Abstract
The understanding into the pathogenesis and treatment of gastric cancer has improved in recent years; however, a number of limitations have delayed the development of effective treatment. Cancer cells can undergo glycolysis and inhibit oxidative phosphorylation in the presence of oxygen (Warburg effect). Previous studies have demonstrated that a rotary cell culture system (RCCS) can induce glycolytic metabolism. In addition, the potential of regulating cancer cells by targeting their metabolites has led to the rapid development of metabolomics. In the present study, human HGC-27 gastric cancer cells were cultured in a RCCS bioreactor, simulating weightlessness. Subsequently, liquid chromatography-mass spectrometry was used to examine the effects of simulated microgravity (SMG) on the metabolism of HGC-27 cells. A total of 67 differentially regulated metabolites were identified, including upregulated and downregulated metabolites. Compared with the normal gravity group, phosphatidyl ethanolamine, phosphatidyl choline, arachidonic acid and sphinganine were significantly upregulated in SMG conditions, whereas sphingomyelin, phosphatidyl serine, phosphatidic acid, L-proline, creatine, pantothenic acid, oxidized glutathione, adenosine diphosphate and adenosine triphosphate were significantly downregulated. The Human Metabolome Database compound analysis revealed that lipids and lipid-like metabolites were primarily affected in an SMG environment in the present study. Overall, the findings of the present study may aid our understanding of gastric cancer by identifying the underlying mechanisms of metabolism of the disease under SMG.
Keywords: HGC-27, SMG, RCCS, metabolism, PE, PC, PS, PA, SM
Introduction
Studies conducted in real and simulated microgravity (SMG) have demonstrated that the physiological functions of living organisms undergo substantial changes in different gravitational conditions (1,2). These changes may have adverse effects, including degenerative changes to bone and muscle, cephalic fluid shifts and electrolyte disturbances, and dysfunction of the immune and cardiovascular system (3,4). Based on these studies, the influence of real and SMG on human macroscopic tissue is relatively clear; however, the influence of these conditions on cells, especially cancer cells, requires further investigation.
The effect of real and SMG on different cancer cells has been previously studied (5–10) and alterations primiarly involved the morphological structure and physiological function. For example, compared with the 2D monolayer structure of cancer cells under normal gravity (NG), cells are more rounded and form complex 3D spheroid structures under SMG (5–7). Furthermore, G2/M and G2+M phases are key mitotic check points, which are increased under SMG (8) and decreased proliferation and increaseed apoptosis are also observed under decreased SMG (9,10).
The influence of metabolism on cancer cells, which functions in the synthesis of various cell substances such as cell membrane, is of interest in the field of cancer. Under physiological conditions, oxidative phosphorylation is the primary process that generates ATP and glycolysis is dominant only when oxygen is low; however, the opposite is observed in tumor cells (11,12). This ability of tumor cells to undergo glycolysis, even under aerobic conditions, is referred to as the Warburg effect (11), and the metabolic reprogramming of several tumor characteristics involved has been previously investigated, including increased ATP synthesis and uncontrolled tumor growth (13). Previous studies have demonstrated that the levels of lactic acid were increased and glucose levels decreased in gastric cancer tissues compared with the control group (14,15). The high levels of lactate may be due to the Warburg effect (11), whereas the low glucose levels may be due to the elevated fructose-6-phosphokinase activity (16), high levels of glucose transporters (17) and increased type II hexokinase activity (18) in gastric cancer tissues. High activity levels of pyruvate kinase and lactate dehydrogenase are associated with cancer proliferation and poor prognosis (19,20), whereas their downregulation in vitro can impair tumor invasion (21,22). Based on these reports, several studies have investigated the influence of normal gravity on physiology and metabolism of cancer (23–25); however, the effect of gravity metabolism on cells, especially gastric cancer cells, is poorly understood.
Previous studies investigating MDA-MB-231 and MCF-7 breast cancer cells have reported changes in energy metabolism and increases in intracellular lactic acid and lactate dehydrogenase activity (26). Metabolomic analysis revealed several metabolic pathways affected and a number of pathways involved in glycometabolism were significantly influenced under SMG, including glycolysis, Kreb's cycle, the pentose phosphate pathway, and the glycerol-phosphate and the malate-aspartate shuttles. The following results were observed: The glycolysis pathway was stimulated by increased levels of glucose-6-phosphate and other glycolytic precursors, such as 3-phosphoglyceraldehyde and 1,3-diphosphoglyceric acid; the Kreb's cycle was activated by the accumulation of acetyl-CoA and the content of acyl-carnitine was decreased under SMG, reflected by increased acyl-CoA levels, which suggests that the β-oxidation pathway was decreased (27). Thus, to demonstrate the effects of SMG on the metabolism of HGC-27 gastric cancer cells in the present study, liquid chromatography-mass spectrometry (LC-MS) was used.
The present study aimed to improve the understanding of the physiology and pathology of gastric cancer cells under SMG. The metabolomics analysis of HGC-27 cells under SMG may promote further understanding of the regularity of tumor occurrence and the development and changes in physiological function, to provide a novel therapeutic strategy for gastric cancer.
Materials and methods
Cell culture
The gastric cancer cell line HGC-27 was purchased from the BeNa Culture Collection. Cells were cultured in RPMI 1640 medium and supplemented with 10% fetal bovine serum (both Thermo Fisher Scientific, Inc), 100 units/ml penicillin and 100 mg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc) and were incubated at 37°C with 5% CO2. The medium was changed three times a week.
Microgravity simulation
The RCCS bioreactor (Synthecon, Inc.) was developed by the National Aeronautics and Space Administration to simulate the effects of microgravity. A total of 12 bioreactors were used (total volume, 55 ml), each equipped with a gas-exchange membrane and incubated at 37°C with 5% CO2. The bottom of the RCCS bioreactor was designed with a silicone membrane and prevented the formation of bubbles (28). Cells in the RCCS bioreactor were grown on cytodex-3 microcarrier beads (Sigma-Alrich; Merck KGaA) to provide a solid support pretreated with phosphate buffered saline (PBS) and 75% ethanol, and stored at 4°C. After washing cytodex-3 microcarrier beads three times with PBS, the beads were added to the rotating culture vessel. Subsequently, 7×106 HGC-27 cells with 250 mg cytodex-3 microcarrier beads were seeded into a 55 ml RCCS bioreactor containing 500 ml of RPMI-1640 complete medium (Gibco; Thermo Fisher Scientific, Inc.), and all air bubbles were removed using a 5 ml syringe. The rotational speed of the high aspect ratio vessel was 0.01 × g. Control groups (NG) of 1 g static cultures were stored in the same incubator at 37°C with 5% CO2. Both groups were cultured in the RCCS bioreactor for 1 and 3 days, respectively.
Sample preparation and treatment
The cell-microcarrier complexes were washed with PBS three times. A total of 4 ml Trypsin (0.25% EDTA) was added at 37°C for 10 min and the cells were dislodged by intermittently tapping the flask against a hard surface. This reaction was terminated by adding 500 ml of RPMI-1640 complete medium followed by centrifugation at 167.7 × g for 5 min at 37°C. The suspension was obtained by filtering the mixture of cells and microcarriers with 70 µm cell sieves. The control group was washed twice using PBS, with the addition of 2–3 ml 0.25% Trypsin-EDTA Solution (Gibco; Thermo Fisher Scientific, Inc.) at 37°C for 5 min. The supernatant was discarded following centrifugation at 167.7 × g for 5 min at 37°C. Cells collected from NG and SMG were stored at −80°C for ~1 week, prior to subsequent experimentation.
A total of 7×106 HGC-27 NG and SMG cells were added to 400 µl cold methanol and the cell pellet was dislodged using a high flux tissue crusher (MP Biomedicals Co., Ltd.) at 4°C. Subsequently, 100 µl of distilled water was added and ultrasonic extraction (Ningbo Xinzhi Biotechnology Co., Ltd.) was performed on ice three times (10 min each time).
Analysis of metabolites using LC- MS
LC-MS was performed using a 100×2.1 mm2 Acquity 1.7 µm C18 column and a ACQUITY Ultra High-Performance Liquid Chromatography system (both Waters Corporation). The following conditions were used; Ionization mode, positive/negative; nitrogen gas temperature, 500°C; nebulizer pressure, 50 psi and flow rate, 0.40 ml/min. Each sample was analyzed six times and the scanning range of mass spectrometry was 50–1,000 m/z and the resolution was 30,000 dpi.
Data processing
MS data matrices were obtained using Progenesis QI software (version 2.0; Waters Corporation). The data matrices were imported into SIMCA-P+ software (version 14.0; Sartorius AG) and unsupervised principal component analysis was used to observe the overall distribution among samples and the degree of dispersion between groups and supervised partial least square discriminant analysis (PLS-DA) was used to distinguish the overall differences of metabolic profiles among groups. For PLS-DA scores, a variable importance (VIP) value >1 was considered to indicate a statistically significant difference, whereas R2Y and Q2>0.5 were considered to indicate a strong prediction capability. The Human Metabolme (HMDB; http://www.hmdb.ca) and the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp) databases were used to identify potential metabolites.
Statistical analysis
Statistical analysis was performed using SPSS software version 21.0 (IBM Corp.). Data are expressed as the mean ± standard deviation for continuous variables and were analyzed using an independent sample t-test or one-way ANOVA, followed by Tukey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference. All experiments were performed in triplicate.
Results
Metabolic alterations of HGC-27 cells under NG and SMG
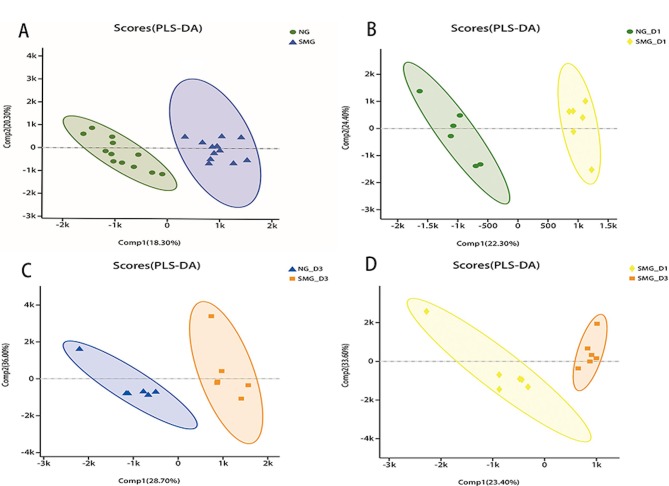
The PLS-DA scores in HGC-27 cells under NG and SMG are presented in Fig. 1. The value of the parameters R2Y and Q2 are >0.5, which suggests that the model was good and that the prediction capability was strong. The greater the separation degree between the two groups, the more significant the classification effect. Comp1 refers to the first principal component interpretation degree and Comp2 refers to the second principal component interpretation degree. The metabolites of NG and SMG cells were grouped, indicating that there was a difference in metabolites between the two groups. A total of 67 differentially regulated metabolites were identified, including 36 upregulated and 31 downregulated metabolites (all P<0.05, VIP>1). Compared with the NG group, phosphatidyl ethanolamine (PE), phosphatidyl choline (PC), arachidonic acid and sphinganine were significantly upregulated in the SMG group, whereas sphingomyelin (SM), phosphatidyl serine (PS), phosphatidic acid (PA), L-Proline, creatine, pantothenic acid, oxidized glutathione (GSSG), adenosine diphosphate (ADP) and adenosine triphosphate (ATP) were significantly downregulated.
Figure 1.
Metabolite-concentration distribution in HGC-27 gastric cancer cells under NG and SMG using PLS-DA. (A) Total NG for 1 and 3 days vs. total SMG for 1 and 3 days. (B) NG for 1 day vs. SMG for 1 day. (C) NG for 3 days vs. SMG for 3 days. (D) SMG for 1 day vs. SMG for 3 days. PLS-DA, partial least squares discriminate analysis; NG, normal gravity; SMG, simulated microgravity; D1, 1 day duration; D3, 3 day duration; Comp, component.
Lipids and lipid-like metabolite alterations of HGC-27 cells in NG and SMG
The HMDB method in LC-MS was used to assess compound classification of HGC-27 cells. Compared with the NG groups, HMDB compound analysis confirmed that glyceropholipids (45.10%) and fatty acids (13.73%) were the most affected by SMG, followed by nucleotides (Fig. 2). Of the 67 metabolites, LC-MS/MS profiling of HGC-27 cells detected 50 lipid molecules, including 31 phospholipids and 7 sphingolipids, which were upregulated in SMG (Table I). These results suggest that these two types of lipids exhibited special biological behaviors in HGC-27 cells under SMG.
Figure 2.
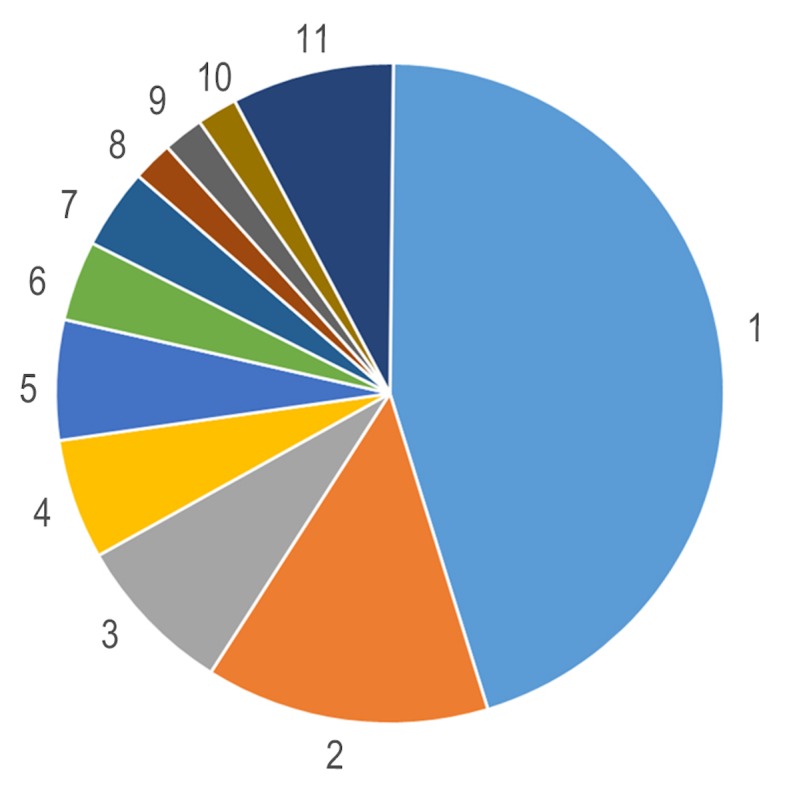
Human Metabolme Database compound classification analysis of the significantly altered metabolites indicating that lipids were the mo st influenced metabolite under simulated microgravity. 1, Glycerophospholipids (45.1%); 2, Fatty Acids (13.73%); 3, Carboxylic acids and derivatives (7.84%); 4, Prenol lipids (5.88%); 5, Sphingolipids (5.88%); 6, Organonitrogen Compounds (3.92%); 7, Organooxygen compounds (3.92%); 8, Alcohols and polyols (1.96%); 9, Carbonyl compounds (1.96%); 10, Flavonoids (1.96%) and 11, Others (7.85%).
Table I.
Quantitated lipid classes and numbers in HGC-27 cells under normal gravity and simulated microgravity.
| Lipid category | Lipid class | Number of lipid species |
|---|---|---|
| Phospholipid | Phosphatidylcholine | 13 |
| Lysophospholipid | 1 | |
| Glycerophosphocholine | 1 | |
| Phosphatidylethanolamine | 12 | |
| Phosphatidylserine | 3 | |
| Phosphatidic acid | 1 | |
| Sphingolipid | Sphingomyelin | 2 |
| Sphinganine | 1 | |
| Oleamide | 1 | |
| Linoleamide | 1 | |
| Dihydroceramide | 1 | |
| Lactosylceramide | 1 | |
| Glycerolipide | Glycinoprenol 9 | 1 |
| Other lipids | Umbelliprenin | 1 |
| Sorbitan stearate | 1 | |
| Pantothenic Acid | 1 | |
| Oleoylcarnitine | 1 | |
| Isobutyryl carnitine | 1 | |
| Glycocholic acid | 1 | |
| Camelliagenin B | 1 | |
| Arachidonic acid | 1 | |
| 8-Hydroxyguanosine | 1 | |
| 3-Hydroxyhexadecanoyl carnitine | 1 | |
| 2-Methylbutyroylcarnitine | 1 | |
| Total lipids | 50 |
A total of 38 significantly affected lipids, particularly phospholipids and sphingolipids, were observed, which characterized the differences between NG and SMG (VIP >1). Detailed information concerning these lipids is presented in Table II. Of these 38 lipids, 25 were upregulated and 13 were downregulated. The two predominant downregulated lipid classes detected in SMG were SM and PS. It was showed that PS [DiMe (11,3)/MonoMe (11,5)], PS [18:3 (9Z,12Z,15Z)/22:1(13Z)], PS [14:1 (9Z)/24:0)], PA[16:0/18:1(11Z)], SM (d18:0/16:1(9Z) and SM (d18: +B5:G321/14:0) levels significantly decreased, whereas Sphinganine, lactosylceramide (d18:1/12:0) and dihydroceramide levels increased under SMG.
Table II.
Identification of the significantly altered metabolites in HGC-27 cells.
| Metabolite | Formula | M/Z | P-value | Regulation | VIP |
|---|---|---|---|---|---|
| SM (d18:+B5:G321/14:0) | C37H75N2O6P | 719,5313 | 8.951×10−6 | Downregulated | 1,5850 |
| SM [d18:0/16:1(9Z)] | C39H79N2O6P | 747,5634 | 2.004×10−2 | Downregulated | 1,8047 |
| PS [DiMe(11,3)/MonoMe(11,5)] | C47H80NO12P | 918,4877 | 1.500×10−8 | Downregulated | 1,1943 |
| PS [18:3(9Z,12Z,15Z)/22:1(13Z)] | C46H82NO10P | 822,5620 | 9.948×10−3 | Downregulated | 1,2988 |
| PS [14:1(9Z)/24:0] | C44H84NO10P | 800,5760 | 1.440×10−7 | Downregulated | 3,0884 |
| PE-NMe2 [14:1(9Z)/16:1(9Z)] | C37H70NO8P | 732,4793 | 2.400×10−8 | Downregulated | 3,9878 |
| PE-NMe [14:1(9Z)/16:1(9Z)] | C36H68NO8P | 718,4649 | 4.190×10−6 | Downregulated | 1,0028 |
| PE (15:0/P-16:0) | C36H72NO7P | 706,4990 | 1.223×10−6 | Upregulated | 1,8137 |
| PE [15:0/24:1(15Z)] | C44H86NO8P | 832,6037 | 4.555×10−3 | Upregulated | 1,3361 |
| PE [15:0/22:5(4Z,7Z,10Z,13Z,16Z)] | C42H74NO8P | 796,5100 | 7.155×10−4 | Upregulated | 1,4220 |
| PE [15:0/22:4(7Z,10Z,13Z,16Z)] | C42H76NO8P | 798,5255 | 2.232×10−2 | Upregulated | 1,3835 |
| PE [15:0/22:2(13Z,16Z)] | C42H80NO8P | 802,5564 | 1.816×10−2 | Downregulated | 2,8050 |
| PE [15:0/22:1(13Z)] | C42H82NO8P | 804,5728 | 7.846×10−5 | Upregulated | 4,1810 |
| PE [15:0/20:2(11Z,14Z)] | C40H76NO8P | 774,5259 | 9.433×10−6 | Downregulated | 8,3376 |
| PE [15:0/18:2(9Z,12Z)] | C38H72NO8P | 746,4948 | 1.153×10−6 | Downregulated | 4,9712 |
| PE [14:0/24:1(15Z)] | C43H84NO8P | 818,5870 | 9.771×10−6 | Upregulated | 1,1963 |
| PE [14:0/20:2(11Z,14Z)] | C39H74NO8P | 757,5542 | 4.356×10−3 | Downregulated | 2,1212 |
| PC [o-16:0/20:4(8Z,11Z,14Z,17Z)] | C44H82NO7P | 812,5771 | 1.832×10−2 | Upregulated | 1,2528 |
| PC [22:5(4Z,7Z,10Z,13Z,16Z)/16:0] | C46H82NO8P | 852,5716 | 4.918×10−2 | Upregulated | 2,0817 |
| PC [22:4(7Z,10Z,13Z,16Z)/14:0] | C44H80NO8P | 826,5566 | 7.767×10−6 | Upregulated | 3,9909 |
| PC [18:4(6Z,9Z,12Z,15Z)/P-18:0] | C44H80NO7P | 810,5616 | 3.320×10−7 | Upregulated | 2,0711 |
| PC [18:4(6Z,9Z,12Z,15Z)/P-16:0] | C42H76NO7P | 782,5270 | 1.608×10−5 | Upregulated | 1,0437 |
| PC [18:1(11Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)] | C48H82NO8P | 876,5722 | 2.134×10−4 | Upregulated | 1,7139 |
| PC [18:0/20:4(5Z,8Z,11Z,14Z)] | C46H84NO8P | 854,5879 | 9.655×10−6 | Upregulated | 1,5839 |
| PC [18:0/18:2(9Z,12Z)] | C44H84NO8P | 830,5881 | 6.867×10−4 | Upregulated | 2,3650 |
| PC [16:1(9Z)/20:3(5Z,8Z,11Z)] | C44H80NO8P | 826,5554 | 1.198×10−4 | Upregulated | 1,5843 |
| PC [16:1(9Z)/16:1(9Z)] | C40H76NO8P | 752,5196 | 2.971×10−2 | Downregulated | 4,8632 |
| PC [16:0/20:5(5Z,8Z,11Z,14Z,17Z)] | C44H78NO8P | 824,5400 | 9.291×10−3 | Upregulated | 3,4401 |
| PC [16:0/20:3(5Z,8Z,11Z)] | C44H82NO8P | 826,5554 | 6.358×10−3 | Upregulated | 1,4770 |
| PC (16:0/18:3(6Z,9Z,12Z)] | C42H78NO8P | 800,5412 | 9.244×10−4 | Upregulated | 2,2188 |
| PA (16:0/18:1(11Z)] | C37H71O8P | 719,4839 | 4.759×10−6 | Downregulated | 1,2779 |
| LysoPC [20:4(8Z,11Z,14Z,17Z)] | C28H50NO7P | 588,3274 | 5.778×10−5 | Upregulated | 1,7989 |
| Sphinganine | C18H39NO2 | 302,3052 | 6.998×10−3 | Upregulated | 2,7342 |
| Oleamide | C18H35NO | 563,5495 | 7.274×10−4 | Upregulated | 31,6443 |
| Linoleamide | C18H33NO | 280,2632 | 1.702×10−2 | Upregulated | 10,9674 |
| Lactosylceramide (d18:1/12:0) | C42H79NO13 | 850,5563 | 5.000×10−9 | Upregulated | 3,5254 |
| Glycerophosphocholine | C8H20NO6P | 258,1098 | 1.328×10−2 | Upregulated | 1,5225 |
| Dihydroceramide | C19H39NO3 | 362,3264 | 2.622×10−2 | Upregulated | 1,9767 |
VIP, variable importance; M/Z, mass-to-charge ratio; SM, sphingomyelin; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PA, phosphatidic acid; LysoPC, lysophosphatidylcholine.
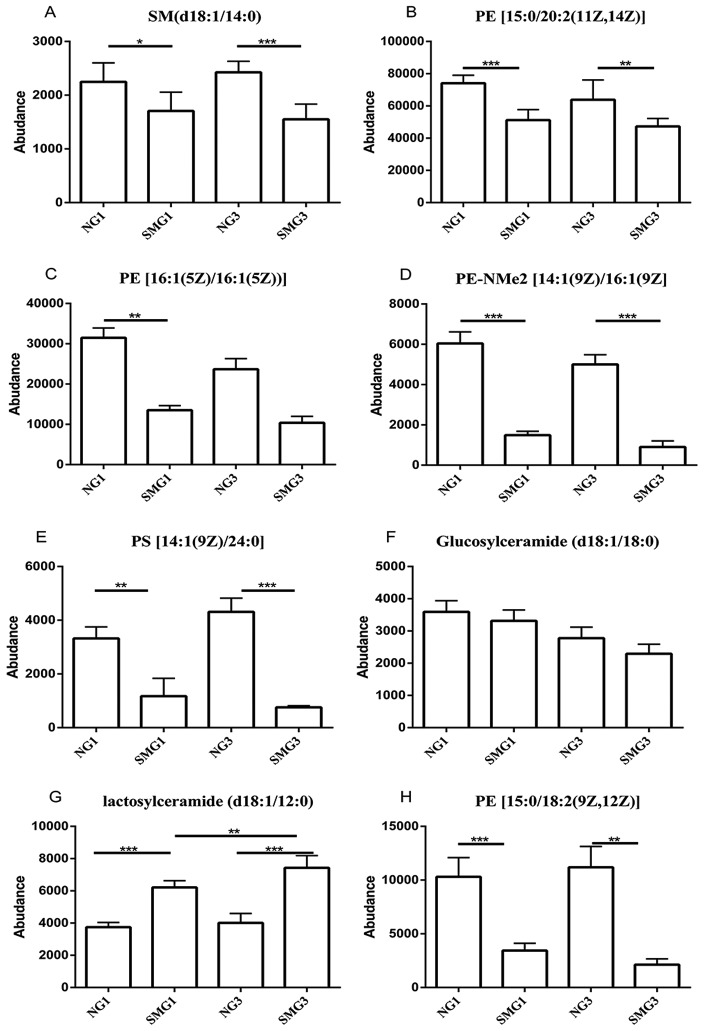
Among the 13 downregulated lipids one class of PS [14:1(9Z)/24:0] (consisting of one chain of myristoleic acid at the C-1 position and one chain of lignoceric acid at the C-2 position; http://www.hmdb.ca/metabolites/HMDB0112319), one class of SM (d18:1/14:0), three classes of PE, [15:0/18:2(9Z,12Z), 15:0/20:2(11Z,14Z), 16:1(5Z)/16:1(5Z) and PE-NMe2 14:1(9Z)/16:1(9Z)] were downregulated after 1 and 3 days of SMG compared with the NG group, (P<0.05; Fig. 3). Glucosylceramide (d18:1/18:0) was downregulated after 1 and 3 days of SMG compared with the NG group, however there was no statistical significance (P>0.05; Fig. 3). Regarding lactosylceramide (d18:1/12:0), there was a significant increase after 3 days of SMG exposure compared with 1 day of SMG (Fig. 3).
Figure 3.
Effects of SMG on the expression of (A) SM (d18:1/14:0), (B) PE [15:0/20:2(11Z,14Z)], (C) PE [16:1(5Z)/16:1(5Z)], (D) PE-NMe2 [14:1(9Z)/16:1(9Z)], (E) PS [14:1(9Z)/24:0], (F) Glucosylceramide (d18:1/18:0), (G) Lactosylceramide (d18:1/12:0) and (H) PE [15:0/18:2(9Z,12Z)]. *P<0.05, **P<0.01 and ***P<0.001. NG, normal gravity; SMG, simulated microgravity; D1, 1 day duration; D3, 3 day duration; PE, phosphatidyl ethanolamine; SM, sphingomyelin.
The influence of fatty acid β-oxidation under SMG
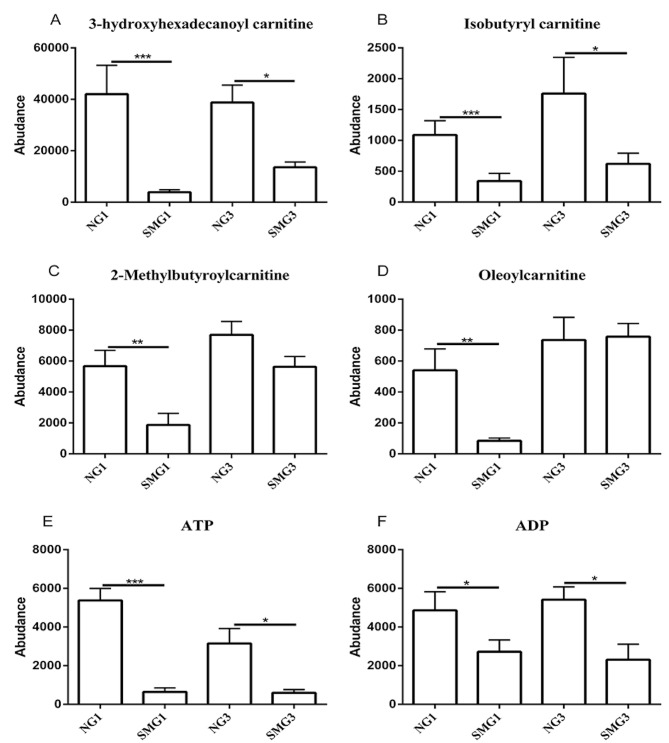
Carnitines, including 3-hydroxyhexadecanoyl carnitine, 2-methylbutyroylcarnitine, isobutyryl carnitine and oleoylcarnitine, serve a role in the oxidation of fatty acids to ATP (29). 3-hydroxyhexadecanoyl carnitine and isobutyryl carnitine were downregulated after 1 and 3 days of SMG compared with the NG group. However, 2-methylbutyroylcarnitine and oleoylcarnitine were only downregulated after 1 day of SMG. Furthermore, ATP and ADP were downregulated after 1 and 3 days of SMG compared with the NG group (Fig. 4).
Figure 4.
Effects of SMG on the expression of (A) 3-hydroxyhexadecanoyl carnitine, (B) Isobutyayl carnitine, (C) 2-Methylbutyroylcarnitine, (D) Oleoylcarbitine, (E) ATP and (F) ADP. *P<0.05, **P<0.01 and ***P<0.001. NG, normal gravity; SMG, simulated microgravity; D1, 1 day duration; D3, 3 day duration; ATP, adenosine triphosphate; ADP, adenosine diphosphate.
KEGG pathway enrichment analysis
Fig. 5 shows the KEGG enrichment pathways under SMG. These pathways could be grouped into six categories: Cellular processes; environmental information processing; genetic information processing; human diseases; metabolism; and organismal systems. The results show that SMG significantly affected multiple pathways and KEGG pathway enrichment analysis showed that the differentially expressed metabolites were primarily associated with: Regulation of the lysosome; ‘FoxO signaling pathway’; ‘leishmaniasis’; ‘GnRH signaling pathway’; ‘platelet activation’; ‘sphingolipid signaling pathway’; ‘Fc γ R-mediated phagocytosis’; ‘long-term depression’; ‘sphingolipid metabolism’; and ‘arginine and proline metabolism’.
Figure 5.
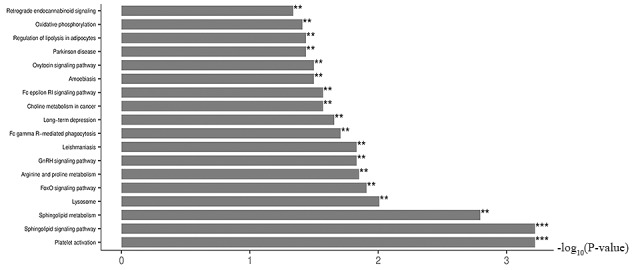
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of the significantly dysregulated metabolites with SMG. x-axis is the -log10(P-value). **P<0.01 and ***P<0.001. FoxO, forkhead box O.
Discussion
Several studies have explored the effects of different durations of NG and SMG on various types of cells, ranging from a few seconds to dozens of days, reporting notable effects on different types of cells, including thyroid, colorectal and breast cancer cells (30–33). In these studies cellular morphology was significantly altered (3D spheroid structure formation and the alteration of the cytoskeleton), the extracellular matrix (ECM) was increased, the cell cycle was modified after 10 and 12 days of SMG (the number of cells in the G1 phase increased, whilst the number of cells in the S and G2/M phases decreased), proliferation capacity was decreased and apoptosis was increased (30,31). The clear alteration of physiological functions of different human cancer cells were observed in our previous study (34). Chen et al (26) reported that different types of tumor can reprogram their metabolic processes to fulfill particular demands after 5 days of SMG, including processes involved in cancer cell proliferation, metastasis, immunological escape and survival, and shifting the aggressive phenotype of cancer cells. This metabolic reprogramming is regarded as a novel characteristic of tumors (35). Chen et al (26) also proposed a novel point to understand the Warburg effect in MDA-MB-231 and MCF-7 breast cancer cells. Glycolysis is often induced by hypoxia; however, glycolysis can also be induced by high levels of adrenomedullin under SMG, which may be a novel mechanism inducing glycolysis. However, to the best of our knowledge, the exact molecular mechanism of this reprogramming in tumor cells remains unknown under SMG.
A previous study reported that 72 h exposure to SMG induces changes in morphology, proliferation, cell cycle distribution and apoptosis in human SGC-7901 gastric cancer cells (36). Thus, the present study investigated whether 1 and 3 days exposure to SMG could influence the metabolites of human HGC-27 gastric cancer cells. Previous studies have demonstrated that SMG influences several metabolic pathways, stimulates lipid metabolism and interrupting the Krebs cycle in human breast cancer cells, osteoblast and oligodendrocyte after 5 d, 110 h and 3 d of SMG, respectively (26,27,37).
The present study demonstrated that SMG significantly affected HGC-27 human gastric cancer cells in a number of processes, in particular lipid metabolism. HGC-27 cells are highly malignant tumor cells (38). Reprograming lipid metabolism is associated with cancer invasion and metastasis (39); however, the influences of lipid metabolism on HGC-27 cells remains unclear. In the present study, HGC-27 lipid metabolites were measured using LC-MS and, to the best of our knowledge, this is the first study to analyze the alteration of lipid metabolic profiles of gastric cancer cells under SMG. As evidenced by LC-MS metabolome analysis, it was revealed the differential expression of 67 metabolites were altered during 1 and 3 days exposure to SMG compared with NG, according to the criteria of a VIP>1 and P<0.05, suggesting several different categories of membrane lipid components were affected by SMG. A total of 50 significantly different lipid metabolites were identified and 1 and 3 days exposure to SMG significantly increased the levels of PE, PC, arachidonic acid and sphinganine, but significantly decreased the levels of SM, PS, PA, pantothenic acid, ADP and ATP (Table I, Fig. 4). These upregulated and downregulated metabolites are associated with cellular processes, biological regulation and metabolic processes. These metabolites, altered in SMG, may have value as markers for the early diagnosis of gastric cancer.
PC, PE, PS, SM and PA are the primary components of membrane phospholipids, which play a significant role in cell membranes (40). A previous study showed that these phospholipids were upregulated in breast cancer tissues (41). The high levels of PE may serve as a biomarker of apoptosis in cancer (42) and PC has been considered as a biomarker of membrane proliferation in ovarian cancer (43). The present study demonstrated that PE and PC levels increased under SMG. Previous studies have indicated that several cancer cell lines, including ML-1 and ONCO-DG1 thyroid carcinoma cells, SGC-7901 gastric carcinoma cells, U251 glioma cells and MDA-MB-231 breast cancer cells, exhibit high levels of apoptosis under SMG after 48, 24, 36, 8 and 72 h of SMG (36,44–47). These high levels of apoptosis may be due to elevated PC levels, which indicates high membrane proliferation. These results are consistent with previous findings that apoptosis in human SGC-7901 gastric cancer cells increase following treatment with SMG after 48 h (36). However, other PEs, such as PE [15:0/18:2(9Z,12Z)], PE [15:0/20:2(11Z,14Z)], PE [16:1(5Z)/16:1(5Z)] and PE-NMe2 [14:1(9Z)/16:1(9Z)] were downegulated after 1 and 3 days of SMG compared with the NG group. It is hypothesized that PE is an key molecule for the synthesis of glycosylphosphatidylinositol anchors, which are glycolipids identified in yeast, protozoa, plants and humans, enabling modified proteins to be tethered to the outer leaflet of the plasma membrane necessary for the immune response, cell-cell communication and embryogenesis (48–51). Failure to attach can lead to dysfunction in the immune response, cell-cell communication and embryogenesis. As levels of PE were decreased in HGC-27 cells in the present study after 1 and 3 days of SMG, an adverse effect, such as gene mutations associated with cell cycle and apoptosis, may occur.
Chang et al (52) and Qian et al (53) observed that the invasive ability of A549 human lung adenocarcinoma and MCF-7 breast cells decreases following treatment with SMG after 72 h. A previous study reported that PS is associated with invasion of colorectal cancer cells (54). In the present study, it was demonstrated that PS [DiMe (11,3)/MonoMe (11,5)], PS [18:3 (9Z,12Z,15Z)/22:1(13Z)] and PS [14:1 (9Z)/24:0)] significantly decreased, suggesting that the the invasive ability of HGC-27 cells was weakened under SMG. This indicates that these phospholipids may have value as biomarkers for metastasis.
SM is located in the plasma membrane of most mammalian cells, which is the primary component of the pulp. The metabolic products of SM, including Cer and sphingosine (Sph), are signalling molecules with biological activity and act as first and/or second messengers controlling the vital activity of cells, such as cell growth, differentiation, aging, and apoptosis (55–57). The present study observed that SM (d18:0/16:1(9Z) and SM (d18: +B5:G321/14:0) levels were decreased, whereas Sphinganine, lactosylceramide (d18:1/12:0) and dihydroceramide levels increased under SMG (Table II). These results suggest that SMG may decrease the formation of sphingomyelin and increase its metabolism. Cer and Sph metabolic products function in the regulation of tumor proliferation and apoptosis, and are negative regulators of proliferation, which can inhibit cell growth and promote apoptosis (58). PE and the metabolic products of SM (Cer and Sph) can contribute to the increase in apoptosis under SMG (59,60). Previous studies have demonstrated a decrease in the proliferation capacity in SGC-7901 gastric cancer cells, U251 glioma cells and A549 lung adenocarcinoma cells, and increased apoptosis after 12, 48 and 72 h of SMG, respectively (36,47,53).
SM metabolism can regulate the function of K-Ras, which was the first human oncogene identified in human cancer and is a member of the Ras superfamily (61). Considering cellular signalling networks in the Ras-Raf-mitogen-activated protein kinase (MAPK) pathways, the role of K-Ras is associated with cell physiological functions, such as proliferation, apoptosis and survival (62). Relevant mutations in pathway components, such as serine/threonine-protein kinase B-raf, epidermal growth factor receptor and neurofibromin, are associated with progression in colon cancer and melanomas (63–65). The present study demonstrated that PA[16:0/18:1(11Z)] levels were decreased after SMG compared with the NG group (Table II). This may be associated with the high levels of phospholipase D2 (PLD2) after 3 days of SMG (66). PLD2 is an enzyme which hydrolyzes PA, promoting local invasion by regulating MT1-matrix metalloproteinase (MMP) (66). Based on the role of PLD2-generated PA in breast cancer metastasis, PLD2 may be regarded as a potential therapeutic target for metastatic breast cancer, and as a plasma membrane target (67). MT1-MMP, tethered to the plasma membrane, is a key protein associated with the migration of cancer cells (67). PLD2 generates small lipid molecules, such as phosphatidic acid, which can regulate the movement of the cytoskeleton, proliferation and migration (68–70). Therefore, it has been suggested that PLD2 and its small lipid molecules can regulate MT1-MMP for invasion and metastasis. However, this result contradicts the data of the present study, which demonstrates that some lipid levels, including PS [DiMe(11,3)/MonoMe(11,5)], PS [18:3(9Z,12Z,15Z)/22:1(13Z)] and PS [14:1(9Z)/24:0], were significantly decreased under SMG, suggesting that the invasive abilities of HGC-27 cells are weakened under SMG. It was hypothesized that the environment of cell proliferation may account for these contradicting results. Furthermore, the different devices used to simulate microgravity, such as random positioning machine and RCCS are speculated to account for the alteration in cell migration; however, further studies are required.
L-Proline, creatine, ADP and ATP levels were decreased after 1 and 3 days of SMG compared with the NG group in the present study. The downregulation of L-Proline in HGC-27 cells under SMG may be due to decreased activation of MMPs and degradation of the microenvironment for the ECM (71), rather than the degradation of collagen, which is regulated under conditions of nutrient stress assoicated with the mTOR signaling pathway, is blocked (72). However, the mTOR pathway participates in the process of apoptosis and autophagy (73).
The present study demonstrated that 3-hydroxyhexadecanoyl carnitine, 2-methylbutyroylcarnitine, isobutyryl carnitine and oleoylcarnitine, associated with β-oxidation, were significantly downregulated under SMG. Creatine is a key component in muscle contraction and can be transported between the mitochondria and cellular ATP utilization sites, acting as a spatial energy buffer (74). The function of creatine in cancer has been studied, reporting that some physiological functions of cancer, such as metabolism, are associated with the creatine kinase system, by regulating ATP provisions (75). The anticancer role of creatine has been demonstrated in vitro and in vivo experimental models (76,77). The downregulation of creatine, ADP and ATP in present study indicates that SMG may influence energy metabolism, an important process in physiological functions, including proliferation, cell cycle, migration and apoptosis after 5 d of expose (26). ATP is produced primarily in the mitochondria. SMG may have interferred with the normal mitochondrial function of HGC-27 gastric cancer cells, which has been illustrated in skeletal muscle tissue and cardiomyocytes (78,79). Mitochondrial function alteration might be an adaptive response for HGC-27 cells to reprogram their metabolism for particular requirements under SMG (80). A previous study reported that glycolysis and the pentose phosphate pathways were dominant, whereas the Krebs cycle was interrupted, which resulted in the downregulation of ATP, after 110 h of SMG (27). Similarly, complex II, a primary protein of the mitochondrial respiratory chain, was decreased and ATP was influenced by a reduction in proton transport when measured by proteomics (27).
Pantothenic acid is associated with cell migration (81). The downregulation of pantothenic acid in the present study may be due to the decreased migration ability, which was consistent with our previous discussion that the invasive ability decreased from downregulating PS [DiMe(11,3)/MonoMe(11,5)], PS [18:3(9Z,12Z,15Z)/22:1(13Z)] and PS [14:1(9Z)/24:0]. Furthermore, the downregulation of pantothenic acid, the key precursor for the biosynthesis of coenzyme A (CoA), can inhibit the synthesis of CoA and, in the Krebs cycle, CoA is a key enzyme for the transformation of α-ketoglutarate to succinyl-CoA (82).
Meanwhile, the KEGG pathway enrichment analysis of the present study revealed that the metabolites associated with the regulation of physiological functions were differentially expressed in SMG compared with NG. When the sphingolipid signaling pathway, sphingolipid metabolism, and arginine and proline metabolism were altered under SMG, their associated physiological functions, such as proliferation, cell cycle, apoptosis and migration, may also have been affected. Therefore, the mechanism of the effect of SMG on these functions in human HGC-27 cells requires further investigation. In addition, KEGG pathway enrichment analysis indicated that the differentially expressed metabolites were primarily associated with the Forkhead box O (FOXO) signaling pathway. FOXO is an important transcription factor that determines cell fate and is associated with a broad range of cellular physiological functions, such as differentiation, apoptosis, cell proliferation, DNA damage and repair and mediating oxidative stress among a wide range of cancers (83,84). The intracellularly initiation pathway of apoptosis induced by the FOXO protein is associated with oxidative stress, which occurs when ROS is released into the cytoplasm (85). The FOXO1 and FOXO3a transcription factors typically actived during oxidative stress causing apoptotic cell injury (86,87). Under other oxidative stresses, FOXO3a translocates from the cytoplasm to the nucleus where it triggers cell death, thus activating the cell apoptosis pathway mediated by the Fas protein (88). In addition, FOXO4 and FOXO3a can mediate cell cycle arrest in mice and cyclin-dependent kinases (CDK) expression through DNA damage (89). The transcription factor, E2F1 forms a complex with FOXO1 and FOXO3a, promoting the transition from G1 to S phase, eventually resulting in cell cycle arrest (90). Notably, knockdown of FOXO3a, in combination with the oncogene c-myc, nuclear factor and p27, results in the initiation of the cell cycle, alteration of malignant mouse cells and activation of carcinogenesis (91).
In summary, the investigation of cancer cells under SMG may provide novel insight into the development of cancer and subsequent changes in physiological function. The present study revealed that human HGC-27 gastric cancer cells have a major effect on lipid metabolism and that SMG may be valuble for cell and cancer research. Ground-based weightlessness simulators like the RCCS, used in the present study, have improved the understanding of how SMG may affect humans. Although the present study is the first to show the changes in metabolic expression of human HGC-27 gastric cancer cells induced by SMG, to the best of our knowledge, the understanding is far from complete. However, the results of the present study may inform future development of novel targets for gastric cancer therapy.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- RCCS
rotary cell culture system
- LC-MS
liquid chromatography-mass spectrometry
- SMG
simulated microgravity
- NG
normal gravity
- PE
phosphatidyl ethanolamine
- PC
phosphatidyl choline
- SM
sphingomyelin
- PS
phosphatidyl serine
- PA
phosphatidic acid
- ECM
extracellular matrix
- Cer
ceramide
- Sph
sphingosine
- PLD2
phospholipase D2
- MMP
matrix metalloproteinase
- FOXO
forkhead box O
Funding
The present study was funded by The Key Program Grants of PLA Planning Technology Research (grant no. SYFD1500128).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
ZC performed the experiments and drafted the initial manuscript. HS, SG, JZ and BL performed the majority of the experiments and statistical analyses. JY, SC, HFY, PS and TZ performed certain experiments and collected the samples. NJ, SG, JZ and HMY interpreted the data. YC designed the study and revised the initial manuscript. All authors read and approved the final manuscript.
Ethical approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Fedotov AA, Akulov SA, Akulova AS. Alterations in cardiovascular system under artificially simulated microgravity: Preliminary study. Conf Proc IEEE Eng Med Biol Soc. 2016;2016:204–206. doi: 10.1109/EMBC.2016.7590675. [DOI] [PubMed] [Google Scholar]
- 2.Atomi Y. Gravitational Effects on human physiology. Subcell Biochem. 2015;72:627–59. doi: 10.1007/978-94-017-9918-8_29. [DOI] [PubMed] [Google Scholar]
- 3.Hughes-Fulford M. Changes in gene expression and signal transduction in microgravity. J Gravit Physiol. 2001;8:P1–4. [PubMed] [Google Scholar]
- 4.Ulbrich C, Wehland M, Pietsch J, Aleshcheva G, Wise P, van Loon J, Magnusson N, Infanger M, Grosse J, Eilles C, et al. The impact of simulated and real microgravity on bone cells and mesenchymal stem cells. Biomed Res Int. 2014;2014:928507. doi: 10.1155/2014/928507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riwaldt S, Bauer J, Wehland M, Slumstrup L, Kopp S, Warnke E, Dittrich A, Magnusson NE, Pietsch J, Corydon TJ, et al. Pathways regulating spheroid formation of human follicular thyroid cancer cells under simulated microgravity conditions: A genetic approach. Int J Mol Sci. 2016;17:528. doi: 10.3390/ijms17040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svejgaard B, Wehland M, Ma X, Kopp S, Sahana J, Warnke E, Aleshcheva G, Hemmersbach R, Hauslage J, Grosse J, et al. Common effects on cancer cells exerted by a randompositioning machine and a 2D clinostat. PLoS One. 2015;10:e0135157. doi: 10.1371/journal.pone.0135157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahana J, Nassef MZ, Wehland M, Kopp S, Krüger M, Corydon TJ, Infanger M, Bauer J, Grimm D. Decreased E-cadherin in MCF-7 human breast cancer cells forming multicellular spheroids exposed to simulated microgravity. Proteomics. 2018;18:e1800015. doi: 10.1002/pmic.201800015. [DOI] [PubMed] [Google Scholar]
- 8.Vidyasekar P, Shyamsunder P, Arun R, Santhakumar R, Kapadia NK, Kumar R, Verma RS. Genome wide expression profiling of cancer cell lines cultured in microgravity reveals significant dysregulation of cell cycle and MicroRNA gene networks. PLoS One. 2015;10:e0135958. doi: 10.1371/journal.pone.0135958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietz C, Infanger M, Romswinkel A, Strube F, Kraus A. Apoptosis induction and alteration of cell adherence in human lung cancer cells under simulated microgravity. Int J Mol Sci. 2019;20(pii):E3601. doi: 10.3390/ijms20143601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YJ, Jeong AJ, Kim M, Lee C, Ye SK, Kim S. Time-averaged simulated microgravity (taSMG) inhibits proliferation of lymphoma cells, L-540 and HDLM-2, using a 3D clinostat. Biomed Eng Online. 2017;16:48. doi: 10.1186/s12938-017-0337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–272. [PubMed] [Google Scholar]
- 12.Nath S, Villadsen J. Oxidative phosphorylation revisited. Biotechnol Bioeng. 2015;112:429–437. doi: 10.1002/bit.25492. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Hu JD, Tang HQ, Zhang Q, Fan J, Hong J, Gu JZ, Chen JL. Prediction of gastric cancer metastasis through urinary metabolomics investigation using GC/MS. World J Gastroenterol. 2011;17:727–734. doi: 10.3748/wjg.v17.i6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JL, Tang HQ, Hu JD, Fan J, Hong J, Gu JZ. Metabolomics of gastric cancer metastasis detected by gas chromatography and mass spectrometry. World J Gastroenterol. 2010;16:5874–5880. doi: 10.3748/wjg.v16.i46.5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis. Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 17.Koukourakis MI, Pitiakoudis M, Giatromanolaki A, Tsarouha A, Polychronidis A, Sivridis E, Simopoulos C. Oxygen and glucose consumption in gastrointestinal adenocarcinomas: Correlation with markers of hypoxia, acidity and anaerobic glycolysis. Cancer Sci. 2006;97:1056–1060. doi: 10.1111/j.1349-7006.2006.00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen PL, Mathupala S, Rempel A, Geschwind JF, Ko YH. A key player in the growth and survival of many cancers and an ideal prospect for therapeutic intervention. Biochim Biophys Acta. 2002;1555:14–20. doi: 10.1016/S0005-2728(02)00248-7. [DOI] [PubMed] [Google Scholar]
- 19.Tech K, Tikunov AP, Farooq H, Morrissy AS, Meidinger J, Fish T, Green SC, Liu H, Li Y, Mungall AJ, et al. Pyruvate kinase inhibits proliferation during postnatal cerebellar neurogenesis and suppresses medulloblastoma formation. Cancer Res. 2017;77:3217–3230. doi: 10.1158/0008-5472.CAN-16-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An J, Zhang Y, He J, Zang Z, Zhou Z, Pei X, Zheng X, Zhang W, Yang H, Li S. Lactate dehydrogenase A promotes the invasion and proliferation of pituitary adenoma. Sci Rep. 2017;7:4734. doi: 10.1038/s41598-017-04366-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Israelsen WJ, Vander Heiden MG. Pyruvate kinase: Function, regulation and role in cancer. Semin Cell Dev Biol. 2015;43:43–51. doi: 10.1016/j.semcdb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Hu L, Chen M, Cao W, Chen H, He T. Pyruvate kinase M2 overexpression and poor prognosis in solid tumors of digestive system: Evidence from 16 cohort studies. Onco Targets Ther. 2016;9:4277–4288. doi: 10.2147/OTT.S106508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jové M, Collado R, Quiles JL, Ramírez-Tortosa MC, Sol J, Ruiz-Sanjuan M, Fernandez M, de la Torre Cabrera C, Ramírez-Tortosa C, Granados-Principal S, et al. A plasma metabolomic signature discloses human breast cancer. Oncotarget. 2017;8:19522–19533. doi: 10.18632/oncotarget.14521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey R, Caflisch L, Lodi A1, Brenner AJ, Tiziani S. Metabolomic signature of brain cancer. Mol Carcinog. 2017;56:2355–2371. doi: 10.1002/mc.22694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navas-Carrillo D, Rodriguez JM, Montoro-García S, Orenes-Piñero E. High-resolution proteomics and metabolomics in thyroid cancer: Deciphering novel biomarkers. Crit Rev Clin Lab Sci. 2017;54:446–457. doi: 10.1080/10408363.2017.1394266. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Yang X, Cui X, Jiang MM, Gui Y, Zhang YN, Luo XD. Adrenomedullin is a key protein mediating rotary cell culture system that induces the effects of simulated microgravity on human breast cancer Cells. Microgravity Sci Technol. 2015;27:417–426. doi: 10.1007/s12217-015-9434-0. [DOI] [Google Scholar]
- 27.Michaletti A, Gioia M, Tarantino U, Zolla L. Effects of microgravity on osteoblast mitochondria: A proteomic and metabolomics profile. Sci Rep. 2017;7:15376. doi: 10.1038/s41598-017-15612-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morabito C, Steimberg N, Mazzoleni G, Guarnieri S, Fanò-Illic G, Mariggiò MA. RCCS bioreactor-based modelled microgravity induces significant changes on in vitro 3D neuroglial cell cultures. Biomed Res Int. 2015;2015:754283. doi: 10.1155/2015/754283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longo N, Frigeni M, Pasquali M. Carnitine transport and fatty acid oxidation. Biochim Biophys Acta. 2016;1863:2422–2435. doi: 10.1016/j.bbamcr.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietsch J, Ma X, Wehland M, Aleshcheva G, Schwarzwälder A, Segerer J, Birlem M, Horn A, Bauer J, Infanger M, Grimm D. Spheroid formation of human thyroid cancer cells in an automated culturing system during the Shenzhou-8 Space mission. Biomaterials. 2013;34:7694–705. doi: 10.1016/j.biomaterials.2013.06.054. [DOI] [PubMed] [Google Scholar]
- 31.Riwaldt S, Pietsch J, Sickmann A, Bauer J, Braun M, Segerer J, Schwarzwälder A, Aleshcheva G, Corydon TJ, Infanger M, Grimm D. Identification of proteins involved in inhibition of spheroid formation under microgravity. Proteomics. 2015;15:2945–2952. doi: 10.1002/pmic.201500067. [DOI] [PubMed] [Google Scholar]
- 32.Arun RP, Sivanesan D, Vidyasekar P, Verma RS. PTEN/FOXO3/AKT pathway regulates cell death and mediates morphogenetic differentiation of Colorectal Cancer Cells under Simulated Microgravity. Sci Rep. 2017;7:5952. doi: 10.1038/s41598-017-06416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopp S, Sahana J, Islam T, Petersen AG, Bauer J, Corydon TJ, Schulz H, Saar K, Huebner N, Slumstrup L, et al. The role of NFκB in spheroid formation of human breast cancer cells cultured on the random positioning machine. Sci Rep. 2018;8:921. doi: 10.1038/s41598-017-18556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen ZY, Guo S, Li BB, Jiang N, Li A, Yan HF, Yang HM, Zhou JL, Li CL, Cui Y. Effect of weightlessness on the 3D structure formation and physiologic function of human cancer cells. Biomed Res Int. 2019;2019:4894083. doi: 10.1155/2019/4894083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marín de Mas I, Aguilar E, Jayaraman A, Polat IH, Martín-Bernabé A, Bharat R, Foguet C, Milà E, Papp B, Centelles JJ, Cascante M. Cancer cell metabolism as new targets for novel designed therapies. Future Med Chem. 2014;6:1791–1810. doi: 10.4155/fmc.14.119. [DOI] [PubMed] [Google Scholar]
- 36.Zhu M, Jin XW, Wu BY, Nie JL, Li YH. Effects of simulated weightlessness on cellular morphology and biological characteristics of cell lines SGC-7901 and HFE-145. Genet Mol Res. 2014;13:6060–6069. doi: 10.4238/2014.August.7.20. [DOI] [PubMed] [Google Scholar]
- 37.Espinosa-Jeffrey A, Nguyen K, Kumar S, Toshimasa O, Hirose R, Reue K, Vergnes L, Kinchen J, Vellis J. Simulated microgravity enhances oligodendrocyte mitochondrial function and lipid metabolism. J Neurosci Res. 2016;94:1434–1450. doi: 10.1002/jnr.23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akagi T, Kimoto T. Human cell line (HGC-27) derived from the metastatic lymph node of gastric cancer. Acta Med Okayama. 1976;30:215–219. [PubMed] [Google Scholar]
- 39.Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015;16:30. doi: 10.1007/s11864-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zalba S, Ten Hagen TL. Cell membrane modulation as adjuvant in cancer therapy. Cancer Treat Rev. 2017;52:48–57. doi: 10.1016/j.ctrv.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HY, Lee KM, Kim SH, Kwon YJ, Chun YJ, Choi HK. Comparative metabolic and lipidomic profiling of human breast cancer cells with different metastatic potentials. Oncotarget. 2016;7:67111–67128. doi: 10.18632/oncotarget.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elvas F, Stroobants S, Wyffels L. Phosphatidylethanolamine targeting for cell death imaging in early treatment response evaluation and disease diagnosis. Apoptosis. 2017;22:971–987. doi: 10.1007/s10495-017-1384-0. [DOI] [PubMed] [Google Scholar]
- 43.Iorio E, Ricci A, Bagnoli M, Pisanu ME, Castellano G, Di Vito M, Venturini E, Glunde K, Bhujwalla ZM, Mezzanzanica D, et al. Activation of phosphatidylcholine cycle enzymes in human epithelial ovarian cancer cells. Cancer Res. 2010;70:2126–2135. doi: 10.1158/0008-5472.CAN-09-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grimm D, Bauer J, Kossmehl P, Shakibaei M, Schöberger J, Pickenhahn H, Schulze-Tanzil G, Vetter R, Eilles C, Paul M, Cogoli A. Simulated microgravity alters differentiation and increases apoptosis in human follicular thyroid carcinoma cells. FASEB J. 2002;16:604–606. doi: 10.1096/fj.01-0673fje. [DOI] [PubMed] [Google Scholar]
- 45.Kossmehl P, Shakibaei M, Cogoli A, Infanger M, Curcio F, Schönberger J, Eilles C, Bauer J, Pickenhahn H, Schulze-Tanzil G, et al. Weightlessness induced apoptosis in normal thyroid cells and papillary thyroid carcinoma cells via extrinsic and intrinsic pathways. Endocrinology. 2003;144:4172–4179. doi: 10.1210/en.2002-0171. [DOI] [PubMed] [Google Scholar]
- 46.Masiello MG, Cucina A, Proietti S, Palombo A, Coluccia P, D'Anselmi F, Dinicola S, Pasqualato A, Morini V, Bizzarri M. Phenotypic switch induced by simulated microgravity on MDA-MB-231 breast cancer cells. Biomed Res Int. 2014;2014:652434. doi: 10.1155/2014/652434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao J, Ma H, Wu L, Cao L, Yang Q, Dong H, Wang Z, Ma J, Li Z. The influence of simulated microgravity on proliferation and apoptosis in U251 glioma cells. In Vitro Cell Dev Biol Anim. 2017;53:744–751. doi: 10.1007/s11626-017-0178-6. [DOI] [PubMed] [Google Scholar]
- 48.Paulick MG, Bertozzi CR. The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for protein. Biochemistry. 2008;47:6991–7000. doi: 10.1021/bi8006324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferguson MA, Homans SW, Dwek RA, Rademacher TW. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science. 1988;239:753–759. doi: 10.1126/science.3340856. [DOI] [PubMed] [Google Scholar]
- 50.Ferguson MA. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J Cell Sci. 1999;112:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- 51.Tsai YH, Liu X, Seeberger PH. Chemical biology of glycosylphosphatidylinositol anchors. Angew Chem Int Ed Engl. 2012;51:11438–11456. doi: 10.1002/anie.201203912. [DOI] [PubMed] [Google Scholar]
- 52.Chang D, Xu H, Guo Y, Jiang X, Liu Y, Li K, Pan C, Yuan M, Wang J, Li T, Liu C. Simulated microgravity alters the metastatic potential of a human lung adenocarcinoma cell line. In Vitro Cell Dev Biol Anim. 2019;49:170–177. doi: 10.1007/s11626-013-9581-9. [DOI] [PubMed] [Google Scholar]
- 53.Qian A, Zhang W, Xie L, Weng Y, Yang P, Wang Z, Hu L, Xu HY, Tian ZC, Shang P. Simulated weightlessness alters biological characteristics of human breast cancer cell line MCF-7. Acta Astronautica. 2008;63:947–958. doi: 10.1016/j.actaastro.2008.01.024. [DOI] [Google Scholar]
- 54.Peng W, Tan S, Xu Y, Wang L, Qiu D, Cheng C, Lin Y, Liu C, Li Z, Li Y, et al. LC-MS/MS metabolome analysis detects the changes in the lipid metabolic profiles of dMMR and pMMR cells. Oncol Rep. 2018;40:1026–1034. doi: 10.3892/or.2018.6510. [DOI] [PubMed] [Google Scholar]
- 55.Toshima K, Nagafuku M, Okazaki T, Kobayashi T, Inokuchi JI. Plasma membrane sphingomyelin modulates thymocyte development by inhibiting TCR-induced apoptosis. Int Immunol. 2019;31:211–223. doi: 10.1093/intimm/dxy082. [DOI] [PubMed] [Google Scholar]
- 56.Hogan PG. Sphingomyelin, ORAI1 channels, and cellular Ca2+ signaling. J Gen Physiol. 2015;146:195–200. doi: 10.1085/jgp.201511479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matanes F, Twal WO, Hammad SM. Sphingolipids as biomarkers of disease. Adv Exp Med Biol. 2019;1159:109–138. doi: 10.1007/978-3-030-21162-2_7. [DOI] [PubMed] [Google Scholar]
- 58.Kohama T, Olivera A, Edsall L, Nagiec MM, Dickson R, Spiegel S. Molecular cloning and functional characterization of murine sphingosine kinase. J Biol Chem. 1998;273:23722–23728. doi: 10.1074/jbc.273.37.23722. [DOI] [PubMed] [Google Scholar]
- 59.Li J, Gray BD, Pak KY, Ng CK. Targeting phosphatidylethanolamine and phosphatidylserine for imaging apoptosis in cancer. Nucl Med Biol. 2019;78:23–30. doi: 10.1016/j.nucmedbio.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Andrieu-Abadie N, Levade T. Sphingomyelin hydrolysis during apoptosis. Biochim Biophys Acta. 2002;1585:126–134. doi: 10.1016/S1388-1981(02)00332-3. [DOI] [PubMed] [Google Scholar]
- 61.van der Hoeven D, Cho KJ, Zhou Y, Ma X, Chen W, Naji A, Montufar-Solis D, Zuo Y, Kovar SE, Levental KR, et al. Sphingomyelin metabolism is a regulator of K-Ras function. Mol Cell Biol. 2018;38(pii):e00373–17. doi: 10.1128/MCB.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cacev T, Radosević S, Spaventi R, Pavelić K, Kapitanović S. NF1 gene loss of heterozygosity and expression analysis in sporadic colon cancer. Gut. 2005;54:1129–1135. doi: 10.1136/gut.2004.053348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17:31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 65.Khoukaz T. Administration of anti-EGFR therapy: A practical review. Semin Oncol Nurs. 2006;22:20–27. doi: 10.1016/j.soncn.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 66.Baek MO, Ahn CB, Cho HJ, Choi JY, Son KH, Yoon MS. Simulated microgravity inhibits C2C12 myogenesis via phospholipase D2-induced Akt/FOXO1 regulation. Sci Rep. 2019;9:14910. doi: 10.1038/s41598-019-51410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Z, Zhang F, He J, Wu P, Tay LWR, Cai M, Nian W, Weng Y, Qin L, Chang JT, et al. Binding of PLD2-generated phosphatidic acid to KIF5B promotes MT1-MMP surface trafficking and lung metastasis of mouse breast cancer cells. Dev Cell. 2017;43:186–197. doi: 10.1016/j.devcel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeiller C, Mebarek S, Jaafar R, Pirola L, Lagarde M, Prigent AF, Némoz G. Phospholipase D2 regulates endothelial permeability through cytoskeleton reorganization and occludin downregulation. Biochim Biophys Acta. 2009;1793:1236–1249. doi: 10.1016/j.bbamcr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 69.Ngo Thai Bich V, Hongu T, Miura Y, Katagiri N, Ohbayashi N, Yamashita-Kanemaru Y, Shibuya A, Funakoshi Y, Kanaho Y. Physiological function of phospholipase D2 in anti-tumor immunity: Regulation of CD8+ T lymphocyte proliferation. Sci Rep. 2018;8:6283. doi: 10.1038/s41598-018-24512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kandori S, Kojima T, Matsuoka T, Yoshino T, Sugiyama A, Nakamura E, Shimazui T, Funakoshi Y, Kanaho Y, Nishiyama H. Phospholipase D2 promotes disease progression of renal cell carcinoma through the induction of angiogenin. Cancer Sci. 2018;109:1865–1875. doi: 10.1111/cas.13609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo L, Cui C, Zhang K, Wang J, Wang Y, Lu Y, Chen K, Yuan J, Xiao G, Tang B, et al. Kindlin-2 links mechano-environment to proline synthesis and tumor growth. Nat Commun. 2019;10:845. doi: 10.1038/s41467-019-08772-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Phang JM, Donald SP, Pandhare J, Liu Y. The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids. 2008;35:681–690. doi: 10.1007/s00726-008-0063-4. [DOI] [PubMed] [Google Scholar]
- 73.Liu W, Wang X, Liu Z, Wang Y, Yin B, Yu P, Duan X, Liao Z, Chen Y, Liu C, et al. SGK1 inhibition induces autophagy-dependent apoptosis via the mTOR-Foxo3a pathway. Br J Cancer. 2017;117:1139–1153. doi: 10.1038/bjc.2017.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wallimann T, Tokarska-Schlattner M, Schlattner U. The creatine kinase system and pleiotropic effects of creatine. Amino Acids. 2011;40:1271–1296. doi: 10.1007/s00726-011-0877-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Campos-Ferraz PL, Gualano B, das Neves W, Andrade IT, Hangai I, Pereira RT, Bezerra RN, Deminice R, Seelaender M, Lancha AH. Exploratory studies of the potential anti-cancer effects of creatine. Amino Acids. 2016;48:1993–2001. doi: 10.1007/s00726-016-2180-9. [DOI] [PubMed] [Google Scholar]
- 76.Martin KJ, Chen SF, Clark GM, Degen D, Wajima M, Von Hoff DD, Kaddurah-Daouk R. Evaluation of creatine analogs as a new class of anticancer agents using freshly explanted human tumor cells. J Natl Cancer Inst. 1994;86:608–613. doi: 10.1093/jnci/86.8.608. [DOI] [PubMed] [Google Scholar]
- 77.Miller EE, Evans AE, Cohn M. Inhibition of rate of tumor growth by creatine and cyclocreatine. Proc Natl Acad Sci USA. 1993;90:3304–3308. doi: 10.1073/pnas.90.8.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoppeler H, Fluck M. Plasticity of skeletal muscle mitochondria: Structure and function. Med Sci Sports Exerc. 2003;35:95–104. doi: 10.1097/00005768-200301000-00016. [DOI] [PubMed] [Google Scholar]
- 79.Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, Hwang DH, Newman JW, Garvey WT. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr. 2009;139:1073–1081. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen WW, Freinkman E, Wang T, Birsoy K, Sabatini DM. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell. 2016;166:1324–1337. doi: 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hutschenreuther A, Birkenmeier G, Bigl M, Krohn K, Birkemeyer C. Glycerophosphoglycerol, Beta-alanine, and pantothenic acid as metabolic companions of glycolytic activity and cell migration in breast cancer cell lines. Metabolites. 2013;3:1084–1101. doi: 10.3390/metabo3041084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leonardi R, Jackowski S. Biosynthesis of pantothenic acid and coenzyme A. EcoSal Plus. 2007;2 doi: 10.1128/ecosalplus.3.6.3.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vurusaner B, Poli G, Basaga H. Tumor suppressor genes and ROS: Complex networks of interactions. Free Radic Biol Med. 2012;52:7–18. doi: 10.1016/j.freeradbiomed.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 84.Gomes AR, Brosens JJ, Lam EW. Resist or die: FOXO transcription factors determine the cellular response to chemotherapy. Cell Cycle. 2008;7:3133–3136. doi: 10.4161/cc.7.20.6920. [DOI] [PubMed] [Google Scholar]
- 85.Hou YQ, Yao Y, Bao YL, Song ZB, Yang C, Gao XL, Zhang WJ, Sun LG, Yu CL, Huang YX, et al. Juglanthraquinone C induces intracellular ROS increase and apoptosis by activating the Akt/Foxo signal pathway in HCC cells. Oxid Med Cell Longev. 2016;2016:4941623. doi: 10.1155/2016/4941623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maiese K, Chong ZZ, Hou J, Shang YC. Erythropoietin and oxidative stress. Curr Neurovasc Res. 2008;5:125–142. doi: 10.2174/156720208784310231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakamura T, Sakamoto K. Forkhead transcription factor FOXO subfamily is essential for reactive oxygen species-induced apoptosis. Mol Cell Endocrinol. 2008;281:47–55. doi: 10.1016/j.mce.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 88.Barthélémy C, Henderson CE, Pettmann B. Foxo3a induces motoneuron death through the Fas pathway in cooperation with JNK. BMC Neurosci. 2004;5(48) doi: 10.1186/1471-2202-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maiese K, Chong ZZ, Li F, Shang YC. Erythropoietin: Elucidating new cellular targets that broaden therapeutic strategies. Prog Neurobiol. 2008;85:194–213. doi: 10.1016/j.pneurobio.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nowak K, Killmer K, Gessner C, Lutz W. E2F-1 regulates expression of FOXO1 and FOXO3a. Biochim Biophys Acta. 2007;1769:244–252. doi: 10.1016/j.bbaexp.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 91.Liu Y, Ao X, Ding W, Ponnusamy M, Wu W, Hao X, Yu W, Wang Y, Li P, Wang J. Critical role of FOXO3a in carcinogenesis. Mol Cancer. 2018;17:104. doi: 10.1186/s12943-018-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.