Highlights
-
•
In cell culture models, low-micromolar doses of alisporivir block SARS-CoV and MERS-CoV replication.
-
•
Combination treatment with alisporivir and ribavirin increases the anti-MERS-CoV activity in cell culture.
-
•
Combination treatment with alisporivir and ribavirin does not protect against SARS-CoV infection in a mouse model.
-
•
Cyclophilin-binding drugs should be explored further in the context of host-directed anti-coronaviral strategies.
Abstract
Currently, there is no registered treatment for infections with emerging zoonotic coronaviruses like SARS- and MERS-coronavirus. We here report that in cultured cells low-micromolar concentrations of alisporivir, a non-immunosuppressive cyclosporin A-analog, inhibit the replication of four different coronaviruses, including MERS- and SARS-coronavirus. Ribavirin was found to further potentiate the antiviral effect of alisporivir in these cell culture-based infection models, but this combination treatment was unable to improve the outcome of SARS-CoV infection in a mouse model. Nevertheless, our data provide a basis to further explore the potential of Cyp inhibitors as host-directed, broad-spectrum inhibitors of coronavirus replication.
1. Main text
The outbreak of the Severe Acute Respiratory Syndrome-coronavirus (SARS-CoV) in 2003 and the continuing circulation of the Middle East respiratory syndrome-coronavirus (MERS-CoV; since 2012) have highlighted the potentially lethal consequences of zoonotic coronavirus infections in humans. Approximately 8000 people were infected during the SARS-CoV epidemic (∼10% mortality; http://www.who.int/csr/sars/en/), while the global MERS tally is now over 1800 laboratory-confirmed cases, with a mortality of about 35% (http://www.who.int/csr/disease/coronavirus_infections/en/). In mid-2015, an air travel-related outbreak in South Korea (186 confirmed cases, 36 deaths, and about 17,000 potentially exposed individuals quarantined) again illustrated the socioeconomic impact of this kind of emerging pathogen, also outside regions in which the virus is endemic (http://www.who.int/csr/don/07-july-2015-mers-korea/en/). The lack of effective methods to prevent or treat coronavirus infections in humans remains a serious public health concern, especially given increasing evidence that SARS-like coronaviruses continue to circulate in bats and may have the potential to readily cross the species barrier and emerge as human pathogens (Ge et al., 2013, Menachery et al., 2015).
Data from cell culture infection models (Chan et al., 2013a, Chan et al., 2013b, de Wilde et al., 2013b, Falzarano et al., 2013a, Kindler et al., 2013, Zielecki et al., 2013) and experiments in rhesus macaques (Falzarano et al., 2013b) and marmosets (Chan et al., 2015) suggested that interferons (IFNs) are potent inhibitors of MERS–CoV replication. The outcome of the clinical use of interferons was variable, with some reports questioning the long-term survival benefits (Al-Tawfiq et al., 2014, Omrani et al., 2014, Shalhoub et al., 2015), whereas others suggested that further investigation is warranted (Khalid et al., 2016), possibly in combination with the use of lopinavir/ritonavir (Kim et al., 2015). Unfortunately, some of the negative outcomes are confounded by the late-stage treatment of critically-ill patients.
Since the development and registration of novel therapeutic compounds is generally time consuming, the repurposing of approved drugs offers one of the few shortcuts to establishing anti-coronavirus therapy. Several FDA-approved compounds have been reported to inhibit MERS-CoV and SARS-CoV replication in cell culture (de Wilde et al., 2014, Dyall et al., 2014, Hart et al., 2013), but their efficacy in animal models remains to be determined. Previously, the FDA-approved drug cyclosporin A (CsA) was shown to inhibit the replication of a variety of viruses (reviewed in Nagy et al., 2011), including coronaviruses (Carbajo-Lozoya et al., 2014, de Wilde et al., 2013b, de Wilde et al., 2011, Kim and Lee, 2014, Pfefferle et al., 2011, Tanaka et al., 2012). CsA targets members of the cyclophilin (Cyp) family, which are peptidyl-proline isomerases (PPIases) that act as chaperones in protein folding and function (Davis et al., 2010). Since the immuno suppressive properties of CsA (Schreiber and Crabtree, 1992) are an undesirable side-effect in the context of antiviral therapy, numerous non-immunosuppressive Cyp inhibitors have been developed. Among these, alisporivir (ALV) showed increased sustained viral response during treatment of chronic hepatitis C virus (HCV) infection in phase III clinical trials, when treatment with ribavirin and pegylated interferon was combined with ALV (Buti et al., 2015, Flisiak et al., 2012, Pawlotsky et al., 2015, Zeuzem et al., 2015).
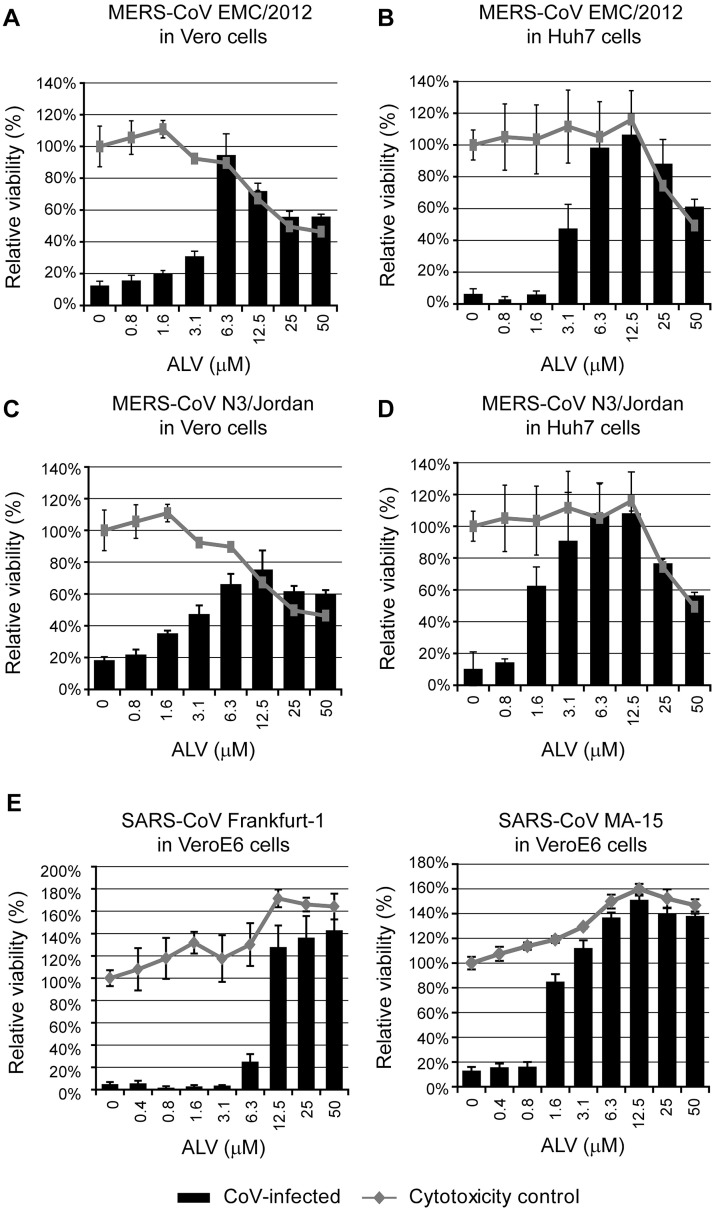
In this study, we have first investigated whether ALV inhibits MERS- and SARS-CoV replication in cell culture. We employed previously described cell-culture based screening assays relying on the rapid cytopathic effect (CPE) observed in coronavirus-infected cells (de Wilde et al., 2013b, de Wilde et al., 2014). First, the viability of MERS-CoV-infected Vero cells (MOI 0.005) treated with increasing ALV concentrations was assessed at 3 days post infection (p.i.). We found that virus-induced CPE was prevented by ALV in a dose-dependent manner (EC50 3.6 μM; Fig. 1 A). A similar efficacy was observed in MERS-CoV-infected Huh7 cells (MOI 0.005) (EC50 3.4 μM; Fig. 1B). The toxicity of ALV treatment was assessed for both cell lines and CC50 values were found to be comparable and 7–13 times higher than the observed EC50 values (26.4 and 43.8 μM, respectively; see Fig. 1A-B). ALV sensitivity was not restricted to the original MERS-CoV isolate (EMC/2012; Zaki et al., 2012) as similar EC50 values of 1.5 and 3.0 μM were determined when using MERS-CoV strain N3/Jordan in Vero (MOI 0.05) and Huh7 cells (MOI 0.005), respectively (Fig. 1C-D). The potential of ALV to inhibit other betacoronaviruses was illustrated by the complete block of SARS-CoV replication in VeroE6 cells. The SARS-CoV field isolate Frankfurt-1 was inhibited with an EC50 of 8.3 μM (CC50 in VeroE6 cells is >50 μM; Fig. 1E; de Wilde et al., 2014) and for the mouse-adapted SARS-CoV strain MA-15 (MOI 0.05; kindly provided by prof. Luis Enjuanes, National Center of Biotechnology (CNB-CSIC), Madrid, Spain) an EC50 of 1.3 μM was determined. The EC50, CC50, and selectivity index (SI) values are summarized in Table 1 .
Fig 1.
MERS-CoV- and SARS-CoV-induced cell death is strongly reduced by low-micromolar concentrations of alisporivir. (a,c) Vero or (b,d) Huh7 cells in 96-well plates were infected with (a,b) MERS-CoV EMC/2012 (MOI 0.005) or (c,d) MERS-CoV N3/Jordan (MOI 0.05 in Vero cells and MOI 0.005 in Huh7 cells) in the presence of 0–50 μM ALV. (e,f) Vero E6 cells in 96-well plates were infected with (e) SARS-CoV isolate Frankfurt-1 (MOI 0.005) or (f) SARS-CoV strain MA-15 (MOI 0.05) in the presence of 0–50 μM ALV. Mock-infected cells that did not receive ALV or solvent were used as a reference for cell viability (their relative viability was set at 100 %). Cells were incubated for 3 days with the exception for MERS-CoV EMC/2012 in Huh7 cells (2 days) and cell viability was monitored using the CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Promega). In addition, the (potential) toxicity of ALV treatment was monitored in parallel in mock-infected cell cultures. Graphs show the results (average and SD) of a representative experiment that was performed in quadruplicate. All virus-cell combinations were tested at least twice. Lines represent relative cell viability in the absence of infection (ALV toxicity control); bars represent relative cell viability after infection and ALV treatment.
Table 1.
Inhibition of SARS-CoV and MERS-CoV infection by ALV treatment.
| Virus | Strain | Cell line | EC50a (μM) | CC50a (μM) | SIb | Read-out |
|---|---|---|---|---|---|---|
| MERS-CoV | EMC/2012 | Vero | 3.6 ± 1.1 | 26.4 ± 1.0 | 7.3 | CPE-based assayc |
| 3.9 ± 1.7 | >20 | >5.1 | Virus yieldd,e | |||
| Huh7 | 3.4 ± 1.0 | 43.8 ± 1.0 | 12.9 | CPE-based assay | ||
| 2.8 ± 1.0 | >20 | >7.1 | Virus yield | |||
| LLC-MK2 | 4.0 ± 1.1 | 14.3 ± 1.8 | 3.6 | Virus yield | ||
| N3/Jordan | Vero | 3.0 ± 1.0 | 26.4 ± 1.0 | 8.8 | CPE-based assay | |
| Huh7 | 1.5 ± 1.0 | 43.8 ± 1.0 | 29.2 | CPE-based assay | ||
| SARS-CoV | Frankfurt-1 | VeroE6 | 8.3 ± 1.0 | >50 | >6.0 | CPE-based assay |
| MA-15 | VeroE6 | 1.3 ± 0.05 | >50 | >38.5 | CPE-based assay |
Data are from two independent laboratories. EC50 and CC50 values were calculated as described previously (de Wilde et al., 2014, Falzarano et al., 2013a). The selectivity index (SI), the relative efficacy of a compound in specifically inhibiting virus replication, was calculated as CC50/EC50. Statistical analyses were performed using the results of at least two independent experiments.
EC50 and CC50 values are means (± SE) from a representative experiment (n = 4) that was repeated at least twice.
SI: Selectivity index, calculated as CC50/EC50.
Data is presented in Fig. 1.
Virus yield is determined by TCID50 assay (data is presented in Fig. S1; Falzarano et al., 2013a).
Experiments performed to independently confirm the antiviral effect of ALV in a second laboratory.
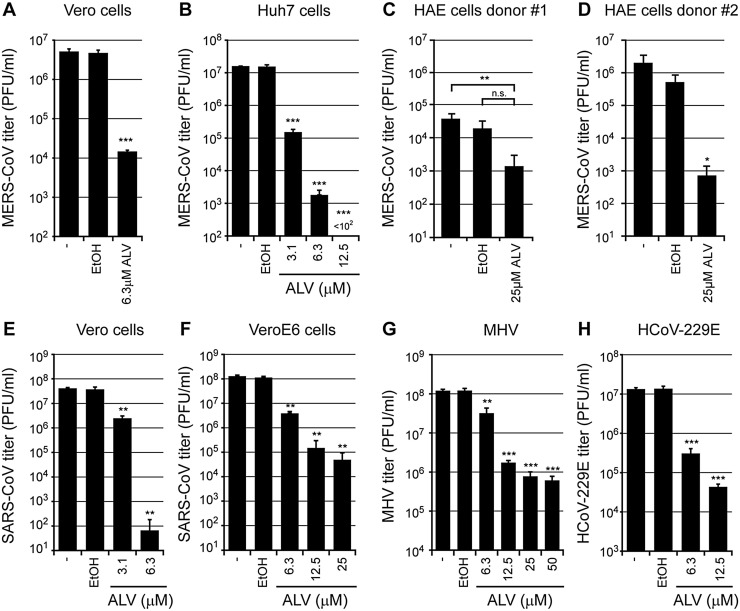
We next established that ALV treatment also reduced the yield of infectious MERS-CoV progeny. Vero and Huh7 cells were infected with MERS-CoV (MOI 0.01) and treated with increasing but non-cytotoxic ALV concentrations (for cell viability data, see Fig. 1; grey lines) from 1 h post infection (p.i.) onward. Progeny virus from MERS-CoV-infected Vero cells was harvested at 48 h p.i. and titrated by plaque assay, which revealed a ∼2 log reduction when using 6.3 μM ALV (Fig. 2 A). This effect was more pronounced in Huh7 cells, as a comparable decrease in progeny titer was already observed when using 3.1 μM ALV (Fig. 2B). Similar results were obtained using fully differentiated primary human airway epithelial (HAE) cells, of which the non-ciliated cells were shown to be the primary target for MERS-CoV infection (Raj et al., 2013). First, air–liquid interface cultures derived from two donors were cultured for 14 days on semi-permeable transwell membranes (van Wetering et al., 2007). Next, these HAE cell cultures were pretreated for 16 h with 25 μM ALV or 0.13% EtOH (vehicle control) and infected with MERS-CoV isolate EMC/2012 (MOI of 2; titer determined on Vero cells). After a 48-h ALV treatment, MERS-CoV progeny titers were reduced by 1 or 3 log, depending on the donor (Fig. 2C-D).
Fig. 2.
Alisporivir inhibits the yield of MERS-CoV, SARS-CoV, MHV, and HCoV-229E from infected cells. (a, b) MERS-CoV EMC/2012-infected (MOI 0.01) (a) Vero and (b) Huh7 cells were treated with 3.1 or 6.3 μM ALV (Vero) or 3.1 to 12.5 μM ALV (Huh7) from 1 h p.i. onward. At 48 h p.i., virus titers in the culture medium were determined by plaque assays as described before (van den Worm et al., 2012). (c,d) HAE cells from two different donors were cultured on semi-permeable 12-well transwell membranes for 14 weeks. About 16 h prior to infection, 25 μM ALV, 0.13% EtOH, or medium was added to the basal side of the cell layer. Subsequently, cells were apically infected with MERS-CoV (MOI of 2; titer determined on Vero cells). After 48 h, virus release from the apical side of the cell layer was determined by harvesting the mucous fluid and subsequent plaque assay. (e, f) SARS-CoV-infected (e) Vero or (f) VeroE6 cells (MOI 0.01) were treated with various concentrations of ALV from 1 h p.i. onwards, and virus titers in the culture medium at 32 h p.i. were determined by plaque assay. (g) 17Cl1 cells infected with MHV-A59 (MOI 0.01) or (h) Huh7 cells infected with HCoV-229E (MOI 0.01) were treated with ALV from 1 h p.i. onwards, and infectious progeny titers were determined at 16 h p.i. and 48 h p.i., respectively. The graphs show the results of one representative experiment (mean ± SD, n = 3). For all experiments, control infections include cells that remained untreated (“-“) or are treated with an amount of EtOH equalling that present at the highest ALV concentration used. Two-sided Student’s t test (Graphpad Prism 7 software) was used to determine the significance of inhibition of virus replication between EtOH-treated and ALV-treated samples (* p < 0.05; ** p < 0.005; *** p < 0.001; n.s. not significant).
In a second laboratory, the ability of ALV to inhibit MERS-CoV replication was assessed independently in Vero, LLC-MK2, and Huh7 cells. Cells were inoculated with MERS-CoV (EMC/2012) at an MOI of 0.001 and medium containing between 0.625 and 20 μM of ALV was added at 1 h p.i. Supernatant was collected on day 3 and levels of infectious virus progeny assessed by TCID50 assay. In parallel, toxicity was assessed using a cell viability assay. On day 3, a 4- to 5-log decrease in infectious progeny was observed at a dose of 10 μM ALV and EC50 values of 3.9, 2.8, and 4.0 μM were calculated for Vero, LLC-MK2, and Huh7 cells, respectively (Fig. S1 and Table 1).
SARS-CoV infection (MOI 0.01) in Vero cells was sensitive to 3.1 μM ALV (Fig. 2E), resulting in a near-complete block when using 6.3 μM ALV. A ∼1.5 log reduction in virus progeny could be achieved in VeroE6 cells using the same dose (Fig. 2F), suggesting that ALV treatment in VeroE6 cells was twice less effective compared to Vero cells. For both SARS-CoV and MERS-CoV, differences in ALV sensitivity in various cell lines were observed. One explanation for such difference would be that ALV uptake differs per cell line. Interestingly, we also observed remarkable difference in e.g. SARS-CoV-induced cell death between VeroE6 and Vero cells, ranging from severe CPE in the former to minimal CPE in the latter after the same incubation period (unpublished observations). This suggests that these related cell lines can respond quite differently to virus infection.
To investigate the potential of ALV as a broad-spectrum coronavirus inhibitor, we assessed the effect of ALV treatment on the replication of two additional coronaviruses, murine hepatitis virus (MHV; strain A59) and human coronavirus 229E (HCoV-229E). Following infection with an MOI of 0.01, media were harvested at 16 h p.i. for MHV on 17Cl1 cells or 48 h p.i. for HCoV-229E on Huh7 cells (Fig. 2G and H). When using an ALV dose of 6.3 μM, MHV progeny titers were decreased by 1 log, while a 25-μM dose resulted in a ∼2-log reduction without showing any signs of toxicity in uninfected cells (cell viability values >85% of untreated 17Cl1 control cells were observed for all ALV concentrations tested; data not shown). HCoV-229E production was reduced to a similar extent (∼1.5 and ∼2-log reduction at 6.3 and 12.5 μM ALV, respectively).
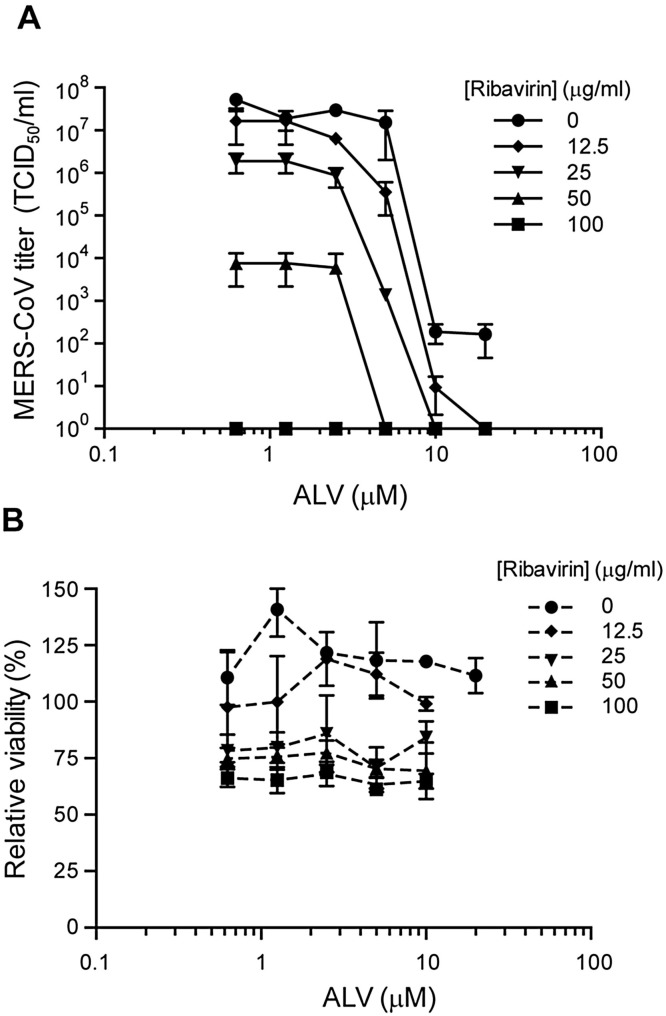
As ribavirin has previously been reported to inhibit MERS-CoV replication (Falzarano et al., 2013a) and ALV and ribavirin have been used together during clinical trials for hepatitis C treatment (Pawlotsky et al., 2015), this combination was tested in LLC-MK2 cells. Combining ribavirin and ALV treatment primarily had an additive effect on antiviral activity, with the exception of the combination of 25 μg/ml ribavirin and 5 μM ALV, which had a synergy value of 1.51 (Fig. 3 A) as calculated using MacSynergy II (Prichard and Shipman, 1990). Increasing concentrations of ribavirin gradually lowered the cell viability measured without affecting cellular morphology when assessed microscopically (Fig. 3B).
Fig. 3.
Additive antiviral effects of combined Ribavirin and Alisporivir treatment in MERS-CoV-infected cell cultures. (a) LLC-MK2 cells infected with MERS-CoV at an MOI of 0.001 were treated with a combination of 0.625 to 20 μM ALV and 12.5 to 100 μg/ml ribavirin from 1 h p.i. onwards. Virus titers in the culture medium at 3 days p.i. were determined by TCID50 as previously described (Falzarano et al., 2013a). (b) In parallel, control cells were treated with the same compound concentrations to determine cytotoxicity with a CellTiter 96 AQueous One solution cell proliferation assay.
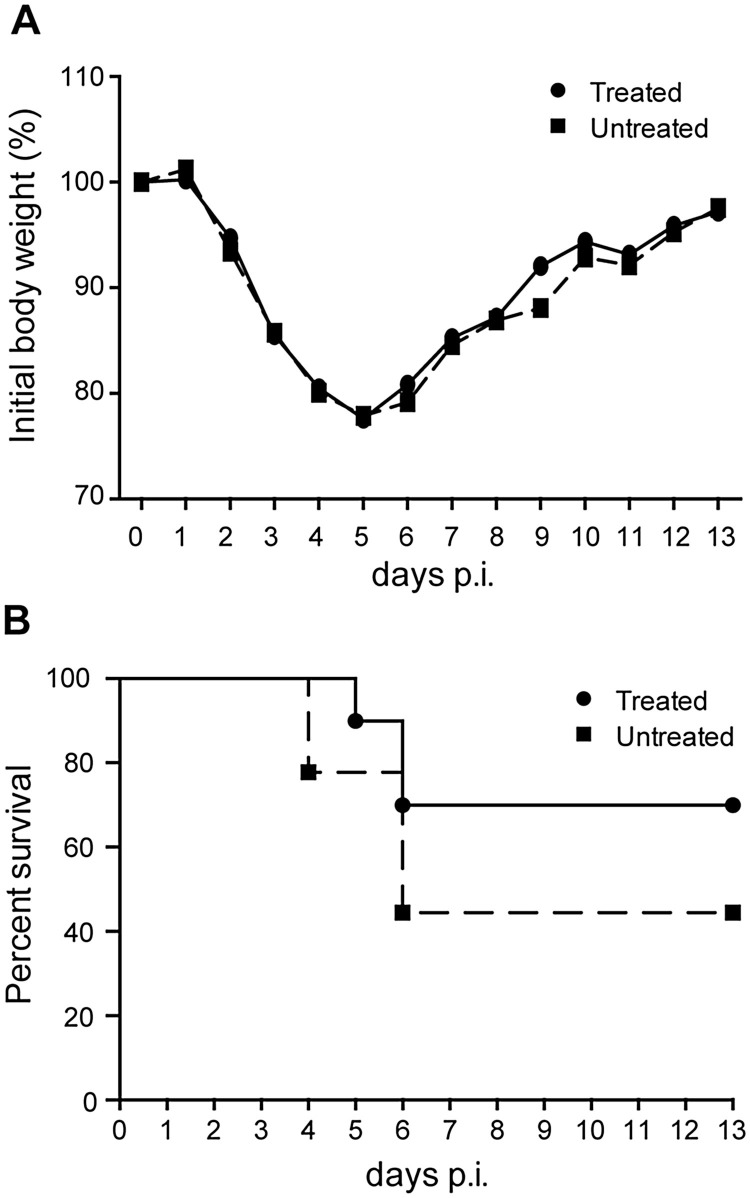
The above findings suggested that a combination approach might have a chance of success in animal models. Therefore, we assessed whether similar inhibitory effects could be observed in a mouse model. We employed the mouse-adapted MA15 strain of SARS-CoV (Roberts et al., 2007) and animals were treated daily with ALV (60 mg/kg) and/or ribavirin (50 mg/kg). Unfortunately, treatment with ALV alone did not enhance survival (data not shown) and combined therapy with ALV and ribavirin did not prevent weight loss (Fig. 4 A) or enhance survival (Fig. 4B).
Fig. 4.
Treatment with Ribavirin and alisporivir does not protect from SARS-CoV infection in a mouse model. (a,b) Combination therapy with alisporivir and ribavirin did not diminish morbidity (a) or mortality (b) in SARS-CoV MA15-infected mice. Six-week-old BALB/c mice were infected with 40,000 PFU of SARS-CoV-MA15, and then treated daily with 60 mg/kg alisporivir and 50 mg/kg ribavirin. Results of two experiments combined, n = 10 for both groups.
Since the ALV concentration required for SARS-CoV inhibition in cell culture was >100-fold higher than that required for inhibition of HCV replication (Coelmont et al., 2010, Paeshuyse et al., 2006, Puyang et al., 2010), the negative outcome of this animal experiment may not be too surprising. Nevertheless, the use of Cyp inhibitors remains a promising and innovative antiviral strategy. This class of drugs can potentially inhibit against broad range of pathogenic viruses, including hepatitis B virus (Phillips et al., 2015; EC50 of 4.1 μM), HCV (Coelmont et al., 2010, Paeshuyse et al., 2006, Puyang et al., 2010; EC50 values between 0.02 and 0.23 μM), and human immunodeficiency virus 1 (Ptak et al., 2008; EC50 values in the low-nanomolar range). We previously reported that also the replication of two arteriviruses, which are distantly related to coronaviruses, can be blocked by the ALV-related non-immunosuppressive CsA analog Debio-064 (de Wilde et al., 2013a).
Our study reveals that ALV is a broad-spectrum coronavirus inhibitor in cell culture, as it inhibits the replication of both alpha- and betacoronaviruses (Figs. 1 and 2 and Carbajo-Lozoya et al., 2014). Further research is needed to elucidate the exact mechanism of action underlying ALV’s interference with coronavirus replication, as well as the involvement of Cyps in the coronavirus replication cycle. For HCV, ALV has been shown to disrupt functional interactions between cyclophilin A (CypA) and viral proteins and/or RNA (Coelmont et al., 2010, Garcia-Rivera et al., 2012, Nag et al., 2012). Interestingly, although ALV has a ∼4 times higher affinity for CypA compared to CsA (unpublished data), the EC50 values for both inhibitors are similar, leaving the possibility that ALV and other Cyp inhibitors target CoV replication independently of CypA.
In a previous study, a 4-log reduction in HCoV-NL63 virus progeny was reported upon treatment of infected CaCo-2 cells with 10 μM ALV (EC50 0.8 μM; Carbajo-Lozoya et al., 2014). The observed variations in EC50 values suggest that different coronaviruses may not be equally sensitive to the drug, although they may also reflect differences in e.g. experimental design, Cyp expression levels, and/or ALV uptake or turnover in different cell lines. The lack of ALV activity in the SARS-CoV animal model suggests that the drug itself may not be suited for the treatment of coronavirus infections. Nevertheless, Cyp inhibitors remain interesting leads for the development of host-directed anti-coronavirus therapy, as well as interesting tools to study the role of Cyps in coronavirus replication in more detail.
Acknowledgements
We thank Dr. Frauke Fischer, Dr. Nikolai Naoumov (Novartis, Switzerland) and Dr. Grégoire Vuagniaux (DebioPharm, Switzerland) for helpful discussions and providing alisporivir. We thank Dr. Michael Cooper and Col. James Cummings (Global Emerging Infections Surveillance and Response Systems, Armed Forces Health Surveillance Branch, Silver Spring, MD, USA) and the Naval Medical Research Unit-3 in Cairo, Egypt, for providing MERS-CoV isolate N3/Jordan, Dr. Ron Fouchier (Erasmus Medical Center Rotterdam, The Netherlands) for sharing MERS-CoV isolate EMC/2012, and Dr. Luis Enjuanes, National Center of Biotechnology (CNB-CSIC), Spain) for providing SARS-CoV strain MA-15. We are grateful to Dr. Pieter Hiemstra (Dept. of Pulmonology, LUMC) for sharing HAE cell cultures, to Renate Verhoosel, Dennis Ninaber, and Diede Oudshoorn for technical assistance. We thank Dr. Dirk Jochmans and Dr. Johan Neyts (KU Leuven, Belgium) for helpful discussions. This research was supported in part by the Council for Chemical Sciences (CW) of the Netherlands Organization for Scientific Research (NWO) through TOP grant 700.57.301, as well as by the Division of Intramural Research (DIR) of the National Institute of Allergy and Infectious Disease (NIAID) and Novartis (Basel, Switzerland). All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Iowa.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2016.11.011.
Contributor Information
Adriaan H. de Wilde, Email: A.H.de_Wilde@lumc.nl.
Eric J. Snijder, Email: E.J.Snijder@lumc.nl.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Al-Tawfiq J.A., Momattin H., Dib J., Memish Z.A. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int. J. Infect. Dis. 2014;20:42–46. doi: 10.1016/j.ijid.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buti M., Flisiak R., Kao J.H., Chuang W.L., Streinu-Cercel A., Tabak F., Calistru P., Goeser T., Rasenack J., Horban A., Davis G.L., Alberti A., Mazzella G., Pol S., Orsenigo R., Brass C. Alisporivir with peginterferon/ribavirin in patients with chronic hepatitis C genotype 1 infection who failed to respond to or relapsed after prior interferon-based therapy: FUNDAMENTAL, a Phase II trial. J. Viral Hepat. 2015;22(7):596–606. doi: 10.1111/jvh.12360. [DOI] [PubMed] [Google Scholar]
- Carbajo-Lozoya J., Ma-Lauer Y., Malesevic M., Theuerkorn M., Kahlert V., Prell E., von Brunn B., Muth D., Baumert T.F., Drosten C., Fischer G., von Brunn A. Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including Alisporivir. Virus Res. 2014;184C:44–53. doi: 10.1016/j.virusres.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Chan K.H., Kao R.Y., To K.K., Zheng B.J., Li C.P., Li P.T., Dai J., Mok F.K., Chen H., Hayden F.G., Yuen K.Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67(6):606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R.W., Chan M.C., Agnihothram S., Chan L.L., Kuok D.I., Fong J.H., Guan Y., Poon L.L., Baric R.S., Nicholls J.M., Peiris J.S. Tropism and innate immune responses of the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 2013;87(12):6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yao Y., Yeung M.L., Deng W., Bao L., Jia L., Li F., Xiao C., Gao H., Yu P., Cai J.P., Chu H., Zhou J., Chen H., Qin C., Yuen K.Y. Treatment with Lopinavir/Ritonavir or interferon-beta1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212(12):1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelmont L., Hanoulle X., Chatterji U., Berger C., Snoeck J., Bobardt M., Lim P., Vliegen I., Paeshuyse J., Vuagniaux G., Vandamme A.M., Bartenschlager R., Gallay P., Lippens G., Neyts J. DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin A induced cis-trans isomerisation in domain II of NS5A. PLoS One. 2010;5(10):e13687. doi: 10.1371/journal.pone.0013687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T.L., Walker J.R., Campagna-Slater V., Finerty P.J., Paramanathan R., Bernstein G., MacKenzie F., Tempel W., Ouyang H., Lee W.H., Eisenmesser E.Z., Dhe-Paganon S. Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases. PLoS Biol. 2010;8(7):e1000439. doi: 10.1371/journal.pbio.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jr., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58(8):4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Martellaro C., Callison J., Munster V.J., Feldmann H. Inhibition of novel beta coronavirus replication by a combination of interferon-alpha2b and ribavirin. Sci. Rep. 2013;3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., de Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P., Brining D., Bushmaker T., Martellaro C., Baseler L., Benecke A.G., Katze M.G., Munster V.J., Feldmann H. Treatment with interferon-alpha2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat. Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flisiak R., Jaroszewicz J., Flisiak I., Lapinski T. Update on alisporivir in treatment of viral hepatitis C. Expert Opin. Invest. Drugs. 2012;21(3):375–382. doi: 10.1517/13543784.2012.658641. [DOI] [PubMed] [Google Scholar]
- Garcia-Rivera J.A., Bobardt M., Chatterji U., Hopkins S., Gregory M.A., Wilkinson B., Lin K., Gallay P.A. Multiple mutations in hepatitis C virus NS5A domain II are required to confer a significant level of resistance to alisporivir. Antimicrob. Agents Chemother. 2012;56(10):5113–5121. doi: 10.1128/AAC.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.Y., Wang L.F., Daszak P., Shi Z.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart B.J., Dyall J., Postnikova E., Zhou H., Kindrachuk J., Johnson R.F., Olinger G.G., Jr., Frieman M.B., Holbrook M.R., Jahrling P.B., Hensley L. Interferon-beta and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2013;95(Pt 3):571–577. doi: 10.1099/vir.0.061911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid I., Alraddadi B.M., Dairi Y., Khalid T.J., Kadri M., Alshukairi A.N., Qushmaq I.A. Acute management and long-term survival among subjects with severe Middle East respiratory syndrome coronavirus pneumonia and ARDS. Respir. Care. 2016;61(3):340–348. doi: 10.4187/respcare.04325. [DOI] [PubMed] [Google Scholar]
- Kim Y., Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. 2014;460–461:180–193. doi: 10.1016/j.virol.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U.J., Won E.J., Kee S.J., Jung S.I., Jang H.C. Combination therapy with lopinavir/ritonavir, ribavirin and interferon-alpha for Middle East respiratory syndrome: a case report. Antivir. Ther. 2015 doi: 10.3851/IMP3002. [DOI] [PubMed] [Google Scholar]
- Kindler E., Jonsdottir H.R., Muth D., Hamming O.J., Hartmann R., Rodriguez R., Geffers R., Fouchier R.A., Drosten C., Muller M.A., Dijkman R., Thiel V. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio. 2013;4(1):e00611–00612. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., Randell S.H., Lanzavecchia A., Marasco W.A., Shi Z.L., Baric R.S. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21(12):1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Robotham J.M., Tang H. Suppression of viral RNA binding and the assembly of infectious hepatitis C virus particles in vitro by cyclophilin inhibitors. J. Virol. 2012;86(23):12616–12624. doi: 10.1128/JVI.01351-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Wang R.Y., Pogany J., Hafren A., Makinen K. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology. 2011;411(2):374–382. doi: 10.1016/j.virol.2010.12.061. [DOI] [PubMed] [Google Scholar]
- Omrani A.S., Saad M.M., Baig K., Bahloul A., Abdul-Matin M., Alaidaroos A.Y., Almakhlafi G.A., Albarrak M.M., Memish Z.A., Albarrak A.M. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect. Dis. 2014;14(11):1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeshuyse J., Kaul A., De Clercq E., Rosenwirth B., Dumont J.M., Scalfaro P., Bartenschlager R., Neyts J. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43(4):761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- Pawlotsky J.M., Flisiak R., Sarin S.K., Rasenack J., Piratvisuth T., Chuang W.L., Peng C.Y., Foster G.R., Shah S., Wedemeyer H., Hezode C., Zhang W., Wong K.A., Li B., Avila C., Naoumov N.V., team V.-s. Alisporivir plus ribavirin, interferon free or in combination with pegylated interferon, for hepatitis C virus genotype 2 or 3 infection. Hepatology. 2015;62(4):1013–1023. doi: 10.1002/hep.27960. [DOI] [PubMed] [Google Scholar]
- Pfefferle S., Schopf J., Kogl M., Friedel C.C., Muller M.A., Carbajo-Lozoya J., Stellberger T., von Dall'armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Zust R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Wessels C., Pohlmann S., Haas J., Drosten C., von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7(10):e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S., Chokshi S., Chatterji U., Riva A., Bobardt M., Williams R., Gallay P., Naoumov N.V. Alisporivir inhibition of hepatocyte cyclophilins reduces HBV replication and hepatitis B surface antigen production. Gastroenterology. 2015;148(2):403–414. doi: 10.1053/j.gastro.2014.10.004. (e407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard M.N., Shipman C., Jr. A three-dimensional model to analyze drug–drug interactions. Antiviral Res. 1990;14(4–5):181–205. doi: 10.1016/0166-3542(90)90001-n. [DOI] [PubMed] [Google Scholar]
- Ptak R.G., Gallay P.A., Jochmans D., Halestrap A.P., Ruegg U.T., Pallansch L.A., Bobardt M.D., de Bethune M.P., Neyts J., De Clercq E., Dumont J.M., Scalfaro P., Besseghir K., Wenger R.M., Rosenwirth B. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob. Agents Chemother. 2008;52(4):1302–1317. doi: 10.1128/AAC.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyang X., Poulin D.L., Mathy J.E., Anderson L.J., Ma S., Fang Z., Zhu S., Lin K., Fujimoto R., Compton T., Wiedmann B. Mechanism of resistance of hepatitis C virus replicons to structurally distinct cyclophilin inhibitors. Antimicrob. Agents Chemother. 2010;54(5):1981–1987. doi: 10.1128/AAC.01236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L., Zaki S.R., Baric R., Subbarao K. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3(1):e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S.L., Crabtree G.R. The mechanism of action of cyclosporin A and FK506. Immunol. Today. 1992;13(4):136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- Shalhoub S., Farahat F., Al-Jiffri A., Simhairi R., Shamma O., Siddiqi N., Mushtaq A. IFN-alpha2a or IFN-beta1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J. Antimicrob. Chemother. 2015;70(7):2129–2132. doi: 10.1093/jac/dkv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Sato Y., Osawa S., Inoue M., Tanaka S., Sasaki T. Suppression of feline coronavirus replication in vitro by cyclosporin A. Vet. Res. 2012;43(1):41. doi: 10.1186/1297-9716-43-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zeuzem S., Flisiak R., Vierling J.M., Mazur W., Mazzella G., Thongsawat S., Abdurakhmanov D., Van Kinh N., Calistru P., Heo J., Stanciu C., Gould M., Makara M., Hsu S.J., Buggisch P., Samuel D., Mutimer D., Nault B., Merz M., Bao W., Griffel L.H., Brass C., Naoumov N.V., Group E.I.S. Randomised clinical trial: alisporivir combined with peginterferon and ribavirin in treatment-naive patients with chronic HCV genotype 1 infection (ESSENTIAL II) Aliment. Pharmacol. Ther. 2015;42(7):829–844. doi: 10.1111/apt.13342. [DOI] [PubMed] [Google Scholar]
- Zielecki F., Weber M., Eickmann M., Spiegelberg L., Zaki A.M., Matrosovich M., Becker S., Weber F. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J. Virol. 2013;87(9):5300–5304. doi: 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Zevenhoven-Dobbe J.C., van der Meer Y., Thiel V., Narayanan K., Makino S., Snijder E.J., van Hemert M.J. Cyclosporin A inhibits the replication of diverse coronaviruses. J. Gen. Virol. 2011;92(Pt 11):2542–2548. doi: 10.1099/vir.0.034983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Li Y., van der Meer Y., Vuagniaux G., Lysek R., Fang Y., Snijder E.J., van Hemert M.J. Cyclophilin inhibitors block arterivirus replication by interfering with viral RNA synthesis. J. Virol. 2013;87(3):1454–1464. doi: 10.1128/JVI.02078-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W., Posthuma C.C., van der Meer Y., Barcena M., Haagmans B.L., Snijder E.J., van den Hoogen B.G. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J. Gen. Virol. 2013;94(Pt 8):1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58(8):4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wetering S., Zuyderduyn S., Ninaber D.K., van Sterkenburg M.A., Rabe K.F., Hiemstra P.S. Epithelial differentiation is a determinant in the production of eotaxin-2 and −3 by bronchial epithelial cells in response to IL-4 and IL-13. Mol. Immunol. 2007;44(5):803–811. doi: 10.1016/j.molimm.2006.04.008. [DOI] [PubMed] [Google Scholar]
- van den Worm S.H., Eriksson K.K., Zevenhoven J.C., Weber F., Zust R., Kuri T., Dijkman R., Chang G., Siddell S.G., Snijder E.J., Thiel V., Davidson A.D. Reverse genetics of SARS-related coronavirus using vaccinia virus-based recombination. PLoS One. 2012;7(3):e32857. doi: 10.1371/journal.pone.0032857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.