Abstract
MicroRNAs are short, endogenous, nonprotein-coding RNAs that are essential for regulation of cellular processes through gene silencing. The miR-34/449 family is conserved in mammalian organisms and generally comprises six homologous genes: miR-34a, miR-34b, miR-34c, miR-449a, miR-449b and miR-449c, at three genomic loci. Strong similarity in the sequence of these miRNAs, particularly at the seed region, predicts robust functional redundancy. A large proportion of the literature on the miR-34/449 family focuses on its role in regulating cell cycle arrest and apoptosis by modulating E2F- and p53-related signaling pathways. A growing subset of the literature reports that the miR-34/449 family is involved in the regulation of immune responses and viral infections, and data suggest the potential for miR-34/446 as a diagnostic and therapeutic target. In this review, we discuss our current understanding of the conservation and transcriptional regulation of the miR-34/449 family and review the literature on its functions in viral infections.
Keywords: miR-34/449 family, Viral infection, Virus replication, Inflammatory responses
1. Introduction
MicroRNAs (miRNAs) encode a type of short, nonprotein-coding RNA that fine-tunes gene expression through post-transcriptional repression (Ambros, 2004; He and Hannon, 2004). The miRNA-mediated repression mechanism has essential regulatory functions in plants and animals. In animals, primary miRNAs (pri-miRNAs) are transcribed from miRNA genes and processed sequentially by two ribonucleases of the RNase III family, Drosha and Dicer, to give rise to approximately 22-bp single-stranded RNAs (Ha and Kim, 2014). Mature miRNAs exert their function by binding the 3ʹ un-translated region (3′UTR) of protein-coding transcripts through a complementary “seed sequence” to reduce target transcript stability or repress protein synthesis (Lai, 2002). A single miRNA can target multiple messenger RNAs (mRNAs), and a single mRNA 3′UTR can contain binding sites for a number of miRNAs (Bartel, 2009). Genes for miRNAs are distributed in different genomic locations, including intergenic regions, introns and exons of protein-coding genes. The majority of miRNA genes are transcribed individually and scattered across the entire genome, whereas a portion of miRNA genes are clustered and transcribed as one long pri-miRNA transcript and processed into individual precursor miRNAs (pre-miRNAs) (Altuvia et al., 2005). According to miRBase, more than a quarter of human miRNA genes are <10 kb away from other miRNA gene loci (Kozomara and Griffiths-Jones, 2014). The genomic distribution of miRNAs within a cluster probably serve to protect them from degradation because the secondary structure of a longer pri-miRNA is complex, with a number of hairpins that stabilize the RNA (Mathelier and Carbone, 2013). Generally, miRNAs within a cluster are paralogous with strong sequence similarity, indicating that they may be the result of genomic duplications (Hertel et al., 2006). The miR-34/449 family generally occupies three different loci in mammalian genomes: miR-34a, miR-34b/c and miR-449a/b/c. The expression of the miR-34/449 family is regulated by p53 and essential for regulation of cell cycle arrest and apoptosis via silencing key related factors, including B-cell CLL/lymphoma 2 (BCL-2), cyclins, CDKs, and E2F (Lize et al., 2011) in different cell types, including virus-infected cells (Wang et al., 2015). Increasing evidence shows that the miR-34 family is an important regulator of inflammatory responses and viral replication. The miR-34/449 family is also used as biomarkers for virus infection. For example, serum expression of miR-34a and its target, heat shock protein 70 (Hsp70), are used as biomarkers for early detection of hepatitis C virus (HCV) infection (Shehata et al., 2017). In this review, we discuss the characteristics and functions of the highly conserved miR-34/449 family and summarize the roles of miR34/449 family in viral infections.
2. Discovery of the miR-34/449 family and genomic organization in humans
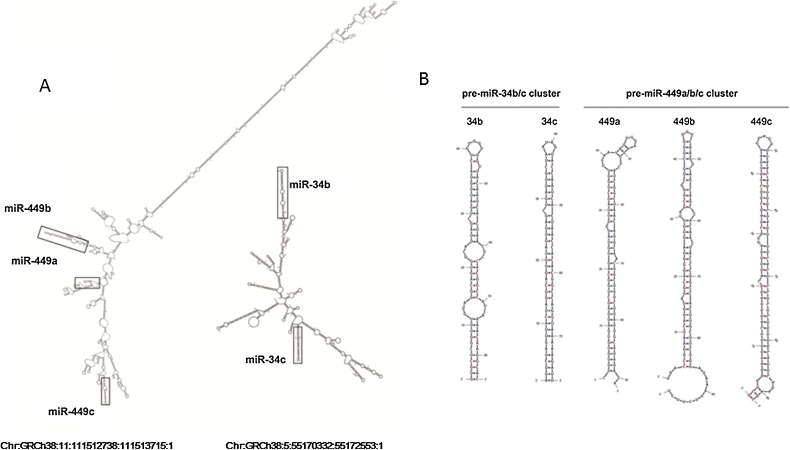
The first human miRNA of the miR-34/449 family to be identified was miR-34a, which was predicted by computational methods (Lim et al., 2003) and validated by microRNP immunoprecipitation from the human retinoblastoma Weri cell line (Dostie et al., 2003). MiR-34b and miR-34c were both cloned and sequenced in 2003 by Houbaviy et al (Houbaviy et al., 2003). miR-449a, miR-449b and miR-449c were identified by separate groups: Xie et al identified miR-449a via bioinformatics and cloning approaches (Xie et al., 2005). miR-449b was predicted and experimentally verified by Cummins et al using the miRAGE method (Cummins et al., 2006). The miR-449c sequence was identified as a miRNA candidate by Berezikov et al using RAKE techniques (Berezikov et al., 2006) and expression was confirmed by Wyman et al (Wyman et al., 2009). miR-34b/c and miR-449a/b/c are classified as two miRNA clusters because miRNAs within these two clusters are co-expressed in testes and located near each other (Bao et al., 2012). The miR-34a gene is on chromosome 1p36.22, and miR-34b/c and miR-449a/b/c clusters are expressed from polycistronic transcripts encoded on chromosomes 11q23.1 and 5q11.2, respectively. The secondary structure of human pri-miR-34b/c and pri-miR-449a/b/c were generated using Mfold, a software using energy minimization criteria to determine optimal suboptimal RNA and DNA secondary structure (Zuker, 2003). The results showed that the secondary structure of pri-miR-449a/b/c places the members of each cluster in close proximity despite a > 3 kb span between pri-miR-449a and pri-miR-449c (Fig. 1 ). Both the pri-miR34-b/c and pri-miR-449a/b/c structures contain a number of other hairpins that may enhance RNA stability (Mathelier and Carbone, 2013). Although the mature sequences of members of the hsa-miR-34/449 family have identical seed sequences (Fig. 2 B), the six trimmed pre-miRNAs are structurally different (Fig. 2). The miR-34/449 family in mammalian genomes has at least one less-conserved paralog, miR-449d, located on the chromosome 29 of Bos taurus (Glazov et al., 2009). Compared with other mammalian miR-34/449 family members, cow miR-449d has two base differences in the seed sequence (Glazov et al., 2009).
Fig. 1.
(A) Secondary structures of hsa-pri-miR-34b/c and miR-449a/b/c clusters predicted by Mfold folding software. (B) Secondary structures of hsa-pre-miR-34a, 34b, 34c, 449a, 449b and 449c.
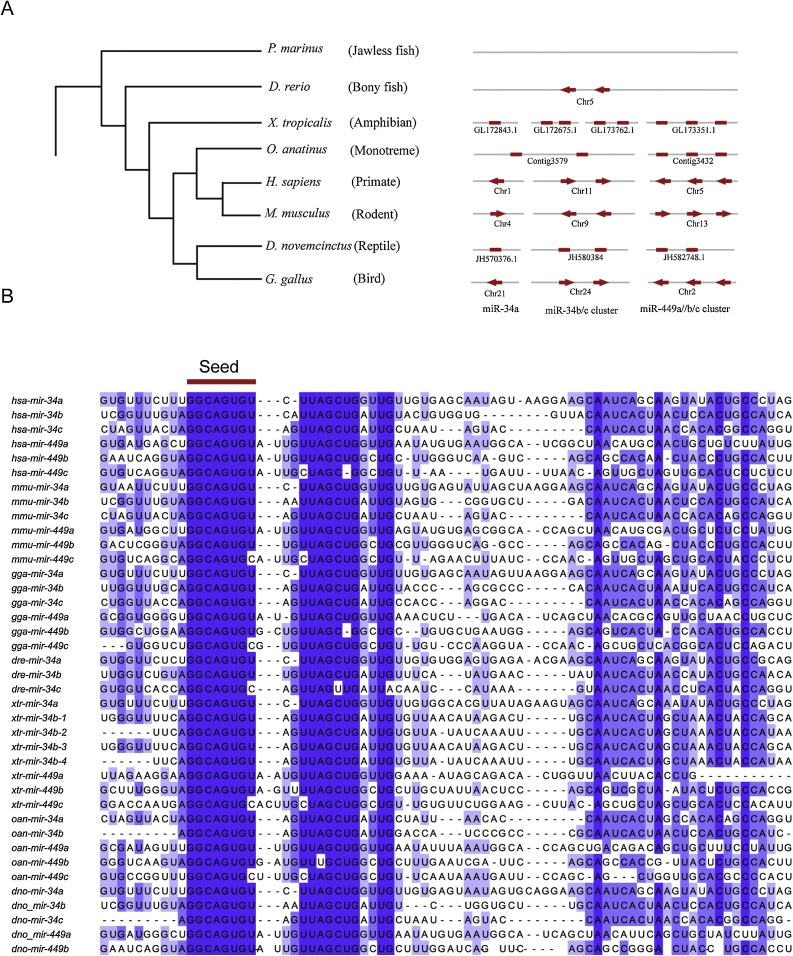
Fig. 2.
(A) Phylogenetic tree of vertebrate species and genomic organization of miR-34/449 family members. (B) Multiple sequence alignment of precursor sequences of miR-34/449 family members. The multiple sequences alignment is obtained using MEGA6 (Tamura et al., 2013) and displayed in Jalview 2.8 (Waterhouse et al., 2009).
3. Comparative genomic analysis of the miR-34/449 family in vertebrates
The starting point of analysis was the collection of precursor sequences of miR-34/449 family members in representative vertebrate species. A combined searching approach was used to identify miR-34/449 family members in multiple genomes from the miRBase Database (http://www.mirbase.org/) and Ensemble Genome Database (http://www.ensembl.org/index.html). For some well-annotated vertebrate genomes, the precursor sequences were retrieved directly from the databases. In addition, BLAST searches were carried out to identify the homologous sequences (Altschul et al., 1997). The representative species of vertebrates analyzed in this study included Gallus gallus (bird), Homo sapiens (primate), Dasypus novemcinctus (reptile), Ornithorhynchus anatinus (ancient mammal), Xenopus tropicalis (amphibian), Danio rerio (bony fish) and Petromyzon marinus (jawless fish). Combined with phylogenetic relationships between selected vertebrate species, we found the miR-34/449 family was conserved in vertebrates but copy numbers and genome organizations varied. As shown in Fig. 2, no members of the miR-34/449 family were discovered in the genome of an ancient vertebrate, the jawless fish P. marines. Only two members of the miR-34/449 family, miR-34b and miR-34c, were discovered in bony fishes such as D. rerio. No miR-34a or miR-449a/b/c was detected in a jawless and a bony fish, suggesting that the emergence of the miR-34a and miR-449a/b/c cluster at the branch led to the tetrapoda. Two miR-34b/c clusters were located in different scaffolds in the X. tropicalis genome, probably due to random duplication. The absence of miR-34c and miR-449c in the genomes of O. anatinus and D. novemcintus was probably because of random deletion. The expansion or shrinking of the miR-34/449 family by random duplication or deletion produced miR-34/449 clusters of different lengths in different organisms. RNA-seq data from H. sapiens and Mus musculus (summarized in miRBase) verified that the 5ʹ arm is highly expressed and more conserved than the 3ʹ arm in the majority of members of the miR-34/449 family.
4. Expression of the miR-34/449 family regulated by viral infection
Similar to protein-coding genes, miRNA genes are transcribed by RNA polymerase II. The transcription of miRNA genes is activated by various cellular processes, including binding of transcription factors to miRNA gene promoter regions and chromatin modifications. The three miR-34/449 family loci have their own transcription units in mammalian genomes. Expression of the miR-34/449 family can be regulated by CpG methylation (Lodygin et al., 2008) or binding of transcription factors, such as p53 (Chang et al., 2007), CREMτ and SOX5 (Bao et al., 2012), SNAIL and ZEB1 (Siemens et al., 2011) and NF-κB (Li et al., 2012) to promoter regions. Therefore, viruses are able to regulate the expression of the miR-34/449 family by modulating the binding of transcription factors to promoters of the gene family. Oncogenic human papillomavirus types 16 (HPV 16) and 18 (HPV 18) reduce miR-34a expression in cervical cancer-related cell lines including the CaSki, SiHa, Hela and C411 cell lines (Wang et al., 2009). The reduction of miR-34 expression is attributed to the expression of viral E6 protein, which destabilizes the tumor suppressor p53, a transcription factor that binds to the promoter of the miR-34a gene to activate expression (Wang et al., 2009). In primary B cells, miR-34a is strongly induced by Epstein–Barr virus infection via increased enrichment of NF-κB to the miR-34a gene promoter. EBV regulation of host gene expression including miRNA genes during latency is largely accomplished by the oncoproteins EBNA-2 and LMP-1 (Kang and Kieff, 2015). Elevated expression of LMP-1 is sufficient to trigger the expression of miR-34a through IKKβ-dependent activation of canonical NF-κB signaling pathways (Forte et al., 2012). In addition, the viral HBx protein encoded by hepatitis B virus strongly represses the expression of miR-34 in HepG2X and/or Hep3BX cells through an unknown mechanism (Ou et al., 2017). As mentioned above, mir-34 family involvement has been demonstrated with shrimp virus, flaviviruses, and a few others but not with all animal viruses. The non-responsive virus-host combinations are summzrized in the Table 1 .
Table 1.
The expression of miR-34/449 family following viral infection.
| Virus | miRNAs | Biological processes | References |
|---|---|---|---|
| HPV16 | miR-34a down-regulation | Promotes cell proliferation | Wang et al. (2009) |
| HPV18 | miR-34a down-regulation | Promotes cell proliferation | Wang et al. (2009) |
| EBV | miR-34a up-regulation | Promotes cell growth | Forte et al. (2012) |
| HBV | miR-34a down-regulation miR-449a down-regulation miR-34c down-regulation |
Promote cell proliferation and induce cell apoptosis Promote virus replication and cell cycle transition and proliferation Repress virus replication and cell proliferation and induce cell apoptosis |
Qu et al. (2017) Zhang et al. (2016) Wang et al. (2015) |
| HCV | miR-449a down-regulation | Promote inflammatory responses | Sarma et al. (2014) |
| DENV | miR-34c down-regulation | Repress virus replication | Smith et al. (2017) |
| WNV | miR-34c down-regulation | Repress virus replication | Smith et al. (2017) |
| JEV | miR-34c down-regulation | Repress inflammatory responses | Smith et al. (2017), Kumari et al. (2016) |
| ALV-J | miR-34b up-regulation | Promote cell proliferation and migration and virus replication | Li et al. (2017) |
| Influenza A virus | miR-34a down-regulation miR-34c up-regulation and miR-449a up-regulation miR-449b up-regulation |
Inhibit cell apoptosis Promote virus replication Promote virus replication |
Fan and Wang (2016) Bakre et al. (2013) Buggele et al. (2013) |
| SARS-CoV | Non-responsive | / | Mallick et al. (2009) |
| Rhinoviruses | Non-responsive | / | Gutierrez et al. (2016) |
| Human Metapneumovirus | Non-responsive | / | Deng et al. (2014) |
| Human Cytomegalovirus | Non-responsive | / |
Meshesha et al. (2012), Wang et al. (2008) |
5. Virus replication regulated by the miR-34/449 family in host cells
The expression of the miR-34/449 family is regulated in host cells after infection with some types of virus. The miR-34/449 family can regulate virus replication by modulating various cellular processes. For instance, miR-449a enhances HBV replication by regulating the expression of two transcription factors. Farnesoid X receptor α (FXRα) is a transcription factor that binds to two motifs within the HBV enhancer II and core promoter regions to enhance HBV transcription and replication (Ramiere et al., 2008). In HepG 2.2.15 cells, increased levels of FXRα induced by miR-449a mimic results in an elevated transcription of the HBV core promoter to around 1.8-fold (Zhang et al., 2016). The cyclic AMP-response element-binding protein 5 (CREB5) is a transcription factor that contains zinc-finger and bZIP DNA-binding domains. CREB5 specifically binds to the cAMP response element (CRE) as a homodimer or a heterodimer with c-Jun or CER-BP1, and acts as a CRE-dependent transactivator (Zu et al., 1993). CREB5 overexpression leads to decreased HBV replication and HBsAg and HBeAg production in Huh7 cells. The 3ʹUTR of CREB5 mRNA has two evolutionarily conserved binding sites of miR-449a. Therefore, increased miR-449a enhances HBV replication by repressing the expression of CREB5 (Zhang et al., 2016). Another member of the miR-34/449 family, miR-34b, represses the expression of the melanoma differentiation-associated gene 5 (MDA5), an activator for the interferon signaling pathway, to facilitate avian leucosis virus subgroup (ALV-J) replication in DF-1 cells (Li et al., 2017). During flavivirus infection, overexpression of miR-34a represses the Wnt signaling pathway via inhibiting the production of CTTNB1, LEF1, WNT1, WNT2 and WNT3. The repressed Wnt signaling pathway promote interferon signaling pathway, finally leading to inhibition of virus replication (Smith et al., 2017). miR-34c and miR-449a also increase influenza virus replication (Bakre et al., 2013). However, the miR-34/449 family also represses virus replication. miR-34c represses HBV DNA replication and viral antigen synthesis in HepG 2.2.15 cells (Wang et al., 2015). miR-449b represses influenza virus replication in A549 cells by targeting histone deacetylase 1 (HDAC1) (Buggele et al., 2013), a protein previously found to be an important repressive component of the interferonβ enhancer (Nusinzon and Horvath, 2006) (Table 1).
6. Cellular processes regulated by the miR-34/449 family after viral infection
The miRNA-mediated silencing machinery plays important roles in regulation of immune responses, cell proliferation and cell apoptosis. The miR-34/449 family is involved in regulation of immune responses after viral infections. miR-34/449 induced by viral infections triggers secretion of diverse inflammatory cytokines or chemokines by regulating different signaling pathways. miR-449a activates the secretion of YKL-40 in HCV-infected liver tissue, an known marker for inflammation in tissues including liver, smooth muscle and cancer cells (Sarma et al., 2012; Volck et al., 1998). miR-449a down-regulation by HCV infection in liver tissue also increases the expression of Notch 1, a key component of the Notch signaling pathway. Influence on the Notch signaling pathway results in nuclear stabilization of p65, which regulates YKL40 expression in concert with CEBPα in response to TNFα (Sarma et al., 2012). In addition, downregulation of miR-449a following HCV infection modulates inflammatory signaling pathways in patients. In HCV-infected patients, miR-449a enhances expression of C-C motif chemokine ligand 2 (CCL2), an inflammatory chemokine upregulated in patients with chronic liver diseases, by targeting IL-6R and JAK1, two key components of the IL-6-mediated signaling cascade (Sarma et al., 2014). Similarly, another member of the miR-34/449 family, miR-34c, silences multiple genes (Notch1, Dll1, JAG1 and Hes1) in Notch signaling pathways. In mouse microglial cells (BV2), overexpressed miR-34c attenuates secretion of inflammatory cytokines TNF and IL-6 induced by JEV infection by regulating the Notch signaling pathway (Kumari et al., 2016) (Table 2 ).
Table 2.
Previously reported target mRNAs of the miR-34/449 family.
| Targets | Virus Type | Reported miRNA regulators | References |
|---|---|---|---|
| Farnesoid X receptor α | HBV | miR-449a | Ramiere et al. (2008) |
| CREB5 | HBV | miR-449a | Zhang et al. (2016) |
| MDA5 | ALV-J | miR-34b | Li et al. (2017) |
| HDAC1 | Influenza virus | miR-449b | Buggele et al. (2013) |
| Notch1 | HCV | mir-449a | Sarma et al. (2012) |
| CCL2 | HCV | miR-449a | Sarma et al. (2014) |
| Notch1 | JEV | miR-34c | Kumari et al. (2016) |
| CTTNB1, LEF1, WNT1-3 | DENV | miR-34a | Smith et al. (2017) |
Following viral infection, the miR-34/449 family can regulate cell proliferation and cell apoptosis. miR-34a, served as an antitumorigenic miRNA, can inhibit virus-infected cell proliferation and induce cell apoptosis. For instance, in influenza A (H1N1) virus infected A549 cells, miR-34a induces cell apoptosis via targeting the Bax protein, a apoptosis regulator (Fan and Wang, 2016). miR-34c, down-regulated by HBV infection, can also induce cell apoptosis in HepG2.2.15 cells (Wang et al., 2015). Some members of the miR-34/449 family are able to regulate cell proliferation via targeting cell cycle related signaling pathways in virus infected cells. For example, in ALV-J infected cells, overexpression of miR-34b significantly increased the number of cells in S phase and reduced the numbers of cells in G2 phase (Li et al., 2017). Conversely, another member of this miRNA family, miR-449a, inhibits cell cycle transition and proliferation in HBV infected cells (Zhang et al., 2016).
7. Concluding remarks
The miR-34/449 family is widely distributed in vertebrate organisms and is important for regulation of virus-host interactions by modulating immune responses and virus replication. The p53 tumor suppressor protein is a transcription factor that regulates the expression of miRNAs. Binding of p53 to promoter regions is crucial for expression of the miR-34/449 family in different cell types. Interestingly, in influenza virus-infected A549 cells, p53 is upregulated (Turpin et al., 2005) but miR-34a is downregulated (Fan and Wang, 2016), indicating that miR-34a expression is independent of p53 in influenza virus-infected cells. In EBV-infected B cells, expression of the miR-34/449 family is tightly associated with NF-κB signaling pathways instead of the p53 signaling pathway (Forte et al., 2012). This evidence demonstrates that in some types of cells, regulation of miR-34/449 family expression following virus infection is independent of the p53 signaling pathway. Members of the conserved miR-34/449 family share the same seed sequences in humans. Therefore, a single protein-coding gene can be targeted by two or more members of the miR-34/449 family (Bao et al., 2012). This finding means that members of this miRNA family have similar functions by silencing the same target. In contrast, members of the miR-34/449 family can have functions that are opposed to the functions of other members of the miRNA family. For instance, served as an antitumorigenic miRNA, miR-34a represses cell proliferation and induces apoptosis in HPV, HBV and Influenza A virus infected cells. However, overexpression of miR-34a was not toxic in B lymphoma cell lines, including EF3D and HTC-116, and inhibition of mir-34a impairs the growth of EBV-transformed cells (Forte et al., 2012). The difference between two opposite characteristics on cell growth is possibly due to the significant difference in the abundance of mRNAs of miR-34a targets. Additionally, in A549 cells, HBV replication is repressed by miR-34c but increased by another member of the family, miR-449a. In closing, the literature reinforces the model that the conserved miR-34/449 family is important in viral infections. The coordinated regulation of viral infection by different members of the miR-34/449 family deepens our understanding of virus-host interactions and also provides new thoughts about diagnosis.
Disclosure of potential conflicts of interest
The author states he has no conflict of interest.
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (Grant No. 2017YFD0501104), the National Natural Science Foundation of China (Grant No. 31772780), the Key Technology R&D Program of Gansu Province of China (Grant No. 1604NKCA045-2), China Agriculture Research System (Grant No. CARS-35).
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia Y., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bakre A., Andersen L.E., Meliopoulos V., Coleman K., Yan X., Brooks P., Crabtree J., Tompkins S.M., Tripp R.A. Identification of host kinase genes required for influenza virus replication and the regulatory role of MicroRNAs. PLoS One. 2013;8(6):e66796. doi: 10.1371/journal.pone.0066796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J., Li D., Wang L., Wu J., Hu Y., Wang Z., Chen Y., Cao X., Jiang C., Yan W., Xu C. MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J. Biol. Chem. 2012;287(26):21686–21698. doi: 10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., van Tetering G., Verheul M., van de Belt J., van Laake L., Vos J., Verloop R., van de Wetering M., Guryev V., Takada S., van Zonneveld A.J., Mano H., Plasterk R., Cuppen E. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16(10):1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggele W.A., Krause K.E., Horvath C.M. Small RNA profiling of influenza A virus-infected cells identifies miR-449b as a regulator of histone deacetylase 1 and interferon beta. PLoS One. 2013;8(9):e76560. doi: 10.1371/journal.pone.0076560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C.J., Arking D.E., Beer M.A., Maitra A., Mendell J.T. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins J.M., He Y., Leary R.J., Pagliarini R., Diaz L.A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Raymond C.K., Roberts B.S., Juhl H., Kinzler K.W., Vogelstein B., Velculescu V.E. The colorectal microRNAome. Proc. Natl. Acad. Sci. U. S. A. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Ptashkin R.N., Wang Q., Liu G., Zhang G., Lee I., Lee Y.S., Bao X. Human metapneumovirus infection induces significant changes in small noncoding RNA expression in airway epithelial cells. Mol. Ther. Nucleic Acids. 2014;3:e163. doi: 10.1038/mtna.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Mourelatos Z., Yang M., Sharma A., Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9(2):180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan N., Wang J. MicroRNA 34a contributes to virus-mediated apoptosis through binding to its target gene Bax in influenza A virus infection. Biomed. Pharmacother. 2016;83:1464–1470. doi: 10.1016/j.biopha.2016.08.049. [DOI] [PubMed] [Google Scholar]
- Forte E., Salinas R.E., Chang C., Zhou T., Linnstaedt S.D., Gottwein E., Jacobs C., Jima D., Li Q.J., Dave S.S., Luftig M.A. The Epstein-Barr virus (EBV)-induced tumor suppressor microRNA MiR-34a is growth promoting in EBV-infected B cells. J. Virol. 2012;86(12):6889–6898. doi: 10.1128/JVI.07056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov E.A., Kongsuwan K., Assavalapsakul W., Horwood P.F., Mitter N., Mahony T.J. Repertoire of bovine miRNA and miRNA-like small regulatory RNAs expressed upon viral infection. PLoS One. 2009;4(7):e6349. doi: 10.1371/journal.pone.0006349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M.J., Gomez J.L., Perez G.F., Pancham K., Val S., Pillai D.K., Giri M., Ferrante S., Freishtat R., Rose M.C., Preciado D., Nino G. Airway secretory microRNAome changes during rhinovirus infection in early childhood. PLoS One. 2016;11(9):e0162244. doi: 10.1371/journal.pone.0162244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hertel J., Lindemeyer M., Missal K., Fried C., Tanzer A., Flamm C., Hofacker I.L., Stadler P.F., Students of Bioinformatics Computer Labs A. The expansion of the metazoan microRNA repertoire. BMC Genom. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray M.F., Sharp P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5(2):351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Kang M.S., Kieff E. Epstein-Barr virus latent genes. Exp. Mol. Med. 2015;47:e131. doi: 10.1038/emm.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari B., Jain P., Das S., Ghosal S., Hazra B., Trivedi A.C., Basu A., Chakrabarti J., Vrati S., Banerjee A. Dynamic changes in global microRNAome and transcriptome reveal complex miRNA-mRNA regulated host response to Japanese Encephalitis Virus in microglial cells. Sci. Rep. 2016;6:20263. doi: 10.1038/srep20263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C. Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30(4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Li J., Wang K., Chen X., Meng H., Song M., Wang Y., Xu X., Bai Y. Transcriptional activation of microRNA-34a by NF-kappa B in human esophageal cancer cells. BMC Mol. Biol. 2012;13:4. doi: 10.1186/1471-2199-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Luo Q., Xu H., Zheng M., Abdalla B.A., Feng M., Cai B., Zhang X., Nie Q., Zhang X. MiR-34b-5p suppresses melanoma differentiation-associated gene 5 (MDA5) signaling pathway to promote avian leukosis virus subgroup j (ALV-J)-infected cells proliferaction and ALV-J replication. Front. Cell. Infect. Microbiol. 2017;7:17. doi: 10.3389/fcimb.2017.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P. Vertebrate microRNA genes. Science. 2003;299(5612):1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lize M., Klimke A., Dobbelstein M. MicroRNA-449 in cell fate determination. Cell Cycle. 2011;10(17):2874–2882. doi: 10.4161/cc.10.17.17181. [DOI] [PubMed] [Google Scholar]
- Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Korner H., Knyazev P., Diebold J., Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7(16):2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- Mallick B., Ghosh Z., Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS One. 2009;4(11):e7837. doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A., Carbone A. Large scale chromosomal mapping of human microRNA structural clusters. Nucleic Acids Res. 2013;41(8):4392–4408. doi: 10.1093/nar/gkt112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshesha M.K., Veksler-Lublinsky I., Isakov O., Reichenstein I., Shomron N., Kedem K., Ziv-Ukelson M., Bentwich Z., Avni Y.S. The microRNA transcriptome of human cytomegalovirus (HCMV) Open Virol. J. 2012;6:38–48. doi: 10.2174/1874357901206010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinzon I., Horvath C.M. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol. Cell. Biol. 2006;26(8):3106–3113. doi: 10.1128/MCB.26.8.3106-3113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q., Wang G., Li B., Li W.F. Decreased miR-34a promotes growth by regulating MAP4K4 in hepatitis B virus related hepatocellular carcinoma. Int. J. Clin. Exp. Med. 2017;10(2):2523–2531. [Google Scholar]
- Ramiere C., Scholtes C., Diaz O., Icard V., Perrin-Cocon L., Trabaud M.A., Lotteau V., Andre P. Transactivation of the hepatitis B virus core promoter by the nuclear receptor FXRalpha. J. Virol. 2008;82(21):10832–10840. doi: 10.1128/JVI.00883-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma N.J., Tiriveedhi V., Crippin J.S., Chapman W.C., Mohanakumar T. Hepatitis C virus-induced changes in microRNA 107 (miRNA-107) and miRNA-449a modulate CCL2 by targeting the interleukin-6 receptor complex in hepatitis. J. Virol. 2014;88(7):3733–3743. doi: 10.1128/JVI.03060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma N.J., Tiriveedhi V., Subramanian V., Shenoy S., Crippin J.S., Chapman W.C., Mohanakumar T. Hepatitis C virus mediated changes in miRNA-449a modulates inflammatory biomarker YKL40 through components of the NOTCH signaling pathway. PLoS One. 2012;7(11):e50826. doi: 10.1371/journal.pone.0050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata R.H., Abdelmoneim S.S., Osman O.A., Hasanain A.F., Osama A., Abdelmoneim S.S., Toraih E.A. Deregulation of miR-34a and its chaperon Hsp70 in hepatitis C virus-induced liver cirrhosis and hepatocellular carcinoma patients. Asian Pac. J. Cancer Prev.: APJCP. 2017;18(9):2395–2401. doi: 10.22034/APJCP.2017.18.9.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens H., Jackstadt R., Hunten S., Kaller M., Menssen A., Gotz U., Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10(24):4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- Smith J.L., Jeng S., McWeeney S.K., Hirsch A.J. A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection. J. Virol. 2017;91(8) doi: 10.1128/JVI.02388-16. e02388-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin E., Luke K., Jones J., Tumpey T., Konan K., Schultz-Cherry S. Influenza virus infection increases p53 activity: role of p53 in cell death and viral replication. J. Virol. 2005;79(14):8802–8811. doi: 10.1128/JVI.79.14.8802-8811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volck B., Price P.A., Johansen J.S., Sorensen O., Benfield T.L., Nielsen H.J., Calafat J., Borregaard N. YKL-40, a mammalian member of the chitinase family, is a matrix protein of specific granules in human neutrophils. Proc. Assoc. Am. Phys. 1998;110(4):351–360. [PubMed] [Google Scholar]
- Wang F.Z., Weber F., Croce C., Liu C.G., Liao X., Pellett P.E. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J. Virol. 2008;82(18):9065–9074. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H.K., McCoy J.P., Banerjee N.S., Rader J.S., Broker T.R., Meyers C., Chow L.T., Zheng Z.M. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15(4):637–647. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang C.M., Jiang Z.Z., Yu X.J., Fan C.G., Xu F.F., Zhang Q., Li L.I., Li R.F., Sun W.S., Zhang Z.H., Liu Y.G. MicroRNA-34c targets TGFB-induced factor homeobox 2, represses cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Oncol. Lett. 2015;10(5):3095–3102. doi: 10.3892/ol.2015.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S.K., Parkin R.K., Mitchell P.S., Fritz B.R., O’Briant K., Godwin A.K., Urban N., Drescher C.W., Knudsen B.S., Tewari M. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One. 2009;4(4):e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Liu H., Xie Z., Deng W., Wu C., Qin B., Hou J., Lu M. Epigenetically regulated miR-449a enhances hepatitis B virus replication by targeting cAMP-responsive element binding protein 5 and modulating hepatocytes phenotype. Sci. Rep. 2016;6:25389. doi: 10.1038/srep25389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y.L., Maekawa T., Nomura N., Nakata T., Ishii S. Regulation of trans-activating capacity of CRE-BPa by phorbol ester tumor promoter TPA. Oncogene. 1993;8(10):2749–2758. [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]