Highlights
-
•
The immunodominant region of the porcine deltacoronavirus S protein, is the C-terminal domain of its S1 subunit.
-
•
Three truncations of the PDCoV S protein were generated, the C and N-terminal domains of the S1 subunit, and S2 subunit.
-
•
Three S protein truncations are capable of inducing PDCoV-neutralizing antibody responses in vivo.
-
•
The C-terminal domains of the S1 subunit-specific antisera showed the most potent PDCoV-neutralizing effect.
Keywords: Porcine deltacoronavirus (PDCoV), Spike glycoprotein, Epitope region, Neutralizing antibody
Abstract
Porcine deltacoronavirus (PDCoV), is an emerging enteropathogenic coronavirus in pigs, that poses a novel threat to swine husbandry worldwide. Crucial to halting PDCoV transmission and infection is the development of effective therapies and vaccines. The spike (S) protein of coronavirus is the major target of host neutralizing antibodies, however the immunodominant neutralizing region in the S protein of PDCoV has not been defined. Here, three truncations of the PDCoV S protein were generated, the N-terminal domain of the S1 subunit (NTD, amino acids (aa) 50–286), the C-terminal domain of the S1 subunit (CTD, aa 278–616), and S2 subunit (aa 601–1087). The proteins were expressed using an E. coli expression system. Polyclonal antisera against the three recombinant proteins were produced in rabbits and mice. All three antisera were able to inhibit PDCoV infection in vitro, as determined by virus neutralization assay, fluorescent focus neutralization assay, and plaque-reduction neutralization. The CTD-specific antisera had the most potent PDCoV-neutralizing effect, indicating that the CTD region may contain the major neutralizing epitope(s) in the PDCoV S protein. Based on these findings, CTD may be a promising target for development of an effective vaccine against PDCoV infection in pigs.
1. Introduction
Porcine deltacoronavirus (PDCoV) was first reported in 2012 in Hong Kong during an investigation of novel coronaviruses (CoVs) (Woo et al., 2012). Thereafter, PDCoV was detected in the United States and isolated from pigs suffering from severe diarrhea in 2014 by Hu et al. in Ohio, USA (Hu et al., 2015; Li et al., 2014; Wang et al., 2014b). Pathogenesis and virulence were subsequently investigated using gnotobiotic and conventionally raised pigs. PDCoV infection is characterized by watery diarrhea and vomiting 1–3 days after infection (Hu et al., 2016; Ma et al., 2015); it has since been detected in swine populations throughout the world, resulting in substantial economic losses (Lee and Lee, 2014; Saeng-Chuto et al., 2017; Song et al., 2015; Suzuki et al., 2018; Wang et al., 2014a).
PDCoV is an enveloped, positive-sense, single-stranded RNA virus belonging to the order Nidovirales, family Coronaviridae, subfamily Coronavirinae, and genus Deltacoronavirus. The PDCoV genome (∼25.4 kb) consists of eight open reading frames (ORFs) and contains six common coronaviral genes in the conserved order: 5’ untranslated region (UTR)-ORF1a-ORF1b-S-E-M-N-3’UTR (Woo et al., 2012). The 5’ORF1a/b comprises two-thirds of the genome and encodes two overlapping viral replicase polyproteins (1a and 1ab), The six following ORFs encode four structural proteins and two strain-specific accessory proteins in the order: spike (S), envelope (E), membrane (M), nonstructural protein 6 (NS6), nucleocapsid (N), nonstructural protein 7 (NS7) (Lee and Lee, 2015).
Among CoV structural proteins, the S glycoprotein is abundantly produced in infected cells and has multiple functions in viral entry and pathogenesis (Li et al., 2017a; Zhang, 2016). The S1 subunit mediates virus binding to cells through its receptor-binding domain (RBD), while the S2 subunit mediates virus-cell membrane fusion. In addition, the S protein is postulated to harbor epitopes that induce neutralizing antibodies (Raj et al., 2013; Zumla et al., 2016). Hain et al. (Hain et al., 2016) generated a recombinant Orf virus (ORFV) that expresses the full-length PEDV S protein. An immunization challenge study in pigs showed that intramuscular inoculation with ORFV-PEDV-S elicited S-specific IgG, IgA, and a neutralizing antibody response. Inoculation with PDCoV S protein may therefore be able to inhibit PDCoV infection and induce neutralizing antibodiess against PDCoV infection.
Several studies have shown that potent neutralizing antibodies against alpha- or beta-CoVs target the RBD region of the S protein (Du et al., 2013a, b; He et al., 2004a; Li et al., 2017a; Yoo and Deregt, 2001). The RBD regions in several CoV genera have been identified. For example, the C-terminus of the S1 domain is the RBD region of transmissible gastroenteritis virus (TGEV) in the genus Alphacoronavirus (Godet et al., 1994) and in severe acute respiratory syndrome coronavirus (SARS-CoV) in the genus Betacoronavirus (Li et al., 2005). In contrast, the RBD regions of murine hepatitis virus and bovine coronavirus, both in the genus Betacoronavirus, are located in the N-terminus of the S1 domain (Peng et al., 2011, 2012). However, the immunodominant neutralizing region associated with delta-CoVs, such as PDCoV, has not been identified.
Recent elucidation of PDCoV spike protein structures by cryo-electron microscopy reveal that the S1 subunit consists of four individually folded domains, designated A, B, C, and D (Xiong et al., 2018). Recently, Li et al. (Li et al., 2018) demonstrated that the porcine aminopeptidase N, previously known to be a functional receptor for transmissible gastroenteritis virus (TGEV), also interacts with the B domain of PDCoV S1 subunit and functions as a major cell entry receptor for PDCoV. Therefore, in this study, we aimed to identify the immunodominant region of PDCoV S protein, and its neutralizing epitopes. Based on previous structural data and the location of the RBD region in the S protein of PDCoV (Li et al., 2018), three truncated S proteins spanning the entire S domain were produced using an E.coli expression system. The constructs were designated NTD (N-terminal domain of the S1 subunit, amino acids (aa) 50-286), CTD (C-terminal domain of the S1 subunit (aa 278-616), and the S2 subunit (aa 601-1087). We purified the recombinant proteins and inoculated rabbits and mice to produce NTD-, CTD-, and S2-specific polyclonal antisera. Sera from NTD-, CTD-, and S2-inoculated mice had PDCoV neutralization activity after the second boost. All antisera, from mice and rabbits, exhibited anti-PDCoV activity in vitro, determined by virus neutralization, fluorescent focus neutralization, and plaque-reduction neutralization assays. Among the three antisera, the CTD-specific sera showed the most potent PDCoV inhibitory effect, indicating that the CTD region may contain the major neutralizing epitope(s) of the PDCoV S protein.
2. Materials and methods
2.1. Ethics statement
All the animal experiments were performed in strict accordance with the guidelines and regulations of the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China. All experiment in this study were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultual University(IACUC#RW2016-090).
2.2. Cells, viruses, and antibodies
Swine testis (ST) cells (ATCC CRL-1746) were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco, USA) with 10 % fetal bovine serum (PAN-Biotech, Germany) at 37 ℃ in a humidified 5 % CO2 atmosphere. The PDCoV strain CHN-SC2015 was isolated from a diarrheal piglet in Sichuan Province (GenBank accession No.MK355396). The virus was propagated in ST cells and DMEM supplemented with 5 μg/mL of trypsin. The titers of CHN-SC2015 strain during several passages were up to 106·64 TCID50 /ml. Horseradish peroxidase (HRP) conjugated goat anti-rabbit IgG and fluorescein isothiocyanate (FITC) labeled goat anti-mouse IgG were purchased from Beijing Biosynthesis Biotechnology Co., Ltd. (Bioss, Beijing, China). HRP conjugated goat anti-pig IgG was purchased from ABclonal Technology Co., Ltd (ABclonal, Wuhan, China). An anti-PDCoV N-protein monoclonal antibody (mAb) and pig anti-PDCoV CHN-SC2015 polyclonal serum were prepared and stored in our laboratory
2.3. Plasmids design and protein expression
The S gene of PDCoV consists of three overlapping regions, the N- and C-terminal domains of the S1 subunit (NTD, aa 50-286 and CTD, aa 278-616), and the S2 domain (aa 601-1087) (Fig. 1 A). Viral RNA was extracted from infected cell supernatants using TRIzol Reagent (Sangon Biotech, China). The RNA was reverse transcribed using a Transcription First Strand cDNA Synthesis kit (Takara) according to the manufacturer's instructions. Gene-specific primers were designed using Primer 5.0 software based on the published CHN-SC2015 sequence (Table 1 ). The three S gene segments were amplified by RT-PCR then each was cloned into a pET32a (+) expression vector, the resulting plasmids are henceforth referred to as pET32a-NTD, pET32a-CTD, and pET32a-S2. The recombinant plasmids were separately transformed into Transetta(DE3) cells, protein expression was induced with 0.8 mM IPTG for 3–6 h. Protein expression was analyzed by SDS-PAGE. The recombinant proteins were purified as previous described (Luo et al., 2017). The concentration of the purified proteins was determined using an enhanced BCA protein assay kit (Beyotime, China).
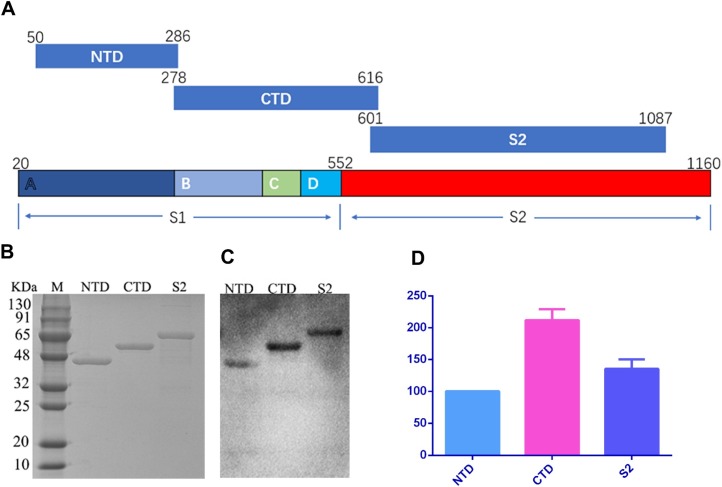
Fig. 1.
Expression of PDCoV S protein segments and comparison of reactivity with pig anti-PDCoV. (A) Schematic showing locations and lengths of the three antigenic S fragments. The S gene was divided into overlapping sections, the N-terminal domain of subunit S1 (NTD, aa 50-286), the C-terminal domain of S1 (CTD, aa 278-616), and S2 (aa 601-1087). The segments were amplified by RT-PCR and then cloned into a pET32a (+) expression vector. Transetta (DE3) cells harboring pET32a-NTD, pET32a-CTD, or pET32a-S2 were induced by IPTG, and cells were collected 4 h after induction. The proteins were purified and diluted to the same concentration (750 μg/ml). (B) SDS-PAGE was performed to analyze protein expression. (C) Expression products were specifically recognized on a western blot using polyclonal pig antisera against PDCoV. (D) Band intensities on the western blot were analyzed using ImageJ. Experiments were performed three times. In each experiment, band intensity values were compared and the intensity of NTD was defined as 100 %.
Table 1.
Oligonucleotide primers used for PCR.
| Primer | Nucleotide sequence (5′–3′)a |
|---|---|
| NTD-F | CGGGATCCATG AATAACTTTGATGTTGGCG |
| NTD-S | CCCTCGAGAGATTTAGGGTCGGAGTAGAAC |
| CTD-F | CGGGATCCATG GATGGGTTCTACTCCGAC |
| CTD-S | CCCTCGAGTGAGGTGTATTGTGCCAGT |
| S2-F | CGGGATCCAATGGCAACAGCCGTTGTCT |
| S2-S | CCCTCGAGGAGCCATTCAAGGTCAACTAGA |
Restriction endonuclease sites: BamHI (italic) and XhoI (underlined).
2.4. Western blot analysis
Equal amounts of purified proteins were separated on a 10 % SDS-PAGE gel, then transferred to a polyvinylidene fluoride membrane (PVDF, BIO-RAD). The membrane was blocked with 2 % bovine albumin V (BSA, Solarbio, China) in PBST (PBS with 0.1 % polysorbate-20), after 2 h the membrane was incubated with pig anti-PDCoV serum (dilution) for 8 h at 4 °C. The membrane was then washed four times with PBST, and incubated with HRP conjugated goat anti-pig IgG (1:5000 in PBST) for 1 h at room temperature. The membrane was washed again four times and the proteins were visualized using enhanced chemiluminescence reagents (ECL; Bio-Rad, USA).
2.5. Antibody production
Rabbits were inoculated with the purified recombinant proteins (NTD, CTD, or S2) for production of polyclonal antibodies, as previously described with some modifications (Luo et al., 2017). Briefly, rabbits were inoculated with 1 mg/rabbit of recombinant protein in Montanide™ Gel 01 PR adjuvant (SEPPIC, France), then boosted three times with the same immunogen and adjuvant at 2-week intervals. Sera was taken from pre-immunized rabbits and then 10 days after each inoculation, sera were heat-inactivated for 1 h at 56 °C and stored at −20 °C.
Female BALB/c mice (6–8-weeks-old) were purchased from Chengdu Dossy Experimental Animal Co., Ltd (Chengdu, China). Mice were divided into 4 groups of 6 mice/group. Three of the groups were subcutaneously injected with 50 μg/mouse of purified protein (NTD, CTD, or S2). The last group was the negative control; each mouse was injected with the same volume of sterile PBS. All animals were boosted twice at two-week intervals. Serum samples were collected each week until 6 weeks after the first immunization. All sera were heat-inactivated for 1 h at 56 °C and stored at −20 °C.
2.6. ELISA
Mouse and rabbit serum titers were assessed by ELISA as previously described with some modifications (Quan et al., 2018). Briefly, 96-well ELISA plates were pre-coated with 1 ug/well of purified PDCoV overnight at 4 °C. Wells were blocked with 5 % non-fat milk for 2 h at 37 °C, then 100 μl of serially diluted sera was aliquoted into each well and plates were incubated for 1 h at 37 °C. Wells were washed four times then incubated with HRP-conjugated anti-mouse IgG or HRP-conjugated anti-rabbit IgG (1:5000 in PBST) for 1 h at 37 °C. Bound antibody was visualized by addition of TMB substrate (3,39,5,59-tetramethylbenzidine) (Invitrogen) for 15 min, the reactions were stopped with 1 N H2SO4. Absorbance at 450 nm was read using microplate absorbance reader (Bio-Rad, USA).
2.7. Virus neutralization assay
A standard micro-neutralization[21] assay was used to quantify the neutralizing activity of the polyclonal serum from each rabbit and mouse group. Briefly, serial dilutions of sera, rabbit or mouse anti-NTD, CTD or S2, were mixed with PDCoV (200 TCID50 in 50 μl) in DMEM. The mixtures were incubated for 1 h at 37 °C to allow for formation of virus-antibody complexes. ST cells grown in 96-well plates were washed 2 times with DMEM supplemented with 5 μg/mL of trypsin, then the virus-antibody mixtures were added to each well. Control sera were treated the same way. After 1 h incubation at 37 °C, unabsorbed virus was removed and cells were overlaid with DMEM supplemented with 2.5 μg/mL of trypsin. After 72 h at 37 °C, the cells incubated with virus and preimmune sera exhibited obvious CPE, the cells incubated with no virus remained healthy. Neutralizing antibody titers were determined by the method of Reed and Muench (Reed and Muench, 1938; Tumpey et al., 2005). Results are expressed as the average of triplicates ± standard deviation.
2.8. Flow cytometry
The binding of mouse anti -NTD, -CTD, and -S2 antibodies to PDCoV-infected ST cells was measured by flow cytometry. Briefly, 2 × 106 ST cells were infected with PDCoV (MOI 0.1) for 24 h, washed twice with PBS and fixed with 4 % formaldehyde in PBS, followed by membrane permeabilization with TritonX-100 for 10 min at 4 °C. The cells were again washed twice with PBS, then incubated with 1 ml of a 1:50 dilution of anti -NTD, -CTD, or -S2 mouse sera for 1 h at 4 °C. Cells were washed then incubated with FITC-goat anti-mouse IgG for 40 min at 4 °C, cells were washed again then analyzed by flow cytometry. Naive mouse antiserum was used as negative control. The flow cytometry data were analyzed using FlowJo software.
2.9. Fluorescent focus neutralization assay (FFN)
The neutralizing activity of the rabbit and mouse anti -NTD, -CTD, and -S2 polyclonal antisera was assessed 4 weeks after the initial inoculation by a FFN assay, as previously described (Okda et al., 2015). The heat-inactivated serum samples were 2-fold serially diluted in DMEM and 200 TCID50 in 50 μl of PDCoV stock was added to each dilution and incubated for 1 h at 37 °C. The virus/serum mixtures were then added to confluent monolayers of ST cells, which had been washed twice with DMEM supplemented with 5 μg/mL trypsin, and incubated for 1.5 h at 37 °C. The overlays were removed and the cells were then incubated for 72 h in DMEM with 2.5 μg/mL of trypsin to allow for replication of non-neutralized virus. Indirect immunofluorescence staining of allowed visualization of PDCoV-infected cells. Endpoint neutralization titers were determined as the highest serum dilution resulting in a 90 % or greater reduction in fluorescent foci relative to PBS-immunized antisera controls.
2.10. Plaque-reduction neutralization test (PRNT)
Plaque-reduction neutralization tests (PRNT) were performed to quantify the titer of PDCoV-neutralizing antibodies in the sera of PDCoV-challenged and unchallenged mice as previously described (Matrosovich et al., 2006). Heat-inactivated sera were two-fold serially diluted from 1:40 to 1:320 in 100 μl of DMEM, to each dilution 50 PFU of PDCoV in 100 μl of DMEM was added, the serum/virus mixtures were then incubated for 1 h at 37 °C. After incubation, 200 μl of each mixture was aliquoted into wells of 6-well plates containing a confluent monolayer of ST cells, plates were incubated for 1.5 h at 37 °C with periodic rocking. After incubation the inoculum was removed and cells were overlaid with 1 × DMEM containing 0.6 % Avicel RC-661(FMC BioPolymer). The plates continued incubating at 37 °C for 3 days, when plaques were easily observed. The monolayers were stained with 1 % crystal violet in 20 % methanol for 30 min at RT, then washed with water. The percent inhibition was calculated as follows: % plaque reduction = (plaque count in virus only sample - plaque count in serum/virus sample) / (plaque count virus only sample) × 100. The PRNT is defined as the reciprocal of the antibody dilution required to reduce the number of plaques by 50 % relative to the control wells
2.11. Statistical analysis
All experimental data were analyzed using GraphPad Prism version 7.0 and expressed as mean ± SD. The differences among the three groups were analyzed using two-way ANOVA. Statistical significance is indicated by* p value < 0.05, ** p value < 0.01, *** p value < 0.001, ****p value < 0.0001.
3. Results
3.1. Preparation and antigenicity analysis of NTD, CTD, and S2
SDS-PAGE analysis showed that the NTD (aa 50-286), CTD (aa 278-616), and S2 (aa 601-1087) fusion proteins were efficiently expressed. The proteins were purified using affinity chromatography, and their concentrations were adjusted to 0.75 mg/ml with PBS (Fig. 1B). After separation using SDS-PAGE and transfer for western blot, the proteins were specifically recognized by pig anti-PDCoV polyclonal antisera (Fig. 1C). Based on band density, the reaction of CTD with the polyclonal antisera was more intense than with the NTD or S2 proteins (Fig. 1D), indicating that the CTD region may be a stronger antigenic site.
3.2. PDCoV neutralizing activity of rabbit polyclonal antisera
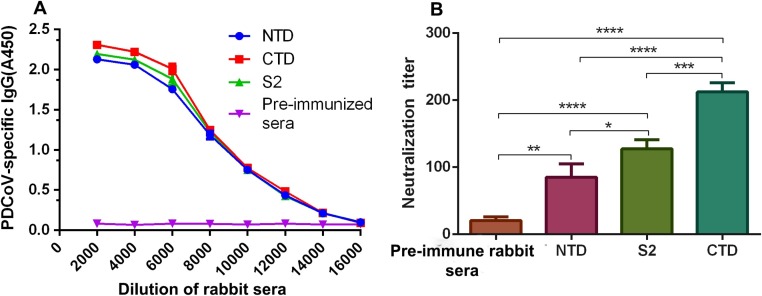
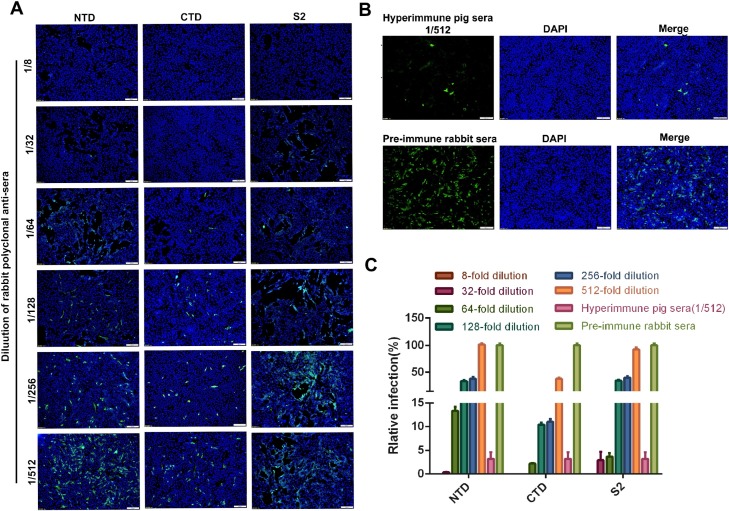
Sera from rabbits inoculated with NTD, CTD, and S2 were tested for neutralizing antibodies against PDCoV by ELISA, virus neutralization (VN), and fluorescent focus neutralization (FFN) assays. As shown in Fig. 2 , all rabbit antisera, anti -NTD, -CTD and -S2, effectively neutralized PDCoV in vitro. Neutralizing antibody titers were 1:88 ± 10, 1:212 ± 11, and 1:125 ± 9.0, respectively. Pre-immune serum exhibited no significant neutralizing effect. The FFN assay (Fig. 3 ) shows that the endpoint neutralizing titer (1:256) of the rabbit polyclonal serum vs. CTD was higher than that for the serum vs. NTD or S2 (1:32 and 1:64, respectively). These results demonstrate that the NTD, CTD, and S2 proteins induced potent anti-PDCoV neutralizing antibody responses in the immunized animals.
Fig. 2.
Detection of antibody responses and evaluation of neutralizing activity for sera of rabbit vaccinated with NTD, CTD and S2, respectively. Sera from 10 days post-last vaccination were used for the detection. (A) ELISA of anti-PDCoV serum IgG levels, data are presented as mean A450 ± SD. (B) PDCoV-neutralizing antibody titers, pre-immune serum was used for a negative control. **P = 0.0031, *P = 0.0378, ****P < 0.0001 for NTD compared to pre-immune serum, S2 and CTD, respectively. ***P = 0.0005 for S2 compared to CTD. The experiment was repeated three times. Statistical differences are indicated by * p value < 0.05, ** p value < 0.01, *** p value < 0.001, **** p value < 0.0001.
Fig. 3.
Comparison of PDCoV-neutralizing antibodies in sera of rabbits inoculated with NTD, CTD, and S2. Sera were collected on day 10 after the final inoculation. (A) FFN assay of the PDCoV-neutralizing activity in each sera. (B) Pre-immune serum was used as negative control and hyperimmune pig anti-serum against PDCoV was used as positive control. (C) Neutralizing activity of the S-specific antisera was quantified by counting virus-infected cells after immunofluorescent staining. Percent infection based on the images in panels A and B, the FITC-positive cells in the control serum was set as 100 %. The total number cells and the PDCoV + cells were counted using three or more pictures for each dilution of sera.
3.3. NTD, CTD, and S2 induce humoral immune responses in mice
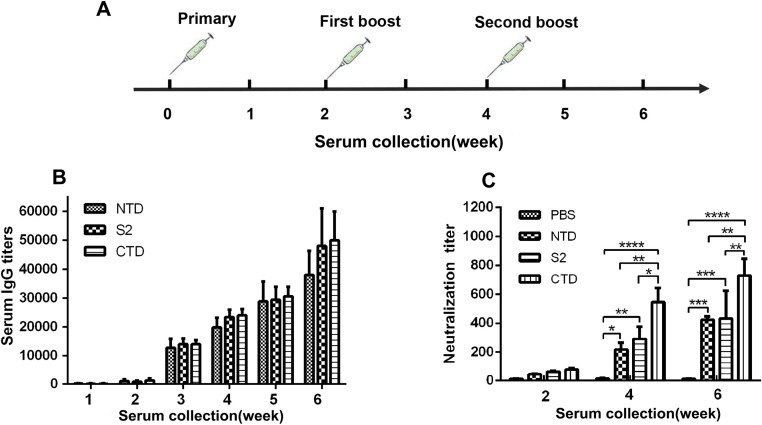
To evaluate the immunogenicity of NTD, CTD, and S2, mice were inoculated by subcutaneous injection with identical concentrations (50 μg/mouse) of the three proteins (Fig. 4 A). Sera were collected every week after the primary inoculation and PDCoV-specific antibodies were detected using ELISA. The results showed that each protein induced PDCoV-IgG antibodies after the first inoculation, and all mice showed a significantly enhanced immune response three weeks after the secondary boost. CTD induced the highest level of IgG antibody (Fig. 4B). The PDCoV neutralizing-activity of the mouse sera was assessed using a virus neutralization (VN) assay. PDCoV neutralization was detected 2 weeks after mice were initially inoculated and the neutralizing titers increased significantly thereafter (Fig. 4C). Sera from the PBS inoculated control group had no significant neutralizing activity. After 4 weeks, each sera effectively neutralized PDCoV infection in vitro; antibody titers were 1:218 ± 38, 1:545 ± 81, and 1: 291 ± 68, respectively (Fig. 4C). More importantly, neutralizing antibody titers induced by CTD were significantly higher than those induced by NTD or S2 at weeks 4 and 6 (Fig. 4C), indicating that CTD contains the main neutralizing epitope in the PDCoV S protein.
Fig. 4.
Kinetics of anti -NTD, -CTD, and -S2 IgG production and PDCoV neutralization. (A) Experimental timeline. Equal doses of antigen were injected on weeks 0, 2, and 4. (B) Time course of PDCoV-specific IgG response in mice upon subcutaneous delivery of NTD, CTD, and S2. Blood samples were collected every week for 6 weeks, and PDCoV-specific antibodies were measured by ELISA. (C) PDCoV-neutralizing titers were measured every two weeks. At week four, *P = 0.0398, **P = 0.0058, ****P < 0.0001 for PBS compared to NTD, CTD and S2, respectively. **P = 0.0016 for NTD compared to CTD, and *P = 0.0105 for S2 compared to CTD. At week six, both ***P = 0.0002 for PBS compared to NTD and S2, and ****P < 0.0001 compared to CTD. **P = 0.0028 for NTD compared to CTD, and **P = 0.0033 for S2 compared to CTD. Statistically significant differences are indicated by* p value < 0.05, ** p value < 0.01, *** p value < 0.001, **** p value < 0.0001.
3.4. Evaluation of the affinity of mouse polyclonal antibodies to PDCoV
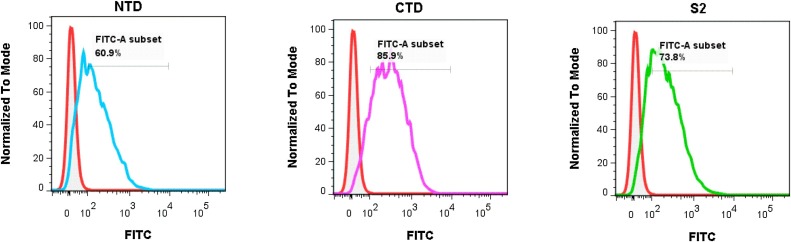
PDCoV infected ST cells were reacted with NTD-, CTD-, and S2-specific mouse antisera, collected at week 4 after the initial inoculation, then with FITC-anti mouse IgG. Antibody binding to PDCoV was evaluated by flow cytometry. As shown in Fig. 5 , PDCoV-specific fluorescence signal was generated by each sera, but the percentage of PDCoV-infected cells bound by the CTD-antisera (85.9 %), was considerably higher than NTD (60.9 %) or S2 (73.8 %) antisera.
Fig. 5.
Evaluation by flow cytometry of the affinity of mouse polyclonal antibodies for PDCoV. PDCoV infected ST cells incubated with anti-NTD, CTD, and S2 polyclonal antisera (collected at week 4), or naive mouse antisera, then with FITC-labelled goat-anti-mouse. The red peaks represent the cells reacted with the negative control serum.
3.5. PDCoV neutralizing activity of mouse polyclonal antisera
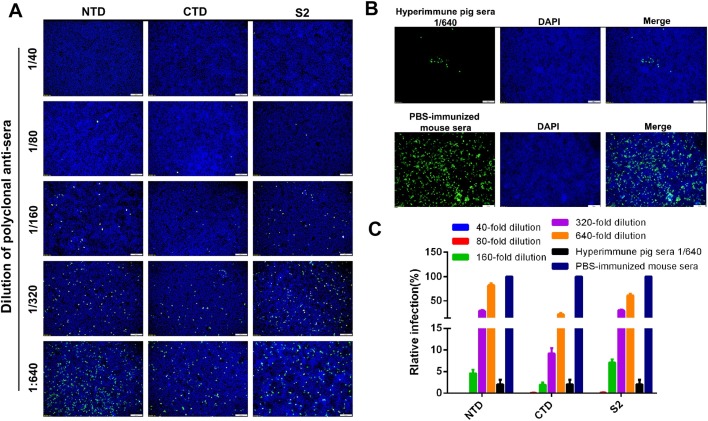
Neutralizing activities of the mouse polyclonal antisera were also tested by fluorescent focus neutralization (FFN) assay. As shown in Fig. 6 , the mouse antisera differed in the extent of neutralization of PDCoV infection. Based on relative amount of infected ST cells, the endpoint neutralizing titer (1:320) of the CTD serum was higher than the NTD or S2 sera (both 1:160). The results suggest that the CTD region may contain the major neutralizing epitope(s) of the PDCoV S protein.
Fig. 6.
PDCoV-neutralizing activity of S-specific mouse polyclonal antisera. Sera collected at week 4 after the primary inoculation were used. (A)The PDCoV neutralizing activity of NTD, CTD, and S2 mouse antisera was determined by fluorescent focus neutralization assay (FFN; >90 % reduction; magnification, 100×). (B) Serum from mice inoculated with PBS was used as negative control and hyperimmune pig anti-PDCoV serum was used as positive control. (C) Neutralizing activity of the S-specific antisera was quantified by counting virus-infected cells after immunofluorescent staining. Percent infection based on the images in panels A and B, the FITC-positive cells in the control serum was set as 100 %. The total number cells and the PDCoV + cells were counted using three or more pictures for each dilution of sera.
3.6. Plaque reduction neutralization test (PRNT)
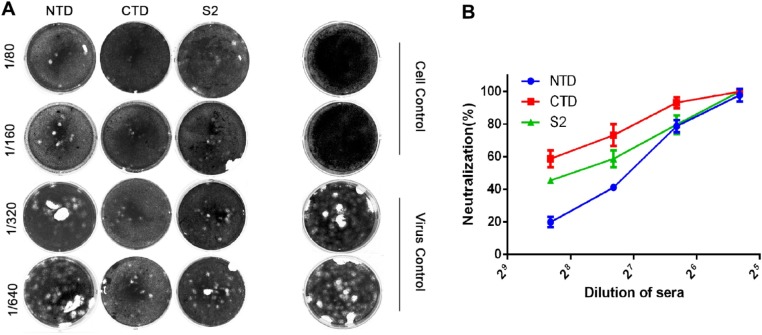
To further demonstrate neutralization activity, a plaque reduction neutralization test (PRNT) was used to evaluate the mouse sera collected at week 4 after the initial inoculation. The PRNT titer (1:320) of the anti-CTD sera was higher than the titers obtained for anti-NTD (1:80) and -S2 sera (1:160) (Fig. 7 ). Of the three protein domains, the CTD may play a more important role in the viral infection process.
Fig. 7.
(A) Plaque reduction neutralization activity of S-specific mouse polyclonal antisera. Sera collected at week 4 after the primary inoculation were used. Approximately 50 PFU of PDCoV were used to infect ST cells in a 12-well plate with or without PDCoV S-specific polyclonal antisera. PDCoV alone and PBS were used as the virus and blank controls, respectively. The images correspond to the neutralizing percentages presented in panel B.
4. Discussion
Several studies have shown that the S glycoprotein of CoVs is primarily involved in virus attachment and cellular entry (Gallagher and Buchmeier, 2001). The S protein of CoVs contains two subunits, S1 and S2. Subunit S1 is located at the N-terminus and binds to cellular receptor(s), whereas S2 is involved in virus-cell membrane fusion (Eckert and Kim, 2001). Given its critical functions in cellular entry, S protein is a high-priority target for vaccine development. S protein-specific serum or secretory neutralizing antibodies may play a key role in protection against coronavirus infection (Okda et al., 2017; Song et al., 2016). Several neutralizing epitopes have been identified in the full-length PEDV S protein, with the majority of epitope(s) mapping to a collagenase equivalent domain (COE) (Okda et al., 2017). Vaccine development has therefore been focused exclusively on the COE epitope. An oral vaccine (pPG-COE-DCpep/L393), a recombinant Lactobacillus casei that expresses DC-targeting peptide fused with a COE protein, was developed by Hou et al. (Hou et al., 2018).
PDCoV, a newly emerged and lethal swine enteric coronavirus, poses a worldwide threat to the pig industry. To date, the immune neutralizing region or epitope of PDCoV remains unknown. Using approaches similar to those used to study PEDV, we successfully identified the immune dominant region(s) of PDCoV S protein. These regions are potentially useful as targets for vaccine or therapeutic agents to control PDCoV infection in pigs.
In order to identify the immune dominant region of PDCoV S protein, we expressed three truncated fragments covering residues 50-1087 of S but excluding the signal peptide, guided by a previous study that used cryo-electron microscopy to analyze the structure of the PDCoV S protein (Xiong et al., 2018). The three recombinant fragments contained the NTD (aa 50-286), CTD (aa 278-616), and S2 (aa 601-1087) regions of the S protein. The purified fragments were specifically recognized on a western blot probed using polyclonal pig antisera against PDCoV. The bands migrated as expected for each protein (NTD, 46 kDa; CTD, 58 kDa; S2, 66 kDa) (Fig. 1C). Interestingly, CTD bound PDCoV at higher a level than did NTD or S2, suggesting that the CTD region may have better antigenicity and contain more or stronger epitopes.
Purified recombinant NTD, CTD and S2 proteins were injected into rabbits to produce specific antisera. All antisera were able to inhibit PDCoV infection in vitro, as determined by VN and FFN assays. Amongst the three antisera, CTD-specific serum was the most effective (Fig. 2, Fig. 3). However, since the neutralizing antibody in these experiments was obtained from a single time point after immunization, the result is not conclusive. To verify the relative efficacies of the antisera, female BALB/c mice were inoculated with purified protein and the corresponding antibodies was evaluated several times after immunization. The inoculated mice showed a significant increase in anti-PDCoV antibody titer after receiving the second immunization (Fig. 4B). As shown in Fig. 4C, anti-CTD antiserum consistently showed the highest neutralizing antibody titers by VN assay across time points. At week 4, mice inoculated with CTD showed a significant increase in neutralizing antibody titers to 1:545 (± 81.7). Similarly, anti-CTD antiserum collected at week 4 also had a markedly higher neutralizing titer (1:320) in FFN and PRNT assays (Fig. 6, Fig. 7). These results suggest that PDCoV CTD has an important role in the viral infection process.
Neutralizing antibody titers in mice inoculated with the RBD regions of MERS-CoV and SARS-CoV reached 1:1000 in VN assays after the second immunization (He et al., 2005; Tai et al., 2017). The three recombinant PDCoV proteins we tested generated somewhat lower neutralizing antibody responses, perhaps because the truncated region of the S protein contains non-neutralizing epitopes that compete with the neutralizing epitopes in eliciting antibody responses (Du et al., 2013a). More precise localization and identification of specific immune dominant epitope(s) of CTD will be needed to test this hypothesis.
The S glycoprotein of CoVs plays a critical role in mediating viral entry and inducing neutralizing antibody responses in infected individuals (Liu et al., 2016; Spaan et al., 1988; Zumla et al., 2016). Previous work demonstrated that an anti-PEDV-S2 mAb recognizing the linear epitope GPRLQPY at the carboxy-terminal of the S2 subunit exhibited neutralizing activity against PEDV (Cruz et al., 2008). Similarly, a neutralizing mAb that specifically recognizes the S1-NTD of MERS-CoV S protein has been identified (Chen et al., 2017). In our study, we also found that regions S2 and S1-NTD can induce neutralizing antibody, consistent with previous reports (Chen et al., 2017; Cruz et al., 2008). In addition, all CoVs use the S1-CTD or S1-NTD regions to interact with cellular receptors or co-receptors for viral attachment. The main functional RBD regions of most α-CoVs and some β-CoVs, including SARS-CoV, MERS-CoV, and HCoV-HKU1, are located in the S1-CTD (Chen et al., 2017; Deng et al., 2016). In contrast, the RBDs of human CoV OC43, MHV, and avian γ-CoV are located in the S1-NTD region. Importantly, the S-RBD region in most CoVs contains critical neutralizing sites that induce potent neutralizing antibodies (Du et al., 2013a; He et al., 2004b; Yoo and Deregt, 2001). Using ten anti-PEDV-S1 mAbs, Li et al.(Li et al., 2017a) showed that six non-overlapping neutralizing and non-neutralizing epitopes exist in the S1 subunit of the PEDV spike protein, and that most of the neutralizing antibodies target the receptor binding domain. The antibodies induced by NTD, CTD, and S2 in our study differed in neutralization capacity against PDCoV in vitro (Fig. 4, Fig. 5, Fig. 6). The superior inhibitory and neutralizing capacity of CTD antiserum suggests that this region may contain the main RBD region or neutralizing epitopes in the PDCoV S protein.
Recently, three research groups demonstrated that pAPN is a major cell entry receptor for PDCoV, and its catalytic domain can interact with S1-CTD (Li et al., 2018; Wang et al., 2018; Zhu et al., 2018). In these studies, APN-knockout IPI-2I cells and ST cells were still susceptible to PDCoV infection. By analyzing the three-dimensional structures of PDCoV S1-CTD, TGEV S1-CTD, and pAPN, Zhu et al. (Zhu et al., 2018) found that the shorter β1–β2 and β3–β4 turns in PDCoV S1-CTD may cause insufficient contact with pAPN compared with TGEV S1-CTD. These data indicate that pAPN may be a major entry receptor for PDCOV, but other unknown PDCOV receptors also exist. Li et al. (Li et al., 2017b) found that MERS-CoV uses both protein and sialoglycan-based receptor determinants, with binding to dipeptidyl peptidase 4 (DPP4) required for entry and cell surface sialic acids (Sias) serving as attachment factors, respectively. Data also show that DPP4 interacts with the CTD region of MERS-CoV S1 subunit (Mou et al., 2013), and the Sia binding site lies within S1-NTD of the MERS-CoV (Li et al., 2017b). In our current study, we showed that the NTD of PDCoV S1 subunit can induce neutralizing antibody responses. Importantly, the sugar-binding capability of PDCoV S1-NTD to mucin has been demonstrated using an enzyme-linked immunosorbent assay (Shang et al., 2017). Therefore, we speculate that one sugar receptor may interact with the NTD region in the S1 Subunit and influence the entry of PDCoV into the cell, just as Sia is used as an additional receptor for MERS-CoV cell entry.
To our knowledge, this is the first report describing the neutralizing regions within the PDCOV S protein. We found that three regions in PDCOV S protein, including NTD (aa 50-286), CTD (aa 278-616), and S2 (aa 601-1087), can induce neutralizing antibody responses, and the specific polyclonal mouse and rabbit sera efficiently inhibit PDCoV entry and infection in ST cells. Because receptor binding domains on CoVs are often targets for potent neutralizing antibodies, we speculate that the CTD and NTD regions may bind to protein and sugar receptors, respectively. Our results provide a foundation for the study of PDCoV receptors and their receptor binding domains. CTD may be more immunogenic than the NTD and S2 regions and may contain the major neutralizing epitope(s) in the PDCoV S protein. Therefore, CTD potentially offers an attractive target for the development of vaccines to prevent PDCoV infection and combat future PDCoV- disease pandemics.
Declaration of Competing Interest
The authors declare that we have no commercial or associative interests that represent a conflict of interest in connection with the work submitted.
Acknowledgements
This study was supported by the National Key Research and Development Program of China (award 2016YFD0500700). We thank Dr. Kwonil Jung from the Ohio State University for critical reading and editing of the manuscript.
References
- Chen Y., Lu S., Jia H., Deng Y., Zhou J., Huang B., Yu Y., Lan J., Wang W., Lou Y., Qin K., Tan W. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg. Microbes Infect. 2017;6(6):e60. doi: 10.1038/emi.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz D.J., Kim C.J., Shin H.J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. 2008;132(1-2):192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F., Ye G., Liu Q., Navid M.T., Zhong X., Li Y., Wan C., Xiao S., He Q., Fu Z.F., Peng G. Identification and comparison of receptor binding characteristics of the spike protein of two porcine epidemic diarrhea virus strains. Viruses. 2016;8(3):55. doi: 10.3390/v8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Kou Z., Ma C., Tao X., Wang L., Zhao G., Chen Y., Yu F., Tseng C.T., Zhou Y., Jiang S. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Kou Z., Ma C., Sun S., Poon V.K., Lu L., Wang L., Debnath A.K., Zheng B.J., Zhou Y., Jiang S. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J. Virol. 2013;87(17):9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68(12):8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain K.S., Joshi L.R., Okda F., Nelson J., Singrey A., Lawson S., Martins M., Pillatzki A., Kutish G.F., Nelson E.A., Flores E.F., Diel D.G. Immunogenicity of a recombinant parapoxvirus expressing the spike protein of Porcine epidemic diarrhea virus. J. Gen. Virol. 2016;97(10):2719–2731. doi: 10.1099/jgv.0.000586. [DOI] [PubMed] [Google Scholar]
- He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. (Baltimore, Md. : 1950) 2005;174(8):4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Jiang X., Jiang Y., Tang L., Xu Y., Qiao X., Min L., Wen C., Ma G., Li Y. Oral immunization against PEDV with recombinant Lactobacillus casei expressing dendritic cell-targeting peptide fusing COE protein of PEDV in piglets. Viruses. 2018;10(3):106. doi: 10.3390/v10030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53(5):1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Saif L.J. Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine deltacoronavirus strain OH-FD22. Arch. Virol. 2016;161(12):3421–3434. doi: 10.1007/s00705-016-3056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2014;2(6):e01191–01114. doi: 10.1128/genomeA.01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015;208:136–145. doi: 10.1016/j.virusres.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li W., Lucio de Esesarte E., Guo H., van den Elzen P., Aarts E., van den Born E., Rottier P.J.M., Bosch B.J. Cell attachment domains of the porcine epidemic diarrhea virus spike protein are key targets of neutralizing antibodies. J. Virol. 2017;91(12):e00273–00217. doi: 10.1128/JVI.00273-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li G., Chen Q., Harmon K.M., Yoon K.J., Schwartz K.J., Hoogland M.J., Gauger P.C., Main R.G., Zhang J. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2014;2(2):e00278–00214. doi: 10.1128/genomeA.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Kenney S.P., Widjaja I., Jung K., Alhamo M.A., van Dieren B., van Kuppeveld F.J.M., Saif L.J., Bosch B.J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. U. S. A. 2018;115(22):E5135–e5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W., Widagdo W., Tortorici M.A., van Dieren B., Lang Y., van Lent J.W.M., Paulson J.C., de Haan C.A.M., de Groot R.J., van Kuppeveld F.J.M., Haagmans B.L., Bosch B.J. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017;114(40):E8508–e8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Ma Y., Yang Y., Zheng Y., Shang J., Zhou Y., Jiang S., Du L., Li J., Li F. Cell entry of porcine epidemic diarrhea coronavirus is activated by lysosomal proteases. J. Biol. Chem. 2016;291(47):24779–24786. doi: 10.1074/jbc.M116.740746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S.X., Fan J.H., Opriessnig T., Di J.M., Liu B.J., Zuo Y.Z. Development and application of a recombinant M protein-based indirect ELISA for the detection of porcine deltacoronavirus IgG antibodies. J. Virol. Methods. 2017;249(1):76–78. doi: 10.1016/j.jviromet.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio. 2015;6(2) doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M., Matrosovich T., Garten W., Klenk H.D. New low-viscosity overlay medium for viral plaque assays. Virol. J. 2006;3:63. doi: 10.1186/1743-422X-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H., Raj V.S., van Kuppeveld F.J., Rottier P.J., Haagmans B.L., Bosch B.J. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87(16):9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okda F., Liu X., Singrey A., Clement T., Nelson J., Christopher-Hennings J., Nelson E.A., Lawson S. Development of an indirect ELISA, blocking ELISA, fluorescent microsphere immunoassay and fluorescent focus neutralization assay for serologic evaluation of exposure to North American strains of Porcine Epidemic Diarrhea Virus. BMC Vet. Res. 2015;11:180. doi: 10.1186/s12917-015-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okda F.A., Lawson S., Singrey A., Nelson J., Hain K.S., Joshi L.R., Christopher-Hennings J., Nelson E.A., Diel D.G. The S2 glycoprotein subunit of porcine epidemic diarrhea virus contains immunodominant neutralizing epitopes. Virology. 2017;509:185–194. doi: 10.1016/j.virol.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U. S. A. 2011;108(26):10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Xu L., Lin Y.L., Chen L., Pasquarella J.R., Holmes K.V., Li F. Crystal structure of bovine coronavirus spike protein lectin domain. J. Biol. Chem. 2012;287(50):41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan K., Zhu Z., Cao S., Zhang F., Miao C., Wen X., Huang X., Wen Y., Wu R., Yan Q., Huang Y., Ma X., Han X., Zhao Q. -Derived outer membrane vesicles deliver Galactose-1-phosphate uridyltransferase and yield partial protection against in mice. J. Microbiol. Biotechnol. 2018;28(12):2095–2105. doi: 10.4014/jmb.1809.09004. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoint. Am. J. Hyg. 1938;27(3):493–497. [Google Scholar]
- Saeng-Chuto K., Lorsirigool A., Temeeyasen G., Vui D.T., Stott C.J., Madapong A., Tripipat T., Wegner M., Intrakamhaeng M., Chongcharoen W., Tantituvanont A., Kaewprommal P., Piriyapongsa J., Nilubol D. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and lao PDR in 2015. Transbound. Emerg. Dis. 2017;64(1):3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Tai W., Du L., Zhou Y., Zhang W., Li F. Cryo-EM structure of porcine delta coronavirus spike protein in the pre-fusion state. J. Virol. 2017;92(4) doi: 10.1128/JVI.01556-17. JVI.01556-01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound. Emerg. Dis. 2015;62(6):575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., Stone S., Drebes D., Greiner L.L., Dvorak C.M.T., Murtaugh M.P. Characterization of anti-porcine epidemic diarrhea virus neutralizing activity in mammary secretions. Virus Res. 2016;226:85–92. doi: 10.1016/j.virusres.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek M.C. Coronaviruses: structure and genome expression. J. Gen. Virol. 1988;69(12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. 6. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Shibahara T., Imai N., Yamamoto T., Ohashi S. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect. Genetics Evol. 2018;61:176–182. doi: 10.1016/j.meegid.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Wang Y., Fett C.A., Zhao G., Li F., Perlman S., Jiang S., Zhou Y., Du L. Recombinant receptor-binding domains of multiple middle east respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J. Virol. 2017;91(1) doi: 10.1128/JVI.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey T.M., García-Sastre A., Taubenberger J.K., Palese P., Swayne D.E., Pantin-Jackwood M.J., Schultz-Cherry S., Solórzano A., Van Rooijen N., Katz J.M., Basler C.F. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J. Virol. 2005;79(23):14933–14944. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Liu Y., Ji C.M., Yang Y.L., Liang Q.Z., Zhao P., Xu L.D., Lei X.M., Luo W.T., Qin P., Zhou J., Huang Y.W. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J. Virol. 2018;92(12):e00318–00318. doi: 10.1128/JVI.00318-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014;20(7):1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg. Infect. Dis. 2014;20(9):1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Tortorici M.A., Snijder J., Yoshioka C., Walls A.C., Li W., McGuire A.T., Rey F.A., Bosch B.J., Veesler D. Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections. J. Virol. 2018;92(4):e01628–01617. doi: 10.1128/JVI.01628-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D., Deregt D. A single amino acid change within antigenic domain II of the spike protein of bovine coronavirus confers resistance to virus neutralization. Clin. Diagn. Lab. Immunol. 2001;8(2):297–302. doi: 10.1128/CDLI.8.2.297-302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016;226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Liu S., Wang X., Luo Z., Shi Y., Wang D., Peng G., Chen H., Fang L., Xiao S. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection. Emerg. Microbes Infect. 2018;7(1):65. doi: 10.1038/s41426-018-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nature reviews. Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]