Abstract
Lung cancer is the most common type of cancer and the leading cause of cancer-associated death worldwide. Malignant pleural effusion (MPE), which is observed in ~50% of advanced non-small cell lung cancer (NSCLC) cases, and most frequently in lung adenocarcinoma, is a common complication of stage III–IV NSCLC, and it can be used to predict a poor prognosis. In the present study, multiple oncogene mutations were detected, including 17 genes closely associated with initiation of advanced lung cancer, in 108 MPE samples using next generation sequencing (NGS). The NGS data of the present study had broader coverage, deeper sequencing depth and higher capture efficiency compared with NGS findings of previous studies on MPE. In the present study, using NGS, it was demonstrated that 93 patients (86%) harbored EGFR mutations and 62 patients possessed mutations in EGFR exons 18–21, which are targets of available treatment agents. EGFR L858R and exon 19 indel mutations were the most frequently observed alterations, with frequencies of 31 and 25%, respectively. In 1 patient, an EGFR amplification was identified and 6 patients possessed a T790M mutation. ALK + EML4 gene fusions were identified in 6 patients, a ROS1 + CD74 gene fusion was detected in 1 patient and 10 patients possessed a BIM (also known as BCL2L11) 2,903-bp intron deletion. In 4 patients, significant KRAS mutations (G12D, G12S, G13C and A146T) were observed, which are associated with resistance to afatinib, icotinib, erlotinib and gefitinib. There were 83 patients with ERBB2 mutations, but only two of these mutations were targets of available treatments. The results of the present study indicate that MPE is a reliable specimen for NGS based detection of somatic mutations.
Keywords: malignant pleural effusion, next generation sequencing, non-small cell lung cancer, mutation
Introduction
Cancer is the first or second leading cause of death in patients aged <70 in 91 of 172 countries and is expected to be the leading cause of cancer-associated death worldwide (1). In China, there has been an increase in the incidence and mortality rates of cancer, and cancer is the leading cause of death, constituting a major public health crisis; ~4,292,000 new cancer cases and ~2,814,000 cancer-associated deaths occurred in China in 2015, with lung cancer being the most common and the leading cause of cancer-associated deaths (2). Non-small cell lung cancer (NSCLC) accounts for 80–85% of lung cancer cases (3), and the majority of patients with NSCLC are diagnosed at an advanced stage with <30% of patients undergoing surgical resection (4).
Large-scale sequencing studies have revealed the complex genomic landscape of NSCLC (5–8) and the genomic differences between lung adenocarcinomas and lung squamous-cell carcinomas (9). In NSCLCs, mutant oncogenes drive proliferation and tumor maintenance, particularly in adenocarcinomas (5). Tumors may be heavily dependent on these driver mutations for survival rather than multiple downstream genetic and epigenetic abnormalities (10). Mutations in EGFR and gene fusion of EML4-anaplastic lymphoma kinase (ALK) are the best studied and actionable genes in lung adenocarcinomas and have been observed in 15–60 and 3–5% of lung cancer cases, respectively (11,12). Other reported oncogenic driver mutations include KRAS, ROS1, BRAF, ERBB2 and MET (8,12). The presence of EGFR mutations and ALK gene fusions in lung cancer are associated with longer median survival times compared with the wild type status (>24 months vs. 5–9 months) (13).
Obtaining a suitable specimen is the first step for accurate gene alteration testing, allowing for histological and molecular analysis, and preventing false-negative results and test failures. The primary sample types currently used for diagnosis are tissues and cytology samples, with surgical resections being the first used for clinical molecular diagnosis (14). However, 50–70% of patients with NSCLCs present with advanced stage cancer at diagnosis and are unresectable (15). It is difficult to obtain an adequate amount of tumor tissue from patients with advanced NSCLC (7). Liquid biopsy is a noninvasive method that overcomes the heterogeneity of tumor tissues, and repeat biopsies and can be obtained throughout the progression of NSCLC, and this may potentially improve early detection and post-treatment monitoring of patients with lung cancer (16,17). Liquid biopsy may be one of the most influential techniques in the field of detecting and monitoring cancer over the next decade (18).
Malignant pleural effusion (MPE), which is observed in ~50% of advanced NSCLC cases and most frequently in lung adenocarcinoma (19,20), is a common manifestation of stage IV NSCLC. For a number of patients, it may be the only specimen available for diagnostic and molecular testing (21,22). With a high tumor content, most MPE samples contain numerous tumor cells, which are sufficient for pathological diagnosis and for determining driver gene status (23–25). MPE samples can be obtained less invasively and repeatedly over time compared with samples from the primary lesion. Furthermore, previous studies have suggested that pleural effusions are a suitable source for enrichment of putative cancer stem cells (26,27).
In 2014, Akamatsu et al (23) examined genetic alterations in 84 patients with pleural effusions using pyro-sequencing and/or capillary electrophoresis, and reverse-transcription polymerase chain reaction (RT-PCR) and their results showed that 24 patients (28.5%) possessed EGFR mutations, including EGFR G719C/S, G719A, exon 19 deletion, T790M, exon 20 insertion, L858R and L861Q mutations, and 4 patients (4.7%) possessed fusion of ALK-EML4. Other studies have used direct sequencing methods, reporting an EGFR mutation rate between 25–68% (25,28–34). The use of high resolution melting (HRM) analysis has demonstrated a 50% EGFR mutation rate (35) in NSCLC. Peptide nucleic acid (PNA) clamping methods have reported an EGFR mutation rate between 39–72% (36–38) in NSCLC. Buttitta et al (39) conducted next generation pyrosequencing in bronchoalveolar lavage, and pleural fluid patients, demonstrating an EGFR mutation rate of 80.5%.
NGS exceeds the limitations of traditional Sanger sequencing, allowing a huge number of independent sequencing reactions to run simultaneously, which has laid the foundation for development of targeted therapies (40). This detection method has the power to identify new single nucleotide variations (SNVs) or indel mutations, and chromosomal rearrangements (41–44). Liu et al evaluated the molecular profiling of lung adenocarcinoma using MPE specimens utilizing NGS, illustrating 22 cases of EGFR mutations within 30 patients (73%), including L858R, G719S, exon 20 insertion and exon 19 deletion, with an average sequencing depth on target of 359X (45).
Although MPE is a potential candidate for molecular testing, its use in multiplexed molecular profiling with NGS has not been fully investigated. In the present study, multiple oncogene mutations were detected, including 17 genes closely associated with advanced lung cancer induction in 108 MPE samples using deep sequencing methods.
Materials and methods
Patient and sample characteristics
Between December 2017 and July 2018, MPE samples from 108 patients (51 males, 56 females, and 1 patient with unknown gender; median age, 68 years; age range, 30–89 years) with pathologically confirmed lung cancer were collected. Written informed consent was obtained from all participants, and the present study was approved by the Institutional Review Board of Affiliated Changzhou No. 2 People's Hospital (Changzhou, China). Pleural effusion samples, 20–50 ml, were obtained at the time of diagnosis. All cell blocks of MPE samples contained tumor cells. Clinical characteristics were obtained from medical records.
Hematoxylin and eosin (HE) staining of MPE samples
The pleural fluid sample was centrifuged for 10 min in a 15-ml centrifuge tube at 268 × g. The supernatant was discarded and 50% ethanol was added to 10 ml, and centrifuged for 10 min at 268 × g. After discarding the supernatant, 10% buffer neutral formaldehyde solution was added to 10 ml, followed by centrifugation for 10 min at 268 × g. The supernatant was discarded and the cell precipitates were removed and wrapped in wrapping paper. Subsequently, 10% buffer neutral formaldehyde solution was added for internal fixation for 10 min, then placed into the automatic dehydrator to dehydrate and transferred into a xylene solution for transparency (15 min at room temperature). The samples were subsequently waxed, embedded and cut into slices of 2–3-µm thickness. HE staining was subsequently performed. Samples were incubated in xylene for 5–10 min once, followed by: 3–5 min in 95% ethanol twice; and 1–2 min in 80% ethanol. Following rinsing, the samples were stained with hematoxylin for 10 min, rinsed and differentiated using 1% ethanol hydrochloride for 2–5 sec. After rinsing with running water for 10–15 min, the samples were stained with 0.5% eosin solution for 3–5 min. Finally, the slides were incubated with 80% ethanol for 1–2 sec, 95% ethanol for 1–2 sec, 100% ethanol for 2–5 sec (repeated twice), xylene for 1–2 min (repeated once). Subsequently, the tissue sections were dried, dropped neutral resins and covered with a cover slide. All the above steps were at room temperature.
DNA extraction and library construction
The tumor DNA was extracted using a human tissue DNA extraction kit (YunYing Medical Technology Co. Ltd.) according to the manufacturer's protocols. DNA was eluted in the elution buffer, and concentration and purity were assessed using a NanoDrop spectrophotometer. DNA was stored at −20°C until use. Library preparation was constructed using the VAHTS Universal DNA Library Prep kit for Illumina® sequencing (Illumina, Inc.). Target enrichment was performed using optimized probes (YunYing Medical Technology Co. Ltd.) that targeted the exons of 17 lung cancer-associated genes and specific introns. Sequencing was performed on an Illumina® NextSeq500 platform (Illumina, Inc.) according to the manufacturer's protocols.
NGS analysis and statistical analysis
The Fastqc software (version 0.11.2; http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and customized python scripts were used for screening FASTQ files, and the adaptor sequences and sequences with Q<30 were removed. Clean reads were mapped to the reference human genome GRCh37-hg19 using Burrows-Wheeler Aligner version 0.7.7. (https://github.com/lh3/bwa). Bam files were then realigned and recalled using GenomeAnalysisTK version 3.5 (https://software.broadinstitute.org/gatk/), which was also used to detect mutations. Somatic mutations with ≥2% mutant allele frequency, and with at least 20 supporting reads were detected using VarScan version 2.3.2 (http://varscan.sourceforge.net/). Pindel version 0.2.5b8 (https://www.sanger.ac.uk/science/tools/pindel) was used for indel detection using default parameters. Structure variation was identified using FACTERA version 1.4.4 with default parameters (https://factera.stanford.edu/). Copy number variations were detected using ONCOCNV version 6.4 with default parameters (http://boevalab.inf.ethz.ch/ONCOCNV/). The amplification refractory mutation system (ARMS)-PCR was performed to detect all mutations with an allele frequency between 1–10%. All statistical analyses were performed in R (version 1.8.1) (46) and RStudio (version 0.99.903) (47).
Results
Patient characteristics
Pleural effusion samples from 108 patients with advanced NSCLC were obtained between December 2017 and July 2018, and patient characteristics are shown in Table I. A total of 51 patients (47.2%) were male and 56 (51.8%) were female, and there was one patient whose gender was unknown. The median age was 68 years (range, 30–89 years), with 11 patients between 30 and 50 years, 51 patients between 50 and 70 years, 44 patients between 70 and 90 years, and 2 patients whose ages were unknown. Adenocarcinoma was the most commonly observed histology, in 60.1% of cases. A total of six representative hematoxylin and eosin stains of MPE samples with the respective controls are shown in the supplemental materials (Fig. S1).
Table I.
Clinical characteristics of patients in this study.
| Characteristic | Number of cases | % |
|---|---|---|
| Sex | ||
| Male | 51 | 47.2 |
| Female | 56 | 51.8 |
| Unknown | 1 | 1 |
| Age, years | ||
| Median (range) | 68 (30–89) | |
| ≥30-<50 | 11 | 10.1 |
| ≥50-<70 | 51 | 47.2 |
| ≥70-<90 | 44 | 40.7 |
| Unknown | 2 | 2 |
| Histology | ||
| Adenocarcinoma | 65 | 60.1 |
| Others unknown | 43 | 39.9 |
Detection of genetic abnormalities
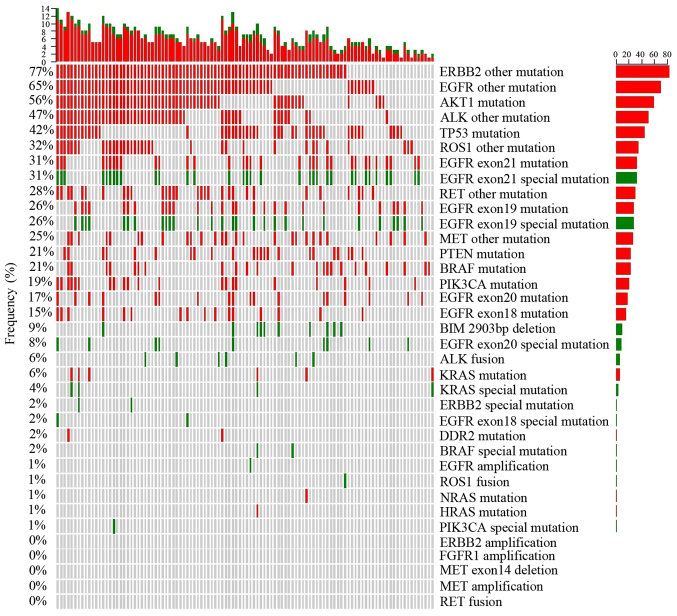
After NGS data processing, the sequencing data of all 108 patient (Table SI) revealed that the average coverage of target regions approached 100%, the average fraction of effective bases on target was ~65%, and the average sequencing depth on target was nearly 3,492X, 10 times greater than in a previous study (45). In the present study, gene mutations were classified into two groups, one group of mutations that can be utilized as targeted therapies and another group of unknown significance at present. Observed genetic abnormalities in the 108 MPE specimens are shown in Fig. 1. Furthermore, ARMS-PCR was performed as described in our previous study (48), was also used to detect all mutations with an allele frequency between 1–10% in MPE samples from the 108 patients involved, to further verify the accuracy of NGS (Figs. S2-S10).
Figure 1.
Plot showing mutations related to certain agents and variants of unknown significance frequency with variant distribution in samples. The frequency of variant-positive samples per gene is shown on the left, and the number of different variants per sample is shown on the upper side. The right side shows the total number of variants per gene. Green indicates variants related to certain therapeutic agents, red indicates variants of unknown significance and gray indicates no variant was detected.
ERBB2 (HER2), a known proto-oncogene located on the long arm of human chromosome 17, is a member of the human EGFR (HER/EGFR/ERBB) family (49). Among the 108 MPE samples, there were only 2 cases (females aged 89 and 62 years old) with insertion of AYVM in M774_A775 in the ERBB2 amino acid sequence, which is sensitive to trastuzumab (Herceptin), ado-trastuzumab emtansine (Kadcyla), and afatinib dimaleate (Gilotrif) treatment. Special ERBB2 mutations are noted in Fig. 1 in green, which indicate that the mutation is associated with a therapeutic agent. In addition to these two cases of ERBB2 alterations, 83 patients (76.8%) were identified as harboring other ERBB2 mutations of unknown present significance, annotated as ‘ERBB2 other mutations’ in Fig. 1. Collectively, these unknown mutations were observed more frequently compared with all other mutations.
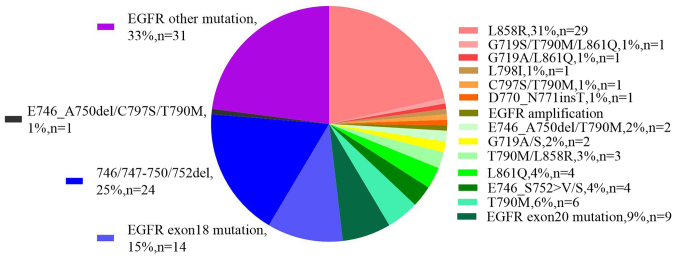
EGFR mutations were first identified as NSCLC driver oncogenes by three independent groups in 2004 (50–52). In the present study, analysis of 108 patients with MPE, demonstrated that 93 patients (86%) harbored EGFR mutations, the breakdown by type of which is shown in Fig. 2 (or Table SII). In particularly, 16 patients harbored mutations in EGFR exon 18, among which, only two women possessed the G719A/G719S alterations. A total of 28 patients (30%) were found to harbor exon 19 alterations, 24 of which were deletion mutations, primarily in the KELREATS sequence, whereas only one deletion was observed in the TSPKANKE sequence. A total of 3 patients exhibited a E746_S752>V mutation and 1 patient had a L747_P753>S alteration on exon 19 of EGFR. A total of 18 abnormalities on exon 20 of EGFR were identified, among which, 1 patient had a L798I mutation, one had a T790M/C797S double alteration (cis relationship), six had a T790M mutation and only one had an insertion of threonine between D770 and N771. A total of 33 patients (35.5%) harbored mutations on exon 21, including L858R (n=29) and L861Q (n=4). There was only one case of EGFR amplification that simultaneously harbored a L858R mutation. Altogether, 62 patients (66.7%) had EGFR mutations that are associated with sensitivity to certain agents, such as icotinib, gefitinib, erlotinib, afatinib and osimertinib. In addition to these mutations, 31 patients (33.3%) possessed EGFR alterations outside of exons 18–21 which do not currently have an available targeted therapy (Fig. 2). It was also observed that 15 patients possessed a wild type EGFR gene among the 108 MPE samples.
Figure 2.
Molecular profiling of EGFR mutation distribution within 93 cases of EGFR positive patients.
AKT1 is a serine-threonine protein kinase that is critical for transmitting growth-promoting signals, most likely via the IGF1 receptor (53). An E17K mutation of AKT1 is associated with AKT1 sensitivity to the inhibitor, uprosertib (53). In the present study the E17K mutation was not observed; however, other alterations of the AKT1 gene were observed in 60 patients (55.6%) (Fig. 1). The TP53 gene is the most frequently mutated gene in cancer, indicating that TP53 serves a crucial tumor suppressor role (54). A total of 45 patients (41.7%) harbored TP53 gene mutations, including SNVs and frameshifts (Fig. 1). BIM is a pro-apoptotic member of the B-cell CLL/lymphoma 2 (BCL2) family of proteins. A frequently observed intronic deletion (2,903 bp) polymorphism in BIM is sufficient to confer intrinsic tyrosine kinase inhibitor (TKI) resistance (55). This intronic deletion was identified in 10 patients (9%) (Fig. 1). BRAF, a human gene which encodes the B-Raf protein, is a member of the Raf kinase family of growth signal transduction protein kinases (56). In the present study, 23 patients (21.3%) harbored BRAF mutations, including two cases of the N581I mutations, which are sensitive to Solafini (57). In 6 patients, KRAS mutations were observed, four of whom had different mutations; G12D, G12S, G13C and A146T. In addition, one mutation occurred in HRAS, and one in NRAS, but these two mutations belonged to variants of unknown significance (Fig. 1). PIK3CA participates in a complex interaction within the tumor microenvironment (58). The present study demonstrated that 21 patients (19.4%) harbored PIK3CA mutations, but only one patient harbored a E545K mutation, which is sensitive to Sirolimus and Ivimos (59) (Fig. 1). PTEN acts as a tumor suppressor gene through its phosphatase activity (60), and in the present study, 23 patients (21.3%) exhibited PTEN mutations (Fig. 1). DDR2 serves a key role in the communication of cells with their microenvironment (61). In the present study, 2 patients harbored DDR2 mutations (Fig. 1). No amplifications of the FGFR1 gene were observed, but 20 patients (18.5%) possessed SNVs in this gene (Fig. 1). There were no MET amplifications or exon 14 skips among the samples, but 27 patients (25%) had SNVs of unknown significance in the MET gene (Fig. 1). Additionally, no RET fusions were detected; however, there were 30 patients (27.8%) with SNVs of unknown significance in the RET gene (Fig. 1).
There was one case of an ROS1-CD74 fusion, where ROS1 intron 33 and CD74 intron 6 were fused in a patient of 34 years old. This fusion resulted in the formation of a new gene, and the corresponding sequences are listed in Table SIII. Meanwhile, 35 patients (32.4%) exhibited SNVs of unknown significance in ROS1 (Fig. 1).
Overall, 6 cases (5.6%) of ALK-EML4 fusions, in 1 man and 5 women, were observed. Table SIII shows the corresponding sequences, in particularly, a man (77 years old) demonstrated gene fusion between ALK exon 20, and EML4 intron 13. Meanwhile, 5 women possessed gene fusion between ALK intron 19, and different EML4 introns (three cases of intron 6, one case of intron 2, and one case of intron 20). A total of 51 patients (47.2%) possessed SNVs in the ALK gene, which have no known significance at present.
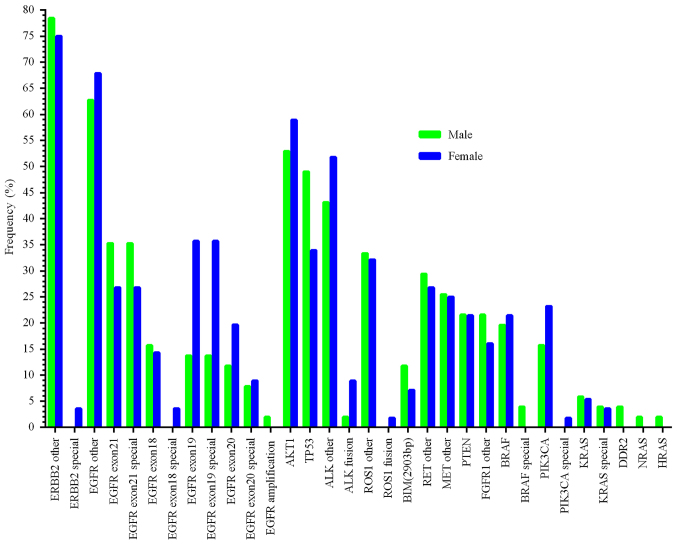
Profiling of mutations between males and females
Among MPE samples from 108 patients, there was only one person without sex or age information, and this patient had mutations of E746_A750 deletion, and T790M in the EGFR gene, as well as TP53 abnormalities (Table SII). Multiplexed molecular profiling in men (n=51), and women (n=56) is shown in Fig. 3. The mutation rate in females was higher than in males in the ERBB2 special mutations, EGFR exon 18 special mutations, EGFR exon 19 special mutations, ALK fusions, ROS1 fusions, and PIK3CA special mutations. The abnormalities in males which were higher compared with females were EGFR exon 21 special mutations, BIM, TP53 mutations, and BRAF special mutations. The rate of EGFR exon 20 special mutations and KRAS special mutations were ~6%, similar between men and women (Fig. 3). In particular, there were 36 female patients (64.3%) harboring EGFR mutations with known significance, higher than in males (50.9%).
Figure 3.
Multiplexed molecular profiling between men (n=51) and women (n=56). ‘Special’ indicates mutations related to certain therapeutic agents.
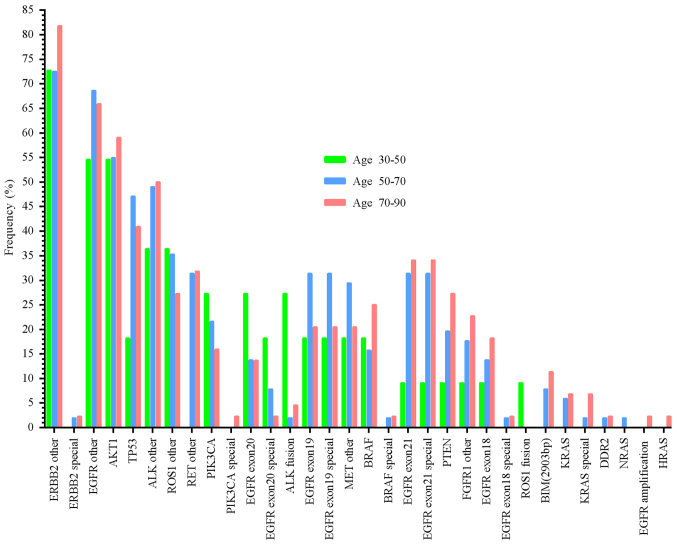
Mutation distributions across different ages
As mentioned above, one patient without age or sex information harbored an E746_A750 deletion, and T790M mutations in the EGFR gene and TP53 abnormalities. Another male patient's age information was not available. The mutation distributions in different ages are presented in Fig. 4. These results demonstrate that some mutation rates increased with age, including ERBB2 special mutations, PIK3CA special mutations, BRAF special mutations, EGFR exon 21 mutations, EGFR exon 21 special mutations, PTEN mutations, FGFR1 other mutations, EGFR exon 18 mutations, EGFR exon 18 special mutations, BIM mutations, KRAS mutations, and KRAS special mutations. In contrast, other mutation frequencies decreased with age, for example the PIK3CA mutations, EGFR exon 20 special mutations and ROS1 fusions. No significant difference in the other mutation rates and age were observed (Fig. 4).
Figure 4.
Mutation distribution across different age groups. ‘Special’ indicates mutations related to certain therapeutic agents.
Discussion
MPE, a common cause of symptoms, hospitalizations and morbidities in patients with disseminated lung cancer, is a cause of the lethal outcomes in various types of cancer originating from the lung (20,62), breast (20), and metastasizes to the pleural cavity (63). Increased tumor burden within the pleural space is associated with reduced overall survival in patients (64–66). However, the reasons why certain patients with pleural tumors develop MPE, whereas others do not remain unknown. In 2010, Stathopoulos et al (67) used two different models to demonstrate that host-derived IL-5 promotes experimental MPE and may be involved in the pathogenesis of human MPE. Stathopoulos et al found that nuclear factor-κB affected tumor progression in a mouse model of MPE by injecting Lewis lung cancer cells directly into the pleural space of syngeneic C57B/6 mice (68).
A number of studies have evaluated the formation of MPE (69–75), and discussed the tumor and host-derived factors likely involved in MPE development, including osteopontin (74), CCL2 (76), VEGF (71), TNF (77), angiopoietins 1/2 (78), interleukin-5 (67), interleukin-6 (79). Wu et al demonstrated for the first time that Toll-like receptor 2 signaling promotes the development of MPE and accelerates the death of mice (72). Some methods and biomarkers have been utilized to improve MPE diagnostic sensitivity and specificity (80–88), for example, utilization of ploidy, agyrophilic nucleolar organizer regions and immunocytochemistry, the combined use of liquid-based cytology, cell block, and carcinoembryonic antigen immunocytochemistry. Han et al reported that low-dose interleukin-2 improved response rate and disease control rate in patients (89). Moreover, combination therapy with lobaplatin and erythromycin has been shown to be a safe and efficient treatment for patients with NSCLC-mediated MPE (90). A previous study demonstrated the low intensity of effusion immune cell PD-L1 expression in these neoplasms, which is associated with poor survival in patients with lung cancer (91).
Varying types of mutations within certain genes may result in proteins with a different conformation, thus providing new insights into mutation-induced drug resistance and/or sensitivity mechanisms, and conformational transitions connecting active and/or inactive states (92–94). Furthermore, these conformations are critical for ligand-independent EGFR activation and downstream signaling (95).
Accurate identification of predictive genetics is crucial not just for patient therapy but also for increasing our knowledge of treatment-induced tumor evolution. Thus far, researchers have illustrated the feasibility of oncogene analysis in MPE patient samples with advanced lung cancer (25,28–38). However, these targeted identification approaches, for example Sanger sequencing, ARMS-PCR, PNA clamping, or HRM analysis are predominantly PCR-based methods, therefore are only suitable for detecting mutations in small regions of DNA. The primary limitation of these methods is that they require multiple PCRs and an adequate amount of DNA. In addition, their sensitivity does not exceed 1:100 (14). Previously published studies have investigated the use of molecular profiling of advanced lung cancer samples with MPE using NGS (39,45). These studies showed that EGFR mutation frequency was 80% (29/36 patients) (39) and 73% (22/30 patients) (45). However, the number of enrolled patients with MPE was not sufficient (30–36 individuals) and the average sequencing depth on target was only 359X (45). In the present study, 108 patients with MPE were recruited between December 2017 and July 2018, and high-quality purified DNA was extracted from cell pellets obtained from pleural effusions. Higher quality specimens, DNA, higher quality libraries, more robust quality methods (for example, sequences with Q<30 were removed; somatic mutations with ≥2% mutant allele frequency, and with at least 20 supporting reads) and controls are required to quantify the NGS results with greater accuracy to reduce false negative and false positive detections. The present study utilized the hybridization capture-based assay (48) and the capture efficiency of YunYing's optimized probes was ~65%, which is superior to Agilent SureSelect, NimbleGen SeqCap EZ, or Illumina TruSeq Capture (96). This higher capture efficiency vastly reduced false negative NGS results. In addition, raw data was screened with a 2% threshold for the mutant allele frequency at every mutant site and most of these sites had at least 10 supporting reads, which greatly reduced the effects of false positives.
Activating mutations in EGFR are the most well-known among lung adenocarcinomas, which are associated with therapeutic agents, and have been described in 15–60% of lung cancer cases in different studies (11,97–103). The two most common mutations of EGFR in lung cancer are exon 19 deletions (60%) and the L858R point mutation (35%) (104–108). A retrospective single-center study reported that the frequency of these specific EGFR mutations was higher in patients with MPE compared with those without MPE in patients with stage IV lung cancer (109). In the present study, 93 patients (86%) with MPE harbored EGFR mutations (Table SII), and molecular profiling of the mutation distribution within these 93 EGFR-positive patients is shown in Fig. 2. Among these 93 patients, 31 (33.3%) presented with mutations holding no known significance, including the rare mutations within exons 18–21 and mutations outside exons 18–21. In total, 29 (31%) patients possessed the L858R mutation, which was the most common mutation. In 2015, Tsai et al (110) reported that the expression of EGFR-L858R in lung cancer cells resulted in the upregulation of CXCR4 using in vitro and in vivo experiments, upregulation of CXCR4 is associated with increased cancer cell invasive ability, promoting subsequent MPE formation. In the present study, the indel mutation in exon 19 was the second most commonly observed mutation among all mutations with 24 patients (25.8%) harboring exon 19 indel mutations. Other studies demonstrated that the EGFR exon 19 deletion in lung cancer cell lines resulted in high basal levels of MYC and HIF-1α expression, which contributed to tumor angiogenesis and may increase the probability of MPE formation (62,111). The results of the present study indicate that higher EGFR mutation frequency was associated with increased tendency for MPE. Overall, in the present study, 62 patients exhibited mutations within EGFR exons 18–21, who were treated with TKIs.
Rearrangements involving the ALK gene are critical events in NSCLC (112). ALK stimulates hypoxia pathways via direct regulation of HIF1α and HIF2α expression, promoting angiogenesis, and subsequently increasing pleural membrane permeability (113). ALK + EML4 gene fusion has been reported in ~5% of patients with MPE using immunohistochemistry, fluorescence in situ hybridization or RT-PCR (114–116). The present study identified 6 patients (5.5%), one male and five females, with the ALK + EML4 gene fusion using NGS.
Pleural metastases are more frequently observed in patients with lung adenocarcinoma compared with patients with wild type KRAS (117,118). Previously, Agalioti et al (119) demonstrated that KRAS-mutant cells disseminated into the pleural cavity recruited splenic mast cells, and CD11b + Gr1+ myeloid cells from bone marrow to the pleural space via CCL2-CCR2 signaling, which promoted the formation of MPE. The analysis of the present study identified 6 patients harboring KRAS mutations, only four of whom exhibited mutations of glycine or alanine, for which there are available treatment agents.
The BIM polymorphism has only been observed in East Asian populations, to the best of our knowledge (120). A 2,903-bp deletion was discovered in NSCLC cancer at a frequency of 12–18% using PCR (120–126). Several studies have reported that the BIM deletion polymorphism confers intrinsic resistance to EGFR-TKIs in cell lines (55,121–124). However, other studies have reported that the BIM deletion polymorphism did not account for intrinsic resistance to EGFR-TKIs (120,125,126), notably for MPE in patients with lung adenocarcinoma (120). The present study also suggested that lung cancer development was not completely dependent upon the BIM pathway. NGS analysis in the present study of MPE samples demonstrated that 10 patients (9.2%) possessed the BIM polymorphism, but the role of this mutation in the development or treatment of lung cancer remains unclear.
In patients with lung cancer, there is a consensus among studies that EGFR mutations are more frequent in female than in male patients (127–131). In the present study, the EGFR mutant frequency in female patients was 64.3% (36/56 patients), higher than in male patients, 50.9% (26/51 patients). This was consistent with previous studies, but EGFR exon 21 special mutations, BIM mutations, TP53 and BRAF special mutations were higher in males than in females in the present study.
Aging is a major risk factor for the development of cancer. The incidence of cancer is positively associated with age and is relatively rare in younger individuals, except for leukemia (132). Aging and cancer either share or diverge with respect to several disease mechanisms (132). Such mechanisms include the role of genomic instability, telomere attrition, epigenetic changes, loss of proteostasis, decreased nutrient sensing and altered metabolism, but also cellular senescence and stem cell function. Cancer cells and aged cells are also fundamentally different, as cancer cells can be thought of as hyperactive cells with advantageous mutations, rapid cell division and increased energy consumption, whereas aged cells are hypoactive with accumulated disadvantageous mutations, cell division inability and a decreased ability for energy production and consumption. A previous study investigating MPE from lung adenocarcinoma demonstrated that fewer Asian patients aged ≤50 years possessed EGFR mutations, but the EGFR mutation types observed were more uncommon (127). Most mutation rates observed in the NGS results in the present study increased with age, but the frequencies of other mutations decreased with age, for example, PIK3CA, EGFR exon 20 special mutation and ROS1 fusions.
The present study has certain limitations. Therapeutic information for the 108 patients was not available. Additionally, there were no control specimens for comparison of pleural effusion.
In conclusion, multiplexed molecular profiling of MPE from lung cancer using NGS was investigated, and due to the higher capture efficiency and deeper sequencing depth of NGS, MPE may be a reliable specimen for NGS detection of somatic mutations using NGS.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81472199) and by the National Natural Science Foundation of Jiangsu Province of China (grant no. BK20141162).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YS and XR designed the study and the experiments. MY contributed to the conception of the study. WW and JY were responsible for data collection and the extraction of DNA. DZ and ZG analyzed the data. YS, XR and MY drafted the manuscript. All authors read, critically revised, and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all participants and the present study was approved by the Institutional Review Board of Affiliated Changzhou No. 2 People's Hospital (Changzhou, China; approval no. 201400101).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Spiro SG, Silvestri GA. One hundred years of lung cancer. Am J Respir Crit Care Med. 2005;172:523–529. doi: 10.1164/rccm.200504-531OE. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network, corp-author. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. discussion 3080. [DOI] [PubMed] [Google Scholar]
- 11.Inamura K. Lung Cancer: Understanding its molecular pathology and the 2015 WHO classification. Front Oncol. 2017;7:193. doi: 10.3389/fonc.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohno T, Nakaoku T, Tsuta K, Tsuchihara K, Matsumoto S, Yoh K, Goto K. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res. 2015;4:156–164. doi: 10.3978/j.issn.2218-6751.2014.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan L, Alexander M, Officer A, MacManus M, Mileshkin L, Jennens R, Herath D, de Boer R, Fox SB, Ball D, Solomon B. Survival difference according to mutation status in a prospective cohort study of Australian patients with metastatic non-small-cell lung carcinoma. Intern Med J. 2018;48:37–44. doi: 10.1111/imj.13491. [DOI] [PubMed] [Google Scholar]
- 14.Han Y, Li J. Sample types applied for molecular diagnosis of therapeutic management of advanced non-small cell lung cancer in the precision medicine. Clin Chem Lab Med. 2017;55:1817–1833. doi: 10.1515/cclm-2017-0112. [DOI] [PubMed] [Google Scholar]
- 15.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaz LA, Jr, Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61:112–123. doi: 10.1373/clinchem.2014.222679. [DOI] [PubMed] [Google Scholar]
- 18.Masters GA, Krilov L, Bailey HH, Brose MS, Burstein H, Diller LR, Dizon DS, Fine HA, Kalemkerian GP, Moasser M, et al. Clinical cancer advances 2015: Annual report on progress against cancer from the American society of clinical oncology. J Clin Oncol. 2015;33:786–809. doi: 10.1200/JCO.2014.59.9746. [DOI] [PubMed] [Google Scholar]
- 19.Froudarakis ME. Pleural effusion in lung cancer: More questions than answers. Respiration. 2012;83:367–376. doi: 10.1159/000338169. [DOI] [PubMed] [Google Scholar]
- 20.Roberts ME, Neville E, Berrisford RG, Antunes G, Ali NJ, BTS Pleural Disease Guideline Group Management of a malignant pleural effusion: British thoracic society pleural disease guideline 2010. Thorax. 2010;65(Suppl 2):ii32–ii40. doi: 10.1136/thx.2010.136994. [DOI] [PubMed] [Google Scholar]
- 21.Chen YL, Lee CT, Lu CC, Yang SC, Chen WL, Lee YC, Yang CH, Peng SL, Su WC, Chow NH, Ho CL. Epidermal growth factor receptor mutation and anaplastic lymphoma kinase gene fusion: Detection in malignant pleural effusion by RNA or PNA analysis. PLoS One. 2016;11:e0158125. doi: 10.1371/journal.pone.0158125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarogoulidis K, Zarogoulidis P, Darwiche K, Tsakiridis K, Machairiotis N, Kougioumtzi I, Courcoutsakis N, Terzi E, Zaric B, Huang H, et al. Malignant pleural effusion and algorithm management. J Thorac Dis. 2013;5(Suppl 4):S413–S419. doi: 10.3978/j.issn.2072-1439.2013.09.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akamatsu H, Koh Y, Kenmotsu H, Naito T, Serizawa M, Kimura M, Mori K, Imai H, Ono A, Shukuya T, et al. Multiplexed molecular profiling of lung cancer using pleural effusion. J Thorac Oncol. 2014;9:1048–1052. doi: 10.1097/JTO.0000000000000203. [DOI] [PubMed] [Google Scholar]
- 24.Agalioti T, Giannou AD, Stathopoulos GT. Pleural involvement in lung cancer. J Thorac Dis. 2015;7:1021–1030. doi: 10.3978/j.issn.2072-1439.2015.04.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai TH, Wu SG, Hsieh MS, Yu CJ, Yang JC, Shih JY. Clinical and prognostic implications of RET rearrangements in metastatic lung adenocarcinoma patients with malignant pleural effusion. Lung Cancer. 2015;88:208–214. doi: 10.1016/j.lungcan.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Tiran V, Stanzer S, Heitzer E, Meilinger M, Rossmann C, Lax S, Tsybrovskyy O, Dandachi N, Balic M. Genetic profiling of putative breast cancer stem cells from malignant pleural effusions. PLoS One. 2017;12:e0175223. doi: 10.1371/journal.pone.0175223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jian G, Songwen Z, Ling Z, Qinfang D, Jie Z, Liang T, Caicun Z. Prediction of epidermal growth factor receptor mutations in the plasma/pleural effusion to efficacy of gefitinib treatment in advanced non-small cell lung cancer. J Cancer Res Clin Oncol. 2010;136:1341–1347. doi: 10.1007/s00432-010-0785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han HS, Eom DW, Kim JH, Kim KH, Shin HM, An JY, Lee KM, Choe KH, Lee KH, Kim ST, et al. EGFR mutation status in primary lung adenocarcinomas and corresponding metastatic lesions: Discordance in pleural metastases. Clin Lung Cancer. 2011;12:380–386. doi: 10.1016/j.cllc.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Wu SG, Gow CH, Yu CJ, Chang YL, Yang CH, Hsu YC, Shih JY, Lee YC, Yang PC. Frequent epidermal growth factor receptor gene mutations in malignant pleural effusion of lung adenocarcinoma. Eur Respir J. 2008;32:924–930. doi: 10.1183/09031936.00167407. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Zhao Y, Wang M, Yap WS, Chang AY. Detection and comparison of epidermal growth factor receptor mutations in cells and fluid of malignant pleural effusion in non-small cell lung cancer. Lung Cancer. 2008;60:175–182. doi: 10.1016/j.lungcan.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 32.Kimura H, Fujiwara Y, Sone T, Kunitoh H, Tamura T, Kasahara K, Nishio K. EGFR mutation status in tumour-derived DNA from pleural effusion fluid is a practical basis for predicting the response to gefitinib. Br J Cancer. 2006;95:1390–1395. doi: 10.1038/sj.bjc.6603428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu SG, Yu CJ, Tsai MF, Liao WY, Yang CH, Jan IS, Yang PC, Shih JY. Survival of lung adenocarcinoma patients with malignant pleural effusion. Eur Respir J. 2013;41:1409–1418. doi: 10.1183/09031936.00069812. [DOI] [PubMed] [Google Scholar]
- 34.Carter J, Miller JA, Feller-Kopman D, Ettinger D, Sidransky D, Maleki Z. Molecular profiling of malignant pleural effusion in metastatic non-small-cell lung carcinoma. The effect of preanalytical factors. Ann Am Thorac Soc. 2017;14:1169–1176. doi: 10.1513/AnnalsATS.201609-709OC. [DOI] [PubMed] [Google Scholar]
- 35.Lin J, Gu Y, Du R, Deng M, Lu Y, Ding Y. Detection of EGFR mutation in supernatant, cell pellets of pleural effusion and tumor tissues from non-small cell lung cancer patients by high resolution melting analysis and sequencing. Int J Clin Exp Pathol. 2014;7:8813–8822. [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Lee OJ, Son SM, Woo CG, Jeong Y, Yang Y, Kwon J, Lee KH, Han HS. EGFR mutation status in lung adenocarcinoma-associated malignant pleural effusion and efficacy of EGFR tyrosine kinase inhibitors. Cancer Res Treat. 2018;50:908–916. doi: 10.4143/crt.2017.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeo CD, Kim JW, Kim KH, Ha JH, Rhee CK, Kim SJ, Kim YK, Park CK, Lee SH, Park MS, Yim HW. Detection and comparison of EGFR mutations in matched tumor tissues, cell blocks, pleural effusions, and sera from patients with NSCLC with malignant pleural effusion, by PNA clamping and direct sequencing. Lung Cancer. 2013;81:207–212. doi: 10.1016/j.lungcan.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 38.Han HS, Lim SN, An JY, Lee KM, Choe KH, Lee KH, Kim ST, Son SM, Choi SY, Lee HC, Lee OJ. Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J Thorac Oncol. 2012;7:355–364. doi: 10.1097/JTO.0b013e31823c4c1b. [DOI] [PubMed] [Google Scholar]
- 39.Buttitta F, Felicioni L, Del Grammastro M, Filice G, Di Lorito A, Malatesta S, Viola P, Centi I, D'Antuono T, Zappacosta R, et al. Effective assessment of egfr mutation status in bronchoalveolar lavage and pleural fluids by next-generation sequencing. Clin Cancer Res. 2013;19:691–698. doi: 10.1158/1078-0432.CCR-12-1958. [DOI] [PubMed] [Google Scholar]
- 40.Biankin AV, Piantadosi S, Hollingsworth SJ. Patient-centric trials for therapeutic development in precision oncology. Nature. 2015;526:361–370. doi: 10.1038/nature15819. [DOI] [PubMed] [Google Scholar]
- 41.Celesti F, Celesti A, Wan J, Villari M. Why deep learning is changing the way to approach NGS data processing: A review. IEEE Rev Biomed Eng. 2018;11:68–76. doi: 10.1109/RBME.2018.2825987. [DOI] [PubMed] [Google Scholar]
- 42.Kamps R, Brandão RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ, Romano A. Next-generation sequencing in oncology: Genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017;18:E308. doi: 10.3390/ijms18020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu YC, Zhou YF, Wang WX, Xu CW, Zhuang W, Du KQ, Chen G. CEP72-ROS1: A novel ROS1 oncogenic fusion variant in lung adenocarcinoma identified by next-generation sequencing. Thorac Cancer. 2018;9:652–655. doi: 10.1111/1759-7714.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Li Y, Liu C, Wang W, Li M, Lv D, Sun G, Chen H, Dong X, Miao Z, et al. Identification of a novel KIF13A-RET fusion in lung adenocarcinoma by next-generation sequencing. Lung Cancer. 2018;118:27–29. doi: 10.1016/j.lungcan.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, Shao D, Deng Q, Tang H, Wang J, Liu J, Guo F, Lin Y, Peng Z, Mao M, et al. Next generation sequencing-based molecular profiling of lung adenocarcinoma using pleural effusion specimens. J Thorac Dis. 2018;10:2631–2637. doi: 10.21037/jtd.2018.04.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R Core Team, corp-author. R Foundation for Statistical Computing; Vienna, Austria: 2014. A language and environment for statistical computing. [Google Scholar]
- 47.RStudio Team, corp-author. RStudio: Integrated Development for R. RStudio, Inc., Boston, MA, 2015. http://www.rstudio.com/ [Google Scholar]
- 48.Bai H, Xia J, Zhao X, Gong Z, Zhang D, Xiong L. Detection of EGFR mutations using target capture sequencing in plasma of patients with non-small-cell lung cancer. J Clin Pathol. 2019;72:379–385. doi: 10.1136/jclinpath-2019-205699. [DOI] [PubMed] [Google Scholar]
- 49.Chmielecki J, Ross JS, Wang K, Frampton GM, Palmer GA, Ali SM, Palma N, Morosini D, Miller VA, Yelensky R, et al. Oncogenic alterations in ERBB2/HER2 represent potential therapeutic targets across tumors from diverse anatomic sites of origin. Oncologist. 2014;20:7–12. doi: 10.1634/theoncologist.2014-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 52.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 53.Hyman DM, Smyth LM, Donoghue MTA, Westin SN, Bedard PL, Dean EJ, Bando H, El-Khoueiry AB, Pérez-Fidalgo JA, Mita A, et al. AKT Inhibition in Solid Tumors With AKT1 Mutations. J Clin Oncol. 2017;35:2251–2259. doi: 10.1200/JCO.2017.73.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J, Yu L, Chen W, Xu Y, Wu M, Todorova D, Tang Q, Feng B, Jiang L, He J, et al. Wild-type p53 promotes cancer metabolic switch by inducing PUMA-dependent suppression of oxidative phosphorylation. Cancer Cell. 2019;35:191–203.e8. doi: 10.1016/j.ccell.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 55.Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, Ariyaratne PN, Takahashi N, Sawada K, Fei Y, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 56.BRAF fusions in clinically advanced non-small cell lung cancer, corp-author. An emerging target for anti-BRAF therapies. J Clin Oncol. 2017;35(Suppl. 15):S9072. doi: 10.1200/JCO.2017.35.15_suppl.9072. [DOI] [Google Scholar]
- 57.Caris Life Sciences, corp-author. BRAF mutations are potentially targetable alterations in a wide variety of solid cancers. https://www.carislifesciences.com/documents/braf-mutations-are-potentially-targetable-alterations-in-a-wide-variety-of-solid-cancers/ [Dec 2;2016 ]; [Google Scholar]
- 58.Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. 2010;347:21–41. doi: 10.1007/82_2010_68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng PK, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, Sengupta S, Wang Z, Bhavana VH, Tran R, et al. Systematic functional annotation of somatic mutations in cancer. Cancer Cell. 2018;33:450–462.e10. doi: 10.1016/j.ccell.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang LH, Wu CF, Rajasekaran N, Shin YK. Loss of tumor suppressor gene function in human cancer: An overview. Cell Physiol Biochem. 2018;51:2647–2693. doi: 10.1159/000495956. [DOI] [PubMed] [Google Scholar]
- 61.Hammerman PS, Sos ML, Ramos AH, Xu C, Dutt A, Zhou W, Brace LE, Woods BA, Lin W, Zhang J, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lui MMS, et al. Malignant pleural effusion from lung cancers with driver mutations. Curr Pulmonol Rep. 2018;7:13–18. doi: 10.1007/s13665-018-0196-1. [DOI] [Google Scholar]
- 63.Davies HE, Mishra EK, Kahan BC, Wrightson JM, Stanton AE, Guhan A, Davies CW, Grayez J, Harrison R, Prasad A, et al. Effect of an indwelling pleural catheter vs. chest tube and talc pleurodesis for relieving dyspnea in patients with malignant pleural effusion: The TIME2 randomized controlled trial. JAMA. 2012;307:2383–2389. doi: 10.1001/jama.2012.5535. [DOI] [PubMed] [Google Scholar]
- 64.Sahn SA, Good JT., Jr Pleural fluid pH in malignant effusions. Diagnostic, prognostic, and therapeutic implications. Ann Intern Med. 1988;108:345–349. doi: 10.7326/0003-4819-108-3-345. [DOI] [PubMed] [Google Scholar]
- 65.Heffner JE, Nietert PJ, Barbieri C. Pleural fluid pH as a predictor of survival for patients with malignant pleural effusions. Chest. 2000;117:79–86. doi: 10.1378/chest.117.1.79. [DOI] [PubMed] [Google Scholar]
- 66.Heffner JE, Heffner JN, Brown LK. Multilevel and continuous pleural fluid pH likelihood ratios for evaluating malignant pleural effusions. Chest. 2003;123:1887–1894. doi: 10.1378/chest.123.6.1887. [DOI] [PubMed] [Google Scholar]
- 67.Stathopoulos GT, Sherrill TP, Karabela SP, Goleniewska K, Kalomenidis I, Roussos C, Fingleton B, Yull FE, Peebles RS, Jr, Blackwell TS. Host-derived interleukin-5 promotes adenocarcinoma-induced malignant pleural effusion. Am J Respir Crit Care Med. 2010;182:1273–1281. doi: 10.1164/rccm.201001-0001OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stathopoulos GT, Zhu Z, Everhart MB, Kalomenidis I, Lawson WE, Bilaceroglu S, Peterson TE, Mitchell D, Yull FE, Light RW, Blackwell TS. Nuclear factor-kappaB affects tumor progression in a mouse model of malignant pleural effusion. Am J Respir Cell Mol Biol. 2006;34:142–150. doi: 10.1165/rcmb.2005-0130OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stathopoulos GT, Psallidas I, Moustaki A, Moschos C, Kollintza A, Karabela S, Porfyridis I, Vassiliou S, Karatza M, Zhou Z, et al. A central role for tumor-derived monocyte chemoattractant protein-1 in malignant pleural effusion. J Natl Cancer Inst. 2008;100:1464–1476. doi: 10.1093/jnci/djn325. [DOI] [PubMed] [Google Scholar]
- 70.Giannou AD, Marazioti A, Spella M, Kanellakis NI, Apostolopoulou H, Psallidas I, Prijovich ZM, Vreka M, Zazara DE, Lilis I, et al. Mast cells mediate malignant pleural effusion formation. J Clin Invest. 2015;125:2317–2334. doi: 10.1172/JCI79840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y, Mathy NW, Lu H. The role of VEGF in the diagnosis and treatment of malignant pleural effusion in patients with nonsmall cell lung cancer (Review) Mol Med Rep. 2018;17:8019–8030. doi: 10.3892/mmr.2018.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu XZ, Zhou Q, Lin H, Zhai K, Wang XJ, Yang WB, Shi HZ. Immune regulation of toll-like receptor 2 engagement on CD4+ T cells in murine models of malignant pleural effusion. Am J Respir Cell Mol Biol. 2017;56:342–352. doi: 10.1165/rcmb.2015-0396OC. [DOI] [PubMed] [Google Scholar]
- 73.Stathopoulos GT, Kalomenidis I. Malignant pleural effusion: Tumor-host interactions unleashed. Am J Respir Crit Care Med. 2012;186:487–492. doi: 10.1164/rccm.201203-0465PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Psallidas I, Stathopoulos GT, Maniatis NA, Magkouta S, Moschos C, Karabela SP, Kollintza A, Simoes DC, Kardara M, Vassiliou S, et al. Secreted phosphoprotein-1 directly provokes vascular leakage to foster malignant pleural effusion. Oncogene. 2013;32:528–535. doi: 10.1038/onc.2012.57. [DOI] [PubMed] [Google Scholar]
- 75.Cui R, Takahashi F, Ohashi R, Yoshioka M, Gu T, Tajima K, Unnoura T, Iwakami S, Hirama M, Ishiwata T, et al. Osteopontin is involved in the formation of malignant pleural effusion in lung cancer. Lung Cancer. 2009;63:368–374. doi: 10.1016/j.lungcan.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 76.Thomas R, Cheah HM, Creaney J, Turlach BA, Lee YC. Longitudinal measurement of pleural fluid biochemistry and cytokines in malignant pleural effusions. Chest. 2016;149:1494–1500. doi: 10.1016/j.chest.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 77.Ho CC, Liao WY, Wang CY, Lu YH, Huang HY, Chen HY, Chan WK, Chen HW, Yang PC. TREM-1 expression in tumor-associated macrophages and clinical outcome in lung cancer. Am J Respir Crit Care Med. 2008;177:763–770. doi: 10.1164/rccm.200704-641OC. [DOI] [PubMed] [Google Scholar]
- 78.Gopinathan G, Milagre C, Pearce OM, Reynolds LE, Hodivala-Dilke K, Leinster DA, Zhong H, Hollingsworth RE, Thompson R, Whiteford JR, Balkwill F. Interleukin-6 stimulates defective angiogenesis. Cancer Res. 2015;75:3098–3107. doi: 10.1158/0008-5472.CAN-15-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yeh HH, Lai WW, Chen HH, Liu HS, Su WC. Autocrine IL-6-induced Stat3 activation contributes to the pathogenesis of lung adenocarcinoma and malignant pleural effusion. Oncogene. 2006;25:4300–4309. doi: 10.1038/sj.onc.1209464. [DOI] [PubMed] [Google Scholar]
- 80.Liu Q, Yu YX, Wang XJ, Wang Z, Wang Z. Diagnostic accuracy of interleukin-27 between tuberculous pleural effusion and malignant pleural effusion: A meta-analysis. Respiration. 2018;95:469–477. doi: 10.1159/000486963. [DOI] [PubMed] [Google Scholar]
- 81.Ye ZJ, Zhou Q, Yin W, Yuan ML, Yang WB, Xiang F, Zhang JC, Xin JB, Xiong XZ, Shi HZ. Interleukin 22-producing CD4+ T cells in malignant pleural effusion. Cancer Lett. 2012;326:23–32. doi: 10.1016/j.canlet.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 82.Sriram KB, Relan V, Clarke BE, Duhig EE, Yang IA, Bowman RV, Lee YC, Fong KM. Diagnostic molecular biomarkers for malignant pleural effusions. Future Oncol. 2011;7:737–752. doi: 10.2217/fon.11.45. [DOI] [PubMed] [Google Scholar]
- 83.Palaoro LA, Blanco AM, Gamboni M, Rocher AE, Rotenberg RG. Usefulness of ploidy, AgNOR and immunocytochemistry for differentiating benign and malignant cells in serous effusions. Cytopathology. 2007;18:33–39. doi: 10.1111/j.1365-2303.2007.00404.x. [DOI] [PubMed] [Google Scholar]
- 84.Nam HS. Malignant pleural effusion: Medical approaches for diagnosis and management. Tuberc Respir Dis (Seoul) 2014;76:211–217. doi: 10.4046/trd.2014.76.5.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Light RW. Pleural effusions. Med Clin North Am. 2011;95:1055–1070. doi: 10.1016/j.mcna.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 86.Jiang B, Wu GP, Zhao YJ, Wang SC. Transcription expression and clinical significance of TTF-1 mRNA in pleural effusion of patients with lung cancer. Diagn Cytopathol. 2008;36:849–854. doi: 10.1002/dc.20926. [DOI] [PubMed] [Google Scholar]
- 87.Woo CG, Son SM, Han HS, Lee KH, Choe KH, An JY, Man Lee K, Lim YH, Lee HC, Lee OJ. Diagnostic benefits of the combined use of liquid-based cytology, cell block, and carcinoembryonic antigen immunocytochemistry in malignant pleural effusion. J Thorac Dis. 2018;10:4931–4939. doi: 10.21037/jtd.2018.07.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang F, Wang J, Zheng X, Hu L, Chen J, Jiang F, Wang Y. Clinical value of jointly detection pleural fluid Midkine, pleural fluid adenosine deaminase, and pleural fluid carbohydrate antigen 125 in the identification of nonsmall cell lung cancer-associated malignant pleural effusion. J Clin Lab Anal. 2018;32:e22576. doi: 10.1002/jcla.22576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han L, Jiang Q, Yao W, Fu T, Zeng Q. Thoracic injection of low-dose interleukin-2 as an adjuvant therapy improves the control of the malignant pleural effusions: A systematic review and meta-analysis base on Chinese patients. BMC Cancer. 2018;18:725. doi: 10.1186/s12885-018-4581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu L, Wang B, Gao M, Zhang Y, Qi Q, Li T, Li C, Wang A, Li Y. Intrapleural combination therapy with lobaplatin and erythromycin for non-small cell lung cancer-mediated malignant pleural effusion. Thorac Cancer. 2018;9:950–955. doi: 10.1111/1759-7714.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tseng YH, Ho HL, Lai CR, Luo YH, Tseng YC, Whang-Peng J, Lin YH, Chou TY, Chen YM. PD-L1 expression of tumor cells, macrophages, and immune cells in non-small cell lung cancer patients with malignant pleural effusion. J Thorac Oncol. 2018;13:447–453. doi: 10.1016/j.jtho.2017.10.034. [DOI] [PubMed] [Google Scholar]
- 92.Li Y, Li X, Ma W, Dong Z. Conformational transition pathways of epidermal growth factor receptor kinase domain from multiple molecular dynamics simulations and bayesian clustering. J Chem Theory Comput. 2014;10:3503–3511. doi: 10.1021/ct500162b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pan AC, Weinreich TM, Shan Y, Scarpazza DP, Shaw DE. Assessing the accuracy of two enhanced sampling methods using EGFR kinase transition pathways: The influence of collective variable choice. J Chem Theory Comput. 2014;10:2860–2865. doi: 10.1021/ct500223p. [DOI] [PubMed] [Google Scholar]
- 94.Sutto L, Gervasio FL. Effects of oncogenic mutations on the conformational free-energy landscape of EGFR kinase. Proc Natl Acad Sci USA. 2013;110:10616–10621. doi: 10.1073/pnas.1221953110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruan Z, Kannan N. Altered conformational landscape and dimerization dependency underpins the activation of EGFR by αC-β4 loop insertion mutations. Proc Natl Acad Sci USA. 2018;115:E8162–E8171. doi: 10.1073/pnas.1803152115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: Key considerations in genomic analyses. Nat Rev Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 97.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 98.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 99.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M, Riely GJ. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang M, Topaloglu U, Petty WJ, Pagni M, Foley KL, Grant SC, Robinson M, Bitting RL, Thomas A, Alistar AT, et al. Circulating mutational portrait of cancer: Manifestation of aggressive clonal events in both early and late stages. J Hematol Oncol. 2017;10:100. doi: 10.1186/s13045-017-0468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Z, Yang JJ, Huang J, Ye JY, Zhang XC, Tu HY, Han-Zhang H, Wu YL. Lung adenocarcinoma harboring EGFR T790M and in trans C797S responds to combination therapy of first- and third-generation EGFR TKIs and shifts allelic configuration at resistance. J Thorac Oncol. 2017;12:1723–1727. doi: 10.1016/j.jtho.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 102.Lin YT, Liu YN, Wu SG, Yang JC, Shih JY. Epidermal growth factor receptor tyrosine kinase inhibitor-sensitive exon 19 insertion and exon 20 insertion in patients with advanced non-small-cell lung cancer. Clin Lung Cancer. 2017;18:324–332.e1. doi: 10.1016/j.cllc.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 103.Ibrahim U, Saqib A, Atallah JP. EGFR exon 18 delE709_T710insD mutated stage IV lung adenocarcinoma with response to afatinib. Lung Cancer. 2017;108:45–47. doi: 10.1016/j.lungcan.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 104.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 105.Chic N, Mayo-de-Las-Casas C, Reguart N. Successful treatment with gefitinib in advanced non-small cell lung cancer after acquired resistance to osimertinib. J Thorac Oncol. 2017;12:e78–e80. doi: 10.1016/j.jtho.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 106.Ganesan P, Ali SM, Wang K, Blumenschein GR, Esmaeli B, Wolff RA, Miller VA, Stephens PJ, Ross JS, Palmer GA, Janku F. Epidermal growth factor receptor P753S mutation in cutaneous squamous cell carcinoma responsive to cetuximab-based therapy. J Clin Oncol. 2016;34:e34–37. doi: 10.1200/JCO.2013.50.3516. [DOI] [PubMed] [Google Scholar]
- 107.Costa DB. Kinase inhibitor-responsive genotypes in EGFR mutated lung adenocarcinomas: Moving past common point mutations or indels into uncommon kinase domain duplications and rearrangements. Transl Lung Cancer Res. 2016;5:331–337. doi: 10.21037/tlcr.2016.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, Lai Z, Markovets A, Vivancos A, Kuang Y, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zou J, Bella AE, Chen Z, Han X, Su C, Lei Y, Luo H. Frequency of EGFR mutations in lung adenocarcinoma with malignant pleural effusion: Implication of cancer biological behaviour regulated by EGFR mutation. J Int Med Res. 2014;42:1110–1117. doi: 10.1177/0300060514539273. [DOI] [PubMed] [Google Scholar]
- 110.Tsai MF, Chang TH, Wu SG, Yang HY, Hsu YC, Yang PC, Shih JY. EGFR-L858R mutant enhances lung adenocarcinoma cell invasive ability and promotes malignant pleural effusion formation through activation of the CXCL12-CXCR4 pathway. Sci Rep. 2015;5:13574. doi: 10.1038/srep13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee JG, Wu R. Erlotinib-cisplatin combination inhibits growth and angiogenesis through c-MYC and HIF-1α in EGFR-mutated lung cancer in vitro and in vivo. Neoplasia. 2015;17:190–200. doi: 10.1016/j.neo.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mertens F, Tayebwa J. Evolving techniques for gene fusion detection in soft tissue tumours. Histopathology. 2014;64:151–162. doi: 10.1111/his.12272. [DOI] [PubMed] [Google Scholar]
- 113.Martinengo C, Poggio T, Menotti M, Scalzo MS, Mastini C, Ambrogio C, Pellegrino E, Riera L, Piva R, Ribatti D, et al. ALK-dependent control of hypoxia-inducible factors mediates tumor growth and metastasis. Cancer Res. 2014;74:6094–6106. doi: 10.1158/0008-5472.CAN-14-0268. [DOI] [PubMed] [Google Scholar]
- 114.Wang Z, Wu X, Han X, Cheng G, Mu X, Zhang Y, Cui D, Liu C, Liu D, Shi Y. ALK gene expression status in pleural effusion predicts tumor responsiveness to crizotinib in Chinese patients with lung adenocarcinoma. Chin J Cancer Res. 2016;28:606–616. doi: 10.21147/j.issn.1000-9604.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu L, Zhan P, Zhou X, Song Y, Zhou X, Yu L, Wang J. Detection of EML4-ALK in lung adenocarcinoma using pleural effusion with FISH, IHC, and RT-PCR methods. PLoS One. 2015;10:e0117032. doi: 10.1371/journal.pone.0117032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhong J, Li X, Bai H, Zhao J, Wang Z, Duan J, An T, Wu M, Wang Y, Wang S, Wang J. Malignant pleural effusion cell blocks are substitutes for tissue in EML4-ALK rearrangement detection in patients with advanced non-small-cell lung cancer. Cytopathology. 2016;27:433–443. doi: 10.1111/cyt.12322. [DOI] [PubMed] [Google Scholar]
- 117.Raparia K, Villa C, Raj R, Cagle PT. Peripheral lung adenocarcinomas with KRAS mutations are more likely to invade visceral pleura. Arch Pathol Lab Med. 2015;139:189–193. doi: 10.5858/arpa.2013-0759-OA. [DOI] [PubMed] [Google Scholar]
- 118.Renaud S, Seitlinger J, Falcoz PE, Schaeffer M, Voegeli AC, Legrain M, Beau-Faller M, Massard G. Specific KRAS amino acid substitutions and EGFR mutations predict site-specific recurrence and metastasis following non-small-cell lung cancer surgery. Br J Cancer. 2016;115:346–353. doi: 10.1038/bjc.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Agalioti T, Giannou AD, Krontira AC, Kanellakis NI, Kati D, Vreka M, Pepe M, Spella M, Lilis I, Zazara DE, et al. Mutant KRAS promotes malignant pleural effusion formation. Nat Commun. 2017;8:15205. doi: 10.1038/ncomms15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu SG, Liu YN, Yu CJ, Yang PC, Shih JY. Association of BIM deletion polymorphism with intrinsic resistance to EGFR tyrosine kinase inhibitors in patients with lung adenocarcinoma. JAMA Oncol. 2016;2:826–828. doi: 10.1001/jamaoncol.2016.0016. [DOI] [PubMed] [Google Scholar]
- 121.Lee JH, Lin YL, Hsu WH, Chen HY, Chang YC, Yu CJ, Shih JY, Lin CC, Chen KY, Ho CC, et al. Bcl-2-like protein 11 deletion polymorphism predicts survival in advanced non-small-cell lung cancer. J Thorac Oncol. 2014;9:1385–1392. doi: 10.1097/JTO.0000000000000238. [DOI] [PubMed] [Google Scholar]
- 122.Cardona AF, Rojas L, Wills B, Arrieta O, Carranza H, Vargas C, Otero J, Corrales-Rodriguez L, Martín C, Reguart N, et al. BIM deletion polymorphisms in Hispanic patients with non-small cell lung cancer carriers of EGFR mutations. Oncotarget. 2016;7:68933–68942. doi: 10.18632/oncotarget.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xia J, Bai H, Yan B, Li R, Shao M, Xiong L, Han B. Mimicking the BIM BH3 domain overcomes resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer. Oncotarget. 2017;8:108522–108533. doi: 10.18632/oncotarget.19411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao M, Zhang Y, Cai W, Li J, Zhou F, Cheng N, Ren R, Zhao C, Li X, Ren S, et al. The Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer. 2014;120:2299–2307. doi: 10.1002/cncr.28725. [DOI] [PubMed] [Google Scholar]
- 125.Lee JK, Shin JY, Kim S, Lee S, Park C, Kim JY, Koh Y, Keam B, Min HS, Kim TM, et al. Primary resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in patients with non-small-cell lung cancer harboring TKI-sensitive EGFR mutations: An exploratory study. Ann Oncol. 2013;24:2080–2087. doi: 10.1093/annonc/mdt127. [DOI] [PubMed] [Google Scholar]
- 126.Isobe K, Hata Y, Tochigi N, Kaburaki K, Kobayashi H, Makino T, Otsuka H, Sato F, Ishida F, Kikuchi N, et al. Clinical significance of BIM deletion polymorphism in non-small-cell lung cancer with epidermal growth factor receptor mutation. J Thorac Oncol. 2014;9:483–487. doi: 10.1097/JTO.0000000000000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wu SG, Chang YL, Yu CJ, Yang PC, Shih JY. Lung adenocarcinoma patients of young age have lower EGFR mutation rate and poorer efficacy of EGFR tyrosine kinase inhibitors. ERJ Open Res. 2017;3:00092–2016. doi: 10.1183/23120541.00092-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu M, Pan X, Xu Y, Wu S, Wu X, Chen B. Methodological comparison of the allele refractory mutation system and direct sequencing for detecting EGFR mutations in NSCLC, and the association of EGFR mutations with patient characteristics. Oncol Lett. 2018;16:1087–1094. doi: 10.3892/ol.2018.8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G, Yang PC. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tseng CH, Chiang CJ, Tseng JS, Yang TY, Hsu KH, Chen KC, Wang CL, Chen CY, Yen SH, Tsai CM, et al. EGFR mutation, smoking, and gender in advanced lung adenocarcinoma. Oncotarget. 2017;8:98384–98393. doi: 10.18632/oncotarget.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kota R, Gundeti S, Gullipalli M, Linga VG, Maddali LS, Digumarti R. Prevalence and outcome of epidermal growth factor receptor mutations in non-squamous non-small cell lung cancer patients. Lung India. 2015;32:561–565. doi: 10.4103/0970-2113.168099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Aunan JR, Cho WC, Soreide K. The biology of aging and cancer: A brief overview of shared and divergent molecular hallmarks. Aging Dis. 2017;8:628–642. doi: 10.14336/AD.2017.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.