Abstract
Background
Effective countermeasures against emerging infectious diseases require an understanding of transmission rate and basic reproduction number (R0). R0 for severe acute respiratory syndrome is generally considered to be >1, whereas that for Middle East respiratory syndrome (MERS) is considered to be <1. However, this does not explain the large-scale outbreaks of MERS that occurred in Kingdom of Saudi Arabia (KSA) and South Korean hospitals.
Aim: To estimate R0 in nosocomial outbreaks of MERS.
Methods
R0 was estimated using the incidence decay with an exponential adjustment model. The KSA and Korean outbreaks were compared using a line listing of MERS cases compiled using publicly available sources. Serial intervals to estimate R0 were assumed to be six to eight days. Study parameters [R0 and countermeasures (d)] were estimated by fitting a model to the cumulative incidence epidemic curves using Matlab.
Findings
The estimated R0 in Korea was 3.9 in the best-fit model, with a serial interval of six days. The first outbreak cluster in a hospital in Pyeongtaek had an R0 of 4.04, and the largest outbreak cluster in a hospital in Samsung had an R0 of 5.0. Assuming a six-day serial interval, the KSA outbreaks in Jeddah and Riyadh had R0 values of 3.9 and 1.9, respectively.
Conclusion
R0 for the nosocomial MERS outbreaks in KSA and South Korea was estimated to be in the range of 2–5, which is significantly higher than the previous estimate of <1. Therefore, more comprehensive countermeasures are needed to address these infections.
Keywords: Nosocomial infection, Basic reproduction number, Epidemiology, Middle east respiratory syndrome coronavirus, Mathematical modelling, South Korea
Introduction
The emergence of infectious diseases associated with Middle East respiratory syndrome (MERS), severe acute respiratory syndrome and Ebola has created unprecedented public health challenges. These challenges are complicated by the lack of basic epidemiological data, which makes it difficult to predict epidemics. Thus, it is important to quantify actual outbreaks as novel infectious diseases emerge. Disease severity and rate of transmission can be predicted by mathematical models using the basic reproduction number (R 0) [1]. For example, R 0 has been used extensively to assess pathogen transmissibility, outbreak severity and epidemiological control [2], [3], [4].
In previous studies, R 0 for MERS has ranged from 0.42 to 0.92 [5], [6], [7], [8], which suggests that the MERS coronavirus (MERS-CoV) has limited transmissibility. However, these studies typically considered community-acquired MERS infections. In this context, nosocomial infections can exhibit different R 0 values as the transmission routes for community-acquired and nosocomial infections often differ [9]. Recent studies have examined large nosocomial outbreaks of MERS-CoV infection in Jeddah and Riyadh within the Kingdom of Saudi Arabia (KSA). One study reported higher nosocomial R 0 values than those from community-acquired infections when using the incidence decay with exponential adjustment (IDEA) model, which yielded values of 3.5–6.7 in Jeddah and 2.0–2.8 in Riyadh [10]. The IDEA model is simple because it does not consider the population-level immune status, which makes it especially useful for modelling emerging infectious diseases in resource-limited settings.
The MERS outbreak in South Korea was associated with nosocomial infections. At that time, the Korea Centre for Disease Control and Prevention (KCDC) assumed that the outbreak had an R 0 <1. Thus, the initial countermeasures were not sufficiently aggressive to prevent the spread of MERS-CoV infection to other hospitals. Therefore, the IDEA model was used to evaluate and compare the MERS R 0 values from the outbreaks in both KSA and South Korean hospitals.
Methods
Data source
KSA data were obtained using a line listing of MERS-CoV cases that was maintained by Andrew Rambaut (updated on 19th August 2015). The line listing was created using data from the KSA Ministry of Health and World Health Organization (WHO) report [10]. Since only 44% of cases in the KSA listing included the onset date, hospitalization dates or reported dates were used instead. The Korean data were obtained from the KCDC. Among the 186 MERS cases, 178 had confirmed onset dates. The eight cases with unknown onset dates were assigned dates based on laboratory confirmation. All cases in KSA and Korea were confirmed based on laboratory findings. Study parameters [R 0 and countermeasures (d)] were estimated by fitting a model to the cumulative incidence epidemic curves using Matlab (Mathworks, Natick, MA, USA).
The data were narrowed down to the nosocomial cases alone. Cases with unknown transmissions were considered to be nosocomial if: (a) the patient was in contact with a healthcare worker and/or hospitalized patients; or (b) the patient was a healthcare worker. Cases were excluded if they could not be verified as nosocomial (e.g. zoonotic transmission, family contact or community infection).
Model
The IDEA model was used to estimate R 0 as reported previously [11], together with publicly available data. The IDEA model is based on the concept that the number of incident cases () in an epidemic generation () can be counted as:
| (1) |
when an outbreak occurs, epidemic control measures can be implemented, which can, in turn, change R 0. Therefore, the relationship between I and R 0 with is defined as follows:
| (2) |
R 0 and d are estimated by fitting from Eq. (2) to the observed cumulative incidence data of MERS using the least-squares data-fitting method. Since the IDEA model is parameterized using epidemic generation time, incidence case counts were aggregated at serial intervals of six, seven and eight days in the present study [10].
Two large outbreaks were considered in each country studied: the outbreaks in Riyadh and Jeddah for KSA; and those in Pyeongtaek St. Mary's Hospital and Samsung Seoul Hospital for South Korea. The term ‘resnorm’ is defined as the norm of the residual, which is the squared 2-norm of the residual; it measures the difference between observed data and the fitted value provided by a model. However, as residuals can be positive or negative, a sum of residuals is not a good measure of overall error in the fit. Therefore, a better measure of error is the sum of the squared residuals (), which is calculated as follows:
| (3) |
The functions to be fit were the given input data (xdata), the observed output data, (ydata) and F(x, xdata), where xdata was an epidemic generation, ydata was the observed cumulative incidence data, and F(x, xdata) was Eq. (2).
Since the generation times and the estimated values differ according to serial interval times, resnorm changes accordingly. Therefore, to compare resnorm with the serial interval time, relative resnorm was defined as follows:
| (4) |
The IDEA model was fitted to the cumulative South Korean MERS-CoV case data from the onset date of the first case to the onset date of the last case. The outbreak start date was defined as 11th May 2015 because that was the symptom onset date for Patient 0, who was the index case and caused the outbreak in the Pyeongtaek hospital. Patient 14 caused the outbreak at the Samsung hospital, and his symptom onset date was 21st May 2015. The last case of the MERS outbreak in South Korea was observed on 4th July 2015. The KSA MERS outbreak model was fitted using the cumulative incidence data from 28th March 2014 to 2nd June 2014 in Jeddah, and from 20th March 2014 to 29th May 2014 in Riyadh.
Ethical considerations
All data used in these analyses were de-identified publicly available data obtained from WHO, the KSA Ministry of Health website or KCDC datasets. As such, these data were deemed to be exempt from institutional review board assessment.
Results
KSA outbreaks were relatively large, with 180 cases (over the course of 67 days) in Jeddah and 142 cases (over the course of 71 days) in Riyadh. The Korean outbreaks involved 186 cases (over the course of 55 days), including 36 cases (over the course of 23 days) in the Pyeongtaek hospital and 91 cases (over the course of 45 days) in the Samsung hospital. Most Korean cases (180) were nosocomial, with the exception of four cases acquired by household transmission and two cases with unknown modes of transmission. In KSA, only two cases involved confirmed zoonotic transmission, while a large number of unknown transmissions (Jeddah: 99 cases; Riyadh: 69 cases) and hospital exposures (Jeddah: 80 cases; Riyadh: 70 cases) were observed (Table I ).
Table I.
Characteristics of selected Middle East respiratory syndrome (MERS) outbreaks in Saudi Arabia and South Korea
| Saudi Arabia |
South Korea |
|||||
|---|---|---|---|---|---|---|
| Jeddah | Riyadh | Total | Pyeongtaek St. Mary's hospital | Samsung Seoul hospital | ||
| Outbreak | Onset date | 28/03/2014 | 20/03/2014 | 11/05/2015 | 15/05/2015 | 25/05/2015 |
| Duration (day) | 67 | 71 | 55 | 23 | 45 | |
| No. of cases | 180 | 142 | 186 | 36 | 91 | |
| Exposure | Nosocomial | 80a | 70a | 180 | 36 | 88 |
| Household | 4 | 0 | 3 | |||
| Zoonotic | 1 | 1 | 0 | 0 | 0 | |
| Unknown | 99 | 69 | 2 | 0 | 0 | |
| Statusb | Healthcare worker | 40 | 8 | 39 | 3 | 15 |
| Patient | 82 | 20 | 36 | |||
| Family or visitor | 63 | 13 | 40 | |||
| Unknown | 140 | 134 | 2 | 0 | 0 | |
| Datec | Onset date | 75 | 66 | 178 | 36 | 85 |
| Hospitalized date | 85 | 79 | 186 | 36 | 91 | |
| Reported date | 180 | 142 | 186 | 36 | 91 | |
Nosocomial cases included healthcare workers and individuals who were in contact with a healthcare worker or hospitalized patients.
The status of cases when they were exposed to MERS.
The number of cases with information for onset date, hospitalization date and reported date of MERS.
The IDEA model was fitted to the daily KSA and Korea MERS-CoV case data according to the onset date. Figure 1 displays the cumulative MERS-CoV case data for the 2014 KSA and the 2015 South Korea MERS outbreaks. The date of symptom onset for Patient 0 was 11th May 2015; however, he was admitted to the Pyeongtaek hospital on 15th May 2015. Therefore, the outbreak was assumed to start on 15th May 2015 via a simulation of the Pyeongtaek hospital outbreak. The outbreak start date for the Samsung hospital was determined to be 25th May 2015, following the same logic (Figure 1).
Figure 1.
Epidemic curves of cumulative cases by selected Middle East respiratory syndrome outbreaks in (a) Saudi Arabia (red circles, Jeddah; blue asterisks, Riyadh) and (b) South Korea (green squares, total; red circles, Pyeongtaek; blue asterisks, Samsung).
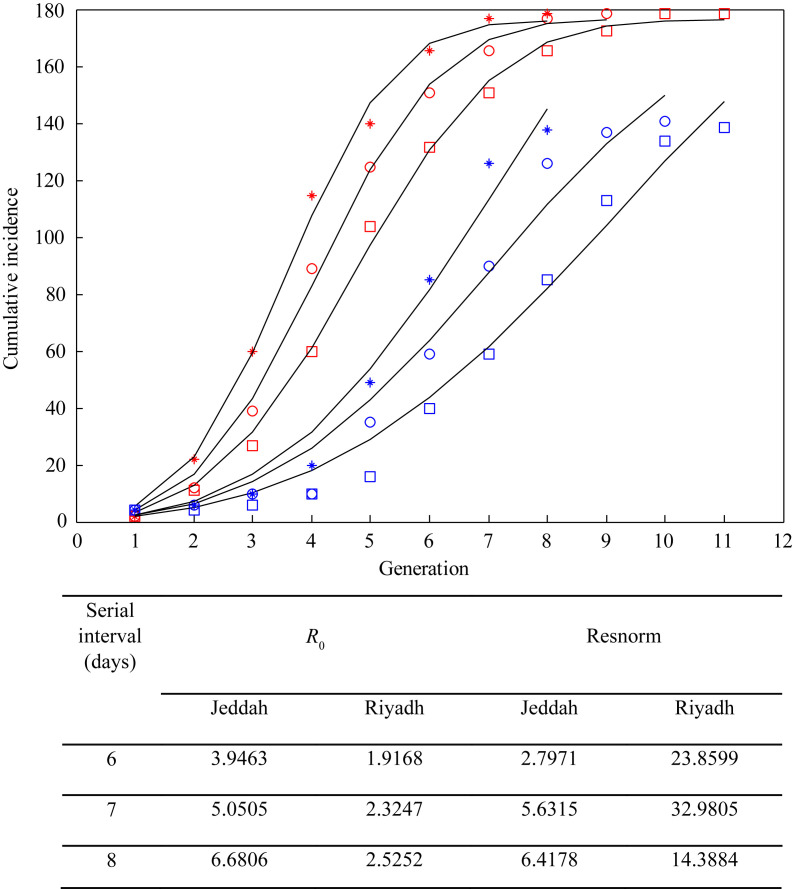
Figure 2 shows the results of the 2014 KSA outbreak. Squares, circles and asterisks represent data aggregation of the number of cases by serial intervals of six, seven and eight days, respectively; the curves represent model fits for best-fit parameters. The estimated R 0 values for Jeddah and Riyadh were in the range of 3.95–6.68 and 1.92–2.52, respectively, using serial intervals of six to eight days. The estimated R 0 values for the Korea MERS outbreak were 3.96, 4.91 and 5.95 for serial intervals of six, seven and eight days, respectively (Figure 3 ). Since most cases were related to nosocomial infections, R 0 for each hospital was also considered. The outbreak in the Samsung hospital was larger than that in the Pyeongtaek hospital (the first Korean outbreak). The Pyeongtaek hospital exhibited best-fit R 0 values of 4.04, 4.23 and 4.39 for serial intervals of six, seven and eight days, respectively, while the Samsung hospital exhibited greater R 0 values of 5.0, 6.8 and 8.11 for serial intervals of six, seven and eight days, respectively. Figure 3 shows that the IDEA model provided well-fitted curves for the cumulative data regarding South Korean MERS symptom-onset dates for all cases.
Figure 2.
Best-fit reproduction number (R0) by serial intervals of Middle East respiratory syndrome in Jeddah and Riyadh, Saudi Arabia, 2014, using the incidence decay with exponential adjustment model. Red squares, Jeddah, six days; red circles, Jeddah, seven days; red asterisks, Jeddah, eight days; blue squares, Riyadh, six days; blue circles, Riyadh, seven days; blue asterisks, Riyadh, eight days.
Figure 3.
Best-fit reproduction number (R0) by serial intervals of Middle East respiratory syndrome in South Korea, 2015, using the incidence decay with exponential adjustment model. Green squares, total, six days; green circles, total, seven days; green asterisks, total, eight days; red squares, Pyeongtaek, six days; red circles, Pyeongtaek, seven days; red asterisks, Pyeongtaek, eight days; blue squares, Samsung, six days; blue circles, Samsung, seven days; blue asterisks, Samsung, eight days.
Although the IDEA model seemed to be appropriate, the original data never fit the model precisely. Therefore, the appropriateness of the model was assessed. Error was evaluated using the relative resnorm to find the best-fit parameters. The results indicated that the best-fit R 0 and serial interval values were 4.9 and seven days for all cases, 4.39 and eight days for the Pyeongtaek hospital, and 5.0 and six days for the Samsung hospital, respectively. d increased with each serial interval because the daily effort of d was aggregated by serial interval.
Discussion
The clusters of MERS-CoV cases in KSA healthcare facilities occurred from late March to late May 2014, while the Korean outbreaks occurred from mid-May to early July 2015. These hospital-based outbreaks exhibited characteristics different from those of community-based outbreaks (higher R 0 values and case fatality rates) [12], [13].
The estimated R 0 is a basic epidemiological variable that is important for selecting appropriate countermeasure efforts. However, an emerging infectious disease often has unknown epidemiology, making it difficult to model mathematically. Several methods have been proposed to address this issue, including the IDEA model. The Richards model can also estimate R 0 using the cumulative daily number of cases and the outbreak turning point (or the peak, ) [14]. In this context, Hsieh used the Richards model to estimate R 0 values for the Korean outbreak as 7.0–19.3. However, the Richards model does not consider any countermeasures implemented during an outbreak; therefore, it can only be used after an outbreak has peaked.
The present study used the IDEA model to estimate R 0 values from the MERS outbreaks in KSA and South Korea. The IDEA model exhibited a good fit: the estimated R 0 values for South Korea were 3.9–8.0, and the best-fit R 0 was 4.9 for a serial interval of seven days. Conversely, R 0 values for Riyadh and Jeddah were 1.9–2.5 and 3.9–6.9, respectively, using serial intervals of six to eight days. Majumder et al. [10] used the IDEA model and estimated very similar R 0 values of 2.0–2.8 for Riyadh and 3.5–6.7 for Jeddah, with serial intervals of six to eight days. However, the estimated R 0 values from the present study were much higher than the previously reported values of <1 for MERS (the threshold for an epidemic) [15]. Regardless, the Korean Government assumed that the outbreak had an R 0 value <1 based on the previous research. The initial criterion for quarantine, therefore, was limited to cases of ‘close contacts’, which were defined as people who were within 2 m of a MERS patient for ≥1 h [16]. These quarantines – established using an incorrectly assumed R 0 – resulted in more MERS patients and greater hospital-to-hospital transmission [16].
A serial interval is the interval between successive cases of an infectious disease. This time period depends on the temporal relationship between the infectiousness of the disease, the clinical onset of the source case, and the incubation period of the receiving case [17]. As MERS becomes infectious with the onset of clinical symptoms, the MERS latency period equals the incubation period. Therefore, the shortest serial interval could be the same as the incubation period, and the longest serial interval could be the sum of the incubation period and the maximum duration of infectiousness. During the Korean MERS outbreak, several super-spreading events occurred because the MERS cases were not isolated immediately upon presentation of clinical symptoms [18]. Thus, these cases contacted susceptible individuals for up to one week after the onset of their clinical symptoms. However, most MERS cases with laboratory confirmation were isolated immediately after onset of clinical symptoms [19], [20]. In this study, as the incubation period was two to 14 days (median: six days), the serial interval was slightly longer than the incubation period. The IDEA model with several serial intervals (four to 12 days) was used and found that intervals of six to eight days provided the best fit. For KSA data, even though the reported date was used instead of the onset date, R 0 was not affected because aggregated data by serial intervals was used in the analysis.
The IDEA model is limited by the fact that d cannot be compared with d of another model. In this context, an increasing d in accordance with increasing serial intervals indicates that the countermeasure efforts are increasing. However, the size of d cannot be compared between two or more models of different outbreaks. Nevertheless, the strength of the IDEA model is its simplicity because R 0 can be estimated using the cumulative number of cases according to the serial interval alone.
In conclusion, the estimated R 0 values from the KSA outbreaks (Riyadh and Jeddah) ranged from 1.9 to 6.9, whereas the estimated values from the South Korean outbreaks ranged from 3.9 to 8.0. Based on these findings, it appears that nosocomial MERS-CoV outbreaks in KSA and South Korea had higher R 0 values than the previously assumed values of <1. Although community-acquired infections are caused by contact, nosocomial infections are caused by a combination of contact and aerosol transmission; therefore, R 0 values for nosocomial infections can be higher than those for community-acquired infections. Hence, more comprehensive countermeasures are needed to address nosocomial MERS infections and prevent spread.
Conflict of interest statement
None declared.
Funding source
This work was supported by the National Cancer Center Grant (NCC-1710141-1) and the Korea National Research Foundation Grant (NRF-2015R1A6A3A01020594).
Data availability
All relevant data are available at http://rambaut.github.io/MERS-Tools/cases2.html.
References
- 1.Chowell G., Sattenspiel L., Bansal S., Viboud C. Mathematical models to characterize early epidemic growth: a review. Phys Life Rev. 2016;18:66–97. doi: 10.1016/j.plrev.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riley S., Fraser C., Donnelly C.A., Ghani A.C., Abu-Raddad L.J., Hedley A.J. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan E.H., Craft D.L., Wein L.M. Emergency response to a smallpox attack: the case for mass vaccination. Proc Natl Acad Sci USA. 2002;99:10935–10940. doi: 10.1073/pnas.162282799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velasco-Hernandez J.X., Gershengorn H.B., Blower S.M. Could widespread use of combination antiretroviral therapy eradicate HIV epidemics? Lancet Infect Dis. 2002;2:487–493. doi: 10.1016/s1473-3099(02)00346-8. [DOI] [PubMed] [Google Scholar]
- 5.Kucharski A.J., Althaus C.L. The role of superspreading in Middle East respiratory syndrome coronavirus (MERS-CoV) transmission. Euro Surveill. 2015;20:14–18. doi: 10.2807/1560-7917.es2015.20.25.21167. [DOI] [PubMed] [Google Scholar]
- 6.Breban R., Riou J., Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013;382:694–699. doi: 10.1016/S0140-6736(13)61492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisman D.N., Leung G.M., Lipsitch M. Nuanced risk assessment for emerging infectious diseases. Lancet. 2014;383:189–190. doi: 10.1016/S0140-6736(13)62123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cauchemez S., Fraser C., Van Kerkhove M.D., Donnelly C.A., Riley S., Rambaut A. Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect Dis. 2014;14:50–56. doi: 10.1016/S1473-3099(13)70304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishiura H., Endo A., Saitoh M., Kinoshita R., Ueno R., Nakaoka S. Identifying determinants of heterogeneous transmission dynamics of the Middle East respiratory syndrome (MERS) outbreak in the Republic of Korea, 2015: a retrospective epidemiological analysis. BMJ Open. 2016;6 doi: 10.1136/bmjopen-2015-009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majumder M.S., Rivers C., Lofgren E., Fisman D. Estimation of MERS-coronavirus reproductive number and case fatality rate for the Spring 2014 Saudi Arabia outbreak: insights from publicly available data. PLOS Curr. 2014 Dec 18;6 doi: 10.1371/currents.outbreaks.98d2f8f3382d84f390736cd5f5fe133c. Edition 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisman D.N., Hauck T.S., Tuite A.R., Greer A.L. An IDEA for short term outbreak projection: nearcasting using the basic reproduction number. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K.M., Ki M., Cho S.I., Hong J.K., Cheong H.K., Kim J.H. Epidemiologic features of the first MERS outbreak in Korea: focus on Pyeongtaek St. Mary's Hospital. Epidemiol Health. 2015;37 doi: 10.4178/epih/e2015041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majumder M.S., Kluberg S.A., Mekaru S.R., Brownstein J.S. Mortality risk factors for Middle East respiratory syndrome outbreak, South Korea, 2015. Emerg Infect Dis. 2015;21:2088–2090. doi: 10.3201/eid2111.151231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh Y.H. 2015 Middle East respiratory syndrome coronavirus (MERS-CoV) nosocomial outbreak in South Korea: insights from modeling. PeerJ. 2015;3 doi: 10.7717/peerj.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J., Chowell G., Jung E. A dynamic compartmental model for the Middle East respiratory syndrome outbreak in the Republic of Korea: a retrospective analysis on control interventions and superspreading events. J Theor Biol. 2016;408:118–126. doi: 10.1016/j.jtbi.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ki M. 2015 MERS outbreak in Korea: hospital-to-hospital transmission. Epidemiol Health. 2015;37 doi: 10.4178/epih/e2015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fine P.E. The interval between successive cases of an infectious disease. Am J Epidemiol. 2003;158:1039–1047. doi: 10.1093/aje/kwg251. [DOI] [PubMed] [Google Scholar]
- 18.Kim S.W., Park J.W., Jung H.D., Yang J.S., Park Y.S., Lee C. Risk factors for transmission of Middle East respiratory syndrome coronavirus infection during the 2015 outbreak in South Korea. Clin Infect Dis. 2017;64:551–557. doi: 10.1093/cid/ciw768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park G.E., Ko J.H., Peck K.R., Lee J.Y., Lee J.Y., Cho S.Y. Control of an outbreak of Middle East respiratory syndrome in a tertiary hospital in Korea. Ann Intern Med. 2016;165:87–93. doi: 10.7326/M15-2495. [DOI] [PubMed] [Google Scholar]
- 20.Cho S.Y., Kang J.M., Ha Y.E., Park G.E., Lee J.Y., Ko J.H. MERS-CoV outbreak following a single patient exposure in an emergency room in South Korea: an epidemiological outbreak study. Lancet. 2016;388:994–1001. doi: 10.1016/S0140-6736(16)30623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are available at http://rambaut.github.io/MERS-Tools/cases2.html.