Highlights
-
•
We report one novel complete genome (CRCoV BJ232) of CRCoV from China.
-
•
CRCoV BJ232 form a separate clade from other Betacoronavirus 1.
-
•
CRCoV-K37 was derived from genetic recombination between CRCoV-BJ232 and BCoV.
-
•
We confirmed the prevalence of CRCoV-BJ232 lineage around the world for a long time.
Keywords: Betacoronavirus, Canine respiratory coronavirus, Genome, Origin, Phylogenetic analysis, Recombination
Abstract
Although canine respiratory coronavirus (CRCoV) is an important respiratory pathogen that is prevalent in many countries, only one complete genome sequence of CRCoV (South Korea strain K37) has been obtained to date. Genome-wide analyses and recombination have rarely been conducted, as small numbers of samples and limited genomic characterization have previously prevented further analyses. Herein, we report a unique CRCoV strain, denoted strain BJ232, derived from a CRCoV-positive dog with a mild respiratory infection. Phylogenetic analysis based on complete genome of all available coronaviruses consistently show that CRCoV BJ232 is most closely related to human coronavirus OC43 (HCoV-OC43) and BCoV, forming a separate clade that split off early from other Betacoronavirus 1. Based on the phylogenetic and SimPlot analysis we propose that CRCoV-K37 was derived from genetic recombination between CRCoV-BJ232 and BCoV. In detail, spike (S) gene of CRCoV-K37 clustered with CRCoV-BJ232. However orf1ab, membrane (M) and nucleocapsid (N) genes were more related to Bovine coronavirus (BCoV) than CRCoV-B232. Molecular epidemic analysis confirmed the prevalence of CRCoV-BJ232 lineage around the world for a long time. Recombinant events among Betacoronavirus 1 may have implications for CRCoV transmissibility. All these findings provide further information regarding the origin of CRCoV.
1. Introduction
Canine respiratory coronavirus (CRCoV), an enveloped positive-stranded RNA virus, belongs to the betacoronavirus genus, subgroup 2a (Erles et al., 2007, Erles et al., 2003). Unlike the enteric canine coronavirus (CCoV) belonging to the alphacoronavirus genus, CRCoV is associated with mild to severe respiratory signs and proposed as an etiological agent of canine infectious respiratory disease (CIRD) (Buonavoglia and Martella, 2007). CRCoV was first detected in 2003 by RT-PCR in the UK from dogs suffering from severe respiratory disease (Erles et al., 2003). Later, the CRCoV-4182 strain was successfully isolated from a dog with respiratory signs and was cultured in the HRT-18 cell line (Erles et al., 2007). To date, data have been made publicly available for only one complete genomic sequence (CRCoV-K37 strain), which was isolated from a South Korea dog (An et al., 2010, Lim et al., 2013).
CRCoV was included in species Betacoronavirus 1, which also contain BCoV, equine coronavirus, HCoV-OC43, porcine hemagglutinating encephalomyelitisvirus (PHEV), human enteric coronavirus (HECoV) and CRCoV (King, 2011). In contract, Canine coronavirus (CCoV) is a member of alphacoronavirus, which include feline coronavirus and porcine transmissible gastroenteritis among others. The virus was distinct from CCoV and only showed 69% nucleotide identity in polymerase and 21% amino acid identity in spike protein (Buonavoglia and Martella, 2007, Erles and Brownlie, 2008).
The evolutionary origin of CRCoV remains uncertain, although successful experimental infection of puppies with BCoV has suggested that CRCoV is of bovine origin and transmitted to dogs from cattle (Kaneshima et al., 2007). Meanwhile, HCoV-OC43 may have emerged after viral transmission from cattle to people (Vijgen et al., 2005). The high nucleotide identity between CRCoV and BCoV, HCoV-OC43 and HECoV indicated that the four viruses have a common ancestor (Erles and Brownlie, 2008), and demonstrated the occurrence of repeated host-species shifts (Vijgen et al., 2006). Although CRCoV strains are frequently circulating in farmed and pet dogs worldwide (Erles and Brownlie, 2005, Kaneshima et al., 2006, Priestnall et al., 2006, Priestnall et al., 2007, Yachi and Mochizuki, 2006), causing respiratory disease, little is known about their genomic evolution and recombination. In the present study, we sequenced and analyzed the first complete genome of CRCoV derived from China (CRCoV-BJ232 strain) and discuss its role in CRCoV evolution.
2. Materials and methods
2.1. Ethics statement
This research was reviewed and approved by the ethics committee of the National Institute for Viral Disease Control and Prevention of the Chinese CDC. All animals were treated strictly according to the guidelines for the Laboratory Animal Use and Care from the Chinese CDC.
2.2. Specimen collection and handling
From December 2013–March 2014, 246 swab specimens (nasopharyngeal and anal swabs) were collected from diseased dogs at the Animal Hospital of China Agricultural University. Most of the dogs showed symptoms of gastrointestinal and/or respiratory tract infections, including coughing, lacrimation, vomiting and diarrhea. All the swab samples were collected by professional veterinarians and were stored at −80 °C.
2.3. CRCoV detection
For molecular detection, total RNA was extracted from swab specimens using a QIAamp Viral RNA Mini Kit (QIAGEN, Germany) according to the manufacturer’s instructions. The RNA was used as a template for reverse transcription-PCR (RT-PCR), and the partial sequences of the S, RNA-dependent RNA polymerase (RdRp), and N genes were amplified using gene-specific primers (Table S1). In detail, reverse transcription was performed by using a SuperScript III kit (Invitrogen, San Diego, CA). The PCR mixture (25 μl) contain 1 μl cDNA, 1 μl forward primer(10 μM), 1 μl reverse primer (10 μM), 12.5 μl Q5® High-Fidelity 2X Master Mix (New England Biolabs) and 9.5 μl nuclease-free water; the mixtures were amplified with 30 cycles at 98 °C for 10s, 52 °C for 30s, and 72 °C for 1 min and final extension step at 72 °C for 2 min in an automated thermal cycler (BIO-RAD, T100 Thermal Cycler).
2.4. Complete genome sequencing of CRCoV
To better characterize the CRCoV discovered here, the CRCoV-positive samples were pretreated and used for random amplification as described previously (Wang et al., 2016); the amplified DNA was used as a template for Illumina HiSeq2500 sequencing (2 × 125 bp, paired-end reads). The sequencing procedure was conducted by Beijing Berry Genomic Company. The full-length genome of CRCoV was assembled using SOAP de novo and was annotated based on the sole annotation of CRCoV K37 strain (JX860640.1).
2.5. Phylogenetic analysis
To determine the evolutionary relationships between the CRCoV discovered here and those identified previously, we collected most of the complete sequences of the Betacoronavirus 1 and conducted nucleotide acid sequence alignments using the MAFFT algorithm. The phylogenetic analyses were conducted based on the nucleotide acid sequences of the orf1ab, S, M, and N genes and complete genome of CRCoV. All the phylogenetic trees were constructed in the PhyML software using the maximum likelihood method, with 500 bootstrap replicates. The following Betacoronavirus 1 genome sequences from the GenBank database were used for the phylogenetic analysis as follows: BCoV reference strain (NC_003045), HECoV (NC_012950), HCoV-OC43 (NC_005147), CRCoV-37 (JX860640), and BCoV strain Quebec (AF220295).
2.6. Recombinant analysis
To confirm the potential recombination events, similarity plots and bootscanning analyses were conducted using the SimPlot software; a sliding window of 1000 nucleotides and 100-nucleotide steps were used as the default settings. In addition, a schematic diagram of the complete CRCoV genome was examined to determine the recombination sites.
2.7. Nucleotide sequence accession number
The complete genome of ChinaBJ232 was deposited in the GenBank database under accession no. KX432213. Other sequences generated in this study, including the partial S, RdRp and complete N gene sequences, were also assigned accession numbers: KX432214-KX432217.
3. Results
3.1. Identification of CRCoV
A total of 246 swab samples were collected in Beijing from December 2013 to March 2014.RT-PCR was performed to detect CRCoV, and 16 (6.5%) out of 246 swabs originating from Chinese dogs tested positive for CRCoV. In addition, five CRCoV-positive samples with high titers were initially processed for complete genome sequencing using deep sequencing technology.
3.2. Complete genomic characterization of the BJ232 strain
Due to the limited specimen size and low viral load, five clinical samples were used for deep sequencing, and only one complete genomic sequence was eventually obtained. The genomic data, formally referred to as “Canine Respiratory Coronavirus strain BJ232” and in this report as “CRCoV-BJ232,” were deposited in GenBank (KX432213). The CRCoV-BJ232 genome isolated in China was found to have 30,868 nucleotides and showed a genomic organization that was highly similar to the Betacoronavirus 1species: 5′-UTR(nt 1–143),ORF1ab (nt 144–21427), ORF2 (nt 21437–22273), HE gene (nt 22285–23559), S gene (nt 23574–27665),E gene (nt 28355–28609), M gene (nt 28624–29316), N gene (nt 29326–30672), and 3′-UTR (nt 30673–30868). Between the S and E gene, three small ORFs were identified: ORF4a (nt 27655–27789), ORF4b (nt 27825–27902) and ORF4c (nt 28039–28293), encoding 4.9-, 2.7- and 12.8-kDa non-structural proteins, respectively, which were analogous to those noted in strain CRCoV-K37. Meanwhile, conventional PCR and Sanger sequencing were conducted to verify the complete genome sequence of CRCoV-BJ232 with 100% nucleotide identity. Thus far, there are only two CRCoV strains with full-length genomic information submitted to GenBank; the previously reported CRCoV-K37 and our current strain, CRCoV-BJ232. Unfortunately, attempts to isolate CRCoV-BJ232 from the five positive samples were unsuccessful; future studies to isolate the virus from more canine samples will allow characterization of its pathogenicity and transmissibility.
3.3. Phylogenetic analysis
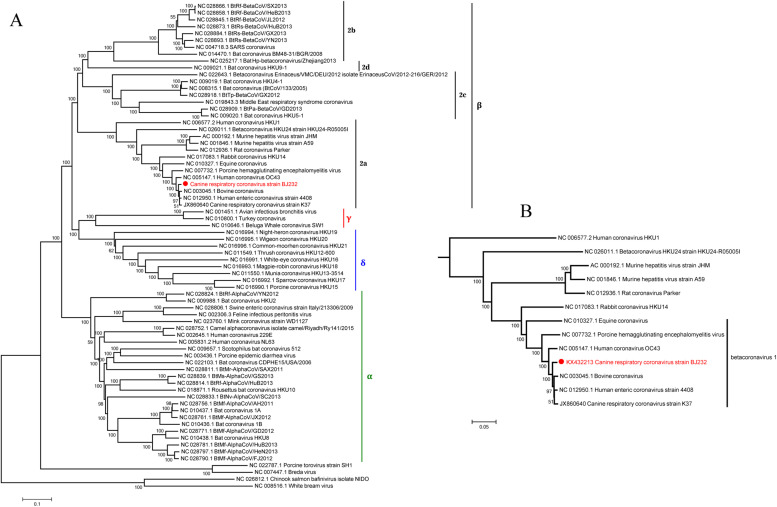
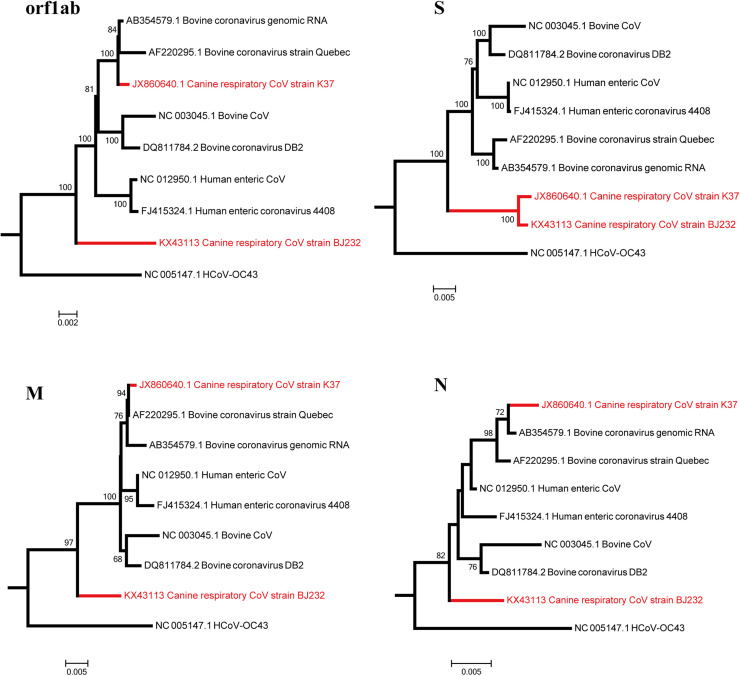
Based on the analysis of whole-genome phylogeny, CRCoV-BJ232 was classified into Betacoronavirus 1 species of betacoronavirus group 2a (Fig. 1 A) and shares a common origin with BCoV, CRCoV-K37, HECoV and HCoV-OC43. Moreover, the latter four viruses clustered together with high branch bootstrap values (Fig. 1B). To investigate the genetic relationships among the Betacoronavirus 1 species, we generated phylogenetic trees based on the sequences oforf1ab, S, M and N genes (Fig. 2 ). Phylogenetic analysis based on different genes consistently show that CRCoV BJ232 is most closely related to HCoV-OC43, forming a separate clade of CRCoV that split off early from other Betacoronavirus 1. However, CRCoV-K37 strain closely clustered with CRCoV-BJ232 strain in S gene, and was more related to BCoV strain Quebec (AF220295.1) in the region of orf1ab, M and N genes. The major genes of the CRCoV-BJ232 strain showed a consistent trend in the evolution, while the phylogenetic topology conflicted in the sub-regions of CRCoV strain K37, especially in the S and orf1ab genes. These findings are consistent with recombination, a phenomenon not uncommon in coronaviruses, implying that the CRCoV K37 strain may have arisen from genetic recombination.
Fig. 1.
Phylogenetic analysis of all available coronaviruses with reference sequence based on complete genome. A, the current taxonomy of the family Coronaviridae; B, the evolution of Betacoronavirus 1 based on complete genomes. StrainBJ232 (accession no. KX432213, indicated by the red dot) was the first CRCoV with complete genome derived from China. The previously reported CRCoV-K37 strain, which was detected in South Korea, was the sole other CRCoV strain with complete genome sequence available. Some other representative strains of the Betacoronavirus genus, subgroup 2a, were also analyzed in PhyML program using the maximum likelihood method, with 500 bootstrap replicates. The numbers at the nodes represent the bootstrap support. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Evolutionary analyses of major coding genes of Betacoronavirus 1. The coding genes used in the phylogenetic analyses included orf1ab, S, M and N. The two CRCoV strains (BJ232 and K37 strains) are indicated in red. All the sequences were analyzed in PhyML program using the maximum likelihood method, with 500 bootstrap replicates.
3.4. Recombinant analysis
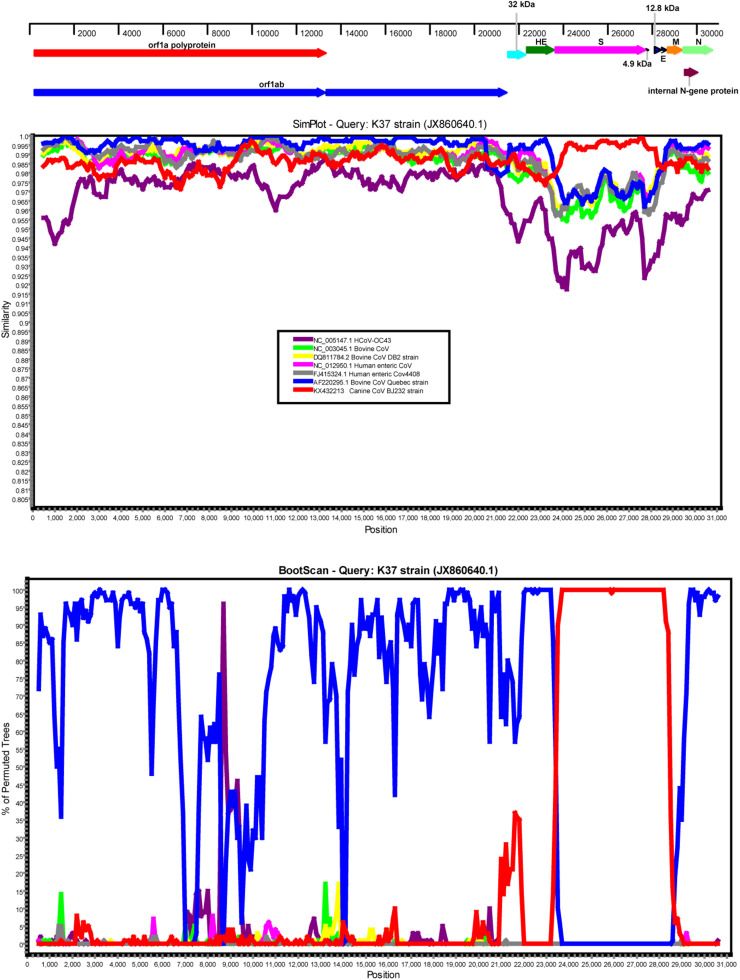
Similarity plots and bootscanning analyses (Fig. 3 ) were performed to confirm the recombination between CRCoV and BCoV. The previously reported CRCoV K37 strain was used as a query sequence, and was compared with HCoV-OC43 (NC.005147), BCoV (BCoV-RefNC_003045, BCoV-DB2 DQ811784, BCoV-Quebec AF220295), HECoV (HECoV-Ref NC_012950, HECoV-4408 FJ415324), and CRCoV (CRCoV-BJ232 KX432213). In the S gene, strain K37 possessed the highest similarity with the BJ232 strain between position 23,842 and 29,072, as expected. However, in the orf1ab, HE, M and N genes, strain K37 was apparently related to BCoV, which further confirmed the occurrence of genetic recombination between CRCoV and BCoV. The consistency in the results of bootscanning and phylogenetic analyses also supported the possibility of recombination in the S gene.
Fig. 3.
Genetic recombination analyses of the complete genome of the South Korea K37 strain. (A) A schematic diagram of the complete genome of the CRCoV-K37 strain. (B) The results of a SimPlot similarity analysis. (C) The results of a bootscanning analysis. The CRCoV-K37 strain was used as the query sequence and was compared with seven other representative strains of BCoV-like viruses, including HCoV-OC43 (NC_005147.1), BCoV (NC_003045.1), BCoV DB2 strain (DQ811784), HECoV (NC_012950.1), HECoV 4408 strain (FJ415324.1), BCoV Quebec strain (AF220295.1), and CRCoV-BJ232 (KX432213). A window size of 1000, a step size of 1000 bp and 100 replicates were used as the default settings.
3.5. Prevalence of recombinant CRCoV worldwide
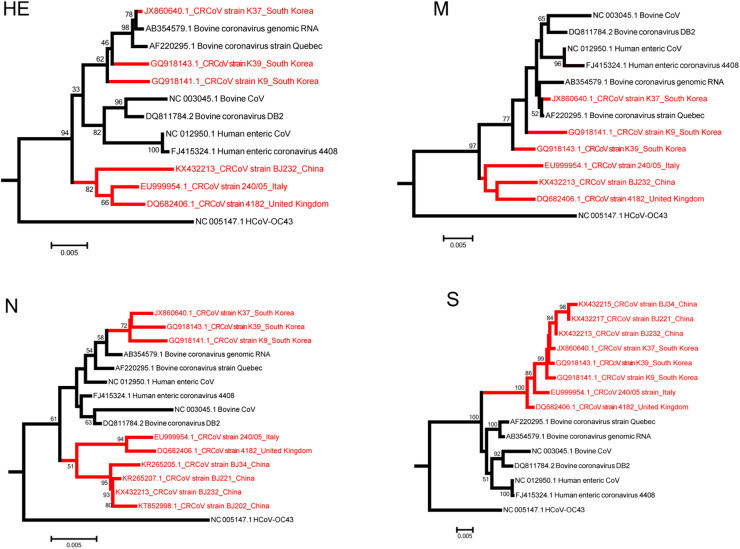
We collected all available CRCoV sequences from the GenBank database and conducted comprehensive phylogenetic analyses separately based on HE, S, M and N genes to understand the epidemiology distribution of the recombinant strain(South Korea strain K37). The BJ232, BJ221, BJ34 and BJ202 strains were collected from China in this study, while the K37, K39 and K9 strains came from South Korea, and the 240/05 and 4182 strains were derived from Italy and the United Kingdom, respectively. As shown in Fig. 4 , the CRCoV strains from China, including the BJ232, BJ221, BJ34 and BJ202 strains, clustered with strains prevalent in Italy and the United Kingdom (the 240/05 and 4182 strains), and the CRCoV strains detected in South Korea (K37, K39 and K9 strains) were all derived from recombinant lineage.
Fig. 4.
Phylogenetic analyses of all available CRCoV strains based on HE, M, N and S genes. The BJ232, BJ221, BJ34 and BJ202 strains come from China in this research,K37, K39 and K9 strains were collected from South Korea, and the 240/05 and 4182 strains were derived from Italy and the United Kingdom, respectively. All the genes were analyzed by PhyML program using the maximum likelihood method, with 500 bootstrap replicates.
4. Discussion
CRCoV is a causative agent for CIRD and a novel pathogen detected in respiratory samples from dogs (Erles and Brownlie, 2008). A serological survey of CRCoV conducted worldwide revealed that antibodies against CRCoV were common in the canine population (Erles et al., 2004, Isaacs et al., 1983, Pratelli et al., 2003, Priestnall et al., 2006). In contrast, corresponding genome-wide analyses have rarely been conducted (An et al., 2010, Lorusso et al., 2009). In the present study, we characterized the first complete genome of CRCoV (named strain BJ232) derived from China, and phylogenetic analyses showed that CRCoV-BJ232occupied a deep branch at the root of members of Betacoronavirus 1, being distinct from HCoV-OC43 and BCoV. Another previously described CRCoV strain, K37, was identified to have arisen from recombination between CRCoV-BJ232 and BCoV. In detail, the CRCoV-K37 strain was related to CRCoV-BJ232 in the S gene, while in the orf1ab, HE, M, and N genes, CRCoV-K37 was noted to possess a close relationship with BCoV.
Recombination is a common phenomenon in coronaviruses and thought to contribute to the emergence of new pathotypes (Gorbalenya, 2008, Wang et al., 2015). Thus far, most of the recombination events of coronaviruses have been reported between species of the same group (Herrewegh et al., 1998, Keck et al., 1988), such as among the bat-associated CoVs (Corman et al., 2014) and 229E-related CoVs (Corman et al., 2015), and the major recombination breakpoint has mainly been within the S gene. Genetic recombinant among alphacoronavirus 1 species frequently occurred, such as ferret coronaviruses (Lamers et al., 2016, Minami et al., 2016). However, few researches about genetic recombination of Betacoronavirus 1 species have been conducted. The discovery of genetic recombination between CRCoV and BCoV confirms the phenomenon, and intraspecies recombination therefore exists for Betacoronavirus 1 species.
Phylogenetic analyses based on all available CRCoV genes showed that the two clusters of CRCoV, including recombinant strain (South Korea K37 strain) and the Chinese BJ232 strain, have been prevalent worldwide for many years. CRCoV and BCoV were closely related in Betacoronavirus 1, located on the outer leaves of phylogenetic tree, and form an independent branch. It was difficult to determine the evolution orientation (transmission from cattle to canine or both from murine), although the latest research supports the murine origins of Betacoronavirus 1 (Lau et al., 2011, Lau et al., 2015). More CRCoV genome sequences are needed to understand the evolutionary relationship between CRCoV and BCoV.
Regarding the pathogenicity and transmissibility of CRCoV, the Chinese BJ232 strain and South Korea K37 strain were both derived from diseased dogs with respiratory infections. Genomic analyses showed that the S genes, which mediate coronavirus attachment, were highly similar in these strains. Because recombination may be a driving force for the formation of pathogenic viruses from less pathogenic virus (Forni et al., 2017), recombinant may play an important role in the pathogenicity and transmissibility of CRCoV. Limitation of this study was that the isolation of CRCoV-BJ232 failed on cell culture. More CRCoV strains were needed to be isolated or rescued using reverse genetic technique to perform experimental inoculation of dogs.
In conclusion, herein, we reported the first complete genome sequence of a CRCoV derived from a diseased dog in China, and confirm that intraspecies recombinant exists for Betacoronavirus 1 species. The results may have implications for the origin and evolution of CRCoV.
Acknowledgments
This work was supported by grants from the National Key Plan for Scientific Research and Development of China (2016YFD0500301) and Megaproject for Infectious Disease Research of China (2014ZX10004001,2013ZX10004601). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have declared that no competing interests exist.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2017.05.006.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- An D.-J., Jeong W., Yoon S.H., Jeoung H.-Y., Kim H.-J., Park B.-K. Genetic analysis of canine group 2 coronavirus in Korean dogs. Vet. Microbiol. 2010;141:46–52. doi: 10.1016/j.vetmic.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonavoglia C., Martella V. Canine respiratory viruses. Vet. Res. 2007;38:355–373. doi: 10.1051/vetres:2006058. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Ithete N.L., Richards L.R., Schoeman M.C., Preiser W., Drosten C., Drexler J.F. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M., Nkrumah E.E., Maganga G.D., Oppong S., Adu-Sarkodie Y. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Brownlie J. Investigation into the causes of canine infectious respiratory disease: antibody responses to canine respiratory coronavirus and canine herpesvirus in two kennelled dog populations. Arch. Virol. 2005;150:1493–1504. doi: 10.1007/s00705-005-0533-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Brownlie J. Canine respiratory coronavirus: an emerging pathogen in the canine infectious respiratory disease complex. Vet. Clin. N. A.: Small Anim. Pract. 2008;38:815–825. doi: 10.1016/j.cvsm.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Toomey C., Brooks H.W., Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003;310:216–223. doi: 10.1016/S0042-6822(03)00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Dubovi E.J., Brooks H.W., Brownlie J. Longitudinal study of viruses associated with canine infectious respiratory disease. J. Clin. Microbiol. 2004;42:4524–4529. doi: 10.1128/JCM.42.10.4524-4529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Shiu K.-B., Brownlie J. Isolation and sequence analysis of canine respiratory coronavirus. Virus Res. 2007;124:78–87. doi: 10.1016/j.virusres.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human Coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E. Nidoviruses ASM Press; Washington, DC: 2008. Genomics and Evolution of the Nidovirales; pp. 15–28. [Google Scholar]
- Herrewegh A.A., Smeenk I., Horzinek M.C., Rottier P.J., de Groot R.J. Feline coronavirus type II strains 79–1683 and 79–1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs D., Flowers D., Clarke J., Valman H., MacNaughton M. Epidemiology of coronavirus respiratory infections. Arch. Dis. Child. 1983;58:500–503. doi: 10.1136/adc.58.7.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneshima T., Hohdatsu T., Satoh K., Takano T., Motokawa, Koyama K., H The prevalence of a group 2 coronavirus in dogs in Japan. J. Vet. Med. Sci. 2006;68:21–25. doi: 10.1292/jvms.68.21. [DOI] [PubMed] [Google Scholar]
- Kaneshima T., Hohdatsu T., Hagino R., Hosoya S., Nojiri Y., Murata M., Takano T., Tanabe M., Tsunemitsu H., Koyama H. The infectivity and pathogenicity of a group 2 bovine coronavirus in pups. J. Vet. Med. Sci. 2007;69:301–303. doi: 10.1292/jvms.69.301. [DOI] [PubMed] [Google Scholar]
- Keck J., Matsushima G.K., Makino S., Fleming J., Vannier D., Stohlman S.A., Lai M. In vivo RNA–RNA recombination of coronavirus in mouse brain. J. Virol. 1988;62:1810–1813. doi: 10.1128/jvi.62.5.1810-1813.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A.M. Elsevier; 2011. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. [Google Scholar]
- Lamers M.M., Smits S.L., Hundie G.B., Provacia L.B., Koopmans M., Osterhaus A.D., Haagmans B.L., Raj V.S. Naturally occurring recombination in ferret coronaviruses revealed by complete genome characterization. J. Gen. Virol. 2016;97:2180–2186. doi: 10.1099/jgv.0.000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Lee P., Tsang A.K., Yip C.C., Tse H., Lee R.A., So L.-Y., Lau Y.-L., Chan K.-H., Woo P.C. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Tsang A.K., Fan R.Y., Luk H.K., Cai J.-P., Chan K.-H., Zheng B.-J., Wang M. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J. Virol. 2015;89:3076–3092. doi: 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.-i., Choi S., Lim J.-A., Jeoung H.-Y., Song J.-Y., Dela Pena R., An D.-J. Complete genome analysis of canine respiratory coronavirus. Genome Announc. 2013;1:00093–100012. doi: 10.1128/genomeA.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A., Desario C., Mari V., Campolo M., Lorusso E., Elia G., Martella V., Buonavoglia C., Decaro N. Molecular characterization of a canine respiratory coronavirus strain detected in Italy. Virus Res. 2009;141:96–100. doi: 10.1016/j.virusres.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami S., Kuroda Y., Terada Y., Yonemitsu K., Van Nguyen D., Kuwata R., Shimoda H., Takano A., Maeda K. Detection of novel ferret coronaviruses and evidence of recombination among ferret coronaviruses. Virus Genes. 2016;52:858–862. doi: 10.1007/s11262-016-1365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Decaro N., Tinelli A., Camero M., Cirone F., Elia G., Cavalli A., Corrente M., Greco G. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J. Virol. Methods. 2003;110:9–17. doi: 10.1016/S0166-0934(03)00081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestnall S.L., Brownlie J., Dubovi E.J., Erles K. Serological prevalence of canine respiratory coronavirus. Vet. Microbiol. 2006;115:43–53. doi: 10.1016/j.vetmic.2006.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestnall S.L., Pratelli A., Brownlie J., Erles K. Serological prevalence of canine respiratory coronavirus in southern Italy and epidemiological relationship with canine enteric coronavirus. J. Vet. Diagon. Invest. 2007;19:176–180. doi: 10.1177/104063870701900206. [DOI] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moës E., Thoelen I., Wollants E., Lemey P., Vandamme A.-M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Lemey P., Maes P., Van Reeth K., Nauwynck H., Pensaert M., Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu D., Shi W., Lu R., Wang W., Zhao Y., Deng Y., Zhou W., Ren H., Wu J. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. MBio. 2015;6:01280–101215. doi: 10.1128/mBio.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhu N., Li Y., Lu R., Wang H., Liu G., Zou X., Xie Z., Tan W. Metagenomic analysis of viral genetic diversity in respiratory samples from children with severe acute respiratory infection in China. Clin. Microbiol. Infect. 2016;22(458):e451–e458. doi: 10.1016/j.cmi.2016.01.006. (e459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachi A., Mochizuki M. Survey of dogs in Japan for group 2 canine coronavirus infection. J. Clin. Microbiol. 2006;44:2615–2618. doi: 10.1128/JCM.02397-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.