Highlights
-
•
Review of the state of the first decade or so of RNA virus metagenomics.
-
•
History, principles, protocols, and major discoveries in RNA virus metagenomics.
-
•
More effort in following up on existing RNA viral discoveries is needed.
Keywords: Metagenomics, RNA virus, Viral discovery, Conjoined circles
Abstract
It is hard to overemphasize the role that metagenomics has had on our recent understanding of RNA virus diversity. Metagenomics in the 21st century has brought with it an explosion in the number of RNA virus species, genera, and families far exceeding that following the discovery of the microscope in the 18th century for eukaryotic life or culture media in the 19th century for bacteriology or the 20th century for virology. When the definition of success in organism discovery is measured by sequence diversity and evolutionary distance, RNA viruses win. This review explores the history of RNA virus metagenomics, reasons for the successes so far in RNA virus metagenomics, and methodological concerns. In addition, the review briefly covers clinical metagenomics and environmental metagenomics and highlights some of the critical accomplishments that have defined the fast pace of RNA virus discoveries in recent years. Slightly more than a decade in, the field is exhausted from its discoveries but knows that there is yet even more out there to be found.
1. Introduction
In 1986, using direct cloning of 16S rDNA genes Norman Pace presciently noted that the method “seems to have no upper limit as to the complexity of the populations” (Pace et al., 1986). Thirty years later, Pace’s maxim only seems ever more true. Deeper and deeper sequencing technologies have enabled the discovery of thousands of viral species. Metagenomic viral discovery can be applied to almost any sample type and novel viruses emerge. Growth in virome and metagenomics papers is outpaced only by that of the viruses discovered (Fig. 1 ). This review is meant to cover the field of modern-day RNA virus metagenomics, where RNA is perhaps best defined as “not DNA”, mostly due to the use of DNA-specific nucleases in library preparations. Many excellent reviews on viral metagenomics already exist, but no specific review focuses on RNA virus metagenomics (Bexfield and Kellam, 2011, Delwart, 2007, Edwards and Rohwer, 2005, Mokili et al., 2012, Rosario and Breitbart, 2011, Tang and Chiu, 2010). Since there is absolutely no way to cover all the samples sequenced or genome organizations found even if one was restricted to a single order or family, the author apologizes in advance if he failed to cite your contribution to the field.
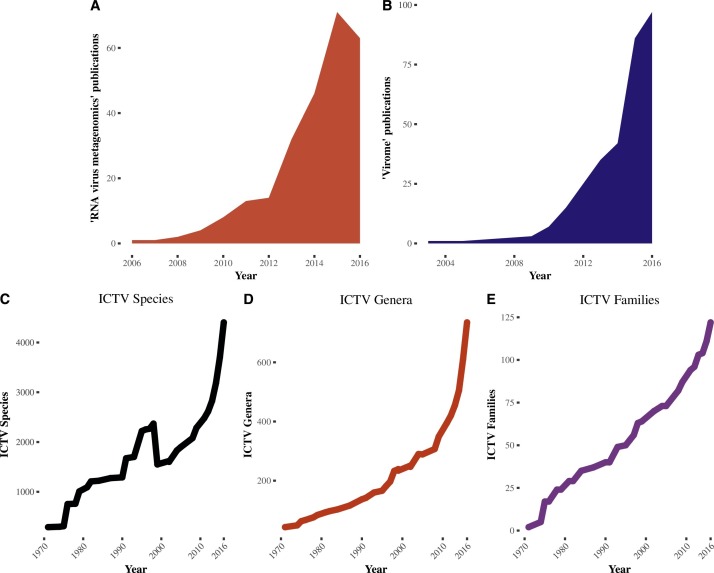
Fig. 1.
Growth everywhere. Publications by year for “RNA virus metagenomics” (A) and “virome” (B) over the past decade using the “Results by Year” graph from Pubmed. Number of viral species (C), genera (D), and families (E) assigned by the ICTV over the past four decades (ICTV, 2017a, ICTV, 2017b). Changepoint analysis with the “at most one change” statistical test was significant at year 1996 for genera and family data and 2015 for species data (“changepoint, ” n.d.).
2. TMRCA of RNA virus metagenomics
2.1. The root
Where do we begin with RNA virus metagenomics? Construction and sequencing of cDNA libraries share most of the features of RNA viral metagenomic protocols and have existed since the late 1970s (Sim et al., 1979, Efstratiadis et al., 1977). One could argue about the meta- or the −genomic nature of the cDNA sequencing example, but the method is nearly unchanged since its introduction. A single organism contains multitudes and now one of the more common ways of discovering new viruses is from searching said cDNA sequencing data (Feng et al., 2008, Shi et al., 2016, Krishnamurthy et al., 2016). Clearly, the depth of sampling has changed in recent years but sequencing even two clones could be considered metagenomics.
RNA virus metagenomics began with a DNA virus and a contaminant. In a prescient 2001 manuscript, Allander et al. added DNase to spin-filtered serum prior to extraction to enrich for viral particles (Allander et al., 2001). Nucleic acids were extracted and separately processed for DNA and RNA libraries and then randomly amplified, enabling the discovery of two bovine parvoviruses from the commercial bovine serum that was used to dilute their viral spike-in controls. Surprisingly, the novel DNA viruses were present in the RNA half of the experiment – the side that was specifically reverse transcribed and amplified in order to test the protocol’s ability to detect GB virus C (Allander et al., 2001).
This experiment is worth focusing on as it foreshadowed several themes in the world of RNA virus metagenomics. The protocol used in the paper is surprisingly similar to that used today (Conceição-Neto et al., 2015). Host or environmental background DNA dwarfs most nucleic acids and sequencing of RNA species circumvents the problem so well that it may be the best way to find high concentration DNA viruses that are being actively transcribed. Even today concerns over whether metagenomics is sufficiently sensitive bedevil its use as a gold standard (Schlaberg et al., 2017, Wylie et al., 2012). Finally, in the world of metagenomics, you do not know what is in your sample until you sequence, and the sensitivity of your assay is perhaps best measured by your ability to recover ubiquitous reagent contaminants (Bhatt et al., 2013, Naccache et al., 2013).
2.2. The early branches
Environmental RNA virus metagenomics arose somewhat later. As recently as 2003, degenerate PCR targeting the RNA-dependent RNA polymerase from picorna-like viruses had shown the potential for viral discovery in marine virus communities with the detection of four new RNA virus families (Culley et al., 2003). Large scale, high-profile efforts such as sailboat-based shotgun sequencing of the Sargasso Sea came two years later and used 0.1–3.0 μm filters that predominantly enriched for DNA from microbial communities (Venter et al., 2004). Three years later, Robert Edwards and Forest Rohwer were still calling for the development of methods that would allow for sequencing of RNA viruses (Edwards and Rohwer, 2005).
Allander’s original method that detected the bovine parvovirus contaminants gave rise to clinical metagenomics in 2005 by discovering a novel human parvovirus (human bocavirus) and detecting a novel human coronavirus (coronavirus HKU1) by screening pooled human nasopharyngeal aspirates (Allander et al., 2005, Woo et al., 2005). Reads to influenza A virus, human orthopneumovirus (RSV), metapneumovirus, human adenovirus, and TTV-like virus were also recovered from the 864 reads. That same year 36,789 colony sequences from DNase-treated RNA extracted from viral concentrates of human feces revealed a preponderance of pepper mild mottle virus and other plant viruses, but no new human viruses (Zhang et al., 2005). The first non-human environmental-focused RNA viral metagenomic survey used DNase-treated extracted nucleic acids from nuclease-treated viral concentrates from coastal communities in British Columbia (Culley et al., 2006). The resultant pure RNA was reverse transcribed and double-stranded cDNA was synthesized, blunt-end ligated, and cloned to reveal nothing, for there was not enough RNA present even in 40–60L of seawater, highlighting the importance of amplification in most any RNA virus metagenomic preparation. After amplification, two novel picorna-like viruses and one novel +ssRNA virus that remains taxonomically unassigned were discovered at high concentrations among the 355 reads.
It is incredible to note that only one year later, researchers were reporting RNA viral metagenomic sorcery of honeybees with hundreds of thousands of reads (Cox-Foster et al., 2007, Koonin, 2007). Next-generation sequencing and rapid bioinformatics gains allowed researchers unbelievable opportunities (Fig. 1) (Delwart, 2007). Due to the short read length of Solexa sequencing at the time, initial viral metagenomic efforts required 454 sequencing, which allowed for hundreds of thousands of reads a few hundred bases long (Rothberg and Leamon, 2008). The result was an array of human viral discoveries, including Merkel cell polyomavirus, human bocavirus 2, salivirus, Lujo arenavirus, Dandenong arenavirus, SFTS bunyavirus, and a variety of new human astroviruses including MLB1 and VA1 through VA5 (Briese et al., 2009, Feng et al., 2008, Finkbeiner et al., 2008a, Finkbeiner et al., 2008b, Greninger et al., 2009, Holtz et al., 2009, Kapoor et al., 2009a, Kapoor et al., 2009b, Koonin, 2007, Li et al., 2009, Meyer et al., 2015, Palacios et al., 2008, Yu et al., 2011). Groups often found the same viruses within days of each other and discrepant names for the same virus were common (Greninger et al., 2009, Holtz et al., 2008, Holtz et al., 2009, Kapoor et al., 2008, Li et al., 2009). Pyrosequencing yielded impressive gains in environmental RNA virus sequencing with profiling of a freshwater lake yielding tens of novel ssRNA and dsRNA viruses (Djikeng et al., 2009). The RNA virus metagenomic age was officially underway.
3. Why has RNA virus metagenomics been so successful?
3.1. Diversity
While early studies of metagenomics lamented the lack of RNA viruses discovered compared to DNA viruses, RNA virus metagenomics appeals to the viral discoverer for a number of reasons. The first is that success in viral metagenomics is often measured by the genetic distance of a newly discovered virus from other known viruses. While finding a new virus in the environment is not necessarily a problem in either DNA or RNA, the low fidelity of RNA polymerases and the sequence space they are capable of sampling, along with the possibility of recombination, lend themselves to new species and genera that are the trophies of metagenomic viral discovery. These properties also favor the metagenomic library preparation process. Sufficient homology to land PCR primers for amplicon-tiling library preparation approaches may not exist, and the number of oligos needed to cover the sequence space of a given RNA virus such as Hepatitis C virus can make hybrid capture approaches cost-prohibitive. De novo assembly methods that allow for the rapid reconstruction of near complete genomes have also dramatically improved over the past decade and can handle widely disparate coverage for each contig (Grabherr et al., 2011, Bankevich et al., 2012). All of these properties often make metagenomics the first method of choice when trying to recover sequence from even a known RNA virus (Greninger et al., 2017a, Ogimi et al., 2017).
3.2. It is not DNA
Another reason for the provision of RNA virus metagenomics is the ease and increased depth and sensitivity associated with removing background DNA. Indeed, one of the difficulties of RNA virus metagenomics is that RNA virus genome sizes are generally measured in the kilobases, but the size and quantity host DNA background measures in the gigabases. If not partitioning sequence selectively across RNA and DNA, a single contaminating host cell can be the equivalent of half a million virions. Removal of host DNA is often required for one to recover complete RNA virus genomes. The library preparation process simply requires a 30-min treatment with DNAse I or Benzonase to remove all background DNA leaving the RNA viruses behind. In addition, RNA viral capsids are measured in nanometers and can be selectively enriched using 0.22 um or 0.45 um filtration. Even if one is interested in DNA viruses, RNA metagenomics allows the recovery of mRNAs that are associated with actively replicating DNA viruses, which may be helpful information when so many DNA viruses can be latent or integrated into host genomes (Perlejewski et al., 2015).
3.3. Sitting right there
Recovery of these RNA viruses is also made easier by the fact that, once host DNA is removed, they can be present at extraordinarily high concentrations in total RNA or constitute a surprisingly large proportion of transcripts in an mRNA library, up to almost 90% of non-ribosomal RNAs in some invertebrates (Li et al., n.d.; Shi et al., 2016). Based on total masses of nucleic acid in virus like particles, it has been estimated that half of marine viruses are RNA viruses due to the larger burst size of eukaryotic RNA viruses (Steward et al., 2013). A particularly instructive example was the discovery of the Lake Sinai virus in honeybees (Runckel et al., 2011). It was originally discovered and assembled metagenomically at such high titer that when researchers went to perform Northern blots, they could see the viral RNA staring back at them after RNA electrophoresis. The author has also previously taken part in studies where the method of screening for known segmented RNA viruses such as rotavirus was to run gel electrophoresis on extracted stool RNA without amplification and look for banding (Greninger et al., 2010, Yu et al., 2012).
3.4. No wet lab necessary
Mining of publicly available transcriptome data has contributed greatly to the discovery of novel RNA viruses (Basler et al., 2005, Schomacker et al., 2004). Indeed, genome-transcriptome studies of eurkaryotes sometimes miss the viruses present in their sequence when they map their RNA-Seq reads to their assembled DNA-Seq genome (Babb et al., 2017, Clarke et al., 2015). RNA viruses are lurking in the unaligned portion of the reads and often can be readily assembled and aligned to NCBI to find many new viruses in a given host (Bekal et al., 2011). Most recently, six novel RNA viruses were discovered in publicly available orb weaver spider transcriptome data (Debat, 2017). Such sequences can be downloaded from the Short Read Archive, assembled, and deposited into NCBI’s Third Party Annotation database (“NCBI Third Party Annotation, ” n.d.). Research parasites of the world can now identify with their obligate intracellular parasitic brethren (Longo and Drazen, 2016).
4. A metastable approach
Rapid increases in sequencing depth achieved with next-generation sequencing have been matched by the increasing ease of library preparation. Original viral sequencing protocols involved the preparation of multiple micrograms of nucleic acid and about a week of work (Thurber et al., 2009, Willerth et al., 2010). Protocols were slowly refined to reduce library preparation to a matter of days (Greninger et al., 2010). The development of transposon-based library creation and amplification have reduced RNA metagenomic library preparation such that most labs perform the process in 12 days, and when coupled to nanopore sequencing the entire sample-to-sequence process can be as little as 6 h (Greninger et al., 2015b, Greninger et al., 2017b, Greninger et al., 2017c).
A typical RNA virus metagenomic protocol entails filtration, nuclease treatment, double-stranded cDNA synthesis, followed by Nextera XT tagmentation and PCR amplification (Alexander L. Greninger et al., 2015c, Hall et al., 2014). The benefit of the protocol is the minimal hands-on time, low cost given the dilutability of the transposase, ease of dual-indexing, and direct compatibility with Illumina sequencers. The protocol can be performed in as little as 4–6 h and is amenable to automation (Greninger et al., 2017b, Greninger et al., 2017c). Variants include the use of additional PCR cycles or amplification prior to tagmentation. However, in the author’s experience these are not necessary given the amplification step present in the tagmentation library preparation protocol and the possibility of extra PCR cycles creating coverage bias (Conceição-Neto et al., 2015). Steps to increase the amount of nucleic acid used as input are helpful for the retrieval of longer reads and reducing PCR jackpotting, although often practitioners move forward with the RNA present. While no doubt the best way to enrich for viral like particles, and perhaps to help define a sequence as likely viral in origin, the use of ultracentrifugation is not necessarily required for viral metagenomics. Rather ultracentrifuge is kept for defined “virome” work, especially where there is a high bacterial background such as feces (Kohl et al., 2015, Temmam et al., 2015, Thurber et al., 2009). Indeed a direct head-to-head comparisons of ultracentrifugation and filtration for viral particle enrichment found the main benefit of ultracentrifugation to be the removal of host DNA background, which is less of a concern for RNA viral metagenomics if extracted nucleic acids are DNAse treated (Kleiner et al., 2015).
5. Metagenomics for the people
As evidenced from the early history, discovery of new etiological agents of human disease drove the adoption of viral metagenomics (Lysholm et al., 2012, Tang and Chiu, 2010). Clinical metagenomics, or direct shotgun sequencing from clinical samples with minimal sample processing, for diagnostic testing has largely been driven by the diversity of RNA viruses that preclude the routine use of single-plex or even multiplex PCR for broad clinical detection. Indeed, current rapid diagnostics followed by 16S or ITS PCR and Sanger sequencing cover most of the actionable and more exotic organisms present in clinical specimens. While metagenomics delivered on the promise of finding novel human viruses, viral discovery in humans has increasingly become a tragic story of patients interacting with the wrong squirrel or tick on the wrong day and most samples sequenced are frankly negative (Hoffmann et al., 2015, McMullan et al., 2012). In part because of the declining number of viral discoveries in humans, clinical metagenomics has taken on the mantle of detecting all known pathogens instead (Naccache et al., 2014, Wood and Salzberg, 2014). Metagenomics has been especially successful at finding known RNA viruses in unexpected sample types (Doan et al., 2016, Naccache et al., 2015). Provided there is sufficient coverage, it can also provide single nucleotide resolution to define strain relatedness for outbreak epidemiology and, where available, antiviral resistance (Barzon et al., 2011, Capobianchi et al., 2013, Greninger et al., 2017b, Greninger et al., 2017c). Furthermore, the vast majority of clinical samples sent for clinical metagenomic sequencing are entirely negative, yielding a greater potential role for “ruling out” an infectious cause through metagenomic testing prior to treating for other diseases such as autoimmune disorders with immunosuppressant medication.
The excitement over using clinical metagenomics for one definitive test for clinical microbiology is tempered by questions around its sensitivity, actionable-ness of the sequence recovered (including cross-contamination), and cost. Since clinical labs are highly unlikely to perform ultracentrifugation, metagenomics likely has the best sensitivity in acellular environments such as cerebrospinal fluid rather than respiratory or stool specimens (Schlaberg et al., 2017). Human cells become an interfering substance and at concentrations much lower than those that interfere with other clinical chemistry measurements (Ranjitkar et al., 2017). Detection of viruses at low concentration such as Zika virus make shotgun metagenomics problematic for a rule-out result in diagnostic virology (Bingham et al., 2016, Landry and St George, 2017, Naccache et al., 2016). As with other culture-independent testing, culture will likely be required to provide antimicrobial sensitivity. While early detection can reduce further diagnostic testing, most RNA viruses in humans have no treatment and are essentially unactionable.
6. Bat-guano crazy about sampling
The ease and ubiquity of RNA virus metagenomics is perhaps best illustrated by the increasingly diverse samples that are sequenced. When sequence divergence is the metric of success, metagenomicists boldly go where no human has gone before. For the purposes of raw discovery, the use of extraordinary mixed samples allow for one-stop shopping and diversify discovery risk, albeit with the drawback that host species cannot be confidently defined. And the continuing need to fund and sell discovery from the basis of security has focused metagenomic efforts on species with high zoonotic pandemic potential (Racaniello, 2016, Temmam et al., 2014). Here, bat guano routinely ranks high on the RNA virus metagenomic list as it checks all three requirements listed above. Bats aggregate a wide array of arthropods, plants, and other animals and produce copious amounts of guano and have been associated with multiple zoonoses. Cameroonian fruit bats, Chinese bats, Myanmar bats, Hungarian bats, French bats, American bats, big brown bats, tricolored bats, and little brown bats have all been profiled showing a vast array of RNA viruses from eukaryotic hosts (Dacheux et al., 2014, Donaldson et al., 2010, Ge et al., 2012, He et al., 2013, He et al., 2017, Kemenesi et al., 2014, Li et al., 2010, Yinda et al., 2017). Bats have been used to put an upper limit – in the hundreds of thousands − of the total number of eukaryotic viruses that remain to be discovered (Anthony et al., 2013). Other insect and arthropod aggregators such as spiders and birds have also yielded many new RNA virus species (Debat, 2017, Ducatez and Guérin, 2015, Shean et al., 2017, Zhou et al., 2015).
The greatest paradigm shifter in recent viral metagenomics work has been the sheer number and diversity of novel RNA viruses present in arthropods and invertebrates. Yong-Zhen Zhang and team at the Chinese CDC have been systematically profiling the transcriptomes of invertebrates to upend our understanding of animal virology. A haul of 112 novel negative-stranded RNA viruses from 70 arthropods surpassed the known diversity of Mononegavirales (Li et al., n.d.). These included the first circular negative stranded RNA viruses ever found, which were part of a larger family of viruses (Chuviridae) that may yet become an order given that it is phylogenetically ancient to existing segmented and unsegmented viruses and was arranged in a number of topologies.
And yet those were just the negative-stranded viruses. When the team increased the number of hosts profiled to 220, they found 1445 novel RNA viruses (Shi et al., 2016). The genomes found illustrated widespread recombination among RNA viruses, viruses with multiple copies of structural genes, loss of structural genes, as well as transfer of genes between virus and host. Indeed, these discoveries so dwarf all previous RNA virus discoveries, especially in the picorna-like superfamily, that the paper itself would be worth reprinting here in full. Three additional novel families of positive-stranded viruses and two novel families of negative stranded viruses were described. Rather than detailing what was found, the surprises are the families in which novel RNA dependent RNA polymerase (RdRp) sequences were not found such as the Arenaviridae, Picornaviridae, Hepeviridae, Paramyxoviridae, Arteriviridae, and Secoviridae. Of course, whole undiscovered taxa basal to these taxa were also recovered. Perhaps most impressively, Zhang’s team finished complete genomes with rapid amplification of cDNA ends (RACE) for each of these viral genomes, a feat that not many metagenomic viral discovers undertake when finding more than a few novel viruses. No doubt evolutionary RNA virologists will have just begun to comb through this data trove before the next ten thousand whole genomes of novel RNA viruses is deposited from peat moss.
Most importantly for veterinarians, RNA virus metagenomics has revealed a number of candidate etiological agents for a variety of animal ailments including avian proventricular dilatation disease, mink shaking syndrome, snake inclusion body disease, python respiratory illness, dairy cow disease, and tilapia die-offs (Kistler et al., 2008, Gancz et al., 2009, Blomström et al., 2010, Stenglein et al., 2014, Stenglein et al., 2012, Uccellini et al., 2014, Hoffmann et al., 2012, Bacharach et al., 2016). Animal feces has proven fruitful hunting for metagenomicists, with canine, feline, porcine, equine, goose, duck, and shrew fecal RNA viromes turning up a plethora of mostly novel picorna-like viruses, albeit with an unclear relationship to disease and perhaps some more closely linked to the invertebrates indicated above (Fawaz et al., 2016, Greninger and Jerome, 2016, Li et al., 2011, Moreno et al., 2017, Nagai et al., 2015, Naoi et al., 2016, Ng et al., 2014b, Phan et al., 2011, Reuter et al., 2012, Sano et al., 2016, Sasaki et al., 2015, Zhang et al., 2014). The recovery of Ancient Northwest Territories cripavirus from 700-year old frozen caribou feces illustrated an incredible stability of encapsidated RNA (Ng et al., 2014a). Screens for fish viruses are still in their infancy and yet the phylogenetic diversity of fish and perhaps their intense exposure to marine RNA viruses may prove them to be the chordate arthropods of viral discovery. Novel members of the Orthomyxoviridae, Picornaviridae, and Reoviridae have recently been described in fish (Bacharach et al., 2016, Reuter et al., 2013, Reuter et al., 2015). Our understanding of reptile and amphibian viral diversity is also still in its early days. The discovery of a non-Old World, non-New World arenavirus in boid snakes with an envelope protein most closely related to filoviruses proved a tantalizing hint toward new variations on old taxonomies (Stenglein et al., 2012). For the basic scientists, a number of novel RNA viruses have been discovered in model organisms such as C. elegans or D. melanogaster (Félix et al., 2011, Webster et al., 2015). Thousands of new viruses have been found in plants through metagenomics, with most awaiting further characterization or even finished assembly (Roossinck et al., 2010). Plant virus metagenomics is probably one of the more fertile areas of growth for viral discovery in the coming years given their incredible biodiversity (Roossinck, 2012).
RNA viral metagenomics has made a number of contributions to our understanding of viral diversity at the far reaches of life. Sequencing of a hot, acidic lake and wastewater revealed a novel ssDNA virus with an RNA virus-like capsid protein, suggesting past recombination between DNA and RNA viruses (Diemer and Stedman, 2012; “RDHV-like virus SF1 replication-like protein and capsid protein genes, complete cds,” 2015). While there are still no definitive RNA viruses of Archaea, viral RNA metagenomics isolated a 5.6 kb contig that contained an capsid-like protein and an RNA-dependent RNA polymerase that differed from the RdRps of viruses from eukaryotes and bacteria (Bolduc et al., 2012). The same can be said of the ciliates, who were previously noted among the eukaryota for having no known RNA viruses (Koonin et al., 2008). Metagenomic sequencing of wastewater after a rainstorm revealed two RNA contigs that were highly suggestive of ciliate RNA viruses in a metagenomic background of Tetrahymena and several viruses best translated in the ciliate genetic code were found in the invertebrate surveys detailed above (Greninger and DeRisi, 2015a, Shi et al., 2016).
Given the incredible growth in bacteriophage diversity from metagenomics, RNA phages were a surprisingly late arrival to the metagenomic discovery party. As recently as 2016, genome sequences from RNA phages were surprisingly few and far between with only 11 ssRNA and 5 dsRNA genomes compared to over 1000 DNA bacteriophage genomes from 494 species (Greninger and DeRisi, 2015b, Krishnamurthy et al., 2016). Mining of metagenomic datasets yielded an additional 122 partial genomes from novel RNA phages covering >100 novel species including the first known RNA phage number of a Gram-positive bacteria (Krishnamurthy et al., 2016). These sequences included multiple unique genomic arrangements as well as ORFs that had no similarity to known proteins. Invertebrate virus sequencing efforts detailed above also found a similar number of levi-like viruses (Shi et al., 2016). Given the many uses of the MS2 phage coat protein for understanding RNA-protein interactions and as a tool for bioengineering RNA affinity purification, the expansion of the RNA phageome will likely create a number interesting tools for molecular biology that can build on MS2.
7. Pity the ICTV
The ridiculous number of virus discoveries in the past several years has put an incredible strain on the whole system of viral taxonomy (Fig. 1C–E). The inclusion of uncharacterized metagenomic viral sequence in taxonomy has long been of some debate at the International Committee on Taxonomy of Viruses (ICTV), the adjudicator of novel families, genera, and species of the viral world. Only this past year did the ICTV codify its existing policy through issue of a consensus statement on the inclusion of metagenomic data in its consideration of taxonomical placement of viruses (Simmonds et al., 2017).
The torrid pace of discoveries has forced the hand of viral discoverers to come up with euphonious names for viruses for which there is almost no biological understanding (ICTV, 2017a, ICTV, 2017b). Convention previously dictated something like location, host, and number, even if the World Health Organization now recommends against ruining the tourist economy along the Ebola River or in Coxsackie, New York (Fukuda et al., 2015). Since sequence similarities govern our understanding of genomic function, there is a temptation to name based on homology and let past discoveries anchor the novel (e.g., picobirnavirus or dicipivirus/cadicivirus/picodicistrovirus) (Woo et al., 2012). Default nomenclature is approaching that of drug manufacturers or therapeutic antibody naming, with alternating consonant-vowel pairs that might have some basis in Latin or relation to the location but obscures the actual place (e.g. avisivirus, aquamavirus, hunnivirus, harkavirus) (Boros et al., 2015, Reuter et al., 2012). Recently, five ancient Chinese states formed the basis for naming novel family lineages (Shi et al., 2016). Detection of ancient recombination in RNA virus sequences is revealing genomic abominations that only a liger could love. What do you get when you cross a picornavirus and a calicivirus? Probably a non-functional, non-replicating piece of RNA, but Picalivirus A–D are still a thing (Greninger and DeRisi, 2015c; Ng et al., 2012). Tombusviridae and Nodaviridae? Tombunodavirus (Grasse and Spring, 2017, Greninger and DeRisi, 2015d).
The inventiveness of nomenclature for novel virus discovery in a space of anarchy combats a steady march of rules and reason by the ICTV (Simmonds et al., 2017). Not even clinically relevant viruses such as human parainfluenza viruses or respiratory syncytial viruses – now human respirovirus, rubulavirus, orthopneumovirus – can resist binomial nomenclature and taxonomical reassignment (Adams et al., 2017). However, if discoverers get out far enough of the rationalization, their original names can take advantage of the movement to have an outsized influence on viral nomenclature (Boros et al., 2015, Runckel et al., 2011, Woo et al., 2012).
8. Remaining frontiers
In a field with seemingly no end to frontiers, it is curious to define even more. No doubt the ‘virtuous cycle’ of metagenomic sequencing will only increase in breadth and depth in the coming years with a concomitant growth of new bioinformatic algorithms leading to discovery of even more new organisms and further decrease in so-called unalignable viral “dark matter” (Fig. 2 ) (Greninger et al., 2015a, Krishnamurthy and Wang, 2017). Turning the crank on RNA virus discovery metagenomics is now the provenance of undergraduate theses (Makhsous et al., 2017, Shean et al., 2017, Zaaijer et al., 2016). Other than sampling more broadly and maintaining the exponential growth in Genbank, what are orthogonal challenges for future RNA virus metagenomic studies?
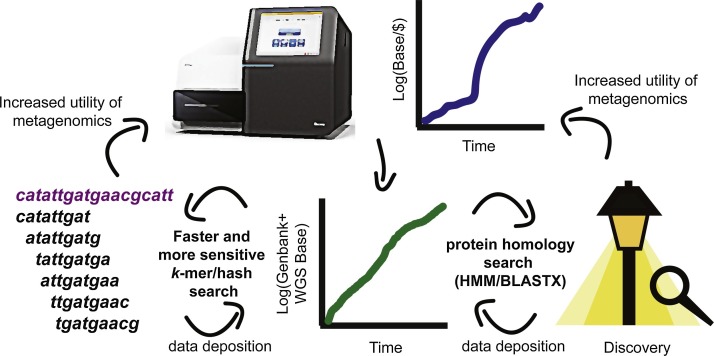
Fig. 2.
The conjoined circles of metagenomic success.
The increased output of modern-day sequencers has led to increased metagenomic sequencing of samples, both environmental and clinical. This in turn has led to an explosion in the NCBI Genbank and WGS databases. New discoveries beget the discovery of more divergent new viruses and organisms as they are now alignable to new references in the database. Rather than searching through gapped alignments, the increased coverage of the Genbank reference database allows for more exact k-mer searching, which allows for faster, more sensitive alignments of reads. This in turn makes metagenomic sequencing more useful, especially in the clinic, which in turn begets more sequencing.
8.1. Easier, faster, better, broader protocols
To start in the wet lab, better methods of host depletion and recovery of full viral genomes are absolutely required. Too many analogies to “finding a needle in a haystack” in the literature necessitate the use of cellulases (Allen et al., 2009, Kowalchuk et al., 2007, Lax and Gilbert, 2015, Lecuit and Eloit, 2014, Naccache et al., 2014, Soueidan et al., 2015). Even if RNA viral transcripts constitute >50% of an arthropod, they rarely do so in mammalian tissue (Feng et al., 2008, Shi et al., 2016). In both clinical metagenomics and complex mixtures there are always low concentration viral sequences, either due to time of sampling, viral biology, or the long tail of ecological abundance, which deeper sequencing alone may not solve. Synthetic biology and programmable nucleases may allow new options for depletion beyond the ribosome although current efficiencies must be improved (Gu et al., 2016, Matranga et al., 2014).
Even though sample preparation methods have become considerably easier in the past decade, they still take several hours of hands-on time and have not routinely been ported to automated liquid handlers. With plunging sequencing costs, library preparation costs for RNA virus metagenomics have taken a front seat. The time and cost of library preparation hamper adoption of metagenomics in the clinical virology lab. Similar to the expanding suite of Cas programmable sequence-guided nucleases, an RNA-directed transposase for adapter tagging followed by one-step, dual-indexed amplification would speed up library preparation as the bulk of time in current preparations is spent on double-stranded cDNA synthesis (Gertz et al., 2012). Such an enzyme might be discovered through the approaches highlighted here, or perhaps through directed evolution of existing DNA transposases (Adey et al., 2010). Another solution might be a one-pot mix of enzymes that performed library preparation in a similar fashion as Gibson cloning.
Even in a host-free environment, current amplification methods do not recover full viral genomes from end-to-end. Instead, the common paradigm is random priming with amplification followed by a follow-up step of RACE to recover ends (Li et al., n.d.; Shi et al., 2016). This second step is highly laborious, especially for segmented RNA viruses, and difficult to execute on viral sequences at low concentration or in complex mixtures. Furthermore, current transposase-based library methods can produce inversions or sequencing artifacts at the end of genome segments or in areas of RNA secondary structure.
As “unbiased” or “agnostic” as metagenomics can be, we still rely on sequencing by synthesis and a four base read-out. The ultimate immunoevasion strategy may be virally-encoded nucleotides (Bryson et al., 2015, Murphy et al., 2013, Weigele et al., 2017). Host and virally-encoded RNA editing already bedevils calling of accurate whole genome sequences in both positive- and negative-strand RNA viruses (Park et al., 2015, Pelet et al., 1991, Piontkivska et al., 2017, Vidal et al., 1990). Adenosine-based modifications of viral genomes have shown effects on viral and host biology, but detecting them requires additional modifications to most sequencing protocols (Gokhale and Horner, 2017, Gonzales-van Horn and Sarnow, 2017, Kennedy et al., 2016, Kennedy et al., 2017). Our understanding of RNA base modifications in viral genomes is still in its infancy. Here, nanopore-based approaches to sequencing may hold the key to new discoveries as they can directly detect modified nucleotides, although current approaches are as dependent on the biology of pore proteins as we are on polymerases (Ayub et al., 2013, Carlsen et al., 2014). The ultimate answer may be direct mass spectrometry-based detection of viral nucleotides (Gooskens et al., 2014, Cobo, 2013). Improvements in sensitivity of nanopore sequencing in terms of sequencing depth and the host depletion strategies highlighted above are required before nanopore-based metagenomics becomes routine or meaningful (Greninger et al., 2015b).
Other than the retroviruses, RNA viruses do not readily provide host linkage information in metagenomics like DNA viruses and phages often do. Linking RNA virus and host has been imputed based on transcript levels or dinucleotide usage, although the latter has recently been shown to more correlated with viral family than host species (Kapoor et al., 2010, Giallonardo et al., 2017, Shi et al., 2016). Even when sequencing discrete organisms such as a singular honeybee, the co-existence of eukaryotic parasites means that imputation of viral host cannot be exact (Runckel et al., 2011). Host promiscuity seems to be a common theme among newly-described RNA viruses (Nunes et al., 2017). Highly-indexed libraries, chemical linkage of RNA species, and physical partitioning through single cell transcriptomics currently provide the best potential solution to the host imputation problem in complex mixtures (Burton et al., 2014, Chow et al., 2015, Turner et al., 2009).
8.2. Experimenting with function
To date, most metagenomics has adopted the 19th century British naturalist approach of cataloging inventory and diversity. This focus of the field has been somewhat punishing for those involved, with burnout of both scientists and reviewers from not fully understanding the why of it all (Canuti and van der Hoek, 2014). This author does not necessarily have a better plan, but perhaps phenotype might be a reasonable start. Mass spectrometry was available to previous generations of biochemists, but they did not necessarily stop to catalog the contents of every fraction, focusing instead on function. This is not to understate the critical impact of the technical training that comes from viral metagenomics, nor to minimize the power of the method. But once we know that viruses are diverse, some concern for function is most likely in order. Although they are now no doubt contributing, viral metagenomicists have previously borrowed liberally from the biochemical functions assigned to strings of sequence without confirmation. Genomics has been the wires that allowed biochemistry to scale (167, 168).
A number of options for functional characterization of these viruses are available but they unfortunately require work. Additional culture models are needed to handle all the new discoveries and genetic engineering methods for more easily establishing cell lines from exotic species are worth pursuing (Ettayebi et al., 2016, Finkbeiner et al., 2012, Stenglein et al., 2012, Bell-Sakyi and Attoui, 2016, Janowski et al., 2017). Focusing discovery on genetically tractable organisms has allowed for relatively rapid functional studies of the new discovered C. elegans Orsay virus (Chen et al., 2017, Jiang et al., 2017). A number of culture-independent methods exist now to characterize the novel viruses and their genes. Biochemical assays for RdRp, proteases, helicases, capsids, and internal ribosome entry sites all exist and synthetic biology allows for easy cloning from a database to test these new proteins found through metagenomics (Ladd Effio et al., 2016, O’Donoghue et al., 2012, Peersen, 2017). Profiling known functions across the new viral species, such as RNA binding strength and sequence specificity of novel RNA phage proteins related to MS2 phage or viral protease specificity and kinetics, might be a place to start (O’Donoghue et al., 2012). Affinity purification-mass spectrometry is a powerful method that allows discovery of viral-host protein–protein interactions in the absence of culture (Jäger et al., 2011, Greninger et al., 2012, Medina et al., 2017, Greene et al., 2016). For vertebrate viruses, serological assays have long been used to confirm roles in disease (O’Sullivan et al., 1997, Bao et al., 2011, Coller et al., 2016).
The ubiquity of these viruses also leads to questions of how we publish in the absence of experiments and what a sequence is worth. The number of novel viruses discovered needed for a high-profile paper has increased by logarithms (Krishnamurthy et al., 2016, Shi et al., 2016). In the absence of particular phenotypic data or wet-lab viral characterization, many authors have turned to Genome Announcements, biorxiv, or simply uploading to Genbank with extra metadata, figuring that sequencing is the most likely future method of both detection and discovery (Debat, 2017, Greninger and DeRisi, 2015e, Greninger and DeRisi, 2015f, Karamendin et al., 2016, Sharman et al., 2016, Sparks et al., 2013). Given the glut of new viruses, expansion of sequencing, and the time it takes to publish, scientists may be more likely to align to your novel virus rather than read about it in a journal and decide to screen for it. This method has allowed for rapid sharing of data and de facto global screens of novel RNA virus prevalence, tropism, and evolution that then lead to something resembling a story.
8.3. New roots
Perhaps the most difficult challenge to spring from sequence gazing at all the novel RNA viruses is our understanding of the origins of RNA viruses. At what point in host evolution does an RNA viral pathogen arrive? And what exactly did the host look like then? RNA virus metagenomics still does not provide the answer. Results from initial RNA virus genomics work suggested picorna-like viruses predated the radiation of the five supergroups of eukaryotic organisms (Koonin et al., 2008). Since then, hints of genetic exchange between hosts and viruses, prominent in DNA virus and bacteriophage genomics, are showing up in RNA viruses (Hughes and Stanway, 2000, Sasaki and Taniguchi, 2008, Shi et al., 2016, Staring et al., 2017). Accounting for each of the domains commonly present in RNA viruses is a challenge for any origin story, though greater knowledge of domain organization and swapping between different RNA viruses may help firm up these dates (Koonin et al., 2015). Biochemical characterization of the incredibly diverse RNA viruses discovered through ongoing metagenomic screens will also test theories of the origin of RNA viruses and better date the origin of different RNA virus clades relative to different epochs of eukaryotic evolution. Recent genetic and proteomic screens of eukaryotic RNA viruses have indicated key reliance on host lipid modifying enzymes that regulate vesicular transport (Greninger et al., 2013, Sasaki et al., 2012, Greninger, 2015, Carette et al., 2011, Marceau et al., 2016, Borawski et al., 2009, Berger et al., 2011, Arita et al., 2013, Nagy and Pogany, 2012, Xu and Nagy, 2016, Salloum et al., 2013). Membrane repurposing is a critical determinant of RNA virus replication which should be added to the “hallmark genes” of RNA viruses, though they currently escape sequence alignment profiling (Koonin and Dolja, 2014). These results have led to a hypothesis in which part of the RNA virus origins involve selfish vesicular transport that likely expanded with the rise of intracellular eukaryotic transport (Greninger, 2015, Koonin et al., 2006, Kuehn and Kesty, 2005). The list of RNA viruses that have acquired a cellular envelope continues to grow (Chen et al., 2015, Feng et al., 2013, McKnight et al., 2017). New concepts in bacterial vesicular transport might be consistent the close evolutionary link between the eukaryotic RNA virus polymerases and bacterial retroelements (Jan, 2017, Kuehn and Kesty, 2005).
9. Make it stop
To put it scientifically, it is nuts that all this and much more have happened in only approximately a decade. If they were not already, RNA viruses have become the star child of evolutionary biologists, whether for day-by-day or eon-by-eon examples of evolution (Koonin, 2007, Koonin et al., 2008, Koonin et al., 2015, Shi et al., 2016, Xue et al., 2017). With our world expanded and our place in it shrunk, there is still much to do regarding viral sequencing for our own quotidian human RNA viruses (Tang et al., 2017). There is but one way to ensure that we find no more viruses. Go sequence something.
Acknowledgements
The author would like to thank Keith Jerome Samia Naccache, Sue Greninger, and Ryan Shean for helpful comments.
References
- Adams M.J., Lefkowitz E.J., King A.M.Q., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M., Sabanadzovic S., Sanfaçon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Gorbalenya A.E., Davison A.J. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2017:2017. doi: 10.1007/s00705-017-3358-5. [DOI] [PubMed] [Google Scholar]
- Adey A., Morrison H.G., Asan, Xun X., Kitzman J.O., Turner E.H., Stackhouse B., MacKenzie A.P., Caruccio N.C., Zhang X., Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T., Emerson S.U., Engle R.E., Purcell R.H., Bukh J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. 2001;98:11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C., Bent A., Charkowski A. Underexplored niches in research on plant pathogenic bacteria. Plant Physiol. 2009;150:1631–1637. doi: 10.1104/pp.109.140004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony S.J., Epstein J.H., Murray K.A., Navarrete-Macias I., Zambrana-Torrelio C.M., Solovyov A., Ojeda-Flores R., Arrigo N.C., Islam A., Khan S.A., Hosseini P., Bogich T.L., Olival K.J., Sanchez-Leon M.D., Karesh W.B., Goldstein T., Luby S.P., Morse S.S., Mazet J.A.K., Daszak P., Lipkin W.I. A strategy to estimate unknown viral diversity in mammals. mBio. 2013;4 doi: 10.1128/mBio.00598-13. (e00598-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita M., Kojima H., Nagano T., Okabe T., Wakita T., Shimizu H. Oxysterol-binding protein family I is the target of minor enviroxime-like compounds. J. Virol. 2013;87:4252–4260. doi: 10.1128/JVI.03546-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayub M., Hardwick S.W., Luisi B.F., Bayley H. Nanopore-based identification of individual nucleotides for direct RNA sequencing. Nano Lett. 2013;13:6144–6150. doi: 10.1021/nl403469r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb P.L., Lahens N.F., Correa-Garhwal S.M., Nicholson D.N., Kim E.J., Hogenesch J.B., Kuntner M., Higgins L., Hayashi C.Y., Agnarsson I., Voight B.F. The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat. Genet. 2017;49:895–903. doi: 10.1038/ng.3852. [DOI] [PubMed] [Google Scholar]
- Bacharach E., Mishra N., Briese T., Zody M.C., Kembou Tsofack J.E., Zamostiano R., Berkowitz A., Ng J., Nitido A., Corvelo A., Toussaint N.C., Abel Nielsen S.C., Hornig M., Del Pozo J., Bloom T., Ferguson H., Eldar A., Lipkin W.I. Characterization of a novel orthomyxo-like virus causing mass die-offs of tilapia. mBio. 2016;7:e00431–00416. doi: 10.1128/mBio.00431-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao C., Guo X., Qi X., Hu J., Zhou M., Varma J.K., Cui L., Yang H., Jiao Y., Klena J.D., Li L., Tao W., Li X., Chen Y., Zhu Z., Xu K., Shen A., Wu T., Peng H., Li Z., Shan J., Shi Z., Wang H. A family cluster of infections by a newly recognized bunyavirus in eastern China, 2007: further evidence of person-to-person transmission. Clin. Infect. Dis. 2011;53:1208–1214. doi: 10.1093/cid/cir732. [DOI] [PubMed] [Google Scholar]
- Barzon L., Lavezzo E., Militello V., Toppo S., Palù G. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 2011;12:7861–7884. doi: 10.3390/ijms12117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler C.F., García-Sastre A., Palese P. A novel paramyxovirus? Emerg. Infect. Dis. 2005;11:108–112. doi: 10.3201/eid1101.040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekal S., Domier L.L., Niblack T.L., Lambert K.N. Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J. Gen. Virol. 2011;92:1870–1879. doi: 10.1099/vir.0.030585-0. [DOI] [PubMed] [Google Scholar]
- Bell-Sakyi L., Attoui H. Virus discovery using tick cell lines. Evol. Bioinform. Online. 2016;12:31–34. doi: 10.4137/EBO.S39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger K.L., Kelly S.M., Jordan T.X., Tartell M.A., Randall G. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J. Virol. 2011;85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bexfield N., Kellam P. Metagenomics and the molecular identification of novel viruses. Vet. J. Lond. Engl. 2011;1997(190):191–198. doi: 10.1016/j.tvjl.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A.S., Freeman S.S., Herrera A.F., Pedamallu C.S., Gevers D., Duke F., Jung J., Michaud M., Walker B.J., Young S., Earl A.M., Kostic A.D., Ojesina A.I., Hasserjian R., Ballen K.K., Chen Y.-B., Hobbs G., Antin J.H., Soiffer R.J., Baden L.R., Garrett W.S., Hornick J.L., Marty F.M., Meyerson M. Sequence-based discovery of bradyrhizobium enterica in cord colitis syndrome. N. Engl. J. Med. 2013;369:517–528. doi: 10.1056/NEJMoa1211115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham A.M., Cone M., Mock V., Heberlein-Larson L., Stanek D., Blackmore C., Likos A. Comparison of test results for zika virus RNA in urine, serum, and saliva specimens from persons with travel-associated zika virus disease – Florida, 2016. MMWR Morb. Mortal. Wkly. Rep. 2016;65:475–478. doi: 10.15585/mmwr.mm6518e2. [DOI] [PubMed] [Google Scholar]
- Blomström A.-L., Widén F., Hammer A.-S., Belák S., Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J. Clin. Microbiol. 2010;48:4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc B., Shaughnessy D.P., Wolf Y.I., Koonin E.V., Roberto F.F., Young M. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated yellowstone hot springs. J. Virol. 2012;86:5562–5573. doi: 10.1128/JVI.07196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borawski J., Troke P., Puyang X., Gibaja V., Zhao S., Mickanin C., Leighton-Davies J., Wilson C.J., Myer V., Cornellataracido I., Baryza J., Tallarico J., Joberty G., Bantscheff M., Schirle M., Bouwmeester T., Mathy J.E., Lin K., Compton T., Labow M., Wiedmann B., Gaither L.A. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 2009;83:10058–10074. doi: 10.1128/JVI.02418-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros Á., Pankovics P., Simmonds P., Pollák E., Mátics R., Phan T.G., Delwart E., Reuter G. Genome analysis of a novel, highly divergent picornavirus from common kestrel (Falco tinnunculus): the first non-enteroviral picornavirus with type-I-like IRES. Infect. Genet. Evol. 2015;32:425–431. doi: 10.1016/j.meegid.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Briese T., Paweska J.T., McMullan L.K., Hutchison S.K., Street C., Palacios G., Khristova M.L., Weyer J., Swanepoel R., Egholm M., Nichol S.T., Lipkin W.I. Genetic detection and characterization of lujo virus, a new hemorrhagic fever–associated arenavirus from southern africa. PLoS Pathog. 2009;5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson A.L., Hwang Y., Sherrill-Mix S., Wu G.D., Lewis J.D., Black L., Clark T.A., Bushman F.D. Covalent modification of bacteriophage T4 DNA inhibits CRISPR-Cas9. mBio. 2015;6:e00648–15. doi: 10.1128/mBio.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J.N., Liachko I., Dunham M.J., Shendure J. Species-level deconvolution of metagenome assemblies with Hi-C-based contact probability maps. G3 Bethesda Md. 2014;4:1339–1346. doi: 10.1534/g3.114.011825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canuti M., van der Hoek L. Virus discovery: are we scientists or genome collectors? Trends Microbiol. 2014;22:229–231. doi: 10.1016/j.tim.2014.02.004. Special Issue: Omics: Fulfilling the Promise. [DOI] [PubMed] [Google Scholar]
- Capobianchi M.R., Giombini E., Rozera G. Next-generation sequencing technology in clinical virology. Clin. Microbiol. Infect. 2013;19:15–22. doi: 10.1111/1469-0691.12056. [DOI] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., Dal Cin P., Dye J.M., Whelan S.P., Chandran K., Brummelkamp T.R. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen A.T., Zahid O.K., Ruzicka J.A., Taylor E.W., Hall A.R. Selective detection and quantification of modified DNA with solid-state nanopores. Nano Lett. 2014;14:5488–5492. doi: 10.1021/nl501340d. [DOI] [PubMed] [Google Scholar]
- Chen Y.-H., Du W., Hagemeijer M.C., Takvorian P.M., Pau C., Cali A., Brantner C.A., Stempinski E.S., Connelly P.S., Ma H.-C., Jiang P., Wimmer E., Altan-Bonnet G., Altan-Bonnet N. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Franz C.J., Jiang H., Jiang Y., Wang D. An evolutionarily conserved transcriptional response to viral infection in Caenorhabditis nematodes. BMC Genomics. 2017;18:303. doi: 10.1186/s12864-017-3689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.-E.T., Winget D.M., White R.A., Hallam S.J., Suttle C.A. Combining genomic sequencing methods to explore viral diversity and reveal potential virus-host interactions. Front. Microbiol. 2015;6:265. doi: 10.3389/fmicb.2015.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T.H., Garb J.E., Hayashi C.Y., Arensburger P., Ayoub N.A. Spider transcriptomes identify ancient large-scale gene duplication event potentially important in silk gland evolution. Genome Biol. Evol. 2015;7:1856–1870. doi: 10.1093/gbe/evv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobo F. Application of MALDI-TOF mass spectrometry in clinical virology: a review. Open Virol. J. 2013;7:84–90. doi: 10.2174/1874357920130927003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller K.E., Berg M.G., Frankel M., Forberg K., Surani R., Chiu C.Y., Hackett J., Dawson G.J. Antibodies to the novel human pegivirus 2 are associated with active and resolved infections. J. Clin. Microbiol. 2016;54:2023–2030. doi: 10.1128/JCM.00515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição-Neto N., Zeller M., Lefrère H., Bruyn P.D., Beller L., Deboutte W., Yinda C.K., Lavigne R., Maes P., Ranst M.V., Heylen E., Matthijnssens J. Modular approach to customise sample preparation procedures for viral metagenomics: a reproducible protocol for virome analysis. Sci. Rep. 2015;5:srep16532. doi: 10.1038/srep16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Foster D.L., Conlan S., Holmes E.C., Palacios G., Evans J.D., Moran N.A., Quan P.-L., Briese T., Hornig M., Geiser D.M., Martinson V., vanEngelsdorp D., Kalkstein A.L., Drysdale A., Hui J., Zhai J., Cui L., Hutchison S.K., Simons J.F., Egholm M., Pettis J.S., Lipkin W.I. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- Culley A.I., Lang A.S., Suttle C.A. High diversity of unknown picorna-like viruses in the sea. Nature. 2003;424:1054–1057. doi: 10.1038/nature01886. [DOI] [PubMed] [Google Scholar]
- Culley A.I., Lang A.S., Suttle C.A. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Dacheux L., Cervantes-Gonzalez M., Guigon G., Thiberge J.-M., Vandenbogaert M., Maufrais C., Caro V., Bourhy H. A preliminary study of viral metagenomics of French bat species in contact with humans: identification of new mammalian viruses. PLoS One. 2014;9:e87194. doi: 10.1371/journal.pone.0087194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debat H.J. An RNA virome associated to the golden orb-Weaver spider Nephila clavipes. bioRxiv. 2017:140814. doi: 10.3389/fmicb.2017.02097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart E.L. Viral metagenomics. Rev. Med. Virol. 2007;17:115–131. doi: 10.1002/rmv.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemer G.S., Stedman K.M. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol. Direct. 2012;7:13. doi: 10.1186/1745-6150-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A., Kuzmickas R., Anderson N.G., Spiro D.J. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One. 2009;4:e7264. doi: 10.1371/journal.pone.0007264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T., Wilson M.R., Crawford E.D., Chow E.D., Khan L.M., Knopp K.A., O’Donovan B.D., Xia D., Hacker J.K., Stewart J.M., Gonzales J.A., Acharya N.R., DeRisi J.L. Illuminating uveitis: metagenomic deep sequencing identifies common and rare pathogens. Genome Med. 2016;8:90. doi: 10.1186/s13073-016-0344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson E.F., Haskew A.N., Gates J.E., Huynh J., Moore C.J., Frieman M.B. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 2010;84:13004–13018. doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez M.F., Guérin J.-L. Identification of a novel coronavirus from guinea fowl using metagenomics. Methods Mol. Biol. Clifton NJ. 2015;1282:27–31. doi: 10.1007/978-1-4939-2438-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R.A., Rohwer F. Viral metagenomics. Nat. Rev. Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Kafatos F.C., Maniatis T. The primary structure of rabbit beta-globin mRNA as determined from cloned DNA. Cell. 1977;10:571–585. doi: 10.1016/0092-8674(77)90090-3. [DOI] [PubMed] [Google Scholar]
- Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R., Neill F.H., Blutt S.E., Zeng X.-L., Qu L., Kou B., Opekun A.R., Burrin D., Graham D.Y., Ramani S., Atmar R.L., Estes M.K. Replication of human noroviruses in stem cell–derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M.-A., Ashe A., Piffaretti J., Wu G., Nuez I., Bélicard T., Jiang Y., Zhao G., Franz C.J., Goldstein L.D., Sanroman M., Miska E.A., Wang D. Natural and experimental infection of caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawaz M., Vijayakumar P., Mishra A., Gandhale P.N., Dutta R., Kamble N.M., Sudhakar S.B., Roychoudhary P., Kumar H., Kulkarni D.D., Raut A.A. Duck gut viral metagenome analysis captures snapshot of viral diversity. Gut Pathog. 2016;8:30. doi: 10.1186/s13099-016-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Hensley L., McKnight K.L., Hu F., Madden V., Ping L., Jeong S.-H., Walker C., Lanford R.E., Lemon S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S.R., Allred A.F., Tarr P.I., Klein E.J., Kirkwood C.D., Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S.R., Kirkwood C.D., Wang D. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol. J. 2008;5:117. doi: 10.1186/1743-422X-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S.R., Zeng X.-L., Utama B., Atmar R.L., Shroyer N.F., Estes M.K. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio. 2012;3:e00159–12. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K., Wang R., Vallat B. Naming diseases: first do no harm. Science. 2015;348 doi: 10.1126/science.348.6235.643. (643-643) [DOI] [PubMed] [Google Scholar]
- Gancz A.Y., Kistler A.L., Greninger A.L., Farnoushi Y., Mechani S., Perl S., Berkowitz A., Perez N., Clubb S., DeRisi J.L., Ganem D., Lublin A. Experimental induction of proventricular dilatation disease in cockatiels (Nymphicus hollandicus) inoculated with brain homogenates containing avian bornavirus 4. Virol. J. 2009;6:100. doi: 10.1186/1743-422X-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li Y., Yang X., Zhang H., Zhou P., Zhang Y., Shi Z. Metagenomic analysis of viruses from the bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 2012;JVI doi: 10.1128/JVI.06671-11. (06671-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J., Varley K.E., Davis N.S., Baas B.J., Goryshin I.Y., Vaidyanathan R., Kuersten S., Myers R.M. Transposase mediated construction of RNA-seq libraries. Genome Res. 2012;22:134–141. doi: 10.1101/gr.127373.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giallonardo F.D., Schlub T.E., Shi M., Holmes E.C. Dinucleotide composition in animal RNA viruses is shaped more by virus family than by host species. J. Virol. 2017;91:e02381–16. doi: 10.1128/JVI.02381-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale N.S., Horner S.M. RNA modifications go viral. PLoS Pathog. 2017;13:e1006188. doi: 10.1371/journal.ppat.1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-van Horn S.R., Sarnow P. Making the mark: the role of adenosine modifications in the life cycle of RNA viruses. Cell Host Microbe. 2017;21:661–669. doi: 10.1016/j.chom.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooskens J., Zevenhoven-Dobbe J.C., Claas E.C., Kroes A.C.M., Posthuma C.C. Mass spectrometry-based comparative sequence analysis for the genetic monitoring of influenza A(H1N1)pdm09 virus. PLoS One. 2014;9:e92970. doi: 10.1371/journal.pone.0092970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasse W., Spring O. ssRNA viruses from biotrophic Oomycetes form a new phylogenetic group between Nodaviridae and Tombusviridae. Arch. Virol. 2017;162:1319–1324. doi: 10.1007/s00705-017-3243-2. [DOI] [PubMed] [Google Scholar]
- Greene T.T., Tokuyama M., Knudsen G.M., Kunz M., Lin J., Greninger A.L., DeFilippis V.R., DeRisi J.L., Raulet D.H., Coscoy L. A Herpesviral induction of RAE-1 NKG2D ligand expression occurs through release of HDAC mediated repression. eLife. 2016:5. doi: 10.7554/eLife.14749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., DeRisi J.L. Draft Genome Sequences of Ciliovirus and Brinovirus from San Francisco Wastewater. Genome Announc. 2015:3. doi: 10.1128/genomeA.00651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., DeRisi J.L. Draft genomes of Leviviridae RNA phages EC and MB recovered from San Francisco wastewater. Genome Announc. 2015:3. doi: 10.1128/genomeA.00652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., DeRisi J.L. Draft Genome Sequence of Picalivirus D Recovered from San Francisco Wastewater. Genome Announc. 2015:3. doi: 10.1128/genomeA.00654-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., DeRisi J.L. Draft genome of tombunodavirus UC1. Genome Announc. 2015:3. doi: 10.1128/genomeA.00655-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., DeRisi J.L. Draft genome sequence of laverivirus UC1, a dicistrovirus-like RNA virus featuring an unusual genome organization. Genome Announc. 2015:3. doi: 10.1128/genomeA.00656-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., DeRisi J.L. Draft Genome Sequences of Marine RNA Viruses SF-1, SF-2, and SF-3 Recovered from San Francisco Wastewater. Genome Announc. 2015:3. doi: 10.1128/genomeA.00653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Jerome K.R. Draft genome sequence of goose dicistrovirus. Genome Announc. 2016:4. doi: 10.1128/genomeA.00068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Runckel C., Chiu C.Y., Haggerty T., Parsonnet J., Ganem D., DeRisi J.L. The complete genome of klassevirus – a novel picornavirus in pediatric stool. Virol. J. 2009;6:82. doi: 10.1186/1743-422X-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Chen E.C., Sittler T., Scheinerman A., Roubinian N., Yu G., Kim E., Pillai D.R., Guyard C., Mazzulli T., Isa P., Arias C.F., Hackett J., Jr., Schochetman G., Miller S., Tang P., Chiu C.Y. A metagenomic analysis of pandemic influenza a (2009 H1N1) infection in patients from north america. PLoS One. 2010;5:e13381. doi: 10.1371/journal.pone.0013381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Knudsen G.M., Betegon M., Burlingame A.L., Derisi J.L. The 3A protein from multiple picornaviruses utilizes the golgi adaptor protein ACBD3 to recruit PI4KIIIβ. J. Virol. 2012;86:3605–3616. doi: 10.1128/JVI.06778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Knudsen G.M., Betegon M., Burlingame A.L., DeRisi J.L. ACBD3 interaction with TBC1 domain 22 protein is differentially affected by enteroviral and kobuviral 3A protein binding. mBio. 2013;4:e00098–00013. doi: 10.1128/mBio.00098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Messacar K., Dunnebacke T., Naccache S.N., Federman S., Bouquet J., Mirsky D., Nomura Y., Yagi S., Glaser C., Vollmer M., Press C.A., Kleinschmidt-DeMasters B.K., Klenschmidt-DeMasters B.K., Dominguez S.R., Chiu C.Y. Clinical metagenomic identification of Balamuthia mandrillaris encephalitis and assembly of the draft genome: the continuing case for reference genome sequencing. Genome Med. 2015;7:113. doi: 10.1186/s13073-015-0235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Naccache S.N., Federman S., Yu G., Mbala P., Bres V., Stryke D., Bouquet J., Somasekar S., Linnen J.M., Dodd R., Mulembakani P., Schneider B.S., Muyembe-Tamfum J.-J., Stramer S.L., Chiu C.Y. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Naccache S.N., Messacar K., Clayton A., Yu G., Somasekar S., Federman S., Stryke D., Anderson C., Yagi S., Messenger S., Wadford D., Xia D., Watt J.P., Van Haren K., Dominguez S.R., Glaser C., Aldrovandi G., Chiu C.Y. A novel outbreak enterovirus D68 strain associated with acute flaccid myelitis cases in the USA (2012-14): a retrospective cohort study. Lancet Infect. Dis. 2015;15:671–682. doi: 10.1016/S1473-3099(15)70093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Pepper G., Shean R.C., Cent A., Palileo I., Kuypers J.M., Schiffer J.T., Jerome K.R. Myeloablation-associated deletion of ORF4 in a human coronavirus 229E infection. Npj Genomic Med. 2017;2:30. doi: 10.1038/s41525-017-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Waghmare A., Adler A., Qin X., Crowley J.L., Englund J.A., Kuypers J.M., Jerome K.R., Zerr D.M. Rule-out outbreak: 24-hour metagenomic next-generation sequencing for characterizing respiratory virus source for infection prevention. J. Pediatr. Infect. Dis. Soc. 2017;6:168–172. doi: 10.1093/jpids/pix019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L., Zerr D.M., Qin X., Adler A.L., Sampoleo R., Kuypers J.M., Englund J.A., Jerome K.R. Rapid metagenomic next-generation sequencing during an investigation of hospital-acquired human parainfluenza virus 3 infections. J. Clin. Microbiol. 2017;55:177–182. doi: 10.1128/JCM.01881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A.L. Picornavirus–host interactions to construct viral secretory membranes. Prog. Mol. Biol. Transl. Sci. 2015;129:189–212. doi: 10.1016/bs.pmbts.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Crawford E.D., O’Donovan B.D., Wilson M.R., Chow E.D., Retallack H., DeRisi J.L. Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17:41. doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.J., Wang J., Todd A.K., Bissielo A.B., Yen S., Strydom H., Moore N.E., Ren X., Huang Q.S., Carter P.E., Peacey M. Evaluation of rapid and simple techniques for the enrichment of viruses prior to metagenomic virus discovery. J. Virol. Methods. 2014;195:194–204. doi: 10.1016/j.jviromet.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Li Z., Yang F., Zheng J., Feng Y., Guo H., Li Y., Wang Y., Su N., Zhang F., Fan Q., Tu C. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel Mammalian viruses. PLoS One. 2013;8:e61950. doi: 10.1371/journal.pone.0061950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Huang X., Zhang F., Tan W., Matthijnssens J., Qin S., Xu L., Zhao Z., Yang L., Wang Q., Hu T., Bao X., Wu J., Tu C. Group a rotaviruses in chinese bats: genetic composition, serology, and evidence for bat-to-human transmission and reassortment. J. Virol. 2017:91. doi: 10.1128/JVI.02493-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B., Scheuch M., Höper D., Jungblut R., Holsteg M., Schirrmeier H., Eschbaumer M., Goller K.V., Wernike K., Fischer M., Breithaupt A., Mettenleiter T.C., Beer M. Novel orthobunyavirus in cattle, Europe, 2011. Emerg. Infect. Dis. 2012;18:469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B., Tappe D., Höper D., Herden C., Boldt A., Mawrin C., Niederstraßer O., Müller T., Jenckel M., van der Grinten E., Lutter C., Abendroth B., Teifke J.P., Cadar D., Schmidt-Chanasit J., Ulrich R.G., Beer M. A variegated squirrel bornavirus associated with fatal human encephalitis. N. Engl. J. Med. 2015;373:154–162. doi: 10.1056/NEJMoa1415627. [DOI] [PubMed] [Google Scholar]
- Holtz L.R., Finkbeiner S.R., Kirkwood C.D., Wang D. Identification of a novel picornavirus related to cosaviruses in a child with acute diarrhea. Virol. J. 2008;5:159. doi: 10.1186/1743-422X-5-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz L.R., Finkbeiner S.R., Zhao G., Kirkwood C.D., Girones R., Pipas J.M., Wang D. Klassevirus 1, a previously undescribed member of the family Picornaviridae, is globally widespread. Virol. J. 2009;6:86. doi: 10.1186/1743-422X-6-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes P.J., Stanway G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 2000;81:201–207. doi: 10.1099/0022-1317-81-1-201. [DOI] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses (ICTV) [WWW Document], n.d. Int. Comm. Taxon. Viruses ICTV. URL https://talk.ictvonline.org/information/w/ictv-information/383/ictv-code (Accessed 7.1.17a).
- International Committee on Taxonomy of Viruses (ICTV) [WWW Document], n.d. Int. Comm. Taxon. Viruses ICTV. URL https://talk.ictvonline.org/taxonomy/p/taxonomy_releases (Accessed 10.13.17b).
- Jäger S., Cimermancic P., Gulbahce N., Johnson J.R., McGovern K.E., Clarke S.C., Shales M., Mercenne G., Pache L., Li K., Hernandez H., Jang G.M., Roth S.L., Akiva E., Marlett J., Stephens M., D’Orso I., Fernandes J., Fahey M., Mahon C., O’Donoghue A.J., Todorovic A., Morris J.H., Maltby D.A., Alber T., Cagney G., Bushman F.D., Young J.A., Chanda S.K., Sundquist W.I., Kortemme T., Hernandez R.D., Craik C.S., Burlingame A., Sali A., Frankel A.D., Krogan N.J. Global landscape of HIV-human protein complexes. Nature. 2011;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan A.T. Outer membrane vesicles (OMVs) of gram-negative bacteria: a perspective update. Front. Microbiol. 2017;8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski A.B., Bauer I.K., Holtz L.R., Wang D. Propagation of astrovirus VA1, a neurotropic human astrovirus, in cell culture. J. Virol. 2017 doi: 10.1128/JVI.00740-17. https://www.ncbi.nlm.nih.gov/pubmed/28701405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Chen K., Sandoval L.E., Leung C., Wang D. An evolutionarily conserved pathway essential for orsay virus infection of caenorhabditis elegans. mBio. 2017:8. doi: 10.1128/mBio.00940-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Victoria J., Simmonds P., Slikas E., Chieochansin T., Naeem A., Shaukat S., Sharif S., Alam M.M., Angez M., Wang C., Shafer R.W., Zaidi S., Delwart E. A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc. Natl. Acad. Sci. 2008;105:20482–20487. doi: 10.1073/pnas.0807979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Li L., Victoria J., Oderinde B., Mason C., Pandey P., Zaidi S.Z., Delwart E. Multiple novel astrovirus species in human stool. J. Gen. Virol. 2009;90:2965–2972. doi: 10.1099/vir.0.014449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Slikas E., Simmonds P., Chieochansin T., Naeem A., Shaukat S., Masroor Alam M., Sharif S., Angez M., Zaidi S., Delwart E. A newly identified bocavirus species in human stool. J. Infect. Dis. 2009;199:196–200. doi: 10.1086/595831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Lipkin W.I., Zaidi S., Delwart E. Use of nucleotide composition analysis to infer hosts for three novel picorna-like viruses. J. Virol. 2010;84:10322–10328. doi: 10.1128/JVI.00601-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamendin K., Kydyrmanov A., Seidalina A., Asanova S., Sayatov M., Kasymbekov E., Khan E., Daulbayeva K., Harrison S.M., Carr I.M., Goodman S.J., Zhumatov K. Complete genome sequence of a novel avian paramyxovirus (APMV-13) isolated from a wild bird in Kazakhstan. Genome Announc. 2016;4:e00167–16. doi: 10.1128/genomeA.00167-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemenesi G., Dallos B., Görföl T., Boldogh S., Estók P., Kurucz K., Kutas A., Földes F., Oldal M., Németh V., Martella V., Bányai K., Jakab F. Molecular survey of RNA viruses in Hungarian bats: discovering novel astroviruses, coronaviruses, and caliciviruses. Vector Borne Zoonotic Dis. Larchmt. 2014;14:846–855. doi: 10.1089/vbz.2014.1637. [DOI] [PubMed] [Google Scholar]
- Kennedy E.M., Bogerd H.P., Kornepati A.V.R., Kang D., Ghoshal D., Marshall J.B., Poling B.C., Tsai K., Gokhale N.S., Horner S.M., Cullen B.R. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19:675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E.M., Courtney D.G., Tsai K., Cullen B.R. Viral epitranscriptomics. J. Virol. 2017:91. doi: 10.1128/JVI.02263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler A.L., Gancz A., Clubb S., Skewes-Cox P., Fischer K., Sorber K., Chiu C.Y., Lublin A., Mechani S., Farnoushi Y., Greninger A., Wen C.C., Karlene S.B., Ganem D., DeRisi J.L. Recovery of divergent avian bornaviruses from cases of proventricular dilatation disease: identification of a candidate etiologic agent. Virol. J. 2008;5:88. doi: 10.1186/1743-422X-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner M., Hooper L.V., Duerkop B.A. Evaluation of methods to purify virus-like particles for metagenomic sequencing of intestinal viromes. BMC Genomics. 2015;16:7. doi: 10.1186/s12864-014-1207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl C., Brinkmann A., Dabrowski P.W., Radonić A., Nitsche A., Kurth A. Protocol for metagenomic virus detection in clinical specimens1. Emerg. Infect. Dis. 2015;21:48–57. doi: 10.3201/eid2101.140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol. Mol. Biol. Rev. MMBR. 2014;78:278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Senkevich T.G., Dolja V.V. The ancient Virus World and evolution of cells. Biol. Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Wolf Y.I., Nagasaki K., Dolja V.V. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V., Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479(60th Anniversary Issue):2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. Metagenomic sorcery and the expanding protein universe. Nat. Biotechnol. 2007;25:540–542. doi: 10.1038/nbt0507-540. [DOI] [PubMed] [Google Scholar]
- Kowalchuk G.A., Speksnijder A.G.C.L., Zhang K., Goodman R.M., Veen van J.A. Finding the needles in the metagenome haystack. Microb. Ecol. . 2007;53:475–485. doi: 10.1007/s00248-006-9201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S.R., Wang D. Origins and challenges of viral dark matter. Virus Res. 2017;239:136–142. doi: 10.1016/j.virusres.2017.02.002. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S.R., Janowski A.B., Zhao G., Barouch D., Wang D. Hyperexpansion of RNA bacteriophage diversity. PLoS Biol. 2016;14:e1002409. doi: 10.1371/journal.pbio.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn M.J., Kesty N.C. Bacterial outer membrane vesicles and the host–pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- Ladd Effio C., Oelmeier S.A., Hubbuch J. High-throughput characterization of virus-like particles by interlaced size-exclusion chromatography. Vaccine. 2016;34:1259–1267. doi: 10.1016/j.vaccine.2016.01.035. [DOI] [PubMed] [Google Scholar]
- Landry M.L., St George K. Laboratory diagnosis of zika virus infection. Arch. Pathol. Lab. Med. 2017;141:60–67. doi: 10.5858/arpa.2016-0406-SA. [DOI] [PubMed] [Google Scholar]
- Lax S., Gilbert J.A. Hospital-associated microbiota and implications for nosocomial infections. Trends Mol. Med. 2015;21:427–432. doi: 10.1016/j.molmed.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Lecuit M., Eloit M. The diagnosis of infectious diseases by whole genome next generation sequencing: a new era is opening. Front. Cell. Infect. Microbiol. 2014:4. doi: 10.3389/fcimb.2014.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Victoria J., Kapoor A., Blinkova O., Wang C., Babrzadeh F., Mason C.J., Pandey P., Triki H., Bahri O., Oderinde B.S., Baba M.M., Bukbuk D.N., Besser J.M., Bartkus J.M., Delwart E.L. A novel picornavirus associated with gastroenteritis. J. Virol. 2009;83:12002–12006. doi: 10.1128/JVI.01241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Victoria J.G., Wang C., Jones M., Fellers G.M., Kunz T.H., Delwart E. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 2010;84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Pesavento P.A., Shan T., Leutenegger C.M., Wang C., Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 2011;92:2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.-X., Shi, M., Tian, J.-H., Lin, X.-D., Kang, Y.-J., Chen, L.-J., Qin, X.-C., Xu, J., Holmes, E.C., Zhang, Y.-Z., n.d. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife 4. 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed]
- Longo D.L., Drazen J.M. Data sharing. N. Engl. J. Med. 2016;374:276–277. doi: 10.1056/NEJMe1516564. [DOI] [PubMed] [Google Scholar]
- Lysholm F., Wetterbom A., Lindau C., Darban H., Bjerkner A., Fahlander K., Lindberg A.M., Persson B., Allander T., Andersson B. Characterization of the viral microbiome in patients with severe lower respiratory tract infections, using metagenomic sequencing. PLoS One. 2012;7:e30875. doi: 10.1371/journal.pone.0030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhsous N., Shean R., Droppers D., Guan J., Jerome K.R., Greninger A. Genome Announc. Press; 2017. Genome Sequences of Three Novel Bunyaviruses, Two Novel Rhabdoviruses, and One Novel Nyamivirus from Washington State Moths. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau C.D., Puschnik A.S., Majzoub K., Ooi Y.S., Brewer S.M., Fuchs G., Swaminathan K., Mata M.A., Elias J.E., Sarnow P., Carette J.E. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C.B., Andersen K.G., Winnicki S., Busby M., Gladden A.D., Tewhey R., Stremlau M., Berlin A., Gire S.K., England E., Moses L.M., Mikkelsen T.S., Odia I., Ehiane P.E., Folarin O., Goba A., Kahn S.H., Grant D.S., Honko A., Hensley L., Happi C., Garry R.F., Malboeuf C.M., Birren B.W., Gnirke A., Levin J.Z., Sabeti P.C. Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol. 2014;15:519. doi: 10.1186/s13059-014-0519-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight K.L., Xie L., González-López O., Rivera-Serrano E.E., Chen X., Lemon S.M. Protein composition of the hepatitis A virus quasi-envelope. Proc. Natl. Acad. Sci. U. S. A. 2017;114:6587–6592. doi: 10.1073/pnas.1619519114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan L.K., Folk S.M., Kelly A.J., MacNeil A., Goldsmith C.S., Metcalfe M.G., Batten B.C., Albariño C.G., Zaki S.R., Rollin P.E., Nicholson W.L., Nichol S.T. A new phlebovirus associated with severe febrile illness in missouri. N. Engl. J. Med. 2012;367:834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- Medina G.N., Knudsen G.M., Greninger A.L., Kloc A., Díaz-San Segundo F., Rieder E., Grubman M.J., DeRisi J.L., de Los Santos T. Interaction between FMDV L(pro) and transcription factor ADNP is required for optimal viral replication. Virology. 2017;505:12–22. doi: 10.1016/j.virol.2017.02.010. [DOI] [PubMed] [Google Scholar]
- Meyer C.T., Bauer I.K., Antonio M., Adeyemi M., Saha D., Oundo J.O., Ochieng J.B., Omore R., Stine O.C., Wang D., Holtz L.R. Prevalence of classic, MLB-clade and VA-clade astroviruses in Kenya and the Gambia. Virol. J. 2015;12:78. doi: 10.1186/s12985-015-0299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokili J.L., Rohwer F., Dutilh B.E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno P.S., Wagner J., Mansfield C.S., Stevens M., Gilkerson J.R., Kirkwood C.D. Characterisation of the canine faecal virome in healthy dogs and dogs with acute diarrhoea using shotgun metagenomics. PLoS One. 2017;12:e0178433. doi: 10.1371/journal.pone.0178433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J., Mahony J., Ainsworth S., Nauta A., Sinderen van D. Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl. Environ. Microbiol. 2013;79:7547–7555. doi: 10.1128/AEM.02229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Third Party Annotation [WWW Document], n.d. URL https://www.ncbi.nlm.nih.gov/genbank/tpa/.
- Naccache S.N., Greninger A.L., Lee D., Coffey L.L., Phan T., Rein-Weston A., Aronsohn A., Hackett J., Delwart E.L., Chiu C.Y. The perils of pathogen discovery: origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J. Virol. 2013;87:11966–11977. doi: 10.1128/JVI.02323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache S.N., Federman S., Veeraraghavan N., Zaharia M., Lee D., Samayoa E., Bouquet J., Greninger A.L., Luk K.-C., Enge B., Wadford D.A., Messenger S.L., Genrich G.L., Pellegrino K., Grard G., Leroy E., Schneider B.S., Fair J.N., Martínez M.A., Isa P., Crump J.A., DeRisi J.L., Sittler T., Hackett J., Miller S., Chiu C.Y. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24:1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache S.N., Peggs K.S., Mattes F.M., Phadke R., Garson J.A., Grant P., Samayoa E., Federman S., Miller S., Lunn M.P., Gant V., Chiu C.Y. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-Generation sequencing. Clin. Infect. Dis. 2015:ciu912. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache S.N., Thézé J., Sardi S.I., Somasekar S., Greninger A.L., Bandeira A.C., Campos G.S., Tauro L.B., Faria N.R., Pybus O.G., Chiu C.Y. Distinct zika virus lineage in salvador, bahia, Brazil. Emerg. Infect. Dis. 2016:22. doi: 10.3201/eid2210.160663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M., Omatsu T., Aoki H., Kaku Y., Belsham G.J., Haga K., Naoi Y., Sano K., Umetsu M., Shiokawa M., Tsuchiaka S., Furuya T., Okazaki S., Katayama Y., Oba M., Shirai J., Katayama K., Mizutani T. Identification and complete genome analysis of a novel bovine picornavirus in Japan. Virus Res. 2015;210:205–212. doi: 10.1016/j.virusres.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012;10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoi Y., Kishimoto M., Masuda T., Ito M., Tsuchiaka S., Sano K., Yamasato H., Omatsu T., Aoki H., Furuya T., Katayama Y., Oba M., Okada T., Shirai J., Mizutani T., Nagai M. Characterization and phylogenetic analysis of a novel picornavirus from swine feces in Japan. Arch. Virol. 2016;161:1685–1690. doi: 10.1007/s00705-016-2834-7. [DOI] [PubMed] [Google Scholar]
- Ng T.F.F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., Oderinde B.S., Wommack K.E., Delwart E. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.F.F., Chen L.-F., Zhou Y., Shapiro B., Stiller M., Heintzman P.D., Varsani A., Kondov N.O., Wong W., Deng X., Andrews T.D., Moorman B.J., Meulendyk T., MacKay G., Gilbertson R.L., Delwart E. Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc. Natl. Acad. Sci. 2014;111:16842–16847. doi: 10.1073/pnas.1410429111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.F.F., Mesquita J.R., Nascimento M.S.J., Kondov N.O., Wong W., Reuter G., Knowles N.J., Vega E., Esona M.D., Deng X., Vinjé J., Delwart E. Feline fecal virome reveals novel and prevalent enteric viruses. Vet. Microbiol. 2014;171:102–111. doi: 10.1016/j.vetmic.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes M.R.T., Contreras-Gutierrez M.A., Guzman H., Martins L.C., Barbirato M.F., Savit C., Balta V., Uribe S., Vivero R., Suaza J.D., Oliveira H., Nunes Neto J.P., Carvalho V.L., da Silva S.P., Cardoso J.F., de Oliveira R.S., da Silva Lemos P., Wood T.G., Widen S.G., Vasconcelos P.F.C., Fish D., Vasilakis N., Tesh R.B. Genetic characterization, molecular epidemiology, and phylogenetic relationships of insect-specific viruses in the taxon Negevirus. Virology. 2017;504:152–167. doi: 10.1016/j.virol.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue A.J., Eroy-Reveles A.A., Knudsen G.M., Ingram J., Zhou M., Statnekov J.B., Greninger A.L., Hostetter D.R., Qu G., Maltby D.A., Anderson M.O., Derisi J.L., McKerrow J.H., Burlingame A.L., Craik C.S. Global identification of peptidase specificity by multiplex substrate profiling. Nat. Methods. 2012;9:1095–1100. doi: 10.1038/nmeth.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan J.D., Allworth A.M., Paterson D.L., Snow T.M., Boots R., Gleeson L.J., Gould A.R., Hyatt A.D., Bradfield J. Fatal encephalitis due to novel paramyxovirus transmitted from horses. Lancet Lond. Engl. 1997;349:93–95. doi: 10.1016/s0140-6736(96)06162-4. [DOI] [PubMed] [Google Scholar]
- Ogimi C., Greninger A.L., Waghmare A.A., Kuypers J.M., Shean R.C., Xie H., Leisenring W.M., Stevens-Ayers T.L., Jerome K.R., Englund J.A., Boeckh M. Prolonged shedding of human Coronavirus in hematopoietic cell transplant recipients: risk factors and viral genome evolution. J. Infect. Dis. 2017;216:203–209. doi: 10.1093/infdis/jix264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N.R., Stahl D.A., Lane D.J., Olsen G.J. The analysis of natural microbial populations by ribosomal RNA sequences. In: Marshall K.C., editor. Advances in Microbial Ecology, Advances in Microbial Ecology. Springer; US: 1986. pp. 1–55. [Google Scholar]
- Palacios G., Druce J., Du L., Tran T., Birch C., Briese T., Conlan S., Quan P.-L., Hui J., Marshall J., Simons J.F., Egholm M., Paddock C.D., Shieh W.-J., Goldsmith C.S., Zaki S.R., Catton M., Lipkin W.I. A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- Park D.J., Dudas G., Wohl S., Goba A., Whitmer S.L.M., Andersen K.G., Sealfon R.S., Ladner J.T., Kugelman J.R., Matranga C.B., Winnicki S.M., Qu J., Gire S.K., Gladden-Young A., Jalloh S., Nosamiefan D., Yozwiak N.L., Moses L.M., Jiang P.-P., Lin A.E., Schaffner S.F., Bird B., Towner J., Mamoh M., Gbakie M., Kanneh L., Kargbo D., Massally J.L.B., Kamara F.K., Konuwa E., Sellu J., Jalloh A.A., Mustapha I., Foday M., Yillah M., Erickson B.R., Sealy T., Blau D., Paddock C., Brault A., Amman B., Basile J., Bearden S., Belser J., Bergeron E., Campbell S., Chakrabarti A., Dodd K., Flint M., Gibbons A., Goodman C., Klena J., McMullan L., Morgan L., Russell B., Salzer J., Sanchez A., Wang D., Jungreis I., Tomkins-Tinch C., Kislyuk A., Lin M.F., Chapman S., MacInnis B., Matthews A., Bochicchio J., Hensley L.E., Kuhn J.H., Nusbaum C., Schieffelin J.S., Birren B.W., Forget M., Nichol S.T., Palacios G.F., Ndiaye D., Happi C., Gevao S.M., Vandi M.A., Kargbo B., Holmes E.C., Bedford T., Gnirke A., Ströher U., Rambaut A., Garry R.F., Sabeti P.C. Ebola virus epidemiology, transmission, and evolution during seven months in Sierra Leone. Cell. 2015;161:1516–1526. doi: 10.1016/j.cell.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peersen O.B. Picornaviral polymerase structure, function, and fidelity modulation. Virus Res. 2017;234:4–20. doi: 10.1016/j.virusres.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelet T., Curran J., Kolakofsky D. The P gene of bovine parainfluenza virus 3 expresses all three reading frames from a single mRNA editing site. EMBO J. 1991;10:443–448. doi: 10.1002/j.1460-2075.1991.tb07966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlejewski K., Popiel M., Laskus T., Nakamura S., Motooka D., Stokowy T., Lipowski D., Pollak A., Lechowicz U., Caraballo Cortés K., Stępień A., Radkowski M., Bukowska-Ośko I. Next-generation sequencing (NGS) in the identification of encephalitis-causing viruses: unexpected detection of human herpesvirus 1 while searching for RNA pathogens. J. Virol. Methods. 2015;226:1–6. doi: 10.1016/j.jviromet.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Phan T.G., Kapusinszky B., Wang C., Rose R.K., Lipton H.L., Delwart E.L. The fecal viral flora of wild rodents. PLoS Pathog. 2011;7:e1002218. doi: 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontkivska H., Frederick M., Miyamoto M.M., Wayne M.L. RNA editing by the host ADAR system affects the molecular evolution of the Zika virus. Ecol. Evol. 2017;7:4475–4485. doi: 10.1002/ece3.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RDHV-like virus SF1 replication-like protein and capsid protein genes, complete cds, 2015.
- Racaniello V. Moving beyond metagenomics to find the next pandemic virus. Proc. Natl. Acad. Sci. 2016;113:2812–2814. doi: 10.1073/pnas.1601512113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P., Greene D.N., Baird G.S., Hoofnagle A.N., Mathias P.C. Establishing evidence-based thresholds and laboratory practices to reduce inappropriate treatment of pseudohyperkalemia. Clin. Biochem. 2017;50(August (12)):663–669. doi: 10.1016/j.clinbiochem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Reuter G., Pankovics P., Knowles N.J., Boros Á. Two closely related novel picornaviruses in cattle and sheep in Hungary from 2008 to 2009, proposed as members of a new genus in the family Picornaviridae. J. Virol. 2012;86:13295–13302. doi: 10.1128/JVI.01142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Boros A., Delwart E., Pankovics P. Novel seadornavirus (family reoviridae) related to banna virus in europe. Arch. Virol. 2013;158:2163–2167. doi: 10.1007/s00705-013-1712-9. [DOI] [PubMed] [Google Scholar]
- Reuter G., Pankovics P., Delwart E., Boros Á. A novel posavirus-related single-stranded RNA virus from fish (Cyprinus carpio) Arch. Virol. 2015;160:565–568. doi: 10.1007/s00705-014-2304-z. [DOI] [PubMed] [Google Scholar]
- Roossinck M.J., Saha P., Wiley G.B., Quan J., White J.D., Lai H., Chavarría F., Shen G., Roe B.A. Ecogenomics: using massively parallel pyrosequencing to understand virus ecology. Mol. Ecol. 2010;19:81–88. doi: 10.1111/j.1365-294X.2009.04470.x. [DOI] [PubMed] [Google Scholar]
- Roossinck M.J. Plant virus metagenomics: biodiversity and ecology. Annu. Rev. Genet. 2012;46:359–369. doi: 10.1146/annurev-genet-110711-155600. [DOI] [PubMed] [Google Scholar]
- Rosario K., Breitbart M. Exploring the viral world through metagenomics. Curr. Opin. Virol. 2011;1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Rothberg J.M., Leamon J.H. The development and impact of 454 sequencing. Nat. Biotechnol. 2008;26:1117–1124. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- Runckel C., Flenniken M.L., Engel J.C., Ruby J.G., Ganem D., Andino R., DeRisi J.L. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One. 2011;6:e20656. doi: 10.1371/journal.pone.0020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salloum S., Wang H., Ferguson C., Parton R.G., Tai A.W. Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog. 2013;9:e1003513. doi: 10.1371/journal.ppat.1003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano K., Naoi Y., Kishimoto M., Masuda T., Tanabe H., Ito M., Niira K., Haga K., Asano K., Tsuchiaka S., Omatsu T., Furuya T., Katayama Y., Oba M., Ouchi Y., Yamasato H., Ishida M., Shirai J., Katayama K., Mizutani T., Nagai M. Identification of further diversity among posaviruses. Arch. Virol. 2016;161:3541–3548. doi: 10.1007/s00705-016-3048-8. [DOI] [PubMed] [Google Scholar]
- Sasaki J., Taniguchi K. Aichi virus 2A protein is involved in viral RNA replication. J. Virol. 2008;82:9765–9769. doi: 10.1128/JVI.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J., Ishikawa K., Arita M., Taniguchi K. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J. 2012;31:754–766. doi: 10.1038/emboj.2011.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Orba Y., Ueno K., Ishii A., Moonga L., Hang’ombe B.M., Mweene A.S., Ito K., Sawa H. Metagenomic analysis of the shrew enteric virome reveals novel viruses related to human stool-associated viruses. J. Gen. Virol. 2015;96:440–452. doi: 10.1099/vir.0.071209-0. [DOI] [PubMed] [Google Scholar]
- Schlaberg R., Chiu C.Y., Miller S., Procop G.W., Weinstock G., Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology, Microbiology Resource Committee of the College of American Pathologists Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch. Pathol. Lab. Med. 2017;141:776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- Schomacker H., Collins P.L., Schmidt A.C. In silico identification of a putative new paramyxovirus related to the Henipavirus genus. Virology. 2004;330:178–185. doi: 10.1016/j.virol.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Sharman M., Kehoe M., Coutts B., Leur J., van Filardo F., Thomas J. Two complete genome sequences of phasey bean mild yellows virus, a novel member of the luteoviridae from Australia. Genome Announc. 2016;4:15–e01569. doi: 10.1128/genomeA.01569-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shean R., Makhsous N., Crawford R., Jerome K.R., Greninger A. Genome Announc. Press; 2017. Draft Genome Sequences of Six Novel Picorna-Like Viruses from Washington State Spiders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Lin X.-D., Tian J.-H., Chen L.-J., Chen X., Li C.-X., Qin X.-C., Li J., Cao J.-P., Eden J.-S., Buchmann J., Wang W., Xu J., Holmes E.C., Zhang Y.-Z. Redefining the invertebrate RNA virosphere. Nature. 2016 doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- Sim G.K., Kafatos F.C., Jones C.W., Koehler M.D., Efstratiadis A., Maniatis T. Use of a cDNA library for studies on evolution and developmental expression of the chorion multigene families. Cell. 1979;18:1303–1316. doi: 10.1016/0092-8674(79)90241-1. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Adams M.J., Benkő M., Breitbart M., Brister J.R., Carstens E.B., Davison A.J., Delwart E., Gorbalenya A.E., Harrach B., Hull R., King A.M.Q., Koonin E.V., Krupovic M., Kuhn J.H., Lefkowitz E.J., Nibert M.L., Orton R., Roossinck M.J., Sabanadzovic S., Sullivan M.B., Suttle C.A., Tesh R.B., van der Vlugt R.A., Varsani A., Zerbini F.M. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- Soueidan H., Schmitt L.-A., Candresse T., Nikolski M. Finding and identifying the viral needle in the metagenomic haystack: trends and challenges. Front. Microbiol. 2015:5. doi: 10.3389/fmicb.2014.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks M.E., Gundersen-Rindal D.E., Harrison R.L. Complete genome sequence of a novel iflavirus from the transcriptome of halyomorpha halys, the brown marmorated stink bug. Genome Announc. 2013;1:e00910–13. doi: 10.1128/genomeA.00910-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staring J., von Castelmur E., Blomen V.A., van den Hengel L.G., Brockmann M., Baggen J., Thibaut H.J., Nieuwenhuis J., Janssen H., van Kuppeveld F.J.M., Perrakis A., Carette J.E., Brummelkamp T.R. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature. 2017;541:412–416. doi: 10.1038/nature21032. [DOI] [PubMed] [Google Scholar]
- Stenglein M.D., Sanders C., Kistler A.L., Ruby J.G., Franco J.Y., Reavill D.R., Dunker F., Derisi J.L. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio. 2012;3 doi: 10.1128/mBio.00180-12. (e00180-00112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein M.D., Jacobson E.R., Wozniak E.J., Wellehan J.F.X., Kincaid A., Gordon M., Porter B.F., Baumgartner W., Stahl S., Kelley K., Towner J.S., DeRisi J.L. Ball python nidovirus: a candidate etiologic agent for severe respiratory disease in python regius. mBio. 2014;5 doi: 10.1128/mBio.01484-14. (e01484-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward G.F., Culley A.I., Mueller J.A., Wood-Charlson E.M., Belcaid M., Poisson G. Are we missing half of the viruses in the ocean? ISME J. 2013;7:672–679. doi: 10.1038/ismej.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P., Chiu C. Metagenomics for the discovery of novel human viruses. Future Microbiol. 2010;5:177–189. doi: 10.2217/fmb.09.120. [DOI] [PubMed] [Google Scholar]
- Tang J.W., Lam T.T., Zaraket H., Lipkin W.I., Drews S.J., Hatchette T.F., Heraud J.-M., Koopmans M.P. INSPIRE investigators, 2017. Global epidemiology of non-influenza RNA respiratory viruses: data gaps and a growing need for surveillance. Lancet Infect. Dis. 2017;17(October (10)):e320–e326. doi: 10.1016/S1473-3099(17)30238-4. Epub 2017 Apr 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmam S., Davoust B., Berenger J.-M., Raoult D., Desnues C. Viral metagenomics on animals as a tool for the detection of zoonoses prior to human infection? Int. J. Mol. Sci. 2014;15:10377–10397. doi: 10.3390/ijms150610377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmam S., Monteil-Bouchard S., Robert C., Pascalis H., Michelle C., Jardot P., Charrel R., Raoult D., Desnues C. Host-Associated metagenomics: a guide to generating infectious RNA viromes. PLoS One. 2015;10:e0139810. doi: 10.1371/journal.pone.0139810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber R.V., Haynes M., Breitbart M., Wegley L., Rohwer F. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- Turner E.H., Ng S.B., Nickerson D.A., Shendure J. Methods for genomic partitioning. Annu. Rev. Genomics Hum. Genet. 2009;10:263–284. doi: 10.1146/annurev-genom-082908-150112. [DOI] [PubMed] [Google Scholar]
- Uccellini L., Ossiboff R.J., de Matos R.E., Morrisey J.K., Petrosov A., Navarrete-Macias I., Jain K., Hicks A.L., Buckles E.L., Tokarz R., McAloose D., Lipkin W.I. Identification of a novel nidovirus in an outbreak of fatal respiratory disease in ball pythons (Python regius) Virol. J. 2014;11:144. doi: 10.1186/1743-422X-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., Fouts D.E., Levy S., Knap A.H., Lomas M.W., Nealson K., White O., Peterson J., Hoffman J., Parsons R., Baden-Tillson H., Pfannkoch C., Rogers Y.-H., Smith H.O. Environmental genome shotgun sequencing of the sargasso sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Vidal S., Curran J., Kolakofsky D. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J. Virol. 1990;64:239–246. doi: 10.1128/jvi.64.1.239-246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C.L., Waldron F.M., Robertson S., Crowson D., Ferrari G., Quintana J.F., Brouqui J.-M., Bayne E.H., Longdon B., Buck A.H., Lazzaro B.P., Akorli J., Haddrill P.R., Obbard D.J. The discovery, distribution, and evolution of viruses associated with drosophila melanogaster. PLoS Biol. 2015;13:e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigele P.R., Müller S., Lee Y.-J., Walsh S., Guan C., Dai N., Correa I. Novel thymidine hypermodifications in viruses encoding a 5-hydroxymethyl-5′-deoxyuridine DNA kinase. FASEB J. 2017;31 (606.6-606.6) [Google Scholar]
- Willerth S.M., Pedro H.A.M., Pachter L., Humeau L.M., Arkin A.P., Schaffer D.V. Development of a low bias method for characterizing viral populations using next generation sequencing technology. PLoS One. 2010;5:e13564. doi: 10.1371/journal.pone.0013564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Chu C., Chan K., Tsoi H., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W., Poon L.L.M., Wong S.S.Y., Guan Y., Peiris J.S.M., Yuen K. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Choi G.K.Y., Huang Y., Teng J.L.L., Tsoi H.-W., Tse H., Yeung M.L., Chan K.-H., Jin D.-Y., Yuen K.-Y. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine picodicistrovirus, in the picornavirus-like superfamily. J. Virol. 2012;86:2797–2808. doi: 10.1128/JVI.05481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie K.M., Mihindukulasuriya K.A., Sodergren E., Weinstock G.M., Storch G.A. Sequence analysis of the human virome in febrile and afebrile children. PLoS One. 2012:7. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Nagy P.D. Enrichment of phosphatidylethanolamine in viral replication compartments via co-opting the endosomal rab5 small GTPase by a positive-Strand RNA virus. PLoS Biol. 2016;14:e2000128. doi: 10.1371/journal.pbio.2000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue K.S., Stevens-Ayers T., Campbell A.P., Englund J.A., Pergam S.A., Boeckh M., Bloom J.D. Parallel evolution of influenza across multiple spatiotemporal scales. eLife. 2017;6:e26875. doi: 10.7554/eLife.26875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C.K., Zell R., Deboutte W., Zeller M., Conceição-Neto N., Heylen E., Maes P., Knowles N.J., Ghogomu S.M., Van Ranst M., Matthijnssens J. Highly diverse population of Picornaviridae and other members of the Picornavirales, in Cameroonian fruit bats. BMC Genomics. 2017;18:249. doi: 10.1186/s12864-017-3632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.-J., Liang M.-F., Zhang S.-Y., Liu Y., Li J.-D., Sun Y.-L., Zhang L., Zhang Q.-F., Popov V.L., Li C., Qu J., Li Q., Zhang Y.-P., Hai R., Wu W., Wang Q., Zhan F.-X., Wang X.-J., Kan B., Wang S.-W., Wan K.-L., Jing H.-Q., Lu J.-X., Yin W.-W., Zhou H., Guan X.-H., Liu J.-F., Bi Z.-Q., Liu G.-H., Ren J., Wang H., Zhao Z., Song J.-D., He J.-R., Wan T., Zhang J.-S., Fu X.-P., Sun L.-N., Dong X.-P., Feng Z.-J., Yang W.-Z., Hong T., Zhang Y., Walker D.H., Wang Y., Li D.-X. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 2011;364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Greninger A.L., Isa P., Phan T.G., Martínez M.A., de la Luz, Sanchez M., Contreras J.F., Santos-Preciado J.I., Parsonnet J., Miller S., DeRisi J.L., Delwart E., Arias C.F., Chiu C.Y. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS One. 2012;7:e49449. doi: 10.1371/journal.pone.0049449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaaijer, S., Columbia University Ubiquitous Genomics 2015 class, Erlich, Y., 2016. Using mobile sequencers in an academic classroom. eLife 5. 10.7554/eLife.14258. [DOI] [PMC free article] [PubMed]
- Zhang T., Breitbart M., Lee W.H., Run J.-Q., Wei C.L., Soh S.W.L., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2005;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Li L., Deng X., Kapusinszky B., Pesavento P.A., Delwart E. Faecal virome of cats in an animal shelter. J. Gen. Virol. 2014;95:2553–2564. doi: 10.1099/vir.0.069674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- changepoint: An R Package for Changepoint Analysis | Killick | Journal of Statistical Software, n.d. 10.18637/jss.v058. i03.
- Zhou H., Zhu S., Quan R., Wang J., Wei L., Yang B., Xu F., Wang J., Chen F., Liu J. Identification and genome characterization of the first sicinivirus isolate from chickens in mainland China by using viral metagenomics. PLoS One. 2015;10:e0139668. doi: 10.1371/journal.pone.0139668. [DOI] [PMC free article] [PubMed] [Google Scholar]