Highlights
-
•
Recent progress in techniques for isolation of hbnAbs has facilitated the study of passive immunization.
-
•
The current effective techniques in broadly neutralizing antibody isolation are reviewed.
-
•
Functional study of neutralizing epitopes provides key knowledge for efficient vaccine design.
Abbreviations: bNAbs, broadly neutralizing antibodies; hbNAbs, human broadly neutralizing antibodies; bnmAbs, broadly neutralizing monoclonal antibodies; Env, envelope
Keywords: Technology, Monoclonal antibody, Phage/yeast display, Neutralization, Single-cell sorting
Abstract
HIV/AIDS has become a worldwide pandemic. Before an effective HIV-1 vaccine eliciting broadly neutralizing monoclonal antibodies (bnmAbs) is fully developed, passive immunization for prevention and treatment of HIV-1 infection may alleviate the burden caused by the pandemic. Among HIV-1 infected individuals, about 20% of them generated cross-reactive neutralizing antibodies two to four years after infection, the details of which could provide knowledge for effective vaccine design. Recent progress in techniques for isolation of human broadly neutralizing antibodies has facilitated the study of passive immunization. The isolation and characterization of large panels of potent human broadly neutralizing antibodies has revealed new insights into the principles of antibody-mediated neutralization of HIV. In this paper, we review the current effective techniques in broadly neutralizing antibody isolation.
1. Introduction
Antibody responses to neutralize human immunodeficiency virus-1 (HIV-1) are mediated by direct binding to viral spikes, which are trimers composed of glycoproteins gp120 and gp41 (Pincus et al., 2017a, Pincus et al., 2017b, Blair et al., 2007, Morris et al., 2000, Micoli et al., 2000, Pegu et al., 2017, Haynes and Mascola, 2017, Liao et al., 2004, Brodine et al., 2003, Ward and Wilson, 2017, Debnath et al., 1994, Moore et al., 1993). GP120 with the attachment of virus particles to the target cells, and the gp41 engages in virus-cell fusion (Durham and Chen, 2016, Desai et al., 2015, Dale et al., 2011, Bieniasz et al., 1997, Rausch et al., 1990). While successful in-vitro, recombinant gp120 has failed to generate cross neutralizing antibodies in vivo possibly due to the natural conformation of gp120 (Prevost, 2017, Wang et al., 2016, Doores et al., 2015, Doores et al., 2010, Zhou et al., 2010, Wu et al., 1997, Watkins et al., 1996, Robert-Guroff et al., 1992). Monoclonal antibodies were first generated in mice in 1975 using a hybridoma technique (Patke et al., 2017, Holzlohner and Hanack, 2017, Bhatia et al., 2016, Pogson et al., 2016, Abusneina and Gauthier, 2016, Milstein, 1999), and were subsequently accepted globally by manufacturers. Now, it is well understood that rodent and murine derived antibodies are immunogenic in humans; when binding to antigens within the human body, the complexes can be recognized as immunogenic agents. To solve this problem, human equivalent forms were generated from rodent or murine antibodies, known as humanized antibodies (Pardi et al., 2017, Wiehe et al., 2017, Rothenberg, 2016, Bhowmick et al., 2016, Hamanoue et al., 2016). In recent years, new techniques have evolved to isolate human monoclonal antibodies against HIV-1 directly which offer an advantage of reduced immunogenicity.
A panel of HIV-1 specific potent broadly neutralizing antibodies has been isolated in recent decades. The first generation antibodies, 2F5, 2G12 and 4E10, were used in combination for passive immunization (Morris et al., 2014, Mehandru et al., 2004, Armbruster et al., 2004). A second generation antibody, IgG b12, was observed to neutralize numerous virus isolates with IC50 in nM level (1 nM is equal to about 0.15ug/ml) for the first time (Li et al., 2017, Bunnik et al., 2010, Ashish et al., 2010, Zwick et al., 2003, Sun et al., 2015). The new generation of potent broadly neutralizing antibodies, including VRC01-class antibodies, NIH45-46, PG9/16,10E8, BG18, IOMA, 35O22, ACS202, can neutralize virus isolates ranging from 73% to 98% with IC50 less than 50 μg/ml, and can neutralize 59%–72% of virus isolates with IC50 less than 1 μg/ml (Morales et al., 2016, Huang et al., 2012, Braibant et al., 2013, Thenin et al., 2012, Ringe et al., 2012, Euler et al., 2011, Davenport et al., 2011, Pancera et al., 2010, Wu et al., 2010, Scheid, 2015, Scheid et al., 2011).
Worth noting, broadly neutralizing antibody 10E8 neutralizes 98% of tested viruses with geometric mean IC50 of 0.22 μg/ml. 10E8 binds to the cell-surface envelope as opposed to phospholipids. The binding of 10E8 and MPER reveals a highly conserved gp41-hydrophobic residue and a critical arginine or lysine just before the transmembrane region. The breadth and potency of 10E8 demonstrates a conserved site for HIV neutralization and potential target for HIV vaccine design (Huang et al., 2012). The highly potent VRC01-class broadly neutralizing antibodies (bnAbs) individually neutralize up to 90% of HIV-1 isolates. VRC01-class antibodies are considered a sub group of bnAbs and have already been isolated from several HIV-1 infected patients. They each neutralize different virus isolates through the interaction with HIV gp120 CD4 binding site. VRC01-class bnAbs are derived from human VH1-2 gene family, which occupies 2% of the human antibody repertoire. Structural studies revealed that these VRC01-class Abs interact with gp120 CD4 binding site by mimicking the CD4 molecule; specifically the secondary structure formed by heavy chain structure and CDR3 in light chain. Additionally, an important deletion in CDRL1 present in many VRC01-class Abs can facilitate the interaction with the CD4 binding site and avoid conflict with glycan at Asn276 (N276) on loop D (Zhou et al., 2010, Georgiev et al., 2014, Wu et al., 2015, Zhou et al., 2013). Next-generation sequencing (NGS)-derived sequencing reveals that VRC01 lineage is comprised of at least six distinct heavy chain clades and five light chain clades. This observation suggests that extraordinary variation in antibody immunity may only occur within a few antibody lineages − or even a single lineage (Wu et al., 2015). Generation of an unmutated common ancestor of VRC01-class antibodies could be a potential mechanism for opposing HIV-1. After stimulation by virus and co-evolution with the virus, the unmutant common ancestor has ability to mature to bnAbs.
Human antibodies can be elicited by B cells to eliminate “foreign immunogens” directly or indirectly. Some antibodies can be produced to neutralize foreign antigens by various mechanisms, while some non-neutralizing antibodies can perform effector functions such as mediating NK cells via antibody-dependent cellular cytotoxicity (ADCC) (Liao et al., 2004, Lin et al., 1998, Thomann et al., 2015, Ito et al., 2009, Mandi et al., 1982). Serum immunoglobulins produced naturally by human immunity have been applied in passive immunotherapy against many infectious diseases since 1990. But this approach suffers a number of problems. Not only have these polyclonal immunoglobulins displayed mixed efficacies, but they also pose the potential threat of blood-borne pathogen transmission. In addition, the resource of serum is limited and expensive. These disadvantages limit the application of polyclonal immunotherapy (Berry, 2017, Stevens et al., 2016, Wang et al., 2007, Sili et al., 2003, Lang et al., 1993). Recent technology developments have made it possible to isolate human antibodies against HIV-1 from both in vivo and in vitro sources. These improved techniques include display methods, Epstein-Barr virus (EBV) immortalization, classical hybridoma procedures, and single-cell sorting followed by molecular cloning.
2. Techniques to make human monoclonal antibodies
2.1. Hybridoma technique, B cell immortalization and microneutralization
In the mid-1970s, hybridoma technology was invented for production of mouse monoclonal antibodies with defined antigen specificities and neutralization activity for application in clinical therapies (Kelso et al., 2016, Hugwil, 2013, Tomita and Tsumoto, 2011, Martin-Lopez et al., 2007, Hencsey et al., 1996, Honda et al., 1990, Köhler and Milstein, 1975). Hybridomas can be generated by effective fusion of B cells and partner cells followed by screening of individual antibody producing cells. Production of monoclonal antibody in individual hybridoma cells can be easily quantified by surface plasmon resonance imaging (Stojanovic et al., 2015). B cell hybridomas can be an important source for screening of monoclonal antibodies. High-throughput screening is used to characterize mouse IgG antibodies; including sub-isotypes, binding activity, and neutralizing activities (Liu et al., 2015, Szafran et al., 2016). Fluorescent antigen can be used to sort antigen binding hybridoma cells from a mixture as opposed to the traditional way of screening using multi-micro well plate screening and limiting dilution. As an extension of this technology, T cell hybridoma was also generated (Krishnan et al., 2015). However, mouse monoclonal antibodies were shown to be problematic in humans due to their immunogenic properties and low effector function. High immunogenicity prevented their application in humans where prolonged dosing was required. Differences in sequence and glycosylation pattern of the Fc region make rodent mAbs poorly effective in mediating effector functions in human (Li et al., 2006). Human B cells proved to be difficult to immortalize using hybridoma technique (Li et al., 2006, Cafri et al., 2013, Park et al., 1997). The primary issue was that mAbs produced by human B cell hybridoma cells could react against self-antigens. In addition, the lack of sufficient maturation outside of the germinal centers and low binding affinities of mAbs derived by hybridoma, meant that only 1.5% of screened clones typically reacted to the target antigen. Due to these problems, antibody humanization was developed as an alternative to generate human format antibodies from rodent derived antibodies (Martin and Rees, 2016, Choi et al., 2015, Olimpieri et al., 2015, Safdari et al., 2013, Tsurushita et al., 2004, Ohtomo et al., 1995). Humanized antibodies are produced by methods of genetic engineering and can potentially reduce the immune response against non-human derived antibodies. This is done by combining the variable (V) region binding domain of a rodent antibody with human antibody constant (C) regions. This kind of chimeric antibody retains the binding specificity with less immunogenicity to humans. In some cases, the chimeric antibodies retain the effects in mediating human complement-dependent cytotoxicity (CDC) or ADCC. It is also possible to produce a humanized antibody without creating a chimeric intermediate first. Once the precise sequences of the desired CDRs are known, these DNA sequences can be directly cloned into antibody expression vectors with human antibody “scaffold”. A wide variety of human antibody expressing vectors has been developed using this method. For example, PDR12 vector is a human IgG1 antibody expressing vector which was originally used for expression of human HIV-1 potent neutralizing antibody IgG b12. Antibody VRC01 was cloned into a different cloning vector described previously by Tiller et al. (Tiller et al., 2008).
Another method to immortalize human memory B cells is by Epstein-Barr virus (EBV) mediated transformation (Lee et al., 2011, Klein, 1996, Straub and Zubler, 1989, Ohashi et al., 2017, McLaughlin et al., 2017, Traggiai et al., 2004). The C3b, EBV-binding receptor positive B cells can be immortalized to generate antibodies for isolation using single cell sorting, or can be cultured with feeder cells for further screening or cloning. The optimized methodology is to activate B cells using Toll-like receptor 9 (TLR9) agonist CpG DNA before or during EBV infection, which has been used to isolate human monoclonal antibodies against various pathogenic viruses (Katsumura et al., 2012, Bass and Darke, 2004, Sun et al., 2017). It has also been reported that human B cells can be immortalized by transforming a retrovirus encoding anti-apoptotic factor B cell lymphoma 6 (BCL6) and BCL-XL in the presence of interleukin 21 (IL-21) and CD40 ligands (Schrader et al., 2012, Rydstrom et al., 2010, Kusam et al., 2009). This process can trigger memory B cells to differentiate into antibody secreting cells. The immortalization of human B cells provides a substantial resource of human B cells for the subsequent screening. After culturing for 1–2 weeks, antibodies can be collected and isolated from the supernatant to perform neutralization against different viruses. Immortalized B cells can also be sorted by antigen using flow-based technology, and the heavy and light chain genes of antibodies secreted by immortalized B cells can then be cloned. The famous 2G12, 2F5 and 4E10 antibodies are generated by this method (Buchacher et al., 1994). However, due to the low efficiency of EBV-induced B cell transformation, this method has limitation in application. Though optimization methods were studied based on EBV-induced B cell transformation, the efficiency still needs a dramatic improve (Sun et al., 2017, Lu et al., 2016, Kwakkenbos et al., 2016). Some reports show that memory B cells can be cultured short-term, and the supernatant can be used directly for screening of neutralization activity in a high throughput neutralization assay (micro-neutralization). A number of famous neutralizing antibodies have been isolated based on these methods including PG9, PG16, PGT121-128 and 10E8 (Huang et al., 2012, Ringe et al., 2012, Walker et al., 2011, Walker et al., 2009, Falkowska et al., 2014).
HIV-1 potent neutralizing antibodies PG9 and PG16 are classic antibodies isolated by microneutralization. These were originally isolated from 30,000 activated memory B cells from one clade A infected donor screened from approximately 1800 characterized HIV infected donors. The memory B cells were cultured at clonal density for the purpose of cloning antibody heavy and light chain pair from each culture well. PG9 neutralized 127 out of 162 viral isolates and PG16 neutralized 119 out of 162 viruses with a median potency (IC50) less than 0.2 μg/ml. 10E8 is an anti-gp41 antibody isolated from 16,500 memory B cells, sorted and cultured with IL-2, IL-21, and CD40-ligand expressing feeder cells. 10E8 neutralized 98% of the tested pseudoviruses with an IC50 below 50 μg/ml, and 72% of the tested viruses with an IC50 values below 1 ug/ml.
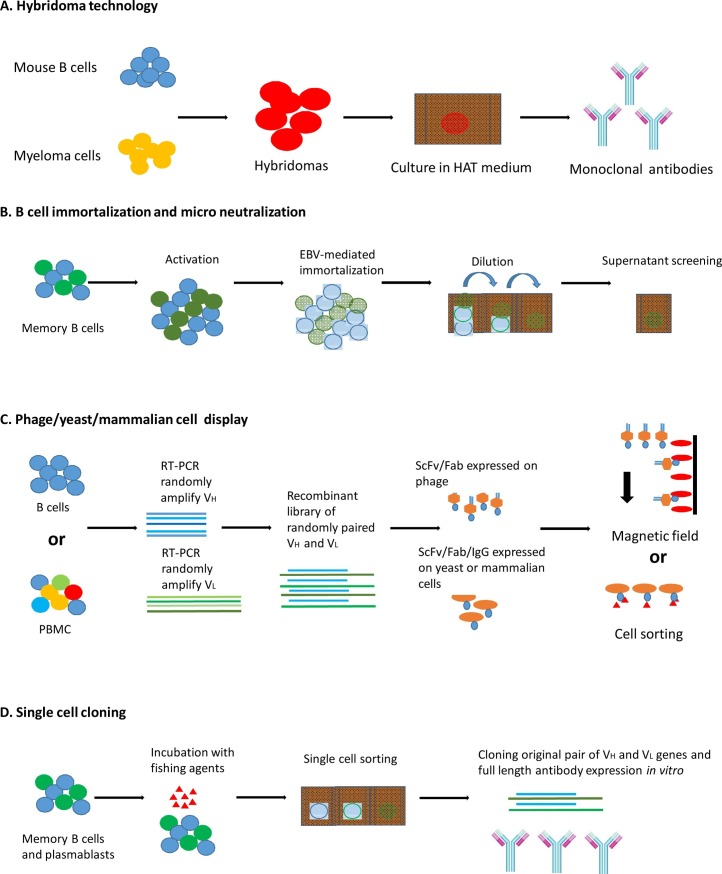
B cell culture and microneutralization can isolate potent broadly neutralizing HIV-1 antibodies. However, the large scale screening of B cells makes this method labor intensive and costly, in addition to low yield; typically ten antigen-specific clones can be isolated out of ten thousands of B cells (Fig. 1 A and B). In current reports, EBV transformation was optimized with efficiency from 0.1%-2% to 7.8%, which enabled the generation of immortalized B cell libraries. However the efficiency still needs to be improved to enable greater B cell survival (Sun et al., 2017).
Fig. 1.
Methods currently used in human monoclonal antibody screening.
2.2. Phage/yeast display
Phage display is a library method which permits screening of antibodies from a large recombinant library (Falkowska et al., 2014, Boots et al., 1997, Chan et al., 1996, Aghebati-Maleki et al., 2017, Rahbarnia et al., 2017, Finlay et al., 2017). Antibodies can be displayed in the form of either single chain variable fragments (ScFv) or antigen-binding fragments (Fab). Recent reports show that single domain antibody can also be displayed in phage display system (Duarte et al., 2016, Kazemi-Lomedasht et al., 2016, Li et al., 2016, Rotman et al., 2015, Tang et al., 2013). Many antibodies against different viruses including rabies virus, severe acute respiratory syndrome (SARS) virus, Ebola virus, yellow fever virus, hepatitis C virus, dengue virus, hepatitis A virus, influenza virus, and HIV, have been successfully isolated using this technology. The principle for phage display technique in isolating antibodies is to construct a phage display recombinant antibody library first. Recombinant antibody library can increase the diversity of antibodies in B cell repertoires, which promotes the screening of antibodies with novel properties (Graus et al., 1998). Antibody libraries are in general constructed by randomly assembling antibody heavy and light chain variable region, and through gene shuffling of the heavy and light chain to further increase the diversity. In summary, phage display creates greater diversity of antibodies and provides rapid and high throughput way of screening.
M43 and m44 are HIV-1 cross-reactive human monoclonal antibodies isolated from a recombinant phage display library by competitive antigen panning (Zhang et al., 2012, Zhang et al., 2008, Zhang et al., 2006, Zhang et al., 2004a, Zhang et al., 2004b). M44 is a gp41 specific cross-reactive antibody, and m43 can recognize both gp120 and gp41. B12 is another potent neutralizing antibody isolated by phage display from recombinant antibody libraries, and it is considered one of the most potent neutralizing antibodies isolated by phage display, as it can neutralize about 40% of known HIV-1 isolates (Li et al., 2017, Mantis et al., 2007, Haussner et al., 2017, Wu et al., 2009). M43, m44 and b12 can be considered standard antibodies isolated by phage display; however the potencies are much lower than antibodies isolated by micro neutralization, such as PG9/16 and/or 10E8.
Phage display selects antibodies in vitro, and antibodies expressed in phage system may not necessarily represent those in mammalian cells due to the difference in protein folding and post transcriptional modifications. It is difficult to draw direct conclusions and gain an understanding of the natural occurrence of antibody heavy and light chain selection based on phage display alone (Zhang et al., 2003). To avoid these limitations, other display methods have been developed to display antibodies on the surface of yeast or mammalian cells, which can be sorted by flow cytometry according to antigen specificity. This is known as yeast surface display, which has become a powerful engineering tool for displaying recombinant proteins on the surface of Saccharomyces cerevisiae via genetic fusion (Traxlmayr and Shusta, 2017, Mei et al., 2017, Andreu and Del Olmo, 2017, Sheehan and Marasco, 2015, Gera et al., 2013, Boder et al., 2012). Yeast surface display is a eukaryotic expression system with the capability to induce post translational modifications on recombinant antibodies. Each yeast cell expresses fusion recombinant proteins which allows for the application of cell sorting for a specific antigen. Yeast display system was not only reported used for isolation of antibodies, but also reported to be used in displaying antigens and other recombinant proteins (Srivastava et al., 2013, Guo et al., 2015). There is a panel of yeast display antigen library which has been constructed, including yeast display H1N1 antigen epitope library, and yeast display SIV antigen epitope library for epitope mapping.
HIV-1 specific antibodies isolated by display techniques are less potent than those isolated by micro neutralization or single B cell sorting and cloning. However in recent studies, phage/yeast display showed great potentials in isolating single domain antibodies from recombinant libraries. Single-domain antibodies are considered a separate class of antibodies composed of antibody fragments consisting of a single monomeric antibody domain. Single-domain antibodies allow a broad range of applications due to their small molecular mass and size, efficient production, and high affinity. Single domain antibodies can be labeled using fluorescent molecules for diagnostic purpose or biotechnological usage. With the conjunction of drug or toxin, single domain antibodies can also be used for therapeutic application. The most well-studied single domain antibodies are antibody heavy-chain variable domains. The DNA sequence will be optimized in order to improve the stability. In addition to heavy-chain variable domains, there are other types of single domain antibodies which have been studied, including CH2 antibodies, and light-chain variable domain antibodies, and some other forms of nanobodies (Duarte et al., 2016, Li et al., 2016, Gong et al., 2016, Louis et al., 2014, Lulf et al., 2014, Bouchet et al., 2012, Bouchet et al., 2011, Boudet et al., 1995).
Mammalian cell display is a powerful method for the isolation of antibodies in scFv format or full length IgG with high affinity. It has been shown that single chain antibodies can be displayed on the surface of human HEK-293T cells and used for affinity maturation (Soga et al., 2015, Tomimatsu et al., 2013, Beerli et al., 2008). Mammalian cell expression system ensures that all of the cellular components will be involved in process of antibody synthesis. Isolation of human antibodies by mammalian cell display can benefit from the advantages of mammalian cell expression system.Mammalian cell display relies on the transfection of antibody expression vectors. In comparison with the yeast display system, mammalian cell expression system allows multiple recombinant protein/antibody delivering vectors in one cell. Thus a further modification, enrichment, and single clone isolation will be needed for panning purposes (Fig. 1C).
2.3. Flow cytometry based single cell sorting and cloning
An important advance in antibody screening and cloning technology is the development of single cell sorting and single cell reverse transcription PCR (Ouisse et al., 2017, Evans et al., 2015, Battye et al., 2000). Human single B cells with different antigen specificities can be sorted by flow cytometer directly, and the original VH and VL pairing of the antibody from single human B cells can be amplified in a high efficient way, with the requirement of relatively few cell numbers. This methodology is quite efficient in obtaining antibody heavy and light chain from extremely rare and highly discrete B cell subpopulations (Fig. 1D). Memory B cells and plasma cells are ideal cell types for the purpose of monoclonal antibody cloning due to the specificity of the cell types. This method is always combined with single cell cloning after flow cytometry based single cell sorting to sort the antigen specific B cells, thus accurate probe design for sorting is essential.
The gp120 or gp41 proteins are not ideal probes, because they are reactive with many HIV-1 antibodies, including non-neutralizing but strong binding antibodies (Wu et al., 2010, Wu et al., 2015, Zhou et al., 2015). One successful example of probe design in fishing potent broadly neutralizing HIV-1 monoclonal antibodies is the agents’ pair of RSC3 and ΔRSC3. RSC3 and ΔRSC3 are computer-assisted designed for sorting of individual B cells expressing CD4bs antibodies. The resurfaced stabilized core 3 (RSC3) is a functional structure core of the CD4 binding site with loop deletion. RSC3 preserved the antigenic structure of the CD4 binding site and eliminated other antigenic regions by substitution with simian immunodeficiency virus (SIV) or non–HIV-1 residues. ΔRSC3 contains one amino acid deletion (at position 371) in RSC3. This single amino acid mutation can knock out the function of CD4 binding site. CD4 binding site antibodies like VRC01 and b12 can bind efficiently with RSC3 but fail to bind with ΔRSC3. Potent broadly neutralizing antibody VRC01 is isolated through the single B cell positive sorting by RSC3 and negative sorting by ΔRSC3. VRC01 neutralized up to 90% of the tested pseudoviruses with an IC50 below 50 μg/ml, and 70% of the tested viruses with an IC50 values below 1 ug/ml (Wu et al., 2010, Wu et al., 2015, McCoy and Burton, 2017).
BG18, IOMA are another two successful bnmAbs isolated by reasonable probe design and single cell sorting/cloning (Gristick et al., 2016, Freund et al., 2017). N-glycans on the trimeric envelope glycoprotein (Env) can be accommodated by broadly neutralizing antibodies. An engineered crystal structures of the HIV-1 Env trimer with an exposed native glycan shield of high-mannose and complex-type N-glycans was used to fishing out the bnmAbs. IOMA, a new CD4-binding site (CD4bs) antibody was thus defined. The heavy chain of IOMA derives from VH1-2*02, which is considered the germline gene of VRC01-class bNAbs, but its light chain lacks the short CDRL3 which defines VRC01-class bNAbs. BG18, BG1, and NC37 are three bNAbs isolated by single IgG+ B cell sorting by using four different fishing agents in four separate sorts: 2CC core, gp140YU2, a 1:1 mixture of gp14092UG37.8 (clade A) + gp140CZA79012 (clade C), and BG505 SOSIP.664 (Sok et al., 2014, Dey et al., 2009, Yang et al., 2000, Sanders et al., 2013, Kovacs et al., 2012). Antibody BG18 (VH4-4 and VL3-25) targeted the glycan-V3 portion of envelope, can neutralize 64% of viruses in a 118-virus panel tested with a geometric mean IC50 of 0.03 mg/ml. Antibodies NC37 (VH1-46/1-2, VK3-20) and BG1 (VH3-49 and Vk1-49) showed geometric mean IC50 of 0.3 and 0.67 mg/ml respectively. NC37 can bind to a quaternary trimer epitope, the core of which overlaps with the CD4bs, and BG1 binds to the V1V2 region.
Antigen baiting and fishing has proven quite successful in isolating HIV-specific human monoclonal IgG expressing B cells and IgM expressing B cells from infected donors using cleverly engineered antigen probes. Flow cytometry based single cell sorting and cloning can also be applied in understanding the natural selection of antibodies by cloning of antibodies in B cells from different infection stage from one donor or B cells in the same stage from different donors. 454 deep sequencing allows for the study of the maturation of a specific antibody population and corresponding B cell subpopulations by the stimulation of virus isolates in progressingstages of infection (Morris et al., 2017, Banerjee et al., 2017, Carter et al., 2015, Karlsson Hedestam et al., 2017, Soto et al., 2016, Stramer et al., 2016, Zhu et al., 2013, Zhu et al., 2012).
2.4. HIV-1 specific bNAbs isolation from immunized animals
The difficulty in eliciting bNAbs in immunized animals has been concluded (Walker et al., 2011, Sanders et al., 2013, Sok et al., 2017). The enormous antigenic diversity of the envelope glycoprotein and the N-linked glycan shield are considered as two major points. Recent study shows that success of rapid elicitation of broadly neutralizing antibodies to HIV-1 by immunization in cows. JR-FL gp120 and BG505 SOSIP are immunized to cows, and strong immune response is observed. And broadly neutralizing antibodies were isolated from peripheral blood mononuclear cells (PBMCs) by sorting with biotinylated BG505 SOSIP. The broadly neutralizing antibody NC-Cow1 displayed a 72% neutralization breadth with a potent median IC50 of 0.028 μg /ml on a 117-virus panel tested.This study gives lights in investigating the generation of antibodies against pathogens that have evolved to avoid human antibody responses in animals.
3. Conclusion
In recent decades, the technology advances have allowed for specific human antibodies to be isolated directly from human B cells and/or from recombinant libraries. These isolated human monoclonal antibodies can be used in therapy because of their safety, efficiency, specificity and tolerance in humans; thus are a powerful tool in fighting against pathogens. Among the developed antibody isolation methods over the last decade, it is difficult to highlight one as the sole preferred method. Different methodology has different advantages. Display techniques basing on recombinant library can be used for isolating single domain antibodies with enlarged library size. But this technique cannot provide us the information of the original pair of heavy chain and light chain of a specific antibody. Single B cell sorting and culturing facilitate the rapid cloning of potent neutralizing antibodies with the requirement of large labor cost. Reasonable design of probes and the development of native-like stabilized trimers have greatly improved efficiency in screening of functional envelope-binding B cells. Thus it is hard to conclude the best way to generate novel bnAbs. Combined methods can be used in screening and isolating potent neutralizing antibodies in order to maximize the advantages of different methods. Meanwhile, the development of novel methodology of broadly neutralizing antibodies isolation and screening are encouraged to broaden our knowledge and enlarge the recourses of antibody classes. A purposed method basing on single cell sorting and viral neutralization instead of fishing agents should provide a direct and efficient way to minimum the workload.
In addition to passive treatment and presentation, another important purpose of isolation and characterization of potent broadly neutralizing antibodies is the functional study of neutralizing epitopes which provides key knowledge for efficient vaccine design. Human broadly neutralizing antibodies serve as a potential source of discovering neutralizing epitopes that can be targeted to fight against many infectious diseases and can thus facilitate the design of the vaccines. Antibody structure studies can provide new insights into the mechanism of recognition of immune evasion epitopes at the atomic level, and how antibodies mature to capture the virus mutant (Morales et al., 2016, Wibmer et al., 2017, Rujas et al., 2016). However in recent reports, few immunogen candidates were reported that can elicit potent broadly neutralizing antibodies in several immunization studies. Native-like HIV-1 envelope trimers BG505 SOSIP.664 and B41 SOSIP.664 are reported to induce neutralizing antibodies against the Tier-2 virus in rabbits, even though the potency is much weaker in comparison to the broadly neutralizing antibodies isolated from human long term non-progressors (Sanders et al., 2015, Bradley et al., 2016).Pre-stimulation of B cells by heterogenous epitopes or antigens showed improved stimulation of B cells in generating potent neutralizing antibodies (Yang et al., 2015). It has also been reported that PGT121-class germline-targeting stabilized trimer immunogens primed PGT121-like responses in PGT121 inferred-germline knock in mice, highlighting the hypothesis of prime-boosting and sequential immunization strategy (Steichen et al., 2016). All these observations highlight the neutralizing antibodies’ role in HIV-1 prevention. Altogether, understanding antibody generation and function mechanisms can help to guide efficient vaccine design and produce effective therapy.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Competing interests
No.
Funding
Not applicable.
Authors’ contributions
The authors claimed no interest.
Acknowledgements
We wish to thank Dr. Mei-yun Zhang, Dr. Zhiwei Chen (The University of Hong Kong), and Dr. Paul Zhou (Institute Pasteur of Shanghai CAS) for helpful discussion.
References
- Abusneina A., Gauthier E.R. Ammonium ions improve the survival of glutamine-starved hybridoma cells. Cell Biosci. 2016;6:23. doi: 10.1186/s13578-016-0092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghebati-Maleki L. Isolation and characterization of anti ROR1 single chain fragment variable antibodies using phage display technique. Hum. Antibodies. 2017;25(1–2):57–63. doi: 10.3233/HAB-170310. [DOI] [PubMed] [Google Scholar]
- Andreu C., Del Olmo M.L. Development of a new yeast surface display system based on Spi1 as an anchor protein. Appl. Microbiol. Biotechnol. 2017;101(1):287–299. doi: 10.1007/s00253-016-7905-x. [DOI] [PubMed] [Google Scholar]
- Armbruster C. Passive immunization with the anti-HIV-1 human monoclonal antibody (hMAb) 4E10 and the hMAb combination 4E10/2F5/2G12. J. Antimicrob. Chemother. 2004;54(5):915–920. doi: 10.1093/jac/dkh428. [DOI] [PubMed] [Google Scholar]
- Ashish Global structure of HIV-1 neutralizing antibody IgG1 b12 is asymmetric. Biochem. Biophys. Res. Commun. 2010;391(1):947–951. doi: 10.1016/j.bbrc.2009.11.170. [DOI] [PubMed] [Google Scholar]
- Banerjee S. Evaluation of a novel multi-immunogen vaccine strategy for targeting 4E10/10E8 neutralizing epitopes on HIV-1 gp41 membrane proximal external region. Virology. 2017;505:113–126. doi: 10.1016/j.virol.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass H., Darke C. Improved efficiency of EBV transformation of B-lymphocytes. Cell Prolif. 2004;37(6):443–444. doi: 10.1111/j.1365-2184.2004.00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye F.L., Light A., Tarlinton D.M. Single cell sorting and cloning. J. Immunol. Methods. 2000;243(1-2):25–32. doi: 10.1016/s0022-1759(00)00225-8. [DOI] [PubMed] [Google Scholar]
- Beerli R.R. Isolation of human monoclonal antibodies by mammalian cell display. Proc. Natl. Acad. Sci. U. S. A. 2008;105(38):14336–14341. doi: 10.1073/pnas.0805942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C.M. Antibody immunoprophylaxis and immunotherapy for influenza virus infection: utilization of monoclonal or polyclonal antibodies? Hum. Vaccin Immunother. 2017 doi: 10.1080/21645515.2017.1363135. Aug 30:0 PMID: 28854120 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia H. Hybridoma cell-culture and glycan profile dataset at various bioreactor conditions. Data Brief. 2016;9:676–678. doi: 10.1016/j.dib.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick M. Humanized monoclonal antibody alemtuzumab treatment in transplant. Exp. Clin. Transplant. 2016;14(1):17–21. [PubMed] [Google Scholar]
- Bieniasz P.D. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16(10):2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair W.S. Identification and characterization of UK-201844: a novel inhibitor that interferes with human immunodeficiency virus type 1 gp160 processing. Antimicrob. Agents Chemother. 2007;51(10):3554–3561. doi: 10.1128/AAC.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder E.T., Raeeszadeh-Sarmazdeh M., Price J.V. Engineering antibodies by yeast display. Arch. Biochem. Biophys. 2012;526(2):99–106. doi: 10.1016/j.abb.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Boots L.J. Anti-human immunodeficiency virus type 1 human monoclonal antibodies that bind discontinuous epitopes in the viral glycoproteins can identify mimotopes from recombinant phage peptide display libraries. AIDS Res. Hum. Retroviruses. 1997;13(18):1549–1559. doi: 10.1089/aid.1997.13.1549. [DOI] [PubMed] [Google Scholar]
- Bouchet J. Inhibition of the Nef regulatory protein of HIV-1 by a single-domain antibody. Blood. 2011;117(13):3559–3568. doi: 10.1182/blood-2010-07-296749. [DOI] [PubMed] [Google Scholar]
- Bouchet J. Single-domain antibody-SH3 fusions for efficient neutralization of HIV-1 Nef functions. J. Virol. 2012;86(9):4856–4867. doi: 10.1128/JVI.06329-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet F. Single peptide and anti-idiotype based immunizations can broaden the antibody response against the variable V3 domain of HIV-1 in mice. Mol. Immunol. 1995;32(7):449–457. doi: 10.1016/0161-5890(95)00007-2. [DOI] [PubMed] [Google Scholar]
- Bradley T. Structural constraints of vaccine-induced tier-2 autologous HIV neutralizing antibodies targeting the receptor-binding site. Cell Rep. 2016;14(1):43–54. doi: 10.1016/j.celrep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braibant M. Cross-group neutralization of HIV-1 and evidence for conservation of the PG9/PG16 epitopes within divergent groups. AIDS. 2013;27(8):1239–1244. doi: 10.1097/QAD.0b013e32835ecb42. [DOI] [PubMed] [Google Scholar]
- Brodine S.K. Diverse HIV-1 subtypes and clinical: laboratory and behavioral factors in a recently infected US military cohort. AIDS. 2003;17(17):2521–2527. doi: 10.1097/00002030-200311210-00016. [DOI] [PubMed] [Google Scholar]
- Buchacher A. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses. 1994;10(4):359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Bunnik E.M. Emergence of monoclonal antibody b12-resistant human immunodeficiency virus type 1 variants during natural infection in the absence of humoral or cellular immune pressure. J. Gen. Virol. 2010;91(Pt 5):1354–1364. doi: 10.1099/vir.0.017319-0. [DOI] [PubMed] [Google Scholar]
- Cafri G. Production of LacZ inducible T cell hybridoma specific for human and mouse gp100(2)(5)(−)(3)(3) peptides. PLoS One. 2013;8(2):e55583. doi: 10.1371/journal.pone.0055583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C.C. HIV-1 neutralizing antibody response and viral genetic diversity characterized with next generation sequencing. Virology. 2015;474:34–40. doi: 10.1016/j.virol.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W. Human recombinant antibodies specific for hepatitis C virus core and envelope E2 peptides from an immune phage display library. J. Gen. Virol. 1996;77(Pt 10):2531–2539. doi: 10.1099/0022-1317-77-10-2531. [DOI] [PubMed] [Google Scholar]
- Choi Y. Antibody humanization by structure-based computational protein design. MAbs. 2015;7(6):1045–1057. doi: 10.1080/19420862.2015.1076600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale B.M. Cell-to-cell transfer of HIV-1 via virological synapses leads to endosomal virion maturation that activates viral membrane fusion. Cell Host Microbe. 2011;10(6):551–562. doi: 10.1016/j.chom.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport T.M. Binding interactions between soluble HIV envelope glycoproteins and quaternary-structure-specific monoclonal antibodies PG9 and PG16. J. Virol. 2011;85(14):7095–7107. doi: 10.1128/JVI.00411-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath A.K. Three-dimensional structure-activity analysis of a series of porphyrin derivatives with anti-HIV-1 activity targeted to the V3 loop of the gp120 envelope glycoprotein of the human immunodeficiency virus type 1. J. Med. Chem. 1994;37(8):1099–1108. doi: 10.1021/jm00034a007. [DOI] [PubMed] [Google Scholar]
- Desai T.M. Fluorescent protein-tagged Vpr dissociates from HIV-1 core after viral fusion and rapidly enters the cell nucleus. Retrovirology. 2015;12:88. doi: 10.1186/s12977-015-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5(5):e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K.J. A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity. Proc. Natl. Acad. Sci. U. S. A. 2010;107(40):17107–17112. doi: 10.1073/pnas.1002717107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K.J. Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope. J. Virol. 2015;89(2):1105–1118. doi: 10.1128/JVI.02905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte J.N. Generation of immunity against pathogens via single-domain antibody-antigen constructs. J. Immunol. 2016;197(12):4838–4847. doi: 10.4049/jimmunol.1600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham N.D., Chen B.K. Measuring t cell-to-T cell HIV-1 transfer, viral fusion, and infection using flow cytometry. Methods Mol. Biol. 2016;1354:21–38. doi: 10.1007/978-1-4939-3046-3_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler Z. Activity of broadly neutralizing antibodies: including PG9, PG16, and VRC01, against recently transmitted subtype B HIV-1 variants from early and late in the epidemic. J. Virol. 2011;85(14):7236–7245. doi: 10.1128/JVI.00196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K. Assurance of monoclonality in one round of cloning through cell sorting for single cell deposition coupled with high resolution cell imaging. Biotechnol. Prog. 2015;31(5):1172–1178. doi: 10.1002/btpr.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40(5):657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay W.J. Phage display: a powerful technology for the generation of high-specificity affinity reagents from alternative immune sources. Methods Mol. Biol. 2017;1485:85–99. doi: 10.1007/978-1-4939-6412-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund N.T. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci. Transl. Med. 2017;9(373) doi: 10.1126/scitranslmed.aal2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev I.S. Antibodies VRC01 and 10E8 neutralize HIV-1 with high breadth and potency even with Ig-framework regions substantially reverted to germline. J. Immunol. 2014;192(3):1100–1106. doi: 10.4049/jimmunol.1302515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gera N., Hussain M., Rao B.M. Protein selection using yeast surface display. Methods. 2013;60(1):15–26. doi: 10.1016/j.ymeth.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Gong X. Specific determination of influenza H7N2 virus based on biotinylated single-domain antibody from a phage-displayed library. Anal. Biochem. 2016;500:66–72. doi: 10.1016/j.ab.2015.09.020. [DOI] [PubMed] [Google Scholar]
- Graus Y.F. Selection of recombinant anti-HuD Fab fragments from a phage display antibody library of a lung cancer patient with paraneoplastic encephalomyelitis. J. Neuroimmunol. 1998;82(2):200–209. doi: 10.1016/s0165-5728(97)00199-9. [DOI] [PubMed] [Google Scholar]
- Gristick H.B. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 2016;23(10):906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Simian immunodeficiency virus infection evades vaccine-elicited antibody responses to V2 region. J. Acquir. Immune Defic. Syndr. 2015;68(5):502–510. doi: 10.1097/QAI.0000000000000530. [DOI] [PubMed] [Google Scholar]
- Hamanoue S. Successful treatment with humanized anti-interleukin-6 receptor antibody (tocilizumab) in a case of AA amyloidosis complicated by familial Mediterranean fever. Mod. Rheumatol. 2016;26(4):610–613. doi: 10.3109/14397595.2014.908810. [DOI] [PubMed] [Google Scholar]
- Haussner C. Peptide paratope mimics of the broadly neutralizing HIV-1 antibody b12. Chembiochem. 2017;18(7):647–653. doi: 10.1002/cbic.201600621. Apr 4. [DOI] [PubMed] [Google Scholar]
- Haynes B.F., Mascola J.R. The quest for an antibody-based HIV vaccine. Immunol. Rev. 2017;275(1):5–10. doi: 10.1111/imr.12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hencsey Z. Effect of medium composition on hybridoma growth and antibody production. Acta Microbiol. Immunol. Hung. 1996;43(4):359–370. [PubMed] [Google Scholar]
- Holzlohner P., Hanack K. Generation of murine monoclonal antibodies by hybridoma technology. J. Vis. Exp. 2017 doi: 10.3791/54832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Ichimori Y., Iwasa S. A human hybrid hybridoma producing a bispecific monoclonal antibody that can target tumor cells for attack by Pseudomonas aeruginosa exotoxin A. Cytotechnology. 1990;4(1):59–68. doi: 10.1007/BF00148811. [DOI] [PubMed] [Google Scholar]
- Huang J. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491(7424):406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugwil A.V. The meaning of the anti-cancer antibody CLN-IgG (Pritumumab) generated by human x human hybridoma technology against the cyto-skeletal protein: vimentin, in the course of the treatment of malignancy. Med. Hypotheses. 2013;81(3):489–495. doi: 10.1016/j.mehy.2013.05.037. [DOI] [PubMed] [Google Scholar]
- Ito A. Defucosylated anti-CCR4 monoclonal antibody exercises potent ADCC-mediated antitumor effect in the novel tumor-bearing humanized NOD/Shi-scid: IL-2Rgamma(null) mouse model. Cancer Immunol. Immunother. 2009;58(8):1195–1206. doi: 10.1007/s00262-008-0632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Karlsson Hedestam G.B. Evolution of B cell analysis and Env trimer redesign. Immunol. Rev. 2017;275(1):183–202. doi: 10.1111/imr.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumura K.R., Maruo S., Takada K. EBV lytic infection enhances transformation of B-lymphocytes infected with EBV in the presence of T-lymphocytes. J. Med. Virol. 2012;84(3):504–510. doi: 10.1002/jmv.23208. [DOI] [PubMed] [Google Scholar]
- Kazemi-Lomedasht F. Production and characterization of novel camel single domain antibody targeting mouse vascular endothelial growth factor. Monoclon. Antib. Immunodiagn. Immunother. 2016;35(3):167–171. doi: 10.1089/mab.2016.0001. [DOI] [PubMed] [Google Scholar]
- Kelso G.F. Impact on monoclonal antibody production in murine hybridoma cell cultures of adenosine receptor antagonists and phosphodiesterase inhibitors. Bioorg. Med. Chem. Lett. 2016;26(2):540–544. doi: 10.1016/j.bmcl.2015.11.075. [DOI] [PubMed] [Google Scholar]
- Klein G. EBV-B cell interactions: immortalization, rescue from apoptosis, tumorigenicity (a short review) Acta Microbiol. Immunol. Hung. 1996;43(2-3):97–105. [PubMed] [Google Scholar]
- Kovacs J.M. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc. Natl. Acad. Sci. U. S. A. 2012;109(30):12111–12116. doi: 10.1073/pnas.1204533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B. Versatility of using major histocompatibility complex class II dextramers for derivation and characterization of antigen-specific, autoreactive T cell hybridomas. J. Immunol. Methods. 2015;426:86–94. doi: 10.1016/j.jim.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusam S. BCL6 cooperates with CD40 stimulation and loss of p53 function to rapidly transform primary B cells. Int. J. Cancer. 2009;125(4):977–981. doi: 10.1002/ijc.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwakkenbos M.J. Corrigendum: generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat. Med. 2016;22(12):1502. doi: 10.1038/nm1216-1502a. [DOI] [PubMed] [Google Scholar]
- Lang A.B. Immunotherapy with human monoclonal antibodies: fragment A specificity of polyclonal and monoclonal antibodies is crucial for full protection against tetanus toxin. J. Immunol. 1993;151(1):466–472. [PubMed] [Google Scholar]
- Lee J.E. MicroRNA signatures associated with immortalization of EBV-transformed lymphoblastoid cell lines and their clinical traits. Cell Prolif. 2011;44(1):59–66. doi: 10.1111/j.1365-2184.2010.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Human antibodies for immunotherapy development generated via a human B cell hybridoma technology. Proc. Natl. Acad. Sci. U. S. A. 2006;103(10):3557–3562. doi: 10.1073/pnas.0511285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. A single-domain antibody-linked Fab bispecific antibody Her2-S-Fab has potent cytotoxicity against Her2-expressing tumor cells. AMB Express. 2016;6(1):32. doi: 10.1186/s13568-016-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Structural analysis of the glycosylated intact HIV-1 gp120-b12 antibody complex using hydroxyl radical protein footprinting. Biochemistry. 2017;56(7):957–970. doi: 10.1021/acs.biochem.6b00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X. Immunogenicity of constrained monoclonal antibody A32-human immunodeficiency virus (HIV) Env gp120 complexes compared to that of recombinant HIV type 1 gp120 envelope glycoproteins. J. Virol. 2004;78(10):5270–5278. doi: 10.1128/JVI.78.10.5270-5278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.J. Effect of interleukin (IL)-12 and IL-15 on activated natural killer (ANK) and antibody-dependent cellular cytotoxicity (ADCC) in HIV infection. J. Clin. Immunol. 1998;18(5):335–345. doi: 10.1023/a:1023290932154. [DOI] [PubMed] [Google Scholar]
- Liu H. A rapid method to characterize mouse IgG antibodies and isolate native antigen binding IgG B cell hybridomas. PLoS One. 2015;10(8):e0136613. doi: 10.1371/journal.pone.0136613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J.M. Binding of HIV-1 gp41-directed neutralizing and non-neutralizing fragment antibody binding domain (Fab) and single chain variable fragment (ScFv) antibodies to the ectodomain of gp41 in the pre-hairpin and six-helix bundle conformations. PLoS One. 2014;9(8):e104683. doi: 10.1371/journal.pone.0104683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Sun Z., Zhang M.-Y. Generation of immortalized human na#xp#ve B cell libraries by optimized EBV transformation. J. Med. Discovery. 2016;2(1):s1. [Google Scholar]
- Lulf S. Structural basis for the inhibition of HIV-1 Nef by a high-affinity binding single-domain antibody. Retrovirology. 2014;11:24. doi: 10.1186/1742-4690-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandi Y., Bakay M., Beladi I. Effect of human adenovirus on antibody-dependent cellular cytotoxicity (ADCC) in chickens. Cell. Immunol. 1982;69(2):395–400. doi: 10.1016/0008-8749(82)90082-x. [DOI] [PubMed] [Google Scholar]
- Mantis N.J. Inhibition of HIV-1 infectivity and epithelial cell transfer by human monoclonal IgG and IgA antibodies carrying the b12 V region. J. Immunol. 2007;179(5):3144–3152. doi: 10.4049/jimmunol.179.5.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A.C., Rees A.R. Extracting human antibody sequences from public databases for antibody humanization: high frequency of species assignment errors. Protein Eng. Des. Sel. 2016;29(10):403–408. doi: 10.1093/protein/gzw018. [DOI] [PubMed] [Google Scholar]
- Martin-Lopez A. Enhanced monoclonal antibody production in hybridoma cells by LPS and Anti-mIgG. Biotechnol. Prog. 2007;23(6):1447–1453. doi: 10.1021/bp070191a. [DOI] [PubMed] [Google Scholar]
- McCoy L.E., Burton D.R. Identification and specificity of broadly neutralizing antibodies against HIV. Immunol. Rev. 2017;275(1):11–20. doi: 10.1111/imr.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin L.P. EBV-Directed T cell therapeutics for EBV-Associated lymphomas. Methods Mol. Biol. 2017;1532:255–265. doi: 10.1007/978-1-4939-6655-4_19. [DOI] [PubMed] [Google Scholar]
- Mehandru S. Neutralization profiles of newly transmitted human immunodeficiency virus type 1 by monoclonal antibodies 2G12: 2F5, and 4E10. J. Virol. 2004;78(24):14039–14042. doi: 10.1128/JVI.78.24.14039-14042.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei M. Application of modified yeast surface display technologies for non-Antibody protein engineering. Microbiol. Res. 2017;196:118–128. doi: 10.1016/j.micres.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Micoli K.J. Requirement of calmodulin binding by HIV-1 gp160 for enhanced FAS-mediated apoptosis. J. Biol. Chem. 2000;275(2):1233–1240. doi: 10.1074/jbc.275.2.1233. [DOI] [PubMed] [Google Scholar]
- Milstein C. The hybridoma revolution: an offshoot of basic research. Bioessays. 1999;21(11):966–973. doi: 10.1002/(SICI)1521-1878(199911)21:11<966::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Moore J.P. Towards a structure of the HIV-1 envelope glycoprotein gp120: an immunochemical approach. Philos. Trans. R Soc. Lond. B Biol. Sci. 1993;342(1299):83–88. doi: 10.1098/rstb.1993.0139. [DOI] [PubMed] [Google Scholar]
- Morales J.F. Fragments of the V1/V2 domain of HIV-1 glycoprotein 120 engineered for improved binding to the broadly neutralizing PG9 antibody. Mol. Immunol. 2016;77:14–25. doi: 10.1016/j.molimm.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C.B. Effectiveness of intranasal immunization with HIV-gp160 and an HIV-1 env CTL epitope peptide (E7) in combination with the mucosal adjuvant LT(R192G) Vaccine. 2000;18(18):1944–1951. doi: 10.1016/s0264-410x(99)00447-8. [DOI] [PubMed] [Google Scholar]
- Morris G.C. MABGEL 1: first phase 1 trial of the anti-HIV-1 monoclonal antibodies 2F5, 4E10 and 2G12 as a vaginal microbicide. PLoS One. 2014;9(12):e116153. doi: 10.1371/journal.pone.0116153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C.D. Differential antibody responses to conserved HIV-1 neutralizing epitopes in the context of multivalent scaffolds and native-like gp140 trimers. MBio. 2017;8(1) doi: 10.1128/mBio.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi A. Reappraisal of EBV in diffuse large B-cell lymphoma (DLBCL): comparative analysis between EBV-positive and −negative DLBCL with EBV-positive bystander cells. Histopathology. 2017 doi: 10.1111/his.13197. [DOI] [PubMed] [Google Scholar]
- Ohtomo T. Humanization of mouse ONS-M21 antibody with the aid of hybrid variable regions. Mol. Immunol. 1995;32(6):407–416. doi: 10.1016/0161-5890(95)00017-9. [DOI] [PubMed] [Google Scholar]
- Olimpieri P.P., Marcatili P., Tramontano A. Tabhu: tools for antibody humanization. Bioinformatics. 2015;31(3):434–435. doi: 10.1093/bioinformatics/btu667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouisse L.H. Antigen-specific single B cell sorting and expression-cloning from immunoglobulin humanized rats: a rapid and versatile method for the generation of high affinity and discriminative human monoclonal antibodies. BMC Biotechnol. 2017;17(1):3. doi: 10.1186/s12896-016-0322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 2010;84(16):8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H. Generation and characterization of a novel fusion partner cell line for the production of human macrophage hybridoma. Hybridoma. 1997;16(6):551–556. doi: 10.1089/hyb.1997.16.551. [DOI] [PubMed] [Google Scholar]
- Patke S. Bisfabs: tools for rapidly screening hybridoma IgGs for their activities as bispecific antibodies. MAbsp. 2017;9(3):430–437. doi: 10.1080/19420862.2017.1281504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegu A. Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol. Rev. 2017;275(1):296–312. doi: 10.1111/imr.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus S.H. Identification of human anti-HIV gp160 monoclonal antibodies that make effective immunotoxins. J. Virol. 2017;91(3) doi: 10.1128/JVI.01955-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus S.H. Design and In vivo characterization of immunoconjugates targeting HIV gp160. J. Virol. 2017;91(3) doi: 10.1128/JVI.01360-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson M. Immunogenomic engineering of a plug-and-(dis)play hybridoma platform. Nat. Commun. 2016;7:12535. doi: 10.1038/ncomms12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost J.et al. Influence of the envelope gp120 Phe 43 cavity on HIV-1 sensitivity to ADCC responses. J. Virol. 2017 doi: 10.1128/JVI.02452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbarnia L. Evolution of phage display technology: from discovery to application. J. Drug Target. 2017;25(3):216–224. doi: 10.1080/1061186X.2016.1258570. [DOI] [PubMed] [Google Scholar]
- Rausch D.M. Peptides derived from the CDR3-homologous domain of the CD4 molecule are specific inhibitors of HIV-1 and SIV infection, virus-induced cell fusion, and postinfection viral transmission in vitro. Implications for the design of small peptide anti-HIV therapeutic agents. Ann. N. Y. Acad. Sci. 1990;616:125–148. doi: 10.1111/j.1749-6632.1990.tb17834.x. [DOI] [PubMed] [Google Scholar]
- Ringe R., Phogat S., Bhattacharya J. Subtle alteration of residues including N-linked glycans in V2 loop modulate HIV-1 neutralization by PG9 and PG16 monoclonal antibodies. Virology. 2012;426(1):34–41. doi: 10.1016/j.virol.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Robert-Guroff M. Cross-neutralization of human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus isolates. J. Virol. 1992;66(6):3602–3608. doi: 10.1128/jvi.66.6.3602-3608.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg M.E. Humanized anti-IL-5 antibody therapy. Cell. 2016;165(3):509. doi: 10.1016/j.cell.2016.04.020. [DOI] [PubMed] [Google Scholar]
- Rotman M. Fusion of hIgG1-Fc to 111In-anti-amyloid single domain antibody fragment VHH-pa2H prolongs blood residential time in APP/PS1 mice but does not increase brain uptake. Nucl. Med. Biol. 2015;42(8):695–702. doi: 10.1016/j.nucmedbio.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Rujas E. Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface. Sci. Rep. 2016;6:38177. doi: 10.1038/srep38177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydstrom K. CD40 is a potential marker of favorable prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Leuk. Lymphoma. 2010;51(9):1643–1648. doi: 10.3109/10428194.2010.492537. [DOI] [PubMed] [Google Scholar]
- Safdari Y. Antibody humanization methods − a review and update. Biotechnol. Genet. Eng. Rev. 2013;29:175–186. doi: 10.1080/02648725.2013.801235. [DOI] [PubMed] [Google Scholar]
- Sanders R.W. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W. HIV-1 VACCINES: HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349(6244):aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F. HIV-specific B cell response in patients with broadly neutralizing serum activity. Science. 2015;350(6265):1175. doi: 10.1126/science.aad7133. [DOI] [PubMed] [Google Scholar]
- Schrader A. Global gene expression changes of in vitro stimulated human transformed germinal centre B cells as surrogate for oncogenic pathway activation in individual aggressive B cell lymphomas. Cell Commun. Signal. 2012;10(1):43. doi: 10.1186/1478-811X-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan J., Marasco W.A. Phage and yeast display. Microbiol. Spectr. 2015;3(1) doi: 10.1128/microbiolspec.AID-0028-2014. (p. AID-0028-2014) [DOI] [PubMed] [Google Scholar]
- Sili U. Large-scale expansion of dendritic cell-primed polyclonal human cytotoxic T-lymphocyte lines using lymphoblastoid cell lines for adoptive immunotherapy. J. Immunother. 2003;26(3):241–256. doi: 10.1097/00002371-200305000-00008. [DOI] [PubMed] [Google Scholar]
- Soga K. Mammalian cell surface display as a novel method for developing engineered lectins with novel characteristics. Biomolecules. 2015;5(3):1540–1562. doi: 10.3390/biom5031540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. U. S. A. 2014;111(49):17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature. 2017;548(7665):108–111. doi: 10.1038/nature23301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. Developmental pathway of the MPER-directed HIV-1-Neutralizing antibody 10E8. PLoS One. 2016;11(6):e0157409. doi: 10.1371/journal.pone.0157409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V. Identification of dominant antibody-dependent cell-mediated cytotoxicity epitopes on the hemagglutinin antigen of pandemic H1N1 influenza virus. J. Virol. 2013;87(10):5831–5840. doi: 10.1128/JVI.00273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen J.M. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity. 2016;45(3):483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N.E. Preserved antiviral adaptive immunity following polyclonal antibody immunotherapy for severe murine influenza infection. Sci. Rep. 2016;6:29154. doi: 10.1038/srep29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic I. Quantification of antibody production of individual hybridoma cells by surface plasmon resonance imaging. Anal. Biochem. 2015;485:112–118. doi: 10.1016/j.ab.2015.06.018. [DOI] [PubMed] [Google Scholar]
- Stramer S.L. Two human immunodeficiency virus Type 2 cases in US blood donors including serologic, molecular, and genomic characterization of an epidemiologically unusual case. Transfusion (Paris) 2016;56(6 Pt 2):1560–1568. doi: 10.1111/trf.13600. [DOI] [PubMed] [Google Scholar]
- Straub C., Zubler R.H. Immortalization of EBV-infected B cells is not influenced by exogenous signals acting on B cell proliferation. Effects of mutant EL-4 thymoma cells and transforming growth factor-beta. J. Immunol. 1989;142(1):87–93. [PubMed] [Google Scholar]
- Sun Z. Reconstitution and characterization of antibody repertoires of HIV-1-infected elite neutralizers. Antiviral Res. 2015;118:1–9. doi: 10.1016/j.antiviral.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Sun Z. Isolation and characterization of HIV-1 envelope glycoprotein specific B cell from immortalized human naive B cell library. J. Gen. Virol. 2017 doi: 10.1099/jgv.0.000706. [DOI] [PubMed] [Google Scholar]
- Szafran A.T. Use of HCA in subproteome-immunization and screening of hybridoma supernatants to define distinct antibody binding patterns. Methods. 2016;96:75–84. doi: 10.1016/j.ymeth.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z. A human single-domain antibody elicits potent antitumor activity by targeting an epitope in mesothelin close to the cancer cell surface. Mol. Cancer Ther. 2013;12(4):416–426. doi: 10.1158/1535-7163.MCT-12-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thenin S. Naturally occurring substitutions of conserved residues in human immunodeficiency virus type 1 variants of different clades are involved in PG9 and PG16 resistance to neutralization. J. Gen. Virol. 2012;93(Pt 7):1495–1505. doi: 10.1099/vir.0.042614-0. [DOI] [PubMed] [Google Scholar]
- Thomann M. In vitro glycoengineering of IgG1 and its effect on Fc receptor binding and ADCC activity. PLoS One. 2015;10(8):e0134949. doi: 10.1371/journal.pone.0134949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329(1–2):112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimatsu K. A rapid screening and production method using a novel mammalian cell display to isolate human monoclonal antibodies. Biochem. Biophys. Res. Commun. 2013;441(1):59–64. doi: 10.1016/j.bbrc.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Tomita M., Tsumoto K. Hybridoma technologies for antibody production. Immunotherapy. 2011;3(3):371–380. doi: 10.2217/imt.11.4. [DOI] [PubMed] [Google Scholar]
- Traggiai E. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 2004;10(8):871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxlmayr M.W., Shusta E.V. Directed evolution of protein thermal stability using yeast surface display. Methods Mol. Biol. 2017;1575:45–65. doi: 10.1007/978-1-4939-6857-2_4. [DOI] [PubMed] [Google Scholar]
- Tsurushita N. Humanization of a chicken anti-IL-12 monoclonal antibody. J. Immunol. Methods. 2004;295(1–2):9–19. doi: 10.1016/j.jim.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Walker L.M. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Optimizing adoptive polyclonal T cell immunotherapy of lymphomas: using a chimeric T cell receptor possessing CD28 and CD137 costimulatory domains. Hum. Gene Ther. 2007;18(8):712–725. doi: 10.1089/hum.2007.028. [DOI] [PubMed] [Google Scholar]
- Wang Y. HIV-1 gp140 epitope recognition is influenced by immunoglobulin DH gene segment sequence. Immunogenetics. 2016;68(2):145–155. doi: 10.1007/s00251-015-0890-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A.B., Wilson I.A. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol. Rev. 2017;275(1):21–32. doi: 10.1111/imr.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins B.A. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J. Virol. 1996;70(12):8431–8437. doi: 10.1128/jvi.70.12.8431-8437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K. Structure and recognition of a novel HIV-1 gp120-gp41 interface antibody that caused MPER exposure through viral escape. PLoS Pathog. 2017;13(1):e1006074. doi: 10.1371/journal.ppat.1006074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe K. Immunodominance of antibody recognition of the HIV envelope V2 region in ig-Humanized mice. J. Immunol. 2017;198(3):1047–1055. doi: 10.4049/jimmunol.1601640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 1997;186(8):1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J. Virol. 2009;83(21):10892–10907. doi: 10.1128/JVI.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Maturation and diversity of the VRC01-Antibody lineage over 15 years of chronic HIV-1 infection. Cell. 2015;161(3):470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Characterization of stable: soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 2000;74(12):5716–5725. doi: 10.1128/jvi.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Identification of non-HIV immunogens that bind to germline b12 predecessors and prime for elicitation of cross-clade neutralizing HIV-1 antibodies. PLoS One. 2015;10(5):e0126428. doi: 10.1371/journal.pone.0126428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Y. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods. 2003;283(1–2):17–25. doi: 10.1016/j.jim.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Zhang M.Y. Identification and characterization of a new cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody. J. Virol. 2004;78(17):9233–9242. doi: 10.1128/JVI.78.17.9233-9242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Y. Identification of a novel CD4i human monoclonal antibody Fab that neutralizes HIV-1 primary isolates from different clades. Antiviral Res. 2004;61(3):161–164. doi: 10.1016/j.antiviral.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Zhang M.Y. Selection of a novel gp41-specific HIV-1 neutralizing human antibody by competitive antigen panning. J. Immunol. Methods. 2006;317(1-2):21–30. doi: 10.1016/j.jim.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Y. Cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody that recognizes a novel conformational epitope on gp41 and lacks reactivity against self-antigens. J. Virol. 2008;82(14):6869–6879. doi: 10.1128/JVI.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Y. Identification and characterization of a broadly cross-reactive HIV-1 human monoclonal antibody that binds to both gp120 and gp41. PLoS One. 2012;7(9):e44241. doi: 10.1371/journal.pone.0044241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329(5993):811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T. Multidonor analysis reveals structural elements: genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39(2):245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T. Structural repertoire of HIV-1-Neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell. 2015;161(6):1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. Somatic populations of PGT135-137 HIV-1-neutralizing antibodies identified by 454 pyrosequencing and bioinformatics. Front. Microbiol. 2012;3:315. doi: 10.3389/fmicb.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts. Proc. Natl. Acad. Sci. U. S. A. 2013;110(43):E4088–97. doi: 10.1073/pnas.1306262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick M.B. A novel human antibody against human immunodeficiency virus type 1 gp120 is V1: V2, and V3 loop dependent and helps delimit the epitope of the broadly neutralizing antibody immunoglobulin G1 b12. J. Virol. 2003;77(12):6965–6978. doi: 10.1128/JVI.77.12.6965-6978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.