 2004). One insertion of 6 aa residues and three deletion sites are present in the S1 domain of bat SL-CoVs, among which two deletion sites (5 and 12–13 aa, respectively) are present in RBD particularly in the receptor binding motif (RBM, 424–494 of Tor2 S1 domain) (Li et al., 2005a) (Fig. 3). These differences may imply that bat SL-CoVs may use a vastly diverse ACE2 or even a different molecule as the entry receptor.
4. Are there other animal reservoirs of SARS-CoV?
4.1. Other animal species that have been investigated for SARS-CoV
Apart from masked palm civets and bats, 29 other animal species from 13 families, 8 animal orders and 2 classes that have been tested for SARS-CoV (Table 4 ). Among them, seven species were shown to harbor the virus under certain circumstances.
Table 4.
Summary of investigations of SARS-CoV in other animal speciesa
| Order | Family | Species | Common name | Methods | Prevalence | Reference |
|---|---|---|---|---|---|---|
| Primates | Cercopithecidae | Macaca mulatta | Rhesus macaque | RT-PCR | 0/20 | Lau et al. (2005) |
| Carnivora | Canidae | Canis familiaris | Dog | Antibody | 0/20 | Chen et al. (2005) |
| Real-time RT-PCR | 0/5 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Nyctereutes procyonoides | Raccoon dog | RT-PCR, virus isolation | 1/1 | Guan et al. (2003) | ||
| RT-PCR | 15/15 | Kan et al. (2005) | ||||
| Vulpes vulpes | Red fox | Real-time RT-PCR | 3/5 | Wang et al., 2005a, Wang et al., 2005b | ||
| Mustelidae | Melogale moschata | Chinese ferret-badger | Antibody | 1/2 | Guan et al. (2003) | |
| Arctonyx collaris | Hog-badger | RT-PCT, antibody | 0/3 | Guan et al. (2003) | ||
| Mustela vison | Mink | Real-time RT-PCR | 0/1 | Wang et al., 2005a, Wang et al., 2005b | ||
| Felidae | Felis catus | Domestic cat | RT-PCT, antibody | 0/4 | Guan et al. (2003) | |
| Real-time RT-PCR | 4/20 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Real-time RT-PCR | 0/13 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Real-time RT-PCR | 0/3 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Antibody | 0/11 | Chen et al. (2005) | ||||
| Artiodactyla | Suidae | Sus scrofa domestica | Pig | Antibody, RT-PCR | 2/108 | Chen et al. (2005) |
| S. scrofa | Wild boar | Real-time RT-PCR | 1/19 | Wang et al., 2005a, Wang et al., 2005b | ||
| Cervidae | Muntiacus reevesi | Chinese muntjac | RT-PCT, antibody | 0/2 | Guan et al. (2003) | |
| Antibody | 0/9 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Bovidae | Bos tarurs | Cattle | Real-time RT-PCR | 0/60 | Chen et al. (2005) | |
| Capra hircas | Goat | Real-time RT-PCR | 0/3 | Wang et al., 2005a, Wang et al., 2005b | ||
| Lagomorpha | Leporidae | Oryctolagus cuniculus | Rabbit | Real-time RT-PCR | 0/6 | Wang et al., 2005a, Wang et al., 2005b |
| Real-time RT-PCR | 0/5 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Lepus sinensis | Chinese hare | RT-PCT, antibody | 0/3 | Guan et al. (2003) | ||
| Rodentia | Castoridae | Castor fiber | Beaver | RT-PCT, antibody | 0/3 | Guan et al. (2003) |
| Muridae | Niviventer fulvescens | Chestnut spiny rat | RT-PCR | 0/12 | Lau et al. (2005) | |
| Rattus rattus flavipectus | Buff-bellied rat | RT-PCR | 0/4 | Lau et al. (2005) | ||
| Rattus sikkimensis | Sikkim rat | RT-PCR | 0/44 | Lau et al. (2005) | ||
| Rattus losea | Lesser rice field rat | Real-time RT-PCR | 1/6 | Wang et al., 2005a, Wang et al., 2005b | ||
| Real-time RT-PCR | 0/16 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Anseriformes | Anatidae | Anas domestica | Duck | Antibody | 0/30 | Chen et al. (2005) |
| Anas platyhynchos | Spotbill duck | Real-time RT-PCR | 0/13 | Wang et al., 2005a, Wang et al., 2005b | ||
| Real-time RT-PCR | 0/9 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Anser anser | Greylag goose | Real-time RT-PCR | 1/10 | Wang et al., 2005a, Wang et al., 2005b | ||
| Real-time RT-PCR | 0/14 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Galliformes | Phasianidae | Gallus domestiaus | Chicken | Antibody | 0/11 | Chen et al. (2005) |
| Gallus gallus | Red jungle fowl | Real-time RT-PCR | 0/46 | Wang et al., 2005a, Wang et al., 2005b | ||
| Francolinus pintadeanus | Chinese francolin | Real-time RT-PCR | 0/31 | Wang et al., 2005a, Wang et al., 2005b | ||
| Real-time RT-PCR | 0/2 | Wang et al., 2005a, Wang et al., 2005b | ||||
| Phasianus colchicus | Common pheasant | Real-time RT-PCR | 0/8 | Wang et al., 2005a, Wang et al., 2005b | ||
| Pavo muticus | Green peafowl | Real-time RT-PCR | 0/2 | Wang et al., 2005a, Wang et al., 2005b | ||
| Columbiformes | Columbidae | Columba livia | Pigeons | Real-time RT-PCR | 0/6 | Wang et al., 2005a, Wang et al., 2005b |
aThis table does not include masked palm civets and bats.
4.1.1. Raccoon dogs
Raccoon dogs (N. procyonoides), belong to Carnivora, Canidae. There are three main subspecises in China: N. p. procyonoides, N. p. ussuriensis, and N. p. orestes. N. p. procyonoides is naturally distributed in the east and the central-south parts of China, including Fujian, Jiangxi, Hubei, Hunan, Jiangsu, Zhejiang, Anhui, Guangxi and Guangdong Provinces. N. p. ussuriensis is naturally distributed in the northeast part of China including Heilongjiang, Liaoning and Jinlin. While N. p. orestes is naturally distributed in the southwest part of China including Yunnan, Guizhou and Sichuan Provinces (Gao et al., 1987). Naturally, raccoon dogs are omnivores and change their food according to the seasonal food availability. Rodents, frogs, lizards, invertebrates, birds and their eggs are often eaten, specially in the early summer. Plants and mushrooms are eaten, while berries and fruits are favored in late summer and autumn. Raccoon dogs are nocturnal and they hibernate in winter. They are strictly monogamous and live in pairs (Kauhala and Saeki, 2004).
Farming of the raccoon dogs started in 1950s in the northeast part of China for fur. The local wild raccoon dogs (N. p. ussuriensis) were domesticated successfully in early 1960s in Jilin and Heilongjiang Provinces (Tong, 1990). In 1988, the farmed raccoon dogs were over 300 million in China (Tong, 1990). Unlike the farmed masked palm civets which were mainly for food and almost disappear after 2004 in China, the farmed raccoon dogs are mainly for fur and are still in very large abundance, specially in northeast China.
Two independent surveys have shown that raccoon dogs from live animal markets harbored SARS-CoV at a very high prevalence (100%) (Guan et al., 2003, Kan et al., 2005). In May 2003, Guan et al. isolated a SARS-CoV-like virus from the feacal sample of a raccoon dog (SZ13) from the Dongmen animal Market, in Shenzhen. The neutralization antibody titer of the serum of the raccoon dog was ≥640 against the SARS-CoV-like virus of a masked palm civet (SZ16) (Guan et al., 2003). In January 2004, Kan et al. investigated 15 raccoon dogs from the Xinyuan animal market, Guangzhou. Of the 15 raccoon dogs, by real-time and nested RT-PCR, 12 were tested positive with both throat and rectal swabs, while 3 tested positive with throat swabs only (Kan et al., 2005). So far there are three spike sequences of raccoon dogs, that of SZ13 (Guan et al., 2003), A030G (Kan et al., 2005), and A031G (Kan et al., 2005). All of them, however, were found to be identical to certain sequences found in masked palm civets. For example, the sequence of SZ13 is identical to that of SZ16 (Guan et al., 2003), and A030G and A031G are identical to A022G (Kan et al., 2005). It remains unclear whether the raccoon dogs were infected by the virus from the masked palm civets or the other way around. It is also possible that both animals were infected by the virus from a hitherto unidentified host. Unlike the situation with masked palm civets, very limited research has been conducted to survey wild or farmed raccoon dog.
4.1.2. Other investigated animals
A survey conducted on 5 January 2004 in a Guangzhou market before the mass culling of masked palm civets demonstrated that red foxes, domestic cats, and Lesser rice field rats may also harbor the virus (Wang et al., 2005a). Real-time RT-PCR revealed that 4 out of 20 cats, 3 out of 5 red foxes, and 1 out of 6 Lesser rice field rats were positive for SARS-CoV. The N and P genes sequences were determined and shown to be identical to those of the human SARS-CoVs. After culling of animals and disinfection of the market place, follow up surveillance was conducted 2 weeks later and only one greylag goose out of 119 animals tested positive for SARS-CoV. At this time cats did not test positive for the virus (Wang et al., 2005a). Also, the same team reported that in 2004, one wild boar out of 102 animals tested positive (Wang et al., 2005a) (Table 4). Since the environment of the animal market was heavily contaminated with SARS-CoV-like viruses during that period (Kan et al., 2005), it remains unknown whether those animals that tested positive by PCR were truly susceptible hosts or simply mechanical carriers of SARS-CoV genetic materials. In this regard, it is worth to note that under experimental conditions, goose was not susceptible to SARS-CoV infection (Swayne et al., 2004).
In another study carried out in a village close to Beijing (Chen et al., 2005), 2 of 108 pigs sampled were positive for the SARS-CoV by antibody analysis and by RT-PCR during a survey which also included 60 cattle, 20 dogs, 11 cats, 11 chickens and 30 ducks. Based on the sequence and available epidemiological data, the authors suggested that the pigs were infected from feed materials contaminated by human SARS patients and were not a natural host of the SARS-CoV.
It needs to be pointed out that there were no standard screening regimens being used for detecting SARS-CoVs in the open markets. During the SARS epidemic and shortly after, many teams were involved into the animal reservoir investigation, and different teams used their own sampling and screening methods. The culling circumstances also added the complexity of the sampling. Therefore, some of the open market data need to be further proven.
4.2. Experimental infection of animals
Experimental data have indicated that SARS-CoV might infect a wide-range of hosts, including masked palm civets (Wu et al., 2005), monkeys (Fouchier et al., 2003, McAuliffe et al., 2004, Qin et al., 2005, Rowe et al., 2004), cats and ferrets (Martina et al., 2003), mice (Glass et al., 2004, Roberts et al., 2005a, Subbarao et al., 2004, Wentworth et al., 2004), pigs and chickens (Weingartl et al., 2004), guinea pigs (Liang et al., 2005), and Golden Syrian Hamsters (Roberts et al., 2005b). However, as most experimental tests were performed to find a suitable animal model for vaccine evaluation, not much is known about transmission of the virus from the inoculated animals. Pigs and chickens, for example, may support SARS-CoV replication to a very limited degree but are not likely to play a role as an amplifying host (Weingartl et al., 2004). A study by Swayne et al. (2004), showed that by intratracheally injecting SARS-CoV into chickens, turkeys, geese, ducks, and quail, failed to cause disease or replicate with all the avian species, suggesting domestic poultry are unlikely the reservoir of the SARS-CoV.
5. Conclusions
Evidence so far indicated that the origin of the 2003 SARS epidemic and the 2004 mild SARS cases was from masked palm civets (Guan et al., 2003, Kan et al., 2005, Song et al., 2005, Wang et al., 2005b). It is still uncertain whether the masked palm civets were infected during trading in animal market or were naturally infected in the wild or under farmed conditions. The very high nt sequence identity among the masked palm civet SARS-CoV-like viruses (>99.6%) clearly suggested that the virus has not resided in the masked palm civet population for a very long time. Nevertheless, it is quite clear that masked palm civets are highly susceptible to the SARS-CoV (Wu et al., 2005, Kan et al., 2005) where the virus can be amplified to very high titers (Kan et al., 2005). Therefore, detailed surveys of the SARS-CoV in masked palm civets will continue to play an important role in the prevention of potential future SARS outbreaks.
Bats, the only flying mammals, constitute about 20% of the mammalian diversity (Kunz and Fenton, 2003). They are distributed widely in the world and most of them are gregarious. Because of unknown mechanisms, they appear to be natural reservoirs of several new and re-emerging viruses, such as Hendra virus, Nipah virus, Menangle virus, lyssavirus and Ebola virus, etc. over the last 20 years (Leroy et al., 2005, Mackenzie et al., 2001). Horseshoe bats have been found to be a natural reservoir of the SARS-CoV-like virus. The genetic diversity of coronaviruses found in bats highlighted our poor understanding of viruses in wild animals. Moreover, bats that harbor the viruses rarely display clinical signs or overt symptoms of pathology. Even though there was little direct evidence to indicate that viruses in bats can cause the severe disease in other domestic animals and in humans, there is an increased possibility of virus variants crossing the species barrier and causing outbreaks in humans as people come into closer contact with wild animals. It is therefore essential to study the viral pathology in the host and to enhance our knowledge and understanding of reservoir host distribution, animal-to-animal and human-to-animal interactions. It is also interesting to find out why bats are such a natural reservoir for so many viruses. It is intriguing to speculate if this is related to the bats’ immune system and/or to their ecology. During the search for the natural reservoir of the SARS-CoV, the investigators also found several new types of coronavirus which are closely related to group 1 and group 2 coronaviruses, further indicating that bats can harbour a much wider spectrum of coronaviruses (Lau et al., 2005, Poon et al., 2005). It is not known whether the coronaviruses in bats would cause a zoonotic disease in humans or in other animals. All these uncertainties underscore the need for further investigations to understand the ecology and pathogenicity of these viruses.
So far the sequence data showed that the average genome identity of the SARS-CoV-like virus from horseshoe bat to the SARS-CoV is about 87–92%. As it was pointed out by Li et al. (2006) that substantial genetic changes in the spike protein of bat SL-CoV are likely to be necessary for the virus to infect humans. Further investigations might reveal viruses with higher identity to SARS-CoV in bats or with lower identity in masked palm civets. However, it is likely that in the emerging path of SARS-CoV, there are still other species missing between horseshoe bats and masked palm civets. One way of revealing possible links and suspects is to look at the ecological circles of both bats and masked palm civets. Alternatively, constant survey of wild animal species for SARS-CoV-like viruses should provide further information on animal reservoirs.
Acknowledgements
The authors thank the supports for the European Commission DISSECT (no. SP22-CT-2004-511060) and EPISARS (no. SP22-CT-2004-511603) contracts, the Programme de Recherche en Réseaux Franco-Chinois (Épidémie du SRAS: de l’émergence au contrôle), and MOST projects (2003CB514118, 2005CB523004, and a special grant for “Animal Reservoir of SARS-CoV”). We would like to thank Dr. Basil M. Arif for scientific editing.
References
- Cavanagh D., Mawditt K., Adzhar A., Gough R.E., Picault J.P., Naylor C.J., Haydon D., Shaw K., Britton P. Does IBV change slowly despite the capacity of the spike protein to vary greatly? Adv. Exp. Med. Biol. 1998;440:729–734. doi: 10.1007/978-1-4615-5331-1_94. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC), 2003. Prevalence of IgG Antibody to SARS-Associated Coronavirus in Animal Traders—Guangdong Province, China, 2003. Morbidity and Mortality Weekly Report 52, 986–987. [PubMed]
- Chen W., Baric R.S. Function of a 5′-end genomic RNA mutation that evolves during persistent mouse hepatitis virus infection in vitro. J. Virol. 1995;69:7529–7540. doi: 10.1128/jvi.69.12.7529-7540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Yan M., Yang L., Ding B., He B., Wang Y., Liu X., Liu C., Zhu H., You B., Huang S., Zhang J., Mu F., Xiang Z., Feng X., Wen J., Fang J., Yu J., Yang H., Wang J. SARS-associated coronavirus transmitted from human to pig. Emerg. Infect. Dis. 2005;11:446–448. doi: 10.3201/eid1103.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Chiu R.W., Chim S.S., Tong Y.K., Fung K.S., Chan P.K., Zhao G.P., Lo Y.M. Tracing SARS-coronavirus variant with large genomic deletion. Emerg. Infect. Dis. 2005;11:168–170. doi: 10.3201/eid1101.040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Duan W.W., Zhu K.M., Li L., Jiang W.X., Xiang X.C. The first incident of virus enteritis in masked civet: its diagnosis and prevention. J. Econ. Anim. 2001;5:34–38. [Google Scholar]
- Enjuanes L., Sune C., Gebauer F., Smerdou C., Camacho A., Anton I.M., Gonzalez S., Talamillo A., Mendez A., Ballesteros M.L., Sanchez C. Antigen selection and presentation to protect against transmissible gastroenteritis coronavirus. Vet. Microbiol. 1992;33:249–262. doi: 10.1016/0378-1135(92)90053-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. Science Press; Beijing: 1987. Fauna Sinica Mammalia Carnivora. p. 8. [Google Scholar]
- Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004;173:4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S., Poon L.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Halpin K., Young P.L., Field H.E., Mackenzie J.S. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- Holmes, K.V., 2001. Coronaviridae and their replication. In: Knipe, D.M., Roizman, B., Howley, P.M., Straus, S.E., Fields, B.N., Griffin, D.E., Martin, M.A., L.R.A. (Eds.), Fields Virology. Lippincott Williams & Wilkins, Philadelphia, Pennsylvania, USA, pp. 1187–1203.
- Hu L.D., Zheng G.Y., Jiang H.S., Xia Y., Zhang Y., Kong X.Y. Mutation analysis of 20 SARS virus genome sequences: evidence for negative selection in replicase ORF1b and spike gene. Acta Pharmacol. Sin. 2003;24:741–745. [PubMed] [Google Scholar]
- Hu W., Bai B., Hu Z., Chen Z., An X., Tang L., Yang J., Wang H., Wang H. Development and evaluation of a multitarget real-time Taqman reverse transcription-PCR assay for detection of the severe acute respiratory syndrome-associated coronavirus and surveillance for an apparently related coronavirus found in masked palm civets. J. Clin. Microbiol. 2005;43:2041–2046. doi: 10.1128/JCM.43.5.2041-2046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Miyazawa T., Nakamura K., Naito R., Inoshima Y., Tung K.C., Lee W.M., Chen M.C., Kuo T.F., Lin J.A., Mikami T. Serosurvey for selected virus infections of wild carnivores in Taiwan and Vietnam. J. Wildl. Dis. 1999;35:578–681. doi: 10.7589/0090-3558-35.3.578. [DOI] [PubMed] [Google Scholar]
- Jiang Z.-G., Li C.-W., Zeng Y. Status of the research on masked palm civets. Chin. J. Zool. 2003;38:120–122. [Google Scholar]
- Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Cui B., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Qi X., Chen K., Du L., Gao K., Zhao Y.T., Zou X.Z., Feng Y.J., Gao Y.F., Hai R., Yu D., Guan Y., Xu J. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Li W., Qu X., Zhang S. Study of the hibernation habit and characteristics of Paguma larvata. J. Hunan Agric. Univ. 1997;23:366–369. [Google Scholar]
- Kauhala K., Saeki M. Raccoon dogs. In: Macdonald D.W., Sillero-Zubiri C., editors. The Biology and Conservation of Wild Canids. Oxford University Press Inc.; New York, USA: 2004. [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kunz T.H., Fenton M.B. University of Chicago Press; Chicago: 2003. Bat Ecology. [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Delicat A., Paweska J.T., Gonzalez J.P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li W., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J. Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. Embo J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Chen Q., Xu J., Liu Y., Lim W., Peiris J.S., Anderson L.J., Ruan L., Li H., Kan B., Di B., Cheng P., Chan K.H., Erdman D.D., Gu S., Yan X., Liang W., Zhou D., Haynes L., Duan S., Zhang X., Zheng H., Gao Y., Tong S., Li D., Fang L., Qin P., Xu W. Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerg. Infect. Dis. 2004;10:1774–1781. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., He C., Lei M., Li S., Hao Y., Zhu H., Duan Q. Pathology of guinea pigs experimentally infected with a novel reovirus and coronavirus isolated from SARS patients. DNA Cell Biol. 2005;24:485–490. doi: 10.1089/dna.2005.24.485. [DOI] [PubMed] [Google Scholar]
- Liu Y.X. Captive breeding of masked palm civet. Chin. J. Zool. 1959;9:425. [Google Scholar]
- Mackenzie J.S., Chua K.B., Daniels P.W., Eaton B.T., Field H.E., Hall R.A., Halpin K., Johansen C.A., Kirkland P.D., Lam S.K., McMinn P., Nisbet D.J., Paru R., Pyke A.T., Ritchie S.A., Siba P., Smith D.W., Smith G.A., van den Hurk A.F., Wang L.F., Williams D.T. Emerging viral diseases of Southeast Asia and the Western Pacific. Emerg. Infect. Dis. 2001;7:497–504. doi: 10.3201/eid0707.017703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.S., Field H.E. Emerging encephalitogenic viruses: lyssaviruses and henipaviruses transmitted by frugivorous bats. Arch. Virol. Suppl. 2004;18:97–111. doi: 10.1007/978-3-7091-0572-6_8. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G., Peiris J.S., Lim W., Osterhaus A.D. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe J., Vogel L., Roberts A., Fahle G., Fischer S., Shieh W.J., Butler E., Zaki S., St Claire M., Murphy B., Subbarao K. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. 2004;330:8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L., Chu D.K., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H., Lau S.K., Woo P.C., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S. Identification of a novel coronavirus in bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Wang J., Wei Q., She M., Marasco W.A., Jiang H., Tu X., Zhu H., Ren L., Gao H., Guo L., Huang L., Yang R., Cong Z., Guo L., Wang Y., Liu Y., Sun Y., Duan S., Qu J., Chen L., Tong W., Ruan L., Liu P., Zhang H., Zhang J., Zhang H., Liu D., Liu Q., Hong T., He W. An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. J. Pathol. 2005;206:251–259. doi: 10.1002/path.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X.X., Hao P., Song X.J., Jiang S.M., Liu Y.X., Wang P.G., Rao X., Song H.D., Wang S.Y., Zuo Y., Zheng A.H., Luo M., Wang H.L., Deng F., Wang H.Z., Hu Z.H., Ding M.X., Zhao G.P., Deng H.K. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280:29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Li W., Yu M., Hao P., Zhang Y., Zhou P., Zhang S., Zhao G., Zhong Y., Wang S., Wang L.F., Shi Z. Full genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J. Gen. Virol. 2006;87:3355–3359. doi: 10.1099/vir.0.82220-0. [DOI] [PubMed] [Google Scholar]
- Roberts A., Paddock C., Vogel L., Butler E., Zaki S., Subbarao K. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 2005;79:5833–5838. doi: 10.1128/JVI.79.9.5833-5838.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Vogel L., Guarner J., Hayes N., Murphy B., Zaki S., Subbarao K. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 2005;79:503–511. doi: 10.1128/JVI.79.1.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Rowe T., Gao G., Hogan R.J., Crystal R.G., Voss T.G., Grant R.L., Bell P., Kobinger G.P., Wivel N.A., Wilson J.M. Macaque model for severe acute respiratory syndrome. J. Virol. 2004;78:11401–11404. doi: 10.1128/JVI.78.20.11401-11404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, M., Li, J., 1997. Raising and management of masked palm civet (P. larvata). J. Shaanxi Normal Univ. (Nat. Sci. ed.) 25 (Suppl.), 157–161.
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., Zheng H.J., Chern S.W., Cheng F., Pan C.M., Xuan H., Chen S.J., Luo H.M., Zhou D.H., Liu Y.F., He J.F., Qin P.Z., Li L.H., Ren Y.Q., Liang W.J., Yu Y.D., Anderson L., Wang M., Xu R.H., Wu X.W., Zheng H.Y., Chen J.D., Liang G., Gao Y., Liao M., Fang L., Jiang L.Y., Li H., Chen F., Di B., He L.J., Lin J.Y., Tong S., Kong X., Du L., Hao P., Tang H., Bernini A., Yu X.J., Spiga O., Guo Z.M., Pan H.Y., He W.Z., Manuguerra J.C., Fontanet A., Danchin A., Niccolai N., Li Y.X., Wu C.I., Zhao G.P. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W.J.M., Brian D., Cavanagh D., de Groot R.J., Enjuanes L., Gorbalenya A.E., Holmes K.V., Masters P., Rottier P., Taguchi F., Talbot P. Coronaviridae. In: Fauquet C.M., Mayo M.A., Maniloff J., Desselberger U., Ball C.A., editors. Virus Taxonomy. Classification and Nomenclature of Viruses. Elsevier Academic Press; London, UK: 2005. pp. 947–964. [Google Scholar]
- Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D.E., Suarez D.L., Spackman E., Tumpey T.M., Beck J.R., Erdman D., Rollin P.E., Ksiazek T.G. Domestic poultry and SARS coronavirus, southern China. Emerg. Infect. Dis. 2004;10:914–916. doi: 10.3201/eid1005.030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y.R. The resources of the precious fur animals in China. In: Tong Y.R., Qian G.C., editors. A Comprehensive Breeding Technique of Chinese Fur Animals. Chinese Agricultural Science and Technology Press; Beijing, China: 1990. pp. 15–24. (chapter 1) [Google Scholar]
- Tu, C., Crameri, G., Kong, X., Chen, J., Sun, Y., Yu, M., Xiang, H., Xia, X., Liu, S., Ren, T., Yu, Y., Eaton, B.T., Xuan, H., Wang, L.F., 2004. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis. 10, 2244–2248. [DOI] [PMC free article] [PubMed]
- Vega V.B., Ruan Y., Liu J., Lee W.H., Wei C.L., Se-Thoe S.Y., Tang K.F., Zhang T., Kolatkar P.R., Ooi E.E., Ling A.E., Stanton L.W., Long P.M., Liu E.T. Mutational dynamics of the SARS coronavirus in cell culture and human populations isolated in 2003. BMC Infect. Dis. 2004;4:32. doi: 10.1186/1471-2334-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Jing H.Q., Xu H.F., Jiang X.G., Kan B., Liu Q.Y., Wan K.L., Cui B.Y., Zheng H., Cui Z.G., Yan M.Y., Liang W.L., Wang H.X., Qi X.B., Li Z.J., Li M.C., Chen K., Zhang E.M., Zhang S.Y., Hai R., Yu D.Z., Xu J.G. Surveillance on severe acute respiratory syndrome associated coronavirus in animals at a live animal market of Guangzhou in 2004. Zhonghua Liu Xing Bing Xue Za Zhi. 2005;26:84–87. [PubMed] [Google Scholar]
- Wang M., Yan M., Xu H., Liang W., Kan B., Zheng B., Chen H., Zheng H., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Liu Y.F., Guo R.T., Liu X.N., Zhan L.H., Zhou D.H., Zhao A., Hai R., Yu D., Guan Y., Xu J. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 2005;11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H.M., Copps J., Drebot M.A., Marszal P., Smith G., Gren J., Andova M., Pasick J., Kitching P., Czub M. Susceptibility of pigs and chickens to SARS coronavirus. Emerg. Infect. Dis. 2004;10:179–184. doi: 10.3201/eid1002.030677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth D.E., Gillim-Ross L., Espina N., Bernard K.A. Mice susceptible to SARS coronavirus. Emerg. Infect. Dis. 2004;10:1293–1296. doi: 10.3201/eid1007.031119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO), 2004. Summary of probable SARS cases with onset of illness from November 1, 2002 to July 31, 2003 (accessed April 21, 2004, at http://www.who.int/csr/sars/country/table2004_04_21/en/index.html).
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Tu C., Xin C., Xuan H., Meng Q., Liu Y., Yu Y., Guan Y., Jiang Y., Yin X., Crameri G., Wang M., Li C., Liu S., Liao M., Feng L., Xiang H., Sun J., Chen J., Sun Y., Gu S., Liu N., Fu D., Eaton B.T., Wang L.F., Kong X. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J. Virol. 2005;79:2620–2625. doi: 10.1128/JVI.79.4.2620-2625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.F., Wang M., Zhang Z.B., Zou X.Z., Gao Y., Liu X.N., Lu E.J., Pan B.Y., Wu S.J., Yu S.Y. An epidemiologic investigation on infection with severe acute respiratory syndrome coronavirus in wild animals traders in Guangzhou. Zhonghua Yu Fang Yi Xue Za Zhi. 2004;38:81–83. [PubMed] [Google Scholar]
- Xu R.H., He J.F., Evans M.R., Peng G.W., Field H.E., Yu D.W., Lee C.K., Luo H.M., Lin W.S., Lin P., Li L.H., Liang W.J., Lin J.Y., Schnur A. Epidemiologic clues to SARS origin in China. Emerg. Infect. Dis. 2004;10:1030–1037. doi: 10.3201/eid1006.030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.Q., Yang Z.C., Yang J.L., Qi S.D. Outbreak of bacterial disease in masked palm civets. J. Tradition. Chin. Vet. Med. 2000;19:36. [Google Scholar]
- Yeh S.H., Wang H.Y., Tsai C.Y., Kao C.L., Yang J.Y., Liu H.W., Su I.J., Tsai S.F., Chen D.S., Chen P.J. Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: molecular epidemiology and genome evolution. Proc. Natl. Acad. Sci. USA. 2004;101(8):2542–2547. doi: 10.1073/pnas.0307904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Roberts R.S., Sims A.C., Deming D., Frieman M.B., Sparks J., Denison M.R., Davis N., Baric R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005;79(23):14909–14922. doi: 10.1128/JVI.79.23.14909-14922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.L., Su X.L., Gao G.C., Zhang W.H. Study of the living habits and hibernation habit of Paguma larvata. Chin. J. Zool. 1991;26:19–22. [Google Scholar]
- Zhang B.L., Su X.L., Gao G.C., Zhang W.H. Study of the distribution and population structure of Paguma larvata. Chin. J. Zool. 1991;26:42–45. [Google Scholar]
- Zhang R. vol. 238. Academy Press; 1999. (Zoogeography of China). p. 477. [Google Scholar]
- Zhao Y. Investigation and treatment of parasites diseases of masked palm civets. Jilin Anim. Husbandry Vet. Med. 2003;12:39. [Google Scholar]
- Zhao Z., Li H., Wu X., Zhong Y., Zhang K., Zhang Y.P., Boerwinkle E., Fu Y.X. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol. Biol. 2004;4:21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris, Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. Technologies for breeding masked palm civets. Sichuan J. Zool. 1994;1994:138–139. [Google Scholar]


