Highlights
-
•
PEDV reverse genetics achieved by targeted RNA recombination, BAC cloning, and in vitro ligation.
-
•
Overview of cell lines assessed for permissiveness for PEDV propagation.
-
•
Biological questions about PEDV addressed and to be addressed by reverse genetics.
-
•
Reverse genetics as a tool for vaccine development.
Keywords: Porcine epidemic diarrhea virus, Reverse genetics, Spike protein, ORF3, Nucleocapsid, Vaccine
Abstract
Emergence of the porcine epidemic diarrhea virus (PEDV) as a global threat to the swine industry underlies the urgent need for deeper understanding of this virus. To date, we have yet to identify functions for all the major gene products, much less grasp their implications for the viral life cycle and pathogenic mechanisms. A major reason is the lack of genetic tools for studying PEDV. In this review, we discuss the reverse genetics approaches that have been successfully used to engineer infectious clones of PEDV as well as other potential and complementary methods that have yet to be applied to PEDV. The importance of proper cell culture for successful PEDV propagation and maintenance of disease phenotype are addressed in our survey of permissive cell lines. We also highlight areas of particular relevance to PEDV pathogenesis and disease that have benefited from reverse genetics studies and pressing questions that await resolution by such studies. In particular, we examine the spike protein as a determinant of viral tropism, entry and virulence, ORF3 and its association with cell culture adaptation, and the nucleocapsid protein and its potential role in modulating PEDV pathogenicity. Finally, we conclude with an exploration of how reverse genetics can help mitigate the global impact of PEDV by addressing the challenges of vaccine development.
1. Introduction
The porcine epidemic diarrhea virus (PEDV) is an enveloped positive-sense RNA virus in the Coronaviridae family (Masters, 2006, Park et al., 2012). The PEDV genome is approximately 28 kb in length, with seven overlapping open reading frames encoding the replicase (ORF1a, 1b) nonstructural proteins and the accessory protein ORF3, as well as the spike (S), envelope (E), membrane (M), and nucleocapsid (N) structural proteins (Kocherhans et al., 2001). The S protein mediates specific receptor binding and membrane fusion to facilitate viral entry and contains major antigenic epitopes for neutralizing antibodies against PEDV (Sun et al., 2007). The E protein is a membrane protein which has been found to play a role in viral budding in other coronaviruses (Ortego et al., 2007), while the M protein plays a role in viral assembly (de Haan et al., 2000). Coronavirus N proteins bind viral RNA, providing structural scaffolding for viral transcription, replication and assembly (McBride et al., 2014). The replicase genes encompassing two-thirds of the genome (ORF1a and ORF1b) encode two polyproteins that are further processed by viral proteases into 16 nonstructural proteins (NSPs 1–16) important for viral replication (Kocherhans et al., 2001). These proteins and ORF3 remain to be fully characterized.
The mechanisms behind PEDV pathogenesis remain largely unknown due to the challenges in propagating field isolates in cell culture and the lack of genetic tools for virus manipulation. Coronaviruses have genomes approximately 30 kb in length, the largest among RNA viruses, which present a significant challenge in engineering vectors for the generation of infectious clones as the distinction between natural and amplification-associated mutations and sequencing errors can be extremely difficult to pin down. In addition, attempts to set up reverse genetics systems have been hampered by the instability and toxicity of viral genomic sequences. Recently, however, a number of reverse genetics techniques have been developed for PEDV, promising to greatly accelerate research into the PEDV replication cycle and pathogenesis. The three techniques used are targeted RNA recombination, the bacterial artificial chromosome (BAC) system, and in vitro ligation. These approaches, along with their strengths and weaknesses, will be discussed in detail in this review, as well as a few alternative methods that have yet to be applied to PEDV. As appropriate cellular substrates are significant factors for successful virus rescue and propagation, we will also present an overview of cell lines that have been studied in the context of PEDV. We will then discuss some topics that can be further explored using reverse genetics technology, namely the role of trypsin in viral propagation, the study of receptors and attachment factors for viral entry, S protein variation and its implications, the significance of ORF3 and N proteins, and how reverse genetics may help overcome challenges in PEDV vaccine design.
2. Reverse genetics of PEDV
2.1. Targeted RNA recombination
Targeted RNA recombination was the first reverse genetics system developed for coronaviruses, being first devised and refined for the generation of recombinant murine hepatitis virus (MHV) (Koetzner et al., 1992, Kuo et al., 2000, van der Most et al., 1992). This technique was subsequently applied to reverse genetics of other coronaviruses such as feline infectious peritonitis virus (FIPV) (Haijema et al., 2003) and PEDV (Li et al., 2013). Considering the complications involved in generating full-length cDNA clones of coronaviruses, this technique eliminates the need to clone the genome in its entirety, including the particularly troublesome replicase genes.
Generally, this technique involves intracellular recombination between full-length viral RNA (typically by infection) and a viral RNA fragment generated by in vitro transcription and containing genetic modifications that result in a selectable phenotype. Such phenotypes have included changes in temperature sensitivity and plaque size, as pioneered by Koetzner et al. (Koetzner et al., 1992). More recently, the use of S proteins that confer a very specific change in host cell tropism has been employed for greater ease in recombinant virus selection (Kuo et al., 2000, Kuo and Masters, 2002). Overall, targeted RNA recombination takes advantage of the high rate of coronavirus RNA recombination during host cell infection (Lai et al., 1985, Liao and Lai, 1992, Makino et al., 1986) as well as strict host tropism for selection of complete recombinant viral genomes.
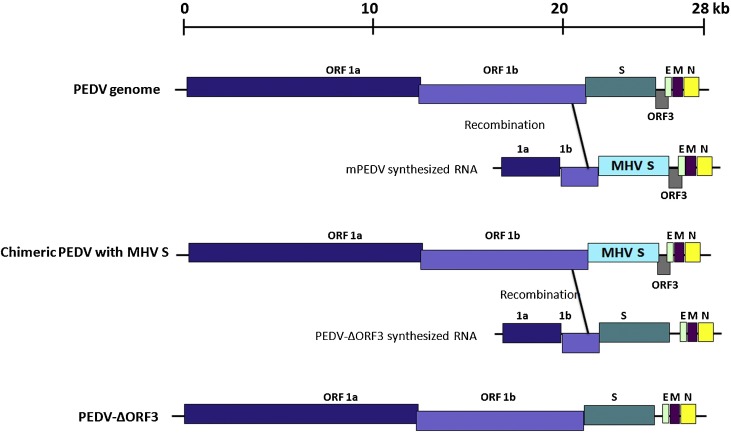
For PEDV, the approach used by Li et al. involved two stages: the generation of a recombinant PEDV with strict murine cell tropism, followed by selection for the final modified PEDV recombinant in Vero cells (Fig. 1 ) (Li et al., 2013). A synthetic donor construct was first prepared encoding (a) the bacteriophage T7 RNA polymerase promoter for RNA transcription, (b) a segment of the ORF1b sequence for targeted recombination, (c) the MHV S protein in place of the PEDV S, and (d) the remaining downstream sequences, including ORF3, E, M and N. Chimeric virus was generated by RNA recombination after electroporation of PEDV-infected Vero cells with transcribed RNA from the construct. Recombinants carrying the MHV S gene (mPEDV) were selected by propagation in murine L cells. For the next stage, a second donor construct encompassing the same genomic region was prepared. This construct carried PEDV S to enable recombinant virus selection in Vero cells, although in practice, it need not be the same S gene as the original virus. Likewise, the remaining genes (ORF3, E, M, N) may be modified and, in the case of ORF3, removed entirely or replaced with a reporter such as luciferase. Electroporation of transcribed RNA into mPEDV-infected L cells followed by selection on Vero yielded the final recombinant virus.
Fig. 1.
Genetic manipulation of the PEDV genome by targeted RNA recombination. An RNA fragment containing the S protein of MHV flanked by upsteam and downstream PEDV sequences was generated by in vitro transcription. Upon electroporation into PEDV-infected cells, recombination occurs between the ORF1b sequences, creating a chimeric PEDV with MHV S. PEDV S was reintroduced back to the genome in a second recombination event with in vitro transcribed RNA encoding PEDV S along with an ORF3 deletion. Figure adapted from Li et al. (Li et al., 2013).
Essentially, this system allows highly flexible modification of the approximately 8 kb structural gene sequence at the 3′ end of the genome. The relatively short cloned donor segment facilitates sequence verification, and use of replication-competent virus as the basis for RNA recombination ensures virus rescue as long as modifications introduced into the donor construct are non-lethal. Swapping of S, ORF3, E, M and/or N genes enables studies into gene product function. And as ORF3 is considered an accessory protein, recombinant PEDVs with reporters in place of ORF3 can be created with this technique, allowing tracking of recombinant virus replication and spread, as well as facilitating quantification of viral growth. Targeted RNA recombination is an elegant solution to obstacles faced in cloning the replicase genes, and remains an option if problems in infectious cDNA cloning appear insurmountable.
The main drawback of this method is that it relies on replicase genes to generate the desired recombinant viruses, preventing modification and hence study of these essential genes. Its dependence on T7 RNA polymerase transcription means that the viral genetic sequence must be carefully assessed for potential cryptic transcription termination signals, and mutations must be carefully implemented to avoid detrimental effect to the virus. Additionally, in vitro selection of the recombinant virus is contingent upon the use of an S gene that is capable of mediating entry into Vero or other known target cells. Viruses carrying mutations that destroy receptor binding or fusion cannot be generated for testing hypotheses regarding cell entry. Similarly, lethal mutations in other regions will prevent recovery of recombinant virus. And even after recombinant viruses have been successfully generated, they may need to be plaque purified and screened, especially in cases where multiple mutations or gene swaps are desired, as a single recombination event is not guaranteed (Lai, 1996). An identification marker at the far 3′ end of the donor sequence can be used for selection (Koetzner et al., 1992), which would require analysis of clones by procedures such as sequencing.
2.2. Infectious cDNA clone generation using the BAC system
Due to the instability and toxicity of the replicase genes when propagated as cDNA in bacteria, it has been extremely challenging to generate a full-length infectious clone for coronaviruses. Application of BACs designed to accommodate large inserts over 300 kb in size (O'Connor et al., 1989, Shizuya et al., 1992) and be maintained at very low plasmid copy numbers per cell (1–2 copies) has enabled engineering of infectious clones for porcine transmissible gastroenteritis virus (TGEV) (Almazan et al., 2000), severe acute respiratory syndrome coronavirus (SARS-CoV) (Almazan et al., 2006, Pfefferle et al., 2009), human CoV OC43 (HCoV-OC43) (St-Jean et al., 2006), FIPV (Balint et al., 2012), Middle East respiratory syndrome CoV (MERS-CoV) (Almazan et al., 2013), and MHV (Fehr et al., 2015). Similarly, we recently reported successful construction of the first PEDV infectious clone using pSMART-BAC (Jengarn et al., 2015).
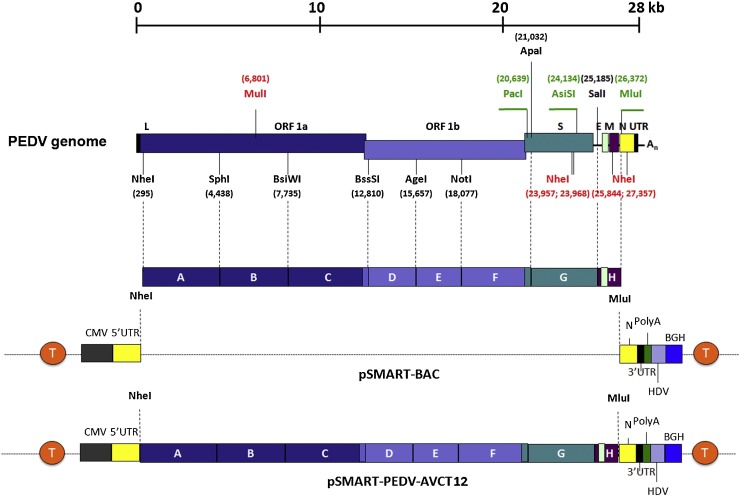
The genome of the PEDV strain AVCT12 was synthesized as eight separate fragments with specific restriction sites for ligation and cloning (Fig. 2 ). Two fragments were combined at a time by three-way ligation into subcloning vectors until the full-length cDNA clone was reconstituted. The expression cassette of this full-length clone comprises, in this order, the CMV promoter, the PEDV genomic sequence flanked by the 5′ and 3′ UTRs, a 25-residue polyA tract, the hepatitis delta virus (HDV) ribozyme cleavage site, and the bovine growth hormone termination and polyA sequence (BGH). Flanking this entire expression cassette are transcription terminator sequences, one at either end, to reduce transcription from cryptic promoters and reduce toxicity to transformed bacteria. This precaution supports the stable maintenance of viral cDNA in the pSMART-BAC system. After the cloning process, however, bacterial transposon sequences (ATGGTACCG) were found to have integrated into the genomic sequence, confounding attempts to rescue the virus. This issue was successfully circumvented by propagating all cloning and subcloning vectors in competent Escherichia coli DH10B and diligent inspection of clone sequences.
Fig. 2.
Construction of a PEDV infectious clone using the BAC system. The AVCT12 genome was divided into small fragments (A–H) and synthesized with native restriction site sequences designated in black, introduced restriction sites in green and deleted restriction sites in red. After fragment assembly, the full-length cDNA was ligated to pSMART-BAC containing transcriptional terminator sequences (T in orange) flanking both directions of the cloning site. Figure adapted from Jengarn et al. (Jengarn et al., 2015).
Virus recovery itself was performed by transfecting the pSMART-BAC-based infectious cDNA clone into the Vero E6-APN cell line for rescue and propagation, although a recent protocol modification to use human embryonic kidney (HEK) 293T cells for transfection prior to transfer to Vero E6-APN has resulted in improved rescue titers (unpublished observations). Rescued virus showed similar growth in cell culture to the parental AVCT12, suggesting that modifications made to facilitate the cloning process did not interfere with in vitro replication.
The major advantage of full-length infectious clones over targeted RNA recombination is that it allows manipulation of the entire PEDV genome. The eight-fragment system enables easy and separate modification of individual genes. Interestingly, the AVCT12 strain in particular has lost expression of its ORF3 gene due to deletion of the region containing the ORF3 start codon. It exhibits strong growth in Vero E6-APN culture, and can support expression of a transgene in place of the ORF3 gene. An AVCT12 construct carrying the fluorescent reporter gene mCherry has been created to facilitate tracking of virus replication and spread.
Although BAC-based infectious clones are extremely powerful and highly desirable tools for manipulation of the PEDV genome, it is not without its challenges. The construct design in itself requires careful planning in the selection of restriction sites for fragment assembly, and should the process necessitate the removal of native restriction sites or introduction of new ones, caution must be taken in their mutagenesis to avoid affecting viral replication. Additionally, we observed insertion of bacterial tranposons into the viral sequence after propagation in bacteria, so regular checking of sequence fidelity at every step during clone construction is critical. This need to recheck viral sequences at each cloning step is a major bottleneck for workflow, especially after the full-length cDNA assembly stage, and having accurate, verified sequence data for the original, infectious virus as a basis for comparison is absolutely vital for cloning success.
2.3. Infectious cDNA clone generation by in vitro ligation
Another approach for minimizing the impact of genomic instability and toxicity in bacteria is to avoid full-length clone propagation in bacteria altogether. With in vitro ligation, multiple fragments of viral cDNA encompassing the entirety of the viral genome are assembled in a stepwise manner in vitro and transcribed as full-length infectious RNA by T7 RNA polymerase. The full-length RNA is then electroporated into cells for virus production. This technique was first developed to simplify preparation of a TGEV molecular clone (Yount et al., 2000), and has subsequently been applied to the reverse genetics of other coronaviruses, namely MHV (Yount et al., 2002), infectious bronchitis virus (IBV) (Fang et al., 2007, Youn et al., 2005), SARS-CoV (Yount et al., 2003), bat SARS-like CoV (Agnihothram et al., 2014, Becker et al., 2008), HCoV-NL63 (Donaldson et al., 2008), and MERS-CoV (Scobey et al., 2013).
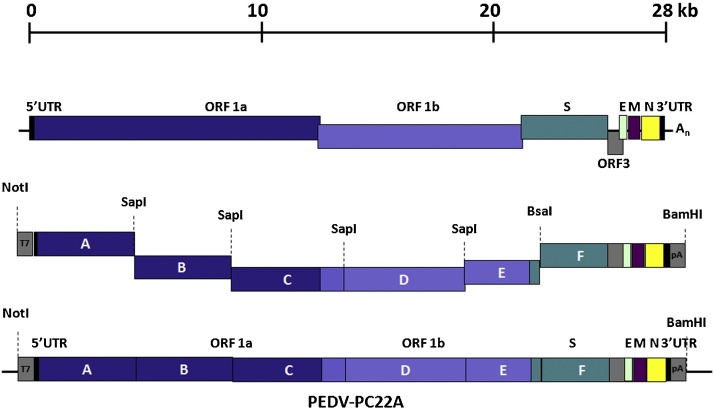
For PEDV, in vitro ligation was used to generate an infectious cDNA clone of the virulent North American strain PC22A (Beall et al., 2016, Oka et al., 2014). Six cDNA fragments spanning the entire PEDV genome were designed with flanking class II restriction endonuclease sites for seamless and specific ligation (see assembly details in Fig. 3 ). Each fragment was inserted into a subcloning vector, with the 5′-most segment ligated to a T7 RNA polymerase promoter. The fragments were subsequently digested, ligated, and transcribed in vitro to generate full-length RNA transcripts of the virus. Infectious RNA was subsequently electroporated into Vero cells for recovery of infectious PEDV. N gene transcripts were also prepared and electroporated along with the genomic RNA as previously done with TGEV (Yount et al., 2000), where N was shown to be critical for efficient virus replication and spread. Such N supplementation has proven to enhance rescue of other coronaviruses as well (Youn et al., 2005, Yount et al., 2003, Yount et al., 2002) but has not been needed for DNA-launched virus production systems like the BAC approach, suggesting the importance of N-mediated encapsidation of coronavirus RNA for transcription initiation.
Fig. 3.
Construction of a PEDV infectious clone using in vitro ligation. The genome of PC22A, a pathogenic strain of PEDV, was divided into six fragments, with the first and the last fragments carrying a T7 RNA polymerase promoter and polyA sequences respectively (indicated in gray boxes). Each fragment was flanked by recognition sites for the type II restriction enzyme SapI, which leaves unique overhangs at both 5′ and 3′ ends and allows directional ligation assembly of full-length cDNA fragments. Figure adapted from Beall et al. (Beall et al., 2016).
The in vitro ligation technique shares many strengths of the BAC system, such as ease in modifying the entire PEDV genome and insertion of alternative genes into the ORF3 expression cassette. Its engineering approach can also be applied to simplify cloning into BAC vectors, by circumventing the stepwise ligation process with a single in vitro ligation reaction. Furthermore, it also offers an alternative to the use of the BAC system by bypassing the need for a stable bacterial vector for the full-length cDNA clone. Instead, the cDNA is maintained as separate segments in smaller subcloning vectors. Toxicity of replicase gene fragments remains an issue even in subcloning vectors however, although it has been determined that splitting the genes at specific regions into separate fragments can help alleviate the problem (Scobey et al., 2013, Yount et al., 2000, Yount et al., 2002). Design of cDNA fragments will need to work around this requirement.
This technique also amplifies certain weaknesses of the targeted RNA recombination approach, due to the sheer length of the RNA transcript. Fragment design will need to take potential transcription termination sites into consideration (Yount et al., 2000). And despite the low error rate of T7 RNA polymerase, specific errors have been associated with the enzyme (Doetsch, 2002, Pomerantz et al., 2006). The correctness of the full-length RNA used as the template for virus production is contingent upon the fidelity of the T7 RNA polymerase, may vary with each experiment, and cannot be verified prior to use.
2.4. Infectious cDNA clone generation in vaccinia vectors
Vaccinia vectors provide another alternative to the use of bacteria for maintenance and propagation of PEDV cDNA. A large DNA virus in the family Poxviridae, the vaccinia virus has a 185 kb genome that is capable of accommodating inserts of up to at least 26 kb in size (Merchlinsky and Moss, 1992). It encodes around 250 genes, containing the necessary replication machinery to enable viral transcription and replication in the cytoplasm. The virus is easy to grow to high titers, and efficiently infects a variety of cell types, both primary cells and cell lines of mammalian and non-mammalian origin, making it powerful as a gene vector for tissue culture. It was first applied to coronavirus reverse genetics by Casais et al., who used vaccinia as a platform for generation of recombinant IBV (Casais et al., 2001). Since then, it has been applied to reverse genetics of MHV (Coley et al., 2005), feline coronavirus (FCoV) (Tekes et al., 2008), and SARS-CoV (van den Worm et al., 2012), as well as the study of HCoV-229E replicase genes (Thiel et al., 2001). While there has been no report of the vaccinia vector being used for PEDV reverse genetics, the advantages it offers are worth consideration.
This approach is based on in vitro ligation of subgenomic cDNA fragments based on native or engineered restriction sites to generate full-length viral cDNA downstream of a T7 RNA polymerase promoter. While assembly steps vary with each report, the cDNA is ultimately ligated at the unique NotI site of the vNotI/tk vaccinia virus DNA (Merchlinsky and Moss, 1992). Some approaches first clone incomplete segments of genomic cDNA into vaccinia (Tekes et al., 2008, van den Worm et al., 2012), although full-length insertions of the 27-kb IBV and 31-kb MHV (Coley et al., 2005) genomes have been successful. The ligation reaction is then transfected into cells infected with fowlpox helper virus, which carries the necessary machinery for initiation of transcription. Recombinant vaccinia viruses are collected, and the DNA purified and used for T7 RNA polymerase-mediated transcription, either by in vitro transcription (Tekes et al., 2008) or intracellularly in the presence of recombinant fowlpox virus expressing T7 RNA polymerase (Casais et al., 2001, van den Worm et al., 2012). In the case of in vitro transcription, full-length coronavirus RNA will need to be electroporated back into cells to generate the final recombinant virus. While multiple steps are necessary for vaccinia vector-based reverse genetics, modifications to recombinant viruses can be achieved by homologous recombination, which, unlike the BAC and in vitro ligation systems described above, does not require reassembly of all component fragments.
2.5. Infectious cDNA clone generation by Gibson assembly
The Gibson assembly method is a relatively recent technology that enables assembly of DNA fragments up to several hundred kb in length (Gibson et al., 2009). DNA fragments are designed with at least 20 overlapping bases at both 5′ and 3′ ends, and are ligated simultaneously in the absence of restriction digestion in a single-step, isothermic reaction. This is accomplished with three key components: (a) a 5′ exonuclease to generate recessed ends, (b) DNA polymerase to fill in the gaps after annealing of sticky ends, and (c) DNA ligase. While Gibson assembly has not been applied to coronavirus infectious clone construction, it has been used to successfully recover infectious clones of viruses such as porcine reproductive and respiratory syndrome virus (PRSSV) (Suhardiman et al., 2015), dengue virus (Siridechadilok et al., 2013), and West Nile virus (Vandergaast et al., 2014). Given that it eliminates the need for restriction site design, it may prove to be a very effective tool to complement in vitro ligation as well as cloning into BAC or vaccinia vectors.
As a ligation method, Gibson assembly does not inherently overcome the issue of sequence instability and toxicity. However, beyond being simply a tool for rapid assembly of cDNA, it can also circumvent the need for bacterial transformation as demonstrated by Suhardiman et al. (Suhardiman et al., 2015) and Siridechadilok et al. (Siridechadilok et al., 2013), where Gibson assembly reactions were directly transfected into cells for virus production. By generating cDNA by RT-PCR from field samples, this approach can be extremely useful for rapid generation of viral clones that preserve viral sequence diversity. However, the complete elimination of bacteria propagation steps also prevents sequence verification and control prior to virus generation. Experimental variation is expected to be greater than the BAC and in vitro ligation methods if assembly of RT-PCR fragments are performed, and as such, this bacteria-free protocol is not recommended for biochemical studies without prior plaque purification and virus sequencing.
3. Cell lines for PEDV cultivation
The development of a cell culture system for PEDV cultivation has been key to enabling PEDV research and is a critical component of reverse genetics rescue of PEDV. Successful in vitro propagation of PEDV was first reported in 1988 using Vero cells (Hofmann and Wyler, 1988), which are derived from kidney epithelial cells of the African green monkey (Yasumura and Kawakita, 1963) and are now used widely in PEDV research. In this study by Hofmann and Wyler, inoculation of Vero cells with field-isolated PEDV of gut origin yielded detectable cytopathic effect involving formation of syncytia containing less than 10 nuclei (Hofmann and Wyler, 1988). After three passages in cell culture, however, Vero infection resulted in much more extensive syncytia (up to 100 nuclei), indicating that a certain degree of adaptation must take place for field isolates to replicate well in culture. Indeed, the success rate of field isolate cultivation can be quite low in Vero cells, and the virus may gradually lose infectivity upon further passaging (Chen et al., 2014). These observations emphasize the need for cell lines allowing for less selective propagation of field isolates. Several traditional and newly established cell lines have, therefore, been examined for their permissiveness for PEDV replication. Table 1, Table 2 provide a representative overview of cell lines that have been tested for PEDV propagation. While cells derived from species other than swine have shown susceptibility to PEDV (Table 1), some porcine cells isolated from different organs were found to be non-permissive (Table 2). One technical reason for the inability of porcine primary cells to support PEDV growth could be the standard practice of adding trypsin to promote replication, as primary cells tend to be hypersensitive to trypsin and detach rapidly, preventing efficient PEDV infection (Hofmann and Wyler, 1988). Mechanistically, this may involve viral entry by field isolates and the question of receptor usage, which will be discussed in detail in a later section. Briefly, while several groups have highlighted porcine aminopeptidase N (pAPN) as the key receptor for cell entry of PEDV (Li et al., 2007, Liu et al., 2015, Nam and Lee, 2010), the considerable number of non-porcine cell lines used for PEDV propagation suggests that other receptors could play a role in infection. Elucidation of the mechanisms underlying infection of field isolates would greatly advance development of optimal cell culture systems for PEDV propagation in vitro, as well as minimize cell culture adaptation that may significantly alter PEDV pathogenesis.
Table 1.
Cells reported as permissive for PEDV propagation in vitro.
| Cell | Cell description | PEDV strain (cell-adapted or field isolates) | Cell maintenance medium | Post-infection medium | Note | Reference |
|---|---|---|---|---|---|---|
| Vero | African green monkey kidney cell line (CCL-81) | V215/78 (field, Germany), Not specified (field, Belgium) |
EMEM, 30 mM HEPES, 10% FBS, 100 IU of penicillin, 100 μg/mL streptomycin | EMEM, 0.3% tryptose phosphate broth, 0.02% yeast extract, 10 μg/mL trypsin | (Hofmann and Wyler, 1988) | |
| KPEDV-9 (cell-adapted) | α-MEM, 5% FBS, antibiotics | α-MEM, 0.3% tryptose phosphate broth, 0.02% yeast extract, 2 μg trypsin | (Oh et al., 2004) | |||
| ISU13-19338E (field, Indiana, US), ISU13-22038 (field, Iowa, US) | MEM, 10% FBS, 2 mM L-glutamine, 0.05 mg/mL gentamicin, 10 units/mL penicillin, 10 μg/mL streptomycin, 0.25 μg/mL amphotericin | MEM, 0.3% tryptose phosphate broth, 0.02% yeast extract, 5 μg/mL trypsin | (Chen et al., 2014) | |||
| PC168, PC170, PC173, PC177, PC179, PC180, PC182, PC21A and PC22A (field, Ohio, Michigan, Illinois and Iowa, US) | DMEM, 5% FBS, 100 unit/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL Fungizone | DMEM, 100 unit/mL penicillin, 100 μg/mL streptomycin, 0.3% tryptose phosphate broth, 10 μg/mL trypsin | (Oka et al., 2014) | |||
| HLJBY (not specified) | DMEM, 10% FBS, 100 units/mL penicillin, 100 units/mL streptomycin | DMEM, 10 μg/mL trypsin | (Meng et al., 2014) | |||
| KPEDV-9 (cell-adapted), SM98LVec (cell-adapted) |
MEM, 10% FBS | MEM, 10 μg/mL trypsin | (Park and Shin, 2014) | |||
| Ohio VBS2 (field, Ohio, US) | Not specified | Maintenance medium, 5 μg/mL trypsin | (Liu et al., 2015) | |||
| YN144 (cell-adapted) | DMEM, 10% FBS | DMEM, 10 μg/mL trypsin | Cells are also permissive to lentivirus pseudotyped with PEDV S | (Wang et al., 2016) | ||
| Vero E6 | African green monkey kidney cell line (CRL-1586) | CV777 (cell-adapted) | DMEM, 10% FBS | DMEM, 2.5 μg/mL of trypsin | Vero E6 shows contact inhibition | (Cao et al., 2015a) |
| Vero- TMPRSS2 |
Vero cells stably expressing TMPRSS2 | MK (cell-adapted) | DMEM, 5% FBS | DMEM, 10% tryptose phosphate broth, 2.5 μg/mL trypsin | (Shirato et al., 2011) | |
| IECs | Swine small intestine epithelial cells (ileum) | CV777 (cell-adapted) | DMEM-F12, 10% FBS | DMEM-F12, 2.5 μg/mL of trypsin | (Cong et al., 2015) | |
| Shaanxi strain (field, China) | DMEM-F12, 5% FBS | Not specified | (Cao et al., 2015b) | |||
| 3D4 | Porcine alveolar macrophage cell line | KPEDV-9 (cell-adapted), SM98LVec (cell-adapted) |
RPMI 1640, 10% FBS, 10 mM HEPES, 1.0 mM sodium pyruvate, 0.1 mM nonessential amino acids | MEM, 10 μg/mL trypsin | (Park and Shin, 2014) | |
| MK-DIEC | Duck intestinal epithelial cell line | Colorado strain (cell-adapted) | 1:1 of DMEM: mammary epithelial growth medium, 70 μg/mL bovine pituitary extract, 5 ng/mL human epidermal growth factor, 5 μg/mL insulin, 0.5 μg/mL hydrocortisone, 2% FBS | DMEM, 0.3% tryptose phosphate broth, 0.02% yeast extract, 1% penicillin/streptomycin, 2.5-10 μg/mL trypsin | Cells endogenously express APN (immunofluorescent staining) | (Khatri, 2015) |
| KSEK6 | Swine epithelial cell line | P5-V (cell adapted) | Information not accessible | Information not accessible | (Kadoi et al., 2002) | |
| IB-RS-2 | Swine kidney cell line | P5-V (cell adapted) | Information not accessible | Information not accessible | (Kadoi et al., 2002) | |
| PK15 | Pig kidney cell line | Ohio VBS2 (field, Ohio, US) | Not specified | Maintenance medium, 5 μg/mL trypsin | (Liu et al., 2015) | |
| YN144 (cell-adapted) | DMEM, 10% FBS | DMEM, 10 μg/mL trypsin | Cells endogenously express APN and are also permissive to lentivirus pseudotyped with PEDV S. | (Wang et al., 2016) | ||
| Huh-7 | Human liver cell line | YN144 (cell-adapted) | DMEM, 10% FBS | DMEM, 10 μg/mL trypsin | Cells endogenously express APN and are also permissive to lentivirus pseudotyped with PEDV S. | (Wang et al., 2016) |
| Ohio VBS2 (field, Ohio, US) | Not specified | Maintenance medium, 5 μg/mL trypsin | (Liu et al., 2015) | |||
| ST | Pig testis cell line | Ohio VBS2 (field, Ohio, US) | Not specified | Maintenance medium, 5 μg/mL trypsin | (Liu et al., 2015) | |
| MRC-5 | Human lung cell line | Ohio VBS2 (field, Ohio, US) | Not specified | Maintenance medium, 5 μg/mL trypsin | (Liu et al., 2015) | |
| Tb1-Lu | Bat lung cell line | Ohio VBS2 (field, Ohio, US) | Not specified | Maintenance medium, 5 μg/mL trypsin | (Liu et al., 2015) | |
| PK15-APN | Pig kidney cells exogenously expressing human or porcine APN | Retrovirus pseudotyped with PEDV S | Not specified | Not specified, no trypsin added | (Liu et al., 2015) | |
| MDCK-APN | Canine kidney cells exogenously expressing human or porcine APN | Retrovirus pseudotyped with PEDV S | Not specified | Not specified, no trypsin added | (Liu et al., 2015) | |
| MARC-145 | African green monkey kidney cell line MA-104 | Colorado/2013 (field, Iowa, US) | DMEM, 10% FBS | DMEM, 0.3% tryptose phosphate broth, 0.02% yeast extract, 2 μg/mL trypsin 250 | (Zhang et al., 2016) |
APN, aminopeptidase N; DMEM, Dulbecco’s modified Eagle’s medium; DMEM-F12, Dulbecco’s modified Eagle’s medium –Ham’s F12 mixture; EMEM, Eagle’s minimal essential medium; MEM, Minimal essential medium; FBS, fetal bovine serum; HEPES, N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid.
Table 2.
Cells reported non-permissive to PEDV in vitro or hypersensitive to trypsin.
| Cell | Cell description | PEDV strain (cell-adapted or field isolates) | Cell maintenance medium | Post-infection medium | Note | Reference |
|---|---|---|---|---|---|---|
| PD5 | Porcine thyroid cell line | V215/78 (field, Germany), Not specified (field, Belgium) |
EMEM, 30 mM HEPES, 10% FBS, 100 IU penicillin, 100 μg/mL streptomycin | EMEM, 0.3% tryptose phosphate broth, 0.2% yeast extract, 10 μg/mL trypsin | No cytopathic effect or viral protein synthesis detected | (Hofmann and Wyler, 1988) |
| PK15 | Porcine kidney cell line | V215/78 (field, Germany), Not specified (field, Belgium) |
EMEM, 30 mM HEPES, 10% FBS, 100 IU penicillin, 100 μg/mL streptomycin | EMEM, 0.3% tryptose phosphate broth, 0.2% yeast extract, 10 μg/mL trypsin | No cytopathic effect or viral protein synthesis detected | (Hofmann and Wyler, 1988) |
| HRT18 | Human rectal tumor cell line | V215/78 (field, Germany), Not specified (field, Belgium) |
EMEM, 30 mM HEPES, 10% FBS, 100 IU penicillin, 100 μg/mL streptomycin | EMEM, 0.3% tryptose phosphate broth, 0.2% yeast extract, 10 μg/mL trypsin | No cytopathic effect or viral protein synthesis detected | (Hofmann and Wyler, 1988) |
| Fetal cells | Porcine primary and secondary cells | V215/78 (field, Germany), Not specified (field, Belgium) |
EMEM, 30 mM HEPES, 10% FBS, 100 IU penicillin, 100 μg/mL streptomycin | EMEM, 0.3% tryptose phosphate broth, 0.2% yeast extract, 10 μg/mL trypsin | Cells damaged by trypsin activity | (Hofmann and Wyler, 1988) |
| Vero 76 | African green monkey kidney cell line (CRL-1587) | PC168, PC170, PC173, PC177, PC179, PC180, PC182, PC21A and PC22A (field, Ohio, Michigan, Illinois and Iowa, US) | DMEM, 5% FBS, 100 unit/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL Fungizone | DMEM, 100 unit/mL penicillin, 100 μg/mL streptomycin, 0.3% tryptose phosphate broth, 10 μg/mL trypsin | Cells rounded by trypsin activity | (Oka et al., 2014) |
| MDBK | Bovine kidney cell line | YN144 (cell-adapted) | DMEM, 10% FBS | DMEM, 10 μg/mL trypsin | No viral protein synthesis detected (immunofluorescent staining) | (Wang et al., 2016) |
| CCL94 | Cat kidney cell line | YN144 (cell-adapted) | DMEM, 10% FBS | DMEM, 10 μg/mL trypsin | No viral protein synthesis detected (immunofluorescent staining) | (Wang et al., 2016) |
| BSR | Hamster kidney cell line | YN144 (cell-adapted) | DMEM, 10% FBS | DMEM, 10 μg/mL trypsin | No viral protein synthesis detected (immunofluorescent staining) | (Wang et al., 2016) |
| MDCK | Canine kidney cell line | YN144 (cell-adapted) | DMEM, 10% FBS | DMEM, 10 μg/mL trypsin | No viral protein synthesis detected (immunofluorescent staining) | (Wang et al., 2016) |
DMEM, Dulbecco’s modified Eagle’s medium; DMEM-F12, Dulbecco’s modified Eagle’s medium –Ham’s F12 mixture; EMEM, Eagle’s minimal essential medium; MEM, Minimal essential medium; FBS, fetal bovine serum; HEPES, N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid.
4. Role of trypsin in PEDV propagation
PEDV propagation in vitro requires considerations beyond selection of an appropriate cell line. While knowledge gained from the study of other coronaviruses, along with trial and error, has helped inform our approaches to PEDV cultivation, the complex factors involved in viral entry into host cells still need to be elucidated before we can develop more robust systems for PEDV cell culture.
One of the key determinants of host cell tropism is the S protein, a 180–220 kDa type I viral glycoprotein that determines viral tropism by mediating host cell entry and membrane fusion. The S protein of the prototype CV777 strain is 1383 amino acids long (Duarte and Laude, 1994), although this length varies among strains as the protein is characterized by both extensive insertion and deletion mutations as well as substitutions.
Typically, type I glycoproteins are expressed as precursor proteins that are activated by proteolytic cleavage into two subunits, the N-terminal and the C-terminal subunits. Proteolytic cleavage is generally programmed by the presence of a cleavage site for host cell proteases such as furin at the interface between the two subunits, as seen with diverse viruses such as influenza virus (Stieneke-Grober et al., 1992), human immunodeficiency virus (HIV) (Hallenberger et al., 1992), and MERS-CoV (Millet and Whittaker, 2014). This cleavage generally releases the fusion peptide, which in the cases of influenza virus and HIV is directly positioned at the N-terminus of the second subunit (Gallaher, 1987, Skehel and Waterfield, 1975). However, coronaviruses appear to have a second cleavage site within S2 that is required for activating the fusion peptide, as first indicated by studies of SARS-CoV S and labeled as S2′ (Belouzard et al., 2009, Madu et al., 2009).
By homology to other type I and coronavirus envelope glycoproteins, the PEDV S protein can be divided into two subunits: the N-terminal S1 and the C-terminal S2. The S1 subunit contains the signal peptide (predicted to be residues 1–20 (Duarte and Laude, 1994)) and receptor binding ectodomain, while the S2 subunit is fusogenic, containing both the fusion peptide and transmembrane domain. However, for many alphacoronaviruses, including PEDV and TGEV, there is no furin-like cleavage site at the predicted interface between S1 and S2 (Duarte and Laude, 1994), raising the possibility that cleavage may occur primarily at the S’ site. Data gleaned from other coronaviruses such as SARS-CoV (Simmons et al., 2005) and MHV (Qiu et al., 2006) suggest that such cleavage may occur during the entry process. Observations from cell culture may not reflect the natural mechanism of PEDV infection, however, as the context during host animal infection is likely different. It is known that coronaviruses tend to be highly species- and tissue-specific, with viral tropism being generally associated with receptor distribution. Enteric tropism, such as that of PEDV, is associated with a trypsin-rich environment, and it is possible that in vivo, PEDV S cleavage is mediated by this less discriminating protease. CV777 isolated from pig intestinal perfusate appeared to present S in cleaved form, with a major 89 kDa band and minor 112 kDa and 129 kDa bands (Egberink et al., 1988). Nevertheless, the exact nature of in vivo cleavage and protease susceptibility remains to be definitively determined.
Given such a context, the first successful cultivation of PEDV was dependent upon the presence of trypsin for continued virus passaging and growth (Hofmann and Wyler, 1988). The Vero-trypsin system is now standard for culturing PEDV in vitro, with regular addition of trypsin required for efficient replication, likely due to trypsin inactivation by Vero cells (Kaverin and Webster, 1995). Robust replication is associated with the formation of extensive syncytia (Hofmann and Wyler, 1988). Trypsin can be functionally replaced by the transmembrane type II serine protease 2 (TMPRSS2), as demonstrated by the propagation of PEDV in Vero cells stably expressing TMPRSS2 (Shirato et al., 2011). This promising approach may allow greater exploration of trypsin-sensitive cell lines for isolating and culturing PEDV.
Interestingly, there have been rare reports of trypsin-independent PEDV strains, such as the highly cell-adapted DR13 (Li et al., 2013, Song et al., 2003) and KPEDV-9 (Kweon et al., 1999, Park et al., 2011), both of which were maintained in Vero cells for over 90 passages. In the absence of trypsin, these viruses do not appear to form syncytia (Li et al., 2013, Park et al., 2014a, Wicht et al., 2014). Similarly, our own experience with the reverse genetics of AVCT12 has revealed that virus rescued in transfected HEK 293T cells in the absence of trypsin are capable of initiating infection of Vero E6-APN without causing syncytia (unpublished observations). These results hint at the possibility that host proteases within the endosome may play a role in cleaving the S protein. Furthermore, the lack of syncytia supports the idea that the S protein is not cleaved during processing and trafficking to the host cell membrane, and is expressed in the inactive form at the cell surface.
Attempts to answer the various questions regarding the nature of PEDV trypsin dependence have been made using reverse genetics as a central tool for hypothesis testing. Recombinant PEDV carrying either S from the trypsin-dependent CV777 strain or the trypsin-independent DR13 were tested for the impact of trypsin on viral entry, with trypsin treatment after receptor binding noticeably increasing relative infection rates of CV777-S virus, and decreasing rates for the DR13-S virus (Wicht et al., 2014). These results suggest that, like SARS-CoV (Matsuyama et al., 2005), PEDV S cleavage by trypsin may occur at the receptor-binding step, possibly due to increased accessibility after receptor binding-induced conformational changes. The significance of receptor binding is supported by similar observations in a study by Park et al. (Park et al., 2011). Trypsin treatment during viral adsorption, but not virus or cell pretreatment, resulted in mild increases in internalized virus and viral titer. This slight change may have been due to the use of KPEDV-9, which has been shown to be trypsin-independent (Kweon et al., 1999, Park et al., 2011, Park et al., 2014a). Interestingly, there is some evidence to suggest that trypsin may play a role during viral egress as well. Electron microscopy of PEDV-infected Vero showed large virion clusters on the cell surface, and increased extracellular viral titers induced by trypsin treatment could be inhibited by leupeptin (Madu et al., 2009).
Reverse genetics-derived PEDV with chimeric S genes have been used to further delineate the nature of trypsin cleavage of PEDV S (Wicht et al., 2014). Trypsin dependence was mapped to the S2 region containing the fusion peptide and first heptad repeat. Of particular interest was the arginine at residue 890, the S’ site directly upstream of the fusion peptide, as arginines or lysines are targets for trypsin digest. The importance of S’ as a cleavage site was underscored when mutation of this region into a furin cleavage site rendered the resultant virus trypsin-independent and capable of replicating to low titers (Li et al., 2015).
For trypsin-independent strains like the cell-adapted DR13, trypsin exposure decreases viral infectivity, strongly indicating changes in S conformation during cell culture adaptation that increase S susceptibility to trypsin and possibly enable cleavage and maturation by host cell proteases (Park et al., 2014a). Cell adaptation may thus result in changes to S structure, with ramifications for S antigenicity in the case of vaccine development, as well as our understanding of the functional structure of S and its mechanism for cellular entry. The ability to preserve natural determinants of entry and replication reinforces the status of reverse genetics as an invaluable tool for studying properties of field isolate-like PEDV.
5. PEDV receptor binding and cell attachment
Despite how integral the elucidation of PEDV S function is to explaining the virus’ tropism and pathogenicity, work focused on direct characterization of the S protein and its receptor usage has been relatively scarce. This is due, in part, to limitations in viral manipulation, although the growing availability of reverse genetics tools is expected to rectify this issue. Thus far, however, we have been relatively reliant upon the homology between PEDV S and that of other coronaviruses to guide our understanding.
Indeed, like TGEV (Delmas et al., 1992, Delmas et al., 1993), PEDV has been shown to utilize pAPN as a receptor for host cell entry (Li et al., 2007, Oh et al., 2003). Initial studies detected a 150 kDa band when probing porcine enterocyte and swine testis (ST) membrane proteins with PEDV in a virus overlay protein binding assay (VOPBA) (Oh et al., 2003). The size of this protein coincided with pAPN, and direct virus–APN binding was found to be dose-dependent and susceptible to competition with anti-pAPN antibodies. Later, expression of a pAPN transgene was shown to enable the non-permissive Madin-Darby canine kidney (MDCK) cell line to support PEDV replication (Li et al., 2007), demonstrating the functional role of APN in PEDV infection. Assessment of pAPN binding to the CHGD-01 strain-derived S1 protein fragments by flow cytometry and ELISA showed strong contribution of the region between amino acids 538–638 in enhancing APN binding. These results suggest that binding function is conferred by the C-terminal domain of S1 (Deng et al., 2016), similar to other coronaviruses such as the APN-binding TGEV and HCoV-229E (Bonavia et al., 2003, Breslin et al., 2003, Godet et al., 1994) and the ACE2-binding HCoV-NL63 and SARS-CoV (Babcock et al., 2004, Lin et al., 2008, Wong et al., 2004). On the other hand, a co-immunoprecipitation study using the strain KNU-0801 by Lee et al. suggested that the N-terminal region of S1 is responsible for mediating pAPN binding, with amino acids at the 25–55 positions resulting in noticeably higher levels of pAPN pulldown (Lee et al., 2011). Nevertheless, the notable differences in S1 fragment expression levels and the lack of comparison with N-terminal truncation fragments leave room for reconciling the results from these two reports. In either case, attempts to characterize S functional domains were limited to binding assays using S1 protein fragments. While these studies provide intriguing insight into the mechanism of S-mediated viral entry, they do not yet demonstrate functional receptor usage by these domains. Manipulation of PEDV S by reverse genetics should enable functional studies in the viral context, supplement our understanding of the binding data, and help account for the differences in experimental results.
Interestingly, while Vero cells are the most common cellular substrate used for isolating or propagating PEDV, the functional receptor has yet to be identified for this cell line. As PEDV appears capable of binding both porcine and human APN (Liu et al., 2015) and APN tissue distribution includes expression on kidney epithelial cells (Dixon et al., 1994), there is a possibility that PEDV may recognize and utilize simian APN. Strangely, VOPBA analysis using the Korean KPEDV-9 strain to probe Vero membrane proteins did not reveal any bands of note, despite the strain being capable of replicating in Vero cells (Oh et al., 2003). Our own work with Vero E6 cells also indicated basal levels of APN hydrolytic activity of leucine p-nitroanilide (Jengarn et al., 2015), although direct comparison with a known APN-negative cell line was not examined.
More recently, however, heparan sulfate has been suggested to act as an attachment factor for PEDV on Vero cells, with ablation of heparan sulfate on the cell surface resulting in reduction of PEDV entry as well as decreased viral titers upon preincubation with heparin (Huan et al., 2015). High affinity for N-acetylneuraminic acid has also been identified by a glycan screen, and treatment of human and bovine mucins with neuraminidase was shown to reduce PEDV S1 binding in a dot blot hybridization assay (Liu et al., 2015). The ability to bind sugars is unsurprising, as this property is shared across viruses in the family Coronaviridae. By homology to other alphacoronaviruses such as TGEV, the sugar binding domain of the PEDV S protein is likely to reside in the N-terminus of the S1 subunit. Indeed, ELISA assessing the binding of S1 fragments to bovine mucin demonstrated stronger sugar binding activity in the N-terminal domain of S1, especially within the first 253 residues (Deng et al., 2016). Whether sugars act as functional receptors for PEDV infection of Vero cells and whether such receptor usage is mediated by the N-terminus of S1 remain to be conclusively demonstrated. Such data will also need to be reconciled with observations of Vero replication by PEDV with massive truncations in the S protein, namely Tottori2/JPN/2014, which has a 194-amino acid deletion from residues 23–213 (by CV777 numbering; Fig. 4 ) and was successfully cultivated in Vero cells (Masuda et al., 2015), and PC177, which has a 197-amino acid deletion from residues 34–227 (CV777 numbering) directly arising from adaptation to Vero cells (Oka et al., 2014).
Fig. 4.
S protein N terminus alignment. N-terminal amino acid sequences of the reference strain CV777 (accession number NP_598310), the S INDEL strain USA/Ohio851 (AHL38184), the N-terminal deletion strain Tottori2/JPN/2014 (BAR92898), and the prototype US virulent strain USA/Colorado/2013 (AGO58924) were aligned using Clustal Omega. Shaded sequences indicate defining insertion and deletion sites. Dashes (−) indicate deletions; asterisks (*), amino acid conservation; colons (:), conservative mutations; dots (.), semi-conservative mutations.
Although work on TGEV S has formed a solid foundation for exploring the function of PEDV S, there are indications that PEDV S has a distinct functional structure. Firstly, the swine testis (ST) cell line, which is known both for its expression of pAPN and permissiveness for TGEV infection, has not been shown to support PEDV replication. Overexpression of pAPN in ST cells, however, resulted in PEDV-associated cytopathic effects and viral spread as shown by staining of PEDV N proteins (Nam and Lee, 2010), suggesting the possible significance of receptor density or other requirements in receptor usage. In contrast, TGEV infection of ST cells overexpressing APN showed decreased production of TGEV virions (Delmas et al., 1995). TGEV S-pseudotyped lentiviruses have also been shown to infect the feline kidney cell line CCL94 (Wang et al., 2016) and the virus uses feline APN as a receptor, unlike PEDV (Tresnan and Holmes, 1998, Tresnan et al., 1996). Given the distinctive properties of PEDV S and the current availability of reverse genetics systems, we expect that more details regarding this protein and its unique functional structure will be forthcoming.
6. PEDV S variants
After the initial isolation of PEDV as the causative agent of a type of porcine diarrhea in Belgium in 1978 (Pensaert and de Bouck, 1978), the virus has been associated with relatively mild outbreaks in Europe and sporadic epidemics in Asia. Emergence of a highly virulent strain in China in 2010 (Sun et al., 2012), however, has been followed by a resurgence in severe PEDV outbreaks, including the first outbreak on the American continent in 2013. Increased surveillance of circulating PEDV strains has resulted in the identification of novel S variants, with most of the variation located in the N-terminus of the S protein. The nature of the evolutionary pressure on this region along with the impact of these variations on viral life cycle and pathogenicity are being gradually elucidated, and widespread application of reverse genetics would further help clarify observations in the field.
Recently sequenced isolates tend to cluster together by both S and whole genome analyses (Lee, 2015), and share a 4-amino acid insertion at residue 58, a 1-amino acid insertion at residue 135 and a 2-amino acid deletion at residue 156 (see USA/Colorado/2013 as the representative sequence, Fig. 4), according to CV777 numbering. These sequences represent the bulk of global epidemic PEDV strains currently detected in circulation.
Among these isolates, however, investigators have also been detecting variants containing a number of insertions and deletions relative to these predominant strains. USA/OH851 was described to show 99% genetic identity at the whole genome level to other strains detected in the United States after the 2013 outbreak, but closer identity to the Chinese CH/HBQX/10 (Zheng et al., 2013) in the S1 region (Wang et al., 2014). This report was followed by isolates from Indiana, Iowa, Minnesota and Ohio (Oka et al., 2014, Vlasova et al., 2014). Closely related strains have also been reported in Europe, namely France (FR/001/2014 to FR/003/2014) (Grasland et al., 2015), Germany (GER/L00719/2014 and GER/L00721/2014) (Hanke et al., 2015), Portugal (Portugal 2015) (Mesquita et al., 2015), Belgium (15V010/BEL/2015) (Theuns et al., 2015) and the Netherlands (NL/GD001/2014, NL/GD002/2014). These strains, currently termed S INDEL strains, have lost the aforementioned insertions and deletions, a throwback to CV777, early isolates and vaccine strains such as SM98-1 and DR13 (see OH851 and CV777 as representative sequences, Fig. 4) despite sequence divergence in the rest of the genome. Survey of the 754 S1 sequences available in the GenBank database indicate greater circulation of strains containing this INDEL pattern than has been noted previously in literature (Table 3 ). These observations are suggestive of recombination events with vaccine strains, and it is certainly curious to note that such recombinants have been preserved in the virus gene pool.
Table 3.
List of strains with S INDEL pattern and their accession numbers in order of collection date.
| Strain | Accession | Collection Date | Collection Location |
|---|---|---|---|
| CV777 | NP_598310 | 1977 | Belgium |
| 83P 5 | BAK32938 | 1982–1983 | Japan |
| CH/S | AEQ55004 | 1986 | China |
| Br1/87 | CAA80971 | 1987 | Great Britain |
| KPEDV-9 | AHJ11040 | 1997 | South Korea |
| Chinju99 | AAN86621 | (2002) | South Korea |
| JS 2004 2 | AAT75298 | 2004 | China |
| DR13 | ABG78322 | (2006) | South Korea |
| LZC | ABM64776 | (2006) | Lanzhou, China |
| LBJ/03 | ABJ51939 | (2006) | Heilongjiang, China |
| DX | ABS72123 | (2007) | Gansu, China |
| HC070225 | AKS26489 | February 2007 | Taiwan |
| virulent DR13 | AFE85962 | 2009 | South Korea |
| Italy/7239/2009 | ALR84908 | January 2009 | Italy |
| MK | AB548624 | (2010) | Japan |
| CH/HBQX/10 | AFV59240 | 2010 | China |
| AVCT12 | BAR73378 | January 2010 | Thailand |
| CH/BJSY/2011 | AFJ97036 | 2011 | China |
| CH/FJND-1/2011 | AER10516 | 2011 | China |
| CH/FJND-2/2011 | AEQ92853 | 2011 | China |
| CH/JL/2011 | AFJ97039 | 2011 | China |
| CH/JLGZL/2011 | AFJ97038 | 2011 | China |
| CH2 | AFL02625 | 2011 | China |
| CH3 | AFL02626 | 2011 | China |
| CH4 | AFL02627 | 2011 | China |
| CH5 | AFL02628 | 2011 | China |
| CH6 | AFL02629 | 2011 | China |
| CH7 | AFL02630 | 2011 | China |
| SC-L | AGU13789 | 2011 | Sichuan, China |
| CH22-JS | AFM55054 | July 2011 | Jiangsu, China |
| Italy/178509/2014 | ALR84952 | July 2011 | Italy |
| Italy/200885/2014 | ALR84953 | July 2011 | Italy |
| HNZZ | AFJ54340 | August 2011 | China |
| CH-HKC-08-2011 | AFR11480 | August 2011 | China |
| CH13-GX | AFM55052 | September 2011 | Guangxi, China |
| CH9-FJ | AFM55051 | September 2011 | Fujian, China |
| AH-M | AHG97559 | October 2011 | Anhui, China |
| CH-STC-12-2011 | ALD19721 | December 2011 | China |
| JS2008 | AGD98685 | (2012) | China |
| CH/FJXM-02/2012 | AFP25201 | 2012 | China |
| CH/AHHF/2012 | AFO42862 | 2012 | China |
| CH/YNKM/2012 | AFO42861 | 2012 | China |
| SD-M | AFX98013 | February 2012 | Shandong, China |
| CH/GD-01/2012 | ALA15706 | March 2012 | Guangdong, China |
| CH/GD-05/2012 | ALA15710 | December 2012 | Guangdong, China |
| CH/GD-06/2012 | ALA15711 | December 2012 | Guangdong, China |
| GDS03 | BAO04590 | 2013 | Guangdong, China |
| GD10 | AKP16759 | 2013 | Guangdong, China |
| GD11 | AKP16760 | 2013 | Guangdong, China |
| GD13 | AKP16762 | 2013 | Guangdong, China |
| KDGN13DJ | AJD09600 | 2013 | South Korea |
| KDJN13_1003SW | AJD09601 | 2013 | South Korea |
| CHM2013 | AJH76957 | 2013 | China |
| CH-GMB-02-2013 | AGO59789 | 2013 | China |
| CH/GD-08/2013 | ALA15733 | January 2013 | Guangdong, China |
| CH/GD-10/2013 | ALA15713 | January 2013 | Guangdong, China |
| CH/GD-15/2013 | ALA15718 | January 2013 | Guangdong, China |
| CH-WTC1-02-2013 | ALD19704 | February 2013 | China |
| CH/GD-11/2013 | ALA15714 | February 2013 | Guangdong, China |
| CH/GD-12/2013 | ALA15715 | February 2013 | Guangdong, China |
| CH-XLC-03-2013 | ALD19703 | March 2013 | China |
| CH/GD-13/2013 | ALA15716 | March 2013 | Guangdong, China |
| USA/Indiana12.83/2013 | AID56667 | June 2013 | Indiana, USA |
| USA/Minnesota52/2013 | AID57081 | June 2013 | Minnesota, USA |
| USA/Iowa23.57/2013 | AID56751 | October 2013 | Iowa, USA |
| USA/Minnesota58/2013 | AID56787 | November 2013 | Minnesota, USA |
| Hawaii/39249/2014 | AKO63237 | November 2013 | Hawaii, USA |
| CH-GLC-2013 | ALV66172 | November 2013 | China |
| CH/GD-17/2013 | ALA15720 | November 2013 | Guangdong, China |
| CH/GD-18/2013 | ALA15721 | December 2013 | Guangdong, China |
| MYZ-1/JPN/2013 | BAT33329 | December 2013 | Miyazaki, Japan |
| USA/Iowa106/2013 | AID57027 | December 2013 | Iowa, USA |
| USA/Iowa107/2013 | AID57033 | December 2013 | Iowa, USA |
| NJ | AIC83850 | (2014) | China |
| ZK_O | BAR73383 | 2014 | Japan |
| GD19 | AKP16768 | 2014 | Guangdong, China |
| GER/L00719/2014 | CDW77213 | 2014 | Germany |
| GER/L00721/2014 | CDW77205 | 2014 | Germany |
| CH/GD-21/2014 | ALA15724 | January 2014 | Guangdong, China |
| CH/GD-22/2014 | ALA15725 | January 2014 | Guangdong, China |
| OH851 | AHL38184 | January 2014 | Ohio, USA |
| USA/Ohio126/2014 | AID57069 | January 2014 | Ohio, USA |
| USA/Minnesota187/2014 | AKJ21701 | January 2014 | Minnesota, USA |
| HUA-PED55 | AJL35174 | January 2014 | Vietnam |
| HUA-PED58 | AJL35175 | January 2014 | Vietnam |
| HUA-PED60 | AJL35176 | January 2014 | Vietnam |
| HUA-PED63 | AJL35177 | January 2014 | Vietnam |
| HUA-PED67 | AJL35178 | January 2014 | Vietnam |
| HUA-PED68 | AJL35179 | January 2014 | Vietnam |
| CH-STNG-2014 | ALV66161 | January 2014 | China |
| KCH-1/JPN/2014 | BAT33317 | March 2014 | Kochi, Japan |
| KCH-2/JPN/2014 | BAT33323 | March 2014 | Kochi, Japan |
| KNU-1406-1 | AIR95864 | March 2014 | South Korea |
| KNU-1406-2 | AIR95870 | March 2014 | South Korea |
| KNU-1406-3 | AIR95871 | March 2014 | South Korea |
| KNU-1406-4 | AIR95872 | March 2014 | South Korea |
| OKY 1/JPN/2014 | BAT33335 | March 2014 | Okayama, Japan |
| SQ2014 | AKP99781 | March 2014 | Jiangsu, China |
| SC1402 | AJW67223 | March 2014 | China |
| CZ2014 | AMB20700 | April 2014 | China |
| EAS1 | AKH45326 | October 2014 | Thailand |
| EAS2 | AKH45332 | October 2014 | Thailand |
| NL/GD001/2014 | AKU46226 | November 2014 | Netherlands |
| CH-SHC-12-2014 | AKN45975 | December 2014 | China |
| CH/GD-26/2014 | ALA15729 | December 2014 | Guangdong, China |
| CH/GD-28/2014 | ALA15731 | December 2014 | Guangdong, China |
| CH/NMG/XLGL | ALJ33145 | December 2014 | China |
| CH/HNYF/14 | ALB08472 | December 2014 | Henan, China |
| FR/001/2014 | AKE47378 | December 2014 | France |
| NL/GD002/2014 | AKU46227 | December 2014 | Netherlands |
| HLJBY | AKJ85723 | (2015) | China |
| 15V010/BEL/2015 | AKC34872 | January 2015 | Belgium |
| CH-HFEC-01-2015 | AKN45968 | January 2015 | China |
| CH-XBC-01-2015 | AKN45979 | January 2015 | China |
| CH-YGC-01-2015 | AKN45980 | January 2015 | China |
| CH-ZWBZa-01-2015 | AKN45982 | January 2015 | China |
| CH-ZWC-01-2015 | AKN45984 | January 2015 | China |
| CH/HNQX-3/14 | ALF39589 | January 2015 | Henan, China |
| CH-JPYC-02-2015 | AKN45971 | February 2015 | China |
| CH/NMG/WLCB | ALJ33144 | February 2015 | China |
| SLO/JH-11/2015 | ALU34112 | September 2015 | Slovenia |
| CH-XDC2-2015 | ALV66166 | March 2015 | China |
Years in parentheses indicate date of sequence submission.
Particularly intriguing is that a number of these recent INDEL strains appear to exhibit decreased virulence relative to circulating non-INDEL strains (Grasland et al., 2015, Lin et al., 2015, Wang et al., 2014, Yamamoto et al., 2015), albeit to differing degrees. Unfortunately, these results are difficult to compare and interpret, due to variations in genomic background, cell culture adaptation, dosage, and animal characteristics. Application of reverse genetics for the generation of these S INDEL mutations would enable analysis of their impact in isogenic viruses under more uniform conditions.
Major deletions in the S gene have also been reported in Korea (MF3809/2008/South Korea) (Park et al., 2014b), Japan (Tottori2/JPN/2014) (Masuda et al., 2015), and the United States (PC177) (Oka et al., 2014). MF3809 is characterized by a 204-amino acid deletion in the C-terminus of S1 (residues 713–916) and exhibited decreasing viral RNA after passaging in Vero cells (Park et al., 2014b). Tottori2, on the other hand, has a 194-amino acid deletion in the N-terminus of S1 and was capable of adapting to Vero culture (Masuda et al., 2015) (Fig. 4). The 197-amino acid deletion in PC177 is, however, understood to have occurred as a result of adaptation to Vero cells, and it is capable of growing to titers similar to non-deletion variants through at least 8 passages (Oka et al., 2014). Such a significant deletion of the S gene has already been observed with TGEV, where a 224-amino acid deletion in the S1 N terminus yielded the antigenically distinct porcine respiratory coronavirus (PRCV) that targets the respiratory tract of swine (Callebaut et al., 1988, Rasschaert et al., 1990). The deleted region encodes the sugar-binding domain (Krempl et al., 1997, Schultze et al., 1996) and is thought to account for the loss of enteric tropism (Ballesteros et al., 1997, Krempl et al., 1997, Laude et al., 1995). It is tempting to speculate that the INDEL mutations and the large S deletions, localized to the N-terminus of S1, may alter receptor or sugar binding domains, affecting efficiency or selectivity of receptor binding and usage. Reverse genetics will be invaluable for fine delineation of the determinants of entry, tropism and pathogenesis encoded in these regions.
7. ORF3 function and cell adaptation
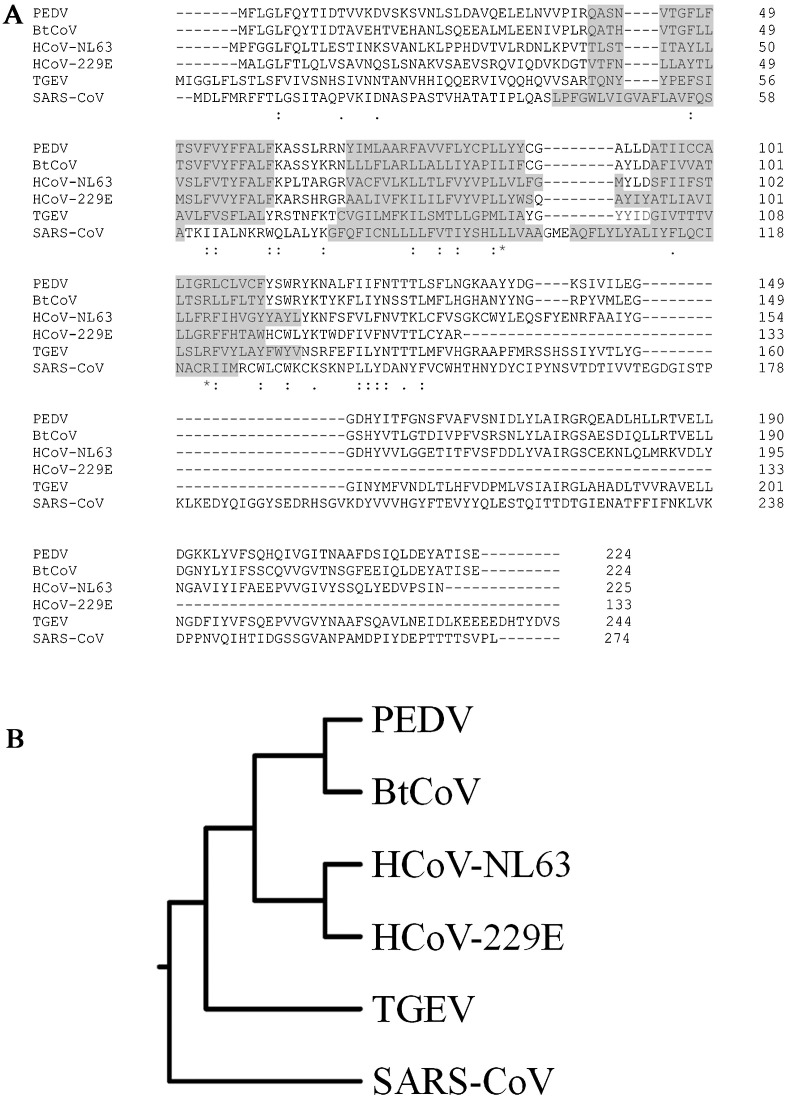
The PEDV ORF3 protein is encoded by subgenomic mRNA located between the S and E genes. It is an accessory protein 224 amino acids in length and is shared between alphacoronaviruses including the bat coronavirus (BtCoV), HCoV-229E, HCoV-NL63 and TGEV (Zeng et al., 2004). PEDV ORF3 is characterized as a transmembrane protein using SwissProt His, comprising at least 3 transmembrane domains (TM): TM1 (Q40–F60), TM2 (Y69–Y88) and TM3 (A95–F111) (ViPR). The alignment of amino acid sequences from different ORF3 proteins and their homologs across the alphacoronavirus genus showed a similar TM pattern (Fig. 5 A) regardless of the low amino acid sequence conservation between them (Fig. 5B).
Fig. 5.
Sequence comparison of ORF3 homologs among coronaviruses. (A) Amino acid alignment of ORF3 and its homologs from PEDV, BtCoV, HCoV-NL63, HCoV-229E, TGEV and SARS-CoV. Shaded sequences indicate transmembrane domains. Dashes (−) indicate deletions; asterisks (*), amino acid conservation; colons (:), conservative mutations; dots (.), semi-conservative mutations. (B) Rooted phylogenetic tree with branch length (UPGMA) of ORF3 and its homologs.
ORF3-like genes of the various coronaviruses are highly variable, with insertions and deletions occasionally resulting in deleted or truncated proteins. Most mutations occur during extensive passaging of the virus in cell culture and some have been correlated with viral attenuation. One study reported attenuation of TGEV virulence after serial passaging in cell culture (Woods, 2001). Sequence analysis of the S gene, which was expected to be the primary determinant of attenuation and cell culture adaptation, revealed mostly identical amino acid sequences between the original and serially passaged viruses. Interestingly, the ORF3 from the serially passaged virus was shorter than that of the virulent original. Similarly, a study comparing attenuated and pathogenic PEDV also found a 51-nucleotide deletion in the ORF3 gene of attenuated vaccine strains, including attenuated DR13, KPEDV-9 and P-5V (Park et al., 2008). In addition, KPEDV-9, which had undergone 93 passages in Vero cells, exhibited reduced pathogenicity in neonatal pigs and was described as safe in pregnant sows (Kweon et al., 1999). Therefore, it has been hypothesized that ORF3 truncation may affect PEDV pathogenesis.
However, not all ORF3 truncations have been associated with reduced pathogenicity. Recently, analysis was performed on the ORF3 of 27 field samples collected from different farms in Fujian, China, between 2010 and 2012 (Chen et al., 2013). These ORF3 exhibited notable heterogeneity and could be divided into 2 groups. Group 1 was composed of 26 viruses with full-length ORF3 (224 amino acids in length) and group 2 had the one strain (P55) with an ORF3 truncation (92 amino acids in length) shared with four selected ORF3 reference sequences (truncated-ORF3 CV777, CH/GSJIII/07, truncated-ORF3 CH/BJ/2011, truncated-ORF3 DBI865, and Zhejiang-08). Despite the truncated ORF3, the virulence of P55 did not differ from that of the isolates in group 1. A different study involved the YN1 field strain, whose ORF3 gene was found to encode an early stop codon at position 145 after serial propagation in Vero cells for 200 passages (Chen et al., 2015). Full-length sequences at passages 15, 30, 60, 90, 144, and 200 had been analyzed, with no changes observed for the NSP2, NSP4–7, NSP10, NSP12 and NSP13 genes during the Vero cell adaptation process. Identified mutations included 9–26 amino acid changes in ORF 1a/b and S, an early termination codon in ORF3, and 1–3 amino acid changes in the E, M and N proteins. The passage 15 virus YN15 had 8 amino acid changes in the ORF3 gene and had already gained the early termination codon. When field-isolated YN1 and cell-adapted YN15 were characterized for pathogenicity in piglets, infected piglets from both groups showed classic and similar signs of infection, with watery diarrhea and emaciation. Therefore, acquisition of a truncated ORF3 by YN1 progeny appears to be more important for cell adaptation than attenuation.
Truncation of ORF3 hints at its dispensability in cell culture. This has been confirmed by reverse genetics, where a recombinant PEDV based on attenuated DR13 (which encodes an ORF3 with a 17-amino acid deletion) was successfully rescued by targeted RNA recombination (Li et al., 2013). Likewise, our own study generated reverse genetics-derived AVCT12, which contains a C-terminal deletion of the S gene resulting in disruption of the ORF3 start codon and absence of the ORF3 protein (Jengarn et al., 2015). While this virus grew to the same titers as the original virus, reverse genetics virus with a restored ORF3 gene could not be rescued. These results suggest that ORF3 may negatively affect viral growth in cell culture. On the other hand, an infectious molecular clone was recently generated based on the virulent PEDV strain PC22A, which possesses an intact ORF3 gene (Beall et al., 2016). And while disruption of the ORF3 with red fluorescent protein did not increase viral titer, both wild-type and ORF3-deleted recombinant viruses replicated to titers around 2–3 log less than recombinant AVCT12. Interestingly, regardless of the presence of ORF3, all inoculated piglets succumbed to illness or were euthanized due to illness at their final time points. The notable differences between these two studies are likely due to viral genetic background—our reverse genetics report focused on the use of an SM98 vaccine-like strain, which may have other adaptations that render ORF3 dispensable, and even detrimental.
The correlation between mutations in the ORF3 gene and cell adaptation of field isolates after cell culture has raised many questions about its function. Based on the relative ORF3 conservation among coronaviruses (Tang et al., 2006), it may be possible to glean clues from other coronaviruses. Table 4 lists the various ORF3 homologs and their known characteristics. The more extensively studied SARS-CoV ORF3a appears to have multiple functions, including ion channel activity (Lu et al., 2006) and induction of apoptosis (Chan et al., 2009). With structural studies revealing the tetramerization and membrane localization possibly indicative of ion channel properties (Lu et al., 2006), ORF3a was tested for ion channel function by injection of ORF3a RNA into Xenopus oocytes. The results suggested that ORF3a forms a potassium-sensitive channel. In a separate study, FRhk-4 cells transfected with siRNA targeting ORF3a prior to infection with SARS-CoV showed a significant decrease in viral release (Chan et al., 2009). Furthermore, mutations interfering with potassium channel activity abolished caspase-dependent apoptosis. Therefore, the ion channel activity of ORF3a may affect SARS-CoV release and its pro-apoptotic properties.
Table 4.
Characteristics of ORF3 homologs in coronaviruses.
| CoV | ORF | Length (aa) |
Function | Subcellular localization |
|---|---|---|---|---|
| SARS | ORF3a | 274 | Ion channel (Lu et al., 2006) and pro apoptotic (Chan et al., 2009) activities | Cytoplasm, plasma membrane (Tan et al., 2004) and Golgi apparatus (Yu et al., 2004) |
| TGEV | ORF3b | 244 | Viral pathogenicity (Wesley and Woods, 2001) | Unknown |
| PEDV | ORF3 | 224 | Ion channel activity (Wang et al., 2012), prolongs S phase and facilitates vesicle formation (Ye et al., 2015) | Cytoplasm and punctuate vesicle-like structures in the cytoplasm (Ye et al., 2015) and unpublished data) |
| HCoV-229E | ORF4a | 133 | Ion channel activity and acts as viroporin (Zhang et al., 2014) | Endoplasmic reticulum/Golgi intermediate compartment (Zhang et al., 2014) |
| HCoV-NL63 | ORF3 | 225 | Structural viral protein (Muller et al., 2010) | Endoplasmic reticulum/Golgi intermediate compartment (Muller et al., 2010) |
A computational model using five different programs predicted four transmembrane regions for PEDV ORF3 (TM1 at Q40–S63, TM2 at R75–I97, TM3 at Y116–Y139, and TM4 at G150–I173), with ORF3 assembling to form a homotetrameric protein complex with a central pore (Wang et al., 2012). This model offers a structural prediction distinct from that predicted by SwissProt His, and this discrepancy will need to be resolved by structural studies in the future. Nevertheless, it is suggestive that PEDV ORF3 may also possess ion channel function.
The first attempt to directly study PEDV ORF3 function involved expression of ORF3 from parental and cell-adapted CV777 in E. coli as a heterodimeric fusion protein (Schmitz et al., 1998). The full-length parental ORF3 was expressed poorly compared to the truncated form from the cell-adapted strain, which the authors suggested may be indicative of a degree of functional toxicity. Expression of CV777 ORF3 in Xenopus laevis and Saccharomyces cerevisiae, however, did reveal potassium ion channel function, along with the importance of residue Y170 for such activity (Wang et al., 2012). In comparison, truncated ORF3 from cell-adapted CV777 lacked this ion channel activity. Virus production was additionally observed to be reduced after infection when cells were treated with siRNA to silence the ORF3 gene, suggesting that ORF3 may affect viral replication like SARS-CoV ORF3a. A more recent study used a Vero cell line stably expressing ORF3 (Vero-ORF3) to assess the protein’s impact on the host cell (Ye et al., 2015). Aside from its cytoplasmic localization, the observed prolongation of the cell cycle S phase and the appearance of large double-membrane vesicles possibly acting as sites of viral replication (Gosert et al., 2002, Pedersen et al., 1999, Snijder et al., 2006) together suggest that ORF3 may help promote viral growth. Indeed, ORF3 increased viral titers of attenuated viruses, although it had no effect on the non-attenuated field isolate, indicating that ORF3 can play a role in viral replication in vitro.
The data so far are conflicting regarding the role of ORF3 and its importance for viral replication and pathogenicity both in vivo and in vitro. This is unsurprising given its nature as an accessory protein, whose role and importance can vary depending upon virus and host cell contexts. It will be essential to accumulate a larger pool of data, and the application of reverse genetics will help control variation within each study, as well as enable analysis of the impact of other viral proteins in conjugation with ORF3 in mediating PEDV adaptation and pathogenicity.
8. N and PEDV pathogenicity
N proteins of coronaviruses carry out multiple functions during the viral life cycle. The most abundant structural protein in virions, the primary function of N is to organize the viral genome, relying on its RNA binding and self-multimerization properties. Additional functions include virus assembly, cell cycle regulation, apoptosis induction, host stress response, translational shutoff and immune system interference (McBride et al., 2014). N has therefore been an attractive target for antiviral research, especially for coronaviruses that are human pathogens such as SARS-CoV and MERS-CoV.
Following the resurgence in severe PEDV epidemics, research on PEDV N has started to gain momentum. Studies on N have focused on its possible roles in PEDV pathogenesis, regulation of host cell environment and immune evasion. Using CV777 infection in porcine intestinal epithelial cells (IECs), Cao et al. demonstrated that active PEDV replication could induce gene expression under the NF-κB promoter in a dose-dependent manner (Cao et al., 2015c). With co-transfection experiments, they identified N, especially the immuno-dominant central region, as responsible for PEDV-dependent NF-κB activation through Toll-like receptors 2, 3 and 9. This is in line with a previous report that N from a PEDV strain isolated in Shaanxi province could activate NF-κB (Xu et al., 2013). In IECs stably transfected with a pGFP-N expression vector, upregulation of NF-κB, along with an endoplasmic reticulum (ER) chaperone GRP78 and IL-8, was observed compared to IECs expressing GFP alone. In contrast, in transfected HEK 293T cells, N derived from strain AJ 1102 inhibited promoter activation of NF-κB, IFN-β and IRF3 (Ding et al., 2014). This study also observed inhibition of Sendai virus-induced IFN-β production and IFN-stimulated gene expression, suggesting a role for N as an interferon antagonist. The authors further demonstrated that N interacts with TBK1 and in turn inhibits TBK1 phosphorylation of IRF3.
Even cellular localization of N is still debatable. Xu et al. suggested that N could localize to the ER, leading to ER stress and cell cycle arrest (Xu et al., 2013). They specifically stated that, in their experimental setup of IECs stably expressing N fused with a C-terminal EGFP, they did not observe N in the nucleolus of the cells. However, using CV777 infection and expression of an N-GFP fusion protein in Vero E6 cells, Shi et al. observed predominant cytoplasmic localization of PEDV N, with a certain fraction in the nucleolus (Shi et al., 2014). They further identified the nucleolar localization and nuclear export signals of N, although the importance of nucleocytoplasmic shuttling of N was not clear.
Together, these results present a likely incomplete and complex story for the roles of N in PEDV pathogenesis and host cell manipulation. These experiments were conducted with different PEDV strains and in various cell lines, possibly giving rise to discrepancies. Another important aspect is the lack of information on how PEDV infection or the viral life cycle is affected should any of these proposed functions of N be altered. Although they can help delineate the role of N in PEDV pathogenicity and identify important molecular features, experiments in the context of transfection should be complemented with rigorous tests for physiological relevance in the context of PEDV infection. For this purpose, reverse genetics platforms for PEDV where mutations of N could be easily introduced are useful tools for addressing these issues.
The reverse genetics platform will also allow further exploration into other biochemical curiosities present in N. For example, N contains three intrinsic disorder regions (IDRs) flanking its structural portions. Unlike other coronaviruses whose IDRs comprise serines and arginines (SR-rich; (Hurst et al., 2009)), IDRs in PEDV N consist of several repeats of asparagines (N-rich). Reports have suggested that the SR-rich IDRs participate in N oligomerization, offer additional RNA binding sites, and allow enough flexibility for the arrangement of viral ribonucleoprotein complex structures (Chang et al., 2009, Luo et al., 2005). Nevertheless, it is unknown whether PEDV’s N-rich IDRs would behave similarly. Experiments with reverse genetics-derived PEDV variants carrying N mutants with IDRs of varying length and amino acid composition could provide very useful insights into virus fitness, replication cycle and virulence.
9. Challenges in PEDV vaccine design
Given the recurrent worldwide outbreaks of PEDV since its first discovery, a number of PEDV vaccines are now commercially available. Vaccine companies in Korea (Komipharm and Daesung), Japan (Nisseiken and Kaketsuken) and China produce PEDV vaccines for the Asian market. And soon after PEDV was introduced into the United States, two commercial vaccines were rapidly developed and granted conditional USDA license to contain the epidemic. The first vaccine developed by Harrisvaccine and Merck, iPED+, uses alphavirus replicon vaccine technology (Vander Veen et al., 2012). The other, marketed by Zoetis, is based on the inactivated/killed virus platform.
Most, if not all, PEDV vaccines are designed for parenteral immunization, which may provoke skepticism among users regarding the efficacy of these vaccines against enteric pathogens like PEDV. It is commonly known that newborn piglets, though immunocompetent, are usually unable to mount primary immune responses in time to prevent disease onset, especially those associated with acute viral infection. They heavily rely on maternal passive immunity obtained from colostrum and milk, as the placentally transferred antibodies during gestation are not sufficient to confer disease protection (Kim et al., 1966, Tlaskalova-Hogenova et al., 1994). Given that PEDV infects its hosts primarily by directly invading enterocytes lining the intestinal villi and the virus does not require systemic spread, it is likely that mucosal immunity, particularly secretory IgA, is responsible for virus neutralization and disease protection as opposed to the IgG predominantly found in sera of sows vaccinated via the parenteral route. Although PEDV-specific IgG may be present in high amounts in colostrum at parturition in sows intramuscularly vaccinated by PEDV live vaccines (Paudel et al., 2014), protection duration tends to be very short due to rapid decline of colostrum production in post-parturient sows (Klobasa et al., 1987).
Several lines of evidence support the notion that oral immunization of pregnant sows with live PEDV is essential for stimulating adequate lactogenic immunity for piglet protection. It has been shown recently that sows orally fed with intestinal homogenates obtained from clinically affected piglets exhibit significantly higher levels of milk anti-PEDV neutralizing antibodies compared to those intramuscularly immunized by the iPED+ vaccine (Scherba et al., 2016). Likewise, Paudel et al. showed that oral administration of live attenuated PEDV strain DR-13 to pregnant sows reduces the mortality rate of nursing piglets (Paudel et al., 2014). A different study demonstrated that sows previously exposed to a mild strain of PEDV can passively transfer sufficient lactogenic immunity to piglets for protection against virulent PEDV exposure (Goede et al., 2015). And, in line with studies in PEDV-infected sows, pregnant sows fed with TGEV also exhibit substantially elevated levels of IgA antibodies in milk that confer passive protection against virulent TGEV to piglets (Bohl et al., 1972, Bohl and Saif, 1975, Chattha et al., 2015). In contrast, sows vaccinated with recombinant subunit vaccines based on the TGEV S protein provide inadequate protective immunity against similar challenge (Park et al., 1998).
Efficacy of live oral vaccines depends largely on the ability of PEDV to infect and replicate in swine enterocytes. Accumulating data seem to suggest that adaptation of PEDV to cell culture affects the efficacy of viruses used as oral vaccines. For instance, sows orally inoculated with parental DR13 displayed higher immune responses when compared to those fed with virus cultured in Vero cells to passages 75 or 90 (Song et al., 2003). Likewise, Chen et al. recently performed virulence experiments by orally inoculating nursing piglets with either a low- (YN15) or high-passage number (YN144) variant of the virulent YN1 strain (Chen et al., 2015). They demonstrated that, while all YN15-infected piglets succumbed to severe diarrhea, those infected with YN144 showed no detectable clinical signs. Immunohistochemical analyses of intestinal samples from YN15- and YN144-infected piglets clearly indicated that YN144 barely infected swine enterocytes. Notably, results from others and our laboratory also suggest that cell-adapted PEDV strains infect and replicate in swine enterocytes less efficiently than in Vero cells (Cong et al., 2015). While it is currently not known exactly how adaptation in cells of non-swine origin can affect the ability of PEDV to initiate infection in swine cells, it is clear that in vivo infection of swine enterocytes is a prerequisite for an effective PEDV vaccine.
To develop such effective PEDV vaccines, not only should the seed vaccine maintain high growth in vitro, but virus adaptation in host cells must also be minimized without compromising attenuation. With these considerations in mind, the reverse genetics systems of PEDV available to-date offer great potential for novel vaccine development. With more insight into differences between field-isolated and cell-adapted variants, it will be possible to genetically manipulate the infectious clone of a high-growth PEDV to switch its tropism preference from monkey (Vero) to swine cells. Investigation of Vero and swine enterocyte entry by recombinant viruses may help identify key residues involved in cell adaptation, which should in turn provide insightful information for augmenting entry of cell-adapted PEDV into intestinal cells. Particularly encouraging is the ease in changing viral tropism by generating the reverse genetics-derived coronavirus bearing chimeric or mutant S genes (Kuo et al., 2000, Menachery et al., 2015, Menachery et al., 2016, Phillips et al., 1999), with Phillips et al. analyzing changes in MHV neurotropism conferred by the S of mildly versus highly neurovirulent strains (Phillips et al., 1999). Similarly, PEDV with chimeric S can be used to identify the determinants of viral tropism at the nucleotide level, paving the way towards the engineering of S to harbor domains necessary for enterocyte entry while maintaining the necessary antigenic sites for eliciting humoral protective immunity.
While it is not clearly elucidated what genetic components of cell-adapted PEDV are responsible for its attenuation, changing tropism to favor swine intestinal cells may affect virulence. If so, based on the well-characterized SARS-CoV candidate vaccines (Netland et al., 2010, Regla-Nava et al., 2015), a number of approaches could be employed to attenuate a potential PEDV vaccine. The 2′O-methyl-transferase (2′O-MTase), NSP16, has been shown to be an attractive target for attenuation of many coronaviruses (Menachery et al., 2014). Though not yet studied in detail, genetic modification of PEDV NSP16 may provide a useful tool for virus attenuation. SARS-CoV with a disrupted or mutated E gene has also been generated and was shown to be highly attenuated in vivo (Netland et al., 2010, Regla-Nava et al., 2015). While it is possible for SARS-CoV with a deleted E gene to regain virulence upon serial passaging, further manipulation of the virus genome such as truncating the NSP1 protein can help maintain the attenuated phenotype of the virus (Jimenez-Guardeno et al., 2015). Indeed, the coronavirus NSP1 has been proposed to play a major role in determining the virulence of many coronaviruses, primarily by interfering with the host innate immune response (Zust et al., 2007). A recent study showing that PEDV NSP1 can disrupt type I interferon signaling by degrading the CREB-binding protein (CBP) (Zhang et al., 2016) supports the possibility of modifying this protein for PEDV attenuation.
10. Concluding remarks
With the advent of reverse genetics systems for the genetic manipulation of PEDV genomes, these technologies promise to revolutionize PEDV research. In this review, we have described PEDV reverse genetic systems as well as past and current applications of this technology towards bettering our understanding of PEDV. Furthermore, we have discussed the many unanswered questions that we deem critical for advancing our understanding of this economically important pathogen, and how reverse genetics may be the tool to provide the much-needed answers. The integration of advances in reverse genetics approaches, animal testing, and novel engineered host cells will be critical for fostering the development of next-generation vaccines to tackle future PEDV and possibly other coronavirus outbreaks.
Acknowledgements
We thank the National Science and Technology Agency, Thailand, and the Betagro Science Center for co-funding (P-11-00087, P-12-01765, P-14-50863) our reverse genetics work described in this review. Ongoing work is supported by the BIOTEC Fellows’ Grant (P-15-51261). We are also grateful to Dr Nanchaya Wanasen for critical reading of the manuscript.
References
- Agnihothram S., Yount B.L., Jr., Donaldson E.F., Huynh J., Menachery V.D., Gralinski L.E., Graham R.L., Becker M.M., Tomar S., Scobey T.D., Osswald H.L., Whitmore A., Gopal R., Ghosh A.K., Mesecar A., Zambon M., Heise M., Denison M.R., Baric R.S. A mouse model for Betacoronavirus subgroup 2c using a bat coronavirus strain HKU5 variant. MBio. 2014;5(2) doi: 10.1128/mBio.00047-14. e00047-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., Gonzalez J.M., Penzes Z., Izeta A., Calvo E., Plana-Duran J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U. S. A. 2000;97(10):5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., Dediego M.L., Galan C., Escors D., Alvarez E., Ortego J., Sola I., Zuniga S., Alonso S., Moreno J.L., Nogales A., Capiscol C., Enjuanes L. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006;80(21):10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., DeDiego M.L., Sola I., Zuniga S., Nieto-Torres J.L., Marquez-Jurado S., Andres G., Enjuanes L. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. MBio. 2013;4(5):e00650–13. doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambrosino D.M. Amino acids 270–510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78(9):4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint A., Farsang A., Zadori Z., Hornyak A., Dencso L., Almazan F., Enjuanes L., Belak S. Molecular characterization of feline infectious peritonitis virus strain DF-2 and studies of the role of ORF3abc in viral cell tropism. J. Virol. 2012;86(11):6258–6267. doi: 10.1128/JVI.00189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros M.L., Sanchez C.M., Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227(2):378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. MBio. 2016;7(1) doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., Pickles R.J., Corti D., Johnston R.E., Baric R.S., Denison M.R. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. U. S. A. 2008;105(50):19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U .S. A. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl E.H., Saif L.J. Passive immunity in transmissible gastroenteritis of swine: immunoglobulin characteristics of antibodies in milk after inoculating virus by different routes. Infect. Immun. 1975;11(1):23–32. doi: 10.1128/iai.11.1.23-32.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl E.H., Gupta R.K., Olquin M.V., Saif L.J. Antibody responses in serum, colostrum, and milk of swine after infection or vaccination with transmissible gastroenteritis virus. Infect. Immun. 1972;6(3):289–301. doi: 10.1128/iai.6.3.289-301.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavia A., Zelus B.D., Wentworth D.E., Talbot P.J., Holmes K.V. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 2003;77(4):2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin J.J., Mork I., Smith M.K., Vogel L.K., Hemmila E.M., Bonavia A., Talbot P.J., Sjostrom H., Noren O., Holmes K.V. Human coronavirus 229E: receptor binding domain and neutralization by soluble receptor at 37 °C. J. Virol. 2003;77(7):4435–4438. doi: 10.1128/JVI.77.7.4435-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut P., Correa I., Pensaert M., Jimenez G., Enjuanes L. Antigenic differentiation between transmissible gastroenteritis virus of swine and a related porcine respiratory coronavirus. J. Gen. Virol. 1988;69(Pt. 7):1725–1730. doi: 10.1099/0022-1317-69-7-1725. [DOI] [PubMed] [Google Scholar]
- Cao L., Ge X., Gao Y., Herrler G., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-beta production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 2015;12:127. doi: 10.1186/s12985-015-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Ge X., Gao Y., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus infection induces NF-kappaB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 2015;96(Pt. 7):1757–1767. doi: 10.1099/vir.0.000133. [DOI] [PubMed] [Google Scholar]
- Cao L., Ge X., Gao Y., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus infection induces NF-kappaB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 2015;96(7):1757–1767. doi: 10.1099/vir.0.000133. [DOI] [PubMed] [Google Scholar]
- Casais R., Thiel V., Siddell S.G., Cavanagh D., Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 2001;75(24):12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.M., Tsoi H., Chan W.M., Zhai S., Wong C.O., Yao X., Chan W.Y., Tsui S.K., Chan H.Y. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41(11):2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83(5):2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattha K.S., Roth J.A., Saif L.J. Strategies for design and application of enteric viral vaccines. Annu. Rev. Anim. Biosci. 2015;3:375–395. doi: 10.1146/annurev-animal-022114-111038. [DOI] [PubMed] [Google Scholar]
- Chen X., Zeng L., Yang J., Yu F., Ge J., Guo Q., Gao X., Song T. Sequence heterogeneity of the ORF3 gene of porcine epidemic diarrhea viruses field samples in Fujian, China, 2010–2012. Viruses. 2013;5(10):2375–2383. doi: 10.3390/v5102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014;52(1):234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhu Y., Wu M., Ku X., Ye S., Li Z., Guo X., He Q. Comparative genomic analysis of classical and variant virulent parental/attenuated strains of porcine epidemic diarrhea virus. Viruses. 2015;7(10):5525–5538. doi: 10.3390/v7102891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley S.E., Lavi E., Sawicki S.G., Fu L., Schelle B., Karl N., Siddell S.G., Thiel V. Recombinant mouse hepatitis virus strain A59 from cloned, full-length cDNA replicates to high titers in vitro and is fully pathogenic in vivo. J. Virol. 2005;79(5):3097–3106. doi: 10.1128/JVI.79.5.3097-3106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Li X., Bai Y., Lv X., Herrler G., Enjuanes L., Zhou X., Qu B., Meng F., Cong C., Ren X., Li G. Porcine aminopeptidase N mediated polarized infection by porcine epidemic diarrhea virus in target cells. Virology. 2015;478:1–8. doi: 10.1016/j.virol.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74(11):4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L'Haridon R., Vogel L.K., Sjostrom H., Noren O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357(6377):417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Sjostrom H., Noren O., Laude H. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 1993;342:293–298. doi: 10.1007/978-1-4615-2996-5_45. [DOI] [PubMed] [Google Scholar]
- Delmas B., Kut E., Gelfi J., Laude H. Overexpression of TGEV cell receptor impairs the production of virus particles. Adv. Exp. Med. Biol. 1995;380:379–385. doi: 10.1007/978-1-4615-1899-0_62. [DOI] [PubMed] [Google Scholar]
- Deng F., Ye G., Liu Q., Navid M.T., Zhong X., Li Y., Wan C., Xiao S., He Q., Fu Z.F., Peng G. Identification and comparison of receptor binding characteristics of the spike protein of two porcine epidemic diarrhea virus strains. Viruses. 2016;8(3) doi: 10.3390/v8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88(16):8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J., Kaklamanis L., Turley H., Hickson I.D., Leek R.D., Harris A.L., Gatter K.C. Expression of aminopeptidase-n (CD 13) in normal tissues and malignant neoplasms of epithelial and lymphoid origin. J. Clin. Pathol. 1994;47(1):43–47. doi: 10.1136/jcp.47.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch P.W. Translesion synthesis by RNA polymerases: occurrence and biological implications for transcriptional mutagenesis. Mutat. Res. 2002;510(1–2):131–140. doi: 10.1016/s0027-5107(02)00258-0. [DOI] [PubMed] [Google Scholar]
- Donaldson E.F., Yount B., Sims A.C., Burkett S., Pickles R.J., Baric R.S. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J. Virol. 2008;82(23):11948–11957. doi: 10.1128/JVI.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte M., Laude H. Sequence of the spike protein of the porcine epidemic diarrhoea virus. J. Gen. Virol. 1994;75(Pt. 5):1195–1200. doi: 10.1099/0022-1317-75-5-1195. [DOI] [PubMed] [Google Scholar]
- Egberink H.F., Ederveen J., Callebaut P., Horzinek M.C. Characterization of the structural proteins of porcine epizootic diarrhea virus, strain CV777. Am. J. Vet. Res. 1988;49(8):1320–1324. [PubMed] [Google Scholar]
- Fang S., Chen B., Tay F.P., Ng B.S., Liu D.X. An arginine-to-proline mutation in a domain with undefined functions within the helicase protein (Nsp13) is lethal to the coronavirus infectious bronchitis virus in cultured cells. Virology. 2007;358(1):136–147. doi: 10.1016/j.virol.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89(3):1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher W.R. Detection of a fusion peptide sequence in the transmembrane protein of human immunodeficiency virus. Cell. 1987;50(3):327–328. doi: 10.1016/0092-8674(87)90485-5. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68(12):8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goede D., Murtaugh M.P., Nerem J., Yeske P., Rossow K., Morrison R. Previous infection of sows with a mild strain of porcine epidemic diarrhea virus confers protection against infection with a severe strain. Vet. Microbiol. 2015;176(1–2):161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 2002;76(8):3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasland B., Bigault L., Bernard C., Quenault H., Toulouse O., Fablet C., Rose N., Touzain F., Blanchard Y. Complete genome sequence of a porcine epidemic diarrhea s gene indel strain isolated in france in december 2014. Genome Announc. 2015;3(3) doi: 10.1128/genomeA.00535-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77(8):4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenberger S., Bosch V., Angliker H., Shaw E., Klenk H.D., Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360(6402):358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- Hanke D., Jenckel M., Petrov A., Ritzmann M., Stadler J., Akimkin V., Blome S., Pohlmann A., Schirrmeier H., Beer M., Hoper D. Comparison of porcine epidemic diarrhea viruses from Germany and the United States, 2014. Emerg. Infect. Dis. 2015;21(3):493–496. doi: 10.3201/eid2103.141165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26(11):2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan C.C., Wang Y., Ni B., Wang R., Huang L., Ren X.F., Tong G.Z., Ding C., Fan H.J., Mao X. Porcine epidemic diarrhea virus uses cell-surface heparan sulfate as an attachment factor. Arch. Virol. 2015;160(7):1621–1628. doi: 10.1007/s00705-015-2408-0. [DOI] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009;83(14):7221–7234. doi: 10.1128/JVI.00440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jengarn J., Wongthida P., Wanasen N., Frantz P.N., Wanitchang A., Jongkaewwattana A. Genetic manipulation of porcine epidemic diarrhoea virus recovered from a full-length infectious cDNA clone. J. Gen. Virol. 2015;96(8):2206–2218. doi: 10.1099/vir.0.000184. [DOI] [PubMed] [Google Scholar]
- Jimenez-Guardeno J.M., Regla-Nava J.A., Nieto-Torres J.L., DeDiego M.L., Castano-Rodriguez C., Fernandez-Delgado R., Perlman S., Enjuanes L. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for the design of a genetically stable vaccine. PLoS Pathog. 2015;11(10):e1005215. doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoi K., Sugioka H., Satoh T., Kadoi B.K. The propagation of a porcine epidemic diarrhea virus in swine cell lines. New Microbiol. 2002;25(3):285–290. [PubMed] [Google Scholar]
- Kaverin N.V., Webster R.G. Impairment of multicycle influenza virus growth in Vero (WHO) cells by loss of trypsin activity. J. Virol. 1995;69(4):2700–2703. doi: 10.1128/jvi.69.4.2700-2703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri M. Porcine epidemic diarrhea virus replication in duck intestinal cell line. Emerg. Infect. Dis. 2015;21(3):549–550. doi: 10.3201/eid2103.141658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.B., Bradley S.G., Watson D.W. Ontogeny of the immune response: i. Development of immunoglobulins in germfree and conventional colostrum-deprived piglets. J. Immunol. 1966;97(1):52–63. [PubMed] [Google Scholar]
- Klobasa F., Werhahn E., Butler J.E. Composition of sow milk during lactation. J. Anim. Sci. 1987;64(5):1458–1466. doi: 10.2527/jas1987.6451458x. [DOI] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23(2):137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetzner C.A., Parker M.M., Ricard C.S., Sturman L.S., Masters P.S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 1992;66(4):1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C., Schultze B., Laude H., Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997;71(4):3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76(10):4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74(3):1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon C.H., Kwon B.J., Lee J.G., Kwon G.O., Kang Y.B. Derivation of attenuated porcine epidemic diarrhea virus (PEDV) as vaccine candidate. Vaccine. 1999;17(20–21):2546–2553. doi: 10.1016/S0264-410X(99)00059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Baric R.S., Makino S., Keck J.G., Egbert J., Leibowitz J.L., Stohlman S.A. Recombination between nonsegmented RNA genomes of murine coronaviruses. J. Virol. 1985;56(2):449–456. doi: 10.1128/jvi.56.2.449-456.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M.C. Recombination in large RNA viruses: coronaviruses. Semin. Virol. 1996;7:381–388. doi: 10.1006/smvy.1996.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Godet M., Bernard S., Gelfi J., Duarte M., Delmas B. Functional domains in the spike protein of transmissible gastroenteritis virus. Adv. Exp. Med. Biol. 1995;380:299–304. doi: 10.1007/978-1-4615-1899-0_48. [DOI] [PubMed] [Google Scholar]
- Lee D.-K., Cha S.-Y., Lee C. The N-terminal region of the porcine epidemic diarrhea virus spike protein is important for the receptor binding. Korean J. Microbiol. Biotechnol. 2011;39(2):140–145. [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12(1):193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.X., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365(1):166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li Z., Zou Y., Wicht O., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS One. 2013;8(8):e69997. doi: 10.1371/journal.pone.0069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wicht O., van Kuppeveld F.J., He Q., Rottier P.J., Bosch B.J. A single point mutation creating a furin cleavage site in the spike protein renders porcine epidemic diarrhea coronavirus trypsin independent for cell entry and fusion. J. Virol. 2015;89(15):8077–8081. doi: 10.1128/JVI.00356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.L., Lai M.M. RNA recombination in a coronavirus: recombination between viral genomic RNA and transfected RNA fragments. J. Virol. 1992;66(10):6117–6124. doi: 10.1128/jvi.66.10.6117-6124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.X., Feng Y., Wong G., Wang L., Li B., Zhao X., Li Y., Smaill F., Zhang C. Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD-ACE2 receptor interaction. J. Gen. Virol. 2008;89(Pt. 4):1015–1024. doi: 10.1099/vir.0.83331-0. [DOI] [PubMed] [Google Scholar]
- Lin C.M., Annamalai T., Liu X., Gao X., Lu Z., El-Tholoth M., Hu H., Saif L.J., Wang Q. Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet. Res. 2015;46:134. doi: 10.1186/s13567-015-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015;89(11):6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U. S. A. 2006;103(33):12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Ye F., Chen K., Shen X., Jiang H. SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization. Biochemistry. 2005;44(46):15351–15358. doi: 10.1021/bi051122c. [DOI] [PubMed] [Google Scholar]
- Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83(15):7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Keck J.G., Stohlman S.A., Lai M.M. High-frequency RNA recombination of murine coronaviruses. J. Virol. 1986;57(3):729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T., Murakami S., Takahashi O., Miyazaki A., Ohashi S., Yamasato H., Suzuki T. New porcine epidemic diarrhoea virus variant with a large deletion in the spike gene identified in domestic pigs. Arch. Virol. 2015;160(10):2565–2568. doi: 10.1007/s00705-015-2522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. U. S. A. 2005;102(35):12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Debbink K., Baric R.S. Coronavirus non-structural protein 16: evasion, attenuation, and possible treatments. Virus Res. 2014;194:191–199. doi: 10.1016/j.virusres.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., Randell S.H., Lanzavecchia A., Marasco W.A., Shi Z.L., Baric R.S. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., Swanstrom J., Sheahan T.P., Pickles R.J., Corti D., Randell S.H., Lanzavecchia A., Marasco W.A., Baric R.S. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U. S. A. 2016 doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Suo S., Zarlenga D.S., Cong Y., Ma X., Zhao Q., Ren X. A phage-displayed peptide recognizing porcine aminopeptidase N is a potent small molecule inhibitor of PEDV entry. Virology. 2014;456–7:20–27. doi: 10.1016/j.virol.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchlinsky M., Moss B. Introduction of foreign DNA into the vaccinia virus genome by in vitro ligation: recombination-independent selectable cloning vectors. Virology. 1992;190(1):522–526. doi: 10.1016/0042-6822(92)91246-q. [DOI] [PubMed] [Google Scholar]
- Mesquita J.R., Hakze-van der Honing R., Almeida A., Lourenco M., van der Poel W.H., Nascimento M.S. Outbreak of porcine epidemic diarrhea virus in Portugal, 2015. Transbound. Emerg. Dis. 2015 doi: 10.1111/tbed.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., van der Hoek L., Voss D., Bader O., Lehmann D., Schulz A.R., Kallies S., Sulima T., Fielding B.C., Drosten C., Niedrig M. Human coronavirus NL63 open reading frame 3 encodes a virion-incorporated N-glycosylated membrane protein. Virol. J. 2010;7:6. doi: 10.1186/1743-422X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam E., Lee C. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Vet. Microbiol. 2010;144(1–2):41–50. doi: 10.1016/j.vetmic.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., DeDiego M.L., Zhao J., Fett C., Alvarez E., Nieto-Torres J.L., Enjuanes L., Perlman S. Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology. 2010;399(1):120–128. doi: 10.1016/j.virol.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Peifer M., Bender W. Construction of large DNA segments in Escherichia coli. Science. 1989;244(4910):1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- Oh J.S., Song D.S., Park B.K. Identification of a putative cellular receptor 150 kDa polypeptide for porcine epidemic diarrhea virus in porcine enterocytes. J. Vet. Sci. 2003;4(3):269–275. [PubMed] [Google Scholar]
- Oka T., Saif L.J., Marthaler D., Esseili M.A., Meulia T., Lin C.M., Vlasova A.N., Jung K., Zhang Y., Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 2014;173(3–4):258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Ceriani J.E., Patino C., Plana J., Enjuanes L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology. 2007;368(2):296–308. doi: 10.1016/j.virol.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Shin H.J. Porcine epidemic diarrhea virus infects and replicates in porcine alveolar macrophages. Virus Res. 2014;191:143–152. doi: 10.1016/j.virusres.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Sestak K., Hodgins D.C., Shoup D.I., Ward L.A., Jackwood D.J., Saif L.J. Immune response of sows vaccinated with attenuated transmissible gastroenteritis virus (TGEV) and recombinant TGEV spike protein vaccines and protection of their suckling pigs against virulent TGEV challenge exposure. Am. J. Vet. Res. 1998;59(8):1002–1008. [PubMed] [Google Scholar]
- Park S.J., Moon H.J., Luo Y., Kim H.K., Kim E.M., Yang J.S., Song D.S., Kang B.K., Lee C.S., Park B.K. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Genes. 2008;36(1):95–104. doi: 10.1007/s11262-007-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Cruz D.J., Shin H.J. Receptor-bound porcine epidemic diarrhea virus spike protein cleaved by trypsin induces membrane fusion. Arch. Virol. 2011;156(10):1749–1756. doi: 10.1007/s00705-011-1044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim H.K., Song D.S., An D.J., Park B.K. Complete genome sequences of a Korean virulent porcine epidemic diarrhea virus and its attenuated counterpart. J. Virol. 2012;86(10):5964. doi: 10.1128/JVI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Cruz D.J., Shin H.J. Clathrin- and serine proteases-dependent uptake of porcine epidemic diarrhea virus into Vero cells. Virus Res. 2014;191:21–29. doi: 10.1016/j.virusres.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Kim S., Song D., Park B. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg. Infect. Dis. 2014;20(12):2089–2092. doi: 10.3201/eid2012.131642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel S., Park J.E., Jang H., Hyun B.H., Yang D.G., Shin H.J. Evaluation of antibody response of killed and live vaccines against porcine epidemic diarrhea virus in a field study. Vet. Q. 2014;34(4):194–200. doi: 10.1080/01652176.2014.973999. [DOI] [PubMed] [Google Scholar]
- Pedersen K.W., van der Meer Y., Roos N., Snijder E.J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 1999;73(3):2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58(3):243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Krahling V., Ditt V., Grywna K., Muhlberger E., Drosten C. Reverse genetic characterization of the natural genomic deletion in SARS-Coronavirus strain Frankfurt-1 open reading frame 7b reveals an attenuating function of the 7b protein in-vitro and in-vivo. Virol. J. 2009;6:131. doi: 10.1186/1743-422X-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.J., Chua M.M., Lavi E., Weiss S.R. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 1999;73(9):7752–7760. doi: 10.1128/jvi.73.9.7752-7760.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz R.T., Temiakov D., Anikin M., Vassylyev D.G., McAllister W.T. A mechanism of nucleotide misincorporation during transcription due to template-strand misalignment. Mol. Cell. 2006;24(2):245–255. doi: 10.1016/j.molcel.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu C., Das Sarma J., Bates P., Weiss S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006;80(12):5768–5776. doi: 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasschaert D., Duarte M., Laude H. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J. Gen. Virol. 1990;71(Pt. 11):2599–2607. doi: 10.1099/0022-1317-71-11-2599. [DOI] [PubMed] [Google Scholar]
- Regla-Nava J.A., Nieto-Torres J.L., Jimenez-Guardeno J.M., Fernandez-Delgado R., Fett C., Castano-Rodriguez C., Perlman S., Enjuanes L., DeDiego M.L. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 2015;89(7):3870–3887. doi: 10.1128/JVI.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherba G., Bromfield C.R., Jarrell V.L., Shipley C.F. Evaluation of responses to both oral and parenteral immunization modalities for porcine epidemic diarrhea virus in production units. J. Swine Health Prod. 2016;24(1):21–28. [Google Scholar]
- Schmitz A., Tobler K., Suter M., Ackermann M. Prokaryotic expression of porcine epidemic diarrhoea virus ORF3. Adv. Exp. Med. Biol. 1998;440:775–780. doi: 10.1007/978-1-4615-5331-1_100. [DOI] [PubMed] [Google Scholar]
- Schultze B., Krempl C., Ballesteros M.L., Shaw L., Schauer R., Enjuanes L., Herrler G. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J. Virol. 1996;70(8):5634–5637. doi: 10.1128/jvi.70.8.5634-5637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D., Grego S., Randell S.H., Baric R.S. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2013;110(40):16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Lv M., Chen J., Shi H., Zhang S., Zhang X., Feng L. Molecular characterizations of subcellular localization signals in the nucleocapsid protein of porcine epidemic diarrhea virus. Viruses. 2014;6(3):1253–1273. doi: 10.3390/v6031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Matsuyama S., Ujike M., Taguchi F. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J. Virol. 2011;85(15):7872–7880. doi: 10.1128/JVI.00464-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuya H., Birren B., Kim U.J., Mancino V., Slepak T., Tachiiri Y., Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. U. S. A. 1992;89(18):8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B., Gomutsukhavadee M., Sawaengpol T., Sangiambut S., Puttikhunt C., Chin-inmanu K., Suriyaphol P., Malasit P., Screaton G., Mongkolsapaya J. A simplified positive-sense-RNA virus construction approach that enhances analysis throughput. J. Virol. 2013;87(23):12667–12674. doi: 10.1128/JVI.02261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J., Waterfield M.D. Studies on the primary structure of the influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 1975;72(1):93–97. doi: 10.1073/pnas.72.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Yang J.S., Oh J.S., Han J.H., Park B.K. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine. 2003;21(17–18):1833–1842. doi: 10.1016/S0264-410X(03)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean J.R., Desforges M., Almazan F., Jacomy H., Enjuanes L., Talbot P.J. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J. Virol. 2006;80(7):3670–3674. doi: 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieneke-Grober A., Vey M., Angliker H., Shaw E., Thomas G., Roberts C., Klenk H.D., Garten W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11(7):2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhardiman M., Kramyu J., Narkpuk J., Jongkaewwattana A., Wanasen N. Generation of porcine reproductive and respiratory syndrome virus by in vitro assembly of viral genomic cDNA fragments. Virus Res. 2015;195:1–8. doi: 10.1016/j.virusres.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.B., Feng L., Shi H.Y., Chen J.F., Liu S.W., Chen H.Y., Wang Y.F. Spike protein region (aa 636789) of porcine epidemic diarrhea virus is essential for induction of neutralizing antibodies. Acta Virol. 2007;51(3):149–156. [PubMed] [Google Scholar]
- Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18(1):161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.C., Zhang J.X., Zhang S.Y., Wang P., Fan X.H., Li L.F., Li G., Dong B.Q., Liu W., Cheung C.L., Xu K.M., Song W.J., Vijaykrishna D., Poon L.L., Peiris J.S., Smith G.J., Chen H., Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80(15):7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78(13):6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G., Hofmann-Lehmann R., Stallkamp I., Thiel V., Thiel H.J. Genome organization and reverse genetic analysis of a type I feline coronavirus. J. Virol. 2008;82(4):1851–1859. doi: 10.1128/JVI.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuns S., Conceicao-Neto N., Christiaens I., Zeller M., Desmarets L.M., Roukaerts I.D., Acar D.D., Heylen E., Matthijnssens J., Nauwynck H.J. Complete genome sequence of a porcine epidemic diarrhea virus from a novel outbreak in Belgium, January 2015. Genome Announc. 2015;3(3) doi: 10.1128/genomeA.00506-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 2001;75(14):6676–6681. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlaskalova-Hogenova H., Mandel L., Trebichavsky I., Kovaru F., Barot R., Sterzl J. Development of immune responses in early pig ontogeny. Vet. Immunol. Immunopathol. 1994;43(1–3):135–142. doi: 10.1016/0165-2427(94)90129-5. [DOI] [PubMed] [Google Scholar]
- Tresnan D.B., Holmes K.V. Feline aminopeptidase N is a receptor for all group I coronaviruses. Adv. Exp. Med. Biol. 1998;440:69–75. doi: 10.1007/978-1-4615-5331-1_9. [DOI] [PubMed] [Google Scholar]
- Tresnan D.B., Levis R., Holmes K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996;70(12):8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R.G., Heijnen L., Spaan W.J., de Groot R.J. Homologous RNA recombination allows efficient introduction of site-specific mutations into the genome of coronavirus MHV-A59 via synthetic co-replicating RNAs. Nucleic Acids Res. 1992;20(13):3375–3381. doi: 10.1093/nar/20.13.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Worm S.H., Eriksson K.K., Zevenhoven J.C., Weber F., Zust R., Kuri T., Dijkman R., Chang G., Siddell S.G., Snijder E.J., Thiel V., Davidson A.D. Reverse genetics of SARS-related coronavirus using vaccinia virus-based recombination. PLoS One. 2012;7(3):e32857. doi: 10.1371/journal.pone.0032857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Veen R.L., Harris D.L., Kamrud K.I. Alphavirus replicon vaccines. Anim. Health Res. Rev. 2012;13(1):1–9. doi: 10.1017/S1466252312000011. [DOI] [PubMed] [Google Scholar]
- Vandergaast R., Hoover L.I., Zheng K., Fredericksen B.L. Generation of West Nile virus infectious clones containing amino acid insertions between capsid and capsid anchor. Viruses. 2014;6(4):1637–1653. doi: 10.3390/v6041637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ViPR, Virus pathogen database and analysis resource: http://www.viprbrc.org/brc/viprDetails.spg?ncbiProteinId=YP_001718613&decorator=corona.
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014;20(10):1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H., Yu W., Xu K., Bian C., Fischer W.B., Schwarz W., Feng L., Sun B. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586(4):384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20(5):917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Deng F., Ye G., Dong W., Zheng A., He Q., Peng G. Comparison of lentiviruses pseudotyped with S proteins from coronaviruses and cell tropisms of porcine coronaviruses. Virol. Sin. 2016;31(1):49–56. doi: 10.1007/s12250-015-3690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R.D., Woods R.D. Partial passive protection with two monoclonal antibodies and frequency of feeding of hyperimmune anti-transmissible gastroenteritis virus (TGEV) serum for protection of three-day-old piglets from a TGEV challenge infection. J. Vet. Diagn. Invest. 2001;13(4):290–296. doi: 10.1177/104063870101300402. [DOI] [PubMed] [Google Scholar]
- Wicht O., Li W., Willems L., Meuleman T.J., Wubbolts R.W., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014;88(14):7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R.D. Efficacy of a transmissible gastroenteritis coronavirus with an altered ORF-3 gene. Can. J. Vet. Res. 2001;65(1):28–32. [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y., Liu H.J., Tong D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164(3–4):212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R., Soma J., Nakanishi M., Yamaguchi R., Niinuma S. Isolation and experimental inoculation of an S INDEL strain of porcine epidemic diarrhea virus in Japan. Res. Vet. Sci. 2015;103:103–106. doi: 10.1016/j.rvsc.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y., Kawakita Y. A line of cells derived from African green monkey kidney. Nippon Rinsho. 1963;21:1209–1210. [Google Scholar]
- Ye S., Li Z., Chen F., Li W., Guo X., Hu H., He Q. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes. 2015;51(3):385–392. doi: 10.1007/s11262-015-1257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn S., Leibowitz J.L., Collisson E.W. In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology. 2005;332(1):206–215. doi: 10.1016/j.virol.2004.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74(22):10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Denison M.R., Weiss S.R., Baric R.S. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 2002;76(21):11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2003;100(22):12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.J., Chen Y.C., Hsiao C.H., Kuo T.C., Chang S.C., Lu C.Y., Wei W.C., Lee C.H., Huang L.M., Chang M.F., Ho H.N., Lee F.J. Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus. FEBS Lett. 2004;565(1–3):111–116. doi: 10.1016/j.febslet.2004.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Yang R.F., Shi M.D., Jiang M.R., Xie Y.H., Ruan H.Q., Jiang X.S., Shi L., Zhou H., Zhang L., Wu X.D., Lin Y., Ji Y.Y., Xiong L., Jin Y., Dai E.H., Wang X.Y., Si B.Y., Wang J., Wang H.X., Wang C.E., Gan Y.H., Li Y.C., Cao J.T., Zuo J.P., Shan S.F., Xie E., Chen S.H., Jiang Z.Q., Zhang X., Wang Y., Pei G., Sun B., Wu J.R. Characterization of the 3a protein of SARS-associated coronavirus in infected vero E6 cells and SARS patients. J. Mol. Biol. 2004;341(1):271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang K., Lv W., Yu W., Xie S., Xu K., Schwarz W., Xiong S., Sun B. The ORF4a protein of human coronavirus 229E functions as a viroporin that regulates viral production. Biochim. Biophys. Acta. 2014;1838(4):1088–1095. doi: 10.1016/j.bbamem.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F.M., Huo J.Y., Zhao J., Chang H.T., Wang X.M., Chen L., Wang C.Q. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus field strains in central China during 2010–2012 outbreaks. Bing Du Xue Bao. 2013;29(2):197–205. [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3(8):e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]