Abstract
The laboratory mouse has proved an invaluable model to identify host factors that regulate the progression and outcome of virus-induced disease. The paradigm is to use single-gene knockouts in inbred mouse strains or genetic mapping studies using biparental mouse populations. However, genetic variation among these mouse strains is limited compared with the diversity seen in human populations. To address this disconnect, a multiparental mouse population has been developed to specifically dissect the multigenetic regulation of complex disease traits. The Collaborative Cross (CC) population of recombinant inbred mouse strains is a well-suited systems-genetics tool to identify susceptibility alleles that control viral and microbial infection outcomes and immune responses and to test the promise of personalized medicine.
Keywords: systems genetics, Collaborative Cross, host factors, genetic mapping, immune response
Highlights
Viral infections are complex traits that are influenced by viral and environmental as well as host factors.
Complete knockouts of genes are rare in humans whereas natural variation at the nucleotide level is abundant. Thus, successful translation from mice to humans is more likely working with natural variation in mouse populations.
The Collaborative Cross is a mouse genetic reference population that is well suited to be utilized to identify networks of host genetics key players that influence complex traits such as viral infections.
Indefinitely reproducible mouse strains with fully sequenced genomes offer the chance for wide collaborations across pathogens. Additionally, it offers the ability to identify cross-pathogen susceptibility or resistance alleles.
Viral Disease Is a Complex Trait
Viral infections pose a major threat to human and animal health, causing significant morbidity and mortality every year. During the past decades, the emergence of several highly pathogenic zoonotic viruses has demonstrated the fragility of the species barrier in protecting human and farm animal populations from pathogens that exist in the animal kingdom. While basic virological research offers the possibility to detect viral strains that are ‘poised’ for emergence and identify mutations that might promote pathogen emergence, accurate prognoses are confounded by our inability to predict disease severity and virulence. Importantly, viruses with identical genome sequences do not always cause the same set of clinical manifestations in humans. Moreover, the complex interplay between environmental, viral, and host genetic factors drives differences in interindividual disease progression, severity, and outcome. These factors change over the course of a lifetime and some, like individual health status, comorbidities, and environmental factors 1, 2, are difficult if not impossible to control. However, perhaps one of the most important key players in the fragile balance of microbial pathogenesis centers around host genetic susceptibility alleles that dramatically influence the course of disease in different individuals. In humans, a growing number of genetic factors like entry receptors, receptor-modifying enzymes, and innate and adaptive immune-related proteins that regulate influenza virus, norovirus, rotavirus, respiratory syncytial virus (RSV), HIV, hepatitis B and C viruses, herpes virus, and other acute and chronic virus disease outcomes have been identified (Table 1 ; more detailed list in [3]).
Table 1.
Genes with Significant Associations with Viral Disease in Humans
| Pathogen | Phenotype | Causal gene | Refs |
|---|---|---|---|
| Dengue virus (DENV) | DENV shock syndrome | MICB, PLCE1 | [73] |
| Epstein–Barr virus (EBV) | EBNA-1 IgG titer | HLA-DRB1, HLA-DQB1 | [74] |
| Hepatitis B virus (HBV) | Chronic infection | HLA-DPA1, HLA-DPB1 | [75] |
| Persistence | INST10 | [76] | |
| Hepatitis C virus (HCV) | Spontaneous clearance | IL28B | [77] |
| Development of hepatocellular carcinoma | TLL1 | [78] | |
| Progression to hepatocellular carcinoma | DEPDC5 | [79] | |
| HIV-1 | Viral load | HLA-B, HLA-C | [80] |
| Viral load control | HCP5 | [81] | |
| Influenza A virus (IAV) | Reduced restriction of viral replication | IFITM3 | [82] |
| Increased incidence and increased risk of viral pneumonia | TNF | [83] | |
| Norwalk virus (NoV) | Resistance | FUT2 | [84] |
| Respiratory syncytial virus (RSV) | Bronchiolitis | SFPA/D | 85, 86 |
| West Nile virus (WNV) | Resistance | CCR5 | [87] |
Accordingly, research on host genetics is a promising tool for understanding susceptibility and virulence patterns in human populations and refining pandemic-preparedness efforts. Furthermore, the discovery of innovative prophylactic or diagnostic and therapeutic treatment options for viral disease that can be leveraged across different host genetic susceptibility patterns can lead to improved personalized medicine.
In this review we discuss historic and new platform strategies designed to unravel the interplay between the complex host and viral genetic determinants that regulate disease severity. Moreover, we discuss recent developments in the field of complex genetics designed to resolve quantitative trait loci (QTLs) (see Glossary) and rapidly identify single candidate genes and alleles that regulate microbial pathogenesis.
The Laboratory Mouse in Viral Disease Research
Animal models offer a strategy to reduce system-wide complexity through standardizing environmental influences without losing the integrity of a functional biological system. By far, most in vivo viral pathogenesis studies are conducted in inbred mouse models. Not only are the husbandry and breeding of mice cost-efficient, but genome sequences, as well as many species-specific immunologic, molecular, and biochemical reagents, are available to the research community. However, disease spectra in inbred mouse models are narrow compared with the diverse spectra noted in outbred populations like humans. For respiratory viral infections, additional phenotypic variations must also be considered, as mice do not sneeze, cough, or develop fever following infection. Rather, they exhibit loss of body weight, reduced respiratory function, and decreased locomotive activity. As human pathogens often replicate less efficiently in mice, it is frequently necessary to use mouse-adapted viral strains selected for increased replication and disease in inbred mouse strains, which may or may not replicate disease symptoms seen in humans [4]. The most commonly used parameters for viral infection intensity in mice are changes in body weight and survival rate. Detailed analysis of disease progression can be undertaken utilizing time-course experiments during which samples of interest are collected and disease kinetics revealed. Most recently, mouse genetic reference populations with diverse genetic backgrounds have been developed that replicate the genetic and disease outcome variability found in outbred populations like humans. These new platforms are further supported by technologies that allow targeted genetic modifications, enabling researchers to study how complex traits regulate microbial pathogenesis.
Genetic Manipulation of the Mouse Genome
Using the mouse as a model organism offers two distinct types of genetic approaches: reverse and forward genetic studies. The most commonly used application in reverse genetic approaches involves the specific ablation of a single gene, either by deleting parts of or the entire gene or by replacing coding exons. Gene trapping by comparison offers the possibility to insert reporter genes into the gene of interest, disrupting its function. Both techniques require the DNA construct of choice to be transfected into mouse embryonic stem cells (mESCs), injection of screened mESC clones into a blastocyst, and transfer of this blastocyst into the uterus of the host animal. Most recently, innovative techniques like transcription activator-like effector nucleases (TALENs) [5] and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) [6] are accelerating the process of targeted genome editing, including the introduction of susceptibility alleles that improve virus replication and pathogenesis in the mouse [7].
Genetically engineered mouse strains have represented the gold standard to investigate the role of specific host genes in regulating disease and immune outcomes following virus infections (e.g. 8, 9). Furthermore, genetically modified strains can be used [10] to understand human monogenic diseases like cystic fibrosis, polycystic kidney disease, and sickle cell disease. Although complete (biallelic) knockout of genes in humans is a relatively rare event, a recent study showed that healthy people have about 100 loss-of-function variants and 20 completely inactivated genes [11]. Consequently, the majority of human diseases are complex traits whose disease outcomes are influenced by multiple genetic factors.
Human genomes are characterized by extensive natural variation, which has accumulated over time through, for example, mutation events (external damage to DNA or internal errors during replication), gene flow, and sexual reproduction, which drives phenotypic interindividual differences across populations. Differences in DNA sequence can modify transcript and protein levels by altering their functional properties, timing, level, and site of expression [12]. Natural variation can be used in forward genetic studies to identify novel genes involved in a variety of quantitative traits and diseases [13].
The occurrence of spontaneous mutations in laboratory mouse strains was the first platform employed for forward genetic approaches [14]. There are various methods to increase the likelihood of mutations by treating male mice with mutagens such as N-ethyl-N-nitrosourea (ENU) [15] or chlorambucil 16, 17, by irradiation 18, 19, and by utilizing transposons such as the sleeping beauty [20] or piggyback [21] system, named according to their transposases, to insert specific DNA sequences. Breeding of those mutated mice allows selection of those with an altered phenotype in the trait of interest. This approach mimics natural genetic variation in humans in the controlled setting of the mouse model and has the power to reveal genomic variation and networks of genes influencing a phenotype rather than analyzing the effect of the absence of a single-gene product, making the translation from mouse to human more likely.
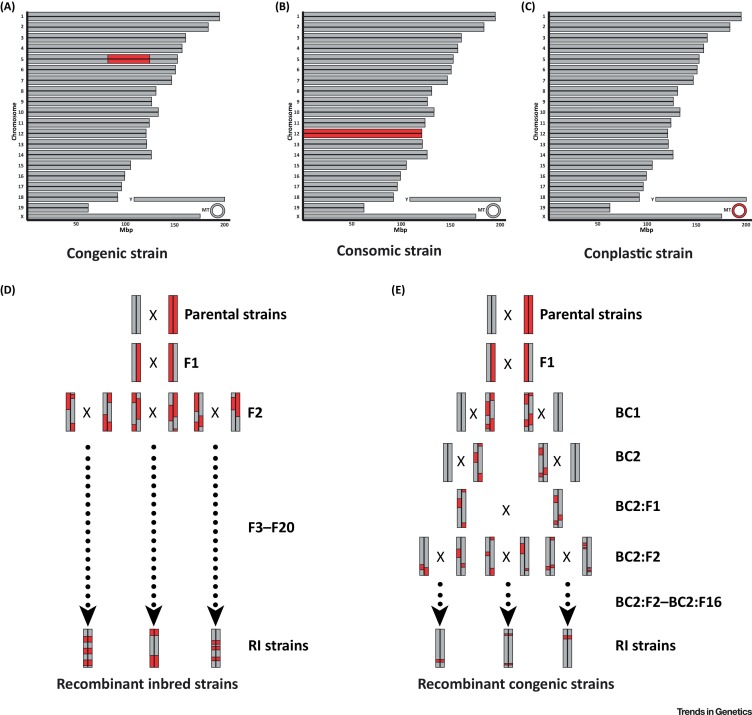
Genetic mapping studies are undertaken to identify the genomic region that causes the altered phenotype of interest. Commonly, QTL analysis is used to reveal genotype–phenotype associations [22]. F2 crosses between an inbred strain carrying the aberrant phenotype of interest and another inbred mouse strain lacking this particular phenotype have been widely performed. The identified chromosomal regions can be large, containing several hundreds of genes. The size of the chromosomal region exclusively depends on the number of recombination breakpoints in the cross and the genetic complexity of the region. Chromosomal locations can be narrowed using consomic, conplastic, congenic, recombinant inbred (RI), or recombinant congenic mouse strains [23] (Figure 1 ).
Figure 1.
Various Genetic Mouse Model Approaches to Narrow Quantitative Trait Locus (QTL) Regions. (A) Congenic mouse strains are produced by transferring a genomic region from an inbred donor strain to an inbred recipient strain through repeated backcrossing. (B) Consomic mouse strains contain an entire chromosome from a donor strain and are generated via backcrossing to the recipient strain. (C) In conplastic mouse strains, the entire mitochondrial DNA is derived from a donor strain and is generated through backcrossing of females from the donor strain to males from the recipient strain. (D) Recombinant inbred mouse strains are generated by crossing two inbred strains to obtain an F1 generation. These F1 mice are crossed to create an F2 generation, which is brother–sister mated for at least 20 generations to achieve mice with a fixed genetic background and equal contributions of the two parental strains. (E) Recombinant congenic strains are produced by crossing two inbred mouse strains. The resulting F1 generation is backcrossed twice (BC1 and BC2) before they are brother–sister mated for an additional 14 generations. The genome composition of the final strains is skewed towards one parental strain in a 7:8 ratio.
The alternative to dealing with large chromosomal regions and the narrowing process is to use mouse genetic reference populations (GRPs). RI stains of mice are popular due to their long-term genetic stability, which helps in integrating data collected in different settings and reproducibility over a long time. The most extensively used mouse GRP is the BXD family of recombinant inbred strains. They are derived from a cross between C57BL/6J and DBA/2J mice [24]. To obtain an F1 generation, C57BL/6J and DBA/2J mice were crossed. The resulting F1 generation is mated to achieve the F2 generation, which is subsequently brother–sister mated for at least 20 generations to generate inbred mouse strains with a fixed genetic architecture. Completely inbred strains are then called BXD strains, which are genotyped once (one animal per line, not every individual) and can be phenotyped indefinitely for every trait of interest. Currently, there are 156 BXD strains available [25]. For decades, recombinant inbred strains such as the BXD family have been used extensively as tools for genetic mapping of Mendelian and quantitative traits. To identify single genes that are responsible for the observed phenotype fine mapping, sequence analysis, expression profiling, and functional studies are typically performed [26]. However, the identification of causal gene variants remains challenging due to the large size and the number of genes under the identified QTL region, coupled with the fact that the parental strains were identical by descent resulting in so-called ‘blind spots’ for genetic mapping. Wild-derived strains other than Mus musculus domesticus need to be employed to cover those spots and increase genetic variation [23]. Various resources have been established to address this issue, among them the heterogeneous stock (HS), which is derived from eight founder strains (A/J, AKR/J, BALBc/J, CBA/J, C3H/HeJ, C57BL/6J, DBA/2J, and LP/J) and maintained through random mating. No inbred mouse lines are created and therefore each mouse exhibits a unique combination of alleles with the goal of containing random variation similar to the human population [27]. The caveat of this mouse population is that every individual mouse needs to be genotyped, which might be too expensive for some researchers. However, genotyping technologies are evolving constantly and costs are decreasing rapidly.
The Collaborative Cross
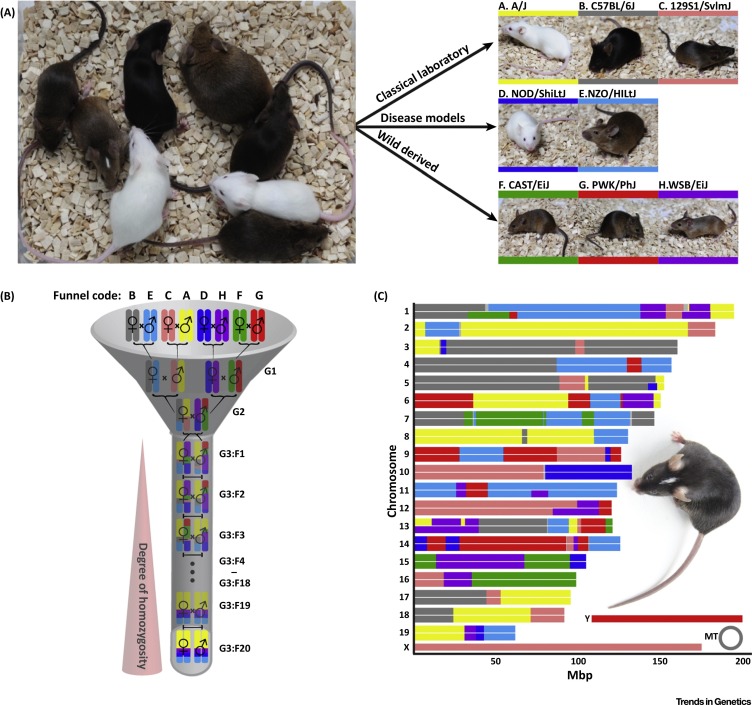
Another mouse GRP that includes other M. musculus subspecies is the CC Mouse Resource (Figure 2 ), which has already been used to successfully identify highly promising candidate genes that are influencing susceptibility or resistance to viral infections (Table 2 ).
Figure 2.
Generation of the Collaborative Cross Resource. (A) The Collaborative Cross panel of recombinant inbred mouse strains is a multiparental population that is derived from eight founder strains. Among these founder strains are classical laboratory mouse strains (A/J, C57BL/B6, 129S1/SvImJ), mouse models for human diseases (NOD/ShiLtJ – type 1 diabetes, NZO/HlLtJ – obesity), and wild-derived mouse strains (CAST/EiJ, PWK/PhJ, WSB/EiJ). Every mouse strain was assigned a letter (A–H) and a particular color that are used by the entire research community. (B) Breeding-funnel design of the Collaborative Cross that guarantees equal distribution of founder alleles to the resulting CC strain. Depicted is the specific breeding funnel for chromosome 19 of the CC strain CC001. (C) Genome architecture of CC001 with founder contributions displayed in their respective colors. Photograph: Klaus Schughart.
Table 2.
QTLs of CC Studies Using Different Viruses
| Pathogen | Phenotype | QTL region | % Variation | Number of genes under QTL | Refs |
|---|---|---|---|---|---|
| SARS-CoV | Vascular cuffing | HrS1: Chr. 3: 18286790–26668414 | 26% | 26 [narrowed to one (Trim55)] | [40] |
| Viral titer | HrS2: Chr. 16: 31583769–36719997 | 22% | 92 (narrowed to 48) | ||
| Eosinophil infiltration | HrS3: Chr. 15: 72103120–75803414 | 26% | 63 (narrowed to 25 – functional change only in Bai1) | ||
| Vascular cuffing | HrS4: Chr. 13: 52822984–54946286 | 21% | 30 (narrowed to nine – Cdhr2) | ||
| D3% weight | HrS5: Chr. 18: 27108062 – 58694005 | 6.6% | 158 [narrowed to one (Ticam2)] | [61] | |
| D4% weight | HrS5: Chr. 18: 27108062–58694005 | 8.5% | |||
| Log titer | HrS5: Chr. 18: 27108062–58694005 | 12.9% | |||
| Hemorrhage | HrS5: Chr. 18: 24762824–7829634 | 6% | |||
| D3% weight | HrS6: Chr. 9: 116476207–telomere | 7% | – | ||
| Log titer | HrS7: Chr. 7: 55169841–11722358 | 12.3% | – | ||
| Log titer | HrS8: Chr. 12: 81649471–108529109 | 5.4% | – | ||
| Hemorrhage | HrS9: Chr. 15: centromere–64.430001 | 9.1% | – | ||
| WNV | Frequency of CD73+ Tregs | HI1: Chr. X: 166 Mb–telomere | – | 43 (narrowed to 22) | [88] |
| Decreased frequency of CXCR3+ Tregs, CXCR3+ CD4+, and CD8+ T cells | HI2: Chr. X: 100–106 Mb | – | 42 (narrowed to 26) | ||
| Increased frequency of ICOS+ Tregs in spleen | HI3: Chr. X: 140–145 Mb | – | 18 (narrowed to 11) | ||
| IAV | D4 weight, log titer, IHC score, D3 clinical, airway inflammation, airway damage | HrI1: Chr. 16: 97.5 Mb–98.2 Mb | 41.67% | Ten (including Mx1) | [39] |
| D4 weight | HrI2: Chr. 7: 89.1 Mb–96.7 Mb | 9.7% | 69 | ||
| Pulmonary edema | HrI3: Chr. 1: 21.7 Mb–29 Mb | 29.73% | 24 | ||
| Airway neutrophils | HrI4: Chr. 15: 77.4 Mb–86.6 Mb | 22.7% | 206 |
To expand genetic variation in GRPs, an innovative strategy for a multiparent population (MPP) of mice was conceptualized in the early 21st century and developed over the next decade [28]. Use of an octoparental crossing scheme between genetically distinct mouse strains was proposed and modifications through the research community were integrated. Breeding of this novel GRP specifically designed for complex genetics started in 2002 [28]. The eight founder strains of the CC include three classical laboratory strains (A/J, C57BL/6J, and 129S1/SvImJ), which have been used extensively in biological research and build the genetic backbone on which most of the knockout mouse strains are generated. Moreover, two mouse models for common human diseases were included (NOD/ShiLtJ for type 1 diabetes and NZO/HlLtJ for obesity) to address research questions of comorbidities. The addition of three wild-derived mouse strains (CAST/EiJ, PWK/PhJ, and WSB/EiJ) not only increased the genetic diversity by adding new alleles that are not present among the classical inbred and disease model strains, but also covered different phylogenetic origins of the mouse species (CAST/EiJ – Mus musculus castaneous, PWK/PhJ – Mus musculus musculus) to encompass 90% of genetic variation present in the M. musculus species [23]. Accordingly, the CC population reaches a level of genetic diversity comparable with the diversity found in the human population.
To guarantee equal contributions of all eight founder strains to each of the resulting CC strains, a specific breeding funnel was elaborated with around 135 unique recombination events and segregating polymorphisms every 100–200 bp [28]. In this way susceptibility alleles are scrambled in new ways, allowing novel allelic combinations leading to an extension of phenotypic range beyond the scope observed in the parental strains. Breeding was performed at three different locations: Oak Ridge National Laboratory in Oak Ridge, TN, which moved to the University of North Carolina at Chapel Hill [29]; the International Livestock Research Institute in Nairobi, Kenya, which moved to the Tel Aviv University in Tel Aviv, Israel [30]; and the Western Australian Institute for Medical Research/Geniad Ltd in Perth, Australia [31]. Although the theoretical plan was perfectly elaborated, hundreds of CC strains became extinct, almost half from problems in male infertility [32].
Prior to the development of the final CC resource, incipient CC lines that were not fully inbred yet (pre-CC lines) were used in genetic mapping studies to provide proof of concept and to show the potential of this newly designed GRP (Box 1 ). Candidate genes for various phenotypes, such as susceptibility to Aspergillus fumigatus infections [33], energy balance traits [34], differences in hematological parameters [35], susceptibility to Klebsiella pneumoniae [36], and neutrophilic inflammation due to house dust mite-induced asthma [37], were successfully identified. Moreover, mapping efforts revealed expression QTLs for extreme host responses to influenza A virus (IAV) infections [38], host response QTLs to IAV [39], and severe acute respiratory syndrome coronavirus (SARS-CoV) pathogenesis [40]. A common observation in all of the studies was that pre-CC lines exhibit an enhanced phenotypic range compared with the variation observed in the eight founder strains or other classical inbred strains and that it was possible to dissect traits that were thought to be inseparably entwined.
Box 1. Ticam2 Plays a Major Role in SARS-CoV Pathogenesis.
As an expansion to their first paper identifying QTLs for SARS pathogenesis in pre-CC mice, the authors utilized an alternative genetic approach to understand host genetic contributions to the course and outcome of SARS-CoV infection [61]. Two pre-CC lines with divergent outcome after SARS-CoV infection were identified. CC003/Unc is resistant to SARS-CoV whereas CC053/Unc is highly susceptible. To dissect host genetic factors that lead to the different outcomes, these two strains were bred and 264 F2 animals generated. Loss of body weight, viral titer in lungs, pulmonary hemorrhage, and histopathological changes were analyzed at multiple time points after infection. Overall, F2 mice exhibited a broader phenotypic range than the parental strains. Five significant QTLs across all analyzed phenotypes were identified, one of which affected multiple SARS-CoV response phenotypes. This QTL (HrS5) on chromosome 18 (27.1–58.6 Mb) was selected for follow-up studies. Integration of different bioinformatics and database approaches led to the identification of Ticam2 as a highly promising candidate gene. Ticam2 is a toll-like receptor (TLR) adapter protein that had not been shown to play an important role in SARS-CoV pathogenesis. Utilizing another tool from the geneticist’s toolbox, Ticam2 knockout mice (Ticam2 −/−) were employed to investigate its effect on disease progression and outcome. Ticam2 −/− mice exhibited significantly more weight loss, similar viral load in lungs on day 4 after infection (however, significantly higher titers on day 2 after infection were reported previously [8]), and similar histopathological findings but significantly increased pulmonary hemorrhage on day 4 after infection. Thus, the authors successfully showed that a screening approach in CC mice in combination with an F2 follow-up study can lead to single candidate genes that can be confirmed using reverse genetic tools. It remains to be determined whether allele swaps can be used to identify the allele driving this disease phenotype.
Alt-text: Box 1
Highlighting another powerful characteristic of the CC, completely new mouse models for spontaneous colitis [41], Ebola-associated hemorrhagic fever [42], novel neurological responses to Theiler’s murine encephalomyelitis virus (TMEV) [43], and persistent West Nile virus (WNV) infection in the brain [44] were discovered, a harbinger of new model systems that may emerge over time.
The Diversity Outbred (DO) population of mice is a GRP complementary to the CC derived from the same set of founder strains. Instead of inbreeding, an outbred population is maintained through random mating, which enhances the mapping resolution even further through the acquisition of even more recombination sites [45]. Early studies utilizing DO mice led to the identification of a specific isoform of Apobec1 contribution to atherosclerosis [46] and sulfotransferases as candidate genes for benzene-induced genotoxicity [47].
Designed specifically for the analysis of complex traits, the CC and DO populations of genetically highly diverse mice provide the first true systems-genetic platform for cumulative and integrated data collection [48]. Systems genetics, as an innovative strategy to investigate the role of host genetics that encompasses diverse molecular omics data [49], also catalyzed the development of a variety of analytic and informatics tools and methods 50, 51, 52, 53 and provides state-of-the-art tools for genetic mapping and candidate gene identification and an opportunity to test the promise of predictive genomic medicine.
From Complex Screens to Candidate Genes: A Recipe for Complex Genetic Studies
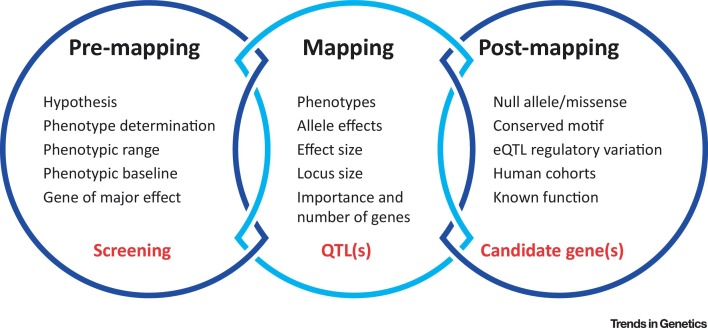
There is increasing evidence for host genetic regulation of viral and microbial pathogenesis in humans (Table 1). Although classical approaches like cell culture experiments or knockout mouse studies have been successfully used to understand infectious disease pathogenesis, the contribution of variation in host genetics can no longer be omitted because it more accurately phenocopies the human condition and is critical to pave the way to personalized medicine. The hurdles for most researchers who are not using complex genetics until now are the lack of expertise in designing complex genetic studies and the inevitable bioinformatics barriers until now. However, complex genetic approaches become exponentially more valuable as data is accumulated and compared across experimental setups and pathogens and combined to gain deeper knowledge. In this review, we provide an experimental framework for the rapid implementation of complex-trait genetic strategies in the laboratory setting, including design of the overall study, best utilization of the CC resource, and ways to narrow down QTLs to pinpoint single candidate genes influencing the trait of interest (reviewed in [54]) (Figure 3 ).
Figure 3.
Schematic of Experimental Approach for Complex Genetic Studies of Viral Infections. eQTL, expression QTL; QTL, quantitative trait loci.
The classic CC hypothesis is that ‘my disease phenotype’ is a complex oligogenic trait regulated by multiple genetic polymorphisms in different genes, which interact to regulate disease outcomes in natural populations. In a second step, the breadth and duration of disease-associated phenotypes is selected and measured in a subset of mouse strains, which typically is either a random selection of CC strains or the eight CC founder strains (RSV – Buntzman 2016, IAV [55]). Analyses of phenotypic baselines are crucial to be able to distinguish between phenotypes that are caused by differences in genome architecture or those that are related to viral pathogenesis. Additionally, multiple sample collection is advised to maximize phenotypic comparisons across the final population. This is important as it is impossible to predict phenotypic outcome as different gene variants segregate in the final population and traits that are closely linked might be broken apart. Another important factor that might complicate experimental design is the impact of known genes of major effect, like Mx1 for IAV [39] or Oas1b for flaviviruses [56]. Selection of CC strains that carry a variation at that locus or even a null allele might help to uncover rather small effects by other modulating genes. Utilizing up to eight different alleles at any given locus in the genome allows the discovery of gene of major effect-dependent and -independent processes (IAV [55], WNV [57]). There is a risk that genetic mapping might exclusively identify the gene of major effect if no attention is paid to these genes while selecting CC strains for the respective study. Genetic mapping offers the possibility to add covariates like batch effects to the calculation and randomization should be achieved whenever possible. Sample collection should anticipate that the CC model is likely to identify CC strains that progress to different stages in disease severity, replicating phenotypes seen in human populations and allowing the identification of susceptibility alleles that regulate disease progression from mild to severe to chronic infections in vivo. This is important as it has been shown that novel disease phenotypes might be discovered in the CC (severe neuroinvasive disease and chronic WNV infection [44]). New models for diseases that are completely unrelated (spontaneous colitis [41], human leukocyte adhesion and recruitment deficiencies [55]) or related to the study design (SARS-CoV [40], Ebola [42]) will generate a panel of variable disease-state mouse models that capture the different phases of disease seen in human populations.
After all phenotype measurements are obtained, genetic mapping is utilized to calculate the likelihood of every position in the genome being associated with the analyzed phenotype, and regions with the highest logarithm of the odds (LOD) score are identified as QTLs. Zooming into the QTL region, bioinformatics tools can be employed to generate allele-effect plots, which show the contribution of each of the eight possible alleles to the QTL. Analyses of allele effects are a good way to determine which parental alleles are driving high and low responses. Afterwards, variations that are unique to the identified strain can be analyzed utilizing the Sanger SNP browseri. Additionally, other resources, like BioGPSii or the gene browser included in Pubmediii, are of great use to gain information on known expression patterns and biological functions. In the line of a systems-genetics approach, transcriptional analyses can be used to identify differentially expressed genes or respective pathways influenced by a particular QTL (WNV [58]). However, these downstream analyses should only be performed on a subset of extreme phenotype samples, as this approach is highly informative and inexpensive. Using different pathogens for the same type of analysis can lead to the discovery of common or unique features across different viral species (IAV and SARS-CoV [59]). Additional tools of the complex genetic toolkit such as diallels of the CC founders and their reciprocal F1 hybrids allow the identification of different types of heritable effects (IAV [60]). Alternatively, once a highly promising candidate gene is identified the use of knockout mouse strains to validate the contribution of this particular gene to the observed phenotype can be used (SARS-CoV 40, 61). The International Knockout Mouse Consortium (IKMC) will soon achieve its goal of having a mouse mutant or a targeted mESC for every gene (http://www.mousephenotype.org), providing a crucial resource for functional annotation and validation of candidate genes. Importantly, it has been shown that the genetic background on which the knockout is created plays an important role 62, 63 and with recent advances of CRISPR/Cas9 technology the development of the identical knockout on different mouse backgrounds may be achievable in a time- and cost-efficient manner. Additionally, CRISPR/Cas9-generated allele swaps between mouse strains offer enhanced specificity, applicability, and translatability compared with complete knockout of genes. Translational aspects can be addressed by comparing identified SNPs with human SNP and gene databases or conserved structural elements and functional motifsiv,v,vi.
Concluding Remarks and Future Directions
The contribution of host genetic factors to the progression and outcome of viral infections has not only been proved but represents a powerful new tool to reveal the complex interplay between novel genes and their polymorphisms and disease severity. Although critical voices were raised in the scientific community [64] as well as in the media, it has been shown that data collected during viral infections in mice and in humans are highly correlated. For example, comparing signature genes derived from transcriptome analyses of IAV-infected CC founder strains, but not classic inbred mouse strains, with infected human volunteers revealed that gene expression in the blood of infected CC mice reproduces much more representative human signature profiles [65].
Human genetic studies are conducted using either candidate-gene approaches or genome-wide association studies (GWASs) [66]. Depending on the research question, both study designs have their own advantages, disadvantages, and limitations. Nevertheless, GWASs revealed many loci that are associated with human diseasevii. However, small numbers of participants, limited sample size, and technical differences in sample collection, and the inability to validate candidate gene–disease associations, make cross-study comparison challenging and reproducibility difficult. Additionally, the low number of accessible human samples often results in lack of statistical significance and/or reproducibility 67, 68. Recommendations on how to enhance the transparency of human GWASs [Strengthening the Reporting of Genetic Association Studies (STREGA)] have been published [69].
However, although human genetic factors influencing infectious disease susceptibility and outcome have been identified, significant difficulties remain in the verification of these factors in humans, mostly due to small effect sizes. Moreover, there are phenotypic traits, especially in the infectious disease context, that are difficult or even impossible to investigate in humans. The only way to unravel the effect of alleles associated with diseases that influence a specific molecular process is to investigate those alleles in controlled experimental settings either individually or in combination [12]. The mouse as the model organism of choice is currently the only mammalian systematic platform that offers not only the resources but also the technology to identify susceptibility and resistance loci for viral infections.
The CC population recombinant inbred mouse panel represents a paradigm shift for microbial pathogenesis studies, immunology, and studies of the role of complex genetic traits in disease. It offers increased mapping resolution compared with classical mouse GRPs and has already been successfully used in the field of infectious diseases, suggesting that an even greater impact exists for the field of immunology [70]. Systems-biology approaches in which a plethora of genetic and genomic (omics) data for the identical experimental condition is collected allows integration of data and offers the possibility of complex modeling and, ultimately, the identification of key factors driving differences in disease progression and outcome [71]. Being a relatively young and novel platform for systems genetics, the CC and its complementary resources have accelerated not only the identification of QTLs from a wide range of phenotypes but also pushed the development of genotyping and databases as well as bioinformatics tools [72]. We would like to highlight that the more that incipient lines of the CC are used in different fields to explore different phenotypes and answer different question, the more valuable the resource itself becomes (see Outstanding Questions). Data generated by laboratories from different fields might be of use for others, shortening their way to obtain results and saving money along the way. In summary, GWASs in mice can help to derive working hypotheses and direct human studies and, accordingly, studies in the two species are highly complementary.
Glossary
- Causal gene variant
the genetic variant that influences a certain trait and explains most of the identified genotype–phenotype association.
- Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)
used to introduce mutations at specific genomic locations (higher cleavage efficiency and versatility).
- Diallel
a defined set of parental strains is bred in every possible combination of mating pairs.
- F2 cross
parental strains are crossed to obtain an F1 (first filial) generation whose offspring is defined as F2 (second filial) generation.
- Forward genetics
unbiased approaches utilizing natural variation in the mouse species for which no prior knowledge of the causative gene is needed.
- Gene flow
introduction of new genetic material from one population to another population, enhancing the overall genetic diversity.
- Gene of major effect
causal gene of a certain trait (e.g., Mendelian traits, monogenic diseases).
- Genetic blind spots
genome regions with extremely low or no genetic diversity, causing difficulties in studies of natural populations and complex traits.
- Genetic mapping
approach in which molecular markers are used to identify the location of one gene or the distances between genes.
- Genome-wide association study (GWAS)
study that seeks to find significant correlations between genetic variants and particular traits.
- Mouse genetic reference populations
a collection of mice with fixed and known genetic architecture.
- Natural variation
random DNA mutations that are caused by mistakes during replication.
- Quantitative trait locus (QTL)
chromosomal region (locus) of variable size that is associated with a certain phenotype (quantitative trait).
- Recombination breakpoint
Genomic locations where chromosomes break before they reattach.
- Reverse genetics
comprises techniques of for the introduction of wanted mutations into the mouse genome. For this hypothesis-driven approach, prior knowledge about the gene of interest is essential.
- Signature gene
a gene or a set of genes that occurs as a result of a specific biological process.
- Transcription activator-like effector nucleases (TALENs)
Transcription activator-like effector nucleases are used to introduce mutations at specific genomic locations (higher precision).
Footnotes
Supplemental information associated with this article can be found online at https://doi.org/10.1016/j.tig.2018.07.005.
Resources
i www.sanger.ac.uk/sanger/Mouse_SnpViewer/rel-1303
ii http://biogps.org/#goto=welcome
iii www.ncbi.nlm.nih.gov/gene/
vii www.ebi.ac.uk/gwas/(see catalog)
Outstanding Questions.
Is direct translation of identified causal genes into humans possible or will it rather be a molecular pathway that is identified in mice to which the homolog in humans needs to be found?
Will it be possible to find treatment options that will have a major effect on disease progression and outcome in humans for a complex network of causal gene variants?
Can interdisciplinary approaches lead to interdisciplinary candidate genes?
Will technology development catch up with the speed and amount of data collection and reveal findings that are already present but not accessible now?
Will it be possible to find panviral susceptibility genes that can be translated into prophylactic or therapeutic treatment options for humans?
Supplemental Information
The following is Supplemental information to this article:
References
- 1.Gautret P. Emerging viral respiratory tract infections – environmental risk factors and transmission. Lancet Infect. Dis. 2014;14:1113–1122. doi: 10.1016/S1473-3099(14)70831-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss R.A., McMichael A.J. Social and environmental risk factors in the emergence of infectious diseases. Nat. Med. 2004;10(12 Suppl):S70–S76. doi: 10.1038/nm1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenney A.D. Human genetic determinants of viral diseases. Annu. Rev. Genet. 2017;51:241–263. doi: 10.1146/annurev-genet-120116-023425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi W.S. The significance of avian influenza virus mouse-adaptation and its application in characterizing the efficacy of new vaccines and therapeutic agents. Clin. Exp. Vaccine Res. 2017;6:83–94. doi: 10.7774/cevr.2017.6.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sommer D. TALEN-mediated genome engineering to generate targeted mice. Chromosome Res. 2015;23:43–55. doi: 10.1007/s10577-014-9457-1. [DOI] [PubMed] [Google Scholar]
- 6.Harms D.W. Mouse genome editing using the CRISPR/Cas system. Curr. Protoc. Hum. Genet. 2014;83:15.7.1–15.7.27. doi: 10.1002/0471142905.hg1507s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cockrell A.S. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol. 2016;2 doi: 10.1038/nmicrobiol.2016.226. 16226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Totura A.L. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. MBio. 2015;6 doi: 10.1128/mBio.00638-15. e00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoggins J.W. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majzoub J.A., Muglia L.J. Knockout mice. N. Engl. J. Med. 1996;334:904–907. doi: 10.1056/NEJM199604043341407. [DOI] [PubMed] [Google Scholar]
- 11.MacArthur D.G. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Justice M.J. Technical approaches for mouse models of human disease. Dis. Models Mech. 2011;4:305–310. doi: 10.1242/dmm.000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter K.W., Crawford N.P. The future of mouse QTL mapping to diagnose disease in mice in the age of whole-genome association studies. Annu. Rev. Genet. 2008;42:131–141. doi: 10.1146/annurev.genet.42.110807.091659. [DOI] [PubMed] [Google Scholar]
- 14.Davisson M.T. Discovery genetics – the history and future of spontaneous mutation research. Curr. Protoc. Mouse Biol. 2012;2:103–118. doi: 10.1002/9780470942390.mo110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell W.L. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc. Natl. Acad. Sci. U. S. A. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell L.B. Chlorambucil effectively induces deletion mutations in mouse germ cells. Proc. Natl. Acad. Sci. U. S. A. 1989;86:3704–3708. doi: 10.1073/pnas.86.10.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flaherty L. Chlorambucil-induced mutations in mice recovered in homozygotes. Proc. Natl. Acad. Sci. U. S. A. 1992;89:2859–2863. doi: 10.1073/pnas.89.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell W.L. X-ray-induced mutations in mice. Cold Spring Harb. Symp. Quant. Biol. 1951;16:327–336. doi: 10.1101/sqb.1951.016.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Russell L.B., Russell W.L. An analysis of the changing radiation response of the developing mouse embryo. J. Cell. Physiol. Suppl. 1954;43(Suppl. 1):103–149. doi: 10.1002/jcp.1030430407. [DOI] [PubMed] [Google Scholar]
- 20.Takeda J. Germline mutagenesis mediated by Sleeping Beauty transposon system in mice. Genome Biol. 2007;8(Suppl. 1):S14. doi: 10.1186/gb-2007-8-s1-s14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L. PiggyBac transposon-based polyadenylation-signal trap for genome-wide mutagenesis in mice. Sci. Rep. 2016;6 doi: 10.1038/srep27788. 27788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abiola O. The nature and identification of quantitative trait loci: a community’s view. Nat. Rev. Genet. 2003;4:911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts A. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm. Genome. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peirce J.L. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams R.W., Williams E.G. Resources for systems genetics. Methods Mol. Biol. 2017;1488:3–29. doi: 10.1007/978-1-4939-6427-7_1. [DOI] [PubMed] [Google Scholar]
- 26.Ermann J., Glimcher L.H. After GWAS: mice to the rescue? Curr. Opin. Immunol. 2012;24:564–570. doi: 10.1016/j.coi.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valdar W. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- 28.Threadgill D.W. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm. Genome. 2002;13:175–178. doi: 10.1007/s00335-001-4001-Y. [DOI] [PubMed] [Google Scholar]
- 29.Chesler E.J. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm. Genome. 2008;19:382–389. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iraqi F.A. The Collaborative Cross, developing a resource for mammalian systems genetics: a status report of the Wellcome Trust cohort. Mamm. Genome. 2008;19:379–381. doi: 10.1007/s00335-008-9113-1. [DOI] [PubMed] [Google Scholar]
- 31.Morahan G. Establishment of “The Gene Mine”: a resource for rapid identification of complex trait genes. Mamm. Genome. 2008;19:390–393. doi: 10.1007/s00335-008-9134-9. [DOI] [PubMed] [Google Scholar]
- 32.Shorter J.R. Male infertility is responsible for nearly half of the extinction observed in the mouse Collaborative Cross. Genetics. 2017;206:557–572. doi: 10.1534/genetics.116.199596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durrant C. Collaborative Cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 2011;21:1239–1248. doi: 10.1101/gr.118786.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathes W.F. Architecture of energy balance traits in emerging lines of the Collaborative Cross. Am. J. Physiol. Endocrinol. Metab. 2011;300:e1124–e1134. doi: 10.1152/ajpendo.00707.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelada S.N. Genetic analysis of hematological parameters in incipient lines of the Collaborative Cross. G3 (Bethesda) 2012;2:157–165. doi: 10.1534/g3.111.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vered K. Susceptibility to Klebsiella pneumonaie infection in Collaborative Cross mice is a complex trait controlled by at least three loci acting at different time points. BMC Genomics. 2014;15:865. doi: 10.1186/1471-2164-15-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutledge H. Genetic regulation of Zfp30, CXCL1, and neutrophilic inflammation in murine lung. Genetics. 2014;198:735–745. doi: 10.1534/genetics.114.168138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottomly D. Expression quantitative trait loci for extreme host response to influenza A in pre-Collaborative Cross mice. G3 (Bethesda) 2012;2:213–221. doi: 10.1534/g3.111.001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferris M.T. Modeling host genetic regulation of influenza pathogenesis in the Collaborative Cross. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gralinski L.E. Genome wide identification of SARS-CoV susceptibility loci using the Collaborative Cross. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogala A.R. The Collaborative Cross as a resource for modeling human disease: CC011/Unc, a new mouse model for spontaneous colitis. Mamm. Genome. 2014;25:95–108. doi: 10.1007/s00335-013-9499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasmussen A.L. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brinkmeyer-Langford C.L. Host genetic background influences diverse neurological responses to viral infection in mice. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-12477-2. 12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham J.B. A mouse model of chronic West Nile virus disease. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svenson K.L. High-resolution genetic mapping using the mouse Diversity Outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smallwood T.L. High-resolution genetic mapping in the Diversity Outbred mouse population identifies Apobec1 as a candidate gene for atherosclerosis. G3 (Bethesda) 2014;4:2353–2363. doi: 10.1534/g3.114.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.French J.E. Diversity Outbred mice identify population-based exposure thresholds and genetic factors that influence benzene-induced genotoxicity. Environ. Health Perspect. 2015;123:237–245. doi: 10.1289/ehp.1408202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Churchill G.A. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 49.Kollmus H. Systems biology and systems genetics – novel innovative approaches to study host-pathogen interactions during influenza infection. Curr. Opin. Virol. 2014;6:47–54. doi: 10.1016/j.coviro.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Mott R. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. U. S. A. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morgan A.P., Welsh C.E. Informatics resources for the Collaborative Cross and related mouse populations. Mamm. Genome. 2015;26:521–539. doi: 10.1007/s00335-015-9581-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gatti D.M. Quantitative trait locus mapping methods for Diversity Outbred mice. G3 (Bethesda) 2014;4:1623–1633. doi: 10.1534/g3.114.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oreper D. Inbred Strain Variant Database (ISVdb): a repository for probabilistically informed sequence differences among the Collaborative Cross strains and their founders. G3 (Bethesda) 2017;7:1623–1630. doi: 10.1534/g3.117.041491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maurizio P.L., Ferris M.T. The Collaborative Cross resource for systems genetics research of infectious diseases. Methods Mol. Biol. 2017;1488:579–596. doi: 10.1007/978-1-4939-6427-7_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leist S.R. Influenza H3N2 infection of the collaborative cross founder strains reveals highly divergent host responses and identifies a unique phenotype in CAST/EiJ mice. BMC Genomics. 2016;17:143. doi: 10.1186/s12864-016-2483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graham J.B. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. MBio. 2015;6:e00493–e00515. doi: 10.1128/mBio.00493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Green R. Oas1b-dependent immune transcriptional profiles of West Nile virus infection in the Collaborative Cross. G3 (Bethesda) 2017;7:1665–1682. doi: 10.1534/g3.117.041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Green R. Identifying protective host gene expression signatures within the spleen during West Nile virus infection in the Collaborative Cross model. Genom. Data. 2016;10:114–117. doi: 10.1016/j.gdata.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Josset L. Annotation of long non-coding RNAs expressed in Collaborative Cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol. 2014;11:875–890. doi: 10.4161/rna.29442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maurizio P.L. Bayesian diallel analysis reveals Mx1-dependent and Mx1-independent effects on response to influenza A virus in mice. G3 (Bethesda) 2017;8:427–445. doi: 10.1534/g3.117.300438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gralinski L.E. Allelic variation in the Toll-like receptor adaptor protein Ticam2 contributes to SARS-coronavirus pathogenesis in mice. G3 (Bethesda) 2017;7:1653–1663. doi: 10.1534/g3.117.041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Threadgill D.W. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 63.Sittig L.J. Genetic background limits generalizability of genotype-phenotype relationships. Neuron. 2016;91:1253–1259. doi: 10.1016/j.neuron.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seok J. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elbahesh H., Schughart K. Genetically diverse CC-founder mouse strains replicate the human influenza gene expression signature. Sci. Rep. 2016;6:26437. doi: 10.1038/srep26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gusareva E.S., Van Steen K. Practical aspects of genome-wide association interaction analysis. Hum. Genet. 2014;133:1343–1358. doi: 10.1007/s00439-014-1480-y. [DOI] [PubMed] [Google Scholar]
- 67.Cantor R.M. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am. J. Hum. Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hayes B. Overview of statistical methods for genome-wide association studies (GWAS) Methods Mol. Biol. 2013;1019:149–169. doi: 10.1007/978-1-62703-447-0_6. [DOI] [PubMed] [Google Scholar]
- 69.Little J. STrengthening the REporting of Genetic Association studies (STREGA) – an extension of the STROBE statement. Eur. J. Clin. Invest. 2009;39:247–266. doi: 10.1111/j.1365-2362.2009.02125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Phillippi J. Using the emerging Collaborative Cross to probe the immune system. Genes Immun. 2014;15:38–46. doi: 10.1038/gene.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bjorkegren J.L.M. Genome-wide significant loci: how important are they? Systems genetics to understand heritability of coronary artery disease and other common complex disorders. J. Am. Coll. Cardiol. 2015;65:830–845. doi: 10.1016/j.jacc.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Srivastava A. Genomes of the mouse Collaborative Cross. Genetics. 2017;206:537–556. doi: 10.1534/genetics.116.198838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khor C.C. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat. Genet. 2011;43:1139–1141. doi: 10.1038/ng.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rubicz R. A genome-wide integrative genomic study localizes genetic factors influencing antibodies against Epstein–Barr virus nuclear antigen 1 (EBNA-1) PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kamatani Y. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat. Genet. 2009;41:591–595. doi: 10.1038/ng.348. [DOI] [PubMed] [Google Scholar]
- 76.Li Y. Genome-wide association study identifies 8p21.3 associated with persistent hepatitis B virus infection among Chinese. Nat. Commun. 2016;7 doi: 10.1038/ncomms11664. 11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rauch A. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. 1345.e1–e7. [DOI] [PubMed] [Google Scholar]
- 78.Matsuura K. Genome-wide association study identifies TLL1 variant associated with development of hepatocellular carcinoma after eradication of hepatitis C virus infection. Gastroenterology. 2017;152:1383–1394. doi: 10.1053/j.gastro.2017.01.041. [DOI] [PubMed] [Google Scholar]
- 79.Miki D. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat. Genet. 2011;43:797–800. doi: 10.1038/ng.876. [DOI] [PubMed] [Google Scholar]
- 80.Fellay J. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.International HIV Controllers Study The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Everitt A.R. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Antonopoulou A. Role of tumor necrosis factor gene single nucleotide polymorphisms in the natural course of 2009 influenza A H1N1 virus infection. Int. J. Infect. Dis. 2012;16:e204–e208. doi: 10.1016/j.ijid.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 84.Lindesmith L. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 85.Lahti M. Surfactant protein D gene polymorphism associated with severe respiratory syncytial virus infection. Pediatr. Res. 2016;51:696–699. doi: 10.1203/00006450-200206000-00006. [DOI] [PubMed] [Google Scholar]
- 86.Lofgren J. Association between surfactant protein A gene locus and severe respiratory syncytial virus infection in infants. J. Infect. Dis. 2002;185:283–289. doi: 10.1086/338473. [DOI] [PubMed] [Google Scholar]
- 87.Glass W.G. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J. Exp. Med. 2006;203:35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Graham J.B. Extensive homeostatic T cell phenotypic variation within the Collaborative Cross. Cell Rep. 2016;21:2313–2325. doi: 10.1016/j.celrep.2017.10.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.