Highlights
-
•
The China lineage strains have become predominant in the Shandong Province.
-
•
Five strains contained the same deletions as the Vietnam/Laos/Thailand lineage strains in the Nsp2 and Nsp3 regions.
-
•
Three strains with a novel pattern of deletions were observed for the first time in the Nsp2 and Nsp3 regions.
Keywords: Porcine deltacoronavirus (PDCoV), Spike protein, Nucleocapsid protein, ORF1a, Deletion
Abstract
Porcine deltacoronavirus (PDCoV) is the etiological agent of acute diarrhoea and vomiting in pigs, threatening the swine industry worldwide. Although several PDCoV studies have been conducted in China, more sequence information is needed to understand the molecular characterization of PDCoV. In this study, the partial ORF1a, spike protein (S) and nucleocapsid protein (N) were sequenced from Shandong Province between 2017 and 2018. The sequencing results for the S protein from 10 PDCoV strains showed 96.7 %–99.7 % nucleotide sequence identity with the China lineage strains, while sharing a lower level of nucleotide sequence identity, ranging from 95.7 to 96.8%, with the Vietnam/Laos/Thailand lineage strains. N protein sequencing analysis showed that these strains showed nucleotide homologies of 97.3%–99.3% with the reference strains. Phylogenetic analyses based on S protein sequences showed that these PDCoV strains were classified into the China lineage. The discontinuous 2 + 3 aa deletions at 400–401 and 758–760 were found in the Nsp2 and Nsp3 coding region in five strains, respectively, with similar deletions having been identified in Vietnam, Thailand, and Laos. Three novel patterns of deletion were observed for the first time in the Nsp2 and Nsp3 regions. Importantly, those findings suggest that PDCoV may have undergone a high degree of variation since PDCoV was first detected in China.
1. Introduction
Porcine deltacoronavirus (PDCoV) was first discovered in 2009 in Hong Kong from swine fecal samples (Woo et al., 2012). PDCoV was recognized as the causative agent of acute diarrhoea and vomiting in pigs (Hu et al., 2015; Song et al., 2015; Wang et al., 2014). Since its appearance, PDCoV has caused significant economic losses for the swine industry worldwide. PDCoV is a member of the genus Deltacoronavirus in the family Coronaviridae and is an enveloped virus that has a positive-sense single-stranded RNA (+ssRNA) genome of 25.4 kb in length (Lee and Lee, 2015; Phan et al., 2018). The PDCoV genome has seven major open reading frames (ORFs). Two overlapping ORFs (ORF1a and ORF1b) encode two replication-associated proteins, which are both autoproteolytically cleaved into 15 nonstructural proteins (Nsp2 to Nsp16) (Wang et al., 2018a; Woo et al., 2010). The remaining ORF encodes the spike protein (S), envelope protein (E), membrane protein (M) and nucleocapsid protein (N). Additionally, three accessory proteins were identified: nonstructural protein 6 (NS6), NS7, and NS7a (Fang et al., 2017, 2016; Luo et al., 2016).
The S protein is the most variable protein among the PDCoV genes, with only 96.0 %–100 % amino acid sequence identity between Chinese and American strains (Zhang et al., 2019b). The S protein plays a pivotal role in the viral entry and stimulates the induction of neutralizing antibodies in the natural host (Chen et al., 2019c; Chiou et al., 2017; Lin et al., 2016; Zhang et al., 2017). N protein is a conservative target for virological detection by PCR (Lee and Lee, 2015). N protein also plays an important role in viral pathogenesis (Chen et al., 2019b; Likai et al., 2019; Shi et al., 2017; Zhang et al., 2015, 2014). PEDV N protein can antagonize beta interferon and interferon-λ production (Ding et al., 2014; Shan et al., 2018). SARS-CoV N protein can bind to DNA in vitro (Chen et al., 2007). hCoV-OC43 N protein interacts with the transcription factor nuclear factor-kappa B (NF-κB) (de Haan and Rottier, 2005). The ORF1a region is the most variable region of the PDCoV genome and substitutions, deletions and insertions have been observed in the Nsp2 and Nsp3 coding region in Vietnam, Thailand, and Laos (Lorsirigool et al., 2017; Wang et al., 2015).
To determine the molecular epidemiology and genetic variations of PDCoV in China, the partial ORF1a, S protein and N protein genes of 10 PDCoV strains from different pig farms located in Shandong Province were sequenced and analysed. This study may provide valuable information for the molecular epidemiology of PDCoV and its emerging variants in China.
2. Materials and methods
2.1. Sample collection
To monitor the prevalence and sequence properties of PDCoV in Shandong Province, China, a total of 58 porcine samples, including 21 faecal samples and 37 intestinal samples, were collected from different commercial swine from September 2017 to December 2018. All samples were stored at −80 °C and were subsequently used for RNA extraction.
2.2. RNA extraction and PDCoV detection
RNAs were extracted from the samples using the RNAeasy mini kit (TaKaRa BIO INC., Dalian, China) according to the manufacturer’s instructions. To detect, differentiate and sequence PDCoV, the One Step RT-PCR kit (TaKaRa Co., Dalian, China) was used to synthesis cDNA.
Ten PDCoV-positive samples were selected for the partial ORF1a, S protein and N protein sequences. The primer sets are listed in Table 1 . TGEV, PEDV and PoRV were detected as described previously (Wang et al., 2018c). The PCR conditions were as follows: an initial PCR activation temperature of 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C∼65 °C for 50 s, and extension at 72 °C for 120 s and another extension at 72 °C for 10 min. The PCR products were ligated into the pMD18-T cloning vector (TaKaRa Co., Dalian, China) and sequenced.
Table 1.
List of primers used in the study.
| Primer name | Primer sequence (5′–3′) | Size (bp) | Target Genes |
|---|---|---|---|
| PDCoV S1-F | 5′-ATGCAGAGAGCTCTATTGATTATGAC-3′ | 1763 bp | S1 |
| PDCoV S1-R | 5′-AACTTGCAAGTACTCCGTCTGAACG-3′ | ||
| PDCoV S2-F | 5′-ATTTTCTCTTTCCGTTCAGACGGAG-3′ | 1750 bp | S2 |
| PDCoV S2-F | 5′-CTACCATTCCTTAAACTTAAAGGACG-3′ | ||
| PDCoV N-F | 5′-ATGGCTGCACCAGTAGTCCCTA-3′ | 1045 bp | N |
| PDCoV N-R | 5′-CTACGCTGCTGATTCCTGCTTTAT-3′ | ||
| PDCoV Nsp2-3F | 5′-TGGCCCGTCTAAACCTGGGGATGTCAT-3′ | 1290 bp | ORF1a/1b |
| PDCoV Nsp2-3R | 5′-ATGGACCTCAGGATCCGGTACTGAGT-3′ | ||
2.3. Nucleotide sequencing and phylogenetic analysis
The partial ORF1a, S protein and N protein sequences of PDCoV were independently used for sequence alignments and phylogenetic analyses. The nucleotide and deduced amino acid sequences were assessed using BioEdit 7.0. All the sequences were aligned using the MEGA5 program, and phylogenetic trees were constructed using the neighbour-joining method. The reliability of the branching orders was evaluated by the bootstrap test (n = 1,000). After nucleotide homology comparison and screening, 29 representative strains were selected from 102 strains for molecular evolutionary analyses (Tables 2 and S1). In addition, these strain sequences have been previously published as reference sequences (Suzuki et al., 2018; Zhang et al., 2019a, c; Zhang et al., 2019d).
Table 2.
PDCoV strains described in this study.
| No. | Strain | Accession No. | Collection date | Collection Country | Length (bp) |
|---|---|---|---|---|---|
| 1 | HKU15-44 | JQ065042 | 2009 | China: Hong Kong | 25430 |
| 2 | USA/Iowa136/2015 | KX022602 | 2015 | USA: Iowa | 25382 |
| 3 | USA/Nebraska145/2015 | KX022605 | 2015 | USA: Nebraska | 25320 |
| 4 | USA/Michigan448/2014 | KR265850 | 2014 | USA: Michigan | 25394 |
| 5 | USA/Indiana453/2014 | KR265851 | 2014 | USA: Indiana | 25394 |
| 6 | USA/Nebraska210/2014 | KR265861 | 2014 | USA: Nebraska | 25404 |
| 7 | OhioCVM1/2014 | KJ769231.1 | 2014 | USA | 25433 |
| 8 | IL/2014/026PDV_P11 | KP981395.1 | 2014 | USA: Illinois | 25422 |
| 9 | USA/Minnesota292/2014 | KR265864 | 2014 | USA: Minnesota | 25395 |
| 10 | YMG/JPN/2014 | LC260044 | 2014 | Japan: Yamagata | 25362 |
| 11 | KNU14-04 | KM820765 | 2012 | South Korea | 25422 |
| 12 | P1_16_BTL_0115/PDCoV/2016/Lao | KX118627 | 2016 | Laos | 25405 |
| 13 | Vietnam/Binh21/2015 | KX834352 | 2015 | Viet Nam | 25406 |
| 14 | Thailand/S5015L/2015 | KU051649 | 2015 | Thailand | 25405 |
| 15 | CH/SXD1/2015 | KT021234 | 2015 | China: Shangxi | 25419 |
| 16 | CHN-JS-2014 | KP757892 | 2014 | China: Jangsu | 25420 |
| 17 | CH/Sichuan/S27/2012 | KT266822 | 2012 | China: Sichuan | 25404 |
| 18 | CHN-AH-2004 | KP757890 | 2004 | China: Anhui | 25420 |
| 19 | CHN-HB-2014 | KP757891 | 2014 | China: Heibei | 25420 |
| 20 | CHN/Tianjin/2016 | KY065120 | 2016 | China: Tianjin | 25413 |
| 21 | CH/JXJGS01/2016 | KY293677 | 2016 | China: Jiangxi | 25438 |
| 22 | CH/Hunan/2014 | KY513724 | 2014 | China: Hunan | 25413 |
| 23 | CH/Jiangsu/2014 | KY513725 | 2014 | China: Jiangsu | 25422 |
| 24 | CHN-HG-2017 | MF095123 | 2017 | China | 25399 |
| 25 | GD | MF431742 | 2015 | China: Guangdong | 25420 |
| 26 | SD | MF431743 | 2014 | China: Shandong | 25414 |
| 27 | CHN/GS/2017/1 | MF642324 | 2017 | China: Gansu | 25420 |
| 28 | CHN-HN-1601 | MG832584 | 2016 | China | 25419 |
| 29 | HNZK-02 | MH708123 | 2018 | China: Henan | 25453 |
3. Results
3.1. Sample screening and genome sequencing
Twelve of fifty-eight (20.68 %) field samples were positive for PDCoV, while the PEDV infection rate was 34.48 % (20/58), and the co-infection rate of PEDV and PDCoV was up to 50.00 % (10/20). 12 PDCoV positive samples were identified from 4 faecal samples and 8 intestinal samples. PRV was identified in 5 of 58 samples. Two of fifty-eight samples were found to be positive for TGEV. PRV/PEDV and TGEV/PEDV co-infections were 15.00 % (3/20) and 5.00 % (1/20), respectively. None of the PDCoV-positive samples were positive for TGEV and PRV. PDCoV/PEDV co-infections were the most common.
To obtain the sequence, the complete S protein, N protein and partial ORF1a genes were amplified in 10 PDCoV-positive samples (Table 1). The 10 strains were named SD01-2018, SD03-2018, SD05-2018, SD06-2018, SD07-2018, SD08-2018, SD09-2018, SD10-2018, SD11-2018, and SD12-2018. The GenBank accession numbers of the complete S protein, N protein and partial ORF1a genes were as follows: MN173803-MN173812, MN173783-MN173792, and MN173793- MN173802, respectively.
3.2. Phylogenetic analysis of the S, N and partial ORF1a/1b genes
3.2.1. Phylogenetic analysis of S protein
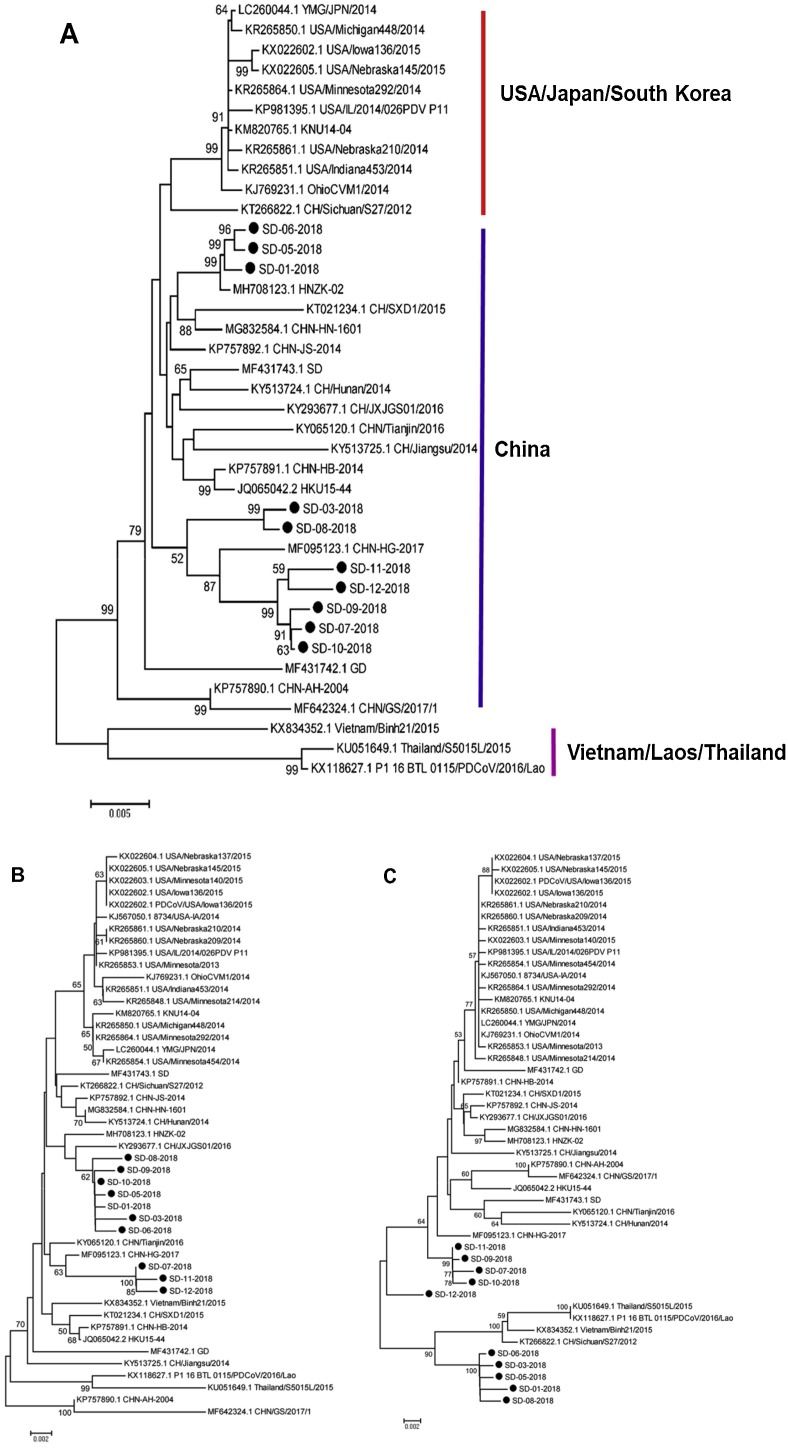
Phylogenetic analysis of the nucleotide sequences of the S protein sequences indicated that all strains worldwide can be categorized into three lineages: the China lineage, USA/Japan/South Korea lineage and Vietnam/Laos/Thailand lineage. All new PDCoVs strains from Shandong in our study belonged to the China lineage (Fig. 1 A).
Fig. 1.
Phylogenetic analyses of PDCoV using the neighbour-joining algorithm and 1000 bootstrap replications in a heuristic search with 29 PDCoV reference strains available in GenBank. A S protein genomic sequence. B N protein sequence. C Partial ORF1a gene.
The sequence alignment results showed that these new strains shared nucleotide sequence homologies of 97.5 %–99.9 % with each other. They also shared up to 96.7 %–99.7 % nucleotide sequence similarity with the representative China lineage strains (HKU15-44, CHN-AH-2004, CHN-HN-2014 and CHN-HG-2017) and 97.5 %–98.6 % with the representative USA/Japan/South Korea lineage strains (OhioCVM1/2014, IL/2014/026PDV_P11, YMG/JPN/2014 and KNU14-04). By contrast, the new strains showed lower 95.7–96.8 % nucleotide sequence identity with the representative Vietnam/Laos/Thailand lineage strains (Vietnam/Binh21/2015, Thailand/S5011/2015 and P1_16_BTL_0115 /2016/Lao).
3.2.2. Phylogenetic analysis of N protein
A phylogenetic tree was constructed using the N protein sequences from the strains isolated from Shandong and the references strains (Fig. 1B). SD07-2018, SD11-2018 and SD12-2018 with CHN-HG-2017 were classified in a group. Seven strains and HNZK-02 and CH/JXJGGS01/2016 were classified in a branch.
The deduced amino acid identity values of N protein sequences were analysed, and shared nucleotide sequence homologies of 98.4 %–99.9 % were found among 10 strains. They shared 97.9–99.3 %, 98.3 %–98.9 %, and 97.3 %–98.9 % sequence identity with the China lineage strains, USA/Japan/South Korea lineage strains and Viet Nam/Laos/Thailand lineage strains, respectively.
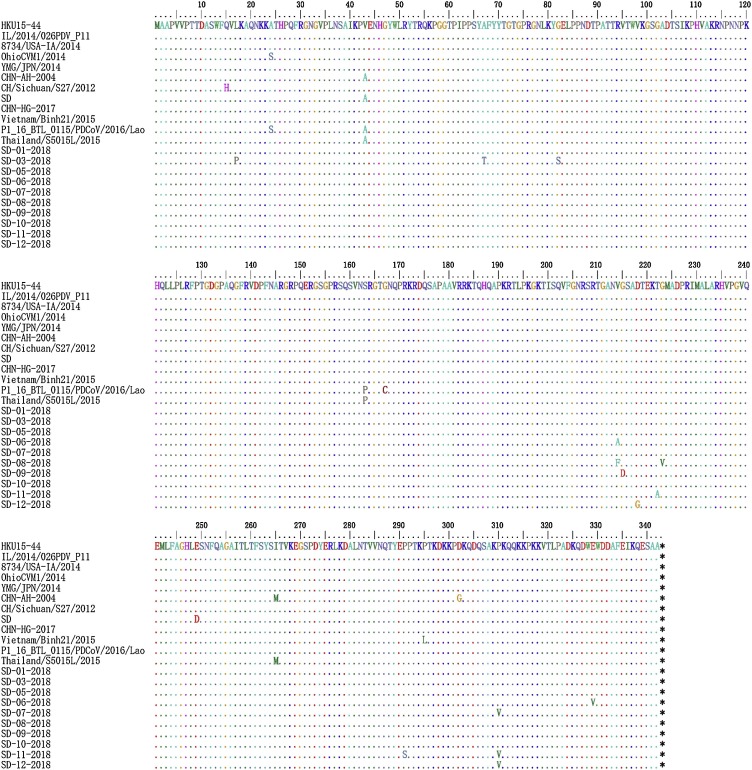
As shown in Fig. 2 , all the PDCoV N protein sequences were the same length (1029 nt) and encoded 342 amino acid residues. Compared to other structural proteins, N protein is the most conserved structural proteins and remains the primary target protein for current diagnostic tests (McBride et al., 2014). Recently, a recombinant N protein-based indirect enzyme-linked immunosorbent assay (ELISA) (rPDCoV-N-ELISA) was established to detect PDCoV IgG antibodies (Su et al., 2016). Similar ELISA test has been done in Taiwan (Hsu et al., 2018). Compared with the reference strain HKU15-44, the following aa changes were detected in CHN-AH-2004, SD, P1_BTL_0115/2016/Lao and Thailand/S5015 L/2015 at 43 V/A, 163S/P, and 265I/M, respectively. Several amino acid mutations were detected in 10 strains at positions 18 L/P, 67Y/T, 82 G/S, 215 G/A/F, 216S/D, 218D/G, 222 G/A, 223 M/V, 291 P/S, 310 P/V, and 329E/V. N protein is a multifunctional protein, the key conserved amino acid site mutations affect their function, for example, the Arg-76 and Tyr-94 residues in the IBV N protein are critical for RNA binding, SARS-CoV N protein also exists Arg-94 and Tyr-122 residues (Tan et al., 2006). PDCOV is difficult to isolate and there are few reverse genetic studies. So far, there have been no related reports that mutations in key amino acids of the N protein will affect the current diagnostic tests.
Fig. 2.
Analysis and comparison of amino acid mutations of the N protein of 10 PDCoV strains in Shandong, China.
3.2.3. Phylogenetic analysis of the partial ORF1a gene
A phylogenetic tree was generated based on the deduced aa sequence of the partial ORF1 gene. SD01-2018, SD03-2018, SD05-2018, and SD08-2018 strains were placed into one subgroup with the Vietnam/Laos/Thailand lineage strain and CH/Sichuan/S27/2012 strain. The other subgroup containing SD07-2018, SD09-2018, SD10-2018 and SD11-2018 was closer to the Chinese strain CHN-HG-2017. The SD12-2018 strain was placed into one subgroup (Fig. 1C).
The partial ORF1a gene of the 10 strains shared 95.9 %–98.7 % nucleotide identity with the China lineage strains (HKU15-44, CHN-AH-2004, CHN-HN-2014 and CHN-HG-2017). These strains shared 96.0 %–97.5 % nucleotide identity with the USA/Japan/South Korea lineage strains (OhioCVM1/2014, IL/2014/026PDV_P11, YMG/JPN/2014 and KNU14-04) and 95.3 %–97.9 % nucleotide identity with the Vietnam/Laos/Thailand lineage strains (Vietnam/Binh21/2015, Thailand/S5011/2015 and P1_16_BTL_0115 /2016/Lao).
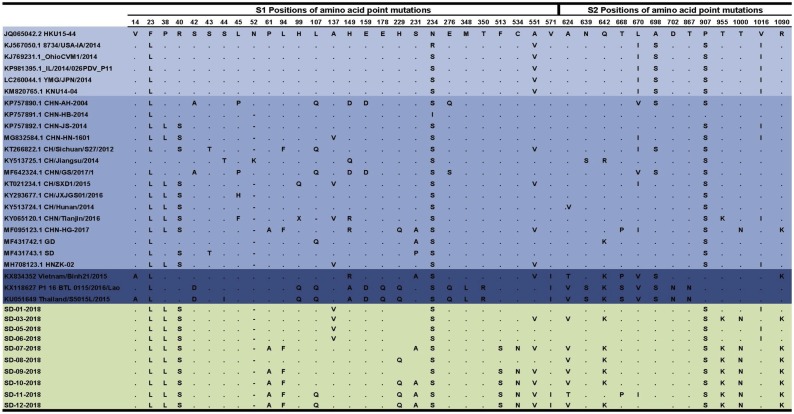
3.3. Amino acid variants in the spike proteins
Compared with the USA/Japan/South Korea lineage and Vietnam/Laos/Thailand lineage strains, the China lineage strains have 3-nt TAA deletions (52 N), leading to the lack of an asparagine in the S gene. This may be the most important feature of the Chinese lineage strains, except for CH/Jiangsu/2014 (Zhang et al., 2019d). Sequence analysis showed that 27 aa mutations were detected in S1 (Fig. 3 ). PDCoV employs host aminopeptidase N (APN) as an entry receptor and interacts with APN via domain B of its S1 protein (residues 298–425) (Li et al., 2018b; Shang et al., 2018). The S1 protein region mutation (residues 229, 231, 234, 276, 348 and 350) may have an impact on virus entry or recognition by neutralizing antibodies.The Vietnam/Laos/Thailand lineage strains had the following unique substitutions: 14 V/A, 42S/D, 99 H/Q,107 H/Q, 149 H/AR, 159E/D, 178E/Q, 229 H/Q, 348 M/L and 350 T/R. The USA/Japan/South Korea lineage strains only had the unique substitutions 23 F/L, 234 N/RS, 551A/V, 670 L/I, 698A/S, 907 P/S and 1016 V/I. Among these 4 strains, SD07-2018, SD09-2018, SD11-2018 and SD12-2018 had another 2 unique substitutions at positions 513 F/S and 534C/N.
Fig. 3.
Analysis and comparison of amino acid mutations of the S protein of 10 PDCoV strains in Shandong, China.
3.4. Novel amino acid deletions or insertions were detected in the partial ORF1a gene
The partial ORF1a gene has the highest genetic diversity in the genomes of the PDCoV strains (Xu et al., 2018). To determine whether the strains in this study possessed the characteristic deletions in the ORF1a gene found in the Thailand PDCoVs previously described, the partial ORF1a sequences containing the Nsp2 and Nsp3 hypervariable region (HVR) from 10 PDCoV strains were sequenced and aligned with the reference isolates, especially those with known pathogenicity, OhioCVM1/2014, IL/2014 /026PDV_P11, YMG/JPN/2014, and CHN-HG- 2017.
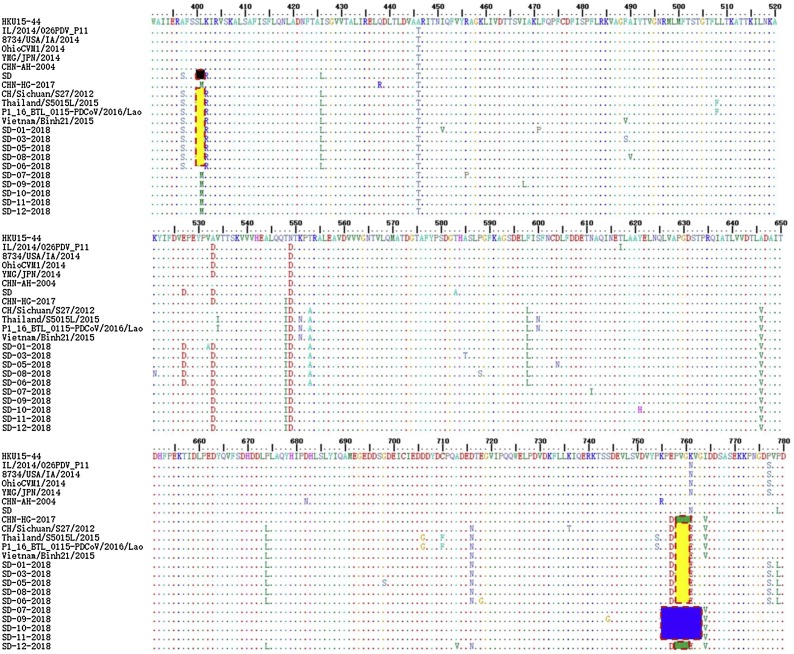
Compared with the OhioCVM1/2014, IL/2014 /026PDV_P11, and YMG/JPN/2014 strains, five strains (SD01-2018, SD03-2018, SD05-2018, SD06-2018 and SD08-2018) in the partial ORF1a gene had the same length of 1255 nucleotides, containing the same two discontinuous deletions of 2 + 3 amino acids at positions 400–401 and 758–760 as the Vietnam/Binh21/2015 and Thailand/S5011/2015 strains, suggesting that these strains are the dominating strain circulating in Shandong Province. Four strains SD07-2018, SD09-2018, SD10-2018 and SD11-2018 in the partial ORF1a gene had a length of 1249 nucleotides, containing a continuous 0 + 7 amino acid deletion at position 755–761. The SD12-2018 strain in the Nsp3 gene had a length of 1261 nucleotides, and a 0 + 3 amino acid deletion at position 758–760. By contrast, no deletions or insertions were detected within the ORF1a gene in the Chinese strains collected early in the study. Additionally, the SD strain contained a 2 + 0 deletion at positions 400–401 in the Nsp2 gene.
As shown in Fig. 4 , five strains (SD01-2018, SD03-2018, SD05-2018, SD06-2018 and SD08-2018) with a 7-aa deletion in ORF1 regions had substitutions identical with those of the Thailand/S5015 L/2015 strain at positions 397A/S, 427I/L, 447A/T, 547 T/I, 550 N/D, 554 T/A, 599 F/L, 647A/V, 670 P/L, 717D/N, and 762 K/E. Additionally, five strains had unique substitutions at positions 529E/D, 778 P/S and 780 P/I. Notably, four strains (SD07-2018, SD09-2018, SD11-2018 and SD12-2018) carried substitutions at position 401 L/M.
Fig. 4.
Amino acid sequence alignments of the partial ORF1gene at 369–791 aa. Two discontinuous amino acid deletions at positions 400–401 (yellow regions) and 758–760 (yellow regions) in Nsp2 and Nsp3 are also observed in SD01-2018, SD03-2018, SD05-2018, SD08-2018, the Vietnam/Laos/Thailand lineage and CH/Sichuan/S27/ 2012 strains. An additional 7 deletions at positions 755–761 (blue regions) in Nsp3 are observed in the SD07-2018, SD09-2018, SD10-2018 and SD11-2018 strains. Three continuous amino acid deletions at positions 758–760 (green regions) in Nsp3 are observed in SD12-2018 and CHN-HG-2017 strains. Two continuous amino acid deletions at positions 401–402 (black regions) in Nsp2 of the SD reference strain.
4. Discussion
Several enteric coronaviruses have been identified from diarrhoeic piglets, such as porcine epidemic diarrhoea virus (PEDV) (Li et al., 2012), transmissible gastroenteritis virus (TGEV) (Xia et al., 2018), Swine acute diarrhea syndrome coronavirus (SADS-CoV), also named as Swine enteric alphacoronavirus (SeACoV) (Fu et al., 2018; Zhou et al., 2018) and PDCoV. Outbreaks of these enteric coronaviruses have been reported in China, causing substantial economic losses in the swine industry (Qing et al., 2016; Zeng et al., 2015; Zhou et al., 2019). In 2014, PDCoV was first detected in China, causing tremendous financial losses in the swine industry, and the virus has rapidly spread nationwide (Dong et al., 2015; Liu et al., 2018; Mai et al., 2018a; Song et al., 2015; Wang et al., 2018b). Thus, the molecular characterization of these PDCoVs is a major focus of Chinese virological research.
Although several studies have shown an obvious increase in the genomic diversity of PDCoV in China (Dong et al., 2016; Liang et al., 2019; Mai et al., 2018b), previous studies have focused mainly on the S genes, while the genetics of the Chinese PDCoVs based on the ORF1 gene are not well characterized. The Vietnam/Laos/Thailand lineage strains had 6-nt (AGTTTG) and 9-nt (GAGCCAGTC) deletions in ORF1a (Le et al., 2018). In 2015, only a few strains with similar deletions have been reported in China, such as the CH/Sichuan/S27/2012 strain with the 6-nt and 9-nt deletions in the ORF1a (Wang et al., 2015). The ORF1a deletion region encoded Nsp2 and Nsp3 proteins. The Nsp3 protein acts as a scaffold protein to interact with itself and bind other viral Nsps or host proteins (Nogales et al., 2012; Yuan et al., 2015). Nsp3 protein contains one or two papain-like protease (PLpro) domain(s) with deubiquitinating (DUB) and deISGylating activities in SARS-CoV and MERS-CoV (Alfuwaires et al., 2017; Neuman, 2016). In MHV, the Nsp3 protein ubiquitin-like domains could interfere with pathways involving ubiquitinylated or ISGylated host targets, thereby leading to the disruption of host anti-viral signal transduction or protein degradation (Chen and Makino, 2004). Nsp3 protein was also recommended as a marker for monitoring coronavirus evolution and for surveying the molecular epidemiology in lineage C betaCoVs (Forni et al., 2016). Additionally, the MERS-CoV Nsp3 Arg911Cys mutation is an example of adaptive evolution(Shokri et al., 2019). In TGEV Nsps 2, 3, and 8 were incorporated into the CoV virions, involving in CoV replication (Nogales et al., 2012).
Mutation and recombination are important mechanisms for PDCoV evolution. The USA/Japan/South Korea lineage strains were characterized by no discontinuous deletions in the Nsp2 and Nsp3 coding regions, while the Vietnam/Laos/Thailand lineage strains showed a discontinuous 5-aa deletion (2aa+3aa) at 400–401 and 758–760 in the Nsp2 and Nsp3 coding regions. These regional deletions were also observed in a Chinese CH/Sichuan/S27/2012 strain. Here, we found 5 novel strains with the same deletion pattern of “2 + 3aa” in the Nsp2 and Nsp3 coding regions. Interestingly, the SD12-2018 and CHN-HG-2017 strains have a continuous “0 + 3aa” in the Nsp3 deletion at 758–760. Additionally, SD07-2018, SD09-2018, SD10-2018 and SD11-2018 strains have a continuous “0 + 7aa” deletion at 755–761 in Nsp3. Two continuous aa deletion in Nsp2 was also identified in a Chinese strain, SD, which has two“2 + 0aa” deletions at position 400–401. Remarkably, natural deletions and insertions occurred in the ORF1a sequence, and these have led to genome size differences among PDCoV strains. However, the role of double deletions in the ORF1 of this virus remains unclear. Thus, the current results indicate that the novel deletion of Nsp2 and Nsp3 may provide another approach to the diagnosis.
The PDCoV S protein is a glycoprotein of approximately 1383 amino acids aa with an apparent molecular mass of 180 kDa. Studies of genomic sequences analyses using S protein genes have shown that PDCoV can be further divided into three lineages: the China lineage, USA/Japan/South Korea lineage and Vietnam/Laos/Thailand lineage Zhang et al., 2019d). The Vietnam/Laos/Thailand lineage has been detected in Southeast Asia (Janetanakit et al., 2016; Lorsirigool et al., 2017, 2016; Saeng-Chuto et al., 2017). The USA/Japan/South Korea lineage has a worldwide distribution (Jang et al., 2017; Nelson et al., 2019; Niederwerder and Hesse, 2018; Perez-Rivera et al., 2019; Suzuki et al., 2018). The China lineage has been the predominant lineage in China since 2014 (Li et al., 2018a; Zhang et al., 2019a). S protein-based phylogenetic trees showed that 7 strains belonging to the China lineage were classified into a minor branch with CHN-HG-2017. The evidence showed that the PDCoV strains of China have genetic diversity.
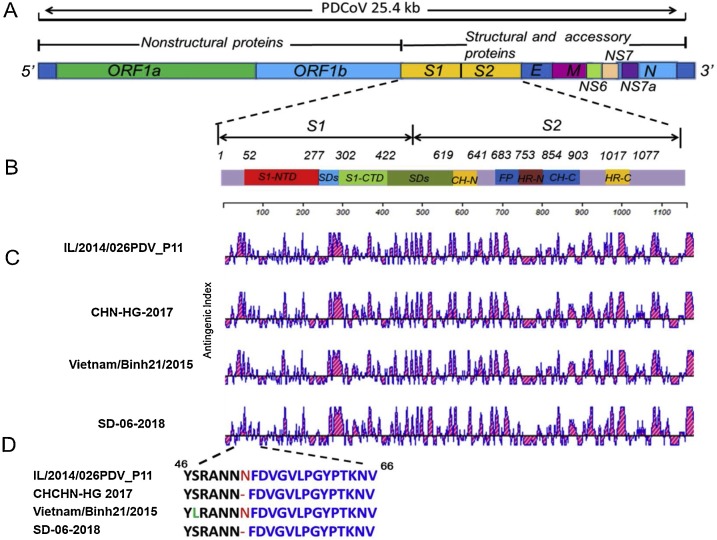
The PDCoV spike (S) protein comprises S1, a receptor-binding subunit, and S2, a membrane fusion subunit. The S1 domain is important for recognizing and binding to cell receptors. Additionally, the S1 domain contains several neutralizing epitopes that stimulate the induction of neutralizing antibodies. The S2 domain is involved in triggering the fusion of the viral envelope and target cell membrane. Therefore, the S protein has been the primary target for the development of vaccines against PDCoV and for determining genetic relatedness among PDCoV isolates. In this study, compared with the USA/Japan/South Korea lineage and Vietnam/Laos/Thailand lineage strains, the 10 strains showed one amino acid (52 N) deletion in the S protein. A comparison of the antigenic index profiles of the S protein among the IL/2014/026PDV_P11, CHN-HG-2017, and Vietnam/Binh21/2015 lineage strains and SD-06-2018 indicated that in SD-06-2018 the deletion region appears to have a higher degree of antigenic change than that of other strains, indicating it may be involved in PDCoV immune escape. S1 NTD has higher sequence variability (Fig. 5 ).
Fig. 5.
Schematic diagrams of the PDCoV genomic structure. (A) Organization of the PDCoV genome. (B) Organization of the S genome and locations of unique amino acids (aa). (C) Comparison of the antigenic index profiles of the S protein among the IL/2014/026PDV_P11 (representative USA/Japan/South Korea lineage strain), CHN-HG-2017 (representative China lineage strain), Vietnam/Binh21/2015 (representative Vietnam/Laos/Thailand lineage strain) and SD-06-2018. (D) Amino acid deletions/insertions are shown with IL/2014/026PDV_P11, CHN-HG-2017, Vietnam/Binh21/2015 and SD-06-2018 strains.
The N protein is the predominant antigen produced in coronavirus-infected cells, making it a major viral target (Kocherhans et al., 2001). The coronavirus N protein is a multifunctional protein involved in virus assembly, translation, apoptosis induction and host innate immune defence. SARS-CoV N protein not only modulates the host cell cycle by regulating cyclin-CDK activity but also inhibits the synthesis of type-1 interferon (1 F N) (Chang et al., 2014; Liao et al., 2005). PDCoV N protein suppressed Sendai virus (SEV)-induced IFN-β production and transcription factor IRF3 activation (Chen et al., 2019a). Furthermore, the N-terminal region (1–246 aa) interacts with pRIG-I and interferes with its function (Chen et al., 2019a). The N protein analysed showed few differences among the three lineage strains. They shared 97.3 %–99.3 % sequence identity with the three lineage strains.
Analysis showed that the N protein and S protein evolutionary rates are inconsistent. N protein is the general target for PDCoV detection, and the genetic extent of variability should be evaluated carefully.
5. Conclusions
In summary, this study provides more information about the deletion and genetic diversity of PDCoV. Phylogenetic analysis revealed that these strains belonged to the Chinese lineage. Meanwhile, new PDCoV strains with different patterns of deletions in Nsp2 and Nsp3 are emerging, and these strains may lead to increased pathogenicity of the virus. Our study provides useful information to prevent PDCoV in China.
Declaration of Competing Interest
The authors declared no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgment
This work was supported by the Wenzhou Basic Agricultural Science and Technology Project (Grant Numbers N20180010 and N20190005).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.197869.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Alfuwaires M., Altaher A., Kandeel M. Molecular dynamic studies of interferon and innate immunity resistance in MERS CoV non-structural protein 3. Biol. Pharm. Bull. 2017;40:345–351. doi: 10.1248/bpb.b16-00870. [DOI] [PubMed] [Google Scholar]
- Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein—forms and functions. Antiviral Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.J., Makino S. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 2004;78:5658–5669. doi: 10.1128/JVI.78.11.5658-5669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Fang P., Wang M., Peng Q., Ren J., Wang D., Peng G., Fang L., Xiao S., Ding Z. Porcine deltacoronavirus nucleocapsid protein antagonizes IFN-beta production by impairing dsRNA and PACT binding to RIG-I. Virus genes. 2019;55:520–531. doi: 10.1007/s11262-019-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Fang P., Wang M., Peng Q., Ren J., Wang D., Peng G., Fang L., Xiao S., Ding Z. Porcine deltacoronavirus nucleocapsid protein antagonizes IFN-beta production by impairing dsRNA and PACT binding to RIG-I. Virus Genes. 2019;55:520–531. doi: 10.1007/s11262-019-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Wang K., Hou Y., Li H., Li X., Yu L., Jiang Y., Gao F., Tong W., Yu H., Yang Z., Tong G., Zhou Y. Genetic evolution analysis and pathogenicity assessment of porcine epidemic diarrhea virus strains circulating in part of China during 2011-2017. Infect. Genet. Evol. 2019;69:153–165. doi: 10.1016/j.meegid.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou H.Y., Huang Y.L., Deng M.C., Chang C.Y., Jeng C.R., Tsai P.S., Yang C., Pang V.F., Chang H.W. Phylogenetic analysis of the spike (S) gene of the new variants of porcine epidemic diarrhoea virus in Taiwan. Transbound. Emerg. Dis. 2017;64:157–166. doi: 10.1111/tbed.12357. [DOI] [PubMed] [Google Scholar]
- de Haan C.A., Rottier P.J. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Zeng S., Sun Q., Chen H., Xiao S. Porcine deltacoronavirus in Mainland China. Emerg. Infect. Dis. 2015;21:2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Yang H., Liu H., Du T., Fang P., Wang D., Chen H., Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Liu X., Hong Y., Wang Y., Dong N., Ma P., Bi J., Wang D., Xiao S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology. 2016;499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Hong Y., Liu X., Dong N., Ma P., Bi J., Wang D., Xiao S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 2017;98:173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Mozzi A., Pozzoli U., Al-Daghri N., Clerici M., Sironi M. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J. Virol. 2016;90:3627–3639. doi: 10.1128/JVI.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Fang B., Liu Y., Cai M., Jun J., Ma J., Bu D., Wang L., Zhou P., Wang H., Zhang G. Newly emerged porcine enteric alphacoronavirus in southern China: identification, origin and evolutionary history analysis. Infect. Genet. Evol. 2018;62:179–187. doi: 10.1016/j.meegid.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T.H., Liu H.P., Chin C.Y., Wang C., Zhu W.Z., Wu B.L., Chang Y.C. Detection, sequence analysis, and antibody prevalence of porcine deltacoronavirus in Taiwan. Arch. Virol. 2018;163:3113–3117. doi: 10.1007/s00705-018-3964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetanakit T., Lumyai M., Bunpapong N., Boonyapisitsopa S., Chaiyawong S., Nonthabenjawan N., Kesdaengsakonwut S., Amonsin A. Porcine Deltacoronavirus, Thailand, 2015. Emerging Infect. Dis. 2016;22:757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G., Lee K.K., Kim S.H., Lee C. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014-2016. Transbound. Emerg. Dis. 2017;64:1364–1370. doi: 10.1111/tbed.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus genes. 2001;23:137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le V.P., Song S., An B.H., Park G.N., Pham N.T., Le D.Q., Nguyen V.T., Vu T.T.H., Kim K.S., Choe S., An D.J. A novel strain of porcine deltacoronavirus in Vietnam. Arch. Virol. 2018;163:203–207. doi: 10.1007/s00705-017-3594-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015;208:136–145. doi: 10.1016/j.virusres.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.L., Zhu L., Ma J.Y., Zhou Q.F., Song Y.H., Sun B.L., Chen R.A., Xie Q.M., Bee Y.Z. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field strains in south China. Virus Genes. 2012;45:181–185. doi: 10.1007/s11262-012-0735-8. [DOI] [PubMed] [Google Scholar]
- Li D., Feng H., Liu Y., Chen Y., Wei Q., Wang J., Liu D., Huang H., Su Y., Wang D., Cui Y., Zhang G. Molecular evolution of porcine epidemic diarrhea virus and porcine deltacoronavirus strains in Central China. Res. Vet. Sci. 2018;120:63–69. doi: 10.1016/j.rvsc.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Kenney S.P., Widjaja I., Jung K., Alhamo M.A., van Dieren B., van Kuppeveld F.J.M., Saif L.J., Bosch B.J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E5135–E5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Li B., Zhang H., Hu H. Complete genome sequences of two porcine deltacoronavirus strains from Henan Province, China. Microbiol. Resour. Announc. 2019;8 doi: 10.1128/MRA.01517-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q.J., Ye L.B., Timani K.A., Zeng Y.C., She Y.L., Ye L., Wu Z.H. Activation of NF-kappaB by the full-length nucleocapsid protein of the SARS coronavirus. Acta Biochim. Biophys. Sin. 2005;37:607–612. doi: 10.1111/j.1745-7270.2005.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likai J., Shasha L., Wenxian Z., Jingjiao M., Jianhe S., Hengan W., Yaxian Y. Porcine deltacoronavirus nucleocapsid protein suppressed IFN-beta production by interfering porcine RIG-I dsRNA-binding and K63-linked polyubiquitination. Front. Immunol. 2019;10:1024. doi: 10.3389/fimmu.2019.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Saif L.J., Marthaler D., Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.J., Zuo Y.Z., Gu W.Y., Luo S.X., Shi Q.K., Hou L.S., Zhong F., Fan J.H. Isolation and phylogenetic analysis of porcine deltacoronavirus from pigs with diarrhoea in Hebei province, China. Transbound. Emerg. Dis. 2018;65:874–882. doi: 10.1111/tbed.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsirigool A., Saeng-Chuto K., Temeeyasen G., Madapong A., Tripipat T., Wegner M., Tuntituvanont A., Intrakamhaeng M., Nilubol D. The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Arch. Virol. 2016;161:2909–2911. doi: 10.1007/s00705-016-2983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsirigool A., Saeng-Chuto K., Madapong A., Temeeyasen G., Tripipat T., Kaewprommal P., Tantituvanont A., Piriyapongsa J., Nilubol D. The genetic diversity and complete genome analysis of two novel porcine deltacoronavirus isolates in Thailand in 2015. Virus Genes. 2017;53:240–248. doi: 10.1007/s11262-016-1413-z. [DOI] [PubMed] [Google Scholar]
- Luo J., Fang L., Dong N., Fang P., Ding Z., Wang D., Chen H., Xiao S. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology. 2016;495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai K., Feng J., Chen G., Li D., Zhou L., Bai Y., Wu Q., Ma J. The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong Province in Southern China. Transbound. Emerg. Dis. 2018;65:166–173. doi: 10.1111/tbed.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai K., Li D., Wu J., Wu Z., Cheng J., He L., Tang X., Zhou Z., Sun Y., Ma J. Complete genome sequences of two porcine deltacoronavirus strains, CHN-GD16-03 and CHN-GD16-05, isolated in Southern China, 2016. Genome Announc. 2018;6 doi: 10.1128/genomeA.01545-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S.W., Zentkovich M.M., Nolting J.M., Bowman A.S. Porcine epidemic diarrhea virus and porcine deltacoronavirus not detected in waterfowl in the North American Mississippi migratory bird flyway in 2013. J. Wildl. Dis. 2019;55:223–226. doi: 10.7589/2018-03-074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res. 2016;135:97–107. doi: 10.1016/j.antiviral.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederwerder M.C., Hesse R.A. Swine enteric coronavirus disease: a review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound. Emerg. Dis. 2018;65:660–675. doi: 10.1111/tbed.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales A., Marquez-Jurado S., Galan C., Enjuanes L., Almazan F. Transmissible gastroenteritis coronavirus RNA-dependent RNA polymerase and nonstructural proteins 2, 3, and 8 are incorporated into viral particles. J. Virol. 2012;86:1261–1266. doi: 10.1128/JVI.06428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rivera C., Ramirez-Mendoza H., Mendoza-Elvira S., Segura-Velazquez R., Sanchez-Betancourt J.I. First report and phylogenetic analysis of porcine deltacoronavirus in Mexico. Transbound. Emerg. Dis. 2019;66:1436–1441. doi: 10.1111/tbed.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan M.V.T., Ngo Tri T., Hong Anh P., Baker S., Kellam P., Cotten M. Identification and characterization of Coronaviridae genomes from Vietnamese bats and rats based on conserved protein domains. Virus Evol. 2018;4 doi: 10.1093/ve/vey035. vey035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Y., Liu J., Huang X., Li Y., Zhang Y., Chen J., Wen X., Cao S., Wen Y., Wu R., Yan Q., Ma X. Immunogenicity of transmissible gastroenteritis virus (TGEV) M gene delivered by attenuated Salmonella typhimurium in mice. Virus Genes. 2016;52:218–227. doi: 10.1007/s11262-016-1296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeng-Chuto K., Lorsirigool A., Temeeyasen G., Vui D.T., Stott C.J., Madapong A., Tripipat T., Wegner M., Intrakamhaeng M., Chongcharoen W., Tantituvanont A., Kaewprommal P., Piriyapongsa J., Nilubol D. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound. Emerg. Dis. 2017;64:3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y., Liu Z.Q., Li G.W., Chen C., Luo H., Liu Y.J., Zhuo X.H., Shi X.F., Fang W.H., Li X.L. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-lambda production by blocking the nuclear factor-kappaB nuclear translocation. J. Zhejiang Univ. Sci. B. 2018;19:570–580. doi: 10.1631/jzus.B1700283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Tai W., Du L., Zhou Y., Zhang W., Li F. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J. Virol. 2018;92 doi: 10.1128/JVI.01556-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Shi H., Sun D., Chen J., Zhang X., Wang X., Zhang J., Ji Z., Liu J., Cao L., Zhu X., Yuan J., Dong H., Wang X., Chang T., Liu Y., Feng L. Nucleocapsid interacts with NPM1 and protects it from proteolytic cleavage, enhancing cell survival, and is involved in PEDV growth. Sci. Rep. 2017;7:39700. doi: 10.1038/srep39700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokri S., Mahmoudvand S., Taherkhani R., Farshadpour F. Modulation of the immune response by Middle East respiratory syndrome coronavirus. J. Cell. Physiol. 2019;234:2143–2151. doi: 10.1002/jcp.27155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Zhou X., Peng Q., Chen Y., Zhang F., Huang T., Zhang T., Li A., Huang D., Wu Q., He H., Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound. Emerg. Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M., Li C., Guo D., Wei S., Wang X., Geng Y., Yao S., Gao J., Wang E., Zhao X., Wang Z., Wang J., Wu R., Feng L., Sun D. A recombinant nucleocapsid protein-based indirect enzyme-linked immunosorbent assay to detect antibodies against porcine deltacoronavirus. J. Vet. Med. Sci. 2016;78:601–606. doi: 10.1292/jvms.15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Shibahara T., Imai N., Yamamoto T., Ohashi S. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect. Genet. Evol. 2018;61:176–182. doi: 10.1016/j.meegid.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg. Infect. Dis. 2014;20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.W., Yue H., Fang W., Huang Y.W. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015;3 doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Su S., Bi Y., Wong G., Gao G.F. Bat-origin coronaviruses expand their host range to pigs. Trends Microbiol. 2018;26:466–470. doi: 10.1016/j.tim.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wang Y., Baloch A.R., Pan Y., Tian L., Xu F., Shivaramu S., Chen S., Zeng Q. Detection and genetic characterization of porcine deltacoronavirus in Tibetan pigs surrounding the Qinghai-Tibet Plateau of China. Transbound. Emerg. Dis. 2018;65:363–369. doi: 10.1111/tbed.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Y., Ji C.J., Zhang X., Xu D.P., Zhang D.L. Infection, genetic and virulence characteristics of porcine epidemic diarrhea virus in northwest China. Infect. Genet. Evol. 2018;62:34–39. doi: 10.1016/j.meegid.2018.04.001. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L., Yang Y., Wang J., Jing Y., Yang Q. Impact of TGEV infection on the pig small intestine. Virol. J. 2018;15:102. doi: 10.1186/s12985-018-1012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Zhong H., Zhou Q., Du Y., Chen L., Zhang Y., Xue C., Cao Y. A highly pathogenic strain of porcine deltacoronavirus caused watery diarrhea in newborn piglets. Virol. Sin. 2018;33:131–141. doi: 10.1007/s12250-018-0003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Chen Z., Song S., Wang S., Tian C., Xing G., Chen X., Xiao Z.X., He F., Zhang L. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J. Biol. Chem. 2015;290:3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S., Zhang H., Ding Z., Luo R., An K., Liu L., Bi J., Chen H., Xiao S., Fang L. Proteome analysis of porcine epidemic diarrhea virus (PEDV)-infected Vero cells. Proteomics. 2015;15:1819–1828. doi: 10.1002/pmic.201400458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi H., Chen J., Shi D., Li C., Feng L. EF1A interacting with nucleocapsid protein of transmissible gastroenteritis coronavirus and plays a role in virus replication. Vet. Microbiol. 2014;172:443–448. doi: 10.1016/j.vetmic.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi H., Chen J., Shi D., Dong H., Feng L. Identification of the interaction between vimentin and nucleocapsid protein of transmissible gastroenteritis virus. Virus Res. 2015;200:56–63. doi: 10.1016/j.virusres.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Liu X., Fang Y., Zhou P., Wang Y., Zhang Y. Detection and phylogenetic analyses of spike genes in porcine epidemic diarrhea virus strains circulating in China in 2016–2017. Virol. J. 2017;14:194. doi: 10.1186/s12985-017-0860-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liang Q., Li B., Cui X., Wei X., Ding Q., Wang Y., Hu H. Prevalence, phylogenetic and evolutionary analysis of porcine deltacoronavirus in Henan province, China. Prev. Vet. Med. 2019;166:8–15. doi: 10.1016/j.prevetmed.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Chen J., Shi D., Shi H., Zhang X., Liu J., Cao L., Zhu X., Liu Y., Wang X., Ji Z., Feng L. Porcine deltacoronavirus enters cells via two pathways: a protease-mediated one at the cell surface and another facilitated by cathepsins in the endosome. J. Biol. Chem. 2019;294:9830–9843. doi: 10.1074/jbc.RA119.007779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.J., Liu D.J., Liu X.L., Ge X.Y., Jongkaewwattana A., He Q.G., Luo R. Genomic characterization and pathogenicity of porcine deltacoronavirus strain CHN-HG-2017 from China. Arch. Virol. 2019;164:413–425. doi: 10.1007/s00705-018-4081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cheng Y., Xing G., Yu J., Liao A., Du L., Lei J., Lian X., Zhou J., Gu J. Detection and spike gene characterization in porcine deltacoronavirus in China during 2016–2018. Infect. Genet. Evol. 2019;73:151–158. doi: 10.1016/j.meegid.2019.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Sun Y., Lan T., Wu R., Chen J., Wu Z., Xie Q., Zhang X., Ma J. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound. Emerg. Dis. 2019;66:687–695. doi: 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.