Highlights
-
•
Nsp16 down-regulates the activities of RIG-I and MDA5 mediated IFN-β and ISRE.
-
•
PEDV nsp10 enhances the inhibitory effect of nsp16 on IFN-β production.
-
•
PEDV nsp16 promotes viral proliferation.
Keywords: PEDV, nsp16, 2'O-MTase, IFN-β, IFIT
Abstract
Type-I IFNs (IFN-I) provide a key mediator of innate antiviral response during virus proliferation. Porcine epidemic diarrhea virus (PEDV), which causes diarrhea in swine of all ages, is a worldwide-distributed alphacoronavirus with economic importance. Here, we screened PEDV RNA modification enzymes involved in regulating antiviral response. Whereas the PEDV nsp13 barely regulates type I IFN, inflammatory cytokines (IL-6, TNF-a) and MHCII, nsp16 and nsp14 (to a lesser extent) down-regulate these antiviral effectors. Importantly, we found nsp16 KDKE tetrad appears to play a key role in interferon inhibition by mutating the D129 catalytic residue. Mechanistically, nsp16 down-regulates the activities of RIG-I and MDA5 mediated IFN-β and ISRE. In turn, the mRNA levels of IFIT family members (IFIT1, IFIT2, IFIT3) was inhibited in cells overexpressing nsp16. In addition, nsp10 enhanced the inhibitory effect of nsp16 on IFN-β. Altogether these results indicate PEDV nsp16 negatively regulates innate immunity to promote viral proliferation. Findings from this study provides novel perspective to advance the understanding in the pathogenesis of PEDV.
1. Introduction
Porcine epidemic diarrhea (PED) is a highly contagious disease of pigs characterized by diarrhea, vomiting, and dehydration in swine of all ages, causing considerable economic losses worldwide each year (Song and Park, 2012; Rui-Qin et al., 2012; Pensaert and De Bouck, 1978). The causative agent of the disease, PED virus (PEDV), is an enveloped, single-stranded positive-sense RNA virus that belongs to a porcine enteropathogenic alphacoronavirus (Kocherhans et al., 2001). The PEDV genome is 28,000 nucleotides (nt) in length, containing seven known open reading frames (ORFs) (Pensaert and Bouck, 1978). The genomic organization is a typical coronavirus with the characteristic gene order. PEDV genome has a 5′ cap and a poly (A) tail, encoding 17 non-structural proteins (nsp1-nsp16, and ORF3) and four structural protein: spike (S), envelope (E), membrane (M) and nucleocapsid (N).
The host innate immune response, predominantly type I interferon (IFN-α and IFN-β), is the first line of defense against pathogens (Tanji and Ip, 2005; Bourgeois et al., 2011). As multi-functional antiviral cytokines, type I interferons can be induced by virus infection, such as Sendai virus (SeV) (Schoggins et al., 2010). Viral RNA is recognized as pathogen-associated molecular pattern (PAMP) by cytoplasmic sensors retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 protein (MDA5) during viral infection (Meylan et al., 2006). Next, RIG-I/MDA-5 binds to the mitochondrial adapter protein MAVS/IPS-1, recruiting TNF receptor-associated factor 3/6 (TRAF3 and TRAF6) (Bradley and Pober, 2001). TRAF3 activates IκB kinase-ε (IKKε) inhibitor and downstream signaling of TANK-binding kinase 1 (TBK1). The transcription factor NF-κB and IRF3 are activated through different mechanisms. Activated IRF-3 and NF-κB enter the nucleus and bind to the IFN-β promoter, thereby activating transcription of IFN-β (Thanos and Maniatis, 1995).
The expression of type I IFN is low in PEDV-infected cells, and PEDV also strongly inhibited RIG-I and poly I:C-mediated IFN-β production (Cao et al., 2015). Accumulating evidence suggests that PEDV has developed a variety of strategies to evade the antiviral activities of IFN. Three PEDV nsps (nsp1, nsp3 and nsp5) have been identified as IFN-β antagonists (Zhang et al., 2016; Jaru-Ampornpan et al., 2016; Wang et al., 2015). PEDV nsp1 mainly disrupts the enhanceosome assembly of IRF3 and CREB-binding protein (CBP), resulting in degradation of CBP in the nucleus (Zhang et al., 2016). PEDV nsp3 encodes papain-like protease 2 (PLP2) with deubiquitinating enzyme activity, which negatively regulates IFN-β expression by removing ubiquitin chains from RIG-I (Jaru-Ampornpan et al., 2016). PEDV nsp5 encoding a 3C-like protease specifically targets NEMO glutamate 231 (Q231) to cleave NEMO residues (Wang et al., 2015). However, the effects and mechanisms of other PEDV non-structural proteins on type I interferons are still being studied extensively.
Coronaviruses are important pathogens causing severe disease in humans and animals. The pathogenesis of these viruses might be related to the inefficient detection by the first line of antiviral response mediated by interferon (Rose et al., 2010; Devaraj et al., 2007; Li et al., 2010; Zhao et al., 2011). In order to evade recognition by the host viral RNA sensor RIG-I or MDA5, some virus encoded several methyltransferases involved in viral RNA capping to carry N7-methylation and 2′O-methylation, similar to the host cell mRNA (Züst et al., 2011a; Daffis et al., 2010; Furuichi and Shatkin, 2000). For example, CoV nsp14 has been reported as a bifunctional enzyme with 3′ -5′ exoribonuclease (ExoN) and N7-MTase activities, and its N7-MTase activity is necessary to viral mRNA cap synthesis and prevents the recognition of viral mRNAs as "non-self" by the host cell (Becares et al., 2016). Furthermore, ribose 2′O-methylation in the cap structure of viral RNA contributes to escape from innate immune recognition. By biochemical assay, it was found that SARS-CoV nsp16 requires nsp10 as a stimulatory factor to execute its 2′O-methyltransferase activity (Chen et al., 2011a). However, the biological activity of PEDV-encoded methyltransferase in cells and their roles in innate immunity are not yet understood.
In this work, we have studied three RNA modification enzymes involved in the formation of cap structures in PEDV: helicase/RNA triphosphatase nsp13, N7 methyltransferase (N7-MTase) nsp14 and 2′O-methyltransferase (2′O-MTase) nsp16 (Ivanov et al., 2004; Chen et al., 2009, 2011b). Our results indicate that nsp14 and nsp16, which are methyltransferase in PEDV, are antagonists of innate immunity, and nsp16 is a more efficient regulator of immune-related genes. In addition, PEDV nsp16 is dependent on the KDKE tetrad to effectively reduce PEDV-induced IFN-β production and promote virus proliferation. Interestingly, the phenomenon not only exists in the coronavirus, but also other virus, such as PRRSV, VSV. It’s speculated that nsp16 is already partially active in absence of nsp10, or other cellular proteins replace the stimulatory factor role of nsp10. PEDV nsp16 appears to be a broad-spectrum anti-innate immune enzyme. Our study provides a novel mechanism by which PEDV restricts interferon production and facilitate virus proliferation, providing a new means for prevention and control of PEDV.
2. Materials and methods
2.1. Cells, virus, and antibody
Human embryonic kidney (HEK) 293 T cells and IPEC-J2 (the intestinal porcine epithelial cell line J2) cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% (V/V) fetal bovine serum (FBS, Biological Industries) and 10 μg/ml streptomycin. PRRSV-permissive PAM cell lines CRL2843-CD163 (3D4/21) cells (Sinha et al., 2012) were cultured in RPMI-1640 medium (Gibco, USA) supplemented with 10% (V/V) FBS plus 100 U/ml penicillin. All cells were cultured in a humidified incubator with 37℃, 5% CO2. The PEDV strain CV777 genotype 1, Sendai virus (SeV) and PRRSV-JXwn06 strain were storage in our laboratory.
Labeled antibodies used in the experiments were purchased from Cell Signaling Technology (CST, Danvers, MA, USA) and Applied Biological Materials Inc (ABM, Vancouver, Canada). A secondary antibody was purchased from Invitrogen (Thermo Fisher Scientific, Waltham, MA, USA). Antibodies to β-actin were purchased from TransGen (Beijing, China).
2.2. Plasmid construction
Total RNAs were extracted from IPEC-J2 cells infected PEDV using TRIzol reagent (TaKaRa, China). First-strand cDNA synthesis was carried out using reverse transcriptase (TaKaRa). Nsp10, nsp13, nsp14 and nsp 16 was synthesized using the specific primers as shown in Table 1 from PEDV strain CV777 genotype 1, and the amplified fragments were cloned into pGEM®-T Easy Vector (Transgen, Beijing).
Table 1.
Primers used for PCR amplification.
| Plasmid name | Primer name | Genbank Number | Sequence of primer(5’-3’) |
|---|---|---|---|
| T-nsp10 | T-nsp10-F | KT323979.1 | ATGGCTGGTAAACAAACA |
| T-nsp10-R | CTACTATTGCATAATGGAC | ||
| T-nsp13 | T-nsp13-F | KT323979.1 | ATGTCTGCAGGGCTTTGTG |
| T-nsp13-R | CTGCAAATCAGACAATTTA | ||
| T-nsp14 | T-nsp14-F | KT323979.1 | ATGGCTAATGAGGGTTGTGG |
| T-nsp14-R | TTGCAAATTGTTACTAAATG | ||
| T-nsp16 | T-nsp16-F | KT323979.1 | ATGGCCAGTGAATGGAAG |
| T-nsp16-R | TCATTTGTTTACGTTGACCAA | ||
| pCMV-nsp10 | pCMV-nsp10-F | KT323979.1 | CCAGTCGACTCTAGAGGATCCATGGCTGGTAAACAAACA |
| pCMV-nsp10-R | CAGGGATGCCACCCGGGATCCCTACTATTGCATAATGGAC | ||
| pCMV-nsp13 | pCMV-nsp13-F | KT323979.1 | CCAGTCGACTCTAGAGGATCCATGTCTGCAGGGCTTTGTG |
| pCMV-nsp13-R | CAGGGATGCCACCCGGGATCCTAACTGCAAATCAGACAATTTA | ||
| pCMV-nsp14 | pCMV-nsp14-F | KT323979.1 | CCAGTCGACTCTAGAGGATCCATGGCTAATGAGGGTTGTGG |
| pCMV-nsp14-R | CAGGGATGCCACCCGGGATCCTAATTGCAAATTGTTACTAAATG | ||
| pCMV-nsp16 | pCMV-nsp16-F | KT323979.1 | CCAGTCGACTCTAGAGGATCCATGGCCAGTGAATGGAAGTGTG |
| pCMV-nsp16-R | CAGGGATGCCACCCGGGATCCTCATTTGTTTACGTTGACCAAATG |
The specific primers pairs (Table 1), harboring common sequence with the vector, were used to amplify the related cDNA encoding nsps gene from clone plasmid, then ligated with pFLAG-CMV2 or pMYC-CMV2 vector by using a one-step clone kit (Vazyme, Nanjing, China). Flag-nsp16 (D129A) expression plasmid was generated by the Fast Mutagenesis System Kit (TransGen Biotech, China).
2.3. Quantitative real-time reverse transcription (RT)-PCR
Treated or untreated cells are collected at the indicated times and total RNAs were extracted using TRIzol reagent (TaKaRa, China). First-strand cDNA was synthesized from purified RNAs of IPEC-J2 cells or HEK293 T cells using a First-Strand Synthesis System (Transgen, Beijing, China) according to the manufacturer’s instructions. The comparatively quantification of gene expression level was analyzed by qRT-PCR that was performed on an ABI 7500 Real-time PCR system (Applied Biosystems, Foster City, CA, USA). The comparative cycle threshold (CT) method was used to calculate the relative gene expression levels according to manufacturer’s protocol (Applied Biosystem). The relative transcript levels of target gene are equal to 2−ΔΔCt threshold method. Data showed the fold increase of mRNA levels. Data are expressed as mean ± SEM representative of three independent experiments. All data presented was relatively quantitative, based on the mRNA level of the endogenous gene β-actin and analyzed using GraphPad Prism 6.0 software. All primer pairs for quantitative real-time PCR are listed in Table 2 .
Table 2.
Primers used for qRT-PCR amplification.
| Primer name | Genbank Number | Sequence of primer(5’-3’) |
|---|---|---|
| PRRSV-N-F | KX286735.1 | GCCTCGTGTTGGGTGGCAGA |
| PRRSV-N-R | CACGGTCGCCCTAATTGAATAGG | |
| PEDV-N-F | JX406145.1 | AACAGCTTCCCAGCGTAGTTGA |
| PEDV-N-R | GAAGTGGCTCTGGATTTGTTCTTC | |
| TNF-α-F | X57321 | GAGATCAACCTGCCCGACT |
| TNF-α-R | CTTTCTAAACCAGAAGGACGTG | |
| IFN-β-F | NM_001003923 | GCAGTATTGATTATCCACGAGA |
| IFN-β-R | TCTGCCCATCAAGTTCCAC | |
| NF-κB-F | X61498.1 | CCCAGCCATTTGCACACCTCAC |
| NF-κB-R | TTCAGAATTGCCCGACCAGTTTTT | |
| IL-6-F | AF518322.1 | AGACAAAATGCTCTTCACCTCTC |
| IL-6-R | CTGGAGGTAGTCCAGGTATATCT | |
| MHC-I-F | AJ131112.2 | CTGGCCCTGACCGGGACT |
| MHC-I-R | ACAGGCCCTGCAGGTAGCTC | |
| MHC-II-F | EU918427.1 | AGGTGCCCGTGGAGCCTG |
| MHC-II-R | ACCTCCGCCCGCTTCTGC | |
| β-actin-F | DQ452569.1 | GAATCCTGCGGCATCCACGA |
| β-actin-R | CTCGTCGTACTCCTGCTTGCT | |
| IFIT1-F | HQ679904.1 | TACATTTCCACTATGGCCGAT |
| IFIT1-R | GGCCTGCTCATAATACTCCA | |
| IFIT2-F | JX070559.1 | AGGAACTAATAGGACACGCTCT |
| IFIT2-R | GATGGCCTTTTCTTCGCACT | |
| IFIT3-F | HQ200930.1 | GCAGCCAAATTTTACCGAGT |
| IFIT3-R | AGTCCTTGGCATATTTCCGTA |
2.4. Immunofluorescence
The procedure for confocal microscopy has been described previously (Su et al., 2018). IPEC-J2 cells were seeded on 12-well plates until the cell density was about 60%˜70%. The cells were transfected with vector or Flag-nsp16 plasmid. At 24 h post-transfection, the cell were infected 2 MOI PEDV. After 24 h, the cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with PBS containing 0.3% Triton X-100 for 10 min at room temperature. After this step, they were blocked for 30 min with 1% bovine serum albumin (BSA) and incubated with anti-PEDV-N mAb at room temperature (RT) in a humid chamber followed by 10 min washing in PBS. Following this, secondary antibody FITC-conjugated anti-mouse IgG (diluted at 1: 200) were used and nuclear DNA was stained with 4′, 6-diamidino-2-phenylindole (DAPI). The PEDV N protein expression was observed with an Olympus confocal microscope. Images were taken at ×100 magnification.
2.5. Luciferase assay
The HEK293 T cells were seeded in 24-well plates and transfected with the Flag-nsp16 or vector plasmids, along with a luciferase reporter (IFN-β-Luc or ISRE-Luc) and the internal control plasmid LacZ. At 12 h post-transfection, the cells were infected with Sendai virus (SeV) or PEDV. The lysed samples were prepared, and the luciferase activity was measured using the multimode microplate reader (Promega) according to the manufacturer’s recommendations. In Fig. 3, Fig. 4, the HEK293 T cells were seeded in 24-well plates and transfected with the Flag-nsp16, Myc-nsp10, IFN-β-luc reporter, the LacZ and poly I:C or Flag-RIG-I(N) or Flag-MDA5(N) (Zhang et al., 2017) for 24 h and then the cells were lysed for luciferase assays and western blotting.
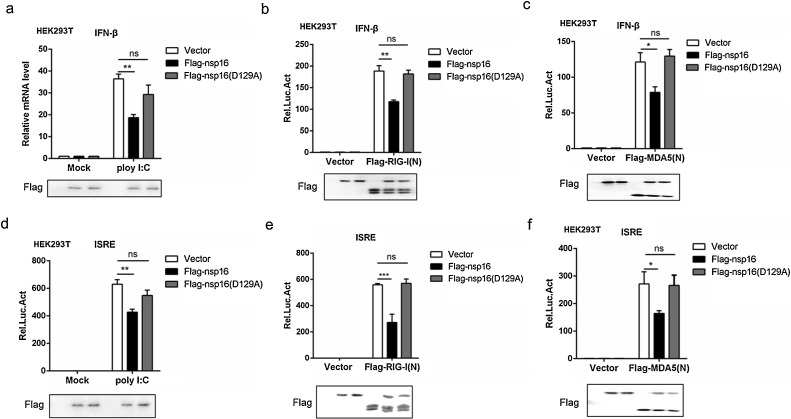
Fig. 3.
Nsp16 plays a negative role in RLR-mediated signal pathway regulation.
(a–c) The Flag-nsp16 or Flag-nsp16 (D129A) or empty vector plasmids together with IFN-β-luc reporter (50 ng), the LacZ (50 ng) and poly I:C (a) Flag-tagged vector or Flag-tagged RIG-I(N) (b) or Flag-tagged MDA5(N) (c) were co-transfected in HEK293 T cells. IFN-β activation was analyzed by the luciferase reporter system. (d–f) ISRE luciferase activity was analyzed in HEK293 T cells co-transfected with Flag-nsp16 or Flag-nsp16 (D129A) or empty vector plasmids together with ISRE-luc reporter (50 ng), the LacZ (50 ng) together with poly I:C (d) Flag-tagged vector or Flag-tagged RIG-I(N) (e) or Flag-tagged MDA5(N) (f).
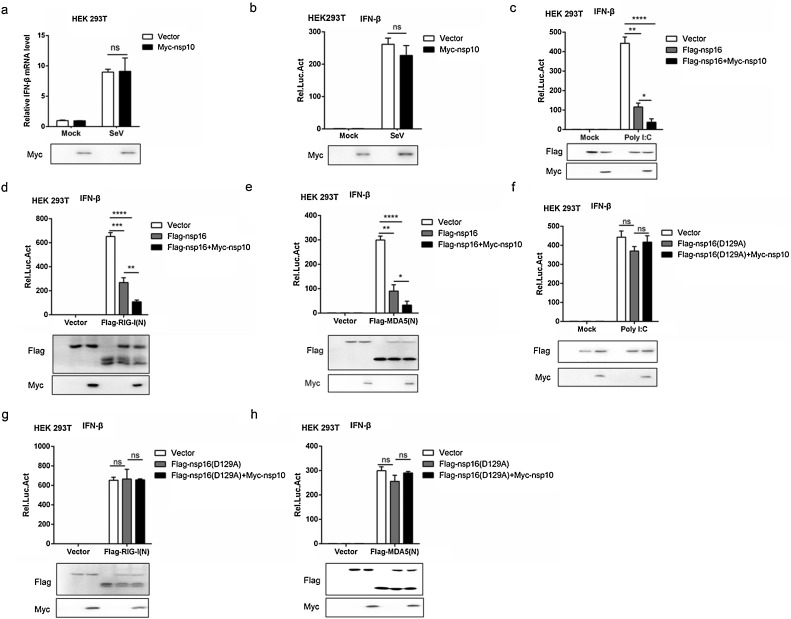
Fig. 4.
Nsp10 enhances the inhibitory effect of nsp16 on type I interferon.
(a) HEK293 T cells were transfected with Myc-nsp10 or Myc-tagged vector. At 24 h post-transfection, the cells were infected with or without 0.1 MOI SeV for 12 h, then IFN-β mRNA levels were analyzed by qRT-PCR. (b) HEK293 T cells were transfected with Myc-nsp10 or Myc-tagged vector together with IFN-β-luc reporter (50 ng), the LacZ (50 ng) and then infected with SeV (MOI = 0.1). After 12 h stimulation, the IFN-β luciferase activity was subsequently analyzed. (c–e) HEK293 T were transfected with Flag-nsp16(WT) or Flag-tagged vector and Myc-nsp10 or Myc-tagged vector together with IFN-β-luc reporter, the LacZ and poly I:C (c) or Flag-RIG-I(N) (d) or Flag-MDA5(N) (e) for 24 h and then the cells were lysed for luciferase assays. (f–h) HEK293 T were transfected with Flag-nsp16(D129A) or Flag-tagged vector and Myc-nsp10 or Myc-tagged vector together with IFN-β-luc reporter, the LacZ and poly I:C (f) or Flag-RIG-I(N) (g) or Flag-MDA5(N) (h) for 24 h and then the cells were lysed for luciferase assays.
2.6. Western blot analyses
Transfected or virus-infected cells were washed twice with cold PBS and lyzed in RIPA buffer (Solarbio, Beijing, China) containing the proteinase inhibitors 20 nM phenylmethanesulfonyl fluoride [PMSF]. Cell lysates were heated in buffer for 10 min and separated with 12% SDS-PAGE. The separated proteins were transferred to the methanol-activated PVDF membrane (Millipore). Membranes were blocked with 5% nonfat dry milk in TBST (0.05% Tween-20) for 1 h and incubated with an antibody against PEDV N (1:2000), PRRSV N (1:5000), β-actin (1:5000) or labeled antibodies (1:5000) for overnight at 4℃, followed by washing and incubation with HRP-conjugated antibody for 1 h at room temperature. Immunodetection was completed using Pierce ECL Western Blotting Substrate (Thermo Scientific).
2.7. Flow cytometry analysis
IPEC-J2 cells were transfected with the Flag-nsp16 or vector plasmid. At 24 h post-transfection, the cells were either mock-infected or infected with 2 MOI PEDV. After 24 h, the cells were harvested and washed for three times with PBS and then treated with 0.25% trypsin at 37 °C for 5 min. Cells were stained with anti-PEDV N antibody, and incubated with goat anti-mouse IgG FITC conjugate (1:200) for 30 min in dark. Fluorescence-activated cell sorting was performed on a FACS LSR II (BD Biosciences, San Jose, CA, USA). A total of 1 × 105 cells was analyzed per run.
2.8. VSV-GFP interferon bioassay
The IPEC-J2 cells were seeded into 6-well plates and transfected with 2 μg Flag-nsp16 or Flag-nsp16 (D129A) plasmid. At 24 h post-transfection, IPEC-J2 cells were infected with 0.1 MOI VSV-GFP virus. After 12 h, the VSV-GFP cells was observed by inverted microscope.
2.9. TCID50 assay
The virus titer was determined using the TCID50 method. IPEC-J2 or 3D4/21 cells were seeded in 96-well plates at a ratio of 1 × 104 cells/well. 100 μl of serially diluted (10−2 to 10-6) viral suspension in 2% DMEM were inoculated in triplicated onto IPEC-J2 or 3D4/21 monolayer cells, and incubated for 1 h at 37 ℃. The normal cells served as mock control. Incubate for 24 h at 37 °C and calculate TCID50 according to the Reed-Muench formula.
2.10. Statistical analyses
Data were subjected to one-way analysis of variance (one-way ANOVA) and expressed as mean ± SEM. Pairwise multiple comparison was conducted to determine which group differed by two-way ANOVA followed by Bonferroni post-tests using Prism 6.0 (GraphPad Software Inc.). P values are indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
3. Results
3.1. PEDV nsp16 is an antagonist of interferon
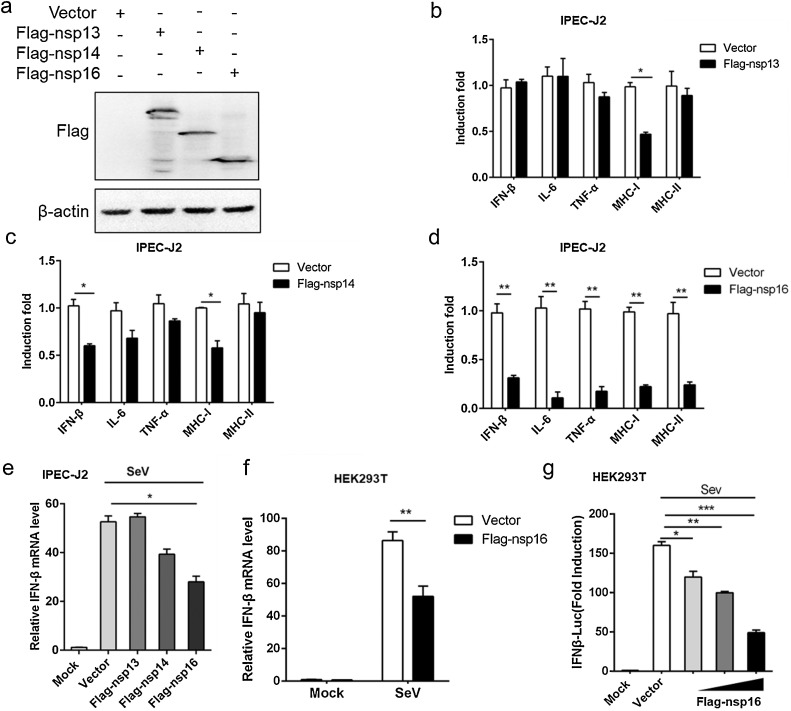
Type I IFN, inflammatory cytokines (IL-6, TNF-α) and major histocompatibility complex class (MHCI, MHCII) play an important role in antiviral defense by directly inducing antiviral effector molecules or indirectly stimulating cell recruitment (Jablons et al., 1989; Beg and Baltimore, 1996; Podlech et al., 1998). To explore whether three RNA modification enzymes (nsp13, nsp14 and nsp16) was involved in the regulation of innate immunity. Eukaryotic expression vectors for three RNA modification enzymes were successfully constructed and expressed (Fig. 1 a). Next, Flag-nsp13, Flag-nsp14, Flag-nsp16 expression plasmids or empty vector were transfected into IPEC-J2 cells, respectively, and then infected with 0.5 M PEDV for 12 h. The results showed that the expression of MHCI mRNA was significantly decreased when Flag-nsp13 was overexpressed in PEDV-infected cells, while nsp13 had no regulatory effect on other immune molecules compared with the control group (Fig. 1b). Meanwhile, experimental data indicated that Flag-nsp14 or Flag-nsp16 expression inhibited PEDV-induced IFN-β production, but nsp16 was more effective than others in inhibiting innate immune-related genes (Fig. 1c, d). These data suggested that nsp16 appears to play a more important role in innate immunity control than nsp14. Later, we investigated the possible involvement of nsp16 in SeV-triggered IFN-β signaling pathways processes. The results showed that nsp16 significantly inhibited the production of IFN-β compared to other non-structural proteins or vector in IPEC-J2 cells (Fig. 1e). The similar result was also observed in 293 T cells (Fig. 1f). Consistently, the luciferase reporter system further confirmed that nsp16 inhibited IFN production in a dose-dependent manner (Fig. 1g). Taken together, these data demonstrated that PEDV nsp16 negatively regulates IFN-β production and antiviral immune responses.
Fig. 1.
PEDV nsp16 is an antagonist of interferon.
(a) IPEC-J2 cells were transfected with pFlag-CMV2 (empty vector) or pFlag-CMV2-nsp13 (Flag-nsp13), pFlag-CMV2-nsp14 (Flag-nsp14), pFlag-CMV2-nsp16 (Flag-nsp16) plasmid respectively, the plasmid expression was detected by western blotting. (b–d) IPEC-J2 cells were transfected with empty vector or Flag-nsp13 (b), Flag-nsp14 (c), Flag-nsp16 (d) plasmid respectively. At 12 h post-transfection, cells were infected with 0.5 MOI PEDV for 12 h. The levels of IFN-β, IL-6, TNF-α, MHCI and MHCII mRNA were analyzed by qRT-PCR. (e) IPEC-J2 cells were transfected with empty vector, Flag-nsp13, Flag-nsp14, Flag-nsp16 plasmid respectively and stimulated with 0.1 MOI SeV or PBS (mock) for 12 h, IFN-β mRNA levels were analyzed by qRT-PCR. (f) HEK293 T cells were transfected with Flag-nsp16 and stimulated with 0.1 MOI SeV for 12 h, IFN-β mRNA levels were analyzed by qRT-PCR. (g) Increasing amounts (100 ng, 300 ng, 500 ng) of Flag-nsp16 or control plasmid together with IFN-β-luc reporter (50 ng), the LacZ (50 ng) were co-transfected in HEK293 T cells. At 12 h post-transfection, cells were infected with SeV (MOI 0.5) or not. After 12 h, the infected cells were collected and IFN-β activation was analyzed by the luciferase reporter system. *P < 0.05**P < 0.01; ***P < 0.001 (analysis of two-way ANOVA followed by Bonferroni post-test). Data are representative of three independent experiments.
3.2. PEDV nsp16 KDKE tetrad necessary for inhibiting IFN induction
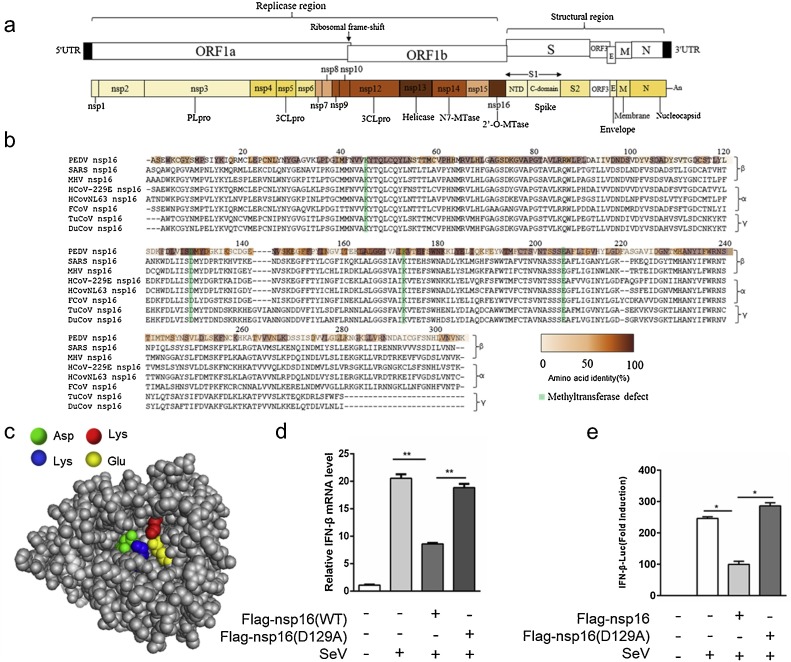
PEDV encoded 16 non-structural proteins, in which nsp16 encoded 2′-O-methyltransferase (Fig. 2 a). It’s known that 2′-O-methyltransferase play a key role in the replication of coronaviruses (Perlman and Netland, 2009; Minskaia et al., 2006a). Next, we attempted to determine the mechanism of PEDV nsp16 inhibiting type I interferon production. Nsp16 is highly conserved in coronavirus. Importantly, the K-d-K-E, an invariant motif within the methyltransferase core required to mediate its activity, is highly conserved among all of the nsp16 sequences detected in the CoV family (Fig. 2b). The crystal structure of SARS coronavirus nsp16 has been resolved (Decroly et al., 2011). PEDV nsp16 3D structure was modelled based on amino acid sequence homology (Fig. 2c). Previous studies found that nsp16 lost the activity of methyltransferase when the tetrad aspartate was mutated to alanine (Feder et al., 2003). PEDV nsp16 mutation (D129A) was unable to limit SeV-induced IFN induction in HEK293 T cells compared to Flag-nsp16 (WT) (Fig. 2d, e). Taken together, these data indicated PEDV nsp16 inhibits PEDV-mediated IFN-β production depending on its KDKE tetrad, indicating that inhibition of type I interferon production by nsp16 is associated with 2′O-MTase enzyme activity. It is worth exploring that PEDV nsp16 also inhibits non-coronavirus mediated IFN-β production in HEK293 T cells and its function depends on KDKE tetrad.
Fig. 2.
The KDKE tetrad is necessary for the ability of PEDV nsp16 to inhibit IFN induction.
(a) Schematic representation of the genomic RNA structure of PEDV. Replicase (ORF1a and ORF1b.), structural regions, and the position of the 5′-capped and 3′ poly(A) tract and the ribosomal frameshift site between ORF1a and ORF1b is shown at the top. Each box represents a protein product, 16 non-structural protein; the envelope proteins, the N protein, and the accessory ORF3 protein; S, spike; E, envelope; M, membrane; N, nucleocapsid; PLpro, papain-like cysteine protease; 3CLpro, the main 3C-like cysteine protease; Hel, helicase; N7-MTase, N7-methyltransferase; 2′-O-MTase, ribose-2′-O-methyltransferase. Colors indicate the level of amino-acid identity with the best-matching protein of other coronaviruses. (b) Sequence comparison of nsp16 and homologs from other viruses. Percent similarity is expressed as a color code from white to brown and shows precise similarity within each element of the matrix. (c) Nsp16 protein 3D structure simulation was performed using Zhang Lab's I-TASSER and PyMOL software. The key amino acid sites that can bind to the SAM substrate are labeled with different colors. (d) The vector, Flag-nsp16 and Flag-nsp16 (D129A) plasmids was transfected in IPEC-J2 cells respectively and then inoculated without or with 0.1 MOI SeV for 12 h, IFN-β mRNA levels were analyzed by qRT-PCR. (e) HEK293 T cells were transfected with Flag-nsp16 or control plasmid together with IFN-β-luc reporter (50 ng), the LacZ (50 ng), and then inoculated without or with 0.1 MOI SeV for 12 h and luciferase activity was subsequently determined. *P < 0.05**P < 0.01 (analysis of two-way ANOVA followed by Bonferroni post-test). Data are representative of three independent experiments.
3.3. PEDV nsp16 plays a negative role in RIG-I-like receptor (RLR) signaling pathway regulation
Virus RNA was always recognized by retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) (Mogensen and Paludan, 2005), and 2′O-methylation in the cap structure of viral RNA play a pivotal role in escaping from innate immune recognition, so we next explore whether nsp16 was involved in the regulation of RLR-mediated IFN-β production. In reporter assays, IFN-β activation was triggered by poly I:C, RIG-I(N) and MDA5(N) respectively (Lin et al., 2016). Compared to the control group, Flag-nsp16 (WT) but not Flag-nsp16 (D129A) reduced the RIG-I ligand poly I:C-induced IFN-β activation (Fig. 3a). Similarly, Flag-nsp16 significantly inhibited RIG-I-, MDA5-induced IFN-β activation, while Flag-nsp16 (D129A) was not (Fig. 3b-c). Consistently, ISRE activation triggered by poly I:C, RIG-I and MDA5 was downregulated by Flag-nsp16 (WT), and Flag-nsp16 (D129A)’s ability to inhibit ISRE activation impaired (Fig. 3d-f). Taken together, these data suggested that nsp16 down regulated RLR-triggered IFN-β signal pathway.
3.4. Nsp10 enhances the inhibitory effect of nsp16 on type I interferon
Previous studies have shown that SARS-CoV nsp16 needs nsp10 as a stimulatory factor to trigger the 2′O-MTase activity (Micka et al., 2015). Next, we examined the effect of PEDV nsp10 expression on IFN-β production. Results demonstrated that nsp10 has no effect on SeV-stimulated IFN β mRNA level in HEK 293 T cells (Fig. 4a). Consistently, the luciferase reporter system further confirmed that nsp10 was not an antagonist of interferon (Fig. 4b). Then, the collective effect of PEDV nsp10 and nsp16 on interferon-mediated innate immunity was examined. Nsp10 significantly enhanced nsp16 inhibition of IFN-β production in poly I:C-, RIG-I- and MDA5-stimulated interferon activation (Fig. 4c-e). However, nsp10 did not alter the role of nsp16 (D129A) in poly I:C-, RIG-I- and MDA5-stimulated interferon activation (Fig. 4f-h). Combined with Fig. 2, PEDV nsp16 relies on the conserved KDKE motif to inhibit coronavirus-induced or non-coronavirus-induced IFNβ production and nsp10 enhances the inhibitory effect of nsp16. So it is hypothesized that nsp16 can exert partially 2′O-MTase activity independently in cells, or that there’re unknown cellular proteins replacing nsp10 to activate nsp16 2′O-MTase activity.
3.5. Nsp16 impacts on PEDV proliferation
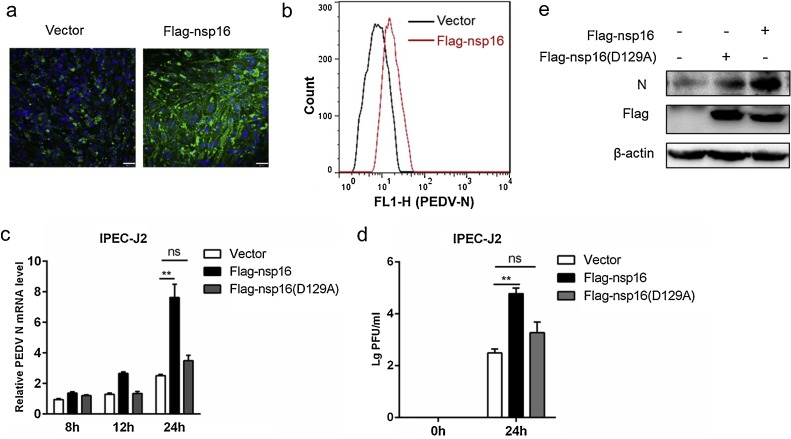
There was growing evidence showed that IFNβ limited viral replication (Taylor and Bresnahan, 2005; Cakebread et al., 2011), so the effect of nsp16 on PEDV proliferation was then investigated. Flag-nsp16 or vector plasmid was transfected into IPEC-J2 cells and inoculated with PEDV for 24 h, and PEDV N protein was detected by immunofluorescence analysis. The result showed nsp16 enhanced PEDV proliferation (Fig. 5 a). In line with that, flow cytometry analysis further confirmed the result (Fig. 5b). Similarly, the overexpression of nsp16 corresponded to an increase in PEDV N gene mRNA whereas Flag-nsp16 (D129A) had no significant effect (Fig. 5c). Meanwhile, the virus titer was significantly higher in Flag-nsp16 plasmid transfected cells compared to the control (Fig. 5d). Western blotting analysis also confirmed nsp16 positively influenced virus replication (Fig.5e). These data collectively indicated that overexpression of nsp16 promotes PEDV proliferation and the effect is related to its methyltransferase activity to a certain extent.
Fig. 5.
Nsp16 can promote PEDV replication.
(a–b) The Flag-nsp16 or Flag-tagged vector plasmids were transfected into IPEC-J2 respectively and then infected with 2 MOI PEDV. At 24 h post-infection, the cells were collected. The PEDV N protein was detected by IFA (a). Analysis of PEDV levels by flow cytometry detection of PEDV N (b). (c) IPEC-J2 transfected with vector or Flag-nsp16 or Flag-nsp16 (D129A) plasmids respectively. At 24 h post-transfection, the cells were either mock-infected or infected with 2 MOI PEDV at the indicated times. The mRNA level of PEDV N were tested by qRT-PCR (c) and the PEDV load was tested by TCID50 (d), and analysis of the PEDV N protein expression levels by western blotting (e). *P < 0.05**P < 0.01 (analysis of two-way ANOVA followed by Bonferroni post-test). Data are representative of three independent experiments.
3.6. Nsp16 promotes virus replication by suppressing cellular antiviral response
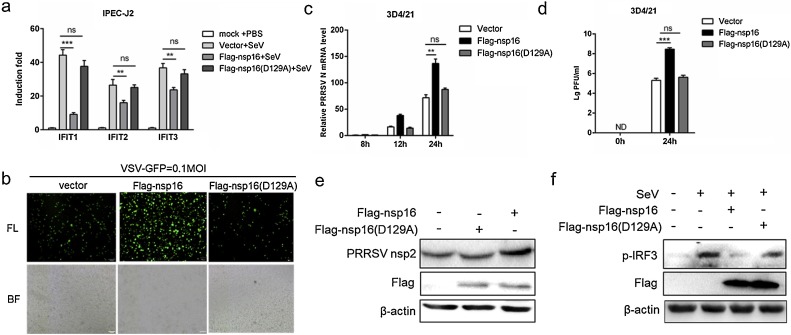
PEDV nsp16 not only inhibits interferon produced by PEDV stimulation, but also non-coronal viruses such as SeV. Next, we attempted to determine if the antiviral effect of nsp16 is extensive. The IFIT (interferon-induced proteins with tetratricopeptide repeats) family is a member of the interferon-stimulated gene that is regulated by viruses or interferons (Fensterl and Sen, 2015), and IFIT proteins has been identified as potent antiviral proteins (Vladimer et al., 2014). Three Sus scrofa IFIT family members (ISG56/IFIT1, ISG54/IFIT2, ISG49/IFIT3) detection primers were designed and the effect of PEDV nsp16 on the IFITs was detected. The results showed that nsp16 overexpression reduced the mRNA level of IFITs, especially IFIT1 (Fig. 6 a). The vesicular stomatitis virus (VSV-GFP) was used to further substantiate the presence of biologically active IFN. Transfection with Flag-nsp16 was beneficial for VSV-GFP infection in IPEC-J2 cells (Fig. 6b). Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) is also a positive-strand RNA virus, which has been found to be frequently mixed with PEDV in recent years. Interestingly, nsp16 also promoted the proliferation of PRRSV in 3D4/21 cells (Fig. 6c-e). Furthermore, the expression of phosphorylated IRF3 in the interferon signaling pathway were also examined by western blotting. The results showed nsp16 inhibited IRF3 activation, while Flag-nsp16 (D129A) was not (Fig.6f). Collectively, these data together reflect that nsp16 promotes virus replication by suppressing cellular antiviral response.
Fig. 6.
Nsp16 promotes virus replication by suppressing cellular antiviral response.
(a) HEK293 T were transfected with Flag-nsp16 or Flag-nsp16 (D129A) plasmid, respectively and then infected with 0.1 MOI SeV for 12 h. Next, cells were harvested, mRNA expression of IFIT, IFIT2, IFIT3 were analyzed by qRT-PCR. (b) HEK293 T were transfected with Flag-nsp16 or Flag-nsp16 (D129A) or Flag-tagged vector and then infected with 0.1 MOI VSV-GFP, fluorescence microscopy imaging examined the proliferation of VSV. (c) 3D4/21 cells transfected with control vector or Flag-nsp16 or Flag-nsp16 (D129A) plasmid, respectively. After 24 h post-transfection, the cells were either mock-infected or infected with PRRSV at an 0.5 MOI at the indicated times. The loads of PRRSV N were tested by qRT-PCR, and PRRSV load was tested by TCID50 (d), and western blotting (e). (f) 3D4/21 cells were transfected with vector or Flag-nsp16 or Flag-nsp16 (D129A) plasmid and then inoculated without or with 0.1 MOI SeV for 12 h, the p-IRF3 protein was detected by western blotting. **P < 0.01 (analysis of two-way ANOVA followed by Bonferroni post-test). Data are representative of three independent experiments.
4. Discussion
Type 1 interferon (IFN-I) and interferon-induced cellular antiviral responses are the first line of defense against pathogen invasion (Rathinam and Fitzgerald, 2011). Studies have found that many viruses including coronaviruses developed a variety of mechanisms to evade innate antiviral immune responses, counteracting the antiviral effects of interferon, thereby establishing a persistent infection. For example, SARS-CoV nsp16 acts as a viral 2′-O-MTase to escape innate immune recognition and facilitate viral replication (Minskaia et al., 2006b; Decroly et al., 2008; Züst et al., 2011b). The latest research found that MERS-CoV nsp16 is essential for IFN resistance and viral pathogenesis (Menachery et al., 2017).
Porcine epidemic diarrhea virus (PEDV) is an important member of the coronavirus. The PEDV mutant strain appeared in China until 2010, and its pathogenicity was significantly enhanced and rapidly spread (Sun et al., 2012). The immunization of sows by passive immunization to protect newborn piglets is currently the main method of preventing PEDV. The existing PEDV commercial vaccine is based primarily on a live attenuated vaccine of the CV777 strain. Although the PEDV vaccine have improved in recent years, its immunoprotective effect is still insufficient (Changhee, 2015). Therefore, it is of great significance to elucidate the molecular pathogenesis of PEDV from the perspective of the interaction between virus and host.
The PEDV genome is 28,000 nucleotides (nt) in length, encoding seven known open reading frames (ORFs) expressed from genomic and subgenomic mRNAs (Duarte et al., 1994). Nsp13, nsp14 and nsp16 are mainly involved in the PEDV capping process and participate in the replication of the viral genome, and their role in the IFN signaling pathway is unclear. Here, we found PEDV nsp16 encoding 2′-O-methyltransferase was an important interferon antagonist, both in PEDV-induced and SeV-induced interferon signaling pathways. PEDV nsp16 inhibited RIG-I- and MDA5-triggered signaling pathways and down-regulated the activity of IFN-β and ISRE promoters depending on the existence of the KDKE motif. Next, we examined the effect of nsp16 on PEDV viral load and found that Flag-nsp16 (WT) but not Flag-nsp16 (D129A) promotes PEDV proliferation. In addition, nsp16 promoted other non-coronavirus proliferation, such as SeV and PRRSV. Furthermore, the expression of nsp16 inhibited IRF3 phosphorylation in cells. The study indicated that PEDV nsp16 appears to be a broad-spectrum anti-innate immune enzyme. The pathogenesis of PEDV might be related to the inefficient detection by the first line of antiviral response mediated by interferon.
Importantly, PEDV nsp10 was not an interferon antagonist, but in the case of co-expression of nsp10 and nsp16, the inhibitory effect on interferon signal transduction was stronger than that of nsp16 alone. Given that CoV nsp10 activates methyltransferase activity of nsp16, the data indicated that inhibition of type I interferon production by nsp16 was associated with its 2′O-MTase enzyme activity. Additionally, PEDV nsp16 relies on the KDKE motif to inhibit virus-induced IFNβ production and improve virus replication. It is hypothesized that nsp16 can exert partially 2′O-MTase activity independently in cells, or that there’re unknown cellular proteins replacing nsp10 to activate nsp16 2′O-MTase activity.
Overall, our study elucidated PEDV nsp16 was an important interferon antagonist and facilitated virus proliferation. However, whether nsp16 has interactions with host partners remains unclear. In addition, the PEDV nsp16 2′O-MTase activity needs to be further demonstrated in vitro and in vivo. The specific mechanism by which nsp16 inhibits the innate immune signaling pathway still needs further exploration.
Funding information
This work was supported by the National Key Research and Development Program of China (2018YFD0500500), and the underprop project of Tianjin Science and Technology in China (16YFZCNC00640). Dr. JH H received the two funding.
Author contributions
Conceived and designed the experiments: JH H. Performed the experiments: PD S, YX S, RQ L, Y L, SR D, and ZX L. Analyzed the data: PD S, and YX S. Contributed reagents/materials: JH H. Wrote the paper: PD S, and JH H.
Conflict of interest
We declare that we have no competing interests.
References
- Becares M., Pascual-Iglesias A., Nogales A., Sola I., Enjuanes L., Zuñiga S.J. Mutagenesis of coronavirus NSP14 reveals its potential role in modulation of the innate immune response. JVI. 2016;90 doi: 10.1128/JVI.03259-15. 03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg A.A., Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- Bourgeois C., Majer O., Frohner I.E., Lesiakmarkowicz I., Hildering K.S., Glaser W., Stockinger S., Decker T., Akira S., Müller M. Conventional dendritic cells mount a type I IFN response against Candida spp. Requiring novel phagosomal TLR7-Mediated IFN-β signaling. J. Immunol. 2011;186:3104–3112. doi: 10.4049/jimmunol.1002599. [DOI] [PubMed] [Google Scholar]
- Bradley J.R., Pober J.S.J.O. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- Cakebread J.A., Xu Y., Grainge C., Kehagia V., Howarth P.H., Holgate S.T., Davies D.E.J., Immunology C. Exogenous IFN-β has antiviral and anti-inflammatory properties in primary bronchial epithelial cells from asthmatic subjects exposed to rhinovirus. J. Allergy Clin. Immunol. 2011;127:1148–1154. doi: 10.1016/j.jaci.2011.01.023. e9. [DOI] [PubMed] [Google Scholar]
- Cao L., Ge X., Gao Y., Herrler G., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-β production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 2015;12:127. doi: 10.1186/s12985-015-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changhee L. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:1–2. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Cai H., Ja Pan, Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., PJPP Tien. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-Methylation by nsp16/nsp10 protein complex. PLoS. 2011;7 doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H.J.N. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2’O)-methyltransferase activity. J. Virol. 2008;82:8071. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.T.K., Baker S.C. Regulation of IRF-3 dependent innate immunity by the PAPAIN-LIKE protease domain of the SARS coronavirus. J. Biol. Chem. 2007;282:32208. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte M., Gelfi J., Lambert P., Rasschaert D., Laude H. Coronaviruses. Springer; 1994. Genome organization of porcine epidemic diarrhoea virus; pp. 55–60. [DOI] [PubMed] [Google Scholar]
- Feder M., Pas J., Wyrwicz L.S., Bujnicki J.M. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2’-O-methyltransferases. Gene. 2003;302:129. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- Fensterl V., Sen G.C. Interferon-induced ifit proteins: their role in viral pathogenesis. J. Virol. 2015;89:2462–2468. doi: 10.1128/JVI.02744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y., Shatkin A.J. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000 doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablons D., Mule J., McIntosh J., Sehgal P., May L., Huang C., Rosenberg S., Lotze M. IL-6/IFN-beta-2 as a circulating hormone. Induction by cytokine administration in humans. J. Immunol. 1989;142:1542–1547. [PubMed] [Google Scholar]
- Jaru-Ampornpan P., Jengarn J., Wanitchang A., Jongkaewwattana A. Porcine epidemic diarrhea virus (PEDV) 3C-like protease-mediated nucleocapsid processing: a possible link to viral cell-culture adaptability. Journal of virology:JVI. 2016 doi: 10.1128/JVI.01660-16. 01660-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23:137. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu Y., Zhang X. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J. Virol. 2010;84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Zhang J., Lin H., Li Z., Sun X., Xin D., Yang M., Sun L., Li L., Wang H.J. Syndecan-4 negatively regulates antiviral signalling by mediating RIG-I deubiquitination via CYLD. Nat. Commun. 2016;7:11848. doi: 10.1038/ncomms11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Mitchell H.D., Dinnon K.H., Leist S.R., Yount B.L., Graham R.L., McAnarney E.T., Stratton K.G., Cockrell A.S. bioRxiv; 2017. MERS-CoV NSP16 Necessary for IFN Resistance and Viral Pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E., Tschopp J., Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Micka L.B., Claire D., Isabelle I., Barbara S., Snijder E.J., Bruno C., Etienne D.J. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS. 2015;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→ 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen T.H., Paludan S.R. Reading the viral signature by Toll-like receptors and other pattern recognition receptors. J. Mol. Med. 2005;83:180–192. doi: 10.1007/s00109-004-0620-6. [DOI] [PubMed] [Google Scholar]
- Pensaert M.B., Bouck P.D. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., De Bouck P. A new coronavirus-like particle associated with diarrhea in swine. J. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlech J., Holtappels R., Wirtz N., Steffens H., Reddehase M. Reconstitution of CD8 T cells is essential for the prevention of multiple-organ cytomegalovirus histopathology after bone marrow transplantation. J. Gen. Virol. 1998;79:2099–2104. doi: 10.1099/0022-1317-79-9-2099. [DOI] [PubMed] [Google Scholar]
- Rathinam V.A., Fitzgerald K.A. Cytosolic surveillance and antiviral immunity. Curr. Opin. Virol. 2011;1:455–462. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose K.M., Elliott R., Martínezsobrido L., Garcíasastre A., Weiss S.R. Murine coronavirus delays expression of a subset of interferon-stimulated genes. J. Virol. 2010;84:5656–5669. doi: 10.1128/JVI.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui-Qin S., Ru-Jian C., Ya-Qiang C., Peng-Shuai L., De-Kun C., Chang-Xu S.J. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Maryline P., Murphy M.Y., Jones C.T., Paul B., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2010;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A., Lin K., Hemann M., Shen H., Beach N.M., Ledesma C., Meng X.J., Wang C., Halbur P.G., Opriessnig T. ORF1 but not ORF2 dependent differences are important for in vitro replication of PCV2 in porcine alveolar macrophages singularly or coinfected with PRRSV. Vet. Microbiol. 2012;158:95. doi: 10.1016/j.vetmic.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Song D., Park B.J. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Shi P., Zhang L., Lu D., Zhao C., Li R., Zhang L., Huang J. The superimposed deubiquitination effect of OTULIN and PRRSV Nsp11 promoted the multiplication of PRRSV. J. Virol.:JVI. 2018 doi: 10.1128/JVI.00175-18. 00175-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R.-Q., Cai R.-J., Chen Y.-Q., Liang P.-S., Chen D.-K., Song C.-X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerging Infect. Dis. 2012;18:161. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji T., Ip Y.T. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 2005;26:193. doi: 10.1016/j.it.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Taylor R.T., Bresnahan W.A.J. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced Beta interferon production. JoV. 2005;79:3873–3877. doi: 10.1128/JVI.79.6.3873-3877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D., Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Vladimer G.I., Górna M.W., Supertifurga G. IFITs: emerging roles as key anti-viral proteins. Front. Immunol. 2014;5:94. doi: 10.3389/fimmu.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S. Porcine epidemic diarrhoea virus 3C-Like protease regulates its interferon antagonism by cleaving NEMO. J. Virol.: JVI. 2015:02514–02515. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.L., Ye H.Q., Liu S.Q., Deng C.L., Li X.D., Shi P.Y., Zhang B.J. West Nile virus NS1 antagonizes interferon-β production by targeting RIG-I and MDA5. JVI. 2017;91 doi: 10.1128/JVI.02396-16. 02396-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Rose K.M., Elliott R., Van R.N., Weiss S.R. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J. Virol. 2011;85:10058–10068. doi: 10.1128/JVI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S. Ribose 2 [prime]-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S.J. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]