Highlights
-
•
By the application of Bayesian phylogeographical analysis, this study demonstrated the spatial- temporal transmission of PEDVs within Korea.
-
•
Of the recent emerged G2a viruses, J3142 strains showed potential recombination breakpoint (376–2,143nt) of S1 gene between KNU1303_Korea strain_G2a (KJ451046) and 45RWVCF0712_Thailand strain_G2b (KF724935).
-
•
The pandemic G2a virus was partial neutralized by the antibodies invoked by the G1- based PED vaccine virus.
Keywords: Novel strains, Porcine epidemic diarrhea virus, South Korea, Piglets, Isolation
Abstract
Since outbreaks of porcine epidemic diarrhea virus (PEDV) in the United States in 2013, explosive outbreaks of PED in South Korea have infected all age groups of pigs in 2014–2015 year. This study analyzed a large collection of the Spike protein coding gene to infer the spatial-temporal diffusion history of PEDV. The studying results suggested that PEDVs in Korea belonged to different genogroups. While classical G1 was continuingly circulating between provinces of Korea, the pandemic G2a were recently introduced from China and USA. By the application of Bayesian phylogeographical analysis, this study demonstrated the spatial-temporal transmission of PEDVs within Korea. Of the recent emerged G2a viruses, J3142 strains showed potential recombination breakpoint (376–2,143nt) of S1 gene between KNU1303_Korea strain_G2a (KJ451046) and 45RWVCF0712_Thailand strain_G2b (KF724935). The pandemic G2a virus was partial neutralized by the antibodies invoked by the G1- based PED vaccine virus.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is an acute contagious diarrhea disease caused in pigs (Song and Park, 2012, Jung et al., 2014). The virus belongs to genus Alpha-coronavirus, family Coronaviridae, which includes other genera; Beta-, Gamma-, and Delta-coronavirus, and has positive-sense single stranded RNA genome with envelope (Woo et al., 2012). PEDV is transmitted mainly through the fecal-oral route, infecting all age groups of pigs but the most severe form of diseases occurs in suckling piglets (Song and Park, 2012). In 2013, PEDV was reported for the first time in North American and now PEDVs are present in all of swine producing countries (Song and Park, 2012, Huang et al., 2013, Mole, 2013). In Korea, PEDV was first reported in 1992 (Kweon et al., 1993), and continuingly circulated and exhibited significant genetic diversity (Choi et al., 2014, Lee and Lee, 2014). PED outbreaks re-occurred in Korea in 2013, however, it was demonstrated that the emerging PEDVs were not variants of old Korean isolates or attenuated vaccine strains (Chung et al., 2015). Though a recent publication characterized the genetic evolution of PEDVs in Korea from 1998 to 2013, to our knowledge, the transmission patterns of the virus in Korea are largely unknown. Using a large dataset collected globally and locally, this study aimed to trace the spatial-temporal dynamics of PEDVs in Korea.
2. Material and methods
2.1. Sample collection
The spike protein coding gene (S gene) was selected for genetic analysis. Aimed at provide a detail about the evolution of PEDV in Korea, we obtained from the previous study 31 partial sequences of the S gene which were collected in years 2002–2005. Additionally, this study sequenced complete S genes from PEDV positive samples which were stored in our laboratory from October 2012 to March 2015. To infer the spatial-temporal diffusion history of PEDV, sequences of the S gene which had known collection date and country of origin were achieved from Genbank. Overall, the final dataset contained 805 sequences originating from Asia (China, Korea, Vietnam, Thailand, Japan, Taiwan), Europe (Belgium, Slovenia, France), and American (USA, Canada, Mexico, Colombia), and covering a sampling period from 1986 to 2015. The details of the dataset are summarized in Supplementary Table S1.
2.2. Bayesian phylogeographic analysis
A Bayesian framework (Lemey et al., 2009) was applied to reconstruct the spatial-temporal diffusion history of PEDVs. In brief, the spatial diffusion of the time-scaled genealogy is modeled as a standard continuous-time Markov chain (CTMC) process over discrete sampling locations. A Bayesian stochastic search variable selection (BSSVS) approach, which allows the exchange rates in the CTMC to be zero with some prior probability, was used to find a parsimonious set of rates explaining the diffusions in the phylogeny. The analysis was performed using BEAST package v1.8.2 under assumptions of (i) a codon based SRD06 nucleotide substitution model, (ii) a constant population size for the coalescent prior, and (iii) the molecular clock model of uncorrelated lognormal distribution. The analysis was run for 100 million chains, sampling every 10,000 generations.
2.3. Potential recombinant origins of the S gene strains
PEDV 411 S complete gene sequences aligned were used by Recombination Detection Program v.4.46 (Tian et al., 2014 and Vlasova et al., 2014); X-Over automated RDP analysis was used to identify recombination points within the PEDV genome.
2.4. Cross serum neutralization assay
In the previous study, it was noted that mutation occurred at SS6 neutralizing epitope of the S gene of an isolate collected from 2013 to 2014 outbreak. This study tested whether PEDV isolated recently was crossed neutralized by serum of pigs which were vaccinated with Korean PED oral vaccine (Attenuated DR13 strain, Green Cross Veterinary Product Co., Ltd., Yong-In, Korea). The serum neutralizing test (SN test) was conducted using the previous method (Song et al., 2007), with some modifications. In brief, sera of pigs (n = 25) were heat inactivated at 56 °C for 30 min and stored at −20°C until use. The sera were serially two- fold diluted. Subsequently either BM3 (a recent PEDV isolate) or DR13 (PED vaccine strain) of 200 TCID50/0.1 ml were mixed at equal volume of the diluted sera. The mixtures were incubated for 1 h at 37 °C. After incubation, 0.1 ml of each virus-serum mixture was infected to monolayer of Vero cells. The presence/absence of CPE were monitored daily for 5 days. The SN titers were expressed as the reciprocal of the highest serum dilution which resulted in the inhibition of CPE.
3. Results and discussions
3.1. Bayesian phylogeography of PEDVs
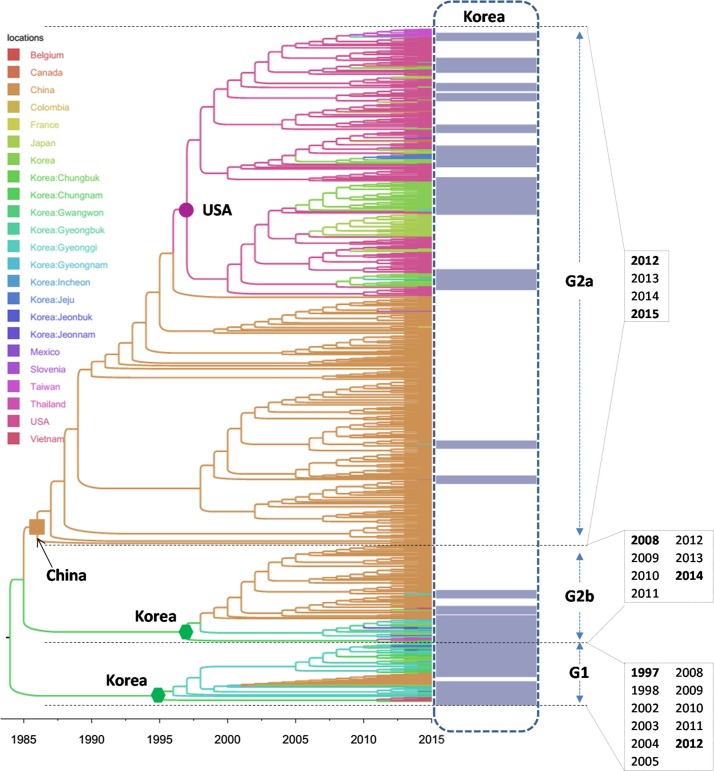
The result of the Bayesian phylogeographic analysis is shown in Fig. 1 . It was obvious that PEDVs are circulating in Korea from the earliest to the latest sequences (years 1997–2015), the virus were classified into genogroup 1 (G1) and genogroup 2 (subgroups 2a and 2b). For the Korean PEDVs of G1, the viruses were found spanning from 1997 until 2012. Meanwhile, the viruses of G2 (G2b and G2a) were detected from 2008 until 2015. As shown in Fig. 1, it was clear that the root of Korean G1 and G2b viruses were inferred to be originated from Korea, while the recent Korean PEDVs were derived from China and USA. The result implied that the transmission histories of PEDVs in Korea were diverse, which reflecting both the continuingly circulation of classical G1 and the introduction of the recent pandemic G2a. The detail of spatial- temporal dynamics of PEDVs between provinces of Korea is shown in Figs. 2–4 .
Fig. 1.
Bayesian time- scaled phylogeny of PEDV with inferred geographical location states. The branches of maximum clade credibility tree were colored according to the most probable location state of their descendent nodes. The color codes are defined in the insert legend. The S gene- based phylogeny clearly divided PEDVs into genogroup 1 (G1) and two subgroups of genogroup 2 (G2a, G2b). For clarity, only the leaves of Korean PEDVs were highlighted. It was obvious that PEDVs circulating in Korea were within G1.
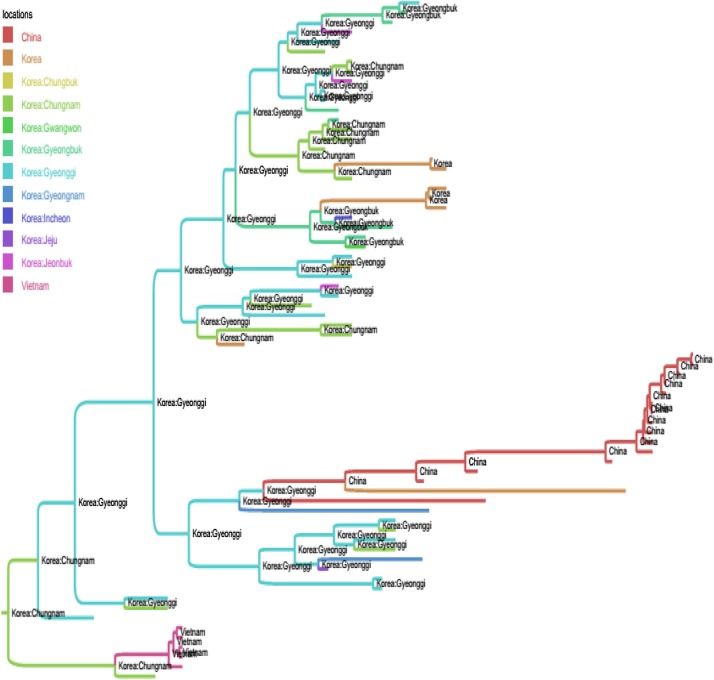
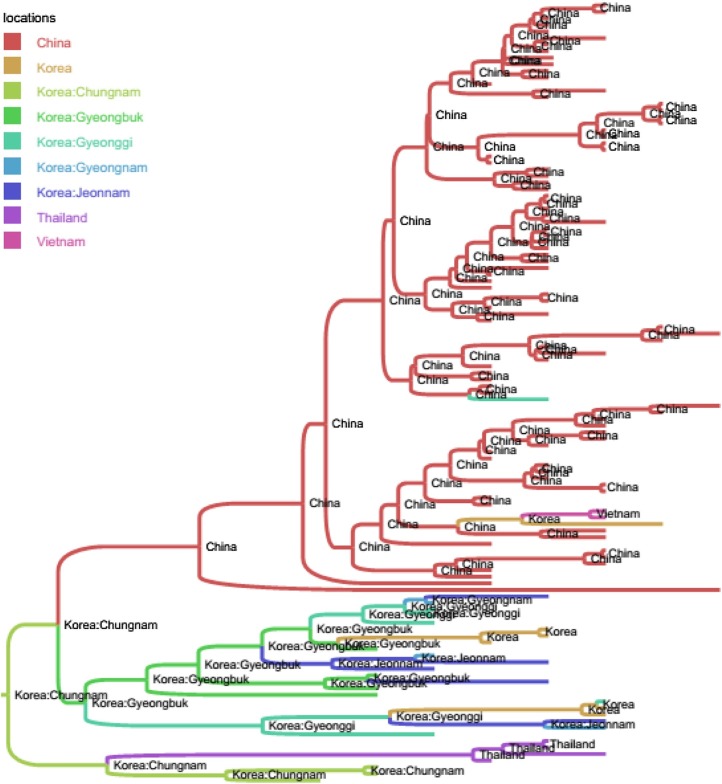
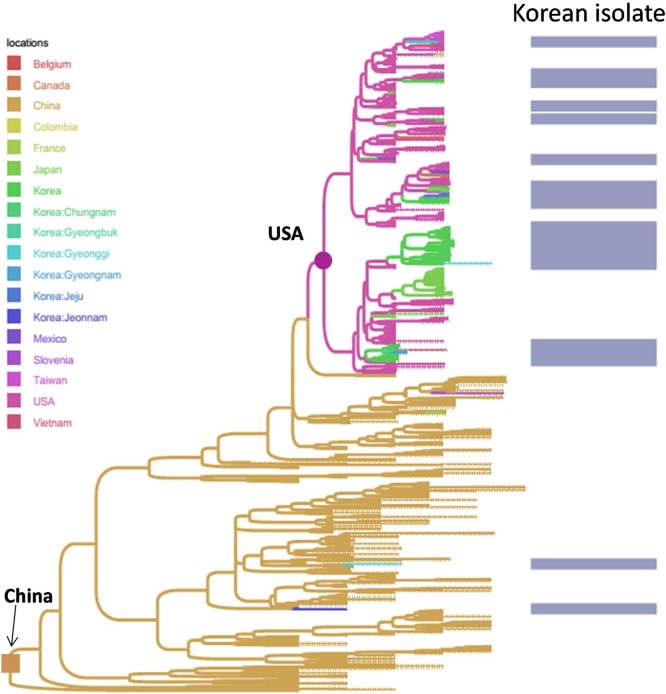
Among the Korean G1 viruses, the result of Fig. 2 suggested the dispersal of the virus between different provinces, of which Gyeonggi was the main source in the viral migration network. Among the Korean G2b viruses (Fig. 3), another province (Chungnam) was predicted to be the initial source for the spreading of virus into Gyeongbuk, and then from Gyeongbuk to Jeonnam. The picture was totally different from the Korean G2a viruses, of which the sources were predicted to be China and USA (Fig. 4).
Fig. 2.
Bayesian time- scaled phylogeny of genogroup 1 of PEDVs. The branches of maximum clade credibility tree were colored according to the most probable location state of their descendent nodes. For clarity, the annotations of leaves were omitted. Shown in every internal nodes of the phylogeny were the inferred locations. It was observed that Gyeonggi was the source for the G1 transmission network within Korea.
Fig. 3.
Bayesian time- scaled phylogeny of sub- genogroup 2b of PEDVs. The branches of maximum clade credibility tree were colored according to the most probable location state of their descendent nodes. For clarity, the annotations of leaves were omitted. Shown in every internal nodes of the phylogeny were the inferred locations. It was observed that Chungnam was predicted to be the initial source for the spreading of virus into several provinces.
Fig. 4.
Bayesian time- scaled phylogeny of sub- genogroup 2a of PEDVs. The branches of maximum clade credibility tree were colored according to the most probable location state of their descendent nodes. For clarity, the annotations of leaves were omitted. It was observed clearly that Korean PEDVs of G2a genetically clustered to the pandemic PEDVs of China and North American strains, and implied for the exotic sources of viral transmission network in Korea.
3.2. Potential recombinant origin of the Korean G2a PEDVs
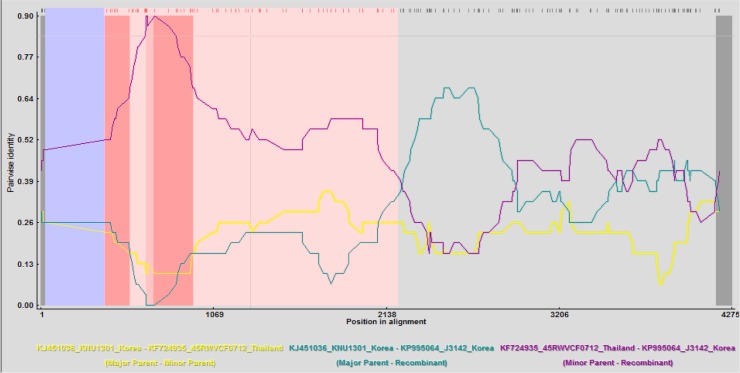
In order to detect potential recombinant strains of J3142 and BM3, we used Recombination Detection Program with 411 references PEDV strains in this study. It has not been found that BM3 has recombinant region. Interestingly, two potential strains of J3142 had recombination which had breakpoint from 376nt (aligned; 391nt) to 2143nt (aligned; 2206nt). One had major patent similarity of 95.3% with KNU1303_Korea strain (KJ451046) which belongs to the subgroup G2a, and the other had minor patent similarity of 96.4% with 45RWVCF0712_Thailand strain (KF724935) which belongs to the subgroup G2b. In this study, J3142 had a variant S1 (Duarte and Laude, 1994, Gallagher and Buchmeier, 2001, Lee et al., 2010) region, suggesting that the recombination between South Korea and Thailand strains had occurred. In Fig. 5 , we used only 411 references for the recombination analysis; thus, it might be possible that in South Korea, there were strains, which are similar to those of Thailand such as 45RWVCF0712, leading to the recombinant event.
Fig. 5.
Identification of potential recombination PEDV strains with reference PEDV strains (n = 411).
3.3. Cross serum neutralization between G1- based vaccine virus and pandemic G2a virus
In the SN test (Table 1 ), 52% (13/25) of the BM3 strain were positive and 88% (22/25) of the DR13 strain were positive. The average titer of the DR13 (14.53 value) was 5 times more than that of the BM3 (2.9 value). The results implied that, the specific antibodies invoked by PED vaccine of G1 inefficiently conferred cross neutralizing activities to the pandemic G2a isolate. Our result was inline with the previous report (Kim et al., 2015) that suggests that limited cross-reactivity was detected between PED vaccine (of genogroup 1) and field viruses (of genogroup 2). The attenuated PEDV vaccines based on the CV777 strains or DR13-derived strains might be antigenically less related to the newly emergent PEDV strains; thus, the development of new vaccines based on current strains are needed (Chen et al., 2014, Chung et al., 2015, Park et al., 2014, Song and Park, 2012, Tian et al., 2013).
Table 1.
Results of serum neutralization (SN) test both BM3 and DR13 strains of PEDV from porcine sera.
| ID | Agea | Sampling site | Collection Date day-month-year | Clinical symptom | PEDV Vaccinnated | BM3 strain |
DR13 strain |
|---|---|---|---|---|---|---|---|
| Mean SN titerb | Mean SN titer | ||||||
| 1 | Suckling | Chungnam, farm DJ | 19−05-2015 | Diarrhea | Yes | 2.67 | 6.67 |
| 2 | 3.33 | 10.67 | |||||
| 3 | 2.33 | 6.67 | |||||
| 4 | 2.67 | 21.33 | |||||
| 5 | Gilt | Gyeongbuk, farm GM |
22−05-2015 | N. K | Yes | <2 | 5.33 |
| 6 | 2.67 | 21.33 | |||||
| 7 | 2.33 | 32 | |||||
| 8 | <2 | 2.33 | |||||
| 9 | Sow | Jeonnam, farm SC |
29−05-2015 | Diarrhea | Yes | <2 | 10.67 |
| 10 | 3.33 | 32 | |||||
| 11 | Sow | Chungnam, farm CS | 03−06-2015 | Severe diarrhea and dehydration | Yes | 5.33 | 21.33 |
| 12 | 4 | 9.33 | |||||
| 13 | Suckling | 2.33 | 21.33 | ||||
| 14 | 2.67 | 21.33 | |||||
| 15 | Suckling | Gyeonggi, Farm NRM |
17−07-2015 | N. K | Yes | 2 | 26.67 |
| 16 | <2 | 13.33 | |||||
| 17 | <2 | 13.33 | |||||
| 18 | <2 | 3.33 | |||||
| 19 | Suckling | Gangwon, farm NFR |
28−07-2015 | N. K | No | <2 | <2 |
| 20 | <2 | 5.33 | |||||
| 21 | <2 | <2 | |||||
| 22 | <2 | <2 | |||||
| 23 | Sow | Chungnam, farm CW | 27−08-2015 | Severe diarrhea, | Yes | <2 | 3.33 |
| 24 | Respiratory disorders | 2 | 18.67 | ||||
| 25 | <2 | 13.33 |
N.K: Not Known.
Samples were sorted into six groups: female (gilt and sow), suckling (30 days), weaned (30–60 days), grower (60–90 days); and finisher (≥90 days).
The mean average of 3 times titer serum neutralizing test for antibodies against PEDV. An antibody titer of ≥2 was considered positive.
In summary, this study suggested that PEDVs in Korea belonged to different genogroups. While classical G1 was continuingly circulating between provinces of Korea, the pandemic G2a were recently introduced from China and USA. By the application of Bayesian phylogeographical analysis, this study demonstrated the spatial- temporal transmission of PEDVs within Korea. Of the recent emerged G2a viruses, J3142 strains showed potential recombination breakpoint (376–2,143nt) of S1 gene between KNU1303_Korea strain_G2a (KJ451046) and 45RWVCF0712_Thailand strain_G2b (KF724935). The pandemic G2a virus was partial neutralized by the antibodies invoked by the G1- based PED vaccine virus.
Conflict of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
The authors would like to thank Hye Jung Yang, and Jung Ah Kim for excellent technical assistance. This study was supported by the BioGreen 21 Program, Rural Development Administration (grant no. PJ011184), and by the Bio-industry Technology Development Program (grant no. 114055031SB010), Ministry of Agriculture, Food and Rural Affairs, South Korea.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2016.06.013.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Virol. 2014;52(1):234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.C., Lee K.K., Pi J.H., Park S.Y., Song C.S., Choi I.S., Lee J.B., Lee D.H., Lee S.W. Comparative genome analysis and molecular epidemiology of the reemerging porcine epidemic diarrhea virus strains isolated in Korea. Infect. Genet. Evol. 2014;26:348–351. doi: 10.1016/j.meegid.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.C., Nguyen V.G., Moon H.J., Lee J.H., Park S.J., Lee G.E., Kim H.K., Noh Y.S., Lee C.H., Goede D. ISolation of porcine epidemic diarrhea virus during outbreaks in South Korea, 2013–2014. Emerg. Infect. Dis. 2015;21(12):2238–2240. doi: 10.3201/eid2112.150437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte M., Laude H. Sequence of the spike protein of the porcine epidemic diarrhoea virus. J. Gen. Virol. 1994;75(5):1195. doi: 10.1099/0022-1317-75-5-1195. [DOI] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-W., Dickerman A.W., Piñeyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.-J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4(5):e00737–00713. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20(4):662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon C., Kwon B., Jung T.S., Kee Y., Hur D., Hwang E., Rhee J., An S. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. J. Vet. Res. 1993;33(2):249–254. [Google Scholar]
- Kim S.-H., Lee J.-M., Jung J., Kim I.-J., Hyun B.-H., Kim H.-I., Park C.-K., Oem J.-K., Kim Y.-H., Lee M.-H., Lee K.-K. Genetic characterization of porcine epidemic diarrhea virus in Korea from 1998 to 2013. Arch. Virol. 2015;160:1055–1064. doi: 10.1007/s00705-015-2353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg. Infect. Dis. 2014;20(7):1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.K., Park C.K., Kim S.H., Lee C.H. Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea. Virus Res. 2010;149(2):175–182. doi: 10.1016/j.virusres.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey A., Rambaut A.J., Drummond M.A. Suchard. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009;5(2009):1–16. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole B. Deadly pig virus slips through US borders. Nature. 2013;499(7459):388. doi: 10.1038/499388a. [DOI] [PubMed] [Google Scholar]
- Park S., Kim S., Song D., Park B. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg. Infect. Dis. 2014;20(12):2089–2092. doi: 10.3201/eid2012.131642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44(2):167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Oh J., Kang B., Yang J.S., Moon H., Yoo H.S., Jang Y., Park B. Oral efficacy of Vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res. Vet. Sci. 2007;82(1):134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Yu Z., Cheng K., Liu Y., Huang J., Xin Y., Li Y., Fan S., Wang T., Huang G. Molecular characterization and phylogenetic analysis of new variants of the porcine epidemic diarrhea virus in Gansu, China in 2012. Viruses. 2013;5(8):1991–2004. doi: 10.3390/v5081991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P.F., Jin Y.L., Xing G., Qv L.L., Huang Y.W., Zhou J.Y. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg. Infect. Dis. 2014;20(10):1735–1738. doi: 10.3201/eid2010.140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014;20(10):1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M. Discovery of seven novel mammalian and avian coronaviruses in Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.