Highlights
-
•
The full-length genome of canine coronavirus type I was determined.
-
•
Sequence analysis showed unique features with respect to canine coronavirus type II.
-
•
By phylogeny, canine coronavirus type I formed a separate cluster.
-
•
The results may contribute to the understanding of the Alphacoronavirus-1 evolution.
Keywords: Dog, Canine coronavirus type I, Genomic analysis
Abstract
Canine coronavirus types I (CCoV-I) and II (CCoV-II) are usually responsible for mild enteritis in dogs. While the CCoV-II genome has been completely sequenced, to date there are no complete genomic sequence data available publicly for CCoV-I. Thus, the aim of the present study was to analyze the full-length genome of a CCoV-I prototype strain that had been recovered from a dog with diarrhea in Italy. CCoV-I strain 23/03 has a genome of 30,000 nucleotides, excluding the 3′ poly(A) tail, displaying the typical Alphacoronavirus-1 organization and the highest genetic relatedness to CCoV-II. However, two distinct features were observed in the CCoV-I genome: (i) the presence of an additional ORF between the spike (S) protein gene and ORF3a; (ii) the diversity of the S protein, which is more closely related to that of feline coronavirus type I and presents a furin cleavage site. The present study may contribute to a better understanding of the Alphacoronavirus-1 evolutionary pattern and may be paradigmatic of how coronaviruses evolve through gene losses, acquisition and exchanges among different members.
1. Introduction
Coronaviruses (CoVs) are large, single-stranded, positive-sense RNA viruses, which are responsible for enteric and/or respiratory disease in mammals and birds. Canine coronavirus (CCoV) is usually responsible for mild enteritis in young dogs (Decaro and Buonavoglia, 2008, Decaro and Buonavoglia, 2011), although fatal disease has been associated to a pantropic variant of the virus (Decaro et al., 2008, Decaro et al., 2010a, Decaro et al., 2012, Marinaro et al., 2010, Zicola et al., 2012, Ntafis et al., 2012). Based on the genetic distance encountered in the spike (S) protein gene (Pratelli et al., 2003), two CCoV genotypes are known, CCoV type I (CCoV-I) and type II (CCoV-II), which are variously distributed worldwide (Decaro et al., 2005, Decaro et al., 2011, Decaro et al., 2013, McElligott et al., 2011; Ntafis et al., 2013, Licitra et al., 2014, Cavalli et al., 2014, Costa et al., 2014). CCoV-II has been found to exist in two different subtypes, CCoV-IIa and CCoV-IIb, the latter being the result of homologous recombination with transmissible gastroenteritis virus of swine (TGEV) (Decaro et al., 2009, Decaro et al., 2010b). Intermediate viruses between CCoV-I and CCoV-II have been also detected (Town and Whittaker, 2012).
CCoV-I and CCoV-II form a unique viral species, Alphacoronavirus-1 (family Coronaviridae, genus Alphacoronavirus), along with feline coronavirus types I (FCoV-I) and II (FCoV-II), TGEV and porcine respiratory coronavirus (PRCoV) (Decaro and Buonavoglia, 2011). An additional ORF, named ORF3, was found in the CCoV-I genome, whereas only its remnants were evident in the genomes of CCoV-II and TGEV, revealing an intriguing evolutionary history within the Alphacoronavirus-1 species (Lorusso et al., 2008).
While the full-length genomes of several strains of CCoV-II have been determined (Decaro et al., 2015), to date there are no complete genomic sequence data available publicly for CCoV-I. Thus, the aim of the present study was to analyze the full-length genome of a CCoV-I prototype strain that had been recovered from a dog with diarrhea in Italy.
2. Materials and methods
2.1. Virus origin
Strain 23/03 was detected during an epidemiological survey for CCoV in Italian dogs with diarrhea (Pratelli et al., 2004). The ill dog, a male German shepherd of 6 weeks of age, belonged to a kennel located in the Apulia region, southern Italy. The feces were collected by a vet directly from the rectal ampulla into a sterile container during the clinical examination of the dog. CCoV-I RNA detection in the specimen was obtained by means of genotype-specific PCR (Pratelli et al., 2004) and real-time RT-PCR (Decaro et al., 2005). Virus isolation attempts using different cell lines of canine and feline origin were unsuccessful, since CCoV-I has not been adapted to the in vitro growth (Decaro and Buonavoglia, 2008, Decaro and Buonavoglia, 2011). The original fecal sample was aliquoted and stored at −70 °C until RNA extraction.
2.2. RNA extraction
An aliquot of the original fecal specimen was clarified by centrifuging at 2500 × g for 10 min. One-hundred-forty microliters of the supernatant were then used for RNA extraction by means of QIAamp® Viral RNA Mini Kit (Qiagen S.p.A., Milan, Italy), following the manufacturer's protocol and the RNA template was stored at −70 °C until its use.
2.3. CCoV detection, quantification and characterization
The RNA extract was subjected to a previously-established TaqMan-based real-time RT-PCR assay for rapid detection and quantification of CCoV RNA (Decaro et al., 2004), with minor modifications. Briefly, a one-step method was adopted using SuperScript® III Platinum® One-Step qRT-PCR Kit (Life Technologies srl, Milan, Italy) and the following 50-μl mixture: 25 μl of master mix, 1 μl of SuperScript® III RT/Platinum Taq Mix, 300 nM of primers CCoV-For and CCoV-Rev, 200 nM of probe CCoV-Pb (Decaro et al., 2004) and 10 μl of template RNA. Duplicates of log10 dilutions of standard RNA were analyzed simultaneously in order to obtain a standard curve for absolute quantification. The thermal profile consisted of reverse transcription at 50 °C for 15 min and activation of Platinum Taq DNA polymerase at 95 °C for 2 min, followed by 45 cycles of denaturation at 95 °C for 15 s, annealing at 48 °C for 30 s and extension at 60 °C for 30 s.
CCoV genotyping was achieved by means of two distinct genotype-specific assays (Decaro et al., 2005) performed by using SuperScript® III Platinum® One-Step qRT-PCR Kit (Life Technologies srl) and the following oligonucleotide sets (final concentrations were 600 and 200 nM for primers and probes, respectively): primer pair CCoVI-F/CCoVI-R and probe CCoVI-Pb for CCoV-I and CCoVII-F/CCoVII-R and probe CCoVII-Pb (Decaro et al., 2005) for CCoV-II. The thermal protocol was as described for CCoV detection except for different annealing temperatures (53 °C and 48 °C for CCoV-I and CCoV-II, respectively).
2.4. RT-PCR amplifications
Overlapping fragments of the genome of CCoV-I strain 23/03 were obtained through RT-PCR reaction carried out using primer sets designed based on the genome sequence of other alphacoronaviruses and the kit SuperScript™ One-Step RT-PCR for Long Templates (Life Technologies srl). Additional RT-PCR assays and subsequent sequencing attempts were performed to close gaps between assembled contigs and to sequence unresolved genomic regions using primers designed on the alignment of the reference Alphacoronavirus strains. The very 5′ and 3′ ends were amplified using 5′ and 3′ RACE System for Rapid Amplification of cDNA Ends (Invitrogen), respectively, following the manufacturer's instructions. The PCR products were detected by electrophoresis through a 1.5% agarose gel and visualization under UV light after ethidium bromide staining.
2.5. Sequence analysis and phylogeny
RT-PCR products were subjected to direct sequencing at the BaseClear B.V. (Leiden, The Netherlands). The sequences were manually edited and analyzed using the Geneious platform (http://www.geneious.com) and the NCBI's (htttp://www.ncbi.nlm.nih.gov) and EMBL's (http://www.ebi.ac.uk) analysis tools. Nucleotide (nt) sequences of the different ORFs were converted into amino acid (aa) sequences and comparative sequence analysis with reference coronavirus sequences was carried out in the full-length genome and encoded structural and nonstructural proteins.
Phylogenetic and molecular evolutionary analyses were conducted using Mega4.1 Beta (Tamura et al., 2007). In order to include in the analysis CCoV-IIb, whose genome has not been completely sequenced, pylogenetic trees were elaborated on a 22,366 genomic sequence (encompassing from the 3′ end of ORF 1a to the 3′ UTR) and on the amino acid (aa) sequences of S, membrane (M), and nucleocapsid (N) proteins using both parsimony and neighbor-joining methods, supplying a statistical support with bootstrapping over 1000 replicates. The following Alphacoronavirus reference strains were used for phylogeny (GenBank accession numbers are indicated in parentheses): CCoV-IIa 1/71 (JQ404409), K378 (KC175340), S378 (KC175341), TN449 (JQ404410), NTU366/F/2008 (GQ477367), CB/05 (KP981644); CCoV-IIb 174/06 (EU856362), 341/05 (EU856361); CCoV A76 (JN856008); FCoV-I Black (EU186072); FCoV-II 79-1146 (DQ010921), 79-1683 (JN634064); TGEV Purdue (DQ811789); PRCoV ISU-1 (DQ811787). The distantly-related Betacoronavirus-1 canine respiratory coronavirus (CRCoV) K37 (JX860640) was used as outgroup.
2.6. Nucleotide sequence accession number
The full-length genome of CCoV-I strain 23/03 was deposited in GenBank under accession number KP849472.
3. Results
3.1. Detection of CCoV-I
By the real-time RT-PCR panels, the fecal sample was confirmed to contain a CCoV-I strain, whose titer was calculated as 6.73 × 106 RNA copies/μl of template. The specimen had no traces of CCoV-II RNA.
3.2. CCoV-I genomic organization
The genome of CCoV-I strain 23/03 has a size of 30,000 nt, excluding the 3′ poly(A) tail, and shows typical Alphacoronavirus-1 organization (Table 1 and Fig. 1 ). The 5′ UTR consists of 313 nt including the leader sequence (L, nt 1–94) and the conserved core 5′-CUAAAC-3′ (nt 95–100) of the transcription regulatory sequence (TRS), which controls the mRNA synthesis through interaction with the viral polymerase during the discontinuous transcription of the negative strand subgenomic RNA of the Nidovirales members (Enjuanes et al., 2000). Similar TRS signals precede each of the 8 putative mRNA encoding for the structural and nonstructural proteins (Table 1). The 3′ end of the viral genome consists of a 274-nt 3′ UTR that is followed by the poly(A) tail. Sequence analysis showed intact structural and non-structural proteins with respect to reference CCoV-II, FCoV-I and FCoV-II genomes. About two-thirds of the viral genome is occupied by the replicase gene encoding for two large polyproteins (pp), pp1a and pp1ab, the latter being synthesized through ribosomal slippage at position 12,327. The polyproteins of the replicase complex are processed by viral proteinases, resulting in several products with different size and function. Sequence comparison with other Alphacoronavirus-1 genomes led to the detection of three putative papain-like proteinase cleavage sites and 11 putative 3C-like proteinase cleavage sites, producing 16 nonstructural proteins (Table 2 ).
Table 1.
Coding potential, putative transcription regulatory sequences and encoded proteins of the CCoV-I 23/03 genome.
| Putative gene |
Putative TRS |
Putative protein |
|||
|---|---|---|---|---|---|
| Gene name | Coding sequence (nt) | Start nt position | TRS sequencea | Protein name | Protein size (aa) |
| ORF1ab | 314–12,327 12,327–20,358 |
90 | UCGAACUAAACGAAAU | Pp1ab | 6681 |
| ORF1a | 314–12,358 | Pp1a | 4014 | ||
| ORF2 | 20,359–24,804 | 20,319 | AUUACUAAACUUUGG | S | 1481 |
| ORF3 | 24,812–25,435 | 24,797 | UUCAUUAAACUCAAA | 3 | 207 |
| ORF3a | 25,447–25,683 | 25,434 | AGAACUAAACAAAUG | 3a | 78 |
| ORF3b | 25,628–25,843 | 3b | 71 | ||
| ORF3c | 25,840–26,595 | 3c | 251 | ||
| ORF4 | 26,561–26,809 | 26,514 | GGUUCUAAACGAAAU | E | 82 |
| ORF5 | 26,820–27,614 | 26,807 | UGAACUAAACAAAAU | M | 264 |
| ORF6 | 27,627–28,769 | 27,611 | AUAACUAAACUUCUA | N | 380 |
| ORF7a | 28,774–29,079 | 28,762 | CGAACUAAACGAAUG | 7a | 101 |
| ORF7b | 29,084–29,725 | 7b | 213 | ||
a The conserved TRS core is underlined.
Fig. 1.

Schematic representation of the genomes of Alphacoronavirus-1 members depicting the genetic differences among the CCoV genotypes. Genes encoding for structural and non-structural proteins are shown in gray and white, respectively. ORF sizes are not drawn to scale. The arrows indicate the transcription regulating sequences preceding each CoV gene.
Table 2.
Putative proteinase cleavage sites of CCoV-I strain 23/03 replicase polyproteins.
| Cleavage product | Polyprotein | Position in polyprotein (aa residues) | Size (aa) |
|---|---|---|---|
| nsp1 | pp1a/pp1ab | 1Met–Gly110 | 110 |
| nsp2 | pp1a/pp1ab | 111Ala–Gly879 | 769 |
| nsp3 | pp1a/pp1ab | 880Gly–Gly2385 | 1506 |
| nsp4 | pp1a/pp1ab | 2386Ser–Gln2875 | 490 |
| nsp5 | pp1a/pp1ab | 2876Ser–Gln3177 | 302 |
| nsp6 | pp1a/pp1ab | 3178Ala–Gln3471 | 294 |
| nsp7 | pp1a/pp1ab | 3472Ser–Gln3554 | 83 |
| nsp8 | pp1a/pp1ab | 3555Ser–Gln3749 | 195 |
| nsp9 | pp1a/pp1ab | 3750Asn–Gln3860 | 111 |
| nsp10 | pp1a/pp1ab | 3861Ala–Gln3995 | 135 |
| nsp11 | pp1a | 3996Ser–Asp4014 | 19 |
| nsp12 | pp1ab | 3996Ser–Gln4924 | 929 |
| nsp13 | pp1ab | 4925Ala–Gln5523 | 599 |
| nsp14 | pp1ab | 5524Ala–Gln6042 | 519 |
| nsp15 | pp1ab | 6043Ser–Gln6381 | 339 |
| nsp16 | pp1ab | 6382Ser–Pro6681 | 300 |
Four structural proteins were detected downstream of the replicase gene, namely the spike (S), small envelope (E), membrane (M) and nucleocapsid (N) proteins. The S protein has a size of 1481 aa, thus being longer than the analogous protein of other Alphacoronavirus-1 members (1451–1457 aa in CCoV-II and FCoV-II, 1457–1464 aa in FCoV-I, 1447–1449 aa in TGEV, 1225 aa in PRCoV). By using the NetNGlyc server (http://www.cbs.dtu.dk/services/NetNGlyc/), 28 N-glycosylation sites were predicted in the CCoV-I 23/03 S protein, whereas 30–33 N-glycosylated Asn residues had been detected in CCoV-II (Sanchez-Morgado et al., 2004; Decaro et al., 2007). At position 802–806, the S protein exhibits a potential cleavage site, represented by the basic aa stretch Arg-Arg-Val-Arg-Arg (RRVRR). This stretch had been also observed in the sequence of the S protein of Elmo/02 (position 801–805), but in this strain an Ala residue has replaced Val at position 803 (Pratelli et al., 2003). With few exceptions (de Haan et al., 2008), other alphacoronaviruses do not share this finding.
The E protein is 82-aa long and does not present any N-glycosylation sites, whereas three N-glycosylated residues have been detected in the 264-aa long M protein, which is in agreement with what has been observed in other FCoV/CCoV strains, with the exception of CCoV-II isolate BGF10 that shows only two glycosylated Asn residues. The N protein of strain CCoV-I 23/03 is 380-aa long product with three potential N-glycosylation sites.
Analogously to CCoV-II and FCoV-I/II, some accessory genes were detected between ORFs 2 (S-protein gene) and 4 (E-protein gene) and downstream of ORF6 (N-protein gene). The S–E intergenic region contains the canonical three ORFs 3a, 3b and 3c, encoding for products with sizes of 78, 71 and 251 aa, respectively, plus an additional accessory protein gene, ORF3, encoding for a putative 206 aa protein, which has been found to be unique to the CCoV-I genome (Lorusso et al., 2008). The 3′ end accessory genes were ORFs 7a and 7b that encodes for 101-aa and 213-aa long proteins, respectively.
3.3. Sequence analysis
Alignment of complete genome sequences of CCoV-I strain 23/03 and reference alphacoronaviruses showed the closest genetic relatedness with CCoV-IIa isolates (83.82–84.98% nt identity), followed by TGEV (82.81%) and FCoV (77.19–77.43%). No comparison was possible with CCoV-IIb since there are no full-length genomes available in the GenBank database for this virus. When the spike protein was analyzed, CCoV-I displayed a higher aa identity to FCoV-I (73.09%) than to CCoV-IIa/IIb (42.74–43.71%), FCoV-II (43.05%) and TGEV (42.83%). Among extant CCoV strains, the closest identity was observed with isolate A76, which has been proven to have a CCoV-I/II recombinant S protein (Regan et al., 2012). The E, M and N proteins of strain 23/03 were all more closely related to the analogous products of CCoV-II and porcine CoV reference strains (Table 3 ).
Table 3.
Percent (%) identities of CCoV-I 23/03 to Alphacoronavirus-1 reference strains in the complete genomic sequence (nucleotide, nt) and structural proteins (amino acid, aa).
| Alphacoronavirus-1 | Strain | Accession number | Identity (%) to CCoV-I 23/03 |
||||
|---|---|---|---|---|---|---|---|
| Full-length genome (nt) | S (aa) | E (aa) | M (aa) | N (aa) | |||
| CCoV-IIa | 1/71 | JQ404409 | 84.96 | 43.52 | 87.80 | 87.50 | 89.01 |
| K378 | KC175340 | 84.82 | 43.52 | 87.80 | 87.50 | 88.74 | |
| S378 | KC175341 | 84.79 | 43.52 | 86.59 | 87.50 | 88.74 | |
| TN449 | JQ404410 | 83.82 | 43.32 | 86.59 | 87.22 | 89.01 | |
| NTU366/F/2008 | GQ477367 | 84.25 | 42.74 | 86.59 | 84.33 | 88.74 | |
| CB/05 | KP981644 | 84.16 | 43.71 | 86.59 | 87.50 | 88.74 | |
| CCoV-IIb | 174/06 | EU856362 | NA | 43.39 | 84.15 | 87.12 | 88.48 |
| 341/05 | EU856361 | NA | 43.13 | 86.59 | 86.74 | 88.74 | |
| CCoV | A76 | JN856008 | 84.62 | 49.18 | 85.37 | 87.50 | 87.96 |
| FCoV-I | Black | EU186072 | 77.39 | 73.09 | 82.93 | 85.66 | 75.33 |
| FCoV-II | 79-1146 | DQ010921 | 77.43 | 43.05 | 82.93 | 84.91 | 75.85 |
| 79-1683 | JN634064 | 77.19 | 43.05 | 86.76 | 77.45 | 75.85 | |
| TGEV | Purdue | DQ811789 | 82.81 | 42.83 | 84.15 | 87.12 | 87.96 |
| PRCoV | ISU-1 | DQ811787 | 80.19 | 40.83 | 84.15 | 88.26 | 86.91 |
NA, sequence not available.
3.4. Phylogeny
In order to include in the analysis the nt sequences available for CCoV-IIb, phylogeny was first constructed on a 23,106-nt fragment spanning from the 3′ end of ORF1a to the very 3′ end of the viral genomes. In the neighbor-joining tree elaborated using these sequences, strain CCoV-I 23/03 clustered separately from extant CCoV/FCoV isolates leading to the formation of an outlier branch (Fig. 2A). The prototype CCoV-I strain formed a separate branch from other analyzed alphacoronaviruses also in the trees elaborated using the M (Fig. 2C) and N (Fig. 2D) proteins, whereas the S protein revealed a clustering with FCoV-I (Fig. 2D). The same tree topologies were obtained through phylogenetic analysis using the maximum-parsimony method (data not shown).
Fig. 2.
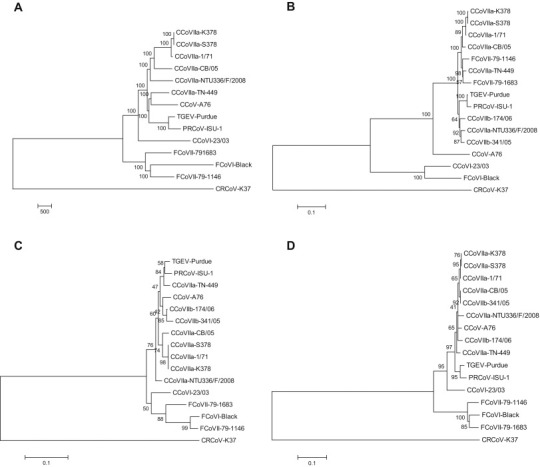
Phylogenetic analysis of CCoV-I strain 23/03 and other members of the species Alphacoronavirus-1. Neighbor-joining trees are based on the 23,106-nt fragment spanning from the 3′ end of ORF3a to the very 3′ end of the viral genome (A), and on the spike (B), membrane (C) and nucleocapsid (N) proteins. For phylogenetic tree construction, the reference strains and GenBank accession numbers are as reported in Table 3. The distantly-related Betacoronavirus-1 canine respiratory coronavirus (CRCoV) K37 (JX860640) was used as outgroup. A statistical support was provided by bootstrapping over 1000 replicates. The scale bars indicate the estimated numbers of nucleotide (A) or amino acid (B–D) substitutions.
4. Discussion
CCoV has progressively emerged as being responsible for moderate to severe enteritis in dogs, with different genotypes and subgenotypes being detected in recent years (Decaro and Buonavoglia, 2008, Decaro and Buonavoglia, 2011). CCoV-II is the oldest genotype, which has been known since 1971 (Binn et al., 1974), whereas CCoV-I was discovered only 30 years later thanks to molecular methods, since this virus has not been adapted to grow in vitro (Pratelli et al., 2003). More recently, two CCoV-II subtypes have been recognized, namely CCoV-IIa and CCoV-IIb, on the basis on the relatedness of the spike protein of the latter virus to that of TGEV (Decaro et al., 2009, Decaro et al., 2010b). In addition, a virulent CCoV-IIa biotype, strain CB/05, has been proven to cause fatal infections and long-lasting lymphopenia in naturally (Buonavoglia et al., 2006, Decaro et al., 2012, Ntafis et al., 2012, Zicola et al., 2012, Pinto et al., 2014) and experimentally (Decaro et al., 2008, Decaro et al., 2011, Marinaro et al., 2010) infected dogs.
To date, while the CCoV-II genome has been fully determined (Decaro et al., 2015), only fragments of the 3′ genomic end are available for CCoV-I. Therefore, in the present study we have carried out the complete genome sequence analysis for this canine pathogen. The results showed that the CCoV-I genome displays some distinct features with respect to CCoV-II and other alphacoronaviruses. The first finding is the presence of an additional accessory protein gene, ORF3, which was located between the S protein gene and ORF3a. This additional gene, which has been recently characterized, was found to be unique to the CCoV-I genome, whereas CCoV-II and TGEV exhibit only 5′ and 3′ end remnants (Lorusso et al., 2008). However, FCoV strains harboring different forms of ORF3 and a CCoV-I N protein gene have been detected, thus proving the circulation of FCoV-I/CCoV-I recombinant viruses (Le Poder et al., 2013). Another finding of the CCoV-I genome is that the S protein gene is closely related to that of FCoV-I, whereas the rest of the genome displays a higher relatedness to CCoV-II. In addition, a furin cleavage site leads to the potential generation of two subunits, S1 and S2. A similar basic motif is present, approximately in the same position, in most beta- and gammacoronaviruses. Cleavage of the CoV S protein has been correlated to cell-fusion activity in vitro but the potential implications in viral pathobiology have not been fully determined (Hingley et al., 1998). In addition to CCoV-I, furin cleavage motifs have been detected in FCoV-I, but even in this case the biological consequences were not completely understood (de Haan et al., 2008). Unlike CCoV-II and FCoV-II that display a similar S protein as a consequence of homologous recombination, CCoV-I does not grow in cell cultures and only few FCoV-I strains have been adapted to grow in vitro. The CoV S protein interacts with cell receptors, thus being responsible for binding of virions to the cell surface (Enjuanes et al., 2000). Thus, the different S proteins between CCoV-I/FCoV-I and CCoV-II/FCoV-II are likely to be responsible for the diverse biological behaviors in cell cultures. The cell receptor for most Alphacoronavirus-1 isolates is the cell surface glycoprotein aminopeptidase N (APN), but there is no evidence for this receptor being used by CCoV-I/FCoV-I (de Haan et al., 2008).
On the basis of the most recent findings, the evolutionary history of Alphacoronavirus-1 members has been tentatively reconstructed, suggesting that CCoV-I and FCoV-I are the ancestral viruses from which TGEV, CCoV-II and FCoV-II have originated through gene losses and recombination events (Lorusso et al., 2008). Our findings corroborate the hypothesis that CCoV-I is the ancestor for CCoV-II, since these viruses exhibit high identity in the entire genome with the exception of the S protein gene, which is markedly diverse, and ORF3, of which only remants are present in the CCoV-II genome.
In conclusion, the full-length sequencing of the CCoV-I genome may contribute to a better understanding of the Alphacoronavirus-1 evolutionary pattern and may be paradigmatic of how CoVs evolve through gene losses, acquisition and exchanges among different members.
Acknowledgements
The authors are grateful to Carlo Armenise, Arturo Gentile and Donato Narcisi for their technical assistance.
References
- Binn L.N., Lazar E.C., Keenan K.P., Huxsoll D.L., Marchwicki B.S., Strano A.J. Recovery and characterization of a coronavirus from military dogs with diarrhea. Proc. Annu. Meet. U. S. Anim. Health Assoc. 1974;78:359–366. [PubMed] [Google Scholar]
- Buonavoglia C., Decaro N., Martella V., Elia G., Campolo M., Desario C., Castagnaro M., Tempesta M. Canine coronavirus highly pathogenic for dogs. Emerg. Infect. Dis. 2006;12:492–494. doi: 10.3201/eid1203.050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli A., Desario C., Kusi I., Mari V., Lorusso E., Cirone F., Kumbe I., Colaianni M.L., Buonavoglia D., Decaro N. Detection and genetic characterization of Canine parvovirus and Canine coronavirus strains circulating in district of Tirana in Albania. J. Vet. Diagn. Invest. 2014;26:563–566. doi: 10.1177/1040638714538965. [DOI] [PubMed] [Google Scholar]
- Costa E.M., de Castro T.X., Bottino, Fde O., Garcia, Rde C. Molecular characterization of canine coronavirus strains circulating in Brazil. Vet. Microbiol. 2014;168:8–15. doi: 10.1016/j.vetmic.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Haijema B.J., Schellen P., Wichgers Schreur P., te Lintelo E., Vennema H., Rottier P.J. Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J. Virol. 2008;82:6078–6083. doi: 10.1128/JVI.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Buonavoglia C. An update on canine coronaviruses: viral evolution and pathobiology. Vet. Microbiol. 2008;132:221–234. doi: 10.1016/j.vetmic.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Buonavoglia C. Canine coronavirus: not only an enteric pathogen. Vet. Clin. North Am. Small Anim. Pract. 2011;38:799–814. doi: 10.1016/j.cvsm.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Pratelli A., Campolo M., Elia G., Martella V., Tempesta M., Buonavoglia C. Quantitation of canine coronavirus RNA in the faeces of dogs by TaqMan RT-PCR. J. Virol. Methods. 2004;119:145–150. doi: 10.1016/j.jviromet.2004.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Ricci D., Elia G., Desario C., Campolo M., Cavaliere N., Di Trani L., Tempesta M., Buonavoglia C. Genotype-specific fluorogenic RT-PCR assays for the detection and quantitation of canine coronavirus type I and type II RNA in faecal samples of dogs. J. Virol. Methods. 2005;130:72–78. doi: 10.1016/j.jviromet.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Elia G., Campolo M., Desario C., Cirone F., Tempesta M., Buonavoglia C. Molecular characterisation of the virulent canine coronavirus CB/05 strain. Virus Res. 2007;125:54–60. doi: 10.1016/j.virusres.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Campolo M., Lorusso A., Desario C., Mari V., Colaianni M.L., Elia G., Martella V., Buonavoglia C. Experimental infection of dogs with a novel strain of canine coronavirus causing systemic disease and lymphopenia. Vet. Microbiol. 2008;128:253–560. doi: 10.1016/j.vetmic.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Campolo M., Lorusso A., Camero M., Elia G., Martella V., Cordioli P., Enjuanes L., Buonavoglia C. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of swine are circulating in dogs. J. Virol. 2009;83:1532–1537. doi: 10.1128/JVI.01937-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Elia G., Martella V., Campolo M., Mari V., Desario C., Lucente M.S., Lorusso E., Kanellos T., Gibbons R.H., Buonavoglia C. Immunity after natural exposure to enteric canine coronavirus does not provide complete protection against infection with the new pantropic CB/05 strain. Vaccine. 2010;28:724–729. doi: 10.1016/j.vaccine.2009.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Elia G., Addie D.D., Camero M., Lucente M.S., Martella V., Buonavoglia C. Recombinant canine coronaviruses in dogs. Europe. Emerg. Infect. Dis. 2010;16:41–47. doi: 10.3201/eid1601.090726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Desario C., Billi M., Mari V., Elia G., Cavalli A., Martella V., Buonavoglia C. Western European epidemiological survey for parvovirus and coronavirus infections in dogs. Vet. J. 2011;187:195–199. doi: 10.1016/j.tvjl.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., von Reitzenstein M., Lucente M.S., Cirone F., Elia G., Martella V., King V.L., Di Bello A., Varello K., Zhang S., Caramelli M., Buonavoglia C. A pantropic canine coronavirus genetically related to the prototype isolate CB/05. Vet. Microbiol. 2012;159:239–244. doi: 10.1016/j.vetmic.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Cordonnier N., Demeter Z., Egberink H., Elia G., Grellet A., Le Poder S., Mari V., Martella V., Ntafis V., von Reitzenstein M., Rottier P.J., Rusvai M., Shields S., Xylouri E., Xu Z., Buonavoglia C. European surveillance for pantropic canine coronavirus. J. Clin. Microbiol. 2013;51:83–88. doi: 10.1128/JCM.02466-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Dowgier G., Elia G., Lanave G., Colaianni M.L., Buonavoglia C. Full-genome sequence of pantropic canine coronavirus. Genome Announc. 2015;3 doi: 10.1128/genomeA.00401-15. pii:e00401-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Brian D., Cavanagh D., Holmes K., Lai M.M.C., Laude H., Masters P., Rottier P., Siddell S., Spaan W.J.M., Taguchi F., Talbot P. Family Coronaviridae. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., Maniloff J., Mayo M.A., McGeoch D.J., Pringle C.R., Wickner R.B., editors. Virus Taxonomy, Classification and Nomenclature of Viruses. Academic Press; New York: 2000. pp. 835–849. [Google Scholar]
- Hingley S.T., Leparc-Goffart I., Weiss R.S. The spike protein of murine coronavirus mouse hepatitis virus strain A59 is not cleaved in primary glial cells and primary hepatocytes. J. Virol. 1998;72:1606–1609. doi: 10.1128/jvi.72.2.1606-1609.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poder S., Pham-Hung d’Alexandry d’Orangiani A.L., Duarte L., Fournier A., Horhogea C., Pinhas C., Vabret A., Eloit M. Infection of cats with atypical feline coronaviruses harbouring a truncated form of the canine type I non-structural ORF3 gene. Infect. Genet. Evol. 2013;20:488–494. doi: 10.1016/j.meegid.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra B.N., Whittaker G.R., Dubovi E.J., Duhamel G.E. Genotypic characterization of canine coronaviruses associated with fatal canine neonatal enteritis in the United States. J. Clin. Microbiol. 2014;52:4230–4238. doi: 10.1128/JCM.02158-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A., Decaro N., Schellen P., Rottier P.J., Buonavoglia C., Haijema B.J., de Groot R.J. Gain, preservation, and loss of a group 1a coronavirus accessory glycoprotein. J. Virol. 2008;82:10312–10317. doi: 10.1128/JVI.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinaro M., Mari V., Bellacicco A.L., Tarsitano E., Elia G., Losurdo M., Rezza G., Buonavoglia C., Decaro N. Prolonged depletion of circulating CD4+ T lymphocytes and acute monocytosis after pantropic canine coronavirus infection in dogs. Virus Res. 2010;152:73–78. doi: 10.1016/j.virusres.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElligott S., Collins P.J., Sleator R.D., Martella V., Decaro N., Buonavoglia C., O'Shea H. Detection and genetic characterization of canine parvoviruses and coronaviruses in southern Ireland. Arch. Virol. 2011;156:495–503. doi: 10.1007/s00705-010-0861-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntafis V., Xylouri E., Mari V., Papanastassopoulou M., Papaioannou N., Thomas A., Buonavoglia C., Decaro N. Molecular characterization of a canine coronavirus NA/09 strain detected in a dog's organs. Arch. Virol. 2012;157:171–175. doi: 10.1007/s00705-011-1141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntafis V., Mari V., Decaro N., Papanastassopoulou M., Pardali D., Rallis T.S., Kanellos T., Buonavoglia C., Xylouri E. Canine coronavirus, Greece. Molecular analysis and genetic diversity characterization. Infect. Genet. Evol. 2013;16:129–136. doi: 10.1016/j.meegid.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L.D., Barros I.N., Budaszewski R.F., Weber M.N., Mata H., Antunes J.R., Boabaid F.M., Wouters A.T., Driemeier D., Brandão P.E., Canal C.W. Characterization of pantropic canine coronavirus from Brazil. Vet. J. 2014;202:659–662. doi: 10.1016/j.tvjl.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Decaro N., Tinelli A., Camero M., Cirone F., Elia G., Cavalli A., Corrente M., Greco G., Buonavoglia D., Gentile M., Tempesta M., Buonavoglia C. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J. Virol. Methods. 2003;110:9–17. doi: 10.1016/S0166-0934(03)00081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Decaro N., Tinelli A., Martella V., Elia G., Tempesta M., Cirone F., Buonavoglia C. Two genotypes of canine coronavirus simultaneously detected in fecal samples of dogs with diarrhea. J. Clin. Microbiol. 2004;42:1797–1799. doi: 10.1128/JCM.42.4.1797-1799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan A.D., Millet J.K., Tse L.P., Chillag Z., Rinaldi V.D., Licitra B.N., Dubovi E.J., Town C.D., Whittaker G.R. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virology. 2012;430:90–99. doi: 10.1016/j.virol.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Morgado J.M., Poynter S., Morris T.H. Molecular characterization of a virulent canine coronavirus BGF strain. Virus Res. 2004;104:27–31. doi: 10.1016/j.virusres.2004.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Town C.D., Whittaker G.R. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virology. 2012;430:90–99. doi: 10.1016/j.virol.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicola A., Jolly S., Mathijs E., Ziant D., Decaro N., Mari V., Thiry E. Fatal outbreaks in dogs associated with pantropic canine coronavirus in France and Belgium. J. Small Anim. Pract. 2012;53:297–300. doi: 10.1111/j.1748-5827.2011.01178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


