Highlights
-
•
A prediction method of coronaviral 3CLpro cleavage sites was proposed to balance the accuracy and false positives.
-
•
Three of the nine putative non-canonical cleavage sites were verified, which are located upstream to nsp4.
-
•
All 11 canonical cleavage sites of MERS-CoV 3CLpro were confirmed and the Michaelis constants were calculated.
Keywords: MERS-CoV, 3C-like protease, Canonical cleavage sites, Non-canonical cleavage sites, Michaelis constants
Abstract
Coronavirus 3C-like protease (3CLpro) is responsible for the cleavage of coronaviral polyprotein 1a/1ab (pp1a/1ab) to produce the mature non-structural proteins (nsps) of nsp4–16. The nsp5 of the newly emerging Middle East respiratory syndrome coronavirus (MERS-CoV) was identified as 3CLpro and its canonical cleavage sites (between nsps) were predicted based on sequence alignment, but the cleavability of these cleavage sites remains to be experimentally confirmed and putative non-canonical cleavage sites (inside one nsp) within the pp1a/1ab awaits further analysis. Here, we proposed a method for predicting coronaviral 3CLpro cleavage sites which balances the prediction accuracy and false positive outcomes. By applying this method to MERS-CoV, the 11 canonical cleavage sites were readily identified and verified by the biochemical assays. The Michaelis constant of the canonical cleavage sites of MERS-CoV showed that the substrate specificity of MERS-CoV 3CLpro is relatively conserved. Interestingly, nine putative non-canonical cleavage sites were predicted and three of them could be cleaved by MERS-CoV nsp5. These results pave the way for identification and functional characterization of new nsp products of coronaviruses.
1. Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) is an enveloped virus carrying a genome of positive-sense RNA (+ssRNA). It was identified as the pathogen of a new viral respiratory disease outbreak in Saudi Arabia in June 2012, named as Middle East Respiratory Syndrome (MERS). MERS-CoV emerged ten years after severe acute respiratory syndrome coronavirus (SARS-CoV) (Zaki et al., 2012) and quickly spread to several countries in Middle East and Europe (Assiri et al., 2013, Tashani et al., 2014). Soon after the first report, the MERS-CoV genome was sequenced and its genomic organization has been elucidated (van Boheemen et al., 2012). This new coronavirus is classified in the lineage C of beta coronavirus, and is close to bat coronavirus HKU4 and HKU5 (de Groot et al., 2013, Lau et al., 2013). Like other coronaviruses (Hussain et al., 2005, Zuniga et al., 2004), MERS-CoV contains a 3′ coterminal, nested set of seven subgenomic RNAs (sgRNAs), enabling translation of at least nine open reading frames (ORFs). The 5′-terminal two thirds of MERS-CoV genome contains a large open reading frame ORF1ab, which encodes polyprotein 1a (pp1a, 4391 amino acids) and polyprotein 1ab (pp1ab, 7078 amino acids), the latter being translated via a −1 ribosomal frameshifting at the end of ORF1a. These two polyproteins were predicted to be subsequently processed into 16 non-structural proteins (nsps) by nsp3, a papain-like protease (PLpro), and nsp5, a 3C-like protease (3CLpro) (Kilianski et al., 2013, van Boheemen et al., 2012).
Protease plays a key role during virus life cycle. It is essential for viral replication by mediating the maturation of viral replicases and thus becomes the target of potential antiviral drugs (Thiel et al., 2003, Ziebuhr et al., 2000). Investigating the cleavage sites of coronavirus proteases and the processing of polyproteins pp1a/1ab will benefit to identify the viral proteins and their potential function for viral replication. Some cleavage sites have been identified and confirmed by previous studies, including three cleavage sites of PLpros of human coronavirus 229E (HCoV 229E), mouse hepatitis virus (MHV), SARS-CoV, MERS-CoV and infectious bronchitis virus (IBV), whose cleavages release the first 3 non-structural proteins (Bonilla et al., 1995, Kilianski et al., 2013, Lim and Liu, 1998, Ziebuhr et al., 2007). The canonical cleavage sites of 3CLpros, the sites between the recognized nsps, have also been characterized, including all sites of MHV, IBV, SARS-CoV and a fraction of sites of HCoV 229E which release the non-structural proteins from nsp4 to nsp16 (Deming et al., 2007, Grotzinger et al., 1996, Liu et al., 1994, Liu et al., 1997, Lu et al., 1995). For 3CLpro of MERS-CoV, two cleavage sites releasing nsp4 to nsp6 have been identified (Kilianski et al., 2013). However, other cleavage sites remain to be characterized.
Furthermore, efforts have been taken to predict these cleavages sites by sequence comparison. Gorbalenya et al. (1989) made the first systematical prediction on IBV pp1a/1ab according to the substrate specificity of 3C protease of picornaviruses. However, two of their predicted cleavage sites within nsp6 of IBV were proved uncleavable (Liu et al., 1997, Ng and Liu, 2000). Gao et al. (2003) developed a software (ZCURVE_CoV) to predict the nsps as well as gene-encoded ORFs of coronaviruses more accurately based on previous studies of 3CLpros cleavage sites of IBV, MHV and HCoV 229E. Later on, non-orthogonal decision trees were used to mine the coronavirus protease cleavage data and to improve the sensitivity and accuracy of prediction (Yang, 2005). However, while these methods focus on the prediction of the canonical cleavage sites and target more and more on prediction accuracy to avoid false positives, potential non-canonical cleavage sites might be neglected. For example, a cleavage site between nsp7 and 8 of MHV strain A59 is not predicted by above methods, but proved to be physiologically important since it produces a shorter nsp7 that can support the growth of MHV carrying a mutation on nsp7–8 cleavage site (Deming et al., 2007). Therefore, the substrate specificities of coronaviruses 3CLpros are complicated. A 3CLpro substrate library of four coronaviruses (HCoV-NL63, HCoV-OC43, SARS-CoV and IBV) containing 19 amino acids × 8 positions variants was constructed by making single amino acid (aa) substitution at each position from P5 to P3′, and their cleavage efficiencies were measured and analyzed to find out the most preferred residues at each position (Chuck et al., 2011). However, the non-canonical cleavage site with less preferred residues of 3CLpro is adopted by coronaviruses (Deming et al., 2007). Thus we speculate that other potential 3CLpro cleavage sites may still exist in coronaviruses.
In order to set up a more moderate and balanced criteria for protease cleavage site identification, we compared six scanning conditions with different stringency to systematically predict the 3CLpro cleavage sites on pp1a/1ab of five coronaviruses including MERS-CoV. As a representative, the cleavability of the predicted cleavage sites of MERS-CoV 3CLpro was analyzed by the recombinant luciferase cleavage assay and the fluorescence resonance energy transfer (FRET) assay. The results showed that all 11 canonical cleavage sites of MERS-CoV pp1a/1ab were cleavable in our experiments and three of nine predicted non-canonical cleavage sites appeared to be cleavable. Our study points out a new direction regarding the prediction and identification of cleavage sites of proteases and contributes to understanding the mechanism of coronaviral polyprotein processing.
2. Materials and methods
2.1. Information collection of coronavirus 3CLpro cleavage sites
The genome sequences of 28 coronaviruses were downloaded from Genebank database and the sequences of the 3CLpro cleavage sites were collected from P4 to P2′ (Tables S1–S4). The substrate profiles of each coronavirus group and the whole Coronavirinae were summarized (Table S5).
2.2. Construction of recombinant 3CLpro expression vectors
The coding sequence of MERS-CoV nsp5 (NC_019843) was synthesized chemically by GenScript and cloned into vectors pET28a and pGEX-6p-1, respectively. The catalytic residue mutation C148A was generated by over lapping PCR with mutagenic primers (Table S6). All the clones and mutations were confirmed by DNA sequencing.
2.3. Expression and purification of recombinant proteins
The expression vectors were transformed into Escherichia coli strain BL21 (DE3). The cells were grown at 37 °C in Lysogeny broth (LB) medium with antibiotics and induced with 0.2 mM isopropylb-d-thiogalactopyranoside (IPTG) at 16 °C for 12 h. The cells were harvested and resuspended in lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.05% NP40, 0.1 mg/ml lysozyme and 1 mM PMSF) at 4 °C. After incubation for 30 min on ice, 10 mM MgCl2 and 10 μg/ml DNase I (Sigma) were added to digest the genomic DNA. The supernatant of cell lysate was applied to affinity chromatography column after centrifugation. The recombinant protein with His-tag was bound with nickel-nitrilotriacetic acid (Ni-NTA) resin (GenScript) and washed with buffer A (50 mM Tris–HCl, pH 7.5, 150 mM NaCl), buffer B (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 20 mM imidazole) and buffer C (50 mM Tris, pH 7.5, 150 mM NaCl, 50 mM imidazole). Proteins were eluted with buffer D (50 mM Tris, pH 7.5, 150 mM NaCl, 250 mM imidazole). GST-tagged protein was bound with GST resin (GenScript), washed with buffer A and eluted with buffer A supplemented with 10 mM reduced glutathione (GSH). The purified proteins were desalted and concentrated by ultrafiltration using 30 kDa amicon ultra 0.5-ml centrifugal filter (Millipore).
2.4. Luciferase-based biosensor assay
All the cleavage sites (eight residues, ranging from P5 to P3′) were inserted into Glo-Sensor 10F linear vector. Comparing to the wild type firefly luciferase (550 aa), Glo-Sensor luciferase has short truncations at both termini with C- and N-part reversed, resulting in the new 234-aa N- and 233-aa C-terminal region respectively. The inserted sequence and the reversed arrangement of the N- and C-terminal regions reduce the luciferase activity dramatically. After the recognition sequence was cut off by nsp5, the luciferase recover its activity and luminescence in the presence of luciferase substrate. A back to front recombinant firefly luciferase inserted with different cleavage sites was expressed when the recombinant plasmids were co-incubated with a cell-free protein expression system extracted from wheat germ (Promega). After incubation for 2 h at 25 °C, nsp5 was added into the system and the whole system was incubated at 30 °C for 1 h. Then, the reaction system was diluted 20 times and mixed thoroughly with equal volume of luciferase substrate. Luciferase luminescence was measured by a luminometer (Promega) after incubation for 5 min at room temperature.
2.5. Peptide-based FRET assay
All the 11 conserved putative recognition sites were designed from P12 to P8′, synthesized and modified with a typical shorter wavelength FRET pair, N-terminal DABCYL and C-terminal Glu-EDANS by GL Biochem (Shanghai). The peptides were completely dissolved in DMSO and the final concentration of DMSO in the reaction system was 1%. 180 μM substrate peptide and 16.3 μM tagged nsp5 were mixed in the solution of 50 mM Tris, pH 7.5, 1 mM EDTA, 50 μM DTT and incubated at 37 °C for 2 h. To calculate kcat/Km, different amounts (7.2–180 μM) of substrate peptides were co-incubated with 16.3 μM nsp5. The reaction system was placed in Giernor black plate and the fluorescence was detected by a microplate reader (Molecular Devices) with Ex/Em (nm/nm) = 340/490. Relative Fluorescence Unit (RFU) was collected every 30 s for 2 h.
2.6. Calculation of Michaelis constants
The initial slope (slope A = RFU/min) was generated from the linear interval of the rising stage. Then, a linear equation was generated using the RFU at plateau (RFUmax) vs. the concentration of substrate. The slope (slope B = RFU/[S]) indicates the RFU change at per unit change of [S]. The initial reaction velocity (V 0 = [S]/min) was calculated through dividing slope A by slope B. The Michaelis–Menten kinetic constants were generated by Lineweaver–Burk plot.
3. Results
The coronavirus 3CLpros and their cleavage sites are evolutionarily conserved among different genera.
To study the genetic diversity and evolution of 3CLpro cleavage sites of coronaviruses pp1a/1ab, 308 primary sequences of 3CLpro cleavage sites (ranging from P4 to P2′) of 28 species of coronaviruses were collected and listed in Tables S1–S4, including the predicted and verified cleavage sites. 11 canonical cleavage sites of each coronavirus were joined end to end to produce a spliced sequence which was then used to produce a phylogenetic tree (Fig. 1A). In addition, the sequences of all coronavirus 3CLpro were used to generate another phylogenetic tree (Fig. 1B). The analyses showed that the phylogenetic distances and taxonomic positions of each virus, in both phylogenetic trees, were mostly consistent with that classified by the International Committee on Taxonomy of Viruses (ICTV) (http://www.ictvonline.org/virusTaxonomy.asp). These results implied that the cleavage sites of coronaviral 3CLpros might co-evolve with 3CLpros, and the genetic diversity of both 3CLpro and its cleavage sites are relatively conserved between different genera of coronaviruses. However, on the phylogenetic tree generated with 3CLpro cleavage sites (Fig. 1A), the members of the genus Gammacoronavirus, although clustered closely, is split into alphacoronaviruses and deltacoronaviruses, suggesting that the cleavage sites of gammacoronaviruses may have undergone recombination events during evolution.
Fig. 1.
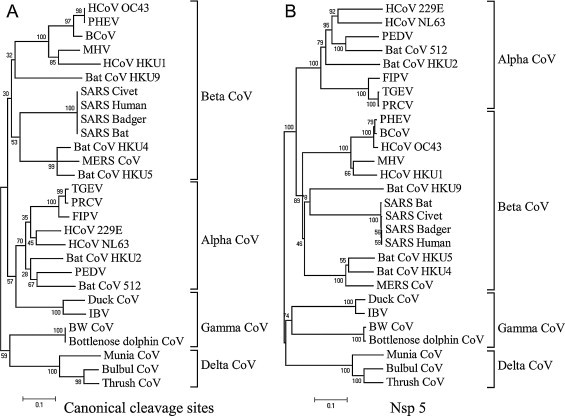
The phylogenetic tree of 26 representative coronaviruses. (A) The tree was generated using an alignment of the joined canonical cleavage sites of 26 coronaviruses. Sequence alignment was performed by ClustalX 2.0, and the tree was built by neighbor-joining method in MEGA 4 (Bootstrap: replication = 1000, random seed = 64,238). (B) The tree was generated by the sequence of nsp5 and the method is the same as described above.
3.1. Setup of the predicting conditions of coronaviruses 3CLpro cleavage sites.
In order to develop an optimized method for cleavage site prediction that can cover all possible cleavage sites with fewer false positives, we have set three levels of criteria (stringent, moderate and mild) for cleavage site prediction. In the stringent rules, 3CLpro cleavage sites only comprise the most preferred residues at each position based on previous description (Chuck et al., 2011). In moderate rules, 3CLpro cleavage sites comprise residues which ever appeared in the cleavage sequences of congeneric coronaviruses at each particular position. As for mild rules, the cleavage sites could comprise any residues ever found in the cleavage sequences of all coronaviruses at each particular position. Because the substrate preference at P4 and P2′ is not strong, we decided to adopt two different lengths of cleavage sequences for prediction, one containing six residues from position P4 to P2′, and the other containing four residues from position P3 to P1′. These two lengths of cleavage sequences, combining with the three different criteria, made up a total of six search conditions for cleavage site predication with decreasing degree of stringency. The canonical cleavage sites of 3CLpro for these seven groups of coronaviruses were summarized in Tables S1–S4 and used to set conditions III to VI. Possible residues at each particular position of 3CLpro cleavage sites were predicted based on all six conditions to make the cleavage site profile of coronaviruses 3CLpro (Table S5). In principle, when condition I was employed, the least number of possible cleavage sites were identified in a scanned sequence, while condition VI predicted the largest number of possible cleavage sites in a scanned sequence.
To the applicability, we applied all the six conditions on five representative coronaviruses, including HCoV 229E from alphacoronavirus, MHV from betacoronavirus lineage A, SARS-CoV from beta coronavirus lineage B, MERS-CoV from betacoronavirus lineage C and IBV from gammacoronavirus. All possible cleavage sites predicted based on each condition were scanned on pp1a/1ab of five representative coronaviruses and the results were summarized in Table 1 . As shown in Table 1, increasing numbers of cleavages sites were found for each coronavirus when conditions from I to VI were applied. The results showed that condition I and II were too strict to cover all 11 canonical cleavages sites; condition V and VI were too loose so as to produce two to three times more than 11 cleavages sites; condition III could only cover the canonical cleavage sites for SARS CoV; only condition IV generates an appropriate number of cleavage sites for all five coronavirus. Therefore, search condition IV was chosen for further analysis of the cleavage sites of MERS-CoV.
Table 1.
The number of cleavage sites in pp1ab of 5 representative coronaviruses predicted by using 6 search conditions.
| HCoV 229E |
MHV |
SARS-CoV |
MERS-CoV |
IBV |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CSa | PSb | CS | PS | CS | PS | CS | PS | CS | PS | |
| Condition Ic | 1 | 0 | 2 | 0 | 2 | 0 | 2 | 0 | 1 | 0 |
| Condition II | 1 | 0 | 4 | 1 | 3 | 0 | 4 | 0 | 2 | 0 |
| Condition III | 11 | 4 | 11 | 5 | 11 | 0 | 11 | 2 | 11 | 3 |
| Condition IV | 11 | 10 | 11 | 14 | 11 | 4 | 11 | 9 | 11 | 5 |
| Condition V | 11 | 9 | 11 | 17 | 11 | 11 | 11 | 12 | 11 | 11 |
| Condition VI | 11 | 15 | 11 | 23 | 11 | 19 | 11 | 19 | 11 | 13 |
Canonical cleavage sites, which are located between recognized nsps.
Putative cleavage sites, which are located inside various nsps.
Six search conditions are designed: conditions I, III and V cover six residues from P4 to P2′; conditions II, IV and VI cover four residues from P3 to P1′. Conditions I and II are set to comprise the most preferred residues at each position; conditions III and IV comprise residues appeared in the cleavage sites of congeneric coronaviruses; conditions V and VI comprise residues appeared in the cleavage sequences of any coronaviruses.
By applying the search condition IV, 9 putative cleavage sites (PSs) as well as 11 canonical cleavage sites (CSs) were predicted (Table 2 ). Although the canonical cleavage sites of MERS-CoV 3CLpro have been predicted by sequence alignment with other coronavirus (van Boheemen et al., 2012), our results suggested that the additional cleavage might occur in the process of MERS-CoV pp1a/1ab processing.
Table 2.
The cleavage site prediction outcomes of MERS-CoV using search condition IV.
| Canonical cleavage sites |
Putative cleavage sites |
||||
|---|---|---|---|---|---|
| Site | Position | Sequence a | Site | Position | Sequence |
| CS4/5 | 3247 | GVLQ↓SG | PS1-1 | 122 | TTLQ↓GK |
| CS5/6 | 3553 | VVMQ↓SG | PS3-1 | 1191 | VLLQ↓GH |
| CS6/7 | 3845 | AAMQ↓SK | PS3-2 | 1278 | DIPQ↓SL |
| CS7/8 | 3928 | SVLQ↓AT | PS3-3 | 1683 | VVLQ↓GL |
| CS8/9 | 4127 | VKLQ↓NN | PS5-1 | 3332 | HAMQ↓GT |
| CS9/10 | 4237 | VRLQ↓AG | PS6-1 | 3580 | IILQ↓AT |
| CS10/12 | 4377 | ALPQ↓SK | PS12-1 | 5076 | NILQ↓AT |
| CS12/13 | 5130 | TTLQ↓AV | PS13-1 | 5591 | VTVQ↓GP |
| CS13/14 | 5908 | YKLQ↓SQ | PS16-1 | 6793 | FKVQ↓NV |
| CS14/15 | 6432 | TKVQ↓GL | |||
| CS15/16 | 6775 | PRLQ↓AS | |||
The “↓” indicates the cleavage position.
3.2. Activity of MERS-CoV 3CLpro in biochemical assays
To verify the activity of MERS-CoV 3CLpro and cleavability of the predicted cleavage sites, the biochemical assay systems of MERS-CoV 3CLpro were established. As shown in Fig. 2A and B, we first expressed and purified MERS-CoV 3CLpro (nsp5) with different tags and mutation: N-terminally GST-tagged nsp5 (Gnsp5, 60.4 kDa), N-terminally His-tagged (34 extra amino acids with 6× His tag and linker provided by vector pET-28a) nsp5 (Hnsp5, 36.9 kDa), Hnsp5 with catalytic residue mutation C148A (Hnsp5m, 36.9 kDa) (Kilianski et al., 2013) and GST tag-GVLQ-nsp5 with C148A mutation and 6× His tag (Gnsp5mH, 61.6 kDa), in which the sequence motif GVLQ represents the last four residues of MERS-CoV nsp4, mimicking the cleavage site of MERS-CoV nsp4/nsp5. In the biochemical assays, the Gnsp5mH with catalytic residue mutation C148A could not undergo self-cleavage at the cleavage site to release GST in incubation for 16 h (Fig. 2C), indicating that the 3CLpro activity of MERS-CoV nsp5 in Gnsp5mH was inactivated by the mutation C148A. Thus, Gnsp5mH was used as protease substrate in the following biochemical assays. To verify the 3CLpro activity of recombinant nsp5s, Gnsp5 and Hnsp5 were incubated with substrate Gnsp5mH for 5 min to 16 h and analyzed by SDS-PAGE (Fig. 2D) and Western blotting, respectively (Fig. 2E). Both Gnsp5 and Hnsp5 showed the proteolysis activity to cleave the substrate Gnsp5mH into two parts: GST (26.0 kDa) and nsp5mH (34.1 kDa), which were confirmed by the correlation of their molecular weight (Fig. 2D and E). However, the 3CLpro activity of Gnsp5 was obviously weaker than that of Hnsp5, which could entirely cleave the substrate Gnsp5mH 2 h post treatment (Fig. 2D and E). These results could be explained by that the larger fusion tag at the N terminus of MERS-CoV 3CLpro significantly reduced the proteolysis activity of 3CLpro, which was consistent with the previous observation (Xue et al., 2007). In the biochemical assays, the relatively lower proteolysis activity of 3CLpro will benefit to observe the influence of different substrates. Therefore, both recombinant Gnsp5 and Hnsp5 were used as MERS-CoV 3CLpro in the following studies.
Fig. 2.
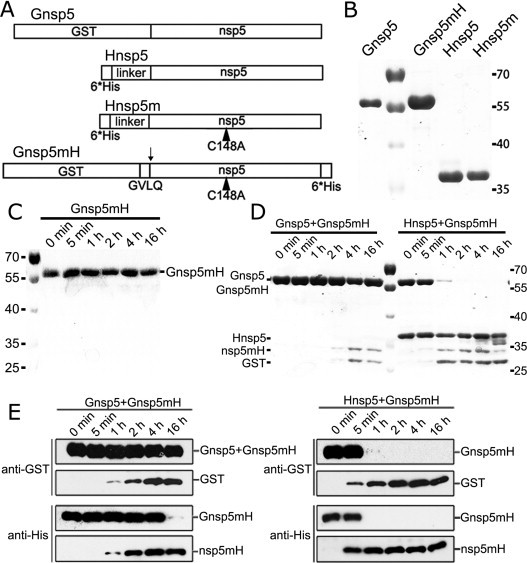
Purification of recombinant nsp5 of MERS-CoV and analysis of substrate cleavage by protein cleavage assays. (A) Diagram of 4 recombinant proteins. The catalytic residue mutation C148A is indicated by a small black triangle. GVLQ (P4-P1) are the last four residues of MERS-CoV nsp4. The insertion of these 4 residues made the N-terminal GST tag cleavable by active nsp5. The cleavage position was indicated by a down arrow. (B) SDS-PAGE analysis of the recombinant proteins. After all of the proteins were purified, they were concentrated to 1 mg/ml. 2 μg Gnsp5mH, Hnsp5, Hnsp5m and 1 μg Gnsp5 were loaded to a 10% SDS PAGE gel and stained with Coomassie brilliant blue. (C) and (D) Gnsp5mH was incubated in 50 mM Tris, pH 7.5, 1 mM EDTA, and 50 μM DTT at 37 °C alone (C) or with Gnsp5 and Hnsp5 (D). The substrate protein was diluted to 0.1 mg/ml. A fraction of the reaction mixture was taken out at each time point (0 min, 5 min, 1 h, 2 h, 4 h, 16 h) and analyzed by 10% SDS-PAGE. Products were detected by CBB staining (D) and Western blot (E).
3.3. Identification of the cleavability of predicted cleavage sites in MERS-CoV pp1a/1ab
To rapidly evaluate the proteolysis activity of MERS-CoV 3CLpro toward the predicted cleavage sites of different substrates, a sensitive luciferase-based biosensor assay was adopted. As shown in Fig. 3A, the canonical cleavage sites (CS) of MERS-CoV nsp4/nsp5 (CS4/5) and nsp5/nsp6 (CS5/6), which were experimentally confirmed in a previous study (Kilianski et al., 2013), were inserted into the inverted and circularly permuted luciferase construct pGlo-10F, in which the N-terminal and C-terminal halves of luciferase gene are separated. The resulting luciferase in translation system in vitro was inactive and could convert into an active luciferase when cleaved by recombinant viral protease at the engineered cleavage sites (such as CS4/5 and CS5/6). In this system, the luciferase signals were detected when incubated with both Gnsp5 and Hnsp5, respectively (Fig. 3B). In contrast, the mutated nsp5 (Hnsp5m) could not convert the inactive luciferase into active form (Fig. 3B). This result indicated that the luciferase-based biosensor assay could be used to evaluate the proteolysis activity of MERS-CoV 3CLpro. Then, the other nine canonical cleavage sites and nine putative cleavage sites composed with 8 aa from MERS-CoV pp1a/1ab were inserted into the luciferase construct pGlo-10F, and the luciferase-based biosensor assays were performed using Hnsp5 and Hnsp5m, respectively. As shown in Fig. 3C, all the 11 canonical cleavage sites of MERS-CoV 3CLpro generated luciferase signal by Hnsp5 at least 6.6 times higher than by the inactive Hnsp5m, indicating that all these canonical sites could be cleaved by MERS-CoV 3CLpro. These results experimentally verified the existence of the 11 predicted canonical cleavage sites. Interestingly, among the nine putative cleavage sites, the luciferase signals of PS1-1, PS3-1 and PS3-3 remarkably increased more than 70 folds when incubated with Hnsp5, indicating that the putative cleavage sites, located inside nsp1 and nsp3 of MERS-CoV respectively, might be cleavable (Fig. 3D). The other 6 predicted putative sites (PS3-2, PS5-1, PS6-1, PS12-1, PS13-1, and PS16-1) showed less than 2.5 folds increase of luciferase signal when they were treated by Hnsp5 comparing with those treated by Hnsp5m (Fig. 3C and D). Due to high sensitivity of the luciferase-based biosensor assay and the fact that the confirmed canonical cleavage sites generated at least 6.6 times increase of luciferase signal, the cleavage signal of these six sites may represent the background level, indicating that they are likely uncleavable per se. These results suggest that previously unrecognized 3CLpro cleavage sites may exist inside the nsps, which were regarded as non-canonical cleavage sites.
Fig. 3.
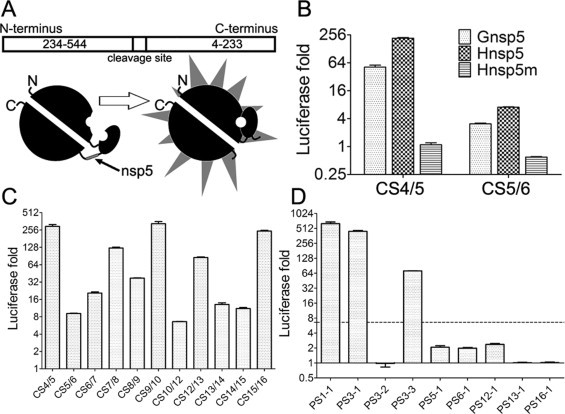
Identification of the cleavability of predicted cleavage sites in recombinant luciferase cleavage assays. (A) Schematic diagram of the recombinant luciferase. (B) Verification of the recombinant luciferase assays. Inactive luciferase was synthesized in the cell-free translation system and the reaction mixture incubated at 25 °C for 2 h. After that, the protein mixture was divided into four parts and incubated with 1.63 μM Gnsp5, Hnsp5, Hnsp5m or H2O, respectively. After incubation for 1 h at 30 °C, the reaction product was diluted 20 times and mixed with equal amount of luciferase substrate. After incubation at room temperature for 5 min, the luciferase luminescence was measured. Luciferase activation fold was calculated through dividing the signal value of the reaction system treated with active Hnsp5 by the one treated with the inactive nsp5 mutant Hnsp5m. (C) The luciferase cleavage assay of predicted 11 canonical cleavage sites and (D) 9 putative cleavage sites. The luciferase expression vector inserted with cleavage sites were added to the wheat germ protein translation mix and incubated at 25 °C for 2 h, and the reaction mixture was divided and treated with Hnsp5 and Hnsp5m, respectively. The dashed line indicates the lowest fold increase of luciferase signal by cleavage of previously confirmed 3CLpro cleavage sites. The data presented here are the mean values ± SD derived from three independent experiments.
3.4. Analysis of the substrate specificity of MERS-CoV 3CLpro
The substrate specificity of coronaviruses 3CLpro is determined by the residues from P4 to P2′ positions of cleavage sites, especially depending on the P1, P2 and P1′ positions, which would benefit the prediction of cleavage site and design the broad-spectrum inhibitors of coronaviruses 3CLpro (Chuck et al., 2011, Hegyi and Ziebuhr, 2002). Previous studies demonstrated that different canonical cleavage sites of some representative coronaviruses are not equally susceptible to proteolysis by recombinant 3CLpro (Fan et al., 2004, Hegyi and Ziebuhr, 2002). To define the susceptibility of the canonical cleavage sites and substrate specificity of MERS-CoV 3CLpro, 20-mer synthetic peptides representing corresponding canonical cleavage sites of MERS-CoV 3CLpro were synthesized and modified with N-terminal DABCYL and C-terminal Glu-EDANS (Fig. 4A). The fluorophore EDANS and quencher DABCYL are widely used in the biochemical assays based on the fluorescence resonance energy transfer (FRET). As shown in Fig. 4B, the peptides represented cleavage sites CS4/5 and CS5/6 were tested to optimize the FRET assay, and the relative fluorescence unit (RFU) folds of both sites significantly increased when incubated with Gnsp5 and Hnsp5. Although the FRET assay system is more costly and less sensitive than the luciferase-based biosensor assay (Fig. 3, Fig. 4), it provides continuous read signals during the process of reaction, which could measure the kinetic characteristic of protease toward different substrates. The initial reaction rate (RFU/min) of all 11 canonical cleavage sites of MERS-CoV were measured and shown in Fig. 4C. The Michaelis constants including kcat, Km, kcat/Km and relative kcat/Km were then calculated (Table 3 ). As shown in Table 3, the substrate specificity of MERS-CoV 3CLpro is relatively conserved with other coronaviruses as previously reported (Fan et al., 2004, Hegyi and Ziebuhr, 2002, Ziebuhr and Siddell, 1999). The relative kcat/Km values of CS4/5 and CS5/6 indicated that the cleavage sites flanking MERS-CoV 3CLpro are converted significantly faster than other sites. The efficient proteolysis at the sites flanking nsp5 implies that the nsp5 (3CLpro) might be released from the polyprotein 1a/1ab at the very early stage of the maturation of viral nsps, which is similar with the HCoV, TGEV, SARS-CoV and MHV (Fan et al., 2004, Hegyi and Ziebuhr, 2002). However, the relative kcat/Km value of CS4/5 is lower than that of CS5/6 (Table 3), which is different from that of the coronaviruses (Fan et al., 2004, Hegyi and Ziebuhr, 2002). This could be explained by that the residue Gly (G) at the P4 of cleavage site between nsp4 and nsp5 of MRES-CoV reduces the protease activity of 3CLpro comparing with the residues Ser (S), Ala (A) and Thr (T) of other coronaviruses (Tables S1-S4) as previous described (Chuck et al., 2011). Whether such disparity plays any role in the replication and pathogenesis of MERS-CoV is unknown.
Fig. 4.
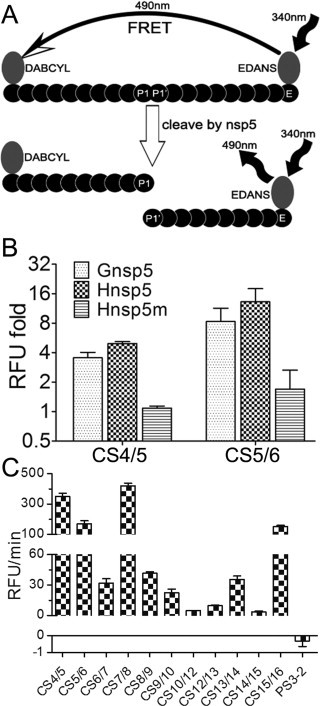
Kinetic analyses of the 11 canonical cleavage sites cleaved by MERS-CoV nsp5 by FRET assays. (A) Diagram of the FRET mechanism. EDANS transfer its 490 nm energy to DABCYL at the excitation of 340 nm, making the emission undetected. After the peptide bond between P1 and P1′ was cut off by nsp5, the separation disabled the energy transferring and the 490 nm emission of EDANS can be detected. (B) 180 μM synthesized peptide was incubated with 16.3 μM tagged nsp5. After incubation for 2 h at 37 °C, the fluorescence (Ex/Em = 340 nm/490 nm) was read by a luminometer. (C) The rate of RFU rise (slope A = RFU/min) at the linear interval right after the reaction began. The data presented here are the mean values ± SD derived from three independent experiments.
Table 3.
The Michaelis constants of the 11 canonical cleavage sites of MERS-CoV 3CLpro.
| kcat (min−1) | Km (μM) | kcat/Km (mM−1 min−1) | kcat/Km (rel) | P valuea | |
|---|---|---|---|---|---|
| CS4/5 | 0.3053 ± 0.05661 | 75.89 ± 17.57 | 4.023 | 1 | – |
| CS5/6 | 0.6811 ± 0.1388 | 88.25 ± 18.19 | 7.717 | 1.9 | 0.015 |
| CS6/7 | 0.2993 ± 0.04865 | 264.7 ± 36.95 | 1.131 | 0.28 | <0.0001 |
| CS7/8 | 2.073 ± 0.5245 | 321.89 ± 97.63 | 6.441 | 1.6 | 0.011 |
| CS8/9 | 0.5161 ± 0.04468 | 423.1 ± 27.32 | 1.220 | 0.30 | <0.0001 |
| CS9/10 | 2.390 ± 0.2397 | 833.8 ± 182.1 | 2.866 | 0.71 | 0.103 |
| CS10/12 | 0.1152 ± 0.02049 | 534.9 ± 91.71 | 0.2154 | 0.053 | <0.0001 |
| CS12/13 | 0.1083 ± 0.002443 | 83.90 ± 3.949 | 1.290 | 0.32 | <0.0001 |
| CS13/14 | 0.1815 ± 0.0200 | 449.7 ± 1.996 | 0.4036 | 0.10 | <0.0001 |
| CS14/15 | 0.05115 ± 0.00878 | 207.2 ± 59.61 | 0.2469 | 0.061 | <0.0001 |
| CS15/16 | 0.3849 ± 0.01126 | 100.7 ± 6.473 | 3.823 | 0.95 | 0.58 |
P value was statistically analyzed by unpaired Students's t-test.
4. Discussion
The processing of viral polyprotein by 3CLpro is essential for the replication of coronaviruses. Besides the 11 canonical cleavage sites of coronaviruses, some additional cleavage sites inside nsps, so-called non-canonical cleavage sites, have also been identified (Deming et al., 2007). Therefore, more non-canonical 3CLpro cleavage sites are to be identified in different coronaviruses. In this study, we designed six search conditions for predicting 3CLpro cleavage sites, among which, the search condition IV provides a feasible way to reveal the potential cleavage sites of 3CLpro within coronaviruses. Based on the genetic diversity of different coronavirus genera (Fig. 1), the scanning condition IV adopted the residues of 3CLpro cleavage sites, which ever appeared in the cleavage sequences of congeneric coronaviruses at position P3 to P1′. In contrast, conditions I, II, III, V and VI were either too restrictive or generated too many false positive outcomes (Table 1). In the suggested condition IV, 4 residues from position P3 to P1′ were applied to the prediction of 3CLpro cleavage site. By measuring the relative protease activities of 3CLpro from different coronavirus genera against 19 amino acids × 8 positions of substrate variants, it is shown that the substrate specificity of position P5, P2′ and P3′ are significantly lower than other positions (Chuck et al., 2011). Therefore, the consideration of six or more residues is unnecessary, which could lead to leave-out of potential cleavage sites (Table 1). Comparing with the previous researches on the prediction and identification of 3CLpro cleavage sites, the scanning condition IV showed its advantages. For example, the two nonexistent putative cleavage sites predicted within nsp6 of IBV (Gorbalenya et al., 1989, Liu et al., 1997, Ng and Liu, 1998) were avoided in our prediction method (data not shown). Notably, the noncanonical cleavage site at the end of MHV nsp7 identified by Deming et al. could be predicted using scanning condition IV.
By using the search condition IV, 9 putative cleavage sites were predicted in MERS-CoV pp1ab in addition to the 11 canonical cleavage sites. The luciferase signal of CS10/12 increased 6.6 fold when treated with nsp5 in the recombinant luciferase cleavage assays, which is the lowest among the 11 canonical cleavage sites (Fig. 3C). Therefore, the 6.6 fold increase of luciferase signal was used arbitrarily as a threshold for judging positive and negative. Among the nine predicted putative cleavage sites, three sites (PS1-1, PS3-1 and PS3-3) showed obviously increasing signals at least 70 times above the background (Fig. 3D) and therefore were regarded as cleavable sites. The increase of signals of other six predicted putative cleavage sites was less than 2.5 times (Fig. 3D). Therefore, they were regarded as non-cleavable sites and thus as false positives from the prediction. Interestingly, the homologous sequence of PS1-1 and PS3-1 are conserved in lineage C of betacoronavirus including MERS-CoV, BatCoV HKU4 and BatCoV HKU5 (Fig. 5A and B). However, PS3-3 is MERS-CoV unique sequence (Fig. 5C). Moreover, the cleavability of a cleavage site in biochemical assays is a necessary but not sufficient condition for its physiological existence in the viral infection. A predicted cleavage site may or may not be accessible by a protease. The 3D structure model of MERS-CoV ADP-ribose-1-monophosphatase (ADRP) domain built by comparative protein modeling and papain like protease (PLpro) domain (Bailey-Elkin et al., 2014) showed that both PS3-1 and PS3-3 are located at the surface of ADRP and PLpro domain, opposite to the enzymatic active centers (Fig. 5D and E), suggesting that these two sites are like approachable by the proteinase. Most recently, the crystal structure of MERS-CoV 3CLpro was determined (Needle et al., 2015). Although PS5-1 is also located at the surface of MERS-CoV 3CLpro, the self-cleavage of MERS-CoV nsp5 was not observed in this study (Fig. 2). Therefore, the threshold we proposed in the luciferase-based biosensor system to exclude the false positive prediction results is reasonable (Fig. 3D). However, further studies are needed to identify the predicted cleavage products from the cells infected by MERS-CoV. Currently, such work with live MERS-CoV is limited in our research facilities due to the biosafety rules, but it can be addressed in collaboration in the future.
Fig. 5.
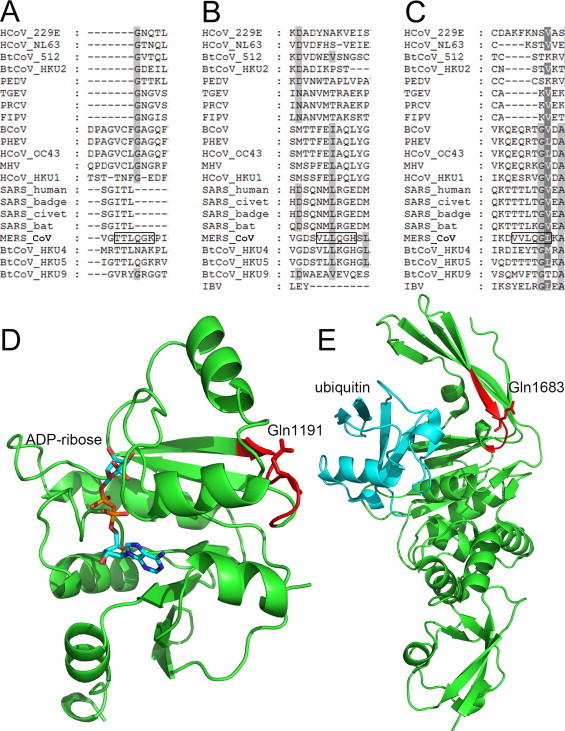
Conservation analysis and the spatial location of the novel noncanonical cleavage sites. The sequence alignment of nsp region covering PS1-1 site (A), PS3-1 site (B) and PS3-3 site (C). The cleavage sites of MERS-CoV were indicated by black boxes. (D) Homology modeled structure of ADRP domain of MERS-CoV (template: 2FAV). ADRP domain was shown as green ribbon. The putative cleavage site was colored in red and the cleavage position Gln was showed by stick. The substrate (ADP-ribose) of ADRP domain was shown by stick and colored by atoms (C: cyan, O: red, N: blue, P: orange). (E) Structure of papain-like protease of MERS-CoV (4RF1). The PLpro domain was shown as cartoon and colored green. The ligand ubiquitin was colored cyan. The putative cleavage site was colored red and the cleavage position Gln was showed by stick.
Notably, the outcomes of the two cleavage assay systems were different. The signal fold change of highly sensitive luciferase-based biosensor assay is dependent on the accumulation of active luciferase cleaved by nsp5 during 1 h (Section 2), while the outcome of the FRET assay is instant relative fluorescence unit (RFU) signal. The RFU/min is the initial speed of the reaction, which reflects but not equals to the efficiency of the cleavage. These differences may be caused by the steric hindrance of the luciferase subunits, the distance between fluorophore and quencher of substrates for FRET assay and substrate solubility. Therefore, the activity observed in the two different systems cannot be compared directly. Based on the characteristic of the two cleavage assay systems, the highly sensitive luciferase-based biosensor assay might be more suitable to high throughput screen the predicted putative cleavage site of protease while the FRET assay better for cleavage kinetic analysis.
According to the Michaelis constants of MERS-CoV, the substrate specificity of MERS-CoV 3CLpro is relatively conserved with other coronaviruses (Fan et al., 2004, Hegyi and Ziebuhr, 2002). Notably, the Pro (P) has been selected as result of evolution at position P2 of cleavage site between nsp10 and nsp12 (CS10/12) of lineage C betacoronavirus, which is not preferred by the 3CLpro based on the previous study (Chuck et al., 2011). However, the relative kcat/Km value of MERS-CoV CS10/12 is 0.053, which is 26.5 fold higher than that of SARS-CoV (Fan et al., 2004). This indicated that the substrate preferences of some cleavage sites could still be varied among different genera of coronaviruses and the proposed scanning condition IV regarding the residues ever appearing in the cleavage sequences of congeneric coronaviruses is reasonable.
In summary, we proposed an optimized search condition for predicting cleavage sites of coronavirus 3CLpro. We verified the 11 canonical cleavage sites of pp1ab in biochemical assays. We further identified three non-canonical cleavage sites in the nsps of MERS-CoV. The results provide clues for possible identification of novel cleavage products of coronavirus nsps and will benefit the studies of the mechanisms of coronavirus replication.
5. Conclusions
Processing of polyprotein 1a/1ab by 3CLpro is essential in coronavirus life cycle. The 3CLpro cleavage site prediction methods established by previous studies are focus on the accuracy, while some noncanonical cleavage sites were missed. In this study, we built a moderate prediction method to balance the accuracy and false positive outcomes. Using this method, 9 putative cleavage sites, in addition to the 11 canonical sites, were predicted in MERS-CoV pp1ab and the cleavability of 3 of them was experimentally confirmed. Interestingly, all these 3 non-canonical cleavage sites are located upstream to nsp4, which is in contrast with previous understanding that the coronavirus 3CL protease only cleaves from nsp4 to nsp16. This suggests a novel role of 3CLpro in coronavirus pp1a/1ab processing. However, the cleavability of these putative cleavage sites needs to be further verified in the viral proteins of MERS-CoV-infected cells. Finally, the catalytic constants of the 11 canonical cleavage sites of MERS-CoV 3CLpro showed its conservation with the cousins in Coronaviridae.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2015.05.018.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., Madani H., Alhakeem R., Al-Tawfiq J.A., Cotten M., Watson S.J., Kellam P., Zumla A.I., Memish Z.A. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369(5):407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Elkin B.A., Knaap R.C., Johnson G.G., Dalebout T.J., Ninaber D.K., van Kasteren P.B., Bredenbeek P.J., Snijder E.J., Kikkert M., Mark B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014;289(50):34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla P.J., Hughes S.A., Pinon J.D., Weiss S.R. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology. 1995;209(2):489–497. doi: 10.1006/viro.1995.1281. [DOI] [PubMed] [Google Scholar]
- Chuck C.P., Chow H.F., Wan D.C., Wong K.B. Profiling of substrate specificities of 3C-like proteases from group 1, 2a, 2b, and 3 coronaviruses. PLoS One. 2011;6(11):e27228. doi: 10.1371/journal.pone.0027228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L., Snijder E.J., Stephens G.M., Woo P.C., Zaki A.M., Zambon M., Ziebuhr J. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D.J., Graham R.L., Denison M.R., Baric R.S. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J. Virol. 2007;81(19):10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 2004;279(3):1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Ou H.-Y., Chen L.-L., Zheng W.-X., Zhang C.-T. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 2003;553(3):451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17(12):4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger C., Heusipp G., Ziebuhr J., Harms U., Suss J., Siddell S.G. Characterization of a 105-kDa polypeptide encoded in gene 1 of the human coronavirus HCV 229E. Virology. 1996;222(1):227–235. doi: 10.1006/viro.1996.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83(Pt 3):595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- Hussain S., Pan J., Chen Y., Yang Y., Xu J., Peng Y., Wu Y., Li Z., Zhu Y., Tien P., Guo D. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79(9):5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilianski A., Mielech A.M., Deng X., Baker S.C. Assessing activity and inhibition of Middle East respiratory syndrome coronavirus papain-like and 3C-like proteases using luciferase-based biosensors. J. Virol. 2013;87(21):11955–11962. doi: 10.1128/JVI.02105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Li K.S., Tsang A.K., Lam C.S., Ahmed S., Chen H., Chan K.H., Woo P.C., Yuen K.Y. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J. Virol. 2013;87(15):8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.P., Liu D.X. Characterisation of a papain-like proteinase domain encoded by ORF1a of the coronavirus IBV and determination of the C-terminal cleavage site of an 87 kDa protein. Adv. Exp. Med. Biol. 1998;440:173–184. doi: 10.1007/978-1-4615-5331-1_22. [DOI] [PubMed] [Google Scholar]
- Liu D.X., Brierley I., Tibbles K.W., Brown T.D. A 100-kilodalton polypeptide encoded by open reading frame (ORF) 1b of the coronavirus infectious bronchitis virus is processed by ORF 1a products. J. Virol. 1994;68(9):5772–5780. doi: 10.1128/jvi.68.9.5772-5780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., Xu H.Y., Brown T.D. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J. Virol. 1997;71(3):1814–1820. doi: 10.1128/jvi.71.3.1814-1820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Lu X., Denison M.R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 1995;69(6):3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needle D., Lountos G.T., Waugh D.S. Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity. Acta Crystallogr. D: Biol. Crystallogr. 2015;71(Pt 5):1102–1111. doi: 10.1107/S1399004715003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.F., Liu D.X. Identification of a 24-kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology. 1998;243(2):388–395. doi: 10.1006/viro.1998.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.F., Liu D.X. Further characterization of the coronavirus infectious bronchitis virus 3C-like proteinase and determination of a new cleavage site. Virology. 2000;272(1):27–39. doi: 10.1006/viro.2000.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashani M., Alfelali M., Barasheed O., Fatema F.N., Alqahtani A., Rashid H., Booy R. Australian Hajj pilgrims’ knowledge about MERS-CoV and other respiratory infections. Virol. Sin. 2014;29(5):318–320. doi: 10.1007/s12250-014-3506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6) doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366(3):965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.R. Mining SARS-CoV protease cleavage data using non-orthogonal decision trees: a novel method for decisive template selection. Bioinformatics. 2005;21(11):2644–2650. doi: 10.1093/bioinformatics/bti404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Ziebuhr J., Schelle B., Karl N., Minskaia E., Bayer S., Siddell S.G., Gorbalenya A.E., Thiel V. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 2007;81(8):3922–3932. doi: 10.1128/JVI.02091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Siddell S.G. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J. Virol. 1999;73(1):177–185. doi: 10.1128/jvi.73.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81(Pt 4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Zuniga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78(2):980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


