Highlights
-
•
Summary of identified viruses associated with Antarctic animals.
-
•
Genomes of Antarctic animals viruses have only been determine in the last five years.
-
•
Limited knowledge of animal virology relative to environmental virology in Antarctica.
Keywords: Penguin, Seal, Petrel, Sharp spined notothen, Antarctica, Wildlife disease
Abstract
The Antarctic, sub-Antarctic islands and surrounding sea-ice provide a unique environment for the existence of organisms. Nonetheless, birds and seals of a variety of species inhabit them, particularly during their breeding seasons. Early research on Antarctic wildlife health, using serology-based assays, showed exposure to viruses in the families Birnaviridae, Flaviviridae, Herpesviridae, Orthomyxoviridae and Paramyxoviridae circulating in seals (Phocidae), penguins (Spheniscidae), petrels (Procellariidae) and skuas (Stercorariidae). It is only during the last decade or so that polymerase chain reaction-based assays have been used to characterize viruses associated with Antarctic animals. Furthermore, it is only during the last five years that full/whole genomes of viruses (adenoviruses, anelloviruses, orthomyxoviruses, a papillomavirus, paramyoviruses, polyomaviruses and a togavirus) have been sequenced using Sanger sequencing or high throughput sequencing (HTS) approaches. This review summaries the knowledge of animal Antarctic virology and discusses potential future directions with the advent of HTS in virus discovery and ecology.
1. Introduction
Among Earth’s oceans, those in the Polar Regions are the smallest and most constrained, the Arctic Ocean encircled by landmasses and the Southern Ocean by the Antarctic Circumpolar Current (ACC). The latter ocean is bounded to its north by the Antarctic Polar Front (APF), a well-known faunal barrier, and has a high degree of endemism among both vertebrates and invertebrates (e.g. Briggs, 1995, Eastman, 2013). Owing to dramatic annual cycles of heat and light, the productivity of the Southern Ocean is highly constrained on a seasonal basis, a characteristic that provides a generally challenging environment for the existence of organisms. Moreover, the high latitude Southern Ocean, i.e. that portion south of the Southern Boundary of the ACC (SBACC), is covered by sea ice for much of the year, in some places the entire year. Most of the birds and seals of a variety of species that inhabit that zone are endemic and resident, the most unvarying species assemblage found in Southern Hemisphere oceans; only a few migrant species augment that assemblage during summer (Ribic and Ainley, 1989). The species comprising this assemblage breed either on Antarctic islands (birds) or the sea ice that surrounds the continent (seals). In contrast, waters north of the SBACC host a much more speciose, seasonally varying seabird and marine mammal assemblage composed of species breeding on low latitude islands bordering the APF (Antarctic and sub-Antarctic) as well as seasonal migrants from more temperate regions (e.g. Ainley et al., 1994, Laws, 1977a, Laws, 1977b, Ribic and Ainley, 1989, Ribic et al., 2011). In accord with trends of diversity varying inversely with latitude, overall diversity of vertebrate species is low in the Southern Ocean, especially south of the SBACC, but populations are immense (e.g. Laws, 1977a, Laws, 1977b).
Inhabiting the pack-ice surrounding Antarctica is a unique assemblage of pagophilic seals, crabeater (Lobodon carcinophaga), leopard (Hydrurda leptonyx), Ross (Ommatophoca rossii) and Weddell seal (Leptonychotes weddellii). Weddell seals colonize near-shore fast-ice regions feeding mainly on fish, while other Antarctic seals remain year round in the pack-ice composed of individual, often compacted floes. Crabeater seals feed principally on krill (Euphausia spp.), and Ross seals mostly on squid. Unlike the other Antarctic seals, leopard seals are solitary and highly predatory feeding on penguins and other seals as well as fish, krill and cephalopods (Siniff, 1981). Southern elephant seals (Mirounga leonina) occupy sub-Antarctic islands for breeding, then migrate south to Antarctica, some hauling out on land for moulting; Antarctic fur seals (Arctocephalus gazella) breed on peri-Antarctic islands, such as Macquarie, South Georgia and South Sandwich, as well as islands of the northern Antarctic Peninsula, around which they also hunt for krill and fish (Siniff et al., 2008).
Confined to pack-ice affected waters south of the SBACC are the Adélie (Pygoscelis adeliae) and emperor (Aptenodytes forsteri) penguins; north of that boundary are three other penguin species: gentoo (P. papua), chinstrap (P. antarctica) and macaroni (Eudyptes chrysolophus) (Williams, 1995). While mostly distributed to the north, populations of chinstrap, gentoo and macaroni penguins breed on islands of the northwestern Antarctic Peninsula, overlapping with Adélie penguins. Gentoo penguins are distributed as far as temperate waters surrounding the Falkland Islands, while macaroni penguins mainly colonize Heard and South Georgia islands in proximity to the APF (Trathan et al., 2016). Colonies of king penguins (A. patagonicus), royal penguins (E. schlegeli) and rockhopper penguins (E. chrysocome) are found on sub-Antarctic islands and do not inhabit the coastal Antarctic continent. Flying birds inhabiting Antarctica and sub-Antarctic regions include skuas, petrels, terns, gulls and Albatross (Brooke, 2004, Murphy, 1936).
In addition to the two penguins endemic to the sea ice zone are the endemic snow (Pagodroma nivea) and Antarctic petrels (Thalassoica antarctica), this pagophilic community increased only in summer by a few other flighted seabirds: skuas, albatross and a few petrel species. The peri-Antarctic and sub-Antarctic islands, and surrounding ice-free ocean, are densely populated by many more petrel and albatross species, with sparse inclusion of skuas and larid species (Brooke, 2004, Murphy, 1936).
2. Early reports of mass mortality in Antarctic animals
Due to its zoogeographical isolation, introduction of pathogens and parasites to these populations of Antarctic wildlife, particularly the endemics of high latitude, may have detrimental effects. Understanding these potential effects required a knowledge of the entities circulating in the ecosystem. This concern drove interest toward detecting viruses, bacteria and parasites among Antarctic animals providing insight to their health. There have been a few reported cases of mass mortality where the disease-causing agent was undetermined. In 1971, several hundred gentoo penguin chicks at a Signey Island colony, South Orkneys, were found dead (MacDonald and Conroy, 1971). Although symptoms were described as similar to puffinosis coronavirus infection, no isolation of the disease agent was possible (Barbosa and Palacios, 2009). In 1972, about 65% of Adélie penguin chicks were reported dead in a colony near Mawson Station. In this case, penguins were found face down and apparently unable to walk or stand properly (Kerry et al., 2009). The cause of this disease was not determined. The only mass mortality reported in seals was the death of over 1500 crabeater seals in a colony around Crown Prince Gustav Channel, Antarctic Peninsula in 1955 (Laws and Taylor, 1957). Interestingly, Weddell seals in the area were unaffected and while the nature of this disease was unknown, a viral infection was suggested. Laws and Taylor (1957) noted that the population of seals in this area was almost ten times higher than normal and predicted that this crowding and partial starvation likely contributed to the effects of the disease.
3. Viruses associated with Antarctic animals
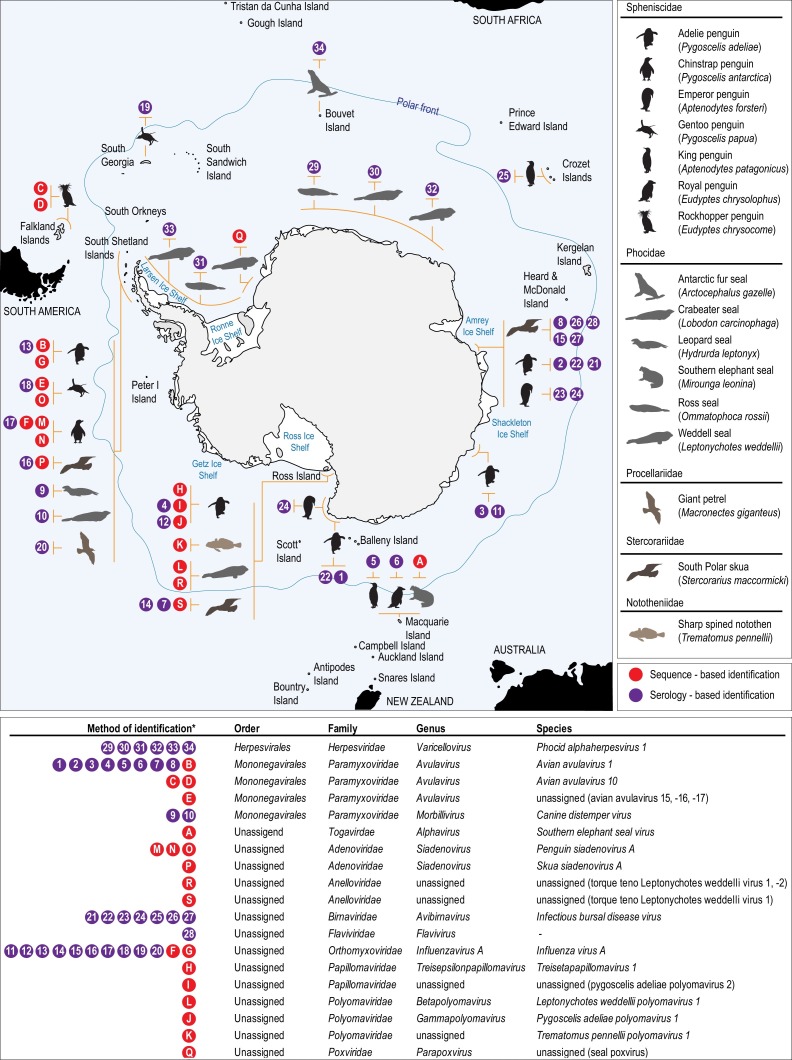
Only within the last ten years has an increase in knowledge of the viral diversity been evident among Antarctic wildlife at a genome level. Early work, beginning in the mid-1970s, on identifying viruses associated with Antarctic animals relied on serology-based assays (Table 1 ). The research then was particularly focused around detecting pathogens posing a risk to animal health due to concerns regarding the impact of increased anthropogenic activity, e.g. research, tourism, on birds and marine mammals (Kerry and Riddle, 2009). During the early period of Antarctic virus research, paramyxoviruses, orthomyxoviruses, birnaviruses, herpesviruses and flaviviruses were serologically detected (Table 1, Fig. 1 ). Subsequently, between 2000 and 2010, probe based assays using polymerase chain reaction (PCR) were used to detect paramyxoviruses, orthomyxoviruses and a poxvirus (Table 2 , Fig. 1). Finally, in the last decade, advancements in high throughput sequencing (HTS) approaches are beginning to have an impact on our knowledge of Antarctic animal virology. For example, in the last five years using viral metagenomic based approaches with HTS, various novel viruses have been identified and characterized at a genomic level (Table 2, Fig. 1). These include adenoviruses, anelloviruses, orthomyxoviruses, papillomaviruses, polyomaviruses and paramyxoviruses.
Table 1.
Summary of Antarctic bird- and mammal-associated viruses detected through serological approaches.
|
Virus taxonomy |
Notes |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID | Order | family | genus | Virus | Host | Method | Sample | Location | Year of collection | Reference |
| 1 | Mononegavirales | Paramyxoviridae | Avulavirus | Avian paramyxovirus | Adelie penguins (Pygoscelis adeliae) | Hemagglutination-inhibition, immunodiffusion tests, morphology |
2/42 serum samples with antibodies to NDV, cloacal swabs |
Wilkes base |
– |
Morgan and Westbury (1981) |
| 2 | Hemagglutination-inhibition, immunodiffusion tests, morphology |
serum, 2 APMV viruses isolated from 550 cloacal swabs |
Peterson Island, Midgley Island, Shirley Island, Cameron Island, d'Urville (Casey station) |
– |
Morgan and Westbury (1981) |
|||||
| 3 | Virus isolation, Haemagglutination test |
Serum samples and cloacal swabs |
Vestfold Hills |
December 1981 |
Morgan and Westbury (1988) |
|||||
| 4 | Indirect ELISA, electron microscopy |
Cloacal swab, serum |
Ross Island |
1978 |
Austin and Webster (1993) |
|||||
| 5 | Royal penguins (Eudyptes chrysolophus) |
Hemagglutination-inhibition, immunodiffusion tests, morphology |
Cloacal samples |
Macquarie Island |
– |
Morgan et al. (1981) |
||||
| 5 | King penguin (Aptenodytes patagonicus) |
Hemagglutination-inhibition, immunodiffusion tests, morphology |
Cloaca samples |
Macquarie Island |
– |
Morgan et al. (1981) |
||||
| 7 | South Polar skua (Stercorarius maccormicki) | Indirect ELISA, electron microscopy |
Serum |
Ross Island |
1978, 1986 |
Austin and Webster (1993) |
||||
| 8 | Haemagglutination-inhibition test |
Serum samples and cloacal swabs |
Davis station |
November/December 1999 |
Miller et al. (2008) |
|||||
| 9 | Morbillivirus | Canine distemper virus | Leopard seal (Hydrurda leptonyx) |
CDV-like antibodies detected through microneutralization test using two CDV strains and a phocine distemper virus isolate |
2/3 serum samples tested positive |
Antarctic peninsula |
1989 |
Bengtson et al. (1991) |
||
| 10 | Crabeater seal (Lobodon carcinophaga) | CDV-like antibodies detected through microneutralization test using two CDV strains and a phocine distemper virus isolate | 35% serum samples tested positive for CDV | Antarctic peninsula | January/March 1989 | Bengtson et al. (1991) | ||||
| 11 | Unassigned | Orthomyxoviridae | Influenzavirus | Influenza A virus | Adelie penguins (Pygoscelis adeliae) | Hemagglutination-inhibition, immunodiffusion tests |
Serum |
Peterson Island (Casey) |
– |
Morgan et al. (1981) |
| 12 | Hemagglutination-inhibition, Neuraminidase-inhibition tests |
Serum |
Ross Island |
1978 |
Austin and Webster (1993) |
|||||
| 13 | Hemagglutination-inhibition test against H1N1, H3N2, H5N1, and H7N2 antigens |
Serum |
Hope Bay |
December–March, 1998, 2001, and 2002 |
Baumeister et al. (2004) |
|||||
| 14 | South Polar skua (Stercorarius maccormicki) | Indirect ELISA, Hemagglutination-inhibition tests |
Serum |
Ross Island |
1978, 1986 |
Austin and Webster (1993) |
||||
| 15 | Capture ELISA |
Serum and cloacal swabs |
Davis station |
November/December 1999 |
Miller et al. (2008) |
|||||
| 16 | Hemagglutination inhibition test against H1N1, H3N2, H5N1, and H7N2 antigens |
Serum |
Potter peninsula and Hope Bay |
December–March, 1998, 2001, and 2002 |
Baumeister et al. (2004) |
|||||
| 17 | Chinstrap penguins (Pygoscelis antarctica) |
Hemagglutination inhibition test against H1N1, H3N2, H5N1, and H7N2 antigens |
Serum |
Potter peninsula |
December-March, 1998, 2001, and 2002 |
Baumeister et al. (2004) |
||||
| 18 | Gentoo penguins (Pygoscelis papua) | Hemagglutination inhibition test against H1N1, H3N2, H5N1, and H7N2 antigens |
Serum |
Potter peninsula |
December-March, 1998, 2001, and 2002 |
Baumeister et al. (2004) |
||||
| 19 | Influenzavirus type A virus antibody ELISA kit |
Bird Island |
1996 |
|||||||
| 20 | Giant petrel (Macronectes giganteus) |
Hemagglutination inhibition test against H1N1, H3N2, H5N1, and H7N2 antigens |
Serum |
Potter peninsula and Harmony peninsula |
December–March, 1998, 2001, and 2002 |
Baumeister et al. (2004) |
||||
| 21 | Birnaviridae | Avibirnavirus | Infectious bursal disease virus | Adelie penguins (Pygoscelis adeliae) | Virus neutralization tests, IBDV serotype 1 and 2 antibodies |
High titer of neutralizing antibodies detected in 2.6% or 133 penguins |
Mawson station |
1995/96 summer |
Gardner et al. (1997) |
|
| 22 | Virus neutralization tests to measure antibody titers to IBDV serotype 1 |
Seroprevalence 7.7%, no significant difference between locations or years. Highly significant titres were obtained from 1.8% of birds |
Mawson coast, Davis Coast, Terra Nova Bay |
November–February 1996-2002 |
Watts et al. (2009) |
|||||
| 23 | Emperor penguin (Aptenodytes forsteri) | Virus neutralization tests, IBDV serotype 1 and 2 antibodies |
High titer of neutralizing antibodies detected in 65.4% of 53 penguins |
Mawson station |
1995/96 summer |
Gardner et al. (1997) |
||||
| 24 | Virus neutralization tests to measure antibody titers to IBDV serotype 1 |
Auster Rookery, Amanda Bay rookery, Cape Washington rookery |
November–February 1996–2001 |
Watts et al. (2009) |
||||||
| 25 | King penguin (Aptenodytes patagonicus) |
Virus neutralization tests, IBDV serotype 1 and 2 antibodies, cough and conjunctivitis clinical signs |
Serum of adults and chicks |
sub-Antarctic Iles Crozet |
November 1996-February 1997 |
Gauthier-Clerc et al. (2002) |
||||
| 26 | South Polar skua (Stercorarius maccormicki) | Antibody neutralization test |
Serum and cloacal swab |
Davis station |
November/December 1999 |
Miller et al. (2008) |
||||
| 27 | Virus neutralization tests to measure antibody titers to IBDV serotype 1 |
Antibodies detected in 11.8% of individuals, Significant difference in prevalence between sample years |
Vestfolds, Davis station |
November–February 1999–2002 |
Watts et al. (2009) |
|||||
| 28 | Flaviviridae | Flavivirus | – | South Polar skua (Stercorarius maccormicki) | Capture ELISA | Serum samples and cloacal swabs | Davis station | November/December 1999 | Miller et al. (2008) | |
| 29 | Herpesvirales | Herpesviridae | Varicellovirus | Phocid alphaherpesvirus 1 | Ross seal (Ommatophoca rossii) |
Indirect ELISA using PhHV-1 as antigen, serum neutralization test |
Serum |
Queen Maud Land |
2001 |
Tryland et al. (2012) |
| 30 | Crabeater seal (Lobodon carcinophaga) | Indirect ELISA using PhHV-1 as antigen, serum neutralization test |
Serum |
Queen Maud Land |
2001 |
Tryland et al. (2012) |
||||
| 31 | Testing for neutralizing antibodies against phocine, feline and canine herspevirus using either microneutralization or by neutralizing peroxidase-linked antibody assay |
Serum |
Weddell Sea |
1990 |
Harder et al. (1991) |
|||||
| 32 | Weddell seal (Leptonychotes weddellii) | Indirect ELISA using PhHV-1 as antigen, serum neutralization test (SNT) |
Serum |
Queen Maud Land |
2001 |
Tryland et al. (2012) |
||||
| 33 | Testing for neutralizing antibodies against phocine, feline and canine herspevirus using either microneutralization or by neutralizing peroxidase-linked antibody assay |
Serum |
Weddell Sea |
1990 |
Harder et al. (1991) and Stenvers et al. (1992) |
|||||
| 34 | Antarctic fur seal (Arctocephalus gazelle) | Indirect ELISA using PhHV-1 as antigen | Serum | Bouvet Island | 2000–2001, 2001–2002 | Tryland et al. (2012) | ||||
CDV: Canine distemper virus.
ELISA: Enzyme-linked immunosorbent assay.
IBDV: Infectious bursal disease virus.
Fig. 1.
Distribution of viruses identified among populations of animals of the Antarctic and high latitude sub-Antarctic. Colored circles denote the method of viral identification. Purple indicates serology based identification, number inside circle corresponding to details provided in Table 1. Red circle indicates sequence based identification, letter inside circle corresponding to details provided in Table 2.
Table 2.
Summary of Antarctic bird- and mammal-associated viruses identified through sequencing based approaches, including respective accession numbers of the partial and full genome sequences.
|
Virus taxonomy |
Notes |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID | Family | Genus | Species & virus | Host | Method | Sample type | Location | Year of collection | Accession # | Reference |
| A | Togaviridae | Alphavirus | Southern elephant seal virus [Southern elephant seal virus (SES virus)] | Southern elephant seals (Mirounga leonina) | Virus cultured in BHK-21 cells virus from blood sucking lice Lepidophthirus macrorhini and used in viral neutralization assay for serology. Negative stain electron microscopy. RT-PCR and Sanger sequencing of capsid protein gene. | Lepidophthirus macrorhini and serum from southern elephant seal virus | Macquarie Island | – | AF315122 HM147990 | Forrester et al. (2012) and La Linn et al. (2001) |
| B | Paramyxoviridae | Avulavirus |
Avian avulavirus 1 [New castle disease virus (NDV)] |
Adélie penguins (Pygoscelis adeliae) |
RT-PCR and real-time PCR targeting F gene of NDV, virus culture, haemagglutination test using antigen against B1 NDV strain, Sanger sequencing. |
Cloacal/tracheal swabs and serum samples |
King George Island |
2006 |
HM143848–HM143850 |
Thomazelli et al. (2010) |
| C | Avian avulavirus 10 [Avian paramyxovirus 10 (APMV10)] | Rockhopper penguins (Eudyptes chrysocome) |
Real-time RT-PCR, hemagglutination assay, binaxNOW influenza A&B test, hemagglutination inhibition assay, ELISA, electron microscopy, Sanger sequencing. |
Cloacal/tracheal swabs and serum samples |
Falkland Islands |
2007 |
HM147142, HM755886–HM755888 (updated following the publication of Goraichuk et al. (2017) |
Miller et al. (2010) |
||
| D | Rockhopper penguins (Eudyptes chrysocome) |
Complete genome sequencing using Illumina and Sanger sequencing. |
Cloacal/tracheal swabs and serum samples |
Falkland Islands |
2007 |
HM147142, HM755886–HM755888 |
Goraichuk et al. (2017) |
|||
| E | Unclassified [Avian paramyxovirus 15, 16, 17 (APMV15, APMV16, APMV17)] | Gentoo penguins (Pygoscelis papua) | Hemagglutination assay, RT-PCR and Sanger sequencing. | Virus isolated from 12 cloacal swabs, 5 confirmed by sequencing | Kopaitic Island | 2014 − 2016 | KY452442–KY452444 | Neira et al. (2017) | ||
| F | Orthomyxoviridae | Influenzavirus A |
Influenzavirus A [Avian Influenza A virus H5N5] |
Chinstrap penguins (Pygoscelis antarctica) |
RT-PCR, HTS, ELISA using nucleoprotein, hemagglutination assay. |
Cloacal/tracheal swabs and serum samples |
Antarctic peninsula |
2015 |
GISAID #s [EPI774530-EPI774536, EPI774538- EPI774539]; [EPI774527- EPI774529] |
Hurt et al. (2016) |
| G | Influenzavirus A [Avian influenza A virus H11N2] | Adélie penguins (Pygoscelis adeliae) | RT-PCR, virus culture, ELISA, whole genome Sanger sequencing. | Cloacal/tracheal swabs and serum samples | Rada Covadonga, Antarctic Peninsula and King George Island | 2013 | KJ729348–KJ729379 | Hurt et al. (2014) | ||
| H | Papillomaviridae | Treisepsilonpapillomavirus |
Treisetapapillomavirus 1 [Pygoscelis adeliae papillomavirus 1] |
Adélie penguins (Pygoscelis adeliae) |
HTS-informed approach, genome recovered by abutting primers, cloned and Sanger sequenced. |
Faeces |
Cape Crozier, Ross Island |
2012/2013 |
KJ173785 |
Varsani et al. (2014) |
| I | Unclassified [Pygoscelis adeliae papillomavirus 2] | Adélie penguins (Pygoscelis adeliae) | HTS-informed approach, genome recovered by abutting primers, cloned and Sanger sequenced. | Cloacal swab | Cape Crozier, Ross Island | 2014 | MF168943 | Van Doorslaer et al. (2017) | ||
| J | Polyomaviridae |
Gammapolyomavirus |
Pygoscelis adeliae polyomavirus 1 [Adelie penguin polyomavirus (AdPyV)] |
Adélie penguins (Pygoscelis adeliae) |
HTS-informed approach, genome recovered by abutting primers, cloned and Sanger sequenced. |
Faeces |
Cape Royds, Ross Island |
2012/2013 |
KP033140 |
Varsani et al. (2015) |
| K |
unassigned |
Trematomus pennellii polyomavirus 1 [Sharp-spined notothenia polyomavirus (SspPyV)] |
Sharp spined notothen (Trematomus pennellii) |
HTS-informed approach, genome recovered by abutting primers, cloned and Sanger sequenced. |
Stomach and liver samples |
Ross Sea |
2012/2013 |
KP768176 |
Buck et al. (2016) |
|
| L |
Betapolyomavirus |
Leptonychotes weddellii polyomavirus 1 [Weddell seal polyomavirus (WsPyV)] |
Weddell seal (Leptonychotes weddellii) |
HTS-informed approach, genome recovered by abutting primers, cloned and Sanger sequenced. |
Kidney |
Ross Sea |
2014 |
KX533457 |
Varsani et al. (2017) |
|
| M | Adenoviridae | Siadenovirus |
Penguin siadenovirus A [Chinstrap penguin adenovirus (CSPAdV)] |
Chinstrap penguins (Pygoscelis antarctica) |
PCR of protein VI and capsid protein hexon genes. |
Lung, liver, kidney, heart, intestine, trachea samples |
King George Island |
2009/2010 |
KC593379–KC593386 |
Lee et al. (2014) |
| N |
Penguin siadenovirus A [Chinstrap penguin adenovirus (CSPAdV)] |
Chinstrap penguins (Pygoscelis antarctica) |
RACE PCR, Sanger sequencing of whole genome. |
Lung, liver, kidney, heart, intestine, trachea samples |
King George Island |
2008–2013 |
KP144329–KP144330 |
Lee et al. (2016) |
||
| O |
Penguin siadenovirus A [Gentoo penguin adenvirus (GPAdV)] |
Gentoo penguins (Pygoscelis papua) |
RACE PCR, Sanger sequencing of whole genome. |
lung, liver, kidney, heart, intestine, trachea, feces |
King George Island |
2008–2013 |
KP279746–KP279747 |
Lee et al. (2016) |
||
| P | Skua siadenovirus A [South polar skua adenovirus 1 (SPSAdV 1)] | South Polar skua (Stercorarius maccormicki) | Nested PCR, RACE PCR, Sanger sequencing of whole genome. | kidney | King George Island | 2007–2009 | HM585353 (full genome) JM585354-HM585358 | Park et al. (2012) | ||
| Q | Poxviridae | Parapoxvirus | Unassigned [Seal poxvirus] | Weddell seal (Leptonychotes weddellii) | Electron microscopy, PCR of B2L gene, sequencing of B2L gene. | Neck skin lesion from a single seal | Queen Maud Land | 2001 | AJ622900 | Tryland et al. (2005) |
| R | Anelloviridae | unassigned | Unassigned [Torque teno Leptonychotes weddelli virus 1,-2(TTLwV1 & TTLwV2)] |
Weddell seal (Leptonychotes weddellii) |
HTS-informed approach, genome recovered by abutting primers, cloned and Sanger sequenced. |
Vaginal, nasal and faecal samples |
Ross Sea |
November-February 2014–2015 |
KY246479–KY246627 |
Fahsbender et al. (2017) |
| S | Unassigned [Torque teno Leptonychotes weddelli virus 1 (TTLwV1) | South Polar skua (Stercorarius maccormicki) | HTS-informed approach, genome recovered by abutting primers, cloned and Sanger sequenced. | Faecal sample | Ross Sea | November/December 2014 | KY246476–KY246478 | Fahsbender et al. (2017) | ||
cDNA: complementary DNA.
ELISA: Enzyme-linked immunosorbent assay.
PCR: Polymerase chain reaction.
HTS: High throughput sequencing.
RACE: Rapid Amplification of cDNA ends.
RT-PCT: Reverse transcription PCR.
3.1. Positive sense RNA viruses
3.1.1. Flaviviridae
Flaviviridae is a family of enveloped positive sense RNA viruses with four genera: Flavivirus, Hepacivirus, Pegivirus and Pestivirus. Their genomes range in length from 8.9–13 kb (Simmonds et al., 2017b). While no genomic information is available for flaviviruses circulating in Antarctic animals, neutralizing antibodies to a flavivirus were detected in the serum of South Polar skuas (Stercorarius maccormicki) around Davis station, East Antarctica (Miller et al., 2008). As lower latitude seabirds are infested with ticks (e.g. Lee and Baust, 1987), which are known to carry flaviviruses, this may indicate the transmission of tick-borne flaviviruses to seabirds, especially given the fact that a flavivirus was isolated from seabird ticks (Ixodes uriae) infecting king penguins on Macquarie Island (Major et al., 2009).
3.1.2. Togaviridae
The family Togaviridae consist of enveloped positive sense RNA viruses with a genome length of about 11–12 kb in length. Togaviruses are classified into two genera, Alphavirus and Rubivirus (Power et al., 2017). While the human pathogenic virus, rubella virus, is the only known member of the single species in the genus Rubivirus to date (species Rubella virus), all animal togaviruses are classified as alphaviruses (Power et al., 2017). The life cycle of alphaviruses requires an arthropod vector, either a mosquito or tick, for transmission to their vertebrate host. The first instance of alphaviruses found in marine mammals was shown in southern elephant seals of Macquarie Island. Initially the seal alphavirus was isolated from the blood-sucking louse (Lepidophthirus macrorhini) that is widespread among southern elephant seals. However, a high seroprevalence of antibodies against the southern elephant seal alphavirus strongly indicated its transmission from lice to seals (La Linn et al., 2001). The full genome of this alphavirus (Southern elephant seal virus) was determined in a later study (Forrester et al., 2012).
3.2. Negative sense RNA viruses
3.2.1. Orthomyxoviridae
Viruses in the family Orthomyxoviridae have enveloped, negative sense RNA genomes that consist of 6–8 segments. Seven genera are established in this family (Influenzavirus A, Influenazavirus B, Influenzavirus C, Influenzavirus D, Isavirus, Quaranjavirus and Thogotovirus) (McCauley et al., 2011). Influenza A virus is the only species in the genus Influenzavirus A, consisting of several pathogenic strains infecting humans, horses, pigs, whales, seals, birds and mink(McCauley et al., 2011). Influenza A virus strains are transmissible to humans and have caused worldwide endemics. This zoonotic nature of influenza viruses has led to extensive research on Influenza A virus. Antarctica continues to provide an interesting environment to study Influenza A viruses, especially in the case of migratory birds. For example, the South Polar skuas that breed on the Antarctic continent in the summer but move north, well into the Northern Hemisphere during the non-breeding season (Weimerskirch et al., 2015), thus act as potential vectors bringing in new variants and reassortants to Antarctica during each breeding season.
The majority of studies screening for influenza have used hemagglutination-inhibition assays to detect antibodies against several common strains of avian influenza virus circulating among Antarctic animals. Antarctic influenza virus research carried out between 1978 and 2002, detected antibodies against several strains of avian influenza virus in South Polar skuas, southern giant petrels (Macronectes giganteus), and Adélie, chinstrap and gentoo penguins from various locations around the Antarctic Peninsula, Ross Island and East Antarctica (Table 1, Fig. 1) (Austin and Webster, 1993, Baumeister et al., 2004, Miller et al., 2008, Morgan and Westbury, 1981). Such research has indicated that influenza virus is highly widespread and prevalent in Antarctic birds. However, the pathogenicity of avian influenza virus in these populations is unknown. No genomic information for Influenza A viruses in Antarctic birds was available until Hurt et al. (2014) used HTS approaches to identify influenza A virus (H11N2) in Adélie penguins around the Antarctic Peninsula. A following study around the same area later identified the strain H5N5 among chinstrap penguins (Hurt et al., 2016).
None of the early studies have detected antibodies against influenza A virus strains in Antarctic pinnipeds, although several studies have looked at crabeater and Weddell seals (Austin and Webster, 1993, McFarlane, 2009).
3.2.2. Paramyxoviridae
Paramyxoviridae is a family of enveloped, non-segmented negative sense RNA viruses in the order Mononegavirales with genomes of ∼15 kb. Paramyxoviruses are divided into seven genera: Aquaparamyxovirus, Ferlavirus, Respirovirus, Morbillivirus, Rubulavirus, Henipavirus, and Avulavirus. The genus Avulavirus includes of 13 formally classified species of avian paramyxoviruses including avian paramyxovirus 1 (AVPM-1) (Afonso et al., 2016). The genus Morbillivirus contains paramyxoviruses infecting mammals.
The majority of research on paramyxoviruses in Antarctica has been based on serological studies using haemagglutination inhibition assay to detect antibodies against paramyxoviruses in serum samples (Table 1). A high prevalence of antibodies to NDV in South Polar skuas has been reported (Miller et al., 2010), whereas low incidences have been found in Adélie and royal penguins around coastal Antarctica and Macquarie Island (Table 1, Fig. 1) (Morgan and Westbury, 1981, Morgan and Westbury, 1988). So far, king, gentoo and rockhopper penguin colonies on Macquarie Island have tested negative for AVPM-1 antibodies (Morgan et al., 1981).
Despite serology-based knowledge of these viruses among Antarctic birds, our understanding of their diversity is extremely limited due to the lack of available genomic data. Partial genome sequences of NDV in Adélie penguins has been obtained using HTS approaches (Thomazelli et al., 2010). Most recently, complete genome sequences of avian paramyxovirus 10 (APMV 10) and three novel avulaviruses (APMV 11, 12, 13) have been determined from rockhopper penguins on the Falkland Islands and gentoo penguins sampled on Kopaitic Island, northern tip of the Antarctic Peninsula (Table 1, Fig. 1) (Goraichuk et al., 2017, Neira et al., 2017).
With the use of sled dogs (Canis familiaris) during the early Antarctic expeditions, concern of morbillivirus infection among Antarctic pinnipeds drove research in monitoring for this virus in seal populations. Antibodies to canine distemper virus (CDV) have been reported in leopard and crabeater seals around the Antarctic Peninsula (Bengtson et al., 1991) and phocine distemper virus (PDV) in Weddell seals from Vestfold Hills, East Antarctica (McFarlane, 2009). With the exception of crabeater seals, both of these studies revealed low antibody titers against CDV and PDV. Several other studies looking at morbilliviruses in Antarctic seals have failed to detect any antibodies against these viruses (Harder et al., 1991, Osterhaus et al., 1988, Stenvers et al., 1992, Yochem et al., 2009). This may suggest morbilliviruses are not persistent in Antarctic seals or perhaps there are diverse morbilliviruses circulating amongst the pinnipeds that cannot be detected using conventional serology assays but likely to be identified using HTS approaches. Unlike avian paramyxoviruses, no genomic data are available for morbilliviruses from Antarctic seals and therefore impossible to tell if there was a spillover event from the canines to the pinnipeds. Sled dogs are no longer allowed in Antarctica.
3.3. Double stranded RNA viruses
3.3.1. Birnaviridae
Viruses in the family Birnaviridae have non-enveloped capsids that encapsidate two linear double stranded segments of RNA, each ∼2.3–3 kb in length. Four genera have been established in this family: Avibirnavirus, Aquabirnavirus, Blosnavirus and Entomobirnavirus (Delmas et al., 2011). Infectious bursal disease virus is the only characterized virus belonging to genus Avibirnavirus. Since its initial identification as virus responsible for a highly infectious disease among chickens, infectious bursal disease virus (IBDV) has widely been isolated from other birds in the poultry industry including ducks and turkeys, however, disease has only been identified in chickens. Between 1995–2002 neutralization assays identified high titers of antibodies against IBDV in Adélie penguin at colonies around Mawson and Davis stations, and Terra Nova Bay, East Antarctica; and emperor penguin at the Auster, Amanda and Cape Washington colonies, also in East Antarctica (Table 1, Fig. 1) (Gardner et al., 1997, Watts et al., 2009). Highest seroprevalence has been detected among emperor penguin colonies with no difference between sampled locations or years (Watts et al., 2009). South Polar skuas around Vestold Hills and Davis Station, East Antarctica, have also had high titer of antibodies against IBDV (Miller et al., 2008, Watts et al., 2009), however, a significant difference in seroprevalence between sampling periods during 1999–2002 was observed. A low titer and prevalence of antibodies to IBDV has been detected in king penguins around Possession Island, among the Crozet Islands along the APF (Gauthier-Clerc et al., 2002). High-titers of IBDV neutralizing antibodies detected in distant populations of penguins and South Polar skua around Antarctica suggests it is unlikely IBDV was introduced through disposal of chicken products around Mawson Station as previously suggested by Gardner et al. (1997). Given that IBDV infection has been commonly detected in other wild avian populations (Hollmén et al., 2000, Kasanga et al., 2008, Ogawa et al., 1998), this virus may be naturally occurring among Antarctic birds (Watts et al., 2009). It is worth noting, however, that IBDV has yet to be isolated from Antarctic birds despite several studies detecting neutralizing antibodies. Therefore, it is difficult to address any questions about the diversity, evolution or transmission of this virus among Antarctic birds.
3.4. Double stranded DNA viruses
3.4.1. Adenoviridae
Adenoviruses are a family of non-enveloped double stranded DNA viruses with a genome length of ∼26–45 kb. The diversification of these viruses is thought to have occurred through several animal hosts including mammals, reptiles, birds, fish and amphibians. The family Adenoviridae has been divided into five genera: Mastadenovirus, Aviadenovirus, Atadenovirus, Siadenovirus, Ichtadenovirus (Harrach et al., 2011).
Most adenovirus research has focused on the implications of human-associated adenoviruses, likely due to the known clinical significance in causing respiratory disease and gastroenteritis. However, the first adenoviruses from Antarctic animals have only recently been identified among South Polar skua (Skua siadenovirus A) (Park et al., 2012), as well as chinstrap, Adélie and gentoo penguins (Penguin siadenovirus A) (Table 2, Fig. 1) (Lee et al., 2014, Lee et al., 2016). This provides important insight to monitoring penguin health in Antarctica, as adenoviruses have been known to cause severe disease among animals.
Whole genomes for these adenoviruses were confirmed using HTS approaches and subsequent phylogenetic analyses of the genomic sequences provide support for the classification of the South Polar skua and penguin adenoviruses in the genus Siadenovirus. Despite this classification, penguin adenovirus genomes are unique in that they lack a putative sialidase gene that is characteristic of other genomes in this genus (Lee et al., 2016).
It is likely that there exits adenoviruses associated with Antarctic seals based on the fact that adenoviruses has been identified in California sea lion (Zalophus californianus), Fur seals (Arctocephalus spp.) and South American sea lion (Otaria flavescens) (Chiappetta et al., 2017, Cortes-Hinojosa et al., 2016, Cortes-Hinojosa et al., 2015, Goldstein et al., 2011, Inoshima et al., 2013).
3.4.2. Herpesviridae
Herpesviridae is a large family of enveloped viruses with a linear, double stranded DNA genome about 120–240 kb in length. This family has been divided into three subfamilies (Alphaherpesvirinae, Betaherpesvirinae and Gammaherpesvirinae). Herpesviruses belonging to two species, based on partial genome sequencing of conserved regions, have been found among pinnipeds in the Northern Hemisphere (Harder et al., 1996): phocid alphaherpesvirus-1 (PhHV-1, species Phocid alphaherpesvirus 1) belonging to the Varicellovirus genus of the Alphaherpesvirinae subfamily and phocid gammaherpesvirus-2 (PhHV-2, Phocid gammaherpesvirus 2) belonging to the Gammaherpesvirinae subfamily. Both PhHV-1 and PhHV-2 have been identified in several non-Antarctic pinniped species around the world from free-ranging populations as well as captive populations in zoos and aquaria (Bellehumeur et al., 2016, Goldstein et al., 2004, Osterhaus et al., 1985).
Among Antarctic pinnipeds, herpesvirus has not been confirmed by molecular methods, however, several studies over the years have shown high levels of PhHV-1 neutralizing antibodies in Antarctic fur seals among sub-Antarctic islands, and Ross, Weddell and crabeater seals off East Antarctica (Table 1, Fig. 1) (Harder et al., 1991, Stenvers et al., 1992, Tryland et al., 2012). Thus, it is highly likely that herpesvirus is widespread and persistent among pinnipeds. However, genomic data is required to confirm this virus among Antarctic pinnipeds as the serological data has only indicated infection of a herpesvirus antigenically similar to PhHV-1.
3.4.3. Papillomaviridae
Papillomaviridae is a large family of non- enveloped, circular, double stranded DNA viruses with ∼7–8 kb genomes and are known to infect skin, squamous and mucosal epithelial cells. All papillomavirus genomes have a very similar organization that can be divided into three regions encoding replication associated and regulatory proteins, structural proteins, and a long control region. While research on human papillomaviruses has been extensive due its clinical significance, relatively few studies have looked at non-human papillomaviruses. Papillomaviruses are found in a range of hosts including mammals, birds, reptiles and fish. It is a well-supported hypothesis that they have co-evolved with their hosts given their diversity and host specificity, with supporting phylogenetic analyses that track diversification of papillomaviruses to the evolution of their host (Bernard et al., 2010, de Villiers et al., 2004).
Two novel papillomavirus, Pygoscelis adeliae papillomavirus 1, −2 (PaPV1, −2), was recently identified in Antarctica from feces and cloacal swab of Adélie penguins at Cape Crozier, Ross Island (Table 2, Fig. 1) using a HTS-informed approach (Van Doorslaer et al., 2017, Varsani et al., 2014). PaPV1 and −2 are related to other avian papillomaviruses, PaPV1 has been assigned to the genus Treisepsilonpapillomavirus whereas PaPV2 is currently unclassified and shares ∼64% genome-wide pairwise identity with PaPV1. These PaPVs are the first papillomaviruses to be discovered in Antarctic animals and are part of the few known avian papillomaviruses.
3.4.4. Polyomaviridae
Polyomaviruses represent a family of non-enveloped, circular, double-stranded DNA viruses with a genome length of 5–6 kb, and infect a range of hosts including mammals, birds, reptiles and fish. This family has four genera: Alphapolyomavirus, Betapolyomavirus, Deltapolyomavirus, and Gammapolyomavirus with three species unassigned to any of these (Moens et al., 2017, Polyomaviridae Study Group of the International Committee on Taxonomy of Viruses et al., 2016). Phylogenetic analyses have shown that avian polyomaviruses cluster together and have been classified under the genus Gammapolyomavirus. Avian polyomaviruses are known to cause inflammatory disease in birds, and can lead to disease of the skin and feathers and mortality in some species. The first polyomavirus identified in Antarctica was found in the feces of Adélie penguins at Cape Royds, Ross Island (Varsani et al., 2015) using a HTS-informed approach. Analysis of this genome shows that it falls in the avian polyomavirus lineage, representing a novel species (Pygoscelis adeliae polyomavirus 1).
Following this, two other polyomaviruses have been identified from Antarctic animals: a polyomavirus from the stomach of a sharp-spined notothen (Trematomus pennellii) (Buck et al., 2016) and most recently from the kidney of a Weddell seal, both sampled in the Ross Sea (Table 2, Fig. 1) (Varsani et al., 2017). The sharp-spined notothen polyomavirus is one of the three polyomaviruses to be identified associated with fish and all three were identified using HTS approaches (Buck et al., 2016).
Polyomavirus sequences have been identified in three other pinniped species: once in a captive Hawaiian monk seal (Neomonachus schauinslandi) (Cortes-Hinojosa et al., 2016), in the placenta of one northern fur seal (Callorhinus ursinus) (Duncan et al., 2013) from Alaska and a stranded free-ranging California sea lion (Zalophus californianus) (Colegrove et al., 2010). However, until recently the genome of California sea lion polyomavirus (CSLPyV) was the only confirmed pinniped polyomavirus. The recently identified Weddell seal polyomavirus is has been proposed to be classified as the species Leptonychotes weddellii polyomavirus 1 (https://talk.ictvonline.org/files/proposals/animal_dna_viruses_and_retroviruses/m/animal_dna_ec_approved/6941).
3.4.5. Poxviridae
Poxviruses are a diverse family of double-stranded DNA viruses with a wide host range among vertebrates and arthropods (Skinner et al., 2011). Poxviruses have been extensively studied for their clinical significance in causing highly pathogenic disease among humans and other animals. While sealpox has yet to be formally classified, studies have identified this virus in populations of harbor seals (Phoca vitulina) (Muller et al., 2003), gray seals (Halichoerus grypus) (Nettleton et al., 1995), Stellar sea lions (Eumetopias jubatus), spotted seals (Phoca largha) (Bracht et al., 2006), and California sea lions (Nollens et al., 2006), causing severe proliferative lesions on the bodies of infected individuals. Partial sequencing has indicated seal poxvirus falls under the parapoxvirus family of which only four species have been classified.
The only case of poxvirus in Antarctica known to date has been the isolation and detection from a skin lesion of a deceased Weddell seal in Queen Maud Land, East Antarctica (Table 2, Fig. 1) (Tryland et al., 2005). Other Weddell seals in the area were analyzed for seal poxvirus, however, all individuals were negative, suggesting poxviruses may not prevalent in this population. Partial sequencing of the Weddell seal parapoxvirus shows it is closely related to harbor and grey seal poxviruses (Tryland et al., 2005).
Recently a seal parapoxvirus was sequenced using HTS from a skin lesion of a grey seal from the Baltic Sea (Gunther et al., 2017). While poxviruses have been identified in several avian species, very little is known about their diversity and host range. Using HTS technology a novel avipoxvirus genome has been sequenced from an African penguin (Spheniscus demersus) (Offerman et al., 2014). It is highly likely that poxviruses will also be recovered from Antarctic penguins through HTS.
3.5. Single stranded DNA viruses
3.5.1. Anelloviridae
Viruses in the family Anelloviridae are non-enveloped, circular single stranded DNA viruses with a genome length of about 2–3.9 kb (Biagini et al., 2011). These viruses have high sequence variability and are highly prevalent in the environment. Despite their ubiquitous nature, the significance of infection and pathogenicity remains unknown. While research has focused on their diversity and significance in humans, anelloviruses have been identified in non-human primates, domesticated animals, rodents and recently in marine mammals. Analysis of lung tissue from a captive California sea lion showing signs of respiratory disease led to discovery of the first anellovirus among pinnipeds, Zalophus californianus anellovirus (ZcAV) (Ng et al., 2009). Since then several novel anellovirus genomes have been recovered in harbor seals (Bodewes et al., 2015, Bodewes et al., 2013). In lung samples of deceased harbor seals along the North American Pacific coast anelloviruses were identified over multiple years demonstrating the persistence of this infection in the population (Ng et al., 2011). Analyses of sub-Antarctic (Arctocephalus tropicalis) and South American fur seal (A. australis) feces also led to the identification of anellovirus sequences (Kluge et al., 2016).
Anelloviruses circulating in the Antarctic ecosystem have recently been shown following detection by HTS and using pairs of abutting primers in the recovery of 152 genomes from vaginal, nasal and faecal samples of Weddell seals in the Ross Sea during the 2014–2015 summer (Fahsbender et al., 2017). Analyses identified two novel anelloviruses, torque teno Leptonychotes weddellii virus (TTLwV-1, TTLwV-2). TTLwV-1 was additionally identified in South Polar skua faecal samples and it was thought that this was as a result of skuas feeding on the placenta and dead carcasses of Weddell seals in the area (Fahsbender et al., 2017).
4. Potential vectors of viruses associated with Antarctic wildlife
Both ecto- and endoparasites have been reported among Antarctic animals. While neither appear to be detrimental to animal health, these organisms may play a significant role as vectors of viruses. Ticks, mites and lice commonly parasitize seals, penguins and other Antarctic birds (Gauthier-Clerc et al., 1998, González-Acuña et al., 2013, McFarlane, 1996). Flaviviruses, orbiviruses, phleboviruses, and nairoviruses have been isolated from seabird ticks (Ixodes uriae) associated with king, rockhopper and royal penguins on Macquarie Island (Major et al., 2009). A novel alphavirus has also been isolated and partially sequenced from lice associated with southern elephant seals of Macquarie Island and the high seroprevalence in the southern elephant seal population showed to this virus strongly suggests its transmission by lice (La Linn et al., 2001). Gastrointestinal parasites, particularly cestode and nematode species, are commonly found in Antarctic seals and penguins. Penguins tend to have a low diversity of parasites and similar profiles have been identified among penguins of the same genus (Diaz et al., 2016, Diaz et al., 2013, Fonteneau et al., 2011, Kleinertz et al., 2014, Vidal et al., 2012). Of the Antarctic seals, gastrointestinal parasites are most prevalent among Weddell and leopard seals. Given that helminth parasites are strongly associated with the diet of the host they infect, this likely explains the higher abundance of parasites among Weddell and leopard seals compared to other Antarctic seals (McFarlane et al., 2009). The potential for endoparasites to transmit viruses to their host has been demonstrated by two genera of plant viruses, nepoviruses and tobraviruses, transmitted by nematodes (Hull, 2014). While our knowledge of parasites in Antarctic animals remains extremely limited and research in this area has been sporadic, developments in molecular technology will undoubtedly have a strong impact toward revealing relationships between organisms and the movement of viruses in the environment.
Recently, Antarctic penguins have been showing signs of disease of unknown pathology, e.g. unexplained incursions of feather loss in Adélie penguins (Grimaldi et al., 2015) in the Ross Sea (2011–2012) but not the years before or after (personal observation). Furthermore, in 2014 observations of an Adélie penguin colony at Hope Bay, Antarctica identified two chicks showing patches without feathers in two sub colonies (Barbosa et al., 2014). Beak and feather disease virus (family Circoviridae) infection in certain psittacines causes feather abnormalities and loss (Pass and Perry, 1984) and hence there is a likelihood that feather loss observed in penguins may be attributed to an unknown circovirus-like agent.
5. Concluding remarks and future directions
Over the last ten years, viral metagenomics has led to a dramatic increase in viral discovery from various environmental and animal samples, for example, Shi et al. (2016) identified ∼1400 novel RNA viruses (from over 200 invertebrate species), Brum et al. (2015) identified ∼5500 distinct dsDNA virus populations (from 43 surface ocean sites worldwide as part of the Tara Oceans expedition) and Paez-Espino et al. (2016) identified ∼120000 partial viral genomes sequences (from ∼3000 geographically diverse samples) using HTS. This together with other studies that have identified large datasets of novel viruses using HTS (e.g. Dayaram et al., 2016, Labonte and Suttle, 2013, Rosario et al., 2015) has shown: 1) the HTS enabled identification of large numbers of previously unknown viruses; 2) we have barely scratched the surface of the viral sequence space and thus their diversity; 3) new taxa will need to be created to classify viruses at a rapid rate to match the pace of virus discovery. All this has opened up discussions on viral classification based on sequence data, either derived from Sanger sequencing or HTS, and lead to a consensus statement by Simmonds et al. (2017a) to incorporate these into current viral taxonomy.
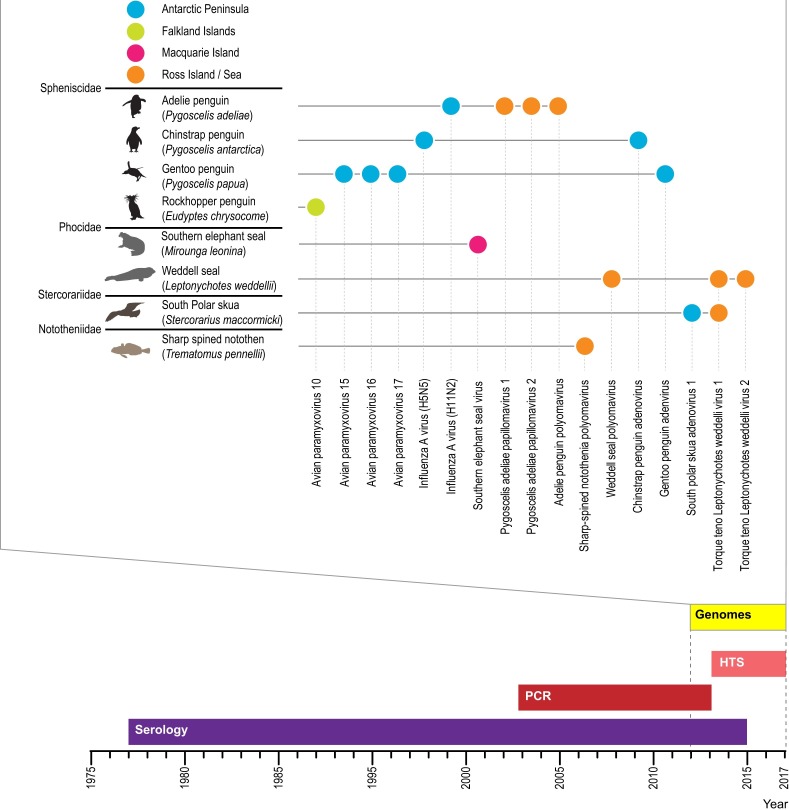
HTS has been used to a large extent in Antarctic environmental virology to study soil (Adriaenssens et al., 2017, Zablocki et al., 2014), lake (Aguirre de Cárcer et al., 2016; Lopez-Bueno et al., 2015, Lopez-Bueno et al., 2009; Yau et al., 2011) and marine (Brum et al., 2017, Miranda et al., 2016) viral ecology. Novel viral genomes from various soils and lake samples (Dziewit and Radlinska, 2016, Kerepesi and Grolmusz, 2017, Meiring et al., 2012, Swanson et al., 2012, Zawar-Reza et al., 2014) have been determined using HTS approaches. In contrast, relatively little is known about viruses associated with Antarctic animals and the associated virus ecology despite the advent of HTS. This perhaps can be attributed to the difficulty in accessing/obtaining animal samples and longitudinal sampling for viral ecology studies. Nonetheless, various studies have used HTS to identify viruses associated with Antarctic animals (Table 2, Fig. 2 ) and we anticipate that the next decade will see a dramatic increase in virology activity and to some extent viral ecology studies of Antarctic animals. Furthermore, it is highly like that large numbers of diverse viruses will be identified using HTS. As sequencing technologies improve, there may be possibility of in-field identification of animal viral pathogens in Antarctica, e.g. using Oxford Nanopore Sequencer (ONS). ONS has been used for metagenomic studies of microbial mats from three lakes in the Antarctic dry valleys (Lakes Fryxell, Lake Vanda and Lake Vida) by Johnson et al. (2017) demonstrating its use in Antarctic field conditions and remote laboratories.
Fig. 2.
Timeline,1975-2017 (present), showing the periods of serology- (purple), PCR- (red) and HTS-based (light red) approaches for viral identification. From 2012 onwards, viral genomes are determined by PCR-, Sanger- and HTS-based approaches. Top panel summarizes the determination of complete genomes by either Sanger sequencing and/or HTS from associated Antarctic animals. Colored circles indicating where they were found: Antarctic Peninsula (dark blue), Falkland Islands (green), Macquarie Island (pink) and Ross Island/sea (orange).
Acknowledgements
ZES is supported through startup funding awarded to AV from The Biodesign Institute and School of Life Sciences, Arizona State University, USA. DGA is supported by National Science Foundation grant PLR-1543541. AV is supported with funds from The Biodesign Institute, Center of Evolution and Medicine, School of Life Science, Arizona State University, USA.
References
- Adriaenssens E.M., Kramer R., Van Goethem M.W., Makhalanyane T.P., Hogg I., Cowan D.A. Environmental drivers of viral community composition in Antarctic soils identified by viromics. Microbiome. 2017;5(1):e83. doi: 10.1186/s40168-017-0301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso C.L., Amarasinghe G.K., Bányai K., Bào Y., Basler C.F., Bavari S., Bejerman N., Blasdell K.R., Briand F.X., Briese T., Bukreyev A., Calisher C.H., Chandran K., Chéng J., Clawson A.N., Collins P.L., Dietzgen R.G., Dolnik O., Domier L.L., Dürrwald R., Dye J.M., Easton A.J., Ebihara H., Farkas S.L., Freitas-Astúa J., Formenty P., Fouchier R.A., Fù Y., Ghedin E., Goodin M.M., Hewson R., Horie M., Hyndman T.H., Jiāng D., Kitajima E.W., Kobinger G.P., Kondo H., Kurath G., Lamb R.A., Lenardon S., Leroy E.M., Li C.X., Lin X.D., Liú L., Longdon B., Marton S., Maisner A., Mühlberger E., Netesov S.V., Nowotny N., Patterson J.L., Payne S.L., Paweska J.T., Randall R.E., Rima B.K., Rota P., Rubbenstroth D., Schwemmle M., Shi M., Smither S.J., Stenglein M.D., Stone D.M., Takada A., Terregino C., Tesh R.B., Tian J.H., Tomonaga K., Tordo N., Towner J.S., Vasilakis N., Verbeek M., Volchkov V.E., Wahl-Jensen V., Walsh J.A., Walker P.J., Wang D., Wang L.F., Wetzel T., Whitfield A.E., Xiè J.T., Yuen K.Y., Zhang Y.Z., Kuhn J.H. Taxonomy of the order Mononegavirales: update 2016. Arch. Virol. 2016;161(8):2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre de Cárcer D., López-Bueno A., Alonso-Lobo J.M., Quesada A., Alcamí A. Metagenomic analysis of lacustrine viral diversity along a latitudinal transect of the Antarctic Peninsula. FEMS Microbiol. Ecol. 2016;92:6. doi: 10.1093/femsec/fiw074. fiw074. [DOI] [PubMed] [Google Scholar]
- Ainley D.G., Ribic C.A., Fraser W.R. Ecological structure among migrant and resident seabirds of the Scotia–Weddell confluence region. J. Anim. Ecology. 1994;63(2):347–364. [Google Scholar]
- Austin F.J., Webster R.G. Evidence of ortho- and paramyxoviruses in fauna from Antarctica. J. Wildl. Dis. 1993;29(4):568–571. doi: 10.7589/0090-3558-29.4.568. [DOI] [PubMed] [Google Scholar]
- Barbosa A., Palacios M.J. Health of Antarctic birds: a review of their parasites, pathogens and diseases. Polar Biol. 2009;32(8):1095–1115. doi: 10.1007/s00300-009-0640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa A., Colominas-Ciuró R., Coria N., Centurión M., Sandler R., Negri A., Santos M. First record of feather-loss disorder in Antarctic penguins. Antarct. Sci. 2014;27(01):69–70. [Google Scholar]
- Baumeister E., Leotta G., Pontoriero A., Campos A., Montalti D., Vigo G., Pecoraro M., Savy V. Serological evidences of influenza A virus infection in Antarctica migratory birds. Int. Congress Ser. 2004;1263:737–740. [Google Scholar]
- Bellehumeur C., Nielsen O., Measures L., Harwood L., Goldstein T., Boyle B., Gagnon C.A. Herpesviruses including novel gammaherpesviruses are widespread among phocid seal species in Canada. J. Wildl. Dis. 2016;52(1):70–81. doi: 10.7589/2015-01-020. [DOI] [PubMed] [Google Scholar]
- Bengtson J.L., Boveng P., Franzen U., Have P., Heide-Jorgensen M.P., Harkonen T.J. Antibodies to canine distemper virus in antarctic seals. Mar. Mamm. Sci. 1991;7(1):85–87. [Google Scholar]
- Bernard H.U., Burk R.D., Chen Z., van Doorslaer K., zur Hausen H., de Villiers E.M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401(1):70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagini P., Bendinelli M., Hino S., Kakkola L., Mankertz A., Niel C., Okamoto H., Raidal S., Teo C.G., Todd D. Family Anellovirdae. In: King A.M.Q., Lefkowitz E., Adams M.J., Carstens E.B., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego, USA: 2011. pp. 331–341. [Google Scholar]
- Bodewes R., Rubio Garcia A., Wiersma L.C., Getu S., Beukers M., Schapendonk C.M., van Run P.R., van de Bildt M.W., Poen M.J., Osinga N., Sanchez Contreras G.J., Kuiken T., Smits S.L., Osterhaus A.D. Novel B19-like parvovirus in the brain of a harbor seal. PLoS One. 2013;8(11):e79259. doi: 10.1371/journal.pone.0079259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., Contreras G.J., Garcia A.R., Hapsari R., van de Bildt M.W., Kuiken T., Osterhaus A.D. Identification of DNA sequences that imply a novel gammaherpesvirus in seals. J. Gen. Virol. 2015;96(5):1109–1114. doi: 10.1099/vir.0.000029. [DOI] [PubMed] [Google Scholar]
- Bracht A.J., Brudek R.L., Ewing R.Y., Manire C.A., Burek K.A., Rosa C., Beckmen K.B., Maruniak J.E., Romero C.H. Genetic identification of novel poxviruses of cetaceans and pinnipeds. Arch. Virol. 2006;151(3):423–438. doi: 10.1007/s00705-005-0679-6. [DOI] [PubMed] [Google Scholar]
- Briggs J.C. Elsevier; 1995. Global Biogeography; p. 14. [Google Scholar]
- Brooke M. Oxford University Press; 2004. Albatrosses and Petrels Across the World. [Google Scholar]
- Brum J.R., Ignacio-Espinoza J.C., Roux S., Doulcier G., Acinas S.G., Alberti A., Chaffron S., Cruaud C., de Vargas C., Gasol J.M., Gorsky G., Gregory A.C., Guidi L., Hingamp P., Iudicone D., Not F., Ogata H., Pesant S., Poulos B.T., Schwenck S.M., Speich S., Dimier C., Kandels-Lewis S., Picheral M., Searson S., Tara Oceans C., Bork P., Bowler C., Sunagawa S., Wincker P., Karsenti E., Sullivan M.B. Ocean plankton: patterns and ecological drivers of ocean viral communities. Science. 2015;348(6237):1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- Brum J.R., Hurwitz B.L., Schofield O., Ducklow H.W., Sullivan M.B. Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 2017;11(2):588. doi: 10.1038/ismej.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck C.B., Van Doorslaer K., Peretti A., Geoghegan E.M., Tisza M.J., An P., Katz J.P., Pipas J.M., McBride A.A., Camus A.C., McDermott A.J., Dill J.A., Delwart E., Ng T.F., Farkas K., Austin C., Kraberger S., Davison W., Pastrana D.V., Varsani A. The ancient evolutionary history of polyomaviruses. PLoS Pathog. 2016;12(4):e1005574. doi: 10.1371/journal.ppat.1005574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappetta C.M., Cibulski S.P., Lima F.E., Varela A.P., Amorim D.B., Tavares M., Roehe P.M. Molecular detection of circovirus and adenovirus in feces of fur seals (Arctocephalus spp.) Ecohealth. 2017;14(1):69–77. doi: 10.1007/s10393-016-1195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrove K.M., Wellehan J.F., Jr., Rivera R., Moore P.F., Gulland F.M., Lowenstine L.J., Nordhausen R.W., Nollens H.H. Polyomavirus infection in a free-ranging California sea lion (Zalophus californianus) with intestinal T-cell lymphoma. J. Vet. Diagn. Invest. 2010;22(4):628–632. doi: 10.1177/104063871002200422. [DOI] [PubMed] [Google Scholar]
- Cortes-Hinojosa G., Gulland F.M., Goldstein T., Venn-Watson S., Rivera R., Waltzek T.B., Salemi M., Wellehan J.F., Jr. Phylogenomic characterization of California sea lion adenovirus-1. Infect. Genet. Evol. 2015;31:270–276. doi: 10.1016/j.meegid.2015.01.024. [DOI] [PubMed] [Google Scholar]
- Cortes-Hinojosa G., Doescher B., Kinsel M., Lednicky J., Loeb J., Waltzek T., Wellehan J.F., Jr. Coinfection of California sea lion adenovirus 1 and a novel polyomavirus in a Hawaiian monk seal (Neomonachus Schauinslandi) J. Zoo Wildl. Med. 2016;47(2):427–437. doi: 10.1638/2014-0252.1. [DOI] [PubMed] [Google Scholar]
- de Villiers E.M., Fauquet C., Broker T.R., Bernard H.U., zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Dayaram A., Galatowitsch M.L., Arguello-Astorga G.R., van Bysterveldt K., Kraberger S., Stainton D., Harding J.S., Roumagnac P., Martin D.P., Lefeuvre P., Varsani A. Diverse circular replication-associated protein encoding viruses circulating in invertebrates within a lake ecosystem. Infect. Genet. Evol. 2016;39:304–316. doi: 10.1016/j.meegid.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Delmas B., Mundt E., Vakharia V.N., Wu J.L. Family Birnaviridae. In: King A.M.Q., Lefkowitz E., Adams M.J., Carstens E.B., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego, USA: 2011. pp. 496–507. [Google Scholar]
- Diaz J.I., Fusaro B., Longarzo L., Coria N.R., Vidal V., Jerez S., Ortiz J., Barbosa A. Gastrointestinal helminths of gentoo penguins (Pygoscelis papua) from stranger point 25 de Mayo/King george island, Antarctica. Parasitol. Res. 2013;112(5):1877–1881. doi: 10.1007/s00436-013-3341-3. [DOI] [PubMed] [Google Scholar]
- Diaz J.I., Fusaro B., Longarzo L., Coria N.R., Vidal V., D'Amico V., Barbosa A. Gastrointestinal helminths of Adélie penguins (Pygoscelis adeliae) from Antarctica. Polar Res. 2016;35(1):e28516. [Google Scholar]
- Duncan C., Goldstein T., Hearne C., Gelatt T., Spraker T. Novel polyomaviral infection in the placenta of a northern fur seal (Callorhinus ursinus) on the Pribilof Islands Alaska, USA. USA. J. Wildl. Dis. 2013;49(1):163–167. doi: 10.7589/2012-04-101. [DOI] [PubMed] [Google Scholar]
- Dziewit L., Radlinska M. Two inducible prophages of an antarctic Pseudomonas sp. ANT_H14 use the same capsid for packaging their genomes—characterization of a novel phage helper-satellite system. PLoS One. 2016;11(7):e0158889. doi: 10.1371/journal.pone.0158889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman J.T. Academic Press; 2013. Antarctic Fish Biology: Evolution in a Unique Environment. [Google Scholar]
- Fahsbender E., Burns J.M., Kim S., Kraberger S., Frankfurter G., Eilers A.A., Shero M.R., Beltran R., Kirkham A., McCorkell R., Berngartt R.K., Male M.F., Ballard G., Ainley D.G., Breitbart M., Varsani A. Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica. Virus Evol. 2017;3(1):vex017. doi: 10.1093/ve/vex017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonteneau F., Geiger S., Marion L., Le Maho Y., Robin J.-P., Kinsella J.M. Gastrointestinal helminths of King penguins (Aptenodytes patagonicus) at Crozet Archipelago. Polar Biol. 2011;34(8):1249–1252. [Google Scholar]
- Forrester N.L., Palacios G., Tesh R.B., Savji N., Guzman H., Sherman M., Weaver S.C., Lipkin W.I. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J. Virol. 2012;86(5):2729–2738. doi: 10.1128/JVI.05591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner H., Kerry K., Riddle M., Brouwer S., Gleeson L. Poultry virus infection in Antarctic penguins. Nature. 1997;387(6630):245. doi: 10.1038/387245a0. [DOI] [PubMed] [Google Scholar]
- Gauthier-Clerc M., Clerquin Y., Handrich Y. Hyperinfestation by ticks Ixodes uriae: A possible cause of death in adult King penguins, a long-lived seabird. Colon Waterbird. 1998;21(2):229–233. [Google Scholar]
- Gauthier-Clerc M., Eterradossi N., Toquin D., Guittet M., Kuntz G., Le Maho Y. Serological survey of the king penguin, Aptenodytes patagonicus in Crozet Archipelago for antibodies to infectious bursal disease, influenza A and Newcastle disease viruses. Polar Biol. 2002;25(4):316–319. [Google Scholar]
- Goldstein T., Mazet J.A., Gulland F.M., Rowles T., Harvey J.T., Allen S.G., King D.P., Aldridge B.M., Stott J.L. The transmission of phocine herpesvirus-1 in rehabilitating and free-ranging Pacific harbor seals (Phoca vitulina) in California. Vet. Microbiol. 2004;103(3–4):131–141. doi: 10.1016/j.vetmic.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Goldstein T., Colegrove K.M., Hanson M., Gulland F.M. Isolation of a novel adenovirus from California sea lions Zalophus californianus. Dis. Aquat. Organ. 2011;94(3):243–248. doi: 10.3354/dao02321. [DOI] [PubMed] [Google Scholar]
- González-Acuña D., Hernández J., Moreno L., Herrmann B., Palma R., Latorre A., Medina-Vogel G., Kinsella M.J., Martín N., Araya K., Torres I., Fernandez N., Olsen B. Health evaluation of wild gentoo penguins (Pygoscelis papua) in the Antarctic Peninsula. Polar Biol. 2013;36(12):1749–1760. [Google Scholar]
- Goraichuk I.V., Dimitrov K.M., Sharma P., Miller P.J., Swayne D.E., Suarez D.L., Afonso C.L. Complete genome sequences of four avian paramyxoviruses of serotype 10 isolated from Rockhopper penguins on the Falkland Islands. Genome Announc. 2017;5(22):00472–00517. doi: 10.1128/genomeA.00472-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi W.W., Hall R.J., White D.D., Wang J., Massaro M., Tompkins D.M. First report of a feather loss condition in Adelie penguins (Pygoscelis adeliae) on Ross Island, Antarctica, and a preliminary investigation of its cause. Emu. 2015;115(2):185–189. [Google Scholar]
- Gunther T., Haas L., Alawi M., Wohlsein P., Marks J., Grundhoff A., Becher P., Fischer N. Recovery of the first full-length genome sequence of a parapoxvirus directly from a clinical sample. Sci. Rep. 2017;7(1):e3734. doi: 10.1038/s41598-017-03997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder T.C., Plotz J., Liess B. Antibodies against European phocine herpesvirus isolates detected in sera of Antarctic seals. Polar Biol. 1991;11(7):509–512. [Google Scholar]
- Harder T.C., Harder M., Vos H., Kulonen K., Kennedy-Stoskopf S., Liess B., Appel M.J., Osterhaus A.D. Characterization of phocid herpesvirus-1 and −2 as putative alpha- and gammaherpesviruses of North American and European pinnipeds. J. Gen. Virol. 1996;77(Pt 1):27–35. doi: 10.1099/0022-1317-77-1-27. [DOI] [PubMed] [Google Scholar]
- Harrach B., Benko M., Both G.W., Brown M.V., Davison A.J., Echavarría M., Hess M., Jones M.S., Kajon A., Lehmkuhl H.D. Family Adenoviridae. In: King A.M.Q., Lefkowitz E.J., Adams M.J., Carstens E.B., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego, CA, USA: 2011. pp. 95–111. [Google Scholar]
- Hollmén T., Franson J.C., Docherty D.E., Kilpi M., Hario M., Creekmore L.H., Petersen M.R. Infectious bursal disease virus antibodies in eider ducks and herring gulls. Condor. 2000;102(3):688–691. [Google Scholar]
- Hull R. Academic Press; Boston: 2014. Plant to Plant Movement, Plant Virology; pp. 669–751. [Google Scholar]
- Hurt A.C., Vijaykrishna D., Butler J., Baas C., Maurer-Stroh S., Silva-de-la-Fuente M.C., Medina-Vogel G., Olsen B., Kelso A., Barr I.G., Gonzalez-Acuna D. Detection of evolutionarily distinct avian influenza a viruses in antarctica. MBio. 2014;5(3):e01098–01014. doi: 10.1128/mBio.01098-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt A.C., Su Y.C., Aban M., Peck H., Lau H., Baas C., Deng Y.M., Spirason N., Ellstrom P., Hernandez J., Olsen B., Barr I.G., Vijaykrishna D., Gonzalez-Acuna D. Evidence for the introduction, reassortment, and persistence of diverse influenza a viruses in Antarctica. J. Virol. 2016;90(21):9674–9682. doi: 10.1128/JVI.01404-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoshima Y., Murakami T., Ishiguro N., Hasegawa K., Kasamatsu M. An outbreak of lethal adenovirus infection among different otariid species. Vet. Microbiol. 2013;165(3–4):455–459. doi: 10.1016/j.vetmic.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Johnson S.S., Zaikova E., Goerlitz D.S., Bai Y., Tighe S.W. Real-Time DNA Sequencing in the antarctic dry valleys using the Oxford nanopore sequencer. J. Biomol. Tech. 2017;28(1):2. doi: 10.7171/jbt.17-2801-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasanga C.J., Yamaguchi T., Wambura P.N., Munang'andu H.M., Ohya K., Fukushi H. Detection of infectious bursal disease virus (IBDV) genome in free-living pigeon and guinea fowl in Africa suggests involvement of wild birds in the epidemiology of IBDV. Virus Genes. 2008;36(3):521–529. doi: 10.1007/s11262-008-0219-z. [DOI] [PubMed] [Google Scholar]
- Kerepesi C., Grolmusz V. The Giant Virus Finder discovers an abundance of giant viruses in the Antarctic dry valleys. Arch. Virol. 2017;162(6):1671–1676. doi: 10.1007/s00705-017-3286-4. [DOI] [PubMed] [Google Scholar]
- Kerry K.R., Riddle M.J. Health of Antarctic wildlife: an introduction. In: Kerry K.R., Riddle M., editors. Health of Antarctic Wildlife. Springer Berlin Heidelberg; Berlin, Heidelberg: 2009. pp. 1–10. [Google Scholar]
- Kerry K., Irvine L., Beggs A., Watts J. Health of Antarctic Wildlife Springer; 2009. An Unusual Mortality Event Among Adélie Penguins in the Vicinity of Mawson Station, Antarctica; pp. 107–112. [Google Scholar]
- Kleinertz S., Christmann S., Silva L.M., Hirzmann J., Hermosilla C., Taubert A. Gastrointestinal parasite fauna of emperor penguins (Aptenodytes forsteri) at the Atka Bay, Antarctica. Parasitol. Res. 2014;113(11):4133–4139. doi: 10.1007/s00436-014-4085-4. [DOI] [PubMed] [Google Scholar]
- Kluge M., Campos F.S., Tavares M., de Amorim D.B., Valdez F.P., Giongo A., Roehe P.M., Franco A.C. Metagenomic survey of viral diversity obtained from feces of subantarctic and South American fur seals. PLoS One. 2016;11(3):e0151921. doi: 10.1371/journal.pone.0151921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Linn M., Gardner J., Warrilow D., Darnell G.A., McMahon C.R., Field I., Hyatt A.D., Slade R.W., Suhrbier A. Arbovirus of marine mammals: a new alphavirus isolated from the elephant seal louse, Lepidophthirus macrorhini. J. Virol. 2001;75(9):4103–4109. doi: 10.1128/JVI.75.9.4103-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte J.M., Suttle C.A. Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J. 2013;7(11):2169–2177. doi: 10.1038/ismej.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws R.M., Taylor R. A mass dying of crabeater seals, Lobodon carcinophagus (Gray) J. Zool. 1957;129(3):315–324. [Google Scholar]
- Laws R.M. Seals and whales of the Southern Ocean. Philos. Trans. R. Soc. Lond. B: Biol. Sci. 1977;279(963):81–96. [Google Scholar]
- Laws R.M. The significance of vertebrates in the Antarctic marine ecosystem. In: Llano G.A., editor. Adaptations Within Antarctic Ecosystem. Gulf Publishing Company; Houston, TX: 1977. pp. 411–438. [Google Scholar]
- Lee R.E., Baust J.G. Cold-hardiness in the antarctic tick, Ixodes uriae. Physiol. Zool. 1987;60(4):499–506. [Google Scholar]
- Lee S.Y., Kim J.H., Park Y.M., Shin O.S., Kim H., Choi H.G., Song J.W. A novel adenovirus in Chinstrap penguins (Pygoscelis antarctica) in Antarctica. Viruses. 2014;6(5):2052–2061. doi: 10.3390/v6052052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Kim J.H., Seo T.K., No J.S., Kim H., Kim W.K., Choi H.G., Kang S.H., Song J.W. Genetic and molecular epidemiological characterization of a novel adenovirus in antarctic penguins collected between 2008 and 2013. PLoS One. 2016;11(6):e0157032. doi: 10.1371/journal.pone.0157032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bueno A., Tamames J., Velazquez D., Moya A., Quesada A., Alcami A. High diversity of the viral community from an Antarctic lake. Science. 2009;326(5954):858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- Lopez-Bueno A., Rastrojo A., Peiro R., Arenas M., Alcami A. Ecological connectivity shapes quasispecies structure of RNA viruses in an Antarctic lake. Mol. Ecol. 2015;24(19):4812–4825. doi: 10.1111/mec.13321. [DOI] [PubMed] [Google Scholar]
- MacDonald D.M., Conroy J.W.H. Virus disease resembling puffinosis in the gentoo penguin (Pygoscelis papua) Br. Antarct. Surv. Bull. 1971;60:80–83. [Google Scholar]
- Major L., Linn M.L., Slade R.W., Schroder W.A., Hyatt A.D., Gardner J., Cowley J., Suhrbier A. Ticks associated with macquarie island penguins carry arboviruses from four genera. PLoS One. 2009;4(2):e4375. doi: 10.1371/journal.pone.0004375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley J.W., Hongo S., Kaverin N.V., Kochs G., Lamb R.A., Matrosovich M.N., Perez D.R., Palese P., Presti R.M., Rimstad E. Family Orthomyxoviridae. In: King A.M.Q., Lefkowitz E., Adams M.J., Carstens E.B., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego, USA: 2011. pp. 749–761. [Google Scholar]
- McFarlane R.A., Norman R.J.d.B., Jones H.I. Springer; 2009. Diseases and Parasites of Antarctic and Sub-Antarctic Seals, Health of Antarctic Wildlife; pp. 57–93. [Google Scholar]
- McFarlane R.A. Some observations on Adelie penguin (Pygoscelis adeliae) mortality in East Antarctica. Avian Pathol. 1996;25(1):187–190. doi: 10.1080/03079459608419134. [DOI] [PubMed] [Google Scholar]
- McFarlane R. Springer; 2009. Health Assessment and Diseases of the Weddell Seal, Leptonochotes Weddelli, in Vestfold Hills, East Antarctica, Health of Antarctic Wildlife; pp. 139–166. [Google Scholar]
- Meiring T.L., Tuffin I.M., Cary C., Cowan D.A. Genome sequence of temperate bacteriophage Psymv2 from Antarctic Dry Valley soil isolate Psychrobacter sp. MV2. Extremophiles. 2012;16(5):715–726. doi: 10.1007/s00792-012-0467-7. [DOI] [PubMed] [Google Scholar]
- Miller G.D., Watts J.M., Shellam G.R. Viral antibodies in south polar skuas around Davis Station, Antarctica. Antarct. Sci. 2008;20(05):455–461. [Google Scholar]
- Miller P.J., Afonso C.L., Spackman E., Scott M.A., Pedersen J.C., Senne D.A., Brown J.D., Fuller C.M., Uhart M.M., Karesh W.B., Brown I.H., Alexander D.J., Swayne D.E. Evidence for a new avian paramyxovirus serotype 10 detected in rockhopper penguins from the Falkland Islands. J. Virol. 2010;84(21):11496–11504. doi: 10.1128/JVI.00822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.A., Culley A.I., Schvarcz C.R., Steward G.F. RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 2016;18(11):3714–3727. doi: 10.1111/1462-2920.13291. [DOI] [PubMed] [Google Scholar]
- Moens U., Calvignac-Spencer S., Lauber C., Ramqvist T., Feltkamp M.C.W., Daugherty M.D., Verschoor E.J., Ehlers B., ICTV Report Consortium ICTV virus taxonomy profile: Polyomaviridae. J. Gen. Virol. 2017;98(6):1159–1160. doi: 10.1099/jgv.0.000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan I.R., Westbury H.A. Virological studies of Adelie Penguins (Pygoscelis adeliae) in Antarctica. Avian Dis. 1981;25(4):1019–1026. [PubMed] [Google Scholar]
- Morgan I.R., Westbury H.A. Studies of viruses in penguins in the Vestfold Hills. Hydrobiologia. 1988;165(1):263–269. [Google Scholar]
- Morgan I.R., Westbury H.A., Caple I.W., Campbell J. A survey of virus infection in sub-antarctic penguins on Macquarie Island, Southern Ocean. Aust. Vet. J. 1981;57(7):333–335. doi: 10.1111/j.1751-0813.1981.tb05839.x. [DOI] [PubMed] [Google Scholar]
- Muller G., Groters S., Siebert U., Rosenberger T., Driver J., Konig M., Becher P., Hetzel U., Baumgartner W. Parapoxvirus infection in harbor seals (Phoca vitulina) from the German North Sea. Vet. Pathol. 2003;40(4):445–454. doi: 10.1354/vp.40-4-445. [DOI] [PubMed] [Google Scholar]
- Murphy R.C. Macmillan; New York: 1936. The Oceanic Birds of South America. [Google Scholar]
- Neira V., Tapia R., Verdugo C., Barriga G., Mor S., Ng T.F.F., Garcia V., Del Rio J., Rodrigues P., Briceno C., Medina R.A., Gonzalez-Acuna D. Novel avulaviruses in penguins, Antarctica. Emerg. Infect. Dis. 2017;23(7):1212–1214. doi: 10.3201/eid2307.170054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton P.F., Munro R., Pow I., Gilray J., Gray E.W., Reid H.W. Isolation of a parapoxvirus from a grey seal (Halichoerus grypus) Vet. Rec. 1995;137(22):562–564. doi: 10.1136/vr.137.22.562. [DOI] [PubMed] [Google Scholar]
- Ng T.F., Suedmeyer W.K., Wheeler E., Gulland F., Breitbart M. Novel anellovirus discovered from a mortality event of captive California sea lions. J. Gen. Virol. 2009;90(Pt 5):1256–1261. doi: 10.1099/vir.0.008987-0. [DOI] [PubMed] [Google Scholar]
- Ng T.F., Wheeler E., Greig D., Waltzek T.B., Gulland F., Breitbart M. Metagenomic identification of a novel anellovirus in Pacific harbor seal (Phoca vitulina richardsii) lung samples and its detection in samples from multiple years. J. Gen. Virol. 2011;92(Pt 6):1318–1323. doi: 10.1099/vir.0.029678-0. [DOI] [PubMed] [Google Scholar]
- Nollens H.H., Jacobson E.R., Gulland F.M., Beusse D.O., Bossart G.D., Hernandez J.A., Klein P.A., Condit R.C. Pathology and preliminary characterization of a parapoxvirus isolated from a California sea lion (Zalophus californianus) J. Wildl. Dis. 2006;42(1):23–32. doi: 10.7589/0090-3558-42.1.23. [DOI] [PubMed] [Google Scholar]
- Offerman K., Carulei O., van der Walt A.P., Douglass N., Williamson A.L. The complete genome sequences of poxviruses isolated from a penguin and a pigeon in South Africa and comparison to other sequenced avipoxviruses. BMC Genomics. 2014;15:e463. doi: 10.1186/1471-2164-15-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Wakuda T., Yamaguchi T., Murata K., Setiyono A., Fukushi H., Hirai K. Seroprevalence of infectious bursal disease virus in free-living wild birds in Japan. J. Vet. Med. Sci. 1998;60(11):1277–1279. doi: 10.1292/jvms.60.1277. [DOI] [PubMed] [Google Scholar]
- Osterhaus A.D., Yang H., Spijkers H.E., Groen J., Teppema J.S., van Steenis G. The isolation and partial characterization of a highly pathogenic herpesvirus from the harbor seal (Phoca vitulina) Arch. Virol. 1985;86(3–4):239–251. doi: 10.1007/BF01309828. [DOI] [PubMed] [Google Scholar]
- Osterhaus A.D., Groen J., De Vries P., UytdeHaag F.G., Klingeborn B., Zarnke R. Canine distemper virus in seals. Nature. 1988;335(6189):403–404. doi: 10.1038/335403a0. [DOI] [PubMed] [Google Scholar]
- Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C. Uncovering earth’s virome. Nature. 2016;7617:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- Park Y.M., Kim J.H., Gu S.H., Lee S.Y., Lee M.G., Kang Y.K., Kang S.H., Kim H.J., Song J.W. Full genome analysis of a novel adenovirus from the South Polar skua (Catharacta maccormicki) in Antarctica. Virology. 2012;422(1):144–150. doi: 10.1016/j.virol.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pass D.A., Perry R.A. The pathology of psittacine beak and feather disease. Aust. Vet. J. 1984;61(3):69–74. doi: 10.1111/j.1751-0813.1984.tb15520.x. [DOI] [PubMed] [Google Scholar]
- Polyomaviridae Study Group of the International Committee on Taxonomy of Viruses, Calvignac-Spencer S., Feltkamp M.C., Daugherty M.D., Moens U., Ramqvist T., Johne R., Ehlers B. A taxonomy update for the family Polyomaviridae. Arch. Virol. 2016;161(6):1739–1750. doi: 10.1007/s00705-016-2794-y. [DOI] [PubMed] [Google Scholar]
- Power A., Huang H., Roehrig J., Strauss E., Weaver S. Togaviridae. In: Maclachlan J.N., Dubovi E.J., Barthold S.W., Swayne D.F., Winton J.R., editors. Fenner's Veterinary Virology. Academic Press; Boston: 2017. pp. 511–524. [Google Scholar]
- Ribic C.A., Ainley D.G. Constancy of seabird species assemblages: an exploratory look. Biol. Oceanogr. 1989;6(2):175–202. [Google Scholar]
- Ribic C.A., Ainley D.G., Glenn Ford R., Fraser W.R., Tynan C.T., Woehler E.J. Water masses, ocean fronts, and the structure of Antarctic seabird communities: putting the eastern Bellingshausen Sea in perspective. Deep Sea Res. Part II. 2011;58(13–16):1695–1709. [Google Scholar]
- Rosario K., Schenck R.O., Harbeitner R.C., Lawler S.N., Breitbart M. Novel circular single-stranded DNA viruses identified in marine invertebrates reveal high sequence diversity and consistent predicted intrinsic disorder patterns within putative structural proteins. Front. Microbiol. 2015;6:696. doi: 10.3389/fmicb.2015.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Lin X.-D., Tian J.-H., Chen L.-J., Chen X., Li C.-X., Qin X.-C., Li J., Cao J.-P., Eden J.-S., Buchmann J., Wang W., Xu J., Holmes E.C., Zhang Y.-Z. Redefining the invertebrate RNA virosphere. Nature. 2016;540(7634):539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Adams M.J., Benko M., Breitbart M., Brister J.R., Carstens E.B., Davison A.J., Delwart E., Gorbalenya A.E., Harrach B., Hull R., King A.M., Koonin E.V., Krupovic M., Kuhn J.H., Lefkowitz E.J., Nibert M.L., Orton R., Roossinck M.J., Sabanadzovic S., Sullivan M.B., Suttle C.A., Tesh R.B., van der Vlugt R.A., Varsani A., Zerbini F.M. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017;15(3):161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Becher P., Bukh J., Gould E.A., Meyers G., Monath T., Muerhoff S., Pletnev A., Rico-Hesse R., Smith D.B., Stapleton J.T., ICTV Report Consortium ICTV virus taxonomy profile: Flaviviridae. J. Gen. Virol. 2017;98(1):2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniff D.B., Garrott R.A., Rotella J.J., Fraser W.R., Ainley D.G. Opinion: projecting the effects of environmental change on Antarctic seals. Antarct. Sci. 2008;20(05):425–435. [Google Scholar]
- Siniff D.B. Seal population-dynamics and ecology. J. R. Soc. N. Z. 1981;11(4):317–327. [Google Scholar]
- Skinner M.A., Buller R.M., Damon I.K., Lefkowitz E.J., McFadden G., McInnes C.J., Mercer A.A., Moyer R.W., Upton C. Family Poxviridae. In: King A.M.Q., Lefkowitz E., Adams M.J., Carstens E.B., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego, USA: 2011. pp. 291–309. [Google Scholar]
- Stenvers O., Plotz J., Ludwig H. Antarctic seals carry antibodies against seal herpesvirus. Arch. Virol. 1992;123(3–4):421–424. doi: 10.1007/BF01317275. [DOI] [PubMed] [Google Scholar]
- Swanson M.M., Reavy B., Makarova K.S., Cock P.J., Hopkins D.W., Torrance L., Koonin E.V., Taliansky M. Novel bacteriophages containing a genome of another bacteriophage within their genomes. PLoS One. 2012;7(7):e40683. doi: 10.1371/journal.pone.0040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomazelli L.M., Araujo J., Oliveira D.B., Sanfilippo L., Ferreira C.S., Brentano L., Pelizari V.H., Nakayama C., Duarte R., Hurtado R., Branco J.O., Walker D., Durigon E.L. Newcastle disease virus in penguins from King George Island on the Antarctic region. Vet. Microbiol. 2010;146(1-2):155–160. doi: 10.1016/j.vetmic.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Trathan P.N., Lynch H.J., Fraser W.R. Changes in penguin distribution over the Antarctic Peninsula and Scotia Arc. Antarct. Environ. Portal. 2016 doi: 10.18124/D43019. [DOI] [Google Scholar]
- Tryland M., Klein J., Nordoy E.S., Blix A.S. Isolation and partial characterization of a parapoxvirus isolated from a skin lesion of a Weddell seal. Virus Res. 2005;108(1–2):83–87. doi: 10.1016/j.virusres.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Tryland M., Nymo I.H., Nielsen O., Nordoy E.S., Kovacs K.M., Krafft B.A., Thoresen S.I., Asbakk K., Osterrieder K., Roth S.J., Lydersen C., Godfroid J., Blix A.S. Serum chemistry and antibodies against pathogens in antarctic fur seals, Weddell seals, crabeater seals, and Ross seals. J. Wildl. Dis. 2012;48(3):632–645. doi: 10.7589/0090-3558-48.3.632. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer K., Ruoppolo V., Schmidt A., Lescroël A., Jongsomjit D., Elrod M., Kraberger S., Stainton D., Dugger K.M., Ballard G., Ainley D.G., Varsani A. Unique genome organization of non-mammalian papillomaviruses provides insights into the evolution of viral early proteins. Virus Evol. 2017;3(2):vex027. doi: 10.1093/ve/vex027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varsani A., Kraberger S., Jennings S., Porzig E.L., Julian L., Massaro M., Pollard A., Ballard G., Ainley D.G. A novel papillomavirus in Adelie penguin (Pygoscelis adeliae) faeces sampled at the Cape Crozier colony, Antarctica. J. Gen. Virol. 2014;95(6):1352–1365. doi: 10.1099/vir.0.064436-0. [DOI] [PubMed] [Google Scholar]
- Varsani A., Porzig E.L., Jennings S., Kraberger S., Farkas K., Julian L., Massaro M., Ballard G., Ainley D.G. Identification of an avian polyomavirus associated with Adelie penguins (Pygoscelis adeliae) J. Gen. Virol. 2015;96(4):851–857. doi: 10.1099/vir.0.000038. [DOI] [PubMed] [Google Scholar]
- Varsani A., Frankfurter G., Stainton D., Male M.F., Kraberger S., Burns J.M. Identification of a polyomavirus in Weddell seal (Leptonychotes weddellii) from the Ross Sea (Antarctica) Arch. Virol. 2017;162(5):1403–1407. doi: 10.1007/s00705-017-3239-y. [DOI] [PubMed] [Google Scholar]
- Vidal V., Ortiz J., Diaz J.I., de Ybanez M.R., Amat M.T., Palacios M.J., Benzal J., Valera F., de la Cruz C., Motas M., Barbosa A. Gastrointestinal parasites in chinstrap penguins from deception island south shetlands, Antarctica. Parasitol. Res. 2012;111(2):723–727. doi: 10.1007/s00436-012-2892-z. [DOI] [PubMed] [Google Scholar]
- Watts J.M., Miller G.D., Shellam G.R. Infectious bursal disease virus and Antarctic birds. In: Kerry K.R., Riddle M., editors. Health of Antarctic Wildlife. Springer Berlin Heidelberg; Berlin, Heidelberg: 2009. pp. 95–105. [Google Scholar]
- Weimerskirch H., Tarroux A., Chastel O., Delord K., Cherel Y., Descamps S. Population-specific wintering distributions of adult south polar skuas over three oceans. Mar. Ecol. Prog. Ser. 2015;538:229–237. [Google Scholar]
- Williams T.D. Oxford University Press; USA: 1995. The Penguins: Spheniscidae, 2. [Google Scholar]
- Yau S., Lauro F.M., DeMaere M.Z., Brown M.V., Thomas T., Raftery M.J., Andrews-Pfannkoch C., Lewis M., Hoffman J.M., Gibson J.A., Cavicchioli R. Virophage control of antarctic algal host-virus dynamics. Proc. Natl. Acad. Sci. U. S. A. 2011;108(15):6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem P.K., Stewart B.S., Gelatt T.S., Siniff D.B. Health assessment of Weddell seals, Leptonychotes weddellii, in McMurdo Sound, Antarctica. In: Kerry K.R., Riddle M., editors. Health of Antarctic Wildlife. Springer Berlin Heidelberg; Berlin, Heidelberg: 2009. pp. 123–138. [Google Scholar]
- Zablocki O., van Zyl L., Adriaenssens E.M., Rubagotti E., Tuffin M., Cary S.C., Cowan D. High-level diversity of tailed phages, eukaryote-associated viruses, and virophage-like elements in the metaviromes of antarctic soils. Appl. Environ. Microbiol. 2014;80(22):6888–6897. doi: 10.1128/AEM.01525-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawar-Reza P., Arguello-Astorga G.R., Kraberger S., Julian L., Stainton D., Broady P.A., Varsani A. Diverse small circular single-stranded DNA viruses identified in a freshwater pond on the McMurdo Ice Shelf (Antarctica) Infect. Genet. Evol. 2014;26:132–138. doi: 10.1016/j.meegid.2014.05.018. [DOI] [PubMed] [Google Scholar]