Abstract
Growing evidence indicated that the gut microbiota was the intrinsic and essential component of the cancer microenvironment, which played vital roles in the development and progression of colorectal cancer (CRC). In our present study, we investigated the alterations of fecal abundant microbiota with real-time quantitative PCR and the changes of indicators of gut mucosal barrier from 53 early-stage CRC patients and 45 matched healthy controls. We found that the traditional beneficial bacteria such as Lactobacillus and Bifidobacterium decreased significantly and the carcinogenic bacteria such as Enterobacteriaceae and Fusobacterium nucleatum were significantly increased in CRC patients. We also found gut mucosal barrier dysfunction in CRC patients with increased levels of endotoxin (LPS), D-lactate, and diamine oxidase (DAO). With Pearson's correlation analysis, D-lactate, LPS, and DAO were correlated negatively with Lactobacillus and Bifidobacterium and positively with Enterobacteriaceae and F. nucleatum. Our present study found dysbiosis of the fecal microbiota and dysfunction of the gut mucosal barrier in patients with early-stage CRC, which implicated that fecal abundant bacteria and gut mucosal barrier indicators could be used as targets to monitor the development and progression of CRC in a noninvasive and dynamic manner.
1. Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-related mortality, accounting for the fifth most commonly diagnosed cancer and the fifth most common cause of death by cancer in China [1]. The overall decline or stabilization in the incidence of CRC was noted in several high-income countries [2]; however, the incidence and mortality rates of CRC in China still showed an increasing trend in the past decades, with approximately 376,300 new cases and 191,000 deaths per year [3]. Recently, many previous studies have verified that the human intestinal microbiota, including microbiome, mycobiome, and virome, was one of the important factors associated with CRC [4–10]. Intestinal microbiota, one part of the tumor microenvironment, has attracted increasing attention, as it can affect CRC growth and spread in many ways. A variety of intestinal commensal bacteria and their metabolites are known for triggering inflammatory cascades and oncogenic signaling, thereby promoting genetic and epigenetic alterations in CRC development [11].
Increasing evidence indicated that intestinal dysbiosis plays vital roles in CRC initiation, progression, and metastasis. Our previous study has found that the structures of the intestinal microbiota altered significantly in CRC patients compared to healthy individuals, which found that Fusobacterium, Porphyromonas, Peptostreptococcus, and Mogibacterium increased in CRC patients significantly [4]. These bacteria might influence CRC risk via cometabolism or metabolic exchange with the host. Wei et al. demonstrated that Ruminococcus obeum and Allobaculum-like bacteria were enriched in the feces of 1,2-dimethyl hydrazine treated rats developing precancerous mucosal lesions [12]. Wang et al. also found that an increase of opportunistic pathogens and a reduction of butyrate-producing bacteria may constitute a major structural imbalance of gut microbiota in these CRC patients [13]. Several bacterial species, such as Fusobacterium nucleatum, Peptostreptococcus anaerobius, and Bacteroides clarus [6, 14–16], have been implicated in the development of CRC, which could promote carcinogenesis upon invasion of host cells. In addition, Coker et al. also revealed CRC-associated mycobiome dysbiosis characterized by altered fungal composition and ecology, signifying that the gut mycobiome might also play a role in CRC development [5]. Recently, an international panel of experts from International Cancer Microbiome Consortium delivered a consensus statement on the role of the human microbiome in carcinogenesis [17], which emphasized that future studies should provide direct evidence to demonstrate the roles of the human commensal microbiome in the aetiopathogenesis of cancer including CRC.
Our present study, enrolling confirmed early-stage CRC subjects and matched healthy controls, aimed at assessing the abundant bacteria in the fecal microbiota with quantitative PCR and evaluating the functions of the gut mucosal barrier, which would demonstrate the altered composition of the fecal microbiota in early-stage CRC patients and host response in an easy and rapid way. These results might be helpful for CRC precise diagnosis and personalized treatment targeting on human microbiota.
2. Methods
2.1. Subjects' Selection
A total of 53 patients who were diagnosed with primary early-stage CRC (aged 46-75 years old) between January 2011 and March 2012 were consecutively recruited from the First Affiliated Hospital, School of Medicine, Zhejiang University, Zhejiang, China. The diagnosis and stages of CRC were based on NCCN clinical practice guidelines in oncology (2010 edition) [18]. None of the patients were on any medications before sample collection. 45 sex-, age-, and body mass index- (BMI-) matched healthy subjects were selected as controls from the same cohorts during a routine physical examination, which were also confirmed by screening colonoscopy and pathology later. The following exclusion criteria were established: obesity (BMI > 30); diabetes; hypertension; family history of CRC; previous colon or rectal surgery; gastrointestinal disorders such as irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD); emergency colonoscopy; known active bacterial, fungal, and viral infections; and the use of probiotics, prebiotics, synbiotics, or antibiotics in the previous month. The study protocol was approved by the Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University (Zhejiang, China). Written informed consent was obtained from all participants prior to the enrollment.
2.2. Sampling
Prior to bowel cleansing for scheduled colonoscopy, fecal samples and blood samples were collected from these participants. We collected approximately 2 g of fresh feces into a sterile plastic cup from these participants and kept these samples in an icebox. Samples for bacterial genomic DNA extraction were transferred immediately to the laboratory and stored at -80°C after preparation within 15 min until use. In addition, blood samples were also collected simultaneously from these participants for intestinal mucosal barrier function analysis. Plasma was also stored at -80°C after preparation within 15 min (Table 1 shows the details of the samples).
Table 1.
Summary information of the samples.
| CRC | Control | |
|---|---|---|
| Sample size | 53 | 45 |
| Age (years): mean ± SD | 52.4 ± 18.8 | 53.7 ± 16.7 |
| Gender (male/female) | 33/20 | 25/20 |
| BMI | 24.8 ± 3.58 | 23.7 ± 3.67 |
2.3. Bacterial Genomic DNA Extraction
Fecal genomic DNA was extracted using a QIAamp® DNA Stool Mini Kit (QIAGEN, Hilden, Germany) according to our previous studies [4, 19]. The amount of bacterial genomic DNA was analyzed using a NanoDrop ND-1000 spectrophotometer; the integrity and size of bacterial genomic DNA were checked by electrophoresis. All bacterial genomic DNA was stored at -80°C for further use.
2.4. Fecal Abundant Bacteria Analysis
For fecal abundant bacteria analysis, real-time quantitative PCR (qPCR) was performed on ABI Prism 7900HT real-time PCR system (Applied Biosystems, Carlsbad, CA) with a Power SYBR Green PCR Master Mix (Takara, Dalian, China) according to the manufacturer's instructions. The bacterial primer sets and the reaction conditions are shown in Table 2 [19, 20]. SDS 2.4 was used for the data analysis. Triplicate repeats were carried out for all reactions in every analysis with a nontemplate included. The quantity of these bacteria was presented as log10 bacteria per gram of feces (wet weight).
Table 2.
Bacterial-specific primer sets for detection of fecal microbiota by qPCR.
| PCR specificity | Primer | Sequence (5′→3′) | Annealing temperature (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| Total bacteria | Uni331F | TCCTACGGGAGGCAGCAGT | 58 | 466 |
| Uni797R | GGACTACCAGGGTATCTATCCTGTT | |||
| Bacteroides-Prevotella group | Bac303F | GAAGGTCCCCCACATTG | 56 | 418 |
| Bac708R | CAATCGGAGTTCTTCG | |||
| Lactobacillus spp. | Lac-F | AGCAGTAGGGAATCTTCCA | 58 | 341 |
| Lac-R | CACCGCTACACATGGAG | |||
| Bifidobacterium spp. | Bifid-F | CTCCTGGAAACGGGTGG | 55 | 550 |
| Bifid-R | GGTGTTCTTCCCGATATCTACA | |||
| Clostridium cluster I | CG1-F | TACCHRAGGAGGAAGCCAC | 63 | 700 |
| CG1-R | GTTCTTCCTAATCTCTACGCAT | |||
| Clostridium cluster XI | CG2-F | ACGCTACTTGAGGAGGA | 58 | 141 |
| CG2-R | GAGCCGTAGCCTTTCACT | |||
| Clostridium cluster XIVab | CG3-F | GAWGAAGTATYTCGGTATGT | 52 | 152 |
| CG3-R | CTACGCWCCCTTTACAC | |||
| Faecalibacterium prausnitzii | Fpra-F | GATGGCCTCGCGTCCGATTAG | 58 | 199 |
| Fpra-R | CCGAAGACCTTCTTCCTCC | |||
| Fusobacterium nucleatum | Fn-F | CAACCATTACTTTAACTCTACCATGTTCA | 60 | 112 |
| Fn-R | GTTGACTTTACAGAAGGAGATTATGTAAAAATC | |||
| Enterobacteriaceae | Eco-F | CATTGACGTTACCCGCGAGAAGAAGC | 63 | 195 |
| Eco-R | CTCTACGAGCTCAAGCTTGC | |||
| Enterococcus spp. | ENco-F | CCCTTATTGTTAGTTGCCATCATT | 61 | 144 |
| ENco-R | ACTCGTTGTACTTCCCATTGT |
2.5. Intestinal Mucosal Barrier Function Analysis
The plasma from each participants was collected for intestinal mucosal barrier function analysis. The parameters of gut mucosal barrier function such as endotoxin (LPS), D-lactate, and diamine oxidase (DAO) were detected by a dry chemical method using the Intestinal Mucosal Barrier Biochemical Index Analysis System (JY-DLT, Beijing Zhongsheng Jinyu Diagnostic Technology Co., Ltd., China) [19, 21, 22]. The experiments were undergone according to the protocols suggested by the manufacturer and conducted within 4 h after plasma extraction.
2.6. Statistical Analysis
The quantitative data of fecal abundant bacteria and other parameters in our study are presented as the mean ± standard deviation (SD), the differences between the two groups were evaluated by Student's t test, and the correlations between variables were tested by Pearson correlation using SPSS version 20.0 (SPSS Inc., Chicago, IL.), respectively. p < 0.05 was considered statistically significant for all analyses.
3. Results
3.1. Characteristics of the Early-Stage CRC Patients
In our present study, these participants were all newly diagnosed early-stage CRC patients in our hospital, who were not treated with antibiotics, prebiotics, probiotics, and synbiotics in the previous month. Before sampling, these participants have been performed a complete physical examination such as colonoscopy and pathology, serological markers detection, and radiological examination. Most of the CRC patients (50/53) were found to be positive in fecal occult blood testing, while several patients (8/53) occurred severe diarrhoea. In our study, the BMI values and the serum liver function, kidney function, lipid levels, blood sugar, and blood pressure in the CRC patients were not different from healthy controls significantly (p > 0.05). However, the serological marker such as carcinoembryonic antigen (CEA) was increased significantly in CRC patients when compared with healthy controls (p < 0.05). Our study demonstrated that no any CRC patients were found to be positive in tumor metastasis.
3.2. Abnormal Fecal Microbiota in CRC Patients
In our present study, we investigated the abundant bacteria in the fecal microbiota by real-time quantitative PCR (Figure 1). The abundant bacteria represented the higher relative abundance of bacteria in the fecal microbiota, which would determine the overall structure and composition of the fecal microbiota. Ten abundant bacteria, which accounted for more than 90% of the total bacteria, were selected to explore the alterations of fecal microbiota in these CRC patients. We found that the copy numbers of the total bacteria were not significantly different between healthy controls and CRC patients. However, the traditional beneficial bacteria such as Lactobacillus and Bifidobacterium decreased significantly in CRC patients. In addition, the gut-indigenous Clostridium included several different clusters, including abundant clusters such as Clostridium cluster I, cluster XI, and cluster XIVb. Our data indicated that Clostridium cluster I was significantly decreased in CRC patients when compared with healthy controls, while Clostridium cluster XI and cluster XIVb were not significantly different between healthy controls and CRC patients. Another bacterium, Enterobacteriaceae, was considered as gut harmful bacteria, which could produce lipopolysaccharide into peripheral blood. We observed that Enterobacteriaceae was significantly increased in CRC patients when compared with healthy controls. F. nucleatum was the confirmed carcinogenic bacterium that associated with CRC development. Consistent with the previous studies, our results demonstrated that F. nucleatum was significantly increased in CRC patients when compared with healthy controls. Our data indicated that dysbiosis of fecal microbiota was found in CRC patients, which might be contributed to CRC initiation, progression, and metastasis.
Figure 1.
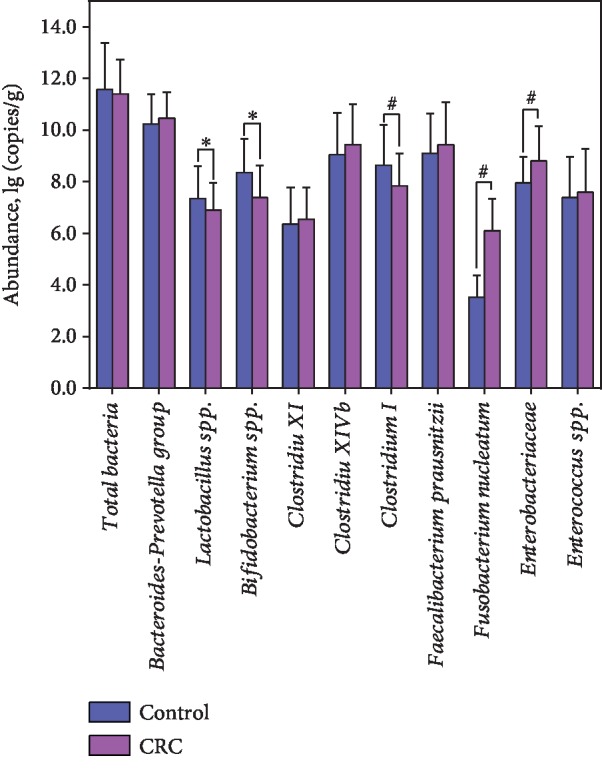
Quantitative real-time PCR analysis of the fecal abundant bacteria in patients with colorectal cancer (log10 copies per gram of fresh feces). ∗p < 0.05; #p < 0.01.
3.3. Changed Intestinal Mucosal Barrier Function
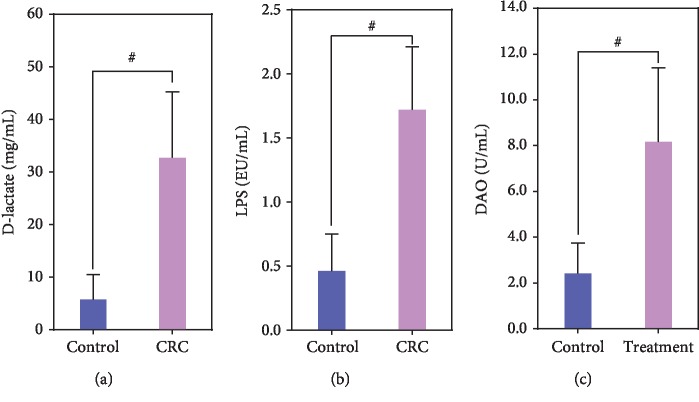
The three parameters such as LPS, D-lactate, and DAO could be used to evaluate the integrity of the gut mucosal barrier. The increased permeability of the gut mucosal barrier was associated with increased systemic and local inflammation, which was closely correlated with the initiation and development of CRC. In our present study, we found obviously disturbed gut mucosal barrier in CRC patients when compared with healthy controls. Our data indicated that plasma D-lactate, a byproduct of bacterial metabolism, increased significantly in CRC patients when compared with healthy controls (p < 0.01; Figure 2(a)). LPS, a structural component of the cell wall of Gram-negative bacteria, was a potent inducer of proinflammatory cytokines that activate a systemic inflammatory response syndrome. Elevated LPS level has been reported to be an early marker of impaired mucosal barrier function. In our study, we also observed that the levels of LPS were significantly higher in the CRC patients than that in the healthy controls (p < 0.01; Figure 2(b)). In addition, DAO, an enzyme which deaminates histamine and polyamines, has its highest activity in the intestinal mucosa in most mammalian species, including humans. DAO activity has been shown to reflect changes associated with the gut mucosal barrier. Our data indicated that significant differences of DAO levels were observed between CRC patients and healthy controls (p < 0.01; Figure 2(c)). Taken together, our results showed that the loss of gut mucosal barrier integrity and function was found in CRC patients, which indicated that the impairment of gut mucosal barrier function might be one of the important factors for the initiation and development of CRC.
Figure 2.
The alterations of the indicators of the gut mucosal barrier in patients with colorectal cancer. (a) D-lactate; (b) LPS; (c) DAO. #p < 0.01.
3.4. Correlations between Differential Bacteria and Indicators of Gut Mucosal Barrier
In our present study, Pearson correlation was used to examine the relationship between differential fecal bacteria and indicators of the gut mucosal barrier (Table 3). We found that the differential fecal bacteria such as Lactobacillus, Bifidobacterium, and Clostridium cluster I correlated with D-Lactate and endotoxin negatively (p < 0.05), while Lactobacillus, but not Bifidobacterium and Clostridium cluster I, correlated with DAO negatively (p < 0.05). In addition, we also found that Fusobacterium nucleatum and Enterobacteriaceae correlated positively with D-Lactate, DAO, and endotoxin (p < 0.05). Among these indicators of gut mucosal barrier, strong correlations (Pearson's r ≥ 0.50) were observed between Bifidobacterium and endotoxin (r = −0.752, p < 0.000), between Clostridium cluster I and endotoxin (r = −0.630, p < 0.000), between Fusobacterium nucleatum and D-Lactate (r = 0.809, p < 0.000), and between Enterobacteriaceae and endotoxin (r = 0.802, p < 0.000).
Table 3.
Pearson's correlation coefficients between differential abundant bacteria and indicators of gut mucosal barrier.
| D-lactate (mg/ml) | DAO (U/ml) | LPS (EU/ml) | ||||
|---|---|---|---|---|---|---|
| r | p | r | p | r | p | |
| Lactobacillus | -0.314 | 0.043 | -0.411 | 0.006 | -0.456 | 0.002 |
| Bifidobacterium | -0.385 | 0.011 | -0.149 | 0.341 | -0.752 | 0.000 |
| Clostridium cluster I | -0.332 | 0.030 | -0.252 | 0.103 | -0.630 | 0.000 |
| Fusobacterium nucleatum | 0.809 | 0.000 | 0.339 | 0.026 | 0.456 | 0.002 |
| Enterobacteriaceae | 0.402 | 0.007 | 0.342 | 0.025 | 0.802 | 0.000 |
4. Discussion
The development of human CRC is influenced by various risk factors, such as diets, genetic, epigenetic, environmental, and metabolic factors, while environmental factors are the predominant trigger for CRC [23]. A growing body of evidence has indicated that human gut microbiota may be an important environmental factor that promotes CRC development [15, 24]. With the advent of high-throughput sequencing platforms “omics” technologies and advanced bioinformatics approaches, the changes of the composition and function of the gut microbiota have been identified in the last decades, and the suspected causative roles and molecular mechanisms of gut microbiota in CRC development have been established by many previous studies. Recently, several bacterial species such as Fusobacterium nucleatum and Peptostreptococcus anaerobius have been confirmed in the CRC development and progression [14, 25]. These fecal bacteria have been considered as one of the most important elements of the tumor microenvironment. Now, fecal candidate bacteria can be used as novel biomarkers with existing methods, such as real-time qPCR and fecal immunochemical test, for noninvasive diagnosis of CRC [7].
With real-time qPCR, we found that the abundant fecal bacteria changed significantly in these early-stage CRC patients, which indicated that the dysbiosis of fecal microbiota might participate in CRC development and progression. In our present study, ten abundant bacteria were investigated to illustrate the alterations of fecal microbiota in CRC patients. In our present study, the two traditional beneficial bacteria such as Lactobacillus and Bifidobacterium (belonged to the lactic acid bacteria category) decreased significantly in these CRC patients, which were consistent with previous study [26]. Mendes et al. has found that the abundance of the genera Lactobacillus and Bifidobacterium increased significantly after microbiota modification by probiotic supplementation [27]. Increasing reports in recent years have shown that these bacteria have demonstrated a host of properties in preventing CRC development by inhibiting initiation or progression through multiple pathways, such as apoptosis, antioxidant DNA damages, immune responses, and epigenetics [28–30]. Although Lactobacillus and Bifidobacterium may only act before carcinogenesis and have little inhibitory impact on established cancer, these two beneficial bacteria were the most frequently used as probiotics to prevent colon carcinogenesis in animal models or patients, which might help regulate the CRC-related tumor microenvironment. Different from Lactobacillus and Bifidobacterium, Enterobacteriaceae was often considered as harmful bacteria in the fecal microbiota, which could induce inflammation in the host gut epithelium [31]. As inflammation has been a well-documented risk factor for various form of cancer [32], it was implicated that Enterobacteriaceae participated actively in the development and progression of CRC. Previous studies showed that the actions of Enterobacteriaceae are similar to the prolonged inflammatory response induced by enterotoxigenic Bacteroides fragilis [33, 34]. Sun et al. has showed significant enrichment of Enterobacteriaceae in the DMH-induced CRC animal model [35]. They also found that Enterobacteriaceae showed higher relative abundance in the early stages of tumor formation, while it was gradually replaced with other bacteria such as Rikenellaceae, Lachnospiraceae, Ruminococcaceae, and Streptococcaceae [35]. Another potential cancer-promoting bacterium, F. nucleatum, was associated with CRC. Previous studies found that F. nucleatum is enriched in both the feces and colonic mucosa of CRC patients [4, 36, 37] and plays important roles in colorectal carcinogenesis [38, 39]. F. nucleatum may promote colorectal tumor growth and inhibit T cell-mediated immune responses against colorectal tumors [40]. Rubinstein et al. also found that F. nucleatum binds E-cadherin on epithelial cells and activates β-catenin signaling, driving epithelial cell proliferation [39]. With quantitative PCR and fecal immunochemical test, Liang and their colleagues found that F. nucleatum alone can discriminate CRC from controls with a sensitivity of 77.7%, and specificity of 79.5%, which can serve as a novel noninvasive diagnostic method for patients with CRC [7]. Mima et al. also found that the amount of F. nucleatum in CRC tissue is associated with shorter survival and may potentially serve as a prognostic biomarker [40]. Bullman et al. observed that treatment of mice bearing a CRC xenograft with the antibiotic metronidazole reduced F. nucleatum load, cancer cell proliferation, and overall tumor growth, which indicated that microbiota modulation as a potential treatment for Fusobacterium-associated colorectal carcinomas [41]. In addition, we also found that Clostridium cluster I decreased significantly in CRC patients when compared with healthy controls. Kostic et al. found that CRC tissues have decreased microbial diversity, including a reduction of certain bacterial genera like Clostridium and Bacteroides [42]. However, the genus Clostridium includes a diverse group of Gram-positive, spore-forming anaerobes [43]. In general, clostridial fermentative metabolism functions by the conversion of hexose sugars to butyrate, acetate, and CO2 [44]. Our study firstly found that Clostridium cluster I was negatively correlated with CRC development. The present study indicated that the abundant bacteria in fecal microbiota participated actively in the development and progression of CRC.
The gut mucosal barrier is a functional unit organized as a multilayer system, and its multiple functions are crucial for maintaining gut homeostasis. The inherent property of the gut to act as a semipermeable barrier is crucial for the maintenance of health. Dysfunction of the gut mucosal barrier leads to increased translocation of commensal bacteria and their metabolites locally and systemically, triggering the inflammatory response. Gut inflammation results in excess production of proinflammatory cytokines that can in turn increase mucosal permeability by altering intercellular tight junction structure and induce apoptosis of intestinal epithelial cells. Numerous scientific evidences showed a significant association between impaired gut mucosal barrier and gastrointestinal/extraintestinal diseases [45–48]. Previous study has found that progression of colorectal neoplasia has been linked to alterations of tumor microenvironment and gut mucosal barrier function, which facilitate the interaction of microbial products with host pathways [11]. D-lactate, LPS, and DAO were three accepted and convenient indicators to evaluate the integrity of the gut mucosal barrier. The increased permeability of the gut mucosal barrier would lead to bacterial translocation that was accompanied by increasing levels of D-lactate, LPS, and DAO, which finally contribute to a number of intestinal diseases such as CRC. D-lactate, a bydroxycarboxylic acid produced by bacterial fermentation, is a useful indicator of increased gut permeability and gut barrier dysfunction. Our present study found that the level of D-lactate was increased significantly in CRC patients, which was correlated with Lactobacillus, Bifidobacterium, and Clostridium cluster I negatively and with F. nucleatum and Enterobacteriaceae positively, confirming the existence of gut mucosal barrier dysfunction. LPS is large molecules found in the outer membrane of Gram-negative bacteria. The increase in LPS is associated with bacterial translocation due to the impairment of intestinal epithelial cell [49]. Previous study has found that gut microbiota dysbiosis alters the intestinal barrier function, increases plasma LPS levels, which promotes endotoxemia, and contributes to the onset and development of CRC [50]. Our study also found that the concentration of LPS increased significantly in CRC patients, which was correlated with Lactobacillus, Bifidobacterium, and Clostridium cluster I negatively and with F. nucleatum and Enterobacteriaceae positively. The results suggest that there was an increased intestinal permeability in these CRC patients. DAO is an enzyme mainly produced in the small intestine involved in the histamine metabolism [51]. Yee et al. has found that an established first-line treatment for patients in T2DM, metformin, inhibits DAO activity [52]. Previous study also found that serum DAO activity decreased step-by-step significantly during anticancer drug therapy in human, which may be to serve as a useful predictor of gastrointestinal toxicity due to anticancer drug [53]. Our present data found that the levels of DAO also increased significantly in CRC patients, which was correlated with Lactobacillus, Bifidobacterium, and Clostridium cluster I negatively and with F. nucleatum and Enterobacteriaceae positively. Taken together, the low levels of D-lactate, LPS, and DAO in normal conditions increased significantly in CRC patients, suggesting a breakdown of the gut mucosal barrier.
In summary, dysbiosis of the fecal microbiota, which was featured by altered abundant fecal bacteria, and dysfunction of the gut mucosal barrier, which was characterized by increased levels of D-lactate, LPS, and DAO, occurred in patients with early-stage CRC. The abundant bacteria in the feces and the indicators of the gut mucosal barrier in the plasma could be used as targets to monitor the development and progression of CRC in the future.
Acknowledgments
The authors thank all of the participants who recruited patients in this study. This present work was funded by the grants from the National Natural Science Foundation of China under Grant Nos. 31700800, 81771724, 81790631, and 31870839 and National S&T Major Project of China under Grant No. 2018YFC2000500 and the Nutrition and Care of Maternal & Child Research Fund Project of Guangzhou Biostime Institute of Nutrition & Care (2019BINCMCF045).
Contributor Information
Li Shao, Email: 0011334@zju.edu.cn.
Zongxin Ling, Email: lingzongxin@zju.edu.cn.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Ethical Approval
All research conformed to the Helsinki Declaration and was approved by the Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University (China), and was implemented in accordance with the approved guidelines.
Consent
Informed written consent was obtained from each of the patients before enrollment.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Authors' Contributions
XL and ZXL conceived and designed the experiments. XL, YWC, LS, and ZXL performed the experiments and analyzed the data. XL and ZXL wrote the paper and edited the manuscript. The final manuscript was read and approved by all authors. Xia Liu and Yiwen Cheng contributed equally to this work.
References
- 1.Chen W., Sun K., Zheng R., et al. Cancer incidence and mortality in China, 2014. Chinese Journal of Cancer Research. 2018;30(1):1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araghi M., Soerjomataram I., Bardot A., et al. Changes in colorectal cancer incidence in seven high-income countries: a population-based study. Lancet Gastroenterol Hepatol. 2019;4(7):511–518. doi: 10.1016/S2468-1253(19)30147-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen W., Zheng R., Baade P. D., et al. Cancer statistics in China, 2015. CA: a Cancer Journal for Clinicians. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Chen W., Liu F., Ling Z., Tong X., Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7(6, article e39743) doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coker O. O., Nakatsu G., Dai R. Z., et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68(4):654–662. doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong S. H., Zhao L., Zhang X., et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153(6):1621–1633.e6. doi: 10.1053/j.gastro.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Liang Q., Chiu J., Chen Y., et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clinical Cancer Research. 2017;23(8):2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 8.Nakatsu G., Li X., Zhou H., et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6(1) doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J., Feng Q., Wong S. H., et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2016;66(1):70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 10.Nakatsu G., Zhou H., Wu W. K. K., et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155(2):529–541.e5. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Irrazabal T., Belcheva A., Girardin S. E., Martin A., Philpott D. J. The multifaceted role of the intestinal microbiota in colon cancer. Molecular Cell. 2014;54(2):309–320. doi: 10.1016/j.molcel.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 12.Wei H., Dong L., Wang T., et al. Structural shifts of gut microbiota as surrogate endpoints for monitoring host health changes induced by carcinogen exposure. FEMS Microbiology Ecology. 2010;73(3):577–586. doi: 10.1111/j.1574-6941.2010.00924.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang T., Cai G., Qiu Y., et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. The ISME Journal. 2012;6(2):320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsoi H., Chu E. S. H., Zhang X., et al. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology. 2017;152(6):1419–1433.e5. doi: 10.1053/j.gastro.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Zeller G., Tap J., Voigt A. Y., et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Molecular Systems Biology. 2014;10(11):p. 766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwong T. N. Y., Wang X., Nakatsu G., et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology. 2018;155(2):383–390.e8. doi: 10.1053/j.gastro.2018.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Scott A. J., Alexander J. L., Merrifield C. A., et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut. 2019;68(9):1624–1632. doi: 10.1136/gutjnl-2019-318556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burt R. W., Barthel J. S., Dunn K. B., et al. NCCN clinical practice guidelines in oncology. Colorectal cancer screening. Journal of the National Comprehensive Cancer Network. 2010;8(1):8–61. doi: 10.6004/jnccn.2010.0003. [DOI] [PubMed] [Google Scholar]
- 19.Xia X., Chen J., Xia J., et al. Role of probiotics in the treatment of minimal hepatic encephalopathy in patients with HBV-induced liver cirrhosis. Journal of International Medical Research. 2018;46(9):3596–3604. doi: 10.1177/0300060518776064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan C., Ling Z., Huang Y., et al. Dysbiosis of intestinal microbiota associated with inflammation involved in the progression of acute pancreatitis. Pancreas. 2015;44(6):868–875. doi: 10.1097/MPA.0000000000000355. [DOI] [PubMed] [Google Scholar]
- 21.Ling Z., Liu X., Guo S., et al. Role of probiotics in mycoplasma pneumoniae pneumonia in children: a short-term pilot project. Frontiers in Microbiology. 2019;9 doi: 10.3389/fmicb.2018.03261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen L., Ao L., Xu H., et al. Poor short-term glycemic control in patients with type 2 diabetes impairs the intestinal mucosal barrier: a prospective, single-center, observational study. BMC Endocrine Disorders. 2019;19(1):p. 29. doi: 10.1186/s12902-019-0354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nistal E., Fernández-Fernández N., Vivas S., Olcoz J. L. Factors determining colorectal cancer: the role of the intestinal microbiota. Frontiers in Oncology. 2015;5:p. 220. doi: 10.3389/fonc.2015.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobhani I., Tap J., Roudot-Thoraval F., et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6(1, article e16393) doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu T., Guo F., Yu Y., et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170(3):548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Keefe S. J. D. Diet, microorganisms and their metabolites, and colon cancer. Nature Reviews Gastroenterology & Hepatology. 2016;13(12):691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendes M. C. S., Paulino D. S., Brambilla S. R., Camargo J. A., Persinoti G. F., Carvalheira J. B. C. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World Journal of Gastroenterology. 2018;24(18):1995–2008. doi: 10.3748/wjg.v24.i18.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong L., Zhang X., Covasa M. Emerging roles of lactic acid bacteria in protection against colorectal cancer. World Journal of Gastroenterology. 2014;20(24):7878–7886. doi: 10.3748/wjg.v20.i24.7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danese S. Inflammatory bowel disease and inflammation-associated colon cancer: partners in crime. Current Drug Targets. 2008;9(5):p. 360. doi: 10.2174/138945008784221134. [DOI] [PubMed] [Google Scholar]
- 30.Nowak A., Paliwoda A., Blasiak J. Anti-proliferative, pro-apoptotic and anti-oxidative activity of Lactobacillus and Bifidobacterium strains: a review of mechanisms and therapeutic perspectives. Critical Reviews in Food Science and Nutrition. 2019;59(21):3456–3467. doi: 10.1080/10408398.2018.1494539. [DOI] [PubMed] [Google Scholar]
- 31.Lupp C., Robertson M. L., Wickham M. E., et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host & Microbe. 2007;2(2):119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Elinav E., Nowarski R., Thaiss C. A., Hu B., Jin C., Flavell R. A. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nature Reviews Cancer. 2013;13(11):759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 33.Housseau F., Sears C. L. Enterotoxigenic Bacteroides fragilis (ETBF)-mediated colitis in Min (Apc+/-) mice: a human commensal-based murine model of colon carcinogenesis. Cell Cycle. 2010;9(1):3–5. doi: 10.4161/cc.9.1.10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangan P. R., Harrington L. E., O'Quinn D. B., et al. Transforming growth factor-β induces development of the TH17 lineage. Nature. 2006;441(7090):231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 35.Sun T., Liu S., Zhou Y., et al. Evolutionary biologic changes of gut microbiota in an 'adenoma-carcinoma sequence' mouse colorectal cancer model induced by 1, 2-dimethylhydrazine. Oncotarget. 2017;8(1):444–457. doi: 10.18632/oncotarget.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn J., Sinha R., Pei Z., et al. Human gut microbiome and risk for colorectal cancer. JNCI: Journal of the National Cancer Institute. 2013;105(24):1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castellarin M., Warren R. L., Freeman J. D., et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kostic A. D., Chun E., Robertson L., et al. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host & Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubinstein M. R., Wang X., Liu W., Hao Y., Cai G., Han Y. W. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/ β-Catenin Signaling via its FadA Adhesin. Cell Host & Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mima K., Nishihara R., Qian Z. R., et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bullman S., Pedamallu C. S., Sicinska E., et al. Analysis ofFusobacteriumpersistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kostic A. D., Gevers D., Pedamallu C. S., et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patakova P., Linhova M., Rychtera M., Paulova L., Melzoch K. Novel and neglected issues of acetone-butanol-ethanol (ABE) fermentation by clostridia: Clostridium metabolic diversity, tools for process mapping and continuous fermentation systems. Biotechnol Adv. 2013;31(1):58–67. doi: 10.1016/j.biotechadv.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 44.Therien J. B., Artz J. H., Poudel S., et al. The physiological functions and structural determinants of catalytic bias in the [FeFe]-hydrogenases CpI and CpII of Clostridium pasteurianum strain W5. Frontiers in Microbiology. 2017;8:p. 1305. doi: 10.3389/fmicb.2017.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scaldaferri F., Pizzoferrato M., Gerardi V., Lopetuso L., Gasbarrini A. The gut barrier: new acquisitions and therapeutic approaches. Journal of Clinical Gastroenterology. 2012;46:S12–S17. doi: 10.1097/MCG.0b013e31826ae849. [DOI] [PubMed] [Google Scholar]
- 46.Weber C. R., Turner J. R. Inflammatory bowel disease: is it really just another break in the wall? Gut. 2007;56(1):6–8. doi: 10.1136/gut.2006.104182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fasano A. Leaky gut and autoimmune diseases. Clinical Reviews in Allergy & Immunology. 2012;42(1):71–78. doi: 10.1007/s12016-011-8291-x. [DOI] [PubMed] [Google Scholar]
- 48.Mu Q., Kirby J., Reilly C. M., Luo X. M. Leaky gut as a danger signal for autoimmune diseases. Frontiers in Immunology. 2017;8:p. 598. doi: 10.3389/fimmu.2017.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cavaillon J. M. Exotoxins and endotoxins: inducers of inflammatory cytokines. Toxicon. 2018;149:45–53. doi: 10.1016/j.toxicon.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez-Sarrias A., Nunez-Sanchez M. A., Avila-Galvez M. A., et al. Consumption of pomegranate decreases plasma lipopolysaccharide-binding protein levels, a marker of metabolic endotoxemia, in patients with newly diagnosed colorectal cancer: a randomized controlled clinical trial. Food & Function. 2018;9(5):2617–2622. doi: 10.1039/C8FO00264A. [DOI] [PubMed] [Google Scholar]
- 51.Zhao L., Luo L., Jia W., et al. Serum diamine oxidase as a hemorrhagic shock biomarker in a rabbit model. PLoS One. 2014;9(8, article e102285) doi: 10.1371/journal.pone.0102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yee S. W., Lin L., Merski M., et al. Prediction and validation of enzyme and transporter off-targets for metformin. Journal of Pharmacokinetics and Pharmacodynamics. 2015;42(5):463–475. doi: 10.1007/s10928-015-9436-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyoshi J., Miyamoto H., Goji T., et al. Serum diamine oxidase activity as a predictor of gastrointestinal toxicity and malnutrition due to anticancer drugs. Journal of Gastroenterology and Hepatology. 2015;30(11):1582–1590. doi: 10.1111/jgh.13004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.