Abstract
Despite the increased molecular knowledge and the diagnostic and therapeutic improvements, the survival of patients with upper aerodigestive tract carcinoma remains poor. The identification of early diagnostic and prognostic biomarkers and the development of molecular models to distinguish patients that will recur and/or develop metastasis after treatment as well as to benefit with target therapies can be important to decrease mortality, improve survival rates and improve the quality of life of these patients. The current study analyzed 21 upper aerodigestive tract carcinomas through array comparative genomic hybridization and methylation-specific multiplex ligation-dependent probe amplification techniques. A number of chromosomal regions and genes were observed with copy number alterations and methylation. A predictive (epi)genomic model that comprises the 3p chromosomal region and WT1, VHL and THBS1 genes was built, highlighting a molecular signature with possible clinical use. The current study may aid in the development of a more individualized patient management and targeted drug design. The power of this genomic and epigenetic model to predict the recurrence and metastasis development should be evaluated and validated in future larger cohort study.
Keywords: copy number alteration, methylation, recurrence and metastasis, upper aerodigestive tract carcinoma, predictive model, head and neck cancer
Introduction
Worldwide, head and neck squamous cell carcinomas (HNSCCs) are highly aggressive tumors in the upper aerodigestive tract affecting more than half a million patients each year (1). So, these tumors that affect the oral cavity, oropharynx, larynx, and hypopharynx are usually described as squamous cell carcinoma of the upper aerodigestive tract, being the patient outcomes strongly associated to tumor stage (2). Patients with advanced stage tumors (stages III and IV) present frequently recurrences (3), which lead to a poor prognosis and quality of life. Cigarette smoking and alcohol consumption have been established as the main risk factors for these carcinomas (4). The Human Papillomavirus (HPV) infection is a well established risk factor for squamous cell carcinoma of the oropharynx but its role remains unclear in oral cavity and laryngeal cancer (5). Surgical intervention, radiotherapy and chemotherapy are the main treatments for HNSCC. The 5-year survival rate for these patients is still about 50% even with treatment advances (6).
These tumors are considered the final stage of a multistep carcinogenic process involving amplifications, deletions, up and downregulation of oncogenes and tumor suppressor genes, responsible for the initiation, promotion and progression of neoplasms (7). In the last years, important progresses in the understanding of molecular biology of upper aerodigestive tract cancer have been achieved, which allowed a better characterization of these tumors; however, the survival of these patients remains without great improvements. Introducing molecular biomarkers as predictive factors to determine which patients will develop recurrences, will fail to response to treatment or will benefit with target therapies may become a great help to improve survival and quality of life of the patients. In this study, we used whole genome copy number alterations (CNAs) and methylation status in order to identify genomic and epigenetic signatures able to predict recurrence/metastasis development in patients with primary upper aerodigestive tract carcinoma treated with curative intent. Our results represent a step further through the identification of clinically significant biomarkers with predictive value for these cancer patients management.
Materials and methods
Study population
The study protocol was approved by the Committee on Ethics in Research of the Faculty of Medicine of the University of Coimbra. All patients provided their written consent to participate in the study after being informed about the research purposes, following the regulations in the Declaration of Helsinki.
The study cohort includes tumor tissue and tissue from surgery resection margin (macroscopically tumor-free tissue) of 21 patients with diagnosis of upper aerodigestive tract carcinoma. The tissue samples were snap-frozen in liquid nitrogen within 30 min after resection and stored at −80°C until use. The patients were recruited between December 2013 and March 2017 from the Department of Otorhinolaryngology-Head and Neck Surgery of Coimbra Hospital and University Centre (CHUC), EPE, Portugal. Diagnosis and staging were performed in accordance with the American Joint Committee on Cancer TNM staging system (8). Patients were followed-up through hospital revisits during routine clinical appointments. The follow-up periods ranged from 6 to 46 months. Details of our study cohort are listed in Table I. For the control group, 7 palatine uvulas from patients diagnosed with sleep apnoea and/or snoring were used (5 males and 2 females, with ages ranging from 31 to 71 years).
Table I.
Clinic-pathologic characteristics of study population (n=21).
| Characteristic | n |
|---|---|
| Sex | |
| Male | 20 |
| Female | 1 |
| Anatomic subsite | |
| Larynx | 8 |
| Hypopharynx | 2 |
| Piriform sinus | 2 |
| Oropharynx | 2 |
| Epiglottis | 1 |
| Supraglottis | 1 |
| Glottis | 1 |
| Hemilarynx | 1 |
| Vocal chord | 1 |
| Pharynx | 1 |
| Palate/left tonsil | 1 |
| Tobacco | |
| Yes | 21 |
| No | 0 |
| Alcohol | |
| Yes | 20 |
| No | 1 |
| Age at diagnosis (Years) | |
| <60 | 12 |
| ≥60 | 9 |
| TNM stage | |
| I | 2 |
| II | 1 |
| III | 2 |
| IV | 16 |
| Treatment | |
| Surgery only | 4 |
| QT alone | 2 |
| RT alone | 2 |
| Surgery + QT | 2 |
| Surgery + RT | 3 |
| Surgery + RT + QT | 1 |
| RT + QT | 3 |
| NA | 4 |
| Vital status | |
| Relapses/Metastasis in follow-up | 8 |
| Dead-Cancer | 8 |
| Dead-non-Cancer | 2 |
QT, chemotherapy; RT, radiotherapy; NA, not available.
DNA extraction and HPV typing
DNA from fresh frozen tissues of patients and controls were extracted using a High Pure PCR Template Preparation Kit (Roche GmbH), according to the manufacturer's instructions. The DNAs were quantified by UV spectrophotometric analysis using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Inc.). All tumor tissue samples were analyzed for HPV infection as previous described (9,10).
Array-CGH analysis
High-resolution whole genome analyses were performed using Agilent SurePrint G3 Human Genome microarray 180 K (Agilent Technologies), according our previous work (2). DNA of tumor samples was labelled with Cy5 by random primer labelling. DNA from controls was labelled with Cy3. Results were analysed using Agilent Genomic Workbench v6.5 software with the following settings: ADM1 as aberration algorithm, threshold of 6.0, moving average 2 Mb. The results are according to Human Genome build 19 and include imbalances with at least three consecutive probes with abnormal log2 ratios.
MS-MLPA analysis
MS-MLPA analyses were performed using MS-MLPA probe set ME002 (MRC-Holland), which can simultaneously detect CNAs in 38 tumor suppressor genes and aberrant methylation patterns in a subset of 25 of these genes. All MS-MLPA reactions were performed according our previous work (11,12). Three controls selected from the previously analyzed control group, without CNAs and methylation values below 20%, as well as a negative control (without DNA), were always included in each MS-MLPA assay. Binning of the raw data and comparative analyses were performed using Coffalyser. NET software. For each probe we determined the specific cutoff values for gain and loss, using 95% confidence intervals as determined on non-cancer subjects. A copy number gain was scored when a value exceeded 1.2 and a copy number loss was scored when a value was lower than 0.8. We considered a gene promoter as methylated when the methylation dosage ratio was ≥0.20, which means that at least 20% of the DNA was methylated. These cut-off values were based on our previous works (9–11).
Statistical analysis
Methylation and CNA data were firstly analysed considering the number of patients that presented genes alterations. The genes that have less than 10% of patients carrying an alteration were discarded. For the remaining genes, 7 regarding methylation and 25 with CNA, a measure of the effect size for discriminating the groups with and without relapses/metastases was calculated. The odds-ratio was used for this purpose. Since the total number of cases is only 21, we selected the four genes that presented the largest odds-ratio to fit a logistic regression, aiming to further study the influence of these genes in the occurrence of relapses/metastaes.
The MS-MLPA data were desribed resorting to a circos plot, which allows by visual inspection, to identify the 3p region as the best candidate as a biomarker of relapses/metastases. We calculate the fraction of deletion and amplification as the quotient between the number of pair bases altered and the number of pair bases composing the 3p arm. We then assessed, with a ROC analysis, this quantitative measure as a potential discrminative variable of the relapses/metastases group.
Finaly, we integrated the genes selected from the methylation and the CNA data with the fraction of deletion in a logistic model. As the number of cases is quite small the classical forms (e.g. ANOVA test, Naguelkerke pseudo R2, Wald test, etc) to evaluate the logistic regression tend to give inconsistent results, instead we adopted a leave-one -out method to compute the accuracy in the cases not entering in the training set and, in turn, to get a generalizability measure.
The analyses were carried out using SPSS v24, R v3.3.2 and Matlab R2018b. The significance level adopt was 0.05.
Results
HPV typification
We did not detect any HPV-positive sample in the 21 patients analyzed.
CNAs detection by array-CGH
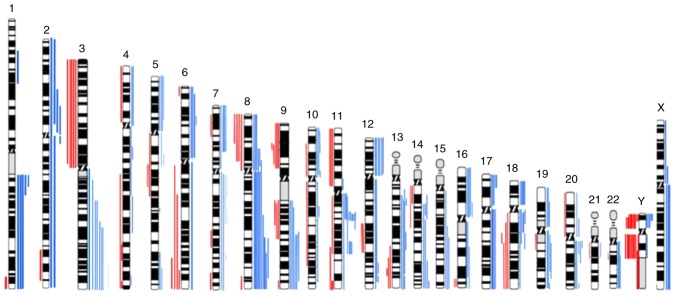
The genomic characterization of tumor samples through whole genome array-CGH revealed several copy number gains and losses in all chromosomes (Fig. 1), being the most frequent copy number gains observed at 3q, 7p, 8q, 9q, 11q13, 12p and 18p and the most frequent copy number losses observed at 3p, 8p 9p and Y (Fig. 1). This aCGH analysis demonstrated that gains of chromosomal segments were more common than losses.
Figure 1.
Ideogram of all copy number alterations detected in 21 upper aerodigestive tract tumors by aCGH. Red bars on the left side of the chromosomes represent copy number losses, blue bars on the right side show copy number gains. Each bar represents one patient. aCGH, Array comparative genomic hybridisation.
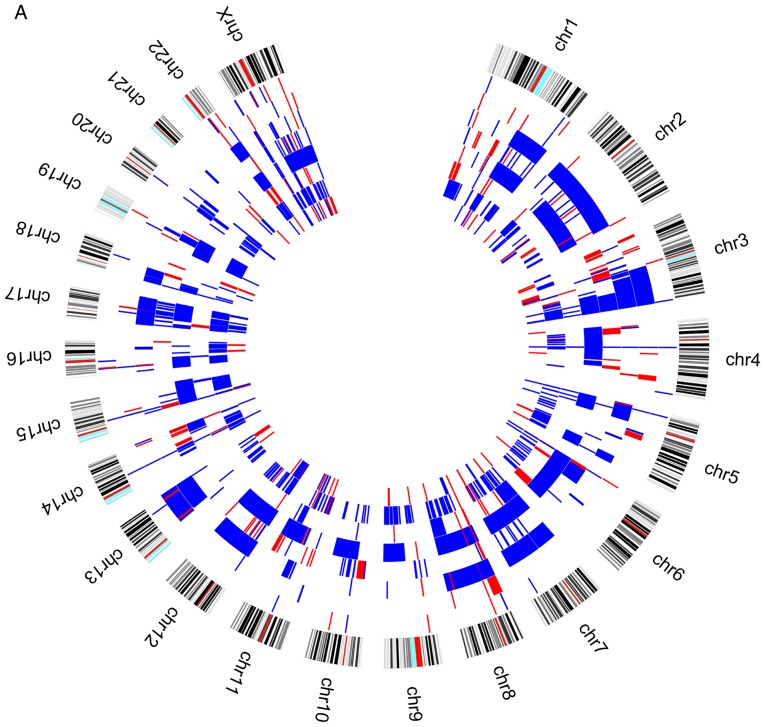
When we analysed the genomic profile of the two groups of patients according to the presence or absence of relapses/metastases during the patients follow-up, we observed that losses at 3p chromosomes are more frequent in patients without relapses/metastases (Fig. 2).
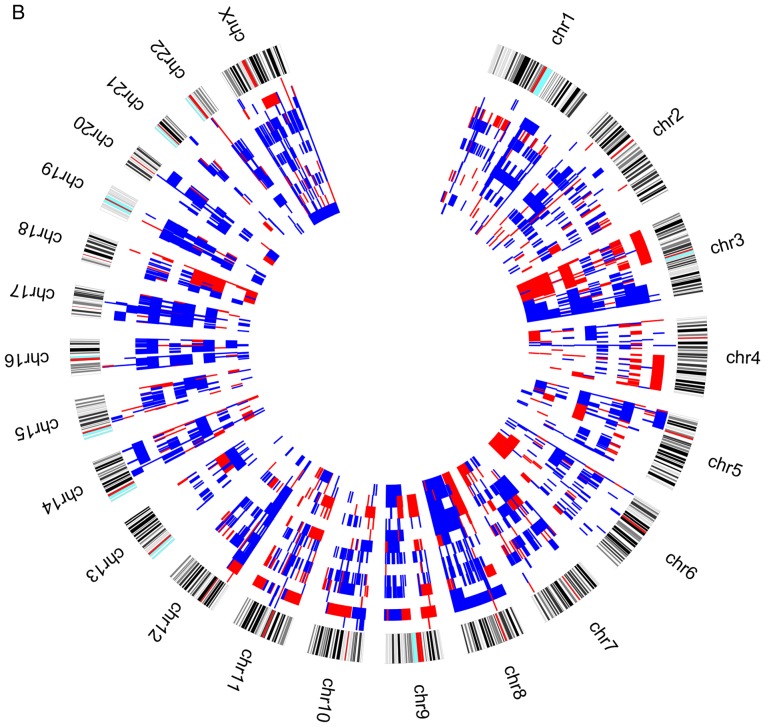
Figure 2.
Circos plot with genomic pattern identified by array-CGH technique for upper aerodigestive tract cancer patients (A) with relapses or metastasis. Blue color represents copy number gains and red copy number losses. Each layer represents a patient and the most outer layer represents the chromosomes. CGH, Comparative Genomic Hybridisation. Circos plot with genomic pattern identified by array-CGH technique for upper aerodigestive tract cancer patients (B) without relapses or metastasis. Blue color represents copy number gains and red copy number losses. Each layer represents a patient and the most outer layer represents the chromosomes. CGH, comparative genomic hybridisation.
A ROC analysis over the fraction of deletion in 3p was performed in order to verify if the genomic differences observed in this chromosome arm could be useful to predict the risk of these patients develop relapses or metastases. This analysis showed that the gains observed in 3p can not discriminate the two groups of patients (AUC=0.510; P=0.942). However, considering the losses in this region, even without statistical significance (P=0.070), the AUC is 0.740 [IC95%(0.529; 0.952)], which suggest that this variable has discriminative power (Fig. 3). This result highlighted 3p losses as a putative biomarker of reduced risk of relapses/metastases development during follow-up of upper aerodigestive tract cancer patients, after the treatment of primary tumor.
Figure 3.
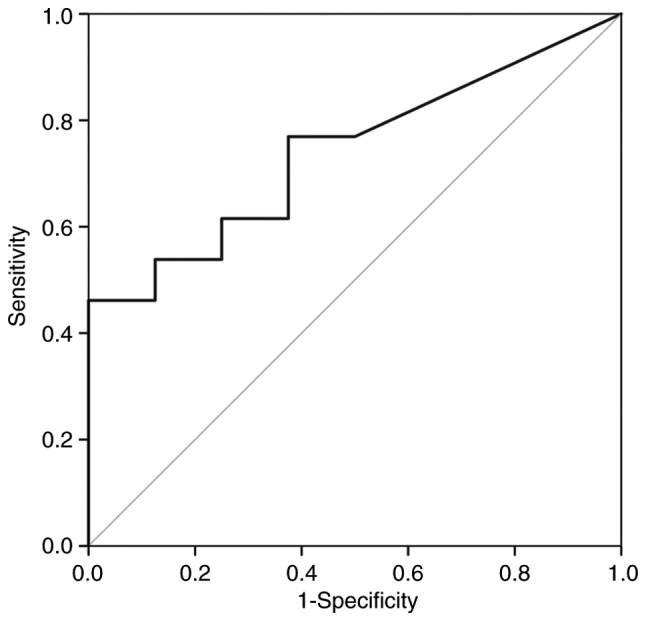
Reciever operator characteristic curve of the fraction of deletion in the short arm of chromosome 3.
The fraction of deletion in 3p was transformed into a bynary variable using as cut point the value that maximizes the Youden index of the ROC anaysis. This variable describing the amount of deletion in 3p is significantly associated with relapses/metastases (P=0.042) and corroborates the previous observation that 3p deletion reduces the chance of relapses/metastases.
CNAs and methylation detection by MS-MLPA
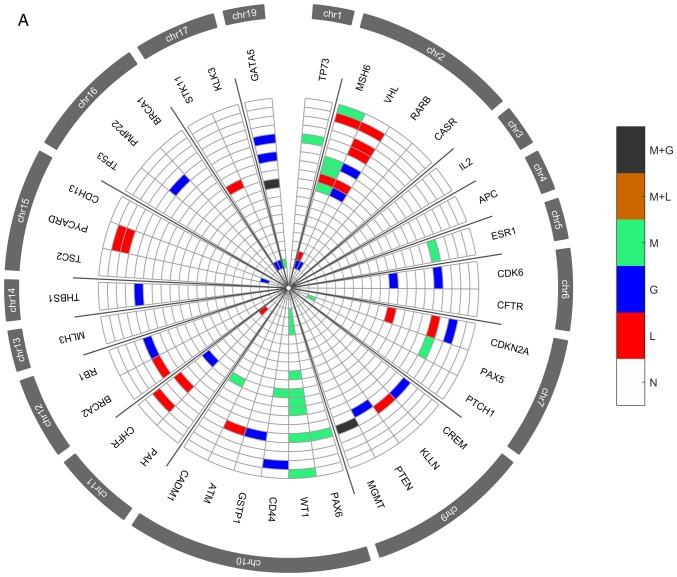
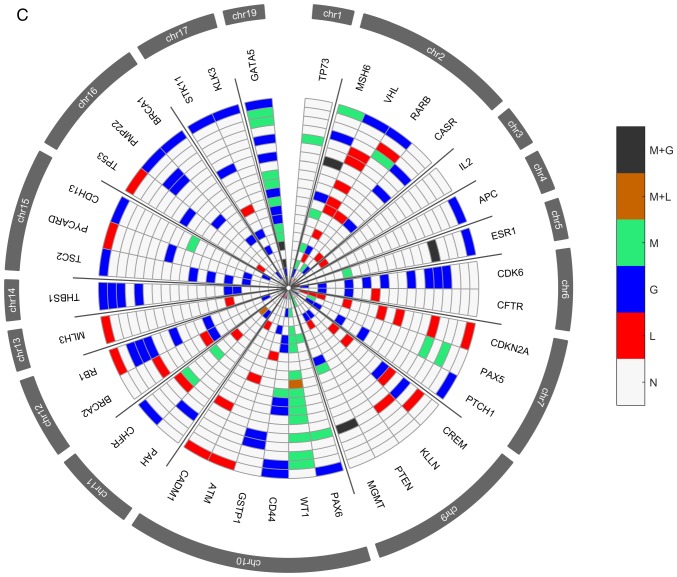
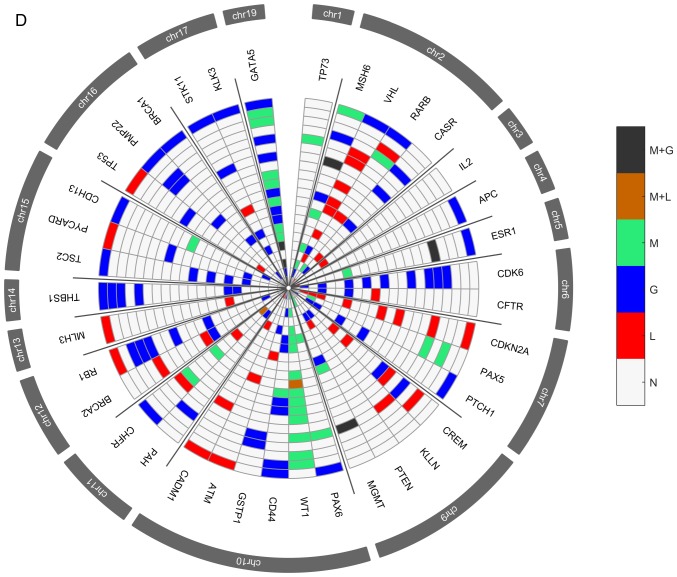
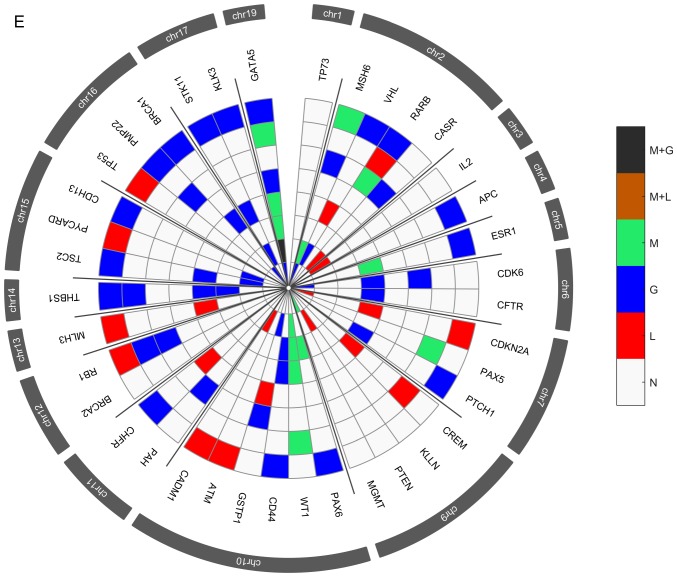
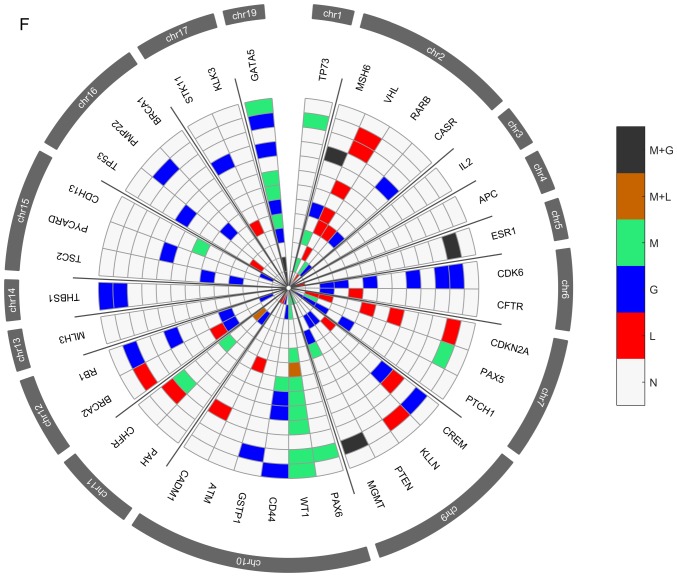
Several genetic and epigenetic alterations were detected in the upper aerodigestive tract tumor and non-tumor tissue samples by MS-MLPA (Fig. 4A and D). The genetic alterations more frequently altered in non-tumor samples were loss of MSH6 (2p16.3), VHL (3p25.3), and CHFR (12q24.33) genes and gain at GATA5 (20q13.33) gene (Fig. 4A-C). The genetic alterations more frequently altered in tumor samples were loss of VHL, CDKN2A (9p21.3), CREM (10p11.21), ATM (11q22.3) and CADM1 (11q23) genes and gain at GATA5, CDK6 (7q21.2), PTCH1 (9q22.32), CD44 (11p13), RB1 (13q14.2), THBS1 (15q14) and TSC2 (16p13.3) genes (Fig. 4D-F). In agreement with array-CGH analysis, the CNV analysis through MS-MLPA assay confirmed the presence of a greater number of gains in relation to losses in these samples.
Figure 4.
Radar chart with analyzed genes for methylation and the respective copy number alterations (A) in all patients with non-tumor upper aerodigestive tract cancer. M + G, methylation and copy number gains; M + L, methylation and copy number losses; M, methylation; G, gains; L, losses; N, Normal. Radar chart with analyzed genes for methylation and the respective copy number alterations (B) in patients with non-tumor upper aerodigestive tract cancer with relapses or metastasis. M + G, methylation and copy number gains; M + L, methylation and copy number losses; M, methylation; G, gains; L, losses; N, Normal. Radar chart with analyzed genes for methylation and the respective copy number alterations (D) in all patients with tumor upper aerodigestive tract cancer. M + G, methylation and copy number gains; M + L, methylation and copy number losses; M, methylation; G, gains; L, losses; N, Normal. Radar chart with analyzed genes for methylation and the respective copy number alterations (C) in patients with non-tumor upper aerodigestive tract cancer without relapses or metastasis. M + G, methylation and copy number gains; M + L, methylation and copy number losses; M, methylation; G, gains; L, losses; N, Normal. Radar chart with analyzed genes for methylation and the respective copy number alterations (E) in patients with tumor upper aerodigestive tract cancer with relapses or metastasis. M + G, methylation and copy number gains; M + L, methylation and copy number losses; M, methylation; G, gains; L, losses; N, Normal. Radar chart with analyzed genes for methylation and the respective copy number alterations (F) in patients with tumor upper aerodigestive tract cancer without relapses or metastasis. M + G, methylation and copy number gains; M + L, methylation and copy number losses; M, methylation; G, gains; L, losses; N, Normal.
In the non-tumor samples, the most frequent methylated genes were WT1 and MSH6 genes (Fig. 4A-C). In the tumor samples, the most frequent methylated genes were WT1, GATA5 and MSH6 genes (Fig. 4D-F). Besides, in the non-tumor samples few genetic and epigenetic alterations were observed, some of those appear in both tumor and non-tumor samples, which may be indicative of dissemination of cells with malignant features.
Only WT1 gene presented in one patient both copy number loss and methylation in the tumor sample. Also, in tumor samples, GATA5, MSH6, ESR1 and MGMT genes presented both copy number gain and methylation (Fig. 4D). While in non-tumor samples, MGMT and GATA5 presented both copy number gain and methylation (Fig. 4A).
After a two step genes reduction process, a statistical model linking upper aerodigestive tract recurrence/metastasis development and the identified genetic and epigenetic profile was build using a logistic regression. The developed model comprises WT1 gene promoter methylation and CNA of VHL and THBS1 genes. The model obtained does not present statistical significance (χ2(5)-8.876; P=0.114) but the value of explained variance is 0.510 and the accuracy (78.9%) is higher than obtained by chance (63.2%). Univariate analysis of the independent variables shows that when WT1 is methylated both in tumor and non-tumor tissues the chance of relapses/metastasis is disminished with odds-ratio equal to 0.33 (P=0.346) and 0.286 (P=0.367), respectively. Regarding CNA, the alteration of the VHL gene reduces the chance of relapses/metastasis (OR=0.167; P=0.173) whereas the alteration of THBS1 gene increases the change of relapses/metastasis (OR=5.5; P=0.146).
Predictive model using genomic and epigenetic data
Taking into account the results obtained by array-CGH and MS-MLPA techniques, the logistic model was adjusted in order to comprise the previously selected genes as well as the 3p chromosome arm. The model obtained was assessed with a leave-one-out method in order to use all the cases but one as the training set. The accuracy to predict the leave-out case was 77.8% that compares to 61.1% if it was by chance. The sensitivity and specificity were 81.8 and 71.4%, respectively. The mean accuracy in the training set was 99.4% and the sensitivity and specificity were 99.5 and 99.1%, respectively.
Discussion
Identification of genome-wide high resolution DNA copy number alterations through array-CGH has been applied to a wide range of tumors including upper aerodigestive tract (2,13–17), being reported commom alterations in multiple chromosome arms, such as losses of 3p and 8p, and gains of 3q, 5p and 8q. However, the pivotal chromosomal alterations and genes that play a central role in upper aerodigestive tract cancer development and progression as well as in recurrence and metastasis development are not still fully understood. The integration of specific biological markers with a role in the clinical management of these tumors together with the TNM staging and histopathological grading need to be explored to improve the diagnosis, prognosis and target therapies design.
In the present study we observed several copy number gains and losses in all chromosomes, which revealed the great genomic complexity that underlies upper aerodigestive tract carcinomas. Gains of chromosomes 3q, 7p, 8, 9q, 11q, 12p,17q and 18p and losses in chromosome 3p, 9p, 11p and Y are the most frequently altered in our cohort (Fig. 1), where are mapped several known and novel putative oncogenes and tumor suppressor genes that can provide a good basis for functional studies with potential to design novel drug targets. Losses in 3p showed to be predictive of reduced risk of relapse/metastasis development during follow-up of our upper aerodigestive tract cancer patients, after the treatment of primary tumor (Figs. 2 and 3). In agreement with this finding, previous published studies had demostrated that 3p losses are an early event that often occur in potential malignant lesions, being included in the Califano genetic progression model for HNSCC (18). The tumor-suppressor genes mapped at 3p responsible for head and neck oncogenesis remains unclear (19).
Additionally, to the genomic characterization of the upper aerodigestive tract carcinoma through array-CGH technology highlighting specific chromosomal alterations, we also identified several genetic and epigenetic alterations both in the tumor and non-tumor tissue samples by MS-MLPA technique (Fig. 4). Some genetic and epigenetic alterations were observed in both tumor and non-tumor samples, sugesting already the presence of cells with molecular malignant features even in tissues without phenotipic manifestation. Considering that in the case of the studied samples, the patients are exposed to the risk factors, namely the carcinogenic action of tobacco smoke often in synergy with alcohol, affecting a large area of tissue, which could be expected that genetic and epigenetic alterations can occur in tumor and macroscopically non-tumor tissue (7). The genetic alterations more frequently altered in tumor samples were loss of VHL (3p25.3), CDKN2A (9p21.3), CREM (10p11.21), ATM (11q22.3) and CADM1 (11q23) genes and gain at GATA5 (20q13.33), CDK6 (7q21.2), CD44 (11p13), PTCH1 (9q22.32), RB1 (13q14.2), THBS1 (15q14) and TSC2 (16p13.3) genes (Fig. 4). In the tumor samples, the most frequent methylated genes were WT1, GATA5 and MSH6 genes (Fig. 4). Tumors encompass a heterogeneous set of cells with different genetic, epigenetic and phenotypic characteristics that can differentially lead to progression, metastasis and drug resistance (20). Nevertheless, cancer treatment is still carried out considering the tumors as a homogenous disease. Nowadays, technologies for interrogating at a single level, the whole genome, transcriptome, epigenome and proteome have suffered great progress allowing to study the intratumor heterogeneity in individual tumors as well as to understand the function and effect of specific cell populations on tumorigenesis, namely, which features could promote tumor initiation, progression or drug resistance (20). So, the great molecular and clinical behavior heterogeneity of upper aerodigestive tract tumors hampers to predict the tumor progression using only the available set of clinical markers; therefore, the development of a prognosis predictive model is a novel and promising strategy to increase the upper aerodigestive tract cancer survival rate and improve the quality of life of the patients, allowing the implementation of precision medicine. The identified genomic and epigenetic signature was used to build a predictive statistical model of recurrence and metastasis development that comprises the 3p chromosomal region and WT1, VHL and THBS1 genes. Methylation of the WT1 gene was observed in the higher number of the patients of our cohort, either in tumor and non-tumor samples, which are in agremment with our previous work in oral squamous cell carcinoma (11). In the 3p chromosome, we verified that specifically the VHL (3p25.3) gene, a tumor suppressor gene with functions related to regulation of genes and control of cell division (21), has a role in our predictive model. A correlation between VHL loss and epithelial-mesenchymal transition in oral squamous cell carcinoma has been suggested, affecting the prognosis of the patients (22). Additionally, gains at THBS1 (15q14) gene were observed in our cohort. This gene is described with a role in stimulation of cancer cell migration and expression of matrix metalloproteases, promoting oral squamous cell invasion (23). Altogether, these results suggest a specific set of chromosomes and genes that seem to have an important role in the development and prediction of relapse/metastasis in the upper aerodigestive tract cancer. This model can help not only in these patients management as well as in the design of targeted therapies. So, our results may improve selection of patients for existing therapies as well as for the development of novel therapies. Improvements in therapies targeting the VHL pathway, namely the VHL-HIF-VEGF axis has been translated into development of therapies with improved clinical response (24,25). Moreover, the upregulation of THBS1, was associated with chemotherapy resistance in breast cancer patients, being shown that THBS1 mediate chemoresistance through the integrin β1/mTOR pathway, which suggest that therapies targeting integrin β1/mTOR pathway may be a promising strategy to overcome chemotherapy resistance (26). Additionally, WT1 protein emerges as a promising tumor antigen for the development of universal cancer vaccines for adjuvant treatment against residual disease and cancer relapses (27). Thus, the identification of prognostic biomarkers to stratify cancer patients into distinct subgroups of clinical outcomes as well as with predictive value for response to novel target therapies is vital.
It is also important to stress some limitations of this study, namely the reduced number of patients analyzed as well as the fact that our cohort presented a relatively reduced clinical follow-up time (range from 6 to 46 months), so, some patients with a molecular profile similar to those with relapse/metastasis could be incorrectly classified only because the patients were not followed up enough time to be diagnosed with recurrence/metastasis. In the future, this predictive model should be tested in larger cohorts of distinct populations of upper aerodigestive tract carcinoma. Further studies and larger follow-up times should be performed to validate the clinical application of this model in the management of these patients.
The clinical application of this genomic and epigenetic predictive model is promising since it is possible to identify newly diagnosed upper aerodigestive tract cancer patients with risk of development of recurrence/metastasis and, in this sense, monitor them closely, avoiding or performing early detection of the recurrences and even provide more aggressive and personalized treatment in order to reduce the morbidity and mortality associated with this disease. We also highlighted in this study some chromosomal regions and genes that can be good candidates for targeted therapy studies.
Taking into account that upper aerodigestive tract cancer has a poor overall survival with tendency to recur, this predictive genomic and epigenetic model for recurrence and metastasis development may pave the way to a more practical and individualized patient management and targeted drug design. Further studies in large cohorts are needed to validate the clinical application of these potential biomarkers and molecular model in the prediction of relapses and metastases development.
Acknowledgements
Not applicable.
Funding
Funding was received from HEALTHY AGING 2020 (grant no. CENTRO-01-0145-FEDER-000012).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
IPR performed the aCGH experiments and wrote the manuscript. FC performed the statistical analysis. MR performed the MS-MLPA experiments. FM, AM and JM collected the samples and performed the critical interpretation of the data from the clinical point of view. IMC and JBM contributed to conception and design of the study and critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Committee on Ethics in Research of the Faculty of Medicine of the University of Coimbra. All patients provided their written consent to participate in the study after being informed about the research purposes, following the regulations in the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Canning M, Guo G, Yu M, Myint C, Groves MW, Byrd JK, Cui Y. Heterogeneity of the head and neck squamous cell carcinoma immune landscape and its impact on immunotherapy. Front Cell Dev Biol. 2019;7:52. doi: 10.3389/fcell.2019.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribeiro IP, Caramelo F, Esteves L, Menoita J, Marques F, Barroso L, Miguéis J, Melo JB, Carreira IM. Genomic predictive model for recurrence and metastasis development in head and neck squamous cell carcinoma patients. Sci Rep. 2017;7:13897. doi: 10.1038/s41598-017-14377-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Visscher AV, Manni JJ. Routine long-term follow-up in patients treated with curative intent for squamous cell carcinoma of the larynx, pharynx, and oral cavity. Does it make sense? Arch Otolaryngol Head Neck Surg. 1994;120:934–939. doi: 10.1001/archotol.1994.01880330022005. [DOI] [PubMed] [Google Scholar]
- 4.de Miguel-Luken MJ, Chaves-Conde M, Carnero A. A genetic view of laryngeal cancer heterogeneity. Cell Cycle. 2016;15:1202–1212. doi: 10.1080/15384101.2016.1156275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torrente MC, Rodrigo JP, Haigentz M, Jr, Dikkers FG, Rinaldo A, Takes RP, Olofsson J, Ferlito A. Human papillomavirus infections in laryngeal cancer. Head Neck. 2011;33:581–586. doi: 10.1002/hed.21421. [DOI] [PubMed] [Google Scholar]
- 6.Thariat J, Vignot S, Lapierre A, Falk AT, Guigay J, Van Obberghen-Schilling E, Milano G. Integrating genomics in head and neck cancer treatment: Promises and pitfalls. Crit Rev Oncol Hematol. 2015;95:397–406. doi: 10.1016/j.critrevonc.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Kujawski M, Rydzanicz M, Sarlomo-Rikala M, Gabriel A, Szyfter K. Chromosome alterations reflect clonal evolution in squamous cell carcinoma of the larynx. Med Sci Monit. 2002;8:BR279–BR282. [PubMed] [Google Scholar]
- 8.7th. Springer; New York, NY: 2010. American Joint Committee on Cancer (AJCC)-TNM staging classification. [Google Scholar]
- 9.Ribeiro IP, Marques F, Caramelo F, Ferrão J, Prazeres H, Julião MJ, Rifi W, Savola S, de Melo JB, Baptista IP, Carreira IM. Genetic imbalances detected by multiplex ligation-dependent probe amplification in a cohort of patients with oral squamous cell carcinoma-the first step towards clinical personalized medicine. Tumour Biol. 2014;35:4687–4695. doi: 10.1007/s13277-014-1614-9. [DOI] [PubMed] [Google Scholar]
- 10.Ribeiro IP, Marques F, Caramelo F, Pereira J, Patrício M, Prazeres H, Ferrão J, Julião MJ, Castelo-Branco M, de Melo JB, et al. Genetic gains and losses in oral squamous cell carcinoma: Impact on clinical management. Cell Oncol (Dordr) 2014;37:29–39. doi: 10.1007/s13402-013-0161-5. [DOI] [PubMed] [Google Scholar]
- 11.Ribeiro IP, Caramelo F, Marques F, Domingues A, Mesquita M, Barroso L, Prazeres H, Julião MJ, Baptista IP, Ferreira A, et al. WT1, MSH6, GATA5 and PAX5 as epigenetic oral squamous cell carcinoma biomarkers-a short report. Cell Oncol (Dordr) 2016;39:573–582. doi: 10.1007/s13402-016-0293-5. [DOI] [PubMed] [Google Scholar]
- 12.Ribeiro IP, Caramelo F, Esteves L, Oliveira C, Marques F, Barroso L, Melo JB, Carreira IM. Genomic and epigenetic signatures associated with survival rate in oral squamous cell carcinoma patients. J Cancer. 2018;9:1885–1895. doi: 10.7150/jca.23239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarmuz-Szymczak M, Pelinska K, Kostrzewska-Poczekaj M, Bembnista E, Giefing M, Brauze D, Szaumkessel M, Marszalek A, Janiszewska J, Kiwerska K, et al. Heterogeneity of 11q13 region rearrangements in laryngeal squamous cell carcinoma analyzed by microarray platforms and fluorescence in situ hybridization. Mol Biol Rep. 2013;40:4161–4171. doi: 10.1007/s11033-013-2496-4. [DOI] [PubMed] [Google Scholar]
- 14.Järvinen AK, Autio R, Haapa-Paananen S, Wolf M, Saarela M, Grénman R, Leivo I, Kallioniemi O, Mäkitie AA, Monni O. Identification of target genes in laryngeal squamous cell carcinoma by high-resolution copy number and gene expression microarray analyses. Oncogene. 2006;25:6997–7008. doi: 10.1038/sj.onc.1209690. [DOI] [PubMed] [Google Scholar]
- 15.Giefing M, Martin-Subero JI, Kiwerska K, Jarmuz M, Grenman R, Siebert R, Szyfter K. Characterization of homozygous deletions in laryngeal squamous cell carcinoma cell lines. Cancer Genet Cytogenet. 2008;184:38–43. doi: 10.1016/j.cancergencyto.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Giefing M, Zemke N, Brauze D, Kostrzewska-Poczekaj M, Luczak M, Szaumkessel M, Pelinska K, Kiwerska K, Tönnies H, Grenman R, et al. High resolution ArrayCGH and expression profiling identifies PTPRD and PCDH17/PCH68 as tumor suppressor gene candidates in laryngeal squamous cell carcinoma. Genes Chromosomes Cancer. 2011;50:154–166. doi: 10.1002/gcc.20840. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Network: Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D. Genetic progression model for head and neck cancer: Implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 19.Lee DJ, Schonleben F, Banuchi VE, Qiu W, Close LG, Assaad AM, Su GH. Multiple tumor-suppressor genes on chromosome 3p contribute to head and neck squamous cell carcinoma tumorigenesis. Cancer Biol Ther. 2010;10:689–693. doi: 10.4161/cbt.10.7.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol. 2018;20:1349–1360. doi: 10.1038/s41556-018-0236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, Zhou X, Wang B, Zhang K, Liu S, Yue K, Zhang L, Wang X. Loss of VHL expression contributes to epithelial-mesenchymal transition in oral squamous cell carcinoma. Oral Oncol. 2014;50:809–817. doi: 10.1016/j.oraloncology.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Pal SK, Nguyen CT, Morita KI, Miki Y, Kayamori K, Yamaguchi A, Sakamoto K. THBS1 is induced by TGFB1 in the cancer stroma and promotes invasion of oral squamous cell carcinoma. J Oral Pathol Med. 2016;45:730–739. doi: 10.1111/jop.12430. [DOI] [PubMed] [Google Scholar]
- 24.Patel PH, Chadalavada RS, Chaganti RS, Motzer RJ. Targeting von Hippel-Lindau pathway in renal cell carcinoma. Clin Cancer Res. 2006;12:7215–7220. doi: 10.1158/1078-0432.CCR-06-2254. [DOI] [PubMed] [Google Scholar]
- 25.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, Srivastava S, Hartman M, Buhari SA, Chan CW, Iau P, Khin LW, Wong A, Tan SH, Goh BC, Lee SC. High expression of intratumoral stromal proteins is associated with chemotherapy resistance in breast cancer. Oncotarget. 2016;7:55155–55168. doi: 10.18632/oncotarget.10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Driessche A, Berneman ZN, Van Tendeloo VF. Active specific immunotherapy targeting the Wilms' tumor protein 1 (WT1) for patients with hematological malignancies and solid tumors: Lessons from early clinical trials. Oncologist. 2012;17:250–259. doi: 10.1634/theoncologist.2011-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.