Highlights
-
•
The impact of BCP mutations on HBV-X biologic activity was analyzed.
-
•
Genotype F wild type and mutant HBV-X induce apoptosis of human hepatocytes.
-
•
HBV-X variants modulate the expression of Bcl-2 family proteins.
-
•
Subgenotypes F1b and F4 HBV-X and variants induce autophagy of human hepatocytes.
Keywords: Hepatitis B virus, Hepatitis B virus X protein, Genotype F, Basal core promoter mutation, Apoptosis, Autophagy
Abstract
The hepatitis B virus X protein (HBV-X) is a multifunctional regulatory protein associated with the pathogenesis of liver disease in chronic HBV infection. Basal core promoter mutations (BCP), associated with the clinical course of chronic HBV infection, affect HBV-X at 130–131 positions. The role of these mutations on HBV-X biological activity remains largely unknown. The aim of this study was to analyze the impact of the presence of different amino acids at 130–131 positions of HBV-X on the biological activity of the protein. Transient expression of wild type and mutant F1b and F4 HBV-X increased cell mortality by the induction of apoptosis in human hepatoma cells. The wild type and mutant HBV-X differentially modulate the expression of pro-apoptotic (Bax) and anti-apoptotic (Bcl-2 and Bcl-X) regulatory proteins of the Bcl-2 family. Furthermore, the expression of HBV-X variants of both subgenotypes induced autophagy of human tumoral hepatocytes. In conclusion, HBV-X variants of the Latin American HBV F genotype promotes human hepatocytes death by the induction of apoptosis and autophagy. The results of this work describe some of the molecular mechanisms by which HBV-X variants contribute to the pathogenesis of liver diseases in the infected liver and help to the biological characterization of genotype F, responsible of the majority of HBV infections in Argentina.
1. Introduction
The hepatitis B virus (HBV) chronically actually infects approximately 257 million people worldwide (WHO, 2017). Chronic infection is associated with the development of severe liver diseases, including cirrhosis and hepatocellular carcinoma (HCC). HBV belongs to the Hepadnaviridae family and can be classified into ten genotypes (A to J) and multiple subgenotypes with characteristic geographic distribution (Kramvis, 2014, Sunbul, 2014). The viral genome consists of a 3.2 kb partially double-stranded DNA with four overlapping open reading frames that codify for: envelope (S/Pre-S1/Pre-S2), core (C/pre-C), polymerase (P) and X (HBV-X) proteins (Norder et al., 2004).
HBV-X is a 154 amino acid multifunctional protein with transcriptional transactivator activity on a number of cellular and viral promoters. It has been associated with the pathogenesis of HBV related diseases, especially in the occurrence of HCC in chronic patients (Chemin and Zoulim, 2009, Kew, 2011). By interacting with various transcription factors or components of signal transduction pathways, HBV-X regulates a wide variety of cellular genes modulating gene transcription, protein degradation, signal transduction, cell proliferation, cell cycle progress, DNA repair, senescence, as well as apoptosis and autophagy (Ma et al., 2011, Tang et al., 2006).
Apoptosis, or programmed cell death, is a highly regulated process that has a vital role in organ development and clearance of diseased or damaged cells. The deregulation of apoptosis is involved in a wide range of pathological processes, including development of HCC (Schattenberg et al., 2011). Numerous studies have shown that HBV-X can modulate programmed cell death with a dual effect, either by inhibiting or stimulating apoptosis. It has been reported that HBV-X induces apoptosis by causing loss of mitochondrial membrane potential (Shirakata and Koike, 2003), inducing Fas ligand gene expression (Yoo and Lee, 2004), increasing Bax translocation to the mitochondria (Kim et al., 2008), activating caspase 8 (Kuo et al., 2012), perturbing intracellular calcium homeostasis (Chami et al., 2003) or by interaction with cellular FLICE inhibitory protein (Kim and Seong, 2003). By contrast, in other studies HBV-X could also inhibit apoptosis by increasing NF-κB activity, activating the PI3 K pathway or inhibiting caspase 3 activity. These controversial results can be explained considering that HBV-X can modulate pro-apoptotic or anti-apoptotic pathways depending on culture conditions, cellular types and protein expression levels (Rawat et al., 2012).
Autophagy, an evolutionaly conserved intracellular catabolic process, involves the formation of a double membrane structure, called autophagosome, which engulfs long-lived cytoplasmic macromolecules and damaged organelles, and delivers them to lysosomes for degradation and recycling (Jackson, 2015). This mechanism plays an important role in maintaining cellular homeostasis, as prevents the accumulation of protein aggregates and damaged organelles in the cell. The dysfunction of autophagy has been implicated in multiple diseases, including neurodegenerative diseases, muscle diseases, cancer, cardiac diseases, and infectious diseases (Levine and Kroemer, 2008). Autophagy has also been implicated in innate and adaptive immune responses against intracellular microbial pathogens (Desai et al., 2015). However, many viruses have evolved mechanisms to use this pathway to benefit their own replications. For example, several single-stranded RNA viruses such as poliovirus, coronavirus, dengue virus, and hepatitis C virus induce the accumulation of autophagic vacuoles and use these membrane vesicles to enhance their replication (Jackson, 2015, Ke and Chen, 2014, Panyasrivanit et al., 2009, Richards and Jackson, 2013). In contrast, other viruses such as herpes simplex virus-1, cytomegalovirus, and Kaposi’s sarcoma herpes virus have evolved mechanisms to suppress autophagy for its own survival (Deretic and Levine, 2009). Furthermore, recent studies showed that HBV could also enhance and use autophagy for its DNA replication, and this enhancement of autophagy might be mediated by HBV-X (Liu et al., 2014, Sir et al., 2010b, Yang et al., 2015, Zhang et al., 2014, Zhou et al., 2016). The persistent activation of autophagy in infected hepatocytes during chronic infection may play an important role in HBV pathogenesis.
Among the naturally occurring mutations associated with the clinical course of chronic HBV infection, appear the basal core promoter (BCP) mutations. The most common at the BCP are A1762T and G1764A, that occur in tandem (Buckwold et al., 1996). This double mutation reduces preCore RNA transcription and hence HBeAg expression. In addition, it moderately increases genome replication through up-regulation of pgRNA levels (Tong and Revill, 2016). BCP mutations are selected during the natural course of chronic HBV infection and its emergence is associated with pathogenesis. Numerous studies have demonstrated that BCP mutations are frequently present in patients with chronic hepatitis, fulminant hepatitis and HCC and, to a lesser extent, in inactive carriers and immunocompromised patients (Liu et al., 2004, Parekh et al., 2003). In addition, there is a high correlation between the occurrence of these mutations and the later development of cirrhosis and HCC in chronic patients (Chen and Yang, 2011, Wei et al., 2017). Due to the overlapping open reading frames in the HBV genome, BCP mutations also affect the X gene, and consequently, HBV-X aminoacidic sequence (Kidd-Ljunggren et al., 1997). The A1762T/G1764A double mutation, involves the K130 M and V131I changes in HBV-X. Despite the numerous studies conducted on the BCP region, linking these mutations to the pathogenesis of liver disease, very few studies have focused on analyzing the role of BCP mutations in relation to HBV-X biological activity. Because of the multiple functions assigned to this protein, amino acid changes at HBV-X might affect its biological activity and, consequently, its role in the pathogenesis of chronic infection.
The aim of this study was to analyze the impact of different amino acids at 130–131 positions of HBV-X on the biological activity of the protein. In particular, the effect of HBV-X variants on cell death by apoptosis and/or autophagy induction in human hepatocytes was studied. Also, apoptotic signal transduction pathways modulated by these protein variants were characterized.
2. Materials and methods
2.1. Samples
Serum samples from patients with chronic subgenotypes F1b and F4 HBV infections were studied (the most prevalent subgenotypes in our region). Samples with known wild type sequence at 1762–1764 (A-G, amino acids K/V in 130–131 of HBV-X) and with mutations at the BCP (T-A, amino acids M/I in 130–131 of HBV-X) were included in the study. Written informed consent to participate in this study was obtained from all patients.
2.2. PCR and cloning
HBV DNA was extracted from 200 μl of serum using QIAamp DNA Mini kit (Qiagen, Germany), according to the manufacturer’s instructions. The X gene was amplified by PCR using the following primers: sense 5′ AGCCACCATGGCTGCTCGGTTGTGC 3′ and antisense 5′ ATTAGGCAGAGGTGAAAAAG 3′. The PCR was performed with 10 μl of DNA in a 50 μl reaction mixture containing 10X buffer, 200 μM dNTPs, 2 mM MgCl2, 25 pmol of each primer and 2.5 U Taq polymerase (Invitrogen, USA). The amplification conditions were: an initial denaturation step at 94 °C for 3 min, then 94 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min for 36 cycles and a final extension at 72 °C for 10 min. The PCR products were inserted into the pGEM-T Easy vector (Promega, USA). The genes were excised from the plasmid with EcoR1 (Promega, USA) and subcloned into the mammalian expression vector pcDNA3.0 (Invitogen, USA) to generate the pcDNA3-X_K/V and pcDNA3-X_M/I plasmids. The sequence and correct orientation of the X gene was confirmed by sequencing analysis (Macrogen Inc., Seoul, Korea) using sense and antisense primers.
2.3. Cell culture
Human hepatoma cell lines HepG2 and Hep3 B were cultured in minimal Eagle’s medium (MEM, GIBCO) supplemented with 10% fetal bovine serum (Natocor), 2 mM glutamine (GIBCO), 0.15% sodium bicarbonate, 1 mM nonessential aminoacids (Gibco) and 1 mM sodium pyruvate (GIBCO). Hepatocellular carcinoma cell line Huh-7 was cultured in Dulbecco’s modified Eagle’s medium (DMEM, Sigma, USA) supplemented with 10% fetal bovine serum, 1 mM nonessential aminoacids, 0.15% sodium bicarbonate and penicillin (100 IU/ml) and streptomycin (100 μg/ml) (GIBCO). Cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2.
2.4. Transfections
Plasmids pcDNA3-X_K/V and pcDNA3-X_M/I containing full length wild type (KV) and mutant (MI) HBV-X of subgenotypes F1b and F4 were used for transfection. Nude vector pCDNA3 was used as control. To perform the transient transfection, cells were grown to 50–60% confluence. Transfections were carried out using X-treme gene HP transfection reagent (Roche, Germany) with a construct concentration of 0.1 μg DNA and reagent:DNA radios of 2:1 for HepG2 and Huh-7 cell lines and 1:1 for Hep3B. Cells were maintained at 37 °C in 5% CO2. After 5 h incubation, transfection mixtures were replaced with fresh medium and incubated for 48 h.
2.5. RT-PCR
Total RNA of transfected cells was isolated with Trizol reagent (Invitrogen, USA), according to the manufacturer’s instructions, and retrotranscribed into cDNA with Random Hexamer Primers (Biodynamics) using Moloney Murine Leukemia Virus Retrotranscriptase enzyme (M-MLV RT, Invitrogen, USA). The cDNA was used as template for amplification of HBV-X mRNA with primers sense 5′ TTGTGCTGCCAACTGGATC 3′ and antisense 5′ GGCAGAGGTGAAAAAGTTGC 3′. The PCR was performed as described in section 2.2. The PCR products were subjected to electrophoresis in 1% agarose gel, stained with ethidium bromide, visualized under UV light and photographed.
2.6. Trypan blue staining
Forty-eight hours post-transfection both adherent and detached cells were harvested, resuspended in culture medium and stained with an equal volume of 0.4% trypan blue solution for 3 min. The number of total, viable (unstained) and dead cells (blue stained) was counted under an inverted microscope (Olympus, Japan) using a hemocytometer, and the percentage of death cells was determined.
2.7. Analysis of DNA fragmentation
DNA fragmentation assays were performed using a phenol/chloroform extraction method modified by Powell and Gannon (2002). Briefly, 48 h post-transfection, approximately 5 × 106 adherent and detached cells were harvested and washed twice with phosphate-buffered saline (PBS). The cell pellet was resuspended in 0.6 ml of lysis buffer (50 mM Tris-HCl pH 8, 100 mM NaCl, 10 mM EDTA pH 8, 0.5% SDS and 100 μg/ml proteinase K) and incubated for 3 h at 55 °C. DNA was extracted with an equal volume of phenol, pH 8.0, followed by extraction with chloroform:isoamyl alcohol (24:1). The aqueous phase was precipitated with 2 vols of ice-cold ethanol and 0.1 vols of 3 M sodium acetate, pH 5.5, at −80 °C for 30 min. The precipitated DNAs were collected by centrifugation at 13,000g for 15 min at 4 °C. They were washed with 70% ethanol, air-dried and resuspended in 30 μl of TE (10 mM Tris-HCl pH 8 and 1 mM EDTA pH 8) buffer. The DNA samples were electrophoresed on 1% agarose gel at 50 V for 3 h. The gels were stained with ethidium bromide, visualized under UV light and photographed.
2.8. Acridine orange and ethidium bromide staining
After transfections, cells were harvested, washed with PBS and resuspended in 1 ml PBS containing 4 μg/ml acridine orange (Sigma, USA) and 4 μg/ml ethidium bromide (Sigma, USA). The cell suspension was immediately dispensed onto slides, viewed under fluorescent microscopy (Leitz Dialux 20, Germany) and photographed. Distinctive fluorescence patterns were observed according to the state of the cells: viable, bright green fluorescent nuclei with an organized structure; early apoptotic, nuclei containing green chromatin were highly condensed or fragmented; late apoptotic, bright orange chromatin with nuclei highly condensed and fragmented; necrotic, diffuse orange nuclei. To calcute the percentage of apoptotic cells for each sample, ten photographs were counted and the percentage of early and late apoptotic cells was determined.
2.9. Western blot analysis
For total protein extraction, adherent and detached cells were harvested, washed with PBS and resuspended in lysis buffer [20 mM Tris, 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, 1 mM PMSF and protease inhibitor cocktail (Sigma, USA)]. The cells were lyzed by 3 freeze/thawing cycles. Lysates were centrifuged and supernatants were harvested. Total protein content was determined using Bradford protein assay reagent (Sigma, USA). Equal amounts of protein were loaded on 12% SDS-polyacrylamide gels and transferred to PVDF Inmobilon-P membranes (Millipore, USA) by electroblotting at 100 V for 60 min. The membranes were blocked in 5% non-fat milk in T-TBS (0.1% Tween-20, 20 mM Tris and 150 mM NaCl, pH 7.6) for 1 h at room temperature, followed by overnight incubation with the primary antibody at 4 °C. The membranes were washed and incubated with the horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Proteins were visualized using an enhanced chemiluminescence (ECL) system (GE Healthcare, UK) by autoradiography. The quantification of the bands was performed using ImageJ analysis software (Wayne Rasband, NIH, United States).
Primary antibodies used: monoclonal antibody anti-Bcl-2 (1:250, Santa Cruz Biotechnology), monoclonal antibody anti-Bax (1:500, Santa Cruz Biotechnology), polyclonal antibody anti-Bcl-XL (1:500, Santa Cruz Biotechnology) and monoclonal antibody anti-β-actin (0.5 μg/ml, Sigma). Secondary antibodies: polyclonal antibody anti-mouse (1:10000 for Bcl-2 and Bax; and 1:30000 for β-actin, Santa Cruz Biotechnology) and polyclonal antibody anti-rabbit (1:10000 for Bcl-XL, Santa Cruz Biotechnology). β-actin detection was used as the total protein loading control.
2.10. Transmission electron microscopy (TEM)
Forty-eight hours after transfection, approximately 5 × 106 cells were scraped, washed twice with PBS and fixed for TEM with 2% glutaraldehyde in phosphate buffer (80 mM Na2HPO4 and 25 mM NaH2PO4, pH 7.4) at 4 °C for 2 h. The fixing solution was removed and the cell pellet was washed with phosphate buffer every 30 min at 4 °C. The cells were post-fixed in 1% osmium tetroxide and embedded in Araldite (Huntsman Advanced Materials). Thin sections (about 1 μm) were stained with toluidine blue and observed by light microscopy in order to select fields. Ultrathin sections were mounted on 200 mesh copper grids and stained with uranyl acetate and lead citrate. The grids containing the sections were observed on a Jeol JEM 1200EX II transmission electron microscope (Servicio Central de Microscopía Electrónica, Universidad Nacional de La Plata) at 80 kV and photographed (ES500W Erlangshen CCD Gatan).
2.11. Statistical analysis
All experiments were performed three times. Statistical significance was determined using a two-tailed Student t test. A value of p < 0.05 was considered to be statistically significant. Results were expressed as mean ± standard deviation. All analyses were performed using GraphPad Prism 5.01 software (GraphPad Software, San Diego, CA, USA).
3. Results
3.1. HBV-X_K/V and HBV-X_M/I are expressed in transfected human hepatoma cells
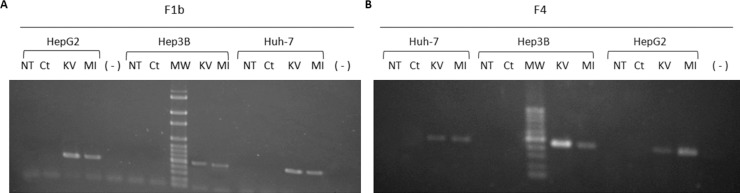
In order to analyze whether hepatoma transfected cells expressed wild type (KV) and mutant (MI) F1b and F4 HBV-X, 48 h post-transfection, the X gene mRNA was amplified by RT-PCR. As is shown in Fig. 1 , HBV-X mRNA was detected in the three cell lines transfected with F1b (Fig. 1A) and F4 (Fig. 1B) HBV-X_K/V and HBV-X_M/I. In contrast, non-transfected cells (NT), control cells transfected with pcDNA3.0 empty vector (Ct), as well as the negative control (−) showed no HBV-X mRNA expression. These findings confirm that as a result of transient transfection, X gene mRNA transcription occurs for both subgenotypes in the three studied cell lines.
Fig. 1.
HBV-X mRNA detection in non-transfected cells (NT), in cells transfected with control plasmid (Ct), F1b (A) and F4 (B) wild type HBV-X (KV) and mutant HBV-X (MI) by RT-PCR. (−): negative control; MW: molecular weight.
3.2. HBV-X_K/V and HBV-X_M/I increase the mortality of hepatoma cells
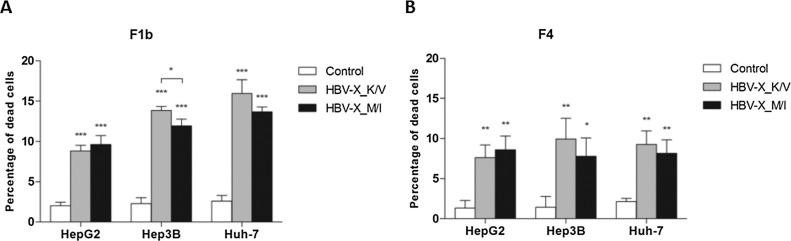
To investigate the effect of HBV-X_K/V and HBV-X_M/I expression on the viability of hepatoma human cells, 48 h after transfection cell mortality was analyzed by the trypan blue exclusion assay. Compared to control transfected cells, the expression of HBV-X_K/V and HBV-X_M/I significantly increased cell mortality in all three cell lines (Fig. 2 ). In cells expressing F1b viral proteins (Fig. 2A), the percentage of dead cells increased 340% and 380% when wild type and mutant HBV-X were expressed in HepG2, 504% and 417% in Hep3B, and 512% and 427% in Huh-7. When comparing the effects induced by the expression of both protein variants in each cell line, the wild type protein induced significantly higher mortality percentages than the mutant HBV-X for Hep3B. Meanwhile, in cells transfected with both F4 HBV-X (Fig. 2B), the increase in mortality was 467% and 542% for HepG2, 590% and 438% for Hep3 B and 343% and 290% for Huh-7, all transfected with wild type and mutant HBV-X, respectively. No significant differences in cell mortality induction were observed between wild type and mutant HBV-X in the three cell lines.
Fig. 2.
HBV-X_K/V and HBV-X_M/I increase mortality in human hepatoma cells. Cells were transfected with a control plasmid, F1b (A) and F4 (B) wild type and mutant HBV-X, and cell mortality was determined by trypan blue exclusion assay. Shown values represent the mean ± standard deviation of three independent experiments. * p < 0.05; ** p < 0.005 and *** p < 0.0001.
Based on the detected increases in cell mortality, the death mechanism induced by wild type and mutant HBV-X expression was analyzed by the examination of apoptosis by different methods.
3.3. HBV-X_K/V and HBV-X_M/I induce apoptotic cell death
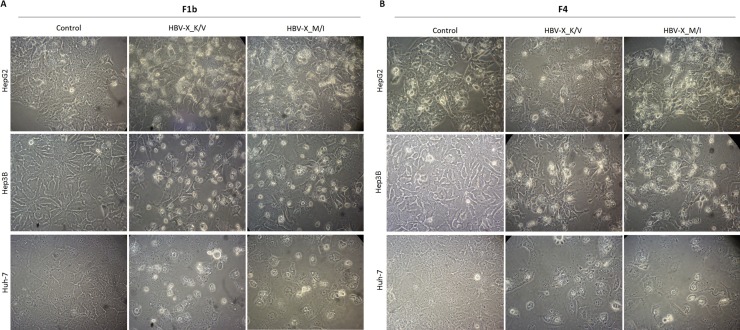
To determine whether HBV-X_K/V and HBV-X_M/I expression induced apoptotic typical morphological changes in hepatoma cells, 48 h after transfection cells were observed under contrast phase microscopy and photographed. The observation of cellular morphology revealed that cells expressing F1b (Fig. 3 A) and F4 (Fig. 3B) HBV-X_K/V and HBV-X_MI showed significant morphological changes, including cell shrinkage, loss of contact with neighboring cells, chromatin condensation and margination to the nuclear membrane, plasma membrane blebbing and apoptotic bodies, all compatible with programmed cell death. Control transfected cells showed the typical morphology of each cell line.
Fig. 3.
HBV-X_K/V and HBV-X_M/I induce typical apoptotic morphological changes in human hepatoma cells. Control, F1b (A) and F4 (B) HBV-X_K/V and HBV-X_M/I transfected cells were observed under contrast phase microscopy and photographed (200 x magnification). Shown results are from one experiment that is representative of three independent similar ones.
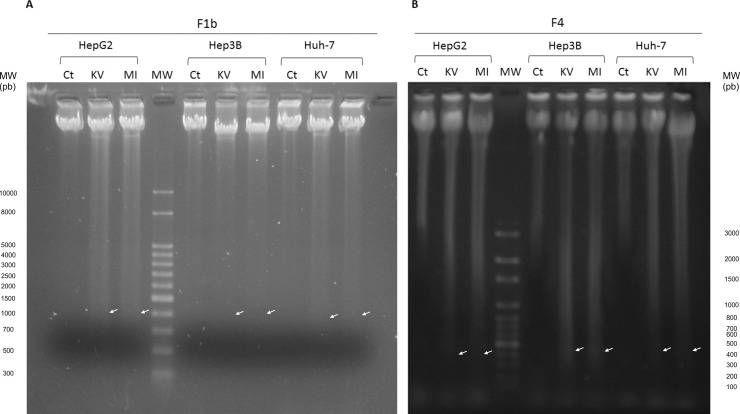
An important feature of apoptotic cell death is the fragmentation of cellular DNA into characteristic oligonucleosomal fragments, multiples of 200 base pairs. As illustrated in Fig. 4 , the expression of F1b (Fig. 4A) and F4 (Fig. 4B) HBV-X_K/V and HBV-X_MI induced DNA fragmentation in the three human hepatoma cells (white arrows), compared to control transfected cells where no DNA ladder was detected. No difference in the intensity of DNA fragmentation was observed when HBV-X_K/V and HBV-X_M/I were compared, in all cases.
Fig. 4.
HBV-X_K/V and HBV-X_M/I induce DNA fragmentation in human hepatoma cells. DNA was extracted from control (Ct), F1b (A) and F4 (B) HBV-X_K/V and HBV-X_M/I transfected cells and electrophoresed on an agarose gel. Arrows show the DNA fragments. MW: molecular weight. Shown results are from one experiment that is representative of three independent similar ones.
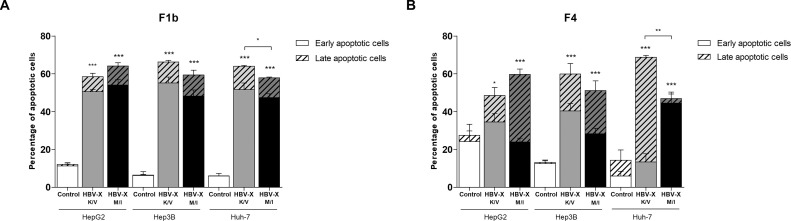
To further confirm the occurrence of apoptosis, transfected cells were stained with acridine orange and ethidium bromide and visualized under fluorescent microscopy. In the three cell lines, a significant decrease in the number of viable cells and a significant increase in early apoptotic, late apoptotic and necrotic cells was observed in F1b and F4 HBV-X_K/V and HBV-X_M/I expressing cells (Supplementary Fig. S1). In cells transfected with F1b viral proteins, the percentage of apoptotic cells (early and late apoptotic cells) increased 376% and 430% in HepG2, 936% and 820% in Hep3 B and 967% and 867% in Huh-7 for wild type and mutant HBV-X, respectively. Meanwhile in cells transfected with F4 HBV-X variants, the apoptotic cells augmented 77% and 117% in HepG2, 357% and 291% in Hep3 B and 380% and 229% in Huh-7 transfected with HBV-X_K/V and HBV-X_M/I, respectively. The comparative analysis between HBV-X_K/V and HBV-X_M/I ability to induce apoptosis revealed that in Huh-7, the induction was significantly higher in cells expressing the wild type protein compared to cells with mutant HBV-X for both subgenotypes (Fig. 5 ).
Fig. 5.
HBV-X_K/V and HBV-X_M/I induce apoptosis in human hepatoma cells. Control, F1b (A) and F4 (B) HBV-X_K/V and HBV-X_M/I transfected cells were stained with acridine orange and ethidium bromide and observed under fluorescent microscopy. The percentage of early, late and total (early + late) apoptotic cells was quantified. Shown values represent the mean ± standard deviation of three independent experiments. * p < 0.05; ** p < 0.005 and *** p < 0.0001.
Taken together, these results demonstrate that F1b and F4 wild type and mutant HBV-X can induce apoptosis in three different human hepatoma cell lines.
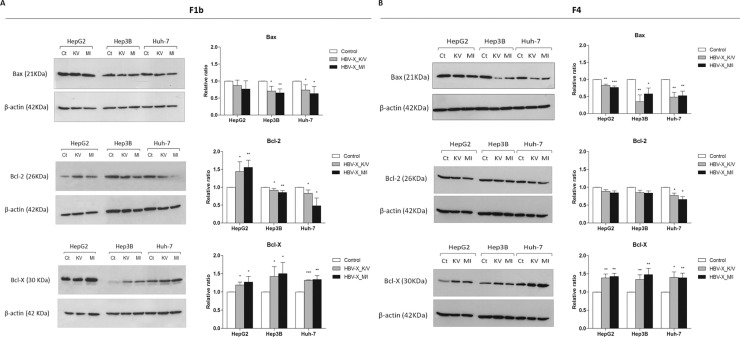
3.4. HBV-X_K/V and HBV-X_M/I modulate the expression of Bcl-2 family proteins
To examine if regulatory apoptotic signal transduction pathways were affected by HBV-X with different amino acids at 130/131 positions, the expression of pro-apoptotic (Bax) and anti-apoptotic (Bcl-2 and Bcl-X) members of the Bcl-2 regulatory family of proteins was analyzed by Western Blot (Fig. 6 ). Bax levels were down-regulated in F1b and F4 HBV-X_K/V and HBV-X_M/I expressing cells. The reduction in Bax expression was greater in HBV-X transfected Hep3 B and Huh-7 compared to HepG2, particularly in those cells that expressed both F4 viral proteins. In cells expressing the F1b viral proteins, the levels of Bcl-2 decreased in transfected Hep3 B and Huh-7, however they significantly increased in HepG2 that expressed wild type and mutant HBV-X. Whereas, in cells transfected with both F4 HBV-X variants Bcl-2 levels were significantly down-regulated in Huh-7 cells. No significant differences in Bcl-2 expression were observed in transfected HepG2 and Hep3B. On the other hand, the expression of Bcl-X showed a significant increase in all F1b and F4 HBV-X transfected cells. The comparative analysis between HBV-X_K/V and HBV-X_M/I ability to modulate Bcl-2 proteins revealed no significant differences in the expression of the pro and anti-apoptotic Bcl-2 proteins in each cell line.
Fig. 6.
HBV-X_K/V and HBV-X_M/I modulate the expression of the regulatory Bcl-2 family proteins. Hepatoma cells were transfected with a control plasmid (Ct), F1b (A) and F4 (B) wild type HBV-X (KV) and mutant HBV-X (MI), and protein expression levels of Bax, Bcl-2 and Bcl-X were analyzed by Western Blot. Protein ratios normalized to β-actin were used to quantify fold changes relative to the control. Shown values represent the mean ± standard deviation of three independent experiments. * p < 0.05; ** p < 0.005 and *** p < 0.0001.
These findings show that F1b and F4 wild type and mutant HBV-X can differentially modulate the expression of pro and anti-apoptotic Bcl-2 proteins in different human hepatoma cell lines, conducing them to apoptotic cell death.
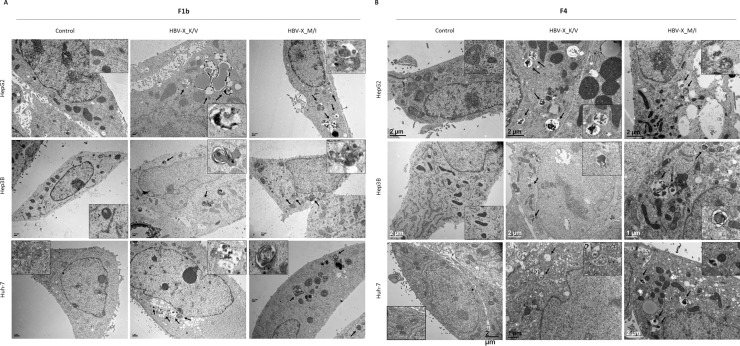
3.5. HBV-X_K/V and HBV-X_M/I induce the formation of autophagic vacuoles
In order to analyze whether HBV-X_K/V and HBV_M/I expression can also induce autophagy in human hepatoma cells, the presence of autophagic vacuoles (autophagosomes and autolysosomes), was detected at the ultrastructural level by transmission electron microscopy. In comparison to control transfected cells whose autophagic vacuoles were rarely observed, an increased number of autophagic vacuoles, including autophagosomes (double-limiting membrane compartments with cytoplasmic material and/or organelles) and autolysosomes (single-limiting membrane compartments containing cytoplasmic materials at various stages of degradation), was detected in F1b (Fig. 7 A) and F4 (Fig. 7B) HBV-X_K/V and HBV_M/I expressing cells in the three hepatoma lines. Additionally, no difference in the type and number of autophagic structures was observed when cells expressing F1b and F4 wild type and mutant HBV-X were compared in each cell line.
Fig. 7.
HBV-X_K/V and HBV-X_M/I induce the formation of autophagic vacuoles. Representative transmission electron micrographs of hepatoma cells transfected with a control vector, F1b (A) and F4 (B) HBV-X_K/V and HBV-X_M/I. Arrows indicate representative autophagic vacuoles. Inset: enlargement of autophagic vacuoles in the electron micrograph.
These results suggest that wild type and mutant HBV-X from the autochthonous genotypes can induce the autophagic process in different human hepatoma cells.
4. Discussion
In the present study, we analyzed the impact of the presence of different amino acids at 130–131 positions of HBV-X on the biological activity of the protein, related to cell death. Particularly, we studied the ability of HBV-X variants to induce apoptosis and autophagy in human hepatocytes, and characterized apoptotic signal transduction pathways of apoptosis regulation affected by different HBV-X variants.
In this work we selected genotype F samples, particularly subgenotypes F1b and F4, the most prevalent subgenotypes in our region (Barbini et al., 2013). These subgenotypes, likely originated in Amerindian populations, are responsible for most of the new acute and chronic infections in our country, and have been scarcely studied so far. Moreover, growing evidence showed a close association of subgenotype F1b with more severe course of chronic HBV infections (Ching et al., 2016, Gounder et al., 2016, Livingston et al., 2007). This study is the first to characterize the biological activity of HBV-X and its variants from local subgenotypes F1b and F4.
To date numerous studies have assessed the impact of HBV-X expression on the modulation of the apoptotic pathway, but the results of these studies have been variable. HBV-X was shown to induce (Chirillo et al., 1997, Kim et al., 2008, Kim and Seong, 2003, Miao et al., 2006), inhibit (Diao et al., 2001, Pan et al., 2001) or have no effect on apoptosis (Madden et al., 2000, Yun et al., 2002). In addition, some studies demonstrated that HBV-X does not directly induce apoptosis in certain cell types, but instead sensitizes these cells to pro-apoptotic stimuli (Su and Schneider, 1997, Wang et al., 2004). Probably the discrepancy about HBV-X apoptotic activities reflect differences in cell types studied and experimental systems used for these analyses. On the other hand, there are evidences that indicate that HBV-X expression levels are also important for the activity of this protein. HBV-X shows low expression levels during the early stages of HBV infection, which contributes to transcriptional activation and virus replication. In contrast, high levels of HBV-X are seen during the late stages of HBV infection. It is possible that HBV-X is able to inhibit apoptosis at an early stage during hepatocyte infection to facilitate HBV replication, and later it can activate apoptosis to contribute to virus spread (Murakami, 2001). The discrepant HBV-X activities reported on apoptosis may reflect concentration dependent effects at different stages of natural HBV infection. However, either stimulation or inhibition of apoptosis could lead to malignant transformation of hepatocytes and development of HCC. Inhibition of apoptosis would allow the accumulation of potentially transforming mutations (Arbuthnot et al., 2000), while enhanced compensatory hepatocyte proliferation, in response to a pro-apoptotic activity, may lead to the selection of premalignant hepatic cells (Koike et al., 1998).
In this work, we demonstrate that F1b and F4 wild type and mutant HBV-X expression has the ability to induce apoptosis in three human hepatoma cell lines. These results are in agreement with previous studies where the viral protein was transiently expressed in HepG2 (Lin et al., 2005, Tang et al., 2012), Hep3 B (Miao et al., 2006) and Huh-7 (Kanda et al., 2004) cells. Therefore, our results confirm previous in vitro studies in relation to HBV-X ability to induce apoptotic hepatocyte death, and add a piece of knowledge into genotype F, one of the less characterized HBV genotypes.
Despite the existence of articles analyzing HBV-X apoptotic activity, very few have studied the impact that mutations and genotypes might have on its biological activity. The only work considering these variables was conducted by Kanda and co-workers (2004), who analyzed whether HBV genotypes A, B, C and D or BCP mutations affected HBV-X-induced apoptosis in Huh-7 cells. Apoptosis studies using different assays revealed no differences in the pro-apoptotic activity of HBV-X, regarding different genotypes and HBV-X variants. However, in the present study, Huh-7 was the only cell line that showed significant differences in the percentage of apoptotic induction between wild type and mutant HBV-X from both analyzed subgenotypes. Quantification of apoptotic cells revealed a significant increase in the percentage of apoptosis in cells expressing HBV-X_K/V in relation to HBV-X_M/I for both subgenotypes. These results suggest that in Huh-7 cells the expression of wild type HBV-X has a greater effect on apoptosis induction than mutant HBV-X. The observed differences between both studies might be associated with the experimental systems and the protein expression levels achieved in the two assays. Additionally, it should be noted that both studies examined different viral genotypes, suggesting that HBV-X biological activity may vary according to the genotype.
The tumor suppressor gene p53 is a transcription factor central in the maintenance of genomic integrity by controlling cell response to stresses, including DNA damage and nucleotide deprivation (Fridman and Lowe, 2003). An increase in p53 leads to the expression of pro-apoptotic proteins, which prompt cells to undergo apoptosis. Several studies showed that HBV-X can interact with p53 inhibiting its pro-apoptotic function, as well as induce hepatocyte apoptosis through a p53-dependent pathway (Truant et al., 1995, Wang et al., 1994). The different human hepatoma cell lines used in this work are characterized by having different p53 status. HepG2 express wild type p53 protein, Hep3 B have a partially deleted gene with no p53 expression and Huh-7 express a mutant form of p53 protein that show higher expression levels and a longer half-life, leading to its accumulation in the cell nucleus (Bressac et al., 1990). According to the results of this work, the fact that both HBV-X variants can induce apoptosis in cells with different p53 phenotypes, suggests that wild type and mutant forms of the protein are able to trigger the apoptotic process independently of the p53 status of each cell line.
Apoptosis can proceed through two signaling pathways: the extrinsic, or death receptor pathway and the intrinsic or mitochondrial pathway. Activation of the intrinsic pathway is regulated by the Bcl-2 family proteins, which include both pro-apoptotic and anti-apoptotic members (Chipuk et al., 2010). In the present study, we demonstrated that wild type and mutant HBV-X differentially regulate the expression of pro and anti-apoptotic Bcl-2 family proteins, finally conducing to apoptotic cell death. These findings revealed the involvement of the intrinsic pathway in HBV-X_K/V and HBV-X_M/I induced apoptosis. Additionally, no difference in the expression of pro and anti-apoptotic proteins was observed when comparing the effects of both HBV-X variants within the same subgenotype, indicating that F1b and F4 HBV-X_K/V and HBV-X_M/I modulate Bcl-2 proteins in a similar way.
The results obtained in this study show that HBV-X variants decreased the abundance of the pro-apoptotic protein Bax, and increased the expression of the anti-apoptotic protein Bcl-X, although the final fact is the induction of apoptosis in the hepatoma cells. It is worth mentioning that the Bcl-2 family proteins include numerous pro and anti-apoptotic members, and the final destiny of a cell, in this case death by apoptosis, depends on the balance of all pro and anti-apoptotic proteins participating in this signaling pathway. Furthermore, the activity of the proteins of the Bcl-2 family also depends on their subcellular localization, on their post-translational modifications and on binding partners. Therefore, these results reflect only a small fraction of the complex network of interactions that exist between the Bcl-2 family proteins. In addition, it must be considered that Bcl-2 proteins are not the only regulators of apoptosis and other downstream regulatory pathways could be differentially activated by the distinct forms of HBV-X.
On the other hand, there is evidence that HBV-X can also induce apoptosis through the extrinsic pathway, upon up-regulation of proteins related to the Fas/FasL signaling pathway, and through activation of caspase 8 (He et al., 2013, Tang et al., 2012). Kuo and co-workers (2012) reported that HBV-X expression in Chang cells induces apoptosis through the simultaneous activation of the extrinsic and intrinsic pathways, through an increase in the expression of pro-apoptotic proteins (Bax and Bad), in the release of cytochrome c and increasing the levels of cleaved caspase 9 (intrinsic pathway) and caspase 8 (extrinsic pathway). In the present study, we demonstrate that in Huh-7 cells the percentage of apoptotic cells induced was higher in HBV-X_K/V expressing cells in relation to HBV-X_M/I expressing ones. However, no significant differences in the expression of pro and anti-apoptotic Bcl-2 proteins were observed between both HBV-X variants in this cell line. It is possible that particularly in Huh-7 cells, HBV-X might also activate the extrinsic apoptotic pathway, and the differences observed in the activation of apoptosis are the result of variations in the levels of induction of the extrinsic pathway between both variants of HBV-X.
On the other hand, it is known that through self-degradation of cellular components, autophagy does not only provide nutrients to maintain vital cellular functions during starvation, but also removes superfluous cells or damaged organelles and invasive pathogens (Levine and Kroemer, 2008). However, many viruses have developed numerous strategies to avoid autophagic degradation or utilize aspects of host autophagy for their own advantage (Desai et al., 2015). One of the characteristic features of autophagy is its dynamic regulation. Cellular autophagic activity is usually low under basal conditions, but can be markedly up-regulated by numerous stimuli, such as viral replication or production of certain viral proteins (Mizushima et al., 2010). Given that the autophagosome is an intermediate structure in a dynamic pathway, the number of autophagosomes observed at any specific time point is a function of the balance between the rate of their generation and the rate of their conversion into autolysosomes. Thus, autophagosome accumulation may represent either autophagy induction or, alternatively, suppression of steps in the autophagy pathway downstream of autophagosome formation.
In the present study, the observation of F1b and F4 HBV-X_K/V and HBV-X_M/I expressing cells by transmission electron microscopy revealed an increase in the number of autophagic vacuoles (autophagosomes and autolysosomes). These results demonstrate the ability of HBV-X to induce autophagy in hepatoma cells. The fact that an increase in the number of both, autophagosomes and autolysosomes was observed, indicates that both HBV-X variants promote autophagy in human hepatoma cells. In addition, no difference in the type and number of the autophagic structures was observed in cells expressing wild type and mutant HBV-X for both subgenotypes, indicating that both protein variants can modulate this process in a similar way. It should be noted that this work is the first to demonstrate that the autochthonous F genotype HBV-X induces autophagy in human hepatocytes.
Discrepant results related to HBV-X autophagic modulation have also been reported. An initial report about the effect of HBV on autophagy suggested that HBV-X sensitized cells to starvation-induced autophagy via upregulation of beclin 1 expression (Tang et al., 2009). Sir and co-workers (2010) demonstrated that HBV can enhance autophagic response in cell cultures, mouse liver, and during natural infection. They proposed that the enhancement of the autophagic response was dependent on HBV-X, through upregulation of PI3KC3. However, this enhancement in the autophagic response was not coupled to an increase of autophagic protein degradation. Recent studies have also demonstrated the induction of HBV-X-mediated autophagy through regulation of the PI3 K/Akt-mTOR pathway and by activating DAPK protein kinase in HepG2 and Chang cells, respectively (Wang et al., 2013, Zhang et al., 2014). On the other hand, Liu et al. (2014) showed that HBV-X expression enhanced the accumulation of autophagic vacuoles and also of proteins commonly degraded through autophagy. The authors proposed that HBV-X suppress rather than stimulated autophagy through a repressive effect on lysosomal function.
Based on these results it is proposed that HBV would have a dual effect on the regulation of the autophagic pathway. On one hand, HBV could induce autophagy to increase viral DNA replication. In this respect, multiples studies have indicated that induction of autophagosomes contributes to virus production, and inhibition in autophagosome formation suppresses virus replication (Sir et al., 2010a, Tian et al., 2011). Additionally, the increase in the number of autophagosomes might play a key role in the acquisition of virus envelope, supporting the idea that autophagosome-derived membranes could be used as a source of membranes for viral envelopment (Li et al., 2011). On the other hand, HBV might also inhibit later stages of autophagy, suppressing lysosomal activity, as a survival mechanism to escape direct destruction and thus avoid the generation of antigenic peptides and/or attenuate antigen presentation.
Thus, HBV-X-mediated activation of autophagy in hepatic cells of chronically infected patients may play an important role in HBV pathogenesis, increasing virus production and maintaining the viability of infected cells. Furthermore, the observation that autophagy is required for HBV DNA replication raises the possibility of targeting this pathway for the treatment of HBV-infected patients.
In conclusion, our study demonstrates that wild type and mutant HBV-X of the most prevalent subgenotypes in our country (F1b and F4), induces cell death by apoptosis and autophagy in human hepatoma cells. Activation of apoptosis in three hepatocyte cell lines with different biologic characteristics is mediated, at least in part, by the modulation of expression levels of pro and anti-apoptotic proteins of the Bcl-2 family. These results support the involvement of the intrinsic apoptotic pathway in HBV-X_K/V and HBV-X_M/I induced apoptosis. The continuous induction of apoptosis and autophagy in hepatocytes expressing HBV-X variants during the course of chronic infections might play an important role in the pathogenesis of HBV, contributing to the development of severe liver diseases and HCC in chronically infected patients.
Even though the precise mechanisms of HBV pathogenesis are not completely understood, the results presented in this study help to describe some of the molecular mechanisms by which different HBV-X variants contribute to the pathogenesis of disease in the infected liver. Furthermore, these results contribute to the biological and molecular characterization of genotype F, indigenous of Latin America, and responsible for the majority of HBV chronic infections in Argentina.
Conflict of interest
The authors declare no conflicts of interest.
Funding
This work was supported by grants from: Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) [PIP2015-0595CO]; Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) [PICT2014-1672] and UBACyT [20020130100505BA 2014-2017].
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.virusres.2017.09.025.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Arbuthnot P., Capovilla A., Kew M. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J. Gastroenterol. Hepatol. 2000;15:357–368. doi: 10.1046/j.1440-1746.2000.02069.x. [DOI] [PubMed] [Google Scholar]
- Barbini L., Elizalde M., Torres C., Campos R. Molecular epidemiology and genetic diversity of hepatitis B virus in Mar del Plata city, Argentina.I nfect. Genet. Evol. 2013;19:152–163. doi: 10.1016/j.meegid.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Bressac B., Galvin K., Liang T., Isselbacher K., Wands J., Ozturk M. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Pnas. 1990;87:1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckwold V.E., Xu Z., Chen M., Yen T.S., Ou J.H. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J. Virol. 1996;70:5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chami M., Ferrari D., Nicotera P., Paterlini-Brechot P., Rizzuto R. Caspase-dependent alterations of Ca2+ signaling in the induction of apoptosis by hepatitis B virus X protein. J. Biol. Chem. 2003;278:31745–31755. doi: 10.1074/jbc.M304202200. [DOI] [PubMed] [Google Scholar]
- Chemin I., Zoulim F. Hepatitis B virus induced hepatocellular carcinoma. Cancer Lett. 2009;286:52–59. doi: 10.1016/j.canlet.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Chen C.-J., Yang H.-I. Natural history of chronic hepatitis B REVEALed. J. Gastroenterol. Hepatol. 2011;26:628–638. doi: 10.1111/j.1440-1746.2011.06695.x. [DOI] [PubMed] [Google Scholar]
- Ching L.K., Gounder P.P., Bulkow L., Spradling P.R., Bruce M.G., Negus S., Snowball M., McMahon B.J. Incidence of hepatocellular carcinoma according to hepatitis B virus genotype in Alaska Native people. Liver Int. 2016;36:1507–1515. doi: 10.1111/liv.13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk J.E., Moldoveanu T., Llambi F., Parsons M.J., Green D.R. The BCL-2 family Reunion. Mol. Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirillo P., Pagano S., Natoli G., Puri P.L., Burgio V.L., Balsano C., Levrero M. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8162–8167. doi: 10.1073/pnas.94.15.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Fang R., Sun J. The role of autophagy in microbial infection and immunity. ImmunoTargets Ther. 2015;4:13–27. doi: 10.2147/ITT.S76720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J., Khine A.A., Sarangi F., Hsu E., Iorio C., Tibbles L.A., Woodgett J.R., Penninger J., Richardson C.D. X protein of hepatitis B virus inhibits fas-mediated apoptosis and is associated with up-regulation of the SAPK/JNK pathway. J. Biol. Chem. 2001;276:8328–8340. doi: 10.1074/jbc.M006026200. [DOI] [PubMed] [Google Scholar]
- Fridman J.S., Lowe S.W. Control of apoptosis by p53. Oncogene. 2003;22:9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- Gounder P.P., Bulkow L.R., Snowball M., Negus S., Spradling P.R., McMahon B.J. Hepatocellular carcinoma risk in alaska native children and young adults with hepatitis B virus: retrospective cohort analysis. J. Pediatr. 2016;178:206–213. doi: 10.1016/j.jpeds.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Zhou G., Qu D., Zhang B., Wang Y., Li D. HBx inhibits proliferation and induces apoptosis via Fas/FasL upregulation in rat renal tubular epithelial cells. J. Nephrol. 2013;26:1033–1041. doi: 10.5301/jn.5000304. [DOI] [PubMed] [Google Scholar]
- Jackson W.T. Viruses and the autophagy pathway. Virology. 2015;479–480:450–456. doi: 10.1016/j.virol.2015.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T., Yokosuka O., Imazeki F., Yamada Y., Imamura T., Fukai K., Nagao K., Saisho H. Hepatitis B virus X protein (HBx)-induced apoptosis in HuH-7 cells: influence of HBV genotype and basal core promoter mutations. Scand. J. Gastroenterol. 2004;39:478–485. doi: 10.1080/00365520310008719. [DOI] [PubMed] [Google Scholar]
- Ke P.Y., Chen S.S.L. Autophagy in hepatitis C virus-host interactions: potential roles and therapeutic targets for liver-associated diseases. World J. Gastroenterol. 2014;20:5773–5793. doi: 10.3748/wjg.v20.i19.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew M.C. Hepatitis B virus X protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2011;26:144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- Kidd-Ljunggren K., Öberg M., Kidd A.H. Hepatitis B virus X gene 1751–1764 mutations: Implications for HBeAg status and disease. J. Gen. Virol. 1997;78:1469–1478. doi: 10.1099/0022-1317-78-6-1469. [DOI] [PubMed] [Google Scholar]
- Kim K.H., Seong B.L. Pro-apoptotic function of HBV X protein is mediated by interaction with c-FLIP and enhancement of death-inducing signal. EMBO J. 2003;22:2104–2116. doi: 10.1093/emboj/cdg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim S.Y., Kim J., Lee H., Choi M., Kim J.K., Ahn J.K. Hepatitis B virus X protein induces apoptosis by enhancing translocation of Bax to mitochondria. IUBMB Life. 2008;60:473–480. doi: 10.1002/iub.68. [DOI] [PubMed] [Google Scholar]
- Koike K., Moriya K., Yotsuyanagi H., Shintani Y., Fujie H., Tsutsumi T., Kimura S. Compensatory apoptosis in preneoplastic liver of a transgenic mouse model for viral hepatocarcinogenesis. Cancer Lett. 1998;134:181–186. doi: 10.1016/s0304-3835(98)00252-3. [DOI] [PubMed] [Google Scholar]
- Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141–150. doi: 10.1159/000360947. [DOI] [PubMed] [Google Scholar]
- Kuo C., Tsai ju, Hwang G.-Y. Apoptosis induced by hepatitis B virus X protein in a CCL13-HBx stable cell line. Oncol. Rep. −132. 2012;28(127) doi: 10.3892/or.2012.1775. [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu Y., Wang Z., Liu K., Wang Y., Liu J., Ding H., Yuan Z. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J. Virol. 2011;85:6319–6333. doi: 10.1128/JVI.02627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N., Chen H.Y., Li D., Zhang S.J., Cheng Z.X., Wang X.Z. Apoptosis and its pathway in X gene-transfected HepG2 cells. World J. Gastroenterol. 2005;11:4326–4331. doi: 10.3748/wjg.v11.i28.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.J., Chen P.J., Lai M.Y., Kao J.H., Chen D.S. Evolution of precore/core promoter mutations in hepatitis B carriers with hepatitis B e antigen seroreversion. J. Med. Virol. 2004;74:237–245. doi: 10.1002/jmv.20176. [DOI] [PubMed] [Google Scholar]
- Liu B., Fang M., Hu Y., Huang B., Li N., Chang C., Huang R., Xu X., Yang Z., Chen Z., Liu W. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014;10:416–430. doi: 10.4161/auto.27286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston S.E., Simonetti J.P., McMahon B.J., Bulkow L.R., Hurlburt K.J., Homan C.E., Snowball M.M., Cagle H.H., Williams J.L., Chulanov V.P. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: preponderance of genotype F. J. Infect. Dis. 2007;195:5–11. doi: 10.1086/509894. [DOI] [PubMed] [Google Scholar]
- Ma J., Sun T., Park S., Shen G., Liu J. The role of hepatitis B virus X protein is related to its differential intracellular localization Cytoplasmic Localization of HBx. Acta Biochim Biophys Sin (Shanghai) 2011;43:583–588. doi: 10.1093/abbs/gmr048. (Advance) [DOI] [PubMed] [Google Scholar]
- Madden C.R., Finegold M.J., Slagle B.L. Expression of hepatitis B virus x protein does not alter the accumulation of spontaneous mutations in transgenic mice. J. Virol. 2000;74:5266–5272. doi: 10.1128/jvi.74.11.5266-5272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Chen G.G., Chun S., Lai P.P.S. Hepatitis B virus X protein induces apoptosis in hepatoma cells through inhibiting Bcl-xL expression. Cancer Lett. 2006;236:115–124. doi: 10.1016/j.canlet.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J. Gastroenterol. 2001;36:651–660. doi: 10.1007/s005350170027. [DOI] [PubMed] [Google Scholar]
- Norder H., Couroucé A.M., Coursaget P., Echevarria J.M., Lee S.D., Mushahwar I.K., Robertson B.H., Locarnini S., Magnius L.O. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- Pan J., Duan L.X., Sun B.S., Feitelson M.A. Hepatitis B virus X protein protects against anti-Fas-mediated apoptosis in human liver cells by inducing NF-kappa B. J. Gen. Virol. 2001;82:171–182. doi: 10.1099/0022-1317-82-1-171. [DOI] [PubMed] [Google Scholar]
- Panyasrivanit M., Khakpoor A., Wikan N., Smith D.R. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J. Gen. Virol. 2009;90:448–456. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- Parekh S., Zoulim F., Ahn S.H., Tsai A., Li J., Kawai S., Khan N., Trepo C., Wands J., Tong S. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J. Virol. 2003;77:6601–6612. doi: 10.1128/JVI.77.12.6601-6612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell R., Gannon F. Purification of DNA by phenol extraction and ethanol precipitation. Oxford Pract. Approach Ser. 2002:1–2. [Google Scholar]
- Rawat S., Clippinger A.J., Bouchard M.J. Modulation of apoptotic signaling by the Hepatitis B Virus X protein. Viruses. 2012;4:2945–2972. doi: 10.3390/v4112945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A.L., Jackson W.T. That which does not degrade you makes you stronger: infectivity of poliovirus depends on vesicle acidification. Autophagy. 2013;9:806–807. doi: 10.4161/auto.23962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattenberg J.M., Schuchmann M., Galle P.R. Cell death and hepatocarcinogenesis: dysregulation of apoptosis signaling pathways. J. Gastroenterol. Hepatol. 2011;26:213–219. doi: 10.1111/j.1440-1746.2010.06582.x. [DOI] [PubMed] [Google Scholar]
- Shirakata Y., Koike K. Hepatitis B virus x protein induces cell death by causing loss of mitochondrial membrane potential. J. Biol. Chem. 2003;278:22071–22078. doi: 10.1074/jbc.M301606200. [DOI] [PubMed] [Google Scholar]
- Sir D., Ann D.K., Ou J.J. Autophagy by hepatitis B virus and for hepatitis B virus. Autophagy. 2010;6:548–549. doi: 10.4161/auto.6.4.11669. [DOI] [PubMed] [Google Scholar]
- Sir D., Tian Y., Chen W-l., Ann D.K., Yen T.-S.B., Ou J.-h.J. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl. Acad. Sci. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F., Schneider R.J. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8744–8749. doi: 10.1073/pnas.94.16.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J. Gastroenterol. 2014;20:5427–5434. doi: 10.3748/wjg.v20.i18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Oishi N., Kaneko S., Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Da L., Mao Y., Li Y., Li D., Xu Z., Li F., Wang Y., Tiollais P., Li T., Zhao M. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- Tang R.X., Kong F.Y., Fan B.F., Liu X.M., You H.J., Zhang P., Zheng K.Y. HBx activates FasL and mediates HepG2 cell apoptosis through MLK3-MKK7-JNKs signal module. World J. Gastroenterol. 2012;18:1485–1495. doi: 10.3748/wjg.v18.i13.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Sir D., Kuo C.-f., Ann D.K., Ou J.-h.J. Autophagy required for hepatitis B virus replication in transgenic mice. J. Virol. 2011;85:13453–13456. doi: 10.1128/JVI.06064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Revill P. Overview of hepatitis B viral replication and genetic variability. J. Hepatol. 2016;64:S4–S16. doi: 10.1016/j.jhep.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R., Antunovic J., Greenblatt J., Prives C., Cromlish J.A. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J. Virol. 1995;69:1851–1859. doi: 10.1128/jvi.69.3.1851-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . WHO; 2017. Hepatitis B. [Google Scholar]
- Wang X.W., Forrester K., Yeh H., Feitelson M.A., Gu J.R., Harris C.C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Hullinger R.L., Andrisani O.M. Sustained activation of p38 mitogen-activated protein kinase and c-Jun N-terminal kinase pathways by Hepatitis B virus X protein mediates apoptosis via induction of Fas/FasL and tumor necrosis factor (TNF) receptor 1/TNF-α expression. Mol. Cell. Biol. 2004;24:10352–10365. doi: 10.1128/MCB.24.23.10352-10365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Guo Q.S., Wang Z.W., Qian H.X. HBx induces HepG-2 cells autophagy through PI3 K/Akt-mTOR pathway. Mol. Cell. Biochem. 2013;372:161–168. doi: 10.1007/s11010-012-1457-x. [DOI] [PubMed] [Google Scholar]
- Wei F., Zheng Q., Li M., Wu M. The association between hepatitis B mutants and hepatocellular carcinoma. Medicine. 2017;96:e6835. doi: 10.1097/MD.0000000000006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Fu Q., Liu C., Li T., Wang Y., Zhang H., Lu X., Sang X., Zhong S., Huang J., Mao Y. Hepatitis B virus promotes autophagic degradation but not replication in autophagosome. Biosci. Trends. 2015;9:111–116. doi: 10.5582/bst.2015.01049. [DOI] [PubMed] [Google Scholar]
- Yoo Y.-G., Lee M.-O. Hepatitis B virus x protein induces expression of fas ligand gene through enhancing transcriptional activity of early growth response factor. J. Biol. Chem. 2004;279:36242–36249. doi: 10.1074/jbc.M401290200. [DOI] [PubMed] [Google Scholar]
- Yun C., Um H.R., Jin Y.H., Wang J.H., Lee M.O., Park S., Lee J.H., Cho H. NF-κB activation by hepatitis B virus X (HBx) protein shifts the cellular fate toward survival. Cancer Lett. 2002;184:97–104. doi: 10.1016/s0304-3835(02)00187-8. [DOI] [PubMed] [Google Scholar]
- Zhang H.T., Chen G.G., Hu B.G., Zhang Z.Y., Yun J.P., He M.L., Lai P.B.S. Hepatitis B virus x protein induces autophagy via activating death-associated protein kinase. J. Viral Hepat. 2014;21:642–649. doi: 10.1111/jvh.12191. [DOI] [PubMed] [Google Scholar]
- Zhou T., Jin M., Ding Y., Zhang Y., Sun Y., Huang S., Xie Q., Xu C., Cai W. Hepatitis B virus dampens autophagy maturation via negative regulation of Rab7 expression. Biosci. Trends. 2016;10:244–250. doi: 10.5582/bst.2016.01049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.