Highlights
-
•
Bats are important reservoir hosts for emerging zoonotic viruses.
-
•
Viruses detected in bats are difficult to isolate using traditional cell lines.
-
•
Bat cell lines provide critical tools to dissect host pathogen interactions.
-
•
Little is known about immune cell populations and their responses in bats.
-
•
Sharing reagents and cell lines will accelerate research and virus discovery.
Keywords: Bats, Chiroptera, Viral reservoirs, Virus isolation, Cell lines, Host-pathogen interaction
Abstract
Bats are natural reservoirs for a variety of emerging viruses that cause significant disease in humans and domestic animals yet rarely cause clinical disease in bats. The co-evolutionary history of bats with viruses has been hypothesized to have shaped the bat-virus relationship, allowing both to exist in equilibrium. Progress in understanding bat-virus interactions and the isolation of bat-borne viruses has been accelerated in recent years by the development of susceptible bat cell lines. Viral sequences similar to severe acute respiratory syndrome corona virus (SARS-CoV) have been detected in bats, and filoviruses such as Marburg virus have been isolated from bats, providing definitive evidence for the role of bats as the natural host reservoir. Although viruses can be readily detected in bats using molecular approaches, virus isolation is far more challenging. One of the limitations in using traditional culture systems from non-reservoir species is that cell types and culture conditions may not be compatible for isolation of bat-borne viruses. There is, therefore, a need to develop additional bat cell lines that correspond to different cell types, including less represented cell types such as immune cells, and culture them under more physiologically relevant conditions to study virus host interactions and for virus isolation. In this review, we highlight the current progress in understanding bat-virus interactions in bat cell line systems and some of the challenges and limitations associated with cell lines. Future directions to address some of these challenges to better understand host-pathogen interactions in these intriguing mammals are also discussed, not only in relation to viruses but also other pathogens carried by bats including bacteria and fungi.
1. Introduction
Bats are an ancient and diverse group of mammals, comprising almost a quarter of all mammalian diversity and inhabiting all continents, other than Antarctica. Phylogenetic analyses based on molecular data classified bats into the suborders Yinpterochiroptera and Yangochiroptera (Teeling et al., 2016; Teeling et al., 2005). The Yinpterochiroptera suborder includes the non echolocating Pteropodidae family and the echolocating Rhinolophoidea family. The Yangochiroptera consist of the remaining echolocating microbat families. The two suborders diverged over 50 million years ago (Lei and Dong, 2016; O'Leary et al., 2013; Simmons et al., 2008). Members of both suborders serve as reservoir or suspected reservoir hosts for a number of zoonotic viruses that are highly pathogenic in humans (Calisher et al., 2006; Moratelli and Calisher, 2015), yet cause no clinical disease in bats that are naturally or experimentally infected. The long co-evolutionary history of bats and their viruses has been speculated to have shaped the unique host-pathogen relationship (Schountz et al., 2017).
Although bats have been linked with a number of viruses, identifying wildlife reservoirs can be a challenging process, requiring extensive ecological sampling. Evidence that a pathogen is permanently maintained in a population, often in the absence of disease, is required to confidently identify its reservoir (Calisher et al., 2006; Haydon et al., 2002). Detection of fragments of viral genomes, combined with serological evidence for viral exposure in multiple individuals of a species are generally the first lines of evidence to associate a host with a pathogen (Halpin et al., 2011; Li et al., 2005; Swanepoel et al., 2007; Young et al., 1996). Additional evidence in the form of virus isolation and the ability to link a reservoir to a spillover event is necessary to confirm the association. Examples of bat viruses that are confirmed or speculated to have ‘jumped’ from bats into humans and agricultural animals include coronaviruses (severe acute respiratory syndrome coronavirus [SARS-CoV], porcine epidemic diarrhea [PEDV] virus), filoviruses (ebolaviruses and Marburg virus), and henipaviruses (Nipah and Hendra viruses) [reviewed by (Moratelli and Calisher, 2015)]. Consistent with the definition of a wildlife reservoir, infection of bats by some of these viruses results in viremia and viral shedding in the absence of clinical disease (Halpin et al., 2011; Schuh et al., 2017). The spillover of some of these viruses to other susceptible hosts, including humans, can result in severe disease and mortality due to the lack of therapeutics and vaccines.
The emergence of zoonotic diseases is increasing globally and mammals, including bats, are major sources of emerging and re-emerging pathogens (Plowright et al., 2015). The drivers of disease emergence include anthropogenic changes to the environment (e.g., agricultural intensification), climate change and the encroachment of human populations into wildlife habitats (Tait et al., 2014). The biological characterization of a newly identified virus often begins with its isolation in cultured cells. Although detection of virus-specific antibodies by serology and the use of molecular methods, such as viral sequencing, are often readily achievable, virus isolation from bats has been far more challenging. For example, bats are implicated as the reservoirs of ebolaviruses based on the detection of viral genome, serological evidence and their confirmed role as a reservoir for the closely related Marburg virus. However, ebolaviruses have yet to be isolated from bats to definitively link them to spillover events (Leroy et al., 2005). Identifying wildlife reservoirs and the viruses they host will require the development of better laboratory tools for virus isolation to study replication kinetics and virus-host interactions. In this perspective, we discuss the importance of reservoir species specific cell lines as tools to study emerging wildlife pathogens with a focus on bats.
2. Current status of cell lines derived from bats
Virus isolation is generally attempted in established immortalized cell lines, such as the type I interferon deficient BHK-21 (baby Syrian hamster kidney cells) (Otsuki et al., 1979) or Vero E6 cell lines (African green monkey kidney cells) (Govorkova et al., 1996). Different primary cells from bats have been studied and some immortalized cell lines have also been generated (Table 1 ). Established cell lines from non-reservoir host species are convenient tools for virus isolation but do not always support propagation or isolation of viruses from wildlife. For example, a bat herpesvirus isolated from cell lines established from the Schreiber’s long-fingered bat, Miniopterus schreibersii, failed to replicate in cells from a variety of other species, including cells from the black flying fox, Pteropus alecto (Zhang et al., 2012). The henipavirus Cedar virus was isolated from primary cells of its putative natural reservoir, P. alecto whereas other cell lines showed no visible cytopathic effect (Marsh et al., 2012). Similarly, the paramyxovirus Menangle virus was isolated using the same P. alecto kidney cell line (Barr et al., 2012).
Table 1.
Some cell types established from bats and range of viruses that have been used in studies with these cells.
| Bat primary cells or cell lines | Origin | Virus/pseudovirus used in the study | Reference |
|---|---|---|---|
| Artibeus jamaicensis primary cells | Embryonic, primary kidney cells | Dengue virus, MERS-CoV | Moreira-Soto et al. (2017); Munster et al. (2016) |
| Carollia perspicillata. | Trachea | Vesicular stomatitis virus (VSV) and Rift-Valley fever virus | Eckerle et al. (2014) |
| Carollia perspicillata – CpKd | Kidney | Bat-associated influenza viruses | Hoffmann et al. (2016) |
| Eidolon helvum | Trachea | VSV and Rift-Valley fever virus | Eckerle et al. (2014) |
| Eidolon helvum – EidNi/41 | Kidney | Bat-associated influenza viruses, Ebolavirus, MERS-CoV, Influenza A virus, O'nyong–nyong virus | Hoffmann et al. (2016), Ng et al. (2015); Cai et al. (2014); Poole et al. (2014); Biesold et al. (2011) |
| Epomops buettikoferi – EpoNi/22.1 | Kidney | Bat-associated influenza viruses, Influenza A virus | Hoffmann et al. (2016); Poole et al. (2014) |
| Eptesicus fuscus – Efk3 | kidney | VSV, porcine epidemic diarrhea virus, herpes simplex virus and MERS-CoV | Banerjee et al. (2016) |
| Hypsignathus monstrosus – hypLu/45.1 | Lung | MERS-CoV, Influenza A virus | Cai et al. (2014); Poole et al. (2014) |
| Hypsignathus monstrosus – hypNi/1.1 | Kidney | Bat-associated influenza viruses, MERS-CoV, Influenza A virus, African Henipavirus | Hoffmann et al. (2016); Cai et al. (2014); Poole et al. (2014);. Kruger et al., (2013) |
| Miniopterus schreibersii primary cells | Lymph node (MsLn), Kidney (MsKi) | Miniopterus schreibersii herpesvirus (MsHV) | Zhang et al. (2012) |
| Myotis daubentonii – MyDauLu/47.1 | Lung | Influenza A virus | Poole et al. (2014) |
| Myotis davidii – MdKi | Kidney | Sendai virus | Liang et al. (2015) |
| Myotis myotis | brain (MmBr), tonsil (MmTo), peritoneal cavity (MmPca), nasal epithelium (MmNep) and nervus olfactorius (MmNol) | Rabies virus, European bat lyssavirus (EBLV – 1 and 2) | He et al. (2014) |
| Myotis velifer incautus (MVI-it) – CRL-6012 | intercapsular tumor | Novel bat gammaherpesvirus | Shabman et al. (2016); Host and Damania (2016) |
| Pipistrellus ceylonicus | Embryonic | Chandipura virus (CHPV), novel adenovirus (BtAdv-RLM) | Mourya et al. (2013) |
| Pipistrellus subflavus – ESU-BSL | Lung | MERS-CoV | Cai et al. (2014) |
| Pteropus alecto | Aorta, bone marrow, brain, foetus, foetal membranes, heart, kidney, liver, lymph nodes, lung, muscle, pharynx, placenta, salivary gland, small intestine, skin, spleen, testes, thymus, uterus | Hendra virus, Pulau virus, Sendai virus, Nipah virus, Hendra virus | Zhou et al. (2013); Crameri et al. (2009); Virtue et al. (2011) |
| Pteropus alecto | Kidney | Influenza A virus, Sendai virus, Pteropine orthoreovirus NB, Nipah virus, Hendra virus | Dlugolenski et al. (2013); Zhou et al. (2013); Virtue et al. (2011) |
| Pteropus alecto | Embryonic | Nipah virus, Hendra virus | Virtue et al. (2011) |
| Rhinolophus affinis | embryonic | VSV | Li et al., (2015) |
| Rhinolophus sinicus | splenocytes | VSV | Li et al., (2015) |
| Rousettus aegyptiacus – RO6E, RO5T | Kidney | MERS-CoV, Influenza A virus, Ebola virus, Marburg virus | Cai et al. (2014); Poole et al. (2014);. Krahling et al. (2010) |
| Rousettus aegyptiacus – RoNi/7.1 | Kidney | Bat-associated influenza viruses, MERS-CoV, Influenza A virus | Hoffmann et al. (2016); Cai et al. (2014); Poole et al. (2014) |
| Tadaria brasiliensis – Tb1Lu | Lung | Bovine leukemia virus, Influenza A virus, mammalian reovirus serotype 3 Dearing | Graves and Ferrer (1976); Poole et al. (2014);. Sandekian et al. (2013) |
| Rousettus aegyptiacus immortalized cells | Foetus | Vaccinia Ankara | Jordan et al. (2009) |
Cell lines have defined characteristics based on their tissue of origin and method of immortalization. For a cell to be suitable for virus propagation, it must bear an appropriate receptor to bind with an incoming virus and it must be permissive. Because viruses differ in their cellular tropisms, working with a few cell lines with epithelial or fibroblastic characteristics limits our ability to isolate or study viruses that may have a tropism towards cell types that are more difficult to immortalize. Current cell lines may lack receptors required by viruses to replicate when cells are infected at low multiplicity of infections. For example, bat cells from various species were resistant to infection with vesicular stomatitis virus (VSV) pseudotypes bearing the SARS-CoV spike protein until the cells were transfected to express the human SARS-CoV receptor, angiotensin-converting enzyme 2 (ACE2) (Hoffmann et al., 2013). Although the lack of appropriate receptors on cell lines makes it difficult to isolate some viruses, this lack of susceptibility to infection can also help us understand the early events in virus infection. For instance, differences in the susceptibility of cells from a variety of bats to filoviruses also led to the identification of unique amino acid substitutions in the filovirus receptor, Neimann-Pick type C1 (NPC1), providing insight into the host range of ebolaviruses (Ng et al., 2015). Similarly, Widagdo et al. have shown that dipeptidyl peptidase 4 (DPP4, CD26), a cellular receptor for MERS-CoV, is expressed in specific cell types in different tissues which also varies between bat species (Widagdo et al., 2017). These studies demonstrate the need to develop cell lines from different organs and bat species and clone them for a variety of cell types as viral receptors may not be equally distributed.
3. Considerations for development of cell lines
Chiroptera consists of over 1200 bat species and different species may have co-evolved with different viruses. Viruses detected in one species, may not infect cells established from a different bat species. Thus, the choice of bat species for generating cell lines capable of supporting a particular virus should be supported by evidence that the species is susceptible to that virus. For example, SARS-CoV-like coronaviruses have been detected in Rhinolophus sinicus (Lau et al., 2005), and this species is the logical candidate for generating cell lines that can support replication and isolation of these viruses.
Because propagation of viruses in cell lines can select for members of the virus population that replicate most efficiently in the chosen cell type, the choice of cell line should be taken into consideration. Propagation of viruses in cell lines other than those of the natural host may lead to accumulation of adaptive mutations or attenuate the virus (Combe and Sanjuan, 2014), which in turn has implications for assessing host-pathogen interactions either in vivo or in vitro. For example, although wild type Marburg virus causes no disease in mice, it can be adapted to mice by serial passage to cause lethal disease (Qiu et al., 2014). Deep sequencing of Marburg virus after adaptation to mice and following propagation in Vero E6 cells has revealed the sequential mutations associated with each passage as the virus adapts to a new host either in vivo or in vitro (Wei et al., 2017). This is also thought to have occurred with Tacaribe virus, which was isolated from artibeus bats and passaged 20 times in newborn mice, resulting in point mutations and deletions (Malmlov et al., 2017; Sayler et al., 2014).
Bats experience a range of body temperatures during different physiological activities (from 4 to 8 °C during hibernation to 41 °C during flight) (O'Shea et al., 2014). Thus, viruses isolated from bats should be studied at different relevant physiological temperatures. Propagation of viruses at temperatures other than the nominal of the reservoir species may also lead to the generation of attenuation mutants; a strategy that has been used for vaccine development for decades. There is evidence that arthropod viruses found in mosquitoes and ticks accumulate mutations when grown at different temperatures (Lemm et al., 1990). Thus, propagation of viruses in susceptible cell lines [reviewed by (Hare et al., 2016)] and at optimized temperatures may minimize mutations and produce viruses similar to wild type virus found in wildlife reservoirs or arthropod vectors [reviewed by (Prescott et al., 2017)].
Controlling for temperature to match temperatures experienced by bats during hibernation is a challenge because in vitro cell culture systems are not viable at low temperatures. Although Miller et al. were able to grow bat cells at temperatures ranging from 37 to 41 °C (Miller et al., 2016), our attempts to cultivate big brown bat cells (Efk3) at lower temperatures of 26–30 °C were not successful (Banerjee and Misra, unpublished). There is evidence that experimental infection of different reptiles and amphibians with chikungunya virus produce varying amounts of viremia depending on the host’s body temperature (Hartwig et al., 2015). There is a need to develop a system to test the replication potential of bat-borne viruses at lower temperatures such as those experienced by bats during torpor and hibernation, rather than only propagating them in cells cultured at 37 °C.
The virus stock that is used to experimentally infect bats is usually generated by propagation in cells of non-bat origin, such as Vero E6 cells. It is unknown whether propagation of a virus in a monkey cell line alters the genotype of the virus. We have previously infected bats with a virus isolated from bat urine and subsequently propagated in bat cells (Baker et al., unpublished). A problem with propagating this virus in bat cells was achieving high virus titre to perform experimental infection studies in bats, perhaps because bat cells appear to have constitutive type I IFN activation (Zhou et al., 2016b). It may be necessary to develop interferon-deficient bat cell lines to propagate bat viruses. Recently, Zhang et al. knocked out an antiviral cell signaling receptor, interferon alpha receptor 2 (IFNAR2) in bat cells using CRISPR/Cas9 technology (Zhang et al., 2017). Similarly, this technology can be used to generate interferon-deficient bat cells in culture.
There are several challenges to establishing primary cells from wildlife species. These challenges range from obtaining healthy tissues to determining the ideal cell types. Primary cells maintain genetic integrity and are therefore most appropriate for the study of natural infection of cells. However, primary cells have finite numbers of cell division before undergoing senescence, thus, it can become problematic for long-term studies. Immortalized cells frequently accumulate chromosomal aberrations over time; therefore, caution must be exercised when maintaining cell stocks. Nonetheless, they are convenient and often receptive to genetic manipulations, including transfection of plasmids and targeted changes (e.g., CRISPR), and often the only cells available that are capable of supporting replication. Thus, the ability to conduct experiments in primary or immortalized cells has advantages and disadvantages.
Primary cells can also harbor latent viruses that can become reactivated during in vitro cultivation when the cells are outside the host and isolated from other components of the immune system that would otherwise control virus replication. Most mammals, including bats harbor persistent viruses [reviewed by (Calisher et al., 2006)], and viruses have been isolated or identified by next generation sequencing (NGS) from bat cell lines (Shabman et al., 2016). Cell lines from any species of bat should therefore only be handled under controlled biosafety conditions and thoroughly tested before being used for experimental studies. This is also a limitation when working with wild-caught bats.
4. Using cell lines to assess the immune response in bats to viral infections
Research in the field of bat immunology steadily increased after bats were implicated as ancestral reservoirs of the coronavirus that caused the SARS outbreak in 2003 (Baker et al., 2013; Lau et al., 2005; Li et al., 2005). Over 200 viruses have been identified in bats and additional new viruses are discovered on a regular basis (Moratelli and Calisher, 2015; Schountz, 2014). Few viruses have been documented to cause disease in bats in addition to rabies and related lyssaviruses (Davis et al., 2012; Davis et al., 2013; McColl et al., 2002). Experimental inoculation of Jamaican fruit bats with high doses of Tacaribe virus caused significant morbidity and mortality similar to natural infection of artibeus bats (Downs et al., 1963). However, the inoculated bats did not transmit the virus. Based on their observations, Cogswell-Hawkinson et al. speculate that Jamaican fruit bats may not be a reservoir host for Tacaribe virus (Cogswell-Hawkinson et al., 2012). Lloviu virus, a filovirus was isolated from dead bats in Spain (Negredo et al., 2011). These examples appear to be exceptions to the usual resistance to viral disease displayed by most bats. Bats have been hypothesized to have evolved unique aspects of their immune systems, such as a strict control of the inflammatory process (Banerjee et al., 2017) and constitutive expression of antiviral interferons (Zhou et al., 2016b) to better survive infections with these viruses. Testing this hypothesis has been possible, in part through the establishment of well characterized cell lines (Schountz et al., 2017).
Cell lines from several bat species including members of both the Yinpterochiroptera (Pteropus alecto, Eidolon helvum and Rousettus aegyptiacus) and Yangochiroptera (Eptesicus fuscus, and Myotis myotis) suborders, have been established (Banerjee et al., 2016; Biesold et al., 2011; Crameri et al., 2009; He et al., 2014; Jordan et al., 2012) (Table 1). These cell lines have been used to cultivate viruses that are speculated or confirmed to have spilled over from bats. These cell lines have also allowed researchers to study the innate immune responses to viruses. Cytokines produced in response to activation of selected innate immune response pathways in bat cells have led to a better understanding of how bats might respond to viral infections in the wild. Bat cells can respond to viruses or viral antigens through the production of antiviral interferons and interferon stimulated genes (Biesold et al., 2011; Omatsu et al., 2008; Zhou et al., 2011; Zhou et al., 2013). The use of bat cell lines has also led to the discovery of viral proteins that counteract the innate immune response in these cells (Virtue et al., 2011). Less work has been performed on the adaptive immune responses of bats using cell lines, in part due to the lack of research on bat T and B cells. However, a recent report used P. alecto cell lines to examine the repertoire of self and Hendra virus epitopes presented by MHC class I molecules (Wynne et al., 2016). More research is needed to establish in vitro and ex vivo cultures of immune cells from representative bat species.
5. Developing bat specific reagents
Establishing cell lines from different species of bats provides the opportunity to perform comparative studies between bat species and with other mammals. Several cell lines have been developed from bats, including some that support the replication of viruses from different families (Table 1). However, to better characterize bat immune responses, we need to develop cell lines or a system where we can continuously establish immune cells in vitro. Current wildlife cell lines are composed primarily of epithelial or fibroblast cell types, which limit the responses generated following infection. These cells may lack many of the receptors and cytokine responses of specialized immune cells. Isolating and developing representative immune cell lines will allow the study of both adaptive and innate immune responses of bats. This also involves developing species and cell-type specific reagents that will allow characterization and culture of immune cells in vitro. Recently, immune cells from black flying foxes have been functionally and phenotypically characterized (Martinez Gomez et al., 2016), along with the establishment of bone-marrow derived dendritic cells and macrophages using black flying fox reagents developed in the laboratory (Zhou et al., 2016a). Systems such as these will further allow exploration of the adaptive immune response in potential reservoir bat species. More efforts are required to generate similar cell types in other bat species. Sharing of resources will play a key role in expanding our understanding of the unique immune response in bats.
Although primary cells are good tools to attempt virus isolation, they have a limited life-span. There are several technologies that can be used to immortalize these primary cells. Immortalized cell lines replicate indefinitely in culture and provide researchers with valuable tools to study viruses and cell-virus interactions. Some of these bat cell lines have been established using existing technology such as simian virus 40 large T-antigen and human hTERT expression systems (Ali and DeCaprio, 2001; He et al., 2009; Xie et al., 2015), whereas others have been generated by using novel technologies such as a bat polyomavirus large T-antigen to immortalize cells (Banerjee et al., 2016) and other bat specific reagents to generate dendritic cells and macrophages (Zhou et al., 2016a). Polyomavirus large T-antigen binds to and attenuates the tumor suppressor protein p53 and proteins of the retinoblastoma tumor suppressor family (pRb, p130 and p107) promoting cell cycle progression. Gamma-herpesviruses have also been used to immortalize cells of the immune system, such as B lymphocytes (Liang et al., 2011). These novel technologies or combination of cell immortalization techniques can be modified for the generation of cell lines from representative cell and tissue types from less studied species of bats and other neglected wildlife reservoirs.
Studying the host-pathogen relationship in wildlife species is often challenging, with in vivo experiments difficult and costly to perform. Developing cell lines will allow preliminary research, which can later be validated in vivo in bats. These cell lines can be used to study or isolate viruses that spillover from animal reservoirs to humans, and pathogens that cause disease in wildlife. The advantages of developing cell lines do not end at studying the immune responses and isolating viruses. These cell lines can be used to propagate isolated viruses to high titre for the development of diagnostic kits and potential vaccines. Studying pathogen-host interactions may also allow us to identify biomarkers associated with viral infection that could be used to predict and prevent spillover to other susceptible hosts. For example, biochemical or hormonal changes linked to increases in viral replication in wild bat populations may be used to predict periods of increased risk of spillover to other susceptible species.
6. Using cell lines to study host responses to pathogens that cause disease in bats
In addition to viruses, bats may also be reservoirs, or even dead-end hosts, for other pathogens such as bacteria (Banskar et al., 2016), fungi or parasites. Bats do not seem to be refractory to disease when it comes to extracellular pathogens such as fungi (Zukal et al., 2014). The white-nose syndrome (WNS) fungus, Pseudogymnoascus destructans, causes significant wing damage at the sites of infection in little brown bats (Myotis lucifugus) (Warnecke et al., 2012; Zukal et al., 2014). Big brown bats (Eptesicus fuscus) seem to be relatively resistant to the fungus (Frank et al., 2014). Identifying, isolating and characterizing the interaction of extracellular and intracellular pathogens with their host can be better facilitated by the availability of bat species specific cell lines.
Research to understand bacterial and fungal infections in bats is growing and has led to the identification of potential pathogenic bacterial species in bats (Avena et al., 2016; Banskar et al., 2016). Cell lines, as previously shown (Nepelska et al., 2017), can be effective tools to dissect the role of these organisms in the overall health of bats. Transcriptomic data from bat cells infected with various pathogens, including bacteria and viruses, will shed light on global pathways that are activated or suppressed in response to these pathogens. These data can be used to develop hypotheses on the overall impact these pathogens might have on bats at an organism level. Recently, a novel Streptomyces species with anti-fungal activity was identified in bats (Hamm et al., 2017). WNS has caused the death of millions of little brown bats in North America (Alves et al., 2014; Knudsen et al., 2013; Zukal et al., 2014) and this new compound amongst others (Boire et al., 2016) might help to curb the spread of WNS. Potential toxic effects of this drug can be tested in uninfected bat cell lines before testing them in live bats or other relevant animal models.
7. Developing in vivo bat models
In addition to cell lines, there is a need for laboratory bred bats with known and tested infection status for ex vivo and in vivo experiments. Establishing a wild-caught bat colony is challenging but achievable with sufficient resources to house and maintain the bats. Although studies using wild-caught outbred bats potentially provide the best representation of what occurs naturally during virus infection, it can be difficult to obtain reproducible data using wild caught animals due to their high genetic diversity. The current infection status and history of viral infections in wild caught bats is also difficult to determine which in turn may affect the outcome and interpretation of experimental infections. There is therefore a need to establish inbred laboratory colonies of bats of known infection status. As much of the knowledge that has been obtained regarding the host responses of bats has been gained from studies using bat cell lines, experimental infections of captive bats provide an opportunity to test whether observations made in vitro also apply in vivo. Studies in captive animals are also necessary to confirm the role of bats as ‘reservoirs’ by carrying out long-term experiments to detect virus shedding and the lack of associated pathology in bats with known infection status (i.e. laboratory bred bats). This was recently attempted with Egyptian rousette bats that were infected with Marburg virus. The bats shed virus between 5 and 19 days post infection (Schuh et al., 2017), but longer term studies might be necessary to further establish bats as reservoirs. Thus there is a need to develop more reliable animal models with known infection status.
8. Conclusion
Although several different cell lines have been established from different species of bats (Table 1), we are still faced with the challenge of isolating several viruses that have been detected in bats. We have detected little brown bat coronavirus sequences (Misra et al., 2009; Subudhi et al., 2017) and others have detected ebolavirus sequences [reviewed by (Olival and Hayman, 2014)] in bats by polymerase chain reaction (PCR), next generation sequencing (NGS) and serology. As discussed, virus isolation is more challenging than nucleic acid detection but with the availability of additional cell lines from various organs and species and technologies such as CRISPR to develop cell lines deficient in components of the immune system, attempts to isolate and propagate viruses in vitro should be more successful.
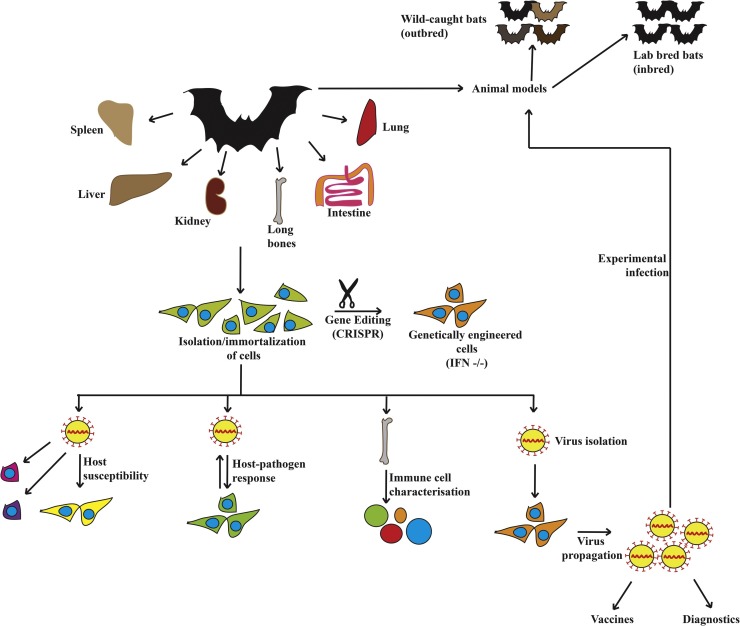
Chiroptera consists of over 1200 species and much remains unknown about their relationship with their microorganisms. Bats are genetically diverse and results from studies done in one bat species may not represent other species of bats. Thus, there is a need to study other species of bats to gain deeper insights into bat-microbe co-evolution, bat immune system and ecological interactions of bats. Bats host more zoonotic viruses per mammalian species (Luis et al., 2013; Olival et al., 2017), even more than rodents, the largest group of mammals. Collaboration between laboratories will be required to develop cell lines from different bat tissues, bat specific reagents and to establish laboratory bat colonies to explore the length and breadth of microbe-host interactions in these ecologically important mammals (Fig. 1 ).
Fig. 1.
There is a need to develop more model cell lines and animal models to decipher the immune responses of bats to viruses. Viruses display tropism towards different cell types and there is a need to generate cell lines representative of the multitude of cell types found in bats to increase the chances of isolating the viruses that they carry. Alternatively, genetically modified cell lines can be generated that do not express antiviral interferons from these cell types for virus isolation. These cells may be used to propagate bat-borne viruses to high titres for ex vivo, in vitro and in vivo experiments. Generating cell lines from a variety of different tissues and bats will allow researchers to explore virus susceptibility and virus-host interactions in these intriguing mammals. Generation of outbred (wild-caught) and inbred (laboratory bred) bat colonies with known infection status will enable systemic immune response studies with relevant viruses isolated from bats. The ability to resist infections with some of these viruses may be an effect of several evolutionary adaptations, which can be better dissected by studying the systemic responses in bats.
Conflicts of interest
None.
Acknowledgements
AB is funded through a Saskatchewan Innovation and Opportunity scholarship and the Department of Veterinary Microbiology at the University of Saskatchewan, Canada. VM is funded through a Discovery Grant awarded by the Natural Sciences and Engineering Research Council of Canada (NSERC).
References
- Ali S.H., DeCaprio J.A. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 2001;11(1):15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
- Alves D.M., Terribile L.C., Brito D. The potential impact of white-nose syndrome on the conservation status of north American bats. PLoS One. 2014;9(9):e107395. doi: 10.1371/journal.pone.0107395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avena C.V., Parfrey L.W., Leff J.W., Archer H.M., Frick W.F., Langwig K.E., Kilpatrick A.M., Powers K.E., Foster J.T., McKenzie V.J. Deconstructing the bat skin microbiome: influences of the host and the environment. Front. Microbiol. 2016;7:1753. doi: 10.3389/fmicb.2016.01753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.L., Schountz T., Wang L.F. Antiviral immune responses of bats: a review. Zoonoses Public Health. 2013;60(1):104–116. doi: 10.1111/j.1863-2378.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Rapin N., Miller M., Griebel P., Zhou Y., Munster V., Misra V. Generation and characterization of Eptesicus fuscus (Big brown bat) kidney cell lines immortalized using the Myotis polyomavirus large T-antigen. J. Virol. Methods. 2016;237:166–173. doi: 10.1016/j.jviromet.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Rapin N., Bollinger T., Misra V. Lack of inflammatory gene expression in bats: a unique role for a transcription repressor. Sci. Rep. 2017;7(1):2232. doi: 10.1038/s41598-017-01513-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banskar S., Bhute S.S., Suryavanshi M.V., Punekar S., Shouche Y.S. Microbiome analysis reveals the abundance of bacterial pathogens in Rousettus leschenaultii guano. Sci. Rep. 2016;6:36948. doi: 10.1038/srep36948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J.A., Smith C., Marsh G.A., Field H., Wang L.F. Evidence of bat origin for Menangle virus, a zoonotic paramyxovirus first isolated from diseased pigs. J. Gen. Virol. 2012;93(Pt 12):2590–2594. doi: 10.1099/vir.0.045385-0. [DOI] [PubMed] [Google Scholar]
- Biesold S.E., Ritz D., Gloza-Rausch F., Wollny R., Drexler J.F., Corman V.M., Kalko E.K., Oppong S., Drosten C., Muller M.A. Type I interferon reaction to viral infection in interferon-competent, immortalized cell lines from the African fruit bat Eidolon helvum. PLoS One. 2011;6(11):e28131. doi: 10.1371/journal.pone.0028131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boire N., Zhang S., Khuvis J., Lee R., Rivers J., Crandall P., Keel M.K., Parrish N. Potent inhibition of pseudogymnoascus destructans, the causative agent of white-nose syndrome in bats, by cold-pressed, terpeneless, valencia orange oil. PLoS One. 2016;11(2):e0148473. doi: 10.1371/journal.pone.0148473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Yu S.Q., Postnikova E.N., Mazur S., Bernbaum J.G., Burk R., Zhang T., Radoshitzky S.R., Muller M.A., Jordan I., Bollinger L., Hensley L.E., Jahrling P.B., Kuhn J.H. CD26/DPP4 cell-surface expression in bat cells correlates with bat cell susceptibility to Middle East respiratory syndrome coronavirus (MERS-CoV) infection and evolution of persistent infection. PLoS One. 2014;9(11):e112060. doi: 10.1371/journal.pone.0112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19(3):531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell-Hawkinson A., Bowen R., James S., Gardiner D., Calisher C.H., Adams R., Schountz T. Tacaribe virus causes fatal infection of an ostensible reservoir host, the Jamaican fruit bat. J. Virol. 2012;86(10):5791–5799. doi: 10.1128/JVI.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combe M., Sanjuan R. Variation in RNA virus mutation rates across host cells. PLoS Pathog. 2014;10(1):e1003855. doi: 10.1371/journal.ppat.1003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crameri G., Todd S., Grimley S., McEachern J.A., Marsh G.A., Smith C., Tachedjian M., De Jong C., Virtue E.R., Yu M., Bulach D., Liu J.P., Michalski W.P., Middleton D., Field H.E., Wang L.F. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One. 2009;4(12):e8266. doi: 10.1371/journal.pone.0008266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A., Gordy P., Rudd R., Jarvis J.A., Bowen R.A. Naturally acquired rabies virus infections in wild-caught bats. Vector Borne Zoonotic Dis. 2012;12(1):55–60. doi: 10.1089/vbz.2011.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.D., Jarvis J.A., Pouliott C., Rudd R.J. Rabies virus infection in Eptesicus fuscus bats born in captivity (naive bats) PLoS One. 2013;8(5):e64808. doi: 10.1371/journal.pone.0064808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugolenski D., Jones L., Tompkins S.M., Crameri G., Wang L.F., Tripp R.A. Bat cells from Pteropus alecto are susceptible to influenza A virus infection and reassortment. Influenza Other Respir Viruses. 2013;7(6):900–903. doi: 10.1111/irv.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs W.G., Anderson C.R., Spence L., Aitken T.H., Greenhall A.H. Tacaribe virus, a new agent isolated from Artibeus bats and mosquitoes in Trinidad, West Indies. Am. J. Trop. Med. Hyg. 1963;12:640–646. doi: 10.4269/ajtmh.1963.12.640. [DOI] [PubMed] [Google Scholar]
- Eckerle I., Ehlen L., Kallies R., Wollny R., Corman V.M., Cottontail V.M., Tschapka M., Oppong S., Drosten C., Muller M.A. Bat airway epithelial cells: a novel tool for the study of zoonotic viruses. PLoS One. 2014;9(1):e84679. doi: 10.1371/journal.pone.0084679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C.L., Michalski A., McDonough A.A., Rahimian M., Rudd R.J., Herzog C. The resistance of a north American bat species (Eptesicus fuscus) to white-nose syndrome (WNS) PLoS One. 2014;9(12):e113958. doi: 10.1371/journal.pone.0113958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorkova E.A., Murti G., Meignier B., de Taisne C., Webster R.G. African green monkey kidney (Vero) cells provide an alternative host cell system for influenza A and B viruses. J. Virol. 1996;70(8):5519–5524. doi: 10.1128/jvi.70.8.5519-5524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves D.C., Ferrer J.F. In vitro transmission and propagation of the bovine leukemia virus in monolayer cell cultures. Cancer Res. 1976;36(11 Pt 1):4152–4159. [PubMed] [Google Scholar]
- Halpin K., Hyatt A.D., Fogarty R., Middleton D., Bingham J., Epstein J.H., Rahman S.A., Hughes T., Smith C., Field H.E., Daszak P., Henipavirus Ecology Research, G Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg. 2011;85(5):946–951. doi: 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm P.S., Caimi N.A., Northup D.E., Valdez E.W., Buecher D.C., Dunlap C.A., Labeda D.P., Lueschow S., Porras-Alfaro A. Western bats as a reservoir of novel streptomyces species with antifungal activity. Appl. Environ. Microbiol. 2017;83(5) doi: 10.1128/AEM.03057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D., Collins S., Cuddington B., Mossman K. The importance of physiologically relevant cell lines for studying virus-host interactions. Viruses. 2016;8(11) doi: 10.3390/v8110297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig A., Bosco-Lauth A., Bowen R. Chikungunya virus in non-mammalian species: a possible new reservoir. New Horiz. Transl. Med. 2015;2(4–5):128. [Google Scholar]
- Haydon D.T., Cleaveland S., Taylor L.H., Laurenson M.K. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg. Infect. Dis. 2002;8(12):1468–1473. doi: 10.3201/eid0812.010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.L., Wu Y.H., He X.N., Liu F.J., He X.Y., Zhang Y. An immortalized goat mammary epithelial cell line induced with human telomerase reverse transcriptase (hTERT) gene transfer. Theriogenology. 2009;71(9):1417–1424. doi: 10.1016/j.theriogenology.2009.01.012. [DOI] [PubMed] [Google Scholar]
- He X., Korytar T., Zhu Y., Pikula J., Bandouchova H., Zukal J., Kollner B. Establishment of myotis cell lines – model for investigation of host-pathogen interaction in a natural host for emerging viruses. PLoS One. 2014;9(10):e109795. doi: 10.1371/journal.pone.0109795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Muller M.A., Drexler J.F., Glende J., Erdt M., Gutzkow T., Losemann C., Binger T., Deng H., Schwegmann-Wessels C., Esser K.H., Drosten C., Herrler G. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS One. 2013;8(8):e72942. doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kruger N., Zmora P., Wrensch F., Herrler G., Pohlmann S. The hemagglutinin of bat-associated influenza viruses is activated by TMPRSS2 for pH-dependent entry into bat but not human cells. PLoS One. 2016;11(3):e0152134. doi: 10.1371/journal.pone.0152134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Host K.M., Damania B. Discovery of a novel bat gammaherpesvirus. mSphere. 2016;1(1) doi: 10.1128/mSphere.00016-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I., Horn D., Oehmke S., Leendertz F.H., Sandig V. Cell lines from the Egyptian fruit bat are permissive for modified vaccinia Ankara. Virus Res. 2009;145(1):54–62. doi: 10.1016/j.virusres.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I., Munster V.J., Sandig V. Authentication of the R06E fruit bat cell line. Viruses. 2012;4(5):889–900. doi: 10.3390/v4050889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen G.R., Dixon R.D., Amelon S.K. Potential spread of white-nose syndrome of bats to the northwest: epidemiological considerations. Northwest Sci. 2013;87(4):292–306. [Google Scholar]
- Krahling V., Dolnik O., Kolesnikova L., Schmidt-Chanasit J., Jordan I., Sandig V., Gunther S., Becker S. Establishment of fruit bat cells (Rousettus aegyptiacus) as a model system for the investigation of filoviral infection. PLoS Negl. Trop. Dis. 2010;4(8):e802. doi: 10.1371/journal.pntd.0000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger N., Hoffmann M., Weis M., Drexler J.F., Muller M.A., Winter C., Corman V.M., Gutzkow T., Drosten C., Maisner A., Herrler G. Surface glycoproteins of an African henipavirus induce syncytium formation in a cell line derived from an African fruit bat, Hypsignathus monstrosus. J. Virol. 2013;87(24):13889–13891. doi: 10.1128/JVI.02458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Dong D. Phylogenomic analyses of bat subordinal relationships based on transcriptome data. Sci. Rep. 2016;6:27726. doi: 10.1038/srep27726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm J.A., Durbin R.K., Stollar V., Rice C.M. Mutations which alter the level or structure of Nsp4 can affect the efficiency of sindbis virus-replication in a host-dependent manner. J. Virol. 1990;64(6):3001–3011. doi: 10.1128/jvi.64.6.3001-3011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Delicat A., Paweska J.T., Gonzalez J.P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438(7068):575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li J., Zhang G., Cheng D., Ren H., Qian M., Du B. Molecular characterization of RIG-I, STAT-1 and IFN-beta in the horseshoe bat. Gene. 2015;561(1):115–123. doi: 10.1016/j.gene.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Paden C.R., Morales F.M., Powers R.P., Jacob J., Speck S.H. Murine gamma-herpesvirus immortalization of fetal liver-derived B cells requires both the viral cyclin D homolog and latency-associated nuclear antigen. PLoS Pathog. 2011;7(9):e1002220. doi: 10.1371/journal.ppat.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y.Z., Wu L.J., Zhang Q., Zhou P., Wang M.N., Yang X.L., Ge X.Y., Wang L.F., Shi Z.L. Cloning, expression, and antiviral activity of interferon beta from the Chinese microbat, Myotis davidii. Virol Sin. 2015;30(6):425–432. doi: 10.1007/s12250-015-3668-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis A.D., Hayman D.T., O'Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R., Mills J.N., Timonin M.E., Willis C.K., Cunningham A.A., Fooks A.R., Rupprecht C.E., Wood J.L., Webb C.T. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. Biol. Sci. 2013;280(1756):20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmlov A., Seetahal J., Carrington C., Ramkisson V., Foster J., Miazgowicz K.L., Quackenbush S., Rovnak J., Negrete O., Munster V., Schountz T. Serological evidence of arenavirus circulation among fruit bats in Trinidad. PLoS One. 2017;12(9):e0185308. doi: 10.1371/journal.pone.0185308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh G.A., de Jong C., Barr J.A., Tachedjian M., Smith C., Middleton D., Yu M., Todd S., Foord A.J., Haring V., Payne J., Robinson R., Broz I., Crameri G., Field H.E., Wang L.F. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012;8(8):e1002836. doi: 10.1371/journal.ppat.1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Gomez J.M., Periasamy P., Dutertre C.A., Irving A.T., Ng J.H., Crameri G., Baker M.L., Ginhoux F., Wang L.F., Alonso S. Phenotypic and functional characterization of the major lymphocyte populations in the fruit-eating bat Pteropus alecto. Sci. Rep. 2016;6:37796. doi: 10.1038/srep37796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl K.A., Chamberlain T., Lunt R.A., Newberry K.M., Middleton D., Westbury H.A. Pathogenesis studies with Australian bat lyssavirus in grey-headed flying foxes (Pteropus poliocephalus) Aust. Vet. J. 2002;80(10):636–641. doi: 10.1111/j.1751-0813.2002.tb10973.x. [DOI] [PubMed] [Google Scholar]
- Miller M.R., McMinn R.J., Misra V., Schountz T., Muller M.A., Kurth A., Munster V.J. Broad and temperature independent replication potential of filoviruses on cells derived from old and new world bat species. J. Infect. Dis. 2016;214(Suppl 3):S297–S302. doi: 10.1093/infdis/jiw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V., Dumonceaux T., Dubois J., Willis C., Nadin-Davis S., Severini A., Wandeler A., Lindsay R., Artsob H. Detection of polyoma and corona viruses in bats of Canada. J. Gen. Virol. 2009;90(Pt 8):2015–2022. doi: 10.1099/vir.0.010694-0. [DOI] [PubMed] [Google Scholar]
- Moratelli R., Calisher C.H. Bats and zoonotic viruses: can we confidently link bats with emerging deadly viruses? Mem. Inst. Oswaldo Cruz. 2015;110(1):1–22. doi: 10.1590/0074-02760150048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Soto A., Soto-Garita C., Corrales-Aguilar E. Neotropical primary bat cell lines show restricted dengue virus replication. Comp. Immunol. Microbiol. Infect. Dis. 2017;50:101–105. doi: 10.1016/j.cimid.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Mourya D.T., Lakra R.J., Yadav P.D., Tyagi P., Raut C.G., Shete A.M., Singh D.K. Establishment of cell line from embryonic tissue of Pipistrellus ceylonicus bat species from India & its susceptibility to different viruses. Indian J. Med. Res. 2013;138(2):224–231. [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Adney D.R., van Doremalen N., Brown V.R., Miazgowicz K.L., Milne-Price S., Bushmaker T., Rosenke R., Scott D., Hawkinson A., de Wit E., Schountz T., Bowen R.A. Replication and shedding of MERS-CoV in Jamaican fruit bats (Artibeus jamaicensis) Sci. Rep. 2016;6:21878. doi: 10.1038/srep21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negredo A., Palacios G., Vazquez-Moron S., Gonzalez F., Dopazo H., Molero F., Juste J., Quetglas J., Savji N., de la Cruz Martinez M., Herrera J.E., Pizarro M., Hutchison S.K., Echevarria J.E., Lipkin W.I., Tenorio A. Discovery of an ebolavirus-like filovirus in europe. PLoS Pathog. 2011;7(10):e1002304. doi: 10.1371/journal.ppat.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepelska M., de Wouters T., Jacouton E., Beguet-Crespel F., Lapaque N., Dore J., Arulampalam V., Blottiere H.M. Commensal gut bacteria modulate phosphorylation-dependent PPARgamma transcriptional activity in human intestinal epithelial cells. Sci. Rep. 2017;7:43199. doi: 10.1038/srep43199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M., Ndungo E., Kaczmarek M.E., Herbert A.S., Binger T., Kuehne A.I., Jangra R.K., Hawkins J.A., Gifford R.J., Biswas R., Demogines A., James R.M., Yu M., Brummelkamp T.R., Drosten C., Wang L.F., Kuhn J.H., Muller M.A., Dye J.M., Sawyer S.L., Chandran K. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. Elife. 2015;4 doi: 10.7554/eLife.11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary M.A., Bloch J.I., Flynn J.J., Gaudin T.J., Giallombardo A., Giannini N.P., Goldberg S.L., Kraatz B.P., Luo Z.X., Meng J., Ni X., Novacek M.J., Perini F.A., Randall Z.S., Rougier G.W., Sargis E.J., Silcox M.T., Simmons N.B., Spaulding M., Velazco P.M., Weksler M., Wible J.R., Cirranello A.L. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science. 2013;339(6120):662–667. doi: 10.1126/science.1229237. [DOI] [PubMed] [Google Scholar]
- O'Shea T.J., Cryan P.M., Cunningham A.A., Fooks A.R., Hayman D.T., Luis A.D., Peel A.J., Plowright R.K., Wood J.L. Bat flight and zoonotic viruses. Emerg. Infect. Dis. 2014;20(5):741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J., Hayman D.T. Filoviruses in bats: current knowledge and future directions. Viruses. 2014;6(4):1759–1788. doi: 10.3390/v6041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546(7660):646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omatsu T., Bak E.J., Ishii Y., Kyuwa S., Tohya Y., Akashi H., Yoshikawa Y. Induction and sequencing of Rousette bat interferon alpha and beta genes. Vet. Immunol. Immunopathol. 2008;124(1-2):169–176. doi: 10.1016/j.vetimm.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki K., Maeda J., Yamamoto H., Tsubokura M. Studies on avian infectious bronchitis virus (IBV). III. Interferon induction by and sensitivity to interferon of IBV. Arch. Virol. 1979;60(3–4):249–255. doi: 10.1007/BF01317496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright R.K., Eby P., Hudson P.J., Smith I.L., Westcott D., Bryden W.L., Middleton D., Reid P.A., McFarlane R.A., Martin G., Tabor G.M., Skerratt L.F., Anderson D.L., Crameri G., Quammen D., Jordan D., Freeman P., Wang L.F., Epstein J.H., Marsh G.A., Kung N.Y., McCallum H. Ecological dynamics of emerging bat virus spillover. Proc. Biol. Sci. 2015;282(1798):20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole D.S., Yu S., Cai Y., Dinis J.M., Muller M.A., Jordan I., Friedrich T.C., Kuhn J.H., Mehle A. Influenza A virus polymerase is a site for adaptive changes during experimental evolution in bat cells. J. Virol. 2014;88(21):12572–12585. doi: 10.1128/JVI.01857-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J., Feldmann H., Safronetz D. Amending Koch's postulates for viral disease: when growth in pure culture leads to a loss of virulence. Antiviral Res. 2017;137:1–5. doi: 10.1016/j.antiviral.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Wong G., Audet J., Cutts T., Niu Y., Booth S., Kobinger G.P. Establishment and characterization of a lethal mouse model for the Angola strain of Marburg virus. J. Virol. 2014;88(21):12703–12714. doi: 10.1128/JVI.01643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandekian V., Lim D., Prud'homme P., Lemay G. Transient high level mammalian reovirus replication in a bat epithelial cell line occurs without cytopathic effect. Virus Res. 2013;173(2):327–335. doi: 10.1016/j.virusres.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Sayler K.A., Barbet A.F., Chamberlain C., Clapp W.L., Alleman R., Loeb J.C., Lednicky J.A. Isolation of Tacaribe virus, a Caribbean arenavirus, from host-seeking Amblyomma americanum ticks in Florida. PLoS One. 2014;9(12):e115769. doi: 10.1371/journal.pone.0115769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T., Baker M.L., Butler J., Munster V. Immunological control of viral infections in bats and the emergence of viruses highly pathogenic to humans. Front. Immunol. 2017;8:1098. doi: 10.3389/fimmu.2017.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schountz T. Immunology of bats and their viruses: challenges and opportunities. Viruses. 2014;6(12):4880–4901. doi: 10.3390/v6124880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A.J., Amman B.R., Jones M.E., Sealy T.K., Uebelhoer L.S., Spengler J.R., Martin B.E., Coleman-McCray J.A., Nichol S.T., Towner J.S. Modelling filovirus maintenance in nature by experimental transmission of Marburg virus between Egyptian rousette bats. Nat. Commun. 2017;8:14446. doi: 10.1038/ncomms14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabman R.S., Shrivastava S., Tsibane T., Attie O., Jayaprakash A., Mire C.E., Dilley K.E., Puri V., Stockwell T.B., Geisbert T.W., Sachidanandam R., Basler C.F. Isolation and characterization of a novel gammaherpesvirus from a microbat cell line. mSphere. 2016;1(1) doi: 10.1128/mSphere.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons N.B., Seymour K.L., Habersetzer J., Gunnell G.F. Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature. 2008;451(7180):818–821. doi: 10.1038/nature06549. [DOI] [PubMed] [Google Scholar]
- Subudhi S., Rapin N., Bollinger T.K., Hill J.E., Donaldson M.E., Davy C.M., Warnecke L., Turner J.M., Kyle C.J., Willis C.K.R., Misra V. A persistently infecting coronavirus in hibernating Myotis lucifugus, the North American little brown bat. J. Gen. Virol. 2017;98(9):2297–2309. doi: 10.1099/jgv.0.000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R., Smit S.B., Rollin P.E., Formenty P., Leman P.A., Kemp A., Burt F.J., Grobbelaar A.A., Croft J., Bausch D.G., Zeller H., Leirs H., Braack L.E., Libande M.L., Zaki S., Nichol S.T., Ksiazek T.G., Paweska J.T., International, S., Technical Committee for Marburg Hemorrhagic Fever Control in the Democratic Republic of, C Studies of reservoir hosts for Marburg virus. Emerg. Infect. Dis. 2007;13(12):1847–1851. doi: 10.3201/eid1312.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait J., Perotto-Baldivieso H.L., McKeown A., Westcott D.A. Are flying-foxes coming to town? Urbanisation of the spectacled flying-fox (Pteropus conspicillatus) in Australia. PLoS One. 2014;9(10):e109810. doi: 10.1371/journal.pone.0109810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E.C., Springer M.S., Madsen O., Bates P., O'Brien S.J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307(5709):580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Teeling E.C., Jones G., Rossiter S.J. Vol. 54. 2016. pp. 25–54. (Phylogeny, Genes, and Hearing: Implications for the Evolution of Echolocation in Bats). [Google Scholar]
- Virtue E.R., Marsh G.A., Baker M.L., Wang L.F. Interferon production and signaling pathways are antagonized during henipavirus infection of fruit bat cell lines. PLoS One. 2011;6(7):e22488. doi: 10.1371/journal.pone.0022488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke L., Turner J.M., Bollinger T.K., Lorch J.M., Misra V., Cryan P.M., Wibbelt G., Blehert D.S., Willis C.K. Inoculation of bats with European Geomyces destructans supports the novel pathogen hypothesis for the origin of white-nose syndrome. Proc. Natl. Acad. Sci. U. S. A. 2012;109(18):6999–7003. doi: 10.1073/pnas.1200374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Audet J., Wong G., He S., Huang X., Cutts T., Theriault S., Xu B., Kobinger G., Qiu X. Deep-sequencing of Marburg virus genome during sequential mouse passaging and cell-culture adaptation reveals extensive changes over time. Sci. Rep. 2017;7(1):3390. doi: 10.1038/s41598-017-03318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo W., Begeman L., Schipper D., Run P.R.V., Cunningham A.A., Kley N., Reusken C.B., Haagmans B.L., Brand J. Tissue distribution of the MERS-coronavirus receptor in bats. Sci. Rep. 2017;7(1):1193. doi: 10.1038/s41598-017-01290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne J.W., Woon A.P., Dudek N.L., Croft N.P., Ng J.H., Baker M.L., Wang L.F., Purcell A.W. Characterization of the antigen processing machinery and endogenous peptide presentation of a bat MHC class I molecule. J. Immunol. 2016;196(11):4468–4476. doi: 10.4049/jimmunol.1502062. [DOI] [PubMed] [Google Scholar]
- Xie X., Pang M., Liang S., Yu L., Zhao Y., Ma K., Kalhoro D.H., Lu C., Liu Y. Establishment and characterization of a telomerase-immortalized canine bronchiolar epithelial cell line. Appl. Microbiol. Biotechnol. 2015;99(21):9135–9146. doi: 10.1007/s00253-015-6794-8. [DOI] [PubMed] [Google Scholar]
- Young P.L., Halpin K., Selleck P.W., Field H., Gravel J.L., Kelly M.A., Mackenzie J.S. Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg. Infect. Dis. 1996;2(3):239–240. doi: 10.3201/eid0203.960315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Todd S., Tachedjian M., Barr J.A., Luo M., Yu M., Marsh G.A., Crameri G., Wang L.F. A novel bat herpesvirus encodes homologues of major histocompatibility complex classes I and II, C-type lectin, and a unique family of immune-related genes. J. Virol. 2012;86(15):8014–8030. doi: 10.1128/JVI.00723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zeng L.P., Zhou P., Irving A.T., Li S., Shi Z.L., Wang L.F. IFNAR2-dependent gene expression profile induced by IFN-alpha in Pteropus alecto bat cells and impact of IFNAR2 knockout on virus infection. PLoS One. 2017;12(8):e0182866. doi: 10.1371/journal.pone.0182866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Cowled C., Todd S., Crameri G., Virtue E.R., Marsh G.A., Klein R., Shi Z., Wang L.F., Baker M.L. Type III IFNs in pteropid bats: differential expression patterns provide evidence for distinct roles in antiviral immunity. J. Immunol. 2011;186(5):3138–3147. doi: 10.4049/jimmunol.1003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Cowled C., Wang L.F., Baker M.L. Bat Mx1 and Oas1, but not Pkr are highly induced by bat interferon and viral infection. Dev. Comp. Immunol. 2013;40(3-4):240–247. doi: 10.1016/j.dci.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Zhou P., Chionh Y.T., Irac S.E., Ahn M., Jia Ng J.H., Fossum E., Bogen B., Ginhoux F., Irving A.T., Dutertre C.A., Wang L.F. Unlocking bat immunology: establishment of Pteropus alecto bone marrow-derived dendritic cells and macrophages. Sci. Rep. 2016;6:38597. doi: 10.1038/srep38597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Tachedjian M., Wynne J.W., Boyd V., Cui J., Smith I., Cowled C., Ng J.H., Mok L., Michalski W.P., Mendenhall I.H., Tachedjian G., Wang L.F., Baker M.L. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proc. Natl. Acad. Sci. U. S. A. 2016;113(10):2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukal J., Bandouchova H., Bartonicka T., Berkova H., Brack V., Brichta J., Dolinay M., Jaron K.S., Kovacova V., Kovarik M., Martinkova N., Ondracek K., Rehak Z., Turner G.G., Pikula J. White-nose syndrome fungus: a generalist pathogen of hibernating bats. PLoS One. 2014;9(5):e97224. doi: 10.1371/journal.pone.0097224. [DOI] [PMC free article] [PubMed] [Google Scholar]