Abstract
Background/Aims
Following lead optimization, a set of substituted imidazopyridines was identified as potent and selective inhibitors of in vitro HCV replication. The particular characteristics of one of the most potent compounds in this series (5-[[3-(4-chlorophenyl)-5-isoxazolyl]methyl]-2-(2,3-difluorophenyl)-5H-imidazo[4,5-c]pyridine or GS-327073), were studied.
Methods
Antiviral activity of GS-327073 was evaluated in HCV subgenomic replicons (genotypes 1b, 1a and 2a), in the JFH1 (genotype 2a) infectious system and against replicons resistant to various selective HCV inhibitors. Combination studies of GS-327073 with other selective HCV inhibitors were performed.
Results
Fifty percent effective concentrations for inhibition of HCV subgenomic 1b replicon replication ranged between 2 and 50 nM and were 100-fold higher for HCV genotype 2a virus. The 50% cytostatic concentrations were ⩾17 μM, thus resulting in selectivity indices of ⩾340. GS-327073 retained wild-type activity against HCV replicons that were resistant to either HCV protease inhibitors or several polymerase inhibitors. GS-327073, when combined with either interferon α, ribavirin, a nucleoside polymerase or a protease inhibitor resulted in overall additive antiviral activity. Combinations containing GS-327073 proved highly effective in clearing hepatoma cells from HCV.
Conclusions
GS-327073 is a potent in vitro inhibitor of HCV replication either alone or in combination with other selective HCV inhibitors.
Abbreviations: RdRp, RNA-dependent RNA polymerase; 2′-C-MeCyt, 2′-C-methylcytidine; CC50, 50% cytotoxic concentration; EC50, 50% effective concentration; HCVcc, cell culture–grown chimeric HCV
Keywords: Non-nucleoside, HCV RdRp inhibitor, Combination, Viral clearance
1. Introduction
An estimated 170–180 million people worldwide are chronically infected with HCV and are thus at increased risk of developing liver cirrhosis, hepatocellular carcinoma, liver failure and end-stage liver disease [1]. The current standard therapy, i.e. pegylated interferon in combination with ribavirin, is associated with significant side-effects; moreover, a sustained virological response is (depending on the genotype) obtained in only 50–60% of the patients [2], [3]. Hence, there is an urgent need for new potent, selective and safe drugs for the treatment of HCV infections [4], [5].
Several potent and selective inhibitors of HCV replication, most of which target the NS3 protease and the NS5B RNA-dependent RNA polymerase (RdRp), have been developed in recent years [4], [6], [7]. Other viral (such as NS4A) [8] and cellular targets (such as cyclophylins) [9] involved in the HCV lifecycle are also being explored. The efficacy of a number of inhibitors has been or is being studied in patients chronically infected with HCV [10].
Both nucleoside and non-nucleoside inhibitors of the HCV RdRp have been reported. Nucleoside HCV polymerase inhibitors act as premature chain terminators following conversion to their 5′-triphosphate metabolite and incorporation in the viral genome. 2′-C-Methylcytidine (2′-C-MeCyt) [11] was the first nucleoside HCV inhibitor to enter clinical studies. Development of this compound has been discontinued in the light of modest antiviral efficacy along with significant gastrointestinal side effects [12]. The efficacy of R1626, a prodrug of 4′-azidocytidine, is currently being evaluated in combination with standard therapy [13]. β-d-2′-Deoxy-2′-fluoro-2′-C-methylcytidine (prodrug R7128) has demonstrated promising efficacy in phase I studies in chronically infected patients [14]. Various non-nucleoside inhibitor classes of the HCV RdRp, including benzimidazoles [15], [16], [17], indoles [18], [19], [20], [21], thiophene-based carboxylic acids [22], [23], phenylalanines [24], [25], dihydropyranones [26], [27], pyranoindoles [28], [29], [30], proline sulfonamides [31], benzothiadiazines [32], [33], [34], [35], [36], benzylidene derivatives [37] and acrylic acids [38], [39] have been reported, but optimization was not further pursued or clinical development has been stopped because of serious adverse effects. Efficacy of the benzofuran non-nucleoside RdRp inhibitor (HCV-796) was demonstrated in infected patients [40], [41] but clinical development was stopped because of potential hepatotoxicity. VCH-759, a new non-nucleoside thiophene-2-carboxylic acid inhibitor of RdRp causes a 2.5 log10 decrease in HCV RNA after 10 days of treatment in HCV-infected patients and was generally well-tolerated in early clinical studies [42].
We recently reported a novel class of substituted imidazopyridines capable of selectively inhibiting HCV replication [43], [44]. The analogue 2-(2-fluorophenyl)-5-[4-bromobenzyl]-5H-imidazo[4,5-c]pyridine served as the starting point for further lead optimization, which resulted in the discovery of a series of highly potent inhibitors of in vitro HCV replication. Here, we report on the anti-HCV activity and the particular characteristics of the activity of an optimized analogue, 5-[[3-(4-chlorophenyl)-5-isoxazolyl]methyl]-2-(2,3-difluorophenyl)-5H-imidazo[4,5-c]pyridine (GS-327073).
2. Materials and methods
2.1. Compounds
The synthesis of the initial lead compound, i.e. 5-[4-bromophenyl)methyl]-2-phenyl-5H-imidazo[4,5-c]pyridine (BPIP), has been reported earlier [43], [45]. The lead optimization process and the synthesis of GS-327073 or (5-[[3-(4-chlorophenyl)-5-isoxazolyl]methyl]-2-(2,3-difluorophenyl)-5H-imidazo[4,5-c]pyridine) will be reported elsewhere. Reference compounds [VX-950, BILN-2061, SCH 503034, 4′-azidocytidine, 2′-C-MeCyt, GSK-4, HCV-796], were synthesized as reported [11], [32], [46], [47], [48], [49], [50], [51], [52]. Ribavirin was purchased from ICN Pharmaceuticals (Costa Mesa, California) and recombinant IFNα 2b (intron® A) was purchased from Schering Plough (Kenilworth, NJ, USA).
2.2. Cell cultures
Huh 7 cells carrying subgenomic HCV replicon I377NS3-3′/wt [Huh 9-13] [53] or I389luc-ubi-neo/NS3-3′/5.1 [Huh 5-2] [54] were kindly provided by R. Bartenschlager, (University of Heidelberg, Heidelberg, Germany). HuH6 replicons [55] have a similar genetic make-up as Huh 5-2 replicons, but (i) replicate in a different cell clone, (ii) carry different adaptive mutations and (iii) replicon replication is not inhibited by IFNγ and relatively independent of cell proliferation. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Merelbeke, Belgium) supplemented with 10% heat-inactivated foetal calf serum (Integro, Zaandam, The Netherlands), 1× MEM non essential amino acids solution without l-glutamine (Gibco), 100 IU of penicillin/ml and 100 μg of streptomycin/ml (Gibco) and 1 mg/ml G418 (Geneticin® Selective Antibiotic, Gibco) for Huh 9-13 and HuH6 or 250 μg/ml G418 for Huh 5-2 cells. Huh-Lunet cells were cultured without G418. Replicons resistant to the protease inhibitors BILN-2061 and VX-950, the nucleoside polymerase inhibitors 4′-azidocytidine and 2′-C-MeCyt, or the non-nucleoside RdRP inhibitor benzothiadiazine (GSK-4) were generated by selective pressure using increasing concentrations of the respective compounds.
2.3. Anti-HCV assay in replicon containing cells
Huh 9-13, HuH6 and Huh 5-2 cells were seeded at a density of 5 × 103 cells per well in 96-well cell culture plates [Huh 9-13, HuH6 (Iwaki, Asahi Techno Glass, Japan)] or in tissue culture treated white 96-well view plates [Huh 5-2 (Packard, Canberra, Canada)] in complete DMEM without G418. Antiviral assays were carried out as reported earlier [56]. Read-out for Huh 5-2 cells was a luciferase assay, quantification of viral RNA in Huh 9-13 and HuH6 was performed by means of real-time PCR. For Huh 5-2 cells, the EC50 was defined as the concentration of compound that reduced the firefly luciferase signal by 50%; for Huh 9-13 and HuH6 cells as the concentration of compound that reduced the amount of HCV RNA by 50%.
2.4. Antiviral assays with HCVcc virus
A J6/JFH chimeric HCV reporter virus (J6/JFH-Rluc2a-FLAG) was constructed [57]. Infectious virus stocks were generated by transfecting in vitro transcribed RNA into Huh-Lunet cells and harvesting culture medium from the transfected cells. To test antiviral activity of compounds against this chimeric 2a virus, Huh-Lunet cells in 96-well plates (5000 cells per well) were infected with the virus (MOI = 0.1) after which serial dilutions of compounds were added. After three days of incubation, medium was removed, cells were washed once with PBS and lysed using a lysis buffer containing 1% Triton X-100. Viral replication was monitored by measuring Renilla luciferase activity according to the manufacturer’s protocol (Promega, Madison, WI), or by RNA extraction and real-time quantitative RT-PCR. Antiviral data were fit to the logistic dose response equation y = a/(1 + (x/b)c) using XLFit software (IDBS, Emmeryville, CA), and EC50 values were calculated from the resulting equations as described previously [58].
2.5. Cytostatic assay
Replicon-containing cells (Huh 5-2, HuH6 or Huh 9-13) were seeded at a density of 5 × 103 cells per well in complete DMEM without G418. The 50% cytotoxic concentration (CC50), which is defined as the concentration that inhibits the proliferation of exponentially growing cells by 50%, was determined using the MTS/PMS method previously described [56].
2.6. Drug combinations
The effects of drug–drug combinations were studied in HuH6 cells, in which replicon replication is largely independent of cell proliferation (which is important when planning combination studies with a cytostatic drug such as ribavirin) [55]. Cells were seeded in complete DMEM without G418 and compounds were added to the cells in the cell culture plates in checkerboard format. Combinations for each pair of compounds were at least performed in three independent experiments. Data were evaluated using the method of Prichard and Shipman [59]. Briefly, the theoretical additive effect is calculated from the dose–response curves of individual compounds by the equation Z = X + Y(1 − X), where X represents the inhibition produced by compound 1 and Y represents that of compound 2. Z represents the effect produced by the combination of compound 1 and compound 2. The theoretical additive surface is subtracted from the actual experimental surface, resulting in a horizontal surface that equals the zero plane when the combination is additive. Data points more than 20% above the zero plane indicate synergistic activity, data points lower than 20% under the zero plane indicate antagonistic activity.
2.7. HCV replicon clearance-rebound assay
Huh 9-13 cells were seeded in 25 cm2 culture flasks at a cell density of 3 × 105 cells per condition; a sample of 1.5 × 105 Huh 9-13 cells was collected in RLT buffer (RNeasy mini kit (Qiagen) [“Clearance 0” sample] and stored at −80 °C until further use. Cells were cultured for 24 h, after which standard cell culture medium containing 1 mg/ml G418 was replaced with medium that contained either no antiviral drug or various concentrations of GS-327073, IFNα 2b, 2′-C-MeCyt or VX-950 (all conditions in the absence of G418). Cells were incubated until 90% confluency was reached. Cells were subsequently subcultured at a density of 3 × 105 per 25 cm2 culture flask containing the same concentration of antiviral drug without G418 (clearance “Cx”) or were subcultured in culture medium containing 1 mg/ml G418 in the absence of antiviral drug (rebound “Rx”). At every subculture step a sample of 1.5 × 105 cells was lysed in RLT buffer and stored at −80 °C. After collection of all samples, RNA was extracted and samples were analyzed by quantitative real-time PCR for their HCV replicon content. Real-time RT-PCR values of all assayed samples were normalized against the “no-drug control” of the same passage. Cells were cultured under clearance and rebound conditions for 3 passages and 4 passages, respectively. Cells that had lost the replicon died in the presence of G418 during the rebound; cells that still contained the replicon were able to survive under G418 pressure.
2.8. Antiviral assays with other viruses
Antiviral assays for herpes simplex virus-1 (KOS strain, TK− KOS ACVr), herpes simplex virus-2 (G strain, herpesviridae), vaccinia virus (poxviridae), vesicular stomatitis virus (rhabdoviridae), coxsackie virus B4 (picornaviridae), respiratory syncytial virus (paramyxoviridae), para-influenza-3 virus (paramyxoviridae), reovirus-1 (reoviridae), sindbis virus (alphaviridae), influenza A virus (H1N1 and H3N2), influenza B virus (orthomyxoviridae), Punta Toro virus (bunyaviridae), HIV-1 (IIIB) and HIV-2 (ROD, retroviridae), YFV-17D (flaviviridae), bovine viral diarrhea virus (BVDV, flaviviridae) and feline coronavirus (coronaviridae) were carried out as reported before [45], [60], [61], [62], [63], [64], [65], [66].
3. Results
3.1. GS-327073 is a potent inhibitor of in vitro HCV replication
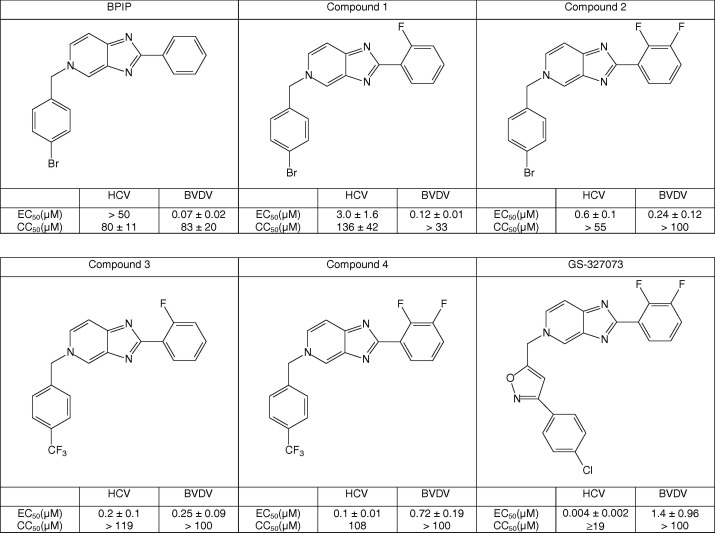
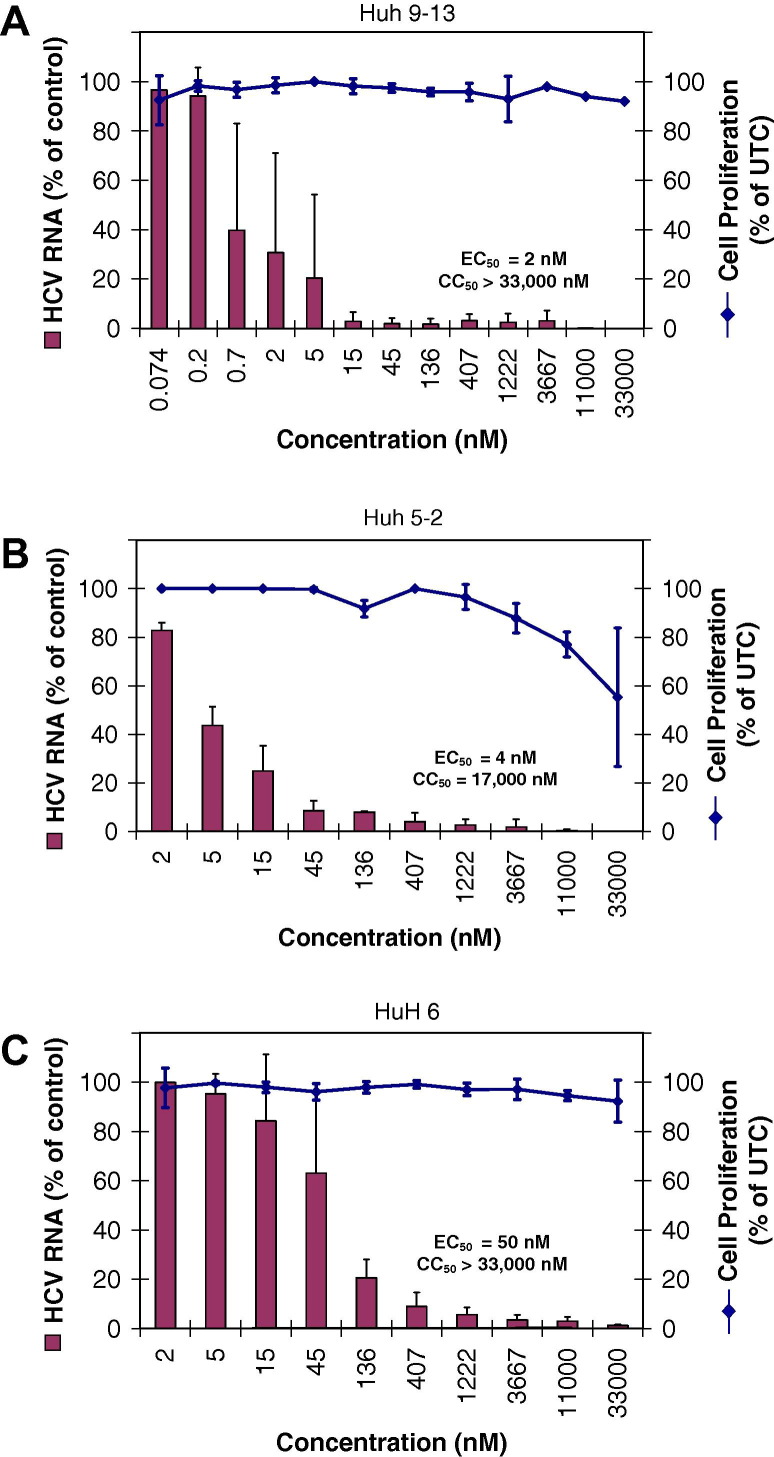
We recently reported the identification of a novel class of 2,5-disubstituted imidazo[4,5-c]pyridines as potent inhibitors of in vitro pestivirus replication [67]. Introduction of a fluorine at position 2 of the phenyl of the lead anti-pestivirus compound 5-[4-bromophenyl)methyl]-2-phenyl-5H-imidazo[4,5-c]pyridine (BPIP) resulted in a molecule that not only exhibited anti-BVDV but also anti-HCV activity (compound 1; EC50 HCV = 3.0 μM; CC50 = 136 μM) (Fig. 1 ) [43]. Introduction of two fluorines on the 2-phenyl ring (2,3-difluoro analogue; compound 2) resulted in a molecule that proved 5-fold more potent against HCV than compound 1 and that retained the anti-BVDV activity. In an attempt to further optimize the anti-HCV activity of this class of compounds, a set of analogues with modifications to the 4-bromo substituent on the 5-benzyl residue (for example the introduction of a trifluoromethyl; compounds 3 and 4) was synthesized [44], resulting in compounds with a 15- to 30-fold increased anti-HCV activity as compared to compound 1. Interestingly, whereas these modifications resulted in an increased anti-HCV activity, there was relatively little effect on BVDV activity. Subsequently, a set of analogues with more extensive modifications at the 5-benzyl position was synthesized (data not shown). This resulted in the identification of a series of analogues with highly potent low nM range anti-HCV activity. One of these, i.e. GS-327073 or 5-[[3-(4-chlorophenyl)-5-isoxazolyl]methyl]-2-(2,3-difluorophenyl)-5H-imidazo[4,5-c]pyridine, was selected for further characterization of the in vitro anti-HCV activity. The compound inhibited HCV (genotype 1) subgenomic replicon replication with mean EC50 values ranging between 2 and 50 nM in a dose-dependent manner (Table 1 , Fig. 2 ). GS-327073 proved markedly more potent than the protease inhibitors BILN-2061, VX-950 and SCH 503034, the nucleoside polymerase inhibitors 2′-C-MeCyt and 4′-azidocytidine and the non-nucleoside polymerase inhibitor GSK-4. GS-327073 inhibited the replication of cell culture–grown chimeric HCV (HCVcc, genotype 2a) designated J6/JFH-Rluc-FLAG in a dose-dependent manner, although less efficiently than the replication of genotype 1b replicon. The antiviral activity of GS-327073 against the J6/JFH-Rluc-FLAG virus was however comparable to the activity of the reference protease or polymerase inhibitors studied.
Fig. 1.
Structural formulae and an overview of some of the major steps in the lead optimization towards GS-327073.
Table 1.
Activity of GS-327073 against various HCV genotype 1b replicons and J6/JFH-Rluc-flag.
| Anti-HCV activity EC50 (μM) |
CC50 (μM) | ||||
|---|---|---|---|---|---|
| Huh 9-13a | Huh 5-2a | HuH6a | J6/JFHb | ||
| GS-327073 | 0.002 ± 0.004 | 0.004 ± 0.002 | 0.05 ± 0.02 | 0.33 ± 0.14 | ⩾17 |
| IFNα 2b (IU/ml) | 0.24 ± 0.17 | 0.10 ± 0.06 | 22 ± 5.4 | 1.9 ± 0.07 | n.d. |
| Ribavirin | n.d. | 38 ± 12 | 28 ± 16 | n.d. | >33 |
| BILN-2061 | 0.01 ± 0.01 | 0.03 ± 0.03 | 0.004 ± 0.002 | n.d. | ⩾22 |
| VX-950 | 0.58 ± 0.10 | 1.1 ± 0.71 | 1.4 ± 0.49 | 0.29 ± 0.11 | ⩾24 |
| SCH 503034 | 0.37 ± 0.26 | 0.93 ± 0.39 | 0.35 ± 0.07 | 0.34c | ⩾7.4 |
| 2′-C-MeCyt | 0.43 ± 0.35 | 2.7 ± 1.5 | 1.02 ± 0.44 | 0.10 ± 0.02 | ⩾26 |
| 4′-Azidocytidine | 2.9 ± 0.87 | 1.4 ± 0.63 | 7.4 ± 3.7 | n.d. | >33 |
| GSK-4 | 0.94 ± 0.62 | 1.8 ± 0.42 | 11 ± 0.81 | n.d. | >33 |
| HCV-796 | 0.08 ± 0.05 | 0.04 ± 0.01 | 0.01 ± 0.003 | <0.1d | >33 |
Data are mean values ± SD from at least three independent experiments.
n.d., not determined.
As determined by a luciferase (Huh 5-2) or RT-qPCR assay (Huh 9-13, HuH6).
As determined by a luciferase assay.
Data from 1 single experiment.
Mean from 2 independent experiments.
Fig. 2.
Dose–response curves for inhibition of HCV replicon replication in (A) Huh 9-13, (B) Huh 5-2 and (C) HuH6 cells by GS-327073.
GS-327073 also inhibited BVDV replication, although about 10-fold less potent than compound 1 (Fig. 1). The compound proved inactive against the yellow fever virus (17D), another member of the Flaviviridae family, as well as against a panel of viruses unrelated to HCV (human immunodeficiency virus 1 and 2, herpes simplex virus 1 and 2, influenza virus A and B, vaccinia virus, vesicular stomatitis virus, respiratory syncitial virus, coxsackie virus, para-influenza virus, reovirus, sindbis virus, feline coronavirus and Punta Toro virus) (data not shown).
3.2. GS-327073 is active against various drug resistant replicons
The activity of GS-327073 was evaluated against a panel of drug-resistant replicons (Table 2 ). GS-327073 retained wild-type activity against replicons resistant to the protease inhibitor BILN-2061 and against BILN-2061/VX-950 double resistant replicons. GS-327073 also retained wild-type activity against replicons resistant to the nucleoside inhibitors 2′-C-MeCyt and 4′-azidocytidine and to the benzothiadiazine GSK-4. The observation that GS-327073 is active against VX-950/BILN-2061res and 2′-C-MeCytres replicon-containing Huh 9-13 was corroborated by the observation that cells containing VX-950/BILN-2061res and 2′-C-MeCytres replicon [that replicated efficiently in the presence of either 5 μg/ml VX-950 or 15 μM 2′-C-MeCyt (and G418 pressure)], died following only one passage in the presence of GS-327073 (300 nM) (data not shown).
Table 2.
In vitro anti-HCV activity of GS-327073 against various protease, nucleoside and non-nucleoside polymerase inhibitor resistant replicons.
| EC50 in μM (fold resistance) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BILN-2061res | VX-950res and BILN-2061res | 2′-C-MeCytres | 4′-Azidocytidineres | GSK-4res | ||||||
| GS-327073 | 0.009 ± 0.005 | (4.5) | 0.004 ± 0.003 | (2.0) | 0.008 ± 0.006 | (4.0) | 0.0072 ± 0.0008 | (3.6) | 0.011 ± 0.001 | (5.5) |
| BILN-2061 | 1.3 ± 0.47 | (130) | 0.80 ± 0.08 | (80) | 0.04 ± 0.03 | (4.0) | 0.014 ± 0.008 | (0.93) | n.d. | |
| VX-950 | 0.31 ± 0.05 | (0.53) | 14 ± 1.4 | (24) | 1.0 ± 0.03 | (1.7) | 0.69 ± 0.15 | (1.2) | n.d. | |
| 2′-C-MeCyt | 0.91 ± 1.1 | (2.1) | 1.0 ± 0.70 | (2.3) | ⩾30 | ⩾81 | 3.4 ± 1.1 | (7.9) | n.d. | |
| 4′-Azidocytidine | n.d. | n.d. | 1.2 ± 0.43 | (0.41) | ⩾16 | (⩾5.5) | n.d. | |||
| GSK-4 | n.d. | n.d. | n.d. | n.d. | >33 | (>35) | ||||
| HCV-796 | n.d. | n.d. | n.d. | n.d. | 0.06 ± 0.03 | (0.75) | ||||
| IFNα 2b (IU/ml) | 1.2 ± 0.83 | (5) | 0.62 ± 0.37 | (0.15) | 0.80 ± 0.46 | (3.3) | 1.4 ± 0.29 | (5.8) | n.d. | |
Data are mean values ± SD from 2 to 5 independent experiments.
n.d., not determined.
3.3. GS-327073 in combination with interferon, protease or polymerase inhibitors results in an additive antiviral activity
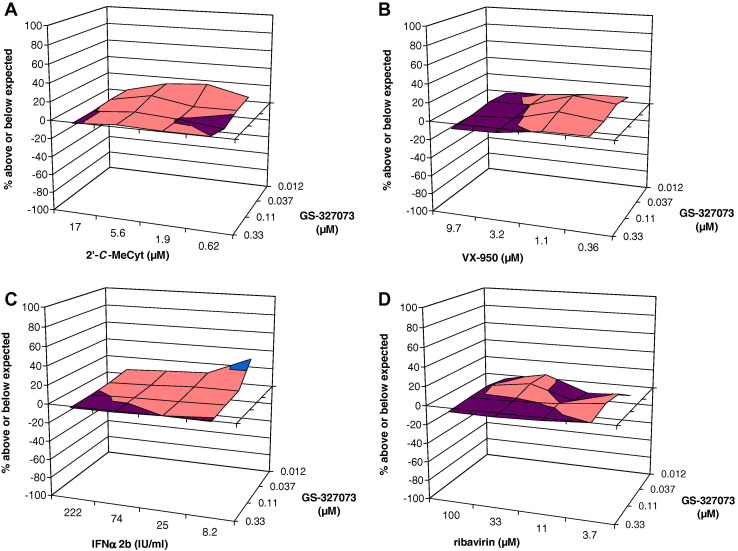
Combination experiments were performed in HuH6 replicon-containing cells in which replicon replication is largely independent of cell proliferation. This is important since the reference drugs 2′-C-MeCyt and ribavirin, at the concentrations used, inhibit host cell proliferation. GS-327073 was combined (in checkerboard format) with either IFNα 2b, VX-950, ribavirin or 2′-C-MeCyt, and data were analyzed using the method of Prichard and Shipman [59]. Each combination resulted overall in an additive antiviral activity (Fig. 3 ). A slight synergistic effect was observed when GS-327073 was combined with low concentrations of IFNα 2b.
Fig. 3.
Anti-HCV activity of the combination of GS-327073 with either (A) 2′-C-MeCyt, (B) VX-950, (C) IFNα 2b or (D) ribavirin. Values between −20% and 0% below expected are labelled in purple. Values between 0% and 20% above expected are labelled in orange. Values between 20% and 40% above expected values are labelled in blue.
3.4. Combinations containing GS-327073 efficiently clear hepatoma cells from their replicon
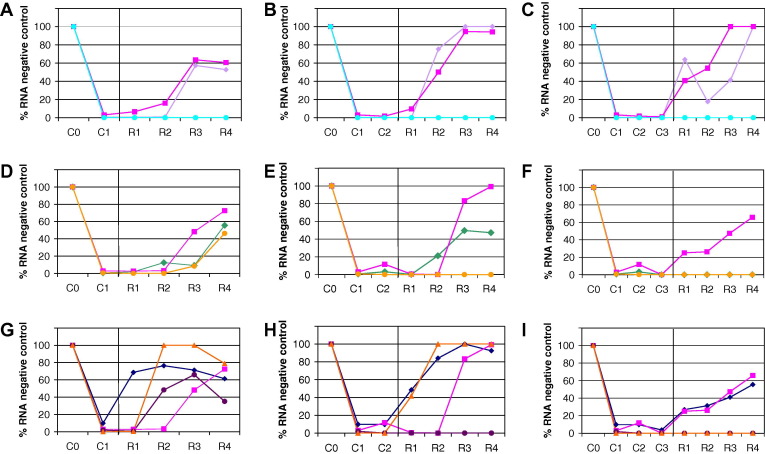
The potential of GS-327073 alone or combined with either a protease inhibitor (VX-950), a nucleoside polymerase inhibitor (2′-C-MeCyt) or IFNα 2b to cure hepatoma cells from their replicon was evaluated. GS-327073 or VX-950, when used at concentrations of five times their EC50, resulted in a rapid and profound drop in replicon level but did not achieve complete clearance of Huh 9-13 cells from their replicon following 3 passages in the presence of either drug (Fig. 4 C). In contrast, the combination GS-327073 (5 × EC50) and VX-950 (5 × EC50) resulted in clearance of Huh 9-13 cells from their replicon following one passage, (Fig. 4A). Combination of GS-327073 (20 × EC50) with either 100 IU/ml IFNα 2b (Fig. 4D–F) or with 2′-C-MeCyt (20 × EC50) (Fig. 4G–I) resulted in clearance of replicon RNA following 2 passages under the combined compound pressure (Fig. 4E and H). Neither 2′-C-MeCyt nor GS-327073 alone (each at 20 × EC50) resulted in clearance, whereas IFNα 2b did so after 3 passages. Furthermore, replicon-containing Huh 9-13 cells surviving rebound conditions (i.e. addition of G418) following GS-327073 or 2′-C-MeCyt treatment did not contain drug resistant replicon (data not shown). In addition, the clearance capacity of the combination of 10 × EC50 of GS-327073 and 10 × EC50 of 2′-C-MeCyt was compared to that of 20 × EC50 of either compounds (Fig. 4G–I). Clearance of replicon from Huh 9-13 cells was observed after 3 passages with the combination at 10 × EC50, whereas either compound alone failed to achieve clearance at 20 × EC50.
Fig. 4.
Clearance of HCV RNA from replicon-containing Huh 9-13 cells following treatment with (A–C) 5 × EC50 of VX-950 (violet diamond), 5 × EC50 GS-327073 (pink square) or the combination of 5 × EC50 GS-327073 and 5 × EC50 VX-950 (green–blue ball); (D–F) 20 × EC50 of IFNα 2b (green diamond), 20 × EC50 GS-327073 (pink square) or the combination of 20 × EC50 GS-327073 and 20 × EC50 IFNα 2b (orange ball); (G–I) 20 × EC50 2′-C-MeCyt (blue diamond), 20 × EC50 GS-327073 (pink square) or the combination of 20 × EC50 GS327073 and 20 × EC50 2′-C-MeCyt (purple ball) or 10 × EC50 GS327073 and 10 × EC50 2′-C-MeCyt (orange triangle). Clearance capacities after one passage in the presence of drug(s) depicted are in graphs A, D and G; after two passages in the presence of drug(s) are depicted in graphs B, E and H; after three passages in the presence of drug(s) are depicted in graphs C, F and I.
3.5. Stability and predicted hepatic clearance
Both in basic (pH 7.3) and in acidic solutions (pH 2.2) GS-327073 remained stable. The NADPH-dependent metabolic stability and predicted hepatic clearance was determined in human, dog and rat hepatic microsomes in vitro according to standard methods. The microsomal half-time for GS-327073 was 236 min in human, 88 min in dog and 96 min in rat hepatic microsomes. The predicted hepatic clearance of GS-327073 was 0.15 L/h/kg in human, 0.36 L/h/kg in dog and 0.63 L/h/kg in rat microsomes.
4. Discussion
We recently reported the anti-pestivirus activity of a novel class of imidazopyridines initially devoid of anti-HCV activity [67]. Introduction of a fluorine at position 2 of the phenyl group of the lead compound resulted in compounds with in vitro anti-HCV activity. Further optimization resulted in the discovery of 2-(2,3-difluorophenyl)-5-[4-(trifluoromethyl)benzyl]-5H-imidazo[4,5-c]pyridine that inhibited HCV replication with an EC50 of about 100 nM [43], [44].
A series of broader modifications to the 5-benzyl group of the substituted 5-benzyl-2-phenyl-5H-imidazo[4,5-c]pyridines was performed. This resulted in a series of molecules with highly potent anti-HCV activity. Within this series, GS-327073 [5-[[3-(4-chlorophenyl)-5-isoxazolyl]methyl]-2-(2,3-difluorophenyl)-5H-imidazo[4,5-c]pyridine], was selected for further characterization. GS-327073 proved particularly effective against genotype 1b replicons (the system used to optimize for antiviral potency) with EC50 values in the low nanomolar range. GS-327073 also inhibited the replication of a genotype 2a cell culture infectious HCVcc (J6/JFH-Rluc-FLAG), but did so less efficiently than for the genotype 1b replicon. The anti-HCV activity of GS-327073 was compared to that of other HCV inhibitors, including ribavirin, the NS3 protease inhibitors BILN-2061, VX-950 and SCH 503034, the nucleoside NS5B polymerase inhibitors 2′-C-MeCyt and 4′-azidocytidine and the non-nucleoside NS5B polymerase benzothiadiazine inhibitor GSK-4. The anti-HCV activity of GS-327073 was superior to that of any of these compounds. GS-327073 retained wild-type antiviral activity against a panel of replicons resistant to various HCV inhibitors, including protease, nucleoside and non-nucleoside polymerase inhibitors. Remarkably, GS-327073 retained a significant antiviral activity against the pestivirus BVDV. GS-327073 proved also cross-resistant in anti-BVDV assay with BPIP, one of the parent compounds in this class of compounds with potent anti-pestivirus activity but no anti-HCV activity. BPIP was shown to target the fingertip of the pestivirus RNA dependent RNA polymerase [45]. It is remarkable that a given non-nucleoside analogue (such as GS-327073) is able to inhibit both pestiviruses and HCV. Representatives of the imidazopyridines are, to the best of our knowledge, the first class of non-nucleoside molecules that are able to inhibit the replication of both pesti- and hepaciviruses, albeit potentially different mechanisms of action may be involved. Detailed studies on the molecular mechanism of action of the imidazopyridines will be reported elsewhere. Replicons resistant to GS-327073 were generated following several months of selective in vitro pressure (data not shown). In comparison to several other STAT-C inhibitors, the genetic barrier to achieve high levels of resistance of GS-327073 is relatively high. This might be explained by the fact that three mutations are required in the RNA-dependent RNA polymerase gene to achieve 50-fold or more resistance.
It is likely that specifically targeted antiviral therapy compounds (STAT-C), in the initial years after they become available for the treatment of HCV infected patients, will be combined with the current standard therapy [68]. At a later stage, two or more potent STAT-C inhibitors with different resistance profiles, may hopefully be combined without further need for interferon and/or ribavirin. It is hence important to obtain information about the antiviral efficacy of particular combinations. For this reason we studied the antiviral activity of GS-327073 when combined with other classes of HCV inhibitors. To this end, two different assay systems were employed. In a first assay, the combined antiviral activity was evaluated in regular antiviral assays, i.e. with a read-out after 3 days of incubation of the replicon containing cells with the antiviral drug(s). All combinations studied resulted in an overall additive antiviral activity, which was in line with our expectation that compounds that do (likely) not interfere with each others’ metabolism or mechanism of action should result in an additive antiviral effect. Combinations of compounds that interfere with each others’ biological activity may either result in a synergistic or antagonistic antiviral activity. For example, the IMP-dehydrogenase inhibitor ribavirin is able to potentiate the antiherpes and anti-HBV activity of purine based deoxynucleoside analogues on one hand, but results in an antagonistic effect on HIV and HCV replication when combined with pyrimidine ribonucleoside analogues [69], [70], [71], [72], [73]. Particular interference of ribavirin with the purine and pyrimidine metabolism is an explanation for these observations. No such antagonistic or synergistic activities were noted when GS-327073 was combined with other selective inhibitors of HCV replication. However, a short antiviral assay may not necessarily predict the antiviral effect of either single compounds or combinations thereof. We, therefore, studied the anti-HCV activity of GS-327073 alone or in combination with either interferon α, a protease or a polymerase HCV inhibitor in “clearance rebound” assays. Replicon containing cells were cultured for one or more passages in the presence of the antiviral drug(s) and in the absence of the selection marker geneticin. Neither drug, at concentrations that were 5- to 20-fold higher than their EC50 values, was able to achieve clearance (except for 3 passages in the presence of 100 IU of interferon α). Surprisingly, and despite the fact that no synergistic activity was noted in a regular 3 day antiviral assay, all GS-327073 containing combinations (5×, 10× and 20 × EC50) efficiently cleared cells from their replicon. Clearance-rebound experiments may possibly have predictive value for estimating the potential of drugs (or combinations thereof) to clear liver cells from replicating virus. Combinations of imidazopyridines with various STAT-C inhibitors (or interferon) may have the potential to achieve rapid viral clearance. Our data also indicate that regular short term antiviral combination assays (in replicon based systems) may not necessarily predict synergistic antiviral effects following long-term culture.
In conclusion, following lead optimization, a series of highly potent HCV inhibitors were developed of which GS-327073 can be considered as a prototype. GS-327073 exhibited favorable pharmacokinetic properties in vitro. Interestingly, several compounds in this series are active against both HCV and pestiviruses. It is currently being studied whether a common mechanism is responsible for both antiviral activities or whether the precise molecular mechanism of action of anti-HCV or anti-pestivirus activity differs.
Acknowledgements
We thank Katrien Geerts for excellent technical assistance. This work is part of the activities of the VIRGIL European Network of Excellence on Antiviral Drug Resistance supported by a grant (LSHM-CT-2004-503359) from the Priority 1 “Life Sciences, Genomics and Biotechnology for Health” Programme in the 6th Framework Programme of the EU. Jan Paeshuyse is supported by a post-doctoral position of the “Fonds voor Wetenschappelijk Onderzoek, Vlaanderen”.
Associate Editor: F. Zoulim
Footnotes
The authors declared that the study was partly funded by Gilead. W.A.L., H.R., L.S.L., S.B., M.P., I.-H.S., E.M., W.Z., are employed by Gilead. D.O. and N.B. were employed by Gilead at the time of preparation of this manuscript.
References
- 1.Craxi A., Laffi G., Zignego A.L. Hepatitis C virus (HCV) infection: a systemic disease. Mol Aspects Med. 2007;29:85–95. doi: 10.1016/j.mam.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 2.Fried M.W., Shiffman M.L., Reddy K.R., Smith C., Marinos C., Goncales L. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 3.Manns M.P., McHutchison J.G., Gordon S.C., Rustgi V.K., Shiffman M., Reindollar R. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 4.Firpi R.J., Nelson D.R. Current and future hepatitis C therapies. Arch Med Res. 2007;38:678–690. doi: 10.1016/j.arcmed.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi N., Takehara T. Antiviral therapy for chronic hepatitis C: past, present, and future. J Gastroenterol. 2006;41:17–27. doi: 10.1007/s00535-005-1740-7. [DOI] [PubMed] [Google Scholar]
- 6.De Francesco R., Migliaccio G. Challenges and successes in developing new therapies for hepatitis C. Nature. 2005;436:953–960. doi: 10.1038/nature04080. [DOI] [PubMed] [Google Scholar]
- 7.Neyts J. Selective inhibitors of hepatitis C virus replication. Antiviral Res. 2006;71:363–371. doi: 10.1016/j.antiviral.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Yang W., Zhao Y., Fabrycki J., Hou X., Nie X., Sanchez A. Selection of replicon variants resistant to ACH-806, a novel hepatitis C virus inhibitor with no cross-resistance to NS3 protease and NS5B polymerase inhibitors. Antimicrob Agents Chemother. 2008;52:2043–2052. doi: 10.1128/AAC.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flisiak R., Dumont J.M., Crabbe R. Cyclophilin inhibitors in hepatitis C viral infection. Expert Opin Investig Drugs. 2007;16:1345–1354. doi: 10.1517/13543784.16.9.1345. [DOI] [PubMed] [Google Scholar]
- 10.Soriano V., Madejon A., Vispo E., Labarga P., Garcia-Samaniego J., Martin-Carbonero L. Emerging drugs for hepatitis C. Expert Opin Emerg Drugs. 2008;13:1–19. doi: 10.1517/14728214.13.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Clark J.L., Hollecker L., Mason J.C., Stuyver L.J., Tharnish P.M., Lostia S. Design, synthesis, and antiviral activity of 2′-deoxy-2′-fluoro-2′-C-methylcytidine, a potent inhibitor of hepatitis C virus replication. J Med Chem. 2005;48:5504–5508. doi: 10.1021/jm0502788. [DOI] [PubMed] [Google Scholar]
- 12.Poordad F., Lawitz E.J., Gitlin N., Rodriguez-Torres M., Box T., Nguyen T. Efficacy and safety of valopicitabine in combination with pegylated interferon and ribavirin in patients with chronic hepatitis C. Hepatology. 2007;46(Suppl.):LB15. [Google Scholar]
- 13.Pockros P.J., Nelson D., Godofsky E., Rodriguez-Torres M., Everson G., Fried M.W. Robust synergistic antiviral effect of R1626 in combination with peginterferon alfa-2a (40 kDa), with or without ribavirin – interim analysis of phase 2a study. Hepatology. 2007;46(Suppl.):167. [Google Scholar]
- 14.Reddy R., Rodriguez-Torres M., Gane E., Robson R., Lalezari J., Everson G.T. Antiviral activity, pharmacokinetics, safety and tolerability of R1728, a novel nucleoside HCV RNA polymerase inhibitor, following multiple, ascending, oral doses in patients with genotype 1 infection who have failed prior interferon therapy. Hepatology. 2007;46(Suppl.):LB9. [Google Scholar]
- 15.Beaulieu P.L. Finger loop inhibitors of the HCV NS5B polymerase: discovery and prospects for new HCV therapy. Curr Opin Drug Discov Dev. 2006;9:618–626. [PubMed] [Google Scholar]
- 16.Hirashima S., Suzuki T., Ishida T., Noji S., Yata S., Ando I. Benzimidazole derivatives bearing substituted biphenyls as hepatitis C virus NS5B RNA-dependent RNA polymerase inhibitors: structure–activity relationship studies and identification of a potent and highly selective inhibitor JTK-109. J Med Chem. 2006;49:4721–4736. doi: 10.1021/jm060269e. [DOI] [PubMed] [Google Scholar]
- 17.Kukolj G., McGibbon G.A., McKercher G., Marquis M., Lefebvre S., Thauvette L. Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J Biol Chem. 2005;280:39260–39267. doi: 10.1074/jbc.M506407200. [DOI] [PubMed] [Google Scholar]
- 18.Beaulieu P.L. Non-nucleoside inhibitors of the HCV NS5B polymerase: progress in the discovery and development of novel agents for the treatment of HCV infections. Curr Opin Investig Drugs. 2007;8:614–634. [PubMed] [Google Scholar]
- 19.Botyanszki J, Roberts CD, Schmitz FU, Gralapp JIM, Griffith RC, Shi D-F, et al. Indole derivates for treating viral infections. EP20060718244(EP1844042) Genelabs Tech Inc [US] 2007.
- 20.Harper S., Avolio S., Pacini B., Di Filippo M., Altamura S., Tomei L. Potent inhibitors of subgenomic hepatitis C virus RNA replication through optimization of indole-N-acetamide allosteric inhibitors of the viral NS5B polymerase. J Med Chem. 2005;48:4547–4557. doi: 10.1021/jm050056+. [DOI] [PubMed] [Google Scholar]
- 21.Harper S., Pacini B., Avolio S., Di Filippo M., Migliaccio G., Laufer R. Development and preliminary optimization of indole-N-acetamide inhibitors of hepatitis C virus NS5B polymerase. J Med Chem. 2005;48:1314–1317. doi: 10.1021/jm049122i. [DOI] [PubMed] [Google Scholar]
- 22.Chan L., Pereira O., Reddy T.J., Das S.K., Poisson C., Courchesne M. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 2: tertiary amides. Bioorg Med Chem Lett. 2004;14:797–800. doi: 10.1016/j.bmcl.2003.10.068. [DOI] [PubMed] [Google Scholar]
- 23.Le Pogam S., Kang H., Harris S.F., Leveque V., Giannetti A.M., Ali S. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J Virol. 2006;80:6146–6154. doi: 10.1128/JVI.02628-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan L., Reddy T.J., Proulx M., Das S.K., Pereira O., Wang W. Identification of N,N-disubstituted phenylalanines as a novel class of inhibitors of hepatitis C NS5B polymerase. J Med Chem. 2003;46:1283–1285. doi: 10.1021/jm0340400. [DOI] [PubMed] [Google Scholar]
- 25.Reddy T.J., Chan L., Turcotte N., Proulx M., Pereira O.Z., Das S.K. Further SAR studies on novel small molecule inhibitors of the hepatitis C (HCV) NS5B polymerase. Bioorg Med Chem Lett. 2003;13:3341–3344. doi: 10.1016/s0960-894x(03)00670-x. [DOI] [PubMed] [Google Scholar]
- 26.Li H., Tatlock J., Linton A., Gonzalez J., Borchardt A., Dragovich P. Identification and structure-based optimization of novel dihydropyrones as potent HCV RNA polymerase inhibitors. Bioorg Med Chem Lett. 2006;16:4834–4838. doi: 10.1016/j.bmcl.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 27.Love R.A., Parge H.E., Yu X., Hickey M.J., Diehl W., Gao J. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J Virol. 2003;77:7575–7581. doi: 10.1128/JVI.77.13.7575-7581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gopalsamy A., Lim K., Ciszewski G., Park K., Ellingboe J.W., Bloom J. Discovery of pyrano[3,4-b]indoles as potent and selective HCV NS5B polymerase inhibitors. J Med Chem. 2004;47:6603–6608. doi: 10.1021/jm0401255. [DOI] [PubMed] [Google Scholar]
- 29.Howe A.Y., Bloom J., Baldick C.J., Benetatos C.A., Cheng H., Christensen J.S. Novel nonnucleoside inhibitor of hepatitis C virus RNA-dependent RNA polymerase. Antimicrob Agents Chemother. 2004;48:4813–4821. doi: 10.1128/AAC.48.12.4813-4821.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howe A.Y., Cheng H., Thompson I., Chunduru S.K., Herrmann S., O’Connell J. Molecular mechanism of a thumb domain hepatitis C virus nonnucleoside RNA-dependent RNA polymerase inhibitor. Antimicrob Agents Chemother. 2006;50:4103–4113. doi: 10.1128/AAC.00365-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gopalsamy A., Chopra R., Lim K., Ciszewski G., Shi M., Curran K.J. Discovery of proline sulfonamides as potent and selective hepatitis C virus NS5b polymerase inhibitors. Evidence for a new NS5b polymerase binding site. J Med Chem. 2006;49:3052–3055. doi: 10.1021/jm060168g. [DOI] [PubMed] [Google Scholar]
- 32.Dhanak D., Duffy K.J., Johnston V.K., Lin-Goerke J., Darcy M., Shaw A.N. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J Biol Chem. 2002;277:38322–38327. doi: 10.1074/jbc.M205566200. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen T.T., Gates A.T., Gutshall L.L., Johnston V.K., Gu B., Duffy K.J. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob Agents Chemother. 2003;47:3525–3530. doi: 10.1128/AAC.47.11.3525-3530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratt J.K., Donner P., McDaniel K.F., Maring C.J., Kati H., Mo T. Inhibitors of HCV NS5B polymerase: synthesis and structure-activity relationships of N-1-heteroalkyl-4-hydroxyquinolon-3-yl-benzothiadiazines. Bioorg Med Chem Lett. 2005;15:1577–1582. doi: 10.1016/j.bmcl.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 35.Rockway T.W., Zhang R., Liu D., Betebenner D.A., McDaniel K.F., Pratt J.K. Inhibitors of HCV NS5B polymerase: synthesis and structure–activity relationships of N-1-benzyl and N-1-[3-methylbutyl]-4-hydroxy-1,8-naphthyridon-3-yl benzothiadiazine analogs containing substituents on the aromatic ring. Bioorg Med Chem Lett. 2006;16:3833–3838. doi: 10.1016/j.bmcl.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 36.Tedesco R., Shaw A.N., Bambal R., Chai D., Concha N.O., Darcy M.G. 3-(1,1-dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)- quinolinones, potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase. J Med Chem. 2006;49:971–983. doi: 10.1021/jm050855s. [DOI] [PubMed] [Google Scholar]
- 37.Lee G., Piper D.E., Wang Z., Anzola J., Powers J., Walker N. Novel inhibitors of hepatitis C virus RNA-dependent RNA polymerases. J Mol Biol. 2006;357:1051–1057. doi: 10.1016/j.jmb.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 38.Pfefferkorn J.A., Greene M.L., Nugent R.A., Gross J., Mitchell M.A., Finzel B.C. Inhibitors of HCV NS5B polymerase. Part 1: evaluation of the southern region of (2Z)-2-(benzoylamino)-3-(5-phenyl-2-furyl)acrylic acid. Bioorg Med Chem Lett. 2005;15:2481–2486. doi: 10.1016/j.bmcl.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 39.Pfefferkorn J.A., Nugent R., Gross R.J., Greene M., Mitchell M.A., Reding M.T. Inhibitors of HCV NS5B polymerase. Part 2: evaluation of the northern region of (2Z)-2-benzoylamino-3-(4-phenoxy-phenyl)-acrylic acid. Bioorg Med Chem Lett. 2005;15:2812–2818. doi: 10.1016/j.bmcl.2005.03.106. [DOI] [PubMed] [Google Scholar]
- 40.Villano S., Howe A., Raible D., Harper D., Speth J., Bichier G. Analysis of HCV NS5B genetic variants following monotherapy with HCV-796, a non-nucleoside polymerase inhibitor, in treatment-naive HCV-infected patients. Hepatology. 2006;44(Suppl.):1127. [Google Scholar]
- 41.Villano S., Raible D., Harper D., Priyamvada C., Bazisotto L., Bichier G. Phase 1 evaluation of antiviral activity of the non-nucleoside inhibitor, HCV-796, in combination with different pegylated interferons in treatment-naive patients with chronic HCV. Hepatology. 2007;46(Suppl.):1302. [Google Scholar]
- 42.Cooper C., Lawitz E.J., Ghali P., Rodriguez-Torres M., Anderson F.H., Lee S.S. Antiviral activity of the non-nucleoside polymerase inhibitor, VCH-759, in chronic Hepatitis C patients:results from a randomized, doubleblind, placebo-controlled, ascending multiple dose study. Hepatology. 2007;46(Suppl.):LB11. [Google Scholar]
- 43.Puerstinger G., Paeshuyse J., De Clercq E., Neyts J. Antiviral 2,5-disubstituted imidazo[4,5-c]pyridines: from anti-pestivirus to anti-hepatitis C virus activity. Bioorg Med Chem Lett. 2007;17:390–393. doi: 10.1016/j.bmcl.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 44.Puerstinger G., Paeshuyse J., Heinrich S., Mohr J., Schraffl N., De Clercq E. Antiviral 2,5-disubstituted imidazo[4,5-c]pyridines: further optimization of anti-hepatitis C virus activity. Bioorg Med Chem Lett. 2007;17:5111–5114. doi: 10.1016/j.bmcl.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 45.Paeshuyse J., Leyssen P., Mabery E., Boddeker N., Vrancken R., Froeyen M. A novel, highly selective inhibitor of pestivirus replication that targets the viral RNA-dependent RNA polymerase. J Virol. 2006;80:149–160. doi: 10.1128/JVI.80.1.149-160.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns CJ, Del Vecchio AM, Bailey TR, Kulkarni BA, Faitg TH, Sherk SR, et al. Benzofuran compounds, compositions and methods for treatment and prophylaxis of hepatitis c viral infections and associated diseases. PCT/US2003/034962(WO/2004/041201) Viropharma Incorporated and Wyeth [US] 2004.
- 47.Eldrup A.B., Allerson C.R., Bennett C.F., Bera S., Bhat B., Bhat N. Structure–activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J Med Chem. 2004;47:2283–2295. doi: 10.1021/jm030424e. [DOI] [PubMed] [Google Scholar]
- 48.Faucher A.M., Bailey M.D., Beaulieu P.L., Brochu C., Duceppe J.S., Ferland J.M. Synthesis of BILN 2061, an HCV NS3 protease inhibitor with proven antiviral effect in humans. Org Lett. 2004;6:2901–2904. doi: 10.1021/ol0489907. [DOI] [PubMed] [Google Scholar]
- 49.Prongay A.J., Guo Z., Yao N., Pichardo J., Fischmann T., Strickland C. Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3-[2(S) -[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization. J Med Chem. 2007;50:2310–2318. doi: 10.1021/jm060173k. [DOI] [PubMed] [Google Scholar]
- 50.Smith D.B., Martin J.A., Klumpp K., Baker S.J., Blomgren P.A., Devos R. Design, synthesis, and antiviral properties of 4′-substituted ribonucleosides as inhibitors of hepatitis C virus replication: the discovery of R1479. Bioorg Med Chem Lett. 2007;17:2570–2576. doi: 10.1016/j.bmcl.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Venkatraman S., Bogen S.L., Arasappan A., Bennett F., Chen K., Jao E. Discovery of (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3-[2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (SCH 503034), a selective, potent, orally bioavailable hepatitis C virus NS3 protease inhibitor: a potential therapeutic agent for the treatment of hepatitis C infection. J Med Chem. 2006;49:6074–6086. doi: 10.1021/jm060325b. [DOI] [PubMed] [Google Scholar]
- 52.Yip Y., Victor F., Lamar J., Johnson R., Wang Q.M., Glass J.I. P4 and P1′ optimization of bicycloproline P2 bearing tetrapeptidyl alpha-ketoamides as HCV protease inhibitors. Bioorg Med Chem Lett. 2004;14:5007–5011. doi: 10.1016/j.bmcl.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Lohmann V., Korner F., Koch J., Herian U., Theilmann L., Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 54.Vrolijk J.M., Kaul A., Hansen B.E., Lohmann V., Haagmans B.L., Schalm S.W. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J Virol Methods. 2003;110:201–209. doi: 10.1016/s0166-0934(03)00134-4. [DOI] [PubMed] [Google Scholar]
- 55.Windisch M.P., Frese M., Kaul A., Trippler M., Lohmann V., Bartenschlager R. Dissecting the interferon-induced inhibition of hepatitis C virus replication by using a novel host cell line. J Virol. 2005;79:13778–13793. doi: 10.1128/JVI.79.21.13778-13793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paeshuyse J., Coelmont L., Vliegen I., Van hemel J., Vandenkerckhove J., Peys E. Hemin potentiates the anti-hepatitis C virus activity of the antimalarial drug artemisinin. Biochem Biophys Res Commun. 2006;348:139–144. doi: 10.1016/j.bbrc.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 57.Paulson MS, Yang H, Shih I, Feng J, Mabery EM, Robinson MF, et al. Comparison of HCV NS3 protease and NS5B polymerase inhibitor activity in 1a, 1b and 2a replicons and 2a infectious virus. Submitted for publication. [DOI] [PubMed]
- 58.Delaney W.E., Edwards R., Colledge D., Shaw T., Torresi J., Miller T.G. Cross-resistance testing of antihepadnaviral compounds using novel recombinant baculoviruses which encode drug-resistant strains of hepatitis B virus. Antimicrob Agents Chemother. 2001;45:1705–1713. doi: 10.1128/AAC.45.6.1705-1713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prichard M.N., Shipman C., Jr. A three-dimensional model to analyze drug–drug interactions. Antiviral Res. 1990;14:181–205. doi: 10.1016/0166-3542(90)90001-n. [DOI] [PubMed] [Google Scholar]
- 60.De Clercq E., Cools M., Balzarini J., Snoeck R., Andrei G., Hosoya M. Antiviral activities of 5-ethynyl-1-beta-d-ribofuranosylimidazole-4- carboxamide and related compounds. Antimicrob Agents Chemother. 1991;35:679–684. doi: 10.1128/aac.35.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keyaerts E., Vijgen L., Pannecouque C., Van Damme E., Peumans W., Egberink H. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antiviral Res. 2007;75:179–187. doi: 10.1016/j.antiviral.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neyts J., Meerbach A., McKenna P., De Clercq E. Use of the yellow fever virus vaccine strain 17D for the study of strategies for the treatment of yellow fever virus infections. Antiviral Res. 1996;30:125–132. doi: 10.1016/0166-3542(96)89697-5. [DOI] [PubMed] [Google Scholar]
- 63.Nunberg J.H., Wright D.K., Cole G.E., Petrovskis E.A., Post L.E., Compton T. Identification of the thymidine kinase gene of feline herpesvirus: use of degenerate oligonucleotides in the polymerase chain reaction to isolate herpesvirus gene homologs. J Virol. 1989;63:3240–3249. doi: 10.1128/jvi.63.8.3240-3249.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pauwels R., Balzarini J., Baba M., Snoeck R., Schols D., Herdewijn P. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 65.Shigeta S., Konno K., Yokota T., Nakamura K., De Clercq E. Comparative activities of several nucleoside analogs against influenza A, B, and C viruses in vitro. Antimicrob Agents Chemother. 1988;32:906–911. doi: 10.1128/aac.32.6.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Witvrouw M., Daelemans D., Pannecouque C., Neyts J., Andrei G., Snoeck R. Broad-spectrum antiviral activity and mechanism of antiviral action of the fluoroquinolone derivative K-12. Antivir Chem Chemother. 1998;9:403–411. doi: 10.1177/095632029800900504. [DOI] [PubMed] [Google Scholar]
- 67.Puerstinger G., Paeshuyse J., Herdewijn P., Rozenski J., De Clercq E., Neyts J. Substituted 5-benzyl-2-phenyl-5H-imidazo[4,5-c]pyridines: a new class of pestivirus inhibitors. Bioorg Med Chem Lett. 2006;16:5345–5349. doi: 10.1016/j.bmcl.2006.07.081. [DOI] [PubMed] [Google Scholar]
- 68.Kwong A.D., Cowherd S., Mueller P. Beyond interferon and ribavirin: antiviral therapies for hepatitis C virus. Drug Discov Today Ther Strateg. 2006;3:211–220. [Google Scholar]
- 69.Baba M., Pauwels R., Balzarini J., Herdewijn P., De Clercq E., Desmyter J. Ribavirin antagonizes inhibitory effects of pyrimidine 2′,3′-dideoxynucleosides but enhances inhibitory effects of purine 2′,3′-dideoxynucleosides on replication of human immunodeficiency virus in vitro. Antimicrob Agents Chemother. 1987;31:1613–1617. doi: 10.1128/aac.31.10.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coelmont L., Paeshuyse J., Windisch M.P., De Clercq E., Bartenschlager R., Neyts J. Ribavirin antagonizes the in vitro anti-hepatitis C virus activity of 2′-C-methylcytidine, the active component of valopicitabine. Antimicrob Agents Chemother. 2006;50:3444–3446. doi: 10.1128/AAC.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pancheva S.N. Potentiating effect of ribavirin on the anti-herpes activity of acyclovir. Antiviral Res. 1991;16:151–161. doi: 10.1016/0166-3542(91)90021-i. [DOI] [PubMed] [Google Scholar]
- 72.Vogt M.W., Hartshorn K.L., Furman P.A., Chou T.C., Fyfe J.A., Coleman L.A. Ribavirin antagonizes the effect of azidothymidine on HIV replication. Science. 1987;235:1376–1379. doi: 10.1126/science.2435003. [DOI] [PubMed] [Google Scholar]
- 73.Ying C., De Clercq E., Neyts J. Ribavirin and mycophenolic acid potentiate the activity of guanine- and diaminopurine-based nucleoside analogues against hepatitis B virus. Antiviral Res. 2000;48:117–124. doi: 10.1016/s0166-3542(00)00121-2. [DOI] [PubMed] [Google Scholar]