Highlights
-
•
Inhibition of VCP by chemical inhibitors decreased WNV infection in a dose-dependent manner.
-
•
Knockdown of endogenous VCP level using siRNA suppressed WNV infection.
-
•
Depletion of VCP levels suppressed WNV infection at the early stages of WNV replication cycle.
-
•
Depletion of VCP levels lowered nascent WNV genomic RNA.
-
•
VCP participates in early stages and viral genomic RNA replication.
Keywords: VCP, WNV replication, Early steps, Genome replication
Abstract
Valosin-containing protein (VCP) is classified as a member of the type II AAA+ ATPase protein family. VCP functions in several cellular processes, including protein degradation, membrane fusion, vesicular trafficking and disassembly of stress granules. Moreover, VCP is considered to play a role in the replication of several viruses, albeit through different mechanisms. In the present study, we have investigated the role of VCP in West Nile virus (WNV) infection. Endogenous VCP expression was inhibited using either VCP inhibitors or by siRNA knockdown. It could be shown that the inhibition of endogenous VCP expression significantly inhibited WNV infection. The entry assay revealed that silencing of endogenous VCP caused a significant reduction in the expression levels of WNV-RNA compared to control siRNA-treated cells. This indicates that VCP may play a role in early steps either the binding or entry steps of the WNV life cycle. Using WNV virus like particles and WNV-DNA-based replicon, it could be demonstrated that perturbation of VCP expression decreased levels of newly synthesized WNV genomic RNA. These findings suggest that VCP is involved in early steps and during genome replication of the WNV life cycle.
1. Introduction
West Nile virus (WNV) belongs to genus the Flavivirus, family Flaviviridae and has an approximately 11 kb positive sense, single-stranded genomic RNA [(+)ssRNA]. The WNV genome encodes three structural proteins (C, prM and E) and seven non-structural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b and NS5) (Brinton, 2014, Fields et al., 2013). Many species of mammals and birds can be infected by WNV (Dauphin et al., 2004, Egberink et al., 2015, Fields et al., 2013, Gamino et al., 2016, Kramer and Bernard, 2001, Lichtensteiger et al., 2003, Read et al., 2005) and infection causes West Nile fever and encephalitis in human and horses (Dauphin et al., 2004, Samuel and Diamond, 2006). WNV was firstly isolated from a Ugandan woman in 1937 (Smithburn et al., 1940, Fields et al., 2013), but has now spread widely to many countries (Fields et al., 2013, Paz, 2015, Troupin and Colpitts, 2016). In the United States, approximately 44,000 cases of WNV infection were reported between 1999 and 2015 (CDC, 2016).
WNV attaches to host cells through the interaction of the viral E protein and cellular receptors on the surface of host cells (Fields et al., 2013). Several attachment receptors of WNV have been reported and include the laminin receptor (Bogachek et al., 2010, Perera-Lecoin et al., 2014, Zaitsev et al., 2014, Zidane et al., 2013), TIM (T cell/transmembrane, immunoglobulin and mucin) and TAM (Tyro3, Axl and Mer) families (Carnec et al., 2016, Morizono and Chen, 2014, Perera-Lecoin et al., 2014), DC-SIGN/L-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin) (Davis et al., 2006, Denizot et al., 2012, Martina et al., 2008, Shimojima et al., 2014) and integrin αvβ3 (Bogachek et al., 2010, Fields et al., 2013, Perera-Lecoin et al., 2014, Smit et al., 2011, Zaitsev et al., 2014). Following attachment, the virus is then internalized into the cytoplasm via clathrin-mediated endocytosis (Brinton, 2014, Chu and Ng, 2004, Fields et al., 2013). WNV particles are delivered to early or intermediate endosomes, which mature into late endosomes, following a conformational change of the viral E protein dimer triggered by the acidic environment in late endosomes. Membrane fusion between viral particles and endosomal membranes then occurs and, thereafter, WNV genomic RNA is released into the cytosol, with subsequent translation and replication (Chu et al., 2006, Chu and Ng, 2004, Fields et al., 2013, Heinz and Allison, 2000, Smit et al., 2011). Host cell membrane rearrangements are induced during replication of flaviviruses, including WNV, to coordinate the processes of genomic RNA replication and virus assembly. Viral genomic RNA replication is thought to occur in endoplasmic reticulum (ER) membrane-derived vesicles (in structures termed vesicle packets) (Gillespie et al., 2010, Kaufusi et al., 2014, Welsch et al., 2009). Encapsidation of nascent viral genomic RNA is achieved by the capsid protein and budding into the ER yielding a viral envelope coated with prM and E proteins (Brinton, 2014, Fields et al., 2013, Suthar et al., 2013, Welsch et al., 2009). The immature virions are transported via the host secretory pathway and virion maturation then occurs in the acidic compartments of the Golgi by cleavage of the prM protein by a furin-like protease (Plevka et al., 2014, Roby et al., 2015, Yu et al., 2008). Mature virions are then released from the infected cells through exocytosis (Fields et al., 2013, Samuel and Diamond, 2006). It has been reported that several cellular pathways and host factors are involved in WNV infection (Ambrose and Mackenzie, 2011, Brinton, 2014, Chahar et al., 2013, Chu and Ng, 2004, Courtney et al., 2012, Fernandez-Garcia et al., 2011, Fields et al., 2013, Gilfoy et al., 2009, Kobayashi et al., 2016a, Krishnan et al., 2008, Ma et al., 2015); however, the role of valosin-containing protein (VCP) has remained controversial.
VCP, also known as CDC48 in Saccharomyces cerevisiae, is well conserved among eukaryotes with orthologues in archaea, protozoa, insects and plants (Meyer et al., 2012, Wolf and Stolz, 2012), and is classified as a member of the type II AAA+ ATPase (adenosine triphosphatase-associated with diverse cellular activities) family (Koller and Brownstein, 1987, Pye et al., 2006, Stolz et al., 2011, Wolf and Stolz, 2012, Xia et al., 2016). VCP is a homohexameric complex composed of six protomers organized as two concentric-rings with a central pore. VCP conformational changes, driven by adenosine triphosphate hydrolysis, acts as a chaperone in protein homeostasis systems, which include ER-associated degradation (ERAD) (Wolf and Stolz, 2012, Xia et al., 2016, Zhong and Pittman, 2006) and mitochondria-associated degradation and autophagy (Bug and Meyer, 2012, Dargemont and Ossareh-Nazari, 2012, Xia et al., 2016, Yamanaka et al., 2012) to prevent accumulation of misfolded-proteins and turnover of certain proteins. Recently, a role of VCP in the disassembly of stress granules (SGs) has also been reported (Buchan et al., 2013, Seguin et al., 2014). Generally, after removal of stress stimuli, SGs are disassembled by VCP and mRNA in the SGs could be restored allowing mRNA translation to proceed. Otherwise, depletion of VCP causes persistence of SGs leading to blockage of mRNA restoration and an arrest of mRNA translation (Buchan et al., 2013). It has also been reported that VCP is involved in chromatin-associated degradation and several nuclear substrates of VCP have been described (Maric et al., 2014, Verma et al., 2011, Wilcox and Laney, 2009). Furthermore, VCP also participates in membrane fusion and vesicular trafficking events (Bug and Meyer, 2012, Meyer et al., 2012, Ramanathan and Ye, 2012, Ritz et al., 2011, Xia et al., 2016). VCP binds to endocytic components, and silencing of VCP leads to a failure of maturation and enlargement of the early endosome (Ramanathan and Ye, 2012).
Interestingly, VCP has also been implicated in the life cycle of several (+)ssRNA viruses. It has been previously reported that VCP facilitates the replication of poliovirus (PV) (Arita et al., 2012). Depletion of VCP caused a reduction of PV infection, whereas, a mutant PV, which has a secretion inhibition-negative phenotype, increases the affinity of binding to VCP and resists VCP-knockdown compared to wild-type PV, suggesting that VCP may play a role in PV replication through cellular secretion pathways (Arita et al., 2012). In addition, other roles for VCP have been described in other picornaviruses (Arita et al., 2012). Although VCP knockdown strongly inhibits PV infection, inhibition of VCP does not affect the replication of Coxsackievirus B3 (Arita et al., 2012), which is also a member of the same genus Enterovirus. In contrast, replication of Aichivirus A, genus Kobuvirus, another member of family Picornaviridae, is enhanced when VCP is depleted (Arita et al., 2012).
A relationship between VCP and Sindbis virus (SINV) replication has also been reported (Panda et al., 2013). VCP is involved in trafficking of the entry receptor of SINV, which is the natural resistance-associated macrophage protein 2 (NRAMP2). Deficiency of VCP suppresses SINV replication through alteration of trafficking routes of NRAMP2 leading to degradation of NRAMP2 by lysosomes.
Studies of infectious bronchitis virus (IBV), family Coronaviridae, have suggested that VCP is engaged in the internalization steps of IBV (Wong et al., 2015). Depletion of VCP using siRNA knockdown, resulted in accumulation of IBV particles in early endosomes as maturation of the endosome and acidification was disrupted. Failure of the acidification of virus-containing endosomes inhibited fusion between the virus envelope and endosomal membrane and prevented IBV exit from the endosomes to the cytosol (Wong et al., 2015)
In the present study, we investigated whether VCP is involved in WNV infection. Specifically, we employed VCP inhibitors and siRNA knockdown to elucidate a potential role of VCP in WNV replication.
2. Methods
2.1. Cell and viruses
Human cervical adenocarcinoma cells, HeLa, were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mM l-glutamine. Human embryonic kidney cells, HEK293T, were grown in high glucose DMEM supplemented with 110 mg/L sodium pyruvate, 2 mM l-glutamine and 5% FBS. African green monkey kidney cells, Vero, were grown with Minimum Essential Media (MEM) supplemented with 5% FBS and 2 mM l-glutamine. Human neuroblastoma cells, SK-N-SH, were grown with Minimum Essential Media (MEM) supplemented with 10% FBS and 2 mM l-glutamine. Cells were grown at 37 °C with 5% supplemented CO2. The mosquito cell line, Aedes albopictus clone C6/36, were grown in MEM supplemented with 10% FBS, 1% non-essential amino acid and 2 mM l-glutamine at 28 °C. WNV New York strain (NY99 6-LP) was propagated in C6/36 at 28 °C. WNV-NY99 6-LP was kindly provided by Dr. Takashima (Laboratory of Public Health, Graduate school of Veterinary Medicine, Hokkaido University, Sapporo, Japan) (Hasebe et al., 2010, Shirato et al., 2004a, Shirato et al., 2004b). Viral titer was measured by plaque assay and stock of viruses were stored at −80 °C until use. All experiments with WNV were performed in the Biosafety level-3 facility at the Research Center for Zoonosis Control, Hokkaido University in accordance with institutional guidelines. Pseudotyped vesicular stomatitis virus (VSV) was provided by Dr. Takada (Research Center for Zoonosis Control, Hokkaido University) (Takada et al., 2007).
2.2. MTT assay
HeLa cells were treated with 5, 10 and 20 μM of Eeyarestatin I (EerI) (Sigma Aldrich, St. Louis, MO) (Wang et al., 2008, Wang et al., 2010) or 12.5, 25 and 50 μM of 3,4-Methylenedioxy-β-nitrostyrene (MDBN) (Abcam, Cambridge, UK) (Chou and Deshaies, 2011) and incubated at 37 °C for 24 h. Thereafter, the treated cells were examined by MTT assay following addition of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and incubated at 37 °C for 1 h, then addition of solubilization solution [10% Triton-X 100 in acidic isopropanol (0.1N HCl)] and incubation at room temperature with shaking for 30 min. Absorbance values were measured at 570 nm and 630 nm using the Model 680 microplate reader (Bio-rad, Hercules, CA).
2.3. Plaque assay
Virus suspensions were diluted in a series of 10-fold dilutions and inoculated onto monolayers of Vero cells. The WNV-inoculated Vero cells were grown with MEM containing 1.25% methyl cellulose, 5% FBS and 2 mM l-glutamine at 37 °C for 4 days. Fixation was done after 4 days using 10% formalin for 10 min at room temperature. The fixed cells were stained with 1% crystal violet in 70% ethanol for 30 min. The number of plaques was counted and the virus titer was determined in plaque forming unit per milliliter (PFU/ml).
2.4. Immunofluorescence assay (IFA)
The WNV-infected cells grown on coverslips were fixed at various times after infection. The infected cells were fixed in 4% paraformaldehyde for 10 min and permeabilized using 0.1% Triton X-100 for 5 min at room temperature. Blocking was performed with 1% bovine serum albumin (BSA) for 30 min before incubation with primary antibody. The cells were incubated with primary antibody (rabbit anti-JEV serum; 1:1500) (Kimura et al., 1994, Kobayashi et al., 2012) that have cross-reactivity with the WNV antigens at 4 °C overnight, followed by incubation with Alexa Fluor 488-conjugated secondary antibody against rabbit IgG (1:2000; Life technologies, Rockville, MD) for 1 h at room temperature. The cells were washed three times with phosphate buffered saline (PBS) before fluorescence microscopy examination. The cells were visualized using an inverted fluorescence microscope (IX70, Olympus, Tokyo, Japan) and images were processed using DP manager software (Olympus).
2.5. Immunoblotting analysis
Cell samples were harvested at the indicated time points using TNE lysis buffer [1% Triton X-100, 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 10% glycerol] and centrifuged at 17,800 × g at 4 °C for 20 min. Only supernatants were collected and mixed with SDS-PAGE sample buffer [0.1 M Tris-HCl (pH 6.8), 3.3% SDS, 11% glycerol]. The samples were separated by 8% polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride filter (Merck Millipore, Billerica, MA). The filters were blocked with 5% skimmed milk for 30 min. The filters were incubated with each primary antibody, including mouse anti-VCP antibody (1:2000; Abcam), mouse anti-envelope protein of West Nile/Kunjin virus (Merck 1:1000; Millipore), rabbit anti-NS3 serum (1:1000), which was prepared from a rabbit immunized by twice intravenous inoculations of synthetic peptides of NS3 (CEREKVYTMDGEYRLRGEER), and mouse anti-actin (1:1000; Merck Millipore). Thereafter, the filters were incubated with secondary antibody, goat anti-mouse IgG antibody conjugated with horseradish peroxidase at 1:10,000 dilution (Biosource International, Camarillo, CA) and washed with TBS [50 mM Tris-HCl (pH 7.5), 150 mM NaCl,] containing 0.05% Tween 20 (TBS-T) three times. Chemiluminescence was detected by Immobilon Western HRP Substrate (Merck Millipore) and visualized with VersaDoc 5000MP (Bio-Rad), and images were analyzed using Quantity One software (Bio-Rad).
2.6. WNV inoculation in the presence of VCP inhibitors
Based on the results of MTT assays, we determined the optimal concentration of EerI (2.5 and 5 μM) and MDBN (6.25 and 12.5 μM) without cytotoxicity in HeLa cells. Multiplicity of infection (MOI) of 1 of WNV NY99 6-LP strain was inoculated into HeLa cells. After 1 h of incubation at 37 °C with rocking, the inocula were removed. Suspensions of either EerI or MDBN diluted in normal cultured medium were added to the WNV-inoculated cells and incubated at 37 °C for 24 h. Thereafter, the inoculated-cells and the supernatants were prepared for IFA, plaque assay and immunoblotting analysis to measure the number of WNV-infected cells, production of infectious WNV and expression of WNV proteins, respectively.
2.7. VCP knockdown
The endogenous VCP was inhibited using siRNA. HeLa cells (2 × 104 cells in 250 μl medium per well) in 48-well plates were transfected with 5 nM of each siRNA targeting VCP, no. (1), (2) and (3), which have the sequences 5′-GAAUAGAGUUGUUCGGAAUTT-3′, 5′-GAACCGUCCCAAUCGGUUATT-3′, and 5′-GGCUCGUGGAGGUAACAUUTT-3′ (Thermo Fisher Scientific, Waltham, MA), respectively, using lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The transfected cells were incubated at 37 °C for 48 h. The expression level of VCP was evaluated using immunoblotting.
2.8. WNV inoculation in siRNA-treated cells
At 48 h post transfection with either siRNA against VCP or control siRNA (Catalogue No: 439084, Thermo Fisher Scientific), HeLa cells were inoculated with WNV NY99 6-LP strain (MOI = 1) and incubated at 37 °C for 1 h with rotation. Thereafter, supernatants of the cells were removed, and normal HeLa culture medium was added to the cells and incubated at 37 °C for 12, 24 and 48 h. The inoculated cells and the supernatants were prepared for IFA, plaque assay and immunoblotting analysis, respectively. To investigate the role of VCP in the early stages of the WNV replicative cycle, the siRNA-transfected cells were inoculated with WNV as described above. After inoculation of WNV, cells were incubated on ice for 1 h, and then washed with PBS five times. The cells were then placed at 37 °C and incubated for 1 h. Thereafter, the inoculated-cells were harvested using trypsin. The detached cells from cell culture plates were centrifuged at 1500 × g for 3 min. After removal of supernatants, total RNA was extracted from cell pellets using Trizol (Thermo Fisher Scientific). Extracted total RNAs were analyzed for WNV genome using real-time reverse transcription PCR (qRT-PCR) analysis.
2.9. Pseudotyped VSV inoculation in siRNA-treated cells
It has been previously reported that VCP knockdown did not affect VSV infection (Panda et al., 2013). Therefore, control experiments using pseudotyped VSV were performed. The pseudotyped VSV encoding GFP was kindly provided by Dr. Takada (Hokkaido University) (Takada et al., 2007). Approximately 30% infectivity of pseudotyped VSV was inoculated into HeLa cells transfected with either siRNA against VCP or control siRNA. The cells were incubated at 37 °C with rotation for 1 h. Thereafter, supernatants of the cells were removed, normal growth media was added to the cells and incubated at 37 °C for 8 h. The percentage positivity following pseudotyped VSV infection was measured by counting the number of GFP-positive cells using an inverted fluorescence microscope (IX70, Olympus, Tokyo, Japan).
2.10. Production of WNV virus-like particles (WNV-VLPs)
WNV-VLPs with reporter DsRed protein were produced following transfection. Three plasmid vectors carrying WNV sequences: pCMV-WNrep-DsRed, pCMV-SVP and pCSXN-C were transfected into HEK293T cells using lipofectamine 2000 (Thermo Fisher Scientific). These plasmids were constructed as follows. A plasmid encoding WNV replicon cDNA, pCMV-WNrep-DsRed encodes the WNV non-structural (NS1-NS5) proteins. Almost all of the sequences encoding structural proteins, including C, prM and E, were deleted and replaced by the gene encoding the DsRed protein. The 3′-terminus of the WNV genome was accomplished by containing sequences enabling ribozyme-mediated post-transcriptional cleavage of the RNA (Kobayashi et al., 2016b). The fragment of prM-E was amplified by PCR from pCAGGS-C-prM-E, which was a gift from Dr. Takashima (Hokkaido University) (Takahashi et al., 2009) as a template, and subcloned into the pCMV vector, and the plasmid was named pCMV-SVP. For the pCSXN-C, the C fragment with restriction sequences of Xho I and Not I (Takara Bio, Kyoto, Japan) were amplified by PCR and inserted into pCSXN-flag which was generated from pCMV-myc (Clontech Laboratories, Mountain View, CA) as previously described (Kobayashi et al., 2013), using Xho I and Not I restriction sites. The plasmid was named as pCXSN-C. The transfected cells were incubated at 37 °C for 72 h. The supernatants from transfected cells were collected and filtered through a 0.45 μm filter (Sigma Aldrich). The WNV-VLPs were concentrated by ultracentifugation at 4 °C, 68,000 × g for 2 h. The supernatant was discarded and only the pellet was collected after ultracentrifugation. The pellet was resuspended with normal HeLa culture medium and the titers of WNV-VLPs were measured by hemagglutination assay as previously described (Makino et al., 2014). The titer of VLPs was calculated as hemagglutination units (HAU)/50 μl based on the highest dilution of VLP suspension causing agglutination of chicken red blood cells.
2.11. Inoculation of WNV-VLPs in VCP knockdown HeLa cells
WNV-VLPs (16 HAU/50 μl) were inoculated into siRNA-treated HeLa cells 48 h post transfection. After 1 h incubation at 37 °C, the inocula were discarded and normal HeLa culture medium was added and incubated for 72 h. Comparison of the quantity of WNV-RNA between VCP knockdown and control was determined using qRT-PCR.
2.12. WNV-VLP production and transfection of pCMV-WNrep-DsRed to siRNA-treated HeLa cells
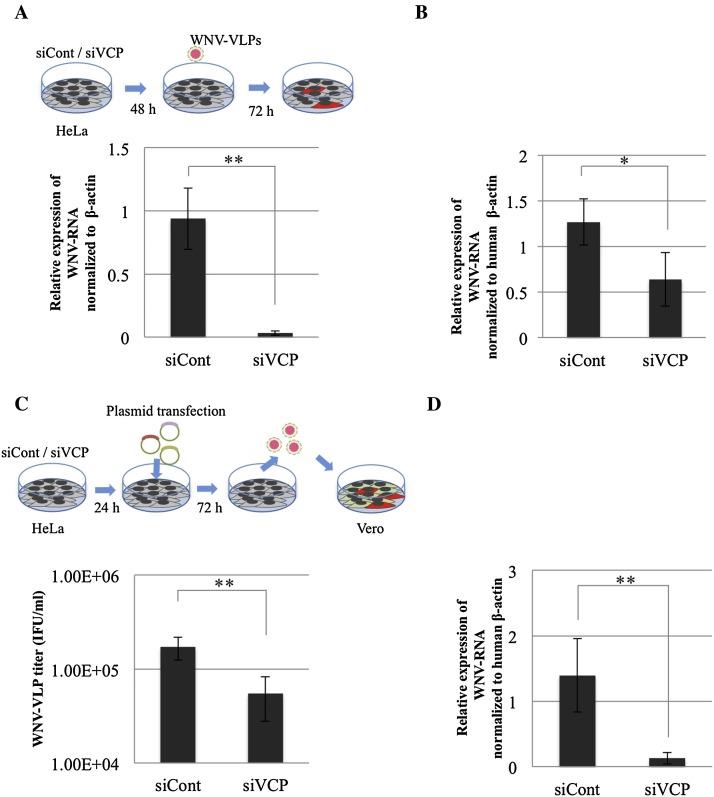
Plasmid transfection to generate WNV-VLPs has been reported to investigate the role of VCP in the late steps of viral life cycle, from genome replication to virus release (Kobayashi et al., 2016a). The results obtained employing the plasmid-encoded VLPs would not be attributable to early steps of WNV infection, including attachment and entry. WNV-VLPs were used to investigate the role of VCP in distinct steps (early and genome replication steps) of WNV infection cycle. At 24 h after siRNA transfection, the plasmid set (pCMV-WNrep-DsRed, pCMV-SVP and pCSXN-C) was transfected into siRNA-treated HeLa cells using FuGENE HD (Promega, Madison, WI). The plasmid transfected-cells were incubated at 37 °C for 72 h. Thereafter, the supernatants from transfected cells were collected and inoculated onto Vero cells monolayers in 10-fold serial dilutions. WNV-VLP titer was calculated as infectious units (IFU)/ml based on the total number of DsRed-positive cells (Fig. 3 C).
Fig. 3.
The role of VCP in distinct steps of WNV life cycle was investigated. (A) WNV-VLP infection after the indicated siRNA treatment, HeLa cells were treated with siRNA against VCP, siVCP (1) or control siRNA (siCont). The indicated siRNA treated-cells were inoculated with WNV-VLPs (16 HAU) at 48 h post transfection and incubated for 72 h. Relative quantification of WNV-RNA normalized to human β-actin from (A) examined by qRT-PCR. Mean ± SD from three independent experiments is shown; ** p < 0.01 (one-way ANOVA). (B) Relative quantification of WNV-RNA expression levels normalized to human β-actin of siRNA-treated cells inoculated with WNV at the early time point of infection. siRNA-treated HeLa cells were inoculated with WNV after 48 h post siRNA transfection. The inoculated cells were incubated on ice for 1 h, followed by washing 5 times with PBS and then transferred to 37 °C. After 1 h incubation, the cells were harvested by trypsin and prepared for qRT-PCR. Mean ± SD from two independent experiments in triplicate is shown; * p < 0.05 (one-way ANOVA). (C) HeLa cells were treated with siRNA against VCP, siVCP (1) or control siRNA (siCont). After 24 h post transfection, the cells were transfected with plasmid set for WNV-VLP production and incubated for 72 h. The culture supernatants were harvested and inoculated on Vero cell monolayers in 10-fold serial dilutions. The viral titers of the harvested supernatants were determined as IFU/ml. Mean ± SD from three independent experiments is shown; ** p < 0.01 (one-way ANOVA). (D) HeLa cells were treated with siRNA, either siVCP (1) or siCont, and then transfected with plasmid containing WNV DNA replicon, pCMV-WNrep-DsRed at 24 h post siRNA treatment and incubated. The transfected cells were harvested at 72 h post transfection of pCMV-WNrep-DsRed. Relative quantification of WNV-RNA normalized to expression of human β-actin was examined by qRT-PCR. Mean ± SD from three independent experiments is shown; ** p < 0.01 (one-way ANOVA).
To investigate the role of VCP in WNV genomic RNA replication, only pCMV-WNrep-DsRed (Kobayashi et al., 2016b) was transfected into the siRNA-treated cells. The procedure and time of transfection are similar to plasmid transfection for VLP production. After 72 h incubation, the total RNAs were extracted and prepared for qRT-PCR.
2.13. Real-time reverse transcription-PCR (qRT-PCR)
Total RNA was isolated by Trizol and chloroform according to the manufacturer’s protocol. The RNA samples were treated with DNase I (Thermo Fisher Scientific) to remove genomic DNA. qRT-PCR was performed with a Brilliant III Ultra-Fast qRT-PCR master mix (Agilent Technologies, Santa Clara, CA) following the manufacturer’s protocol. The oligonucleotide primers and fluorescent probe targeting the 3′UTR of WNV, 5′-AAGTTGAGTAGACGGTGCTG-3′ and 5′-AGACGGTTCTGAGGGCTTAC-3′, WNV probe, FAM-5′-GCTCAACCCCAGGAGGACTGG-3′-BHQ, were used for detection of WNV-RNA. A TaqMan Gene expression assays kit corresponding to human β-actin (Thermo Fisher Scientific) was used as an endogenous control. The expression level of viral RNA was normalized to the expression of human β-actin.
2.14. Statistical analysis
The statistical significance was calculated using one-way ANOVA.
3. Results
3.1. WNV infection is inhibited in the presence of VCP inhibitors
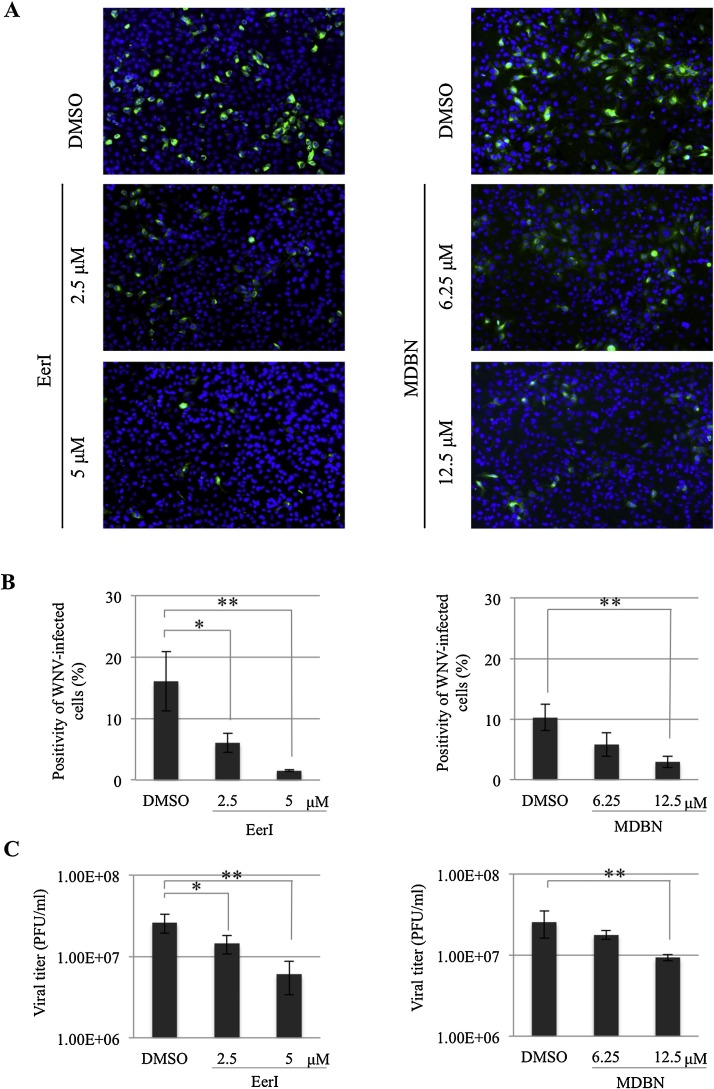
To determine if VCP is involved in WNV infection, the effect of VCP inhibitors, both EerI (Wang et al., 2008, Wang et al., 2010) and MDBN (Chou and Deshaies, 2011) at concentrations without cytotoxicity were assayed in WNV infection. The cytotoxicity of either EerI or MDBN treatment in HeLa cells was examined using a MTT assay (Supplementary Fig. 1). Each VCP inhibitor was added to HeLa cells at 1 h.p.i. with WNV. WNV-inoculated cells and cultured supernatants were harvested at 24 h.p.i. The number of WNV-infected cells was examined by IFA, and this revealed that the number of WNV-infected cells was significantly decreased in a dose-dependent manner in the presence of either EerI or MDBN (Fig. 1A and B). Viral titers of supernatants from WNV-inoculated cells were also measured by plaque assay. Consistently, this demonstrated that viral titers of supernatants from WNV-inoculated cells were significantly decreased in a dose-dependent manner in the presence of either EerI or MDBN (Fig. 1C). We also confirmed the inhibitory effects of EerI in WNV infection in a different cell line, human neuroblastoma SK-N-SH cells. Inhibition of VCP by EerI both decreased the percentage of WNV-infected cells and viral titer in SK-N-SH cells (Supplementary Fig. 2). These findings suggest that VCP may play a role in WNV infection.
Fig. 1.
WNV infection is inhibited in the presence of VCP inhibitors. (A) WNV infection in the presence of either EerI (left panels) or MDBN (right panels). HeLa cells were inoculated with WNV (MOI = 1) and then treated with EerI or MDBN at 1 h.p.i. Cells were harvested at 24 h.p.i. and stained with anti-JEV antibody (Kimura et al., 1994, Kobayashi et al., 2012) that has cross reactivity with WNV antigen (green). Cell nuclei were counterstained with DAPI (blue). (B) Positivity of WNV-infected cells from (A). Mean ± SD from triplicate experiments is shown; * p < 0.05, ** p < 0.01 (one-way ANOVA). (C) The culture supernatants from (A) were collected at 24 h.p.i. and the viral titers of the harvested supernatants were examined by plaque assay. Mean ± SD from three independent experiments is shown; * p < 0.05, ** p < 0.01 (one-way ANOVA).
3.2. WNV infection is inhibited by knockdown of VCP
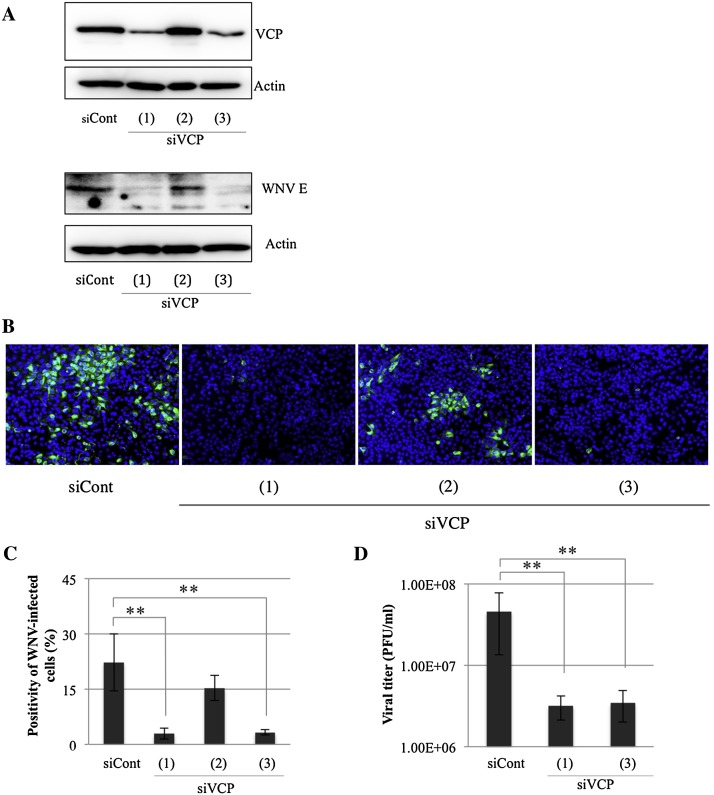
To confirm that the inhibition of WNV infection was caused by perturbation of VCP activity, small interfering RNAs (siRNAs) were employed to deplete endogenous VCP. HeLa cells were transfected with either of three siRNAs targeting three different regions of the VCP gene [siVCP (1), (2) and (3)] or a control siRNA (siCont) and then inoculated with WNV 48 h post transfection and incubated for 24 h. The expression level of VCP after silencing was confirmed by immunoblotting. Reverse transfection of siVCP (1) and (3) for 48 h strongly decreased expression levels of endogenous VCP in HeLa cells (Fig. 2A). Furthermore, depletion of endogenous VCP reduced expression levels of WNV-E protein at 24 h.p.i. of WNV (Fig. 2A). Thereafter, we examined siRNA targeting of VCP [(1) and (3)] which significantly reduced the percentage of WNV-infected cells (Fig. 2B and C). However, siRNA (2) failed to knockdown endogenous VCP as shown in the immunoblotting and IFA results (Fig. 2A–C). We further measured viral titers in supernatants of WNV-inoculated HeLa cells treated by the siRNAs against VCP. Plaque assays revealed that the viral release was significantly inhibited by siRNA treatment [siCVP (1) and (3)] (Fig. 2D). We also examined the effect of siRNA against VCP on WNV infection by IFA at different time points (12, 24 and 48 h). A decrease in the immunofluorescence signals between cells transfected with control and VCP siRNAs was detected (Supplementary Fig. 3). These results indicate that a depletion of VCP significantly inhibits WNV infection. In contrast, VCP-knockdown did not affect infection by pseudotyped VSV (Supplementary Fig. 4).
Fig. 2.
WNV infection is inhibited in VCP knockdown cells. (A) HeLa cells were treated with either siRNA against VCP [siVCP (1), (2) and (3)] or control siRNA (siCont). The siRNA-treated cells were inoculated with WNV (MOI = 1) at 48 h.p.i. The inoculated cells were harvested at 24 h.p.i. The expression of endogenous VCP protein and WNV envelope protein after treatment with the indicated siRNA were examined by immunoblotting with mouse anti-VCP antibody and mouse anti-WNV/Kunjin envelope protein. The expression of actin was examined after reprobing as an endogenous control. (B) WNV-infected cells from (A), after 24 h incubation with WNV, the cells were harvested and examined by immunofluorescence assay. WNV-infected cells were stained with anti-JEV antibody (green) and cell nuclei were counterstained with DAPI (blue). (C) Positivity of WNV-infected cells from (B). Mean ± SD from three independent experiments is shown; ** p < 0.01 (one-way ANOVA). (D) The culture supernatants from (A) were collected at 24 h.p.i. and the viral titers of the harvested supernatants were determined using plaque assay. Mean ± SD from three independent experiments is shown; ** p < 0.01 (one-way ANOVA).
3.3. VCP participates in the early and genome replication steps during the WNV life cycle
We next investigated the specific role of VCP in the life cycle of WNV. The intracellular life cycle of WNV is divided into two major steps, early and late (Fernandez-Garcia et al., 2011, Kaufmann and Rossmann, 2011). The early step consists of viral attachment, entry and uncoating (Jiang et al., 2010, Kaufmann and Rossmann, 2011), while the late step involves genome translation, genome replication, viral assembly and release (Kobayashi et al., 2016a). WNV-VLPs were employed to determine whether VCP participates in the early or late replication steps (Kobayashi et al., 2014). WNV-VLPs are unable to produce progeny virions because of the absence of WNV structural protein coding sequences in their genome. Therefore, our results are independent of the assembly and virion-releasing steps (Hasebe et al., 2010, Scholle et al., 2004). Thus, WNV- VLPs allow the determination of whether VCP plays a role in either an early step or during the genomic replication of the WNV life cycle. WNV-VLPs were inoculated into both VCP and control siRNA-transfected cells and monitored by expression of DsRed-encoded in the WNV-VLP replicon. qRT-PCR demonstrated that the quantity of WNV-RNA in VCP-knockdown (siVCP (1)) cells was significantly lower than that in control siRNA-treated cells (Fig. 3A). These results suggest that VCP knockdown significantly inhibits infection of WNV-VLPs through an inhibition of early step and/or genome replication steps of the WNV life cycle.
To confirm the role of VCP in the early stages of WNV replication, the siRNA-treated cells were inoculated with WNV and viral RNA was investigated at an early time point of WNV infection, at 2 h post infection. The result revealed that silencing of VCP (siVCP (1)) significantly decreased the quantity of WNV-RNA at 2 h post inoculation of WNV compared to control siRNA treated-cells (Fig. 3B). This suggests that VCP plays a role in the early step of WNV replication cycle, including attachment or entry into cells.
To examine whether VCP plays a role in the late step of the WNV life cycle, three plasmids encoding WNV sequences (pCMV-C, pCMV-SVP and pCMV-WNrep-DsRed) were co-transfected into either VCP-knockdown (siVCP (1)) or control siRNA-treated cells. The plasmid transfection protocol for VLP production has been previously employed to investigate the role of host factor(s) in the late stages of viral infection, including genome replication and virus release (Kobayashi et al., 2016a). The components of the VLP are transfected into cells, therefore, the results obtained employing the plasmid-encoded VLPs were not associated with the early steps of WNV replication (Kobayashi et al., 2016a). At 72 h after plasmid transfection, we examined the WNV-VLP titers in the supernatants from plasmid transfected-cells. The titer of WNV-VLPs was significantly decreased in VCP-knockdown (siVCP (1)) cells compared with control siRNA-treated cells (Fig. 3C). These data suggest that VCP is also involved in the genome replication and/or assembly and release steps of the WNV life cycle.
To confirm that VCP is important for WNV genome replication, the WNV DNA-based replicon pCMV-WNrep-DsRed was introduced into both VCP knockdown and control siRNA-treated cells. This replicon consists of coding sequences of WNV non-structural proteins, while almost all of the sequences encoding WNV structural proteins were deleted and replaced by sequences encoding DsRed. The replicon can replicate and translate to synthesize viral genomic RNA and the non-structural proteins of WNV, respectively. However, it is not capable of producing viral progeny because of the absence of structural protein sequences of WNV. The results of qRT-PCR of the plasmid-transfected cells at 72 h post transfection demonstrated a 10-fold reduction of synthesized WNV-RNA in the VCP-knockdown cells (siVCP (1)) compared to control siRNA-treated cells (Fig. 3D). Taken together, these data indicate that VCP plays a role in the genome replication of the WNV life cycle.
4. Discussion
In the present study, we have investigated the roles of VCP during WNV infection. We found that perturbation of endogenous VCP using a potent VCP inhibitor or siRNA targeting VCP significantly inhibited WNV infection. We could confirm that the expression of endogenous VCP in HeLa cells is depleted after 48 h siRNA transfection [with siVCP (1) and (3)], while another siRNA siVCP (2) did not silence expression of endogenous VCP. Silencing of endogenous VCP, by siVCP (1) and (3), demonstrated that depletion of VCP significantly suppressed WNV infection. This finding suggests that VCP is required for WNV infection and, thereafter, we employed the most potent siRNA [siVCP (1)] to investigate the roles of VCP in WNV replication. A previous report indicated that silencing of endogenous VCP and nuclear protein localization 4 (NPL4), a VCP cofactor, did not inhibit WNV infection, whereas depletion of ubiquitin fusion degradation 1-like (UFD1L) and p47, other cofactors of VCP, suppressed WNV infection (Krishnan et al., 2008). We suggest that the differences between this previous report and the present results may be related to the use of different strains of WNV or could be related to the silencing efficiency of the siRNA employed in the experiments. In addition, control experiments from the present study using pseudotyped VSV demonstrated that depletion of VCP did not suppress pseudotyped VSV infection. These results suggest that VCP plays a role in WNV infection specifically and inhibition of WNV infection in VCP knockdown cells is not a consequence of cellular cytotoxicity.
Employing VLPs, we demonstrated that perturbation of VCP suppressed the infectivity of WNV-VLPs (Fig. 3A). This indicates that VCP is potentially involved in either the early steps or during genome replication of WNV. Using plasmid transfection to generate WNV-VLPs, to bypass early steps of the WNV life cycle, we next examined whether VCP plays a role in the late steps (from genome replication until virus release) of the viral life cycle. Knockdown of VCP reduced the yield of WNV-VLPs compared to control siRNA-treated cells (Fig. 3C) and this finding suggests that VCP also participates in the late steps of WNV life cycle. Taken together, we hypothesized that VCP may be implicated in the genome replication steps of WNV. Therefore, a WNV DNA-based replicon was employed to clarify whether VCP is required for genome replication of WNV. Depletion of VCP significantly decreased expression levels of synthesized WNV-RNA (Fig. 3D) and this finding indicates that VCP is engaged in WNV genomic RNA replication.
It has been previously reported that VCP was found to be localized in the cytosol, ER and nucleus, and can play a role in several cellular processes (Arita et al., 2012, Meyer et al., 2012, Meyer and Weihl, 2014, Yamanaka et al., 2012). The possible mechanism(s) of VCP involvement in WNV infection may be based on the localization and physiological function of VCP. Functional roles of VCP in the replication of the WNV-related virus in the family Flaviviridae, hepatitis C virus (HCV) have been reported (Yi et al., 2016). VCP knockdown significantly decreased expression of HCV RNA levels and VCP was found to be colocalized with the HCV replication complex. It is thus possible that this function of VCP in the HCV life cycle is also required for WNV genome replication, however, no direct evidence currently exists for an interaction between WNV replicase components and VCP and further investigations are required.
Apart from during the genome replication of the WNV life cycle, VCP might potentially be involved in other steps. The present study demonstrates that VCP may also function in the early steps, during either attachment or entry, of the viral life cycle (Fig. 3B). The results of an entry assay revealed that silencing of endogenous VCP caused a significant reduction in the expression levels of WNV-RNA compared to control siRNA-treated cells. This suggests that VCP may also play a role in either the binding or entry steps of the WNV life cycle. A role for VCP in early stages of viral infection has previously been reported for coronavirus and Sindbis virus (Panda et al., 2013, Wong et al., 2015). Depletion of VCP inhibited coronavirus infection through a failure in the maturation of virus-loaded endosomes leading to accumulation of coronavirus particles in the early endosomal compartment (Wong et al., 2015). Studies on Sindbis virus indicated that VCP functioned as a regulator of viral entry as knockdown of VCP caused an alteration of trafficking and resulted in the degradation of Sindbis virus entry receptor (Panda et al., 2013). However, the possible function of VCP on early stages of WNV replication has not been investigated and will require further study.
In conclusion, our findings suggest that VCP is required for replication of WNV at a number of different stages of the viral life cycle, thus, VCP potentially represents a candidate for the therapeutic inhibition of WNV infection.
Competing financial interests
The authors declare no competing financial interests.
Author contributions statement
P. Wallaya, S.K., M.S., Y.O and H.S. conceived the experiments, P. Wallaya and S.K. conducted the experiments and analysed the data. P. Wallaya, S.K., M.S., Y.O., and H.S. contributed reagents/materials/analysis tools. P. Wallaya, S.K., M.C., W.H., Y.O. and H.S. wrote the paper. All authors reviewed the manuscript.
Acknowledgements
We thank Dr. Takashima for donating the WNV NY99 6-LP strain and Dr. Takada for providing pseudotyped VSV. This work was supported by the Program for Leading Graduate Schools “Fostering Global Leaders in Veterinary Science for Contributing to One Health”, and grants (16H06429, 16K21723, 16H06431) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2016.11.029.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Ambrose R.L., Mackenzie J.M. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 2011;85(6):2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita M., Wakita T., Shimizu H. Valosin-containing protein (VCP/p97) is required for poliovirus replication and is involved in cellular protein secretion pathway in poliovirus infection. J. Virol. 2012;86(10):5541–5553. doi: 10.1128/JVI.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogachek M.V., Zaitsev B.N., Sekatskii S.K., Protopopova E.V., Ternovoi V.A., Ivanova A.V., Kachko A.V., Ivanisenko V.A., Dietler G., Loktev V.B. Characterization of glycoprotein E C-end of West Nile virus and evaluation of its interaction force with alphaVbeta3 integrin as putative cellular receptor. Biochemistry (Mosc) 2010;75(4):472–480. doi: 10.1134/s0006297910040115. [DOI] [PubMed] [Google Scholar]
- Brinton M.A. Replication cycle and molecular biology of the West Nile virus. Viruses. 2014;6(1):13–53. doi: 10.3390/v6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., Kolaitis R.M., Taylor J.P., Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153(7):1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bug M., Meyer H. Expanding into new markets–VCP/p97 in endocytosis and autophagy. J. Struct. Biol. 2012;179(2):78–82. doi: 10.1016/j.jsb.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC), 2016. West Nile virus disease cases and deaths reported to CDC by year and clinical presentation, 1999–2015. (Accessed 18.08.16).
- Carnec X., Meertens L., Dejarnac O., Perera-Lecoin M., Hafirassou M.L., Kitaura J., Ramdasi R., Schwartz O., Amara A. The phosphatidylserine and phosphatidylethanolamine receptor CD300a binds dengue virus and enhances infection. J. Virol. 2016;90(1):92–102. doi: 10.1128/JVI.01849-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahar H.S., Chen S., Manjunath N. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology. 2013;436(1):1–7. doi: 10.1016/j.virol.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.F., Deshaies R.J. Quantitative cell-based protein degradation assays to identify and classify drugs that target the ubiquitin-proteasome system. J. Biol. Chem. 2011;286(19):16546–16554. doi: 10.1074/jbc.M110.215319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J.J., Ng M.L. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 2004;78(19):10543–10555. doi: 10.1128/JVI.78.19.10543-10555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J.J., Leong P.W., Ng M.L. Analysis of the endocytic pathway mediating the infectious entry of mosquito-borne flavivirus West Nile into Aedes albopictus mosquito (C6/36) cells. Virology. 2006;349(2):463–475. doi: 10.1016/j.virol.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Courtney S.C., Scherbik S.V., Stockman B.M., Brinton M.A. West nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response. J. Virol. 2012;86(7):3647–3657. doi: 10.1128/JVI.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargemont C., Ossareh-Nazari B. Cdc48/p97, a key actor in the interplay between autophagy and ubiquitin/proteasome catabolic pathways. Biochim. Biophys. Acta. 2012;1823(1):138–144. doi: 10.1016/j.bbamcr.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Dauphin G., Zientara S., Zeller H., Murgue B. West Nile: worldwide current situation in animals and humans. Comp. Immunol. Microbiol. Infect. Dis. 2004;27(5):343–355. doi: 10.1016/j.cimid.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Davis C.W., Nguyen H.Y., Hanna S.L., Sánchez M.D., Doms R.W., Pierson T.C. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J. Virol. 2006;80(3):1290–1301. doi: 10.1128/JVI.80.3.1290-1301.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizot M., Neal J.W., Gasque P. Encephalitis due to emerging viruses: CNS innate immunity and potential therapeutic targets. J. Infect. 2012;65(1):1–16. doi: 10.1016/j.jinf.2012.03.019. [DOI] [PubMed] [Google Scholar]
- Egberink H., Addie D.D., Boucraut-Baralon C., Frymus T., Gruffydd-Jones T., Hartmann K., Horzinek M.C., Hosie M.J., Marsilio F., Lloret A., Lutz H., Pennisi M.G., Radford A.D., Thiry E., Truyen U., Möstl K., Diseases E.A.B.o.C. West Nile virus infection in cats: ABCD guidelines on prevention and management. J. Feline Med. Surg. 2015;17(7):617–619. doi: 10.1177/1098612X15588453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Garcia M.D., Meertens L., Bonazzi M., Cossart P., Arenzana-Seisdedos F., Amara A. Appraising the roles of CBLL1 and the ubiquitin/proteasome system for flavivirus entry and replication. J. Virol. 2011;85(6):2980–2989. doi: 10.1128/JVI.02483-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B.N., Knipe D.M., Howley P.M. 6th ed. vol. 2. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2013. (Fields Virology). [Google Scholar]
- Gamino V., Escribano-Romero E., Blázquez A.B., Gutiérrez-Guzmán A.V., Martín-Acebes M., Saiz J.C., Höfle U. Experimental north american west nile virus infection in the red-legged partridge (Alectoris rufa) Vet. Pathol. 2016;53(3):585–593. doi: 10.1177/0300985815612554. [DOI] [PubMed] [Google Scholar]
- Gilfoy F., Fayzulin R., Mason P.W. West Nile virus genome amplification requires the functional activities of the proteasome. Virology. 2009;385(1):74–84. doi: 10.1016/j.virol.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie L.K., Hoenen A., Morgan G., Mackenzie J.M. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 2010;84(20):10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasebe R., Suzuki T., Makino Y., Igarashi M., Yamanouchi S., Maeda A., Horiuchi M., Sawa H., Kimura T. Transcellular transport of West Nile virus-like particles across human endothelial cells depends on residues 156 and 159 of envelope protein. BMC Microbiol. 2010;10:165. doi: 10.1186/1471-2180-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz F.X., Allison S.L. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 2000;55:231–269. doi: 10.1016/S0065-3527(00)55005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Weidner J.M., Qing M., Pan X.B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P.Y., Block T.M., Guo J.T. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J. Virol. 2010;84(16):8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B., Rossmann M.G. Molecular mechanisms involved in the early steps of flavivirus cell entry. Microbes Infect. 2011;13(1):1–9. doi: 10.1016/j.micinf.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufusi P.H., Kelley J.F., Yanagihara R., Nerurkar V.R. Induction of endoplasmic reticulum-derived replication-competent membrane structures by West Nile virus non-structural protein 4B. PLoS One. 2014;9(1):e84040. doi: 10.1371/journal.pone.0084040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Kimura-Kuroda J., Nagashima K., Yasui K. Analysis of virus-cell binding characteristics on the determination of Japanese encephalitis virus susceptibility. Arch. Virol. 1994;139(3–4):239–251. doi: 10.1007/BF01310788. [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Orba Y., Yamaguchi H., Kimura T., Sawa H. Accumulation of ubiquitinated proteins is related to West Nile virus-induced neuronal apoptosis. Neuropathology. 2012;32(4):398–405. doi: 10.1111/j.1440-1789.2011.01275.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Suzuki T., Igarashi M., Orba Y., Ohtake N., Nagakawa K., Niikura K., Kimura T., Kasamatsu H., Sawa H. Cysteine residues in the major capsid protein, Vp1, of the JC virus are important for protein stability and oligomer formation. PLoS One. 2013;8(10):e76668. doi: 10.1371/journal.pone.0076668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Orba Y., Yamaguchi H., Takahashi K., Sasaki M., Hasebe R., Kimura T., Sawa H. Autophagy inhibits viral genome replication and gene expression stages in West Nile virus infection. Virus Res. 2014;191:83–91. doi: 10.1016/j.virusres.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Suzuki T., Kawaguchi A., Phongphaew W., Yoshii K., Iwano T., Harada A., Kariwa H., Orba Y., Sawa H. Rab8b regulates transport of west nile virus particles from recycling endosomes. J. Biol. Chem. 2016;291(12):6559–6568. doi: 10.1074/jbc.M115.712760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Yoshii K., Hirano M., Muto M., Kariwa H. A novel reverse genetics system for production of infectious west nile virus using homologous recombination in mammalian cells. J. Virol. Methods. 2016 doi: 10.1016/j.jviromet.2016.11.006. [DOI] [PubMed] [Google Scholar]
- Koller K.J., Brownstein M.J. Use of a cDNA clone to identify a supposed precursor protein containing valosin. Nature. 1987;325(6104):542–545. doi: 10.1038/325542a0. [DOI] [PubMed] [Google Scholar]
- Kramer L.D., Bernard K.A. West Nile virus infection in birds and mammals. Ann. N. Y. Acad. Sci. 2001;951:84–93. doi: 10.1111/j.1749-6632.2001.tb02687.x. [DOI] [PubMed] [Google Scholar]
- Krishnan M.N., Ng A., Sukumaran B., Gilfoy F.D., Uchil P.D., Sultana H., Brass A.L., Adametz R., Tsui M., Qian F., Montgomery R.R., Lev S., Mason P.W., Koski R.A., Elledge S.J., Xavier R.J., Agaisse H., Fikrig E. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455(7210):242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtensteiger C.A., Heinz-Taheny K., Osborne T.S., Novak R.J., Lewis B.A., Firth M.L. West Nile virus encephalitis and myocarditis in wolf and dog. Emerg. Infect. Dis. 2003;9(10):1303–1306. doi: 10.3201/eid0910.020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Dang Y., Wu Y., Jia G., Anaya E., Zhang J., Abraham S., Choi J.G., Shi G., Qi L., Manjunath N., Wu H. A CRISPR-based screen identifies genes essential for West-Nile-virus-induced cell death. Cell Rep. 2015;12(4):673–683. doi: 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino Y., Suzuki T., Hasebe R., Kimura T., Maeda A., Takahashi H., Sawa H. Establishment of tracking system for West Nile virus entry and evidence of microtubule involvement in particle transport. J. Virol. Methods. 2014;195:250–257. doi: 10.1016/j.jviromet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Maric M., Maculins T., De Piccoli G., Labib K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 2014;346(6208):1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina B.E., Koraka P., van den Doel P., Rimmelzwaan G.F., Haagmans B.L., Osterhaus A.D. DC-SIGN enhances infection of cells with glycosylated West Nile virus in vitro and virus replication in human dendritic cells induces production of IFN-alpha and TNF-alpha. Virus Res. 2008;135(1):64–71. doi: 10.1016/j.virusres.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Meyer H., Weihl C.C. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci. 2014;127(Pt 18):3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H., Bug M., Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 2012;14(2):117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Morizono K., Chen I.S. Role of phosphatidylserine receptors in enveloped virus infection. J. Virol. 2014;88(8):4275–4290. doi: 10.1128/JVI.03287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D., Rose P.P., Hanna S.L., Gold B., Hopkins K.C., Lyde R.B., Marks M.S., Cherry S. Genome-wide RNAi screen identifies SEC61A and VCP as conserved regulators of Sindbis virus entry. Cell Rep. 2013;5(6):1737–1748. doi: 10.1016/j.celrep.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz S. Climate change impacts on West Nile virus transmission in a global context. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370(1665) doi: 10.1098/rstb.2013.0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera-Lecoin M., Meertens L., Carnec X., Amara A. Flavivirus entry receptors: an update. Viruses. 2014;6(1):69–88. doi: 10.3390/v6010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plevka P., Battisti A.J., Sheng J., Rossmann M.G. Mechanism for maturation-related reorganization of flavivirus glycoproteins. J. Struct. Biol. 2014;185(1):27–31. doi: 10.1016/j.jsb.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye V.E., Dreveny I., Briggs L.C., Sands C., Beuron F., Zhang X., Freemont P.S. Going through the motions: the ATPase cycle of p97. J. Struct. Biol. 2006;156(1):12–28. doi: 10.1016/j.jsb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Ramanathan H.N., Ye Y. The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res. 2012;22(2):346–359. doi: 10.1038/cr.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R.W., Rodriguez D.B., Summers B.A. West Nile virus encephalitis in a dog. Vet. Pathol. 2005;42(2):219–222. doi: 10.1354/vp.42-2-219. [DOI] [PubMed] [Google Scholar]
- Ritz D., Vuk M., Kirchner P., Bug M., Schütz S., Hayer A., Bremer S., Lusk C., Baloh R.H., Lee H., Glatter T., Gstaiger M., Aebersold R., Weihl C.C., Meyer H. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 2011;13(9):1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby J.A., Setoh Y.X., Hall R.A., Khromykh A.A. Post-translational regulation and modifications of flavivirus structural proteins. J. Gen. Virol. 2015;96(Pt 7):1551–1569. doi: 10.1099/vir.0.000097. [DOI] [PubMed] [Google Scholar]
- Samuel M.A., Diamond M.S. Pathogenesis of West Nile Virus infection: a balance between virulence, innate and adaptive immunity, and viral evasion. J. Virol. 2006;80(19):9349–9360. doi: 10.1128/JVI.01122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle F., Girard Y.A., Zhao Q., Higgs S., Mason P.W. trans-Packaged West Nile virus-like particles: infectious properties in vitro and in infected mosquito vectors. J. Virol. 2004;78(21):11605–11614. doi: 10.1128/JVI.78.21.11605-11614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin S.J., Morelli F.F., Vinet J., Amore D., De Biasi S., Poletti A., Rubinsztein D.C., Carra S. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ. 2014;21(12):1838–1851. doi: 10.1038/cdd.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M., Takenouchi A., Shimoda H., Kimura N., Maeda K. Distinct usage of three C-type lectins by Japanese encephalitis virus: DC-SIGN, DC-SIGNR, and LSECtin. Arch. Virol. 2014;159(8):2023–2031. doi: 10.1007/s00705-014-2042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kimura T., Mizutani T., Kariwa H., Takashima I. Different chemokine expression in lethal and non-lethal murine West Nile virus infection. J. Med. Virol. 2004;74(3):507–513. doi: 10.1002/jmv.20205. [DOI] [PubMed] [Google Scholar]
- Shirato K., Miyoshi H., Goto A., Ako Y., Ueki T., Kariwa H., Takashima I. Viral envelope protein glycosylation is a molecular determinant of the neuroinvasiveness of the New York strain of West Nile virus. J. Gen. Virol. 2004;85(Pt 12):3637–3645. doi: 10.1099/vir.0.80247-0. [DOI] [PubMed] [Google Scholar]
- Smithburn K.C., Hughes T.P., Burke A.W., Paul J.H. A Neurotropic Virus Isolated from the Blood of a Native of Uganda. Am. J. Trop. Med. Hyg. 1940;s1–20(20):471–492. [Google Scholar]
- Smit J.M., Moesker B., Rodenhuis-Zybert I., Wilschut J. Flavivirus cell entry and membrane fusion. Viruses. 2011;3(2):160–171. doi: 10.3390/v3020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Hilt W., Buchberger A., Wolf D.H. Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 2011;36(10):515–523. doi: 10.1016/j.tibs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Suthar M.S., Diamond M.S., Gale M. West Nile virus infection and immunity. Nat. Rev. Microbiol. 2013;11(2):115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- Takada A., Ebihara H., Feldmann H., Geisbert T.W., Kawaoka Y. Epitopes required for antibody-dependent enhancement of Ebola virus infection. J. Infect. Dis. 2007;196(Suppl. 2):S347–S356. doi: 10.1086/520581. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Ohtaki N., Maeda-Sato M., Tanaka M., Tanaka K., Sawa H., Ishikawa T., Takamizawa A., Takasaki T., Hasegawa H., Sata T., Hall W.W., Kurata T., Kojima A. Effects of the number of amino acid residues in the signal segment upstream or downstream of the NS2B-3 cleavage site on production and secretion of prM/M-E virus-like particles of West Nile virus. Microbes Infect. 2009;11(13):1019–1028. doi: 10.1016/j.micinf.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Troupin A., Colpitts T.M. Overview of west nile virus transmission and epidemiology. Methods Mol. Biol. 2016;1435:15–18. doi: 10.1007/978-1-4939-3670-0_2. [DOI] [PubMed] [Google Scholar]
- Verma R., Oania R., Fang R., Smith G.T., Deshaies R.J. Cdc48/p97 mediates UV-dependent turnover of RNA pol II. Mol. Cell. 2011;41(1):82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li L., Ye Y. Inhibition of p97-dependent protein degradation by Eeyarestatin I. J. Biol. Chem. 2008;283(12):7445–7454. doi: 10.1074/jbc.M708347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Shinkre B.A., Lee J.G., Weniger M.A., Liu Y., Chen W., Wiestner A., Trenkle W.C., Ye Y. The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS One. 2010;5(11):e15479. doi: 10.1371/journal.pone.0015479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5(4):365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox A.J., Laney J.D. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat. Cell Biol. 2009;11(12):1481–1486. doi: 10.1038/ncb1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D.H., Stolz A. The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim. Biophys. Acta. 2012;1823(1):117–124. doi: 10.1016/j.bbamcr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Wong H.H., Kumar P., Tay F.P., Moreau D., Liu D.X., Bard F. Genome-wide screen reveals valosin-containing protein requirement for coronavirus exit from endosomes. J. Virol. 2015;89(21):11116–11128. doi: 10.1128/JVI.01360-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D., Tang W.K., Ye Y. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene. 2016;583(1):64–77. doi: 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Sasagawa Y., Ogura T. Recent advances in p97/VCP/Cdc48 cellular functions. Biochim. Biophys. Acta. 2012;1823(1):130–137. doi: 10.1016/j.bbamcr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Yi Z., Fang C., Zou J., Xu J., Song W., Du X., Pan T., Lu H., Yuan Z. Affinity purification of the hepatitis C virus replicase identifies valosin-containing protein, a member of the ATPases associated with diverse cellular activities family, as an active virus replication modulator. J. Virol. 2016;90(21):9953–9966. doi: 10.1128/JVI.01140-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Zhang W., Holdaway H.A., Li L., Kostyuchenko V.A., Chipman P.R., Kuhn R.J., Rossmann M.G., Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- Zaitsev B.N., Benedetti F., Mikhaylov A.G., Korneev D.V., Sekatskii S.K., Karakouz T., Belavin P.A., Netesova N.A., Protopopova E.V., Konovalova S.N., Dietler G., Loktev V.B. Force-induced globule-coil transition in laminin binding protein and its role for viral-cell membrane fusion. J. Mol. Recognit. 2014;27(12):727–738. doi: 10.1002/jmr.2399. [DOI] [PubMed] [Google Scholar]
- Zhong X., Pittman R.N. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 2006;15(16):2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- Zidane N., Ould-Abeih M.B., Petit-Topin I., Bedouelle H. The folded and disordered domains of human ribosomal protein SA have both idiosyncratic and shared functions as membrane receptors. Biosci. Rep. 2013;33(1):113–124. doi: 10.1042/BSR20120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.