Highlights
-
•
An overview of the interactions of PEDV and its target cells during the initial stage of infection.
-
•
A description of the multidomain structure of the spike (S) protein.
-
•
A summary of observations on aminopeptidase N as the PEDV protein receptor.
-
•
An overview with new data on the significance of the N-terminal S domain in sialic acid binding.
-
•
A summary of the requirements for proteolytic activation of the fusion function of the S protein.
Keywords: Porcine epidemic diarrhea virus, PEDV, PED, Virus, Coronavirus, Spike, Virus entry, Sialic acid, Sialic acid binding, Receptor interaction, Membrane fusion, Proteolytic activation, Virus tropism
Abstract
Porcine epidemic diarrhea virus (PEDV), a coronavirus discovered more than 40 years ago, regained notoriety recently by its devastating outbreaks in East Asia and the Americas, causing substantial economic losses to the swine husbandry. The virus replicates extensively and almost exclusively in the epithelial cells of the small intestine resulting in villus atrophy, malabsorption and severe diarrhea. Cellular entry of this enveloped virus is mediated by the large spike (S) glycoprotein, trimers of which mediate virus attachment to the target cell and subsequent membrane fusion. The S protein has a multidomain architecture and has been reported to bind to carbohydrate (sialic acid) and proteinaceous (aminopeptidase N) cell surface molecules. PEDV propagation in vitro requires the presence of trypsin(-like) proteases in the culture medium, which capacitates the fusion function of the S protein. Here we review the current data on PEDV entry into its host cell, including therein our new observations regarding the functional role of the sialic acid binding activity of the S protein in virus infection. Moreover, we summarize the recent progress on the proteolytic activation of PEDV S proteins, and discuss factors that may determine tissue tropism of PEDV in vivo.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is the causative agent of porcine epidemic diarrhea (PED), an enteric disease affecting pigs of all ages. The disease is characterized by acute watery diarrhea, dehydration and vomiting, with high mortality in neonatal piglets. Devastating outbreaks of PED in East Asia (since 2010) and in North America (since 2013) have revitalized the research into this porcine coronavirus that was first identified in 1978 (Stevenson et al., 2013). PEDV primarily replicates in the villous enterocytes of the small intestine. Its entry into host cells is mediated by the spike (S) glycoprotein that is exposed on the virion surface. This key entry factor is considered the main determinant of viral host and tissue tropism. Moreover, the S protein is highly immunogenic and the main target for neutralizing antibodies. Understanding this protein’s function will thus aid the design of strategies against this enteric swine coronavirus and is fundamental to our understanding of its epidemiology and pathogenesis. In this review, following a brief and general introduction on PEDV, we will describe the structure and function of the spike glycoprotein. In particular, we will report the generation of a recombinant PEDV virus harboring a large deletion in the S protein’s N-terminal region for studies to assess the role of sialic acid binding activity of PEDV S in infection. Finally we will discuss the mechanism by which the S protein is proteolytically activated for membrane fusion.
1.1. Coronaviruses infecting swine
PEDV is a member of the Coronaviridae family (subfamily Coronavirinae, family Coronaviridae, order Nidovirales). This family of viruses comprises a large group of enveloped viruses with a positive-sense RNA genome of up to 32 kilobases. Coronaviruses infect a broad range of mammalian and avian hosts and can cause respiratory, enteric, hepatic and neurological disease. Pathogenic coronaviruses are found in farm animals as well as in humans and have demonstrated potential to cross the host-species barrier. Two zoonotic coronaviruses – the severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV) – have emerged over the last two decades, both causing severe and often fatal respiratory disease in humans. Coronaviruses have recently been subdivided into four genera: Alphacoronavirus, Betacoronavirus (lineages A–D), Gammacoronavirus and Deltacoronavirus (de Groot et al., 2013). Pathogenic viruses in each genus include transmissible gastroenteritis virus (TGEV), human coronavirus (HCoV) 229E and HCoV-NL63 (α-CoV), mouse hepatitis virus (MHV, β-CoV, lineage A), SARS-CoV (β-CoV, lineage B), MERS-CoV (β-CoV, lineage C), avian infectious bronchitis virus (IBV, γ-CoV) and porcine deltacoronavirus (PDCoV, δ-CoV). In swine five coronaviruses have been identified, representing three of the four genera. PEDV, TGEV and the natural TGEV deletion mutant porcine respiratory virus (PRCoV) belong to the Alphacoronavirus genus. TGEV mainly infects epithelial cells from the small intestine and causes enteritis and fatal diarrhea in piglets; it is clinically indistinguishable from PEDV. Unlike TGEV, PRCoV mostly infects epithelial cells of the respiratory tract and alveolar macrophages causing a mild or often subclinical respiratory disease. The porcine hemagglutinating encephalomyelitis virus (PHEV) belongs to the Betacoronavirus genus; it targets respiratory and neuronal tissues and causes vomiting, wasting disease and neurological disorders in seronegative piglets (Straw et al., 2006). The recently identified PDCoV of the Deltacoronavirus genus has enteric tropism causing mild to moderate disease in young piglets (Jung et al., 2015).
1.2. PEDV epidemiology
PED was not detected in swine until the 1970s. The first PED outbreak in swine was recognized in England in 1971. Seven years later the etiological agent was identified as a coronavirus and officially named as PEDV (Pensaert and De Bouck, 1978). PED was prevalent throughout Europe causing sporadic, localized outbreaks in the 1980s, 1990s and in subsequent years (Martelli et al., 2008). PED was first reported in Asia in 1982 and since then it has had an increasingly great economic impact on the Asian swine industry. Particularly since 2010, devastating outbreaks have been reported in China and other Asian countries causing up to 100% mortality in suckling piglets (Li et al., 2012, Puranaveja et al., 2009, Sun et al., 2012). PEDV entered the United States (US) for the first time in April 2013 and this virulent strain rapidly spread across the US to 36 states, as well as to other countries in North- and South-America, including Canada (Pasick et al., 2014), Mexico (Vlasova et al., 2014), the Dominican Republic, Colombia and Peru (Ojkic et al., 2015, Oka et al., 2014). A less virulent PEDV strain has been detected in the US characterized by small genomic insertions and deletions (S INDEL strain) in the viral spike glycoprotein. Since 2014, PEDV has reemerged in Europe including Germany (Hanke et al., 2015), Italy, Austria (Steinrigl et al., 2015), The Netherlands, Belgium (Theuns et al., 2015), Portugal (Mesquita et al., 2015), France (Grasland et al., 2015) and Ukraine (Dastjerdi et al., 2015).
1.3. PEDV pathogenesis
PEDV mainly infects and replicates in villous enterocytes of the small intestine (duodenum, jejunum and ileum) (Debouck et al., 1981, Ducatelle et al., 1981, Ducatelle et al., 1982). Infection results in destruction of the intestinal epithelium with subsequent villus shortening causing watery diarrhea that lasts for about a week. Other clinical symptoms include vomiting, anorexia and fever. Pigs of all ages are susceptible, but symptoms are most severe in suckling piglets of less than one week old with mortality rates often approaching 100%. Fatality rates in weaned pigs are much lower (1–3%) while mortality has not been observed among fattening pigs (Hou et al., 2007). Many studies indicate that PEDV does not replicate outside the intestinal tract, though PEDV was detected in a recent study by RT-PCR and IHC in other organs of experimentally infected piglets including lung, liver, kidney and spleen (Park and Shin, 2014).
2. Structure, function and antigenicity of the PEDV S protein
The PEDV S protein is the key protein responsible for virus entry into the target cell. It mediates the essential functions of receptor binding and subsequent fusion of the viral and cellular membranes during cell entry, thereby releasing the viral nucleocapsid into the cytoplasm. The PEDV S protein is a ±1383-residues long glycoprotein of 180–200 kilodalton in size. Trimers of these S proteins form the club-shaped, ±20 nm long projections (spikes) on the virion surface that provide the coronavirus its typical crown-like appearance on electron micrographs (Pensaert and De Bouck, 1978). Like other CoV spike proteins, PEDV S is a type I membrane glycoprotein with an N-terminal signal peptide, a large extracellular region, a single transmembrane domain and a short cytoplasmic tail (Fig. 1 ). The ectodomain of coronavirus spike proteins can be divided into two domains with distinct functions: the N-terminal S1 subunit responsible for receptor binding and the C-terminal membrane anchored S2 domain responsible for membrane fusion. The border between the S1 (residues 1–726, CV777 numbering [GB: AF353511]) and S2 (residues 727–1383) subunit can be deduced from the sequence alignment with alphacoronavirus S proteins (e.g. type I feline and canine coronaviruses) of which the S1-S2 junction is clearly demarcated by the presence of a furin protease cleavage site (de Haan et al., 2008). Recent elucidation of the cryo-EM structure of a betacoronavirus spike trimer demonstrated a modular S1 protein architecture with four discrete domains (Walls et al., 2016), two of which may interact with host receptors and hence serve as a receptor binding domain for betacoronaviruses. Two independently folding domains have been assigned in the S1 subunit of alphacoronaviruses that can interact with host cell surface molecules: an N-terminal domain (TGEV S residues 1–244) that can exhibit sialic acid binding activity, and a more C-terminal domain (TGEV S residues 506–655) that can interact with a protein receptor (Reguera et al., 2012). Relative to S1, the S2 subunit is more conserved in amino acid sequence and contains the typical structural features found in class I viral fusion proteins including a hydrophobic fusion peptide (FP), heptad repeat regions (HR1 and HR2) and a C-terminal trans membrane (TM) region (Fig. 1). As for all class I fusion proteins, membrane fusion is started by the exposure and insertion of the FP into the target cell membrane, after which the fusion protein jackknives such that the antiparallel HR1 and HR2 regions form a stable six-helix bundle structure. During this transition, the fusion peptide and the transmembrane domain come into close proximity enabling the fusion of the viral and cellular membrane (Eckert and Kim, 2001). A key feature of the coronavirus S protein is the proteolytic priming of the fusion potential by host proteases that can occur at the S1/S2 junction as well as adjacent to the fusion peptide within the S2 region at the S2′ position (Belouzard et al., 2009, Yamada and Liu, 2009). The PEDV spike proteins on the virion are uncleaved, yet they undergo this proteolytic requirement for fusion during the entry stage (Wicht et al., 2014b Shirato et al., 2011a, Shirato et al., 2011b).
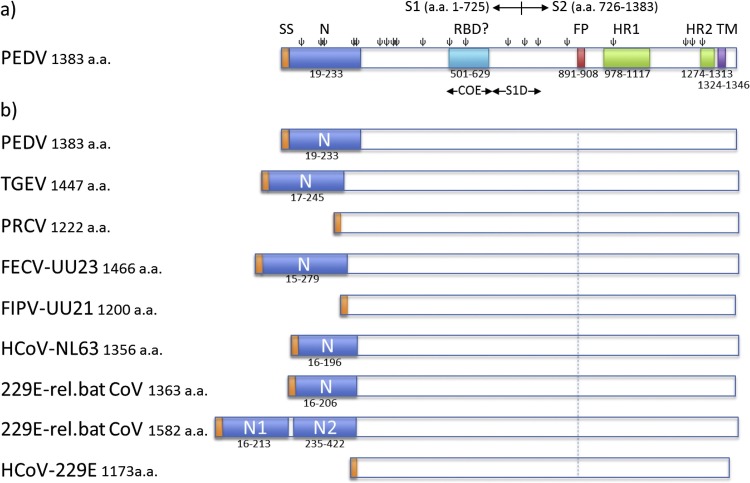
Fig. 1.
Schematic presentation of spike proteins of PEDV and related alphacoronaviruses. a) The PEDV S protein. The S1 and S2 subunits, the signal peptide (SP, residues 1–18), the N-terminal domain (N, residue 19–233), regions containing neutralizing epitopes (COE, residues 499–638 and S1D, residues 636–789), the fusion peptide (FP; residues 891–908), two heptad repeat regions (HR1, residues 978–1117 and HR2, residues 1274–1313), the transmembrane domain (TM, residues 1324–1346; predicted by TMHMM server) and predicted N-glycosylation sites (ψ; predicted by the NetNGlyc server) are indicated. b) N-terminal domains (N) in the spike proteins of PEDV and related alphacoronaviruses. Schematic representations of the spike proteins are shown of a number of viruses belonging to Alphacoronavirus genus including PEDV strain CV777 (GB: AAK38656.1), TGEV strain Purdue P115 (GB: ABG89325.1), PRCoV strain ISU-1 (GB: ABG89317.1), FECV strain UU23 (GB: ADC35472.1), FIPV strain UU21 (GB: ADL71466.1), HCoV-NL63 (GB: YP_003767.1), HCoV-229E related bat CoV with one N domain (GB: ALK28775.1), HCoV-229E related bat CoV with two N domains (GB: ALK28765.1) and HCoV-229E strain inf-1 (GB: NP_073551.1). Spike proteins are drawn to scale and aligned at the position of the conserved fusion peptide indicated by the dashed line. Signal sequence is indicated in orange.
Exerting critical functions in cell entry, the coronavirus S protein is the main target for humoral immunity. A recent study demonstrated that immunization of pregnant sows with the PEDV S1 protein could provide passive immunity against PEDV to suckling piglets through colostrum and milk (Oh et al., 2014). The S protein ectodomain contains at least two neutralizing B-cell epitopes (Chang et al., 2002, Cruz et al., 2006, Zhang et al., 2016). One of the neutralizing epitopes maps to a domain homologous to the collagenase resistant fragment (CO-26 K) of the TGEV S protein and was coined ‘CO-26 K equivalent’ (COE) domain for PEDV (residues 499–638). The CO-26 K fragment in TGEV S harbors the receptor binding domain to which most neutralizing antibodies are directed (Godet et al., 1994). Analogous to TGEV, immunization of mice with the PEDV S COE region elicited virus neutralizing antibodies (Chang et al., 2002). The S1D region (residues 636–789) in PEDV S contains another neutralizing epitope and spans the S1-S2 junction. Two non-neutralizing B-cell epitopes (SS2: 748-YSNIGVCK-755 and SS6: 764-LQDGQVKI-771) have been identified within this region, just downstream of the predicted S1-S2 junction (Sun et al., 2008) (Fig. 1a).
3. PEDV receptor usage
Receptor interaction plays a crucial role in the cell and tissue tropism of coronaviruses as well as in pathogenesis and cross-species transmission. Coronaviruses can bind to a wide range of proteinaceous and carbohydrate cell surface molecules via their spike proteins. PEDV has been reported to utilize the aminopeptidase N protein (APN, also known as CD13) as a functional receptor (Li et al., 2007). APN serves as a receptor for multiple alphacoronaviruses, including the canine coronavirus type II, feline coronavirus type II, TGEV and PRCoV and human coronavirus 229E (HCoV-229E) (Delmas et al., 1992, Yeager et al., 1992). It is, however, not a universal receptor for these viruses as the human coronavirus NL63 (HCoV-NL63) uses angiotensin converting enzyme 2 (ACE2) for its entry (Hofmann et al., 2005). APN is a 150 kDa glycosylated type II transmembrane protein that is highly abundant on the apical membrane of mature enterocytes (Mina-Osorio, 2008). It was termed ‘moonlighting enzyme’ referring to its multiple biological functions, including peptide metabolism, cell motility and adhesion (Mina-Osorio, 2008). Several lines of evidence support the view that APN is a functional receptor for PEDV: i) in a virus overlay protein binding assay a ∼150 kDa protein was identified as a PEDV binding protein in swine testis (ST) cells and enterocytes, and this interaction could be blocked by antibodies against porcine APN (pAPN); ii) APN overexpression in otherwise non-permissive cell lines (i.c. Madin Darby canine kidney and ST cells) conferred susceptibility of these cells to PEDV infection (Nam and Lee, 2010); iii) PEDV S1 was shown to biochemically interact with soluble APN in a dot-blot assay and with cell surface expressed APN in a FACS assay (Liu et al., 2015 Deng et al., 2016); iv) preincubation of cells with pAPN antibodies or a pAPN-binding peptide prior to inoculation interfered with PEDV infection (Li et al., 2007, Meng et al., 2014), and v) transgenic mice expressing pAPN appeared susceptible to PEDV infection (Park et al., 2015). The APN binding domain was recently shown to reside within a domain in the C-terminal half of PEDV S1 (residues 477–629), corresponding to the RBD of TGEV S1 (residues 505–655). Intriguingly, though suggested to share the same receptor, the two aromatic residues within the extending loops of the TGEV RBD known to be critical for APN binding are not conserved in the PEDV RBD, indicating a different mode of receptor interaction. African green monkey Vero cell lines are historically used for isolation and propagation of PEDV strains. Yet, these cells do not express APN as inferred from mass spectrometry analyses of the Vero cell proteome (Guo et al., 2014, Li et al., 2007, Shirato et al., 2011a, Shirato et al., 2011b, Zeng et al., 2015) and the lack of mRNA transcripts in these cells (our own data, not shown), indicating that other receptors may be involved in PEDV entry into these cells.
PEDV exhibits promiscuous binding to cellular receptors (Liu et al., 2015). Unlike TGEV, PEDV was shown to interact with and utilize human APN as a functional receptor. Moreover, PEDV can infect multiple cell lines in vitro from different species besides porcine including bat and primate (human and non-human). Intriguingly, the genome sequence and genome organization of PEDV are more related to bat alphacoronaviruses than to other viruses from the same genus suggesting that interspecies transmission between bats and pigs, perhaps via an intermediate host, may have occurred (Woo et al., 2012). The ability of PEDV to infect cells of multiple species suggests that the virus – similar to MERS-CoV – utilizes evolutionarily conserved host cell components as receptors thereby increasing the potential for cross-species and potentially zoonotic transmission (Gallagher and Perlman, 2013, Raj et al., 2013). Apart from its interaction with APN, PEDV is able to bind sialic acids (Deng et al., 2016, Liu et al., 2015). Several coronaviruses from the alpha-, beta- and gammacoronavirus genus have been shown to bind to sialic acids, a feature variably required for virus infection in cell culture depending on the particular (Schwegmann-Weßels and Herrler, 2006). This lectin activity resides within the N-terminal domain of their spike protein. Notably, betacoronaviruses within the lineage A1 including HCoV-OC43, bovine coronavirus (BCoV) and PHEV use (acetylated) sialic acid as a primary receptor. Other coronaviruses can bind to sialoglycoconjugates in addition to their primary protein receptor. The APN-recruiting TGEV S1 can bind to sialic acids (N-acetylneuraminic acid [Neu5Ac] and N-glycolylneuraminic acid [Neu5Gc]) via its N-terminal region (residues 21–244) (Krempl et al., 1997, Reguera et al., 2012, Schultze et al., 1996). The sialic acid binding activity appears dispensable for TGEV infection in cell culture but enhances infection under shorter inoculation conditions (Schwegmann-Wessels et al., 2011). PEDV was shown to have hemagglutination (HA) activity using rabbit erythrocytes, but this HA activity was only observed after pretreatment of the virus with trypsin or neuraminidase (Park et al., 2010). PEDV lacks the HE protein that is responsible for the HA activity of beta-coronaviruses like MHV, HCoV-OC43 and BCoV. Because in other hemagglutinating coronaviruses lacking an HE protein such as IBV and TGEV the HA activity resides in the S protein, in PEDV this activity is also likely to occur in this protein. Chang et al. demonstrated that PEDV S1 could bind to bovine and porcine mucins as shown by dot-blot hybridization assay. Furthermore, they used a glycan array screen to identify which type(s) of sugar was the most favored by PEDV S1, observing that Neu5Ac shows the highest binding affinity (Liu et al., 2015). The sialic acid binding activity of PEDV was shown very recently to occur in the N-terminal region of its spike protein (residues 1–320), analogous to other sialic acid-binding coronaviruses (Deng et al., 2016). However, it is unknown whether sialic acid binding by the S protein can facilitate entry of PEDV into cells.
4. Structural and functional assessment of the PEDV spike protein N-domain
4.1. N-domains in spike proteins of alphacoronaviruses
For a number of alphacoronaviruses the loss of the S protein N-domain correlates with a loss of enteric tropism (Fig. 1b). i) The clearest example of this phenomenon is TGEV that displays a dual tropism targeting the respiratory and enteric tract. On the other hand, PRCoV, a natural deletion variant of TGEV lacking the S protein N-domain, lost its enteric tropism and mainly replicates in the respiratory tract (Cox et al., 1990, Sanchez et al., 1992); ii) feline coronaviruses come in two pathotypes, the clinically mild or asymptomatic enteric FECV and the highly pathogenic, systemically replicating FIPV. FIPV is thought to originate from FECV by mutation and seems to have lost its tropism for enterocytes (Desmarets et al., 2013). Sequencing analysis of multiple FECV and FIPV field strains indicates that, contrary to enteric FECV field strains, some FIPV strains (e.g. strains UU16 and UU21, Fig. 1b) have a large deletion in the N-domain; iii) clinical isolates of HCoV-229E encode a relatively short spike protein of 1173 residues in length due to the apparent lack of the N-domain (Farsani et al., 2012). Recently, bat coronaviruses related to HCoV-229E have been isolated from hipposiderid bats, some of them encoding unusual large spike glycoproteins of 1571–1582 residues in length (Corman et al., 2015). Intriguingly, PSI-BLAST analysis indicates that these 229E-related bat coronaviruses carry two consecutive N-domain homologs in the first 425 residues part of their S protein − which we designated N1 and N2 − with 32% and 35% amino acid sequence identity to the N-domain of the HCoV-NL63 S protein, respectively (Fig. 1b). Other 229E-related bat coronaviruses isolated from bats of the same genus carry shorter spike proteins (±1362 a.a.) that harbor a single N-domain (Fig. 1b). The HCoV-229E and a related alpaca coronavirus lacking an N-domain in its spike protein have respiratory tropism and are not generally associated with gastro-enteritis (Esper et al., 2010), whereas most bat coronaviruses have enteric tropism (Drexler et al., 2014). Together the above examples suggest a role of the alphacoronavirus spike N-domain for replication in the intestinal tract. In case of TGEV, the loss of enteric tropism was specifically associated with the loss of the sialic binding activity that resides in this domain. A TGEV mutant with a four amino acids deletion in its S protein N-domain that resulted in a loss of hemagglutinating activity displayed reduced enteropathogenicity (Krempl et al., 1997). How sialic acid binding activity in this N-terminal spike domain influences enteric tropism of these alphacoronaviruses remains to be resolved. It has been hypothesized i) that surface-bound sialoglycoconjugates may protect the virus from hostile conditions in the stomach, ii) that virion binding to mucins may prevent the loss of viruses by intestinal peristalsis, and iii) that binding to mucins may allow the viruses to pass the thick mucus barrier thereby gaining access to their intestinal target cells (Krempl and Herrler, 2001). In the following paragraphs ($4.2–$4.4) we describe our recent data regarding the role of the N-terminal domain and the sialic-acid binding activity of the S protein in PEDV infection in cell culture.
4.2. The caDR13-PEDV spike N-domain is dispensable for virus propagation in cell culture
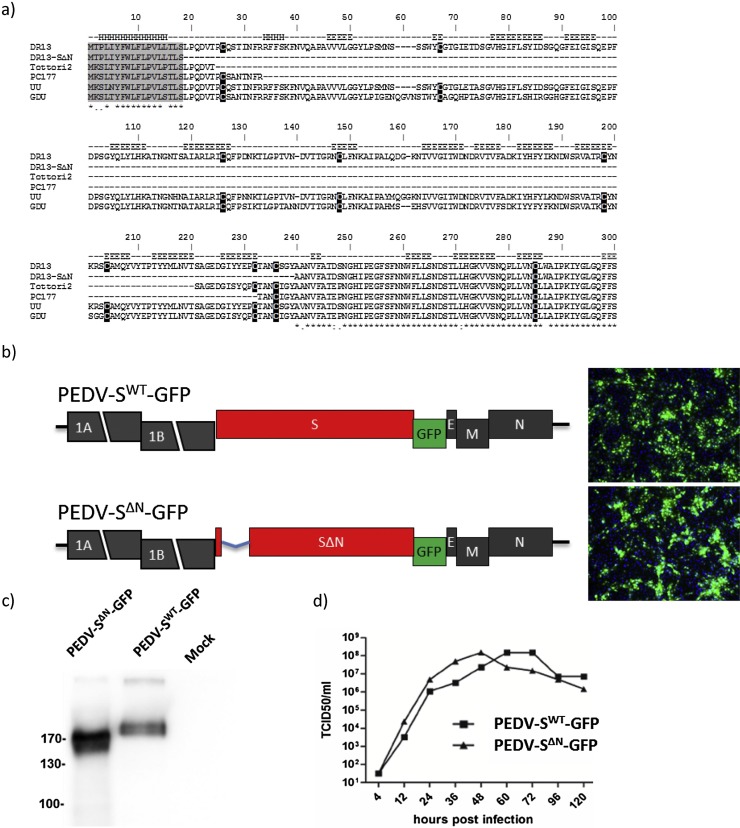
Since N-domain deletions had been observed in spike proteins of natural variants of different alphacoronaviruses, we attempted to generate a recombinant PEDV with a similar deletion using our previously described reverse genetics system based on the cell culture adapted PEDV DR13 strain (caDR13) (Li et al., 2013). We designed a PEDV recombinant with a 215 residues long N-terminal spike deletion (residues 19–233) analogous to the deletion observed in the spike protein of the FIPV strain UU21 (GB: ADC35472.1). The deletion was introduced into the previously described pPEDV-ΔORF3/GFP transfer vector and recombinant viruses were generated using targeted recombination (Li et al., 2013). Recombinant viruses, designated PEDV-SWT-GFP and PEDV-SΔN-GFP, could be rescued, plaque purified and their identity was confirmed by Sanger sequencing and Western blot analysis (data not shown and Fig. 2 c). Propagation of both viruses was compared in Vero cells. Intriguingly, PEDV-SΔN-GFP exhibited growth characteristics similar to that of PEDV-SWT-GFP indicating that deletion of the N-domain did not impair propagation of the PEDV caDR13 strain in vitro (Fig. 2d).
Fig. 2.
Generation and characterization of a recombinant PEDV (PEDV-SΔN) with a large 215-residues long deletion in the N-terminal region of the S protein. a) Amino acid sequence alignment of the N-terminal region of the spike proteins of PEDV variants. The N-terminal region (residues 1–294) of the spike protein of the cell culture adapted DR13 strain (caDR13; GB: JQ023162.1) was aligned with the corresponding spike protein sequences of the caDR13 PEDV-SΔN recombinant described in this study and two earlier described PEDV variants with deletions in the N-domain − PC177 (GB: AKR05184.1) and Tottori2 (GB: BAR92898.1) − as well as of the PEDV-GDU strain (GB: KU985229) and PEDV-UU strain (GB: KU985229). Alphahelical (‘H’) and betasheet ('E') secondary structural elements were predicted using the JPRED4 server (http://www.compbio.dundee.ac.uk/jpred/). Conserved residues and gaps are indicated in the alignment using the ‘*’ and ‘-‘ symbols, respectively. Signal peptide and cysteine residues are marked in grey and black, respectively. b) Schematic representation of the recombinant PEDV genomes carrying the wildtype spike (SWT) gene (PEDV-SWT-GFP) or the SΔN gene (PEDV-SΔN-GFP). For both viruses, the S proteins carried a C-terminal Flag-tag peptide (VQDYKDDDDK) and the ORF3 gene was substituted by the GFP gene (Wicht et al., 2014b). Left panel displays pictures of Vero cells infected with PEDV-SWT-GFP or PEDV-SΔN-GFP taken by fluorescence microscopy. c) Western-blot analysis of the S protein of recombinant PEDV-SWT-GFP and PEDV-SΔN-GFP. Recombinant viruses propagated in Vero (CCL81) cells in the absence of trypsin were semi-purified by sedimentation through a 20% (w/w) sucrose cushion, subjected to Western blot analysis, after which S proteins were detected with an anti-Flag monoclonal antibody (Sigma). d) Multi-step growth kinetics of PEDV-SWT-GFP and PEDV-SΔN-GFP. Vero cells were inoculated with recombinant PEDV (MOI = 0.01) for 3 h in the absence of trypsin, after which the inoculum was replaced by fresh culture medium, following a previously described procedure (Wicht et al., 2014b). Virus infectivity in culture medium was determined at different times p.i. (4, 12, 24, 36, 48, 60, 72, 96 or 120 h p.i.) by a quantal assay on Vero cells from which TCID50 values were calculated.
4.3. Sialic acid binding capacity of S protein differs among PEDV strains
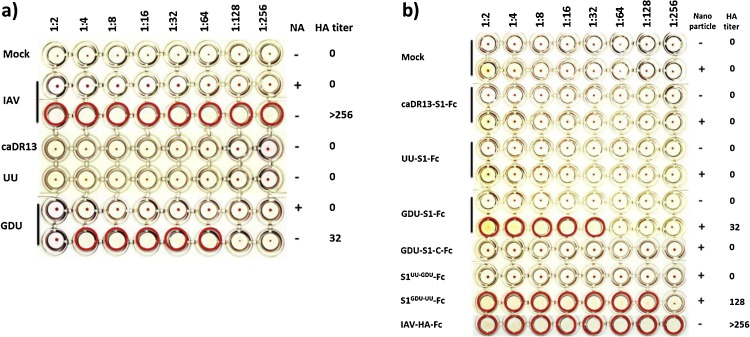
We assessed the sialic acid binding activity of different strains of PEDV using an HA assay. Surprisingly, viruses of different strains displayed remarkable differences in their ability to hemagglutinate human erythrocytes. Apart from the cell culture adapted DR13 strain we used two recent PEDV field isolates from China (strain GDU) and The Netherlands (strain UU, S INDEL type). PEDV-GDU was able to agglutinate human erythrocytes. Hemagglutination was dependent on sialic acids since pretreatment of the erythrocytes with bacterial neuraminidase abolished hemagglutination of PEDV-GDU as well as of influenza virus A (IAV) that was taken along as a positive control. In contrast to PEDV-GDU, the PEDV-caDR13 as well as the PEDV-UU strain did not show detectable HA activity (Fig. 3 a). We confirmed the strain dependent HA activity using a recombinant S1 protein-based HA assay. S1 proteins of different PEDV strains C-terminally fused to the human IgG Fc portion (S1-Fc) were expressed in HEK–293 T cells and subsequently affinity-purified by using protein A sepharose beads as described before (Raj et al., 2013). The PEDV-GDU S1-Fc − expressed as dimers due to the disulphide-linkage of the Fc moiety − was not able to hemagglutinate human erythrocytes presumably due to a too low avidity. To improve the sialic acid binding avidity and increase the sensitivity of the hemagglutination assay, the S1-Fc proteins were complexed via their Fc portion with protein A-coupled, 200nm-sized nanoparticles. We demonstrated that these GDU-S1-Fc loaded nanoparticles were able to hemagglutinate erythrocytes. This indicates that the multivalent interactions between these S1-Fc loaded nanoparticles and sialic acids increases the avidity of binding to erythrocytes. Multimerization on nanoparticles may hence provide a general and versatile method to increase the avidity of receptor-ligand interactions. The similarly multimerized caDR13- or UU-S1-Fc proteins, however, still did not exhibit HA activity, consistent with the observed lack of virus-mediated hemagglutination. Most of the differences (51 out of 69 residues) between the S1 subunits of the hemagglutinating GDU and the non-hemagglutinating UU strain are located in the first 249 residues of S1. In order to map the sialic acid binding domain within the S1 subunit we constructed a S1 chimera (fused to Fc) in which the S1 N-domain (residues 1–249) of the GDU strain was replaced by that (residues 1–246) of the UU strain (S1UU−GDU-Fc), as well as the reciprocal chimera (S1GDU−UU-Fc). Only the S1GDU−UU-Fc protein was able to agglutinate erythrocytes demonstrating that the sialic acid binding activity occurs within the N-terminal 249 residues of the S1 protein (Fig. 3b).
Fig. 3.
PEDV virions of different strains vary in their hemagglutination activity. a) Virions of the PEDV-GDU strain but not of the PEDV-UU or PEDV-caDR13 strain are able to agglutinate human erythrocytes. The HA assay was performed according to a previously described procedure (Park et al., 2010) and the influenza A virus (A/California/07/2009) was taken along as a positive control. Two-fold serial dilutions of viruses (start dilution 1 × 107 TCID50/ml) were made in 50 μl phosphate buffered saline supplemented with 0.1% bovine serum albumin. Neuraminidase (from Arthrobacter ureafaciens, Sigma, cat.no. 10269611001) treated (10 mU ml−1 at 37 °C for 2 h) or mock treated human erythrocytes were washed with phosphate buffered saline. 50 μl erythrocyte suspension (0.5%) was mixed with 50 ul of each virus dilution in V-shaped 96 wells plates and incubated for 2 h on ice after which the wells were photographed. Virus dilutions are indicated above the plate. Wells positive for hemagglutination are encircled in red. b) The Fc-tagged S1 protein of the PEDV-GDU strain but not that of the PEDV-UU or PEDV-caDR13 strain is able to agglutinate human erythrocytes. To enhance the sensitivity of the S1-based hemagglutination assay, we premixed 5 μg of S1-Fc proteins with 1 ul of protein A-coupled, 200nm-sized nanoparticles (nano-screenMAG-Protein A beads; Chemicell GmbH, cat.no. 4503-1) to increase the avidity of S1-Fc proteins for sialic acids on the erythrocyte surface. The influenza A virus hemagglutinin glycoprotein ectodomain (A/California/07/2009, GB: ACP41953.1) fused to the human Fc portion (IAV-HA-Fc) was taken along as a positive control. The start dilution of IAV-HA-Fc and PEDV-S1-Fc was 5 μg and two-fold serial dilutions of virus-nanoparticle mixtures were tested as decribed above.
4.4. Sialic acid binding activity of the PEDV S protein facilitates cell entry
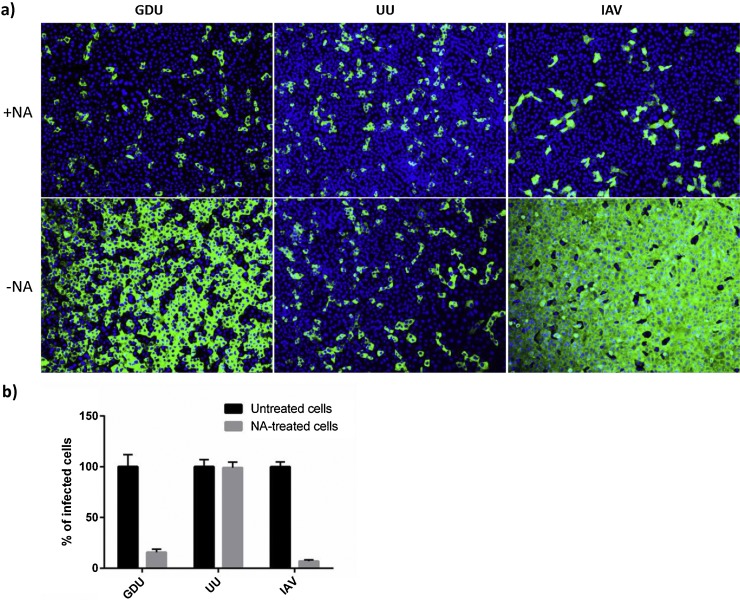
To assess the role of cell surface exposed sialic acids in PEDV infection, Vero cells were pretreated with neuraminidase prior to inoculation with different PEDV strains (Fig. 4 ). The sialic acid dependent influenza A virus (IAV) was taken along as a positive control. The neuraminidase pretreatment greatly reduced infection (6.5 ± 1.1 fold reduction) of the PEDV-GDU strain, to a similar extent as seen for IAV. No effect of neuraminidase pretreatment was observed on infection with the PEDV-UU strain. These results are in accordance with the HA activity of both PEDV strains and indicate a strain-dependent binding and receptor recruitment of sialic acid-containing glycoproteins or glycolipids by PEDV spike proteins during virus entry of Vero cells.
Fig. 4.
Dependency of PEDV on cell surface sialic acids for infection of Vero cells varies between strains. a) Vero cells were pretreated with PBS or PBS containing neuraminidase (100 mU ml−1; Sigma, cat.no. 10269611001) at 37 °C for 2 h. Cells were subsequently inoculated with PEDV-GDU, PEDV-UU or influenza A virus (IAV-WSN) (Baggen et al., 2016) at a multiplicity of infection of 5, 1 or 5, respectively, for 1 h at 37 °C in the presence of trypsin. Inoculum was removed and cells were washed thrice with fresh medium and further incubated at 37 °C in medium supplemented with 1% fetal calf serum. At 14 h p.i. cells were fixed and infected cells were visualized by immunofluorescence staining using a mouse monoclonal antibody detecting PEDV nucleocapsid protein (BioNote, Republic of Korea). b) Percentage of infected cells (relative to PBS-treated) was calculated by counting the infected cells in 10 x microscopic fields. Statistical significance was assessed by unpaired one-tailed Student’s test (* = P<0.01). The infection experiments were repeated three times, and representative images are shown.
4.5. Functional role of the PEDV N-domain
Our data and those of others indicate that the S protein N-domain of at least some PEDV strains is dispensable for replication of PEDV in vitro. The recombinant PEDV-caDR13 mutant lacking a large part (215 a.a., residues 19–233) of its S protein N-terminal region replicated with equal efficiency in Vero cells as the parental virus. Slightly smaller deletions in the N-terminal portion of the PEDV S protein have been documented for the PC177 (197 a.a., residues 34–230) as well as the Tottori2 strain (194 a.a., residues 23–216) (Oka et al., 2014, Masuda et al., 2015). Whereas our caDR13-based recombinant virus was generated by rational design using reverse-genetics, the PC177 deletion mutant most likely arose as a result of adaptation to Vero cells (Oka et al., 2014). That this deletion mutant was apparently selected during passaging suggests that the absence of the N-domain might even have provided an advantage under the used conditions. Intriguingly, the deletion in the spike protein of the Tottori2 strain was already detected in RNA sequenced directly from samples of PEDV-positive pigs showing mild clinical disease, suggesting that this domain is also dispensable in vivo and may reduce virus pathogenicity (Masuda et al., 2015). Yet, the dispensability of this spike domain for virus replication may vary among PEDV strains depending on their ability to recruit sialic acids as a receptor for entry.
Our observations demonstrate that the sialic acid binding activity of the S protein facilitates infection by PEDV. The sensitivity to neuraminidase treatment of PEDV-GDU both in infection and in the HA assay points to sialic acids being used as auxiliary receptors. Different variants of PEDV exhibit different sialic acid binding potential. Contrary to the GDU strain, virions of the caDR13 and UU strain as well as their S1 proteins lacked the ability to hemagglutinate erythrocytes. Consistently, removal of cell surface sialic acids from Vero cells did not affect infection by these two viruses (Fig. 4 and data not shown). The highly passaged caDR13 strain has accumulated numerous mutations in its spike protein some of which may have changed its cellular receptor binding properties. The recent clinical strains GDU and UU have been passaged in cell culture for 24 and 27 passages, respectively, yet their spike N-domain sequences are identical to those of the original isolates (data not shown), excluding cell culture adaptation artifacts. The inability of the UU virus to agglutinate erythrocytes and its independence on cell surface sialic acids in infection indicates that functional receptor recruitment of cell surface sialic acids is not a universal feature of all PEDV strains. Variations in sialic acid binding activity of the S1 proteins of different PEDV strains were also observed using bovine mucin by Deng et al., with a recently isolated chinese PEDV variant showing stronger binding affinity than the prototype Belgium strain CV777 (Deng et al., 2016). We mapped the sialic acid binding activity to the first 246 residues of the S protein by exchanging the N-domains of GDU and UU S1 proteins in DR13-based viruses. Recombinant viruses carrying such chimeric spike proteins with opposite sialic acid dependency might clarify the contribution of sialic acid binding activity to virus propagation in vitro but also in vivo.
The GDU spike protein has high homology (∼99% amino acid sequence identity) to the spike protein of the original highly virulent US strains whereas the UU spike protein (∼94% identity to original US strain) is of the less virulent S INDEL type (Vlasova et al., 2014, Wang et al., 2014, Lin et al., 2015). Most of the variation in the spike proteins of the original virulent US strains and the S INDEL strains maps to the N-terminal region of the S protein (Wang et al., 2014, Lee et al., 2010, Vlasova et al., 2014, Hanke et al., 2015). All amino acid insertions and deletions that characterize the S INDEL strains occur within this region. The coronavirus spike protein is highly immunogenic and the main target for neutralizing antibodies. Differences in neutralizing titers of antisera raised against S proteins of different PEDV subtypes correlated with variation in these spike proteins (Wang et al., 2015). Antigenic variation in the N-domain is consistent with a functional relevance of this domain of the S protein in vivo and may have provided the virus evolutionary advantage in the evasion from adaptive immune responses. The inter-strain variation in sialic acid dependence observed for PEDV has also been seen for viruses of other virus families, including enterovirus 68, human norovirus and T3 reovirus (Baggen et al., 2016, Rydell et al., 2009, Dermody et al., 1990) though the significance of this polymorphism is unknown. Clearly, further studies are needed in vitro and in vivo to functionally assess differences in sialic acid binding activity of S proteins and their consequences for virus infection and pathogenesis.
5. Requirements for proteolytic spike protein activation
5.1. PEDV propagation in cell culture depends on supplemental trypsin
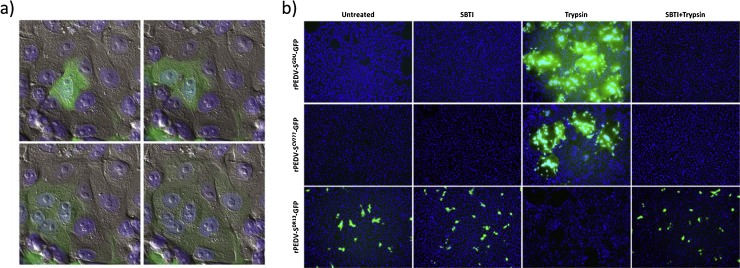
As first demonstrated by Hofmann and Wyler, propagation of PEDV in cultured cells strictly requires the supplementation of trypsin to the culture medium (Fig. 5 ) (Hofmann and Wyler, 1988). Yet, cell culture adaptation of PEDV may result in a trypsin-independent propagation phenotype, as illustrated by the cell-passaged DR13 strain. Analysis of PEDV recombinants carrying spike proteins of trypsin-dependent and trypsin-independent viruses in an isogenic background demonstrated that the differences in trypsin dependency were determined by the spike protein (Fig. 5b) (Wicht et al., 2014b, Shirato et al., 2010). Moreover, inclusion of trypsin after the inoculation stage appeared to be required for cell–cell fusion and syncytia formation (Fig. 5a) (Hofmann and Wyler, 1988, Wicht et al., 2014b). These observations suggest a role of the trypsin in activation of the spike protein’s membrane fusion potential. In addition to cell entry, proteolysis is also required for release of progeny virus from the infected cell (Shirato et al., 2010).
Fig. 5.
Contrasting trypsin-dependency of recombinant PEDV carrying spike proteins from different PEDV isolates. a) PEDV requires proteolysis to activate membrane fusion. The pictures show a PEDV-infected cell fusing with neighbouring cells upon trypsin addition. Snapshots from a time-lapse movie of an infected cell culture taken at 3, 16, 27 and 45 min after addition of trypsin to the culture medium are shown in the following order: upper left, upper right, lower left and lower right, respectively. Pictures represent an overlay of fluorescence (virus encoded-mEGFP labelled cells [green] and Hoechst 33342 stained nuclei [blue]) and DIC images). b) Recombinant viruses encoding spike proteins of PEDV-CV777, PEDV-GDU and the cell-culture adapted DR13 isolate (GB: AF353511, AFP81695.1, JQ023162.1, respectively) were generated in an isogenic background by targeted RNA recombination, as described before (Wicht et al., 2014b). In all recombinant viruses the ORF3 gene was replaced by that of GFP. Vero cells were inoculated with recombinant viruses (MOI 0.1) in the absence or presence of soybean trypsin inhibitor (SBTI) or trypsin, or in the presence of both, as indicated. The cells were fixed at 15 h post infection, nuclei were stained with DAPI (blue) and nuclei and infected cells (GFP; green) were visualized by fluorescence microscopy.
5.2. Cleavage required to activate spike fusion function
Fusion of the coronavirus envelope membrane with a host cell membrane is driven by conformational changes in the spike protein. These conformational changes are irreversible and hence tightly regulated in time and space in order to prevent premature activation of the fusion protein. Conformational changes in the spike protein can be initiated by receptor binding as well as by acidic pH. Similar to other class I viral fusion proteins, the coronavirus spike protein requires proteolytic processing to activate its fusion potential. The spikes of a number of coronaviruses, particularly within the beta- and gammacoronavirus genera, are cleaved during biogenesis in the infected cell at S1/S2 junction by the subtilisin-like proprotein convertase furin(Belouzard et al., 2012). However, the spike protein of PEDV and many other alphacoronaviruses is presented on the virion surface in an uncleaved form. Recently, a second, more universal cleavage site (S2′) has been proposed within the S2 subunit located just upstream of the fusion peptide for some beta- and gammacoronavirus spike proteins including that of MERS-CoV, SARS-CoV, MHV and IBV (Belouzard et al., 2012, Burkard et al., 2014, Heald-Sargent and Gallagher, 2012, Millet and Whittaker, 2014). This cleavage is thought to occur at the cell surface or in the endosome compartment during virus-cell entry. PEDV is an excellent model virus to study proteolytic activation of coronavirus spike proteins, given its unique requirement for supplemental trypsin(-like) proteases for infection in cell culture (68). We and others have shown that this proteolytic activation by trypsin proteases only occurred after the receptor binding stage (Millet and Whittaker, 2014, Shirato et al., 2011b, Wicht et al., 2014b). Treatment of virions or cells with trypsin prior to receptor binding did not rescue infectivity (Wicht et al., 2014a). This indicates that the trypsin cleavage site within the PEDV S protein is inaccessible on the virion, in contrast to other class I viral fusion proteins including influenza virus hemagglutinin which can be proteolytically primed at any stage after folding. The dependency on virus-cell interaction for exposure of the S protein cleavage site might prevent premature triggering of the S protein fusion machinery in the protease-rich intestine. Introduction of a furin cleavage site in the PEDV S protein by a single valine-to-arginine substitution (V888R) at a position N-terminal of the predicted fusion peptide yielded a mutant virus exhibiting trypsin-independent membrane fusion (Li et al., 2015). This observation further supports the hypothesis that cleavage just upstream of the fusion peptide is a general and essential requirement for activation of the CoV spike protein’s membrane fusion function (Belouzard et al., 2009, Yamada and Liu, 2009, Millet and Whittaker, 2014).
5.3. Cleavage as tropism determinant
Replication of PEDV seems to be restricted to the enterocytes of the intestinal epithelium. Multiple factors may determine virus tropism including in particular the availability of functional receptors and fusion-activating proteases. Alterations in cleavage requirements can have a profound effect on tissue tropism and pathogenicity of viruses. The highly pathogenic phenotype of avian influenza viruses is largely determined by the acquisition of a multibasic cleavage site in the HA protein which switches processing of the hemagglutinin protein by tissue resident trypsin-like proteases to the ubiquitously expressed furin-like proteases (Millet and Whittaker, 2014, Yamada and Liu, 2009). The strict requirement of PEDV for supplemental trypsin proteases during its cell entry and release are likely met in vivo by intestine-resident proteases. Gastric and pancreatic proteases or proteases locally expressed by intestinal epithelial cells may facilitate these processes essential for PEDV infection in the animal host (Zamolodchikova, 2012) and hence limit its tropism to the enteric tract.
Acknowledgements
We thank Bas Kolpa and Gilbert van Hagen from the veterinary practice De Oosthof for their help in acquiring the Dutch PEDV-UU isolate. We are indebted to the National Natural Science Foundation of China for financial support (31272572). Berend-Jan Bosch was supported by an ESCMID (European Society of Clinical Microbiology and Infectious Diseases) Research Grant (2015). We thank Mark Bakkers and dr. Raoul de Groot for technical support regarding the nanoparticle-based hemagglutination assay technology.
References
- Baggen J., Thibaut H.J., Staring J., Jae L.T., Liu Y., Guo H., Slager J.J., de Bruin J.W., van Vliet A.L., Blomen V.A., Overduin P., Sheng J., de Haan Xander C.A., de Vries E., Meijer A., Rossmann M.G., Brummelkamp T.R., van Kuppeveld F.J. Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1399–1404. doi: 10.1073/pnas.1524498113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J., Bosch B.J., de Haan C.A. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Bae J., Kang T., Kim J., Chung G., Lim C., Laude H., Yang M., Jang Y. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M., Nkrumah E.E., Maganga G.D., Oppong S., Adu-Sarkodie Y., Vallo P., da Silva Filho L.V., Leroy E.M., Thiel V., van der Hoek L., Poon L.L., Tschapka M., Drosten C., Drexler J.F. Evidence for an ancestral association of human Coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E., Pensaert M.B., Callebaut P., Vandeun K. Intestinal replication of a porcine respiratory Coronavirus closely related antigenically to the enteric transmissible gastroenteritis virus. Vet. Microbiol. 1990;23:237–243. doi: 10.1016/0378-1135(90)90154-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz D.J.M., Kim C., Shin H. Phage-displayed peptides having antigenic similarities with porcine epidemic diarrhea virus (PEDV) neutralizing epitopes. Virology. 2006;354:28–34. doi: 10.1016/j.virol.2006.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastjerdi A., Carr J., Ellis R.J., Steinbach F., Williamson S. Porcine epidemic diarrhea virus among farmed pigs, Ukraine. Emerg. Infect. Dis. 2015;21:2235–2237. doi: 10.3201/eid2112.150272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck P., Pensaert M., Coussement W. The pathogenesis of an enteric infection in pigs, experimentally induced by the coronavirus-like agent, CV 777. Vet. Microbiol. 1981;6:157–165. [Google Scholar]
- Delmas, B., Gelfi, J., L'Haridon, R., Sjöström, H., Laude, H., 1992. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. [DOI] [PMC free article] [PubMed]
- Deng F., Ye G., Liu Q., Navid M.T., Zhong X., Li Y., Wan C., Xiao S., He Q., Fu Z.F. Identification and comparison of receptor binding characteristics of the spike protein of two porcine epidemic diarrhea virus strains. Viruses. 2016;8:55. doi: 10.3390/v8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody T.S., Nibert M.L., Bassel-Duby R., Fields B.N. A sigma 1 region important for hemagglutination by serotype 3 reovirus strains. J. Virol. 1990;64:5173–5176. doi: 10.1128/jvi.64.10.5173-5176.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarets L., Theuns S., Olyslaegers D., Dedeurwaerder A., Vermeulen B.L., Roukaerts I., Nauwynck H.J. Establishment of feline intestinal epithelial cell cultures for the propagation and study of feline enteric coronaviruses. Vet. Res. 2013;44:10.1186. doi: 10.1186/1297-9716-44-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatelle R., Coussement W., Pensaert M., Debouck P., Hoorens J. In vivo morphogenesis of a new porcine enteric coronavirus, CV 777. Arch. Virol. 1981;68:35–44. doi: 10.1007/BF01315165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatelle R., Coussement W., Debouck P., Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets: II. Electron microscopic study. Vet. Pathol. 1982;19:57–66. doi: 10.1177/030098588201900109. [DOI] [PubMed] [Google Scholar]
- Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Esper F., Ou Z., Huang Y.T. Human coronaviruses are uncommon in patients with gastrointestinal illness. J. Clin. Virol. 2010;48:131–133. doi: 10.1016/j.jcv.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsani S.M.J., Dijkman R., Jebbink M.F., Goossens H., Ieven M., Deijs M., Molenkamp R., van der Hoek L. The first complete genome sequences of clinical isolates of human coronavirus 229E. Virus Genes. 2012;45:433–439. doi: 10.1007/s11262-012-0807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T., Perlman S. Public health: broad reception for coronavirus. Nature. 2013;495:176–177. doi: 10.1038/495176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (Coronavirus) spike protein. J. Virol. 1994;68:8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasland, B., Bigault, L., Bernard, C., Quenault, H., Toulouse, O., Fablet, C., Rose, N., Touzain, F., Blanchard, Y., 2015. Complete genome sequence of a porcine epidemic diarrhea s gene indel strain isolated in france in december 2014. Genome Announc 3, 10.1128/genomeA. 00535-15 [DOI] [PMC free article] [PubMed]
- Guo D., Zhu Q., Zhang H., Sun D. Proteomic analysis of membrane proteins of vero cells: exploration of potential proteins responsible for virus entry. DNA Cell Biol. 2014;33:20–28. doi: 10.1089/dna.2013.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke D., Jenckel M., Petrov A., Ritzmann M., Stadler J., Akimkin V., Blome S., Pohlmann A., Schirrmeier H., Beer M., Hoeper D. Comparison of porcine epidemic diarrhea viruses from Germany and the United States, 2014. Emerg. Infect. Dis. 2015;21:493–496. doi: 10.3201/eid2103.141165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X.L., Yu L.Y., Liu J. Development and evaluation of enzyme-linked immunosorbent assay based on recombinant nucleocapsid protein for detection of porcine epidemic diarrhea (PEDV) antibodies. Vet. Microbiol. 2007;123:86–92. doi: 10.1016/j.vetmic.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., Saif L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C., Herrler G. Sialic acid binding activity of transmissible gastroenteritis coronavirus affects sedimentation behavior of virions and solubilized glycoproteins. J. Virol. 2001;75:844–849. doi: 10.1128/JVI.75.2.844-849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C., Schultze B., Laude H., Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997;71:3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.K., Park C.K., Kim S.H., Lee C. Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea. Virus Res. 2010;149:175–182. doi: 10.1016/j.virusres.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Ge J., Li Y. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365:166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li Z., Zou Y., Wicht O., van Kuppeveld F.J.M., Rottier P.J.M., Bosch B.J. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS One. 2013;8:e69997. doi: 10.1371/journal.pone.0069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wicht O., van Kuppeveld F.J.M., He Q., Rottier P.J.M., Bosch B. A single point mutation creating a furin cleavage site in the spike protein renders porcine epidemic diarrhea Coronavirus trypsin independent for cell entry and fusion. J. Virol. 2015;89:8077–8081. doi: 10.1128/JVI.00356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Annamalai T., Liu X., Gao X., Lu Z., El-Tholoth M., Hu H., Saif L.J., Wang Q. Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet. Res. 2015;46 doi: 10.1186/s13567-015-0278-9. (134-015-0278-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015;89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli R., Lavazza A., Nigrelli A.D., Merialm G., Alborali L.G., Pensaert M.B. Epidemic of diarrhoea caused by porcine epidemic diarrhoea virus in Italy. Vet. Rec. 2008;162:307–310. doi: 10.1136/vr.162.10.307. [DOI] [PubMed] [Google Scholar]
- Masuda T., Murakami S., Takahashi O., Miyazaki A., Ohashi S., Yamasato H., Suzuki T. New porcine epidemic diarrhoea virus variant with a large deletion in the spike gene identified in domestic pigs. Arch. Virol. 2015;160:2565–2568. doi: 10.1007/s00705-015-2522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Suo S., Zarlenga D.S., Cong Y., Ma X., Zhao Q., Ren X. A phage-displayed peptide recognizing porcine aminopeptidase N is a potent small molecule inhibitor of PEDV entry. Virology. 2014;456:20–27. doi: 10.1016/j.virol.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita J.R., Hakze-van der Honing R., Almeida A., Lourenco M., van der Poel W.H., Nascimento M.S. Outbreak of porcine epidemic diarrhea virus in Portugal, 2015. Transbound Emerg. Dis. 2015;62:586–588. doi: 10.1111/tbed.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2014 doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina-Osorio P. The moonlighting enzyme CD13: old and new functions to target. Trends Mol. Med. 2008;14:361–371. doi: 10.1016/j.molmed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam E., Lee C. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Vet. Microbiol. 2010;144:41–50. doi: 10.1016/j.vetmic.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Lee K., Choi H., Lee C. Immunogenicity and protective efficacy of recombinant S1 domain of the porcine epidemic diarrhea virus spike protein. Arch. Virol. 2014;159:2977–2987. doi: 10.1007/s00705-014-2163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic D., Hazlett M., Fairles J., Marom A., Slavic D., Maxie G., Alexandersen S., Pasick J., Alsop J., Burlatschenko S. The first case of porcine epidemic diarrhea in Canada. Can. Vet. J. 2015;56:149–152. [PMC free article] [PubMed] [Google Scholar]
- Oka T., Saif L.J., Marthaler D., Esseili M.A., Meulia T., Lin C., Vlasova A.N., Jung K., Zhang Y., Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 2014;173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Shin H. Porcine epidemic diarrhea virus infects and replicates in porcine alveolar macrophages. Virus Res. 2014;191:143–152. doi: 10.1016/j.virusres.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Cruz D.J., Shin H.J. Trypsin-induced hemagglutination activity of porcine epidemic diarrhea virus. Arch. Virol. 2010;155:595–599. doi: 10.1007/s00705-010-0620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Park E., Yu J., Rho J., Paudel S., Hyun B., Yang D., Shin H. Development of transgenic mouse model expressing porcine aminopeptidase N and its susceptibility to porcine epidemic diarrhea virus. Virus Res. 2015;197:108–115. doi: 10.1016/j.virusres.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasick J., Berhane Y., Ojkic D., Maxie G., Embury-Hyatt C., Swekla K., Handel K., Fairles J., Alexandersen S. Investigation into the role of potentially contaminated feed as a source of the first-Detected outbreaks of porcine epidemic diarrhea in Canada. Transbound. Emerg. Dis. 2014;61:397–410. doi: 10.1111/tbed.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M., De Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranaveja S., Poollperm P., Lertwatcharasarakul P., Kesdaengsakonwut S., Boonsoongnern A., Urairong K., Kitikoon P., Choojai P., Kedkovid R., Teankum K., Thanawongnuwech R. Chinese-like strain of porcine epidemic diarrhea virus, Thailand. Emerg. Infect. Dis. 2009;15:1112–1115. doi: 10.3201/eid1507.081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J., Santiago C., Mudgal G., Ordoño D., Enjuanes L., Casasnovas J.M. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8:e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydell G.E., Nilsson J., Rodriguez-Diaz J., Ruvoen-Clouet N., Svensson L., Le Pendu J., Larson G. Human noroviruses recognize sialyl Lewis x neoglycoprotein. Glycobiology. 2009;19:309–320. doi: 10.1093/glycob/cwn139. [DOI] [PubMed] [Google Scholar]
- Sanchez C.M., Gebauer F., Sune C., Mendez A., Dopazo J., Enjuanes L. Genetic evolution and tropism of transmissible gastroenteritis Coronaviruses. Virology. 1992;190:92–105. doi: 10.1016/0042-6822(92)91195-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B., Krempl C., Ballesteros M.L., Shaw L., Schauer R., Enjuanes L., Herrler G. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J. Virol. 1996;70:5634–5637. doi: 10.1128/jvi.70.8.5634-5637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Weßels C., Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 2006;23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Wessels C., Bauer S., Winter C., Enjuanes L., Laude H., Herrler G. The sialic acid binding activity of the S protein facilitates infection by porcine transmissible gastroenteritis coronavirus. Virol. J. 2011;8:435. doi: 10.1186/1743-422X-8-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Maejima M., Hirai A., Ami Y., Takeyama N., Tsuchiya K., Kusanagi K., Nunoya T., Taguchi F. Enhanced cell fusion activity in porcine epidemic diarrhea virus adapted to suckling mice. Arch. Virol. 2010;155:1989–1995. doi: 10.1007/s00705-010-0790-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Matsuyama S., Ujike M., Taguchi F. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J. Virol. 2011;85:7872–7880. doi: 10.1128/JVI.00464-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Maejima M., Matsuyama S., Ujike M., Miyazaki A., Takeyama N., Ikeda H., Taguchi F. Mutation in the cytoplasmic retrieval signal of porcine epidemic diarrhea virus spike (S) protein is responsible for enhanced fusion activity. Virus Res. 2011;161:188–193. doi: 10.1016/j.virusres.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrigl A., Fernández S.R., Stoiber F., Pikalo J., Sattler T., Schmoll F. First detection, clinical presentation and phylogenetic characterization of Porcine epidemic diarrhea virus in Austria. BMC Vet. Res. 2015;11:1. doi: 10.1186/s12917-015-0624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Straw, B.E., Zimmerman, J., D'Allaire, S., Taylor, D., 2006. Diseases of swine 9 th edition. Diseases of Swine 9th Edition, 387–395
- Sun D., Feng L., Shi H., Chen J., Cui X., Chen H., Liu S., Tong Y., Wang Y., Tong G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008;131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Cai R., Chen Y., Liang P., Chen D., Song C. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuns, S., Conceicao-Neto, N., Christiaens, I., Zeller, M., Desmarets, L.M., Roukaerts, I.D., Acar, D.D., Heylen, E., Matthijnssens, J., Nauwynck, H.J., 2015. Complete genome sequence of a porcine epidemic diarrhea virus from a novel outbreak in belgium, january 2015. Genome Announc 3, 10.1128/genomeA. 00506-15. [DOI] [PMC free article] [PubMed]
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, may 2013–February 2014. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B., Frenz B., Rottier P.J., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016 doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen J., Shi D., Shi H., Zhang X., Yuan J., Jiang S., Feng L. Immunogenicity and antigenic relationships among spike proteins of porcine epidemic diarrhea virus subtypes G1 and G2. Arch. Virol. 2015 doi: 10.1007/s00705-015-2694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicht O., Burkard C., de Haan C.A., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Identification and characterization of a proteolytically primed form of the murine coronavirus spike proteins after fusion with the target cell. J. Virol. 2014;88:4943–4952. doi: 10.1128/JVI.03451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicht O., Li W., Willems L., Meuleman T.J., Wubbolts R.W., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014;88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Liu D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009;83:8744–8758. doi: 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager, C.L., Ashmun, R.A., Williams, R.K., Cardellichio, C.B., Shapiro, L.H., Look, A.T., Holmes, K.V., 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. [DOI] [PMC free article] [PubMed]
- Zamolodchikova T. Serine proteases of small intestine mucosa—localization, functional properties, and physiological role. Biochemistry (Moscow) 2012;77:820–829. doi: 10.1134/S0006297912080032. [DOI] [PubMed] [Google Scholar]
- Zeng S., Zhang H., Ding Z., Luo R., An K., Liu L., Bi J., Chen H., Xiao S., Fang L. Proteome analysis of porcine epidemic diarrhea virus (PEDV)‐infected Vero cells. Proteomics. 2015;15:1819–1828. doi: 10.1002/pmic.201400458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yao Y., Gao X., Wang Y., Jia X., Xiao Y., Wang T., Li X., Tian K., 2016, Development of a Neutralizing Monoclonal Antibody Against Porcine Epidemic Diarrhea Virus S1 Protein, Monoclonal Antibodies in Immunodiagnosis and Immunotherapy [DOI] [PubMed]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Middle east respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus study group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Haijema B.J., Schellen P., Wichgers Schreur P., te Lintelo E., Vennema H., Rottier P.J. Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J. Virol. 2008;82:6078–6083. doi: 10.1128/JVI.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]