Highlights
-
•
Porcine deltacoronavirus (PDCoV) is a novel swine enteropathogenic coronavirus.
-
•
The etiology and molecular epidemiology of PDCoV are described.
-
•
The cell culture for PDCoV isolation and propagation are demonstrated.
-
•
The disease mechanisms and pathogenesis of PDCoV are reviewed.
Keywords: Porcine deltacoronavirus, Pathogenesis, Review, Disease, Virus, Pigs
Abstract
Porcine deltacoronavirus (PDCoV) (family Coronaviridae, genus Deltacoronavirus) is a novel swine enteropathogenic coronavirus that causes acute diarrhea/vomiting, dehydration and mortality in seronegative neonatal piglets. PDCoV diarrhea was first reported in the US in early 2014, concurrently with co-circulation of porcine epidemic diarrhea virus (PEDV) (family Coronaviridae, genus Alphacoronavirus). The origin of PDCoV in pigs and also its sudden emergence or route of introduction into the US still remains unclear. In the US, since 2013–2014, the newly emerged PDCoV and PEDV have spread nationwide, causing a high number of pig deaths and significant economic impacts. The current US PDCoV strains are enteropathogenic and infect villous epithelial cells of the entire small and large intestines although the jejunum and ileum are the primary sites of infection. Similar to PEDV infections, PDCoV infections also cause acute, severe atrophic enteritis accompanied by transient viremia (viral RNA) that leads to severe diarrhea and/or vomiting, followed by dehydration as the potential cause of death in nursing piglets. At present, differential diagnosis of PDCoV, PEDV, and transmissible gastroenteritis virus (TGEV) is essential to control viral diarrheas in US swine. Cell culture-adapted US PDCoV (TC-PDCoV) strains have been isolated and propagated by us and in several other laboratories. TC-PDCoV strains will be useful to develop serologic assays and to evaluate if serial cell-culture passage attenuates TC-PDCoV as a potential vaccine candidate strain. A comprehensive understanding of the pathogenesis and epidemiology of epidemic PDCoV strains is currently needed to prevent and control the disease in affected regions and to develop an effective vaccine. This review focuses on the etiology, cell culture isolation and propagation, molecular epidemiology, disease mechanisms and pathogenesis of PDCoV infection.
1. Introduction
Porcine deltacoronavirus (PDCoV), a member of the genus Deltacoronavirus in the family Coronaviridae of the order Nidovirales, causes acute diarrhea, vomiting, dehydration and mortality in neonatal piglets (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015). PDCoV was first reported in pigs in Hong Kong in 2012 (Woo et al., 2012). Among a total of 7140 rectal, tracheal, or cloacal swab samples from wild Asian leopard cats, bats, wild birds, domestic cats, cattle, chickens, dogs, pigs, rodents, monkeys, and humans in China and Hong Kong in 2007–2011, DCoVs were detected only in pigs and wild birds, with a 10.1% positive rate for PDCoV in the 169 swine rectal swab samples tested (Woo et al., 2012). However, it appears that DCoVs were present in rectal swabs of small mammals, such as Asian leopard cats and Chinese ferret badgers, at live-animal markets in China since 2005–2006 (Dong et al., 2007). PDCoV diarrhea was first reported in the US in early 2014 (Li et al., 2014, Marthaler et al., 2014b, Wang et al., 2014a, Wang et al., 2014b), concurrently with porcine epidemic diarrhea virus (PEDV) (family Coronaviridae, genus Alphacoronavirus), which causes similar, but more severe clinical disease in neonatal piglets (Cima, 2013, Stevenson et al., 2013). Recent studies reported that PDCoV CHN-AH-2004 and CH/Sichuan/S27/2012 strains, genetically related to the current US PDCoV strains, were previously present in diarrheic piglets in China in 2004 and 2012, respectively, prior to the first outbreak of PDCoV infection in the US in early 2014 (Dong et al., 2015, Wang et al., 2015b).
The family Coronaviridae can be genetically divided into the four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Woo et al., 2012). Bats are the projected host for the gene source of Alphacoronaviruses and Betacoronaviruses, while birds are the suspect host for Gammacoronaviruses and Deltacoronaviruses (Woo et al., 2012). The other swine enteropathogenic coronavirus, transmissible gastroenteritis virus (TGEV), also belongs to Alphacoronavirus. Similar epidemiological, clinical, and pathological features among PDCoV, PEDV and TGEV have complicated their diagnosis, requiring differential laboratory tests (Jung and Saif, 2015). In the US, since 2013–2014, the newly emerged PEDV and PDCoV have spread nationwide and affected pigs alone or jointly, causing a high number of pig deaths and significant economic impacts and hindering the control of both viral infections in farms dually affected by these two viruses (Anon., 2014, Jung and Saif, 2015). At present, differential diagnosis of PEDV, PDCoV, and TGEV is critical to control the coronaviral epidemic diarrheas in US pig farms.
Cultivable, cell culture-adapted PDCoV (TC-PDCoV) strains include the two isolates, USA/IL/2014 and Michigan/8977/2014, isolated at the USDA National Veterinary Services Laboratories (Chen et al., 2015b, Ma et al., 2015) and TC-PDCoV strain OH-FD22 isolated in our lab (Hu et al., 2015). The TC-PDCoV OH-FD22 has been serially passaged >90 times on LLC-PK cells, with high sustained infectious titers (Hu et al., 2015). The TC-PDCoV strains are useful for PDCoV pathogenesis studies, virological and serological assays, and vaccine development. This review focuses on the current understanding of the etiology, molecular epidemiology, disease mechanisms and pathogenesis of PDCoV, and cell culture isolation and propagation of the virus.
2. Etiology
2.1. PDCoV structure and genome
PDCoV is enveloped and pleomorphic with diameter of 60–180 nm, excluding the projections (Ma et al., 2015). PDCoV has a single-stranded positive-sense RNA genome of approximately 25.4 kb in size (excluding the poly A-tail) that encodes four structural proteins, namely, spike (S), envelope (E), membrane (M), and nucleocapsid (N), and four nonstructural proteins (Lee and Lee, 2014, Li et al., 2014, Ma et al., 2015, Marthaler et al., 2014a, Wang et al., 2015b). The PDCoV genome organization and arrangement consist of: 5′ untranslated region, open reading frame 1a/1b (ORF1a/1b), S, E, M, nonstructural protein 6 (NS6), N, nonstructural protein 7 (NS7), and 3′ UTR. Although general characteristics of the structural and nonstructural proteins of coronaviruses and their roles in viral replication have been identified (Jung and Saif, 2015, Saif et al., 2012), the detailed functions and roles of structural and nonstructural proteins of PDCoV in host cells are unknown. A recent study reported multifunctional roles of PDCoV N protein similar to those of other coronaviruses. The N protein was identified in both the nucleus and cytoplasm of stably N-expressing porcine kidney cells, suggesting RNA synthesis or ribosome biogenesis through its interactions with ribosomal subunits or nucleolar proteins (Lee and Lee, 2015).
2.2. Antigenic cross-reactivity between PDCoV and PEDV/TGEV
There was no cross-reactivity of PDCoV USA/IL/2014 strain with antibodies to either PEDV or TGEV, as determined by indirect immunofluorescence assays of virus-infected cells using a rabbit antiserum against synthetic M protein of PDCoV, a PEDV monoclonal antibody (SD6-29) targeting the N protein, and a TGEV polyclonal antiserum (Chen et al., 2015b). This finding was also supported by other reports (Jung et al., 2015c, Ma et al., 2015), showing no cross-reactivity of pig hyperimmune or convalescent antiserum to PDCoV OH-FD22 or CVM1 strains with PEDV, as determined by immunohistochemical staining of frozen or formalin-fixed intestinal tissues of PEDV-infected pigs. However, a recent study reported evidence of antigenic cross-reactivity between PDCoV Michigan/8977/2014 strain and PEDV, possibly sharing at least one conserved or similar epitope on their N proteins, as determined by enzyme-linked immunosorbent assay (ELISA) and western blot using monoclonal PEDV and PDCoV N-specific antibodies, whereas no cross-reactivity was detected when virus neutralization, indirect immunofluorescence, and immunostaining assays were conducted on either virus-infected cells or intestinal tissues using pig hyperimmune antisera to PEDV or PDCoV (Ma et al., 2016).
3. Cell culture for isolation and propagation of PDCoV
LLC porcine kidney (LLC-PK) and swine testicular (ST) cells support the isolation and serial propagation of PDCoV in cell cultures supplemented with exogenous protease trypsin or pancreatin (Hu et al., 2015). Trypsin contributed to a significant increase in PDCoV growth after several passages in LLC-PK cells. However, even without supplemental trypsin, PDCoV replicated in LLC-PK cells, but without cytopathic effects (CPE) and at relatively lower virus titers compared to the trypsin supplemented cell cultures (Hu et al., 2015). Thus, trypsin is not essential, but is beneficial for the propagation of PDCoV in LLC-PK cells. The cell-culture conditions required for the propagation of PDCoV in ST cells differ from those in LLC-PK cells. Trypsin did not support PDCoV propagation in ST cells. In contrast, PDCoV could be serially propagated in ST cells supplemented with pancreatin or small intestinal contents from gnotobiotic (Gn) piglets (Hu et al., 2015). The addition of trypsin and pancreatin in PDCoV-inoculated LLC-PK and ST cells, respectively, resulted in similar CPE that consisted of enlarged, rounded, and densely granular cells that occurred singly or in clusters and then, cell shrinkage and detachment as a result of apoptotic cell death (Jung et al., 2016, Hu et al., 2015). The roles of trypsin or pancreatin in cell entry and release of PDCoV virions in LLC-PK or ST cells remain unknown.
4. Molecular epidemiology
4.1. Discovery of deltacoronaviruses
Coronaviruses in the genus Deltacoronavirus have been identified recently in a variety of wild small mammals and avian species (Dong et al., 2007, Woo et al., 2009a, Woo et al., 2009b, Woo et al., 2012). In 2006, novel coronaviruses were detected in feces of wild Asian leopard cats (Prionailurus bengalensis) and Chinese ferret badgers (Melogale moschata) during a virological surveillance for severe acute respiratory syndrome (SARS) coronavirus in southern China. The isolates were genetically similar to each other and commonly had low amino acid similarity and unique phylogeny distinct from other coronavirus groups (Dong et al., 2007). The two isolates were subsequently shown genetically to belong to Deltacoronavirus (Woo et al., 2012). Another molecular epidemiology study was conducted in dead wild birds for a 7-month period (December 2006 to June 2007) in Hong Kong (Woo et al., 2009b). Three novel avian coronaviruses (BuCoV HKU11, ThCoV HKU12, and MuCoV HKU13) were identified and proposed as a novel coronavirus subgroup 3C under the group 3 coronaviruses (Woo et al., 2009b). Recently, the three isolates were further identified genetically to belong to Deltacoronavirus (King et al., 2012).
A coronavirus surveillance study was conducted in a variety of domestic and wild animal species from 2007 to 2011, and the presence of DCoVs in the feces of domestic pigs was first reported in 2012 (Woo et al., 2012). Seven DCoVs in pigs and wild birds were identified, which were designated porcine coronavirus HKU15 and the avian coronavirueses, white-eye coronavirus HKU16, sparrow coronavirus HKU17, magpie robin coronavirus HKU18, night heron coronavirus HKU19, wigeon coronavirus HKU20, and common moorhen coronavirus HKU21 (Woo et al., 2012). This study first verified that PDCoV was present in the feces of pigs in Mainland China and Hong Kong in 2007–2011, with a positive rate of 10.1%. Two Chinese PDCoV complete genome sequences were generated from two different isolates, HKU15-44 and HKU15-155 (GenBank accession no. JQ065042 and JQ065043, respectively) (Woo et al., 2012). The helicase, S and N genes of PDCoV HKU15-44 and HKU15-155 were closely related to those of the coronaviruses isolated from the Asian leopard cats and Chinese ferret badgers [nucleotide (nt) similarity ≥99.8%] (Woo et al., 2012) (Fig. 1 A), implying the potential interspecies transmission of a DCoV between these wild small mammals and pigs. Further molecular surveillance studies are needed to define the potential role of the small mammals as an intermediate host of PDCoV and the mechanisms of interspecies transmission of DCoVs between the small mammals and domestic pigs or wild birds, and also between domestic pigs and wild birds.
Fig. 1.
Phylogenetic analyses of the S gene nucleotide sequences (A) and complete genomes (B) of some representative porcine and avian deltacoronaviruses. Reference sequences obtained from GenBank are indicated by strain names and accession numbers. The trees were constructed using the distance-based neighbor-joining method of the software MEGA6.06 (http://www.megasoftware.net). Bootstrap analysis was carried out on 1000 replicate data sets, and values are indicated adjacent to the branching points. Scale bars represent 0.1 nucleotide substitutions per site.
4.2. Emergence of PDCoV infection in US swine in february 2014
On February 11, 2014, the Ohio Department of Agriculture first announced the presence of PDCoV in US swine. In early February 2014, the Animal Disease Diagnostic Laboratory at the Ohio Department of Agriculture received 42 fecal or intestinal samples from diarrheic sows and piglets on 5 Ohio farms, and 39 (92.9%) were positive for PDCoV by RT-PCR (Wang et al., 2014a). The PDCoV HKU15-OH1987 strain identified (GenBank accession no. KJ462462) had a 99% nt identity to PDCoV HKU15-44 and HKU15-155 (Wang et al., 2014a) (Fig. 1B), implying a possible importation of a Chinese PDCoV into US swine. During a similar period, the University of Minnesota Veterinary Diagnostic Laboratory also received fecal swabs from diarrheic pigs. The causative agent of the diarrhea was identified as the PDCoV SDCV/USA/Illinois121/2014 strain (GenBank accession no: KJ481931.1) that also had approximately 99% nt identity to the two Hong Kong PDCoV strains (Marthaler et al., 2014a). The Iowa State University Veterinary Diagnostic Laboratory also identified another US PDCoV strain USA/IA/2014/8734 (GenBank accession no: KJ567050) that had 98.9% nt identity to the HKU15-44 strain and 99.2% nt identity to the HKU15-155 strain (Li et al., 2014). PDCoVs were further detected in other US states (Minnesota, South Dakota, Nebraska, Illinois, Indiana, Michigan, Kentucky, Pennsylvania, and Ohio) (Wang et al., 2014b). Of the 435 samples tested, 109 (25%) were positive for PDCoV by RT-PCR, and they shared high nt similarity (≥99.8%) with each other and also 98.9–99.2% nt similarity with PDCoV HKU15-44 and HKU15-155 (Wang et al., 2014b). As of March 2016, PDCoV has been detected in 18 US states according to a report of the USDA Animal and Plant Health Inspection Service (www.aphis.usda.gov/animal-health/secd).
4.3. Evidence of presence of PDCoV in US swine prior to the first detection in february 2014
Several retrospective studies were conducted to determine the earliest date when PDCoV existed in US swine. A retrospective study using real-time RT-PCR was performed on swine samples that were submitted to the Iowa State University Veterinary Diagnostic Laboratory for investigation of enteric diseases between October 2012 and December 2013 (Sinha et al., 2015). A total of 5 samples were positive for PDCoV RNA among the 1734 clinical samples from 18 states tested. PDCoV RNA was detected in fecal samples collected on August 19th 2013 from Minnesota, August 20th and August 27th from Iowa, and August 29th 2013 from Illinois (Sinha et al., 2015). Based on these data, PDCoV has been present in US swine since at least August 2013. Another retrospective study using an indirect ELISA for the detection of PDCoV IgG antibodies demonstrated their prevalence in US pigs (Thachil et al., 2015). A total of 968 serum samples collected between 2006 and 2014 were tested. The PDCoV IgG antibodies were first detected in the archival serum samples collected in 2010 (Thachil et al., 2015). However, the origin of US PDCoV in US swine remains unclear.
4.4. Epidemiology of PDCoV in other countries
To date, PDCoV also has been reported in Canada, Korea, China, and Thailand. Epidemic diarrhea related to PDCoV was first detected in 6 Ontario farms in Canada in March 2014 (Marthaler et al., 2014b). In April 2014, PDCoV (KUN14-04 strain, Genbank accession no. KM820765) was first identified in feces of diarrheic piglets in South Korea. The Korean strain had nt identities of 98.8–99.0% to HKU15-44 and HKU15-155 and 99.6–99.8% to US PDCoV strains (Lee and Lee, 2014). A recent survey conducted in South Korea reported that of 691 diarrheic fecal samples collected on 59 pig farms in January 2014 to March 2015, only 2 samples from 1 pig farm (March 2015) were PDCoV-positive by RT-PCR (Lee et al., 2016). Two Korean PDCoV strains SL2 and SL5 isolated in the study were also closely related to US PDCoV strains, but they appeared to genetically differ from the previous Korean strain KNU14-04 isolated in 2014 (Lee et al., 2016). The first identification of PDCoV HKU15-44 and HKU15-155 strains in swine in Hong Kong, China for the period 2007–2011 were documented in 2012 (Woo et al., 2012). Since then, PDCoV has been identified in diarrheic pigs in mainland China (Dong et al., 2015, Song et al., 2015, Wang et al., 2015b). A study also found PDCoV RNA in diarrheic pig samples collected in 2004 in China (Dong et al., 2015). The prevalence of PDCoV in mainland China was high (>30%), and co-infections with PDCoV and PEDV (51%) were also common (Dong et al., 2015, Song et al., 2015). All PDCoV strains reported from China shared high nt identities (≥98.9%) with each other and with the other PDCoV strains found globally (Dong et al., 2015, Song et al., 2015) (Fig. 1B), implying a possible global circulation of a single genotype. PDCoV strains with multiple mutation or deletion sites in their S, nonstructural, or 3′ UTR genes have been also found in China, whereas the mutations were not found in the genomes of US PDCoV strains (Wang et al., 2015a, Wang et al., 2015b). In June 2015, a PDCoV-related diarrhea outbreak was identified on a pig farm in Thailand (Janetanakit et al., 2016). PDCoV strains detected in Thailand shared the highest nt identities (≥98.4%) with the Chinese PDCoV strain CHN-AH-2004 (Janetanakit et al., 2016).
5. Transmission
Like PEDV and TGEV (Jung and Saif, 2015, Saif et al., 2012), the fecal–oral route may be the main means of PDCoV transmission. Since aerosolized PEDV remains infectious (Alonso et al., 2014), whether aerosolized PDCoV is also infectious should be investigated. Like PEDV transmission, diarrheal feces and/or vomitus and other contaminated fomites, such as transport trailers (Lowe et al., 2014) and feed (Dee et al., 2014), may be major transmission sources of the virus. Another possible reservoir for PDCoV includes carriers, such as older pigs with subclinical infection, similar to PEDV infection in weaned pigs (Jung et al., 2015a).
6. Disease mechanisms and pathogenesis of PDCoV
6.1. Tissue tropism of PDCoV
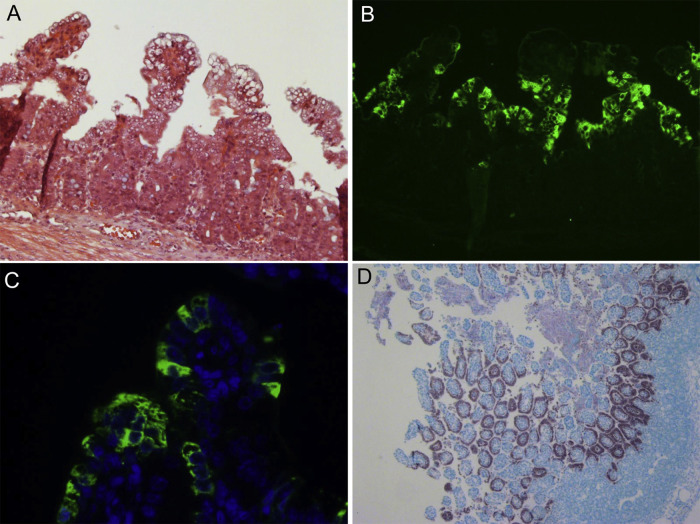
Like PEDV and TGEV (Jung and Saif, 2015, Saif et al., 2012), PDCoV is cytolytic, and infected enterocytes rapidly undergo acute necrosis, leading to marked villous atrophy in the small but not in the large intestine (Fig. 2 A) (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015). Like PEDV (Jung and Saif, 2015) and TGEV (Kim et al., 2000), PDCoV may not induce apoptotic death of enterocytes in the small intestine of infected pigs (Jung et al., 2016). PDCoV antigens (Fig. 2B and C) and nucleic acids (Fig. 2D) are observed mainly in villous enterocytes of the small (duodenum to ileum) and large intestines (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015). As tested at post-inoculation days (PID) 3–7, PDCoV antigens were detected mainly in the villous epithelium of the atrophied mid-jejunum to ileum of experimentally infected pigs and to a lesser extent, in duodenum, proximal jejunum and cecum/colon (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015). Occasionally, a few PDCoV antigen-positive cells are detected in the intestinal crypts (Jung et al., 2016). Porcine small intestinal villous enterocytes express large amounts of aminopeptidase N (APN), a 150-kDa glycosylated transmembrane protein, identified as the cellular receptor for PEDV and TGEV (Delmas et al., 1992, Li et al., 2007). Whether APN also functions as the cellular receptor for PDCoV or if other cellular receptors are involved in cell entry of this virus is unknown. In addition, a few PDCoV antigen-positive cells were also detected in the intestinal lamina propria, Peyer’s patches, and mesenteric lymph nodes (MLN) during the early stages of infection (Hu et al., 2016, Jung et al., 2016). As tested at PID 23–24 after experimentally infected pigs had recovered from clinical disease and ceased fecal virus shedding, larger numbers of PDCoV antigen-positive cells were detected in these tissues (Hu et al., 2016). The PDCoV antigen-positive cells might include antigen-presenting cells, such as macrophages.
Fig. 2.
Histopathology and localization of porcine deltacoronavirus (PDCoV) antigens by immunofluorescence (IF) staining and PDCoV nucleic acids by in situ hybridization in the small intestine of gnotobiotic pigs inoculated with US PDCoV strain OH-FD22. (A) Hematoxylin and eosin–stained jejunum of an inoculated pig at post-inoculation day (PID) 3 (48–51 h after onset of clinical signs), showing acute diffuse, severe atrophic enteritis, with diffuse, moderate vacuolation of enterocytes lining the epithelium of atrophied villi. Original magnification, ×200. (B) IF staining of a serial section of the jejunum (Panel A) of inoculated pig at PID 3, showing that the epithelial cells lining atrophied villi are positive for PDCoV antigen (green staining). Original magnification, ×200. (C) IF staining (higher magnification) of formalin-fixed, paraffin-embedded jejunum of the inoculated pig at PID 3, showing localization of PDCoV antigens (green staining) in the cytoplasm of villous epithelial cells. Nuclei were stained with blue-fluorescent 4′, 6-diamidino-2-phenylindole dihydrochloride. Hyperimmune pig antiserum to PDCoV strain OH-FD22 was used for IF staining. Original magnification, ×600. (D) In situ hybridization of formalin-fixed, paraffin-embedded jejunum of the inoculated pig at PID 3, indicating that the epithelial cells lining the atrophied villi are PDCoV RNA-positive (black staining). Nitroblue tetrazolium/5-bromocresyl-3-indolylphosphate, methyl green counterstain. Original magnification ×80.
PDCoV antigens were not detected in other organs, such as stomach, lung, heart, tonsil, spleen, liver and kidney (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015). However, by qRT-PCR, PDCoV RNA could be detected in low to moderate quantities in multiple organs, possibly due to the effect of viremia (Chen et al., 2015b, Ma et al., 2015). Lung tissues of orally infected pigs were negative for PDCoV antigen (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c). In our preliminary study, four 10- to 14-day-old Gn pigs orally and nasally inoculated with TC-PDCoV OH-FD22 at LLC-PK cell-culture passages 5 (P5) or 80 (P80) were negative for PDCoV RNA in nasal swabs and bronchoalveolar lavage fluids at PID 3–10, although the inoculated pigs were positive for both clinical disease and fecal virus shedding for the period, suggesting lack of PDCoV replication in the respiratory tract. However, further studies are still needed to investigate whether extra-intestinal replication of PDCoV occurs.
6.2. Pathophysiology
Based on clinical observations in experimentally infected Gn or conventional nursing pigs, like PEDV and TGEV (Jung and Saif, 2015, Saif et al., 2012), PDCoV also induces acute, watery diarrhea, frequently accompanied by acute, mild to moderate vomiting, ultimately leading to dehydration, loss of body weight, lethargy, and death (Jung et al., 2015c, Ma et al., 2015). Similar to PEDV infection (Jung et al., 2014), in a controlled experimental setting, clinical pigs exhibited dehydration, loss of bodyweight, and lethargy, but frequently maintained their appetite until sudden death (Ma et al., 2015). Among the different clinical signs identified in Gn or conventional pigs, only diarrhea could be observed in experimentally infected conventional nursing pigs (Chen et al., 2015b), indicating that a variety of disease outcomes are expected based on different experimental conditions. However, conventional pigs appeared to be more susceptible to PDCoV infection, compared to Gn pigs, under similar experimental conditions, such as when the same virus and inoculation dose and age of pigs were used (Ma et al., 2015). Based on field observations in US swine (Anon., 2014), PDCoV caused a number of deaths (up to a 40% mortality rate) among suckling pigs, a lower rate than that found in PEDV infections. The clinical impact and disease severity of PDCoV in the field may be less than that of PEDV (Anon., 2014). On the other hand, a PDCoV-related diarrhea outbreak in a breeding farm in China resulted in a higher mortality rate (more than 80%) among suckling piglets (Song et al., 2015).
Like PEDV and TGEV (Jung and Saif, 2015, Saif et al., 2012), diarrhea induced by PDCoV may be a consequence of malabsorption due to massive loss of absorptive enterocytes. Functional disorders of infected enterocytes may also contribute to the malabsorptive diarrhea. Like PEDV (Ducatelle et al., 1982), mild vacuolation observed in the infected colonic epithelial cells may interfere with the vital reabsorption of water and electrolytes (Jung et al., 2015c). As a consequence of extensive loss of enterocytes, brush border membrane-bound digestive enzymes such as disaccharidases (lactase, sucrase, and maltase), leucine APN, and alkaline phosphatase were significantly decreased in the small intestine of diarrheic piglets after PEDV infection (Coussement et al., 1982, Jung et al., 2006), resulting in maldigestive diarrhea. Because of the similar pathological features between PDCoV and PEDV, PDCoV infection may also result in maldigestive diarrhea. Dehydration is exacerbated by vomiting, but the mechanisms by which vomiting is induced in PDCoV infection are poorly understood. Whether PDCoV infection induces metabolic acidosis, similar to hyperkalemia and acidosis in acute TGEV and PEDV infections (Jung and Saif, 2015, Saif et al., 2012), needs to be studied.
6.3. Onset of diarrhea and fecal virus shedding and peak viral titers
Based on experimental findings (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015), the onset of diarrhea after PDCoV infection appears to be acute, which coincided mostly with or was detected 1–2 days later than the detection of viral RNA in feces. Fecal viral RNA titers peaked 1–3 days later. The findings of these studies are summarized in Table 1 . Five 14-day-old Gn pigs inoculated with 8.8 or 11.0 log10 GE of two field PDCoV strains OH-FD22 and OH-FD100 all showed severe diarrhea and/or vomiting at post-inoculation hours (PIH) 21–24, which coincided with the detection of viral RNA in feces (Jung et al., 2015c). Six 14-day-old Gn pigs inoculated with 10.1-10.8 log10 GE [≈11 log10 50% tissue culture infectious dose (TCID50) or 9 log10 plaque forming units (PFU) per pig (Hu et al., 2015)] of the TC-PDCoV OH-FD22 P5, P20, and P40 all showed severe diarrhea and/or vomiting at PID 2, which also coincided with the detection of viral RNA in feces (Hu et al., 2016). In this study, fecal viral RNA titers peaked, ranging from 7.8 to 8.9 log10 GE/ml, at PID 2–5 and then decreased gradually thereafter (Hu et al., 2016).
Table 1.
Summary of clinical signs and fecal virus RNA shedding in gnotobiotic (Gn) or conventional pigs experimentally inoculated with US porcine deltacoronavirus (PDCoV).
| Pig, age (days) at inoculation | PDCoV strain, oral inoculation dose | Onset of diarrhea (PIH or PID)b | Onset of fecal virus shedding (PIH or PID)b | Peak viral RNA titer in feces (gram) or rectal swab fluids (ml) | Duration of diarrhea (days) | Duration of fecal virus shedding | Reference |
|---|---|---|---|---|---|---|---|
| Gn, 14 | Wild-type OH-FD22 or OH–FD100, 8.8–11.0 log10 GE/piga | PIH 21-24 | PIH 21-24 | NDc | 6 | PID 1 to 20 | Jung et al., 2015c, Hu et al., 2016 |
| Conventional, 5 | Cell culture-adapted USA/IL/2014 (P11), 3 × 104 TCID50/piga | PID 5 | PID 4 or 5 | NDc | NDc | NDc | Chen et al., 2015b |
| Gn, 10 | Cell culture-adapted Michigan/8977/2014 (P15), 106 PFU/piga | PID 3 | PID 1 or 2 | NDc | NDc | NDc | Ma et al., 2015 |
| Conventional, 10 | PID 1 | PID 1 or 2 | 6-7 log10 RNA copies/gram at PID 7 | 7–10 | PID 1 to 21 | ||
| Gn, 14 | Cell culture-adapted OH-FD22 at cell-culture passages 5 (P5), 20 (P20), and 40 (P40), 10.1–10.8 log10 GE/piga | PID 2 | PID 2 | 7.8–8.9 log10 GE/ml at PID 2 to 5 | 5–7 | PID 2 to 20 (for the strain OH-FD22 P20) | Hu et al., 2016 |
GE, genome equivalents; TCID50, 50% tissue culture infectious dose; PFU, plaque forming unit; P11 or P15, PDCoV cell-culture passages 11 or 15.
PIH, postinoculation hour; PID, postinoculation day.
ND, not determined.
Another study using conventional 5-day-old pigs and a cell culture-adapted PDCoV USA/IL/2014 strain (P11) reported the onset of diarrhea at PID 5 in 5 of 5 pigs orally inoculated with 3 × 104 TCID50/pig of the virus, which was 1 day later or coincided with the detection of viral RNA in feces at PID 4 (3/5 pigs tested) or 5 (2/5 pigs tested) (Chen et al., 2015b). Fecal viral RNA peaked at approximately PID 5–7 (Chen et al., 2015b). In another study using another strain genetically similar to the USA/IL/2014 strain, four 10-day-old conventional and three 10-day-old Gn pigs orally inoculated with 106 PFU (≈109 genomic RNA copies) of a cell culture-adapted PDCoV Michigan/8977/2014 strain (P15) all exhibited moderate to severe diarrhea at PID 1 (conventional pig) and PID 3 (Gn pig), respectively (Ma et al., 2015). Fecal shedding occurred at PID 1 (1/4 pigs tested) or PID 2 (3/4 pigs tested) in conventional pigs, whereas fecal shedding was detected at PID 1 (1/3 pigs tested) or PID 2 (2/3 pigs tested) in Gn pigs prior to the onset of diarrhea (Ma et al., 2015). In the infected conventional pigs, fecal viral RNA titers peaked, ranging from 6 to 7 log10 RNA copies/gram, on PID 7 and then decreased gradually thereafter (Ma et al., 2015).
6.4. Duration of diarrhea and fecal virus shedding
Based on experimental findings (Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015), persisting diarrhea in infected nursing pigs is observed for approximately 5–10 days. Prolonged fecal virus RNA shedding was also evident in PDCoV-infected nursing pigs. After recovery from disease, pigs continued to shed PDCoV RNA in the feces. The findings of these studies are summarized in Table 1. The Gn pig inoculated with 8.8 log10 GE of the field PDCoV strain OH-FD22 developed diarrhea at PIH 21–24, which was consistently observed until PID 7 (Jung et al., 2015c). Four 10-day-old conventional pigs orally inoculated with 106 PFU of the Michigan/8977/2014 strain all exhibited diarrhea at PID 1, which then persisted for another 6–9 days (Ma et al., 2015). The Gn pigs inoculated with 10.1–10.8 log10 GE of the TC-PDCoV OH-FD22 P5, P20, and P40 all showed diarrhea at PID 2, which then persisted for another 4–6 days (Hu et al., 2016). Hu et al. (2016) reported that similar to prolonged fecal virus RNA shedding (until PID 20) observed in the wild-type PDCoV OH-FD22-inoculated pigs, fecal virus shedding in the TC-PDCoV OH-FD22 P5 or P20-inoculated pigs was detectable until PID 18 or 20, respectively (Hu et al., 2016). The TC-PDCoV OH-FD22 P40-inoculated pig also had PDCoV RNA-positive feces until PID 15 (Hu et al., 2016). One of the four 10-day-old conventional pigs (25%) inoculated with the Michigan/8977/2014 strain had detectable PDCoV RNA in the feces until PID 21 (Ma et al., 2015).
6.5. Intestinal replication of PDCoV during disease progression
There is little information on intestinal replication of PDCoV during disease progression in pigs of varying ages. In previous experimental infection studies, most of the infected Gn or conventional suckling pigs were euthanized at PID 3–7 (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015). Thus, the earlier, pathological characteristics related to PDCoV infection at PID 1–3 are unknown. In experimental PEDV infections in seronegative suckling pigs, during the incubation period, i.e. at approximately PIH 12–24, PEDV antigen-positive cells were seen throughout the small intestine and as many as 30–100% of the absorptive epithelial cells were positive (Debouck et al., 1981; Madson et al., 2015), consistent with fecal virus shedding in subclinical pigs. After that, during the clinical period, moderate to large numbers of PEDV antigen-positive cells were observed throughout the small intestine (Debouck et al., 1981; Jung et al., 2015a, Madson et al., 2015). Whether PDCoV also replicates similarly in the small intestine of nursing pigs, needs to be studied in well-controlled experimental settings with large numbers of pigs and multiple time-points. More information on the pathogenesis of PDCoV in pigs at earlier time-points (PIDs 1–2) is needed to fully understand disease progression. Relative to experimental PEDV infections (Jung et al., 2015a, Jung et al., 2014, Madson et al., 2015), PDCoV-infected pigs also appeared to shed less PDCoV RNA and had lower peak shedding titers in the feces (Chen et al., 2015b, Jung et al., 2015c), implying a potentially lower replication rate of PDCoV in the gastrointestinal tract of pigs. Studies are also needed to investigate if this might contribute to the lower mortality of PDCoV-infected nursing pigs, as compared to PEDV infections.
6.6. Age-dependent variation to PDCoV infection
Based on field observations (Anon., 2014, Song et al., 2015), PDCoV caused a number of deaths (up to a 40% mortality rate in US and >80% in China) among suckling pigs. Similarly, PED is the most devastating in nursing piglets causing 100% morbidity and 50–100% mortality (Jung and Saif, 2015, Saif et al., 2012). Several mechanisms by which PEDV infection induces greater disease severity in nursing versus weaned pigs, have been defined (Annamalai et al., 2015, Jung et al., 2015a). Compared to 9-day-old nursing pigs that began to show severe clinical disease, villous atrophy and fecal virus shedding at PID 1, a longer incubation period of PEDV was evident before 26-day-old weaned pigs shed fecal virus RNA (by 1 more day) or developed lesions and clinical disease (by 2 more days) (Annamalai et al., 2015, Jung et al., 2015a). There were significantly decreased functional natural killer cells in the ileum and blood of nursing piglets, compared to weaned pigs, which may have contributed to the greater susceptibility of nursing pigs to PEDV infection (Annamalai et al., 2015). Studies are needed to confirm if nursing pigs also have higher susceptibility to PDCoV infection. Anatomical and physiological factors that may contribute to the longer recovery of nursing pigs from PED include: 1) the slower turnover of enterocytes (5–7 days) in neonatal piglets compared to 2–3 days in 3-week-old weaned pigs (Moon et al., 1973); and 2) the anatomically underdeveloped large intestine of nursing piglets that may increase their vulnerability to dehydration, compared to weaned pigs (Jung et al., 2015a). There was also a lack of crypt stem cells and lower numbers of proliferating crypt cells in the small intestine of nursing pigs, compared to weaned pigs (Jung et al., 2015a). This could lead to the slower turnover of enterocytes in nursing vs weaned pigs, contributing to a slower recovery from PED in nursing piglets and the greater susceptibility of nursing pigs to PED (diarrhea and dehydration). Because of the similar pathological features between PDCoV and PEDV, studies are needed to confirm if PDCoV infection may also induce greater disease severity and deaths of nursing piglets vs. weaned pigs by mechanisms similar to those contributing factors to PEDV infection.
6.7. Acute viremia
Viremia where viral RNA in serum ranged from 100.5–103 TCID50/mL was detected in conventional 5-day-old pigs (15/15; 100%) inoculated with the USA/IL/2014 strain at PID 4 (incubation period) and PID 5–7 (clinical period) (Chen et al., 2015b). Similarly, viral RNA in serum ranged from 3.4–4.1 log10 copies of PDCoV RNA/mL in 19-day-old Gn pigs inoculated with the Ohio wild-type field CVM1 strain at PID 1–3 (clinical period) (Ma et al., 2015). In another study, viral RNA in serum ranged from 4.8–5.2 log10 GE/mL in Gn piglets (6/6; 100%) inoculated with the US TC-PDCoV OH-FD22 during the clinical period at PID 3 (Hu et al., 2016). The early, severe diarrhea/vomiting might be accompanied by acute, transient viremia, but no one has yet confirmed the presence of infectious virus in the serum. Similar to PEDV infection of nursing pigs that was accompanied by early viremia (viral RNA, up to 100%), viremia in PDCoV-infected pigs might also be related to the severity of atrophic enteritis and structural alteration of tight and adherens junctions in the jejunal and ileal villous epithelium in the small intestine (Jung et al., 2015b, Jung et al., 2014).
6.8. Immune responses to PDCoV
There is a dearth of information on the innate and adaptive immune responses to PDCoV. After PDCoV infection, infiltration of macrophages, lymphocytes, eosinophils, and neutrophils were observed in the lamina propria of the small intestine (Chen et al., 2015b, Ma et al., 2015, Wang et al., 2015a). Our study reported the development of PDCoV-specific antibodies in serum of PDCoV-infected pigs (Hu et al., 2016). The 14-day-old Gn pigs orally inoculated with the original OH-FD22 and TC-PDCoV OH-FD22 (P5, P20, and P40) had detectable serum IgG, IgA and virus neutralization (VN) antibodies at PID 14. Thereafter, PDCoV-specific IgG, IgA and VN antibody titers increased and remained high at the end of experiment, PID 23/24, when the pigs were fully recovered from clinical disease and fecal virus RNA shedding. Similarly, in PEDV CV777-infected pigs, serum PEDV-specific IgG antibody was first detected at approximately PID 7–12 (de Arriba et al., 2002).
6.9. Virulence of high cell culture-passaged PDCoV (P80)
The TC-PDCoV OH-FD22 has been serially passaged >P90 on LLC-PK cells. Our study confirmed the enteropathogenicity of TC-PDCoV OH-FD22 P5, P20, and P40 in Gn pigs (Hu et al., 2016). The virulence appeared to be similar to that of the parent wild-type virus, as evident by severe diarrhea/vomiting, atrophic enteritis, and high levels of serum IgG, IgA and VN antibodies in inoculated Gn pigs. Our pilot study also revealed that two 10-day-old Gn pigs inoculated orally with 10.2 log10 GE of the TC-PDCoV OH-FD22 P80 (passage 80) developed delayed (by 1–3 days), but severe clinical signs with high viral RNA shedding titers in the feces, ranging from 7.7 to 8.8 log10 GE/ml at PIDs 5 and 6, which is similar to those in Gn pigs inoculated with the original field strain OH-FD22 or TC-PDCoV OH-FD22 (cell passages 5–40). Therefore, the TC-PDCoV OH-FD22 needs to be further passaged to evaluate if higher cell-culture passaged virus becomes attenuated, because attenuation of the virulence of PEDV strains has been induced through high cell-culture passages (93rd–144th) (Chen et al., 2015a, Kweon et al., 1999, Sato et al., 2011, Song et al., 2003).
6.10. Co-infections with PDCoV and other enteric viruses
The severity of clinical disease caused by PDCoV in nursing piglets may be affected by co-infections with other viruses including rotavirus (RV) and PEDV. However, the impact of dual PDCoV and PEDV or rotavirus infection on the disease outcome in pigs needs to be delineated. A survey conducted in the US reported that of 293 samples collected from diarrheic pigs in the US and Canada in early 2014, 89 (30%) were PDCoV-positive by RT-PCR (Marthaler et al., 2014b). Of the 89 PDCoV-positive samples, 69 (78%) were positive for PEDV, group A RV (RVA), RVB, or RVC. Co-infections with PDCoV and RVC were the most common [52 (58%) samples], and 29 (29/89; 33%) were also positive for PEDV. Our lab also identified that seven (41.2%) of 42 PDCoV-positive samples, collected on different farms with diarrhea outbreaks in Ohio and Indiana, were also positive for PEDV (3 samples) or RVC (4 samples) by RT-PCR (Hu et al., 2015). In mainland China, a surveillance study based on nested RT-PCR results reported that of 356 porcine diarrheic samples collected in Jiangxi during 2012–2015, 120 (33.7%) were PDCoV-positive; 231 (64.9%) were PEDV-positive; and 281(78.9%) were positive for either PEDV or PDCoV (Song et al., 2015). Co-infections with PDCoV and PEDV (19.7%) were common in diarrheic pigs in Jiangxi, China. Another study tested by RT-PCR reported that of 215 diarrheic samples collected in Anhui, Guangxi, Hubei, and Jiangsu during 2004–2014, 14 (6.51%) were positive for PDCoV, 110 (51.2%) were positive for PEDV, and 5 (2.3%) were positive for TGEV. Of the 14 PDCoV-positive samples, 7 (50%) were also positive for PEDV; and 2 were co-infected with PEDV, TGEV, and PDCoV.
7. Lesions
7.1. Gross lesions
In infected nursing pigs, similar to PEDV and TGEV infections (Jung and Saif, 2015, Saif et al., 2012), gross lesions are limited to the gastrointestinal tract and are characterized by thin and transparent intestinal walls (proximal jejunum to colon) with accumulation of large amounts of yellow fluid in the intestinal lumen (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015). The stomach is filled with curdled milk. The transparency and fragility of affected intestines appeared milder, as compared to PEDV infections.
7.2. Histological lesions
In infected nursing pigs, similar to PEDV and TGEV infections (Jung and Saif, 2015, Saif et al., 2012), histological lesions are characterized by acute, multifocal to diffuse, mild to severe atrophic enteritis in the proximal jejunum to ileum (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015, Wang et al., 2015a), occasionally accompanied by mild vacuolation of superficial epithelial cells in the cecum and colon that coincided with a few PDCoV antigen-positive cecal and colonic epithelial cells (Jung et al., 2015c). No villous atrophy or histologic lesions were evident in the remainder of the small intestine, duodenum (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c), which coincided with few PDCoV antigen-positive duodenal epithelial cells. During acute infection, vacuolated enterocytes or massive cell exfoliation were seen on the tips or the entire villi in the jejunum and ileum. Atrophied villi are frequently fused and covered with a degenerated or regenerated flattened epithelium. Infiltration of inflammatory cells, such as macrophages, lymphocytes, neutrophils, and eosinophils, was evident in the lamina propria (Chen et al., 2015b, Ma et al., 2015, Wang et al., 2015a). No lesions were seen in other organs, such as stomach, lung, spleen, liver, kidney, and MLN of infected pigs, all of which were also negative for PDCoV antigen (Chen et al., 2015b, Hu et al., 2016, Jung et al., 2015c, Ma et al., 2015). However, one study found an acute, focal, mild degeneration or necrosis of gastric epithelial cells in the gastric pits of stomach (Ma et al., 2015).
8. Conclusions
PDCoV is a novel enteropathogenic coronavirus in pigs. The disease related to PDCoV was first reported in the US in early 2014, when PEDV infections were also epidemic. However, since 2005–2006, DCoVs already had been identified in rectal swabs of wild small mammals (Asian leopard cats and Chinese ferret badgers), domestic pigs, and wild birds. Moreover, PDCoV (CHN-AH-2004 and CH/Sichuan/S27/2012 strains), genetically related to the current US PDCoV strains, has been present in Chinese diarrheic pigs since 2004, prior to the first outbreak of PDCoV infection in US swine. Although birds are thought to be the suspect host for Deltacoronaviruses, the detailed genetic relatedness of PDCoV with other DCoV isolates from wild birds and other mammal species should be studied further to clarify the origin of PDCoV in pigs and also the source of the sudden emergence of US PDCoV in US swine.
US PDCoV strains are enteropathogenic and infect villous epithelial cells of the entire small and large intestines, although the jejunum and ileum are the primary sites of infection. Similar to PEDV (alphacoronavirus) infections, PDCoV (deltacoronavirus) infections also cause acute, severe atrophic enteritis accompanied by viremia (viral RNA) that leads to severe diarrhea and/or vomiting, followed by dehydration as the potential cause of death in nursing piglets. Differential diagnosis of PEDV, PDCoV and TGEV is essential to control viral diarrheas in US pig farms. Currently, cultivable, US TC-PDCoV strains have been isolated and propagated in several laboratories. TC-PDCoV strains will be useful to evaluate if high level cell-culture passaged viruses become attenuated and can be potential vaccine candidate strains. Relative to PEDV infections, causing similar, but more severe clinical disease and lesions, the lack of comprehensive understanding of the pathogenic characteristics of PDCoV and effective preventive (PDCoV vaccine) and therapeutic interventions has impeded biosecurity and development of methods for prevention and control of PDCoV in US swine. It is also critical to understand the pathogenic aspects of PDCoV infection distinct from those of PEDV, possibly clarifying why PDCoV infection results in distinctively lower mortality rates in nursing pigs, as compared to the high mortality usually observed among PEDV-infected seronegative nursing pigs.
Conflict of interest
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper. The authors have no conflict of interest to declare.
Acknowledgements
Salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. This paper was in part supported by a grant from the OARDC SEEDS Grants Program, Grant OHOA1536 (Jung K, PI).
Contributor Information
Kwonil Jung, Email: jung.221@osu.edu.
Linda J. Saif, Email: saif.2@osu.edu.
References
- Alonso C., Goede D.P., Morrison R.B., Davies P.R., Rovira A., Marthaler D.G., Torremorell M. Evidence of infectivity of airborne porcine epidemic diarrhea virus and detection of airborne viral RNA at long distances from infected herds. Vet. Res. 2014;45:73. doi: 10.1186/s13567-014-0073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai T., Saif L.J., Lu Z., Jung K. Age-dependent variation in innate immune responses to porcine epidemic diarrhea virus infection in suckling versus weaned pigs. Vet. Immunol. Immunopathol. 2015;168:193–202. doi: 10.1016/j.vetimm.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon USDA to require reports of PED. J. Am. Vet. Med. Assoc. 2014;244:1234. [PubMed] [Google Scholar]
- Chen F., Zhu Y., Wu M., Ku X., Ye S., Li Z., Guo X., He Q. Comparative genomic analysis of classical and variant virulent parental/attenuated strains of porcine epidemic diarrhea virus. Viruses. 2015;7:5525–5538. doi: 10.3390/v7102891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P., Stafne M., Thomas J., Arruda P., Burrough E., Madson D., Brodie J., Magstadt D., Derscheid R., Welch M., Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cima G. Viral disease affects U.S. pigs: porcine epidemic diarrhea found in at least 11 states. J. Am. Vet. Med. Assoc. 2013;243:30–31. [PubMed] [Google Scholar]
- Coussement W., Ducatelle R., Debouck P., Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. I. Histological and histochemical studyI. Vet. Pathol. 1982;19:46–56. doi: 10.1177/030098588201900108. [DOI] [PubMed] [Google Scholar]
- de Arriba M.L., Carvajal A., Pozo J., Rubio P. Isotype-specific antibody-secreting cells in systemic and mucosal associated lymphoid tissues and antibody responses in serum of conventional pigs inoculated with PEDV. Vet. Immunol. Immunopathol. 2002;84:1–16. doi: 10.1016/S0165-2427(01)00386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouck P., Pensaert M., Coussement W. The pathogenesis of an enteric infection in pigs, experimentally induced by the coronavirus-like agent, CV 777. Vet. Microbiol. 1981;6:157–165. [Google Scholar]
- Dee S., Clement T., Schelkopf A., Nerem J., Knudsen D., Christopher-Hennings J., Nelson E. An evaluation of contaminated complete feed as a vehicle for porcine epidemic diarrhea virus infection of naive pigs following consumption via natural feeding behavior: proof of concept. BMC Vet. Res. 2014;10:176. doi: 10.1186/s12917-014-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L’Haridon R., Vogel L.K., Sjostrom H., Noren O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L., Zhang S.Y., Peiris J.S., Smith G.J., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007;81:6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatelle R., Coussement W., Debouck P., Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. II. Electron microscopic study. Vet. Pathol. 1982;19:57–66. doi: 10.1177/030098588201900109. [DOI] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Saif L.J. Experimental infection of gnotobiotic pigs with the cell culture-adapted porcine deltacoronavirus strain OH-FD22. Vet. Microbiol. 2016 doi: 10.1007/s00705-016-3056-8. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetanakit T., Lumyai M., Bunpapong N., Boonyapisitsopa S., Chaiyawong S., Nonthabenjawan N., Kesdaengsakonwut S., Amonsin A. Porcine deltacoronavirus, Thailand, 2015. Emerg. Infect. Dis. 2016;22:757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Ahn K., Chae C. Decreased activity of brush border membrane-bound digestive enzymes in small intestines from pigs experimentally infected with porcine epidemic diarrhea virus. Res. Vet. Sci. 2006;81:310–315. doi: 10.1016/j.rvsc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20:662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Annamalai T., Lu Z., Saif L.J. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 2015;178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Eyerly B., Annamalai T., Lu Z., Saif L.J. Structural alteration of tight and adherens junctions in villous and crypt epithelium of the small and large intestine of conventional nursing piglets infected with porcine epidemic diarrhea virus. Vet. Microbiol. 2015;177:373–378. doi: 10.1016/j.vetmic.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Eyerly B., Lu Z., Chepngeno J., Saif L.J. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg. Infect. Dis. 2015;21:650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet. Microbiol. 2016;182:57–63. doi: 10.1016/j.vetmic.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Kim O., Tai J.H., Chae C. Transmissible gastroenteritis virus induces apoptosis in swine testicular cell lines but not in intestinal enterocytes. J. Comp. Pathol. 2000;123:64–66. doi: 10.1053/jcpa.2000.0386. [DOI] [PubMed] [Google Scholar]
- King A., Adams M.J., Carstens E.B., Lefkowitz E.J. Elsevier Academic Press; San Diego, CA, USA: 2012. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; pp. 785–795. [Google Scholar]
- Kweon C.H., Kwon B.J., Lee J.G., Kwon G.O., Kang Y.B. Derivation of attenuated porcine epidemic diarrhea virus (PEDV) as vaccine candidate. Vaccine. 1999;17:2546–2553. doi: 10.1016/S0264-410X(99)00059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Complete genome characterization of Korean Porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2014;2:e01191–14. doi: 10.1128/genomeA.01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015;208:136–145. doi: 10.1016/j.virusres.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Chung H.C., Nguyen V.G., Moon H.J., Kim H.K., Park S.J., Lee C.H., Lee G.E., Park B.K. Detection and phylogenetic analysis of porcine deltacoronavirus in Korean swine farms, 2015. Transbound. Emerg. Dis. 2016 doi: 10.1111/tbed.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.X., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365:166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Chen Q., Harmon K.M., Yoon K.J., Schwartz K.J., Hoogland M.J., Gauger P.C., Main R.G., Zhang J. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2014;2:e00278–14. doi: 10.1128/genomeA.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Gauger P., Harmon K., Zhang J., Connor J., Yeske P., Loula T., Levis I., Dufresne L., Main R. Role of transportation in spread of porcine epidemic diarrhea virus infection, United States. Emerg. Infect. Dis. 2014;20:872–874. doi: 10.3201/eid2005.131628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6:e00064. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Oglesbee M., Krakowka S., Neihaus A., Wang G., Jia A., Song H., Li J. Two-way antigenic cross-activity between porcine epidemic diarrhea virus and porcine deltacoronavirus. Vet. Microbiol. 2016;186:90–96. doi: 10.1016/j.vetmic.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson D.M., Arruda P.H., Magstadt D.R., Burrough E.R., Hoang H., Sun D., Bower L.P., Bhandari M., Gauger P.C., Stevenson G.W., Wilberts B.L., Wang C., Zhang J., Yoon K.J. Characterization of porcine epidemic diarrhea virus isolate US/Iowa/18984/2013 infection in 1-day-old cesarean-derived colostrum-deprived piglets. Vet. Pathol. 2015;53:44–52. doi: 10.1177/0300985815591080. [DOI] [PubMed] [Google Scholar]
- Marthaler D., Jiang Y., Collins J., Rossow K. Complete genome sequence of strain SDCV/USA/Illinois121/2014, a porcine deltacoronavirus from the United States. Genome Announc. 2014;2:e00218–14. doi: 10.1128/genomeA.00218-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler D., Raymond L., Jiang Y., Collins J., Rossow K., Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg. Infect. Dis. 2014;20:1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H.W., Norman J.O., Lambert G. Age dependent resistance to transmissible gastroenteritis of swine (TGE). I. Clinical signs and some mucosal dimensions in small intestine. Can. J. Comp. Med. 1973;37:157–166. [PMC free article] [PubMed] [Google Scholar]
- Saif L.J., Pensaert M.P., Sestak K., Yeo S.G., Jung K. Diseases of Swine. 10th ed. Wiley-Blackwell Iowa State University; 2012. Coronaviruses; pp. 501–524. [Google Scholar]
- Sato T., Takeyama N., Katsumata A., Tuchiya K., Kodama T., Kusanagi K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes. 2011;43:72–78. doi: 10.1007/s11262-011-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A., Gauger P., Zhang J., Yoon K.J., Harmon K. PCR-based retrospective evaluation of diagnostic samples for emergence of porcine deltacoronavirus in US swine. Vet. Microbiol. 2015;179:296–298. doi: 10.1016/j.vetmic.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Yang J.S., Oh J.S., Han J.H., Park B.K. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine. 2003;21:1833–1842. doi: 10.1016/S0264-410X(03)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound. Emerg. Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Thachil A., Gerber P.F., Xiao C.T., Huang Y.W., Opriessnig T. Development and application of an ELISA for the detection of porcine deltacoronavirus IgG antibodies. PLoS One. 2015;10:e0124363. doi: 10.1371/journal.pone.0124363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg. Infect. Dis. 2014;20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Hayes J., Sarver C., Byrum B., Zhang Y. Porcine deltacoronavirus: histological lesions and genetic characterization. Arch. Virol. 2015;161:171–175. doi: 10.1007/s00705-015-2627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.W., Yue H., Fang W., Huang Y.W. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from mainland China. Genome Announc. 2015;3:e00945–15. doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Huang Y., Yuen K.Y. Coronavirus diversity: phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lai K.K., Huang Y., Lee P., Luk G.S., Dyrting K.C., Chan K.H., Yuen K.Y. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]