Abstract
A number of studies suggest an association between miRNAs and diffuse large B-cell lymphoma (DLBCL). The present study aimed to investigate the prognostic value of microRNA (miR-150) in primary gastrointestinal (PGI)-DLBCL, by assessing the association between miR-150 expression and clinicopathological characteristics in patients with PGI-DLBCL. A total of 84 patients diagnosed with PGI-DLBCL were recruited and both tumor and adjacent non-tumor tissue samples were collected. miR-150 expression was assessed via reverse transcription-quantitative (RT-q)PCR analysis. The results demonstrated that miR-150 expression was significantly lower in PGI-DLBCL tissues compared with adjacent non-tumor tissues. Furthermore, receiver operating characteristic (ROC) curve analysis indicated that the optimal cut-off value of miR-150 for predicting survival was 8.965 with high sensitivity (79.8%) and specificity (77.1%). Patients were divided into two groups according to this cut-off value, as follows: High (n=18) and low expression (n=66) groups. Low miR-150 expression was significantly associated with clinical stage, International Prognostic Index (IPI), Eastern Cooperative Oncology Group status and use of rituximab. RT-qPCR analysis demonstrated that miR-150 expression was significantly lower in patients with high IPI scores compared with patients with low IPI scores. Downregulated miR-150 expression was significantly associated with shorter overall survival (OS) time and progression-free survival (PFS) time in patients with PGI-DLBCL. Furthermore, miR-150 level and IPI score were identified as two risk factors for OS and PFS. The diagnostic value of miR-150 was evaluated via ROC curve analysis, with an area under the curve value of 0.882. Taken together, the results of the present study suggest that miR-150 is a potential diagnostic marker of PGI-DLBCL, and may also serve as a useful prognostic factor for survival outcomes in patients with PGI-DLBCL.
Keywords: biomarker, miRNA, miR-150, PGI-DLBCL, prognosis
Introduction
Gastrointestinal (GI) lymphomas account for ~5–20% of all cases of extranodal lymphomas (1). However, primary (P)GI lymphoma is considered rare, accounting for only 1–4% of all types of GI cancer (2). In the last decades, the incidence of PGI has exhibited an increasing trend (3). PGI may present throughout the gastrointestinal tract; most commonly in the stomach, followed by the small intestine, ileocecal region and rectum (4,5). Diffuse large B-cell lymphoma (DLBCL) is the most common pathological subtype of GI lymphoma, followed by mucosa-associated lymphoid tissue (MALT) lymphoma (5). DLBCL is typically diagnosed at an advanced stage due to non-specific symptoms, such as abdominal pain, loss of appetite and unintentional weight loss (6). However, the pathological and neoplastic molecular mechanisms underlying the progression of PGI-DLBCL are yet to be elucidated. Increasing evidence suggest that the aberrant expression of microRNAs (miRNAs/miRs) may regulate cancer progression. Thus, miRNAs may serve as promising diagnostic and prognostic biomarkers for different types of cancer (7–9). Previous studies have reported that miRNAs serve as tumor suppressor genes or oncogenes in the context of DLBCL (10–14). However, to the best of our knowledge, the molecular mechanism underlying the influence of miRNAs in the pathogenesis of PGI-DLBCL is not yet fully understood.
miRNAs are a conserved class of long non-coding RNAs (~19–24 nucleotides in length) and regulate posttranscriptional gene repression via direct binding to the 3′-untranslated region of downstream mRNAs (15,16). miRNAs play a key role in several biological pathways, including cell proliferation, stress resistance, cell apoptosis and fat metabolism (7,13). miR-150 influences the development of different types of solid tumors, such as colorectal, lung, breast, pancreatic and esophageal cancer (17–21). For example, downregulation of miR-150 in patients with esophageal squamous-cell carcinoma is associated with poor prognosis (19). Conversely, high miR-150 expression has been reported to promote clonogenicity and cell proliferation, while suppressing apoptosis in breast cancer cells (22). Furthermore, high miR-150 expression in patients with non-small cell lung cancer (NSCLC) is associated with shorter overall survival (OS) time, suggesting the prognostic relevance of upregulation of miR-150 in NSCLC tissues (23). Increased miR-150 expression has also been reported in other carcinomas, such as thyroid and prostate cancer (24,25).
Furthermore, miR-150 has been demonstrated to be associated with hematopoietic specificity in malignant lymphoma (26). Monticelli et al (27) studied miRNA expression profiles of hematopoietic lineage cells and reported low miR-150 expression in progenitor B- and T-cells, but high miR-150 expression in mature cells. Notably, downregulation of miR-150 was demonstrated to be associated with poor prognosis in chronic lymphocytic leukemia (28). In a related clinical study, significantly repressed miR-150 expression in Burkitt lymphoma tissues was indicated to inhibit the proliferation of Raji cells, suggesting its potential role as a novel therapeutic target (29). Conversely, Gebauer et al (30) found that the increased expression of miR-150 was observed in malt marginal zone lymphoma, which indicates that miR-150 may represent a potential treatment for MALT lymphoma.
Despite the aforementioned contradictory results, miR-150 expression level is associated with the progression of lymphoma and influences survival outcomes. However, to the best of our knowledge, the association between miR-150 and PGI-DLBCL has not yet been fully investigated. Thus, the present study aimed to determine whether miR-150 expression significantly influenced the pathogenesis and prognosis of patients with PGI-DLBCL.
Materials and methods
Study participants
The present study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute (Tianjin, China) and written informed consent was obtained from all patients prior to the study start. A total of 84 PGI-DLBCL tissues and paired adjacent non-tumor tissues were collected from 44 men and 40 women (mean age, 54 years; age range, 35–70 years) who underwent radical surgery at the Tianjin Medical University Cancer Institute and Hospital, between January 2011 to December 2015. Tissue samples were stored at −80°C prior to subsequent experimentation. All patients were randomly selected, with a balanced distribution of age and sex, no selection bias and no external validation. Diagnosis of PGI-DLBCL was confirmed by pathologists (Department of Pathology, Tianjin Medical University Cancer Institute and Hospital) and based on morphological, immunophenotypic and molecular tests, according to the World Health Organization (WHO) classification criteria (31). The inclusion criteria were as follows: Patients were diagnosed with PGI-DLBCL according to the WHO classification criteria, patients did not receive radiotherapy or chemotherapy prior to surgery, no previous tumors or associated tumors were reported and complete clinical data and long-term follow-up data were available. All patients were staged according to the Lugano staging system for gastrointestinal non-Hodgkin's lymphoma (32).
RNA and cDNA preparation and Reverse transcription (RT)-PCR
Total RNA was extracted from 10-µm-thick formalin-fixed paraffin-embedded PGI-DLBCL tissues and adjacent non-tumor tissue sections using TRIzol® reagent (Invitrogen, Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The concentrations and purity of total RNA were measured using NanoDrop 2000 (NanoDrop Technologies; Thermo Fisher Scientific, Inc.) and total RNA was reverse transcribed into cDNA using the cDNA synthesis kit (Takara Bio, Inc.), according to the manufacturer's protocol, and subsequently stored at −20°C. Subsequently, qPCR was performed using the CM9600 Sequence Detection System (Bio-Rad Laboratories, Inc.). According to the manufacturer's instructions, DNA was amplified with 10 µl of kit-supplied QuantiTect SYBR Green RT-PCR Master mix, 0.4 µl of each primer (10 µM), 2 µl of cDNA (50 ng RNA), and 7.2 µl of double distilled H2O (all Applied Biosystems; Thermo Fisher Scientific, Inc.). The following primer sequences were used for qPCR: miR-150 forward, 5′-TCTCCCAACCCTTGTACCAGTG-3′ and reverse, 5′-TCTCCCAACCCTTGTACCAGTG-3′; and U6 forward, 5′-GCGCGTCGTGAAGCGTTC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′. The following thermocycling conditions were used for qPCR: initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 sec and annealing at 60°C for 1 min. Relative miRNA levels were quantified using the 2−ΔΔCq method and normalized to U6 snRNA (RNU6B; Invitrogen; Thermo Fisher Scientific, Inc.). All experiments were performed in triplicate.
Immunohistochemical staining and scoring
Expression of B-cell lymphoma 6 (BCL-6), CD10 and multiple myeloma antigen 1 (MUM1) was determined via immunohistochemical staining. The slices were stored at room temperature for 60 min, then soaked twice in xylene for 10 min and rehydrated in decreasing concentrations of ethanol for 5 min at a time (100, 95, 85, 75 and 50%), prior to incubation in pure water for 5 min. Following deparaffinization and rehydration, the sections were incubated with 3% H2O2 for 5–10 min at room temperature to inhibit endogenous peroxidase activity, and subsequently washed with PBS 3 times. The slices were immersed in 0.01 mol citrate buffer, heated to boiling in a medical microwave oven and baked for 8 min. When the buffer temperature dropped to room temperature, the slices were soaked in PBS twice for 5 min. Subsequently, 10% normal goat serum (Cell Signaling Technology, Inc.) was used to block the samples at room temperature for 60 min. Tissue sections were incubated with primary antibodies (all 1:100) directed against BCL-6 (cat. no. ZM-0011; OriGene Technologies, Inc.), CD10 (cat. no. ZM0283; OriGene Technologies, Inc.) and MUM1 (cat. no. ZA-0583; OriGene Technologies, Inc.) overnight at 4°C. Subsequently incubated with secondary antibodies (cat. no. 31926; Cell Signaling Technology, Inc.) and incubated at room temperature for 30 min. The slides were stained with 3,3′-diaminobenzidine at room temperature for 15 min. The specimens were visualized under a light microscope (magnification, ×40).
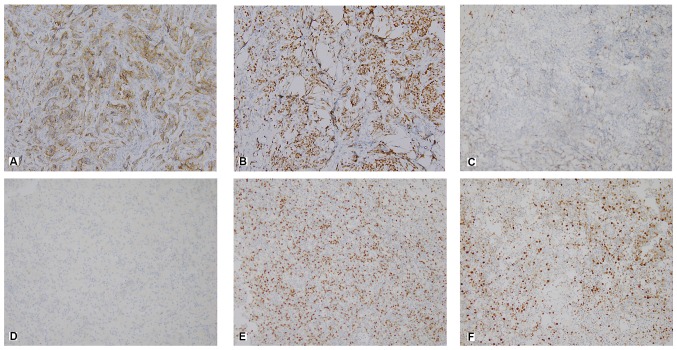
A semi-quantitative scoring system was used to assess the percentage of positive tumor cells and the staining intensity. The experimental group and the control group were photographed, and the relevant results were obtained by using Image Pro Plus image analysis software (version 7.0, Meyer Instruments, Inc.). Staining intensity was scored as follows: 0, no staining; 1+, weak staining; 2+, moderate staining; and 3+, strong staining (Fig. 1).
Figure 1.
Representative histological and immunohistochemical images of patients with primary gastrointestinal diffuse large B-cell lymphoma, in the GCB and non-GCB subgroups (magnification, ×400). Immunostaining for (A) CD10, (B) BCL-6 and (C) MUM-1 in the GCB group. Immunostaining for (D) CD10, (E) BCL-6 and (F) MUM-1 in the non-GCB group. GCB, germinal center B-cell like; BCL-6, B-cell lymphoma 6; MUM-1, multiple myeloma antigen 1.
Treatment and response assessment
According to the National Comprehensive Cancer Network treatment guidelines (33), all patients were treated with CHOP [cyclophosphamide (750 mg/m2, day 1), doxorubicin (50 mg/m2, day 1), vincristine (1.4 mg/m2, day 1) and prednisone (100 mg, days 1–5)] or CHOP-like regimen with rituximab (375 mg/m2 at day 1) (R-CHOP). Patients were treated for 6–8 cycles (each cycle, 21 days). The treatment response at the end of every two cycles was assessed based on results from computed tomography, magnetic resonance imaging, color ultrasound or 18F-fluorodeoxyglucose positron emission tomography, according to the International Working Group response criteria as either: Complete response, partial response, stable disease and progressive disease (34).
Statistical analysis
Statistical analysis was performed using SPSS (version 20.0; IBM Corp.) and GraphPad Prism (version 7.0; GraphPad Software, Inc.) software. Data were presented as the mean ± standard deviation. The association between miR-150 expression and clinicopathological characteristics was assessed using Pearson's χ2 test. The paired Student's t-test was used to compare the miR-150 expression between tumor and adjacent non-tumor tissue samples, while unpaired Students' t-test was used to assess miR-150 expression in the International Prognostic Index (IPI) subgroups. Survival curves were generated using the Kaplan-Meier method and compared using the log-rank test. OS time was measured from the date of diagnosis to the date of mortality or the last follow-up. Progression-free survival (PFS) time was measured from the date of diagnosis to the date of disease progress, recurrence, mortality or last follow-up. In the multivariate and univariate analyses, the prognostic value of miR-150 in patients with PGI-DLBCL was evaluated using Cox regression. Receiver operating characteristic (ROC) curve analysis was performed and the area under the curve (AUC) was measured to assess miR-150 specificity and sensitivity. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-150 expression and clinicopathological characteristics in PGI-DLBCL
Data on the association between miR-150 expression and clinicopathological characteristics in patients with PGI-DLBCL are presented in Table I. Of the 84 assessed patients, 63 (75.00%) exhibited stomach involvement, 47 (55.95%) had advanced disease (clinical stages IIE-IV) and 65 (77.38%) presented with a good performance status (0–1). All tissue samples were stained with CD10, BCL6 and MUM1, which classified 31 patients (36.90%) into the germinal center B-cell like (GCB) subgroup and 53 patients (63.10%) into the non-GCB subgroup. Elevated lactate dehydrogenase (LDH) expression was observed in 31 patients (36.90%), while B symptoms (Systemic symptoms such as: i) Fever of unknown origin exceeding 38°C for 3 consecutive days; ii) unexplained weight loss of >10% within 6 months; iii) night sweats) were identified in 60 patients (71.43%) and low IPI scores (0–2) were observed in 41 patients (48.81%). A total of 54 patients (64.29%) were treated with the R-CHOP regimen. In all, lymph node and bone marrow metastasis were observed in 32 (38.10%) and 21 (25.00%) patients, respectively. According to receiver operating characteristic (ROC) curve analysis, the most notable cut-off point for miR-150 was 8.965, with 79.8% sensitivity and 77.1%, specificity, respectively. Patients were divided into two subgroups according to this cut-off value, as follows: Low miR-150 group (≤8.965; n=66) and high miR-150 group (>8.965; n=18). Low miR-150 expression was significantly associated with clinical stage (P=0.029), IPI score (P=0.025), ECOG score (P=0.002) and treatment regime (P=0.049). However, no significant association was observed between low miR-150 expression and sex, age, tumor origin, pathological type, LDH level, B symptoms, lymph node metastasis and bone marrow metastasis.
Table I.
Association between miR-150 expression and clinicopathological characteristics in patients with primary gastrointestinal diffuse large B-cell lymphoma (n=84).
| miR-150 expression level | ||||
|---|---|---|---|---|
| Characteristic | Patients, n=84 | Low, n=66 | High, n=18 | P-value |
| Sex | 0.761 | |||
| Male | 44 | 34 | 10 | |
| Female | 40 | 32 | 8 | |
| Age, years | 0.969 | |||
| ≤60 | 33 | 26 | 7 | |
| >60 | 51 | 40 | 11 | |
| Tumor origin | 0.759 | |||
| Stomach | 63 | 50 | 13 | |
| Intestinal | 21 | 16 | 5 | |
| Pathological type | 0.844 | |||
| GCB | 31 | 24 | 7 | |
| Non-GCB | 53 | 42 | 11 | |
| Lugano staging status | 0.029a | |||
| I–II | 37 | 25 | 12 | |
| IIE-IV | 47 | 41 | 6 | |
| IPI score | 0.025a | |||
| 0–2 | 41 | 28 | 13 | |
| 3–5 | 43 | 38 | 5 | |
| ECOG score | 0.002a | |||
| 0–1 | 65 | 56 | 9 | |
| 2–4 | 19 | 10 | 9 | |
| B symptoms | 0.274 | |||
| Negative | 24 | 17 | 7 | |
| Positive | 60 | 49 | 11 | |
| LDH level | 0.064 | |||
| Normal | 53 | 45 | 8 | |
| Elevated | 31 | 21 | 10 | |
| Treatment | 0.047a | |||
| CHOP | 30 | 20 | 10 | |
| R-CHOP | 54 | 46 | 8 | |
| Lymph node metastasis | 0.309 | |||
| Negative | 52 | 39 | 13 | |
| Positive | 32 | 27 | 5 | |
| Bone marrow metastasis | 0.759 | |||
| Negative | 63 | 49 | 14 | |
| Positive | 21 | 17 | 4 | |
P<0.05. miR, microRNA; GCB, germinal center B-cell like; IPI, International Prognostic Index; ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase; CHOP, cyclophosphamide, doxorubicin, vincristine and prednisoneor; R-CHOP, CHOP-like regimen with rituximab; B symptoms, systemic symptoms such as: i) Fever of unknown origin exceeding 38°C for 3 consecutive days; ii) unexplained weight loss of >10% within 6 months; iii) night sweats.
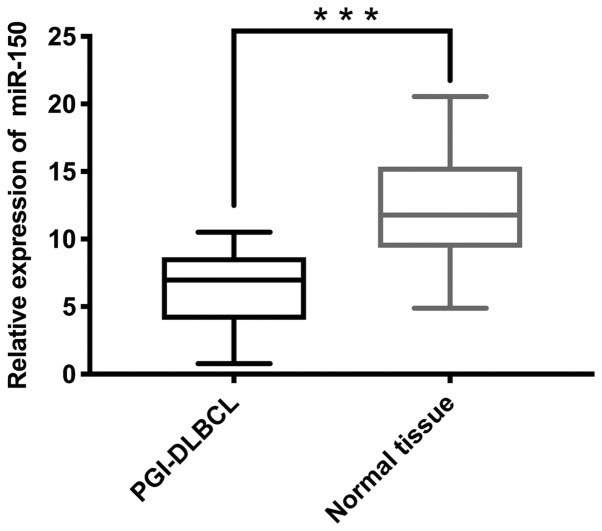
miR-150 expression is significantly lower in PGI-DLBCL tissues compared with non-tumor tissues
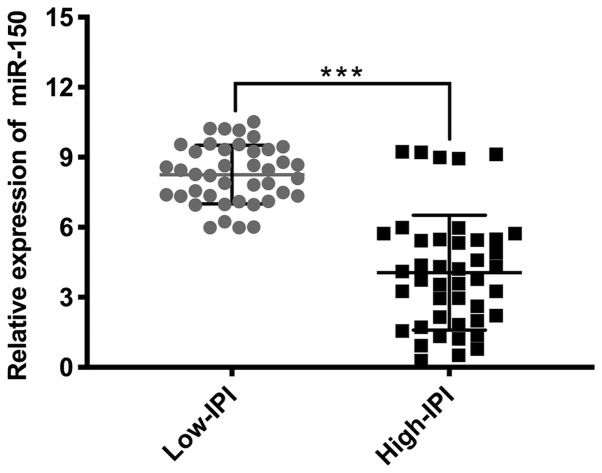
The mean miR-150 expression in tumor tissues was 6.11 and the median was 5.74 (minimum, 0.29; maximum, 10.52), while the average miR-150 expression in adjacent non-tumor tissues was 12.00 and the median was 11.45 (minimum, 2.23; maximum, 20.56). RT-qPCR analysis demonstrated significantly lower miR-150 expression in PGI-DLBCL tissues compared with adjacent non-tumor tissues (P<0.001; Fig. 2). Subgroup analysis was subsequently performed after patients with PGI-DLBCL were divided into two groups: Low IPI score (0–2) and high IPI score (3–5). miR-150 expression was significantly elevated in patients with low-IPI scores compared with patients in the high-IPI group (P<0.001; Fig. 3).
Figure 2.
miR-150 expression in PGI-DLBCL tissues and adjacent non-tumor tissues. Reverse transcription-quantitative PCR analysis demonstrated that miR-150 expression was significantly lower in PGI-DLBCL tissues compared with adjacent non-tumor tissues. All results were normalized to the internal reference gene U6. ***P<0.001. miR, microRNA; PGI-DLBCL, primary gastrointestinal diffuse large B-cell lymphoma.
Figure 3.
miR-150 expression in the IPI subgroups. Reverse transcription-quantitative PCR analysis demonstrated that miR-150 expression was significantly lower in the high-IPI subgroup compared with the low-IPI subgroup. All results were normalized to the internal reference gene U6. ***P<0.001. miR, microRNA; IPI, International Prognostic Index.
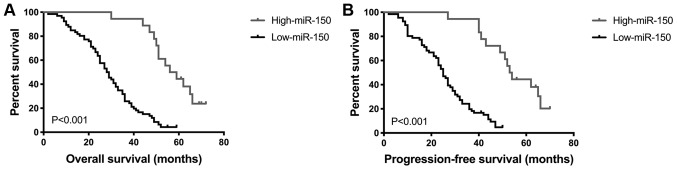
Low miR-150 expression is associated with shorter OS and PFS time
Survival analysis of all 84 patients with PGI-DLBCL was performed using the Kaplan-Meier method, in order to determine the prognostic value of miR-150 in PGI-DLBCL. OS and PFS time were significantly shorter in patients with low miR-150 expression compared with patients with high miR-150 expression (both P<0.001; Fig. 4). Univariate analysis demonstrated that the prognosis of PGI-DLBCL was significantly associated with miR-150 level, treatment and IPI score (Table II). Multivariate analysis indicated that miR-150 level and IPI score were two prognostic indicators of OS and PFS time in patients with PGI-DLBCL (miR-150 OS, P=0.015 and PFS, P=0.015; IPI OS, P=0.004 and PFS, P=0.002; Table III). Taken together, these results suggest that miR-150 may be used as a prognostic marker in patients with PGI-DLBCL.
Figure 4.
Kaplan-Meier analysis of overall survival time and progression-free survival time according to miR-150 expression. Downregulation of miR-150 expression was associated with a poorer (A) overall survival time and (B) progression-free survival time of patients with primary gastrointestinal diffuse large B-cell lymphoma. P<0.001. miR, microRNA.
Table II.
Univariate analysis of the prognostic characteristics for primary gastrointestinal diffuse large B-cell lymphoma.
| Characteristic | OS, HR (95% CI) | P-value | PFS, HR (95% CI) | P-value |
|---|---|---|---|---|
| miR-150 expression (Low vs. high) | 1.761 (1.101–2.814) | 0.018a | 1.873 (1.170–2.998) | 0.009a |
| Treatment (CHOP vs. R-CHOP) | 1.972 (1.251–3.108) | 0.003a | 2.704 (1.314–3.274) | 0.002a |
| IPI score (0–2 vs. 3–5) | 1.984 (1.236–3.185) | 0.005a | 2.054 (1.279–3.300) | 0.003a |
| ECOG (0–1 vs. 2–4) | 1.298 (0.822–2.049) | 0.263 | 1.358 (0.860–2.143) | 0.189 |
| Lugano staging status (I–II vs. IIE-IV) | 1.165 (0.737–1.842) | 0.513 | 1.230 (0.778–1.944) | 0.376 |
P<0.05. OS, overall survival; PFS, progression-free survival; HR, hazard ratio; CI, confidence interval; miR, microRNA; IPI, International Prognostic Index; ECOG, Eastern Cooperative Oncology Group; CHOP, cyclophosphamide, doxorubicin, vincristine and prednisoneor; R-CHOP, CHOP-like regimen with rituximab.
Table III.
Multivariate analysis of the independent prognostic characteristics for primary gastrointestinal diffuse large B-cell lymphoma.
| Characteristic | OS, HR (95% CI) | P-value | PFS, HR (95% CI) | P-value |
|---|---|---|---|---|
| miR-150 expression (Low vs. high) | 2.043 (1.147–3.638) | 0.015a | 2.074 (1.154–3.730) | 0.015a |
| IPI score (0–2 vs. 3–5) | 2.030 (1.262–3.264) | 0.004a | 2.082 (1.295–3.346) | 0.002a |
| Treatment (CHOP vs. R-CHOP) | 1.305 (0.552–3.085) | 0.544 | 1.499 (0.633–3.549) | 0.357 |
P<0.05. OS, overall survival; PFS, progression-free survival; HR, hazard ratio; CI, confidence interval; miR, microRNA; IPI, International Prognostic Index; CHOP, cyclophosphamide, doxorubicin, vincristine and prednisoneor; R-CHOP, CHOP-like regimen with rituximab.
miR-150 has a potential diagnostic value for PGI-DLBCL
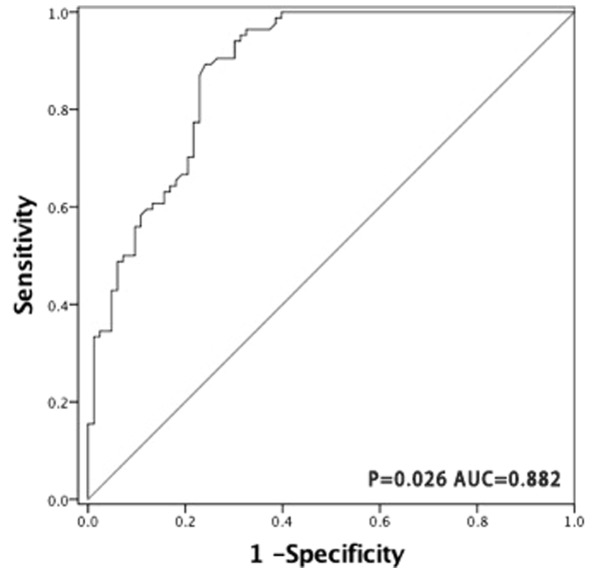
The ROC curve was generated by dividing all tumor and non-tumor tissue samples into one class, in order to determine whether miR-150 may be used as a biomarker to distinguish PGI-DLBCL from adjacent non-tumor tissues. The AUC was 0.882 (P=0.026; Fig. 5), suggesting that miR-150 has a potential diagnostic value for patients with PGI-DLBCL.
Figure 5.
Receiver operating characteristic curve analysis was performed to distinguish the diagnostic value of miR-150 between tumor and adjacent non-tumor tissue samples. miR-150 demonstrated high sensitivity (79.8%) and specificity (77.1%) in the diagnosis of primary gastrointestinal diffuse large B-cell lymphoma, with an optimal cut-off value of 8.965. miR, microRNA.
Discussion
PGI lymphoma originates in the stomach, with or without peri-gastric or abdominal lymph node invasion (35). DLBCL is the most frequently diagnosed histopathological subtype of PGI lymphoma; however, PGL-DLBCL accounts for ~2% of all lymphomas (36). Thus, studies specifically focused on PGI-DLBCL are limited. Although the pathogenesis of PGI-DLBCL remains unclear, its increased incidence may be associated with genetic and epigenetic molecular mechanisms (37,38). Previous studies have demonstrated that the imbalance of miRNA expression seems to be a common feature in several types of cancer. Emerging evidence suggests that these miRNAs act as promising biomarkers and therapeutic targets (39), and thus may be used as a tool to develop novel anticancer cell strategies.
A previous study have implicated aberrant miR-150 expression in tumorigenesis. Additionally, miR-150 may have a potential therapeutic role against cancer by regulating the expression of oncogenes and/or tumor suppressor genes (40). A number of studies have demonstrated dynamic changes in miR-150 expression in several solid tumors, and also during the development of hematological malignancies. For example, Zhang et al (41) reported that miR-150 regulates NSCLC progression by inducing the proliferation and migration of NSCLC cells. Furthermore, malignant hematopoiesis and leukemia development are associated with aberrant miR-150 expression (42,43). Elevated miR-150 expression was demonstrated to be associated with decreased postoperative survival of patients with gastric cancer (44). However, to the best of our knowledge, little is known about the expression pattern and clinical significance of miR-150 in PGI-DLBCL. Thus, the present study aimed to investigate miR-150 expression in patients with PGI-DLBCL. Taken together, the current results demonstrated that miR-150 may influence the pathogenesis of PGI-DLBCL and act as a key independent prognostic factor for this disease.
The results of the present study indicated that miR-150 expression was significantly lower in PGI-DLBCL tissues compared with adjacent non-tumor tissues. Furthermore, miR-150 expression was also decreased in the high-IPI subgroup compared with the low-IPI subgroup. Statistical analysis demonstrated that low miR-150 expression was significantly associated with stage (IIE-IV), IPI score (3–5), ECOG status (2–4) and treatment (R-CHOP). ROC curve analysis indicated that the optimal cut-off concentration of miR-150 was 8.965, with an AUC value of 0.882, and high sensitivity (79.8%) and specificity (77.1%). The present results suggest that miR-150 expression level may help distinguish between patients with PGI-DLBCL and healthy individuals. The OS and PFS time were significantly shorter in patients with low miR-150 expression compared with those with high miR-150 expression. Furthermore, miR-150 was demonstrated to be an independent prognostic factor during both univariate and multivariate analyses.
The IPI score is the most commonly used prognostic evaluation tool in DLBCL, and the prognosis of patients with a high IPI score is generally poor, while that of patients with a low IPI score is relatively favorable (33,45). In the present study, statistical analysis demonstrated significant differences between the miR-150 and IPI subgroups, suggesting that both may be used as a powerful index to determine the prognosis of patients. Furthermore, a previous study reported that the prognosis of patients with DLBCL, with GCB expression profiles is more favorable compared with patients with non-GCB (46). However, the present study failed to demonstrate a significant difference between miR-150 expression level and pathological subtype. Regarding treatment, a significant association was observed between miR-150 expression and the R-CHOP treatment regime, which is consistent with a previous finding that the CHOP regimen with rituximab (CD20 antibody) significantly improves the prognosis of patients (47).
A number of limitations exist within the present study. First, inadequate samples containing other extranodal lymphomas were obtained from patients, limiting assessment to only PGI-DLBCL specimens. This made it difficult to determine the effect of miR-150 expression in other extranodal lymphomas. Secondly, although the present study demonstrated the ability of miR-150 to predict the prognosis of patients with PGI-DLBCL, due to population size limitations, it failed to independently verify the sample settings and the existence of the same expression results in different samples. Future studies should aim to increase the population size and include patients with other extranodal lymphomas, in order to overcome these shortcomings and verify the current results.
Taken together, the results of the present study indicate that that miR-150 serves a vital role in the pathogenesis and progression of PGI-DLBCL, whereby low miR-150 expression is associated with poor predicted clinical outcomes. Furthermore, miR-150 expression was investigated in PGI-DLBCL tissues, which consolidated its role as a valuable diagnostic and prognostic biomarker, and potential therapeutic target in patients with PGI-DLBCL.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- AUC
area under the curve
- BCL-6
B-cell lymphoma 6
- ECOG
Eastern Cooperative Oncology Group
- GCB
germinal center B-cell like
- IPI
International Prognostic Index
- LDH
lactate dehydrogenase
- MALT
mucosa-associated lymphoid tissue
- miRNA
microRNA
- miR-150
microRNA-150
- MUM1
multiple myeloma antigen 1
- NSCLC
non-small cell lung cancer
- NSCLC
non-small cell lung cancer
- OS
overall survival
- PFS
progression-free survival
- PGI-DLBCL
primary gastrointestinal diffuse large B-cell lymphoma
- ROC
receiver operating characteristic
- RT-qPCR
reverse transcription-quantitative PCR
Funding
The present study was funded by the National Natural Science Foundation of China (grant nos. 81100337 and 81470283).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HaZ and YW designed the study and reviewed the final manuscript. XinW performed the experiments, analyzed the experimental data and drafted the initial manuscript. PG, LC and YK helped perform the experiments. XiaW, ZZ, YY and HY acquired patient information and specimens. HuZ, XL, LQ, LL, QZ, ZQ and TD helped acquire the specimens. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute (Tianjin, China) and written informed consent was provided by all patients prior to the study start. All procedures were performed in accordance with the ethical standards of the Institutional Review Board and The Declaration of Helsinki, and its later amendments or comparable ethical standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Peng JC, Zhong L, Ran ZH. Primary lymphomas in the gastrointestinal tract. J Dig Dis. 2015;16:169–176. doi: 10.1111/1751-2980.12234. [DOI] [PubMed] [Google Scholar]
- 2.Wang T, Gui W, Shen Q. Primary gastrointestinal non-Hodgkin's lymphoma: Clinicopathological and prognostic analysis. Med Oncol. 2010;27:661–666. doi: 10.1007/s12032-009-9265-1. [DOI] [PubMed] [Google Scholar]
- 3.Cogliatti SB, Schmid U, Schumacher U, Eckert F, Hansmann ML, Hedderich J, Takahashi H, Lennert K. Primary B-cell gastric lymphoma: A clinicopathological study of 145 patients. Gastroenterology. 1991;101:1159–1170. doi: 10.1016/0016-5085(91)90063-Q. [DOI] [PubMed] [Google Scholar]
- 4.Koch P, del Valle F, Berdel WE, Willich NA, Reers B, Hiddemann W, Grothaus-Pinke B, Reinartz G, Brockmann J, Temmesfeld A, et al. Primary gastrointestinal non-Hodgkin's lymphoma: I. Anatomic and histologic distribution, clinical features, and survival data of 371 patients registered in the German multicenter study GIT NHL 01/92. J Clin Oncol. 2001;19:3861–3873. doi: 10.1200/JCO.2001.19.18.3861. [DOI] [PubMed] [Google Scholar]
- 5.Papaxoinis G, Papageorgiou S, Rontogianni D, Kaloutsi V, Fountzilas G, Pavlidis N, Dimopoulos M, Tsatalas C, Xiros N, Economopoulos T. Primary gastrointestinal non-Hodgkin's lymphoma: A clinicopathologic study of 128 cases in Greece. A hellenic cooperative oncology group study (HeCOG) Leuk Lymphoma. 2006;47:2140–2146. doi: 10.1080/10428190600709226. [DOI] [PubMed] [Google Scholar]
- 6.Suresh B, Asati V, Lakshmaiah KC, Babu G, Lokanatha D, Jacob LA, Lokesh KN, Rudresh AH, Rajeev LK, Smitha S, et al. Primary gastrointestinal diffuse large B-cell lymphoma: A prospective study from South India. South Asian J Cancer. 2019;8:57–59. doi: 10.4103/sajc.sajc_52_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 8.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Zhu FQ, Zeng L, Tang N, Tang YP, Zhou BP, Li FF, Wu WG, Zeng XB, Peng SS. MicroRNA-155 downregulation promotes cell cycle arrest and apoptosis in diffuse large B-cell lymphoma. Oncol Res. 2016;24:415–427. doi: 10.3727/096504016X14685034103473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang H, Shen J, Zheng Z, Luo X, Gao R, Zhuang X. MicroRNA-146a rs2910164 polymorphism and the risk of diffuse large B cell lymphoma in the Chinese Han population. Med Oncol. 2014;31:306. doi: 10.1007/s12032-014-0306-z. [DOI] [PubMed] [Google Scholar]
- 12.Berglund M, Hedström G, Amini RM, Enblad G, Thunberg U. High expression of microRNA-200c predicts poor clinical outcome in diffuse large B-cell lymphoma. Oncol Rep. 2013;29:720–724. doi: 10.3892/or.2012.2173. [DOI] [PubMed] [Google Scholar]
- 13.Ivey KN, Srivastava D. microRNAs as developmental regulators. Cold Spring Harb Perspect Biol. 2015;7:a008144. doi: 10.1101/cshperspect.a008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larrabeiti-Etxebarria A, Lopez-Santillan M, Santos-Zorrozua B, Lopez-Lopez E, Garcia-Orad A. Systematic review of the potential of MicroRNAs in diffuse large B cell lymphoma. Cancers (Basel) 2019;11(pii):E144. doi: 10.3390/cancers11020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 16.Koscianska E, Krzyzosiak WJ. Current understanding of the role of microRNAs in spinocerebellar ataxias. Cerebellum Ataxias. 2014;1:7. doi: 10.1186/2053-8871-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi N, Cao J, Song Y, Shen J, Liu W, Fan J, He J, Shi Y, Zhang X, Lu N, et al. A microRNA signature predicts survival in early stage small-cell lung cancer treated with surgery and adjuvant chemotherapy. PLoS One. 2014;9:e91388. doi: 10.1371/journal.pone.0091388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang W, Xu P, Wang H, Niu Z, Zhu D, Lin Q, Tang L, Ren L. MicroRNA-150 suppresses triple-negative breast cancer metastasis through targeting HMGA2. Onco Targets Ther. 2018;11:2319–2332. doi: 10.2147/OTT.S161996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokobori T, Suzuki S, Tanaka N, Inose T, Sohda M, Sano A, Sakai M, Nakajima M, Miyazaki T, Kato H, Kuwano H. miR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104:48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, Zhang P, Wang F, Zhang H, Yang J, Peng J, Liu W, Qin H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61:1447–1453. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 21.Lee KH, Lee JK, Choi DW, Do IG, Sohn I, Jang KT, Jung SH, Heo JS, Choi SH, Lee KT. Postoperative prognosis prediction of pancreatic cancer with seven microRNAs. Pancreas. 2015;44:764–768. doi: 10.1097/MPA.0000000000000346. [DOI] [PubMed] [Google Scholar]
- 22.Huang S, Chen Y, Wu W, Ouyang N, Chen J, Li H, Liu X, Su F, Lin L, Yao Y. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PLoS One. 2013;8:e80707. doi: 10.1371/journal.pone.0080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin QW, Sun XF, Yang GT, Li XB, Wu MS, Zhao J. Increased expression of microRNA-150 is associated with poor prognosis in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:842–846. [PMC free article] [PubMed] [Google Scholar]
- 24.Dezhong L, Xiaoyi Z, Xianlian L, Hongyan Z, Guohua Z, Bo S, Shenglei Z, Lian Z. miR-150 is a factor of survival in prostate cancer patients. J BUON. 2015;20:173–179. [PubMed] [Google Scholar]
- 25.Dettmer MS, Perren A, Moch H, Komminoth P, Nikiforov YE, Nikiforova MN. MicroRNA profile of poorly differentiated thyroid carcinomas: New diagnostic and prognostic insights. J Mol Endocrinol. 2014;52:181–189. doi: 10.1530/JME-13-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Wang X, Zhang Y, Wang D, Gao H, Wang L, Gao S. Prognostic role of microRNA-150 in various carcinomas: A meta-analysis. Onco Targets Ther. 2016;9:1371–1379. doi: 10.2147/OTT.S97969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szurián K, Csala I, Piurkó V, Deák L, Matolcsy A, Reiniger L. Quantitative miR analysis in chronic lymphocytic leukaemia/small lymphocytic lymphoma-proliferation centres are characterized by high miR-92a and miR-155 and low miR-150 expression. Leuk Res. 2017;58:39–42. doi: 10.1016/j.leukres.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Wang M, Yang W, Li M, Li Y. Low expression of miR-150 in pediatric intestinal Burkitt lymphoma. Exp Mol Pathol. 2014;96:261–266. doi: 10.1016/j.yexmp.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 30.Gebauer N, Kuba J, Senft A, Schillert A, Bernard V, Thorns C. MicroRNA-150 Is up-regulated in extranodal marginal zone lymphoma of MALT type. Cancer Genomics Proteomics. 2014;11:51–56. [PubMed] [Google Scholar]
- 31.Tomonaga M. Outline and direction of revised WHO classification of tumors of haematopoietic and lymphoid tissues. Rinsho Ketsueki. 2009;50:1401–1406. (In Japanese) [PubMed] [Google Scholar]
- 32.Huang J, Jiang W, Xu R, Huang H, Lv Y, Xia Z, Sun X, Guan Z, Lin T, Li Z. Primary gastric non-Hodgkin's lymphoma in Chinese patients: Clinical characteristics and prognostic factors. BMC Cancer. 2010;10:358. doi: 10.1186/1471-2407-10-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horwitz SM, Zelenetz AD, Gordon LI, Wierda WG, Abramson JS, Advani RH, Andreadis CB, Bartlett N, Byrd JC, Fayad LE, et al. NCCN guidelines insights: Non-Hodgkin's lymphomas, version 3.2016. J Natl Compr Canc Netw. 2016;14:1067–1079. doi: 10.6004/jnccn.2016.0117. [DOI] [PubMed] [Google Scholar]
- 34.Cheson BD, Pfistner B, Juweid ME, Gascoyne RD, Specht L, Horning SJ, Coiffier B, Fisher RI, Hagenbeek A, Zucca E, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25:579–586. doi: 10.1200/JCO.2006.09.2403. [DOI] [PubMed] [Google Scholar]
- 35.Ferreri AJ, Montalban C. Primary diffuse large B-cell lymphoma of the stomach. Crit Rev Oncol Hematol. 2007;63:65–71. doi: 10.1016/j.critrevonc.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Dawson IM, Cornes JS, Morson BC. Primary malignant lymphoid tumours of the intestinal tract. Report of 37 cases with a study of factors influencing prognosis. Br J Surg. 1961;49:80–89. doi: 10.1002/bjs.18004921319. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura S, Iida M. Molecular genetics in primary gastrointestinal lymphomas. Fukuoka Igaku Zasshi. 2008;99:123–130. (In Japanese) [PubMed] [Google Scholar]
- 38.Nakamura S, Matsumoto T, Nakamura S, Jo Y, Fujisawa K, Suekane H, Yao T, Tsuneyoshi M, Iida M. Chromosomal translocation t(11;18)(q21;q21) in gastrointestinal mucosa associated lymphoid tissue lymphoma. J Clin Pathol. 2003;56:36–42. doi: 10.1136/jcp.56.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: Opportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T, et al. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25:1324–1334. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Lin J, Ye Y, Oba T, Gentile E, Lian J, Wang J, Zhao Y, Gu J, Wistuba II, et al. Serum MicroRNA-150 predicts prognosis for early-stage non-small cell lung cancer and promotes tumor cell proliferation by targeting tumor suppressor gene SRCIN1. Clin Pharmacol Ther. 2018;103:1061–1073. doi: 10.1002/cpt.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Y, Jiang X, Chen J. The role of miR-150 in normal and malignant hematopoiesis. Oncogene. 2014;33:3887–3893. doi: 10.1038/onc.2013.346. [DOI] [PubMed] [Google Scholar]
- 43.Fayyad-Kazan H, Bitar N, Najar M, Lewalle P, Fayyad-Kazan M, Badran R, Hamade E, Daher A, Hussein N, ElDirani R, et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J Transl Med. 2013;11:31. doi: 10.1186/1479-5876-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, Ogawa R, Harata K, Fujii Y. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537–542. [PubMed] [Google Scholar]
- 45.Song JL, Wei XL, Zhang YK, Hao XX, Huang WM, Wei Q, Wei YQ, Feng R. The prognostic value of the international prognostic index, the national comprehensive cancer network IPI and the age-adjusted IPI in diffuse large B cell lymphoma. Zhonghua Xue Ye Xue Za Zhi. 2018;39:739–744. doi: 10.3760/cma.j.issn.0253-2727.2018.09.007. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Imhoff GW, Boerma EJ, van der Holt B, Schuuring E, Verdonck LF, Kluin-Nelemans HC, Kluin PM. Prognostic impact of germinal center-associated proteins and chromosomal breakpoints in poor-risk diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:4135–4142. doi: 10.1200/JCO.2006.05.5897. [DOI] [PubMed] [Google Scholar]
- 47.Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R, Morel P, Van Den Neste E, Salles G, Gaulard P, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:235–242. doi: 10.1056/NEJMoa011795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.