Abstract
Numerous viruses of human or animal origin can spread in the environment and infect people via water and food, mostly through ingestion and occasionally through skin contact. These viruses are released into the environment by various routes including water run‐offs and aerosols. Furthermore, zoonotic viruses may infect humans exposed to contaminated surface waters. Foodstuffs of animal origin can be contaminated, and their consumption may cause human infection if the viruses are not inactivated during food processing. Molecular epidemiology and surveillance of environmental samples are necessary to elucidate the public health hazards associated with exposure to environmental viruses. Whereas monitoring of viral nucleic acids by PCR methods is relatively straightforward and well documented, detection of infectious virus particles is technically more demanding and not always possible (e.g. human norovirus or hepatitis E virus). The human pathogenic viruses that are most relevant in this context are nonenveloped and belong to the families of the C aliciviridae, A denoviridae, H epeviridae, P icornaviridae and R eoviridae. Sampling methods and strategies, first‐choice detection methods and evaluation criteria are reviewed.
Keywords: food borne virus, faecal‐oral transmission, nonenveloped virus, gastroenteritis, hepatitis molecular detection
Short abstract
Virus hazards from food, water and the environment, their reservoirs and routes of transmission; Sampling methods and sampling strategies thereof, including the first choice test methods, and criteria for data evaluation are described.
Introduction: main food and environmental virus hazards
Food and environmental virology mostly studies viruses that can be transmitted through water, sewage, soil, air, fomites (objects capable of transmitting microbial pathogens) or food (Bidawid et al., 2009). Most such viruses are enteric viruses transmitted via the faecal–oral route. Infected humans can excrete large amounts of human pathogenic viruses; animal and plant material as well as other excreta and secreta can also carry high viral loads (Breitbart et al., 2003; Zhang et al., 2006; de Roda Husman & Bartram, 2008). Viruses transmitted via the faecal–oral route are generally nonenveloped and thus very stable in the environment (Rzeżutka & Cook, 2004) and include major aetiological agents, some of which are thought to be emerging zoonotic pathogens. These viruses cannot always be effectively eliminated by current methods of sewage treatment (Vantarakis & Papapetropoulou, 1999; Thompson et al., 2003; Van Heerden et al., 2003; Van den Berg et al., 2005) and consequently cause viral contamination of the environment from treated as well as untreated wastewater. Other examples of indirect routes are run‐off from manure used in agriculture. There is also direct faecal contamination of the environment from humans and animals, for example by bathers or by defecation of free‐range or wild animals onto soil or surface waters. The resulting viral contamination of sea and coastal water, rivers and other surface waters, groundwaters, and irrigated vegetables and fruit is associated with subsequent risks of reintroduction of the viral pathogens into human and animal populations (Yates et al., 1985; Metcalf et al., 1995; Muscillo et al., 1997; Koopmans et al., 2002; La Rosa et al., 2007). Human exposure to even low levels of these pathogenic viruses in the environment, such as norovirus (NoV), can cause infection and disease (Lindesmith et al., 2003; Teunis et al., 2008). Individuals with an impaired immune system, including children, the elderly, pregnant women and people with HIV/AIDS, are more susceptible to such infections, and the disease outcome may be more severe. This is the case, for example, for rotavirus (RV), which is a more serious problem for young children in developing than in developed countries (Havelaar & Melse, 2003). Genetic susceptibility may also play a role in the susceptibility to infection, as in the case of NoV and the ABO histo‐blood group receptor genotype (Hutson et al., 2002).
Environmentally transmitted viruses include major aetiological agents of mild diseases such as gastroenteritis as well as agents of more severe diseases such as meningitis and hepatitis. Most of these viruses belong to the families Adenoviridae, Caliciviridae, Hepeviridae Picornaviridae and Reoviridae (Dubois et al., 1997; Muscillo et al., 2001; Lodder & de Roda Husman, 2005). The major enteric virus families include one or several types and variants of virus; the different groups may differ as concerns persistence, pathogenicity and infectivity. Some of these viruses, such as hepatitis E virus (HEV) (the sole member of the Hepeviridae), are thought to be zoonotic pathogens. New human pathogenic viruses that may also be transmitted via the environment emerge frequently (McKinney et al., 2006). Enteric viruses are predominantly transmitted via the faecal–oral route and are present in wastewater; therefore, such water is a potential source of infection if not treated or used appropriately (Gantzer et al., 1998; Baggi et al., 2001; Asano & Cotruvo, 2004). These agents are adapted to the hostile environment of the gut and in most cases, can persist for a very long time in water, soil or food matrices (Raphael et al., 1985; Richards, 2001; Le Cann et al., 2004; Van Zyl et al., 2006; Espinosa et al., 2008; Hansman et al., 2008).
Caliciviruses: major viral causes of gastroenteritis
NoV and sapovirus (SaV) are the most important human agents of diarrhoea worldwide (Patel et al., 2009). NoVs are the leading cause of food‐borne outbreaks of acute gastroenteritis and the most common cause of sporadic infectious gastroenteritis affecting people of all age group (Green, 2007; Patel et al., 2008, 2009). SaVs are mainly associated with sporadic acute gastroenteritis in young children (Hansman et al., 2007a; Khamrin et al., 2007; Monica et al., 2007) and are less commonly involved than NoV in epidemic gastroenteritis (Green, 2007), although some outbreaks have been described (Johansson et al., 2005; Hansman et al., 2007b, c). The burden of calicivirus (including NoV) has been clearly documented in numerous geographical areas worldwide (Hall et al., 2005; EFSA, 2009; Scallan et al., 2011).
NoVs and SaVs are icosaedric nonenveloped viruses with an ssRNA (+) genome of between 7.3 and 8.3 kb. They are both classified within the family of the Caliciviridae, as the genera Norovirus and Sapovirus, each subdivided into five genogroups (Karst et al., 2003) and several serotypes. Three genogroups (GI, GII and GIV) containing more than 20 genotypes of NoV are known to infect human beings, and the intra‐genotype nucleotide diversity can be as high as 15% (Zheng et al., 2006). Most human infections are caused by GI and GII, whereas GIII affects swine. In the case of SaV, at least four distinct genogroups containing a number of genotypes and variants can infect humans (Farkas et al., 2004). Thus, NoV and SaV detection can be difficult owing to the large number of genogroups and genotypes; furthermore, currently available detection methods are not sufficiently powerful, and indeed, the prevalence of uncommon NoV variants is probably underestimated (La Rosa et al., 2008).
NoV is believed to be transmitted mainly by person‐to‐person contact or by aerosols after projectile vomiting (Marks et al., 2000, 2003). Consumption of food or water contaminated by faecal matter or vomitus (Marks et al., 2000, 2003; Rutjes et al., 2006), and exposure to contaminated surfaces or fomites, are also the sources of infection (Wu et al., 2005; D'Souza et al., 2006). The ease with which NoV is transmitted and spread is mainly because of its infectious dose being low – fewer than 10 virus particles are required for the infection (Teunis et al., 2008) – high resistance to disinfection (Duizer et al., 2004a; Jimenez & Chiang, 2006; Whitehead & McCue, 2009) and possible long‐term stability and persistence in the environment (Wu et al., 2005; D'Souza et al., 2006).
The most common cause of NoV food‐borne outbreaks is the consumption of shellfish, fresh produce and ready‐to‐eat food contaminated by infected, but possibly asymptomatic, food handlers (Daniels et al., 2000; Cannon & Vinjé, 2008; Lamhoujeb et al., 2008). The long‐term stability and persistence of NoV on contaminated surfaces used in food preparation areas also make a substantial contribution to disease transmission (Cheesbrough et al., 2000; Evans et al., 2002; Kuusi et al., 2002; Taku et al., 2002; Clay et al., 2006; D'Souza et al., 2006; Mattison et al., 2007; Lamhoujeb et al., 2008, 2009). Moreover, NoV is resistant to many industrial food preservation methods and can survive chilling, freezing, acidification, reduced water activity and modified atmosphere packaging (Baert et al., 2009).
NoV has also been documented as a water‐borne pathogen, and numerous outbreaks have originated from sewage‐polluted drinking water (Nygård et al., 2003; Maunula et al., 2005; Hewitt et al., 2007; ter Waarbeek et al., 2010) and recreational water (Hoebe et al., 2004; Maunula et al., 2004; Sartorius et al., 2007). This may be a consequence of its suspected resistance to wastewater treatment (Lodder & de Roda Husman, 2005; Van den Berg et al., 2005; da Silva et al., 2007; La Rosa et al., 2009; Nordgren et al., 2009; Skraber et al., 2009) in addition to its survival ability in aquatic settings (Kadoi & Kadoi, 2001; Allwood et al., 2003; Bae & Schwab, 2008). Additionally, shellfish grown and harvested in wastewater‐polluted water can concentrate NoV, which may be inadequately eliminated by standard depuration procedures (Muniain‐Mujika et al., 2002): the consequence is outbreaks of gastroenteritis after consumption of shellfish (Le Guyader et al., 2006a; Le Guyader et al., 2008; Webby et al., 2007).
Hepatitis A virus: prevalent in developing countries
Hepatitis A virus (HAV) is an icosaedric nonenveloped virus species with an ssRNA (+) genome of approximately 7.5 kb and is classified in the family of the Picornaviridae, genus Hepatovirus. Approximately 1.4 million people worldwide become infected with HAV annually (Issa & Mourad, 2001). The incidence of infection varies between regions of the world, with the highest rate in developing countries where sewage treatment and hygiene practices can be poor. Conversely, the number of reported cases of HAV infection has declined substantially in countries with effective programmes of immunization with a licensed vaccine. For example, in the USA, the number of cases has been reduced by 92% to an infection rate as low as one case per 100 000 persons per year (Daniels et al., 2009); similar situations now also apply to other countries including Canada, Australia, Japan and New Zealand (Jacobsen & Koopman, 2004).
HAV can, via sewage discharge, contaminate soil, food crops and natural watercourses (Bosch, 1998; Cook & Rzeżutka, 2006). Consequently, food (Pebody et al., 1998; Hutin et al., 1999; Lees, 2000; Dentinger et al., 2001; Nygård et al., 2001; Greening, 2006) and drinking water (Divizia et al., 2004; Tallon et al., 2008) are considered major vehicles of HAV transmission to humans. In an epidemiological investigation, 6.5% of acute cases of hepatitis A were identified as food‐ or water‐borne; however, this figure is probably an underestimate, because a considerable proportion of cases (~68%) remain uncharacterized (Daniels et al., 2009).
HAV is able to survive in several environments, notably in water, food and soil (Rzeżutka & Cook, 2004). Water is considered to be the most important source of infectious virus because it can survive for long periods in this environment. For example, the virus can survive for up to 60 days in tap water (Enriquez et al., 1995), over 6 weeks in river water (Springthorpe et al., 1993), over 8 weeks in groundwater (Sobsey et al., 1989) and even up to 30 weeks in sea water (Crance et al., 1998). HAV is also able to survive in various types of soil and remains infectious after 12 weeks (Sobsey et al., 1989).
Adenoviruses: some serotypes cause gastroenteritis in children
Adenovirus (AdV) is an icosaedric nonenveloped virus with a dsDNA genome 28–45 kb long. They are classified as members of the Adenoviridae family, genus Mastadenovirus, which includes 20 known species: three bovine, five human and three porcine. Fifty‐one serotypes of human AdV (hAdV) in six subgroups (A‐F) have been described (Wold & Horwitz, 2007). hAdV serotypes 40/41, included in Group F, are the major causes of gastroenteritis in young children and are readily spread by the faecal–oral route. They are sensitive to chemical disinfection but are more resistant to the effects of UV light than other enteric viruses (Thurston‐Enriquez et al., 2003). hAdV is shed from the gut on a long‐term basis regardless of the site of initial infection, although the mechanism has not been fully clarified in humans (Calcedo et al., 2009; Echavarria, 2009; Roy et al., 2009). A limited number of probable water‐borne outbreaks of hAdV have been reported, particularly in association with conjunctivitis and swimming pools (Martone et al., 1980). Chlorination failures are often cited as a major factor in outbreaks.
Enteroviruses: common viral causes of gastroenteritis
The genus Enterovirus (EV) comprises spherical nonenveloped viruses, with an ssRNA (+) genome of 7.2–8.5 kb, in the family of the Picornaviridae. Four species have been distinguished (A, B, C and D) within which the serotypes are known by their traditional names: human EV (hEV) A includes some coxsackievirus A strains; hEV B contains coxsackievirus A9, coxsackievirus B1‐6 and most of the echoviruses; and hEV C contains polioviruses 1–3 and some coxsackievirus A strains. The more recently identified hEVs have been given individual numbers, from EV68, and are classified amongst all four species (Stanway et al., 2005).
These viruses may replicate in the respiratory tract and the gut and can be transmitted through aerosols and by the respiratory route or via the faecal–oral route. Many infections are asymptomatic, and as few as one in 100 may result in clinical illness. The wide range of diseases includes classical poliomyelitis, aseptic meningitis, cardiac disease, hand, foot‐and‐mouth disease, conjunctivitis and rashes. A common clinical picture is self‐limiting fever, malaise, muscle aches and headache; diarrhoea and vomiting are present only as a part of more generalized systemic illness. Clinical illness in temperate climates is more common in the summer months; all age groups are affected, and immunity to one serotype does not protect against infection with other serotypes (Moore et al., 1984). The serotypes of echoviruses and coxsackieviruses then circulate and dominate within communities change over time, and there is molecular drift within serotypes (Savolainen et al., 2001). hEVs can be found in all aquatic matrices reflecting their widespread occurrence in populations (Sellwood et al., 1981; Hovi et al., 1996; Sedmark et al., 2003). However, transmission of hEV infection through an aquatic route has been difficult to confirm as the number of asymptomatic infections is so large and the transmission by close personal contact so common.
HEV: zoonotic transmission as an emerging problem
HEV is a small, spherical and nonenveloped ssRNA (+) virus of approximately 7.2 kb. It is classified within the family of the Hepeviridae, genus Hepevirus. HEV is a major cause of acute human hepatitis in regions with inadequate water supplies and poor sanitary conditions (Purcell & Emerson, 2001; Guthmann et al., 2006), and there is an increasing evidence of locally acquired HEV infections in industrialized countries (Zanetti et al., 1999; Widdowson et al., 2003; Buti et al., 2004; Mansuy et al., 2004; Ijaz et al., 2005; Waar et al., 2005). HEV sequences worldwide can be classified into four major genotypes (1–4) (Lu et al., 2006). The relatively conserved genotypes 1 and 2 circulate primarily in humans causing the majority of HEV infections including all epidemics in Asia and Africa countries and also in Mexico. By contrast, for genotypes 3 and 4, only isolated cases of human infection have been described and only in more industrialized countries including the USA, Japan, China and countries in Europe. Although four genotypes of HEV exist, there only seems to be one serotype present (Zhou et al., 2003; Herremans et al., 2007; Mushahwar, 2008). Previously, HEV infections in industrialized countries were believed to be travel related, but recently an increasing number of indigenous HEV cases have been reported (Zanetti et al., 1999; Widdowson et al., 2003; Mansuy et al., 2004; Lu et al., 2006; Borgen et al., 2008). Serological studies have reported the presence of HEV antibodies in a variety of animal species, notably cows, cats, dogs and rodents. However, HEV RNA has not been detected in these species, and the validity of the assays used is seldom well established owing to the lack of positive reference samples: consequently, these results must be interpreted with caution (Bouwknegt et al., 2007). The presence of HEV has been reported in food, water and animals including pigs (Rutjes et al., 2009a). In several animal species, HEV genotype 3 and 4 sequences have been detected, with pigs being the animal most frequently involved in countries formerly designated as nonendemic for HEV. HEV RNA has also been detected in wild boar in several countries (Takahashi et al., 2004; de Deus et al., 2008; Martelli et al., 2008; Adlhoch et al., 2009), in Sika deer (Tei et al., 2003), in roe deer (Reuter et al., 2009), in red deer (Rutjes et al., 2010) and in mongoose (Nakamura et al., 2006). Furthermore, a human HEV genotype 1 strain was detected in workhorses in Egypt (Saad et al., 2007).
The non‐travel‐related HEV infections in industrialized countries may be of zoonotic origin. Sequences of the swine HEV genotype 3 and 4 strains closely related to human strains have been isolated in many countries worldwide (van der Poel et al., 2001; Huang et al., 2002; Clemente‐Casares et al., 2003; Lu et al., 2006; Rutjes et al., 2007; Reuter et al., 2009), suggesting that pigs may be the reservoir of the indigenous infections in these countries. More direct evidence of zoonotic food‐borne transmission of genotype 3 was obtained when four cases of hepatitis E could be linked directly to eating raw deer meat: identical HEV strains were found in the deer meat consumed and the patients (Tei et al., 2003; Li et al., 2005).
RV, astrovirus and other agents of gastroenteritis: water‐borne pathogens affecting mostly children
Viruses of the genus Rotavirus are icosahedral nonenveloped nonturreted virions with a triple capsid structure and a segmented dsRNA genome of approximately 18.5 kb. They are classified in the Reoviridae family, and there are five major groups (A‐E) (Estes & Kapikian, 2007). Group A RV (GARV) is associated with a large majority of human RV infections and represents the major cause of child mortality because of diarrhoea worldwide (Parashar et al., 2006; Sánchez‐Padilla et al., 2009). GARV is also widespread in wild and domestic animal species, and it has been suggested that zoonotic transmission plays a substantial role in the introduction of novel strains into the human population (Cook et al., 2004; Bányai et al., 2009). Within GARV, at least 19 G‐ and 27 P‐types can be distinguished on the basis of sequence diversity of the genes encoding the two outer capsid proteins (VP7 and VP4) (Matthijnssens et al., 2008; Van Doorn et al., 2009). The recent introduction of vaccines for human use may lead to the emergence of novel RV genotypes or the re‐emergence of older strains, particularly from animal reservoirs, and such strains could displace those currently predominating (Cook et al., 2004; Iturriza‐Gómara et al., 2004; Kang et al., 2005; Steyer et al., 2008).
RV persist similarly in polluted and nonpolluted fresh water (Hurst & Gerba, 1980) and even when subjected to light exposure, which can seriously affect the stability and viability of other enteric RNA viruses, for example astrovirus (Fujioka & Yoneyama, 2002; Lytle & Sagripanti, 2005). Inactivation of virus infectivity in different types of water has been consistently found to correlate with higher temperatures (John & Rose, 2005).
The genus Mamastrovirus (AstV) includes spherical nonenveloped viruses with an ssRNA (+) genome of between 6.8 and 7 kb. They are members of the Astroviridae family. There are six species affecting bovines, felines, mink, ovines, porcines and humans (HAstV). HAstV is a common cause of gastroenteritis in children and also in the elderly and immunocompromised individuals (Herrmann et al., 1991; Guix et al., 2002; Mendez & Arias, 2007). Eight genotypes of HAstVs have been described to date and are classified into genogroup A (HAstV‐1 to 5 and HAstV‐8) and genogroup B (HAstV‐6 and 7) (Gabbay et al., 2007). HAstVs have been occasionally found associated with gastroenteritis outbreaks involving possible water‐borne or food‐borne transmission (Leclerc et al., 2002; Maunula et al., 2004; Smith et al., 2006; Domínguez et al., 2008; Scarcella et al., 2009), and their presence in seafood has been discussed and may depend on rainfall conditions (Le Cann et al., 2004; Riou et al., 2007). Recently, the possible zoonotic transmission of astroviruses from cows was proposed (Kapoor et al., 2009).
Other viruses, such as kobuvirus, aichivirus, picobirnavirus and torovirus, are also found in the environment, but further epidemiological studies and wide‐ranging investigations of diagnostic spectra are needed to document their distribution in the environment and impact on food safety and health.
Shedding of pathogenic viruses into the environment
Zoonotic transmission
One of the main routes of transmission of viruses to humans is zoonotic, associated with the consumption of contaminated products of animal origin, or during food manipulation by infected handlers. The other most frequent cause of virus‐contaminated foods is contact with faecal‐polluted waters (Fig. 1). Inadequately treated drinking water, consumption of crops contaminated after being irrigated with wastewater or fertilized with sewage and ingestion of shellfish grown in polluted waters are, therefore, common causes of food‐borne viral infection of people (Bosch, 1998). Several factors affect the contamination of shellfish, vegetables, berries, fruits and herbs. Climatic variables such as season, tidal cycles, rainfall and flooding have all been implicated in viral contamination of the environment (Le Guyader et al., 2000; Griffin et al., 2003; Suffredini et al., 2008; Guillois‐Bécel et al., 2009). Likewise, good livestock, agriculture and manufacturing practices are absolutely necessary to minimize the risk of viral contamination of food. Inappropriate irrigation practices, wastewater treatment and reuse, sewage overflows, and wastewater releases from polluted sources are the direct causes of viral environmental contamination and food‐borne outbreaks (Le Guyader et al., 2000; Griffin et al., 2003; Jiménez‐Clavero et al., 2003; Choi et al., 2004; Suffredini et al., 2008; Guillois‐Bécel et al., 2009) (Fig. 1). Shellfish grown in areas close to intensive farming, or waste treatment plants, present a high risk of enteric virus carriage (Le Guyader et al., 2000; Ley et al., 2002).
Figure 1.
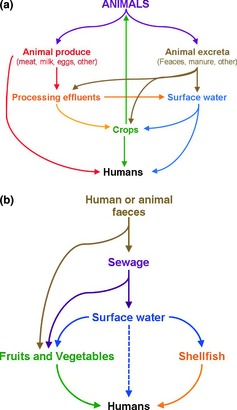
Contamination routes for environmental virus hazards (a) of animal origin and (b) in foods. (a) Contamination routes of environmental virus hazards of animal origin. Zoonotic route of contamination from the original source (animal) to humans. (b) Environmental virus contamination of foods. Contamination from original source to humans using food and water as a route of transmission.
There has been increasing concern about the effects on human and animal health of pathogenic viruses in animal manure. In recent years, outbreaks of food‐borne diseases associated with the consumption of animal products have received much attention, leading to consumer concern about the safety of the food supply. The health risk associated with animal operations depends on diverse factors. The most important is related to the animal species being reared and the concentration of pathogenic microorganisms in animal manure. Some viruses survive both for long periods and despite treatment, and their ability to remain infectious in the environment until ingested by a human or animal host is an added concern. However, it has been difficult to determine the role of livestock in most water‐borne virus outbreaks because both humans and various wildlife species can shed the same viruses and thereby serve as sources of infection or contamination. EVs are shed in faeces and, consequently, are disseminated through contaminated soil and water; therefore, any other animal species grazing in the same pastures and/or drinking from the same water sources as infected livestock are likely to be exposed. Consequently, they may be contaminated by the same or closely related virus variants and therefore present a high risk of further disseminating the virus (Ley et al., 2002; Jiménez‐Clavero et al., 2005).
Most pathogenic viruses emerging in human populations are of animal origin (Taylor et al., 2001). There is a large spectrum of transmission modes for zoonotic viruses with domestic animal or wildlife reservoirs. They can be direct or indirect (Kruse et al., 2004) and include transmission by contaminated food, water, air and soil (Fig. 1). Meat can be contaminated by excreta during processing, but may also have been contaminated earlier because of infection of the living animal. The risk of food‐borne infection depends on the virus infection route, the level of contamination and the extent of inactivation during food processing. Livestock industries produce large amounts of residues that can cause substantial environmental problems. Indeed, accidental or deliberate spills, overuse of fertilizer and emissions of incorrectly, or incompletely, treated animal wastes are the major environmental risks (Jongbloed & Lenis, 1998; Jiménez‐Clavero et al., 2005). Cook et al. (2004) estimated that contamination of arable land with animal RV in spread animal waste used as fertilizer may be considerable, and similarly substantial contamination is plausible or even likely for other viruses shed in large numbers in animal excreta. As expected, detection of animal viruses in contaminated waters (groundwater, lakes, rivers, estuaries, runoffs and animal watering tanks from farms, etc.) is much more frequent in areas of intensive than less active farming (Jiménez‐Clavero et al., 2005). The modes and the levels of environmental contamination with viruses differ for the different types of viruses and animal species.
Occupational exposure
The working environment and procedures can be sources of viral dissemination. However, the difficulties associated with evidencing cases and relating them to possible exposure make it very complex to assess the risk of infection. Health care facilities are the most extensively studied occupational settings. In such facilities, blood‐borne viruses, including human immunodeficiency virus (HIV), hepatitis B virus and hepatitis C virus, can be transmitted mainly by accidents with infected needles or sharp objects (Davanzo et al., 2008). Air‐borne viruses such as the influenza virus, respiratory syncytial virus, AdV, rhinovirus, coronavirus, measles, rubella, mumps viruses and parvovirus B19 are also easily spread (Aitken & Jeffries, 2001). Viral agents transmitted via the faecal–oral route, such as RV, hAdV 40 and 41 and NoV, are frequently associated with nosocomial and health care–related infections spread by contamination of air, hands and surfaces (Lopman et al., 2004). Workers involved in sewage treatment and reuse for agricultural and industrial purposes can be exposed to enteric viruses. Seroepidemiological surveys show that workers in wastewater treatment plants (Clark et al., 1985; Heng et al., 1994; De Serres & Laliberte, 1997; Weldon et al., 2000; Divizia et al., 2008) and in spray irrigation activities (Katzenelson et al., 1976; WHO, 2006) are at higher risk than the general population, in terms of enteric and hepatic infections. Veterinary and zootechnical jobs can also expose workers to zoonotic viruses through contact with manure and inhalation of aerosols generated by activities such as washing and cleaning (Cook et al., 2004). Serological studies indicate that workers in the intensive animal husbandry sector may be exposed to zoonotic viruses, notably H1 swine influenza virus (Olsen et al., 2002). Workers in these fields of activity may therefore possibly have a role in species‐jumping from animal to human populations (Baker & Gray, 2009).
Environmental matrices containing human pathogenic viruses
Human pathogenic viruses are excreted and secreted by humans into their environment through faeces, urine, saliva, sweat and tears (de Roda Husman & Bartram, 2008). The principal matrices, which can be contaminated with human viruses and represent potential sources of infection, are water, sewage, sludge, manure, air, hard surfaces, crops such as fruit and vegetables, shellfish and animal products. The range of complexity in the structure and electrostatic charge of these matrices and of the viruses is such that their interactions are extremely diverse, with corresponding differences as concerns virus inactivation and removal. In general, virus survival is influenced by parameters such as moisture, temperature, association with solids and exposure to UV.
Water and sewage
Surface waters can readily become contaminated with viruses. In the EU, guidelines for sewage discharge (Directive 91/271/EEC) concerning urban wastewater treatment were adopted in 1991 to protect the water environment from the adverse effects of discharges of urban wastewater and from certain industrial discharges. This is an important standard as it not only regulates the conditions of discharge according to the inhabitant equivalent but also stipulates requirements for corresponding collection and treatment facilities. However, the reduction values required for discharges from urban wastewater treatment plants are evaluated according to chemical and biochemical parameters, including biochemical oxygen demand, chemical oxygen demand, total suspended solids and total phosphorus and nitrogen; they do not address highly stable pathogens, like viruses. In sludge (solids remaining after wastewater treatment), viruses may be present and constitute a potential hazard.
Drinking water is abstracted from surface water in many countries and treated by sedimentation, filtration and/or disinfection, which, if done effectively, can produce a virus‐free end product, although this may be dependent on the quality of the source water (Rutjes et al., 2009b; Teunis et al., 2009; Lodder et al., 2010). The European Directive concerning quality of water intended for human consumption is Directive 98/83/EC. Monitoring should provide information about the organoleptic and microbiological quality of the water supplied as well as information concerning the effectiveness of drinking water treatment (particularly disinfection). This directive includes microbiological limits based on bacterial standards, but viruses are not considered in any of the current directives.
Manure
Manure can be defined as urine and faecal material produced by animals housed in artificial environments, such as farms and zoos. It may also contain straw bedding, is often stored for long periods and is used as a fertilizer on agricultural land. In general, enteric viruses including caliciviruses, HAV and HEV are considered to be stable in faeces (Rzeżutka & Cook, 2004). After dispersion of viruses into the environment, the inactivation rates differ substantially between types of virus and inactivation is faster in liquid manure (mixture of urine and water with less bedding material) than in solid manure. Enteric viruses can survive for a very long time (even years) at temperatures below 5 °C and especially in the absence of UV light. There is good evidence that inactivation of viruses in the environment is less effective if they are absorbed onto or embedded within suspended solid matter that is not dried out. Viruses like HAV, NoV and HEV can resist complete inactivation in the environment for a very long time (Pesaro et al., 1995).
Air and hard surfaces
The importance of air‐borne spreading of enteric viruses is not well defined, unlike water‐borne or food‐borne spreading. This is largely owing to the difficulties in identifying this transmission route for single cases or outbreaks. The air‐borne transmission of viruses is dependent on the likelihood of material containing viruses to form aerosols and on the survival of viruses in the air. Enteric viruses can be aerosolized by, for example, violent vomiting (as associated with NoV) (Marks et al., 2000), toilet flushing (Barker & Jones, 2005), spray irrigation (Petterson et al., 2001) and various processes at wastewater treatment plants (Carducci et al., 1995). Some enteric viruses can cause infection by ocular contact or by inhalation and virus catchment by mucus and subsequent swallowing. Nevertheless, the most common mechanism of dissemination is the deposition of aerosol particles on surfaces, particularly food, vegetation and clothes. Surfaces such as door handles, banisters for staircases, flushing handles on toilets, toys, telephones, drinking cups and fabrics have all been implicated in the transmission of enteric viruses (Barker & Jones, 2005; Gallimore et al., 2008). Faecal material or vomit may contaminate these surfaces, and the viruses contained may then be ingested following direct contact or transfer from hands (Boone & Gerba, 2007). The characteristics of the material and the virus contribute to determining the survival rate (Abad et al., 1994; Vasickova et al., 2010). The detection of virus on a large variety of surfaces, like tables, door knobs, walls, toilets seats, thermometers, toys, cotton cloth, carpets, bed covers, gloves, drinking glasses, paper (Boone & Gerba, 2007) has helped to explain the routes of transmission of NoV (Wu et al., 2005; Boxman et al., 2009a), RV (Ansari et al., 1988) and rhinovirus (Ansari et al., 1991) in localized cases and outbreaks.
Food
Food and food environments are a major source of viral transmission to humans (Koopmans et al., 2002; Koopmans & Duizer, 2004). Food‐borne viral outbreaks are reported worldwide every year and are associated with a wide variety of foods (e.g. Verhoef et al., 2008; Kuo et al., 2009; Robesyn et al., 2009; Vivancos et al., 2009). The viruses most frequently involved in food‐borne infections are NoV and HAV, but other viruses, particularly human RV, hEV, HEV and AstV, are also transmitted by food. For NoV and HAV, person‐to‐person spread is the most common transmission route. Secondary spread of these viruses after introduction by, for example, food‐borne contamination is common and often results in larger, prolonged outbreaks (WHO and FAO, 2008). Estimates of the proportion of viral illnesses attributed to food are in the range of around 5% for HAV to 12–47% for NoV. However, all currently available estimates of food‐borne illnesses make assumptions and use extrapolations from different data sources (Scallan et al., 2011). Nevertheless, all essentially conclude that viruses are an important cause of food‐borne illness (WHO and FAO, 2008; Scallan et al., 2011). The incidence of outbreaks of food‐borne viral disease has increased considerably during the last decades, possibly due to the rapid globalization of the food market, the increase in personal travel and food transportation, and the profound changes in food consumption habits (Rodríguez‐Lázaro et al., 2009).
Food products can be contaminated at various points along the food supply chain. This can be because of poor practice in primary production and/or misuse of natural and environmental resources (Appleton, 2000), e.g. the irrigation of vegetables with polluted water – including contamination through roots owing to drop irrigation (Urbanucci et al., 2009) – contact with human faeces or faecally soiled materials and poor hygiene practice by food handlers during the harvest of fresh produce. Furthermore, contamination may arise by inappropriate practices during processing or at the point of sale/consumption (Boxman et al., 2009b). Also, there may be cross‐contamination from polluted working instruments or surfaces, which have been contaminated previously by infected food handlers or contaminated food items (D'Souza et al., 2006; Boxman et al., 2009b; Dreyfuss, 2009). In addition, shellfish, fresh produce or ready‐to‐eat foods may be contaminated with human excreta, either directly or indirectly, and viral food‐borne outbreaks may also originate from zoonotic viruses intrinsically present in food consumed. This has been demonstrated for HEV in raw meat and liver from wild boar and deer (Matsuda et al., 2003; Tei et al., 2003; Takahashi et al., 2004). Moreover, the potential for food‐borne transmission is a concern with every new emerging infection, even for viruses that are primarily respiratory, for example, the highly pathogenic avian influenza virus. Indeed, infectious avian influenza virus has been cultured from frozen exported meat, raising the issue of possible dissemination of such viruses via the food chain (WHO and FAO, 2008).
Foods commonly implicated in outbreaks are those that are minimally processed, such as shellfish or fresh produce, although ready‐to‐eat foods that have been contaminated by an infected food handler are also involved. Traditionally, bivalve mollusc shellfish such as oysters, mussels, clams and cockles have been considered as a principal source of food‐borne virus that may subsequently be disseminated (Pintó et al., 2009). Filter‐feeding shellfish can concentrate viruses from polluted water: the filtration can lead to concentrations in shellfish 100–1000 times higher than that in the surrounding water (Carter, 2005). In addition, specific binding of NoV to the shellfish epithelia has been observed, and this may impede the release of virus during shellfish depuration (Le Guyader et al., 2006b; Maalouf et al., 2011). Fresh produce has high water content – absorbed from groundwater during growth – and may be eaten raw and without peeling, both procedures that may remove external contamination. Viruses can survive on their surface once harvested (Carter, 2005) and can remain infectious for several days or weeks and even during commercial and household storage for periods of up to 5 weeks (Bosch et al., 2006). However, any food that has been manipulated by foodhandlers and is not (or insufficiently) subjected to subsequent preservation and/or cooking is susceptible to be a source of transmission of enteric viruses.
Virus survival in foods can be affected by diverse factors. Kott & Fishelson (1974) found that poliovirus persisted longer on tomato and lettuce plants in phosphate‐buffered saline than in oxidation pond effluent, possibly due to microbial activity in such effluents. Also, natural irradiation in combination with natural antiviral substances generally present in fruit may greatly reduce virus infectivity (Konowalchuk & Speirs, 1978). However, natural or added constitutes in food such as fat, salt and sucrose may protect viruses against inactivation by heating or high hydrostatic pressure (Kovač et al., 2010). Conversely, components like acids and various components of fruit juices may enhance the rate of viral inactivation (Kovač et al., 2010).
Sampling strategies
Surveillance of food and environmental virus hazards
For successful public health intervention regarding food and environmental virus hazards, the early and accurate identification of infectious viral agents is of primary importance. The ability to identify quickly the causative viral pathogen of an emerging viral epidemic markedly increases the chances of success of any countermeasures for containment, prevention and control of the possible disease. Surveillance of environmental viruses can underpin the detection of both cases and outbreaks by identifying an increase in frequency of disease above its background incidence (Centers for Disease Control and Prevention, 2001) and by estimating disease impact. In addition, surveillance can help generate hypotheses and stimulate research, evaluating prevention and control measures and facilitating planning.
Many countries and international organizations, notably the World Health Organization (WHO) and the European Centre for Disease Prevention and Control (ECDC), and international research projects have devoted considerable energy to developing integrated surveillance networks; these networks are for tracking environmental viruses including food‐ and water‐borne viral pathogens such as NoV, RV and EV and for providing information about the viruses’ genetic structure and geographical distribution and about the host populations and environmental matrix involved. Recent advances in molecular biology, including DNA chip technology and whole‐genome sequencing technologies, continuously improve diagnostic power to detect and characterize a wide range of pathogens and their variants. Public health surveillance systems for outbreak detection can establish the relative value of different approaches for the detection of outbreaks at the earliest stages and provide the information needed to improve their efficacy. However, substantial costs can be incurred in developing, enhancing and managing these surveillance systems and investigating false alarms (Wagner et al., 2001). Furthermore, the overall economic benefits of surveillance systems for early detection and response to outbreaks have not been clearly established.
Sampling methods
A rational sampling plan is essential for the analysis of human pathogenic viruses, which may be present in small quantities and distributed heterogeneously in matrices; the plan should be established on a risk‐based approach (Andrews & Hammack, 2003; Food Standard Agency, 2004a, b). Consequently, a sample or subsamples must represent the original matrix (e.g. water and food), and the sampling process (including the storage and transportation) must not alter the condition of the sample and thus not affect the subsequent analysis (Food Standard Agency, 2004a, b). Other aspects that also must be considered when developing a sampling programme are the characteristics of the matrix to be analysed (nature: solid, semi‐solid, viscous or liquid; type: food, water or environmental sample; composition: rich in fat, protein or plant contents such as tannins; and amount: scarce or abundant), and the subsequent analytical method to be used (cell culture, immunological or molecular). If, for example, a sampling plan for a pâté factory is required, a balanced approach needs to be based on the observation that a sample suitable for public health (for example 25 g of a pâté) might not be suitable for subsequent analysis using a molecular method because of the heterogeneous nature and composition of the matrix. Any inadequacy concerning one of the aspects will affect the validity of the final analytical result.
Various international bodies, such as the International Organisation for Standardisation (ISO), the European Committee for the Normalisation (CEN) and the European Food Safety Authority (EFSA), and national bodies, such as the U.S. Department of Health and Human Services (USDHHS), have defined principles and/or standards for the sampling of foods and water. For example, ISO has established a series of standards for sampling (ISO 5667 series, ISO 18593:2004; ISO 8066:2004; ISO 24276:2006; ISO 7002:1986; ISO 17604:2003); however, there is no specific mention of sampling for human enteric pathogenic viruses in any of these standards. The CEN/ISO ad hoc expert committee for viruses in food ‘CEN/TC 275/WG6/TAG4’ is currently working on the first international standard for a horizontal method for the detection of HAV and NoV in food. However, the sampling process is not included in this planned standard, and the committee has decided to examine the ISO 6887 series for suitability. Similarly, the FDA's Bacteriological Analytical Manual (BAM) includes a general protocol for ‘food sampling and preparation of sample homogenate’ (Andrews & Hammack, 2003), in which the scientific basis for sampling only uses previously published bacteriological criteria (ICMSF, 1986, 2002), despite the BAM having defined a specific protocol for the detection and quantification of HAV (Goswami, 2001).
A large number of studies are related to viral food‐ and water‐borne outbreaks, sporadic cases or studies using samples collected to determine the presence of different enteric viruses in food or the environment or to evaluate new methods for the detection of viruses in diverse matrices (Supporting Information, Tables S1 and S2). Several important lessons can be learnt from these studies. First, there is an evident lack of harmonization in the sample size, and therefore, a serious risk in the representativeness of the sampling strategies used. This is most important as most of those studies are related to viral diarrhoeal outbreaks: the consequences may include the true aetiological agent of the gastroenteritis not being found, or the infectious dose being under‐ or overestimated. In these studies, sizes of samples used were extremely diverse, ranging from 50 μL to 3000 L (i.e. an almost 108‐fold difference) for water and from 1.5–200 g for food samples. Second, there is a lack of homogeneity in the selection of the animal tissues or part of the sample tested once the sample is collected. This also can affect the detection of human pathogenic viruses. For example, different shellfish tissues can be tested for human enteric viruses (i.e. the whole shellfish, the mantle, the gills, the stomach or the digestive diverticula). However, it has been demonstrated that the efficiency of recovery can differ substantially between types of sample and even that the virus may not be detectable in some (Wang et al., 2008). In a study evaluating different tissues of naturally contaminated oysters to identify the most suitable for the detecting virus, the percentages of samples positive were different for the whole oyster (0.7%), mantle (2.2%), gills (14.7%), stomach (13.9%) and the digestive diverticula (13.2%), and the detection was not possible when the adductor muscles were tested (Wang et al., 2008). Another important factor is the ambiguous use of individual or pooled samples for foodstuffs, especially in the case of shellfish. This affects directly both the representativeness and analytical sensitivity of the final results. For example, de Roda Husman et al. (2007) observed that pooling digestive glands of several oysters never resulted in a positive signal, whereas RT‐PCR testing of the individual digestive glands of single oysters revealed the presence of virus RNA. This indicates that pooling can affect the final results negatively and even can produce false negative results owing to the simple mechanism of reducing the size of each individual sample used in the pool. This can be of great relevance to public health. Conversely, the use of individual samples can also affect the representativeness of the population studied. A balanced approach to difficult food matrices may therefore be to analyse a representative number of individual samples; however, this could greatly increase both the cost and the time required for the analyses and even may be unfeasible in the field. Two other important aspects also have to be considered: the period of time from the sampling to the start of the analysis in the laboratory and the conditions of storage of the sample during that period. These issues can be of particular importance if complex matrixes are analysed, as the stability of the virus may be compromised. However, they are usually not rigorously addressed during sampling, and most studies do not provide the relevant details. Even where this information is provided, the lack of uniformity is again evident. Samples are sometimes stored frozen (Loisy et al., 2000; Schvoerer et al., 2000, 2001; Donaldson et al., 2002), refrigerated at 4 °C (Pina et al., 2001; La Rosa et al., 2007), at room temperature (Beuret et al., 2002) or kept on ice (Noble & Fuhrman, 2001; Katayama et al., 2008).
Sample representativeness
Representativeness expresses the degree to which sample data accurately and precisely reflect a characteristic or variable at a sampling point. Representativeness is a qualitative factor, which is largely dependent on the appropriate design of the sampling programme. The representativeness criterion is best satisfied by making certain that sampling locations are selected suitably and that a sufficient number of samples are collected. The sampling strategy must be unbiased, sufficient (i.e. it summarizes all relevant information about the parent population, which contained the sample, but ignoring any sample‐specific information), efficient (i.e. the more the statistical values for various samples cluster around the true value and the lower the sampling error, the greater the efficiency) and consistent (the larger the sample, the closer the statistic should be to its true value) (Jarman, 1984).
Transport and storage
After sampling is completed, samples should to be transported to the laboratory facilities as soon as possible. For example, the AFNOR method XP T 90‐451 ‘Recherche des entérovirus’ in water (AFNOR, 1990) states that after in situ concentration by filtration, the sample cartridge should be removed and enclosed aseptically such that the filtration device must not be left completely dry; thereafter, samples should be transported to the laboratory within 24 h at a suitable temperature. On the other hand, the ISO method 19458 ‘Water quality – Sampling for microbiological analysis’ (ISO, 2006), although not specific for mammalian virus, states that viruses should be transported and stored for a period of 24–72 h, at a temperature of 5 ± 3 °C. The guidelines ‘Standard Methods for the examination of Water and Wastewater’ (Eaton et al., 2005) states that samples cannot be held more than 2 h at temperatures of 25 °C or 48 h at 2–10 °C; samples have to be stored at −70 °C if not processed in this time frame. Dahling & Wright (1984) also indicate that samples stored at −70 °C are stable without virus loss for up to 4 days. Mocé i Llivina (2004) tested the stability of EV at −70 °C and demonstrated that they could infect cells after 11 months of storage at this temperature when adsorbed to cellulose ester membranes. In conclusion, transport and storage should be performed as quickly as possible, at a controlled temperature (5 ± 3 °C). In this temperature range, samples can be stored for up to 48 h. If this time cannot be respected, the samples should be frozen at −70 °C.
It is of utmost importance that laboratory personnel recognize that the safe and efficient transportation of any infectious substance is in the interest of public health generally. The packaging of infectious substances for transport must therefore be designed to minimize the risk of damage during transport. Sending or transporting infectious viruses should respect the ‘Guidance on regulations for the Transport of Infectious Substances 2009–2010’ (WHO, 2008). Different forms of transportation (road, rail, sea and air) of infectious substances have different safety requirements and therefore their own international convention or code based on UN Model Regulations. As far as laboratory personnel are concerned, their responsibility lies in ensuring that the goods are packaged according to WHO regulations. Some countries have their own national regulations; when this is not the case, International Guidelines should be followed.
Safety in the laboratory
HAV and NoV are both classed as Hazard Group 2, with a vaccine currently being available for HAV. HEV is classed as Hazard Group 3 in some countries, and therefore, any intentional use of this virus in laboratories in those countries must be performed strictly in containment level 3 facilities (CL3). However, the handling of pathogenic viruses must conform with any specific national recommendations: for example, in the case of HEV, the classification differs between countries and various international bodies. Indeed, the WHO and USA recommendations for this organism is biosafety level (BSL) 2, the Spanish recommendation is generally BSL 3 but not with all BSL 3 precautions as there is no evidence of aerosol contamination, and the British recommendation is BSL 3. This should be borne in mind when sending a sample likely to contain a virus to another laboratory. Only laboratories with the available CL3 facilities should handle any package suspected of containing a CL3 microorganism. Guidance should be sought from a national body, which provides advice on best practice procedures for the safe handling and containment of Hazard Group 2, 3 and 4 organisms. Note that many national guidelines are based on EU or international guidelines. If no national regulatory body of this type exists in a country, international or European guidelines, such as the WHO Laboratory Biosafety Manual 2nd Ed. (WHO, 2003), should be followed.
Detection and identification of food and environmental virus hazards
Detection of viruses in food and environmental samples is challenging because of the large variety and complexity of samples, the possible heterogeneous distribution of a small number of viruses and the presence of components that may inhibit or interfere with virus detection (Goyal, 2006). A general flow chart for the analytical process (from sampling to final identification and characterization) for the detection of human enteric viruses is given in Fig. 2. It is necessary to separate and concentrate viruses from environmental materials before performing tests for detection (Sair et al., 2002). As no standard procedure or systematic approach evaluating the adsorption of viruses onto different substrates has yet been developed, it is difficult to draw conclusions about the mechanisms involved in virus adsorption (Jin & Flury, 2002); consequently, establishing appropriate separation and concentration processes is even more demanding. Whatever the method used, the final concentrate should not be cytotoxic to cell cultures used in infectivity assays and should be free of any inhibitors, which may be co‐extracted or co‐concentrated from environmental samples (Goyal, 2006). A variety of biological and chemical substances that are present in environmental matter or are used during sample processing have been found to act as inhibitors, including polysaccharides, haeme, phenol and cations (Atmar, 2006). Known PCR inhibitors in shellfish extracts include glycogen and acidic polysaccharides (Schwab et al., 1998).
Figure 2.
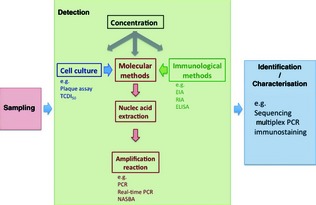
Schematic diagram of the analytical process of detection and identification of environmental virus hazards. TCDI 50, median tissue culture infective dose assay; EIA, enzymatic immunoassay; RIA, radioimmunoassay; ELISA, enzyme‐linked immunosorbent assay; NASBA, nucleic acid sequence–based amplification.
For virological analysis of aerosols, the key issue is sample collection and preparation for the different detection procedures (mainly based on cell culture and/or molecular techniques). The sample size is generally 1–3 m3 of air. Various approaches have been developed, based on the property of air‐borne particles of attaching to every surface with which they enter into contact (Verreault et al., 2008). There are three different principles underlying the most commonly used air samplers: membrane filtration, impact on solid surfaces followed by elution, or impingement in a liquid medium. The eluates produced can be further concentrated (Verreault et al., 2008). Other methods for the virological analysis of aerosols include cyclone or electrostatic precipitators, and in recent years, the fear of bioterrorism has triggered assessments of various new methodologies (including mass spectrometry) able to identify dangerous species in the air. However, it is unlikely that such techniques will be suitable for routine environmental analysis in the near future, and furthermore, they require the establishment of very large databases of environmental samples.
To elucidate the fate of virus dispersed through air, surface monitoring should be also performed, because larger droplets tend to settle out. Surface sampling is most extensively used in health care settings and in food production to assess not only viral contamination but also the efficacy and correct application of disinfection procedures. For hard surfaces, a defined surface area (i.e. 10 or 36 cm2) should be swabbed; the swab is then eluted, and the elute is processed as a liquid sample. Alternative methods are contact plates, which can be similarly eluted.
Concentration of viruses
The aim of concentrating virus is to collect most of the virus present in the sample in a minimal volume (Cliver, 2008); this small sample can then be used for virus detection by molecular, immunological or cell culture–based methods (Fig. 2). Protocols for the concentration of viruses in water samples are generally based on four steps (Croci et al., 2008): adsorption of viruses to a filter; elution of adsorbed viruses using a protein‐rich buffer; reconcentration of viruses by flocculation, precipitation or filtration, and extraction of viruses, for example with chloroform. In solid samples (including foodstuffs), sample processing often starts with a washing step (in the case of fresh produce) or a homogenization step (in the case of, for example, shellfish); the virus is concentrated after this first step (Rodríguez‐Lázaro et al., 2007; Croci et al., 2008). If appropriate, a minimal volume of a diluent can be added to favour dissociation of the virus from the solid matter but avoiding interference with subsequent virus concentration/extraction. For dispersion of the sample in the diluent, a suitable mixing technique is required. The following step is the removal of food solids from the extract by, for example, filtration or differential centrifugation. Concentration methods appropriate for a wide variety of matrices include adsorption elution, differential precipitation, ultracentrifugation and ultrafiltration (Rodríguez‐Lázaro et al., 2007).
Detection methods used for human enteric viruses
Various approaches can be used to detect human enteric viruses in concentrated samples. They range from direct observation by electron microscopy to the detection of cytopathic effects in specific cell lines and of indirect diagnostic signals using immunological or molecular methods (Fig. 2).
Direct observation by electron microscopy is a laborious, painstaking and time‐consuming task, is also subjective, and has a limited sensitivity (Atmar & Estes, 2001). The observation of cytopathic effects produced in specific cell lines is not always possible as some enteric viruses, notably NoV and HEV cannot be propagated in mammalian cell lines. Even when possible, this is not a simple or cost‐effective technique. It may also require the adaptation of the virus before it can grow effectively (Pintó & Bosch, 2008). There are immunological tests such as enzymatic immunoassay, radioimmunoassay or enzyme‐linked immunosorbent assay (ELISA), and many are commercially available for the main enteric viruses. However, their analytical sensitivity is still too poor for effective testing of environmental samples.
To overcome these various limitations and disadvantages, molecular techniques are now being used routinely in viral laboratories, and real‐time quantitative PCR (q‐PCR) has become the method of choice for the detection of enteric viruses. This approach has been reinforced by the recommendation of the international ISO/CEN committee CEN/TC275/WG6/TAG 4 that real‐time PCR should serve as the basis for the forthcoming international standards for the detection of NoV and HAV (Lees and CEN WG6 TAG4, 2010). A large number of scientific studies using molecular methods for the detection of enteric viruses have already been published (see Table S3 for a representative list of the published references).
q‐PCR is a molecular technique that allows the quantification of the amount of the target template (i.e. specific virus) initially present in a sample (Heid et al., 1996). Other major advantages of this technique include the closed‐tube format that reduces the risk of carry‐over contamination, the wide dynamic range of quantification and the possibilities for automation (Rodríguez‐Lázaro et al., 2007). However, q‐PCR also suffers from some limitations. The volume used in the amplification reaction is very small; therefore, only concentration methods that can deliver a very small volume of the resulting nucleic acid solution (i.e. in the microlitre range) from a realistic food or environmental sample can be used. In addition, the quality of the nucleic acids is an important factor that directly affects the analytical sensitivity of the assay, and diverse compounds present in samples can inhibit the amplification reaction. The standardization of inhibition tests would help overcome this limitation once appropriate synthetic standards become available (La Rosa et al., 2010). Finally, definitive international standardization efforts are required to guarantee effective implementation in the real‐life analytical contexts.
Other detection options include the combination of cell culture or immunological methods and a molecular technique. The combination of a cell culture step and subsequent detection by a molecular technique such as RT‐PCR or nucleic acid sequence–based amplification (NASBA) reduces the incubation periods and also allows the detection of viruses that grow without causing cytopathic effects (Table S3) (Dubois et al., 2002; Duizer et al., 2004b).
Index viruses
Classic microbiological indicators such as faecal coliforms (Escherichia coli and enterococci) are the most commonly used indicators to evaluate both the level of faecal contamination and also efficiencies of the elimination of pathogens by water purification processes. However, the adequacy of these bacterial markers to indicate the presence and concentration of human viruses and protozoa cysts has been questioned in recent years (Lipp et al., 2001; Tree et al., 2003). EV, evaluated as cultivable enteric viruses, is the sole viral measure that has been included in past regulations. Results obtained by applying molecular techniques have shown that the presence of EVs does not significantly correlate with the presence of other pathogenic viruses that may be more abundant. Diverse groups of bacteriophages have also been suggested as indicators of viral contamination; this would allow in theory the use of simple assays for the detection of infectious viruses (Savichtcheva & Okabe, 2006; Love et al., 2008), although their presence does not clearly correlate with the presence of specific viral pathogens (Formiga‐Cruz et al., 2003).
The improvement in molecular technologies for detecting viruses present in water and food has focused attention on new groups of DNA viruses that may be quantified with cost‐effective molecular assays and are excreted in large quantities by the populations of widely divergent geographical areas. hAdV are often being detected in the environment (He & Jiang, 2005; Van Heerden et al., 2005a; Katayama et al., 2008; Muscillo et al., 2008) and have been proposed along with human polyomaviruses as a molecular index of viral contamination of human origin (Puig et al., 1994; Pina et al., 1998; Bofill‐Mas et al., 2000). Testing for hAdV is of interest for two different reasons: both to assess the presence of this human pathogen itself and also as a more general indicator. Most of the population is seropositive for the most common AdV and also for the human polyomaviruses JCPyV and BKPyV. The presence of these viruses in water therefore presents only a low risk for healthy immunocompetent populations (Bofill‐Mas et al., 2001). Specific animal AdV or polyomaviruses have been also proposed as microbial source tracking tools (Hundesa et al., 2006, 2009).
hAdV and JCPyV have been found in 98% of the sewage samples analysed from widely diverse geographical areas around the world (Bofill‐Mas et al., 2000), with concentrations of about 105–107 genome equivalents (GE) L−1. The concentrations are generally higher for hAdV than for JCPyV. These viruses have also been commonly found in river water and have been used as a marker for the evaluation of the efficiency with which water treatment plants eliminate virus (Bofill‐Mas et al., 2006; Albinana‐Gimenez et al., 2009a).
q‐PCR methods have been developed for the detection of hAdV in sewage, shellfish, river water and drinking water (Puig et al., 1994; Pina et al., 1998; Formiga‐Cruz et al., 2002; Albinana‐Gimenez et al., 2009b) and in sea water (Calgua et al., 2008). hAdV has also shown to be very stable in the environment and resistant to water treatments (Thompson et al., 2003; Mena & Gerba, 2009). A very high proportion of environmental or shellfish samples presenting human viral pathogens contain AdV (Formiga‐Cruz et al., 2002); they are the most abundant viruses, as assessed by PCR, and are regularly found in faecal contamination. In a study using q‐PCR, hAdV was detected in 100% of the urban sewage samples analysed at concentrations of 104–105 GE mL−1, and these viruses were still present in treated effluents at concentrations of 102–103 GE L−1. The biosolids generated accumulated 102–105 AdV GE g−1. JCPyV also were quantified, and the concentrations found were 103–104 GE mL−1 in urban sewage, 102–103 GE L−1 in treated effluent and 103 GE g−1 in the biosolids generated (Bofill‐Mas et al., 2006).
The application of index viruses in future regulations on the microbiological quality of water should be a step forward for improving the control of the environment, food and water. However, this would require further studies, including epidemiological studies, for the definition of acceptable values of index viruses and to identify where such values would be appropriate.
Evaluation and interpretation of test results
One of the major differences between the study of the presence and enumeration of bacteria and that of viruses in food and in the environment is the availability of a “gold standard” method for detection. Classical culture‐based techniques are considered the gold standard for the detection of bacteria, but the situation is exactly the opposite for the detection of viruses, since no accepted standard method exists. The lack of a defined and consensus standard method for the detection and quantification of viruses is hindering and slowing the adaptation of quantitative viral risk assessment (QVRA) models for food and food environments. Therefore, the establishment and application of a common and validated method for virus detection would make a large contribution to the effective harmonization of QVRA studies. The combination of cell culture and PCR generally produces higher viral counts than those resulting from cell culture methods (i.e. plaque‐forming units or TCID50) and could be considered a de facto standard (Havelaar & Rutjes, 2008).
Validity of molecular detection methods
The reliability of the results produced by molecular techniques is undermined by the lack of standard methods for the detection of viruses in environmental samples and the wide diversity of viruses, matrices, assays and recovery efficiencies described. Molecular techniques, if used with the appropriate quality controls, could allow substantial progress in the control of viral contamination of environment and food. These quality controls must include at least one negative and one positive reaction control, one negative and one positive process control and an internal or external amplification control (Hoorfar et al., 2004; Costafreda et al., 2006; Rodríguez‐Lázaro et al., 2007; Pintó & Bosch, 2008; D'Agostino et al., 2011; Diez‐Valcarce et al., 2011a, b; Martínez‐Martínez et al., 2011) (Table 1). Controls for the estimation of the efficiency of the concentration and/or extraction procedures are also very important. Several approaches have suggested the use of nonpathogenic virus surrogates, with similar structural characteristics and which are not present naturally in the samples to be tested. As examples, Mengo virus MC0 (Costafreda et al., 2006) and feline calicivirus and murine NoV‐1 (Cannon et al., 2006) have been proposed as appropriate surrogates for HAV and human NoV, respectively.
Table 1.
Analytical controls for (RT) real‐time PCR‐based detection of viral hazards in food matrices
| Process controls |
| Processing Positive Control (PPC): A negative sample spiked with sufficient viral target and processed throughout the entire protocol. A positive signal should be obtained indicating that the entire process was correctly performed |
| Processing Negative Control (PNC): A negative sample spiked with sufficient amount of nontarget or water and processed throughout the entire protocol. A negative signal should be obtained, indicating the lack of contamination throughout the entire process. For example, the inclusion of encapsidated RNA (or DNA) or bacteriophages |
| Environmental Control: A tube containing the master mixture or water left open in the PCR set‐up room to detect possible contaminating nucleic acids in the environment |
| Amplification controls |
| Positive PCR control: A viral template known to contain the target sequence. Positive amplification indicates that amplification was performed correctly. It could be used a natural virus or chimerical nucleic acids |
| Negative PCR control (or No Template Control ‐NTC‐ or Reagent Control or Blank): Including all reagents used in the amplification except the template nucleic acids. Usually, water is added instead of the template. A negative signal indicates the absence of specific contamination in the amplification assay |
| External Amplification control (EAC): An aliquot of a solution of control DNA, containing a defined quantity or copy number, added to an aliquot of the nucleic acid of the extracted sample and analysed in a separate reaction tube. A positive signal indicates that the sample nucleic acid extract did not contain any inhibitory substances |
| Internal Amplification Control (IAC): Chimerical nontarget nucleic acid added to the master mix to be co‐amplified with the same primer set as the viral target but with an amplicon size visually distinguishable or different internal sequence region from the target amplicon. The amplification of the IAC both in the presence and in the absence of the target indicates that the amplification conditions are adequate |
Adapted from Rodríguez‐Lázaro et al. (2007), Pintó & Bosch (2008), Bosch et al. (2011) and D'Agostino et al. (2011).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Negative results obtained using correctly designed and controlled PCR assays can provide robust evidence for the absence of pathogens or indicators in the samples analysed with strong implications for risk assessment. Such negative results from well standardized and highly sensitive PCR assays may be acceptable and may facilitate the implementation of potential regulations requiring the absence of pathogens from defined sample volumes, as is the case for food or water safety criteria. More studies are needed to evaluate the significance of positive results, because the differing sensitivities of diverse techniques, like infectivity assays if available, do not allow a definitive evaluation of the infectious capability of the viral genomes detected. Also, if viral measures are considered for regulations concerning the microbiological quality of bathing water or other environmental samples, epidemiological studies would be needed to establish acceptable limits for index viruses.
Infectious particles vs. PCR GE: implications for public health
Viral infectivity is defined as the capacity of viruses to enter the host cell and exploit its resources to replicate and produce progeny infectious viral particles (Black, 1996; Rodríguez et al., 2009), which may lead to infection and subsequent disease in the human host. Therefore, the information required in risk assessment studies is the number of viral particles with infective capacity. Obviously, cell culture–based methods are the soundest methodologies for the estimation of the number of infective particles. However, as indicated earlier, there are no available culture models for some of the most significant food and environmental virus hazards, notably human NoV, HEV and even wild‐type HAV. In these cases, only molecular methods are available, but although RTq‐PCR is a quantitative and sensitive tool, it cannot distinguish between infective and noninfective viruses (Richards, 1999). This limits its usefulness for public health purposes. The ratio between GE and infectious particles has been reported to increase with the time, is strongly dependent upon water and climatic conditions and virus type, and can vary from 70 : 1 to 50 000 : 1 for EV in natural surface water (Rutjes et al., 2005) and in artificial ground and surface waters (de Roda Husman et al., 2009). For example, wastewater can contain up to 1500 GE HAstV L−1 but do not show any infective capacity. To overcome this limitation, several different approaches based on (RT) PCR have been assessed (reviewed in Rodríguez et al., 2009; see Table 2 for examples). However, it is unclear whether any direct PCR method can satisfactorily assess viral infectivity.
Table 2.
Molecular‐based methods used for assessing viral infectivity
| Method | Treatment | Detection | Type of sample | Target virus | References |
|---|---|---|---|---|---|
| Molecular | Proteinase and RNase | RT‐PCR | Cell culture | FCV HAV, MNoV, poliovirus 1, | Nuanualsuwan & Cliver (2002, 2003); Baert et al. (2008) |
| Proteinase and RNase | qNASBA | Stool samples and cell culture | NoV, FCV | Lamhoujeb et al. (2008, 2009) | |
| RNase protection assay | qRT‐PCR | Stool samples and cell culture | NoV, FCV | Topping et al. (2009) | |
| 5’ NTR RT‐PCR | Cell culture | HAV | Bhattacharya et al. (2004); Li et al. (2002, 2004) | ||
| Long target region (LTR) qRT‐PCR | Cell culture | HAV, poliovirus 1, F‐specific RNA phages | Li et al. (2002); Simonet & Gantzer (2006a, b) | ||
| Cell culture + molecular | Attachment to cell monolayer | RT‐PCR | Cell culture | HAV, poliovirus 1, FCV | Nuanualsuwan & Cliver (2003) |
| Virus replication in cell culture (ICC: integrated cell culture) | RT‐PCR | Different types of water, sewage effluent, faecal specimens and cell culture | AdV, AstV, EV, poliovirus, RV, HAV, MS2 | Blackmer et al. (2000); Chapron et al. (2000); Jiang et al. (2004); Ko et al. (2003, 2005); Lee & Kim (2002); Lee & Jeong (2004); Li et al. (2009); Nuanualsuwan & Cliver (2003); Reynolds et al. (1996); Shieh et al. (2008) | |
| Immunological +molecular | Antibody capture | RT‐PCR | Different types of water, faecal samples and cell culture | HAV, NoV, poliovirus 1, FCV | Gilpatrick et al. (2000); Myrmel et al. (2000); Schwab et al. (1996) |
| Immunomagnetic separation | qRT‐PCR | Artificially contaminated groundwater | HAV | Abd El Galil et al. (2004) |
RT‐PCR, reverse transcriptase PCR; qRT‐PCR, reverse transcriptase real‐time PCR; qNASBA, real‐time nucleic acid sequence–based amplification; FCV, feline calicivirus; mNoV, murine NoV.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Risk assessment
As stated earlier, QVRA is theoretically a powerful statistical tool for the estimation of the probability of a viral infection or disease based on exposure of the human host to the viral hazard and for establishing the dose–response relationship (Haas, 1983; Haas et al., 1993). Consequently, QVRA has been used for exposure to various virus hazards in different environmental matrices, mostly for aquatic environments (e.g. Van Heerden et al., 2005b).
In general, the risk analysis framework (FAO and WHO, 2006) consists of hazard identification, exposure assessment, hazard characterization and risk characterization, which should identify and preferably quantify the risk. In the case of QVRA for environmental exposure, this framework reads as follows: (1) hazard identification: the identification of viral agents that may be present in a particular environmental matrix and are capable of causing adverse health effects; (2) exposure assessment: quantitative evaluation of the likely intake of viral agents via exposure to environmental sources; (3) hazard characterization: quantitative evaluation of the nature of the adverse effects associated with the viral agents that may be present in the environment one is exposed to and; (4) risk characterization: the integration of hazard identification, exposure assessment and hazard characterization into a risk estimate of the likelihood and the severity of the adverse effects in a given population with attendant uncertainties.
Various viral characteristics, as described in this paper, are important determinants of the risk of infection or disease upon exposure: numbers (or dose), infectivity and pathogenicity to humans. Application of QVRA has been rendered difficult by the lack of culturing systems and low environmental levels of viruses that present a possible public health risk but cannot be typed or quantified. Moreover, standardized methods for quantification of virus hazards in different environmental matrixes and dose–response models for the main environmental virus hazards are not available. For reliable quantification of viruses in food and environmental matter, various factors need to be determined: the detection efficiency of the assay used, the controls appropriate for accurately measuring both the true concentration and the release of virus into the environment, and the contamination of the food (Pintó & Bosch, 2008; Pintó et al., 2009). This is of the utmost importance for unculturable viruses, such as HEV and human NoV, for which only molecular quantitative detection methods are available. The raw numbers of GE, which are the data generated by such methods, must be corrected for the efficiency of the concentration and nucleic acid extraction steps and the capacity of the enzyme involved in the amplification‐based detection. A formula for the estimation of exposure to viruses in food matrices has been proposed by Havelaar & Rutjes (2008).
Following exposure assessment, hazard characterization is possible using dose–response models, which describe the relationship between virus particles detected and the probability of disease. Viral dose–response models are based on three basic biological assumptions: single hit, independent action and random distribution (FAO and WHO, 2006). Using these assumptions, three different models can be applied to environmental virus hazards (Haas, 1983; Teunis & Havelaar, 2000; Zwietering & Havelaar, 2006). For example, Pintó et al. (2009) estimated the relationship between HAV numbers in frozen coquina shellfish involved in two hepatitis outbreaks and the risk for human health. However, for HAV, immunity needs to be taken into account. Similarly, for human NoV that only induces short‐lived immunity, risk assessment should also take into account the observation that a proportion of the population is resistant to infection with NoV genogroup GI (Hutson et al., 2002; Lindesmith et al., 2003; Rockx et al., 2005) or GII (Thorven et al., 2005; Larsson et al., 2006).
The viral risk can thus be estimated from the information obtained from an exposure assessment and the dose–response relationship (Zwietering & Havelaar, 2006). In addition, the estimation of the disease incidence can be also extrapolated to estimates of disease burden and costs (Havelaar & Rutjes, 2008). Published risk assessments for environmental viruses mainly concern water‐borne or food‐borne exposure, but other routes may be considered as well. For food‐borne viruses, the EU research project ‘Integrated monitoring and control of food‐borne viruses in European food supply chains’ (KBBE 213178; VITAL; http://www.eurovital.org) has been launched to develop pro‐active integrated monitoring and risk management strategies for the control of viral contamination of food supply chains. Moreover, a network of food and environmental virologists, under COST Action 929, ENVIRONET (http://www.cost929-environet.org), has been established to improve our knowledge and the role of the environment and food in the transmission of enteric viral disease.
Concluding remarks and recommendations
Environmental virus hazards are increasingly recognized as a cause of illness in all age groups. Caliciviruses (NoV), AdV, EV, RV, HAV and HEV are the most common causes of illness because of environmental exposure. The major routes of exposure to environmental viruses involve human or animal faeces, surface water or sewage, especially irrigation waters in relation to crops, and fresh and noncooked produce along the food chain, and in particular bivalve molluscs, which filter feed in virus‐contaminated waters. In addition to the risks associated with the contamination of environmental or food matrixes with viruses of human origin, there are also pathogenic viruses that are zoonotic, i.e. of animal origin and transmitted from animals.
Education of populations at risk should give particular attention to describing potential virus contamination routes, especially for those working with water, sewage, faeces and food. Education about risks is also important for health care workers and consumers. The most important preventive measures include the improvement of hygienic conditions during harvesting, processing and handling of potentially contaminated environmental matter. Legislation on handling and treatment of water, sewage and foods should be adapted as needed to reduce the risk of environmental virus contamination. The systems for sewage treatment and the codes of practice for agricultural use of sewage and surface water should be reviewed to address these issues.
Methods related to virus purification and detection of viral particles should be improved such that survival of human pathogenic viruses in the environment can be followed reliably. In parallel, techniques should be further developed for effective virus inactivation and decontamination of environmental materials suspected to pose a risk. When human disease is caused by environmental exposure to viruses, and also for the assessment of virus contamination in environmental matter, virus monitoring is required, and it may be beneficial to implement a virus surveillance strategy. Unfortunately, this is not straightforward. Samples must represent the environmental matter being studied, and tests for specific virus hazards may need specific sampling and sample processing techniques. Safe and efficient transport and laboratory practices are of utmost importance for laboratory workers and the outcomes of prevention and control measures.
The development of a suitable detection technique for a virus in an environmental sample requires a targeted specific approach. This generally starts with the separation and concentration of the virus. Appropriate concentration methods include adsorption elution, differential precipitation, ultracentrifugation and ultrafiltration. Then, various virus identification methods can be used; possible methods range from classical techniques like cell culture and electron microscopy to molecular techniques like RT‐PCR and microarrays, and combinations may also be used. Development of a general method that can be applied to different matrices is difficult and, indeed, may not be feasible. Nevertheless, the CEN/TC 275‐ Food Analysis, Horizontal Methods; Working Group 6, Technical Advisory Group 4 (CENTAG4) is pursuing efforts for the development of such horizontal methods for detection of viruses in foods.
To evaluate the extent of environmental virus contaminations, it can be helpful to test for particular index viruses, whose presence correlates with the presence of other pathogenic, viruses that may be more abundant. Because of their wide applicability and high level of sensitivity and specificity, molecular techniques are most commonly used for the detection of environmental virology. Powerful molecular techniques can be extremely valuable if appropriate controls are used. However, for estimation of the true virus hazard, the detection of GE, which is the output of molecular techniques, has to be related to the quantity of infectious particles present.
To estimate the probability of a viral infection, the statistical tool QVRA can be used. This involves virus hazard identification, exposure assessment, hazard characterization and risk characterization. Satisfactory exposure assessment requires a reliable quantification of the virus present in the environmental material. For reliable quantification of virus in environment, the detection efficiency of the assay used must be determined, and appropriate controls must be employed to determine accurately the true concentration and release of virus in the environment. In conclusion, the study of environmental virus hazards is extremely important to estimate the public health risks associated with viruses.
Supporting information
Table S1. Sampling methods used for detection viral hazards in food matrices.
Table S2. Sampling methods used for detection viral hazards in water samples.
Table S3. Detection methods for viral hazards in different environmental matrices.
Acknowledgements
This review was written as a part of COST action 929 ENVIRONET (http://www.cost929-environet.org), by its Working Group 1, ‘Current and Emerging Issues’ and under grant agreement no. KBBE‐213178 (EU VII FP project VITAL).
References
- Abad FX, Pinto RM & Bosch A (1994) Survival of enteric viruses on environmental fomites. Appl Environ Microbiol 60: 3704–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd El Galil KH, El Sokkary MA, Kheira SM, Salazar AM, Yates MV, Chen W & Mulchandani A (2004) A combined IMS‐molecular beacon RT‐PCR assay for detection of hepatitis A from environmental samples. Appl Environ Microbiol 70: 4371–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlhoch C, Wolf A, Meisel H, Kaiser M, Ellerbrok H & Pauli G (2009) High HEV prevalence in four different wild boar populations in East and West Germany. Vet Microbiol 139: 270–278. [DOI] [PubMed] [Google Scholar]
- AFNOR (1990) AFNOR XP T 90‐451 Recherche des entérovirus. AFNOR, Dartford. [Google Scholar]
- Aitken C & Jeffries DJ (2001) Nosocomial spread of viral disease. Clin Microbiol Rev 14: 528–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albinana‐Gimenez N, Miagostovich M, Calgua B, Huguet JM, Matia L & Girones R (2009a) Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking water‐treatment plants. Water Res 43: 2011–2019. [DOI] [PubMed] [Google Scholar]
- Albinana‐Gimenez N, Clemente‐Casares P, Calgua B, Courtois S, Huguet JM & Girones R (2009b) Comparison of methods for the quantification of human adenoviruses, Polyomavirus JC and Norovirus in source and drinking water. J Virol Methods 158: 104–109. [DOI] [PubMed] [Google Scholar]
- Allwood PB, Malik YS, Hedberg CW & Goyal SM (2003) Survival of F‐specific RNA coliphage, feline calicivirus, and Escherichia coli in water: a comparative study. Appl Environ Microbiol 69: 5707–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews WH & Hammack TS (2003) Food Sampling and Preparation of Sample Homogenate. Bacteriological Analytical Manual (BAM). US Food and Drug Administration, US Department of Health and Human Services, Silver Spring, MD. [Google Scholar]
- Ansari SA, Sattar SA, Springthorpe VS, Wells GA & Tostowaryk W (1988) Rotavirus survival on human hands and transfer of infectious virus to animate and nonporous inanimate surfaces. J Clin Microbiol 26: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari SA, Springthorpe VS, Sattar SA, Rivard S & Rahman M (1991) Potential role of hands in the spread of respiratory viral infections: studies with human parainfluenza virus 3 and rhinovirus 14. J Clin Microbiol 29: 2115–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton H (2000) Control of food‐borne viruses. Br Med Bull 56: 172–183. [DOI] [PubMed] [Google Scholar]
- Asano T & Cotruvo JA (2004) Groundwater recharge with reclaimed municipal wastewater health and regulatory considerations. Water Res 38: 1941–1951. [DOI] [PubMed] [Google Scholar]
- Atmar RL (2006) Molecular methods of virus detection in foods Viruses in Foods. Food Microbiology and Food Safety Series (Goyal SM. ed.), pp. 121–149. Springer, New York, NY. [Google Scholar]
- Atmar RL & Estes MK (2001) Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin Microbiol Rev 14: 15–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J & Schwab KJ (2008) Evaluation of murine norovirus, feline calicivirus, poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl Environ Microbiol 74: 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baert L, Wobus CE, Van Coillie E, Thackray LB, Debevere J & Uyttendaele M (2008) Detection of murine norovirus 1 by using plaque assay, transfection assay, and real‐time reverse transcription‐PCR before and after heat exposure. Appl Environ Microbiol 74: 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baert L, Debevere J & Uyttendaele M (2009) The efficacy of preservation methods to inactivate foodborne viruses. Int J Food Microbiol 131: 83–94. [DOI] [PubMed] [Google Scholar]
- Baggi F, Demarta A & Peduzzi R (2001) Persistence of viral pathogens and bacteriophages during sewage treatment: lack of correlation with indicator bacteria. Res Microbiol 152: 743–751. [DOI] [PubMed] [Google Scholar]
- Baker WS & Gray GC (2009) A review of published reports regarding zoonotic pathogen infection in veterinarians. J Am Vet Med Assoc 234: 1271–1278. [DOI] [PubMed] [Google Scholar]
- Bányai K, Martella V, Molnár P, Mihály I, Van Ranst M & Matthijnssens J (2009) Genetic heterogeneity in human G6P[14] rotavirus strains detected in Hungary suggests independent zoonotic origin. J Infect 59: 213–215. [DOI] [PubMed] [Google Scholar]
- Barker J & Jones MV (2005) The potential spread of infection caused by aerosol contamination of surfaces after flushing a domestic toilet. J Appl Microbiol 99: 339–347. [DOI] [PubMed] [Google Scholar]
- Beuret C, Kohler D, Baumgartner A & Luthi TM (2002) Norwalk‐like virus sequences in mineral waters: one‐year monitoring of three brands. Appl Environ Microbiol 68: 1925–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya SS, Kulka M, Lampel KA, Cebula TA & Goswami BB (2004) Use of reverse transcription and PCR to discriminate between infectious and non‐infectious hepatitis A virus. J Virol Methods 116: 181–187. [DOI] [PubMed] [Google Scholar]
- Bidawid S, Bosch A, Cook N, Greening G, Taylor M & Vinjé J (2009) Editorial. Food Environ Virol 1: 1–2. [Google Scholar]
- Black JG (1996) Microbiology: Principles and Applications, 3rd edn Prentice Hall, Upper Saddle River, NJ. [Google Scholar]
- Blackmer F, Reynolds KA, Gerba CP & Pepper IL (2000) Use of integrated cell culture‐PCR to evaluate the effectiveness of poliovirus inactivation by chlorine. Appl Environ Microbiol 66: 2267–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill‐Mas S, Pina S & Girones R (2000) Documenting the epidemiologic patterns of polyomaviruses in human populations studying their presence in urban sewage. Appl Environ Microbiol 66: 238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill‐Mas S, Formiga‐Cruz M, Clemente‐Casares P, Calafell F & Girones R (2001) Potential transmission of human polyomaviruses through the intestinal tract after exposure of virions or viral DNA. J Virol 75: 10290–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill‐Mas S, Albinana‐Gimenez N, Clemente‐Casares P, Hundesa A, Rodiguez‐Manzano J, Allard A, Calvo M & Girones R (2006) Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl Environ Microbiol 72: 7894–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone SA & Gerba CP (2007) Significance of fomites in the spread of respiratory and enteric disease. Mini‐review. Appl Environ Microbiol 73: 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgen K, Herremans T, Duizer E, Vennema H, Rutjes S, Bosman A, de Roda‐Husman AM & Koopmans M (2008) Non‐travel related HEV genotype 3 infections in The Netherlands; A case series 2004‐2006. BMC Infect Dis 8: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A (1998) Human enteric viruses in the water environment: a minireview. Int Microbiol 1: 191–196. [PubMed] [Google Scholar]
- Bosch A, Pintό RM & Abad FX (2006) Survival and transport of enteric viruses in the environment Viruses in Foods. Food Microbiology and Food Safety Series (Goyal SM, ed.), pp. 151–187. Springer, New York, NY. [Google Scholar]
- Bosch A, Sanchez G, Abbaszadegan M et al (2011) Analytical methods for virus detection in water and food. Food Anal Methods 4: 4–13. [Google Scholar]
- Bouwknegt M, Lodder‐Verschoor F, van der Poel WH, Rutjes SA & de Roda Husman AM (2007) Hepatitis E virus RNA in commercial porcine livers in The Netherlands. J Food Prot 70: 2889–2895. [DOI] [PubMed] [Google Scholar]
- Boxman IL, Dijkman R, te Loeke NA, Hägele G, Tilburg JJ, Vennema H & Koopmans M (2009a) Environmental swabs as a tool in norovirus outbreak investigation, including outbreaks on cruise ships. J Food Prot 72: 111–119. [DOI] [PubMed] [Google Scholar]
- Boxman I, Dijkman R, Verhoef L, Maat A, van Dijk G, Vennema H & Koopmans M (2009b) Norovirus on swabs taken from hands illustrate route of transmission: a case study. J Food Prot 72: 1753–1755. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P & Rohwer F (2003) Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185: 6220–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buti M, Clemente‐Casares P, Jardi R, Formiga‐Cruz M, Schaper M, Valdes A, Rodriguez‐Frias F, Esteban R & Girones R (2004) Sporadic cases of acute autochtonous hepatitis E in Spain. J Hepatol 41: 126–131. [DOI] [PubMed] [Google Scholar]
- Calcedo R, Vandenbeghe LH, Roy S, Somanathan S, Wang L & Wilson JM (2009) Host immune responses to chronic adenovirus infections in human and nonhuman primates. J Virol 83: 2623–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calgua B, Mengewein A, Grünert A, Bofill‐Mas S, Clemente‐Casares P, Hundesa A, Wyn‐Jones P, López‐Pila JM & Girones R (2008) Development and application of a one‐step low cost procedure to concentrate viruses from seawater samples. J Virol Methods 153: 79–83. [DOI] [PubMed] [Google Scholar]
- Cannon JL & Vinjé J (2008) Foodborne viruses Viruses in the Environment (Palombo EA. & Kirkwood CD, eds), pp. 45–75. Research Signpost, Kerala, India. [Google Scholar]
- Cannon JL, Papafragkou E, Park GW, Osborne J, Jaykus LA & Vinjé J (2006) Surrogates for the study of norovirus stability and inactivation in the environment: a comparison of murine norovirus and feline calicivirus. J Food Prot 69: 2761–2765. [DOI] [PubMed] [Google Scholar]
- Carducci A, Arrighi S, Simonini A & Ruschi A (1995) Detection of coliphage and enteroviruses in sewage and aerosol from an activated sludge wastewater treatment plant. Lett Appl Microbiol 21: 207–209. [DOI] [PubMed] [Google Scholar]
- Carducci A, Tozzi E, Rubulotta E, Casini B, Cantiani L, Muscillo M, Rovini E & Pacini R (2000) Assessing airborne biological hazard from urban wastewater treatment. Water Res 34: 1173–1178. [Google Scholar]
- Carter MJ (2005) Enterically infecting viruses: pathogenicity, transmission and significance for food and waterborne infection. J Appl Microbiol 98: 1354–1380. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2001) Updated guidelines for evaluating public health surveillance systems: recommendations from the guidelines working group. MMWR Recomm Rep 50: RR‐13. [PubMed] [Google Scholar]
- Chapron CD, Ballester NA, Fontaine JH, Frades CN & Margolin AB (2000) Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture‐nested PCR procedure. Appl Environ Microbiol 66: 2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesbrough JS, Green J, Gallimore CI, Wright PA & Brown DW (2000) Widespread environmental contamination with Norwalk‐like viruses (NLV) detected in a prolonged hotel outbreak of gastroenteritis. Epidemiol Infect 125: 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Song I, Stine S, Pimentel J & Gerba C (2004) Role of irrigation and wastewater reuse: comparison of subsurface irrigation and furrow irrigation. Water Sci Technol 50: 61–68. [PubMed] [Google Scholar]
- Clark CS, Linneman CC & Gartside PS (1985) Serologic survey of rotavirus, Norwalk agent and Pototheca wickeramii in wastewater workers. Am J Public Health 75: 83–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay S, Maherchandani S, Malik YS & Goyal SM (2006) Survival on uncommon fomites of feline calicivirus, a surrogate of noroviruses. Am J Infect Control 34: 41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente‐Casares P, Pina S, Buti M, Jardi R, Martín M, Bofill‐Mas S & Girones R (2003) Hepatitis E virus epidemiology in industrialized countries. Emerg Infect Dis 9: 448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliver DO (2008) Historic overview of food virology Food‐Borne Viruses: Progress and Challenges (Koopmans MPG, Cliver DO. & Bosch A, eds), pp. 1–28. ASM Press, Washington, DC. [Google Scholar]
- Cook N & Rzeżutka A (2006) Hepatitis viruses Emerging Foodborne Pathogens (Motarjemi Y. & Adams M, eds), pp. 282–308. Woodhead Publishing Limited, Cambridge. [Google Scholar]
- Cook N, Bridger J, Kendall K, Iturriza‐Gómara M, El‐Attar L & Gray J (2004) The zoonotic potential of rotavirus. J Infect 48: 289–302. [DOI] [PubMed] [Google Scholar]
- Costafreda MI, Bosch A & Pintó RM (2006) Development, evaluation, and standardization of a real‐time TaqMan reverse transcription‐PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl Environ Microbiol 72: 3846–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crance JM, Gantzer C, Schwartzbrod L & Deloince R (1998) Effect of temperature on the survival of hepatitis A virus and its capsidal antigen in synthetic seawater. Env Tox Water Quality 13: 89–92. [Google Scholar]
- Croci L, Dubois E, Cook N, de Medici D, Schultz AC, China B, Rutjes SA, Hoorfar J & Van der Poel WHM (2008) Current methods for extraction and concentration of enteric viruses from fresh fruit and vegetables: towards international standards. Food Anal Methods 1: 73–84. [Google Scholar]
- da Silva AK, Le Saux JC, Parnaudeau S, Pommepuy M, Elimelech M & Le Guyader FS (2007) Evaluation of removal of noroviruses during wastewater treatment, using real‐time reverse transcription‐PCR: different behaviors of genogroups I and II. Appl Environ Microbiol 73: 7891–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino M, Cook N, Rodriguez‐Lazaro D & Rutjes S (2011) Nucleic acid amplification‐based methods for detection of enteric viruses: definition of controls and interpretation of results. Food Environ Virol 3: 55–60. [Google Scholar]
- Dahling D & Wright B (1984) Process and transport of environmental virus samples. Appl Environ Microbiol 47: 1272–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels NA, Bergmire‐Sweat DA, Schwab KJ et al (2000) A foodborne outbreak of gastroenteritis associated with Norwalk‐like viruses: first molecular traceback to deli sandwiches contaminated during preparation. J Infect Dis 181: 1467–1470. [DOI] [PubMed] [Google Scholar]
- Daniels D, Grytdal S & Wasley A (2009) Surveillance for acute viral hepatitis – United States, 2007. Centers for Disease Control and Prevention (CDC). MMWR Surveill Summ 58: 1–27. [PubMed] [Google Scholar]
- Davanzo E, Frasson C, Morandin M & Trevisan A (2008) Occupational blood and body fluid exposure of university health care workers. Am J Infect Control 36: 753–756. [DOI] [PubMed] [Google Scholar]
- de Deus N, Peralta B, Pina S, Allepuz A, Mateu E, Vidal D, Ruiz‐Fons F, Martin M, Gortazar C & Segales J (2008) Epidemiological study of hepatitis E virus infection in European wild boars (Sus scrofa) in Spain. Vet Microbiol 129: 163–170. [DOI] [PubMed] [Google Scholar]
- de Roda Husman AM & Bartram J (2008) Global supply of virus safe drinking‐water Human Viruses in Water (Bosch A, ed.), pp. 127–162. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- de Roda Husman AM, Lodder‐Verschoor F, van den Berg HHJL, Le Guyader FS, van Pelt H, van der Poel WHM & Rutjes SA (2007) Rapid virus detection procedure for molecular tracing of shellfish associated with disease outbreaks. J Food Prot 70: 967–974. [DOI] [PubMed] [Google Scholar]
- de Roda Husman AM, Lodder WJ, Rutjes SA, Schijven JF & Teunis PF (2009) Long‐term inactivation study of three enteroviruses in artificial surface and groundwaters, using PCR and cell culture. Appl Environ Microbiol 75: 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Serres G & Laliberte G (1997) Hepatitis A among workers from a wastewater treatment plant during a small community outbreak. Occup Environ Med 54: 60–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentinger CM, Bower WA, Nainan OV, Cotter SM, Myers G, Dubusky LM, Fowler S, Salehi ED & Bell BP (2001) An outbreak of hepatitis A associated with green onions. J Infect Dis 183: 1273–1276. [DOI] [PubMed] [Google Scholar]
- Diez‐Valcarce M, Kovač K, Cook N, Rodríguez‐Lázaro D & Hernández M (2011a) Construction and analytical application of internal amplification controls (IAC) for detection of Food Supply Chain‐Relevant Viruses by Real‐Time PCR‐Based Assays. Food Anal Methods 4: 437–445. [Google Scholar]
- Diez‐Valcarce M, Cook N, Hernández M & Rodríguez‐Lázaro D (2011b) Analytical application of a sample process control in detection of foodborne viruses. Food Anal Methods doi: 10.1007/s12161-011-9262-9. [DOI] [Google Scholar]
- Divizia M, Gabrieli R, Donia D et al (2004) Waterborne gastroenteritis outbreak in Albania. Water Sci Technol 50: 57–61. [PubMed] [Google Scholar]
- Divizia M, Cencioni B, Palombi L & Panà A (2008) Sewage workers: risk of acquiring enteric virus infections including hepatitis A. New Microbiol 31: 337–341. [PubMed] [Google Scholar]
- Domínguez A, Torner N, Ruíz L et al (2008) Aetiology and epidemiology of viral gastroenteritis outbreaks in Catalonia (Spain) in 2004‐2005. J Clin Virol 43: 126–131. [DOI] [PubMed] [Google Scholar]
- Donaldson KA, Griffin DW & Paul JH (2002) Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real‐time RTPCR. Water Res 36: 2505–2514. [DOI] [PubMed] [Google Scholar]
- Dreyfuss MS (2009) Is norovirus a foodborne or pandemic pathogen? An analysis of the transmission of norovirus‐associated gastroenteritis and the roles of food and food handlers. Foodborne Pathog Dis 6: 1219–1228. [DOI] [PubMed] [Google Scholar]
- D'Souza DH, Sair A, Williams K, Papafragkou E, Jean J, Moore C & Jaykus L (2006) Persistence of caliciviruses on environmental surfaces and their transfer to food. Int J Food Microbiol 108: 84–91. [DOI] [PubMed] [Google Scholar]
- Dubois E, LeGuyader F, Haugarreau L, Kopecka H, Cormier M & Pommepuy M (1997) Molecular epidemiological survey of rotaviruses in sewage by reverse transcriptase seminested PCR and restriction fragment length polymorphism assay. Appl Environ Microbiol 63: 1794–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E, Agier C, Traore O, Hennechart C, Merle G, Cruciere C & Laveran H (2002) Modified concentration method for the detection of enteric viruses on fruits and vegetables by reverse transcriptase‐polymerase chain reaction or cell culture. J Food Prot 65: 1962–1969. [DOI] [PubMed] [Google Scholar]
- Duizer E, Bijkerk P, Rockx B, De Groot A, Twisk F & Koopmans M (2004a) Inactivation of caliciviruses. Appl Environ Microbiol 70: 4538–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MP & Estes MK (2004b) Laboratory efforts to cultivate noroviruses. J Gen Virol 85: 79–87. [DOI] [PubMed] [Google Scholar]
- Eaton AD, Clesceri LS, Rice EW, Greenberg AE & Franson MAH (2005) Standard Methods for the Examination of Water and Wastewater, 21st edn American Public Health Association, Washington, DC. [Google Scholar]
- Echavarria M (2009) Adenoviruses Principles and Practice of Clinical Virology, 6th edn (Zuckerman AJ, Banatvala JE, Griffiths P, Schoub B. & Mortimer P, eds), pp. 463–488. John Wiley & Sons, Chichester. [Google Scholar]
- EFSA (2009) The community summary report on food‐borne outbreaks in the European Union in 2007. EFSA J 271: 1–127. [Google Scholar]
- Enriquez CE, Hurst CJ & Gerba CP (1995) Survival of enteric adenoviruses 40 and 41 in tap, sea, and waste water. Water Res 29: 2548–2553. [Google Scholar]
- Espinosa AC, Mazari‐Hiriart M, Espinosa R, Maruri‐Avidal L, Méndez E & Arias CF (2008) Infectivity and genome persistence of rotavirus and astrovirus in groundwater and surface water. Water Res 42: 2618–2628. [DOI] [PubMed] [Google Scholar]
- Estes MK & Kapikian AZ (2007) Rotaviruses Fields Virology, 5th edn (Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B. & Straus SE, eds), pp. 1917–1974. Lippincot William & Wilkins, Philadelphia, PA. [Google Scholar]
- Evans MR, Meldrum R, Lane W, Gardner D, Ribeiro CD, Gallimore CI & Westmoreland D (2002) An outbreak of viral gastroenteritis following environmental contamination at a concert hall. Epidemiol Infect 129: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T, Zhong WM, Jing Y, Huang PW, Espinosa SM, Martinez N, Morrow AL, Ruiz‐Palacios GM, Pickering LK & Jiang X (2004) Genetic diversity among sapovirus. Arch Virol 149: 1309–1323. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organisation & World Heath Organisation (2006) Working principles for risks analysis for application in the framework of the Codex Alimentarius Procedural Manual of the Codex Alimentarius Commission, 16th edn, p. 103 Food and Agriculture Organisation and World Heath Organisation, Geneva, Switzerland. [Google Scholar]
- Food Standard Agency (2004a) Practical Sampling Guidance for Food Standards and Feeding Stuffs. Part 1: Overall Objectives of Sampling. Food Standard Agency, London. [Google Scholar]
- Food Standard Agency (2004b) Practical Sampling Guidance for Food Standards and Feeding Stuffs. Part 2: Food Standards Sampling. Food Standard Agency, London. [Google Scholar]
- Formiga‐Cruz M, Tofiño‐Quesada G, Bofill‐Mas S et al (2002) Distribution of human virus contamination in shellfish from different growing areas in Greece, Spain, Sweden and the United Kingdom. Appl Environ Microbiol 68: 5990–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formiga‐Cruz M, Allard AK, Conden‐Hansson AC et al (2003) Evaluation of potential indicators of viral contamination in shellfish with applicability to diverse geographical areas. Appl Environ Microbiol 69: 1556–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka R & Yoneyama BS (2002) Sunlight inactivation of human enteric viruses and fecal bacteria. Water Sci Technol 46: 291–295. [PubMed] [Google Scholar]
- Gabbay YB, Linhares AC, Cavalcante‐Pepino EL, Nakamura LS, Oliveira DS, Da Silva LD, Mascarenhas JDP, Oliveira CS, Monteiro TAF & Leite JPG (2007) Prevalence of human astrovirus genotypes associated with acute gastroenteritis among children in Belém, Brazil. J Med Virol 79: 530–538. [DOI] [PubMed] [Google Scholar]
- Gallimore CI, Taylor C, Gennery AR, Cant AJ, Galloway A, Xerry J, Adigwe J & Gray JJ (2008) Contamination of the hospital environment with gastroenteric viruses: comparison of two pediatric wards over a winter season. J Clin Microbiol 46: 3112–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantzer C, Maul A, Audic JM & Schwartzbrod L (1998) Detection of infectious enteroviruses, enterovirus genomes, somatic coliphages, and Bacteroides fragilis phages in treated wastewater. Appl Environ Microbiol 64: 4307–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpatrick SG, Schwab KJ, Estes MK & Atmar RL (2000) Development of an immunomagnetic capture reverse transcription‐PCR assay for the detection of Norwalk virus. J Virol Methods 90: 69–78. [DOI] [PubMed] [Google Scholar]
- Goswami WW (2001) Detection and Quantitation of Hepatitis A Virus in Shellfish by the Polymerase Chain Reaction. Bacteriological Analytical Manual (BAM). US Food and Drug Administration, US Department of Health and Human Services, Silver Spring, MD. [Google Scholar]
- Goyal SM (2006) Methods of virus detection in foods Viruses in Foods (Goyal SM, ed.), pp. 101–119. Food Microbiology and Food Safety Series. Springer, New York. [Google Scholar]
- Green KY (2007) Caliciviridae: the noroviruses Fields Virology (Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B. & Straus SE, eds), pp. 949–979. Lippincott‐Raven Publishers, Philadelphia, PA. [Google Scholar]
- Greening GE (2006) Human and animal viruses in food (including taxonomy of enteric viruses) Viruses in Foods (Goyal SM, ed.), pp. 5–42. Springer, New York, NY. [Google Scholar]
- Griffin DW, Donaldson KA, Paul JH & Rose JB (2003) Pathogenic human viruses in coastal waters. Clin Microbiol Rev 16: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillois‐Bécel Y, Couturier E, Le Saux JC et al (2009) An oyster‐associated hepatitis A outbreak in France in 2007. Euro Surveill 14: 19144. [PubMed] [Google Scholar]
- Guix S, Caballero S, Villena C, Bartolome R, Latorre C, Rabella N, Simo M, Bosch A & Pinto RM (2002) Molecular epidemiology of astrovirus infection in Barcelona, Spain. J Clin Microbiol 40: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmann JP, Klovstad H, Boccia D et al (2006) A large outbreak of hepatitis E among a displaced population in Darfur, Sudan, 2004: the role of water treatment methods. Clin Infect Dis 42: 1685–1691. [DOI] [PubMed] [Google Scholar]
- Haas CN (1983) Estimation of risk due to low doses of microorganisms: a comparison of alternative methodologies. Am J Epidemiol 118: 573–582. [DOI] [PubMed] [Google Scholar]
- Haas CN, Rose JB, Gerba C & Regli S (1993) Risk assessment of virus in drinking water. Risk Anal 13: 545–552. [DOI] [PubMed] [Google Scholar]
- Hall G, Kirk MD, Becker N, Gregory JE, Unicomb L, Millard G, Stafford R, Lalor K & OzFoodNet Working Group (2005) Estimating foodborne gastroenteritis, Australia. Emerg Infect Dis 11: 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman GS, Ishida S, Yoshizumi S, Miyoshi M, Ikeda T, Oka T & Takeda N (2007a) Recombinant sapovirus gastroenteritis, Japan. Emerg Infect Dis 13: 786–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman GS, Oka T, Okamoto R et al (2007b) Human sapovirus in clams, Japan. Emerg Infect Dis 13: 620–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman GS, Saito H, Shibata C, Ishizuka S, Oseto M, Oka T & Takeda N (2007c) Outbreak of gastroenteritis due to sapovirus. J Clin Microbiol 45: 1347–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman GS, Oka T, Li TC, Nishio O, Noda M & Takeda N (2008) Detection of human enteric viruses in Japanese clams. J Food Prot 71: 1689–1695. [DOI] [PubMed] [Google Scholar]
- Havelaar AH & Melse JM (2003) Quantifying public health risks The WHO Guidelines for Drinking‐Water Quality: A Burden of Disease Approach. Rijksinstituut voor Volksgezondheid en Milieu (RIVM), Bilthoven, The Netherlands. [Google Scholar]
- Havelaar AH & Rutjes SA (2008) Risk assessment of viruses in food: opportunities and challenges Food‐Borne Viruses: Progress and Challenges (Koopmans MPG, Cliver DO. & Bosch A, eds), pp. 221–236. ASM Press, Washington, DC. [Google Scholar]
- He J & Jiang S (2005) Quantification of enterococci and human adenoviruses in environmental samples by real‐time PCR. Appl Environ Microbiol 71: 2250–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ & Williams PM (1996) Real time quantitative PCR. Genome Res 6: 986–994. [DOI] [PubMed] [Google Scholar]
- Heng BH, Goh KT, Doraisingham S & Quek GH (1994) Prevalence of hepatitis A virus infection among sewage workers in Singapore. Epidemiol Infect 17: 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herremans M, Bakker J, Duizer E, Venneam H & Koopmans MPG (2007) Use of serological assays for diagnosis of hepatitis E virus genotype 1 and 3 infections in a setting of low endemicity. Clin Vaccine Immunol 14: 562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JE, Taylor DN, Echeverria P & Blacklow NR (1991) Astroviruses as a cause of gastroenteritis in children. N Engl J Med 324: 1757–1760. [DOI] [PubMed] [Google Scholar]
- Hewitt J, Bell D, Simmons GC, Rivera‐Aban M, Wolf S & Greening GE (2007) Gastroenteritis outbreak caused by waterborne norovirus at a New Zealand ski resort. Appl Environ Microbiol 73: 7853–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe CJ, Vennema H, de Roda Husman AM & van Duynhoven YT (2004) Norovirus outbreak among primary schoolchildren who had played in a recreational water fountain. J Infect Dis 189: 699–705. [DOI] [PubMed] [Google Scholar]
- Hoorfar J, Malorny B, Abdulmawjood A, Cook N, Wagner M & Fach P (2004) Practical considerations in the design of internal amplification controls for diagnostic PCR assays. J Clin Microbiol 42: 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovi T, Stenvik M & Rosenlew M (1996) Relative abundance of enterovirus serotypes in sewage differs from that in patients: clinical and epidemiological implications. Epidemiol Infect 116: 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FF, Haqshenas G, Guenette DK, Halbur PG, Schommer SK, Pierson FW, Toth TE & Meng XJ (2002) Detection by reverse transcription‐PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J Clin Microbiol 40: 1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundesa A, Maluquer de Motes C, Bofill‐Mas S, Albinana‐Gimenez N & Girones R (2006) Identification of human and animal adenoviruses and polyomaviruses for determination of sources of fecal contamination in the environment. Appl Environ Microbiol 72: 7886–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundesa A, Maluquer de Motes C, Albinana‐Gimenez N, Rodrigues‐Manzano J, Bofill‐Mas S, Suñen E & Girones R (2009) Development of a qPCR assay for the quantification of porcine adenoviruses as an MST tool for swine fecal contamination in the environment. J Virol Meth 158: 130–135. [DOI] [PubMed] [Google Scholar]
- Hurst CJ & Gerba CP (1980) Stability of simian rotavirus in fresh and estuarine water. Appl Environ Microbiol 39: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin YJF, Pool V, Cramer EH et al (1999) A multistate, foodborne outbreak of hepatitis A. N Engl J Med 340: 595–602. [DOI] [PubMed] [Google Scholar]
- Hutson AM, Atmar RL, Graham DY & Estes MK (2002) Norwalk virus infection and disease is associated with ABO histo‐blood group type. J Infect Dis 185: 1335–1337. [DOI] [PubMed] [Google Scholar]
- ICMSF (1986) Microorganisms in Foods 2: Sampling for Microbiological Analysis: Principles and Specific Applications. University of Toronto Press, Toronoto, Canada. [Google Scholar]
- ICMSF (2002) Microorganisms in Foods 7: Microbiological Testing in Food Safety Management. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- Ijaz S, Arnold E, Banks M et al (2005) Non‐travel‐associated hepatitis E in England and Wales: Demographic, clinical, and molecular epidemiological characteristics. J Infect Dis 192: 1166–1172. [DOI] [PubMed] [Google Scholar]
- International Organisation for Standardisation (2006) ISO 19458:2006 Water Quality – Sampling for Microbiological Analysis. International Organisation for Standardisation, Geneva, Switzerland. [Google Scholar]
- Issa IA & Mourad FH (2001) Hepatitis A: an updated overview. J Med Liban 49: 61–65. [PubMed] [Google Scholar]
- Iturriza‐Gómara M, Kang G & Gray J (2004) Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol 31: 259–265. [DOI] [PubMed] [Google Scholar]
- Jacobsen KH & Koopman JS (2004) Declining hepatitis A seroprevalence: a global review and analysis. Epidemiol Infect 132: 1005–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman B (1984) Underpriviledged areas: validation and distribution of scores. Br Med J 289: 1587–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YJ, Liao GY, Zhao W, Sun MB, Qian Y, Bian CX & Jiang SD (2004) Detection of infectious hepatitis A virus by integrated cell culture/strand‐specific reverse transcriptase‐polymerase chain reaction. J Appl Microbiol 97: 1105–1112. [DOI] [PubMed] [Google Scholar]
- Jimenez L & Chiang M (2006) Virucidal activity of a quaternary ammonium compound disinfectant against feline calicivirus: a surrogate for norovirus. Am J Infect Control 34: 269–273. [DOI] [PubMed] [Google Scholar]
- Jiménez‐Clavero MA, Fernández C, Ortiz JA, Pro J, Carbonell G, Tarazona JV, Roblas N & Ley V (2003) Teschoviruses as indicators of fecal contamination of water. Appl Environ Microbiol 69: 6311–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez‐Clavero MA, Escribano‐Romero E, Mansilla C, Gómez N, Córdoba L, Roblas N, Ponz F, Ley V & Sáiz JC (2005) Survey of bovine enterovirus in biological and environmental samples by a highly sensitive real‐time reverse transcription‐PCR. Appl Environ Microbiol 71: 3536–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y & Flury M (2002) Fate and transport of viruses in porous media. Adv Agron 77: 39–102. [Google Scholar]
- Johansson PJ, Bergentoft K, Larsson PA, Magnusson G, Widell A, Thorhagen M & Hedlund KO (2005) A nosocomial sapovirus‐associated outbreak of gastroenteritis in adults. Scand J Infect Dis 37: 200–204. [DOI] [PubMed] [Google Scholar]
- John DE & Rose JB (2005) Review of factors affecting microbial survival in groundwater. Environ Sci Technol 39: 7345–7356. [DOI] [PubMed] [Google Scholar]
- Jongbloed AW & Lenis NP (1998) Environmental concerns about animal manure. J Anim Sci 76: 2641–2648. [DOI] [PubMed] [Google Scholar]
- Kadoi K & Kadoi BK (2001) Stability of feline caliciviruses in marine water maintained at different temperatures. New Microbiol 24: 17–21. [PubMed] [Google Scholar]
- Kang G, Kelkar SD, Chitambar SD, Ray P & Naik T (2005) Epidemiological profile of rotaviral infection in India: challenges for the 21st century. J Infect Dis 192: S120–S126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Li L, Victoria J, Oderinde B, Mason C, Pandey P, Zaidi S & Delwart E (2009) New species of astroviruses in human stool. J Gen Virol 90: 2965–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Wobus CE, Lay M, Davidson J & Virgin HW (2003) STAT1‐dependent innate immunity to a Norwalk‐like virus. Science 299: 1575–1578. [DOI] [PubMed] [Google Scholar]
- Katayama H, Haramoto E, Oguma K, Yamashita H, Tajima A, Nakajima H & Ohgaki S (2008) One‐year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res 42: 1441–1448. [DOI] [PubMed] [Google Scholar]
- Katzenelson E, Buium I& Shuval HI (1976) Risk of communicable disease infection associated with wastewater irrigation in agricultural settlements. Science 194: 944–946. [DOI] [PubMed] [Google Scholar]
- Khamrin P, Maneekarn N, Peerakome S, Tonusin S, Malasao R, Mizuguchi M, Okitsu S & Ushijima H (2007) Genetic diversity of noroviruses and sapoviruses in children hospitalized with acute gastroenteritis in Chiang Mai, Thailand. J Med Virol 79: 1921–1926. [DOI] [PubMed] [Google Scholar]
- Ko G, Cromeans TL & Sobsey MD (2003) Detection of infectious adenovirus in cell culture by mRNA reverse transcription‐PCR. Appl Environ Microbiol 69: 7377–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko G, Cromeans TL & Sobsey MD (2005) UV inactivation of adenovirus type 41 measured by cell culture mRNA RT‐PCR. Water Res 39: 3643–3649. [DOI] [PubMed] [Google Scholar]
- Konowalchuk J & Speirs JI (1978) Antiviral effect of commercial juices and beverages. Appl Environ Microbiol 35: 1219–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M & Duizer E (2004) Foodborne viruses: an emerging problem. Int J Food Microbiol 90: 23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M, von Bonsdorff CH, Vinjé J, de Medici D & Monroe S (2002) Foodborne viruses. FEMS Microbiol Rev 26: 187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kott H & Fishelson L (1974) Survival of enteroviruses on vegetables irrigated with chlorinated oxidation pond effluents. Isr J Technol 12: 290–297. [Google Scholar]
- Kovač K, Diez‐Valcarce M, Hernandez M, Raspor P & Rodríguez‐Lázaro D (2010) High hydrostatic pressure as emergent technology for the elimination of foodborne viruses. Trends Food Sci Technol 21: 558–568. [Google Scholar]
- Kruse H, Kirkemo AM & Handeland K (2004) Wildlife as a source of zoonotic infection. Emerg Infect Dis 10: 2067–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo HW, Schmid D, Jelovcan S, Pichler AM, Magnet E, Reichart S & Allerberger F (2009) A foodborne outbreak due to norovirus in Austria, 2007. J Food Prot 72: 193–196. [DOI] [PubMed] [Google Scholar]
- Kuusi M, Nuorti JP, Maunula L, Minh NN, Ratia M, Karlsson J & von Bonsdorff CH (2002) A prolonged outbreak of Norwalk‐like calicivirus (NLV) gastroenteritis in a rehabilitation centre due to environmental contamination. Epidemiol Infect 129: 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G, Fontana S, Di Grazia A, Iaconelli M, Pourshaban M & Muscillo M (2007) Molecular identification and genetic analysis of norovirus genogroups I and II in water environments: comparative analysis of different reverse transcription‐PCR assays. Appl Environ Microbiol 73: 4152–4161 [Erratum in: Appl Environ Microbiol 2007, 73: 6329]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G, Pourshaban M, Iaconelli M & Muscillo M (2008) Detection of genogroup IV noroviruses in environmental and clinical samples and partial sequencing through rapid amplification of cDNA ends. Arch Virol 153: 2077–2083. [DOI] [PubMed] [Google Scholar]
- La Rosa G, Pourshaban M, Iaconelli M & Muscillo M (2009) Quantification of Norovirus genogroups I and II in environmental and clinical samples using Taqman Real‐Time RT‐PCR. Food Environ Virol 1: 15–22. [Google Scholar]
- La Rosa G, Iaconelli M, Pourshaban M & Muscillo M (2010) Detection And Molecular Characterization Of Noroviruses From Five Sewage Treatment Plants In Central Italy. Water Res 44: 1777–1784. [DOI] [PubMed] [Google Scholar]
- Lamhoujeb S, Fliss I, Ngazoa SE & Jean J (2008) Evaluation of the persistence of infectious human noroviruses on food surfaces by using real‐time nucleic acid sequence‐based amplification. Appl Environ Microbiol 74: 3349–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamhoujeb SFI, Ngazoa SE & Jean J (2009) Molecular study of the persistence of infectious human norovirus on food‐contact surfaces. Food Environ Virol 1: 51–56. [Google Scholar]
- Larsson MM, Rydell GE, Grahn A, Rodríguez‐Diaz J, Akerlind B, Hutson AM, Estes MK, Larson G & Svensson L (2006) Antibody prevalence and titer to norovirus (genogroup II) correlate with secretor (FUT2) but not with ABO phenotype or Lewis (FUT3) genotype. J Infect Dis 194: 1422–1427. [DOI] [PubMed] [Google Scholar]
- Le Cann P, Ranarijaona S, Monpoeho S, Le Guyader F & Ferre V (2004) Quantification of human astroviruses in sewage using real‐time RT‐PCR. Res Microbiol 155: 11–15. [DOI] [PubMed] [Google Scholar]
- Le Guyader F, Haugarreau L, Miossec L, Dubois E & Pommepuy M (2000) Three‐year study to assess human enteric viruses in shellfish. Appl Environ Microbiol 66: 3241–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guyader FS, Bon F, DeMedici D et al (2006a) Detection of multiple noroviruses associated with an international gastroenteritis outbreak linked to oyster consumption. J Clin Microbiol 44: 3878–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guyader FS, Loisy F, Atmar RL, Hutson AM, Estes MK, Ruvoen‐Clouet N, Pommepuy M & Le Pendu J (2006b) Norwalk virus‐specific binding to oyster digestive tissues. Emerg Infect Dis 12: 931–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guyader FS, Le Saux JC, Ambert‐Balay K, Krol J, Serais O, Parnaudeau S, Giraudon H, Delmas G, Pommepuy M, Pothier P & Atmar RL (2008) Aichi virus, norovirus, astrovirus, enterovirus, and rotavirus involved in clinical cases from a French oyster‐related gastroenteritis outbreak. J Clin Microbiol 12: 4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc H, Schwartzbrod L & Dei‐Cas E (2002) Microbial agents associated with waterborne diseases. Crit Rev Microbiol 28: 371–409. [DOI] [PubMed] [Google Scholar]
- Lee HK & Jeong YS (2004) Comparison of total culturable virus assay and multiplex integrated cell culture‐PCR for reliability of waterborne virus detection. Appl Environ Microbiol 70: 3632–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH & Kim SJ (2002) Detection of infectious enteroviruses and adenoviruses in tap water in urban areas in Korea. Water Res 36: 248–256. [DOI] [PubMed] [Google Scholar]
- Lees D (2000) Viruses and bivalve shellfish. Int J Food Microbiol 59: 81–116. [DOI] [PubMed] [Google Scholar]
- Lees D & CEN WG6 TAG4 (2010) International standardisation of a method for detection of human pathogenic viruses in molluscan shellfish. Food Env Virol 2: 146–155. [Google Scholar]
- Ley V, Higgins J & Fayer R (2002) Bovine enteroviruses as indicators of fecal contamination. Appl Environ Microbiol 68: 3455–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JW, Xin ZT, Wang XW, Zheng JL & Chao FH (2002) Mechanisms of inactivation of hepatitis A virus by chlorine. Appl Environ Microbiol 68: 4951–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JW, Xin ZT, Wang XW, Zheng JL & Chao FH (2004) Mechanisms of inactivation of hepatitis A virus in water by chlorine dioxide. Water Res 38: 1514–1519. [DOI] [PubMed] [Google Scholar]
- Li TC, Chijiwa K, Sera N et al (2005) Hepatitis E virus transmission from wild boar meat. Emerg Infect Dis 11: 1958–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Gu AZ, He M, Shi H‐C & Yang W (2009) UV inactivation and resistance of rotavirus evaluated by integrated cell culture and real‐time RT‐PCR assay. Water Res 43: 3261–3269. [DOI] [PubMed] [Google Scholar]
- Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, Stewart P, LePendu J & Baric R (2003) Human susceptibility and resistance to Norwalk virus infection. Nat Med 9: 548–553. [DOI] [PubMed] [Google Scholar]
- Lipp EK, Farrah SA & Rose JB (2001) Assessment of microbiological fecal pollution and human enteric pathogens in a coastal community. Mar Pollut Bull 42: 286–293. [DOI] [PubMed] [Google Scholar]
- Lodder WJ & de Roda Husman AM (2005) Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Appl Environ Microbiol 71: 1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder WJ, van den Berg HHJL, Rutjes SA & de Roda‐Husman AM (2010) Presence of enteric viruses in source waters for drinking water production in the Netherlands. Appl Environ Microbiol 76: 5965–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisy F, Le Cann P, Pommepuy M & Le Guyader FS (2000) An improved method for the detection of Norwalk‐like caliciviruses in environmental samples. Lett Appl Microbiol 31: 411–415. [DOI] [PubMed] [Google Scholar]
- Lopman BA, Reacher MH, Vipond IB, Hill D, Perry C, Halladay T, Brown DW, Edmunds WJ & Sarangi J (2004) Epidemiology and cost of nosocomial gastroenteritis, Avon, England, 2002‐2003. Emerg Infect Dis 10: 1827–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Vinjé J, Khalil SM, Murphy J, Lovelace GL & Sobsey MD (2008) Evaluation of RT‐PCR and reverse line blot hybridization for detection and genotyping F+ RNA coliphages from estuarine waters and molluscan shellfish. J Appl Microbiol 104: 1203–1212. [DOI] [PubMed] [Google Scholar]
- Lu L, Li C & Hagedorn CH (2006) Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Microbiol 16: 5–36. [DOI] [PubMed] [Google Scholar]
- Lytle CD & Sagripanti JL (2005) Predicted inactivation of viruses of relevance to biodefense by solar radiation. J Virol 79: 14244–14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maalouf H, Schaeffer J, Parnaudeau S, Le Pendu J, Atmar RL, Crawford SE & Le Guyader FS (2011) Strain‐dependent norovirus bioaccumulation in oysters. Appl Environ Microbiol 10: 3189–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy JM, Peron JM, Abravanel F, Poirson H, Dubois M, Miedouge M, Vischi F, Alric L, Vinel JP & Izopet J (2004) Hepatitis E in the south west of France in individuals who have never visited an endemic area. J Med Virol 74: 419–424. [DOI] [PubMed] [Google Scholar]
- Marks PJ, Vipond IB, Carlisle D, Deakin D, Fey RE & Caul EO (2000) Evidence for airborne transmission of Norwalk‐like virus (NLV) in a hotel restaurant. Epidemiol Infect 124: 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PJ, Vipond IB, Regan FM, Wedgwood K, Fey RE & Caul EO (2003) A school outbreak of Norwalk‐like virus: evidence for airborne transmission. Epidemiol Infect 131: 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli F, Caprioli A, Zengarini M, Marata A, Fiegna C, Di Bartolo I, Ruggeri FM, Delogu M & Ostanello F (2008) Detection of hepatits E virus (HEV) in a demographic managed wild boar (Sus scrofa scrofa) population in Italy. Vet Microbiol 126: 74–81. [DOI] [PubMed] [Google Scholar]
- Martínez‐Martínez M, Diez‐Valcarce M, Hernández M & Rodríguez‐Lázaro D (2011) Design and application of nucleic acid standards for quantitative detection of enteric viruses by real‐time PCR. Food Environ Virol 3: 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone WJ, Hierholzer JC, Keenlyside RA et al (1980) An outbreak of adenovirus type 3 disease at a private recreation centre swimming pool. Am J Epidemiol 111: 229–237. [DOI] [PubMed] [Google Scholar]
- Matsuda H, Okada K, Takahashi K & Mishiro S (2003) Severe hepatitis E virus infection after ingestion of uncooked liver from a wild boar. J Infect Dis 188: 944. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Rahman M et al (2008) Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol 153: 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K, Karthikeyan K, Abebe M, Malik N, Sattar SA, Farber JM & Bidawid S (2007) Survival of calicivirus in foods and on surfaces: experiments with feline calicivirus as a surrogate for norovirus. J Food Prot 70: 500–503. [DOI] [PubMed] [Google Scholar]
- Maunula L, Kalso S, Von Bonsdorff CH & Ponka A (2004) Wading pool water contaminated with both noroviruses and astroviruses as the source of a gastroenteritis outbreak. Epidemiol Infect 132: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunula L, Miettinen IT & von Bonsdorff CH (2005) Norovirus outbreaks from drinking water. Emerg Infect Dis 11: 1716–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney KR, Gong YY & Lewis TG (2006) Environmental transmission of SARS at Amoy Gardens. J Environ Health 68: 26–30. [PubMed] [Google Scholar]
- Mena KD & Gerba CP (2009) Waterborne adenovirus. Rev Environ Contam Toxicol 198: 133–167. [DOI] [PubMed] [Google Scholar]
- Mendez E & Arias CF (2007) Astroviruses Fields Virology, 5th edn (Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B. & Straus SE, eds), pp. 981–1000. Lippincot William & Wilkins, Philadelphia, PA. [Google Scholar]
- Metcalf TG, Melnick JL & Estes MK (1995) Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology‐a trip of over 50 years. Annu Rev Microbiol 49: 461–487. [DOI] [PubMed] [Google Scholar]
- Mocé i Llivina L (2004) Avenços metodològics en la detecció de virus entèrics en Aigües. PhD Thesis, Universitat de Barcelona, Barcelona, Spain. [Google Scholar]
- Monica B, Ramani S, Banerjee I et al (2007) Human caliciviruses in symptomatic and asymptomatic infections in children in Vellore, South India. J Med Virol 79: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Kaplan MH, McPhee J, Bregman DJ & Klein SW (1984) Epidemiologic, clinical and laboratory features of coxsackievirus B1‐B5 infections in the United States, 1970–79. Public Health Rep 99: 515–522. [PMC free article] [PubMed] [Google Scholar]
- Muniain‐Mujika I, Girones R, Tofino‐Quesada G, Calvo M & Lucena F (2002) Depuration dynamics of viruses in shellfish. Int J Food Microbiol 77: 125–133. [DOI] [PubMed] [Google Scholar]
- Muscillo M, Carducci A, La Rosa G, Cantiani L & Marianelli C (1997) Enteric virus detection in Adriatic seawater by cell culture, polymerase chain reaction and polyacrylamide gel electrophoresis. Wat Res 31: 1980–1984. [Google Scholar]
- Muscillo M, La Rosa G, Marianelli C, Zaniratti S, Capobianchi MR, Cantiani L & Carducci A (2001) A new RT‐PCR method for the identification of reoviruses in seawater samples. Water Res 35: 548–556. [DOI] [PubMed] [Google Scholar]
- Muscillo M, Pourshaban M, Iaconelli M, Fontana S, Di Grazia A, Manzara S, Fadda G, Santangelo R & La Rosa G (2008) Detection and quantification of human adenoviruses in surface waters by nested PCR, TaqMan real‐time PCR and cell culture assays. Water Air Soil Pollut 191: 1–4.32214526 [Google Scholar]
- Mushahwar IK (2008) Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J Med Virol 80: 646–658. [DOI] [PubMed] [Google Scholar]
- Myrmel M, Rimstad E & Wasteson Y (2000) Immunomagnetic separation of a Norwalk‐like virus (genogroup I) in artificially contaminated environmental water samples. Int J Food Microbiol 62: 17–26. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Takahashi K, Taira K, Taira M, Ohno A, Sakugawa H, Arai M & Mishiro S (2006) Hepatitis E virus infection in wild mongoose in Okinawa, Japan: demonstration of anti‐HEV antibodies and a full‐genome nucleotide sequence. Hepatol Res 34: 137–140. [DOI] [PubMed] [Google Scholar]
- Noble RT & Fuhrman JA (2001) Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia 460: 175–184. [Google Scholar]
- Nordgren J, Matussek A, Mattsson A, Svensson L & Lindgren PE (2009) Prevalence of norovirus and factors influencing virus concentrations during one year in a full‐scale wastewater treatment plant. Water Res 43: 1117–1125. [DOI] [PubMed] [Google Scholar]
- Nuanualsuwan S & Cliver DO (2002) Pretreatment to avoid positive RT‐PCR results with inactivated viruses. J Virol Methods 104: 217–225. [DOI] [PubMed] [Google Scholar]
- Nuanualsuwan S & Cliver DO (2003) Capsid functions of inactivated human picornaviruses and feline calicivirus. Appl Environ Microbiol 69: 350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygård K, Andersson Y, Lindkvist P et al (2001) Imported rocket salad partly responsible for increased incidence of hepatitis A cases in Sweden, 2000–2001. Euro Surveil 6: 151–153. [DOI] [PubMed] [Google Scholar]
- Nygård K, Torven M, Ancker C, Knauth SB, Hedlund KO, Giesecke J, Andersson Y & Svensson L (2003) Emerging genotype (GGIIb) of norovirus in drinking water, Sweden. Emerg Infect Dis 9: 1548–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen CW, Brammer L, Easterday BC, Arden N, Belay E, Baker I & Cox NJ (2002) Serologic evidence of H1 swine influenza virus infection in swine farm residents and employees. Emerg Infect Dis 8: 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar UD, Gibson CJ, Bresse JS & Glass RI (2006) Rotavirus and severe childhood diarrhea. Emerg Infect Dis 12: 304–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J & Parashar UD (2008) Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis 14: 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MM, Hall AJ, Vinje J & Parashar UD (2009) Noroviruses: a comprehensive review. J Clin Virol 44: 1–8. [DOI] [PubMed] [Google Scholar]
- Pebody RG, Leino T, Ruutu P, Kinnunen L, Davidkin I, Nohynek H & Leinikki P (1998) Foodborne outbreaks of hepatitis A in a low endemic country: an emerging problem? Epidemiol Infect 120: 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaro F, Sorg I & Metzler A (1995) In situ inactivation of animal viruses and a coliphage in nonaerated liquid and semiliquid animal wastes. Appl Environ Microbiol 62: 92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterson SR, Ashbolt NJ & Sharma A (2001) Microbial risks from wastewater irrigation of salad crops: a screening‐level risk assessment. Water Environ Res 73: 667–672. [DOI] [PubMed] [Google Scholar]
- Pina S, Puig M, Lucena F, Jofre J & Girones R (1998) Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl Environ Microbiol 64: 3376–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina S, Buti M, Jardí R, Clemente‐Casares P, Jofre J & Girones R (2001) Genetic analysis of hepatitis A virus strains recovered from the environment and from patients with acute hepatitis. J Gen Virol 82: 2955–2963. [DOI] [PubMed] [Google Scholar]
- Pintó RM & Bosch A (2008) Rethinking virus detection in food Foodborne Viruses: Progress and Challenges (Koopmans M, Cliver DO. & Bosch A, eds), pp. 171–188. ASM Press, Washington, DC. [Google Scholar]
- Pintó RM, Costafreda MI & Bosch A (2009) Risk assessment in shellfish‐borne outbreaks of Hepatitis A. Appl Environ Microbiol 75: 7350–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Jofre J, Lucena F, Allard A, Wadell G & Girones R (1994) Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl Environ Microbiol 60: 2963–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell RH & Emerson SU (2001) Hepatitis E virus. Rev Med Virol 13: 145–154. [DOI] [PubMed] [Google Scholar]
- Raphael RA, Sattar SA& Springthorpe VS (1985) Long‐term survival of human rotavirus in raw and treated river water. Can J Microbiol 31: 124–128. [DOI] [PubMed] [Google Scholar]
- Reuter G, Fodor D, Forgách P, Kátai A & Szucs G (2009) Characterization and zoonotic potential of endemic hepatitis E virus (HEV) strains in humans and animals in Hungary. J Clin Virol 44: 277–281. [DOI] [PubMed] [Google Scholar]
- Reynolds KA, Gerba CP & Pepper IL (1996) Detection of infectious enteroviruses by an integrated cell culture‐PCR procedure. Appl Environ Microbiol 62: 1424–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GP (1999) Limitations of molecular biological techniques for assessing the virological safety of foods. J Food Prot 62: 691–697. [DOI] [PubMed] [Google Scholar]
- Richards GP (2001) Enteric virus contamination of foods through industrial practices: a primer on intervention strategies. J Ind Microbiol Biotechnol 27: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou P, Le Saux JC, Dumas F, Caprais MP, Le Guyader SF & Pommepuy M (2007) Microbial impact of small tributaries on water and shellfish quality in shallow coastal areas. Water Res 41: 2774–2786. [DOI] [PubMed] [Google Scholar]
- Robesyn E, De Schrijver K, Wollants E, Top G, Verbeeck J & Van Ranst M (2009) An outbreak of hepatitis A associated with the consumption of raw beef. J Clin Virol 44: 207–210. [DOI] [PubMed] [Google Scholar]
- Rockx BH, Vennema H, Hoebe CJ, Duizer E & Koopmans MP (2005) Association of histo‐blood group antigens and susceptibility to norovirus infections. J Infect Dis 191: 749–754. [DOI] [PubMed] [Google Scholar]
- Rodríguez RA, Pepper IL & Gerba CP (2009) Application of PCR‐based methods to assess the infectivity of enteric viruses in environmental samples. Appl Environ Micoribiol 75: 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Lázaro D, Lombard B, Smith HV et al (2007) Trends in analytical methodology in food safety and quality: monitoring microorganisms and genetically modified organisms. Trends Food Sci Technol 18: 306–319. [Google Scholar]
- Rodríguez‐Lázaro D, Cook N, D'Agostino M & Hernandez M (2009) Current challenges in molecular diagnostics in food microbiology Global Issues in Food Science and Technology (Barbosa‐Cánovas G, Mortimer A, Colonna P, Lineback D, Spiess W. & Buckle K, eds), pp. 211–223. Elsevier, Maryland Heights, MO. [Google Scholar]
- Roy S, Vandenbeghe LH, Kryazhimskiy S et al (2009) Isolation and characterisation of adenoviruses persistently shed from the gastrointestinal tract of non‐human primates. PLoS Pathog 5: e1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjes SA, Italiaander R, van den Berg HH, Lodder WJ & de Roda Husman AM (2005) Isolation and detection of enterovirus RNA from large‐volume water samples by using the NucliSens miniMAG system and real‐time nucleic acid sequence‐based amplification. Appl Environ Microbiol 71: 3734–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjes SA, Lodder‐Verschoor F, van der Poel WHM, van Duijnhoven YT & de Roda Husman AM (2006) Detection of noroviruses in foods: a study on virus extraction procedures in foods implicated in outbreaks of human gastroenteritis. J Food Prot 69: 1949–1956. [DOI] [PubMed] [Google Scholar]
- Rutjes SA, Lodder WJ, Bouwknegt M & de Roda Husman AM (2007) Increased hepatitis E virus prevalence on Dutch pig farms from 33 to 55% by using appropriate internal quality controls for RT‐PCR. J Virol Methods 143: 112–116. [DOI] [PubMed] [Google Scholar]
- Rutjes SA, Lodder WJ, Lodder‐Verschoor F, van den Berg HH, Vennema H, Duizer E, Koopmans M & de Roda Husman AM (2009a) Sources of hepatitis E virus genotype 3 in The Netherlands. Emerg Infect Dis 15: 381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjes SA, Lodder WJ, Docters van Leeuwen A & de Roda Husman AM (2009b) Detection of infectious rotavirus in naturally contaminated source waters for drinking water production. J Appl Microbiol 107: 97–105. [DOI] [PubMed] [Google Scholar]
- Rutjes SA, Lodder‐Verschoor F, Lodder WJ, Van der Giessen J, Reesink H, Bouwknegt M & de Roda‐Husman AM (2010) Seroprevalence and molecular detection of hepatitis E virus in wild boar and red deer in The Netherlands. J Virol Methods 168: 197–206. [DOI] [PubMed] [Google Scholar]
- Rzeżutka A & Cook N (2004) Survival of human enteric viruses in the environment and food. FEMS Microbiol Rev 28: 441–453. [DOI] [PubMed] [Google Scholar]
- Saad MD, Hussein HA, Bashandy MM, Kamel HH, Earhart KC, Fryauff DJ, Younan M & Mohamed AH (2007) Hepatitis E virus infection in work horses in Egypt. Inf Gen Evol 7: 368–373. [DOI] [PubMed] [Google Scholar]
- Sair AI, D'Souza DH, Moe CL & Jaykus LA (2002) Improved detection of human enteric viruses in foods by RT‐PCR. J Virol Methods 100: 57–69. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Padilla E, Grais RF, Guerin PJ, Steele AD, Burny ME & Luquero FJ (2009) Burden of disease and circulating serotypes of rotavirus infection in sub‐Saharan Africa: systematic review and meta‐analysis. Lancet Infect Dis 9: 567–576. [DOI] [PubMed] [Google Scholar]
- Sartorius B, Andersson Y, Velicko I et al (2007) Outbreak of norovirus in Vastra Gotaland associated with recreational activities at two lakes during August 2004. Scand J Infect Dis 39: 323–331. [DOI] [PubMed] [Google Scholar]
- Savichtcheva O & Okabe S (2006) Alternative indicators of fecal pollution: relations with pathogens and conventional indicators, current methodologies for direct pathogen monitoring and future application perspectives. Water Res 40: 2463–2476. [DOI] [PubMed] [Google Scholar]
- Savolainen C, Hovi T & Mulders MN (2001) Molecular epidemiology of echovirus 30 in Europe: succession of dominant sublineages within a major genotype. Arch Virol 146: 521–537. [DOI] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL & Griffin PM (2011) Foodborne illness acquired in the United States: major pathogens. Emerg Infect Dis 17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcella C, Carasi S, Cadoria F et al (2009) An outbreak of viral gastroenteritis linked to municipal water supply, Lombardy, Italy, June 2009. Euro Surveill 14: 1–3. [DOI] [PubMed] [Google Scholar]
- Schvoerer E, Bonnet F, Dubois V, Cazaux G, Serceau R, Fleury HJ & Lafon ME (2000) PCR detection of human enteric viruses in bathing areas, waste waters and human stools in Southwestern France. Res Microbiol 151: 693–701. [DOI] [PubMed] [Google Scholar]
- Schvoerer E, Ventura M, Dubos O, Cazaux G, Serceau R, Gournier N, Dubois V, Caminade P, Fleury HJ & Lafon ME (2001) Qualitative and quantitative molecular detection of enteroviruses in water from bathing areas and from a sewage treatment plant. Res Microbiol 152: 179–186. [DOI] [PubMed] [Google Scholar]
- Schwab KJ, Deleon R & Sobsey MD (1996) Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl Environ Microbiol 62: 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab KJ, Neill FH, Estes MK, Metcalf TG & Atmar RL (1998) Distribution of norwalk virus within shellfish following bioaccumulation and subsequent depuration by detection using RT‐PCR. J Food Prot 61: 1674–1680. [DOI] [PubMed] [Google Scholar]
- Sedmark G, Bina D & MacDonald J (2003) Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage from Milwaukee, Wisconsin, collected Aug 1994 to Dec 2002. Appl Environ Microbiol 69: 7181–7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellwood J, Dadswell JV & Slade JS (1981) Viruses in sewage as an indicator of their presence in the community. J Hyg Camb 86: 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh YC, Wong CI, Krantz JA & Hsu FC (2008) Detection of naturally occurring enteroviruses in waters using direct RT‐PCR and integrated cell culture‐RT‐PCR. J Virol Methods 149: 184–189. [DOI] [PubMed] [Google Scholar]
- Simonet J & Gantzer C (2006a) Degradation of the poliovirus 1 genome by chlorine dioxide. J Appl Microbiol 100: 862–870. [DOI] [PubMed] [Google Scholar]
- Simonet J & Gantzer C (2006b) Inactivation of poliovirus 1 and F‐specific RNA phages and degradation of their genomes by UV irradiation at 254 nanometers. Appl Environ Microbiol 72: 7671–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skraber S, Ogorzaly L, Helmi K, Maul A, Hoffman L, Cauchie HM & Gantzer C (2009) Occurrence and persistence of enteroviruses, noroviruses and F‐specific RNA phages in natural wastewater biofilms. Water Res 43: 4780–4789. [DOI] [PubMed] [Google Scholar]
- Smith A, Reacher M, Smerdon W, Adak GK, Nichols G & Chalmers RM (2006) Outbreaks of waterborne infectious intestinal disease in England and Wales, 1992‐2003. Epidemiol Infect 134: 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobsey MD, Shields PA, Hauchman FH, Hazard RL & Caton LW III (1989) Survival and transport of hepatitis A virus in soils, groundwater and wastewater. Water Sci Technol 10: 97–106. [Google Scholar]
- Springthorpe VS, Loh CL, Robertson WJ & Sattar SA (1993) In situ survival of indicator bacteria, MS‐2 phage and human pathogenic viruses in river water. Water Sci Technol 27: 413–420. [Google Scholar]
- Stanway G, Brown F, Christian P et al (2005) Picornaviridae. Virus Taxonomy – 8th Report of the International Committee on Taxonomy Viruses (Fauquet CM, Mayo MA, Maniloff J, Desselberger U. & Bull LA, eds), pp. 757–778. Elsevier Academic Press, London. [Google Scholar]
- Steyer A, Poljsak‐Prijatelj M, Barlic‐Maganja D & Marin J (2008) Human, porcine and bovine rotaviruses in Slovenia: evidence of interspecies transmission and genome reassortment. J Gen Virol 89: 1690–1698. [DOI] [PubMed] [Google Scholar]
- Suffredini E, Corrain C, Arcangeli G et al (2008) Occurrence of enteric viruses in shellfish and relation to climatic‐environmental factors. Lett Appl Microbiol 47: 467–474. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Kitajima N, Abe N & Mishiro S (2004) Complete or near‐complete nucleotide sequence of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology 330: 501–505. [DOI] [PubMed] [Google Scholar]
- Taku A, Gulati BR, Allwood PB, Palazzi K, Hedberg CW & Goyal SM (2002) Concentration and detection of caliciviruses from food contact surfaces. J Food Prot 65: 999–1004. [DOI] [PubMed] [Google Scholar]
- Tallon LA, Love DC, Moore ZS & Sobsey MD (2008) Recovery and sequence analysis of hepatitis A virus from spring water implicated in an outbreak of acute viral hepatitis. Appl Environ Microbiol 74: 6158–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LH, Latham SM & Woolhouse MEJ (2001) Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 356: 983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei S, Kitajima N, Takahashi K & Mishiro S (2003) Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362: 371–373. [DOI] [PubMed] [Google Scholar]
- ter Waarbeek HL, Dukers‐Muijrers NH, Vennema H & Hoebe CJ (2010) Waterborne gastroenteritis outbreak at a scouting camp caused by two norovirus genogroups: GI and GII. J Clin Virol 47: 268–272. [DOI] [PubMed] [Google Scholar]
- Teunis PF & Havelaar AH (2000) The Beta Poisson dose‐response model is not a single‐hit model. Risk Anal 20: 513–520. [DOI] [PubMed] [Google Scholar]
- Teunis PF, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J & Calderon RL (2008) Norwalk virus: how infectious is it? J Med Virol 80: 1468–1476. [DOI] [PubMed] [Google Scholar]
- Teunis PFM, Rutjes SA, Westrell T & de Roda‐Husman AM (2009) Characterization of drinking water treatment for virus risk assessment. Water Res 43: 395–404. [DOI] [PubMed] [Google Scholar]
- Thompson SS, Jackson JL, Suva‐Castillo M, Yanko WA, El Jack Z, Kuo J, Chen CL, Williams FP & Schnurr DP (2003) Detection of infectious human adenoviruses in tertiary‐treated and ultraviolet‐disinfected wastewater. Water Environ Res 75: 163–170. [DOI] [PubMed] [Google Scholar]
- Thorven M, Grahn A, Hedlund KO, Johansson H, Wahlfrid C, Larson G & Svensson L (2005) A homozygous nonsense mutation (428G–>A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J Virol 79: 15351–15355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston‐Enriquez JA, Haas CN, Jacangelo J & Gerba CP (2003) Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl Environ Microbiol 69: 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping JR, Schnerr H, Haines J et al (2009) Temperature inactivation of Feline calicivirus vaccine strain FCV F‐9 in comparison with human noroviruses using an RNA exposure assay and reverse transcribed quantitative real‐time polymerase chain reaction‐A novel method for predicting virus infectivity. J Virol Methods 156: 89–95. [DOI] [PubMed] [Google Scholar]
- Tree JA, Adams MR & Lees DN (2003) Chlorination of indicator bacteria and viruses in primary sewage effluent. Appl Environ Microbiol 69: 2038–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanucci A, Myrmel M, Berg I, von Bonsdorff CH & Maunula L (2009) Potential internalisation of caliciviruses in lettuce. Int J Food Microbiol 135: 175–178. [DOI] [PubMed] [Google Scholar]
- Van den Berg H, Lodder W, van der Poel WHM, Vennema H & de Roda Husman AM (2005) Genetic diversity of noroviruses in raw and treated sewage water. Res Microbiol 156: 532–540. [DOI] [PubMed] [Google Scholar]
- van der Poel WHM, Verschoor F, van de Heide R, Herrera MI, Vivo A, Kooreman M & de Roda‐Husman AM (2001) Hepatitis E virus sequences in swine related to sequences in humans, the Netherlands. Emerg Infect Dis 7: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorn LJ, Kleter B, Hoefnagel E, Stainier I, Poliszczak A, Colau B & Quint W (2009) Detection and genotyping of human rotavirus VP4 and VP7 genes by reverse transcriptase PCR and reverse hybridization. J Clin Microbiol 47: 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heerden J, Ehlers MM, Van Zyl WB & Grabow WOK (2003) Incidence of adenoviruses in raw and treated water. Water Res 37: 3704–3708. [DOI] [PubMed] [Google Scholar]
- Van Heerden J, Ehlers MM, Heim A & Grabow WOK (2005a) Prevalence, quantification and typing of adenoviruses detected in river and treated drinking water in South Africa. J Appl Microbiol 99: 234–242. [DOI] [PubMed] [Google Scholar]
- Van Heerden J, Ehlers MM, Vivier JC & Grabow WO (2005b) Risk assessment of adenoviruses detected in treated drinking water and recreational water. J Appl Microbiol 99: 926–933. [DOI] [PubMed] [Google Scholar]
- Van Zyl WB, Page NA, Grabow WO, Steele AD & Taylor MB (2006) Molecular epidemiology of group A rotaviruses in water sources and selected raw vegetables in southern Africa. Appl Environ Microbiol 72: 4554–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantarakis A & Papapetropoulou M (1999) Detection of enteroviruses, adenoviruses and hepatitis A viruses in raw sewage and treated effluents by nested‐PCR. Water Air Soil Pollut 114: 85–93. [Google Scholar]
- Vasickova P, Pavlik I, Verani M & Carducci A (2010) Issues concerning survival of viruses on surfaces. Food Environ Virol 2: 24–34. [Google Scholar]
- Verhoef L, Boxman IL, Duizer E, Rutjes SA, Vennema H, Friesema IH, de Roda Husman AM & Koopmans M (2008) Multiple exposures during a norovirus outbreak on a river‐cruise sailing through Europe, 2006. Euro Surveill 13: 18899. [PubMed] [Google Scholar]
- Verreault D, Moineau S & Duchaine C (2008) Methods for sampling of airborne viruses. Microbiol Mol Biol Rev 72: 413–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos R, Shroufi A, Sillis M, Aird H, Gallimore CI, Myers L, Mahgoub H & Nair P (2009) Food‐related norovirus outbreak among people attending two barbeques: epidemiological, virological, and environmental investigation. Int J Infect Dis 13: 629–635. [DOI] [PubMed] [Google Scholar]
- Waar K, Herremans MM, Vennema H, Koopmans MP & Benne CA (2005) Hepatitis E is a cause of unexplained hepatitis in The Netherlands. J ClinVirol 33: 145–149. [DOI] [PubMed] [Google Scholar]
- Wagner MM, Tsui FC, Espino JU, Dato VM, Sittig DF, Caruana RA, McGinnis LF, Deerfield DW, Druzdzel MJ & Fridsma DB (2001) The emerging science of very early detection of disease outbreaks. J Pub Health Mgmt Pract 26: 51–59. [DOI] [PubMed] [Google Scholar]
- Wang D, Wu Q, Yao L, Wei M, Kou X & Zhang J (2008) New target tissue for food‐borne virus detection in oysters. Lett Appl Microbiol 47: 405–409. [DOI] [PubMed] [Google Scholar]
- Webby RJ, Carville KS, Kirk MD et al (2007) Internationally distributed frozen oyster meat causing multiple outbreaks of norovirus infection in Australia. Clin Infect Dis 44: 1026–1031. [DOI] [PubMed] [Google Scholar]
- Weldon M, Vanegdom MJ, Hendrick KA et al (2000) Prevalence of antibody to hepatitis A virus in drinking water workers and wastewater workers in Texas from 1996 to 1997. J Occup Environ Med 42: 821–826. [DOI] [PubMed] [Google Scholar]
- Whitehead K & McCue KA (2009) Virucidal efficacy of disinfectant actives against feline calicivirus, a surrogate for norovirus, in a short contact time. Am J Infect Control 38: 26–30. [DOI] [PubMed] [Google Scholar]
- WHO (2003) Laboratory Biosafety Manual, 2nd edn World Health Organisation, Geneva, Switzerland. [Google Scholar]
- WHO (2006) Guidelines for the safe use of wastewater, excreta and greywater. Volume 2: Wastewater use in agriculture. World Health Organisation, Geneva, Switzerland. [Google Scholar]
- WHO (2008) WHO/HSE/EPR/2008.10 Guidance on regulations for the transport of infectious substances 2009–2010. World Health Organisation; Geneva, Switzerland. [Google Scholar]
- WHO & FAO (2008) Viruses in Food: Scientific Advice to Support Risk Management Activities. World Health Organization and Food and Agriculture Organization, Geneva, Switzerland. [Google Scholar]
- Widdowson MA, Jaspers WJ, Van der Poel WH, Verschoor F, De Roda‐Husman AM, Winter HL, Zaaijer HL & Koopmans M (2003) Cluster of cases of acute hepatitis associated with hepatitis E virus infection acquired in the Netherlands. Clin Inf Dis 36: 29–33. [DOI] [PubMed] [Google Scholar]
- Wold WSM & Horwitz MS (2007) Adenoviruses Fields Virology, 5th edn (Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B. & Straus SE, eds), pp. 2395–2436. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- Wu HM, Fornek M, Schwab KJ, Chapin AR, Gibson K, Schwab E, Spencer C & Henning K (2005) A norovirus outbreak at a long‐term‐care facility: the role of environmental surface contamination. Infect Control Hosp Epidemiol 26: 802–810. [DOI] [PubMed] [Google Scholar]
- Yates MV, Gerba CP & Kelley LM (1985) Virus persistence in groundwater. Appl Environ Microbiol 49: 778–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti AR, Schlauder GG, Romano L, Tanzi E, Fabris P, Dawson GJ & Mushahwar IK (1999) Identification of a novel variant of hepatitis E virus in Italy. J Med Virol 57: 356–360. [DOI] [PubMed] [Google Scholar]
- Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SWL, Hibberd ML, Liu ET, Rohwer F & Ruan YJ (2006) RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 4: 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI & Monroe SS (2006) Norovirus classification and proposed strain nomenclature. Virology 346: 312–323. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Purcell RH & Emerson SU (2003) An ELISA for putative neutralizing antibodies to hepatitis E virus detects antibodies to genotype 1, 2, and 4. Vaccine 22: 2578–2585. [DOI] [PubMed] [Google Scholar]
- Zwietering MH & Havelaar A (2006) Dose‐response relationships and food‐borne disease Food Consumption and Disease Risk (Potter ME, ed.), pp. 422–439. Woodhead Publishing, Cambridge. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Sampling methods used for detection viral hazards in food matrices.
Table S2. Sampling methods used for detection viral hazards in water samples.
Table S3. Detection methods for viral hazards in different environmental matrices.


