Highlights
-
•
Recombination accounts for the emergence of IBV variants.
-
•
Strain ck/CH/LGX/130530 is originated from recombination events between a pathogenic tl/CH/LDT3/03- and H120-like strain.
-
•
Recombination event occurred in the 5′ end of the Gene 1 region did not result in changes in the genotype, serotype, and protectotype of the IBV.
-
•
The replicase gene of IBV ck/CH/LGX/130530 isolate is associated with viral pathogenicity.
Keywords: Infectious bronchitis coronavirus, Recombination, Serotype, Pathogenicity, Vaccination-challenge test
Abstract
An infectious bronchitis coronavirus, designated as ck/CH/LGX/130530, was isolated from an IBV strain H120-vaccinated chicken in this study. Analysis of the S1 gene showed that isolate ck/CH/LGX/130530 was a tl/CH/LDT3/03-like virus, with a nucleotide sequence similarity of 99%. However, a complete genomic sequence analysis showed that ck/CH/LGX/130530 was more closely related to a Massachusetts type strain (95% similarity to strain H120) than to the tl/CH/LDT3/03 strain (86%), suggesting that recombination might have occurred during the origin of the virus. A SimPlot analysis of the complete genomic sequence confirmed this hypothesis, and it showed that isolate ck/CH/LGX/130530 emerged from a recombination event between parental IBV H120 strain and pathogenic tl/CH/LDT3/03-like virus. The results obtained from the pairwise comparison and nucleotide similarity showed that the recombination breakpoint was located in the nsp14 gene at nucleotides 17055–17083. In line with the high S1 gene sequence similarity, the ck/CH/LGX/130530 isolate was serotypically close to that of the tl/CH/LDT3/03 strain (73% antigenic relatedness). Furthermore, vaccination with the LDT3-A vaccine, which was derived from the tl/CH/LDT3/03 strain by serial passaging in chicken eggs, provided good protection against challenge with the tl/CH/LDT3/03 strain, in contrast to the poor protection offered with the H120 vaccine. Interestingly, isolate ck/CH/LGX/130530 exhibited low pathogenicity toward specific-pathogen-free chickens compared with the nephropathogenic tl/CH/LDT3/03 strain, which was likely due to natural recombination in the 5′ 17-kb region of the genome. Our results also indicate that the replicase gene of IBV isolate ck/CH/LGX/130530 is associated with viral pathogenicity.
1. Introduction
Infectious bronchitis virus (IBV), a member of the Gammacoronaviridae, order Nidovirales, is an important pathogen that infects domestic chickens of all ages, causing an acute, highly contagious respiratory disease (Cavanagh and Gelb Jr., 2008). The IBV genome contains a single positive-strand RNA molecule, which is about 27.6 kb long and which has a cap at its 5′ end and a poly (A) tail at its 3′ end (Boursnell et al., 1987). The genome of IBV comprises ten open reading frames (ORFs), and the first 20 kb of the genome is made up of ORF1, which is a replicase gene. The replicase has two ORFs, 1a and 1b, and 1b is produced as a 1ab polyprotein by a −1 ribosomal frame-shifting mechanism. ORF1 encodes 15 non-structural proteins associated with RNA replication and transcription. The IBV genome encodes four major structural proteins: the spike (S) glycoprotein, the small envelope (E) protein, the membrane (M) glycoprotein, and the nucleocapsid (N) protein (Cavanagh and Gelb, 2008). In addition, IBV has other genes that encode for proteins interspersed among structural genes, namely 3a, 3b, 5a, and 5b (Boursnell et al., 1987).
Numerous serotypes have been described for IBV thus far, as frequent point mutations occur in the S1 domain of the S gene of the virus. Another important feature of IBV, as well as other coronaviruses, is the high rate of homologous and non-homologous RNA–RNA recombination that has been demonstrated to occur among selected and unselected markers during the course of infection (Masters, 2006). RNA recombination in coronaviruses is thought to result from a copy-choice mechanism (Kirkegaard and Baltimore, 1986). In this scheme, the viral polymerase, with its nascent RNA strand intact, detaches from one template and resumes elongation at the identical position, or a similar position, on another template. Although strong selective pressures are able to create the appearance of local clustering of recombinational hot spots (Banner et al., 1990), the sites of recombination are considered to be random (Banner and Lai, 1991). Because homologous recombination can occur between genomic and subgenomic RNAs, with the latter providing a source of donor and acceptor templates that become more numerous as a function of proximity to the 3′ end of the genome, the rate of recombination increases from the 5′ to 3′ end of the coronavirus genome, (Masters, 2006).
Accumulating evidence suggests that recombination is an inherent component in IBV evolution, and it is believed that recombination events can occur in the field under the following conditions: when there are extremely large numbers of chickens, especially when kept at high density; when the virus is easily spread; and when there is a co-circulation of serotypes, including proof of co-infection with more than one serotype in a given flock (Cavanagh, 2007). The circumstantial evidence for recombination, derived from gene sequence comparisons, has shown that the recombination events had occurred a very long time ago. However, it was shown that recombination in the S1 domain of the S protein of the IBV genome is a mechanism that leads to the emergence of new IBV strains, and it could be responsible for changes in viral pathogenicity and replication in a single generation (Hewson et al., 2014). In general, it is difficult to ascertain what effect recombination has on the serotype, pathogenicity, virulence, and tissue tropism of recombinant strains using only gene sequence analyses. Serotyping and animal studies, paired with differential analyses of genes, genomic sequencing, and phylogenetic analyses, will be essential to provide insights into the altered characteristics of recombinant IBV strains and their effect on poultry health.
2. Materials and methods
2.1. Epidemiological background and virus isolation
An IBV strain, designated as ck/CH/LGX/130530, was isolated from tracheal swabs of diseased broilers suspected of having an IBV infection in Guangxi province, China, in 2013. The flock investigated in this study contained about 15,000 broilers. One-day-old chickens were vaccinated against IBV with the live attenuated H120 vaccine, and 15-day-old chickens boosted with Ma5 live vaccines. Some of the chickens showed early clinical signs, including listlessness, huddling, and ruffled feathers, when they were 20 days old. On day 25, some of the diseased chickens began to die, and gross examinations showed mild to severe tracheitis, nephritis, and proventriculitis. The morbidity rate was about 7%, and the mortality rate was 3%. The clinical signs of the chickens tended to disappear gradually after about day 35.
Tracheal swabs were collected from 30 chickens in the diseased flock on day 23 and centrifuged at 6000 × g for 10 min. The supernatants were pooled and inoculated into five 9-day-old embryonated specific-pathogen-free (SPF) eggs via the allantoic cavity (0.2 ml per egg). The eggs were candled daily, and embryos deaths that occurred within 24 h of inoculation were considered to be non-specific. Allantoic fluids were collected at 96 h post-inoculation, pooled together, and clarified by centrifugation at 3000 × g for 5 min. The clear supernatant was used for further passaging. The samples were blind passaged four times, and characteristic IBV lesions, such as dwarfing, stunting, or curling of embryos, were observed at the fourth passage level. Allantoic fluids of the inoculated embryos were collected for reverse transcription-PCR (RT-PCR) amplification as previously described (Liu and Kong, 2004).
In addition, a pathogenic IBV strain, tl/CH/LDT3/03, and two vaccine strains, H120 and LDT3-A; the LDT3 was derived from the tl/CH/LDT3/03 strain by serial passaging in chicken eggs, were used in the virus cross-neutralization and vaccination-challenge tests in this study. The pathogenic strain tl/CH/LDT3/03 and vaccine strain LDT3-A were also used for complete genomic sequencing. Embryo-propagated viral stocks of these viruses were produced by inoculating the virus into embryonated SPF chicken eggs via the allantoic cavity, and the infectious allantoic fluid was collected 48 h post-inoculation as previously described (Liu and Kong, 2004). The titers of the four viruses, ck/CH/LGX/130530, tl/CH/LDT3/03, H120, and LDT3-A, were determined by inoculating 10-fold dilutions into groups of five 10-day-old embryonated chicken eggs. The median embryo infectious dose (EID50) was calculated using the method of Reed and Muench (1938).
All experiments were conducted using standard procedures with the formal approval of the Ethical and Animal Welfare Committee of Harbin Veterinary Research Institute, China.
2.2. RNA extraction, gene amplification, and sequencing
Genomic RNA was extracted from virus-infected allantoic fluid with the RNAiso Plus kit (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions and stored at −80 °C until further use. Twenty overlapping primers (Zhang et al., 2015) were used to amplify the genomes of the isolate ck/CH/LGX/130530, strain tl/CH/LDT3/03 and the LDT3-A vaccine strain. All gene fragments were amplified using the RT-PCR kit (TaKaRa) according to the manufacturer’s instructions. The 3′/5′-termini of the three IBV strains were determined as previously described (Zhang et al., 2015) using the 3′/5′ RACE kit (TaKaRa) according to the manufacturer’s instructions. The RT-PCR products were sequenced directly and/or cloned into the pMD18-T vector (TaKaRa), and three independent clones were sequenced for each amplicon.
2.3. Sequence analysis
The assembly of contiguous sequences of ck/CH/LGX/130530, tl/CH/LDT3/03 and LDT3-A was performed with GeneDoc software (Ammayappan and Vakharia, 2009). Both the S1 gene and complete genomic sequences of IBV isolate ck/CH/LGX/130530 were used for BLAST searching of the National Center for Biotechnology Information (NCBI) database. The tl/CH/LDT3/03 and partridge/GD/S1/42003 strains, as well as the LDT3-A vaccine, were used as one group of parental viruses for sequence comparisons. In addition, Massachusetts type H120 and Mass 41 strains were selected and used as another group of parental viruses for sequence comparisons. Similarity and breakpoint analyses of the complete genomic sequence of ck/CH/LGX/130530 were aligned with those of tl/CH/LDT3/03, LDT3-A, partridge/GD/S1/42003, H120, and Mass 41, and the multiple alignment results were introduced into SimPlot version 3.5.1 to identify likely recombination breakpoints (Lole et al., 1999). SimPlot analyses were performed using a 1000 bp window with a 100 bp step. IBV strain Mass 41 was used as the query strain. To confirm regions in which recombination events occurred in the genome of the ck/CH/LGX/130530 isolate, pairwise comparisons of the complete genomic sequence of ck/CH/LGX/130530 were performed with those of tl/CH/LDT3/03, LDT3-A, H120, and M41, and the nucleotide similarities of the different corresponding gene fragments were calculated. In addition, pairwise comparisons of the sequence of the 3′ 7.0 kb region of ck/CH/LGX/130530 were performed with those of partridge/GD/S1/42003, tl/CH/LDT3/03, and LDT3-A to elucidate whether the tl/CH/LDT3/03-like sequence of isolate ck/CH/LGX/130530 was derived from the pathogenic tl/CH/LDT3/03 or attenuated LDT3-A vaccine strains.
2.4. Confirmation of the recombination in the natural condition
In order to confirm the predictive recombination event that occurred in the genomes of the ck/CH/LGX/130530 had a natural origin, and was not a methodological artifact originated during the inoculation in eggs, five of the 30 tracheal swabs which were used for virus isolation were selected and used for amplification of the switch site and flanking sequences independently with primers Rec5′ (5′-GTGGTATATTGGTTGTCATGCGC-3′) and Rec3′ (5′-CAACAAGCCCTTCCGGTGTAAC-3′). One of the amplicon was sequenced directly.
2.5. GenBank accession numbers
The complete genomic sequences of IBV isolate ck/CH/LGX/130530, strain tl/CH/LDT3/03 and the LDT3-A vaccine strain were deposited into GenBank (accession numbers KP343691, KT852992 and KR608272, respectively).
2.6. Virus cross-neutralization tests
Virus neutralization tests were performed using anti-sera against strains ck/CH/LGX/130530, tl/CH/LDT3/03, and H120 to determine their antigenic relationship. For serotyping, reciprocal β virus neutralization tests were performed using constant (102 EID50) viral titers and diluted serum against ck/CH/LGX/130530, tl/CH/LDT3/03, or H120 in SPF chicken embryos. The end-point of each serum sample was calculated using the methods of Reed and Muench (1938). Cross-reactivity R values were calculated according to the formula published by Archett and Horsfall (1950).
2.7. Pathogenicity to 1-day-old SPF chickens
Three groups of 10 1-day-old SPF layer chickens were placed in separate isolators. Birds in groups 1 and 2 were challenged by intraocular/intranasal route at one-day-old with 105 EID50 of the isolate ck/CH/LGX/130530 and strain tl/CH/LDT3/03, respectively. Birds in group 3 were mock inoculated with the same volume of sterile allantoic fluid. Blood samples were collected on days 4, 8, 12, 16, 20 and 24 post-challenge from all birds. Chicks were monitored daily for clinical signs, such as tracheal rales, nasal discharge, watery eye, and wheezing. The clinical signs from all the birds were counted by three people over a 2-min period. Morbidity and mortality were recorded daily for 24 days following IBV challenge. Gross lesions were also carefully examined and recorded for the dead chickens. The kidneys of the dead chickens infected with IBV strain tl/CH/LDT3/03 were subjected to immunohistochemistry (IHC) for the detection of IBV antigen using monoclonal antibody 6D10 (Han et al., 2013) directed against the nucleoprotein as previously described (de Wit et al., 2011a, de Wit et al., 2011b, Xu et al., 2015).
2.8. Protection provided by vaccination of H120 and LDT3-A against challenge of isolate ck/CH/LGX/130530
Sixty one-day-old SPF chickens were divided into six groups. One-day-old birds in groups 1 and 3 were vaccinated with 104 EID50 of the H120 vaccine via the intranasal route. Birds in group 2 were vaccinated with the same dose of the LDT3-A vaccine. Birds in groups 4, 5, and 6 were mock inoculated with the same volume of sterile allantoic fluid. Blood samples were collected on 4, 8, 12, 16, and 20 days of age from birds in all groups. At 20 days of age, birds in groups 1, 2, and 5 were challenged with 1 × 105 EID50 of the ck/CH/LGX/130530 isolate via the intraocular/intranasal route. Birds in groups 3 and 4 were challenged with 1 × 105 EID50 of pathogenic tl/CH/LDT3/03 (Liu et al., 2005). Birds in group 6 were mock-inoculated with sterile allantoic fluid and served as the negative control. Tracheal swabs and blood samples were collected on days 4, 8, 12, and 16 post-challenge from all birds. The tracheal swabs were used for virus re-isolation, and the blood samples were used to examine antibodies against IBV.
2.9. Virus re-isolation and antibody detection
The tracheal swabs were centrifuged individually at 6000 × g for 10 min, and each of the supernatant samples was inoculated into two to five 9-day-old SPF embryonated eggs via the allantoic cavity (0.2 ml per egg). The eggs were candled daily until 7 days post-inoculation and then examined for characteristic IBV lesions, such as dwarfing, stunting, or curling of embryos. Allantoic fluid from two of the inoculated embryos was collected, pooled, and used for RT-PCR amplification as previously described (Liu and Kong, 2004). A positive sample was recorded if specific lesions were observed and the RT-PCR amplification was positive. Sera were examined for antibodies against IBV using a commercial enzyme-linked immunosorbent assay (ELISA) kit (IDEXX, Portland, ME, USA) according to the manufacturer’s instructions.
3. Results
3.1. Molecular characteristics of the IBV ck/CH/LGX/130530 isolate, tl/CH/LDT3/03 strain and the LDT3-A vaccine
Each of the sequences of the IBV isolate ck/CH/LGX/130530, strain tl/CH/LDT3/03 and the LDT3-A vaccine were assembled into a complete genome. The size of the ck/CH/LGX/130530 and LDT3-A genomes were 27,613, 27,670 and 27,687 nucleotides, respectively, excluding the poly-A tail at the 3′ ends of the genomes. The genome organization and arrangement of the three IBV strains are in the order of: 5′ untranslated region (UTR), open reading frame 1a/1b (ORF1a/1b), S, ORF3, M, ORF5, N, and 3′ UTR.
The results of the BLAST search showed that the S1 gene of isolate ck/CH/LGX/130530 was closely related to two strains, tl/CH/LDT3/03 and partridge/GD/S1/42003 (99% similarity), which were both isolated in Guangdong province, China, in 2003. This result indicated that the IBV ck/CH/LGX/130530 isolate has a tl/CH/LDT3/03 genotype. In contrast, the S1 gene of this isolate had less than 85% similarity to that of the H120 vaccine strain. Interestingly, the results of the BLAST search using the complete genomic sequence showed that ck/CH/LGX/130530 was closely related to a Massachusetts type strain (95% similarity to H120 strain); however, it showed less than 86% nucleotide similarity to Massachusetts H120, indicating that a recombination event likely occurred in the genome of the ck/CH/LGX/130530 isolate.
3.2. The genomic sequence of the IBV ck/CH/LGX/130530 isolate is mosaic
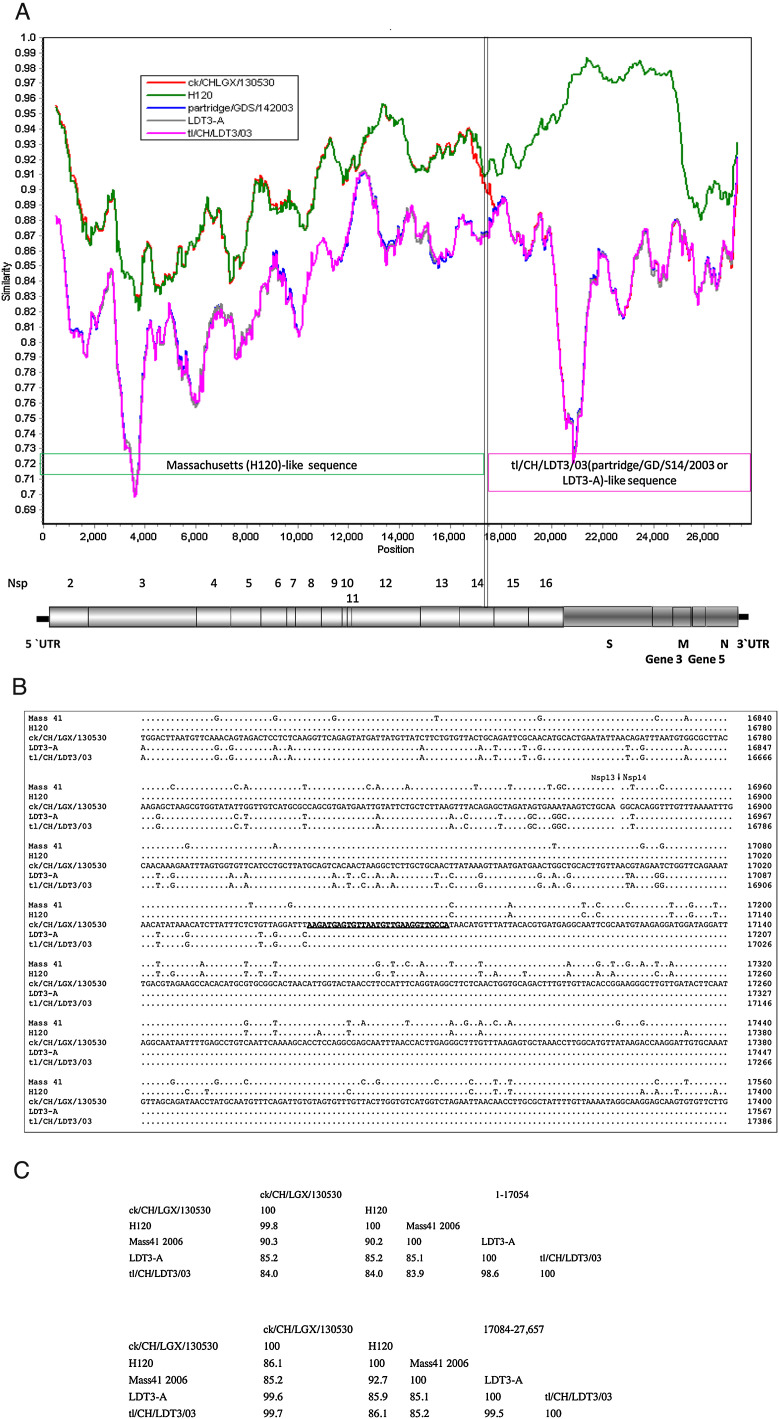
To identify the recombination events that possibly occurred in the genome of the IBV ck/CH/LGX/130530 isolate, similarity plots were performed using strains H120, tl/CH/LDT3/03I, LDT3-A, and partridge/GD/S14/2003 as representatives of the two main groups of IBVs, while the M41 strain served as a query. As illustrated in Fig. 1 A, a similarity plot identified a crossover event in the nsp14 of Gene 1 region.
Fig. 1.
Recombination analysis of the IBV ck/CH/LGX/130530 isolate. Similarity plot using Mass 41 as the query sequence (A). The dotted lines show the deduced recombination breakpoints. The hollow arrows show the different fragments and their colors are the same as those of the parental viruses. The numbers show the nucleotide positions of the corresponding fragments in the genome of the ck/CH/LGX/130530 isolate. Multiple sequence alignment of the predicted breakpoint and flanking sequences among IBV Mass 41, H120, ck/CH/LGX/130530, tl/CH/LDT3/03 and LDT3-A strains (B). The numbers on the right of each alignment show the nucleotide positions in the genome of each virus. The sequences of ck/CH/LGX/130530 are listed, and only the nucleotides differing from those of ck/CH/LGX/130530 are depicted. The region where the template switches (breakpoint) have taken place is underlined. The deleted nucleotides are indicated by a -. GenBank accession numbers are in bold. The GenBank accession numbers are Mass 41 (AY851295), H120 (FJ888351), and partridge/GD/S14/2003 (AY646283). Percentages of nucleotide sequence identity among Mass 41, H120, ck/CH/LGX/130530, tl/CH/LDT3/03 and LDT3-A strains (C). The percentages of nucleotide sequence identity of the corresponding gene fragments are indicated.
To obtain the precise regions of the possible crossover points involved in the recombination events, the genomic sequence of the ck/CH/LGX/130530 isolate was carefully pairwise compared with those of the H120, M41, tl/CH/LDT3/03 and LDT3-A viruses. As shown in Fig. 1B, a recombination breakpoint (nt 17,055–17,083) was located in the nsp14 gene of isolate ck/CH/LGX/130530. In addition, data regarding nucleotide similarities using the corresponding gene fragments supported the aforementioned results (Fig. 1C).
Our results demonstrated that the Massachusetts-like sequences in the genome of the ck/CH/LGX/130530 isolate were closely related to those of the H120 vaccine strain, rather than those of the pathogenic M41 strain. In contrast, of the 21 mutations between the pathogenic tl/CH/LDT3/03 and the LDT3-A vaccine strains in the 3′ 7.0 kb region sequences, ck/CH/LGX/130530 shared 16 mutations with the pathogenic tl/CH/LDT3/03 strain and only five with the LDT3-A strain (Table 1 ), suggesting that this region of ck/CH/LGX/130530 was more similar to that of the pathogenic tl/CH/LDT3/03 strain than the LDT3-A vaccine.
Table 1.
Pairwise comparisons of nucleotide sequences of the 3′ 7.0-kb region of IBV isolate ck/CH/LGX/130530 with those of partridge/GD/S14/2003, pathogenic tl/CH/LDT3/03, and LDT3-A vaccine strains.
| Strain | Genome position |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 20437 | 20741 | 21365 | 21641 | 22953 | 23034 | 23128 | 23508 | 23743 | 23848 | 24346 | 24436 | 24587 | 24994 | 25039 | 25308 | 25601 | 25727 | 26110 | 26445 | 26742 | |
| S |
E |
M |
5a | 5b |
N |
||||||||||||||||
| 57 | 361 | 985 | 1261 | 2573 | 2654 | 2748 | 3118 | 3363 | 3469 | 124 | 214 | 67 | 474 | 519 | 110 | 50 | 176 | 191 | 509 | 806 | |
| Partridge/GD/S14/2003 | T | G | C | G | C | C | T | C | T | G | G | C | C | T | C | C | C | C | C | C | C |
| tl/CH/LDT3/03 | T | G | C | G | C | C | T | C | T | G | G | C | C | T | C | C | C | C | C | C | C |
| ck/CH/LGX/130530 | T | T | C | G | C | T | T | C | T | G | G | C | C | A | C | C | T | C | C | C | T |
| LDT3-A | A | T | T | T | T | T | C | T | C | T | T | A | T | A | T | T | T | T | T | T | T |
We compared the nucleotide sequences of the 3′ 7.0-kb region of IBV isolate ck/CH/LGX/130530 with those of Partridge/GD/S14/2003, pathogenic tl/CH/LDT3/03, and LDT3-A vaccine strains. Of the 21 mutations, 16 were the same in the pathogenic tl/CH/LDT3/03 and ck/CH/LGX/130530 strain (in gray); in contrast, only five mutations were the same in the vaccine LDT3-A strain and the ck/CH/LGX/130530 isolate. Nucleotide positions correspond to those in the sequence of the IBV LDT3-A vaccine genome. GenBank accession numbers are the same as those shown in Fig. 1.
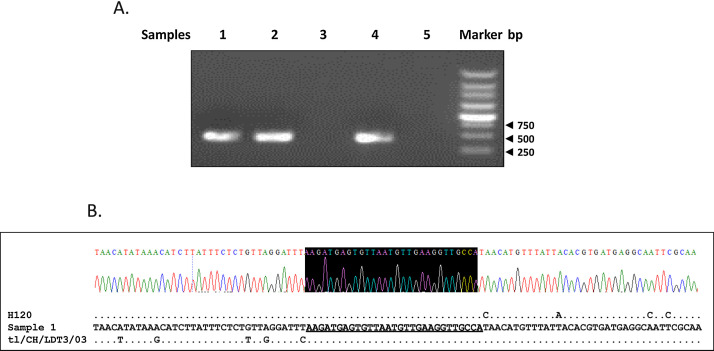
In addition, three of the five tracheal swabs which had been used for virus isolation were positive for RT-PCR amplification (Fig. 2 A). The predictive switch site and flanking sequences were same as those of IBV isolate ck/CH/LGX/130530 (Fig. 2B). Taken together, our results clearly indicated that the ck/CH/LGX/130530 isolate is a naturally recombinant virus that emerged from a recombination event between pathogenic tl/CH/LDT3/03- and H120-like viruses.
Fig. 2.
Identification of the recombination from the collected tracheal swab samples of the chickens showing clinical signs. Five out of 30 tracheal swab samples were identified directly by RT-PCR individually using primers Rec5′ and Rec3′, and 3 were shown to be positive (Samples 1, 2 and 4) (A). The PCR production of sample 1 was sequenced directly and switch site (in black) and flanking sequences were shown (B). The sequences of sample 1 were compared with those of H120 and tl/CH/LDT3/03 strains. The sequences of sample 1 are listed, and only the nucleotides differing from those of ck/CH/LGX/130530 are depicted. The predictive switch site and flanking sequences were same as those of IBV isolate ck/CH/LGX/130530.
3.3. Isolate ck/CH/LGX/130530 has the same serotype as tl/CH/LDT3/03
The ck/CH/LGX/130530 isolate was serologically characterized with the predicted parental viruses H120 and tl/CH/LDT3/03 (LDT3-A was used) to determine the antigenic relationship among the three strains. End-points were calculated by the Reed and Muench (1938) method. Serum against the H120 strain did not neutralize the ck/CH/LGX/130530 isolate and vice versa, which indicated that the ck/CH/LGX/130530 isolate is distinctly related to strain H120 (Table 2 ). In contrast, the ck/CH/LGX/130530 isolate has the same serotype as the LDT3-A strain, with an R value of 0.73.
Table 2.
Titers were obtained using reciprocal β virus neutralization tests (diluted serum, constant virus doses).
| Virus | Serum |
||
|---|---|---|---|
| ck/CH/LGX/130530 | tl/CH/LDT3/03 | H120 | |
| ck/CH/LGX/130530 | 337.8 | 181 | <2 |
| tl/CH/LDT3/03 | 181 | 181 | <2 |
| H120 | <2 | <2 | 147 |
3.4. Isolate ck/CH/LGX/130530 exhibits low pathogenicity toward SPF chickens
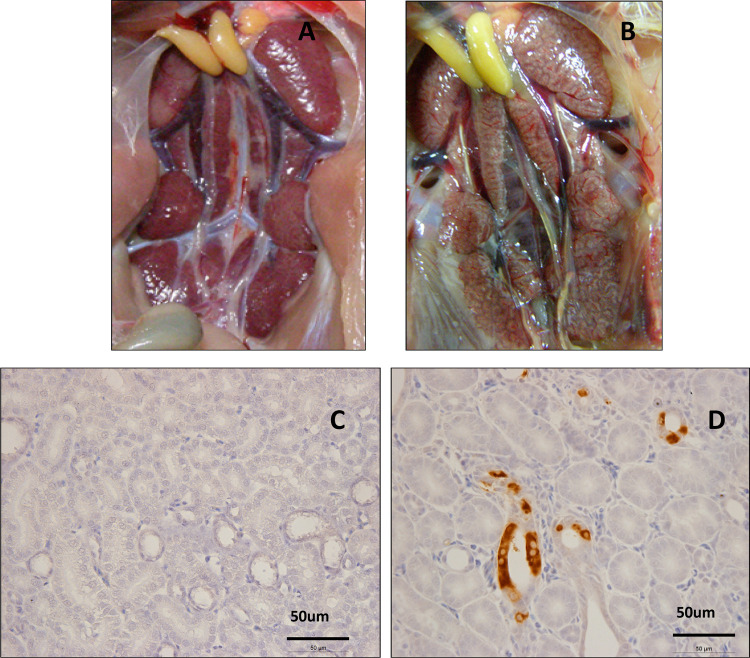
To determine its pathogenicity, ten 1-day-old SPF chickens were challenged with the IBV ck/CH/LGX/130530 isolate. As illustrated in Table 3 , only 3 out of 10 SPF chickens challenged with isolate ck/CH/LGX/130530 showed mild clinical signs of an IB-like disease between 3 and 7 days after challenge. In contrast, all the chickens challenged with strain tl/CH/LDT3/03 showed obvious clinical signs and 7 out of 10 died between 4 and 9 days after challenge, with gross lesions (nephritis) mainly confined to the kidneys (Fig. 3 A). Furthermore, obvious IHC IBV positive cells were seen in the kidneys of those dead chickens (Fig. 3B), with accompanying nephritis, which identified the strain tl/CH/LDT3/03 as nephropathogenic strains. No cilical signs had been observed in the chickens of control group. Serum had not converted at 4 dpi in chickens inoculated with any of the IBVs (Table 3), however antibodies were detected in birds after 8 and 16 days after challenged with tl/CH/LDT3/03 and ck/CH/LGX/130530, respectively. We did not observe seroconversion in any chickens of the control group.
Table 3.
Pathogenicity to 1-day-old SPF chickens.
| Group | Morbidity | Mortality | Antibody responsea |
|||||
|---|---|---|---|---|---|---|---|---|
| 4db | 8d | 12d | 16d | 20d | 24d | |||
| ck/CH/LGX/130530 | 3/10 | 0/10 | 0/10 | 1/10 | 4/10 | 9/10 | 10/10 | 10/10 |
| tl/CH/LDT3/03 | 10/10 | 7/10 | 0/10 | 4/6 | 3/3 | 3/3 | 3/3 | 3/3 |
| Negative control | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
Number that seroconverted/number inoculated.
Days after challenge.
Fig. 3.
Pathogenicity of the isolate ck/CH/LGX/130530 to SPF chickens. The kidney of SPF chicken which was mock inoculated with sterile allantoic fluid (A). The renal lesions of the kidney caused by inoculation with isolate ck/CH/LGX/130530 (B). Kidneys are swollen, tubules and ureters are distended, and uric acid crystals are present. Representation of the type of immunohistochemical staining of IBV seen in the kidneys of chickens mocked inoculated with sterile allantoic fluid (C) and infected chickens (D). Images were taken at 50 × magnification.
3.5. The H120 vaccine does not provide sufficient protection against IBV strains ck/CH/LGX/130530 and tl/CH/LDT3/03
All of the chickens vaccinated with either the H120 or LDT3-A vaccines did not show clinical signs and mortality after challenge with the ck/CH/LGX/130530 isolate. However, nine and five of the ten birds vaccinated with the H120 vaccine were positive for virus re-isolation at 4 and 8 days post-challenge with the ck/CH/LGX/130530 isolate, respectively. In contrast, only one bird in the LDT3-A vaccinated group was positive for virus re-isolation from the respiratory tract at 4 days after ck/CH/LGX/130530 challenge, and all birds were negative thereafter (Table 4 ). The challenge virus was detected in all birds at days 4 and 8 post-challenge with the ck/CH/LGX/130530 isolate.
Table 4.
Results of the vaccination-challenge tests.
| Groupa | Vaccination strain | Challenge strain | Morbidity | Mortality | Antibody responseb |
Virus recoveryc |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vaccinated |
Challenged |
4dd | 8d | 12d | 16d | |||||||||||||
| 4d | 8d | 12d | 16d | 20d | 4d | 8d | 12d | 16d | 20d | |||||||||
| 1 | H120 | ck/CH/LGX/130530 | 0/10 | 0/10 | 0/10 | 1/10 | 6/10 | 10/10 | 10/10 | 10/10 | 10/10 | 10/10 | 10/10 | 10/10 | 9/10 | 5/10 | 2/10 | 0/10 |
| 2 | LDT3-A | ck/CH/LGX/130530 | 0/10 | 0/10 | 0/10 | 1/10 | 5/10 | 9/10 | 10/10 | 10/10 | 10/10 | 10/10 | 10/10 | 10/10 | 1/10 | 0/10 | 0/10 | 0/10 |
| 3 | H120 | tl/CH/LDT3/03 | 10/10 | 2/10 | 0/10 | 1/10 | 5/10 | 10/10 | 10/10 | 10/10 | 8/8 | 8/8 | 8/8 | 8/8 | 10/10 | 8/8 | 1/8 | 0/8 |
| 4 | – | tl/CH/LDT3/03 | 10/10 | 3/10 | – | – | – | – | – | 0/10 | 7/7 | 7/7 | 7/7 | 7/7 | 10/10 | 7/7 | 0/7 | 0/7 |
| 5 | – | ck/CH/LGX/130530 | 2/10 | 0/10 | – | – | – | – | – | 0/10 | 9/10 | 10/10 | 10/10 | 10/10 | 10/10 | 10/10 | 3/10 | 0/10 |
| 6 | – | – | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
One-day-old birds in groups 1 and 3 were vaccinated with 104 EID50 of the H120 vaccine, while birds in group 2 were vaccinated with the LDT3-A vaccine. Twenty-day-old birds in groups 1, 2, and 5 were challenged with 105 EID50 of the ck/CH/LGX/130530 isolate, while birds in groups 3 and 4 were challenged with the IBV isolate tl/CH/LDT3/03. Birds in group 6 were not exposed to any viruses and served as negative controls.
Number that seroconverted/number inoculated.
Two procedures were used for virus recovery after challenge, as described previously (Zhang et al., 2015). First, embryos that were inoculated with individual nasopharyngeal swab samples were observed for lesions. Second, RT-PCR using a pair of oligonucleotide primers, N(−) and N(+), was conducted on RNA recovered from the allantoic fluid of the same eggs. The results from the two procedures were identical. Numbers indicate the number of birds that were positive for virus recovery/number challenged.
Days after challenge.
To evaluate the ability of the H120 vaccine to protect against tl/CH/LDT3/03, ten H120-vaccinated chickens were challenged with the IBV strain tl/CH/LDT3/03. The results demonstrated that all of the chickens developed clinical signs and that two chickens died at 5 and 6 days post-challenge. Meanwhile, all chickens were positive for virus re-isolation 4 and 8 days post-challenge, suggesting that H120 vaccination could not provide protection against the IBV strain tl/CH/LDT3/03. In accordance with our previous results (Liu et al., 2005), the IBV strain tl/CH/LDT3/03 was highly pathogenic to SPF chickens, as it resulted in 100% morbidity and 30% mortality (Table 4). In contrast, only a small proportion of chickens (20%) developed mild clinical signs at about 4–10 days after ck/CH/LGX/130530 challenge. All challenged chickens survived through the entire observation period of 25 days. None of the chicks challenged with the ck/CH/LGX/130530 isolate showed seroconversion at 4 days post-challenge, but antibodies were detected by ELISA in nearly all of the birds after 8 days post-challenge.
4. Discussion
Recombination events between different strains of IBV could result in the emergence of several novel strains, and the detection of some novel strains has caused considerable concern. It had been reported that the Canadian Qu_mv strain originated from a recombination event between Massachusetts and Arkansas-like strains (Smati et al., 2002). California 99-like viruses presumably originated from multiple recombination events between Connecticut, Massachusetts, and Arkansas vaccines (Mondal and Cardona, 2007). In the course of our continuous surveillance of IBV in China, several natural recombinant IBV strains have been isolated and identified. These IBV strains were shown to have originated from recombination events between field and vaccine strains, different types of vaccine strains, or different types of field strains (Liu et al., 2013a, Liu et al., 2013b, Liu et al., 2014, Zhang et al., 2015). Some of the recombinant strains were shown to have undergone multiple recombination events (Liu et al., 2013b, Liu et al., 2014, Zhang et al., 2015). In addition, other reports in China also confirmed our results (Zhao et al., 2013, Feng et al., 2014, Zhou et al., 2014), indicating that recombination played an important role in the course of IBV evolution in China. In this study, we isolated and identified an IBV isolate, ck/CH/LGX/130530, that likely originated from a recombination event between a Massachusetts-type H120-like strain and a field tl/CH/LDT3/03-like strain, as shown by SimPlot analyses, pairwise comparisons, and similarity comparisons using corresponding gene fragments.
The tl/CH/LDT3/03 strain was first isolated in 2003 in Guangdong province, China (Liu et al., 2005), which was close to Guangxi province where ck/CH/LGX/130530 was isolated. The tl/CH/LDT3/03-like viruses were also isolated and shown to be one of the major types of viruses circulating in H120-vaccinated chicken flocks in southern China, including Guangxi province, in recent years (Luo et al., 2012, Feng et al., 2014). The company at which ck/CH/LGX/130530 was isolated had more than a 20-year history of chicken breeding, and it had used the H120 vaccine against IBV for more than 15 years. It has been showed that the H120 vaccine strain may persist in various internal organs of chickens for 163 days or longer (Cavanagh and Gelb, 2008). Therefore, recombination events between the H120 vaccine and field strains may lead to the creation of a new virus, such as the ck/CH/LGX/130530 isolated in the present study, although we cannot exclude the possibility that this isolate was introduced from other regions. However, previous studies showed that the viruses of tl/CH/LDT3/03 genotype had been isolated in the chicken flocks of the company (Li et al., 2012, Li et al., 2013) although it was not the flock in which ck/CH/LGX/130530 had been isolated, implicating that recombination event had been likely occurred in this company. The results showed that H120-like sequences in the genome of the ck/CH/LGX/130530 isolate had less than 99.8% similarity to those of the H120 vaccine, and similarly, the tl/CH/LDT3/03-like sequences of the ck/CH/LGX/130530 isolate had about 99.7% similarity to those of the tl/CH/LDT3/03 strain. These results imply that there has been a recent recombination event, causing near homogenization of certain genomic regions between two highly diverged viruses. Moreover, the occurrence of such events suggests an important role for recombination between a pathogenic field IBV strain and the vaccine strain.
If the extensive use of the H120 vaccine in nearly all chicken flocks in China could provide good protection against tl/CH/LDT3/03 challenge, this would serve as evidence against the aforementioned conclusion that the ck/CH/LGX/130530 isolate originated from a recombination event between H120-like and tl/CH/LDT3/03-like viruses, as there would be only a small chance of co-infection with two different serotype viruses in a given flock, which is believed to be an important prerequisite for recombination between IBV strains in the field (Cavanagh, 2007). However, we found that the tl/CH/LDT3/03 challenge virus could be re-isolated from 100% of the H120-vaccinated SPF chickens at 4 and 8 days post-challenge, indicating that the tl/CH/LDT3/03 virus could replicate and be shed via the oropharynx from the H120-vaccinated chickens at least eight days after infection. Therefore, it is possible that a tl/CH/LDT3/03-like virus and the H120 vaccine strain could co-exist in a certain chicken flock, which would make such a recombination event possible.
The S1 gene of the ck/CH/LGX/130530 isolate showed greater than 99% similarity to that of tl/CH/LDT3/03, indicating that the two IBV strains share the same genotype. It is believed that the S1 domain of IBV is the main antigenic viral protein containing neutralization epitopes and accounts for serotypical variations (Cavanagh, 2003). This can largely account for the observed results that the ck/CH/LGX/130530 isolate was shown to be serotypically close to tl/CH/LDT3/03 by a cross-neutralization test. In addition, vaccination with the LDT3-A vaccine, which was derived from a tl/CH/LDT3/03 strain by serial passaging in chicken eggs, provided good protection against the ck/CH/LGX/130530 challenge, indicating that they belonged to the same protectotype (de Wit et al., 2011a, de Wit et al., 2011b). Consequently, the recombination event that possibly occurred in the ck/CH/LGX/130530 isolate did not result in changes in the genotype, serotype, and protectotype, as compared to those of one of its parental viruses, tl/CH/LDT3/03, as the extent of the similarity between the ck/CH/LGX/130530 isolate and its other parental virus, the H120 strain, was greater than 99.8% in the 5′ end of the Gene 1 region, which is 17,000 nt long.
The results in this and our previous study (Liu et al., 2005) showed that IBV tl/CH/LDT3/03 was a highly nephropathogenic strain; however, the virulence of the IBV isolate ck/CH/LGX/130530 toward SPF chickens was low in this study. The nucleotide sequences of the ck/CH/LGX/130530 isolate from genome position 17,055 to the 3′ end of the genome were shown to be derived from a tl/CH/LDT3/03-like virus (as it shared more than 99.7% nucleotide identity in this region); this region included a part of nsp14, and complete nsp15 and nsp16 of the Gene 1 region, the M, N, and S genes, Gene3, Gene5, and the 3′ UTR. The remaining sequences were likely derived from the attenuated H120 vaccine strain. It was previously demonstrated that the accessory protein genes of IBV contributed to virulence (Shen et al., 2003, Youn et al., 2005). However, replacing the complete replicase gene of the pathogenic IBV M41 strain with that of a non-pathogenic Beaudette-derived gene demonstrated that the IBV replicase gene is a determinant of pathogenicity (Armesto et al., 2009). This was also observed in another coronavirus, which led to the belief that changes within the replicase gene, especially the RNA-dependent RNA polymerase that constitutes the core of viral replicases, can change the replicase fidelity, as in the vast majority of cases, the alterations in replication fidelity decreased viral fitness and attenuated virulence (Smith et al., 2014, Smith et al., 2015). Our results confirmed that the replicase gene was associated with viral pathogenicity although this observation for the ck/CH/LGX/130530 isolate needs to be verified by future reverse genetic and animal studies.
Acknowledgements
This work was supported by grants from the China Agriculture Research Systerm (No. CARS-41-K12), National “Twelfth Five-Year” Plan for Science & Technology Support (2015BAD12B03), and Special Fund for Agro-scientific Research in the Public Interest (No. 201303033).
References
- Ammayappan A., Vakharia V.N. Complete nucleotide analysis of the structural genome of the infectious bronchitis virus strain Md27 reveals its mosaic nature. Viruses. 2009;1:1166–1177. doi: 10.3390/v1031166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archett I., Horsfall F.L. Persistant antigenic variation of influenza A viruses after complete neutralization with heterologous immune serum. J. Exp. Med. 1950;92:441–458. doi: 10.1084/jem.92.5.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armesto M., Cavanagh D., Britton P. The replicase gene of avian coronavirus infectious bronchitis virus is a determinant of pathogenicity. PLoS One. 2009;4:E7384. doi: 10.1371/journal.pone.0007384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner L.R., Keck J.G., Lai M.M. A clustering of RNA recombination sites adjacent to a hypervariable region of the peplomer gene of murine coronavirus. Virology. 1990;175:548–555. doi: 10.1016/0042-6822(90)90439-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner L.R., Lai M.M. Random nature of coronavirus RNA recombination in the absence of selection pressure. Virology. 1991;185:441–445. doi: 10.1016/0042-6822(91)90795-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E., Brown T.D., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus. Avian Pathol. 2003;32:567–582. doi: 10.1080/03079450310001621198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D., Gelb J. Infectious bronchitis. In: Saif Y.M., Fadly A.M., Glisson J.R., McDougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th ed. Wiley-Blackwell Publishing; Iowa: 2008. pp. 117–135. [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- de Wit J.J., Cook J.K., van der Heijden H.M. Infectious bronchitis virus variants: a review of the history: current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J.J., Nieuwenhuisen-van Wilgen J., Hoogkamer A., van de Sande H., Zuidam G.J., Fabri T.H. Induction of cystic oviducts and protection against early challenge with infectious bronchitis virus serotype D388 (genotype QX) by maternally derived antibodies and by early vaccination. Avian Pathol. 2011;40:463–471. doi: 10.1080/03079457.2011.599060. [DOI] [PubMed] [Google Scholar]
- Feng K., Xue Y., Wang F., Chen F., Shu D., Xie Q. Analysis of S1 gene of avian infectious bronchitis virus isolated in southern China during 2011–2012. Virus Genes. 2014;49:292–303. doi: 10.1007/s11262-014-1097-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Zhao F., Shao Y., Liu X., Kong X., Song Y., Liu S. Fine level epitope mapping and conservation analysis of two novel linear B-cell epitopes of the avian infectious bronchitis coronavirus nucleocapsid protein. Virus Res. 2013;171:54–64. doi: 10.1016/j.virusres.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson K.A., Noormohammadi A.H., Devlin J.M., Browning G.F., Schultz B.K., Ignjatovic J. Evaluation of a novel strain of infectious bronchitis virus emerged as a result of spike gene recombination between two highly diverged parent strains. Avian Pathol. 2014;43:249–257. doi: 10.1080/03079457.2014.914624. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Mo M.L., Huang B.C., Fan W.S., Wei Z.J., Wei T.C., Li K.R., Wei P. Continuous evolution of avian infectious bronchitis virus resulting in different variants co-circulating in Southern China. Arch. Virol. 2013;158:1783–1786. doi: 10.1007/s00705-013-1656-0. [DOI] [PubMed] [Google Scholar]
- Li M., Wang X.Y., Wei P., Chen Q.Y., Wei Z.J., Mo M.L. Serotype and genotype diversity of infectious bronchitis viruses isolated during 1985–2008 in Guangxi, China. Arch. Virol. 2012;157:467–474. doi: 10.1007/s00705-011-1206-6. [DOI] [PubMed] [Google Scholar]
- Liu S., Chen J., Chen J., Kong X., Shao Y., Han Z., Feng L., Cai X., Gu S., Liu M. Isolation of avian infectious bronchitis coronavirus from domestic peafowl (Pavo cristatus) and teal (Anas) J. Gen. Virol. 2005;86:719–725. doi: 10.1099/vir.0.80546-0. [DOI] [PubMed] [Google Scholar]
- Liu S., Kong X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and nonvaccinated flocks in China. Avian Pathol. 2004;33:321–327. doi: 10.1080/0307945042000220697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Shao Y., Ma H., Sun C., Zhang X., Li C., Han Z., Yan B., Kong X., Liu S. Comparative analysis of four Massachusetts type infectious bronchitis coronavirus genomes reveals a novel Massachusetts type strain and evidence of natural recombination in the genome. Infect. Genet. Evol. 2013;14:29–38. doi: 10.1016/j.meegid.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ma H., Xu Q., Sun N., Han Z., Sun C., Guo H., Shao Y., Kong X., Liu S. Characterization of a recombinant coronavirus infectious bronchitis virus with distinct S1 subunits of spike and nucleocapsid genes and a 3’ untranslated region. Vet. Microbiol. 2013;162:429–436. doi: 10.1016/j.vetmic.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xu Q., Han Z., Liu X., Li H., Guo H., Sun N., Shao Y., Kong X. Origin and characteristics of the recombinant novel avian infectious bronchitis coronavirus isolate ck/CH/LJL/111054. Infect. Genet. Evol. 2014;23:189–195. doi: 10.1016/j.meegid.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Qin J., Chen F., Xie Q., Bi Y., Cao Y., Xue C. Phylogenetic analysis of the S1 glycoprotein gene of infectious bronchitis viruses isolated in China during 2009–2010. Virus Genes. 2012;44:19–23. doi: 10.1007/s11262-011-0657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Shen S., Wen Z.L., Liu D.X. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene functional characterization of the 3b protein in pathogenesis and replication. Virology. 2003;311:16–27. doi: 10.1016/S0042-6822(03)00117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Case J.B., Blanc H., Isakiv O., Shomron N., Vignuzzi M., Denison M.R. Mutations in coronavirus nonstructural protein 10 decrease virus replication fidelity. J. Virol. 2015 doi: 10.1128/JVI.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smati R., Silim A., Guertin C., Henrichon M., Marandi M., Arella M., Merzouki A. Molecular characterization of three new avian infectious bronchitis virus (IBV) strains isolated in Québec. Virus Genes. 2002;25:85–93. doi: 10.1023/A:1020178326531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Sexton N.R., Denison M.R. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Ann. Rev. Virol. 2014;1:111–132. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
- Mondal S.P., Cardona C.J. Genotypic and phenotypic characterization of the California 99 (Cal99) variant of infectious bronchitis virus. Virus Genes. 2007;34:327–341. doi: 10.1007/s11262-006-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Han Z., Wang Q., Zhang T., Gao M., Zhao Y., Shao Y., Li H., Kong X., Liu S. Emergence of novel nephropathogenic infectious bronchitis viruses currently circulating in Chinese chicken flocks. Avian Pathol. 2015 doi: 10.1080/03079457.2015.1118435. Accepted. [DOI] [PubMed] [Google Scholar]
- Youn S., Collisson E.W., Machamer C.E. Contribution of trafficking signals in the cytoplasmic tail of the infectious bronchitis virus spike protein to virus infection. J. Virol. 2005;21:13209–13217. doi: 10.1128/JVI.79.21.13209-13217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Han Z., Xu Q., Wang Q., Gao M., Wu W., Shao Y., Li H., Kong X., Liu S. Serotype shift of a 793/B genotype infectious bronchitis coronavirus by natural recombination. Infect. Genet. Evol. 2015;32:377–387. doi: 10.1016/j.meegid.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Zou N., Wang F., Guo M., Liu P., Wen X., Cao S., Huang Y. Analysis of a QX-like avian infectious bronchitis virus genome identified recombination in the region containing the ORF 5a, ORF 5b, and nucleocapsid protein gene sequences. Virus Genes. 2013;46:454–464. doi: 10.1007/s11262-013-0884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Tang M., Jiang Y., Chen X., Shen X., Li J., Dai Y., Zou J. Complete genome sequence of a novel infectious bronchitis virus strain circulating in China with a distinct S gene. Virus Genes. 2014;49:152–156. doi: 10.1007/s11262-014-1063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]