Graphical abstract
Keywords: Evolution, Virus, Hepatitis, Zoonotic, Homologues
Abstract
Hepatitis viruses are major threats to human health. During the last decade, highly diverse viruses related to human hepatitis viruses were found in animals other than primates. Herein, we describe both surprising conservation and striking differences of the unique biological properties and infection patterns of human hepatitis viruses and their animal homologues, including transmission routes, liver tropism, oncogenesis, chronicity, pathogenesis and envelopment. We discuss the potential for translation of newly discovered hepatitis viruses into preclinical animal models for drug testing, studies on pathogenesis and vaccine development. Finally, we re-evaluate the evolutionary origins of human hepatitis viruses and discuss the past and present zoonotic potential of their animal homologues.
Introduction
Approximately 1.3 million people die annually from viral hepatitis worldwide.1 These deaths are predominantly associated with cirrhosis and hepatocellular carcinoma (HCC) resulting from chronic infections with hepatitis B virus (HBV; 887,000 deaths) and hepatitis C virus (HCV; 399,000 deaths),2 as well as hepatitis and liver failure resulting from acute infections with hepatitis A virus (HAV; 11,000 deaths) and hepatitis E virus (HEV; 44,000 deaths).1 Worldwide, approximately 5% of people infected with HBV are simultaneously infected with hepatitis delta virus (HDV).[1], [3]
Key point.
Human hepatitis viruses have ancient zoonotic origins.
The recent discoveries of novel hepatitis viruses from animals allow revisiting the enigmatic evolutionary origins of human hepatitis viruses. In chapter 1 of this review, we discuss the diverse animal homologues of all human hepatitis viruses that were discovered over the last decades. In chapter 2, we analyse hepatitis virus evolution based on the unique genomic and morphologic properties of human and non-human hepatitis viruses. In chapter 3, we discuss the level of evolutionary conservation of the characteristic infection patterns of human hepatitis viruses and review the ability to translate the recent virus discoveries into tractable animal models. Finally, in chapter 4, we discuss the evolutionary origins of human hepatitis viruses in the context of a plethora of newly discovered animal homologues. In this chapter, we also evaluate the potential of animal homologues for past and present cross-species transmission and compare macro-evolutionary patterns of the different hepatitis virus families.
Chapter 1: Milestones towards the understanding of the evolutionary origins of hepatitis viruses
In the following section, we outline the path towards the discovery of human hepatitis viruses and provide detail on the huge expansion in the number of animal homologues discovered during the last decade.
The discovery of human hepatitis viruses
The path towards the discovery of human hepatitis viruses started with the differentiation between 2 forms of transmissible jaundice. Infectious hepatitis linked to epidemic outbreaks of faecal-orally transmitted jaundice (termed hepatitis A) was differentiated from a parenterally transmitted jaundice with a relatively longer incubation period (termed hepatitis B).[4], [5] Following the discoveries of the causative HAV and HBV in 1973 and 1970,[6], [7] the HBV-associated HDV was discovered in 1980.8 In parallel, it was noted that most cases of post-transfusion hepatitis were neither linked to infection with HAV, nor HBV.9 However, it was not until 1989 that the causative HCV was finally described.10 HEV was discovered in 1983 as the cause of a predominantly water-borne acute non-A, non-B hepatitis.11 The discoveries of human hepatitis viruses were followed by milestones of major clinical relevance, including tools for reliable diagnosis (e.g., described in[12], [13]), the development of direct-acting antiviral treatments against HCV and preventive vaccinations against HAV and HBV14 (Fig. 1 ). Notably, scientific progress was not evenly distributed among human hepatitis viruses. The clinical relevance of HBV and HCV likely contributed to the enormous achievements attained continuously for these 2 viruses (Fig. 1).
Fig. 1.
Selected milestones in hepatitis virus research. References include for HAV,[6], [19], [25], [26], [28], [73], [107], [219], [220], [221] for HBV,[7], [30], [67], [15], [16], [17], [18], [32], [33], [34], [35], [222], [223], [224], [225] for HCV,[9], [10], [21], [37], [38], [71], [214], [42], [43], [44], [45], [226], [227], [228], [229], [230] for HDV[8], [47], [67], [91], [146], [231], [232], [233] and for HEV.[11], [48], [50], [51], [54], [56], [59], [74], [133], [234], [235], [236], [237], [238] Due to space constraints, only selected milestones are shown. Animal pictograms represent host orders in which distinct hepatitis viruses were detected. DAA, direct-acting antiviral; HAV, hepatitis A virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitis delta virus; HEV, hepatitis E virus; miR-122, micro RNA-122; NRTI, nucleoside reverse transcriptase inhibitor; NTCP, sodium taurocholate co-transporting polypeptide; RBV, ribavirin.
A new era of virus discovery
The first discoveries of non-human hepatitis viruses were ground breaking, yet sporadic. In 1978, a genetically distant relative of HBV, the woodchuck hepatitis virus (WHV), was identified in American woodchucks, a marmot-like rodent species.15 The discovery of WHV was followed by the discovery of duck hepatitis B virus (DHBV), HBV variants in apes, and a divergent HBV species termed woolly monkey hepatitis B virus (WMHBV).[16], [17], [18] For hepatitis viruses other than HBV, knowledge on potential non-human hosts remained scarce. In the 1990s, a distinct HAV type was detected in macaques19 and HEV was recovered from swine.20 In 1995, a non-human primate (NHP) virus distantly related to HCV, termed GB virus-B (GBV-B) was isolated from a laboratory tamarin.21 The tamarin had been inoculated with the serum of a patient with hepatitis, but since GBV-B was never detected in humans, it was most likely a tamarin virus.22
Over the last decades there has been an explosive expansion of the recognized viral diversity in animals, driven by novel sequencing techniques and an unprecedented focus on zoonotic pathogens which followed the identification of highly pathogenic viruses such as Ebola virus and SARS-coronavirus in bats.[23], [24] A plethora of HAV-related viruses were recovered from various mammalian species during 2015–2018, including predominantly bats and rodents, but also tree shrews, seals and marsupials.[25], [26], [27], [28], [29] HBV-related viruses were detected in bats during 2013–2015[30], [31] and in a domestic cat in 2018.32 A distinct HBV species termed capuchin monkey hepatitis B virus (CMHBV) was described in 2018.33 Endogenous and exogenous viruses distantly related to HBV were detected in reptiles, fish and amphibians.[34], [35], [36], [37] HCV-related viruses were detected in horses in 2012, and evidence for sporadic spill-over infections of the horse-associated viruses into dogs and donkeys was found.[38], [39], [40], [41] Soon afterwards, highly diverse HCV-related viruses were detected in bats and rodents,[42], [43] in cattle,[44], [45] and in black-and-white colobus monkeys.46 Hepatitis D-like agents were recently detected in ducks and snakes.[47], [48] Zoonotic HEV genotypes were discovered in wild boars, camelids, rabbits and rats.[49], [50], [51], [52], [253] In addition, divergent HEV-related viruses were described in bats, ferrets, rodents, birds, and fish.[53], [54], [55], [56], [57], [58], [59] In sum, homologues of all human hepatitis viruses exist in diverse animals. Apart from the detection of zoonotic HEV strains, none of the recent studies into viral diversity in animals revealed any direct ancestor of human hepatitis viruses, as outlined below.
Key point.
Diverse animals host hepatitis viruses, but ongoing zoonotic transmission is documented only for hepatitis E virus.
Chapter 2: Conservation and de novo emergence of unique properties of hepatitis viruses
Human hepatitis viruses are assigned to diverse virus families and genera (Table 1 ). Namely, they belong to the families Picornaviridae, genus Hepatovirus (HAV), Hepadnaviridae, genus Orthohepadnavirus (HBV), Flaviviridae, genus Hepacivirus (HCV), and Hepeviridae, genus Orthohepevirus (HEV). The genus Deltavirus (HDV) is unassigned to any virus family. In general, human and non-human hepatitis virus homologues resemble each other in major genomic properties such as structure of the genomic nucleic acid, open reading frame (ORF) composition, genome length and presence and type of noncoding regions (Table 1, Fig. 2 ). Nonetheless, there are striking differences among viruses from the same family or genus. Hypothetically, de novo emergence of genomic features and recombination events during evolution may have contributed to the rise of viruses eventually pathogenic to humans. A prototypic example for micro-evolutionary events within animal reservoirs enabling efficient human infection is the recombination event leading to the functional tetherin antagonist in chimpanzee-associated ancestors of the HIV-1 group M.60
Table 1.
Properties of human hepatitis viruses.
| HAV | HBV | HCV | HDV | HEV | |
|---|---|---|---|---|---|
| Virus family, genus | Picornaviridae, Hepatovirus |
Hepadnaviridae, Orthohepadnavirus |
Flaviviridae, Hepacivirus |
Unassigned, Deltavirus | Hepeviridae, Orthohepevirus |
| Genome type | Positive-sense linear ssRNA | Circular, partially dsDNA (full length negative-sense, partial positive-sense), replication via reverse transcription | Positive-sense linear ssRNA | Viroid-like, negative-sense circular ssRNA | Positive-sense linear ssRNA |
| Approx. genome length (nt) | 7,500 | 3,200 | 9,600 | 1,700 | 7,200 |
| Virion diameter (nm) | 27–32 | 42 | 55–65 | 36–43 | 30–34 |
| Envelope | No/quasi-enveloped | Yes | Yes | Yes | No/quasi-enveloped |
| Course of infection | Acute61 | Acute/chronic (children 30–90%; adults <5%)62 | Acute/chronic (80–85%)63 |
Acute/chronic (>80% if superinfection)64 |
Acute/chronic (<1%)65 |
| Predominant transmission routes | Mainly faecal-oral, parenteral | Vertical, parenteral, sexual |
Parenteral | Parenteral, sexual | Faecal-oral, food-borne, parenteral |
| Cellular receptor | Unknown66 | NTCP, heparan sulfate proteoglycans[67], [68] | CD81, SR-B1, LDL receptor, claudin-1, occludine[69], [70], [71] | NTCP, heparan sulfate proteoglycans[67], [72] | Unknown |
dsDNA, double-stranded DNA; nt, nucleotide; NTCP, sodium taurocholate co-transporting polypeptide; ssRNA, single-stranded RNA.
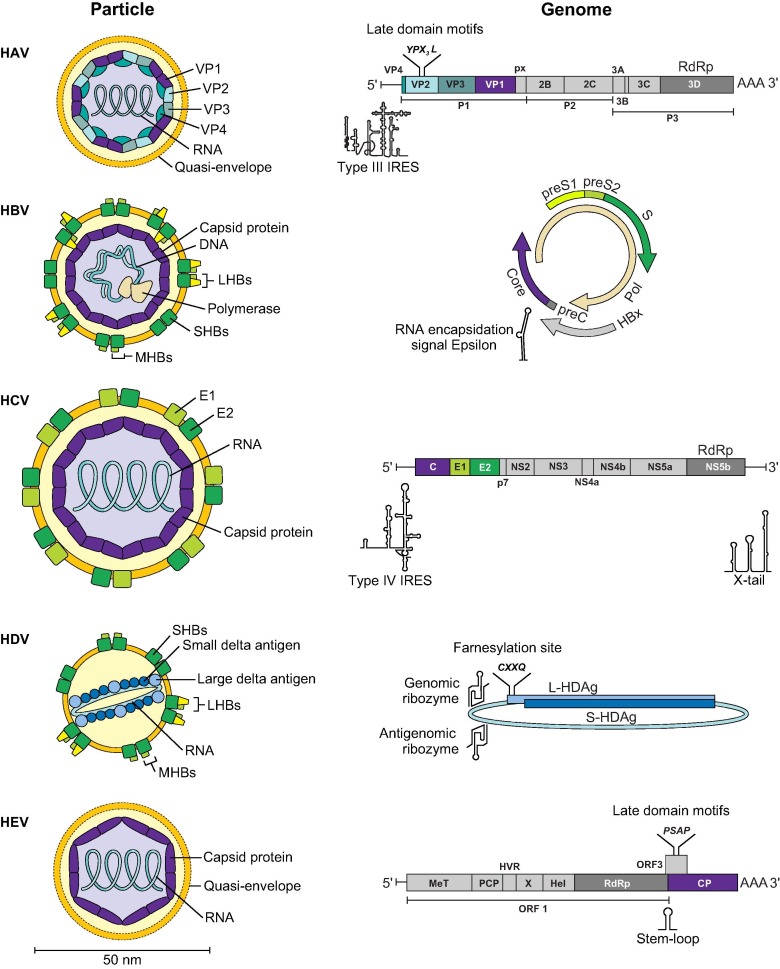
Fig. 2.
Virus particles and genome characteristics of human hepatitis viruses. C, core protein; CP, capsid protein; E, envelope protein; HAV, hepatitis A virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitis delta virus; HEV, hepatitis E virus; Hel, Helicase; HVR, hypervariable region; IRES, Internal ribosomal entry site; LHBs, large surface protein; L-HDAg, large hepatitis delta antigen; MHBs, medium surface protein; MeT, methyltransferase; NS, non-structural protein; ORF, open reading frame; PCP, papain-like cysteine protease; Pol, polymerase; RdRp, RNA-dependent RNA polymerase; S, surface protein; SHBs, small surface protein; S-HDAg, small hepatitis delta antigen; VP, viral protein; X, X-domain/ADP-ribose-binding module. Important secondary structures are depicted below genomes.
Envelopment might not be conserved among animal hepatitis viruses
Recent studies revealed that HAV and HEV, thought previously to be non-enveloped viruses, exist in an enveloped form in blood, referred to as quasi-envelopment (Fig. 2).[73], [74] In contrast to enveloped viruses, there is no hint of virus-encoded proteins in the quasi-envelope of HAV and HEV, which raises questions on how quasi-enveloped viruses enter susceptible cells (summarised in75). Interestingly, some non-primate HAV- and HEV-related viruses (henceforth, hepatoviruses and hepeviruses) lack critical genome structures involved in quasi-envelopment. This includes the apparent absence of a C-terminal VP1 capsid protein extension termed pX in some HAV-related bat viruses25 and the lack of short amino acid motifs termed late domain motifs in several divergent hepeviruses from diverse mammals and birds.29 In contrast, HBV-related viruses (henceforth, hepadnaviruses) are enveloped viruses, yet fish were recently found to host a non-enveloped Hepadnaviridae sister family, termed Nackednaviridae.36 The discovery of nackednaviruses suggested that envelopment emerged de novo in hepadnaviruses capable of infecting mammals and ultimately humans.36 A hallmark of HCV infection is the formation of lipoviral particles, which incorporate both host-derived lipoproteins and virus-derived glycoproteins.76 Interestingly, equine hepacivirus capsid proteins associate with intracellular lipid components, suggesting similarities between the replication of HCV and non-human HCV-related viruses (henceforth, hepaciviruses).77 However, whether the formation of lipoviral particles is evolutionarily conserved among non-human hepaciviruses remains unknown.
On the one hand, it is thus possible that envelopment and quasi-envelopment are not conserved among animal hepatitis viruses. On the other hand, ancestral animal hepatitis viruses may exploit unknown strategies for envelopment that differ from those found in human viruses.29
Key point.
The de novo emergence of virus properties such as envelopment and recombination events may have contributed to the rise of viruses eventually pathogenic for humans
Recombination events occurred during hepatitis virus evolution
Recombination events among viruses can occur when 1 cell is co-infected with 2 different viruses that interact during replication. For all human hepatitis viruses, recombination has been exhaustively described.[78], [79], [80], [81], [82], [83] The plethora of recently discovered viruses has enabled revisiting the occurrence of recombination events during the genealogy of human hepatitis viruses and their animal homologues.
For hepatoviruses there is evidence for recombination in the coding sequence of very distantly related viruses associated with different host orders, which hints at a broad host range.29 In addition, variations in the 5′-genome ends of animal hepatoviruses harbouring the internal ribosome entry site (IRES) hint at ancient recombination events involving different viral families.25 For hepadnaviruses, the mosaic genome structure of extinct and extant HBV genotypes suggests that recombination has been a major micro-evolutionary feature of HBV evolution.[80], [190] Recombination among viruses associated with different host species was also described among primate and bird hepadnaviruses.84 Whether inter-host recombination events also occurred during the genealogy of the newly identified bat and cat hepadnaviruses demands further investigation. For non-human hepaciviruses, recombination events among diverse hosts have not been unambiguously proven yet.85 However, as with hepatoviruses, hints for ancient recombination events are found in the 5′-genome ends of non-primate hepaciviruses, likely involving viruses belonging to different genera.42 For hepeviruses, there is evidence for recombination involving different host orders, similar to hepatoviruses.29 Importantly, the camelid-associated HEV genotypes 7 and 8 show evidence of recombination at the boundary of ORFs encoding non-structural and structural proteins.29 Whether recombination events in these HEV strains contribute to their zoonotic potential is an intriguing question. Notably, recombination events likely enabled the rise of the family Hepeviridae per se, as the different hepevirus ORFs are derived from diverse alphavirus- and astrovirus-like ancestors.86
To summarise, recombination was likely a frequent event during the genealogy of hepatitis viruses. Whether transmission to humans is a consequence of recombination events in animal reservoirs requires further elucidation.
Distinct features of HDV
HDV is not a complete virus but a subviral agent with a very short, circular RNA genome which can replicate autonomously within a cell but requires the surface proteins of HBV for cell release and uptake (Fig. 2).87 Of all human hepatitis viruses, the least is known about the origin of HDV. HDV has been hypothesised to have originated from plant viroids, via recombination with cellular mRNA, or from RNA intermediates of HBV.[88], [89], [90] However, these theories are conflicting and not supported by experimental evidence. Interestingly, HDV-like agents were recently detected in a pool of oropharyngeal and cloacal samples of ducks and in various tissues of snakes.[47], [48] The apparent lack of detectable hepadnaviruses in these preliminary investigations seems consistent with the lack of a predicted large delta antigen containing the C-terminal farnesylation signal required for interaction with the HBV envelope (Fig. 2).[91], [92] On the one hand, this may indicate that the unique association of HDV with HBV is not evolutionarily conserved. On the other hand, divergent helper viruses and divergent mechanisms of envelopment cannot be excluded based on present knowledge. Although snake and duck HDV-like agents are genetically clearly distinct from human HDV, they show typical HDV genome properties, including the ORF encoding the small delta antigen, genomic and antigenomic ribozymes mediating autocatalytic cleavage and an extremely high degree of genomic self-complementarity of the circular genome. The detection of these divergent viruses in ancient vertebrates strongly suggests that other HDV-like viruses exist, which might include ancestors of human HDV.
Chapter 3: Evolutionary conservation of infection patterns
The comparison of hepatitis virus infection patterns among human and non-human hosts might reveal unique viral properties that eventually enabled human infection. For example, chronic courses of viral infections in animal reservoirs were found to be a strong predictor of human-to-human transmissibility after zoonotic introduction into humans.93 Here, we compare transmission routes, organ tropism, disease outcomes, receptor usage and immune evasion strategies among human and non-human hepatitis viruses.
Conservation of transmission routes among hepatitis viruses
Human hepatitis viruses differ in their transmission routes. HAV and HEV are mainly transmitted through the enteric route. Similarly, faecal shedding has been observed invariably in non-human hosts of hepatoviruses and hepeviruses (Table 2 ). In contrast, HBV and HDV are transmitted via blood and other body fluids, including semen and vaginal secretions.[87], [94] In addition, there is a high risk of perinatal transmission of HBV.95 Like in humans, both perinatal and horizontal HBV transmission has been described for gibbons.96 For non-primate hosts, vertical hepadnavirus transmission is known to be effective in rodents and birds.[97], [98] Based on a higher prevalence in female tent-making bats compared to males, predominantly sexual virus transmission has been suggested for the Tent-making bat HBV (TBHBV)254. Transmission routes for other bat or cat hepadnaviruses remain unknown.[30], [32] HCV is a blood-borne virus. However, perinatal transmission rates are much lower for HCV than HBV and sexual transmission is rare.[63], [99] HCV is mainly transmitted via blood transfusions, sharing of equipment in injecting drug use and reuse of injection needles in healthcare,[1], [63] which raises questions on the transmission mode of HCV in scattered prehistoric human populations. Here, fights, use of weapons or tools as well as cultural or religious practices such as tattooing, circumcision, acupuncture or scarification might have enabled virus transmission.100 Transmission routes among non-human hepaciviruses are unclear. In small mammals, transmission of hepaciviruses through biting and scratching associated with mating and territorial behaviour is conceivable.101 In horses, parenteral transmission of equine hepacivirus has been suggested,[102], [103] possibly including sexual transmission and veterinary practices.[41], [102] Transmission via human-aided routes would suggest a relatively recent spread of this virus among equines worldwide and is consistent with the low virus diversity found in equines.[38], [41], [102], [104] Similar transmission routes are conceivable for cattle hepaciviruses.[44], [45]
Table 2.
Evolutionary conservation of infection patterns.
|
Hallmarks of human infection |
Hallmarks of infection in non-human animals |
||
|---|---|---|---|
| Naturally or experimentally infected with autochthonous virus | Experimentally infected with human virus | ||
| HAV | Long faecal shedding; Quasi-envelopment; Never chronic but may cause protracted infections of up to one year; Faecal-oral transmission |
Bat, rodent, hedgehog, shrew: Acute infection; Faecal shedding; Hepatotropism25 Woodchuck: Mild fever; Faecal shedding; Hepatotropism105 |
Chimpanzee: Faecal shedding; Quasi-envelopment; Acute hepatitis[73], [106], [107] New and Old World monkeys: Faecal shedding; Hepatotropism[108], [109], [110] Guinea pig: Faecal shedding; Hepatotropism; Subclinical disease111 |
| HBV | Oncogenic (HCC); Age-dependent chronicity rate; Vertical, parenteral, sexual transmission; Acute, fulminant courses in adults |
New World primates: Acute and chronic; Hepatitis; Vertical transmission[17], [33] Old World primates: Acute and chronic; Horizontal and vertical transmission; Hepatitis[96], [112], [113] Bats: Apparently acute and chronic; Sexual transmission[30], [254] Woodchuck: Acute and chronic; Hepatitis; Oncogenic; Hepatotropism114 (summarised in115) Squirrels: Hepatitis116; Oncogenic117 Cat: Viremia32 Duck: Acute and chronic118; Hepatitis; Vertical transmission; Not oncogenic; No exclusive hepatotropism (summarised in[115], [119]) |
Chimpanzee: Acute and chronic; Hepatitis; Oncogenic (summarised in[120], [121]) Tupaia: Acute and chronic; Hepatitis122; Low viremia (summarised in115) |
| HCV | Oncogenic (HCC); High chronicity rate; Parenteral transmission |
Cattle: Apparently acute and chronic; Hepatotropism[44], [45] Horse: Apparently acute and chronic; Hepatotropism102 Rat: Hepatotropism123 Bank vole: Hepatotropism42 Marmoset: Acute and chronic; Hepatitis126 Tamarin: Acute hepatitis; High viral titres127 |
Chimpanzee: Acute and chronic; Hepatitis; Oncogenic[124], [125](summarised in121) Tupaia: Mild hepatitis; Acute and chronic; Oncogenic128 |
| HDV | HBV-dependent infection; Co-infection with HBV: Mild to severe acute disease; Superinfection with HBV: Mostly chronic, worsens disease outcome compared to HBV mono-infection |
Snake: No hepatotropism; Apparently independent of HBV48 |
Chimpanzee: Hepatotropism; HBV co-infection and superinfection; Co-infection: Mild disease; Superinfection: Acute severe disease, >50% subclinical chronic HDV infection[129], [130] (summarised in131); Woodchuck: Uses WHV envelope; Hepatotropism; Acute and chronic (summarised in131) |
| HEV | Low chronicity rate; Quasi-envelopment; Mainly faecal-oral transmission; Zoonotic |
Rabbit: Faecal shedding; Acute and chronic hepatitis; Viremia132 Rat: Faecal shedding; Apparent hepatotropism; Mild hepatitis[133], [134] Ferret: Faecal shedding; Viremia; Acute and chronic hepatitis135 Bat: Faecal shedding; Viremia54 Swine: Faecal shedding; Viremia; Acute and chronic[136], [137] Moose: Faecal shedding; Mild hepatitis138 Chicken: Hepatitis-splenomegaly syndrome59 Camel: Faecal shedding; Viremia[51], [52] |
Chimpanzee: Severe acute hepatitis139 Macaque: Faecal shedding; Viremia; Hepatitis[140], [141] Swine: Faecal shedding; Mild hepatitis142 |
HAV, hepatitis A virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitits delta virus; HEV, hepatitis E virus; HCC, hepatocellular carcinoma; WHV, woodchuck hepatitis virus.
In sum, virus transmission seems to be evolutionary conserved among enteric hepatitis viruses and their homologues, while transmission routes of non-human hepadnaviruses and hepaciviruses are poorly understood.
Determinants of hepatotropism are poorly understood
For HAV and HEV, little is known about the viral and host factors that promote liver tropism in humans. In bat hepatoviruses, but in none of the other non-primate hepatoviruses, almost equal amounts of virus were detectable in the liver and spleen.25 However, whether this is due to sequestered macrophages containing hepatovirus genetic material, as observed during experimental infections of mice with HAV,143 or due to extrahepatic viral replication remains to be determined.
For HBV and HDV, after low-specific binding to heparin-sulfate proteoglycans (summarised in144), hepatocyte entry is mediated via the sodium taurocholate co-transporting polypeptide (NTCP for the human receptor, Ntcp for the homologous receptor in animals).67 These liver-specific receptors likely govern the hepatotropism of HBV and HDV.67 Interestingly, the avian hepadnavirus DHBV does not show a strict hepatotropism (summarised in119). This is potentially explained by the usage of a receptor that differs from that used by HBV. Notably, even among mammalian hepadnaviruses, Ntcp usage might not be conserved, as illustrated by the apparent lack of hNTCP usage of some bat hepadnaviruses and WHV.[30], [145] In the snake delta-like agent, the apparent lack of hepatotropism may be associated with the absence of a detectable hepadnavirus (Table 2).48
For HCV, an important, yet not exclusive factor determining hepatotropism in humans is its interaction with the liver-specific micro RNA-122 (miR-122).146 All known non-human hepaciviruses contain at least one miR-122 binding site in their 5′-genome ends, which may contribute to their apparently conserved hepatotropism.[147], [148]
Liver tropism is thus a widely, yet not perfectly conserved attribute of the animal homologues of human hepatitis viruses. Because the factors determining hepatotropism of human hepatitis viruses are not entirely understood, systems allowing experimental infections with the newly discovered animal viruses will hopefully provide urgently needed insights into the determinants of viral hepatotropism.
Chronic courses of infection occur in diverse hepatitis virus hosts
Prehistoric human populations were small and scattered, raising the question of how hepatitis viruses survived in these populations. This is illustrated by the disappearance of HAV in isolated populations due to the livelong immunity HAV infections engender (summarised in25). In contrast to prehistoric humans, other hepatovirus hosts such as small mammals were abundant and widespread, which presumably aided viral survival in these populations.25
In contrast to HAV, both HBV and HCV establish chronic infections in humans, which presumably favoured virus maintenance in prehistoric human populations and among NHPs. Chronic hepatitis virus infections are commonly defined as infections that persist for more than 6 months for HBV and HCV, and 3 months for HEV.[149], [150], [151] The chronicity rate varies drastically among human hepatitis viruses. Chronic HEV infections are very rare (<1% of infections) and almost exclusively occur in immunocompromised patients.152 In contrast, chronic HCV infections are very frequent at up to 80–85% (Table 1).63 For HBV, the chronicity rate varies depending on the age at which the infection occurs, with rates of up to 90% for infected neonates, 30% for children aged 1–5 years and less than 5% for older children and adults (summarised in62).
Interestingly, chronic courses of infection are not unique for human HBV and HCV. Similar to human HBV infection, the chronicity rate of the rodent-associated WHV is age-dependent. While animals infected as newborns generally develop chronic infections, WHV usually causes acute self-limiting infections in animals infected at older ages (summarised in115). Chronic courses of hepadnavirus infections also occur in ducks, squirrels, and NHPs (Table 2).[30], [96], [112], [113], [117], [118], [153] Whether chronic hepadnavirus infections occur in bats is unclear.30
Among non-human hepaciviruses, studies reporting chronic infections are scarce. Chronic hepacivirus infections with prolonged viremia for more than 6 months were sporadically observed in experimentally infected horses and naturally infected cattle.[44], [102], [154] The chronicity rates in horses and cattle thus seem to be much lower than in human HCV infections.[44], [102], [103] For other non-human hepaciviruses, whether chronic infections occur remains unknown. Similarly, chronic courses of hepevirus infections in non-human hosts are largely unknown. Nonetheless, an apparent chronic HEV infection in wild boars and prolonged viral shedding in experimentally infected immunocompromised domestic swine, rabbits, and ferrets suggest that chronicity is not limited to human HEV infections.[132], [135], [137], [155]
To conclude, many questions regarding chronic infections in animal hepatitis viruses are still unanswered. Translational animal models to assess therapeutics for chronic hepatitis B and E, and investigating viral pathogenesis will require some correlate of chronic infection, which may be conceivable given the evolutionary conservation of chronicity among several animal hepadnaviruses and hepaciviruses.
Key point.
Chronicity may have aided hepatitis B and hepatitis C virus survival in scattered prehistoric human populations.
Mechanisms leading to HCC differ among hepatitis viruses
Chronic hepatitis virus infections can result in severe disease outcomes, including cirrhosis and HCC.156 In humans, chronic HBV infections result in HCC in 15–40% of cases157 and chronic HCV infections in approximately 2.5% of cases.158
Interestingly, woodchucks infected with WHV and ground squirrels infected with the genetically related ground squirrel hepatitis virus are at a high risk of developing HCC when infected at birth (summarised in159) (Table 2). While oncogenesis in woodchucks is mainly caused by N-myc gene activation via targeted insertion of hepadnaviral DNA into host DNA, oncogenesis in ground squirrels is associated with an activation of c-myc genes.160 For human HBV, the mechanisms of oncogenesis remain poorly understood. HBV DNA fragments can integrate into the hepatocyte genome during viral replication.161 However, in contrast to rodent hepadnaviruses, no specific integration site associated with oncogenesis has been identified. Accumulation of integration events into genes that may enhance oncogenesis, such as those encoding the telomerase reverse transcriptase (TERT), a histone H3 lysine 4 methyltransferase (MLL4), and cyclin E1 (CCNE1) have been described,[162], [163] but their contribution to oncogenesis is unclear. Furthermore, integration sites in tumour cells seem to be enhanced in sites critical for chromosome stability, such as in proximity to telomeres.163 In addition, the viral HBx protein promotes oncogenesis through complex interactions with the host cell, such as altering the expression of host oncogenes and tumour suppressors, stimulating cell-cycle entry by activating cyclins and cyclin-dependent kinase pathways and blocking apoptosis (summarised in164). In humans, HBx also interacts with the structural maintenance of chromosomes (Smc) complex Smc5/6 and stabilises the extrachromosomal cccDNA (covalently closed circular DNA) produced during HBV replication.165 Interestingly, HBx presumably emerged de novo in mammals during hepadnavirus evolution.37 Although the role of the HBx protein in non-human hepadnavirus infections is unknown, conservation of the HBx gene in mammalian orthohepadnaviruses suggests that HBx-mediated oncogenesis may be conceivable in diverse mammals. In concordance, ducks infected with avian hepadnaviruses lacking an HBx do not develop HCC (summarised in115).
In contrast to HBV, integration of viral genetic material into the host genome does not occur during HCV infections. Here, the core protein and non-structural proteins NS3 and NS5A promote HCC development by altering the expression of host genes involved in diverse oncogenic pathways, including proteins involved in cell-cycle control (such as p53, p21, cyclins) and apoptosis (summarised in164). Strikingly, HCC development has not been observed in any non-human hepacivirus host yet. Hypothetically, oncogenesis might either be unique to human HCV infections, or oncogenesis in animals has not been observed due to the relatively short life spans of the investigated animals, e.g. mice and rats infected with rat hepaciviruses.166 In contrast to these rodents, some bat species have a comparably long life span of up to 30 years.167 However, whether bats can get chronically infected with hepaciviruses and if this infection leads to oncogenesis is unknown. In addition, it has been hypothesised that bats may have a lower risk of developing cancer than other mammals, potentially because of their unique physiological and immunological properties.168 In horses and cattle, chronic courses of hepacivirus infections have been described and it would be intriguing to study whether these large and relatively long-lived animals develop HCC. However, costly long-term infection studies and longitudinal epidemiological investigations will be needed to address this question.
If and how animals infected with hepatitis viruses develop HCC is not well understood. HCC can be induced in mice, but this requires genetic engineering of the mouse genome, the use of chemotoxic agents, injection of tumour cells or xenograft approaches (summarised in169). Both HBV and HCV can cause chronic infections and HCC in chimpanzees, but using chimpanzees as animal models is strongly restricted for ethical reasons. An essential step towards an animal model for oncogenic hepatitis is the identification of a tractable, long-living host with oncogenic mechanisms similar to human HBV and HCV infections.
Receptor usage differs across hepatitis virus homologues
A crucial step during viral infection and a major factor limiting cross-species transmission is the interaction of viral proteins with host cell membrane structures enabling viral attachment and entry into cells. Strikingly, the cellular receptor molecules of HAV and HEV are still unknown. Recent data revealed that contrary to textbook knowledge, TIM1 is not essential for HAV entry into hepatocytes or epithelial cells.[66], [170]
HBV host specificity seems to be determined by a few essential amino acids of the cellular receptor NTCP/Ntcp. Interestingly, human HBV can use the Ntcp of great apes and New World monkeys, but 1 specific amino acid exchange in the Ntcp is needed for a cross-species transmission of human HBV to Old World monkeys.[33], [171] Similarly, 1 amino acid exchange in the woodchuck Ntcp enables an efficient infection with HBV.172 However, additional unidentified host-specific co-factors might be essential for viral entry and infection. For example, while HDV infection of mouse hepatocytes can be enabled by alteration of just 3 amino acid residues in the murine Ntcp, HBV infection is not possible in these animals.173 In addition, HBV infection can be enabled in pig and macaque hepatocytes expressing the human NTCP, but not in similarly engineered hepatocytes of mouse, rat, and dog origin.174 Strikingly, HBV can readily infect primary hepatocytes of the Asian tree shrew,175 which implies that both the HBV cellular receptor and potential co-factors are conserved in this animal.
For HCV, a complex interaction of various receptor molecules has been described[69], [70], [71] (Table 1). Like HBV, the host range of HCV seems to be limited by its receptors. Interestingly, rodents can be infected with HCV following modifications of either the host or the virus. On the host side, human liver-chimeric mice susceptible to HCV provide promising opportunities to circumvent host specificity.176 On the viral side, only 3 amino acid exchanges within the viral envelope proteins of HCV have been shown to allow infection of mouse hepatocytes in vitro, but additional adaptations might be necessary for efficient replication in vivo.177 Among non-human hepaciviruses, entry mechanisms have not been well studied, but the inability of the equine hepacivirus to infect human cells in vitro suggests a high level of host specificity.154 Recently detected hepaciviruses might provide exciting new opportunities for animal models, as exemplified by a rat hepacivirus capable of infecting immunocompetent laboratory mice and rats.166 Notably, any tractable animal model of human hepatitis would greatly benefit from susceptibility to human viruses or recombinants thereof, which has rarely been achieved.
Viral immune evasion strategies differ in host specificity
In addition to viral entry, cross-species transmission is limited by host immune responses and the ability of viruses to evade these responses after host switching. Among other mechanisms, HAV and HCV evade the human innate immune response by cleavage of the mitochondrial antiviral signalling protein (MAVS). MAVS is an essential part of interferon pathways induced by viral double-stranded RNA.178 MAVS is cleaved by the HAV protease precursor 3ABC 178 and the HCV protease NS3/4A179 at different sites. Strikingly different levels of host specificity are evident in HAV- and HCV-mediated MAVS cleavage. HAV-mediated MAVS cleavage seems to be rather host-specific based on the inability of human HAV to cleave murine MAVS.143 The ability of non-human hepatoviruses to cleave cognate and heterologous MAVS has not been studied yet and requires further investigation. Interestingly, for hepaciviruses, MAVS cleavage via the NS3/4A protease is conserved among equine, bat, rodent, and primate hepaciviruses,[178], [180], [181] hence reflecting a conserved immune evasion strategy. Strikingly, human MAVS can be cleaved by all of these diverse non-human hepaciviruses,181 hinting at a less host-specific mechanism compared to HAV.
In conclusion, both virus-receptor interactions and host immune responses can restrict cross-species transmission. Understanding the underlying mechanisms is essential for adapting potential novel animal models to human hepatitis viruses.
Key point.
Some biological properties, such as hepatotropism, receptor usage or mechanisms leading to HCC differ among human hepatitis viruses and ancestral viruses carried by animals other than primates.
Key point.
Animal homologues of hepatitis B and C viruses provide novel opportunities for in vivo studies of viral pathogenesis.
Chapter 4: Evolutionary origins of hepatitis viruses
The plethora of newly discovered viruses provides insights into the genealogy of human hepatitis viruses.182 Here, we discuss hypotheses on the when, the whence, and the where of human hepatitis virus evolution.
The age of human hepatitis viruses is underestimated
For all human hepatitis viruses, projections of the most recent common ancestors (MRCA) based on extant viruses yield surprisingly similar results in the range of several thousand years before present.[183], [184], [185] Hypothetically, environmental factors such as the formation of large human populations, rearing of livestock and changes in land use might have contributed to the rise of hepatitis viruses in humans and explain the similarities of calculated MRCA. However, projections of ancient MRCA need to be treated with caution, because of the technical constraints of bioinformatic programmes, recombination events, and an inevitable sampling bias. These limitations are best illustrated by variations of the projected origins of HBV, which vary by several orders of magnitude.[186], [187], [188] In addition, ancient HBV sequences from mummies and human remains revealed that HBV strains closely related to contemporary strains already occurred in the Neolithic, strongly suggesting that HBV is much older than previously thought.[189], [190], [191] The avian-associated hepadnaviruses were dated to have emerged around 6,000 years ago and thus again in a similar range as those yielded by projections of human-associated hepatitis virus MRCA.186 Again, the limitations of such projections became evident when endogenous hepadnavirus elements were identified in the genomes of birds and reptiles.[37], [192], [193], [194], [195] According to these molecular fossils, hepadnaviruses must have occurred in the Early Mesozoic > 200 million years ago (mya), predating the rise of mammals (Fig. 3 A) and again exceeding the MRCA from bioinformatic calculations by several orders of magnitude.196
Key point.
Hepatitis viruses are much older than previously thought, with hepatitis B virus ancestors predating the rise of mammals.
Fig. 3.
Hepatitis virus homologues in non-human hosts. (A) Simplified phylogenetic tree of mammalian orders, Aves, Reptilia, Amphibia, Osteichthyes, Condrichthyes, and Insecta (Phylogeny adapted from[196], [239]). Squares indicate orders for which homologues of human hepatitis viruses were found; black, same virus genus as human representative; grey, viruses distantly related to human hepatitis viruses. Host taxa for which no homologous hepatitis viruses were found are shown without pictograms and are coloured in grey. Numbers next to nodes show approximate time of divergence in million years according to.[196], [239] (B) GenBank entries of human hepatitis viruses and their homologues per mammalian host order given in percent of all entries of the search term. Host orders with less than 0.1% of entries are summarised as others. Host orders with less than 0.5% of entries are magnified 10-fold to the right. GenBank search terms and final entries per mammalian host taxon were: for HAV (January 17, 2018): “Hepatovirus [Organism] AND host [All Fields]”, Human, n = 3,186, Rodentia, n = 90, Eulipotyphla, n = 49, Chiroptera, n = 32, NHP, n = 6, Scandentia, n = 1, and Carnivora, n = 1; for HBV (January 19, 2018): “Hepadnaviridae [Organism] AND host [All Fields]”, Human, n = 68,867, NHP, n = 156, Aves, n = 87, Chiroptera, n = 73, Rodents, n = 25 and Cetartiodactyla, n = 10; for HCV (January 18, 2018): “Hepacivirus [Organism] AND host [All Fields]”, Human, n = 132,645, NHP, n = 491, Perissodactyla, n = 456, Rodentia, n = 182, Carnivora, n = 25, Cetartiodactyla, n = 18 and Chiroptera, n = 12; for HDV (January 17, 2018): “Deltavirus [Organism] AND host [All Fields]”, Human, n = 912; for HEV (January 17, 2018): “Hepeviridae [Organism] AND host [All Fields]”, Human, n = 7,601, Cetartiodactyla, n = 3,152, Lagomorpha, n = 456, Rodentia, n = 316, Aves, n = 155, Carnivora, n = 76, Non-human primates, n = 16, Chiroptera, n = 10, Scandentia, n = 2, Eulipotyphla, n = 1 and Perissodactyla, n = 1. (C) Patristic distance (maximum percentage pairwise amino acid distance) per host order calculated in MEGA7240 using translation alignments generated in Geneious 9.1.8.241 HAV: A GenBank search with the term “Hepatovirus” was performed on January 18, 2018, and all sequences longer than 6,000 nucleotides were selected. Duplicates, cell culture-adapted strains, or viruses isolated from experimentally infected animals were excluded from the dataset. Sorting according to host orders resulted in: Human, n = 101, NHP, n = 5, Chiroptera, n = 4, Rodentia, n = 7 and Eulipotyphla, n = 5 sequences for translation alignments of complete polyproteins. Non-homologous regions (px/3A) were subsequently deleted. For HBV, HCV and HDV, GenBank searches as performed for 3B were used. HBV: All sequences with complete polymerase CDS from non-human hosts were selected. Complete polymerase CDS of reference sequences from242 and the capuchin monkey hepatitis B virus (CMHBV, Acc. No. KY703886) were used. Sorting according to host orders resulted in Human, n = 37, NHP, n = 92, Chiroptera, n = 60 and Rodentia, n = 5 sequences. HCV: All sequences with complete polyprotein CDS from non-human hosts were selected. Complete polyprotein CDS of reference sequences from (https://hcv.lanl.gov/components/sequence/HCV/search/searchi.html) and GBV-B (Acc. No. NC001655) were used. Sorting according to host orders resulted in Human, n = 178, NHP, n = 4, Chiroptera, n = 5, Rodentia, n = 15, Perissodactyla, n = 19, Cetartiodactyla, n = 10 sequences. HDV: Sequences with complete HDAg CDS were selected resulting in a total of 158 human-derived sequences. HEV: A GenBank search with the term “Hepeviridae” was performed on January 25, 2018, and all sequences longer than 6,000 nucleotides were selected. Duplicates, cell culture-adapted strains, or viruses isolated from experimentally infected animals were excluded from the dataset. Sorting according to host orders resulted in Human, n = 165, Chiroptera, n = 3, Rodentia, n = 18, Cetartiodactyla, n = 93, Aves, n = 11, Carnivora, n = 13 and Lagomorpha, n = 17 sequences. CDS, coding sequences; HAV, hepatitis A virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitis delta virus; HEV, hepatitis E virus; NHP, non-human primates. Bars are coloured according to host taxon.
Ancient origins of non-human hepatitis viruses
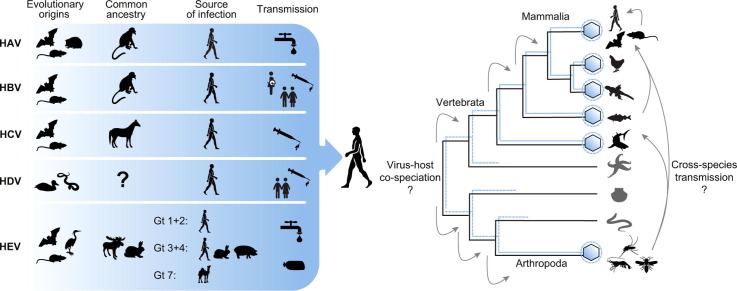
Interestingly, hepatitis viruses are found in diverse vertebrate taxa (Fig. 3A), hinting at a long evolutionary association between vertebrates and hepatitis viruses. The common ancestors of vertebrates date back to the Ordovician, approximately 450 mya, from where they diversified into fish, amphibians, reptiles, birds, and mammals. Notably, all of these vertebrate taxa harbour viruses with some genetic relationship to human hepatitis viruses. Remarkably, viruses and viral elements with genetic or structural relationships to hepatoviruses, hepadnaviruses, hepaciviruses, and hepeviruses also exist in arthropods,[197], [198], [199], [200] which are evolutionarily ancient animals that evolved approximately 500 mya.201 These recent observations further corroborate an ancient evolutionary origin of hepatitis viruses.
The genetic diversity of viruses in a given host taxon provides insights into the time of virus evolution within that host. In public databases, the number of HAV, HBV, and HCV genomic sequences originating from humans highly exceeds the number of sequences from their non-human homologues (Fig. 3B). In contrast, the majority of HEV sequences originates from even-toed ungulates, including swine, camels, cattle, deer, sheep, and moose. This is consistent with the high impact of these zoonotic viruses on human health. Interestingly, despite the comparably low number of genomic sequences available for animal homologues of HAV, HBV, and HCV, the genetic diversity of non-human viruses generally exceeds that of the human-associated viruses (Fig. 3C). This is especially conspicuous for viruses hosted by small mammals, hinting at a longer association of small mammals with hepatitis viruses compared to humans. The comparably low diversity in human hepatitis viruses may additionally be linked to evolutionary pressures favouring certain genotypes,202 the rapid expansion of the human population potentially favouring certain viruses over others, and to extinction events best illustrated by the recent findings of an extinct HBV genotype in human remains.[190], [191]
In conclusion, hepatitis viruses are presumably much older than previously thought. The comparison of hepatitis virus diversity in different host taxa suggests that evolution in non-human hosts generally preceded the introduction of hepatitis viruses into the human lineage.
Complex macro-evolutionary events during hepatitis virus evolution
Key point.
Both cross-species transmission and co-speciation likely shaped hepatitis virus evolution.
Hypothetically, hepatitis viruses may have accompanied speciation of their hosts. Indeed, similarities in the topologies of relatively old vertebrate lineages such as fish, amphibians and birds and their hepato-, hepadna- and hepaciviruses may hint at virus-host co-speciation.[36], [203] In addition, evidence for co-speciation exists for the New World monkey hepadnaviruses CMHBV and WMHBV.33 However, cross-species transmission must also have occurred during the evolution of hepatitis viruses. Apart from the zoonotic HEV genotypes, examples of cross-species transmission events include hepatoviruses in marsupials that are of likely rodent origin,28 horse-associated hepaciviruses in dogs and donkeys,[38], [41] gorillas infected with chimpanzee hepadnaviruses, orangutans infected with gibbon hepadnaviruses,119 and rat hepeviruses in shrews.204 In addition, the tentative HBV genotype J likely emerged from a recombination event involving human- and gibbon-associated parental strains, hinting at cross-species viral transmission from gibbons to humans (summarised in80). Evidence for cross-species transmission events in hepatitis viruses might indicate that humans or their primate precursors acquired hepatitis viruses from another animal host.
It remains unclear when this hypothetical host switching into primates occurred. Nonetheless, a relatively recent introduction of hepatitis viruses into humans or their primate precursors is strongly supported by the lack of genetic diversity among primate viruses compared to non-primate hepatitis virus homologues, and by the complete absence of human viruses clustering in basal positions in phylogenetic reconstructions (Fig. 4 A-E).
Fig. 4.
Phylogenetic relationships of human hepatitis viruses and their vertebrate homologues. Phylogenies are based on amino acid alignments for hepatoviruses, hepaciviruses, deltaviruses, and hepeviruses and a nucleotide alignment for hepadnaviruses generated using MAFFT243 in Geneious 9.1.8.241 Phylogenies were generated using a WAG substitution model (for amino acid sequences) or an HKY substitution model (for nucleotide sequences) excluding all ambiguous data or gaps. Hepadnavirus and deltavirus phylogenies were generated using a Maximum Likelihood algorithm and 1,000 bootstrap replicates in MEGA7.240 For all other virus genera, Bayesian phylogenies were generated using MrBayes V3.1.244 Outgroups were: Hepatoviruses, Avian encephalomyelitis virus, accession number (Acc. No.) NC003990; Hepadnaviruses, Bluegill hepadnavirus, Acc. No. NC030445; Hepaciviruses, Wenling shark virus, Acc. No. KR902729; Hepeviruses, Piscihepevirus, Acc. No. HQ731075. The deltavirus phylogeny was calculated without an outgroup. Bayesian phylogenies were run for 2 million generations, sampled every 100 steps. After an exclusion of 5,000 of the total 20,000 trees as burn-in, final trees were annotated with TreeAnnotator from the BEAST package and visualised with FigTree.245 Bayesian posterior probability support of grouping above 0.9 or bootstrap support above 90% is highlighted by filled circles at nodes. The scale bar indicates genetic distance. Animal pictograms are coloured according to their host taxa: Black and red, Primates; Orange, Rodentia; Light green, Lagomorpha; Pink, Scandentia; Yellow, Perissodactyla; Light brown, Eulipotyphla; Light blue, Chiroptera; Dark blue, Carnivora; Dark green, Cetartiodactyla; Grey, Marsupialia; Turquois, Aves; Dark brown, Reptilia. (A) For hepatoviruses, the complete P1 domain was used. Acc. No. of representative sequences were: M59286, KT452691, KT877158, KT452714, KT452730, KR703607, KT452742, KT452735, AB279735, AB279732, AY644670, AY644676, AF268396, AB020564, D00924, KT452685, KT452658, MG181943. (B) For hepadnaviruses, complete genomes were used. Acc. No. of representative sequences were: MH307930 NC024444, FN594748, EU594434, AB032432, FJ798097, FM209516, LT992443, AF193863, FM209514, GU563556, AB194951, AB368296, FJ899779, AB036920, AB516393, AF046996, NC24445, NC004107, AB486012, FJ023659. (C) For hepaciviruses, domains encoding the structural proteins C-E1-E2-p7 were used. Acc. No. of representative sequences were: KC411777, JQ434004, D00944, GU814265, M62321, Y12083, Y13184, D17763, EF108306, KC796074, KC411806, KC796077, U22304, KJ950938, KC551800, KP641127. (D) For deltaviruses, complete S-HDAg coding regions were used. Acc. No. of representative sequences were: X04451, X60193, L22063, AF018077, AJ584848, AJ584847, AJ584844, AJ584849, MH988742, MH824555. (E) For hepeviruses, complete ORF 2 sequences encoding the viral capsid were used. Acc. No. of representative sequences were: AB197673, DQ279091, AB573435, AF082843, AP003430, KJ013415, KJ496143, KT818608, KX387865, AB602441, M73218, M74506, KF951328, KR905549 GU345042, AY535004, JQ001749. Gt, Genotype; HAV, hepatitis A virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitis delta virus; HEV, hepatitis E virus; ORF, open-reading frame; S-HDAg, small form HDV antigen.
Interestingly, the frequency of predicted cross-species transmission events varies among the different hepatitis virus families. While cross-species transmission may be relatively frequent in the enterically transmitted hepato- and hepeviruses, it seems to be comparably infrequent in blood-borne hepadna- and hepaciviruses.[29], [36], [205] Possible explanations for this observation include potentially easier transmission of highly stable enterically transmitted viruses compared to viruses likely transmitted exclusively through contact with body fluids. Additionally, different levels of host specificity due to receptor interactions or viral immune evasion strategies may play a role, as outlined earlier.
The macro-evolutionary patterns of hepatitis virus are thus complex and likely include both co-speciation and cross-species transmission events. Apart from certain HEV strains, no recent zoonotic event is evident in the phylogenetic reconstructions of human hepatitis viruses.
A recent NHP origin of human hepatitis viruses is unlikely
Since genetic relatedness of hosts facilitates cross-species transmission of viruses206 one could assume human hepatitis virus origins in NHPs, similar to the origins of HIV in monkeys and apes.207 Indeed, relatives of HAV, HBV, and HCV are found in NHPs (Fig. 4A). However, for HAV and HCV these detections are sporadic and the viruses are not genetically diversified in NHPs22 (Fig. 4A/C). In addition, hepaciviruses from NHPs do not share a recent common ancestor with human HCV (Fig. 4C), suggesting a non-recent host switch into NHPs from an unknown source. Among primate HBV, diversified strains from Old World apes are intermixed with human strains and recombination events among these viruses hint at past cross-species transmission.119 However, the relatively large genetic diversity of human viruses compared to viruses from Old World apes may hint at an origin of HBV in the human stem lineage.[33], [208] Interestingly, both the WHMBV and the CMHBV surface proteins can use the hNTCP for cell entry, suggesting zoonotic potential of these divergent HBV species from New World monkeys. Nonetheless, the phylogenetic relationships of primate HBV suggest that neither CMHBV, nor WMHBV are direct ancestors of human HBV.33 Hence, our current knowledge on primate HAV, HBV and HCV diversity does not support a recent origin of human viruses in NHP. Thus, studies investigating hepatitis virus diversity in diverse NHP species and NHP remains are urgently needed.
The complex where and whence of human hepatitis viruses
Small mammals such as bats and rodents are particularly relevant animal reservoirs of human pathogens.[209], [210] Their important role as hosts for zoonotic pathogens has been linked to their species richness, abundance, worldwide distribution, the synanthropic behaviour of some species, the ability of bats to fly, to gregarious populations of some bat species and to unique immunological properties of bats.[209], [211], [212] Interestingly, highly diversified homologues of HAV, HBV, HCV, and HEV were detected in these 2 mammalian orders[25], [28], [30], [42], [43], [54], [213], [214] (Fig. 4), hinting at the importance of small mammals during hepatitis virus evolution.
Key point.
Small mammals were important hosts during the evolution of human hepatitis viruses.
One can assume that the diversity of viruses is highest in areas of prolonged circulation. Based on this assumption, the origins of human hepatitis viruses can be mainly projected to the Old World (Fig. 5 ).[80], [100] In addition, diverse viruses assigned to HAV and HBV are found in Old World NHPs which hints at a relatively long association of hepatitis viruses with primates in the Old World compared to the New World. However, divergent HBV and HDV genotypes occur in American natives, namely HBV genotypes F and H and HDV genotype 3.[187], [215] Prior to the novel animal delta-like agents, it was not possible to root the phylogenetic tree of HDV. Now, upon inclusion of the novel snake and duck delta-like agents, surprising similarities are found in the sister relationship of the New World HBV and HDV genotypes and Old World genotypes (Fig. 4B,D). Notably, the existence of these divergent New World genotypes does not necessarily suggest ancient origins of HBV and HDV in the New World, because these genotypes show only limited genetic diversity compared to the Old World genotypes. It seems plausible that the ancestors of New World HBV and HDV genotypes were introduced to the New World during the peopling of the Americas via the Beringian land bridge approximately 15–23 thousand years ago,216 before then going extinct in the Old World.33
Fig. 5.
Global distribution of hepatitis virus genotypes. Distribution of the most prevalent genotypes were adapted and simplified from246 for HAV, from[247], [248] for HBV, from[249], [250] for HCV, from87 for HDV and from185 for HEV. NHP genotypes are shown as pictograms and represent HAV genotype 5 in Africa, HAV genotypes 4 and 6 in Asia,246 Chimpanzee HBV and Gorilla HBV in Africa and Orangutan HBV and Gibbon HBV genotypes in Asia.[119], [251] HAV, hepatitis A virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitis delta virus; HEV, hepatitis E virus; NHP, non-human primate.
The evolutionary ancestors of human hepatitis viruses are unknown. Presumably, a zoonotic introduction into humans or their primate precursors occurred non-recently in the Old World. Further evidence elucidating the genealogy of human hepatitis viruses can be expected in the coming years.
Key point.
The direct ancestors of human hepatitis viruses so far remain unknown.
Concluding remarks
Major progress has been made in the study of human hepatitis viruses during the last 60 years. However, crucial aspects of hepatitis virus evolution and pathogenesis remain poorly understood. At the same time, properties we took for certain have been called into question, such as the predominant association of hepatitis viruses with human hosts, the determinants of hepatotropism and the distinction of typically either enveloped or non-enveloped viruses. In this review, we discussed pathogenesis, evolutionary origins and zoonotic risks of human hepatitis viruses in light of recent discoveries of a plethora of animal homologues.
It is anticipated that the newly discovered animal viruses will help to answer crucial questions regarding the pathogenesis of hepatitis viruses, e.g., whether the existence of quasi-enveloped particles of HAV and HEV is evolutionarily conserved, and which viral determinants are involved in oncogenesis and the potential to cause chronic infections. Although hepatotropism is not fully understood even for the well-investigated HAV, HCV and HEV, the recent detection of HDV-like agents in animals apparently showing both a broad organ tropism and a lack of detectable helper viruses emphasises the need to investigate the bases underlying the unique biological properties of human hepatitis viruses.[47], [48]
The evolutionary origins of human hepatitis viruses have remained enigmatic. The recent discoveries of highly diverse animal homologues hint at the evolutionary origins of all human hepatitis viruses in non-human hosts. These origins may be rather ancient, and involve old vertebrate lineages such as reptiles, amphibians, birds, fish and potentially even arthropods[198], [199], [200] (Fig. 6 A). Among mammals, small mammals such as bats and rodents harbour particularly diversified hepatitis viruses. One may speculate that arthropod-borne hepatitis virus precursors may have been passed to insectivorous small mammals via the blood-borne route or by ingestion of insects.[25], [217] These hosts hypothetically acted as a point of entry for ancient viruses carried by arthropods or lower vertebrates into mammals. In small mammals, viruses may then have diversified and eventually have been transmitted directly or via intermediate hosts to humans.[29], [119] It is also conceivable that several independent introductions of hepatitis viruses into different vertebrate lineages from arthropods, lower vertebrates and yet unknown sources have occurred (Fig. 6B). Presumably, hepatitis virus evolution combined both long-term virus-host associations and cross-species transmission events, such as those projected for other viruses.[29], [34], [36], [119], [203], [218] Future discoveries of viruses will identify missing links in hepatitis virus evolution and may identify direct ancestors of human hepatitis viruses.
Fig. 6.
Evolutionary origins of hepatitis viruses. (A) Evolutionary origins, ancestors and sources of human infections. Animal pictograms represent host taxa. (B) Hypotheses for the evolution of hepatitis-like viruses among diverse animals. Black lines, simplified bilateria phylogeny from252 including selected taxa. Black pictograms, animal taxa comprising hosts of hepatitis viruses and related viruses. Grey pictograms, representatives of selected animal taxa. From bottom to top: phylum Nematoda, superphylum Lophotrochozoa, phylum Echinodermata. Blue lines: hypothesised virus evolution. Gt, genotype; HAV, hepatitis A virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitis delta virus; HEV, hepatitis E virus.
Finally, there is an increasing drive towards the elimination of viral hepatitis B and C as aspired by the Word Health Organization.1 Notably, a prerequisite for the eradication of hepatitis viruses in humans is the absence of an animal reservoir. The discovery of hepatitis virus homologues in non-human hosts might imply that reintroductions into the human population after successful eradication and cessation of vaccination in the case of HBV may be possible. This is best illustrated by the ability of the surface proteins of the bat-borne TBHBV to confer viral entry into human hepatocytes, hinting at the zoonotic potential of this bat hepadnavirus.30
In summary, the last decades have yielded unprecedented insight into the evolution of human hepatitis viruses, refuting their conceptualisation as predominantly human pathogens in all cases. Beyond enhancing our understanding of viral evolution, the diverse animal homologues of human hepatitis viruses allow conceptualisation of new animal models for preclinical testing of urgently needed therapeutics for chronic hepatitis B and C, and for determining the many unknowns of hepatitis virus pathogenesis.
Financial support
This study was supported by a stipend from the German Center for Infection Research to AR and by funding from the German Research Foundation (DFG, DR 810/1-1) to JFD.
Conflict of interest
The authors declare no conflicts of interest that pertain to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We thank Dieter Glebe for critical reading of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhep.2018.11.010.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.(WHO) WHO. Global hepatitis report, 2017. https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/
- 2.(WHO) WHO. Hepatitis C fact sheet. 2017; http://www.who.int/news-room/fact-sheets/detail/hepatitis-c.
- 3.Romeo R., Del Ninno E., Rumi M., Russo A., Sangiovanni A., de Franchis R. A 28-year study of the course of hepatitis Delta infection: a risk factor for cirrhosis and hepatocellular carcinoma. Gastroenterology. 2009;136:1629–1638. doi: 10.1053/j.gastro.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 4.Krugman S., Giles J.P., Hammond J. Infectious hepatitis. Evidence for two distinctive clinical, epidemiological, and immunological types of infection. JAMA. 1967;200:365–373. doi: 10.1001/jama.200.5.365. [DOI] [PubMed] [Google Scholar]
- 5.Maccallum F.O. Homologous serum hepatitis. Proc R Soc Med. 1946;39:655–657. doi: 10.1177/003591574603901013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feinstone S.M., Kapikian A.Z., Purceli R.H. Hepatitis A: detection by immune electron microscopy of a viruslike antigen associated with acute illness. Science. 1973;182:1026–1028. doi: 10.1126/science.182.4116.1026. [DOI] [PubMed] [Google Scholar]
- 7.Dane D.S., Cameron C.H., Briggs M. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet. 1970;1:695–698. doi: 10.1016/s0140-6736(70)90926-8. [DOI] [PubMed] [Google Scholar]
- 8.Rizzetto M., Canese M.G., Gerin J.L., London W.T., Sly D.L., Purcell R.H. Transmission of the hepatitis B virus-associated delta antigen to chimpanzees. J Infect Dis. 1980;141:590–602. doi: 10.1093/infdis/141.5.590. [DOI] [PubMed] [Google Scholar]
- 9.Feinstone S.M., Kapikian A.Z., Purcell R.H., Alter H.J., Holland P.V. Transfusion-associated hepatitis not due to viral hepatitis type A or B. N Engl J Med. 1975;292:767–770. doi: 10.1056/NEJM197504102921502. [DOI] [PubMed] [Google Scholar]
- 10.Choo Q.L., Kuo G., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 11.Balayan M.S., Andjaparidze A.G., Savinskaya S.S., Ketiladze E.S., Braginsky D.M., Savinov A.P. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology. 1983;20:23–31. doi: 10.1159/000149370. [DOI] [PubMed] [Google Scholar]
- 12.Drosten C., Weber M., Seifried E., Roth W.K. Evaluation of a new PCR assay with competitive internal control sequence for blood donor screening. Transfusion. 2000;40:718–724. doi: 10.1046/j.1537-2995.2000.40060718.x. [DOI] [PubMed] [Google Scholar]
- 13.Drexler J.F., Kupfer B., Petersen N., Grotto R.M., Rodrigues S.M., Grywna K. A novel diagnostic target in the hepatitis C virus genome. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung R.T., Baumert T.F. Curing chronic hepatitis C–the arc of a medical triumph. N Engl J Med. 2014;370:1576–1578. doi: 10.1056/NEJMp1400986. [DOI] [PubMed] [Google Scholar]
- 15.Summers J., Smolec J.M., Snyder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason W.S., Seal G., Summers J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanford R.E., Chavez D., Brasky K.M., Burns R.B., 3rd Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaudin M., Wolstenholme A.J., Tsiquaye K.N., Zuckerman A.J., Harrison T.J. The complete nucleotide sequence of the genome of a hepatitis B virus isolated from a naturally infected chimpanzee. The Journal of general virology. 1988;69:1383–1389. doi: 10.1099/0022-1317-69-6-1383. [DOI] [PubMed] [Google Scholar]
- 19.Nainan O.V., Margolis H.S., Robertson B.H., Balayan M., Brinton M.A. Sequence analysis of a new hepatitis A virus naturally infecting cynomolgus macaques (Macaca fascicularis) J Gen Virol. 1991;72:1685–1689. doi: 10.1099/0022-1317-72-7-1685. [DOI] [PubMed] [Google Scholar]
- 20.Meng X.J., Halbur P.G., Shapiro M.S., Govindarajan S., Bruna J.D., Mushahwar I.K. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J Virol. 1998;72:9714–9721. doi: 10.1128/jvi.72.12.9714-9721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simons J.N., Pilot-Matias T.J., Leary T.P., Dawson G.J., Desai S.M., Schlauder G.G. Identification of two flavivirus-like genomes in the GB hepatitis agent. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson B.H. Viral hepatitis and primates: historical and molecular analysis of human and nonhuman primate hepatitis A, B, and the GB-related viruses. J Viral Hepat. 2001;8:233–242. doi: 10.1046/j.1365-2893.2001.00295.x. [DOI] [PubMed] [Google Scholar]
- 23.Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 24.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 25.Drexler J.F., Corman V.M., Lukashev A.N., van den Brand J.M., Gmyl A.P., Brunink S. Evolutionary origins of hepatitis A virus in small mammals. Proc Natl Acad Sci U S A. 2015;112:15190–15195. doi: 10.1073/pnas.1516992112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anthony S.J., St Leger J.A., Liang E., Hicks A.L., Sanchez-Leon M.D., Jain K. Discovery of a novel hepatovirus (phopivirus of seals) related to human hepatitis. A Virus. mBio. 2015;6 doi: 10.1128/mBio.01180-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu J.M., Li L.L., Zhang C.Y., Lu S., Ao Y.Y., Gao H.C. A novel hepatovirus identified in wild woodchuck Marmota himalayana. Sci Rep. 2016;6:22361. doi: 10.1038/srep22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Oliveira Carneiro I, Sander A.L., Silva N., Moreira-Soto A., Normann A., Flehmig B. A novel marsupial hepatitis A virus corroborates complex evolutionary patterns shaping the genus Hepatovirus. J Virol. 2018;92(13):e00082-18. doi: 10.1128/JVI.00082-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sander A.L., Corman V.M., Lukashev A.N., Drexler J.F. Evolutionary Origins of Enteric Hepatitis Viruses. Cold Spring Harb Perspect Med. 2018;8(12) doi: 10.1101/cshperspect.a031690. pii: a031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drexler J.F., Geipel A., Konig A., Corman V.M., van Riel D., Leijten L.M. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. 2013;110:16151–16156. doi: 10.1073/pnas.1308049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He B., Zhang F., Xia L., Hu T., Chen G., Qiu W. Identification of a novel Orthohepadnavirus in pomona roundleaf bats in China. Arch Virol. 2015;160:335–337. doi: 10.1007/s00705-014-2222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aghazadeh M., Shi M., Barrs V.R., McLuckie A., Lindsay S., Jameson B. A novel hepadnavirus identified in an immunocompromised domestic cat in Australia. Viruses. 2018;10:269. doi: 10.3390/v10050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Carvalho Dominguez Souza B.F., Konig A., Rasche A., de Oliveira Carneiro I., Stephan N., Corman V.M. A novel hepatitis B virus species discovered in capuchin monkeys sheds new light on the evolution of primate hepadnaviruses. J Hepatol. 2018;68:1114–1122. doi: 10.1016/j.jhep.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 34.Dill J.A., Camus A.C., Leary J.H., Di Giallonardo F., Holmes E.C., Ng T.F. Distinct viral lineages from fish and amphibians reveal the complex evolutionary history of hepadnaviruses. J Virol. 2016;90:7920–7933. doi: 10.1128/JVI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hahn C.M., Iwanowicz L.R., Cornman R.S., Conway C.M., Winton J.R., Blazer V.S. Characterization of a novel hepadnavirus in the white sucker (Catostomus commersonii) from the Great Lakes Region of the United States. J Virol. 2015;89:11801–11811. doi: 10.1128/JVI.01278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauber C., Seitz S., Mattei S., Suh A., Beck J., Herstein J. Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe. 2017;22 doi: 10.1016/j.chom.2017.07.019. 387–399 e386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suh A., Weber C.C., Kehlmaier C., Braun E.L., Green R.E., Fritz U. Early mesozoic coexistence of amniotes and hepadnaviridae. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burbelo P.D., Dubovi E.J., Simmonds P., Medina J.L., Henriquez J.A., Mishra N. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol. 2012;86:6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapoor A., Simmonds P., Gerold G., Qaisar N., Jain K., Henriquez J.A. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyons S., Kapoor A., Schneider B.S., Wolfe N.D., Culshaw G., Corcoran B. Viraemic frequencies and seroprevalence of non-primate hepacivirus and equine pegiviruses in horses and other mammalian species. The Journal of General Virology. 2014;95:1701–1711. doi: 10.1099/vir.0.065094-0. [DOI] [PubMed] [Google Scholar]
- 41.Walter S., Rasche A., Moreira-Soto A., Pfaender S., Bletsa M., Corman V.M. Infection patterns and recent evolutionary origins of equine hepaciviruses in donkeys. J Virol. 2017;91 doi: 10.1128/JVI.01711-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drexler J.F., Corman V.M., Muller M.A., Lukashev A.N., Gmyl A., Coutard B. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quan P.L., Firth C., Conte J.M., Williams S.H., Zambrana-Torrelio C.M., Anthony S.J. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci U S A. 2013;110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corman V.M., Grundhoff A., Baechlein C., Fischer N., Gmyl A., Wollny R. Highly divergent hepaciviruses from African cattle. J Virol. 2015;89:5876–5882. doi: 10.1128/JVI.00393-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baechlein C., Fischer N., Grundhoff A., Alawi M., Indenbirken D., Postel A. Identification of a novel hepacivirus in domestic cattle from Germany. J Virol. 2015;89:7007–7015. doi: 10.1128/JVI.00534-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauck M., Sibley S.D., Lara J., Purdy M.A., Khudyakov Y., Hyeroba D. A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild Old World primate. J Virol. 2013;87:8971–8981. doi: 10.1128/JVI.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wille M., Netter H.J., Littlejohn M., Yuen L., Shi M., Eden J.-S. A divergent hepatitis D-like agent in birds. Viruses. 2018;10 doi: 10.3390/v10120720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hetzel U., Szirovicza L., Smura T., Prahauser B., Vapalahti O., Kipar A. Identification of a novel deltavirus in Boa constrictor. BioRxiv. 2018 doi: 10.1101/429753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi M., Nishizawa T., Sato H., Sato Y., Jirintai, Nagashima S. Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J Gen Virol. 2011;92:902–908. doi: 10.1099/vir.0.029470-0. [DOI] [PubMed] [Google Scholar]
- 50.Zhao C., Ma Z., Harrison T.J., Feng R., Zhang C., Qiao Z. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J Med Virol. 2009;81:1371–1379. doi: 10.1002/jmv.21536. [DOI] [PubMed] [Google Scholar]
- 51.Woo P.C., Lau S.K., Teng J.L., Tsang A.K., Joseph M., Wong E.Y. New hepatitis E virus genotype in camels, the Middle East. Emerg Infect Dis. 2014;20:1044–1048. doi: 10.3201/eid2006.140140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rasche A., Saqib M., Liljander A.M., Bornstein S., Zohaib A., Renneker S. Hepatitis E virus infection in dromedaries, North and East Africa, United Arab Emirates, and Pakistan, 1983–2015. Emerg Infect Dis. 2016;22:1249–1252. doi: 10.3201/eid2207.160168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raj V.S., Smits S.L., Pas S.D., Provacia L.B., Moorman-Roest H., Osterhaus A.D. Novel hepatitis E virus in ferrets, the Netherlands. Emerg Infect Dis. 2012;18:1369–1370. doi: 10.3201/eid1808.111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drexler J.F., Seelen A., Corman V.M., Fumie Tateno A., Cottontail V., Melim Zerbinati R. Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J Virol. 2012;86:9134–9147. doi: 10.1128/JVI.00800-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johne R., Heckel G., Plenge-Bonig A., Kindler E., Maresch C., Reetz J. Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis. 2010;16:1452–1455. doi: 10.3201/eid1609.100444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Batts W., Yun S., Hedrick R., Winton J. A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii) Virus Res. 2011;158:116–123. doi: 10.1016/j.virusres.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 57.de Souza W.M., Romeiro M.F., Sabino-Santos G., Jr., Maia F.G.M., Fumagalli M.J., Modha S. Novel orthohepeviruses in wild rodents from Sao Paulo State, Brazil. Virology. 2018;519:12–16. doi: 10.1016/j.virol.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang B., Li W., Zhou J.H., Li B., Zhang W., Yang W.H. Chevrier’s field mouse (Apodemus chevrieri) and Pere David’s Vole (Eothenomys melanogaster) in China carry orthohepeviruses that form two putative novel genotypes within the species orthohepevirus C. Virol Sin. 2018;33:44–58. doi: 10.1007/s12250-018-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Payne C.J., Ellis T.M., Plant S.L., Gregory A.R., Wilcox G.E. Sequence data suggests big liver and spleen disease virus (BLSV) is genetically related to hepatitis E virus. Vet Microbiol. 1999;68:119–125. doi: 10.1016/s0378-1135(99)00067-x. [DOI] [PubMed] [Google Scholar]
- 60.Sauter D., Schindler M., Specht A., Landford W.N., Munch J., Kim K.A. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lemon S.M., Ott J.J., Van Damme P., Shouval D. Type A viral hepatitis: a summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J Hepatol. 2018;68:167–184. doi: 10.1016/j.jhep.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 62.Paganelli M., Stephenne X., Sokal E.M. Chronic hepatitis B in children and adolescents. J Hepatol. 2012;57:885–896. doi: 10.1016/j.jhep.2012.03.036. [DOI] [PubMed] [Google Scholar]
- 63.Webster D.P., Klenerman P., Dusheiko G.M. Hepatitis C. Lancet. 2015;385:1124–1135. doi: 10.1016/S0140-6736(14)62401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pascarella S., Negro F. Hepatitis D virus: an update. Liver Int. 2011;31:7–21. doi: 10.1111/j.1478-3231.2010.02320.x. [DOI] [PubMed] [Google Scholar]
- 65.Pischke S., Behrendt P., Bock C.T., Jilg W., Manns M.P., Wedemeyer H. Hepatitis E in Germany–an under-reported infectious disease. Deutsches Arzteblatt Int. 2014;111:577–583. doi: 10.3238/arztebl.2014.0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Das A., Hirai-Yuki A., Gonzalez-Lopez O., Rhein B., Moller-Tank S., Brouillette R. TIM1 (HAVCR1) is not essential for cellular entry of either quasi-enveloped or naked hepatitis A virions. mBio. 2017;8 doi: 10.1128/mBio.00969-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1 doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schulze A., Gripon P., Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology. 2007;46:1759–1768. doi: 10.1002/hep.21896. [DOI] [PubMed] [Google Scholar]
- 69.Catanese M.T., Ansuini H., Graziani R., Huby T., Moreau M., Ball J.K. Role of scavenger receptor class B type I in hepatitis C virus entry: kinetics and molecular determinants. J Virol. 2010;84:34–43. doi: 10.1128/JVI.02199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamamoto S., Fukuhara T., Ono C., Uemura K., Kawachi Y., Shiokawa M. Lipoprotein receptors redundantly participate in entry of hepatitis C virus. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 72.Lamas Longarela O., Schmidt T.T., Schoneweis K., Romeo R., Wedemeyer H., Urban S. Proteoglycans act as cellular hepatitis delta virus attachment receptors. PLoS One. 2013;8 doi: 10.1371/journal.pone.0058340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feng Z., Hensley L., McKnight K.L., Hu F., Madden V., Ping L. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Takahashi M., Tanaka T., Takahashi H., Hoshino Y., Nagashima S., Jirintai Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. J Clin Microbiol. 2010;48:1112–1125. doi: 10.1128/JCM.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng Z., Hirai-Yuki A., McKnight K.L., Lemon S.M. Naked viruses that aren’t always naked: quasi-enveloped agents of acute hepatitis. Annu Rev Virol. 2014;1:539–560. doi: 10.1146/annurev-virology-031413-085359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Catanese M.T., Uryu K., Kopp M., Edwards T.J., Andrus L., Rice W.J. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci U S A. 2013;110:9505–9510. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tanaka T., Kasai H., Yamashita A., Okuyama-Dobashi K., Yasumoto J., Maekawa S. Hallmarks of hepatitis C virus in equine hepacivirus. J Virol. 2014;88:13352–13366. doi: 10.1128/JVI.02280-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lemon S.M., Murphy P.C., Shields P.A., Ping L.H., Feinstone S.M., Cromeans T. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991;65:2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen X., Zhang Q., He C., Zhang L., Li J., Zhang W. Recombination and natural selection in hepatitis E virus genotypes. J Med Virol. 2012;84:1396–1407. doi: 10.1002/jmv.23237. [DOI] [PubMed] [Google Scholar]
- 80.Locarnini S., Littlejohn M., Aziz M.N., Yuen L. Possible origins and evolution of the hepatitis B virus (HBV) Semin Cancer Biol. 2013;23:561–575. doi: 10.1016/j.semcancer.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 81.Kalinina O., Norder H., Mukomolov S., Magnius L.O. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol. 2002;76:4034–4043. doi: 10.1128/JVI.76.8.4034-4043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin C.C., Lee C.C., Lin S.H., Huang P.J., Li H.P., Chang Y.S. RNA recombination in Hepatitis delta virus: identification of a novel naturally occurring recombinant. J Microbiol Immunol Infect. 2017;50:771–780. doi: 10.1016/j.jmii.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 83.Sy B.T., Nguyen H.M., Toan N.L., Song L.H., Tong H.V., Wolboldt C. Identification of a natural intergenotypic recombinant hepatitis delta virus genotype 1 and 2 in Vietnamese HBsAg-positive patients. J Viral Hepat. 2015;22:55–63. doi: 10.1111/jvh.12228. [DOI] [PubMed] [Google Scholar]
- 84.Yang J., Xi Q., Deng R., Wang J., Hou J., Wang X. Identification of interspecies recombination among hepadnaviruses infecting cross-species hosts. J Med Virol. 2007;79:1741–1750. doi: 10.1002/jmv.20983. [DOI] [PubMed] [Google Scholar]
- 85.Theze J., Lowes S., Parker J., Pybus O.G. Evolutionary and phylogenetic analysis of the hepaciviruses and pegiviruses. Genome Biol Evol. 2015;7:2996–3008. doi: 10.1093/gbe/evv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kelly A.G., Netzler N.E., White P.A. Ancient recombination events and the origins of hepatitis E virus. BMC Evol Biol. 2016;16:210. doi: 10.1186/s12862-016-0785-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hughes S.A., Wedemeyer H., Harrison P.M. Hepatitis delta virus. Lancet. 2011;378:73–85. doi: 10.1016/S0140-6736(10)61931-9. [DOI] [PubMed] [Google Scholar]
- 88.Robertson H.D. How did replicating and coding RNAs first get together? Science. 1996;274:66–67. doi: 10.1126/science.274.5284.66. [DOI] [PubMed] [Google Scholar]
- 89.Taylor J.M. Host RNA circles and the origin of hepatitis delta virus. World J Gastroenterol. 2014;20:2971–2978. doi: 10.3748/wjg.v20.i11.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cunha C., Tavanez J.P., Gudima S. Hepatitis delta virus: a fascinating and neglected pathogen. World J Virol. 2015;4:313–322. doi: 10.5501/wjv.v4.i4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Polson A.G., Bass B.L., Casey J.L. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 92.Otto J.C., Casey P.J. The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J Biol Chem. 1996;271:4569–4572. doi: 10.1074/jbc.271.9.4569. [DOI] [PubMed] [Google Scholar]
- 93.Geoghegan J.L., Senior A.M., Di Giallonardo F., Holmes E.C. Virological factors that increase the transmissibility of emerging human viruses. Proc Natl Acad Sci U S A. 2016;113:4170–4175. doi: 10.1073/pnas.1521582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.MacLachlan J.H., Cowie B.C. Hepatitis B virus epidemiology. Cold Spring Harb Perspect Med. 2015;5 doi: 10.1101/cshperspect.a021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gentile I., Borgia G. Vertical transmission of hepatitis B virus: challenges and solutions. Int J Womens Health. 2014;6:605–611. doi: 10.2147/IJWH.S51138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Noppornpanth S., Haagmans B.L., Bhattarakosol P., Ratanakorn P., Niesters H.G., Osterhaus A.D. Molecular epidemiology of gibbon hepatitis B virus transmission. J Gen Virol. 2003;84:147–155. doi: 10.1099/vir.0.18531-0. [DOI] [PubMed] [Google Scholar]
- 97.Tagawa M., Robinson W.S., Marion P.L. Duck hepatitis B virus replicates in the yolk sac of developing embryos. J Virol. 1987;61:2273–2279. doi: 10.1128/jvi.61.7.2273-2279.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kulonen K., Millman I. Vertical transmission of woodchuck hepatitis virus. J Med Virol. 1988;26:233–242. doi: 10.1002/jmv.1890260303. [DOI] [PubMed] [Google Scholar]
- 99.Terrault N.A., Dodge J.L., Murphy E.L., Tavis J.E., Kiss A., Levin T.R. Sexual transmission of hepatitis C virus among monogamous heterosexual couples: the HCV partners study. Hepatology. 2013;57:881–889. doi: 10.1002/hep.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simmonds P. The origin of hepatitis C virus. Curr Top Microbiol Immunol. 2013;369:1–15. doi: 10.1007/978-3-642-27340-7_1. [DOI] [PubMed] [Google Scholar]
- 101.Schmid J., Rasche A., Eibner G., Jeworowski L., Page R.A., Corman V.M. Ecological drivers of Hepacivirus infection in a neotropical rodent inhabiting landscapes with various degrees of human environmental change. Oecologia. 2018;188:289–302. doi: 10.1007/s00442-018-4210-7. [DOI] [PubMed] [Google Scholar]
- 102.Pfaender S., Cavalleri J.M., Walter S., Doerrbecker J., Campana B., Brown R.J. Clinical course of infection and viral tissue tropism of hepatitis C virus-like nonprimate hepaciviruses in horses. Hepatology. 2015;61:447–459. doi: 10.1002/hep.27440. [DOI] [PubMed] [Google Scholar]
- 103.Ramsay J.D., Evanoff R., Wilkinson T.E., Jr., Divers T.J., Knowles D.P., Mealey R.H. Experimental transmission of equine hepacivirus in horses as a model for hepatitis C virus. Hepatology. 2015;61:1533–1546. doi: 10.1002/hep.27689. [DOI] [PubMed] [Google Scholar]
- 104.Lyons S., Kapoor A., Sharp C., Schneider B.S., Wolfe N.D., Culshaw G. Nonprimate hepaciviruses in domestic horses, United kingdom. Emerg Infect Dis. 2012;18:1976–1982. doi: 10.3201/eid1812.120498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu J.M., Li L.L., Xie G.C., Zhang C.Y., Ao Y.Y., Duan Z.J. Experimental infection of Marmota monax with a novel hepatitis A virus. Arch Virol. 2018;163:1187–1193. doi: 10.1007/s00705-018-3715-z. [DOI] [PubMed] [Google Scholar]
- 106.Emerson S.U., Tsarev S.A., Govindarajan S., Shapiro M., Purcell R.H. A simian strain of hepatitis A virus, AGM-27, functions as an attenuated vaccine for chimpanzees. J Infect Dis. 1996;173:592–597. doi: 10.1093/infdis/173.3.592. [DOI] [PubMed] [Google Scholar]
- 107.Dienstag J.L., Feinstone S.M., Purcell R.H., Hoofnagle J.H., Barker L.F., London W.T. Experimental infection of chimpanzees with hepatitis A virus. J Infect Dis. 1975;132:532–545. doi: 10.1093/infdis/132.5.532. [DOI] [PubMed] [Google Scholar]
- 108.LeDuc J.W., Lemon S.M., Keenan C.M., Graham R.R., Marchwicki R.H., Binn L.N. Experimental infection of the New World owl monkey (Aotus trivirgatus) with hepatitis A virus. Infect Immun. 1983;40:766–772. doi: 10.1128/iai.40.2.766-772.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Amado L.A., Marchevsky R.S., de Paula V.S., Hooper C., Freire Mda S., Gaspar A.M. Experimental hepatitis A virus (HAV) infection in cynomolgus monkeys (Macaca fascicularis): evidence of active extrahepatic site of HAV replication. Int J Exp Pathol. 2010;91:87–97. doi: 10.1111/j.1365-2613.2009.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maynard J.E., Lorenz D., Bradley D.W., Feinstone S.M., Krushak D.H., Barker L.F. Review of infectivity studies in nonhuman primates with virus-like particles associated with MS-1 hepatitis. Am J Med Sci. 1975;270:81–85. doi: 10.1097/00000441-197507000-00012. [DOI] [PubMed] [Google Scholar]
- 111.Hornei B., Kammerer R., Moubayed P., Frings W., Gauss-Muller V., Dotzauer A. Experimental hepatitis A virus infection in guinea pigs. J Med Virol. 2001;64:402–409. doi: 10.1002/jmv.1065. [DOI] [PubMed] [Google Scholar]
- 112.Norder H., Ebert J.W., Fields H.A., Mushahwar I.K., Magnius L.O. Complete sequencing of a gibbon hepatitis B virus genome reveals a unique genotype distantly related to the chimpanzee hepatitis B virus. Virology. 1996;218:214–223. doi: 10.1006/viro.1996.0181. [DOI] [PubMed] [Google Scholar]
- 113.Warren K.S., Heeney J.L., Swan R.A., Heriyanto, Verschoor E.J. A new group of hepadnaviruses naturally infecting orangutans (Pongo pygmaeus) J Virol. 1999;73:7860–7865. doi: 10.1128/jvi.73.9.7860-7865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Coffin C.S., Michalak T.I. Persistence of infectious hepadnavirus in the offspring of woodchuck mothers recovered from viral hepatitis. J Clin Invest. 1999;104:203–212. doi: 10.1172/JCI5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dandri M., Petersen J. Animal models of HBV infection. Best Pract Res Clin Gastroenterol. 2017;31:273–279. doi: 10.1016/j.bpg.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 116.Feitelson M.A., Millman I., Halbherr T., Simmons H., Blumberg B.S. A newly identified hepatitis B type virus in tree squirrels. Proc Natl Acad Sci U S A. 1986;83:2233–2237. doi: 10.1073/pnas.83.7.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Testut P., Renard C.A., Terradillos O., Vitvitski-Trepo L., Tekaia F., Degott C. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J Virol. 1996;70:4210–4219. doi: 10.1128/jvi.70.7.4210-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Omata M., Uchiumi K., Ito Y., Yokosuka O., Mori J., Terao K. Duck hepatitis B virus and liver diseases. Gastroenterology. 1983;85:260–267. [PubMed] [Google Scholar]
- 119.Rasche A., Souza B., Drexler J.F. Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr Opin Virol. 2016;16:86–94. doi: 10.1016/j.coviro.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 120.Wieland S.F. The chimpanzee model for hepatitis B virus infection. Cold Spring Harb Perspect Med. 2015;5 doi: 10.1101/cshperspect.a021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lanford R.E., Walker C.M., Lemon S.M. The chimpanzee model of viral hepatitis: advances in understanding the immune response and treatment of viral hepatitis. ILAR J. 2017;58:172–189. doi: 10.1093/ilar/ilx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ruan P., Yang C., Su J., Cao J., Ou C., Luo C. Histopathological changes in the liver of tree shrew (Tupaia belangeri chinensis) persistently infected with hepatitis B virus. Virol J. 2013;10:333. doi: 10.1186/1743-422X-10-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Firth C., Bhat M., Firth M.A., Williams S.H., Frye M.J., Simmonds P. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio. 2014;5:e01933–01914. doi: 10.1128/mBio.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fernandez J., Taylor D., Morhardt D.R., Mihalik K., Puig M., Rice C.M. Long-term persistence of infection in chimpanzees inoculated with an infectious hepatitis C virus clone is associated with a decrease in the viral amino acid substitution rate and low levels of heterogeneity. J Virol. 2004;78:9782–9789. doi: 10.1128/JVI.78.18.9782-9789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bukh J., Apgar C.L., Engle R., Govindarajan S., Hegerich P.A., Tellier R. Experimental infection of chimpanzees with hepatitis C virus of genotype 5a: genetic analysis of the virus and generation of a standardized challenge pool. J Infect Dis. 1998;178:1193–1197. doi: 10.1086/515683. [DOI] [PubMed] [Google Scholar]
- 126.Iwasaki Y., Mori K., Ishii K., Maki N., Iijima S., Yoshida T. Long-term persistent GBV-B infection and development of a chronic and progressive hepatitis C-like disease in marmosets. Front Microbiol. 2011;2:240. doi: 10.3389/fmicb.2011.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bukh J., Apgar C.L., Yanagi M. Toward a surrogate model for hepatitis C virus: An infectious molecular clone of the GB virus-B hepatitis agent. Virology. 1999;262:470–478. doi: 10.1006/viro.1999.9941. [DOI] [PubMed] [Google Scholar]
- 128.Amako Y., Tsukiyama-Kohara K., Katsume A., Hirata Y., Sekiguchi S., Tobita Y. Pathogenesis of hepatitis C virus infection in Tupaia belangeri. J Virol. 2010;84:303–311. doi: 10.1128/JVI.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Negro F., Bergmann K.F., Baroudy B.M., Satterfield W.C., Popper H., Purcell R.H. Chronic hepatitis D virus (HDV) infection in hepatitis B virus carrier chimpanzees experimentally superinfected with HDV. J Infect Dis. 1988;158:151–159. doi: 10.1093/infdis/158.1.151. [DOI] [PubMed] [Google Scholar]
- 130.Ponzetto A., Negro F., Popper H., Bonino F., Engle R., Rizzetto M. Serial passage of hepatitis delta virus in chronic hepatitis B virus carrier chimpanzees. Hepatology. 1988;8:1655–1661. doi: 10.1002/hep.1840080631. [DOI] [PubMed] [Google Scholar]
- 131.Gerin J.L. Animal models of hepatitis delta virus infection and disease. ILAR J. 2001;42:103–106. doi: 10.1093/ilar.42.2.103. [DOI] [PubMed] [Google Scholar]
- 132.Han J., Lei Y., Liu L., Liu P., Xia J., Zhang Y. SPF rabbits infected with rabbit hepatitis E virus isolate experimentally showing the chronicity of hepatitis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Johne R., Plenge-Bonig A., Hess M., Ulrich R.G., Reetz J., Schielke A. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol. 2010;91:750–758. doi: 10.1099/vir.0.016584-0. [DOI] [PubMed] [Google Scholar]
- 134.Li T.C., Yoshizaki S., Ami Y., Suzaki Y., Yasuda S.P., Yoshimatsu K. Susceptibility of laboratory rats against genotypes 1, 3, 4, and rat hepatitis E viruses. Vet Microbiol. 2013;163:54–61. doi: 10.1016/j.vetmic.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 135.Li T.C., Yang T., Yoshizaki S., Ami Y., Suzaki Y., Ishii K. Ferret hepatitis E virus infection induces acute hepatitis and persistent infection in ferrets. Vet Microbiol. 2016;183:30–36. doi: 10.1016/j.vetmic.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 136.Schlosser J., Eiden M., Vina-Rodriguez A., Fast C., Dremsek P., Lange E. Natural and experimental hepatitis E virus genotype 3-infection in European wild boar is transmissible to domestic pigs. Vet Res. 2014;45:121. doi: 10.1186/s13567-014-0121-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schlosser J., Vina-Rodriguez A., Fast C., Groschup M.H., Eiden M. Chronically infected wild boar can transmit genotype 3 hepatitis E virus to domestic pigs. Vet Microbiol. 2015;180:15–21. doi: 10.1016/j.vetmic.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 138.Lin J., Karlsson M., Olofson A.S., Belak S., Malmsten J., Dalin A.M. High prevalence of hepatitis e virus in Swedish moose–a phylogenetic characterization and comparison of the virus from different regions. PLoS One. 2015;10 doi: 10.1371/journal.pone.0122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arankalle V.A., Ticehurst J., Sreenivasan M.A., Kapikian A.Z., Popper H., Pavri K.M. Aetiological association of a virus-like particle with enterically transmitted non-A, non-B hepatitis. Lancet. 1988;1:550–554. doi: 10.1016/s0140-6736(88)91351-7. [DOI] [PubMed] [Google Scholar]
- 140.Tsarev S.A., Emerson S.U., Tsareva T.S., Yarbough P.O., Lewis M., Govindarajan S. Variation in course of hepatitis E in experimentally infected cynomolgus monkeys. J Infect Dis. 1993;167:1302–1306. doi: 10.1093/infdis/167.6.1302. [DOI] [PubMed] [Google Scholar]
- 141.Erker J.C., Desai S.M., Schlauder G.G., Dawson G.J., Mushahwar I.K. A hepatitis E virus variant from the United States: molecular characterization and transmission in cynomolgus macaques. J Gen Virol. 1999;80:681–690. doi: 10.1099/0022-1317-80-3-681. [DOI] [PubMed] [Google Scholar]
- 142.Halbur P.G., Kasorndorkbua C., Gilbert C., Guenette D., Potters M.B., Purcell R.H. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J Clin Microbiol. 2001;39:918–923. doi: 10.1128/JCM.39.3.918-923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hirai-Yuki A., Hensley L., McGivern D.R., Gonzalez-Lopez O., Das A., Feng H. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science. 2016;353:1541–1545. doi: 10.1126/science.aaf8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Glebe D., Bremer C.M. The molecular virology of hepatitis B virus. Semin Liver Dis. 2013;33:103–112. doi: 10.1055/s-0033-1345717. [DOI] [PubMed] [Google Scholar]
- 145.Konig A., Doring B., Mohr C., Geipel A., Geyer J., Glebe D. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J Hepatol. 2014;61:867–875. doi: 10.1016/j.jhep.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 146.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 147.Yu Y., Scheel T.K.H., Luna J.M., Chung H., Nishiuchi E., Scull M.A. miRNA independent hepacivirus variants suggest a strong evolutionary pressure to maintain miR-122 dependence. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Simmonds P., Becher P., Bukh J., Gould E.A., Meyers G., Monath T. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol. 2017;98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Terrault N.A., Lok A.S.F., McMahon B.J., Chang K.M., Hwang J.P., Jonas M.M. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67:1560–1599. doi: 10.1002/hep.29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kamar N., Izopet J., Dalton H.R. Chronic hepatitis e virus infection and treatment. J Clin Exp Hepatol. 2013;3:134–140. doi: 10.1016/j.jceh.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Westbrook R.H., Dusheiko G. Natural history of hepatitis C. J Hepatol. 2014;61:S58–68. doi: 10.1016/j.jhep.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 152.Kamar N., Izopet J., Pavio N., Aggarwal R., Labrique A., Wedemeyer H. Hepatitis E virus infection. Nat Rev Dis Primers. 2017;3:17086. doi: 10.1038/nrdp.2017.86. [DOI] [PubMed] [Google Scholar]
- 153.Hu X., Margolis H.S., Purcell R.H., Ebert J., Robertson B.H. Identification of hepatitis B virus indigenous to chimpanzees. Proc Natl Acad Sci U S A. 2000;97:1661–1664. doi: 10.1073/pnas.97.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Scheel T.K., Kapoor A., Nishiuchi E., Brock K.V., Yu Y., Andrus L. Characterization of nonprimate hepacivirus and construction of a functional molecular clone. Proc Natl Acad Sci U S A. 2015;112:2192–2197. doi: 10.1073/pnas.1500265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cao D., Cao Q.M., Subramaniam S., Yugo D.M., Heffron C.L., Rogers A.J. Pig model mimicking chronic hepatitis E virus infection in immunocompromised patients to assess immune correlates during chronicity. Proc Natl Acad Sci U S A. 2017;114:6914–6923. doi: 10.1073/pnas.1705446114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Global Burden of Disease Liver Cancer C. Akinyemiju T., Abera S., Ahmed M., Alam N., Alemayohu M.A. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lok A.S. Chronic hepatitis B. N Engl J Med. 2002;346:1682–1683. doi: 10.1056/NEJM200205303462202. [DOI] [PubMed] [Google Scholar]
- 158.Ghouri Y.A., Mian I., Rowe J.H. Review of hepatocellular carcinoma: epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16:1. doi: 10.4103/jcar.JCar_9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mason W.S. Animal models and the molecular biology of hepadnavirus infection. Cold Spring Harb Perspect Med. 2015;5 doi: 10.1101/cshperspect.a021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hansen L.J., Tennant B.C., Seeger C., Ganem D. Differential activation of myc gene family members in hepatic carcinogenesis by closely related hepatitis B viruses. Mol Cell Biol. 1993;13:659–667. doi: 10.1128/mcb.13.1.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Seeger C., Mason W.S. Molecular biology of hepatitis B virus infection. Virology. 2015;479–480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sung W.K., Zheng H., Li S., Chen R., Liu X., Li Y. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 163.Zhao L.H., Liu X., Yan H.X., Li W.Y., Zeng X., Yang Y. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7:12992. doi: 10.1038/ncomms12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mesri E.A., Feitelson M.A., Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Decorsiere A., Mueller H., van Breugel P.C., Abdul F., Gerossier L., Beran R.K. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–389. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 166.Trivedi S., Murthy S., Sharma H., Hartlage A.S., Kumar A., Gadi S.V. Viral persistence, liver disease, and host response in a hepatitis C-like virus rat model. Hepatology. 2017 doi: 10.1002/hep.29494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wilkinson G.S., South J.M. Life history, ecology and longevity in bats. Aging Cell. 2002;1:124–131. doi: 10.1046/j.1474-9728.2002.00020.x. [DOI] [PubMed] [Google Scholar]
- 168.Wang L.F., Walker P.J., Poon L.L. Mass extinctions, biodiversity and mitochondrial function: are bats 'special' as reservoirs for emerging viruses? Curr Opin Virol. 2011;1:649–657. doi: 10.1016/j.coviro.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Brown Z.J., Heinrich B., Greten T.F. Mouse models of hepatocellular carcinoma: an overview and highlights for immunotherapy research. Nat Rev Gastroenterol Hepatol. 2018;15:536–554. doi: 10.1038/s41575-018-0033-6. [DOI] [PubMed] [Google Scholar]
- 170.Feigelstock D., Thompson P., Mattoo P., Zhang Y., Kaplan G.G. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J Virol. 1998;72:6621–6628. doi: 10.1128/jvi.72.8.6621-6628.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Muller S.F., Konig A., Doring B., Glebe D., Geyer J. Characterisation of the hepatitis B virus cross-species transmission pattern via Na+/taurocholate co-transporting polypeptides from 11 New World and Old World primate species. PLoS One. 2018;13 doi: 10.1371/journal.pone.0199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fu L., Hu H., Liu Y., Jing Z., Li W. Woodchuck sodium taurocholate cotransporting polypeptide supports low-level hepatitis B and D virus entry. Virology. 2017;505:1–11. doi: 10.1016/j.virol.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 173.He W., Cao Z., Mao F., Ren B., Li Y., Li D. Modification of three amino acids in sodium taurocholate cotransporting polypeptide renders mice susceptible to infection with hepatitis D virus in vivo. J Virol. 2016;90:8866–8874. doi: 10.1128/JVI.00901-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lempp F.A., Wiedtke E., Qu B., Roques P., Chemin I., Vondran F.W.R. Sodium taurocholate cotransporting polypeptide is the limiting host factor of hepatitis B virus infection in macaque and pig hepatocytes. Hepatology. 2017;66:703–716. doi: 10.1002/hep.29112. [DOI] [PubMed] [Google Scholar]
- 175.Kock J., Nassal M., MacNelly S., Baumert T.F., Blum H.E., von Weizsacker F. Efficient infection of primary tupaia hepatocytes with purified human and woolly monkey hepatitis B virus. J Virol. 2001;75:5084–5089. doi: 10.1128/JVI.75.11.5084-5089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bissig K.D., Wieland S.F., Tran P., Isogawa M., Le T.T., Chisari F.V. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120:924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.von Schaewen M., Dorner M., Hueging K., Foquet L., Gerges S., Hrebikova G. Expanding the host range of hepatitis C virus through viral adaptation. mBio. 2016;7 doi: 10.1128/mBio.01915-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yang Y., Liang Y., Qu L., Chen Z., Yi M., Li K. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci U S A. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Horner S.M., Gale M., Jr. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19:879–888. doi: 10.1038/nm.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Parera M., Martrus G., Franco S., Clotet B., Martinez M.A. Canine hepacivirus NS3 serine protease can cleave the human adaptor proteins MAVS and TRIF. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Anggakusuma Brown RJ, Banda D.H., Todt D., Vieyres G., Steinmann E. Hepacivirus NS3/4A proteases interfere with MAVS signaling in both their cognate animal hosts and humans: implications for zoonotic transmission. J Virol. 2016;90:10670–10681. doi: 10.1128/JVI.01634-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Morse S.S., Mazet J.A., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kulkarni M.A., Walimbe A.M., Cherian S., Arankalle V.A. Full length genomes of genotype IIIA Hepatitis A virus strains (1995–2008) from India and estimates of the evolutionary rates and ages. Infect Genet Evol. 2009;9:1287–1294. doi: 10.1016/j.meegid.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 184.Forni D., Cagliani R., Pontremoli C., Pozzoli U., Vertemara J., De Gioia L. Evolutionary analysis provides insight into the origin and adaptation of HCV. Front Microbiol. 2018;9:854. doi: 10.3389/fmicb.2018.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Forni D., Cagliani R., Clerici M., Sironi M. Origin and dispersal of Hepatitis E virus. Emerg Microbes Infect. 2018;7:11. doi: 10.1038/s41426-017-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhou Y., Holmes E.C. Bayesian estimates of the evolutionary rate and age of hepatitis B virus. J Mol Evol. 2007;65:197–205. doi: 10.1007/s00239-007-0054-1. [DOI] [PubMed] [Google Scholar]
- 187.Paraskevis D., Angelis K., Magiorkinis G., Kostaki E., Ho S.Y., Hatzakis A. Dating the origin of hepatitis B virus reveals higher substitution rate and adaptation on the branch leading to F/H genotypes. Mol Phylogenet Evol. 2015;93:44–54. doi: 10.1016/j.ympev.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 188.Zehender G., Ebranati E., Gabanelli E., Sorrentino C., Lo Presti A., Tanzi E. Enigmatic origin of hepatitis B virus: an ancient travelling companion or a recent encounter? World J Gastroenterol. 2014;20:7622–7634. doi: 10.3748/wjg.v20.i24.7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kahila Bar-Gal G., Kim M.J., Klein A., Shin D.H., Oh C.S., Kim J.W. Tracing hepatitis B virus to the 16th century in a Korean mummy. Hepatology. 2012;56:1671–1680. doi: 10.1002/hep.25852. [DOI] [PubMed] [Google Scholar]
- 190.Muhlemann B., Jones T.C., Damgaard P.B., Allentoft M.E., Shevnina I., Logvin A. Ancient hepatitis B viruses from the Bronze Age to the Medieval period. Nature. 2018;557:418–423. doi: 10.1038/s41586-018-0097-z. [DOI] [PubMed] [Google Scholar]
- 191.Krause-Kyora B., Susat J., Key F.M., Kuhnert D., Bosse E., Immel A. Neolithic and medieval virus genomes reveal complex evolution of hepatitis B. eLife. 2018;7 doi: 10.7554/eLife.36666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Suh A., Brosius J., Schmitz J., Kriegs J.O. The genome of a Mesozoic paleovirus reveals the evolution of hepatitis B viruses. Nat Commun. 2013;4:1791. doi: 10.1038/ncomms2798. [DOI] [PubMed] [Google Scholar]
- 193.Cui J., Holmes E.C. Endogenous hepadnaviruses in the genome of the budgerigar (Melopsittacus undulatus) and the evolution of avian hepadnaviruses. J Virol. 2012;86:7688–7691. doi: 10.1128/JVI.00769-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu W., Pan S., Yang H., Bai W., Shen Z., Liu J. The first full-length endogenous hepadnaviruses: identification and analysis. J Virol. 2012;86:9510–9513. doi: 10.1128/JVI.01164-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gilbert C., Meik J.M., Dashevsky D., Card D.C., Castoe T.A., Schaack S. Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc Biol Sci R Soc. 2014;281:20141122. doi: 10.1098/rspb.2014.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Foley N.M., Springer M.S., Teeling E.C. Mammal madness: is the mammal tree of life not yet resolved? Philos Trans R Soc Lond B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang X., Ren J., Gao Q., Hu Z., Sun Y., Li X. Hepatitis A virus and the origins of picornaviruses. Nature. 2015;517:85–88. doi: 10.1038/nature13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gong Z., Han G.Z. Insect retroelements provide novel insights into the origin of hepatitis B viruses. Mol Biol Evol. 2018;35(9):2254–2259. doi: 10.1093/molbev/msy129. [DOI] [PubMed] [Google Scholar]
- 199.Shi M., Lin X.D., Tian J.H., Chen L.J., Chen X., Li C.X. Redefining the invertebrate RNA virosphere. Nature. 2016 doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 200.Shi M., Lin X.D., Vasilakis N., Tian J.H., Li C.X., Chen L.J. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the flaviviridae and related viruses. J Virol. 2016;90:659–669. doi: 10.1128/JVI.02036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rota-Stabelli O., Daley A.C., Pisani D. Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr Biol. 2013;23:392–398. doi: 10.1016/j.cub.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 202.Tedder R.S., Bissett S.L., Myers R., Ijaz S. The ‘Red Queen’ dilemma--running to stay in the same place: reflections on the evolutionary vector of HBV in humans. Antivir Ther. 2013;18:489–496. doi: 10.3851/IMP2655. [DOI] [PubMed] [Google Scholar]
- 203.Shi M., Lin X.D., Chen X., Tian J.H., Chen L.J., Li K. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 204.Guan D., Li W., Su J., Fang L., Takeda N., Wakita T. Asian musk shrew as a reservoir of rat hepatitis E virus, China. Emerg Infect Dis. 2013;19:1341–1343. doi: 10.3201/eid1908.130069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hartlage A.S., Cullen J.M., Kapoor A. The strange, expanding world of animal hepaciviruses. Annu Rev Virol. 2016;3:53–75. doi: 10.1146/annurev-virology-100114-055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Longdon B., Brockhurst M.A., Russell C.A., Welch J.J., Jiggins F.M. The evolution and genetics of virus host shifts. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tebit D.M., Arts E.J. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis. 2011;11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 208.Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82:693–712. doi: 10.1099/0022-1317-82-4-693. [DOI] [PubMed] [Google Scholar]
- 209.Luis A.D., Hayman D.T., O'Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Han B.A., Schmidt J.P., Bowden S.E., Drake J.M. Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci U S A. 2015;112:7039–7044. doi: 10.1073/pnas.1501598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wu Z., Lu L., Du J., Yang L., Ren X., Liu B. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome. 2018;6:178. doi: 10.1186/s40168-018-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kapoor A., Simmonds P., Scheel T.K., Hjelle B., Cullen J.M., Burbelo P.D. Identification of rodent homologs of hepatitis C virus and pegiviruses. mBio. 2013;4:e00216–00213. doi: 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Alvarado-Mora M.V., Romano C.M., Gomes-Gouvea M.S., Gutierrez M.F., Carrilho F.J., Pinho J.R. Dynamics of hepatitis D (delta) virus genotype 3 in the Amazon region of South America. Infect Genet Evol. 2011;11:1462–1468. doi: 10.1016/j.meegid.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 216.Nielsen R., Akey J.M., Jakobsson M., Pritchard J.K., Tishkoff S., Willerslev E. Tracing the peopling of the world through genomics. Nature. 2017;541:302–310. doi: 10.1038/nature21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Junglen S., Drosten C. Virus discovery and recent insights into virus diversity in arthropods. Curr Opin Microbiol. 2013;16:507–513. doi: 10.1016/j.mib.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Pybus O.G., Theze J. Hepacivirus cross-species transmission and the origins of the hepatitis C virus. Curr Opin Virol. 2016;16:1–7. doi: 10.1016/j.coviro.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 219.Provost P.J., Hilleman M.R. Propagation of human hepatitis A virus in cell culture in vitro. Proc Soc Exp Biol Med. 1979;160:213–221. doi: 10.3181/00379727-160-40422. [DOI] [PubMed] [Google Scholar]
- 220.Najarian R., Caput D., Gee W., Potter S.J., Renard A., Merryweather J. Primary structure and gene organization of human hepatitis A virus. Proc Natl Acad Sci U S A. 1985;82:2627–2631. doi: 10.1073/pnas.82.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mao J.S., Dong D.X., Zhang H.Y., Chen N.L., Zhang X.Y., Huang H.Y. Primary study of attenuated live hepatitis A vaccine (H2 strain) in humans. J Infect Dis. 1989;159:621–624. doi: 10.1093/infdis/159.4.621. [DOI] [PubMed] [Google Scholar]
- 222.Blumberg B.S., Alter H.J., Visnich S. A “new” antigen in leukemia sera. JAMA. 1965;191:541–546. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 223.Greenberg H.B., Pollard R.B., Lutwick L.I., Gregory P.B., Robinson W.S., Merigan T.C. Effect of human leukocyte interferon on hepatitis B virus infection in patients with chronic active hepatitis. N Engl J Med. 1976;295:517–522. doi: 10.1056/NEJM197609022951001. [DOI] [PubMed] [Google Scholar]
- 224.Galibert F., Mandart E., Fitoussi F., Tiollais P., Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 225.Szmuness W., Stevens C.E., Harley E.J., Zang E.A., Oleszko W.R., William D.C. Hepatitis B vaccine: demonstration of efficacy in a controlled clinical trial in a high-risk population in the United States. N Engl J Med. 1980;303:833–841. doi: 10.1056/NEJM198010093031501. [DOI] [PubMed] [Google Scholar]
- 226.Hoofnagle J.H., Mullen K.D., Jones D.B., Rustgi V., Di Bisceglie A., Peters M. Treatment of chronic non-A, non-B hepatitis with recombinant human alpha interferon. A preliminary report. N Engl J Med. 1986;315:1575–1578. doi: 10.1056/NEJM198612183152503. [DOI] [PubMed] [Google Scholar]
- 227.Yanagi M., Purcell R.H., Emerson S.U., Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci U S A. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bacon B.R., Gordon S.C., Lawitz E., Marcellin P., Vierling J.M., Zeuzem S. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sherman K.E., Flamm S.L., Afdhal N.H., Nelson D.R., Sulkowski M.S., Everson G.T. Response-guided telaprevir combination treatment for hepatitis C virus infection. N Engl J Med. 2011;365:1014–1024. doi: 10.1056/NEJMoa1014463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rizzetto M., Canese M.G., Arico S., Crivelli O., Trepo C., Bonino F. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut. 1977;18:997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kos A., Dijkema R., Arnberg A.C., van der Meide P.H., Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 233.Wang K.S., Choo Q.L., Weiner A.J., Ou J.H., Najarian R.C., Thayer R.M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature. 1986;323:508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 234.Khuroo M.S. Study of an epidemic of non-A, non-B hepatitis. Possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am J Med. 1980;68:818–824. doi: 10.1016/0002-9343(80)90200-4. [DOI] [PubMed] [Google Scholar]
- 235.Reyes G.R., Yarbough P.O., Tam A.W., Purdy M.A., Huang C.C., Kim J.S. Hepatitis E virus (HEV): the novel agent responsible for enterically transmitted non-A, non-B hepatitis. Gastroenterol Jpn. 1991;26:142–147. doi: 10.1007/BF02779285. [DOI] [PubMed] [Google Scholar]
- 236.Meng X.J., Purcell R.H., Halbur P.G., Lehman J.R., Webb D.M., Tsareva T.S. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Husain M.M., Aggarwal R., Kumar D., Jameel S., Naik S. Effector T cells immune reactivity among patients with acute hepatitis E. J Viral Hepat. 2011;18:e603–608. doi: 10.1111/j.1365-2893.2011.01489.x. [DOI] [PubMed] [Google Scholar]
- 238.Kamar N., Izopet J., Tripon S., Bismuth M., Hillaire S., Dumortier J. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N Engl J Med. 2014;370:1111–1120. doi: 10.1056/NEJMoa1215246. [DOI] [PubMed] [Google Scholar]
- 239.Inoue J.G., Miya M., Lam K., Tay B.H., Danks J.A., Bell J. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol Biol Evol. 2010;27:2576–2586. doi: 10.1093/molbev/msq147. [DOI] [PubMed] [Google Scholar]
- 240.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Pourkarim M.R., Amini-Bavil-Olyaee S., Kurbanov F., Van Ranst M., Tacke F. Molecular identification of hepatitis B virus genotypes/subgenotypes: revised classification hurdles and updated resolutions. World J Gastroenterol. 2014;20:7152–7168. doi: 10.3748/wjg.v20.i23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 245.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Cristina J., Costa-Mattioli M. Genetic variability and molecular evolution of hepatitis A virus. Virus Res. 2007;127:151–157. doi: 10.1016/j.virusres.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 247.Littlejohn M., Locarnini S., Yuen L. Origins and evolution of hepatitis B virus and hepatitis D virus. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol. 2014;20:5427–5434. doi: 10.3748/wjg.v20.i18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Messina J.P., Humphreys I., Flaxman A., Brown A., Cooke G.S., Pybus O.G. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Murphy D.G., Sablon E., Chamberland J., Fournier E., Dandavino R., Tremblay C.L. Hepatitis C virus genotype 7, a new genotype originating from central Africa. J Clin Microbiol. 2015;53:967–972. doi: 10.1128/JCM.02831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Utsumi T., Wahyuni R.M., Lusida M.I., Yano Y., Priambada N.P., Amin M. Full genome characterization and phylogenetic analysis of hepatitis B virus in gibbons and a caretaker in Central Kalimantan, Indonesia. Arch Virol. 2015;160:685–692. doi: 10.1007/s00705-014-2323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ramulu H.G., Raoult D., Pontarotti P. The rhizome of life: what about metazoa? Front Cell Infect Microbiol. 2012;2:50. doi: 10.3389/fcimb.2012.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sridhar S., Yip C.C.Y., Wu S., Cai J., Zhang A.J., Leung K.H. Rat Hepatitis E Virus as Cause of Persistent Hepatitis after Liver Transplant. Emerging Infect Dis. 2018;24:2241–2250. doi: 10.3201/eid2412.180937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Hiller T., Rasche A., Brändel S.D., König A., Jeworowski L., Teague O'Mara M. Host Biology and Anthropogenic Factors Affect Hepadnavirus Infection in a Neotropical Bat. EcoHealth. 2019 doi: 10.1007/s10393-018-1387-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.