Abstract
Ecological niche modeling (ENM) is widely employed in ecology to predict species’ potential geographic distributions in relation to their environmental constraints and is rapidly becoming the gold-standard method for disease risk mapping. However, given the biological complexity of disease systems, the traditional ENM framework requires reevaluation. We provide an overview of the application of ENM to disease systems and propose a theoretical framework based on the biological properties of both hosts and parasites to produce reliable outputs resembling disease system distributions. Additionally, we discuss the differences between biological considerations when implementing ENM for distributional ecology and epidemiology. This new framework will help the field of disease ecology and applications of biogeography in the epidemiology of infectious diseases.
Keywords: ecological niche modeling, host-space, disease biogeography, disease risk, parasites
Highlights
Infectious diseases greatly impact human health, biodiversity, and global economies, highlighting the need to understand and predict their distributions.
Ecological niche modeling (ENM) was not originally designed to explicitly reconstruct complex biological phenomena such as diseases or parasitism, requiring a reevaluation of the traditional framework.
We provide an integrative ENM framework for disease systems that considers suitable host availability, parasite ecologies, and different scales of modeling.
Disease transmission is driven by factors related to parasite availability and host exposure and susceptibility, which can be incorporated in ENM frameworks.
Challenges and Opportunities to Map Disease Risk
The recent rise of emerging infectious diseases (EIDs) (see Glossary) [1] has increased the burden of infectious diseases and negatively impacted the global economy 2, 3, 4, 5. Approximately 60% of emerging human diseases are caused by pathogenic parasites of animal origin (zoonoses), particularly wildlife [6]. As human activities intensify, contact with wildlife and exposure to novel parasites increase, potentially driving zoonotic disease emergence 1, 7. Given the threat that EIDs pose to human populations, understanding the underlying drivers of parasite geographic distribution and their spillover to humans is particularly relevant for epidemiologists, public-health practitioners, and policy makers [9].
Ecological niche modeling (ENM) has proven useful to forecast the distribution of a vast number of organisms 10, 11, 12, 13 and is increasingly employed to predict parasite distributions locally and globally 14, 15, 16, 17. Despite great strides made in the implementation of ENM to forecast complex biological phenomena such as disease systems [18], traditional frameworks may render biologically unrealistic predictions and thus must be revised, as we show in this review. We provide an overview of the current state of disease ENM and propose a framework based on the biological properties of both parasites and hosts to produce reliable outputs resembling disease systems distributions. Specifically, our theoretical framework: (i) addresses the selection of an appropriate modeling approach and highlights the importance of including biologically sound predictor variables; (ii) proposes the concept of a microscale parasitic niche defined by host traits to identify relevant parasite–host associations; and (iii) integrates traditional parasite ENM with the proposed microscale niche to better understand geographic distributions and improve fine-scale predictions of disease transmission risk.
ENM and Biotic Interactions
ENM estimates the distributions of species by linking their geographic occurrence with their environmental constraints, often utilizing correlative approaches (detailed explanations in [18]). A plethora of algorithms is available to perform ENM (e.g., MaxEnt, Regression Trees) and methods for the development of accurate models have been described at length (see 19, 20 for comparisons). Ecological interactions are hypothesized to affect species distributions only locally (i.e., the Eltonian noise hypothesis [10]) and are usually considered irrelevant in traditional coarse-scale ENM applications 10, 21. However, growing evidence suggests that biotic interactions may have a larger role in shaping broad-scale species distributions, especially under changing conditions (e.g., climate change) 10, 22, 23, 24. For example, continental-scale distributions of the North American warbler are better explained when coupling biotic interactions (e.g., vegetation requirements) with abiotic factors (climate data) [23]. Biotic interactors can be included in ENM by incorporating interacting species as predictor variables (preprocessing), restricting the distribution of the focal species to regions where interactions may occur (post-processing), and linking demographic population models to the final ENM 10, 25, 26, 27.
Modeling Disease Systems
The two most common disease distribution modeling methods are black-box and component-based approaches (Table 1 ; [18]). Black-box approaches model the overall geographic distribution of a final manifestation of host–parasite interactions (e.g., disease outbreaks), assuming that this outcome summarizes all biotic interactions involved in transmission 28, 29, 30. This approach provides a pragmatic framework to generalize disease distributions and is useful when transmission dynamics are poorly understood and data are limited, which is frequently the case for EIDs [31]. However, the black-box oversimplification may be perilous as it neglects ecological complexity (e.g., the identity of key host species for transmission). Alternatively, component-based approaches consider the individual ecologies of all species involved in disease transmission (e.g., parasites, hosts) 16, 17, 32. This approach allows the identification of host species and prioritization of areas for disease surveillance and control. Component-based approaches require in-depth knowledge of the disease system (e.g., the identity and ecologies of relevant species, transmission cycle), which may not be readily available, especially for emerging diseases (e.g., Middle East respiratory syndrome) [18]. Obtaining the necessary information on disease systems can be labor intensive, time consuming, and economically unfeasible [18]. Choosing between these two approaches for disease distribution modeling should be done in accordance with the research question, data availability, and implicit assumptions 18, 22.
Table 1.
Applications of ENM Frameworks to Describe and Predict Disease Distributions
| Modeling approach | Description | Advantage | Limitation |
|---|---|---|---|
| Black box | Considers disease system as an epiphenomenon, using disease cases as occurrences to calibrate the model; for examples see 28, 29, 30 | Useful when information on the disease system is limited or for exploratory analysis to identify potential areas for disease surveillance | Ecologically relevant information on disease transmission is limited; location of disease cases may not represent site of infection, limiting transmission risk estimates. Sampling biases amplify inaccuracies in model predictions. |
| Componentbased | Considers key species involved in disease system (e.g., hosts, vectors, parasite); for examples see 16, 17, 32 | Useful in designing evidence-based control strategies and detailed identification of potential transmission areas to allocate resources for surveillance | Deep understanding of the natural history of the disease is necessary (e.g., all hosts are known). Exclusion of key species when modeling a disease system may underestimate risk. Assumes that the parasite is uniformly distributed across the landscape (e.g., a parasite can be found across its host’s geographic range). Data for model calibration may not be available. |
Traditional ENM Framework Limitations for Disease Systems
Data Quality and Availability
A major challenge of the application of ENM to disease systems is the lack of reliable, high-quality disease occurrence repositories. This has led to the widespread use of black-box modeling for disease outbreaks 28, 29, 30 and component-based modeling of hosts only 17, 32. The use of outbreak data to model disease distributions raises methodological issues. Disease data are generally aggregated at coarse politicoadministrative levels (e.g., province, country), losing crucial information on the local natural history of the disease 1, 33. Additionally, the geographic site of infection and the associated uncertainty are generally not reported and may instead refer to the health-care facility where it is diagnosed [1], potentially misleading the identification of ecological conditions favoring disease occurrence. This could be further complicated by centralized health-care infrastructures and scarce epidemiological resources, resulting in predictions of the site of diagnosis instead of the site of infection and parasite persistence [34]. Lack of information on the ecology of a disease system may hinder proper identification of the parasite species (or strain in the case of viruses) causing the disease and/or the hosts involved in their transmission. This is particularly true for EIDs [31], posing a significant challenge to the prediction of geographic distributions in an ENM framework.
Host–Parasite Interactions
At least two interacting species – a parasite and a host 35, 36 – are present in a disease system. These systems vary in complexity as some parasites can infect multiple hosts, potentially requiring the presence of keystone host species for their transmission (e.g., vectors) and maintenance (e.g., reservoir hosts). Here we define ‘parasites’ broadly to encompass all organisms capable of causing disease (i.e., pathogens), including microparasites (e.g., viruses, bacteria, fungi, protozoa) and macroparasites (e.g., flatworms, nematodes). Similarly, we broadly define ‘hosts’ to include arthropod vectors and vertebrate reservoir hosts of parasites.
Since biotic interactions lie at the core of disease systems, neglecting interacting species and their role in parasite dynamics (maintenance, reproduction, and transmission) may lead to failure to forecast disease distributions (Figure 1 ). Parasite transmission is strongly influenced by interactions among infected and susceptible hosts, which can be altered by host behavior and demography 6, 37, 38. For example, parasite transmission was found to be related to spider monkey (Ateles hybridus) grooming activity [39] and to aggregation behavior during hibernation in bat colonies of multiple species [40].
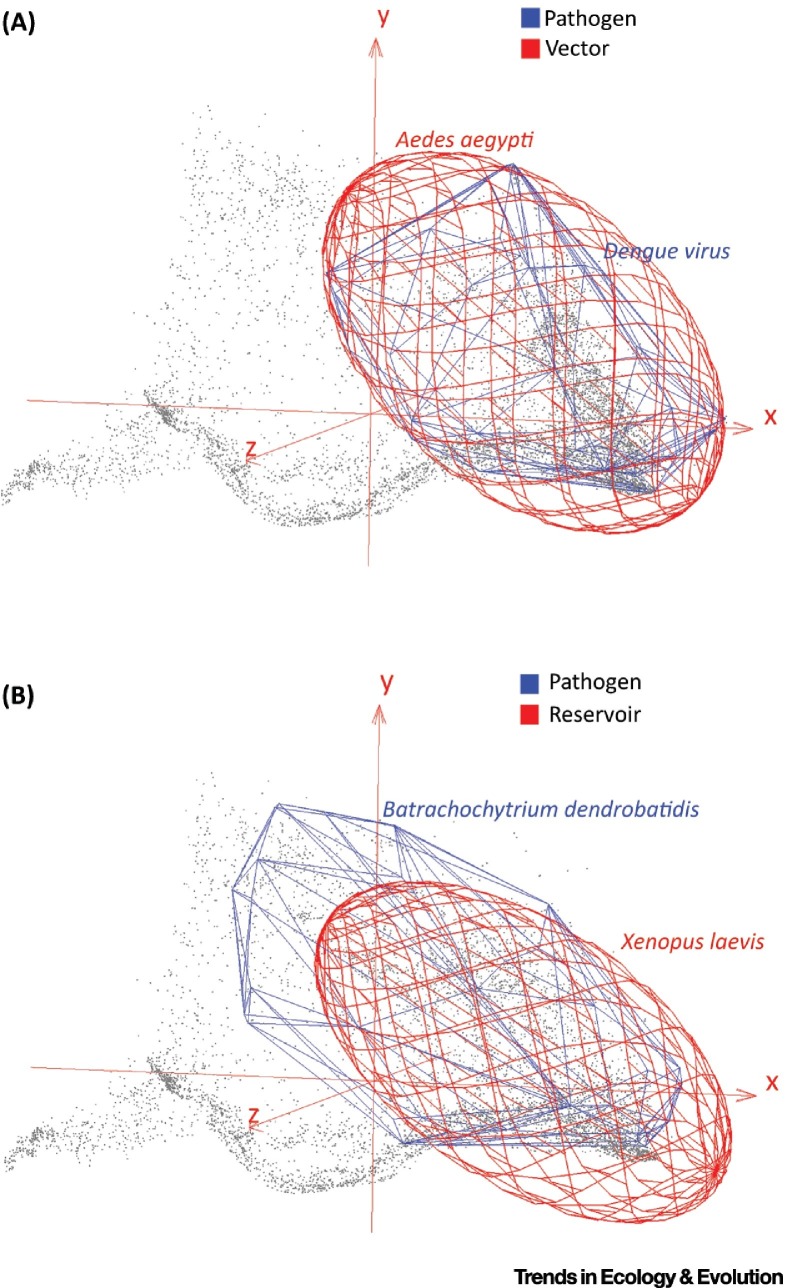
Figure 1.
Host versus Parasite Ranges. Gray points represent a hypervolume of 15 satellite-derived global bioclimatic variables as described by the first three axes of a principal components analysis (PCA); red ellipsoids and blue polyhedra are 3D representations of n-dimensional hypervolumes of host and parasite niches, respectively. (A) Parasite matching the niche of the host. The niche of dengue virus (blue polyhedron) coincides with that of its vector, the mosquito Aedes aegypti (red ellipsoid), suggesting a coevolutionary history. In this case, modeling of the host would be a good proxy of the potential distribution of the parasite. (B) Parasite does not match the niche of the host. The niche of the amphibian chytrid fungus (Batrachochytrium dendrobatidis, blue polyhedron) coincides only partially with that of its main reservoir, the African clawed frog (Xenopus laevis, red ellipsoid). Niche dissimilarity may suggest that this host species may not be the natural reservoir of the parasite. Modeling only the host would underestimate the potential distribution of the parasite. Data sources: environmental variables [89]; species occurrences represent global compendiums for A. aegypti[8], dengue virus [90], X. laevis[91], and B. dendrobatidis92, 93.
Transmission dynamics can be further altered by the structure of the ecological community 9, 41, 42, 43, 44. For example, host species living at higher population densities with smaller body sizes and shorter generation spans were more likely to be competent reservoirs for multihost vector-borne diseases [37]. Likewise, host species diversity was found to alter transmission by decreasing host density (i.e., dilution effect) and increasing contact rates between host species (i.e., amplification effect) in a rodent-borne disease [45]. Host immunity and parasite–parasite interactions may also shape disease distributions as they may facilitate or limit transmission 6, 7, 41, 46. For instance, decreased competence of the bacterium Rickettsia conorii was observed in dogs previously infected by other Rickettsia species [47]. Given the complexity of these interactions, traditional single-species ENM approaches could fail to accurately predict disease distributions and transmission risk, particularly at finer scales. However, traditional approaches may be sufficient for some disease systems, especially if they are simple or well understood (e.g., dengue). Therefore, appropriate selection of the approach will depend on data availability and the question at hand.
Parasite Occurrence versus Disease Expression
A common assumption of disease ENM is that predicted host distributions and disease presence are equivalent 17, 32. This should be considered with caution since susceptible hosts may occur where parasites are absent, and even when infection occurs, disease may be absent 7, 48. For example, flying foxes (Pteropus medius) are necessary hosts for Nipah virus persistence; however, this virus can be absent in areas where flying fox populations are present [48]. Therefore, host presence should be considered only as ‘vessels’ available for parasite introduction, establishment, and spread (Box 1 ). Likewise, parasites are generally assumed to be homogeneously distributed across the host’s range (uniform prevalence; Figure 1A) and fine-scale mechanisms underlying parasite transmission (e.g., host movement, behavior, demographics) are usually not considered.
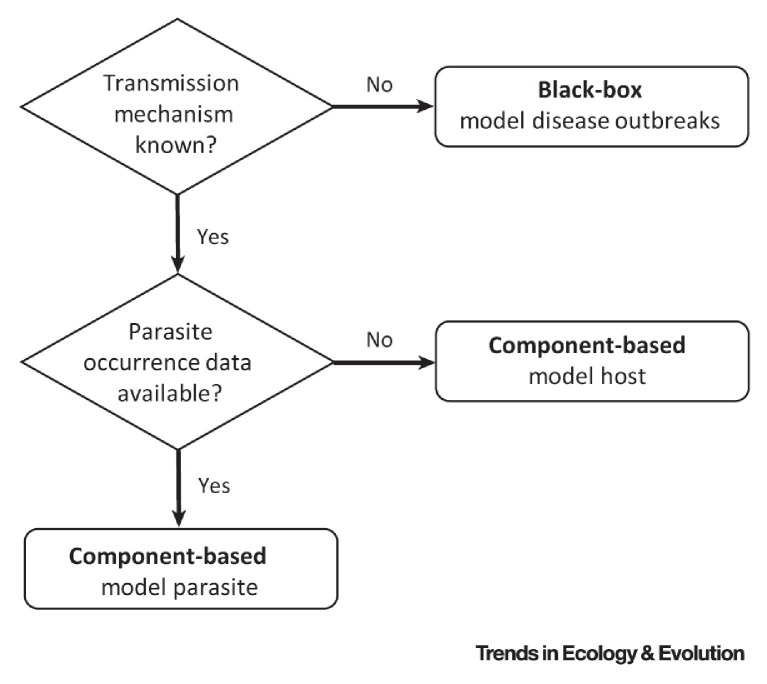
Figure I.
Modeling Approach Selection for Disease Ecological Niche Modeling (ENM). The appropriate selection of the modeling approach (black box vs component based) for diseases will depend on data availability and knowledge of disease transmission dynamics.
Box 1. Guidelines for ENM to Predict Disease Distributions.
The predictive power and biological realism of ENM forecasts of diseases is likely to improve by the inclusion of biotic interactors 10, 43. However, reliable parasite, or disease, occurrence records and information on disease natural history may be lacking, posing an exceptional challenge for disease distribution modeling. Knowing the data limitations of a disease system is crucial for predictor variable and evaluation metric selection and the incorporation of biotic interactors [24]. Understanding of the biological meaning, assumptions, and units of outputs is equally important for proper model interpretation. We discuss the most frequently encountered scenarios in disease ENM and how biotic interactors should be included accordingly (Figure I).
-
(1)
Available occurrence data and known transmission mechanism: When parasite occurrence data are available, these should be preferred over disease outbreak data to minimize spatiotemporal uncertainty. However, if reliable site of infection data are available these should ideally be used. In this case, component-based approaches focused on modeling the geographic potential of the parasite can be implemented. Information on host species (e.g., distribution, abundance) can be incorporated as predictor variables (preprocessing) to complement abiotic variables. However, model outputs should be interpreted with caution as proper definition of units may be difficult [24].
-
(2)
Unavailable occurrence data and known transmission mechanism: Given the reliance of parasites on host species, component-based modeling of hosts could be used to identify suitable areas for parasite persistence. That is, estimated host distributions are used as a proxy for potential parasite distributions. Selection of host species (e.g., identity, number) for modeling will depend on their role in the disease system as well as the nature of the system itself. If parasite persistence depends on interactions between different host species, stacked or joint host distribution models (post-processing) can be used, assuming parasite presence is equal or more likely in areas where all of its hosts are found than where only one host species is found [24]. Likewise, seasonal factors capable of affecting transmission (e.g., rainfall, migration patterns) should be accounted for whenever possible. However, excessive use of abiotic and biotic variables could generate over-fit and complex models, which may be difficult to parameterize and interpret.
-
(3)
Unavailable occurrence data and unknown transmission mechanism: This situation warrants black-box modeling [18]. The point-radius method can be used to mitigate geographic uncertainty inherent to human disease data [91]. Additionally, to reduce uncertainty in environmental dimensions, outlying disease cases reported in areas of inconsistent environmental conditions (e.g., imported cases) can be removed. Due to the temporal lag between infection and disease expression, temporal uncertainty of exposure should be considered to ensure that environmental variables match disease reports.
Alt-text: Box 1
Outcomes of host–parasite interactions are highly variable, ranging from no apparent negative effects on the host (e.g., asymptomatic or subclinical infection) to host mortality 49, 50, 51. A review of mammal–virus associations reported that the vast majority of infected mammal species were asymptomatic (224 of 312 mammal–virus pairs) [52]. Hantavirus infections in North America can result in hantavirus pulmonary syndrome, which is often fatal in humans while having no discernible impact on deer mice (Peromyscus maniculatus), its primary host [46]. Host immunity, genetics, and physiology also play important roles in disease expression, varying among individuals 6, 49, 53 and populations [54]. The generalist amphibian chytrid fungus (Batrachochytrium dendrobatidis) can cause disease in some amphibian species but not others; thus, mapping a single host would underestimate the parasite’s geographic distribution (Figure 1B).
Environmental Predictors
Selection of ecologically relevant predictor variables is necessary to generate reliable modeling outputs and should be supported by the biology of the species and the spatiotemporal scale at hand 26, 55. Variables directly affecting a species’ physiology are preferred since their relationships with its geographic distribution are assumed to be stable across spatiotemporal scales 26, 56. Indirect variables may be employed as proxies for direct variables, although these should be avoided if they are correlated with factors driving the demography, dispersal, or distribution of biotic interactors [26]. For example, in the tropics, altitude could serve as a proxy for temperature and has been used to predict the distribution of mosquito-borne diseases, since vector distribution is restricted by low temperatures. However, elevation can be a confounding factor not related to the species’ physiology, as compared with temperature, and in general, should be avoided.
In disease systems, the effects (direct/indirect) of abiotic variables depend on the parasite’s ecology and relationship with its hosts. Parasite life cycles range from having free-living stages to being completely restricted within a host. Leptospira bacteria (the causative agent of leptospirosis) are capable of persisting in humid soils and waterlogged environments [57]. In this case, environmental variables such as precipitation or the presence of seasonal water bodies are more likely to have direct effects. Conversely, parasites unable to persist outside their hosts, like rabies viruses [58], are likely to be influenced by environmental variables (e.g., climate) indirectly. Hence, host availability may directly affect the maintenance of host-restricted parasites. The nature of the parasite–host association will determine the ecological relevance of environmental variables and how these should be employed to model parasite distributions.
ENM Implementation for Disease Control
Epidemiological strategies to control diseases focus on regulating parasite transmission from a source population (usually wildlife or domestic hosts) to a target population (usually humans or domestic animals) 38, 59, 60. Component-based ENM (details in Modeling Disease System section) can be used to identify areas where potential disease sources and target populations overlap, allowing informed interventions 60, 61. This requires a comprehensive understanding of the natural history of the disease system to properly identify host species acting as sources and spreaders of infection [38]. Misidentification of the host and parasite species involved in epidemics and spillover events, or their functional roles in disease maintenance, may lead to ineffective or counterproductive control measures with potential social and economic costs 6, 38, 61.
In single-host systems (Figure 2 A), disease control strategies should target regions where source populations overlap with the target population (Figure 2B; 38, 61). This can rapidly become complicated in multihost systems as changes in the host community may impact parasite maintenance or transmission (Figure 2C; see 41, 61). Further complications arise when hosts act as bridges facilitating parasite transmission between spatially disjoint host populations (Figure 2C,D) [38]. For example, wild birds associated with wetlands and aquatic environments, such as shorebirds (Charadriiformes; gulls, terns, and waders) and waterfowl (Anseriformes; ducks, geese, and swans), constitute the host reservoir for avian influenza (Figure 2C). Since shorebirds and livestock are spatially disjunct (Figure 2D), strategies aimed at this group only would not stop influenza transmission from waterfowl to livestock and consequent human infection, despite them being part of the reservoir.
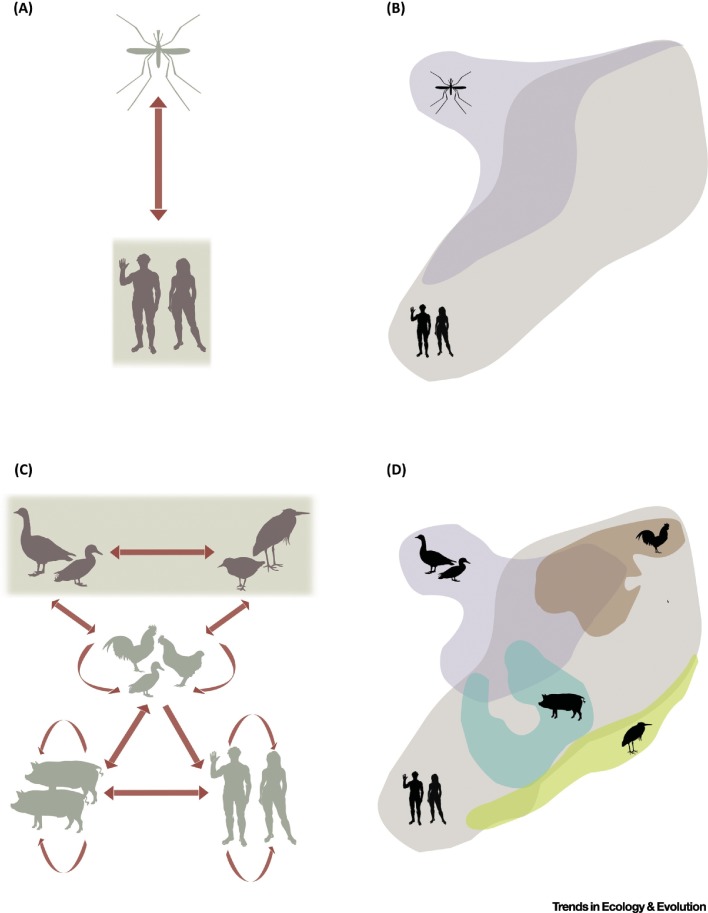
Figure 2.
Disease System Components. (A) Single-host–single-vector disease system (e.g., dengue fever). Only the host and vector are necessary to sustain parasite transmission. In this case, a vector is the only possible source of infection. (B) Component-based ecological niche modeling (ENM) that considers the distributions of the vector (purple polygon) and the target population (light-gray polygon) to identify geographic areas where the two may overlap as a proxy of disease transmission risk (dark-shaded area). Control and prevention strategies should focus on overlapping regions. (C) Multihost disease system (e.g., avian influenza). Multiple waterfowl and shorebird species constitute the natural hosts of the parasite (shaded area). The parasite, however, can infect other species (domestic or wild). In this example, interspecific transmission (spillover) of the parasite among wild and domestic bird species, and among bird and mammal species, has resulted in human infection. (D) In this scenario, ENM should consider overlap among species to identify transmission mechanisms with greater propensity to threaten human health. Here, domestic mammals play a critical role in parasite spillover, and control strategies should aim to reduce overlap between birds, both wild and domestic, and domestic mammals to reduce transmission risk to humans. Animal silhouette source: [94].
Niches in Host-Space
Appropriate selection and inclusion of biotic interactors in parasite ENM requires prior identification of suitable host species (Figure 3 A); that is, host species that possess characteristics supporting parasite survival, reproduction, and transmission and are therefore essential for parasite persistence 35, 36, 62, 63. For a given parasite, different suitable host species must share particular traits enabling its establishment and persistence 9, 35, 63. Here, we propose an adaptation of the niche concept that considers host traits as microscale abiotic and biotic dimensions of parasite niches, defined here as host-space (H-space in Figure 3B).
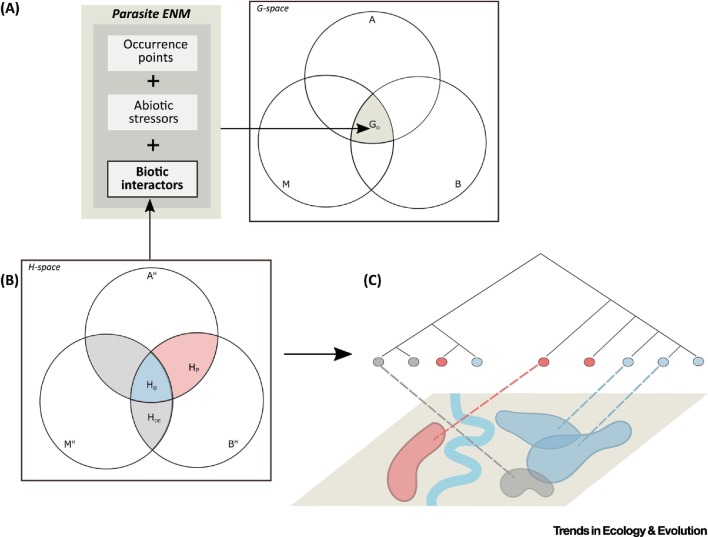
Figure 3.
Graphic Representation of Niches in Host-Space (BAM-H) as a Framework Complementary to Traditional Parasite Ecological Niche Modeling (ENM). (A) Traditional parasite ENM generates estimates of species’ occupied geographic space (GO) using data on species occurrence, abiotic stressors (e.g., climate), and biotic interactors as inputs. (B) Adaptation of the BAM framework to host-space (BAM-H). BH represents the set of dynamically linked biotic factors favoring parasite persistence and AH comprises the set of suitable host traits for pathogen establishment and persistence, while MH represents host populations available for transmission dispersal. HO, occupied host breadth; HP, potential host breadth; HDE, dead-end hosts unable to sustain parasite maintenance (i.e., sink populations). (C) Theoretical example of how the host-space concept can be linked to host phylodynamics (tree) and ecologies to further our understanding of parasite distributions across landscapes (tan rectangle below). Blue represents suitable host populations where the parasite is present (HO) and red represents suitable host populations where the parasite is not found (HP), which may be separated by geographic barriers such as rivers (thick blue line), while gray represents dead-end hosts (HDE).
Under this proposed approach, associations between parasites and host populations can be summarized by adapting the traditional biotic, abiotic, and movement (BAM) framework (Box 2 ) to host-space. We refer to this adapted framework as BAM-H (Figure 3B). Here, BH represents the set of dynamically linked (biotic) factors favoring persistence in hosts where bidirectional effects with parasite load can be observed (i.e., affected by parasite abundance) 26, 64, such as immune response (similar to predator–prey interactions) 46, 49, 54 or coinfection (similar to facilitation/competition) 65, 66, 67. HIV infection, for example, facilitates the establishment of other parasitic organisms including viruses, bacteria [65], and protozoa [68]. AH comprises the set of physical and chemical (e.g., body temperature, pH, presence of cell receptors in the membrane) host traits representing suitable conditions for establishment and persistence with generally unidirectional effects on parasite load 6, 26, 36, 41, 64. For example, rabies virus can survive only in hosts with a specific body temperature range (∼4–39 °C) [58], resulting in a predominance of mammalian hosts (∼37 °C) but not birds (∼40 °C). Similarly, SARS coronavirus cannot infect cells lacking angiotensin-converting enzyme 2 (ACE2), its entry receptor [69]. Last, MH represents the set of host species populations that have been accessible for the parasite to disperse (i.e., transmit). Transmission between individuals is essential to guarantee parasite maintenance [also expressed as basic reproductive number (R0) > 1] in a host population (i.e., intraspecies transmission) or community (i.e., interspecies transmission). Although dispersal-related parasite traits (e.g., free living vs host restricted) are important determinants of transmissibility, parasite mobility can be further constrained by host demography and ecology 18, 70; however, these traits may be less relevant for parasites with environmental reservoirs (e.g., anthrax). For example, parasite persistence may not be possible if the host population size is too small (similar to Allee effects). Social contact, grooming rate, or burrowing behavior limits the transmission of parasites (e.g., fleas in small mammals) [71]. Factors constraining transmission can operate across multiple spatial scales, ranging from limited dispersal between individuals in a population (e.g., decreased host population size for density-dependent transmission) to barriers between host populations (e.g., geographic barriers).
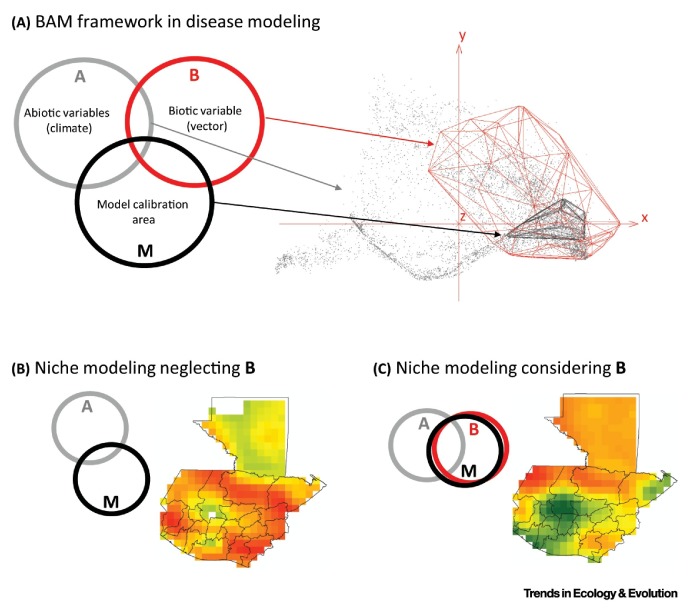
Figure II.
Applying the Biotic, Abiotic, and Movement (BAM) Framework to Real-World Situations. (A) Transferring the theoretical representation of the BAM framework (left) to an empirical ecological niche modeling (ENM) application using real-world data to reconstruct the geography of dengue virus in Guatemala (right). Here, the biotic component of the parasite, B, is denoted by the fundamental niche of the dengue vector, the mosquito Aedes aegypti (red polyhedron) [8]. A (gray points) summarizes global bioclimatic variables [89] condensed in three principal components. Finally, the dispersal potential, M, was restricted to Guatemala as the area of interest (black polyhedron). (B,C) Model predictions of disease distributions are affected by the inclusion of biotic interactions within the ENM framework.
Box 2. BAM: A Simple Framework to Represent Species Distributions.
The BAM framework represents species’ geographic distributions by summarizing the interaction of three factors: dynamically linked biotic interactors (B), unlinked abiotic stressors (A), and dispersal capacity (M) (Figure II). Areas where all three of these conditions are met (B ∩ A ∩ M) represent the species’ actual distribution and a proxy of the species’ realized niche. Traditional ENM applications consider the B component to have negligent effects (the Eltonian noise hypothesis [10]) when modeling species’ geographic distributions under the BAM framework. However, biotic interactions play a critical role in parasitic relationships in nature, so they should be considered with caution in disease ecology.
Implementing a traditional ENM framework [i.e., A ∩ M (Figure IIB)] to map the distribution of dengue virus in Guatemala provides different predictions than a model accounting for biotic interactions [B ∩ A ∩ M (Figure IIC)]. Adding information on vector abundance, immunity of hosts, and behavior, among other variables, would add complexity to the final risk estimation but may provide a more complete history of the plausible manifestation of the disease in the area of interest.
Alt-text: Box 2
BH ∩ AH defines a parasite’s potential host breadth (HP) (Figure 3B,C): host populations that the parasite could theoretically infect and persist in, in the absence of dispersal or demographic barriers 36, 72, 73. BH ∩ AH ∩ MH determines a parasite’s occupied host breadth (HO) (Figure 3B,C): the subset of potential host species it can infect considering dispersal limitations at different scales (e.g., geographic barriers, demographic constraints). HO constitutes the suite of host populations that the parasite effectively occupies. Parasites may be absent in suitable hosts due to local parasite extinction, seasonality (e.g., host migration patterns, precipitation patterns needed for parasite transmission), dispersal, or transmission limitations (e.g., geographic barriers, low host density, host immunity) and in the case of economically important hosts (e.g., livestock) due to disease management control (e.g., vaccination, disease control programs). Transient infections could also occur in dead-end hosts (HDE) (Figure 3B,C), unsuitable hosts limiting their persistence (R 0 ≤ 1; i.e., sink populations). Our approach is complementary to traditional parasite ENM and classic transmission models. By identifying parasite host breadth, BAM-H would allow the proper identification of relevant biotic interactors that inform parasite ENM and should therefore be used jointly (Figure 3).
Closely related host species tend to share ecological, physiological, and immunological traits, making them more likely to share parasites 36, 74, 75, 76. The identification of closely related hosts (i.e., sharing similar traits) could help to identify potential reservoirs and predict potential spillover (HP), analogous to predicting suitable geographic areas (novel hosts) for species invasion (parasite spillover) in invasion ecology. Parasite sharing among closely related host species (phylogenetic clustering) could be interpreted as parasite niche conservatism in host-space [as observed in kissing bug species (Triatoma sp.) in North and Central America] [77]. Potential hosts (HP) would therefore be more likely to be closely related to known hosts. Conversely, parasite sharing among distantly related host species (phylogenetic overdispersion) implies that parasite sharing among known hosts is driven by factors other than host relatedness, important to consider when determining HP. Such factors may include broad physiological tolerances (large AH) or increased transmissibility (MH) between host populations with overlapping geographic ranges (Figure 3C 36, 75). Furthermore, parasites may experience expansions in their occupied host breadth (HO) following landscape alterations or shifts in their geographic distributions 75, 78 (see Box 3 for applications).
Box 3. Improving Epidemiological Surveillance: Using Ecophylogenetics to Estimate Host Breadth.
Epidemiological surveillance aims to identify and monitor disease-causative agents and to use this information to inform public health policy. However, how can we possibly achieve these goals when we do not know which areas or host species should be targeted for surveillance? Ecophylogenetics provide a basis to disentangle the relative contributions of host relatedness (phylogeny) and host ecology (e.g., interaction networks, demography) to parasite-sharing patterns among different host populations (Figure 3C [78]). Integrating phylogenetics with community ecology could help in understanding patterns of host phylogenetic clustering and overdispersion across spatiotemporal scales. This holds great potential for the development of new tools to predict parasite host breadths (HP and HO in our proposed framework), a key component of understanding disease transmission dynamics at the landscape level 56, 77. Additionally, ecophylogenetics may provide insights on potential spillover events that could result in disease emergence, a highly relevant topic under global change and increased international movement of species leading to shifts in host communities and translocation of parasites to new areas and hosts 79, 80. Adoption of the host-space framework under ecophylogenetics could improve epidemiological surveillance, allowing early targeting of potentially suitable hosts (HP) 62, 63.
Current initiatives such as the Global Mammal Database [81], the Malaria Atlas Project [82], and COMADRE [83] could lay the groundwork to implement the host-space framework and serve as a platform to integrate data from different sources similar to large-scale biodiversity repositories (e.g., the Global Biodiversity Information Facility [84]). Coupling ecophylogenetics with ENM could increase our understanding of how external environmental conditions (e.g., climate) and host traits explain disease distributions, although these approaches have rarely been applied until recently [78]. Nevertheless, although the theoretical basis to target potential hosts via ecophylogenetics exists, further work is required to successfully implement this approach.
Alt-text: Box 3
From Disease Distributions to Risk Mapping
Despite widespread recognition of the need for risk assessments to ensure successful disease intervention strategies, definitions of risk remain inconsistent. These definitions seem to confound the different processes contributing to risk, hindering proper quantification and comparisons between assessments [85]. A recently proposed framework aimed to disentangle the underlying mechanisms of risk by decomposing it into three processes: parasite availability (hazard), contact with parasites (exposure), and likelihood of infection (susceptibility) [85].
Our proposed integrative ENM framework combines traditional parasite ENM with the host-space concept, allowing a more comprehensive estimate of the potential geographic distribution of diseases across scales, and could therefore be implemented to estimate hazard (parasite availability). However, we must note that this is only one component driving disease risk for a target population (Box 3). Several often interacting factors such as behavior [59], nutrition [86], immune history [47], and social status 33, 87, 88 are critical for successful parasite maintenance. When possible, these factors should be considered and incorporated into ENM frameworks to enrich risk assessments. Exposure can be incorporated by overlaying the geographic distribution and densities of the target population (e.g., humans) [14]. Susceptibility factors could be added by including information on socioeconomic (e.g., GDP, age) or cultural (e.g., taboo systems, traditional practices) factors influencing exposure to hazard. An example of susceptibility factors increasing exposure is the traditional funeral practices involving the touching and kissing of dead bodies that contributed to the spread of Ebola in the 2014 West African outbreak [59].
Disease risk mapping still faces considerable challenges. Gathering information of parasite occurrence data, in both animals and humans, can suffer from logistic (e.g., sampling in remote areas, ethical human-subject-research regulations) and ecological (e.g., low prevalence, latency) limitations. Parasite detection may be limited by the choice of clinical screening method. Serology tests report past infection whereas PCR or deep-sequencing methods detect parasites present at the moment of collection. Data on susceptibility factors are limited and their effects are not always understood, hindering proper quantification of susceptibility. We believe that a next frontier in disease risk mapping should focus on overcoming these limitations. Investment in active surveillance efforts in wildlife and human populations, as well as new technologies and tools (Box 3) for parasite detection and identification techniques, may improve our ability to collect more reliable disease occurrence data. Interdisciplinary approaches integrating ecology and social sciences may further our understanding on how biological and socioeconomic factors interact to influence disease risk.
Concluding Remarks
ENM is a powerful tool to better understand the distributional ecology of diseases. We described how biotic interactions make disease systems more ecologically complex than the traditional biodiversity studies for which ENM was designed. Limited knowledge on disease natural history (e.g., transmission mechanism, host species involved) may considerably change modeling assumptions, resulting in ecologically unrealistic outputs. Here, we present a new framework – the host-space niche – that is complementary to traditional parasite ENM, which should improve the integration of parasite–host interactions. This host-space niche framework will help in identifying relevant biotic interactors and understanding disease distributions across landscapes.
Finally, we point out that risk can be defined only when a target population is identified. Risk depends on multiple interacting factors including parasite presence, exposure, and the susceptibility of the target population. We emphasize the need for a clear and uniform definition of risk as well as a unified methodological framework to quantify it. Quantification of disease transmission risk is also important for strategic allocation of resources for public health and the conservation of endangered host populations.
The ideas presented here should encourage discussion towards a comprehensive methodological framework to quantify and map disease distributions and risk that are based on ecological and epidemiological theory (see Outstanding Questions). However, challenges remain, particularly ensuring that disease occurrence data reflect the site of infection and the biological realism of model assumptions. Increased epidemiological surveillance and data sharing via online repositories will facilitate the establishment of a renovated field of disease mapping based on ecological theories. We provide guidelines (Box 1) to estimate the geographic distribution of diseases via ENM by accounting for data limitations and different ecological scales. The incorporation of biotic interactors into the models will allow more realistic estimates of disease distributions that could help to guide cost-effective disease control efforts.
Outstanding Questions.
How can disease occurrence data be improved? Besides spatial and temporal biases, disease occurrence data may also be skewed by uneven sampling of host species (e.g., reservoirs) and reports of the locality of development of symptoms or the locality of the diagnosis, misrepresenting the locality of infection. Active surveillance efforts targeting parasite sampling in host taxa could help to reduce these biases, increase understanding of parasite dispersal, and improve occurrence reporting.
Can host traits be used to predict parasite host breadth? We have proposed a niche-conservatism approach to identify potential hosts given their phylogenetic relatedness with known hosts. Host traits essential for parasite establishment can be determined in a more mechanistic manner through experimental testing of parasite physiological niches and may be validated in laboratory essays.
How can risk factors be integrated into ENM frameworks to quantify disease transmission risk? Behavioral and clinical surveillance provides insights on factors influencing exposure and susceptibility to infective parasites. However, an interdisciplinary approach is necessary to adequately generate data-driven models useful for policy making. Integration of quantitative and qualitative approaches remains a challenge in ENM disease risk mapping, which usually neglects the human dimension of disease transmission.
Acknowledgments
E.E.J. and C.Z-T. were supported by the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT program. L.E.E. was supported by a Global Change Center, Virginia Tech, seed grant. The authors thank Robert P. Anderson and Peter J. Galante, for insightful comments and inputs on earlier versions of this manuscript. Comments from two anonymous reviewers and the editor were extremely helpful and greatly improved the content of the manuscript.
Glossary
- Allee effect
correlation between population size or density and the per capita growth rate (mean individual fitness) of a population.
- Basic reproductive number (R0)
expected number of secondary cases caused by a single infectious individual in a population.
- Biotic, abiotic, and movement (BAM) framework
simplified representation of a species’ geographic distribution determined by the intersection of suitable abiotic conditions (A), biotic interactions (B), and the species’ dispersal capacity (M).
- Bridge host
host population capable of facilitating transmission between two otherwise geographically disconnected maintenance and target populations.
- Coinfection
simultaneous infection of a host by multiple parasite species.
- Disease system
set of species, including parasites, susceptible hosts, and vectors, involved in the maintenance, transmission, and expression of a disease.
- Ecological niche modeling (ENM)
computational method used to predict species geographic distributions by combining factors related to a species’ environmental requirements with those related to occurrence and dispersal.
- Ecophylogenetics
field in biology that focuses on the study of ecological patterns in biological communities (e.g., community assembly, species co-occurrence) explained by the evolutionary relationships among coexisting species.
- Eltonian noise hypothesis
ecological hypothesis stating that local-scale ecological interactions have negligible effects on a species’ geographic distribution.
- Emerging infectious disease (EID)
diseases that have increased in incidence or geographic range, found in novel hosts or caused by newly evolved parasites.
- Exposure
likelihood of contact between a target population and hazards. The degree of exposure will depend on the contact rate, the parasite’s transmission mechanism, and the nature of a contact event.
- Hazard
relative number of available parasites at a given space and time acting as potential sources of harm (e.g., disease outbreak) to a target population.
- Host
a living organism that can be infected by a parasite or any other infectious agent under natural conditions.
- Host breadth
the range of different host populations that a parasite is known to (occupied host breadth) or could potentially (potential host breadth) infect and persist in.
- Maintenance
ability of a host or group of host species to keep a parasite circulating within an epidemiologically connected group of individuals over the long term.
- Niche
set of abiotic (e.g., physical, environmental) and biotic (e.g., interactions with other species) conditions that allow a species’ persistence in a given area when accounting for its dispersal ability.
- Niche conservatism
retention of niche-related ecological traits over time, frequently among related species.
- Parasite
an organism dependent on a different organism (host) for its survival and reproduction that may or may not cause negative effects on its host.
- Pathogen
a parasite, usually a microorganism (e.g., bacterium, virus) capable of causing disease in its host.
- Phylogenetic clustering
pattern observed in ecological community structure when driven by environmental filtering where species within a community are more closely related than expected by chance.
- Phylogenetic overdispersion
pattern observed in ecological community structure when driven by competition where species within a community are more distantly related than expected by chance.
- Reservoir
habitat in which a parasite can grow, reproduce, and survive. Reservoirs are typically considered to be biotic (e.g., hosts); however, they can also be abiotic.
- Risk
likelihood of an adverse event (e.g., disease outbreak) occurring in a target population because of exposure to a hazard.
- Risk factor
factor capable of facilitating or limiting risk by modifying either hazard or exposure.
- Spillover
process in which a pathogen is transmitted into a novel host species, mainly referring to the transmission of zoonotic diseases to humans.
- Susceptibility
possibility of a given exposure to hazard resulting in harm; also termed vulnerability by some authors.
- Target population
population that is the focus of a study or public health intervention.
- Vector
a living organism, typically invertebrate, acting as intermediary in the transmission of a parasite from a source to a target population.
- Zoonoses
diseases that can be transmitted between humans and animals under natural conditions.
References
- 1.Allen T. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 2017;8:1124. doi: 10.1038/s41467-017-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan V.Y. Pandemic risk: how large are the expected losses? Bull. World Health Organ. 2018;96:129. doi: 10.2471/BLT.17.199588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pike J. Economic optimization of a global strategy to address the pandemic threat. Proc. Natl. Acad. Sci. U. S. A. 2014;111:18519–18523. doi: 10.1073/pnas.1412661112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC . US Department of Health and Human Services; 2016. Cost of the Ebola Epidemic. [Google Scholar]
- 5.Cunningham A.A. One Health, emerging infectious diseases and wildlife: two decades of progress? Philos. Trans R. Soc. Lond. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2016.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plowright R.K. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15:502–510. doi: 10.1038/nrmicro.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estrada-Peña A. Effects of environmental change on zoonotic disease risk: an ecological primer. Trends Parasitol. 2014;30:205–214. doi: 10.1016/j.pt.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Kraemer M.U. The global compendium of Aedes aegypti and Ae albopictus occurrence. Sci. Data. 2015;2:150035. doi: 10.1038/sdata.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gervasi S.S. The context of host competence: a role for plasticity in host–parasite dynamics. Trends Parasitol. 2015;31:419–425. doi: 10.1016/j.pt.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araújo C.B. The importance of biotic interactions in species distribution models: a test of the Eltonian noise hypothesis using parrots. J. Biogeogr. 2014;41:513–523. [Google Scholar]
- 11.Martínez-Freiría F. Contemporary niche contraction affects climate change predictions for elephants and giraffes. Divers. Distrib. 2016;22:432–444. [Google Scholar]
- 12.Jezkova T. Range and niche shifts in response to past climate change in the desert horned lizard Phrynosoma platyrhinos. Ecography. 2016;39:437–448. doi: 10.1111/ecog.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuckerberg B. Novel seasonal land cover associations for eastern North American forest birds identified through dynamic species distribution modelling. Divers. Distrib. 2016;22:717–730. [Google Scholar]
- 14.Pigott D.M. Mapping the zoonotic niche of Ebola virus disease in Africa. Elife. 2014;3 doi: 10.7554/eLife.04395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barro A.S. Redefining the Australian anthrax belt: modeling the ecological niche and predicting the geographic distribution of Bacillus anthracis. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackburn J.K. Modeling the ecological niche of Bacillus anthracis to map anthrax risk in Kyrgyzstan. Am. J. Trop. Med. Hyg. 2017;96:550–556. doi: 10.4269/ajtmh.16-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samy A.M. Leishmaniasis transmission: distribution and coarse-resolution ecology of two vectors and two parasites in Egypt. Rev. Soc. Bras. Med. Trop. 2014;47:57–62. doi: 10.1590/0037-8682-0189-2013. [DOI] [PubMed] [Google Scholar]
- 18.Peterson A.T. JHU Press; 2014. Mapping Disease Transmission Risk: Enriching Models Using Biogeography and Ecology. [Google Scholar]
- 19.Elith J. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- 20.Elith J., Graham C.H. Do they? How do they? Why do they differ? On finding reasons for differing performances of species distribution models. Ecography. 2009;32:66–77. [Google Scholar]
- 21.Fraterrigo J.M. Local-scale biotic interactions embedded in macroscale climate drivers suggest Eltonian noise hypothesis distribution patterns for an invasive grass. Ecol. Lett. 2014;17:1447–1454. doi: 10.1111/ele.12352. [DOI] [PubMed] [Google Scholar]
- 22.Ilsøe S.K. Global variation in woodpecker species richness shaped by tree availability. J. Biogeogr. 2017;44:1824–1835. [Google Scholar]
- 23.Sanín C., Anderson R.P. A framework for simultaneous tests of abiotic, biotic, and historical drivers of species distributions: empirical tests for North American wood warblers based on climate and pollen. Am. Nat. 2018;192:E48–E61. doi: 10.1086/697537. [DOI] [PubMed] [Google Scholar]
- 24.Atauchi P.J. Species distribution models for Peruvian plantcutter improve with consideration of biotic interactions. J. Avian Biol. 2018;49:jav–01617. [Google Scholar]
- 25.Descombes P. Simulated shifts in trophic niche breadth modulate range loss of Alpine butterflies under climate change. Ecography. 2016;39:796–804. [Google Scholar]
- 26.Anderson R.P. When and how should biotic interactions be considered in models of species niches and distributions? J. Biogeogr. 2017;44:8–17. [Google Scholar]
- 27.Mod H.K. Biotic interactions boost spatial models of species richness. Ecography. 2015;38:913–921. [Google Scholar]
- 28.Pigott D.M. Mapping the zoonotic niche of Marburg virus disease in Africa. Trans. R. Soc. Trop. Med. Hyg. 2015;109:366–378. doi: 10.1093/trstmh/trv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyakarahuka L. Ecological niche modeling for filoviruses: a risk map for Ebola and Marburg virus disease outbreaks in Uganda. PLoS Curr. 2017 doi: 10.1371/currents.outbreaks.07992a87522e1f229c7cb023270a2af1. Published online September 5, 2017. https://dx.doi.org/10.1371/currents.outbreaks.07992a87522e1f229c7cb023270a2af1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quiner C.A., Nakazawa Y. Ecological niche modeling to determine potential niche of Vaccinia virus: a case only study. Int. J. Health Geogr. 2017;16:28. doi: 10.1186/s12942-017-0100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soucy J.-P.R. High-resolution ecological niche modeling of Ixodes scapularis ticks based on passive surveillance data at the northern frontier of Lyme disease emergence in North America. Vector Borne Zoonotic Dis. 2018;18:235–242. doi: 10.1089/vbz.2017.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baak-Baak C.M. Ecological niche model for predicting distribution of disease-vector mosquitoes in Yucatán State, México. J. Med. Entomol. 2017;54:854–861. doi: 10.1093/jme/tjw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farnham A. Legionnaires’ disease incidence and risk factors, New York, New York, USA, 2002–2011. Emerg. Infect. Dis. 2014;20:1795. doi: 10.3201/eid2011.131872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson A.T. Mapping transmission risk of Lassa fever in West Africa: the importance of quality control, sampling bias, and error weighting. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luis A.D. Network analysis of host–virus communities in bats and rodents reveals determinants of cross‐species transmission. Ecol. Lett. 2015;18:1153–1162. doi: 10.1111/ele.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olival K.J. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostfeld R.S. Life history and demographic drivers of reservoir competence for three tick-borne zoonotic pathogens. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caron A. Bridge hosts, a missing link for disease ecology in multi-host systems. Vet. Res. 2015;46:83. doi: 10.1186/s13567-015-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rimbach R. Brown spider monkeys (Ateles hybridus): a model for differentiating the role of social networks and physical contact on parasite transmission dynamics. Philos. Trans R. Soc. Lond. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2014.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langwig K.E. Host and pathogen ecology drive the seasonal dynamics of a fungal disease, white-nose syndrome. Proc. Biol. Sci. 2015;282 doi: 10.1098/rspb.2014.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson P.T. Why infectious disease research needs community ecology. Science. 2015;349 doi: 10.1126/science.1259504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandes R.S. Culex quinquefasciatus from Rio de Janeiro is not competent to transmit the local Zika virus. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostfeld R.S. Tick-borne disease risk in a forest food web. Ecology. 2018;99:1562–1573. doi: 10.1002/ecy.2386. [DOI] [PubMed] [Google Scholar]
- 44.Young H.S. Context-dependent effects of large-wildlife declines on small-mammal communities in central Kenya. Ecol. Appl. 2015;25:348–360. doi: 10.1890/14-0995.1. [DOI] [PubMed] [Google Scholar]
- 45.Luis A.D. Species diversity concurrently dilutes and amplifies transmission in a zoonotic host–pathogen system through competing mechanisms. Proc. Natl. Acad. Sci. U. S. A. 2018;115:7979–7984. doi: 10.1073/pnas.1807106115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehmer E.M. Evaluating the impacts of coinfection on immune system function of the deer mouse (Peromyscus maniculatus) using Sin Nombre virus and Bartonella as model pathogen systems. J. Wildl. Dis. 2018;54:66–75. doi: 10.7589/2017-01-015. [DOI] [PubMed] [Google Scholar]
- 47.Levin M. Effects of homologous and heterologous immunization on the reservoir competence of domestic dogs for Rickettsia conorii (israelensis) Ticks Tick Borne Dis. 2014;5:33–40. doi: 10.1016/j.ttbdis.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Epstein J. Nipah virus ecology and infection dynamics in its bat reservoir, Pteropus medius, in Bangladesh. Int. J. Infect. Dis. 2016;53:20–21. [Google Scholar]
- 49.Soares M.P. Disease tolerance and immunity in host protection against infection. Nat. Rev. Immunol. 2017;17:83. doi: 10.1038/nri.2016.136. [DOI] [PubMed] [Google Scholar]
- 50.Casadevall A., Pirofski L.-A. What is a host? Incorporating the microbiota into the damage-response framework. Infect. Immun. 2015;83:2–7. doi: 10.1128/IAI.02627-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pirofski L.-A., Casadevall A. What is infectiveness and how is it involved in infection and immunity? BMC Immunol. 2015;16:13. doi: 10.1186/s12865-015-0076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levinson J. Targeting surveillance for zoonotic virus discovery. Emerg. Infect. Dis. 2013;19:743. doi: 10.3201/eid1905.121042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rasmussen A.L. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nedelec Y. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 2016;167:657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 55.Fourcade Y. Paintings predict the distribution of species, or the challenge of selecting environmental predictors and evaluation statistics. Glob. Ecol. Biogeogr. 2018;27:245–256. [Google Scholar]
- 56.Cizauskas C.A. Parasite vulnerability to climate change: an evidence-based functional trait approach. R. Soc. Open Sci. 2017;4:160535. doi: 10.1098/rsos.160535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thibeaux R. Seeking the environmental source of leptospirosis reveals durable bacterial viability in river soils. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McElhinney L.M. Effects of carcase decomposition on rabies virus infectivity and detection. J. Virol. Methods. 2014;207:110–113. doi: 10.1016/j.jviromet.2014.06.024. [DOI] [PubMed] [Google Scholar]
- 59.Pandey A. Strategies for containing Ebola in West Africa. Science. 2014;346:991–995. doi: 10.1126/science.1260612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dicko A.H. Using species distribution models to optimize vector control in the framework of the tsetse eradication campaign in Senegal. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10149–10154. doi: 10.1073/pnas.1407773111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viana M. Assembling evidence for identifying reservoirs of infection. Trends Ecol. Evol. 2014;29:270–279. doi: 10.1016/j.tree.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han B.A. Undiscovered bat hosts of filoviruses. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han B.A. Rodent reservoirs of future zoonotic diseases. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7039–7044. doi: 10.1073/pnas.1501598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson R.P. A framework for using niche models to estimate impacts of climate change on species distributions. Ann. N. Y. Acad. Sci. 2013;1297:8–28. doi: 10.1111/nyas.12264. [DOI] [PubMed] [Google Scholar]
- 65.Bruchfeld J. Tuberculosis and HIV coinfection. Cold Spring Harb. Perspect. Med. 2015;5 doi: 10.1101/cshperspect.a017871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J.Y. HCV and HIV co-infection: mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2014;11:362. doi: 10.1038/nrgastro.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Susi H. Co-infection alters population dynamics of infectious disease. Nat. Commun. 2015;6:5975. doi: 10.1038/ncomms6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cota G.F. Leishmania–HIV co-infection: clinical presentation and outcomes in an urban area in Brazil. PLoS Negl. Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Escobar L.E., Craft M.E. Advances and limitations of disease biogeography using ecological niche modeling. Front. Microbiol. 2016;7:1174. doi: 10.3389/fmicb.2016.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Young H.S. Drivers of intensity and prevalence of flea parasitism on small mammals in East African savanna ecosystems. J. Parasitol. 2015;101:327–335. doi: 10.1645/14-684.1. [DOI] [PubMed] [Google Scholar]
- 72.Hoyt J.R. Widespread bat white-nose syndrome fungus, northeastern China. Emerg. Infect. Dis. 2016;22:140. doi: 10.3201/eid2201.151314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Millán J. Patterns of exposure of Iberian wolves (Canis lupus) to canine viruses in human-dominated landscapes. Ecohealth. 2016;13:123–134. doi: 10.1007/s10393-015-1074-8. [DOI] [PubMed] [Google Scholar]
- 74.Huang S. Phylogenetically related and ecologically similar carnivores harbour similar parasite assemblages. J. Anim. Ecol. 2014;83:671–680. doi: 10.1111/1365-2656.12160. [DOI] [PubMed] [Google Scholar]
- 75.Clark N.J. Climate, host phylogeny and the connectivity of host communities govern regional parasite assembly. Divers. Distrib. 2018;24:13–23. [Google Scholar]
- 76.Krasnov B.R. Trait-based and phylogenetic associations between parasites and their hosts: a case study with small mammals and fleas in the Palearctic. Oikos. 2016;125:29–38. [Google Scholar]
- 77.Ibarra-Cerdeña C.N. Phylogeny and niche conservatism in North and Central American triatomine bugs (Hemiptera: Reduviidae: Triatominae), vectors of Chagas’ disease. PLoS Negl. Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fountain-Jones N.M. Towards an eco-phylogenetic framework for infectious disease ecology. Biol. Rev. 2018;93:950–970. doi: 10.1111/brv.12380. [DOI] [PubMed] [Google Scholar]
- 79.Mier-y-Teran-Romero L. Mosquitoes on a plane: disinsection will not stop the spread of vector-borne pathogens, a simulation study. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith K. Summarizing US wildlife trade with an eye toward assessing the risk of infectious disease introduction. Ecohealth. 2017;14:29–39. doi: 10.1007/s10393-017-1211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stephens P.R. Global mammal parasite database version 2.0. Ecology. 2017;98:1476. doi: 10.1002/ecy.1799. [DOI] [PubMed] [Google Scholar]
- 82.Malaria Atlas Project . 2018. Malaria Atlas Project. https://map.ox.ac.uk/ [Google Scholar]
- 83.Salguero-Gómez R. COMADRE: a global database of animal demography. J. Anim. Ecol. 2016;85:371–384. doi: 10.1111/1365-2656.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Global Biodiversity Information Facility . 2018. Global Biodiversity Information Facility. https://www.gbif.org/ [Google Scholar]
- 85.Hosseini P.R. Does the impact of biodiversity differ between emerging and endemic pathogens? The need to separate the concepts of hazard and risk. Philos. Trans R. Soc. Lond. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2016.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kinung’hi S.M. Coinfection of intestinal schistosomiasis and malaria and association with haemoglobin levels and nutritional status in school children in Mara region, northwestern Tanzania: a cross-sectional exploratory study. BMC Res. Notes. 2017;10:583. doi: 10.1186/s13104-017-2904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qi X. The effects of socioeconomic and environmental factors on the incidence of dengue fever in the Pearl River Delta, China, 2013. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arthur R.F. Contact structure, mobility, environmental impact and behaviour: the importance of social forces to infectious disease dynamics and disease ecology. Philos. Trans R. Soc. Lond. B Biol. Sci. 2017;372 doi: 10.1098/rstb.2016.0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vega G.C. MERRAclim, a high-resolution global dataset of remotely sensed bioclimatic variables for ecological modelling. Sci. Data. 2017;4:170078. doi: 10.1038/sdata.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Messina J.P. A global compendium of human dengue virus occurrence. Sci. Data. 2014;1:140004. doi: 10.1038/sdata.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Global Biodiversity Information Facility . 2018. Xenopus laevis. [DOI] [Google Scholar]
- 92.Global Biodiversity Information Facility . 2018. Batrachochytrium dendrobatidis. [DOI] [Google Scholar]
- 93.Global Bd-Mapping Project . 2018. Global Bd Database. http://www.bd-maps.net. [Google Scholar]
- 94.PhyloPic . 2018. PhyloPic. http://www.phylopic.org. [Google Scholar]