Highlights
-
•
PDCoV infection dynamics and appropriate sample collection are reviewed.
-
•
Virological methods for PDCoV detection are discussed.
-
•
Serological methods for PDCoV detection are discussed.
-
•
Global prevalence of PDCoV in swine population is described.
-
•
Genetic analyses of global PDCoV are discussed.
Keywords: Porcine deltacoronavirus, PDCoV, Diagnostics, Prevalence, Genetic evolution
Abstract
Porcine deltacoronavirus (PDCoV) was first reported in Hong Kong, China in 2012 and reported in United States swine in February 2014. PDCoV has subsequently been detected in South Korea, mainland China, and Thailand. PDCoV has been experimentally confirmed to cause diarrhea in inoculated pigs and need to be differentially diagnosed from porcine epidemic diarrhea virus and transmissible gastroenteritis virus in the field. Rapid diagnosis is critical for the implementation of efficient control strategies against PDCoV. Developing high-quality diagnostic methods and understanding PDCoV infection dynamics to collect appropriate specimens at the appropriate time window are important to obtain reliable diagnostic results. Among the virological methods, PDCoV-specific RT-PCR remains the method of choice for the detection of PDCoV; immunohistochemistry combined with hematoxylin and eosin staining has also been commonly used to examine histopathological lesions caused by PDCoV. Serological assays can provide information about previous exposure to PDCoV and also determine antibody responses to infection or vaccination. Prevalence of PDCoV is lower compared to that of PEDV. However, among PDCoV-positive samples, co-infection with other enteric pathogen e.g. PEDV is common. It is also important to understand molecular epidemiology of PDCoV and genetic relationships of global PDCoVs. This review discusses PDCoV infection dynamics and appropriate sample collection for diagnostic testing, the commonly used virological and serological methods for PDCoV diagnosis, prevalence and genetic evolution of PDCoVs.
1. Introduction
Coronaviruses (CoVs) belong to the subfamily Coronavirinae in the family Coronaviridae within the order Nidovirales. Four genera have thus far been described in the subfamily Coronavirinae and these include: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Masters and Perlman, 2013). Five porcine CoVs have been recognized: transmissible gastroenteritis virus (TGEV), porcine respiratory coronavirus (PRCV), and porcine epidemic diarrhea virus (PEDV) in the Alphacoronavirus genus; porcine hemagglutinating encephalomyelitis virus (PHEV) in the Betacoronavirus genus; and porcine deltacoronavirus (PDCoV) in the Deltacoronavirus genus.
Porcine deltacoronavirus was first detected in pig samples collected in 2009 in Hong Kong during a molecular surveillance study (Woo et al., 2012). But the clinical significance of PDCoV was not addressed in that study. In February 2014, emergence of PDCoV in U.S. swine was reported and the virus rapidly spread to multiple states in the U.S. (Li et al., 2014, Marthaler et al., 2014a, Marthaler et al., 2014b, Wang et al., 2014a, Wang et al., 2014b, Wang et al., 2014c). Shortly thereafter, PDCoV was detected in the South Korean swine population (Lee and Lee, 2014). Recently PDCoV has also been detected in mainland China and Thailand (Chen et al., 2015a, Janetanakit et al., 2016, Song et al., 2015, Wang et al., 2015). PDCoV was reported to be associated with naturally infected clinical cases that were presented with severe diarrhea, vomiting, and dehydration in piglets (Janetanakit et al., 2016, Li et al., 2014, Song et al., 2015, Wang et al., 2014a) together with histopathological lesions typical for atrophic enteritis (Wang et al., 2016). Experimental infection studies have confirmed that conventional and gnotobiotic piglets inoculated with PDCoV developed mild to severe diarrhea, gross and microscopic intestinal lesions (Chen et al., 2015b, Jung et al., 2015, Ma et al., 2015).
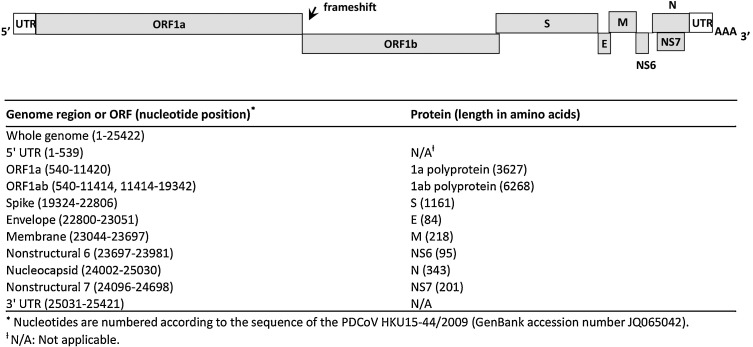
PDCoV is an enveloped, single-stranded, positive-sense RNA virus with a genome of appropriately 25 kb in length. The PDCoV genome organization and nucleotide locations are depicted in Fig. 1 . The genome arrangements are in the order of: 5′ untranslated region (UTR), open reading frame 1a/1b (ORF1a/1b), spike (S), envelope (E), membrane (M), nonstructural protein 6 (NS6), nucleocapsid (N), nonstructural protein 7 (NS7), and 3′ UTR. The functions of PDCoV individual proteins have not been elucidated. But according to studies on other CoVs, the replicase polyproteins 1a (pp1a) and pp1ab are generally cleaved by virus-encoded proteases into 16 non-structural proteins involved in viral transcription and replication (Masters and Perlman, 2013). Among the structural proteins, the S glycoprotein of CoVs generally functions in receptor binding, cell membrane fusion and entry; in addition, the S protein is postulated to harbor epitopes to induce neutralizing antibodies. In regards to molecular characterization of PDCoV, the whole genome sequences and/or S and N gene sequences have been frequently used for phylogenetic analysis (Homwong et al., 2016, Janetanakit et al., 2016, Lee et al., 2016, Song et al., 2015, Wang et al., 2014a). The S, M and N protein genes have also been targeted for the development of virological and serological diagnostic assays for PDCoV (Chen et al., 2015b, Ma et al., 2015, Marthaler et al., 2014b, Song et al., 2015, Su et al., 2015, Thachil et al., 2015, Wang et al., 2014a).
Fig. 1.
Schematic diagrams of PDCoV genome organization. The PDCoV entire genome organization is depicted at the top. The 5′ untranslated region (UTR), ORFs 1a and 1b encoding replicase polyproteins, spike (S), envelope (E), membrane (M), nonstructural protein 6 (NS6), nucleocapsid (N), nonstructural protein 7 (NS7) genes, and 3′ UTR are shown, with the ribosomal frameshift site indicated. The nucleotide locations of each ORF in the PDCoV genome are depicted at the bottom.
Clinical symptoms of PDCoV infection can include diarrhea, dehydration, variable vomiting and mortality in neonatal piglets; these clinical manifestations are similar to other swine enteric pathogens such as PEDV and TGEV. Thus specific laboratory diagnostic testing is imperative to differentiate PDCoV from PEDV and TGEV infection. This paper reviews PDCoV infection dynamics and appropriate sample collection for diagnostic testing as well as the commonly used virological and serological methods for PDCoV diagnosis. In addition, this article provides information known thus far on PDCoV prevalence and genetic evolution.
2. PDCoV infection dynamics and appropriate sample collection for testing
Several experimentally infected studies have investigated PDCoV pathogenesis in conventional and gnotobiotic (Gn) pigs (Chen et al., 2015b, Jung et al., 2015, Ma et al., 2015). Details of the disease mechanisms and pathogenesis of PDCoV have been reviewed in another paper (Jung et al., 2016). Here we focus on summarizing the PDCoV infection dynamics in pigs to help understand how to collect appropriate clinical samples at the appropriate time for diagnostic testing.
In the studies performed by Jung et al. (2015) and Ma et al. (2015), 10-19-day-old Gn pigs were inoculated with PDCoV-positive intestinal contents or cell culture-adapted PDCoV isolates. Diarrhea and/or vomiting were observed starting from 24 h post inoculation (hpi) or 72 hpi and diarrhea persisted through 72–120 hpi when pigs were euthanized and necropsied (Jung et al., 2015, Ma et al., 2015). Fecal viral RNA was detected from 24 hpi and lasted through 72–120 hpi (Jung et al., 2015, Ma et al., 2015). Because pigs still had diarrhea and shed virus at the termination of these studies (72–120 hpi), eventual duration of diarrhea and PDCoV shedding was not determined in Gn pigs in these studies (Jung et al., 2015, Ma et al., 2015).
In 5-day-old conventional pigs experimentally inoculated with 3 × 104 TCID50 of cell culture-adapted PDCoV USA/IL/2014 isolate, onset of diarrhea occurred on 5 days post inoculation (DPI) and lasted through 7 DPI when pigs were necropsied; viral RNA shedding in fecal swabs was detected from 2 to 7 DPI (Chen et al., 2015b). The study was terminated at 7 DPI in efforts to adequately capture gross and microscopic lesions caused by PDCoV infection; eventual duration of diarrhea and PDCoV shedding was not determined (Chen et al., 2015b).
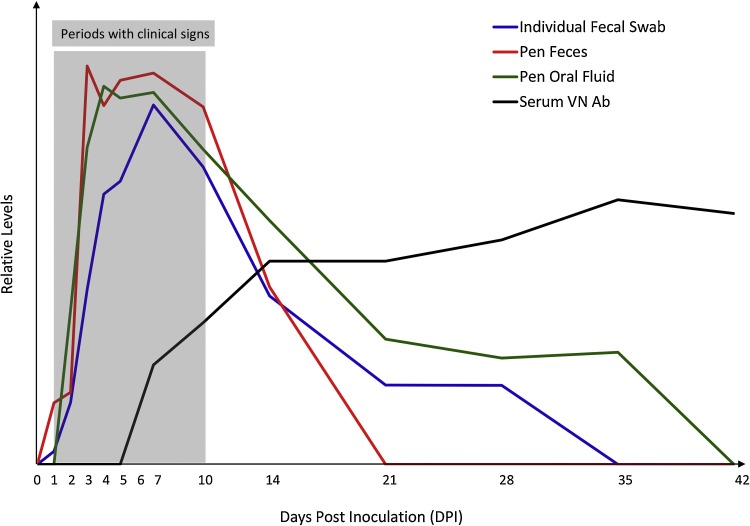
In another study, four 10-day-old conventional pigs were orally inoculated with 5 ml of virus containing 106 PFU of PDCoV Michigan/8977/2014 isolate and monitored through 21 DPI (Ma et al., 2015). Diarrhea occurred on 1 DPI and persisted for 7–10 days and all four pigs recovered from disease on 10 DPI. Fecal viral RNA shedding was detected on 1–2 DPI, peaked on 7 DPI, gradually decreased after 10 DPI, but was still detectable from 1 out of 4 pigs on 21 DPI (Ma et al., 2015). My group has recently performed an experimental infection study where fifteen 3-week-old conventional pigs were orally inoculated with 105 TCID50 of PDCoV USA/IL/2014 isolate and monitored through 42 DPI with viral RNA shedding levels examined in individual fecal swabs (fecal swabs were collected from each pig at each time point), pen-based feces (two feces were collected at each time point from the floor of the room housing all PDCoV-inoculated pigs), and pen-based oral fluids (two oral fluids were collected at each time point from the room housing all PDCoV-inoculated pigs) (unpublished data). A schematic diagram presenting PDCoV infection dynamics in nursery pigs is summarized in Fig. 2 . Clinical signs generally did not last over 10 days. Viral RNA shedding peaked on 7 DPI and gradually decreased thereafter no matter in individual fecal swabs, pen-based feces, or pen-based oral fluids. However, duration of viral RNA shedding in these specimen types are different. PDCoV RNA was detected through at least 14 DPI (negative on 21 DPI) in pen-based feces, at least 28 DPI (negative on 35 DPI) in individual fecal swabs, and at least 35 DPI (negative on 42 DPI) in pen-based oral fluids. PDCoV shedding patterns under field conditions have not been reported.
Fig. 2.
Schematic diagram of PDCoV infection dynamics in 3-week-old weaned pigs. Time periods with clinical signs, average virus shedding levels (viral RNA as determined by PDCoV M gene-based real-time RT-PCR) in individual fecal swabs, pen-based feces, and pen-based oral fluids, and average neutralizing antibody levels in serum are indicated.
Based on aforementioned experimental studies, recommendations on sample collections for PDCoV diagnostic testing are provided below.
-
•
Collection of fecal samples and oral fluids. PDCoV-infected pigs shed high-levels of virus during the acute infection periods (1–10 DPI); individual fecal swabs, pen-based feces, and/or pen-based oral fluids can be collected for PDCoV PCR or virus isolation (VI) testing. Oral fluids and individual fecal swabs appear to be better specimen types than pen-based feces for PCR testing when PDCoV infection is already over 10-14 days.
-
•
Serum samples for PDCoV PCR and VI testing or not? PDCoV infection can induce acute viremia (Chen et al., 2015b, Ma et al., 2015) but viremia is transient and at low-level; thus serum is not the best choice for PDCoV PCR or VI testing.
-
•
Tissue sample collections for PDCoV testing. For sick (clinical) pigs euthanized and necropsied during the acute infection phase (1–10 DPI), the small intestines especially the jejunum and ileum sections can be collected freshly for PDCoV PCR and VI testing or can be collected in 10% buffered formalin for histology and immunohistochemistry (IHC) examinations. Low-levels of PDCoV RNA has been detected in other non-enteric tissues by PCR but PDCoV antigen has not been consistently detected in those non-enteric tissues by IHC or immunofluorescence staining (Chen et al., 2015b, Jung et al., 2015, Ma et al., 2015); therefore, non-enteric tissues are not the best choice for PDCoV diagnostic testing.
-
•
Feed or environmental samples for PDCoV testing. A number of PEDV studies have shown that contaminated trailers and feed/ingredients can serve as a vehicle to transmit PEDV (Bowman et al., 2015a, Dee et al., 2014a, Lowe et al., 2014, Pasick et al., 2014, Pillatzki et al., 2015). Since PDCoV probably has the similar transmission routes to PEDV, environmental samples and feed are occasionally submitted for PDCoV PCR testing.
-
•
Samples for PDCoV antibody testing. No information has been published regarding dynamics of antibody responses against PDCoV infection. According to our unpublished data, PDCoV-inoculated weaned pigs started to develop low-level virus neutralizing (VN) antibody in serum on 7 DPI and VN antibody titers increased from 14 DPI and were maintained through the end of the study (42 DPI; Fig. 2). For PEDV, serum, oral fluid, colostrum and milk samples can be used for antibody detection; it is assumed that these specimens can also be used for PDCoV antibody detection although more research is definitely needed in these areas.
3. PDCoV diagnostic methods
PDCoV diagnostic methods can be divided into two categories: virological and serological methods. Virological methods include detection of virus particles (electron microscopy), detection of viral nucleic acid (various RT-PCRs and in situ hybridization), detection of viral antigen (immunofluorescence staining and immunohistochemistry), and detection of viable virus (virus isolation and swine bioassay). Serological assays can be used to determine previous exposure to a virus, to determine kinetics of antibody response to virus infection, and to evaluate efficacy of vaccines. The most commonly used serological assays include indirect fluorescent antibody (IFA) assay, virus neutralization (VN) test or fluorescent focus neutralization (FFN) test, enzyme linked immunosorbent assays (ELISAs), and fluorescent microsphere immunoassays (FMIA) although some of these assays have not been well validated for the detection of PDCoV antibodies. Table 1 summarizes the current virological and serological methods for PDCoV detection.
Table 1.
Summary of virological and serological methods for PDCoV detection.
| Assay Category | Assay | Appropriate Samples | Comment |
| Detection of virus particles | Electron microscopy | Feces, intestine, virus isolates | Valuable for diagnosing unknown or new viruses. Low sensitivity and time-consuming procedures. Not specific for PDCoV. Not suitable for routine diagnostic testing. |
| Detection of viral RNA | Pan-CoV RT-PCR | Samples with relatively high concentrations of virus | Not specific for PDCoV and needs confirmation by sequencing. Time consuming. Not suitable for routine diagnostic testing. |
| Standard RT-PCR for PDCoV | Feces, rectal swabs, intestinal tissues or contents, oral fluid, feed, environmental samples, etc. | Less sensitive compared to rRT-PCR. Time consuming. Not a high-throughput assay. Needs special attention when running nested PCR to avoid cross contamination. | |
| Singleplex rRT-PCR for PDCoV | A rapid, sensitive and high-throughput assay with quantification capability. | ||
| Multiplex rRT-PCR for PDCoV and PEDV and/or TGEV | A rapid, sensitive and high-throughput assay with quantification capability. Can detect and differentiate PDCoV from other pathogens such as PEDV and TGEV. | ||
| RT-iiPCR for PDCoV | Feces, rectal swab, oral fluid | Comparable sensitivity to rRT-PCR. Portable device useful for on-site detection. | |
| PDCoV in situ hybridization | Intestine tissues | Can be used to confirm virus distribution within tissue lesions. | |
| Detection of viral antigen | Immunofluorescence staining | Intestine tissues; virus-infected cell culture | Can be used to confirm PDCoV VI results or virus distribution within tissue lesions. |
| IHC | Intestine tissues | Can be used to confirm virus distribution within tissue lesions. | |
| Detection of viable virus | Virus isolation | Feces, intestinal tissues or contents | Detects live virus. Not very sensitive and low success rate. |
| Swine bioassay | Feed, environmental samples, disinfectant-treated samples, etc. | Uesful to assess whether a sample contains infectious virus especially when virus isolation is unsuccessful in the sample. | |
| Detection of virus-specific antibody | VN/FFN | Serum, colostrum, milk | Detect PDCoV Ab with neutralizing activity. Cannot distinguish Ab isotypes. |
| IFA | Serum | Detect PDCoV Ab. Not reflecting neutralizing activity. Has the capability to determine Ab isotypes if needed. Not a high-throughput assay. | |
| ELISA | Serum, colostrum, milk, oral fluid, feces | Detect PDCoV Ab. Not reflecting neutralizing activity. Has the capability to determine Ab isotypes if needed. A high-throughput assay. | |
| FMIA | Serum | Capability to detect Ab against multiple proteins or pathogens. |
3.1. Virological methods for PDCoV detection
3.1.1. Electron microscopy (EM)
Electron microscopy allows direct visualization of virus particles. Two EM techniques are commonly used in diagnostic laboratories: negative-stain EM for detection of virus particles in a fluid matrix; ultrathin-section EM for detection of virus particles in fixed tissues or cells. Based on characteristic morphology and size of virus particles observed under EM, viruses can be assigned to appropriate family, e.g. coronavirus-like particles were observed in some feces during initial investigation of diarrheic cases caused by PDCoV. Although EM cannot identify viruses to the species level, identification to the family level can still facilitate next-step testing to achieve definite diagnosis. However, EM generally is less sensitive and needs presence of sufficient amount of virus (about 105–6 virions per milliliter) in examined specimens. In addition, EM requires expensive equipment and highly skilled microscopist. EM is not a tool routinely used for PDCoV diagnostic testing.
3.1.2. Various polymerase chain reaction (PCR)-based assays for PDCoV detection
Polymerase chain reaction (PCR) is a technique that can in vitro amplify specific nucleic acid sequences and produce billion copies of target sequences after 35–40 cycles of three-step process (denaturation, annealing, and extension) within a few hours. Various PCRs including pan-coronavirus RT-PCR and PDCoV-specific RT-PCRs in different formats have been used to detect PDCoV RNA in a variety of samples. Table 1 comments on the properties of these RT-PCRs and Table 2 summarizes PDCoV-specific RT-PCRs regarding the target gene, sequences of primers and probes, limit of detection, and reference papers.
Table 2.
Summary of PDCoV-specific RT-PCR assays.
| RT-PCR Method | Target Gene | Primer or Probe | Sequences (5′-3′) | Limit of Detection | Reference |
|---|---|---|---|---|---|
| Standard One-step RT-PCR | M | primer 67F | ATCCTCCAAGGAGGCTATGC | Not defined | Wang et al. (2014a) |
| primer 560R | GCGAATTCTGGATCGTTGTT | ||||
| N | primer 41F | TTTCAGGTGCTCAAAGCTCA | Not defined | Wang et al. (2014a) | |
| primer 735R | GCGAAAAGCATTTCCTGAAC | ||||
| Standard Nested RT-PCR | N | Outer primer F | TGCTACCTCTCCGATTCCCA | Not defined | Song et al. (2015) |
| Outer primer R | ATCCTGTTTGTCTGCTGGCA | ||||
| Inner primer F | GACACTGAGAAGACGGGTATGG | ||||
| Inner primer R | TAGTTGGTTTGGTAGGTGGCTC | ||||
| Real-time singleplex one-step RT-PCR | M | Forward primer | ATCGACCACATGGCTCCAA | 2 RNA copies per reaction | Marthaler et al. (2014b) |
| Reverse primer | CAGCTCTTGCCCATGTAGCTT | ||||
| Probe | FAM-CACACCAGTCGTTAAGCAT GGCAAGCT-BHQ | ||||
| M | PDCoV-F | CGACCACATGGCTCCAATTC | 0.056 TCID50per reaction | Chen et al. (2015b), Zhang et al. (2016) | |
| PDCoV-R | CAGCTCTTGCCCATGTAGCTT | ||||
| PDCoV-P | FAM-CACACCAGT/ZEN/CGTTAA GCATGGCAAGC-IABkFQ | ||||
| N | Forward primer | CGCTTAACTCCGCCATCAA | Not defined | Ma et al. (2015) | |
| Reverse primer | TCTGGTGTAACGCAGCCAGTA | ||||
| Probe | 6FAM-CCCGTTGAAAACC-MGB | ||||
| Real-time duplex PEDV/PDCoV RT-PCR | M | PEDV rF | GGTTGTGGCGCAGGACA | 7 RNA copies per reaction for PEDV; 14 RNA copies per reaction for PDCoV | Zhang et al. (2016) |
| PEDV rR | CGGCCCATCACAGAAGTAGT | ||||
| PEDV rP | FAM-CATTCTTGG/ZEN/TGGTCT TTCAATCCTGA-IABkFQ | ||||
| M | PDCoV rF | TGAGAGTAGACTCCTTGCAGGGA | |||
| PDCoV rR | GAGAATTGGAGCCATGTGGT | ||||
| PDCoV rP | NED-TGTACCCATTGGATCCATAA-MGB | ||||
| RT-iiPCR | M | PDCoV iiF | GAGAGTAGACTCCTTGCAGGGATTAT | 9 RNA copies per reaction | Zhang et al. (2016) |
| PDCoV iiR | GCTTGCCATGCTTAACGACTG | ||||
| PDCoV iiP | FAM-AATGCACCTCCATGTACC-MGB | ||||
3.1.2.1. Pan-coronavirus (Pan-CoV) RT-PCR
Coronaviruses can infect a variety of host species and there are also quite genetic diversities among coronaviruses. However, a pan-coronavirus one-step RT-PCR targeting a conserved region (251 bp) of polymerase gene has been developed that can detect most if not all of the coronaviruses (Moes et al., 2005). Electron microscopy together with pan-coronavirus RT-PCR followed by genetic sequencing played an important role in identifying PEDV and PDCoV during the early stage of investigating diarrheic swine cases in the United States (US) (Li et al., 2014, Stevenson et al., 2013). However, pan-coronavirus RT-PCR is not PEDV- or PDCoV-specific and genetic sequencing of the amplified product is needed to determine virus identity. This property together with the relatively lower sensitivity make pan-coronavirus RT-PCR not suitable for routine PDCoV diagnostic testing.
3.1.2.2. Standard PDCoV-specific RT-PCR
Several standard (also called conventional or gel-based) one-step RT-PCR or nested RT-PCR assays have been developed for the specific detection of PDCoV (Song et al., 2015, Wang et al., 2014a). These RT-PCR assays target a conserved region of PDCoV M or N genes (Table 2). Following PCR amplification, electrophoresis on agarose gels and visualization of amplicons under UV light are generally performed to determine the RT-PCR results. The limit of detection of these standard PDCoV RT-PCR assays was not defined in the papers (Song et al., 2015, Wang et al., 2014a). Due to two steps of reaction set up, nested RT-PCR is more prone to contamination and special attention needs to be paid when performing nested RT-PCR. The standard RT-PCR assays especially the nested RT-PCR assays have not been widely used by veterinary diagnostic laboratories for PDCoV detection; most of the diagnostic labs use real-time RT-PCR for PDCoV detection.
3.1.2.3. Real-time RT-PCR for PDCoV detection
A number of probe-based real-time RT-PCR (rRT-PCR) assays have been developed for the specific detection of PDCoV (Chen et al., 2015b, Ma et al., 2015, Marthaler et al., 2014b, Zhang et al., 2016). These rRT-PCR assays target the conserved regions of PDCoV M or N genes with the limit of detection (LOD) of 2 viral RNA copies per reaction for the assay described by Marthaler et al. (2014b) and LOD of 0.056 TCID50 per reaction for the assay described by Chen et al. (2015b) and Zhang et al. (2016) (Table 2). Because of the similarity in clinical symptoms caused by PDCoV, PEDV and TGEV, there is a value to simultaneously detect and distinguish these swine enteric coronaviruses. Zhang et al. (2016) described a duplex real-time PEDV/PDCoV RT-PCR targeting M genes for simultaneous detection and differentiation of PDCoV from PEDV. The LOD of this duplex rRT-PCR is 7 RNA copies per reaction for PEDV and 14 RNA copies per reaction for PDCoV (Table 2). The performance of this duplex PEDV/PDCoV rRT-PCR on testing 170 clinical samples (86 fecal swabs, 24 feces, 19 intestines, and 41 oral fluids) was comparable to that of singleplex PDCoV rRT-PCR and singleplex PEDV rRT-PCR (Zhang et al., 2016). Some commercial multiplex rRT-PCR assays for simultaneous detection and differentiation of PDCoV, PEDV and TGEV have been developed (e.g. EZ-PED/TGE/PDCoV MPX 1.0, Tetracore Inc., Rockville, MD) and used by some veterinary diagnostic laboratories for routine diagnostic testing.
3.1.2.4. Reverse transcription-insulated isothermal PCR (RT-iiPCR) for PDCoV detection
In recent years, a point-of-need PCR detection platform integrating the insulated isothermal PCR (iiPCR) technology and a field-deployable device (POCKIT™ Nucleic Acid Analyzer) has been developed for automatic detection and interpretation of PCR results within one hour (Tsai et al., 2012). A PDCoV RT-iiPCR has recently been developed and validated for diagnostic testing (Zhang et al., 2016). This PDCoV RT-iiPCR assay has a limit of detection of 9 RNA copies per reaction (Table 2) and has diagnostic performance comparable to PDCoV real-time RT-PCRs (Zhang et al., 2016). The PDCoV RT-iiPCR is a potentially useful tool for on-site detection.
3.1.2.5. Key points about PDCoV-specific RT-PCRs
Among aforementioned PDCoV-specific RT-PCR assays, real-time RT-PCRs have been most commonly used by veterinary diagnostic laboratories and remain the method of choice for PDCoV detection. Some key points about PDCoV rRT-PCRs are listed below.
-
•
A variety of specimen types such as feces, rectal swabs, intestinal tissues or contents, oral fluid, environmental samples, feed and so on can be tested for presence of PDCoV RNA by rRT-PCR. Magnetic bead-based reagents and automated extraction on various magnetic particle processors such as Thermo KingFisher, Qiagen BioSprint, and Life Technologies MagMAX Express have been used in many laboratories for high throughput nucleic acid extraction. But nucleic acid extraction procedures need to be optimized based on specimen types.
-
•
Proper controls should be included in nucleic acid extraction and PCR reaction setup. For example, positive extraction control (a PDCoV-positive sample that undergoes nucleic acid extraction and PCR reaction), negative extraction control (phosphate buffered saline that undergoes nucleic acid extraction and PCR reaction), positive amplification control (PDCoV-positive RNA for PCR reaction), and negative amplification control (nuclease-free water for PCR reaction) are generally included. In addition, an internal positive control (IPC) such as Xeno™ RNA is recommended to be included. An IPC can be added to every reaction either at the nucleic acid extraction or at the PCR reaction setup step. The rRT-PCR master mix contains primers and probes for the IPC target, thus theoretically the IPC should amplify in every reaction. The absence of IPC amplification may indicate presence of PCR inhibitors in the sample. If both PDCoV and IPC fail to amplify in a sample, PDCoV PCR result is inconclusive and additional testing or resubmission of the sample is recommended.
-
•
When rRT-PCR assays reach their limit of detection, interpretation of PCR results could be a challenge.
-
•
rRT-PCR has many advantages such as rapid turnaround time between sample receipt and obtaining results, high sensitivity and high specificity, high throughput, multiplexing capability, and capability for genomic quantitation. However, PCR detects viral nucleic acid and does not distinguish live from dead virus in a sample.
3.1.3. Immunofluorescence staining, immunohistochemistry and in situ hybridization
The immunofluorescence staining is to detect viral antigen using virus-specific monoclonal or polyclonal antibodies that are either fluorophore-conjugated or unconjugated (if unconjugated, a fluorophore-conjugated secondary antibody is needed); fluorescent staining of viral antigens is visualized under a fluorescent microscope. The immunofluorescence staining has been conducted on PDCoV-infected cell culture for confirmation of virus isolation (Chen et al., 2015b, Hu et al., 2015), or performed on cryosections of tissues for determining the location of viral antigen in tissues (Jung et al., 2015).
Immunohistochemistry is to detect viral antigen in formalin-fixed paraffin-embedded tissues using virus-specific monoclonal or polyclonal antibodies followed by an enzyme-linked secondary antibody and chemical substrate; IHC can be visualized under a light microscope. PDCoV IHC has been described in some studies (Chen et al., 2015b, Ma et al., 2015).
For PDCoV immunofluorescence staining and IHC, either M peptide-specific rabbit antiserum or polyclonal antiserum against PDCoV generated in pigs has been used in the published papers (Chen et al., 2015b, Hu et al., 2015, Jung et al., 2015, Ma et al., 2015). Recently, PDCoV N protein-specific monoclonal antibody has been developed by South Dakota State University and made commercially available through the Medgene labs (Brookings, SD, http://medgenelabs.com) that can be used for PDCoV immunofluorescence staining and/or IHC.
In situ hybridization (ISH) is to detect viral nucleic acid present in fixed tissues using a labeled complementary DNA, RNA or modified nucleic acid strand (e.g. probe). Different than PCR approach where viral nucleic acid in a sample is amplified before detection, ISH detects viral nucleic acid that is not going through an amplification process. So far, only one published paper has described PDCoV detection using ISH approach (Jung et al., 2015).
The gross and histological evaluation of tissues is an important diagnostic method to correlate with ancillary tests such as PCR. However, the gross and microscopic lesions caused by PDCoV infection are not pathognomonic. Immunofluorescence staining, IHC and/or ISH along with hematoxylin and eosin examination can confirm lesions caused by PDCoV and also reveal locations of viral antigen/nucleic acid within lesions.
3.1.4. Virus isolation (VI)
Obtaining a PDCoV isolate that can efficiently grow in cell culture is critical for pathogenesis study, development of diagnostic assays, and vaccine development. The National Veterinary Services Laboratories (NVSL) under USDA has isolated PDCoV USA/IL/2014 strain and Michigan/8977/2014 strain in swine testicular (ST, ATCC CRL-1746) cell line (Chen et al., 2015b, Ma et al., 2015); however, detailed isolation protocols have not been published. To present, only one published paper has described primary PDCoV isolation from clinical samples in cell cultures in detail (Hu et al., 2015). They reported successful isolation and propagation of PDCoV OH-FD22 strain in LLC-PK1 cell line (ATCC CCL-101) and ST cell line. PDCoV cytopathic effects (CPE) in LLC-PK1 and ST cells are similar and are characterized as enlarged, rounded, granular cells followed by cell detachment (Hu et al., 2015). It was found that PDCoV OH-FD22 replicated in LLC-PK1 cells in the absence of trypsin, but virus titer was about 10,000-fold lower than that with trypsin (5 log10 PFU/ml vs 9 log10 PFU/ml); also, no visible CPE was observed without trypsin (Hu et al., 2015). Addition of small intestinal contents (SIC) from Gn pigs together with trypsin (SIC + trypsin) or addition of pancreatin alone to the culture medium resulted in PDCoV titer of 7 log10 PFU/ml which was about 100-fold lower than that with trypsin (9 log10 PFU/ml) (Hu et al., 2015). Therefore, the optimized culture conditions for PDCoV OH-FD22 propagation in LLC-PK1 cells are advanced MEM (Gibco, USA) supplemented with 1% antibiotic-antimycotic (Gibo), 1% nonessential amino acids (Gibo), 1% HEPES (Gibo), and 10% trypsin (Gibco) (Hu et al., 2015). Interestingly, it was found that trypsin did not support PDCoV OH-FD22 propagation in ST cells; in contrast, PDCoV propagated efficiently in ST cells when 10% Gn pig SIC or 1% pancreatin was added (Hu et al., 2015). The optimized culture conditions for PDCoV OH-FD22 propagation in ST cells are advanced MEM supplemented with 1% antibiotic-antimycotic, 1% HEPES, and 1% pancreatin (Sigma, USA) (Hu et al., 2015). The mechanisms of beneficial effects of trypsin or pancreatin on PDCoV propagation in LLC-PK1 or ST cells remain unknown. However, it is noteworthy that the PDCoV USA/IL/2014 strain and Michigan/8977/2014 strain propagate well in ST cells grown in MEM containing 5 μg/ml trypsin 250 (BD Biosciences) or DMEM containing 0.2 μg/ml TPCK-trypsin (Invitrogen) (Chen et al., 2015b, Ma et al., 2015).
Hu et al. attempted to isolate virus from 10 PDCoV PCR-positive clinical samples (4 fecal samples and 6 intestinal content samples) but virus was only successfully isolated from one intestinal content; VI from fecal samples was unsuccessful (Hu et al., 2015). In fact, the low success rate of PDCoV VI is similar to what has been observed for PEDV VI. In two studies (Chen et al., 2014, Oka et al., 2014 Oka et al., 2014), PEDV VI was successful in 2 out of 50 and 9 out of 88 PEDV PCR-positive samples, respectively; in addition, PEDV was only successfully isolated from intestinal homogenates or contents but not from fecal samples in those two studies. However, in a recent study, 3 PEDVs were successfully isolated in cell culture from feces (Chen et al., 2016a). In another study, PEDV VI from 68 clinical samples was unsuccessful; but after one clinical sample (intestinal homogenate) was inoculated into pigs, PEDV was successfully isolated from fresh intestinal homogenates and cecum contents collected from the inoculated pigs (Chen et al., 2016b). It is speculated that high concentration of virus and immediate VI attempts on the fresh samples are the key to success of VI in cell culture. This approach can be considered for PDCoV VI as well when there is a difficulty to isolate PDCoV directly from clinical samples.
3.1.5. Swine bioassay
Due to low success rate of PDCoV VI in cell culture, currently PDCoV VI is not a reliable assay to determine whether infectious virus is present in a clinical specimen or if ‘X’ treatment will inactivate the virus. For PEDV, swine bioassay has been commonly used to assess infectivity of PEDV present in various samples, including feed and feed ingredients after disinfectant or other treatments (Bowman et al., 2015b, Dee et al., 2014b, Dee et al., 2015). Notably, some samples were PEDV VI negative after treatment but swine bioassay positive (Dee et al., 2015), emphasizing the importance and necessity of performing swine bioassay to reliably determine virus infectivity of a sample. Based on some PEDV study, a neonatal pig bioassay is more sensitive than a weaned pig bioassay to assess PEDV infectivity and a 7-day duration would be appropriate for PEDV swine bioassay (Thomas et al., 2015). There have not been reports on PDCoV swine bioassay, but experience and knowledge learned from PEDV studies could possibly be applied to PDCoV.
3.2. Serological methods for PDCoV detection
3.2.1. Indirect fluorescent antibody (IFA) assay
The first step of IFA assay is to prepare PDCoV-infected ST or LLC-PK1 cell plates and get plates fixed at the appropriate time points. Then serum samples to be tested are added to the plates to bind to the virus antigen followed by addition of fluorophore-conjugated anti-swine secondary antibody and visualization under a fluorescence microscope. IFA assay can be used for detection of PDCoV antibody in a qualitative manner (determination as positive or negative) or in a quantitative manner (determination of antibody titer). To determine PDCoV antibody titer, test serums are serially two-fold diluted and incubated with PDCoV-infected cell plate(s); the reciprocal of the highest dilution of serum that still gives positive fluorescent staining is considered as the antibody titers. Theoretically, IFA antibody isotype can be determined by using different types of secondary antibody; however, for most occasions, IFA assay is used to determine IgG antibody. IFA assay is not automated and is subjective with respect to result reading and reporting. Dynamics of PDCoV IFA antibody production and duration has not been well defined and correlations of PDCoV IFA antibody to other serological assays are unknown at this point.
3.2.2. Virus neutralization (VN) assays
For VN test, heat-inactivated serum samples are serially two-fold diluted and incubated with a fixed amount of PDCoV (generally 100–200 TCID50) at 37 °C for 1–2 h. Subsequently, the virus-serum mixture is added to ST or LLC-PK1 cells and incubated at 37 °C. The endpoint determination of neutralizing antibody can be based on viral CPE development (conventional VN test) or based on fluorescence staining (this format of VN test is also called fluorescent focus neutralization [FFN] assay). CPE-based VN test needs to wait until a viral CPE has fully developed in the virus control wells and generally takes 3-4 days. Sometimes serum cytotoxicity can be mistaken as viral CPE, making it difficult to interpret the results. For FFN assay, the cell plates inoculated with virus-serum mixture are generally incubated for 24–48 h and then stained with a FITC-conjugated monoclonal antibody against PDCoV N protein (Medgene labs). The reciprocal of the highest serum dilution resulting in >90% reduction of fluorescent staining as compared to the negative serum control is considered as the neutralizing antibody titer of the serum sample. For another closely-related swine enteric coronavirus, PEDV, virus neutralization assay in the FFN format has been frequently used (Okda et al., 2015, Thomas et al., 2015). VN assays (both conventional CPE-based and FFN) are labor intensive, requiring manual reading and interpretation of the results. VN assays detect neutralizing antibodies but do not determine the antibody isotypes. So far, there have been no publications describing dynamics of PDCoV VN antibody production and duration. We have some preliminary data showing that PDCoV VN antibody was detected from 7 DPI through 42 DPI (the end of the study) in PDCoV-inoculated weaned pigs (unpublished). But correlations of PDCoV VN antibody to other serological assays have not been determined yet.
VN or FFN assays detect functional antibodies with neutralizing activity. In addition to measuring neutralizing antibody levels in serum, FFN assay has been optimized to quantify PEDV neutralizing antibody levels in colostrum and milk to reflect lactogenic immunity (Clement et al., 2016, Okda et al., 2015). Logistically, FFN assay can be adapted to measure PDCoV neutralizing antibody in colostrum and milk although no data is available yet in published literatures.
3.2.3. Enzyme linked immunosorbent assays (ELISAs)
Several indirect ELISAs have been developed for the detection of antibodies against PDCoV and these include a eukaryotic expressed PDCoV S1 protein-based ELISA (Thachil et al., 2015), a prokaryotic expressed PDCoV N protein-based ELISA (Su et al., 2015), and a PDCoV whole virus-based ELISA (Ma et al., 2016). The PDCoV S1 protein-based ELISA was validated using 210 field serum samples and had a diagnostic sensitivity of 90.6% and a diagnostic specificity of 94.8% when the clinical exposure history and PDCoV PCR testing on fecal samples were used to reflect the true PDCoV serological status of those farms from which serum samples were collected (Thachil et al., 2015). The PDCoV N protein-based ELISA was validated using 62 serum samples and had a sensitivity of 100% and a specificity of 90.4% when compared to the Western blot results on the same 62 serum samples (Su et al., 2015). The PDCoV whole virus-based ELISA was validated using a few immune sera generated from Gn pigs that were immunized intramuscularly twice with inactivated PDCoV; diagnostic sensitivity and specificity were not provided in the paper (Ma et al., 2016). The optical density (OD) of ELISA is determined using ELISA plate reader and the sample-to-positive (S/P) ratios are calculated accordingly if S/P ratios are used to determine ELISA results. ELISA is considered a relatively objective assay. ELISA is a high throughput assay and is suitable for testing a large number of samples. In addition, ELISA can be used to measure antibody isotypes (e.g. IgG, IgA, and IgM) in various specimen types. For example, PEDV ELISAs have been used to detect IgG and/or IgA antibodies in serum, oral fluid, and feces samples (Bjustrom-Kraft et al., 2016, Gerber et al., 2014, Gerber and Opriessnig, 2015). No publications have described dynamics of PDCoV antibody production and antibody duration as measured by PDCoV ELISAs.
3.2.4. Fluorescent microsphere immunoassay (FMIA)
The FMIA is a microsphere (bead)-based fluidic assay that has been increasingly used for serologic testing of animal infectious diseases (Christopher-Hennings et al., 2013, Langenhorst et al., 2012). For the FMIA, up to 100 color-coded bead sets (fluorescent microspheres) can be used and each bead set can be conjugated to individual antigens for the detection of antibodies in biological samples. One apparent advantage of FMIA is that it allows simultaneous detection of antibodies to multiple pathogens present in a single sample. A FMIA based on PEDV N protein has recently been developed for the detection of PEDV antibody in serum samples (Okda et al., 2015). The FMIA for the detection of PDCoV antibody has not been reported but it is expected that the FMIA technology can be successfully applied to detect PDCoV antibodies.
3.2.5. Antigenic cross-reactivity among swine enteric coronaviruses?
It is important to understand the antigenic relationships among porcine enteric coronaviruses (PDCoV, PEDV and TGEV) so that specific immunoassays for each virus can be developed. Ma et al. (2016) reported that the conserved or similar epitopes on N proteins of PEDV and PDCoV could cause two-way antigenic cross-reactivity of the two viruses. However, the pig PDCoV antisera did not cross-neutralize PEDV and the pig PEDV antisera did not cross-neutralize PDCoV either (Ma et al., 2016). In addition, the PDCoV IFA, S1 protein-based ELISA, and N protein-based ELISA assays specifically recognized antibodies against PDCoV and did not cross-react to antisera against PEDV or TGEV (Chen et al., 2015b, Ma et al., 2016, Su et al., 2015, Thachil et al., 2015).
Lin et al. (2015) reported that pig hyperimmune antisera against TGEV Miller strain cross-reacted with PEDV by cell culture immunofluorescence (CCIF) assay (similar to IFA assay described above) but pig antisera against PEDV did not cross-react with TGEV by CCIF assay; this one-way antigenic cross-reactivity by CCIF assay could be due to one epitope on the N-terminal region of PEDV/TGEV (Lin et al., 2015). It must be emphasized that pig antisera against PEDV did not cross-neutralize TGEV (regardless of TGEV Purdue strain or Miller strain) and vice versa (Lin et al., 2015). In addition, the PEDV whole virus-based ELISA and S1 protein-based ELISAs did not cross-react to pig antisera against PDCoV, PRCV, or TGEV Purdue strain in Chen et al. (2016b) study where pig antisera against TGEV Miller strain were not included. The PEDV N protein-based ELISAs also did not cross-react to pig antisera against TGEV and PRCV (Okda et al., 2015).
4. Prevalence of PDCoV infection and genetic evolution of PDCoV
4.1. History and prevalence of PDCoV infection in different countries
PDCoV has so far been detected in Hong Kong, the United States, Canada, South Korea, mainland China, and Thailand (Janetanakit et al., 2016, Lee and Lee, 2014, Li et al., 2014, Marthaler et al., 2014b, Song et al., 2015, Wang et al., 2014a, Woo et al., 2012).
4.1.1. In Hong Kong and mainland China
In a large-scale animal agent surveillance study conducted by a Hong Kong research group, a total of 7140 samples (tracheal swabs, rectal swabs, cloacal swabs, or nasopharyngeal aspirates, etc.) collected from multiple host species during a 53-month period (February 2007 to June 2011) were tested by a RT-PCR targeting conserved region of deltacoronavirus polymerase gene; among 169 pig samples, 17 pigs were positive (10.06% positive) and that was the first report of presence of deltacoronavirus in domestic pigs (Woo et al., 2012).
Several publications have reported detection of PDCoV RNA in domestic pigs in mainland China (Chen et al., 2015a, Dong et al., 2015, Song et al., 2015, Wang et al., 2015). Distribution and prevalence of PDCoV in China have not been determined nationwide but some local prevalence data have been reported. In one study, among 356 samples (170 fecal samples from sows or piglets less than 10 days old and 186 intestinal samples from piglets less than 10 days old) that were collected during November 2012 to March 2015 from diarrheic pigs on 51 farms in Jiangxi province, China and tested by PCRs, 120 (33.71%) samples were PDCoV positive, 231 (64.89%) were PEDV positive, 0 were positive for TGEV, and 70 (19.66%) were positive for both PDCoV and PEDV (co-infected); of the 120 PDCoV-positive samples, 70 (58.33%) were co-infected with PEDV (Song et al., 2015). In another study, 215 intestinal or fecal samples collected during 2004–2014 from diarrheic piglets in four provinces (Anhui, Guangxi, Hubei and Jiangsu) of China were tested by PCRs; among them, 14 (6.51%) were PDCoV positive, 110 (51.2%) were PEDV positive, 5 (2.3%) were TGEV positive, and 2 (0.9%) were co-infected with PDCoV, PEDV and TGEV; of the 14 PDCoV-positive samples, 7 (50%) were co-infected with PEDV (Dong et al., 2015). These studies suggest that PDCoV prevalence in Chinese swine is lower compared to PEDV infection and PDCoV co-infection with PEDV is common. Interestingly, one study also showed that PDCoV RNA was detected as early as in 2004 from diarrheic pig samples collected in Anhui province, China (Dong et al., 2015). This is the earliest date that PDCoV RNA was detected in pigs worldwide.
4.1.2. In the U.S. and Canada
In February 2014, the Ohio Department of Agriculture first announced the detection of PDCoV in U.S. swine after investigating the etiological agents for diarrheic sows and piglets on 5 Ohio farms (Wang et al., 2014a). Subsequently, the Veterinary Diagnostic Laboratories in Iowa and Minnesota reported detection of PDCoV in diarrheic pig samples collected from Iowa and Illinois (Li et al., 2014, Marthaler et al., 2014a). PDCoV was also detected in 6 Ontario farms in Canada in mid-March 2014 (Marthaler et al., 2014b). It remains unclear when PDCoV was introduced into the U.S., but a PCR-based retrospective testing of archived diagnostic samples at the Iowa State University Veterinary Diagnostic Laboratory revealed that PDCoV RNA could be detected as early as August 2013 in pig samples collected from Minnesota, Iowa and Illinois (Sinha et al., 2015). A retrospective serological study using an indirect PDCoV S1 protein-based ELISA suggested the presence of PDCoV IgG antibody in 4 archived serum samples collected in 2010 from U.S. pigs (Thachil et al., 2015).
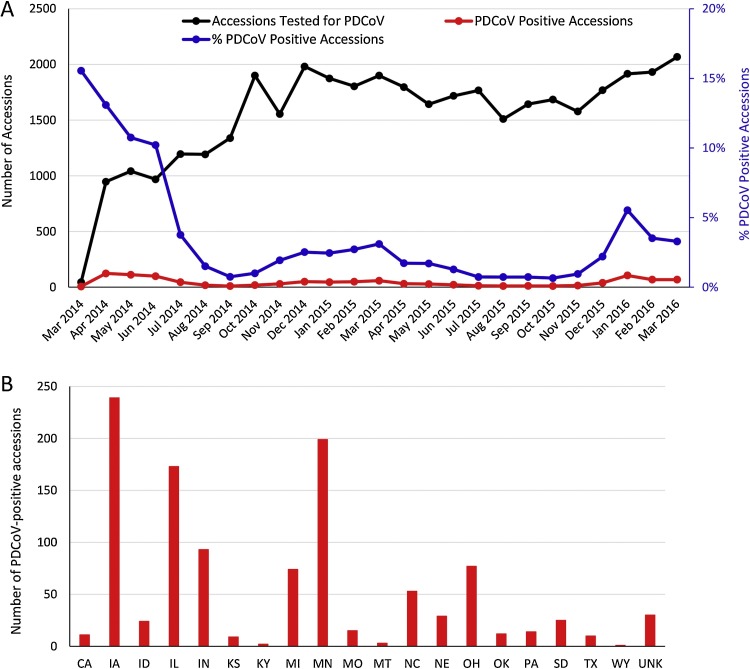
The prevalence of PDCoV in U.S. swine was evaluated in two studies performed in the early stage of PDCoV emergence in the US. In one study, 293 porcine samples collected during January 6 to February 27, 2014 from 7 U.S. states (Ohio, Illinois, Minnesota, Nebraska, Michigan, South Dakota, and Missouri) and Canada were tested by PCRs (Marthaler et al., 2014b). Of 293 samples, 89 (30%) were PDCoV positive and were from Ohio, Illinois, Minnesota, and Nebraska. Of the 89 PDCoV-positive samples, 69 (78%) were co-infected with PEDV, porcine rotavirus A, porcine rotavirus B, or porcine rotavirus C; PDCoV and porcine rotavirus C co-infection was most common (52/89 [58%]) while PDCoV and PEDV co-infection was 29/89 [33%] (Marthaler et al., 2014b). In another study, 435 samples collected during February 7 to April 9, 2014 from 10 U.S. states (Minnesota, South Dakota, Nebraska, Illinois, Indiana, Michigan, Kentucky, Pennsylvania, Maryland and Ohio) were tested by PCRs (Wang et al., 2014b). Of 435 samples, 109 (25%) samples from 9 aforementioned states except Maryland were PDCoV positive; of the 109 PDCoV-positive samples, 19 (17%) were co-infected with PEDV (Wang et al., 2014b). Now PDCoV has been present in U.S. swine for over two years since its emergence in February 2014, what is the current prevalence status of PDCoV in U.S. swine? The weekly-updated USDA Swine Enteric Coronavirus Disease (SECD) Situation Report (www.aphis.usda.gov/animal-health/secd), which compiles PDCoV and PEDV PCR testing results from 17 participating National Animal Health Laboratory Network (NAHLN) laboratories, provides a more accurate and thorough description on swine enteric coronavirus status in U.S. swine. Some key points on PDCoV prevalence based on the report data up to the end of March 2016 are summarized below.
-
•
From March 2014 to March 2016, a total of 38,774 biological accessions (not samples; one accession/case could include multiple samples) were tested for PDCoV by PCR and 1092 (2.8%) accessions were PDCoV positive. Distributions of biological accessions tested and the numbers as well as the percentages of PDCoV-positive accessions by month are shown in Fig. 3 A. During April-September 2014, approximately 1000–1500 accessions were tested per month; from October 2014 to March 2016, roughly 1500–2000 accessions were tested per month. The percentage of PDCoV-positive accessions ranged from 15.6%-10.2% during March-June 2014; the positive rates dropped to 3.8% in July 2014 and then were kept at low levels ( < 3%) from Aug 2014 to Dec 2015 although slight fluctuation was observed; in January 2016, the PDCoV-positive accession rate increased to 5.5%, consistent with the clinical observations that PDCoV broke on multiple farms in January 2016.
-
•
Distributions of 1092 PDCoV-positive accessions by state from where the samples were collected during March 2014-March 2016 are shown in Fig. 3B. The top 7 states that have been PDCoV positive are Iowa (239 positive accessions), Minnesota (199 positive accessions), Illinois (173 positive accessions), Indiana (93 positive accessions), Ohio (77 positive accessions), Michigan (74 positive accessions), and North Carolina (53 positive accessions).
-
•
From June 5, 2014, the USDA SECD reports have also included information on premises. During the periods of June 2014 to March 2016, a total of 160, 2523, and 141 premises in the U.S. have been confirmed positive by PCR for PDCoV, PEDV, and PDCoV & PEDV co-infection, respectively. These data indicate that in U.S. swine, prevalence of PDCoV has been much lower than that of PEDV; among PDCoV-positive premises, co-infection with PEDV is common. The confirmed positive premises for SECD (including PEDV, PDCoV, and PDCoV & PEDV co-infection) are further categorized into each production class: 360 SECD-positive Nursery premises, 726 SECD-positive Wean-to-Finish premises, 128 SECD-positive Farrow-to-Finish premises, 786 SECD-positive Finisher premises, 413 SECD-positive Sow/Breeding premises, and 407 SECD-positive premises with Unknown age information.
Fig. 3.
Biological accessions tested for PDCoV by RT-PCR in the US. (A) Distributions of biological accessions tested and the numbers as well as the percentages of PDCoV-positive accessions by month. (B) Distributions of PDCoV-positive accessions by state from where the samples were collected during March 2014-March 2016. Figures were drawn based on the data available at www.aphis.usda.gov/animal-health/secd.
4.1.3. In South Korea
In South Korea, PDCoV (KOR/KNU14-04/2014 isolate, GenBank accession no. KM820765) was first identified in April 2014 in fecal samples of diarrheic pigs (Lee and Lee, 2014). Subsequently, in a survey study, 681 fecal samples collected from pigs showing signs of diarrhea on 59 farms during the period of January 2013 to March 2015 were tested by PDCoV RT-PCR and only 2 samples collected from a 600-scale sow farm in March 2015 were positive for PDCoV (SL2 and SL5 isolates) (Lee et al., 2016). Co-infection of PDCoV with PEDV, TGEV, porcine rotavirus A, or Kobuvirus was not identified in that survey study (Lee et al., 2016).
4.1.4. In Thailand
In June 2015, acute diarrhea in piglets, gilts and sows on a swine farm was observed and testing 30 samples from the affected swine farm revealed that 26 samples were positive for PDCoV by RT-PCR; none of the 30 samples was positive for PEDV, TGEV, porcine rotavirus A, B, C, PRRSV, or circovirus (Janetanakit et al., 2016). No other prevalence data of PDCoV in Thailand is available at this point.
4.2. Genetic analyses of global PDCoVs
As of the end of March 2016, there are 50 complete PDCoV genomes available in GenBank. These include 2 from Hong Kong, 7 from the mainland China, 1 from South Korea, 2 from Thailand, and 38 from the U.S. with collection dates ranging from May 24, 2004 to Jun 30, 2015 (Table 3 ).
Table 3.
List of global PDCoVs with whole genome sequences available in GenBank through March 2016.
| Strain name | Country | Collection date | GenBank # | Reference | Note: nucleotide deletion or insertionsa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HKU15-44/2009 | China/Hong Kong | Year 2009 | JQ065042 | Woo et al. (2012) | |||||||||
| HKU15-155/2010 | China/Hong Kong | Year 2010 | JQ065043 | Woo et al., 2012) | 3-nt AAT del in S gene resulting in one aa N del; 3-nt TTA del in 3′UTR | ||||||||
| CHN/CHN-AN-2004 | China/Mainland | 24-May-2004 | KP757890 | Dong et al., 2015) | 3-nt TAA del in 3′UTR | ||||||||
| CHN/CH-Sichuan-S27/2012 | China/Mainland | Year 2012 | KT266822 | Wang et al., 2015) | 6-nt TTTGAA del in ORF1a between nt 1738 and 1745; 9-nt CCGGTTGGT del in ORF1a between nt 2810 and 2820; 3-nt AAT del in S gene resulting in one aa N del | ||||||||
| CHN/CHN—HN-2014 | China/Mainland | 24-Nov-2014 | KT336560 | Unpublished | 3-nt AAT del in S gene resulting in one aa N del | ||||||||
| CHN/CHN-JS-2014 | China/Mainland | 20-Dec-2014 | KP757892 | Dong et al. (2015) | 3-nt AAT del in S gene resulting in one aa N del | ||||||||
| CHN/CHN—HB-2014 | China/Mainland | 26-Dec-2014 | KP757891 | Dong et al. (2015) | 3-nt AAT del in S gene resulting in one aa N del | ||||||||
| CHN/CH-SXD1/2015 | China/Mainland | 20-Mar-2015 | KT021234 | Chen et al. (2015a) | 3-nt AAT del in S gene resulting in one aa N del | ||||||||
| CHN/CHJXNI2/2015 | China/Mainland | Mar-2015 | KR131621 | Song et al. (2015) | 3-nt AAT del in S gene resulting in one aa N del | ||||||||
| KOR/KNU14-04/2014 | South Korea | Apr-2014 | KM820765 | Lee and Lee (2014) | |||||||||
| THA/S5011/2015 | Thailand | 10-Jun-2015 | KU051641 | Janetanakit et al. (2016) | 3-nt CCT del in 5′UTR; 1-nt A del in 5′UTR; 6-nt TTTGAA del in ORF1a between nt 1738 and 1745; 9-nt CCGGTTGGT del in ORF1a between nt 2810 and 2820; 1-nt C ins in 3′UTR | ||||||||
| THA/S5015L/2015 | Thailand | 30-Jun-2015 | KU051649 | Janetanakit et al. (2016) | 3-nt CCT del in 5′UTR; 1-nt A del in 5′UTR; 6-nt TTTGAA del in ORF1a between nt 1738 and 1745; 9-nt CCGGTTGGT del in ORF1a between nt 2810 and 2820; 1-nt C ins in 3′UTR | ||||||||
| USA/Minnesota/2013 | USA | 14-Oct-2013 | KR265853 | Homwong et al. (2016) | |||||||||
| USA/Illinois121/2014 | USA | 4-Jan-2014 | KJ481931 | Marthaler et al. (2014a) | |||||||||
| USA/Illinois133/2014 | USA | 8-Jan-2014 | KJ601777 | Marthaler et al. (2014b) | |||||||||
| USA/Illinois134/2014 | USA | 8-Jan-2014 | KJ601778 | Marthaler et al. (2014b) | |||||||||
| USA/Illinois136/2014 | USA | 11-Jan-2014 | KJ601779 | Marthaler et al. (2014b) | |||||||||
| USA/Ohio137/2014 | USA | 26-Jan-2014 | KJ601780 | Marthaler et al. (2014b) | |||||||||
| USA/Ohio/OH1987/2014 | USA | 31-Jan-2014 | KJ462462 | Wang et al. (2014a) | |||||||||
| USA/Nebraska209/2014 | USA | 5-Feb-2014 | KR265860 | Homwong et al. (2016) | |||||||||
| USA/Nebraska210/2014 | USA | 5-Feb-2014 | KR265861 | Homwong et al. (2016) | |||||||||
| USA/Minnesota159/2014 | USA | 11-Feb-2014 | KR265859 | Homwong et al. (2016) | |||||||||
| USA/Illinois/IL2768/2014 | USA | 12-Feb-2014 | KJ584355 | Wang et al. (2014b) | |||||||||
| USA/Indiana/IN2847/2014 | USA | 13-Feb-2014 | KJ569769 | Wang et al. (2014c) | |||||||||
| USA/PA3148/2014 | USA | 18-Feb-2014 | KJ584358 | Wang et al. (2014b) | |||||||||
| USA/Iowa/IA8734/2014 | USA | 20-Feb-2014 | KJ567050 | Li et al. (2014) | |||||||||
| USA/SD3424/2014 | USA | 20-Feb-2014 | KJ584356 | Wang et al. (2014b) | |||||||||
| USA/Nebraska/NE3579/2014 | USA | 21-Feb-2014 | KJ584359 | Wang et al. (2014b) | |||||||||
| USA/Illinois272/2014 | USA | 23-Feb-2014 | KR265856 | Homwong et al. (2016) | |||||||||
| USA/Illinois273/2014 | USA | 23-Feb-2014 | KR265857 | Homwong et al. (2016) | |||||||||
| USA/OhioCVM1/2014 | USA | 1-Mar-2014 | KJ769231 | Ma et al. (2015) | |||||||||
| USA/Minnesota442/2014 | USA | 6-Mar-2014 | KR265847 | Homwong et al., 2016) | |||||||||
| USA/Kentucky/KY4813/2014 | USA | 7-Mar-2014 | KJ584357 | Wang et al. (2014b) | |||||||||
| USA/Minnesota214/2014 | USA | 14-Mar-2014 | KR265848 | Homwong et al. (2016) | |||||||||
| USA/Minnesota292/2014 | USA | 14-Mar-2014 | KR265864 | Homwong et al. (2016) | |||||||||
| USA/Michigan8977/2014 | USA | 17-Mar-2014 | KM012168 | Unpublished | |||||||||
| USA/Michigan/MI6148/2014 | USA | 18-Mar-2014 | KJ620016 | Wang et al. (2014b) | |||||||||
| USA/Ohio444/2014 | USA | 26-Mar-2014 | KR265862 | Homwong et al. (2016) | |||||||||
| USA/Ohio445/2014 | USA | 27-Mar-2014 | KR265863 | Homwong et al. (2016) | |||||||||
| USA/Michigan447/2014 | USA | 2-Apr-2014 | KR265849 | Homwong et al. (2016) | |||||||||
| USA/Michigan448/2014 | USA | 2-Apr-2014 | KR265850 | Homwong et al. (2016) | |||||||||
| USA/Illinois449/2014 | USA | 21-Apr-2014 | KR265852 | Homwong et al. (2016) | |||||||||
| USA/NorthCarolina452/2014 | USA | 6-May-2014 | KR265858 | Homwong et al. (2016) | |||||||||
| USA/Ohio/OH11846/2014 | USA | 7-May-2014 | KT381613 | Wang et al. (2016) | |||||||||
| USA/Indiana453/2014 | USA | 13-May-2014 | KR265851 | Homwong et al. (2016) | |||||||||
| USA/Minnesota454/2014 | USA | 21-May-2014 | KR265854 | Homwong et al. (2016) | |||||||||
| USA/Minnesota455/2014 | USA | 21-May-2014 | KR265855 | Homwong et al. (2016) | |||||||||
| USA/Illinois/2014/026PDV | USA | May-2014 | KP981395 | Chen et al. (2015b) | |||||||||
| USA/Iowa459/2014 | USA | 5-Jun-2014 | KR265865 | Homwong et al. (2016) | |||||||||
| USA/Arkansas61/2015 | USA | 24-Mar-2015 | KR150443 | Homwong et al. (2016) | |||||||||
Nucleotide deletions or insertions are based on comparison to the PDCoV HKU15-44/2009 strain. Nucleotide positions are based on PDCoV HKU15-44/2009.
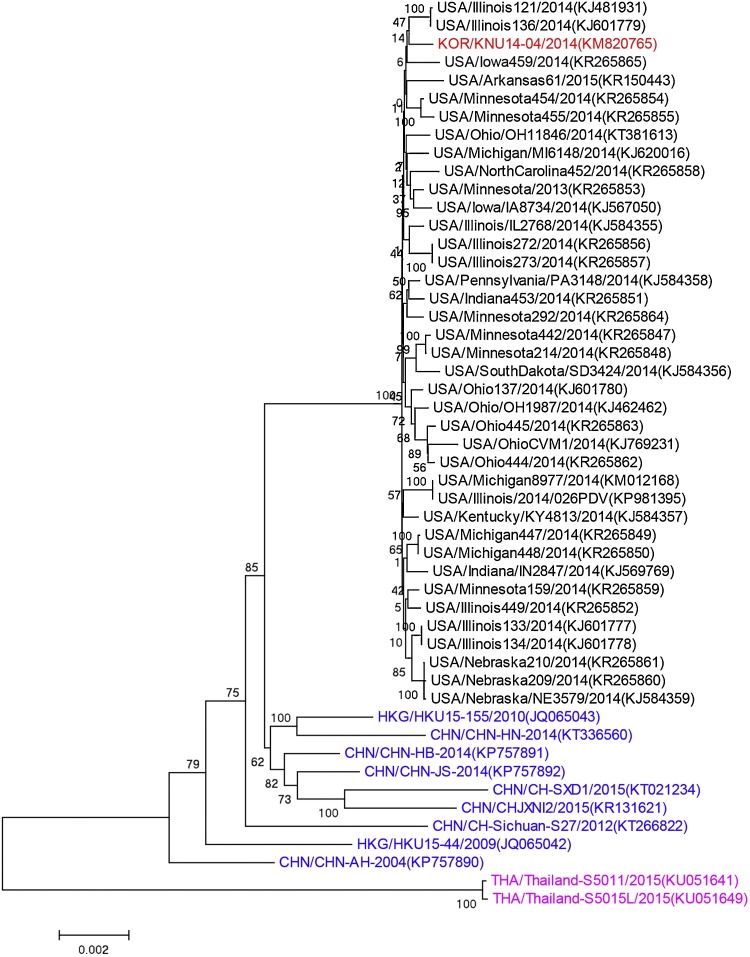
Phylogenetic analysis of 50 PDCoV whole genome sequences indicated that 38 U.S. PDCoVs and 1 Korean PDCoV clustered together, PDCoVs from Hong Kong and mainland China clustered separately from U.S. and Korean PDCoVs, 2 Thailand PDCoVs formed another separate cluster (Fig. 4 ). Phylogenetic trees based on PDCoV ORF1ab, S, M and N genes gave similar results to the tree based on the whole genomes (data not shown). Six Chinese isolates HKU15-155, HN-2014, HB-2014, JS-2014, SXD1, and CHJXNI2 appeared to be more closely related to each other than to the Chinese isolates Sichuan-S27, HKU15-44 and AH-2004.
Fig. 4.
Phylogenetic analysis of 50 global PDCoV complete genome sequences. The tree was constructed using the distance-based neighbor-joining method of the software MEGA6.06. Bootstrap analysis was carried out on 1000 replicate data sets, and values are indicated adjacent to the branching points. The US, Korean, Chinese, and Thailand PDCoVs are indicated by black, red, blue and purple font color, respectively.
Pairwise comparison of nucleotide identities of 50 global PDCoV is summarized in Table 4 . When all 50 sequences were compared, nucleotide identity data at the whole genome level and individual genes suggested that the S gene had more genetic diversity compared to other genes. The 50 sequences were further divided into 3 categories for pairwise comparisons: 9 Chinese isolates (including Hong Kong and mainland China), U.S. & Korean isolates (39 sequences), and 2 Thailand isolates. Among the U.S. & Korean PDCoVs, high nucleotide identity was observed (99.6-99.9% at the whole genome level and 99.5-99.9% for S gene), suggesting that the U.S. & Korean PDCoVs likely have the same origin; also, it appears that the U.S. PDCoVs have not undergone significant genetic changes during the periods October 14, 2013 to March 24, 2015 when these U.S. PDCoV samples were collected. Among 9 Chinese PDCoV isolates, nucleotide identities ranged from 98.6-99.5% and the genetic diversity among Chinese PDCoVs was higher than that observed among U.S. & Korean PDCoVs. The 2 Thailand PDCoVs were highly similar to each other but the Thailand PDCoVs were not very closely related to Chinese, U.S. or Korean PDCoVs.
Table 4.
Pairwise comparison of nucleotide identity of 50 global porcine deltacoronaviruses by different geness.
| Viruses | Gene, nucleotide identity% |
|||||||
|---|---|---|---|---|---|---|---|---|
| Whole genome | ORF1ab | S | E | M | NS6 | N | NS7 | |
| Among all global PDCoVs (50 seq) | 97.1–99.9 | 97.3–100 | 95.4–100 | 98.4–100 | 98–100 | 97.8–100 | 96.9–100 | 97.3–100 |
| Among Chinese strains (Hong Kong & mainland) (9 seq) | 98.6–99.5 | 98.6–99.6 | 97.3–99.6 | 98.8–99.6 | 99−99.6 | 98.2–99.6 | 98.3–99.9 | 98.3–99.8 |
| Among US & Korean strains (39 seq) | 99.6–99.9 | 99.7–99.9 | 99.5–99.9 | 99.2–100 | 99.5–100 | 99.6–100 | 99.1–99.8 | 98.6–100 |
| Among Thailand strains (2 seq) | 99.9 | 99.9 | 99.9 | 100 | 100 | 100 | 100 | 100 |
| Chinese (9 seq) vs US & Korean strains (39 seq) | 98.7–99.2 | 98.7–99.4 | 97.9–98.9 | 98.4–99.6 | 98.6–99.3 | 98.5–100 | 98.1–99.3 | 98.3–99.6 |
| Chinese (9 seq) vs Thailand strains (2 seq) | 97.1–97.7 | 97.3–97.9 | 95.4–96.8 | 99.2–100 | 98.6–99 | 97.8–98.9 | 97.1–97.8 | 97.5–98.3 |
| US & Korean (39 seq) vs Thailand strains (2 seq) | 97.2–97.3 | 97.4–97.5 | 96.1–96.3 | 99.2–100 | 98−98.4 | 98.2–98.9 | 96.9–97.2 | 97.3–97.6 |
When 38 U.S. PDCoVs and 1 Korean PDCoV (KNU14-04) were compared, no deletions or insertions were identified among them. In contrast, Chinese and Thailand PDCoVs have shown some deletions or insertions (Table 3). Specifically, when compared to the Chinese HKU15-44 and US & Korean PDCoVs, the Chinese strains HKU15-155, HN-2014, JS-2014, HB-2014, SXD1, and CHJXNI2 all had the same 3-nt (AAT) deletion in the S gene between nt 19473 and 19477, resulting in deletion of one deduced amino acid Asparagine (Asn or N). The 2 Thailand PDCoVs had a 6-nt (TTTGAA) deletion in ORF1a between nt 1738 and 1745 resulting in amino acid changes from SSLKI to SRI as well as a 9-nt (CCGGTTGGT) deletion in ORF1a between nt 2810 and 2820 resulting in amino acid changes from PEPVGKV to PDDV, when compared to Chinese HKU15-44 and U.S. & Korean PDCoVs. Interestingly, the Chinse Sichun-S27 PDCoV harbored both the 3-nt deletion in the S gene and the 6-nt & 9-nt deletions in the ORF1a. The biological significances of these deletions remain unknown. The evolutionary rates of PDCoVs are estimated to be 3.8 × 10−4 substitutions/site/year at the whole genome level and 2.0 × 10−3 substitutions/site/year for the spike gene (3483 nt) (Homwong et al., 2016).
Although the earliest detection of PDCoV was in 2004 in mainland China, it is difficult to decipher the evolutionary relationships of PDCoVs from different countries and to definitely infer the origin country of PDCoV at this point with limited sequence data. However, the currently available sequence data support the inference that PDCoV in South Korea was from U.S. although the routes of introducing PDCoV from U.S. to South Korea have not been identified. Not only for PDCoV, other infectious diseases such as PED have emerged or reemerged in multiple countries in recent years. Without knowing introduction routes of the pathogens between countries, it still remains a serious challenge to efficiently control spread of the emerging or reemerging diseases between countries or continents.
How DCoV originated in or was transmitted to domestic pigs remains unclear. In 2007, Dong et al. (2007) reported a previously unrecognized coronavirus lineage detected from wild Asian leopard cats and Chinese ferret badgers from samples collected in southern China in 2006. In 2009, Woo et al. (2009b) reported identification of three novel avian coronaviruses in wild birds (bulbul CoV HKU11, thrush CoV HKU12, and munia CoV HKU13). These three novel avian CoVs and CoVs identified from Asian leopard cats and Chinese ferret badgers all belong to the newly established Deltacoronavirus genus (Woo et al., 2010, Woo et al., 2012). In 2012, identification of seven additional novel deltacoronaviruses in pigs and birds were reported (Woo et al., 2012). Among deltacoronaviruses, PDCoV is most genetically similar to CoVs in Asian leopard cats based on available helicase, spike and nucleocapsid gene sequences (Woo et al., 2012). It is unknown how DCoVs were transmitted between small mammals (such as Asian leopard cats and Chinese ferret badgers) and domestic pigs. A model of CoV evolution has been proposed in which bats are the gene source of Alphacoronavirus and Betacornoavirus whereas birds are the gene source of Gammacoronavirus and Deltacoronavirus (Woo et al., 2009a, Woo et al., 2012). It is likely that DCoVs are transmitted from wild birds to small mammals and/or domestic pigs. More surveillance and sequence data are needed in order to understand interspecies transmission of DCoVs between wild birds and small mammals, between wild birds and domestic pigs, and/or between small mammals and domestic pigs.
5. Conclusion
Recent outbreaks of PDCoV in multiple countries have caused significant economic losses. Rapid diagnosis is critical for the implementation of efficient control strategies against PDCoV. Understanding PDCoV infection dynamics and collecting appropriate specimens at the appropriate time window are also important to obtain reliable diagnostic results. A number of virological and serological methods have been developed and used for PDCoV diagnostic testing. Among the virological methods, PDCoV-specific RT-PCR remains the method of choice for the detection of PDCoV; IHC combined with hematoxylin and eosin staining has also been commonly used to examine histopathological lesions caused by PDCoV; success rate of virus isolation in cell cultures has been low. Serological assays can provide information about previous exposure to PDCoV and also determine antibody responses to infection or vaccination when vaccine(s) are available. So far, kinetics of antibody responses to PDCoV infection and duration of PDCoV antibodies have not been studied in detail; correlations of various serological assays for the detection of PDCoV antibodies have not been established. PDCoV has been detected in several countries e.g. China, US, South Korea and Thailand. Prevalence of PDCoV in these countries is lower compared to that of PEDV. However, among PDCoV-positive samples, co-infection with other enteric pathogen e.g. PEDV is common. PDCoV RNA has been detected in samples collected in 2004 in mainland China. PDCoVs circulating in China have shown some different genetic characteristics as evidenced by phylogenetic and comparative sequence analyses. In contrast, PDCoVs detected in the U.S. and South Korea are genetically closely related and have not shown remarkable genetic changes. PDCoVs detected in Thailand are not very closely related to Chinese, US and Korean PDCoVs. The origin of PDCoV has not been clearly identified. It remains to be determined whether global PDCoVs have the similar pathogenicity.
Acknowledgments
I thank faculty, staff and graduate students at the Iowa State University Veterinary Diagnostic Laboratory who have participated in porcine deltacoronavirus diagnostics and research. I also thank all individuals who have published porcine deltacoronavirus work in the literatures that make this review article possible.
References
- Bjustrom-Kraft J., Woodard K., Gimenez-Lirola L.G., Rotolo M., Wang C., Sun Y., Lasley P., Zhang J., Baum D., Gauger P., Main R., Zimmerman J. Porcine epidemic diarrhea virus (PEDV) detection and antibody response in commercial growing pigs. BMC Vet. Res. 2016;12:99. doi: 10.1186/s12917-016-0725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A.S., Krogwold R.A., Price T., Davis M., Moeller S.J. Investigating the introduction of porcine epidemic diarrhea virus into an Ohio swine operation. BMC Vet. Res. 2015;11:38. doi: 10.1186/s12917-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A.S., Nolting J.M., Nelson S.W., Bliss N., Stull J.W., Wang Q., Premanandan C. Effects of disinfection on the molecular detection of porcine epidemic diarrhea virus. Vet. Microbiol. 2015;179(3–4):213–218. doi: 10.1016/j.vetmic.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014;52(1):234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhu Y., Wu M., Ku X., Yao L., He Q. Full-Length genome characterization of chinese porcine deltacoronavirus strain CH/SXD1/2015. Genome Announc. 2015;3(5) doi: 10.1128/genomeA.01284-15. e01284-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P.C., Stafne M.R., Thomas J.T., Madson D.M., Huang H., Zheng Y., Li G., Zhang J. Pathogenesis comparison between the United States porcine epidemic diarrhea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J. Gen. Virol. 2016;97:1107–1121. doi: 10.1099/jgv.0.000419. [DOI] [PubMed] [Google Scholar]
- Chen Q., Thomas J.T., Gimenez-Lirola L.G., Hardham J.M., Gao Q., Gerber P.F., Opriessnig T., Zheng Y., Li G., Gauger P.C., Madson D.M., Magstadt D.R., Zhang J. Evaluation of serological cross-reactivity and cross-neutralization between the United States porcine epidemic diarrhea virus prototype and S-INDEL-variant strains. BMC Vet. Res. 2016;12(1):70. doi: 10.1186/s12917-016-0697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher-Hennings J., Araujo K.P., Souza C.J., Fang Y., Lawson S., Nelson E.A., Clement T., Dunn M., Lunney J.K. Opportunities for bead-based multiplex assays in veterinary diagnostic laboratories. J. Vet. Diagn. Invest. 2013;25(6):671–691. doi: 10.1177/1040638713507256. [DOI] [PubMed] [Google Scholar]
- Clement T., Singrey A., Lawson S., Okda F., Nelson J., Diego D., Nelson E., Christopher-Hennings J. Measurement of neutralizaing antibodies against porcine epidemic diarrhea virus in sow serum, colostrum, and milk samples and piglet serum samples after feedback. J. Swine Heal. Prod. 2016;24:1–10. [Google Scholar]
- Dee S., Clement T., Schelkopf A., Nerem J., Knudsen D., Christopher-Hennings J., Nelson E. An evaluation of contaminated complete feed as a vehicle for porcine epidemic diarrhea virus infection of naive pigs following consumption via natural feeding behavior: proof of concept. BMC Vet. Res. 2014;10(1):176. doi: 10.1186/s12917-014-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee S., Neill C., Clement T., Christopher-Hennings J., Nelson E. An evaluation of a liquid antimicrobial (Sal CURB(R)) for reducing the risk of porcine epidemic diarrhea virus infection of naive pigs during consumption of contaminated feed. BMC Vet. Res. 2014;10:220. doi: 10.1186/s12917-014-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee S., Neill C., Clement T., Singrey A., Christopher-Hennings J., Nelson E. An evaluation of porcine epidemic diarrhea virus survival in individual feed ingredients in the presence or absence of a liquid antimicrobial. Porcine Health Manage. 2015;1:9. doi: 10.1186/s40813-015-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L., Zhang S.Y., Peiris J.S., Smith G.J., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007;81(13):6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 2015;21(12):2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber P.F., Opriessnig T. Detection of immunoglobulin (Ig) A antibodies against porcine epidemic diarrhea virus (PEDV) in fecal and serum samples. MethodsX. 2015;2:368–373. doi: 10.1016/j.mex.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber P.F., Gong Q., Huang Y.W., Wang C., Holtkamp D., Opriessnig T. Detection of antibodies against porcine epidemic diarrhea virus in serum and colostrum by indirect ELISA. Vet. J. 2014;202(1):33–36. doi: 10.1016/j.tvjl.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homwong N., Jarvis M.C., Lam H.C., Diaz A., Rovira A., Nelson M., Marthaler D. Characterization and evolution of porcine deltacoronavirus in the United States. Prev. Vet. Med. 2016;123:168–174. doi: 10.1016/j.prevetmed.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53(5):1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetanakit T., Lumyai M., Bunpapong N., Boonyapisitsopa S., Chaiyawong S., Nonthabenjawan N., Kesdaengsakonwut S., Amonsin A. Porcine deltacoronavirus, Thailand, 2015. Emerg. Infect. Dis. 2016;22(4):757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., Saif L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21(4):650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016 doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenhorst R.J., Lawson S., Kittawornrat A., Zimmerman J.J., Sun Z., Li Y., Christopher-Hennings J., Nelson E.A., Fang Y. Development of a fluorescent microsphere immunoassay for detection of antibodies against porcine reproductive and respiratory syndrome virus using oral fluid samples as an alternative to serum-based assays. Clin. Vaccine Immunol. 2012;19(2):180–189. doi: 10.1128/CVI.05372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Complete genome characterization of korean porcine deltacoronavirus strain KOR/KNU14-04/ Genome Announc. 2014;2(6) doi: 10.1128/genomeA.01191-14. e01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Chung H.C., Nguyen V.G., Moon H.J., Kim H.K., Park S.J., Lee C.H., Lee G.E., Park B.K. Detection and phylogenetic analysis of porcine deltacoronavirus in korean swine farms, 2015. Transbound. Emerg. Dis. 2016 doi: 10.1111/tbed.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Chen Q., Harmon K.M., Yoon K.J., Schwartz K.J., Hoogland M.J., Gauger P.C., Main R.G., Zhang J. Full-Length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2014;2(2) doi: 10.1128/genomeA.00278-14. e00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Gao X., Oka T., Vlasova A.N., Esseili M.A., Wang Q., Saif L.J. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J. Virol. 2015;89(6):3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Gauger P., Harmon K., Zhang J., Connor J., Yeske P., Loula T., Levis I., Dufresne L., Main R. Role of transportation in spread of porcine epidemic diarrhea virus infection, United States. Emerg. Infect. Dis. 2014;20(5):872–874. doi: 10.3201/eid2005.131628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio. 2015;6(2):e00064. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Oglesbee M., Krakowka S., Niehaus A., Wang G., Jia A., Song H., Li J. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine deltacoronavirus. Vet. Microbiol. 2016;186:90–96. doi: 10.1016/j.vetmic.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Jiang Y., Collins J., Rossow K. Complete genome sequence of strain SDCV/USA/Illinois121/2014, a porcine deltacoronavirus from the United States. Genome Announc. 2014;2(2) doi: 10.1128/genomeA.00218-14. e00218-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Raymond L., Jiang Y., Collins J., Rossow K., Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg. Infect. Dis. 2014;20(8):1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. Coronaviridae. In: Knipe D.M., Howley P.M., editors. Field Virology. sixth ed. Wolters Kluwer; Lippincott Williams & Wilkins: 2013. pp. 825–858. [Google Scholar]
- Moes E., Vijgen L., Keyaerts E., Zlateva K., Li S., Maes P., Pyrc K., Berkhout B., van der Hoek L., Van Ranst M. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect. Dis. 2005;5(1):6. doi: 10.1186/1471-2334-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Saif L.J., Marthaler D., Esseili M.A., Meulia T., Lin C.M., Vlasova A.N., Jung K., Zhang Y., Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 2014;173(3–4):258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okda F., Liu X., Singrey A., Clement T., Nelson J., Christopher-Hennings J., Nelson E.A., Lawson S. Development of an indirect ELISA, blocking ELISA, fluorescent microsphere immunoassay and fluorescent focus neutralization assay for serologic evaluation of exposure to North American strains of Porcine Epidemic Diarrhea Virus. BMC Vet. Res. 2015;11:180. doi: 10.1186/s12917-015-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasick J., Berhane Y., Ojkic D., Maxie G., Embury-Hyatt C., Swekla K., Handel K., Fairles J., Alexandersen S. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound. Emerg. Dis. 2014;61(5):397–410. doi: 10.1111/tbed.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillatzki A., Gauger P., Madson D., Burrough E., Zhang J., Chen Q., Magstadt D., Arruda P., Stevenson G., Yoon K.J. Experimental inoculation of neonatal piglets with feed naturally contaminated with porcine epidemic diarrhea virus (PEDV) J. Swine Health Prod. 2015;23(6):317–320. [Google Scholar]
- Sinha A., Gauger P., Zhang J., Yoon K.J., Harmon K. PCR-based retrospective evaluation of diagnostic samples for emergence of porcine deltacoronavirus in US swine. Vet. Microbiol. 2015;179(3–4):296–298. doi: 10.1016/j.vetmic.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-Length genome sequence analysis. Transbound. Emerg. Dis. 2015;62(6):575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25(5):649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Su M., Li C., Guo D., Wei S., Wang X., Geng Y., Yao S., Gao J., Wang E., Zhao X., Wang Z., Wang J., Wu R., Feng L., Sun D. A recombinant nucleocapsid protein-based indirect enzyme-linked immunosorbent assay to detect antibodies against porcine deltacoronavirus. J. Vet. Med. Sci. 2015 doi: 10.1292/jvms.15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thachil A., Gerber P.F., Xiao C.T., Huang Y.W., Opriessnig T. Development and application of an ELISA for the detection of porcine deltacoronavirus IgG antibodies. PLoS One. 2015;10(4):e0124363. doi: 10.1371/journal.pone.0124363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.T., Chen Q., Gauger P.C., Gimenez-Lirola L.G., Sinha A., Harmon K.M., Madson D.M., Burrough E.R., Magstadt D.R., Salzbrenner H.M., Welch M.W., Yoon K.J., Zimmerman J.J., Zhang J. Effect of porcine epidemic diarrhea virus infectious doses on infection outcomes in naive conventional neonatal and weaned pigs. PLoS One. 2015;10(10):e0139266. doi: 10.1371/journal.pone.0139266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.L., Wang H.T., Chang H.F., Tsai C.F., Lin C.K., Teng P.H., Su C., Jeng C.C., Lee P.Y. Development of TaqMan probe-based insulated isothermal PCR (iiPCR) for sensitive and specific on-site pathogen detection. PLoS One. 2012;7(9):e45278. doi: 10.1371/journal.pone.0045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014;20(7):1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg. Infect. Dis. 2014;20(9):1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhang Y., Byrum B. Complete genome sequence of porcine Coronavirus HKU15 strain IN2847 from the United States. Genome Announc. 2014;2(2) doi: 10.1128/genomeA.00291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.W., Yue H., Fang W., Huang Y.W. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from mainland China. Genome Announc. 2015;3(5) doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hayes J., Sarver C., Byrum B., Zhang Y. Porcine deltacoronavirus: histological lesions and genetic characterization. Arch. Virol. 2016;161:171–175. doi: 10.1007/s00705-015-2627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234(10):1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lai K.K., Huang Y., Lee P., Luk G.S., Dyrting K.C., Chan K.H., Yuen K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009;83(2):908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Tsai Y.L., Lee P.A., Chen Q., Zhang Y., Chiang C.J., Shen Y.H., Li F.C., Chang H.G., Gauger P.C., Harmon K.M., Wang H.T. Evaluation of two singleplex reverse transcription-Insulated isothermal PCR tests and a duplex real-time RT-PCR test for the detection of porcine epidemic diarrhea virus and porcine deltacoronavirus. J. Virol. Methods. 2016;234:34–42. doi: 10.1016/j.jviromet.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]