Highlights
-
•
To gain insights into mechanisms of PEDV-pAPN interactions, the present study aimed at identifying the domain that is critical for PEDV binding.
-
•
Results showed PEDV infection was restricted to pAPN domain VII expressing NIH3T3 cells.
-
•
PEDV harvested from pAPN or domain VII expressing NIH3T3 cells was induced indirect plaques in Vero cells.
-
•
Our results demonstrate that PEDV recognizes pAPN and that the main interactive point is lodged within domain VII of the pAPN.
Keywords: Porcine epidemic diarrhea virus, Porcine APN, Pathogenesis, Binding and entry, Domain VII
Abstract
Porcine epidemic diarrhea virus (PEDV) infects swine intestinal cells causing enteric disease. Research has shown that the entry into these cells is through porcine aminopeptidase N (pAPN) receptor. To gain insights into mechanisms of PEDV-pAPN interactions, the present study aimed at identifying the domain that is critical for PEDV binding. To this end, NIH3T3 cell lines constitutively expressing pAPN or pAPN mutants were generated. The mutants were; domain VII deletion mutant and domains IV–VI deletion mutant. In the latter, domain VII was linked to the transmembrane segment through domain III. Results showed PEDV infection was restricted to pAPN and pAPN domain VII expressing NIH3T3 cells. Further, reducing PEDV titre 10 fold resulted in 37.8% decrease in foci indicating positive correlation. A time course test at 12, 24, 36, 48 and 60 h showed that foci increased 6 fold in the overall time range. Also, PEDV harvested from pAPN or domain VII expressing NIH3T3 cells was induced indirect plaques in Vero cells confirming successful entry and replication. Collectively, our results demonstrate that PEDV recognizes pAPN and that the main interactive point is lodged within domain VII of the pAPN. These findings are important for therapeutic development as well as creating a platform for future studies on PEDV.
1. Introduction
Coronaviruses (CoVs) are enveloped, positive-stranded RNA viruses potentially pathogenic in respiratory and intestinal tracts of humans, animals and birds (David and Kathryn, 2007, Susan and Navas-Martin, 2005). Analyses by serological cross-reactivity and genome phylogeny distinguish these viruses into four genera which include Alpha, Beta, Gamma and Delta-CoV (de Groot et al., 2011). Studies have speculated Alpha and Beta-CoVs to have originated from bats whereas Gamma and Delta-CoVs gene source is the birds (Woo et al., 2012). PEDV belongs to the genus Alpha-CoV (de Groot et al., 2011).
Host specificity, tissue tropism and virulence of CoVs are dictated by interactions between the spike glycoproteins of the viruses and cellular glycoproteins on surface receptors. Receptor binding and cell entry is essential steps for virus entry into host cell, which leads to internalization of the viruses (Park et al., 2014, Liu et al., 2015). An envelope-anchored spike protein mediates coronavirus entry into cells (Liu et al., 2015). The spike ectodomain consists of a receptor-binding subunit, S1, and a membrane fusion subunit, S2. Two domains in S1, calls S1-NTD and S1-CTD, can potentially function as receptor-binding domains (Li, 2012, Li, 2015) In some CoVs, entry mechanisms progress via sialic acid interactions with spike and/or hemagglutinin esterase glycoproteins (Haijema et al., 2003, Kuo et al., 2000, Phillips et al., 1999, Sanchez et al., 1999). The spike protein of Murine hepatitis virus (MHV) binds carcinoembryonic cell adhesion molecule 1a for cell penetration while human (HCoV-NL63) and severe acute respiratory syndrome (SARS)-CoV recognize human angiotensin-converting enzyme 2 (hACE-2) on host cells (Dveksler et al., 1991, Dveksler et al., 1993). Aminopeptidase N (APN) serves as receptor for feline (FCoV), canine (CCoV), Transmissible gastroenteritis virus (TGEV), PEDV as well as human (HCoV -229E) (Delmas et al., 1992, Tresnan et al., 1996, Yeager et al., 1992, Kolb et al., 1998, Li, 2015, Liu et al., 2015).
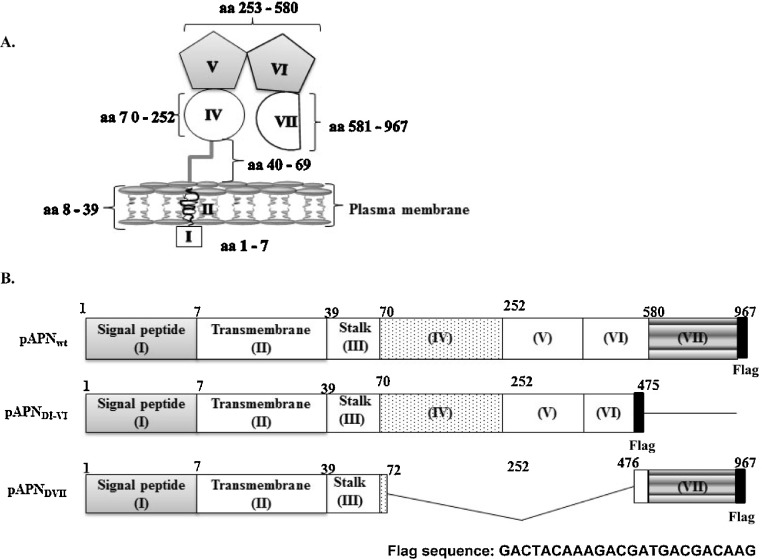
APN/CD13 is a 150 kDa, zinc dependent metalloprotease consisting of 967 amino acids (Rawlings and Barrett, 1995). It is ubiquitously present in diverse organs, tissues and cell types (Luan and Xu, 2007). It is expressed as a glycosylated homodimer on the surface of epithelial cells of liver, intestine, kidney and respiratory tract fibroblasts and leukocytes (Kenny and Maroux, 1982, Lendeckel et al., 2000, Nam and Lee, 2010). Additionally, APN is found on endothelial cells such as cerebral astrocytes as well as pericytes and synaptic membrane in the central nervous system (Kay et al., 1994, Soichiro et al., 1983, Mina-Osorio et al., 2008). APN cleaves the N-terminal amino acids from peptides. In addition, APN plays role as a receptor and signaling molecule (Mina-Osorio et al., 2008). A seven domain organization has been described for APN structure. The first domain constitutes a short cytoplasmic tail on the N-terminal end (amino acids [aa] 1–7). Domain II (aa 8–39) consists of the transmembrane segment and links to a stalk forming domain III (aa 40–70). Domain IV spans from aa 71 to 252. The peptidase activity is housed by domains V and VI (aa 253–580). Located on the C terminus is domain VII consisting of aa 581–967 (Sjostrom et al., 2000).
Previous investigations by virus overlay protein binding assay showed that PEDV bound pAPN in permissive cells and it was also shown that anti-pAPN blocked this binding (Oh et al., 2003). Further, expression of pAPN in non-permissive Madin-Darby canine kidney cells resulted in susceptibity of these cells to PEDV (Li et al., 2009). However, PEDV fails to infect native porcine APN expressing swine testis cells which are highly susceptible to TGEV (Hofmann and Wyler, 1988). In the current study, by constitutive expression of pAPN in non-susceptible NIH3T3 cells, we have sought to annotate the previous findings stipulating pAPN receptor function for PEDV. In addition, this study aimed at identification of using pAPN protein domain(s) that are critical for its binding and entry.
2. Materials and methods
2.1. Cell lines and virus
Vero cells were cultured in alpha minimum essential medium ([α-MEM] Gibco). Mouse embryonic fibroblasts (NIH3T3) and human embryonic kidney (HEK 293T) cells were cultured in Dulbecco's Modified Eagle Medium ([DMEM] Gibco). All growth media were supplemented with 10% fetal bovine serum ([FBS] Gibco). The PEDV strain used in this study was Korean isolate.
2.2. Porcine aminopeptidase (pAPN) and mutants plasmid constructs
The clones used in this study are illustrated (Fig. 1 B). Three clones were designed and denoted as follows; (1) wild type pAPN (pAPNwt), (2) domain VII deletion mutant of pAPN (pAPNDI-VI) and (3) domains IV–VI deletion pAPN mutant (pAPNDVII). These were amplified by PCR (Horton, 1993) using pfu polymerase (Solgent). Primers were designed using GenBank sequences (accession number HQ824547) and all antisense primer sequences were fused in frame to Flag epitope sequence. The wild type pAPN was amplified using sense 5′-CCC AAG CTT ACC ATG GCC AAG GGA TTC TAC-3′ and anti-sense 5′-CCC CTC GAG TCA CTT GTC GTC ATC GTC TTT GTA GTC GCT GTG CTC TAT GAA CCA-3′ primers. Mutants pAPNDI-VI PCR primers were sense 5′ - CCC AAG CTT ACC ATG GCC AAG GGA TTC TAC-3′ and anti-sense 5′ - CCC CTC GAG TCA CTT GTC GTC ATC GTC TTT GTA GTC GCT GTG CTC TAT GAA CCA-3′ while pAPNDVII was amplified using 5′ - CGG TAC CGC GCC TCG GTT ATC AGG ATG CTC-3′ and 5′- CCC CTC GAG TCA CTT GTC GTC ATC GTC TTT GTA GTC GCT GTG CTC TAT GAA CCA-3′ sense and anti-sense primers respectively. The cDNAs encoding pAPNwt and mutants were cloned into the Hind III and Xho I sites of pcDNA 3.0 (Invitrogen).
Fig. 1.
Schematic representation of pAPN domains. (A) Conformation of domains of pAPN on cell surface. Domain I, made up of 7 aa, constitutes a short cytoplasmic tail. Domain II is the transmembrane domain (aa 8–39). Protruding from the plasma membrane is domain III known as stalk (40–69). Domain IV spans from aa 70–252. The aminopeptidase activity is within domains V and VI (aa 253–580). From aa 581 to 967 is domain VII. (B) Three pAPN clones used in the study are illustrated. The domains are numbered I–VII with the amino acids positions indicated above (1–967). Solid lines show deleted domain(s). The wild type (pAPNwt) is the full length of pAPN. The middle one (pAPNDI-VI) was a domain VII deletion mutant and at the bottom, pAPNDVII, domains IV through VI were deleted.
2.3. Transient expression of pAPN
HEK 293T cells were transfected using plasmid constructs containing pAPN clone or mutants cDNA using Fugene HD (Roche) according to the manufacturer. At 24 h after transfection, cells were assayed for pAPN expression by immunoblotting using mouse anti-pAPN polysera and anti-Flag antibody (Santa-Cruz).
2.4. Flow cytometry
Extracellular expression of pAPN was determined by detection of Flag epitope. Cells were harvested using cells dissociation solution (Sigma). These were washed two times in 2 ml of 1 X phosphate buffered saline (PBS), centrifuged at 300 × g for 5 min and supernatants were discarded. Cells were resuspended in 100 μl of staining buffer (1 X PBS containing 2% FBS) and surface antigens were stained with anti-Flag antibodies. After 2 times wash with 2 ml of 1 X PBS and re-suspension in 100 μl staining buffer, cells were incubated with phycoerythrin (PE) conjugated secondary antibody (BD Biosciences) and analyzed by flow cytometry (FACS Calibur, Becton-Dickinson).
2.5. Stable cell lines
NIH3T3 cells were transfected with pcDNA3.0 with or without pAPNwt or mutants using Fugene HD (Roche) as suggested by the manufacturer and 24 h after transfection, cells were subjected to 400 μg/ml G418 selection pressure. The cell lines generated were designated as follows; NIH3T3 stably harboring only vector (NIH3T3pcDNA3.0), expressing pAPNwt (NIH3T3-pAPNwt), expressing pAPNDI-VI (NIH3T3-pAPNDI-VI) and pAPNDVII mutant (NIH3T3-pAPNDVII), respectively. After antibiotic selection, the cells were analyzed for expression by western blot.
2.6. Virus propagation and plaque assay
The KPEDV-9 was propagated in Vero cells using serum free α-MEM in the presence of 10 μg/ml trypsin (Sigma) as described (Oh et al., 2003). For the quantitation of virus, plaque assay was performed as described (Dulbecco, 1952). Briefly, Vero cells (2.5 × 105 cells/ml) were seeded in a 6-well plate 24 h prior to inoculation. Cells were washed with 1 ml of 1X PBS and 200 μl of inoculum was added. After 1 h adsorption, inoculum was removed and cells were overlaid with serum free α-MEM containing 0.9% methylcellulose and 10 μg/ml trypsin. These were incubated at 37 °C with 5% CO2 until plaques formed. The overlay medium was discarded and cells washed with 1 ml 1X PBS then fixed with 2 ml of fixation solution (5% methanol and 10% acetic acid) for 30 min at room temperature. Cells were washed with 1 ml of 1X PBS and incubated with 1 ml of 1% crystal violet for 20 min at room temperature. The dye was then discarded and cells washed were gently with distilled water then air-dried. A plaque count was conducted to deduce the number of plaque forming units (pfu) per ml of virus suspension. This virus culture technique was adapted for inoculation of NIH3T3 cells with PEDV at a multiplicity of infection (m.o.i.) of 0.06–0.6. The inocula were removed after 1 h adsorption and replaced with fresh medium. After various time periods after inoculation, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Cells were blocked with 4% skim milk for 30 min at room temperature and detection of viral antigens was performed using mouse anti-PEDV polysera for 1 h at 37 °C. After 3 times wash with 200 μl of 1 X PBS, cells were incubated with horseradish peroxidase (HRP) - conjugated anti-mouse IgG (Santa-Cruz) for 1 h, 37 °C. The cells were washed 3 times with 1 X PBS. Reaction was visualized by addition of 3,3′-diaminobenzidine chromogenic substrate (Vector Lab).
2.7. Indirect plaque assay
PEDV titer attained in NIH3T3 cell types was analyzed indirectly in Vero cells as previously described (Yu et al., 2010). NIH3T3 cell lines expressing or not expressing surface pAPNwt or mutants were seeded at a density of 2.5 × 105 per ml. At 24 h post inoculation (p.i.) with PEDV at m.o.i. 0.6, virus was harvested after by 3 cycles of freezing-thawing. The supernatants were re-inoculated into Vero cells along with PEDV control inoculation. Since dead or defective virus particle are incapable of forming plaques, virus viability in NIH3T3 cell lines was verified by plaque assay.
3. Results
3.1. Surface expression of pAPN
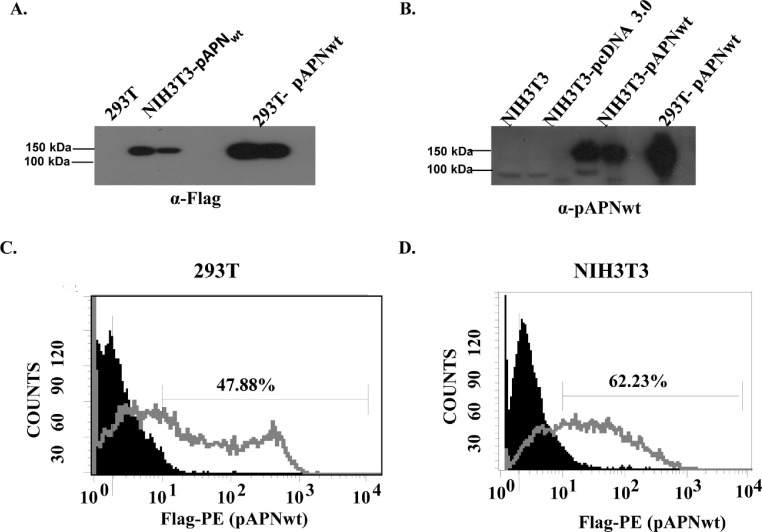
Western blot assays using anti-Flag antibody and mouse anti-pAPN polysera revealed pAPNwt was expressed at approximately 150 kDa in both 293T and NIH3T3 cells (Fig. 2 A and B). Additionally, surface expression was confirmed by flow cytometry using anti-Flag and P.E. labeling. The results confirmed 47.88% (293T) and 62.23% (NIH3T3) surface expression (Fig. 2C and D).
Fig. 2.
Expression of pAPN. (A) Western blot analysis showed pAPNwt fusion protein detected using anti-Flag antibody. Western blot analysis using anti-Flag antibody and mouse anti-pAPN polysera revealed pAPNwt expression bands at approximately 150 kDa. (B) Western blot analysis revealed pAPNwt expression band detected using anti-pAPNwt polysera. Approximately 150 kDa pAPNwt expression confirmed. (C) Flow cytometry confirmation of extracellular expression of pAPNwt transiently expressing 293T cells. (D) Confirmation of stable extracellular expression of pAPNwt in NIH3T3.
3.2. PEDV infection in NIH3T3-pAPNwt cell lines
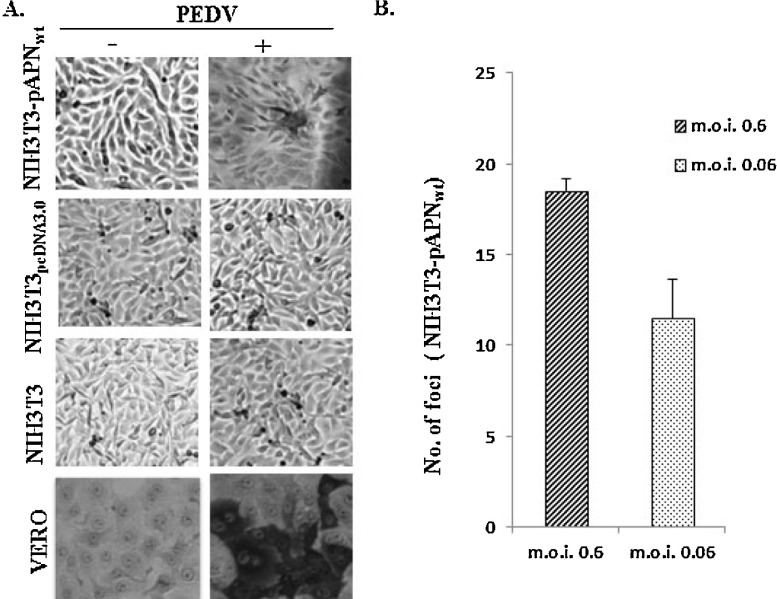
To investigate susceptibility of NIH3T3-pAPNwt, PEDV was inoculated into NIH3T3-pAPNwt, wild type NIH3T3 and NIH3T3pcDNA3.0. Although the typical cytopathic effects (CPE) such as cell rounding and syncytia formation were not observed in NIH3T3- pAPNwt cells, detection of PEDV antigens in fixed cells showed stained foci, but not in wild type NIH3T3 cells nor in NIH3T3pcDNA3.0 (Fig. 3 A). This result confirmed that pAPNwt is important for PEDV infection. Further, PEDV was serially diluted by a factor of 10 and inoculated to NIH3T3-pAPNwt. Comparison of PEDV at m.o.i. 0.6 and 0.06 showed that number of foci was positively correlated to concentration (Fig. 3B). The number of foci formed in NIH3T3-pAPNwt at an m.o.i of 0.6 reduced by 37.8% at an m.o.i. of 0.06. Although NIH3T3-pAPNwt was susceptible to PEDV, the number of progeny virions produced was much lower than those in Vero cells.
Fig. 3.
pAPNwt is important for PEDV entry. (A) Immunocytochemistry examinations of various cell lines inoculated with PEDV showed PEDV antigens detected in pAPNwt expressing NIH3T3 as well as Vero control cells but not pAPNwt deficient cells. (B) Correlation between foci count and virus titer was determined. PEDV with predetermined titer in Vero cells was inoculated at m.o.i. of 0.6 and 0.06. The error bars represent standard deviations of the mean values.
3.3. Expression of pAPNDI-VI and pAPNDVII
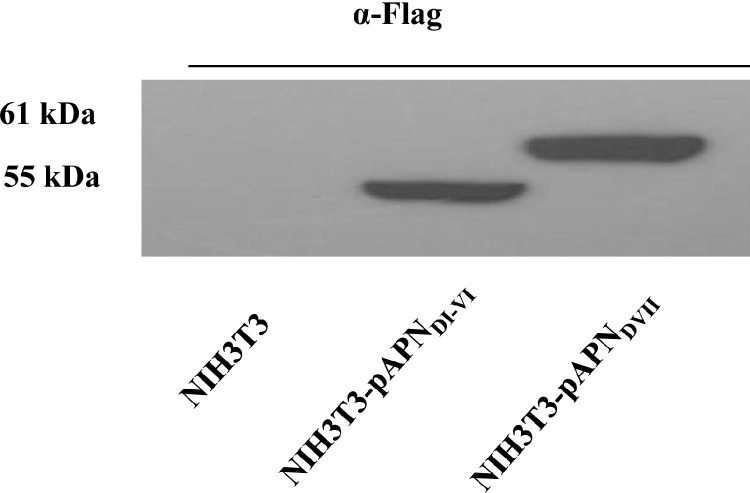
To identify pAPNwt protein domain(s) important for PEDV binding, deletion mutants were generated and their stable expression in NIH3T3 was verified by western blotting. As shown in Fig. 4 , pAPNDVII and pAPNDI-VI expression was detected at 61 kDa and 55 kDa, respectively.
Fig. 4.
Expression of pAPN mutants. Constitutive expression of pAPNDI-VI and pAPNDVII in NIH3T3 cells was determined by western blot analysis. Loading from the left is NIH3T3 control, NIH3T3-pAPNDI-VI and NIH3T3-pAPNDVII lysates. Detection was done through Flag tags.
3.4. Susceptibility of NIH3T3-pAPNwt and mutants cell lines to PEDV
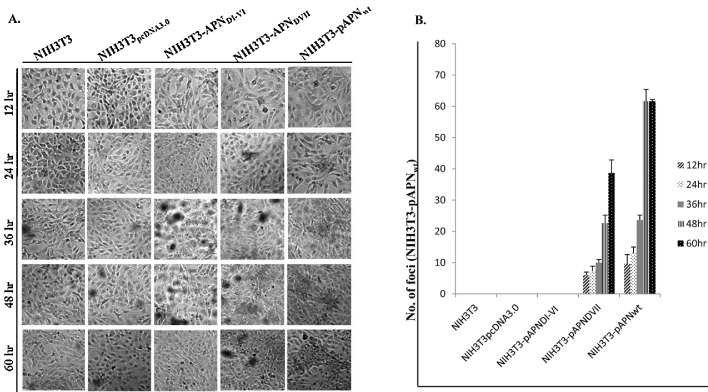
Susceptibility assays showed infection in pAPN domain-VII expressing cells but virus entry was abrogated in absence of seventh domain (pAPNDI-VI) as shown in (Fig. 5 A). Also, there was no observable infection of both NIH3T3 wild type and NIH3T3pcDNA3.0 controls. Analysis of infection between 12 and 60 h p.i. showed that the average number of foci increased more than 6 times in NIH3T3-pAPNDVII. Similarly, PEDV induced foci increased more than 6 fold over time (12 h–60 h) in pAPNwt expressing cells. Also notable was the increase in foci spread through time especially in NIH3T3-pAPNwt (Fig. 5B).
Fig. 5.
PEDV replicates in NIH3T3 upon entry. (A) PEDV infected NIH3T3 cell lines expressing or non-expressing pAPNwt/mutants. Immuncytochemistry procedures performed to detect presence of PEDV antigens through chromogenic foci formation. The foci were evaluated through a time range of 12–60 h. (B) Quantitation of foci between 12 and 60 h in NIH3T3, NIH3T3-pAPNwpcDNA3.0, NIH3T3- pAPNDI-VI, NIH3T3-pAPNDVII and NIH3T3- pAPNwt cells. The error bars represent standard deviations of the mean values.
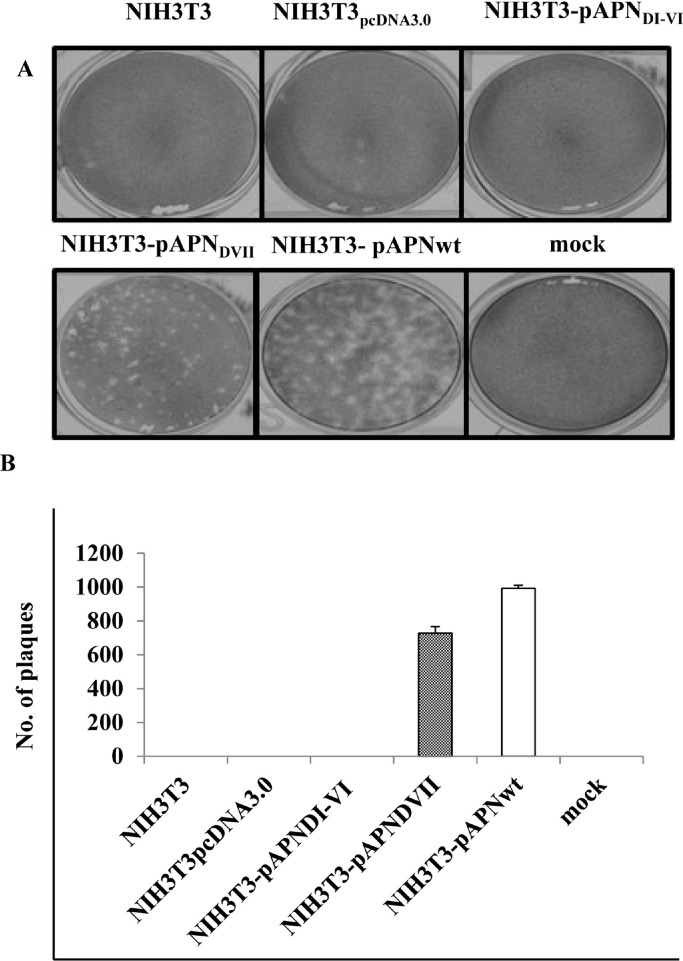
3.5. Infectivity of progeny virion from NIH3T3-pAPNwt and mutants cell lines
Lysates of PEDV or mock-infected NIH3T3 cell lines were re-inoculated in Vero cells for the indirect plaque assay. NIH3T3-pAPNwt and NIH3T3-pAPNDVII lysates induced plaques but NIH3T3, NIH3T3pcDNA3.0 and NIH3T3-pAPNDI-VI lysates did not (Fig. 6 A). On quantification, NIH3T3-pAPNwt had more plaque forming particles (992 pfu/ml) on average while NIH3T3-pAPNDVII lysate had 725 pfu/ml (Fig. 6B).
Fig. 6.
PEDV can be recovered from NIH3T3pAPNVII and NIH3T3pAPNwt cell lines. (A) Image showing plaques assay in Vero cells inoculated with NIH3T3, NIH3T3-pAPNwpcDNA3.0, NIH3T3- pAPNDI-VI, NIH3T3-pAPNDVII and NIH3T3- pAPNwt lysates. (B) Graph of means of plaques formed by the respective lysates. The error bars represent standard deviations of the mean values.
4. Discussion
The hallmark of coronaviruses is the spike glycoprotein which is their functional attachment molecule to the surface of host cells. As of fact and characteristic of all viruses, the attachment involves definite interactions with precise receptors which determine host specificity and tissue tropism (Sandrine et al., 2012). The extensively studied coronavirus prototype, MHV is able to penetrate the hepatocytes through the biliary glycoprotein on the surface of the liver and cause hepatitis (Godfraind and Coutelier, 1998). In alphacoronaviruses, pAPN on the microvillar membrane of the small intestine is the known gateway for TGEV infections in swine (Delmas et al., 1992, Delmas et al., 1994) while human APN (CD13) is the attachment receptor for HCoV-229E which targets infections to lungs where it has been associated with common cold or severe pneumonia in immunocompromised persons (Lachance et al., 1998, Pene et al., 2003). APN/CD13 is a monomeric or homodimeric type II membrane-bound zinc-dependent metalloproteinase, which cleaves neutral or basic amino acids from the N-terminus of oligopeptides and can release neutral and basic amino acid from the N-terminal of peptides (Mou et al., 2009). It is widely expressed on various cells such as epithelial cells of the intestine and kidney, hepatocytes, osteoclasts, endometrial cells, fibroblasts, endothelial cells, bone marrow stromal cells and on neuronal synaptic membranes. APN is involved in many physiology and pathology processes such as hydrolysis of nutrients, inactivation of bioactive peptides, inflammatory bowel diseases, rheumatoid arthritis and cancer (Bank et al., 2008, Yamashita et al., 2007, Shimizu et al., 2002). In 2006, the 3D structure of APN has been studied according to the co-crystal complex of APN and bestatin by Ito (Ito et al., 2006). Based on this report on overall structure and subunits of APN, it is composed of consists of 26 α-helices and 26 β-strands and can be divided into four domains, an N-terminal β-domain (Met1-Asp193), a catalytic domain (Phe194-Gly444), a middle β-domain (Thr445-Trp546), and a C-terminal α-domain (Ser547-Ala870). They also reported the C-terminal α-domain might be important as the active site. More recently, Shan et al. reported the protein structure on pAPN by SWISS-MODEL web server (Shan et al., 2015). They reported pAPN contains small intra-cellular domain and large extra cellular domain divided into 3 subdomains named SPA, SPB and SPC.
In virology, it has been reported that APN is involved in binding of corona viruses and mediating cytomegalovirus infection and antigen presentation (Shahwan et al., 2013, Regan et al., 2012, Li et al., 2007, Kasman, 2005). It has been established that TGEV binds the pAPN receptor on the surface of swine enterocytes (Delmas et al., 1992, Delmas et al., 1994). Additionally, TGEV along with CCoV, TGEV and HCoV-229E exhibit cross-binding activities to feline APN (fAPN), which is the major receptor for feline infectious peritonitis virus and it has been demonstrated that these viruses can utilize this receptor to infect fAPN-expressing BHK21 and NIH3T3 cells (Tresnan et al., 1996, Tusell and Holmes, 2006, Tusell et al., 2007). PEDV was also reported to interact with pAPN in porcine enterocytes (Li et al., 2009), and is capable of infecting MDCK and ST cells expressing the pAPN receptor (Oh et al., 2003, Nam and Lee, 2010). More recently, Li et al. reported pAPN is important receptor for PEDV infection in swine small intestine (IECs). They found higher expression increased the susceptibility by PEDV, and this was confirmed by siRNA inhibition results. They also found PEDV enters via apical plasma membrane and released from there in polarized epithelial cells (Cong et al., 2015). Other group reported that porcine and human APN serve as efficient receptor for PEDV infection (Li, 2015). They found although TGEV Spike protein bound porcine APN much more tightly than human APN, PEDV Spike protein efficiently bound both porcine and human APN. They also reported that Neu5Ac as the PEDV coreceptor. Very recently, Shirato et al. reported pAPN is not functional receptor for PEDV but promotes the its infection through protease activity (Shirato et al., 2016). Taken together it is acceptable that pAPN as the PEDV receptor still little bit controversial.
Because PEDV infects porcine enterocytes causing intestinal lesions that are indistinguishable from those caused by TGEV (Ducatelle et al., 1982, Hooper and Haelterman, 1969), it is conceivable that the two viruses may recognize the same receptor. Curiously, TGEV readily infects porcine primary cells whereas attempts to isolate PEDV in both primary and secondary fetal porcine cells have not been successful (Hofmann and Wyler, 1988, Kusanagi et al., 1992). This ambiguity is further compounded by the fact that Vero cells, with no surface pAPN, are susceptible to PEDV (Hofmann and Wyler, 1988). To clarify these doubts, the current study employed the NIH3T3 cells which are non-susceptible to PEDV infection. By constitutive expression of pAPNwt on these cells, the receptor function would be established by the conversion of the cells into a susceptible cell line (NIH3T3-pAPNwt).
Our results demonstrated that there was extracellular expression of pAPNwt with a surface presence of 62.23% by flow cytometry. Whether this surface receptor would allow PEDV entry was investigated by infection of the cells along with experimental controls, NIH3T3wt and empty vector harboring cells (NIH3T3pcDNA3.0). Results of our analyses showed that although cytopathic effects such as cell rounding and or syncytia formation were not vividly evident, PEDV antigens were detectable immunocytochemistry. In addition, the number of foci formation was positively correlated with the virus titer and the time allowed for virus replication. This development could only be attributed to the pAPNwt because the wild type NIH3T3 and NIH3T3pcDNA3.0 cells were not permissive to PEDV infection. Since NIH3T3 cells being fibroblasts, have a morphology that may not allow close association between cells, spaces in between cells may have resulted in the lack of syncytia or plaque formation in NIH3T3. In addition, PEDV has already been adapted to Vero cells and it is assumable that NIH3T3 may be an unfamiliar environment for PEDV replication. Indeed, even new isolates of PEDV are able to show distinct signs of CPE in Vero cells only after several passages (Pan et al., 2010), which could probably be an adaptation period.
As the functional domain study on pAPN, Shan et al. reported that SPC region on pAPN might be putative functional domain for PEDV infection (Shan et al., 2015). They found MDCK cells expressing only SPC region successfully infected by PEDV and even shown cytopathic effects (CPE). They also reported that PEDV titer in MDCK cell expressing only SPC region generated same number of virus in pAPN expressing MDCK and Vero cells. In our study, to determine the specific interactive point of pAPNwt with PEDV, domain mutants were expressed stably in NIH3T3 cells. The cloning strategy of the mutants followed cues that coronaviruses identify certain intermittent motifs on fAPN which are amino acids (aa) 288–290, aa 732–746 (called R1), and aa 764–788 (called R2) which determine their tropism (Tusell et al., 2007). Thus, two mutants; (David and Kathryn, 2007) expressing domains I–VI of pAPNwt and (Susan and Navas-Martin, 2005) expressing domain VII of pAPNwt were constructed and designated pAPND1-VI and pAPNDVII, respectively. NIH3T3 stable cell lines expressing these mutants were assayed for their susceptibility to PEDV and similar to TGEV (Delmas et al., 1994), PEDV successfully infected pAPNDVII expressing cells but that was abrogated in the absence of the seventh domain (pAPNDI-VI). Also noteworthy, PEDV harvested from NIH3T3-pAPNwt and NIH3T3-pAPNDVII induced plaques on Vero cell monolayers indicating virus entry and propagation in those cells. Taken together, all our data confirmed that PEDV replicated in both NIH3T3-pAPNwt and NIH3T3-pAPNDVII suggesting successful entry into the cell lines. We therefore concluded that domain VII of pAPN is critical for PEDV entry. Although structural domain analysis on pAPN used in this study is little bit different from those from Shan et al. study, but functional binding domain from both studies are identical. Especially, our results promising because we studied stable expression of wild type and deletion mutants from stable cell lines expressing those proteins. Even if our studies confirmed with stable cell lines but still don’t know how those mutations on pAPN affected on its stability, 3-D structure and especially function. Still more studies required to answer on those questions.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and future Planning (grant number 2015R1A2A2A01003785).
References
- Bank U., Bohr U.R., Reinhold D., Lendeckel U., Ansorge S., Malfertheiner P., Tager M. Inflammatory bowel diseases: multiple benefits from therapy with dipeptidyl- and alanyl-aminopeptidase inhibitors. Front. Biosci. 2008;13:3699–3713. doi: 10.2741/2960. [DOI] [PubMed] [Google Scholar]
- Cong Y., Li X., Bai Y., Lv X., Herrler G., Enjuanes L., Zhou X., Qu B., Meng F., Cong C., Ren X., Li G. Porcine aminopeptidase N mediated polarized infection by porcine epidemic diarrhea virus in target cells. Virol. 2015;478:1–8. doi: 10.1016/j.virol.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David E.W., Kathryn V.H. Coronavirus binding and entry. In: Volker T., editor. Coronavirues: Molecular and Cellular Biology. Caister Academic Press; Norfolk, UK: 2007. pp. 3–31. [Google Scholar]
- de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. Coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic; London, United Kingdom: 2011. pp. 806–828. [Google Scholar]
- Delmas B., Gelfi J., L’Haridon R., Vogel L.K., Sjostrom H., Noren O., Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Int. J. Sci. Nat. 1992;357:417–419. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Kut E., Syostrom H., Noren O., Laude H. Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase-N that is distinct from the enzymatic site. J. Virol. 1994;68:5216–5224. doi: 10.1128/jvi.68.8.5216-5224.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatelle R., Coussement W., Debouck P., Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. II. Electron microscopic study. Vet. Pathol. 1982;19:57–66. doi: 10.1177/030098588201900109. [DOI] [PubMed] [Google Scholar]
- Dulbecco R. Production of plaques in monolayer tissue cultures by single particles of an animal virus. Proc. Nat. Acad. Sci. 1952;38:747–752. doi: 10.1073/pnas.38.8.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dveksler G.S., Pensiero M.N., Cardellichio C.B., Williams R.K., Jiang G.S., Holmes K.V., Dieffenbach C.W. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dveksler G.S., Dieffenbach C.W., Cardellichio C.B., McCuaig K., Pensiero M.N., Jiang G.S., Beauchemin N., Holmes K.V. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J. Virol. 1993;67:1–8. doi: 10.1128/jvi.67.1.1-8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfraind C., Coutelier J.P. Morphological analysis of mouse hepatitis virus A59-induced pathology with regard to viral receptor expression. Histol. Histopathol. 1998;13:181–199. doi: 10.14670/HH-13.181. [DOI] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77:4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper B.E., Haelterman E.O. Lesions of the gastrointestinal tract of pigs infected with transmissible gastroenteritis. Can. J. Comp. Med. 1969;33:29–36. [PMC free article] [PubMed] [Google Scholar]
- Horton R.M. SOEing together tailor-made genes. In: White B.A., editor. vol. 15. Humana Press Inc.; Totowa, NJ: 1993. pp. 251–261. (PCR Protocols: Current Methods and Applications). [Google Scholar]
- Ito K., Nakajima Y., Onohara Y., Takeo M., Nakashima K., Matsubara F., Ito T., Yoshimoto T. Crystal structure of aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli and conformational change of methionine 260 involved in substrate recognition. J. Biol. Chem. 2006;281(44):33664–33676. doi: 10.1074/jbc.M605203200. [DOI] [PubMed] [Google Scholar]
- Kasman L.M. CD13/aminopeptidase N and murine cytomegalovirus infection. Virol. 2005;334(1):1–9. doi: 10.1016/j.virol.2005.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay B., Kenny A.J., Turner A.J. Localization of aminopeptidase N and dipeptidyl peptidase IV in pig striatum and in neuronal and glial cell cultures. Eur. J. Neurosci. 1994;6:531–537. doi: 10.1111/j.1460-9568.1994.tb00297.x. [DOI] [PubMed] [Google Scholar]
- Kenny A.J., Maroux S. Topology of microvillar membrance hydrolases of kidney and intestine. Physiol. Rev. 1982;62(1):91–128. doi: 10.1152/physrev.1982.62.1.91. [DOI] [PubMed] [Google Scholar]
- Kolb A.F., Hegyi A., Maile J., Heister A., Hagemann M., Siddell S.G. Molecular analysis of the coronavirus-receptor function of aminopeptidase N. Adv. Exp. Med. Biol. 1998;440:61–67. doi: 10.1007/978-1-4615-5331-1_8. [DOI] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J.M. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusanagi K., Kuwahara H., Katoh T., Nunoya T., Ishikawa Y., Samejima T., Tajima M. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J. Vet. Med. Sci. 1992;54:313–318. doi: 10.1292/jvms.54.313. [DOI] [PubMed] [Google Scholar]
- Lachance C., Arbour N., Cashman N.R., Talbot P.J. Involvement of aminopeptidase N N (CD13) in infection of human neural cells by human coronavirus 229E. J. Virol. 1998;72:6511–6519. doi: 10.1128/jvi.72.8.6511-6519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendeckel U., Kähne T., Riemann D., Neubert K., Arndt M., Reinhold D. The role of membrane peptidases in immune functions. Adv. Exp. Med. Biol. 2000;477:1–24. doi: 10.1007/0-306-46826-3_1. [DOI] [PubMed] [Google Scholar]
- Li B.X., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virol. 2007;365(1):166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.X., Ma G.P., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Bing Du Xue Bao. 2009;25:220–225. [PubMed] [Google Scholar]
- Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J. Virol. 2012;86(5):2856–2858. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015;89(11):6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y., Xu W. The structure and main functions of aminopeptidase N. Curr. Med. Chem. 2007;14:639–647. doi: 10.2174/092986707780059571. [DOI] [PubMed] [Google Scholar]
- Mina-Osorio P., Winnicka B., O'Conor C., Grant C.L., Vogel L.K., Rodriguez-Pinto D., Holmes K.V., Ortega E., Shapiro L.H. CD13 is a novel mediator of monocytic/endothelial cell adhesion. J. Leukoc. Biol. 2008;84:448–459. doi: 10.1189/jlb.1107802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou J., Fang H., Jing F., Wang Q., Liu Y., Zhu H., Shang L., Wang X., Xu W. Design, synthesis and primary activity evaluation of l-arginine derivatives as amino-peptidase N/CD13 inhibitors. Bioorg. Med. Chem. 2009;17(13):4666–4673. doi: 10.1016/j.bmc.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam E., Lee C. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Vet. Microbiol. 2010;144(1–2):41–50. doi: 10.1016/j.vetmic.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.S., Song D.S., Park B.K. Identification of a putative cellular receptor 150 kDa polypeptide for porcine epidemic diarrhea virus in porcine enterocytes. J. Vet. Sci. 2003;4:269–275. [PubMed] [Google Scholar]
- Pan X., Li W., Zhou Q., Wang D., Bi Y., Chen F., Song Y. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol. J. 2010;9:195. doi: 10.1186/1743-422X-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Cruz D.J., Shin H.J. Clathrin- and serine proteases-dependent uptake of porcine epidemic diarrhea virus into Vero cells. Virus Res. 2014;191:21–29. doi: 10.1016/j.virusres.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pene F., Merlat A., Vabret A., Rozenberg F., Buzyn A., Dreyfus F., Cariou A., Freymuth F., Lebon P. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 2003;37:929–932. doi: 10.1086/377612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.J., Chua M.M., Lavi E., Weiss S.R. Pathogenesis of chimeric MHV-4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 1999;73:7752–7760. doi: 10.1128/jvi.73.9.7752-7760.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N., Barrett A. Families of aspartic peptidases, and those of unknown catalytic mechanism. Meth. Enzymol. 1995;248:105–120. doi: 10.1016/0076-6879(95)48009-9. [DOI] [PubMed] [Google Scholar]
- Regan A.D., Millet J.K., Tse L.P., Chillag Z., Rinaldi V.D., Licitra B.N., Dubovi E.J., Town C.D., Whittaker G.R. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virol. 2012;430(2):90–99. doi: 10.1016/j.virol.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M., Izeta A., Sanchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Duran J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrine B., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahwan K., Hesse M., Mork A.K., Herrler G., Winter C. Sialic acid binding properties of soluble coronavirus spike (S1) proteins: differences between infectious bronchitis virus and transmissible gastroenteritis virus. Viruses. 2013;5(8):1924–1933. doi: 10.3390/v5081924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Z., Yin J., Wang Z., Chen P., Li Y., Tang L. Identification of the functional domain of the porcine epidemic diarrhoea virus receptor. J. Gen. Virol. 2015;96(9):2656–2660. doi: 10.1099/vir.0.000211. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Tani K., Hase K., Ogawa H., Huang L., Shinomiya F., Sone S. CD13/aminopeptidase N-induced lymphocyte involvement in inflamed joints of patients with rheumatoid arthritis. Arth. Rheum. 2002;46(9):2330–2338. doi: 10.1002/art.10517. [DOI] [PubMed] [Google Scholar]
- Shirato K., Maejima M., Islam M.T., Miyazaki A., Kawase M., Matsuyama S., Taguchi F. Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhoea virus, but promotes its infectivity via aminopeptidase activity. J. Gen. Virol. 2016 doi: 10.1099/jgv.0.000563. [DOI] [PubMed] [Google Scholar]
- Sjostrom H., Noren O., Olsen J. Structure and function of aminopeptidase N. Adv. Exp. Med. Biol. 2000;477:25–34. doi: 10.1007/0-306-46826-3_2. [DOI] [PubMed] [Google Scholar]
- Soichiro M., Song I., Morita A., Erickson R.H., Kim Y.S. Distribution and biosynthesis of aminopeptidase n and dipeptidyl aminopeptidase IV in rat small intestine. Biochim. Biophys. Acta—Gen. Subj. 1983;761:66–75. doi: 10.1016/0304-4165(83)90363-x. [DOI] [PubMed] [Google Scholar]
- Susan R.W., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresnan D.B., Levis R., Holmes K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996;70:8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusell S.M., Holmes K.V. Molecular interactions of group 1 coronaviruses with feline APN. Adv. Exp. Med. Biol. 2006;581:289–291. doi: 10.1007/978-0-387-33012-9_49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusell S.M., Schittone S.A., Holmes K.V. Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants of viral host range. J. Virol. 2007;81:1261–1273. doi: 10.1128/JVI.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M., Kajiyama H., Terauchi M., Shibata K., Ino K., Nawa A., Mizutani S., Kikkawa F. Involvement of aminopeptidase N in enhanced chemosensitivity to paclitaxel in ovarian carcinoma in vitro and in vivo. Int. J. Cancer. 2007;120(10):2243–2250. doi: 10.1002/ijc.22528. [DOI] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Int. J. Sci. Nat. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Shin B., Park E.S., Yang S., Choi S., Kangm M., Rho J. Protein arginine methyltransferase 1 regulates herpes simplex virus replication through ICP27 RGG-box methylation. Biochem. Biophys. Res. Commun. 2010;391:322–328. doi: 10.1016/j.bbrc.2009.11.057. [DOI] [PubMed] [Google Scholar]