Abstract
Avian infectious bronchitis virus (IBV) is causing major economic losses to the poultry industry. The analysis of the S1 gene has been used to determine IBV genotype. The aim of this study was genotyping of IBVs circulating among the Iranian broiler flocks in the period between 2015 to 2017. Trachea samples from 278 broiler flocks were collected from broiler farms in eight provinces of Iran. After Real-time RT-PCR, IBV-positive samples were further characterized based on S1 gene. The results of the Real-time RT-PCR showed that 52.16% of flocks were IBV positive. Four genotypes were detected and the frequency of occurrence rates of IS-1494-like, 793/B, QX and Massachusetts IBV genotypes were 70.34%, 19.31%, 7.58% and 2.75%, respectively. Sequence analysis revealed that nucleotide identities within IS-1494-like group ranged between 98.86–100%, while each of the QX, Massachusetts and 793/B groups were 98.05–100%, 98.20–100% and 93.29–100% respectively. These results show that the IS-1494-like IBV is the dominant IBV genotype in Iran. Proper control strategies are essential to overcoming the high frequency of occurrence of IS-1494-like IBV. The phylogenetic relationship of the strains with respect to different sequences and geographical regions displayed complexity and diversity. Further studies are needed and should include the isolation and full-length molecular characterization of IBV in Iran.
Keywords: Avian infectious bronchitis, Iran, Genotyping, Phylogenetic analysis, Spike
1. Introduction
The Avian infectious bronchitis virus (IBV) continues to be one of the most economically-impactful diseases in poultry despite intensive control (Jackwood, 2012). The virus affects mainly the respiratory tract, causing tracheal rales, sneezing, coughing, reduced weight gain and mortality, particularly in broiler chickens (Cook et al., 2012). The etiologic agent is a gamma coronavirus. IBV is an enveloped virus approximately 120 nm in diameter with a single-stranded, non-segmented, positive sense RNA genome. The RNA genome of 27.6 kb has at least 10 open reading frames (ORF), from 5′ to 3′ are as follows: 5′-1a-1b-S (S1,S2)-3a,b,c(E)-M-5a,b-N-Poly(A)-3′. The genome encodes four main structural proteins (spike glycoprotein (S), small envelope protein (E), membrane glycoprotein (M), and nucleocapsid protein (N)) and numerous non-structural proteins (Liu et al., 2009). The S1 spike glycoprotein carries virus-neutralizing and serotype-specific antigenic determinants, which can evolve rapidly especially within the three hypervariable regions (HVR) (Abro et al., 2012). Variations in the S1 protein have led to the emergence of numerous variants over time, some of which persist and spread, while others disappear after a short time. (Jackwood, 2012). Improving vaccination efficiency and the understanding of IBV evolution requires knowledge of the IBV variants in the region (Najafi et al., 2016). In Iran, several IBV genotypes have been reported; the first isolation of IBV in Iranian chicken flocks was reported in 1994 (Aghakhan et al., 1994). Later, several Iranian researchers identified the 793/B genotype (Seyfi Abad Shapouri et al., 2004). Genotypes of IBV strains isolated in Iran during 2014–2015 were classified into seven distinct phylogenetic groups (Massachusetts (Mass), 793/B, IS-1494, IS-720, QX, IR-1, and IR-2) (Najafi et al., 2016). Currently, Ma5, H120, and attenuated 793/B like IBV-based vaccination strategies are applied for IB control on poultry farms in Iran. Despite the use of vaccines, IBV still occurs in Iranian poultry farms. Vaccine failures are often associated with the emergence of antigenic variants that differ from the vaccine viruses (Huang and Wang, 2006), thus, continuous surveillance programs are necessary to control IB. The present study was conducted to evaluate the frequency of occurrence of different IBV genotypes circulating in Iranian poultry farms in the period 2015–2017.
2. Material and methods
2.1. Sample collection
Trachea samples (10 tracheas from each flock) from 278 broiler flocks were collected from eight provinces of Iran from March 2015 to February 2017. The investigated broiler chicks showed signs of respiratory problems. Samples from 35 flocks were obtained from each of the 7 provinces of Golestan, Kerman, Ardabil, Isfahan, Ghazvin, Kurdestan, Razavi Khorasan, and 33 flocks from Khuzestan province. All flocks received the IBV vaccines (Massachusetts + 793/B type vaccine). Geographical locations of these provinces are shown in Fig. 1 .
Fig. 1.
Geographical locations of Iranian provinces from where IBV strains were obtained.
2.2. RNA extraction and cDNA synthesis
Ten tracheas from each flock (n = 278) were pooled together and homogenized. RNA was extracted from the pooled samples using RNA easy mini kit (Qiagen) as recommended by the supplier. The cDNA was synthesized using a RevertAid First Strand cDNA synthesis Kit (Thermo Scientific) (Seger et al., 2016).
2.3. Real time PCR, genotyping and phylogenetic analysis
Real-time PCR assay was done in order to amplify a conserved sequence within the 5′-untranslated region (UTR) of the IBV genome described by Callison et al. (Callison et al., 2006). The amplification reaction was performed using a nested PCR assay targeting the hypervariable region 3 of spike gene (393 bp) sequences on cDNA of all positive samples in Real-time PCR step. SX1 (5′-CACCTAGAGGTTTGYTWGCATG-3′) and SX2 (5′-TCCACCTCTATAAACACCYTTAC-3′) primer set were used in first reaction while SX3 (5′-TAATACTGGYAATTTTTCAGATGG-3′) and SX4 (5′ AATACAGATTGCTTACAACCACC-3′) primers were employed in the second round (Worthington et al., 2008). The cycling conditions for both first and second rounds of PCRs were an initial denaturation step at 94 °C for 2 min and 35 cycles of denaturation at 94 °C for 15 s, annealing at 58 °C for 30 s, and polymerization at 72 °C for 30 s. We sent all positive samples for sequencing. The AccuPrep® PCR purification Kit (Bioneer Co., Korea) was used for the purification of the PCR products. Sequencing was performed with the primers (both directions) (Bioneer Co., Korea). Chromatograms were evaluated with CromasPro (CromasPro Version 1.5). After sequences editing, we did NCBI BLAST on the results and did primary identifications. In the following, we draw the phylogenetic tree. Because of the high number of sequences in tree (Data has not been shown), the identical sequences have been removed in each group and finally selected 51 sequences for drawing the final tree and submission in gene bank. Phylogenetic tree for the S1 glycoprotein was generated using the neighbor-joining method with 1000 bootstrap replicates to assign confidence levels to branches using MEGA 7 software package by Kimura-2-parameter model (Kumar et al., 2016). Sequences reported in this paper were added to the GenBank database under accession number (MF322802-MF322852).
3. Results
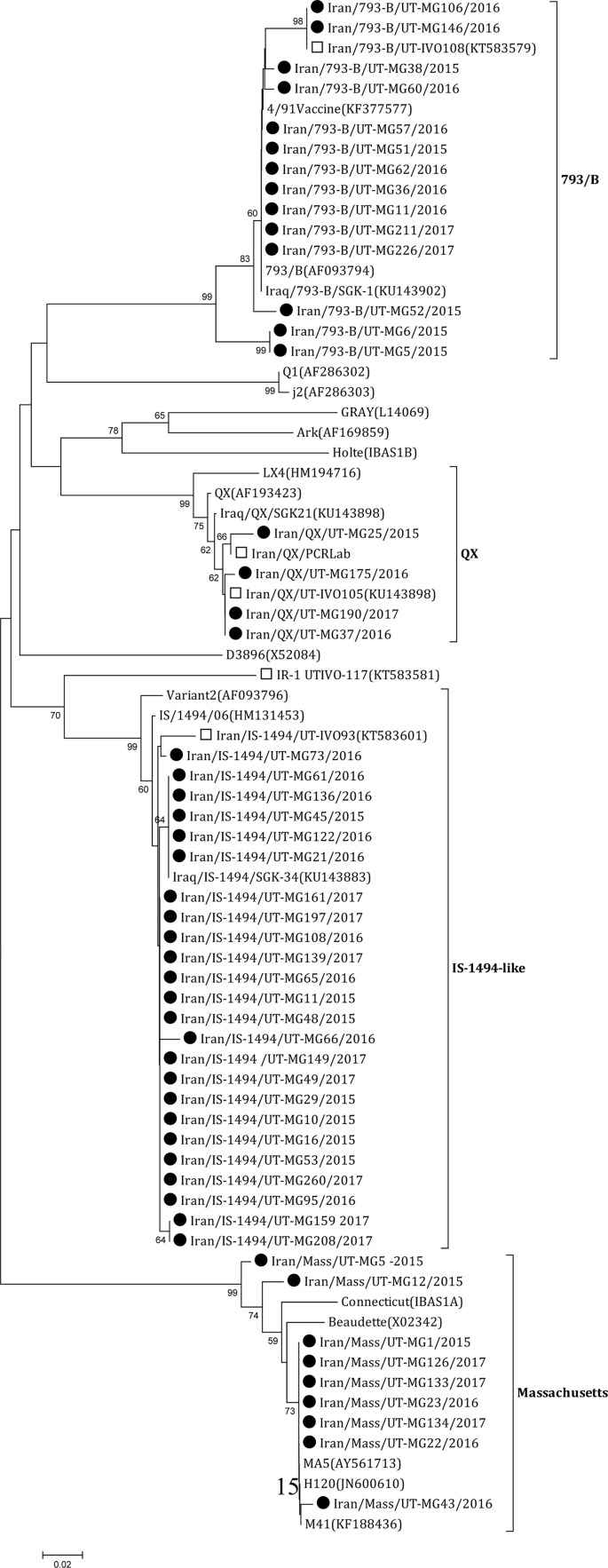
From 2015 to 2017, 278 flocks were examined; of these, 145 (52.16%) were found to be IBV-positive. Based on phylogenetic analysis, the strains were clustered in four distinct groups (Fig. 2 ). The distributions of different IBV genotypes expressed as a percentage of total IBV detected is shown in Table 1 . The most frequently occurring type was IS-1494-like IBV with 70.34% frequency of occurrence rate; 793/B was the second most common type with 19.31% frequency of occurrence. The third and fourth most frequently detected IBV types were Massachusetts and QX with rates of 7.58% and 2.75% respectively (Fig. 3 ). QX IBV was not detected in Isfahan, Kurdistan, Golestan, and Khuzestan provinces. The highest frequently occurring of IBV was detected in Isfahan province (68.57%) and the lowest in Khuzestan province (39.39%). Sequence similarity among IS-1494-like IBV strains of this study varied between 98.86% to 100% and they had more than 98.47% nucleotide similarity to IS/1494/06 (HM131453). Some of our strains were identical to Iraq/IS-1494/SGK-34 (KU143883). Nucleotide identity among 793/B viruses of the present study ranged from 93.29% to 100%. Half of them were totally identical to 4/91 vaccine (KF377577), and others shared homologies between 95.65% to 97.71% to the vaccine (Table 2 ). QX type IBVs of this study shared 98.05% to 100% homologies. IBVs related to the Massachusetts strain had sequence similarities between 98.20% to 100%. They shared more than 98.86% similarity to M41 (KF188436). Massachusetts type viruses of this study share 98.2% similarity to IBV/Brazil/SGO/0116 and MDL15-3697 respectively. The average similarity of Iranian IS-1494-like, QX, Massachusetts, and 793/B to H120 vaccine strain are 79.86%, 74.5%, 99.1%, and 71.68%, respectively (Fig. 4 & Table 2). The similarity of mentioned IBV genotypes to 4/91 IBV vaccine strain are 80%, 81.3%, 72%, and 97.82%, respectively (Fig. 5 & Table 2).
Fig. 2.
Phylogenetic tree based on a partial sequence of the S1 gene, showing the relationship between the Iranian strains and other IBV strains. The neighbor-joining method was used with the Kimura2-parameter substitution model and 1000 bootstrap replicates to assign confidence level to the branches of the phylogenetic tree. Strains detected in the current study are indicated by black circles while Iranian previously identified strains marked by white squares. The vertical lines are for spacing branches and labels. The scale bar represents the distance unit between sequence pairs.
Table 1.
IBV genotype distribution (Percentage;%), given as proportional percentages, for each of eight provinces investigated during 2015–2017.
| Golestan | Kerman | Ardabil | Isfahan | Qazvin | Kurdistan | Razavi Khorasan | Khuzestan | |
|---|---|---|---|---|---|---|---|---|
| IS-1494 (Var2) | 87.5 | 73.68 | 55 | 66.66 | 78.94 | 63.15 | 73.33 | 69.23 |
| 793/B | 6.25 | 10.52 | 35 | 29.16 | 10.52 | 15.78 | 13.33 | 30.76 |
| Massachusetts | 6.25 | 10.52 | 5 | 4.16 | 5.26 | 21.05 | 6.66 | 0 |
| QX | 0 | 5.26 | 5 | 0 | 5.26 | 0 | 6.66 | 0 |
Fig. 3.
The frequently occurring of IBV genotype circulating in Iranian broiler farms during 2015–2017.
Table 2.
Percent identity of partial nucleotide sequences of the S1 glycoprotein genes of some Iranian IBVs to those of IBV reference strains.
 |
Fig. 4.
Percent identity of partial S1 gene sequences of some IBVs from the current study to that of the H120 strain.
Fig. 5.
Percent identity of partial S1 gene sequences of some IBVs from the current study to that of the 4/91 vaccine strain.
4. Discussion
The avian infectious bronchitis virus (IBV) first isolated in 1937 can be destructive and cause severe economic losses in the poultry industry worldwide (De Wit et al., 2011). Because of mutations or recombination events or both occurring during virus replication, dozens of serotypes and genotypes of IBV have been detected around the world (Cook et al., 2012). The first isolation of IBV in Iran was reported by Aghakhan et al. (1994). Genotyping of IBV strains isolated in Iran were classified into seven distinct phylogenetic groups (Massachusetts, 793/B, IS-1494, IS-720, QX, IR-1, and IR-2) (Najafi et al., 2016).
In the present study, IS-1494–like IBV was the most dominant type, followed by the 793/B type. Massachusetts and QX type IBVs were the third and fourth most frequently detected types, respectively. In comparison to our last survey (2014–2015) and other reports from Iran the IR1, IS-720, and IR-2 IB genotypes have disappeared (Fig. 6 ) (Najafi et al., 2016).
Fig. 6.
Comparison of the frequency of IBV genotypes during 2015–2017 and 2014-1015 (Najafi et al., 2016)circulating in Iranian broiler farms during 2015–2017.
Although IS-1494-like IBV was first reported in Israel in 2006, Egypt/Beni-Suef/01 was isolated in Egypt, with 99% similarity to IS/1494/06 of the Israeli variant 2 isolates, in 2001 (Gelb et al., 2005). Since then IS-1494-like IBV type was detected in Jordan, Turkey and other Middle East countries (Ganapathy et al., 2015). In Iran, IS-1494-like IBV was detected in 2010 for the first time and was reported as the second most frequently occurring IBV from 2010 to 2014 (Hosseini et al., 2015), thereafter it turned out to be the most dominant type in the molecular study done from 2014 to 2015 (Najafi et al., 2016). Based on the data, not only is IS-1494-like IBV type still the predominant IBV type, its appearance has remarkably increased up to two times more than that reported in the previous study (Najafi et al., 2016). Since the IS-1494-like IBV type vaccines are not used in Iran and because current vaccines and vaccination strategy do not protect completely against IS-1494-like, it is highly likely to become more widespread (Habibi et al., 2017).
793/B IBV type was first found in 1998 in Iran (Vasfi-Marandi and Bozorgmehrifard, 2001). According to the last two epidemiological data, the frequency of occurrence was 8.4% from 2010 to 2014, rising up to 21% from 2014 to 2015 (Najafi et al., 2016). According to our results, its presence is still high (19.33%) despite vaccination. As most 793/B IBVs detected in this study have the same nucleotide sequences with those of vaccine, the high frequency of occurrence probably reflects re-isolation of 793/B like IB vaccine strains. Like previous data, some 793/B type viruses were detected which differ from the vaccine viruses.
QX type IBV was first isolated by Bozorgmehri-Fard et al. in Iran, the incidence was 9.6% from 2010 to 2014 and 10% from 2014 to 2015 (Najafi et al., 2016).The present study shows a reduction in the frequency of occurrence rate of QX IBV, which may reflect the efficacy of combined Massachusetts and 793/B type IB vaccines. Epidemiological analysis of IBV in Italy and Spain indicated a causal relationship between Massachusetts plus 793/B vaccination and a decreased prevalence of QX (Franzo et al., 2016).
Kahya et al. detected IS-1494-06 like IBV strain in turkey, 2011 (Kahya et al., 2013). S1 analysis of Turkish IBVs (2014–2015) showed 17.2% of the sequences were closely related to MA5 vaccine strain, 49.5% to H120, and 48.9% of those were similar to the IS-1494-like IBV (Yilmaz et al., 2016). Mahmood et al. (2011) detected 793/B and Sul/01/09 type IBV strain in the North of Iraq (2008–2010) (Mahmood et al., 2011). In early 2011, Iraqi QX (IBV/CH/Kurdistan-Iraq/7266-10/2011) was isolated (Amin et al., 2012).The percentage of IS-1494-like, 793/B, QX, and DY12-2 genotypes in five Iraqi governorates was 46.87%, 40.62%, 9.37% and 3.12% in 2014, respectively (Seger et al., 2016). The presence of antibody against Massachusetts (M41), Arkansas, Connecticut, D-274, D-1466, and 4/91 IBV strains have been detected (Ahmed et al., 2007, Munir et al., 2012). Of 39 isolates obtained from 243 Oman backyard poultry in 2012, 66.67% of samples showed the greatest homology to genotype 793/B, 5.12% related closely to M41, 5.12% clustered with D274 and 2.56% belonged to IS-1494-like (Al-Shekaili et al., 2015). In another study from 2009 to 2014, the prevalence of 793/B, IS-1494-like, and Massachusetts genotypes were reported to be 63.7%, 21.2%, and 3%, respectively in Oman (Ganapathy et al., 2015). In summary, IS-1494-like and 793/B are the most prevalent IBV genotypes circulating in our region such as Oman, Iraq, and Turkey.
Due to intensive trading and uncontrolled movement of inhabitants and animals across borders, approximately the same distribution of IBV genotypes exists. The updated data from continuing molecular surveillance of IBV in Iran completes the puzzle of circulating IBV genotypes in the region. The study helps us to monitor the IBV strains to see whether new variants have emerged and we can evaluate the efficacy of our national IB vaccination program. Further, it is necessary to develop a vaccine in order to control the rising spread of the IS-1494-like IBV type.
Conflict of interest
All authors declare that they have no conflict of interest.
Acknowledgments
We would like to thank Ghalyanchi lab experts (Mr. Behrooz Asadi & Mr. Ahmad Vahedi), and Iranian Veterinary Organization lab experts (Dr. Hossein Maghsoudloo and Dr. Hamed Abdollahi) for their technical supports. Research Council, University of Tehran, financially (Grant No. 28692/6/14) supported this project.
References
- Abro S.H., Ullman K., Belák S., Baule C. Bioinformatics and evolutionary insight on the spike glycoprotein gene of QX-like and Massachusetts strains of infectious bronchitis virus. Virol. J. 2012;9(1):1. doi: 10.1186/1743-422X-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghakhan S., Abshar N., Fereidouni S.R.N., Marunesi C., Khodashenas M. Studies on avian viral infections in Iran. Arch. Razi. 1994;44:1–10. [Google Scholar]
- Ahmed Z., Naeem K., Hameed A. Detection and seroprevalence of infectious bronchitis virus strains in commercial poultry in Pakistan. Poult. Sci. 2007;86(7):1329–1335. doi: 10.1093/ps/86.7.1329. [DOI] [PubMed] [Google Scholar]
- Al-Shekaili T., Baylis M., Ganapathy K. Molecular detection of infectious bronchitis and avian metapneumoviruses in Oman backyard poultry. Res. Vet. Sci. 2015;99:46–52. doi: 10.1016/j.rvsc.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin O., Valastro V., Salviato A., Drago A., Cattoli G., Monne I. Circulation of QX-like infectious bronchitis virus in the Middle East. Vet. Rec. 2012;171(21) doi: 10.1136/vr.100896. 530–530. [DOI] [PubMed] [Google Scholar]
- Callison S.A., Hilt D.A., Boynton T.O., Sample B.F., Robison R., Swayne D.E., Jackwood M.W. Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J. Virol. Methods. 2006;138(1–2):60–65. doi: 10.1016/j.jviromet.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J.K., Jackwood M., Jones R. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41(3):239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- De Wit J., Cook J.K., Van der Heijden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40(3):223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo G., Tucciarone C.M., Blanco A., Nofrarías M., Biarnés M., Cortey M., Majó N., Catelli E., Cecchinato M. Effect of different vaccination strategies on IBV QX population dynamics and clinical outbreaks. Vaccine. 2016;34(46):5670–5676. doi: 10.1016/j.vaccine.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy K., Ball C., Forrester A. Genotypes of infectious bronchitis viruses circulating in the Middle East between 2009 and 2014. Virus Res. 2015;210:198–204. doi: 10.1016/j.virusres.2015.07.019. [DOI] [PubMed] [Google Scholar]
- Gelb J., Jr, Weisman Y., Ladman B., Meir R. S1 gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from the United States and Israel (1996 to 2000) Avian Pathol. 2005;34(3):194–203. doi: 10.1080/03079450500096539. [DOI] [PubMed] [Google Scholar]
- Habibi M., Karimi V., Langeroudi A., Ghafouri S., Hashemzadeh M., Farahani R., Maghsoudloo H., Abdollahi H., Seifouri P. Combination of H120 and 1/96 avian infectious bronchitis virus vaccine strains protect chickens against challenge with IS/1494/06 (variant 2)-like infectious bronchitis virus. Acta Virol. 2017;61(2):150–160. doi: 10.4149/av_2017_02_04. [DOI] [PubMed] [Google Scholar]
- Hosseini H., Fard M.H.B., Charkhkar S., Morshed R. Epidemiology of avian infectious bronchitis virus genotypes in Iran (2010–2014) Avian Dis. 2015;59(3):431–435. doi: 10.1637/11091-041515-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Huang Y.-P., Wang C.-H. Development of attenuated vaccines from Taiwanese infectious bronchitis virus strains. Vaccine. 2006;24(6):785–791. doi: 10.1016/j.vaccine.2005.08.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W. Review of infectious bronchitis virus around the world. Avian Dis. 2012;56(4):634–641. doi: 10.1637/10227-043012-Review.1. [DOI] [PubMed] [Google Scholar]
- Kahya S., Coven F., Temelli S., Eyigor A., Carli K.T. Presence of IS/1494/06 genotype-related infectious bronchitis virus in breeder and broiler flocks in Turkey. Ankara universitesi veteriner fakültesi dergisi. 2013;60(1):27–31. [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.-l., Su J.-l., Zhao J.-x., Zhang G.-z. Complete genome sequence analysis of a predominant infectious bronchitis virus (IBV) strain in China. Virus Genes. 2009;38(1):56–65. doi: 10.1007/s11262-008-0282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood Z.H., Sleman R.R., Uthman A.U. Isolation and molecular characterization of Sul/01/09 avian infectious bronchitis virus, indicates the emergence of a new genotype in the Middle East. Vet. Microbiol. 2011;150(1):21–27. doi: 10.1016/j.vetmic.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S., Hussain M., Farooq U., Jamal Q., Afreen M., Bano K., Khan J., Ayaz S., Kim K.Y., Anees M. Quantification of antibodies against poultry haemagglutinating viruses by haemagglutination inhibition test in Lahore. Afr. J. Microbiol. Res. 2012;6(21):4614–4619. [Google Scholar]
- Najafi H., Langeroudi A.G., Hashemzadeh M., Karimi V., Madadgar O., Ghafouri S.A., Maghsoudlo H., Farahani R.K. Molecular characterization of infectious bronchitis viruses isolated from broiler chicken farms in Iran, 2014–2015. Arch. Virol. 2016;161(1):53–62. doi: 10.1007/s00705-015-2636-3. [DOI] [PubMed] [Google Scholar]
- Seger W., GhalyanchiLangeroudi A., Karimi V., Madadgar O., Marandi M.V., Hashemzadeh M. Genotyping of infectious bronchitis viruses from broiler farms in Iraq during 2014–2015. Arch. Virol. 2016;161(5):1229–1237. doi: 10.1007/s00705-016-2790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfi Abad Shapouri M., Mayahi M., Assasi K., Charkhkar S. A survey of the prevalence of infectious bronchitis virus type 4/91 in Iran. Acta Vet. Hung. 2004;52(2):163–166. doi: 10.1556/AVet.52.2004.2.4. [DOI] [PubMed] [Google Scholar]
- Vasfi-Marandi M., Bozorgmehrifard M. Isolation and identification of infectious bronchitis viruses in chickens between 1997 and 2000 in Iran. J. Facul. Vet. Med. Univ. Tehran. 2001;56(3):119–124. [Google Scholar]
- Worthington K.J., Currie R., Jones R.C. A reverse transcriptase-polymerase chain reaction survey of infectious bronchitis virus genotypes in Western Europe from 2002 to 2006. Avian Pathol. 2008;37(3):247–257. doi: 10.1080/03079450801986529. [DOI] [PubMed] [Google Scholar]
- Yilmaz H., Altan E., Cizmecigil U.Y., Gurel A., Ozturk G.Y., Bamac O.E., Aydin O., Britton P., Monne I., Cetinkaya B. Phylogeny and S1 gene variation of infectious bronchitis virus detected in broilers and layers in Turkey. Avian Dis. 2016;60(3):596–602. doi: 10.1637/11346-120915-Reg.1. [DOI] [PubMed] [Google Scholar]