Abstract
Emerging viruses are a major threat to human health. Recent outbreaks have emphasized the urgent need for new antiviral treatments. For several pathogenic viruses, considerable efforts have focused on vaccine development. However, during epidemics infected individuals need to be treated urgently. High-throughput screening of clinically tested compounds provides a rapid means to identify undiscovered, antiviral functions for well-characterized therapeutics. Repurposed drugs can bypass part of the early cost and time needed for validation and authorization. In this review we describe recent efforts to find broad spectrum antivirals through drug repurposing. We have chosen several candidates and propose strategies to understand their mechanism of action and to determine how resistance to antivirals develops in infected cells.
Keywords: Virus, Antivirals, Drug repurposing
1. Introduction
Viruses are a constant threat to humans, animals and plants. There are hundreds of viruses that can cause disease in humans but there is no treatment for most of them. Emerging or reemerging viruses are a public health concern (Howard and Fletcher, 2012). Some of these pathogens are the Rift Valley fever virus (RVFV), dengue virus (DENV), West Nile virus (WNV), yellow fever virus (YFV), Ebola virus (EBOV), SARS- and MERS-CoV, zika virus (ZIKV), Crimean Congo hemorrhagic fever virus (CCHFV), severe fever with thrombocytopenia syndrome virus (SFTSV), chikungunya virus (CHIKV) and influenza A virus (IAV). Recent outbreaks of these viruses are summarized in Table 1 . International travel and migrations, globalization of commerce, technology and industry, agriculture development or climate changes are favoring the emergence and reemergence of viruses that could easily spread and potentially become a pandemic (Jones et al., 2008). A clear example of the potential of an emerging virus to become a pandemic is the emergence of human immunodeficiency virus (HIV) in the 1980s (http://aidsinfo.unaids.org/) (Sharp and Hahn, 2011). There is no cure for HIV, although antiretroviral treatment can control the virus (Barre-Sinoussi et al., 2013). In the case of influenza virus there are both vaccines and antiviral drugs. However, these viruses constantly change and antiviral resistance emerges (To et al., 2015). In 2013, human infections with a new avian influenza virus in China caused considerable concern due to the pandemic potential of this virus (Su et al., 2015). The experts consider that the virus could gain the ability to spread among people, triggering a global epidemic (Su et al., 2015; Tanner et al., 2015).
Table 1.
Recent outbreaks of pathogenic human viruses.
| Virus | Year | Number of cases | Country/Territory | Total number of territories |
|---|---|---|---|---|
| Zika virus | (2013-2018) | >220,000 | Americas, Africa, Asia | 85 |
| Rift Valley fever virus (RVFV) | (2013-2018) | >1,000 | Africa, Saudi Arabia and Yemen | 31 |
| Ebola virus | (2014-2018) | >28,900 | West Africa (Liberia, Guinea, Sierra Leone, Nigeria, and Mali) | 6 |
| Middle East respiratory syndrome coronavirus (MERS) | (2012-2017) | >2,200 | Middle East, North Africa, Europe, the United States of America, and Asia | 27 |
| Severe acute respiratory syndrome coronavirus (SARS-CoV) | (2002-2003) | >8,000 | North America, South America, South Africa, Europe, and Asia | 30 |
| Influenza virus | Annual epidemics | 3 to 5 million cases per year | Worldwide | |
| Chikungunya virus (CHIKV) | (2013-2018) | > 2 million | Americas, Africa, Asia, Europe, the Indian subcontinent and Pacific Islands in Oceania | 112 |
| Dengue virus | Annual epidemics | 390 million dengue infections per year | Africa, the Americas, the Eastern Mediterranean, South-East Asia and the Western Pacific. | 111 |
| Yellow fever virus (YFV) | (2013-2018) | >15,000 | 33 countries in Africa and 11 countries in South America | 44 |
| West Nile virus (WNV) | (2013-2018) | >12,000 Since 2013: Around 2500 cases per year | North and South America, Europe, Africa, the Middle East, and West Asia and Oceania | 83 |
| Crimean-Congo hemorrhagic fever virus (CCHFV) | (2000-2015) | >1,000 | Africa, Asia, Eastern and Southern Europe and Central Asia | 42 |
| Severe fever with thrombocytopenia syndrome virus (SFTSV) | (2009-2016) | >1300 Since 2016: >1500 cases per year | South Korea, Japan, China and United Arab Emirates | 4 |
Sources: European Centre for Disease Prevention and Control. ECDC. (https://ecdc.europa.eu/en/threats-and-outbreaks) U.S. Centers for Disease Control and Protection. CDC. (https://www.cdc.gov/outbreaks/index.html) Chinese Center for Disease Control and Prevention (China CDC). (http://www.chinacdc.cn/en/aboutus/orc_9362/201810/t20181008_194504.html) World Health Organization (WHO). (https://www.who.int/csr/don/en/).
Due to all these reasons, we urgently need new, efficient, broad-spectrum antivirals (Howard and Fletcher, 2012; Schor and Einav, 2018). The traditional strategy in drug development of one virus one drug, based in the development of molecules that specifically target a viral protein, has been very successful for certain viruses like HIV or hepatitis C virus (HCV) (Barre-Sinoussi et al., 2013; Li and De Clercq, 2017). In fact, most approved antiviral treatments target a viral protein, and thus are usually quite virus specific. However, direct acting antivirals (DAA) have some drawbacks.
The development of new therapeutic compounds is time and resources consuming (DiMasi et al., 2016). The whole process from the discovery of a new drug that can potentially treat a specific disease or condition to its approval to be used in humans can take several years (on average 12 to 16 years) (Nosengo, 2016). The first stage, drug discovery, can take between 3 to 6 years. Then preclinical studies, usually in animal models, are carried out. These studies that are aimed to test the efficiency and safety of a molecule in a suitable model before it is tested in humans can go on for about 3 years. If a molecule has shown promising results up to here, then clinical trials in humans are started. These trials, from phase I to phase III, usually last about 5 years. Finally, if a molecule successfully passes all the process it needs to get the approval from the appropriate agency. At the end it is estimated that only about 5% of the candidate molecules successfully pass the whole process. Adding to this the cost of drug development, about 1–2 billion dollars or more in some cases (DiMasi et al., 2016), the one virus one drug strategy is not the most cost efficient approach. In addition, the use of DAAs can easily lead to the appearance of drug resistant viruses due to the elevated mutation rate of viruses (Iyidogan and Anderson, 2014; Menendez-Arias et al., 2014; Perales et al., 2015).
To circumvent these limitations other strategies aim to develop molecules that target host cellular factors that are needed for viral replication. Although viruses can also find ways to use alternative routes to replicate in a host, emergence of resistant viruses is less likely (Lin and Gallay, 2013; Zeisel et al., 2013). In addition these approaches focused in host cellular pathways needed for viral replication can favor the development of broad spectrum antivirals. There are families of viruses that share certain cellular pathways. Thus, drugs that inhibit a specific cellular target could work for several viruses. As a drawback drugs that inhibit a host cell target have higher chances of producing undesirable side effects in the patient compared to drugs that target a viral protein.
Drug repurposing or repositioning is an alternative approach that can lead to less costly and faster approval of new treatments for viral infections (Mercorelli et al., 2018). A clear advantage of drug repositioning over traditional drug development is that since the repositioned drug has already passed a significant number of tests, its safety is known and the risk of failure is reduced. Although repositioned drugs can most likely skip phase I clinical trials, they still need to go through phase II and III trials to test their efficacy for the new condition (Nosengo, 2016). In some instances these drugs might have lost their patents, as is the case of generic medicines. In these cases, when a new formulation or a new medical use exists for an “old” drug, second medical use patents can be obtained for them. On the other hand, increasing implication of practicing physicians and researchers at academic centers in clinical trials can also help in the repurposing of these drugs (Nosengo, 2016). Additionally, there are thousands of drugs that have already passed clinical trials that confirm their safe use in humans. However, for some reasons, they lack or have poor efficiency to treat the condition under scrutiny and have not reached the market. These collections of drugs are another source of potential antivirals. Several drugs in the market have been tested for their potential use as broad spectrum antiviral treatments (Table 2 ). Many of these compounds have shown promising results in preclinical studies. Below we summarize the main results and antiviral mechanisms of selected candidates for repositioning as broad spectrum antiviral drugs.
Table 2.
Drug repurposing in antiviral discovery: Promising candidates.
Abbreviations: CCHFV Crimean Congo hemorrhagic fever virus; CHIKV chikungunya virus; HCMV human cytomegalovirus; DENV dengue virus; EBOV Ebola virus; HAV hepatitis A virus; HBV hepatitis B virus; HCV hepatitis C Virus; HIV human immunodeficiency virus; HPV human papilloma virus; HSV herpes simplex virus; huAdV human adenovirus; JEV japanesse encephalitis virus; LASV Lassa virus; MARV Marburg virus; MERS-CoV Middle East respiratory syndrome coronavirus; RVFV Rift Valley fever virus; SARS-CoV severe acute respiratory syndrome coronavirus; VSV vesicular stomatitis virus; VV vaccinia virus; WNV West Nile virus; YFV yellow fever virus; ZIKV zika virus.
2. Digoxin and other cardiac glycosides
Digoxin is a cardiac glycoside or cardiotonic steroid that has been used for treating certain heart conditions including heart failure, atrial fibrillation, atrial flutter, and paroxysmal atrial tachycardia (Gheorghiade et al., 2006). Cardiac glycosides comprise a large family of natural compounds that share a common steroid structure. Digoxin was first isolated from foxglove plant, digitalis lanata, in 1930, although foxglove plant extracts have been used from ancient times to treat heart conditions (Gheorghiade et al., 2006). Digoxin has been shown to block the Na+/K+ ATPase, raising intracellular Na + levels. The increase in intracellular Na + leads to an increment in intracellular Ca2+ due to inhibition of Na+/Ca2+ exchanger. Elevated intracellular Ca2+ ultimately leads to an increased contractibility of the cardiac muscle (Smith, 1988). Inhibition of the Na+/K+ ATPase has been shown to have an effect in multiple signaling cascades through modulation of neighboring tyrosine kinases, including src, ultimately affecting gene expression (Nesher et al., 2007). Thus, cardiac glycosides are in the spotlight for drug repurposing.
Some studies have shown the potential use of digoxin and other cardiac glycosides to treat cancer (Prassas and Diamandis, 2008). Cardiac glycosides can induce apoptosis in diverse cancer cells by multiple mechanisms including inhibition of Na+/K+ ATPase, suppression of nuclear factor-kappaB and inhibition of topoisomerase II (Ishida et al., 2016; Vaklavas et al., 2011). By performing a chemical screen with an insect cell-based reporter system, the cardiac glycoside digoxin was identified as a specific inhibitor of the retinoic acid receptor-related orphan nuclear receptor (RORγt) transcriptional activity (Huh et al., 2011), which is required for induction of IL-17 transcription and for the manifestation of TH17-dependent autoimmune disease in mice. This study indicated that digoxin and its derivatives could be used as therapeutic agents that attenuate inflammatory lymphocyte function and autoimmune disease.
Recent studies have shown that digoxin and other cardiac glycosides can have a potential use as antivirals. Wong and colleagues reported the suppression of HIV-1 replication by digoxin (Wong et al., 2013). Digoxin was shown to accomplish its antiretroviral effects by two mechanisms: 1) inducing oversplicing of HIV-1 RNA which reduces the accumulation of viral RNAs encoding for structural proteins needed for new virion assembly, and 2) selectively inhibiting the expression of HIV-1 regulatory protein Rev. These studies showed that digoxin exerted its anti-HIV effect by altering the function of a subset of SR proteins, a conserved family of serine and arginine-rich proteins involved in RNA splicing. A screening of a drug library for compounds inhibiting late steps of HIV-1 replication cycle also identified several members of the cardiac glycoside family (Laird et al., 2014).
The cardiac glycosides digoxin and ouabain have also been shown to impair replication of human cytomegalovirus (HCMV) and herpes simplex virus (HSV) that belong to the family of dsDNA viruses Herpesviridae (Dodson et al., 2007; Hartley et al., 2006; Kapoor et al., 2012). Inhibition happens at an early post-entry step and produces a decrease in the expression of immediate-early proteins IE1 and IE2. Similarly, digoxin and digitoxin have been reported to suppress replication of human adenovirus (HAdV), another dsDNA virus, in cell cultures (Grosso et al., 2017). Although the precise mechanism of action is not fully clear, these drugs altered viral mRNA processing, blocking replication before viral DNA synthesis.
Some cardiac glycosides like procillaridin A, bufallin, covallatoxin and digitoxin are able to inhibit hepatitis B virus (HBV) in cell cultures (Okuyama-Dobashi et al., 2015). In this study, however, digoxin did not show an anti-HBV effect. Digoxin also inhibits alphaviruses (single-strand positive-sense RNA viruses) like chikungunya, Ross River virus and Sindbis virus, as well as the unrelated mammalian orthoreovirus (polysegmented double-strand RNA virus) and vesicular stomatitis virus from the Rhabdoviridae family (negative-sense RNA virus) (Ashbrook et al., 2016). In this study digoxin was shown to impair chikungunya infection at an early post-entry step.
Some coronaviruses like feline infectious peritonitis virus, murine hepatitis virus, and MERS-CoV are inhibited by ouabain and bufalin (Burkard et al., 2015). The antiviral effect of these cardiac glycosides was observed only when the drug was added prior to infection. Infection was not affected when the drugs were added 2 h post-infection, suggesting that for these viruses the drugs were acting during the entry step.
3. Sunitinib and other tyrosine kinase inhibitors
Sunitinib is a small molecule that inhibits multiple tyrosine kinase receptors like vascular endothelial growth factor receptor VEGFR, platelet-derived growth factor receptors PDGFRα and PDGFRβ, fibroblast growth factor receptor 1, and epidermal growth factor receptor (Mendel et al., 2003; Sun et al., 2003). Sunitinib has been approved by the FDA for the treatment of some cancers (Atkins et al., 2006).
Recent studies have shown that sunitinib may also have broad spectrum antiviral activity. Intracellular trafficking of viruses relies on the endocytic and exocytic cellular pathways. These processes usually require signal transduction, suggesting that kinase inhibitors may have antiviral activity by blocking the endocytic or exocytic pathways. Binding of the HCV core protein to adaptor protein 2 (AP-2) μ2 subunit (AP2M1) is essential for HCV assembly. Phosphorylation of AP2 by adaptor associated kinase (AKK1) and cycling G-associated kinase (GAK) regulate this interaction (Neveu et al., 2012, 2015). It has also been shown that AP1 and AP2 co-traffic with HCV viral particles within the cell (Bekerman et al., 2017). These studies showed that sunitinib prevents HCV entry and assembly, apparently through an inhibitory effect on AAK1 and GAK, with no effect on HCV RNA replication. Erlotinib, another anticancer drug, also inhibits HCV entry and assembly. Similarly, sunitinib and/or erlotinib can restrict infection by DENV and EBOV in vitro and in a murine animal model and have potent in vitro antiviral activity against ZIKV, WNV, CHIKV, Junin virus (JUNV) and respiratory syncytial virus (RSV) (Bekerman et al., 2017; Pu et al., 2018). Sunitinib has also been shown to inhibit HCMV infection in cell culture (Cai et al., 2014) and HIV-1 infection of resting CD4 T cells (Guo et al., 2013).
Although these studies showed the potential antiviral activity of sunitinib, this drug can also inhibit protein kinase R (PKR) and 2′5′oligoadenylate synthetase (OAS)/RNase L system that act as antiviral effectors in response to type I interferons (Jha et al., 2013). The capacity of sunitinib to block these two innate immunity pathways could hamper its potential use as antiviral. The development of more selective GAK inhibitors could bypass this hurdle. In this sense, a screening of a library based on a bicyclic, heteroaromatic flat scaffold designed to discover novel ligands of GAK, led to the identification of a hit compound based on a isothiazolo[4,3-b]pyridine scaffold (Kovackova et al., 2015). Some isothiazolo[4,3-b]pyridine derivatives with low nanomolar affinity for GAK exhibited inhibitory activity against HCV in cell culture, acting at the level of viral entry and assembly (Kovackova et al., 2015).
4. Chloroquine and its analogs
Chloroquine (CQ) is a weak base 4-aminoquinolone derivative that can easily enter the cells and accumulate in acidic compartments like endosomes, lysosomes or trans-Golgi network vesicles raising their pH (Thome et al., 2013). In the clinic CQ has been used for the treatment of non-resistant malaria (Slater, 1993; White, 1996). Additionally CQ has been proven to have anti-inflammatory properties, and has been approved for the clinical management of some autoimmune diseases such as rheumatoid arthritis and lupus erythematosus (Rainsford et al., 2015).
Many viruses use acidic organelles at some point of their replication cycle. This has prompted the study of CQ and its hydroxyl analogue hydroxychloroquine (HCQ) as potential antiviral drugs. CQ and HCQ have been tested and proved to have in vitro, and in some instances in vivo, antiviral activity against several viruses, as described below.
Two main mechanisms of action have been described for the antiviral activity of CQ (Al-Bari, 2017). On the one hand, many viruses use acidic endocytic vesicles, like endosomes or lysosomes, to enter the host cell. The acidification of these vesicles triggers conformational changes in the envelope proteins of the virus that induce the fusion of the viral and cellular membranes and the delivery of the virion content into the cell. CQ raises the pH of these vesicles impeding the conformational changes needed for viral entry or uncoating and blocking the infection. CQ has also been shown to impair maturation of some viral proteins in the Golgi network by blocking the action of pH-dependent enzymes, like proteases or glycosyltransferases, which are needed for the maturation process. The antiretroviral effect of CQ in cell cultures has been shown to occur through the inhibition of the glycosylation of HIV viral particles (Savarino et al., 2004), while the antiviral activity of CQ against SARS-CoV seems to be related to the poor glycosylation of the SARS-CoV receptor ACE2 (Vincent et al., 2005). CQ has been reported to inhibit DENV replication by blocking the cleavage of the prM protein that occurs in acidic compartments (Boonyasuppayakorn et al., 2014; Randolph et al., 1990).
CQ and some of its derivatives have been reported to have strong antiviral activity against SARS-CoV and MERS-CoV in different cell lines with IC50 ranging from 3.0 to 8.8 μM (de Wilde et al., 2014; Dyall et al., 2014; Keyaerts et al., 2004; Vincent et al., 2005). However, in a SARS-CoV mouse model CQ treatment did not reduce virus titer in the lungs (Barnard et al., 2006)
Some studies have reported inhibitory effect of CQ against influenza A viruses (IAV) H3N2 and H1N1 in vitro at concentrations similar to doses used for malaria treatment (Ooi et al., 2006). However, the susceptibility of IAV to CQ seems to depend on the pH requirements of the viruses and the electrostatic potential of hemagglutinin 2 (Di Trani et al., 2007). In a mouse model of infection with IAV, CQ treatment dramatically increased the survival of infected mice when administered therapeutically but not when administered as a prophylactic treatment (Yan et al., 2013). Furthermore, a randomized, double-blinded placebo controlled clinical trial did not protect against influenza infection (Paton et al., 2011).
Dengue virus and WNV replication has also been reported to be blocked by CQ in cell culture (Boonyasuppayakorn et al., 2014; Farias et al., 2014). A recent study has demonstrated the efficacy of CQ in the prophylactic and therapeutic treatment of Aotus monkeys inoculated with DENV (Farias et al., 2015). However, a double-blinded, placebo-controlled clinical trial in Vietnam failed to show a beneficial impact of CQ treatment in the outcome of the infection except for a modest reduction in the percentage of patients that developed dengue hemorrhagic fever in the CQ-treated arm (Tricou et al., 2010). Chloroquine also exhibits antiviral activity against ZIKV, another member of the Flaviviridae family, in different cell lines with an IC50 around 9.8–14.2 μM (Delvecchio et al., 2016). Additionally, some studies have demonstrated that chloroquine can protect mice against ZIKV infection (Li et al., 2017; Shiryaev et al., 2017).
CQ has also been reported to inhibit CHIKV in cell cultures in a dose dependent manner when added at an early point during infection (Sourisseau et al., 2007). Although in this study the therapeutic index of the drug in cell cultures was low, the results pointed at the potential use of CQ for the treatment of CHIKV infection. However, a clinical trial in the French Reunion Island during an outbreak of CHIKV to test the efficacy and safety of CQ treatment did not show evidence of a significant antiviral effect in CQ treated patients compared to placebo treated patients (De Lamballerie et al., 2008). This lack of antiviral effect could be due to the high viral load in the serum of patients during the acute phase of the infection. The rather narrow therapeutic index of CQ leaves little room for a dose adjustment that could allow the use of CQ as a therapeutic treatment in acute chikungunya infection. Nonetheless, in cell cultures, where the viral titer is lower, CQ strongly inhibits CHIKV infection at concentrations achieved in plasma of individuals on CQ treatment for malaria, which suggest that CQ could be used as a prophylactic treatment during chikungunya outbreaks to prevent transmission (Khan et al., 2010). However, a recent study in non-human primates showed that chloroquine treatment exacerbated the disease and/or suppressed the antiviral immunity in the chloroquine treated macaques compared to the placebo group (Roques et al., 2018).
CQ and HCQ were both shown to inhibit HIV replication in monocytic and T cell lines as well as in monocytes and primary T cells at a post-transcriptional step (Sperber et al., 1993; Tsai et al., 1990). CQ can also inhibit other viruses like CCHFV (Ferraris et al., 2015), hepatitis A virus (HAV) (Bishop, 1998) and EBOV (Madrid et al., 2013) in cell lines. Additionally, CQ has been shown to protect mice against a challenge of a deadly dose of EBOV (Madrid et al., 2013).
In cell culture CQ has shown potent antiviral activity against a broad range of viruses and in some cases efficacy in animal model of infection, as discussed above. However, clinical trials aimed to test the efficacy of CQ as antiviral has shown very limited antiviral effect. The rather narrow therapeutic index of chloroquine might be behind the limited antiviral effect of chloroquine in clinical trials. The development of chloroquine derivatives with lower toxicity could improve their effectiveness for the treatment of viral infections. The immunomodulatory activity of CQ can also contribute to the control of viral diseases reducing the excessive release of cytokines and other pro-inflammatory mediators.
5. Cyclosporin A and its derivatives
Cyclophilin A (CypA) is a peptidylprolyl isomerase that is expressed in the cytosol (Schmid, 1995). Cyclosporin A (CsA) is a cyclic undecapeptide with immune suppressive activity that mainly targets cellular cyclophilins (Cyps) (Handschumacher et al., 1984; Schreiber, 1991). The Cyp-CsA complex inhibits the phosphatase activity of calcineurin needed for the nuclear translocation of the nuclear factor of activated T cells (NFAT), which eventually leads to the block of transcription of cytokines and inhibition of T cell activation (Matsuda and Koyasu, 2000). CsA is one of the drugs approved by the FDA for immunosuppressive therapy to avoid rejection in organ transplants (Hartono et al., 2013).
CsA has been reported to have antiviral activity against a wide range of viruses including human papilloma virus (HPV) (Bienkowska-Haba et al., 2009), vesicular stomatitis virus (VSV) (Bose et al., 2003), vaccinia virus (VV) (Damaso and Moussatche, 1998), HIV-1 (Franke et al., 1994; Thali et al., 1994; Wainberg et al., 1988), and HCV (Kaul et al., 2009; Nakagawa et al., 2004; Yang et al., 2008). Although the immunosuppressive activity of CsA does not make it a good candidate for antiviral treatment, some CsA derivatives, like alisporivir, NIM811 and SCY-635, with reduced immunosuppressive properties that retain the ability to bind cyclophilin have been developed and proved to conserve the antiviral activity (Flisiak et al., 2008; Ma et al., 2016; Paeshuyse et al., 2006; Watashi et al., 2014). The replication of certain viruses like HIV, HCV, HPV, VSV, VV or influenza virus relies on their interaction with cyclophilins at certain steps of their replication cycle. Thus, the antiviral activity of cyclosporine A (CsA) and some of its non-immunosuppressive analogs against these viruses has been shown to be related to its ability to bind cellular cyclophilins and inhibiting the interaction with the viral proteins (Bienkowska-Haba et al., 2009; Bose et al., 2003; Damaso and Moussatche, 1998; Franke et al., 1994; Kaul et al., 2009; Nakagawa et al., 2004; Thali et al., 1994; Wainberg et al., 1988; Yang et al., 2008).
CsA also blocks the replication of diverse coronaviruses (de Wilde et al., 2018, 2011; Pfefferle et al., 2011). The nucleocapsid protein of SARS-CoV specifically binds CypA (Luo et al., 2004), and CypA has been found to be incorporated into the SARS-CoV particle (Neuman et al., 2008). However, siRNA knock-down of cellular CypA and CypB, the main targets of CsA, did not have an effect in the infectivity of this virus, suggesting that either these cyclophilins are not required for viral replication or that the remaining levels of cellular cyclophilins are enough to support normal replication (de Wilde et al., 2011).
Additionally CsA and some CsA analogs have been shown to inhibit HBV entry into cells (Watashi et al., 2014). However, this inhibitory activity was independent on the binding to CypA or calcineurin, and correlated to the ability of CsA to inhibit the transporter activity of sodium taurocholate cotransporting polypeptide (NTCP), a membrane protein that has been proposed to be the HBV receptor.
CypA has been reported to interact with influenza A virus matrix protein M1 suppressing viral replication in cell cultures and animal models (Li et al., 2016; Liu et al., 2009). The role of CypA in the viral cycle is unclear. Although it seems to act at several steps of the viral life cycle, the isomerase activity of CypA does not seem to be implicated (Liu et al., 2009). Overexpression of CypA inhibits M1 translocation into the nucleus (Liu et al., 2009) while depletion of CypA accelerated the replication of the virus (Liu et al., 2012b). Additionally CypA has been shown to increase degradation of M1 though the ubiquitin proteasome system (Liu et al., 2012b). The effect of CsA on influenza A virus infection has also been investigated. CsA-treated mice were administered a dose of influenza A virus that would be lethal for untreated mice, but they survived (Schiltknecht and Ada, 1985). CsA has also been reported to inhibit the propagation of several strains of influenza A virus in cell cultures blocking a late step of the replication cycle by mechanisms that might implicate CypA-dependent and –independent pathways (Hamamoto et al., 2013; Liu et al., 2012a; Ma et al., 2016).
Globally these results point at non-immunosuppressive CsA analogs as promising broad-spectrum antivirals. The most advanced CsA analogue is alisporivir or Debio 025 that has been evaluated in phase II and phase III clinical trials with HCV infected patients (ClinicalTrials.gov Identifier: NCT01215643 and NCT02753699) with promising results on their safety and efficacy. Further studies to evaluate the efficacy of CsA analogs with reduced immunosuppressive activity as antivirals in clinical trials are assured.
6. Silver nanoparticles
The antibacterial effect of silver metal has been known from ancient times (Barillo and Marx, 2014). Recent advances in nanotechnology have allowed the introduction of silver nanoparticles (AgNPs) for biomedical applications (Burdusel et al., 2018). AgNPs can be prepared by several techniques, including physical, chemical and biological methods. Depending on the size and the technique used to prepare the nanoparticles their properties and toxicity can vary (Thorley and Tetley, 2013; Ullah Khan et al., 2018).
AgNPs have been proven to have broad antimicrobial activity against Gram positive and Gram negative bacteria (Kim et al., 2007; Morones et al., 2005). In addition, some studies have shown that AgNPs also have antiviral activity against a broad range of viruses including herpes simplex virus (Baram-Pinto et al., 2009), influenza virus (Papp et al., 2010; Xiang et al., 2013, 2011), hepatitis B virus (Lu et al., 2008), HIV-1 (Elechiguerra et al., 2005; Lara et al., 2010a, b; Sun et al., 2005), Rift Valley fever virus (Borrego et al., 2016), dengue virus (Murugan et al., 2016; Sujitha et al., 2015) and Tacaribe virus (Speshock et al., 2010).
The antiviral mechanism of action of AgNPs is not well defined. AgNPs are thought to interfere with virus-cell attachment and entry into the target cell or by directly inducing structural changes in the virion turning it non-infective (Elechiguerra et al., 2005; Xiang et al., 2013). Additionally, AgNPs can enter the cell and exert their antiviral activity by interfering with cellular proteins needed for viral replication or by direct interaction with viral proteins. Silver ions (Ag+) can react with thiols and phosphate groups present on proteins and nucleic acids inhibiting different steps of the replication cycle (Lara et al., 2010a). AgNPs formulated as topical vaginal gels could be used to prevent the transmission of sexually transmitted infections like HIV and HSV. Studies in an in vitro system using human cervical tissue cultures that simulate in vivo conditions have shown encouraging results inhibiting HIV transmission (Lara et al., 2010b).
Although the use of AgNPs for biomedical applications seems promising, further studies are needed to better understand their potential toxicity and long term effects on human health and in the environment. The toxicity of AgNPs is highly related to the release of Ag+, which directly depends on the size of the nanoparticles. The smaller the size the higher the amount of Ag+ released. Ag+ has been reported to alter mitochondrial function (Chappell and Greville, 1954; Kone et al., 1988). Moreover, exposure of cells to AgNPs induce the production of high levels of reactive oxygen species (ROS) and JNK and p53 activation leading to mitochondria-dependent cell apoptosis (Hsin et al., 2008). Therefore, further studies are needed to better understand the mode of action of AgNPs, their cell specificity and toxicological issues in order to generate new and more effective compounds as well as the use in combination with other drugs in the treatment of different viral diseases.
7. Imaging for studying antivirals
High Content Imaging (HCI) is a popular and basic tool of early drug discovery in multiple disease research areas (Carpenter, 2007). HCI was developed with the contribution of major advances in robotics, imaging and automated image processing (Moffat et al., 2017). HCI is already a fundamental technology for antiviral screening and validation (Bernatchez et al., 2018; Hoenen, 2017; Lowen et al., 2018; Mudhasani et al., 2014; Panchal et al., 2010; Tan et al., 2014).
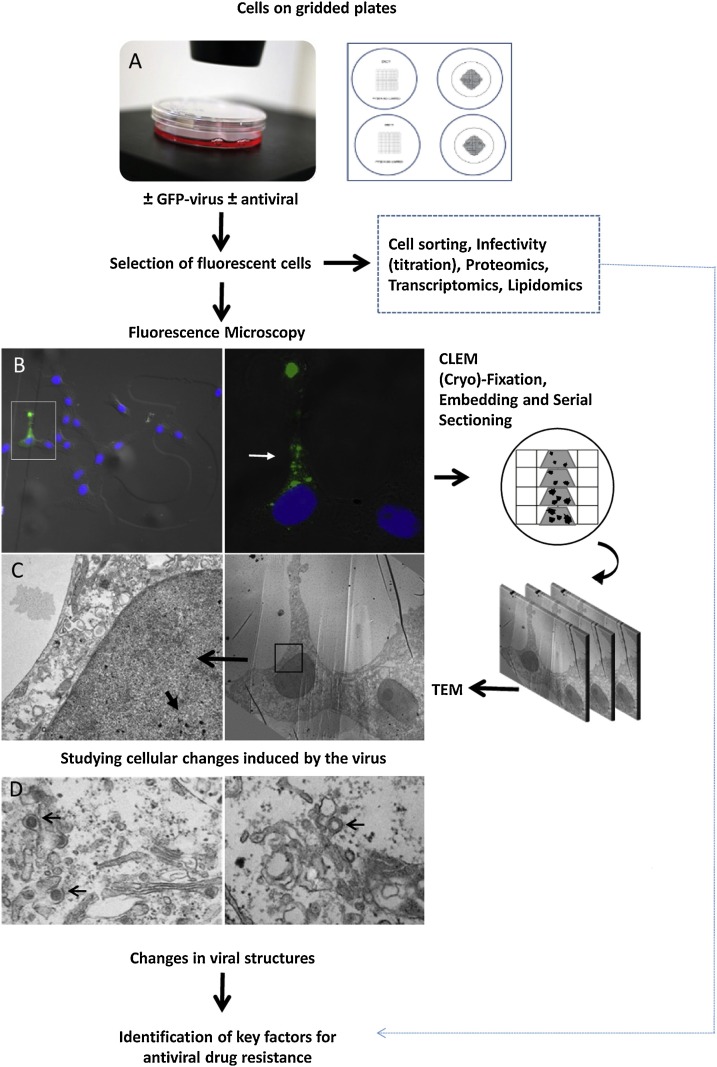
Moreover, recent developments in light and electron microscopy are having a significant impact in virology. In particular, live-cell microscopy, super-resolution microscopy, correlative light and electron microscopy (CLEM) and 3D imaging are now fundamental tools for studying virus-cell interactions (Bykov et al., 2016; Fernandez de Castro et al., 2017; Francis and Melikyan, 2018; Risco et al., 2014; Witte et al., 2018). Microscopy is also a powerful tool for studying antivirals. With CLEM, scientists can study events at the level of single cells in culture (Fernandez de Castro et al., 2014; Hellstrom et al., 2015; Tenorio et al., 2018) and infected animals (Lowen et al., 2018). CLEM is starting to show its potential for testing antivirals because changes in viral structures assembled in cells where the treatment was effective and in those where the antiviral drug failed can be now studied in detail (Berger et al., 2014; Lowen et al., 2018; Martinez et al., 2014).
Cell sorting and CLEM in combination with proteomics, transcriptomics and lipidomics, will be exceptionally informative in identifying new targets for antiviral drugs, to characterize the mechanism of action of old and new antiviral compounds and to understand how resistance to antivirals is developed in cells (Fig. 1 ). The strategy for these studies can be as follows: Duplicated cell cultures infected with a fluorescent virus and treated in the presence or absence of an antiviral drug are processed either by CLEM or cell sorting. Fluorescence microscopy will reveal interesting features in single cells that are then processed for ultrastructural analysis. Electron microscopy will show how viral structures are affected by the antiviral drug. In parallel, key cell factors involved in cell resistance to infection or in viral escape to the antiviral drug will be identified by comparing fluorescent and non-fluorescent cells separated by cell sorting and studied by proteomics, transcriptomics and lipidomics. For these studies, cells that differentiate into tissues and 3D organoids (Liu et al., 2013; Takebe et al., 2015; Walters et al., 2013) will provide more physiologically relevant cellular systems to study virus infection and to test antivirals.
Fig. 1.
Experimental strategy to study the mechanism of action of antiviral drugs in single cells and to investigate the origin of antiviral drug resistance.
8. Conclusions and perspectives
Emerging viruses are a major threat to human health and the rate at which new pathogenic viruses are emerging has accelerated in the past fifty years. The frequent outbreaks have highlighted the urgent need for new antiviral treatments. Generally, antiviral drug development has focused on targeting viral components to block virus entry, replication, morphogenesis and propagation, or on the modulation of the host immune response (Zhu et al., 2015). Drug repurposing is an important alternative. In this review we have chosen several repurposed drugs with proven antiviral activity. With different mechanisms of action, digoxin, sunitinib, chloroquine, cyclosporine A and silver nanoparticles are promising candidates for broad spectrum antivirals that could be used in a combined antiviral therapy. Drug repurposing is now facilitated by a number of resources (Pollastri and Campbell, 2011), such as pathogen target bioinformatics resources, public data repositories of screening data, structural biology resources and compound collections, such as the library of small molecules from the National Institutes of Health (http://www.nihclinicalcollection.com) (Ashbrook et al., 2016; Cao et al., 2015) or the Canadian DrugBank (https://www.drugbank.ca/), a large drug database commonly used by computational drug repositioning methods. DrugBank contains FDA approved drugs and those in clinical trials and is frequently updated with new information. With new and powerful screening assays and prediction tools, in silico structure-based screening of these large chemical libraries is identifying potential inhibitors of viral infections including repurposed compounds that target a variety of viral proteins and host factors (Abu Bakar and Ng, 2018; Barrows et al., 2016; Dowall et al., 2016; Martinez et al., 2015; Yuan et al., 2017). Another strategy that will surely reveal more promising candidates is based on comparing data from databases of pathogens (Brown and Patel, 2017; Sharma et al., 2015) with databases of drugs, e.g., drug information from the National Library of Medicine (https://www.nlm.nih.gov/learn-about-drugs.html). Imaging studies combined with functional assays will show the localization and intracellular effects of these new inhibitors (Panchal et al., 2010). Combination therapies with more than one drug against more than one target will be necessary to minimize problems of resistance to repurposed antiviral drugs.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by a research grant from the Spanish Ministry of Economy, Industry and Competitiveness (BIO2015-68758-R, MINECO-FEDER to CR). BP was recipient of a post-doctoral contract from the Severo Ochoa Center of Excellence Program.
References
- Abu Bakar F., Ng L.F.P. Nonstructural proteins of alphavirus-potential targets for drug development. Viruses. 2018;10 doi: 10.3390/v10020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bari M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017;5 doi: 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashbrook A.W., Lentscher A.J., Zamora P.F., Silva L.A., May N.A., Bauer J.A., Morrison T.E., Dermody T.S. Antagonism of the sodium-potassium ATPase impairs Chikungunya virus infection. MBio. 2016;7 doi: 10.1128/mBio.00693-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins M., Jones C.A., Kirkpatrick P. Sunitinib maleate. Nat. Rev. Drug Discov. 2006;5:279–280. doi: 10.1038/nrd2012. [DOI] [PubMed] [Google Scholar]
- Baram-Pinto D., Shukla S., Perkas N., Gedanken A., Sarid R. Inhibition of herpes simplex virus type 1 infection by silver nanoparticles capped with mercaptoethane sulfonate. Bioconjug. Chem. 2009;20:1497–1502. doi: 10.1021/bc900215b. [DOI] [PubMed] [Google Scholar]
- Barillo D.J., Marx D.E. Silver in medicine: a brief history BC 335 to present. Burns. 2014;40(Suppl. 1):S3–S8. doi: 10.1016/j.burns.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P.K., Sidwell R.W. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir. Chem. Chemother. 2006;17:275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- Barre-Sinoussi F., Ross A.L., Delfraissy J.F. Past, present and future: 30 years of HIV research. Nat. Rev. Microbiol. 2013;11:877–883. doi: 10.1038/nrmicro3132. [DOI] [PubMed] [Google Scholar]
- Barrows N.J., Campos R.K., Powell S.T., Prasanth K.R., Schott-Lerner G., Soto-Acosta R., Galarza-Munoz G., McGrath E.L., Urrabaz-Garza R., Gao J., Wu P., Menon R., Saade G., Fernandez-Salas I., Rossi S.L., Vasilakis N., Routh A., Bradrick S.S., Garcia-Blanco M.A. A screen of FDA-Approved drugs for inhibitors of zika virus infection. Cell Host Microbe. 2016;20:259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekerman E., Neveu G., Shulla A., Brannan J., Pu S.Y., Wang S., Xiao F., Barouch-Bentov R., Bakken R.R., Mateo R., Govero J., Nagamine C.M., Diamond M.S., De Jonghe S., Herdewijn P., Dye J.M., Randall G., Einav S. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Invest. 2017;127:1338–1352. doi: 10.1172/JCI89857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Romero-Brey I., Radujkovic D., Terreux R., Zayas M., Paul D., Harak C., Hoppe S., Gao M., Penin F., Lohmann V., Bartenschlager R. Daclatasvir-like inhibitors of NS5A block early biogenesis of hepatitis C virus-induced membranous replication factories, independent of RNA replication. Gastroenterology. 2014;147:1094–1105. doi: 10.1053/j.gastro.2014.07.019. e1025. [DOI] [PubMed] [Google Scholar]
- Bernatchez J.A., Yang Z., Coste M., Li J., Beck S., Liu Y., Clark A.E., Zhu Z., Luna L.A., Sohl C.D., Purse B.W., Li R., Siqueira-Neto J.L. Development and validation of a phenotypic high-content imaging assay for assessing the antiviral activity of small-molecule inhibitors targeting zika virus. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00725-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska-Haba M., Patel H.D., Sapp M. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N.E. Examination of potential inhibitors of hepatitis A virus uncoating. Intervirology. 1998;41:261–271. doi: 10.1159/000024948. [DOI] [PubMed] [Google Scholar]
- Boonyasuppayakorn S., Reichert E.D., Manzano M., Nagarajan K., Padmanabhan R. Amodiaquine, an antimalarial drug, inhibits dengue virus type 2 replication and infectivity. Antiviral Res. 2014;106:125–134. doi: 10.1016/j.antiviral.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego B., Lorenzo G., Mota-Morales J.D., Almanza-Reyes H., Mateos F., Lopez-Gil E., de la Losa N., Burmistrov V.A., Pestryakov A.N., Brun A., Bogdanchikova N. Potential application of silver nanoparticles to control the infectivity of Rift Valley fever virus in vitro and in vivo. Nanomedicine. 2016;12:1185–1192. doi: 10.1016/j.nano.2016.01.021. [DOI] [PubMed] [Google Scholar]
- Bose S., Mathur M., Bates P., Joshi N., Banerjee A.K. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J. Gen. Virol. 2003;84:1687–1699. doi: 10.1099/vir.0.19074-0. [DOI] [PubMed] [Google Scholar]
- Brown A.S., Patel C.J. A standard database for drug repositioning. Sci. Data. 2017;4:170029. doi: 10.1038/sdata.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdusel A.C., Gherasim O., Grumezescu A.M., Mogoanta L., Ficai A., Andronescu E. Biomedical applications of silver nanoparticles: an up-to-Date overview. Nanomaterials (Basel) 2018;8 doi: 10.3390/nano8090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C., Verheije M.H., Haagmans B.L., van Kuppeveld F.J., Rottier P.J., Bosch B.J., de Haan C.A. ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells. J. Virol. 2015;89:4434–4448. doi: 10.1128/JVI.03274-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov Y.S., Cortese M., Briggs J.A., Bartenschlager R. Correlative light and electron microscopy methods for the study of virus-cell interactions. FEBS Lett. 2016;590:1877–1895. doi: 10.1002/1873-3468.12153. [DOI] [PubMed] [Google Scholar]
- Cai H., Kapoor A., He R., Venkatadri R., Forman M., Posner G.H., Arav-Boger R. In vitro combination of anti-cytomegalovirus compounds acting through different targets: role of the slope parameter and insights into mechanisms of Action. Antimicrob. Agents Chemother. 2014;58:986–994. doi: 10.1128/AAC.01972-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Forrest J.C., Zhang X. A screen of the NIH Clinical Collection small molecule library identifies potential anti-coronavirus drugs. Antiviral Res. 2015;114:1–10. doi: 10.1016/j.antiviral.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E. Image-based chemical screening. Nat. Chem. Biol. 2007;3:461–465. doi: 10.1038/nchembio.2007.15. [DOI] [PubMed] [Google Scholar]
- Chappell J.B., Greville G.D. Effect of silver ions on mitochondrial adenosine triphosphatase. Nature. 1954;174:930–931. doi: 10.1038/174930b0. [DOI] [PubMed] [Google Scholar]
- Chapuis A.G., Paolo Rizzardi G., D’Agostino C., Attinger A., Knabenhans C., Fleury S., Acha-Orbea H., Pantaleo G. Effects of mycophenolic acid on human immunodeficiency virus infection in vitro and in vivo. Nat. Med. 2000;6:762–768. doi: 10.1038/77489. [DOI] [PubMed] [Google Scholar]
- Damaso C.R., Moussatche N. Inhibition of vaccinia virus replication by cyclosporin A analogues correlates with their affinity for cellular cyclophilins. J. Gen. Virol. 1998;79(Pt 2):339–346. doi: 10.1099/0022-1317-79-2-339. [DOI] [PubMed] [Google Scholar]
- De Lamballerie X., Boisson V., Reynier J.C., Enault S., Charrel R.N., Flahault A., Roques P., Le Grand R. On chikungunya acute infection and chloroquine treatment. Vector Borne Zoonotic Dis. 2008;8:837–839. doi: 10.1089/vbz.2008.0049. [DOI] [PubMed] [Google Scholar]
- de Wilde A.H., Zevenhoven-Dobbe J.C., van der Meer Y., Thiel V., Narayanan K., Makino S., Snijder E.J., van Hemert M.J. Cyclosporin A inhibits the replication of diverse coronaviruses. J. Gen. Virol. 2011;92:2542–2548. doi: 10.1099/vir.0.034983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Pham U., Posthuma C.C., Snijder E.J. Cyclophilins and cyclophilin inhibitors in nidovirus replication. Virology. 2018;522:46–55. doi: 10.1016/j.virol.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvecchio R., Higa L.M., Pezzuto P., Valadao A.L., Garcez P.P., Monteiro F.L., Loiola E.C., Dias A.A., Silva F.J., Aliota M.T., Caine E.A., Osorio J.E., Bellio M., O’Connor D.H., Rehen S., de Aguiar R.S., Savarino A., Campanati L., Tanuri A. Chloroquine, an endocytosis blocking agent, inhibits zika virus infection in different cell models. Viruses. 2016;8 doi: 10.3390/v8120322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Trani L., Savarino A., Campitelli L., Norelli S., Puzelli S., D’Ostilio D., Vignolo E., Donatelli I., Cassone A. Different pH requirements are associated with divergent inhibitory effects of chloroquine on human and avian influenza A viruses. Virol. J. 2007;4:39. doi: 10.1186/1743-422X-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M.S., Zachariah M., Harris E. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology. 2002;304:211–221. doi: 10.1006/viro.2002.1685. [DOI] [PubMed] [Google Scholar]
- DiMasi J.A., Grabowski H.G., Hansen R.W. Innovation in the pharmaceutical industry: new estimates of R&D costs. J. Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Dodson A.W., Taylor T.J., Knipe D.M., Coen D.M. Inhibitors of the sodium potassium ATPase that impair herpes simplex virus replication identified via a chemical screening approach. Virology. 2007;366:340–348. doi: 10.1016/j.virol.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall S.D., Bewley K., Watson R.J., Vasan S.S., Ghosh C., Konai M.M., Gausdal G., Lorens J.B., Long J., Barclay W., Garcia-Dorival I., Hiscox J., Bosworth A., Taylor I., Easterbrook L., Pitman J., Summers S., Chan-Pensley J., Funnell S., Vipond J., Charlton S., Haldar J., Hewson R., Carroll M.W. Antiviral screening of multiple compounds against ebola virus. Viruses. 2016;8 doi: 10.3390/v8110277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jr., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elechiguerra J.L., Burt J.L., Morones J.R., Camacho-Bragado A., Gao X., Lara H.H., Yacaman M.J. Interaction of silver nanoparticles with HIV-1. J. Nanobiotechnol. 2005;3:6. doi: 10.1186/1477-3155-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias K.J., Machado P.R., de Almeida Junior R.F., de Aquino A.A., da Fonseca B.A. Chloroquine interferes with dengue-2 virus replication in U937 cells. Microbiol. Immunol. 2014;58:318–326. doi: 10.1111/1348-0421.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias K.J., Machado P.R., Muniz J.A., Imbeloni A.A., da Fonseca B.A. Antiviral activity of chloroquine against dengue virus type 2 replication in Aotus monkeys. Viral Immunol. 2015;28:161–169. doi: 10.1089/vim.2014.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de Castro I., Sanz-Sanchez L., Risco C. Metallothioneins for correlative light and electron microscopy. Methods Cell Biol. 2014;124:55–70. doi: 10.1016/B978-0-12-801075-4.00003-3. [DOI] [PubMed] [Google Scholar]
- Fernandez de Castro I., Fernandez J.J., Barajas D., Nagy P.D., Risco C. Three-dimensional imaging of the intracellular assembly of a functional viral RNA replicase complex. J. Cell. Sci. 2017;130:260–268. doi: 10.1242/jcs.181586. [DOI] [PubMed] [Google Scholar]
- Ferraris O., Moroso M., Pernet O., Emonet S., Ferrier Rembert A., Paranhos-Baccala G., Peyrefitte C.N. Evaluation of Crimean-Congo hemorrhagic fever virus in vitro inhibition by chloroquine and chlorpromazine, two FDA approved molecules. Antiviral Res. 2015;118:75–81. doi: 10.1016/j.antiviral.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flisiak R., Horban A., Gallay P., Bobardt M., Selvarajah S., Wiercinska-Drapalo A., Siwak E., Cielniak I., Higersberger J., Kierkus J., Aeschlimann C., Grosgurin P., Nicolas-Metral V., Dumont J.M., Porchet H., Crabbe R., Scalfaro P. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- Francis A.C., Melikyan G.B. Live-cell imaging of early steps of single HIV-1. Infection. Viruses. 2018;10 doi: 10.3390/v10050275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke E.K., Yuan H.E., Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Gheorghiade M., van Veldhuisen D.J., Colucci W.S. Contemporary use of digoxin in the management of cardiovascular disorders. Circulation. 2006;113:2556–2564. doi: 10.1161/CIRCULATIONAHA.105.560110. [DOI] [PubMed] [Google Scholar]
- Grosso F., Stoilov P., Lingwood C., Brown M., Cochrane A. Suppression of adenovirus replication by cardiotonic steroids. J. Virol. 2017;91 doi: 10.1128/JVI.01623-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Xu X., Rasheed T.K., Yoder A., Yu D., Liang H., Yi F., Hawley T., Jin T., Ling B., Wu Y. Genistein interferes with SDF-1- and HIV-mediated actin dynamics and inhibits HIV infection of resting CD4 T cells. Retrovirology. 2013;10:62. doi: 10.1186/1742-4690-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto I., Harazaki K., Inase N., Takaku H., Tashiro M., Yamamoto N. Cyclosporin A inhibits the propagation of influenza virus by interfering with a late event in the virus life cycle. Jpn. J. Infect. Dis. 2013;66:276–283. doi: 10.7883/yoken.66.276. [DOI] [PubMed] [Google Scholar]
- Handschumacher R.E., Harding M.W., Rice J., Drugge R.J., Speicher D.W. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- Hartley C., Hartley M., Pardoe I., Knight A. Ionic Contra-Viral Therapy (ICVT); a new approach to the treatment of DNA virus infections. Arch. Virol. 2006;151:2495–2501. doi: 10.1007/s00705-006-0824-x. [DOI] [PubMed] [Google Scholar]
- Hartono C., Muthukumar T., Suthanthiran M. Immunosuppressive drug therapy. Cold Spring Harb. Perspect. Med. 2013;3 doi: 10.1101/cshperspect.a015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom K., Vihinen H., Kallio K., Jokitalo E., Ahola T. Correlative light and electron microscopy enables viral replication studies at the ultrastructural level. Methods. 2015;90:49–56. doi: 10.1016/j.ymeth.2015.04.019. [DOI] [PubMed] [Google Scholar]
- Henry S.D., Metselaar H.J., Lonsdale R.C., Kok A., Haagmans B.L., Tilanus H.W., van der Laan L.J. Mycophenolic acid inhibits hepatitis C virus replication and acts in synergy with cyclosporin A and interferon-alpha. Gastroenterology. 2006;131:1452–1462. doi: 10.1053/j.gastro.2006.08.027. [DOI] [PubMed] [Google Scholar]
- Hoenen T. Luciferase-expressing Ebolaviruses as tools for screening of antivirals. Methods Mol. Biol. 2017;1628:189–194. doi: 10.1007/978-1-4939-7116-9_14. [DOI] [PubMed] [Google Scholar]
- Howard C.R., Fletcher N.F. Emerging virus diseases: can we ever expect the unexpected? Emerg. Microbes Infect. 2012;1:e46. doi: 10.1038/emi.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin Y.H., Chen C.F., Huang S., Shih T.S., Lai P.S., Chueh P.J. The apoptotic effect of nanosilver is mediated by a ROS- and JNK-dependent mechanism involving the mitochondrial pathway in NIH3T3 cells. Toxicol. Lett. 2008;179:130–139. doi: 10.1016/j.toxlet.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Huh J.R., Leung M.W., Huang P., Ryan D.A., Krout M.R., Malapaka R.R., Chow J., Manel N., Ciofani M., Kim S.V., Cuesta A., Santori F.R., Lafaille J.J., Xu H.E., Gin D.Y., Rastinejad F., Littman D.R. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida J., Konishi M., Ebner N., Springer J. Repurposing of approved cardiovascular drugs. J. Transl. Med. 2016;14:269. doi: 10.1186/s12967-016-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyidogan P., Anderson K.S. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses. 2014;6:4095–4139. doi: 10.3390/v6104095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha B.K., Dong B., Nguyen C.T., Polyakova I., Silverman R.H. Suppression of antiviral innate immunity by sunitinib enhances oncolytic virotherapy. Mol. Ther. 2013;21:1749–1757. doi: 10.1038/mt.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Cai H., Forman M., He R., Shamay M., Arav-Boger R. Human cytomegalovirus inhibition by cardiac glycosides: evidence for involvement of the HERG gene. Antimicrob. Agents Chemother. 2012;56:4891–4899. doi: 10.1128/AAC.00898-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul A., Stauffer S., Berger C., Pertel T., Schmitt J., Kallis S., Zayas M., Lohmann V., Luban J., Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R., Klichko V., Margolis D. Ex vivo modeling of the effects of mycophenolic acid on HIV infection: considerations for antiviral therapy. AIDS Res. Hum. Retroviruses. 2005;21:116–124. doi: 10.1089/aid.2005.21.116. [DOI] [PubMed] [Google Scholar]
- Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Santhosh S.R., Tiwari M., Lakshmana Rao P.V., Parida M. Assessment of in vitro prophylactic and therapeutic efficacy of chloroquine against Chikungunya virus in vero cells. J. Med. Virol. 2010;82:817–824. doi: 10.1002/jmv.21663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Kuk E., Yu K.N., Kim J.H., Park S.J., Lee H.J., Kim S.H., Park Y.K., Park Y.H., Hwang C.Y., Kim Y.K., Lee Y.S., Jeong D.H., Cho M.H. Antimicrobial effects of silver nanoparticles. Nanomedicine. 2007;3:95–101. doi: 10.1016/j.nano.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Kone B.C., Kaleta M., Gullans S.R. Silver ion (Ag+)-induced increases in cell membrane K+ and Na+ permeability in the renal proximal tubule: reversal by thiol reagents. J. Membr. Biol. 1988;102:11–19. doi: 10.1007/BF01875349. [DOI] [PubMed] [Google Scholar]
- Kovackova S., Chang L., Bekerman E., Neveu G., Barouch-Bentov R., Chaikuad A., Heroven C., Sala M., De Jonghe S., Knapp S., Einav S., Herdewijn P. Selective inhibitors of cyclin g associated kinase (GAK) as anti-hepatitis C agents. J. Med. Chem. 2015;58:3393–3410. doi: 10.1021/jm501759m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.C., Chuang Y.C., Chang C.P., Lin Y.S., Perng G.C., Wu H.C., Hsieh S.L., Yeh T.M. Minocycline suppresses dengue virus replication by down-regulation of macrophage migration inhibitory factor-induced autophagy. Antiviral Res. 2018;155:28–38. doi: 10.1016/j.antiviral.2018.05.002. [DOI] [PubMed] [Google Scholar]
- Laird G.M., Eisele E.E., Rabi S.A., Nikolaeva D., Siliciano R.F. A novel cell-based high-throughput screen for inhibitors of HIV-1 gene expression and budding identifies the cardiac glycosides. J. Antimicrob. Chemother. 2014;69:988–994. doi: 10.1093/jac/dkt471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara H.H., Ayala-Nunez N.V., Ixtepan-Turrent L., Rodriguez-Padilla C. Mode of antiviral action of silver nanoparticles against HIV-1. J. Nanobiotechnology. 2010;8:1. doi: 10.1186/1477-3155-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara H.H., Ixtepan-Turrent L., Garza-Trevino E.N., Rodriguez-Padilla C. PVP-coated silver nanoparticles block the transmission of cell-free and cell-associated HIV-1 in human cervical culture. J. Nanobiotechnology. 2010;8:15. doi: 10.1186/1477-3155-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leela S.L., Srisawat C., Sreekanth G.P., Noisakran S., Yenchitsomanus P.T., Limjindaporn T. Drug repurposing of minocycline against dengue virus infection. Biochem. Biophys. Res. Commun. 2016;478:410–416. doi: 10.1016/j.bbrc.2016.07.029. [DOI] [PubMed] [Google Scholar]
- Lemaitre M., Guetard D., Henin Y., Montagnier L., Zerial A. Protective activity of tetracycline analogs against the cytopathic effect of the human immunodeficiency viruses in CEM cells. Res. Virol. 1990;141:5–16. doi: 10.1016/0923-2516(90)90052-k. [DOI] [PubMed] [Google Scholar]
- Li G., De Clercq E. Current therapy for chronic hepatitis C: the role of direct-acting antivirals. Antiviral Res. 2017;142:83–122. doi: 10.1016/j.antiviral.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chen C., Wong G., Dong W., Zheng W., Li Y., Sun L., Zhang L., Gao G.F., Bi Y., Liu W. Cyclophilin A protects mice against infection by influenza A virus. Sci. Rep. 2016;6:28978. doi: 10.1038/srep28978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhu X., Ji X., Quanquin N., Deng Y.Q., Tian M., Aliyari R., Zuo X., Yuan L., Afridi S.K., Li X.F., Jung J.U., Nielsen-Saines K., Qin F.X., Qin C.F., Xu Z., Cheng G. Chloroquine, a FDA-approved drug, prevents zika virus infection and its associated congenital microcephaly in mice. EBioMedicine. 2017;24:189–194. doi: 10.1016/j.ebiom.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Gallay P. Curing a viral infection by targeting the host: the example of cyclophilin inhibitors. Antiviral Res. 2013;99:68–77. doi: 10.1016/j.antiviral.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Sun L., Yu M., Wang Z., Xu C., Xue Q., Zhang K., Ye X., Kitamura Y., Liu W. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell. Microbiol. 2009;11:730–741. doi: 10.1111/j.1462-5822.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- Liu X., Zhao Z., Li Z., Xu C., Sun L., Chen J., Liu W. Cyclosporin A inhibits the influenza virus replication through cyclophilin A-dependent and -independent pathways. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhao Z., Xu C., Sun L., Chen J., Zhang L., Liu W. Cyclophilin A restricts influenza A virus replication through degradation of the M1 protein. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yang R., He Z., Gao W.Q. Generation of functional organs from stem cells. Cell Regen. (Lond.) 2013;2:1. doi: 10.1186/2045-9769-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen R.G., Bocan T.M., Kane C.D., Cazares L.H., Kota K.P., Ladner J.T., Nasar F., Pitt L., Smith D.R., Soloveva V., Sun M.G., Zeng X., Bavari S. Countering zika virus: the USAMRIID response. Adv. Exp. Med. Biol. 2018;1062:303–318. doi: 10.1007/978-981-10-8727-1_21. [DOI] [PubMed] [Google Scholar]
- Lu L., Sun R.W., Chen R., Hui C.K., Ho C.M., Luk J.M., Lau G.K., Che C.M. Silver nanoparticles inhibit hepatitis B virus replication. Antivir. Ther. (Lond.) 2008;13:253–262. [PubMed] [Google Scholar]
- Luo C., Luo H., Zheng S., Gui C., Yue L., Yu C., Sun T., He P., Chen J., Shen J., Luo X., Li Y., Liu H., Bai D., Shen J., Yang Y., Li F., Zuo J., Hilgenfeld R., Pei G., Chen K., Shen X., Jiang H. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem. Biophys. Res. Commun. 2004;321:557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Li F., Musharrafieh R.G., Wang J. Discovery of cyclosporine A and its analogs as broad-spectrum anti-influenza drugs with a high in vitro genetic barrier of drug resistance. Antiviral Res. 2016;133:62–72. doi: 10.1016/j.antiviral.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid P.B., Chopra S., Manger I.D., Gilfillan L., Keepers T.R., Shurtleff A.C., Green C.E., Iyer L.V., Dilks H.H., Davey R.A., Kolokoltsov A.A., Carrion R., Jr., Patterson J.L., Bavari S., Panchal R.G., Warren T.K., Wells J.B., Moos W.H., Burke R.L., Tanga M.J. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis D., Heredia A., Gaywee J., Oldach D., Drusano G., Redfield R. Abacavir and mycophenolic acid, an inhibitor of inosine monophosphate dehydrogenase, have profound and synergistic anti-HIV activity. J. Acquir. Immune Defic. Syndr. 1999;21:362–370. [PubMed] [Google Scholar]
- Martinez M.G., Snapp E.L., Perumal G.S., Macaluso F.P., Kielian M. Imaging the alphavirus exit pathway. J. Virol. 2014;88:6922–6933. doi: 10.1128/JVI.00592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J.P., Sasse F., Bronstrup M., Diez J., Meyerhans A. Antiviral drug discovery: broad-spectrum drugs from nature. Nat. Prod. Rep. 2015;32:29–48. doi: 10.1039/c4np00085d. [DOI] [PubMed] [Google Scholar]
- Matsuda S., Koyasu S. Mechanisms of action of cyclosporine. Immunopharmacology. 2000;47:119–125. doi: 10.1016/s0162-3109(00)00192-2. [DOI] [PubMed] [Google Scholar]
- Mendel D.B., Laird A.D., Xin X., Louie S.G., Christensen J.G., Li G., Schreck R.E., Abrams T.J., Ngai T.J., Lee L.B., Murray L.J., Carver J., Chan E., Moss K.G., Haznedar J.O., Sukbuntherng J., Blake R.A., Sun L., Tang C., Miller T., Shirazian S., McMahon G., Cherrington J.M. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin. Cancer Res. 2003;9:327–337. [PubMed] [Google Scholar]
- Menendez-Arias L., Alvarez M., Pacheco B. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance. Curr. Opin. Virol. 2014;8:1–9. doi: 10.1016/j.coviro.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Mercorelli B., Palu G., Loregian A. Drug Repurposing for Viral Infectious Diseases: How Far Are We? Trends Microbiol. 2018;26:865–876. doi: 10.1016/j.tim.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis M., Kleinschmidt M.C., Doerr H.W., Cinatl J., Jr. Minocycline inhibits West Nile virus replication and apoptosis in human neuronal cells. J. Antimicrob. Chemother. 2007;60:981–986. doi: 10.1093/jac/dkm307. [DOI] [PubMed] [Google Scholar]
- Mishra M.K., Basu A. Minocycline neuroprotects, reduces microglial activation, inhibits caspase 3 induction, and viral replication following Japanese encephalitis. J. Neurochem. 2008;105:1582–1595. doi: 10.1111/j.1471-4159.2008.05238.x. [DOI] [PubMed] [Google Scholar]
- Moffat J.G., Vincent F., Lee J.A., Eder J., Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- Morones J.R., Elechiguerra J.L., Camacho A., Holt K., Kouri J.B., Ramirez J.T., Yacaman M.J. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16:2346–2353. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- Mudhasani R., Kota K.P., Retterer C., Tran J.P., Whitehouse C.A., Bavari S. High content image-based screening of a protease inhibitor library reveals compounds broadly active against Rift Valley fever virus and other highly pathogenic RNA viruses. PLoS Negl. Trop. Dis. 2014;8:e3095. doi: 10.1371/journal.pntd.0003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan K., Aruna P., Panneerselvam C., Madhiyazhagan P., Paulpandi M., Subramaniam J., Rajaganesh R., Wei H., Alsalhi M.S., Devanesan S., Nicoletti M., Syuhei B., Canale A., Benelli G. Fighting arboviral diseases: low toxicity on mammalian cells, dengue growth inhibition (in vitro), and mosquitocidal activity of Centroceras clavulatum-synthesized silver nanoparticles. Parasitol. Res. 2016;115:651–662. doi: 10.1007/s00436-015-4783-6. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Sakamoto N., Enomoto N., Tanabe Y., Kanazawa N., Koyama T., Kurosaki M., Maekawa S., Yamashiro T., Chen C.H., Itsui Y., Kakinuma S., Watanabe M. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem. Biophys. Res. Commun. 2004;313:42–47. doi: 10.1016/j.bbrc.2003.11.080. [DOI] [PubMed] [Google Scholar]
- Nesher M., Shpolansky U., Rosen H., Lichtstein D. The digitalis-like steroid hormones: new mechanisms of action and biological significance. Life Sci. 2007;80:2093–2107. doi: 10.1016/j.lfs.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates 3rd J.R., Wuthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu G., Barouch-Bentov R., Ziv-Av A., Gerber D., Jacob Y., Einav S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu G., Ziv-Av A., Barouch-Bentov R., Berkerman E., Mulholland J., Einav S. AP-2-associated protein kinase 1 and cyclin G-associated kinase regulate hepatitis C virus entry and are potential drug targets. J. Virol. 2015;89:4387–4404. doi: 10.1128/JVI.02705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534:314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- Okuyama-Dobashi K., Kasai H., Tanaka T., Yamashita A., Yasumoto J., Chen W., Okamoto T., Maekawa S., Watashi K., Wakita T., Ryo A., Suzuki T., Matsuura Y., Enomoto N., Moriishi K. Hepatitis B virus efficiently infects non-adherent hepatoma cells via human sodium taurocholate cotransporting polypeptide. Sci. Rep. 2015;5:17047. doi: 10.1038/srep17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi E.E., Chew J.S., Loh J.P., Chua R.C. In vitro inhibition of human influenza A virus replication by chloroquine. Virol. J. 2006;3:39. doi: 10.1186/1743-422X-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeshuyse J., Kaul A., De Clercq E., Rosenwirth B., Dumont J.M., Scalfaro P., Bartenschlager R., Neyts J. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43:761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- Pan Q., de Ruiter P.E., Metselaar H.J., Kwekkeboom J., de Jonge J., Tilanus H.W., Janssen H.L., van der Laan L.J. Mycophenolic acid augments interferon-stimulated gene expression and inhibits hepatitis C Virus infection in vitro and in vivo. Hepatology. 2012;55:1673–1683. doi: 10.1002/hep.25562. [DOI] [PubMed] [Google Scholar]
- Panchal R.G., Kota K.P., Spurgers K.B., Ruthel G., Tran J.P., Boltz R.C., Bavari S. Development of high-content imaging assays for lethal viral pathogens. J. Biomol. Screen. 2010;15:755–765. doi: 10.1177/1087057110374357. [DOI] [PubMed] [Google Scholar]
- Papp I., Sieben C., Ludwig K., Roskamp M., Bottcher C., Schlecht S., Herrmann A., Haag R. Inhibition of influenza virus infection by multivalent sialic-acid-functionalized gold nanoparticles. Small. 2010;6:2900–2906. doi: 10.1002/smll.201001349. [DOI] [PubMed] [Google Scholar]
- Paton N.I., Lee L., Xu Y., Ooi E.E., Cheung Y.B., Archuleta S., Wong G., Wilder-Smith A. Chloroquine for influenza prevention: a randomised, double-blind, placebo controlled trial. Lancet Infect. Dis. 2011;11:677–683. doi: 10.1016/S1473-3099(11)70065-2. [DOI] [PubMed] [Google Scholar]
- Perales C., Quer J., Gregori J., Esteban J.I., Domingo E. Resistance of hepatitis C virus to inhibitors: complexity and clinical implications. Viruses. 2015;7:5746–5766. doi: 10.3390/v7112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Schopf J., Kogl M., Friedel C.C., Muller M.A., Carbajo-Lozoya J., Stellberger T., von Dall’Armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Zust R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Wessels C., Pohlmann S., Haas J., Drosten C., von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri M.P., Campbell R.K. Target repurposing for neglected diseases. Future Med. Chem. 2011;3:1307–1315. doi: 10.4155/fmc.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prassas I., Diamandis E.P. Novel therapeutic applications of cardiac glycosides. Nat. Rev. Drug Discov. 2008;7:926–935. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- Pu S.Y., Xiao F., Schor S., Bekerman E., Zanini F., Barouch-Bentov R., Nagamine C.M., Einav S. Feasibility and biological rationale of repurposing sunitinib and erlotinib for dengue treatment. Antiviral Res. 2018;155:67–75. doi: 10.1016/j.antiviral.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainsford K.D., Parke A.L., Clifford-Rashotte M., Kean W.F. Therapy and pharmacological properties of hydroxychloroquine and chloroquine in treatment of systemic lupus erythematosus, rheumatoid arthritis and related diseases. Inflammopharmacology. 2015;23:231–269. doi: 10.1007/s10787-015-0239-y. [DOI] [PubMed] [Google Scholar]
- Randolph V.B., Winkler G., Stollar V. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology. 1990;174:450–458. doi: 10.1016/0042-6822(90)90099-d. [DOI] [PubMed] [Google Scholar]
- Risco C., de Castro I.F., Sanz-Sanchez L., Narayan K., Grandinetti G., Subramaniam S. Three-dimensional imaging of viral infections. Annu. Rev. Virol. 2014;1:453–473. doi: 10.1146/annurev-virology-031413-085351. [DOI] [PubMed] [Google Scholar]
- Roques P., Thiberville S.D., Dupuis-Maguiraga L., Lum F.M., Labadie K., Martinon F., Gras G., Lebon P., Ng L.F.P., de Lamballerie X., Le Grand R. Paradoxical effect of chloroquine treatment in enhancing Chikungunya virus infection. Viruses. 2018;10 doi: 10.3390/v10050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino A., Lucia M.B., Rastrelli E., Rutella S., Golotta C., Morra E., Tamburrini E., Perno C.F., Boelaert J.R., Sperber K., Cauda R. Anti-HIV effects of chloroquine: inhibition of viral particle glycosylation and synergism with protease inhibitors. J. Acquir. Immune Defic. Syndr. 2004;35:223–232. doi: 10.1097/00126334-200403010-00002. [DOI] [PubMed] [Google Scholar]
- Schiltknecht E., Ada G.L. In vivo effects of cyclosporine on influenza A virus-infected mice. Cell. Immunol. 1985;91:227–239. doi: 10.1016/0008-8749(85)90046-2. [DOI] [PubMed] [Google Scholar]
- Schmid F.X. Protein folding. Prolyl isomerases join the fold. Curr. Biol. 1995;5:993–994. doi: 10.1016/s0960-9822(95)00197-7. [DOI] [PubMed] [Google Scholar]
- Schor S., Einav S. Repurposing of kinase inhibitors as broad-spectrum antiviral drugs. DNA Cell Biol. 2018;37:63–69. doi: 10.1089/dna.2017.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S.L. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science. 1991;251:283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- Sharma D., Priyadarshini P., Vrati S. Unraveling the web of viroinformatics: computational tools and databases in virus research. J. Virol. 2015;89:1489–1501. doi: 10.1128/JVI.02027-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Hahn B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 2011;1 doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev S.A., Mesci P., Pinto A., Fernandes I., Sheets N., Shresta S., Farhy C., Huang C.T., Strongin A.Y., Muotri A.R., Terskikh A.V. Repurposing of the anti-malaria drug chloroquine for Zika Virus treatment and prophylaxis. Sci. Rep. 2017;7:15771. doi: 10.1038/s41598-017-15467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater A.F. Chloroquine: mechanism of drug action and resistance in Plasmodium falciparum. Pharmacol. Ther. 1993;57:203–235. doi: 10.1016/0163-7258(93)90056-j. [DOI] [PubMed] [Google Scholar]
- Smith T.W. Digitalis. Mechanisms of action and clinical use. N. Engl. J. Med. 1988;318:358–365. doi: 10.1056/NEJM198802113180606. [DOI] [PubMed] [Google Scholar]
- Sourisseau M., Schilte C., Casartelli N., Trouillet C., Guivel-Benhassine F., Rudnicka D., Sol-Foulon N., Le Roux K., Prevost M.C., Fsihi H., Frenkiel M.P., Blanchet F., Afonso P.V., Ceccaldi P.E., Ozden S., Gessain A., Schuffenecker I., Verhasselt B., Zamborlini A., Saib A., Rey F.A., Arenzana-Seisdedos F., Despres P., Michault A., Albert M.L., Schwartz O. Characterization of reemerging chikungunya virus. PLoS Pathog. 2007;3:e89. doi: 10.1371/journal.ppat.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber K., Kalb T.H., Stecher V.J., Banerjee R., Mayer L. Inhibition of human immunodeficiency virus type 1 replication by hydroxychloroquine in T cells and monocytes. AIDS Res. Hum. Retroviruses. 1993;9:91–98. doi: 10.1089/aid.1993.9.91. [DOI] [PubMed] [Google Scholar]
- Speshock J.L., Murdock R.C., Braydich-Stolle L.K., Schrand A.M., Hussain S.M. Interaction of silver nanoparticles with Tacaribe virus. J. Nanobiotechnology. 2010;8:19. doi: 10.1186/1477-3155-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Bi Y., Wong G., Gray G.C., Gao G.F., Li S. Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J. Virol. 2015;89:8671–8676. doi: 10.1128/JVI.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujitha V., Murugan K., Paulpandi M., Panneerselvam C., Suresh U., Roni M., Nicoletti M., Higuchi A., Madhiyazhagan P., Subramaniam J., Dinesh D., Vadivalagan C., Chandramohan B., Alarfaj A.A., Munusamy M.A., Barnard D.R., Benelli G. Green-synthesized silver nanoparticles as a novel control tool against dengue virus (DEN-2) and its primary vector Aedes aegypti. Parasitol. Res. 2015;114:3315–3325. doi: 10.1007/s00436-015-4556-2. [DOI] [PubMed] [Google Scholar]
- Sun L., Liang C., Shirazian S., Zhou Y., Miller T., Cui J., Fukuda J.Y., Chu J.Y., Nematalla A., Wang X., Chen H., Sistla A., Luu T.C., Tang F., Wei J., Tang C. Discovery of 5-[5-fluoro-2-oxo-1,2- dihydroindol-(3Z)-ylidenemethyl]-2,4- dimethyl-1H-pyrrole-3-carboxylic acid (2-diethylaminoethyl)amide, a novel tyrosine kinase inhibitor targeting vascular endothelial and platelet-derived growth factor receptor tyrosine kinase. J. Med. Chem. 2003;46:1116–1119. doi: 10.1021/jm0204183. [DOI] [PubMed] [Google Scholar]
- Sun R.W., Chen R., Chung N.P., Ho C.M., Lin C.L., Che C.M. Silver nanoparticles fabricated in Hepes buffer exhibit cytoprotective activities toward HIV-1 infected cells. Chem. Commun. (Camb.) 2005:5059–5061. doi: 10.1039/b510984a. [DOI] [PubMed] [Google Scholar]
- Szeto G.L., Brice A.K., Yang H.C., Barber S.A., Siliciano R.F., Clements J.E. Minocycline attenuates HIV infection and reactivation by suppressing cellular activation in human CD4+ T cells. J. Infect. Dis. 2010;201:1132–1140. doi: 10.1086/651277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T., Enomura M., Yoshizawa E., Kimura M., Koike H., Ueno Y., Matsuzaki T., Yamazaki T., Toyohara T., Osafune K., Nakauchi H., Yoshikawa H.Y., Taniguchi H. Vascularized and Complex Organ Buds from Diverse Tissues via Mesenchymal Cell-Driven Condensation. Cell Stem Cell. 2015;16:556–565. doi: 10.1016/j.stem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Takhampunya R., Ubol S., Houng H.S., Cameron C.E., Padmanabhan R. Inhibition of dengue virus replication by mycophenolic acid and ribavirin. J. Gen. Virol. 2006;87:1947–1952. doi: 10.1099/vir.0.81655-0. [DOI] [PubMed] [Google Scholar]
- Tan K.H., Ki K.C., Watanabe S., Vasudevan S.G., Krishnan M. Cell-based flavivirus infection (CFI) assay for the evaluation of dengue antiviral candidates using high-content imaging. Methods Mol. Biol. 2014;1138:99–109. doi: 10.1007/978-1-4939-0348-1_7. [DOI] [PubMed] [Google Scholar]
- Tanner W.D., Toth D.J., Gundlapalli A.V. The pandemic potential of avian influenza A(H7N9) virus: a review. Epidemiol. Infect. 2015;143:3359–3374. doi: 10.1017/S0950268815001570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenorio R., Fernandez de Castro I., Knowlton J.J., Zamora P.F., Lee C.H., Mainou B.A., Dermody T.S., Risco C. Reovirus sigmaNS and muNS proteins remodel the endoplasmic reticulum to build replication neo-organelles. MBio. 2018;9 doi: 10.1128/mBio.01253-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M., Bukovsky A., Kondo E., Rosenwirth B., Walsh C.T., Sodroski J., Gottlinger H.G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- Thome R., Lopes S.C., Costa F.T., Verinaud L. Chloroquine: modes of action of an undervalued drug. Immunol. Lett. 2013;153:50–57. doi: 10.1016/j.imlet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Thorley A.J., Tetley T.D. New perspectives in nanomedicine. Pharmacol. Ther. 2013;140:176–185. doi: 10.1016/j.pharmthera.2013.06.008. [DOI] [PubMed] [Google Scholar]
- To K.K., Zhou J., Chan J.F., Yuen K.Y. Host genes and influenza pathogenesis in humans: an emerging paradigm. Curr. Opin. Virol. 2015;14:7–15. doi: 10.1016/j.coviro.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricou V., Minh N.N., Van T.P., Lee S.J., Farrar J., Wills B., Tran H.T., Simmons C.P. A randomized controlled trial of chloroquine for the treatment of dengue in Vietnamese adults. PLoS Negl. Trop. Dis. 2010;4:e785. doi: 10.1371/journal.pntd.0000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W.P., Nara P.L., Kung H.F., Oroszlan S. Inhibition of human immunodeficiency virus infectivity by chloroquine. AIDS Res. Hum. Retroviruses. 1990;6:481–489. doi: 10.1089/aid.1990.6.481. [DOI] [PubMed] [Google Scholar]
- Ullah Khan S., Saleh T.A., Wahab A., Khan M.H.U., Khan D., Ullah Khan W., Rahim A., Kamal S., Ullah Khan F., Fahad S. Nanosilver: new ageless and versatile biomedical therapeutic scaffold. Int. J. Nanomedicine. 2018;13:733–762. doi: 10.2147/IJN.S153167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaklavas C., Chatzizisis Y.S., Tsimberidou A.M. Common cardiovascular medications in cancer therapeutics. Pharmacol. Ther. 2011;130:177–190. doi: 10.1016/j.pharmthera.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainberg M.A., Dascal A., Blain N., Fitz-Gibbon L., Boulerice F., Numazaki K., Tremblay M. The effect of cyclosporine A on infection of susceptible cells by human immunodeficiency virus type 1. Blood. 1988;72:1904–1910. [PubMed] [Google Scholar]
- Walters M.S., Gomi K., Ashbridge B., Moore M.A., Arbelaez V., Heldrich J., Ding B.S., Rafii S., Staudt M.R., Crystal R.G. Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir. Res. 2013;14:135. doi: 10.1186/1465-9921-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watashi K., Sluder A., Daito T., Matsunaga S., Ryo A., Nagamori S., Iwamoto M., Nakajima S., Tsukuda S., Borroto-Esoda K., Sugiyama M., Tanaka Y., Kanai Y., Kusuhara H., Mizokami M., Wakita T. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP) Hepatology. 2014;59:1726–1737. doi: 10.1002/hep.26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N.J. The treatment of malaria. N. Engl. J. Med. 1996;335:800–806. doi: 10.1056/NEJM199609123351107. [DOI] [PubMed] [Google Scholar]
- Witte R., Andriasyan V., Georgi F., Yakimovich A., Greber U.F. Concepts in Light Microscopy of Viruses. Viruses. 2018;10 doi: 10.3390/v10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R.W., Balachandran A., Ostrowski M.A., Cochrane A. Digoxin suppresses HIV-1 replication by altering viral RNA processing. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D.X., Chen Q., Pang L., Zheng C.L. Inhibitory effects of silver nanoparticles on H1N1 influenza A virus in vitro. J. Virol. Methods. 2011;178:137–142. doi: 10.1016/j.jviromet.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Xiang D., Zheng Y., Duan W., Li X., Yin J., Shigdar S., O’Connor M.L., Marappan M., Zhao X., Miao Y., Xiang B., Zheng C. Inhibition of A/Human/Hubei/3/2005 (H3N2) influenza virus infection by silver nanoparticles in vitro and in vivo. Int. J. Nanomedicine. 2013;8:4103–4113. doi: 10.2147/IJN.S53622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Zou Z., Sun Y., Li X., Xu K.F., Wei Y., Jin N., Jiang C. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23:300–302. doi: 10.1038/cr.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Robotham J.M., Nelson H.B., Irsigler A., Kenworthy R., Tang H. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Chan J.F., den-Haan H., Chik K.K., Zhang A.J., Chan C.C., Poon V.K., Yip C.C., Mak W.W., Zhu Z., Zou Z., Tee K.M., Cai J.P., Chan K.H., de la Pena J., Perez-Sanchez H., Ceron-Carrasco J.P., Yuen K.Y. Structure-based discovery of clinically approved drugs as Zika virus NS2B-NS3 protease inhibitors that potently inhibit Zika virus infection in vitro and in vivo. Antiviral Res. 2017;145:33–43. doi: 10.1016/j.antiviral.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Zeisel M.B., Lupberger J., Fofana I., Baumert T.F. Host-targeting agents for prevention and treatment of chronic hepatitis C - perspectives and challenges. J. Hepatol. 2013;58:375–384. doi: 10.1016/j.jhep.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Zhu J.D., Meng W., Wang X.J., Wang H.C. Broad-spectrum antiviral agents. Front. Microbiol. 2015;6:517. doi: 10.3389/fmicb.2015.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink M.C., Uhrlaub J., DeWitt J., Voelker T., Bullock B., Mankowski J., Tarwater P., Clements J., Barber S. Neuroprotective and anti-human immunodeficiency virus activity of minocycline. JAMA. 2005;293:2003–2011. doi: 10.1001/jama.293.16.2003. [DOI] [PubMed] [Google Scholar]